Abstract
Mutations of the IGH variable region in patients with chronic lymphocytic leukemia are associated with a favorable prognosis, whereas cytogenetic complexity (≥3 unrelated aberrations) and translocations have been associated with an unfavorable prognosis. While mutational status of IGHV is stable, cytogenetic aberrations frequently evolve. However, the relationships of these features as prognosticators at diagnosis are unknown. We examined the CpG-stimulated metaphase cytogenetic features detected within 1 year of diagnosis of chronic lymphocytic leukemia and correlated these features with outcome and other clinical features including IGHV mutational status. Of 329 untreated patients, 53 (16.1%) had a complex karyotype, and 85 (25.8%) had a translocation. The median time to first treatment (TFT) was 47 months. In univariable analyses, significant risk factors for shorter TFT (P<0.05) were Rai stage 3-4, 2-microglobulin >3.5 mg/L, log-transformed white blood cell count, unmutated IGHV, a complex karyotype, a translocation, and trisomy 8, del(11q) or del(17p) detected by fluorescence in situ hybridization. In multivariable analysis, there was a significant effect modification of IGHV status on the relationship between translocation and TFT (P=0.002). In IGHV-mutated patients, those with a translocation had an over 3.5 times higher risk of starting treatment than those without a translocation (P<0.001); however, among IGHV-unmutated patients, a translocation did not significantly increase the risk of starting treatment (hazard ratio 1.00, P=0.99). The effect of Rai stage 3-4, log-transformed white blood cell count and complex karyotype remained statistically significant, whereas that of del(17p) did not (P=0.51). In summary, the presence of a translocation in IGHV-mutated patients appeared to negate the improved prognosis of mutated IGHV, but the presence of a translocation did not have an effect on TFT in high-risk IGHV-unmutated patients.
Introduction
Chronic lymphocytic leukemia (CLL) has a varied clinical course. Some patients have an indolent disease with a lifespan similar to that of their peers without the disease. Other patients have rapid disease progression, require treatment early in the course of the disease, are less responsive to therapy, or exhibit short remission after therapy and have a relatively short overall survival. Factors that have prognostic significance include clinical variables such as gender, age and Rai/Binet stage, and biological factors such as IGHV mutational status, ZAP70 status and genetic abnormalities as detected by fluorescence in situ hybridization (FISH). IGHV mutational status is a significant and independent prognostic factor that does not change over the course of the disease. Patients with unmutated IGHV have a worse prognosis than those with mutated IGHV.1-3
The karyotype of the patient is another genetic factor that influences disease course and response to therapy. The complexity of the karyotype is prognostic with three or more unrelated abnormalities predictive of a more adverse prognosis.4-12 Furthermore, a complex karyotype has been associated with unmutated IGHV, CD38 expression and deletion of 17p or mutated TP53.5,9,11,13-16 Additional studies showed that in addition to complex karyotypes, translocations, particularly unbalanced translocations, were associated with a poor prognosis, reflected as both an increased risk of requirement for treatment and a shortened overall survival.5,9,17-19 Translocations have also been associated with a complex karyotype and with deletions of 17p, and in some cases with del(11q),9 but not in others.20While these studies are informative, the relationship of translocations to mutated versus unmutated IGHV detected early in the course of the disease is unclear.
Traditionally metaphase cytogenetic studies of CLL have been minimally informative, as CLL cells do not readily divide spontaneously in culture, nor do they respond to traditional B-cell mitogens. The advent of CpG oligodeoxynucleotide stimulation of CLL cells has facilitated detection of cytogenetic abnormalities in up to 80% of CLL patients.17,21-26 As a result of this increased detection of karyotypic abnormalities, the significance of translocations and of a complex karyotype at disease diagnosis and during disease progression can be assessed more accurately. As CLL progresses, the karyotype frequently evolves.27,28 Therefore, the significance of karyotypic complexity and/or translocations at disease presentation is not clear. We examined CpG-stimulated karyotypes completed within 1 year of diagnosis of CLL and correlated the presence of a translocation and of complexity with disease prognosticators such as IGHV mutational status, sex, age, Rai stage, FISH abnormalities and with time to first treatment (TFT).
Methods
Patients and samples
All patients had immunophenotypically defined B-cell CLL as outlined by the International Workshop on CLL Revised CLL Guidelines.29 Peripheral blood or bone marrow was collected from patients after having obtained written informed consent in accordance with the Declaration of Helsinki and under a protocol approved by the institutional review board of The Ohio State University (Columbus, OH, USA).
Cytogenetic analyses
Cells were stimulated with CpG oligodeoxynucleotides and analyzed according to standard laboratory procedures (described in detail in the Online Supplementary Methods). FISH using probes for D13S319, D12Z3, ATM, TP53, BCL6, MYC, MYB (Abbott Molecular, Des Plaines, IL, USA) and SEC63 (Empire Genomics, Buffalo, NY, USA) were done according to the manufacturers’ recommendations.
Patients’ inclusion criteria
Patients with CLL who had cytogenetic analyses between December 2006 and December 2013 and were within 1 year of diagnosis were identified from The Ohio State University Wexner Medical Center databases. Balanced and unbalanced translocations and insertions were identified. Inversions were considered as balanced translocations, and “adds” as unbalanced translocations. A complex karyotype was defined as ≥3 independent aberrations on metaphase cytogenetics. The association of complex karyo type defined as ≥5 independent aberrations with outcome was also examined. Baseline characteristics were obtained from the patients’ charts. White blood cell (WBC) count and 2-microglobulin concentration, determined within 90 days of diagnosis were used.
Statistical analyses
The patients’ demographic and genetic characteristics were described using mean and standard deviation or median and range for continuous variables, and frequency and percentage for categorical variables. Associations between translocations or cytogenetic complexity with FISH abnormalities were tested using Fisher exact tests. Cases with both balanced and unbalanced translocations were classified together for statistical analyses.
TFT was calculated from the date of diagnosis until the date of first treatment or last follow-up, censoring patients who had not started treatment at the date last seen; patients who died prior to starting treatment were censored at the date last seen. Kaplan-Meier curves were used to estimate TFT probability, and Cox proportional hazard models were used to examine the association between potential risk factors and TFT. Table 1 shows variables considered for modeling. WBC counts were log-transformed.
Significant risk factors from univariable Cox models were included in the multivariable Cox model. Due to missing values in IGHV mutation status (n=37, 11%), WBC count (n=83, 25%), and 2-microglobulin (n=146, 44%), a multiple imputation procedure was applied to obtain combined results from 15 multiply imputed datasets. Using stepwise backward selection, variables that reached statistical significance after adjusting for all other covariates remained in the final model. To evaluate for potential effect modification, pairwise interactions among all variables in the final model were further tested, and TFT curves were generated.
Among patients who started chemoimmunotherapy, overall survival from the start of therapy was determined. Univariable analyses were performed testing for risk factors associated with overall survival, but multivariable analyses were precluded because of the limited number of deaths. The analyses were conducted using Stata 14, SAS 9.4 and the statistical significance was set as P<0.05.
Results
Patients’ characteristics
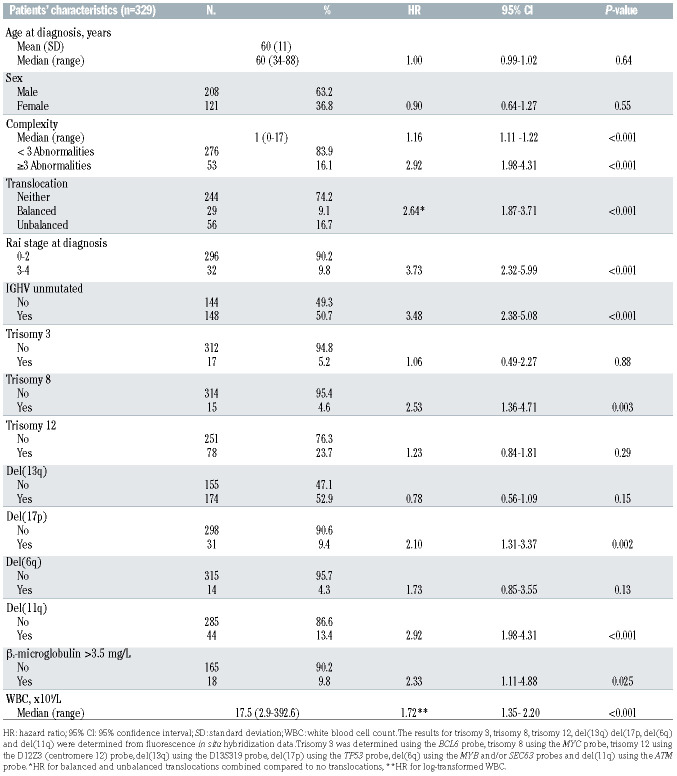
Diagnostic samples meeting the above criteria were identified for 329 patients. The patients’ clinical data are presented in Table 1. The median age of the patients was 60 years (range, 34-88), 36.8% were female, and 90.2% had Rai stage 0-2.
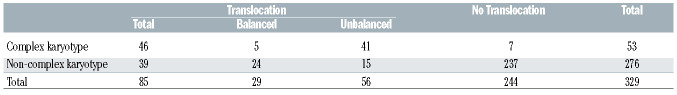
Unbalanced translocations are more frequent in complex karyotypes, but balanced translocations are more frequent in non-complex karyotypes
Translocations occurred in 85 (25.8%) patients: 29 with balanced, 48 with unbalanced and eight with both a balanced and an unbalanced translocation. Defining complexity as ≥3 unrelated aberrations, 16.1% of patients had complex karyotypes, while 7.6% had ≥5 aberrations. The distributions of karyotype complexity, defined as ≥3 aberrations, and the presence of a translocation are shown in Table 2. Of the 85 cases with a translocation, 46 also had a complex karyotype and 39 did not. Unbalanced translocations were more frequent in cases with a complex karyo type (41/46), while balanced translocations were more frequent in non-complex cases (24/39). No translocation was identified in 244 cases; 237 of these had non-complex karyotypes.
Translocations and complex karyotypes occur more frequently in unmutated IGVH chronic lymphocytic leukemia
IGHV data were available for 292 patients; data were not available for 37 patients (Table 3). Unmutated IGHV was found in 148 patients; 51 (34.5%) had a translocation, and 97 (65.5%) did not have a translocation; 31 (20.9%) had complex karyotypes (≥3 aberrations), while 117 (79.1%) had non-complex karyotypes. Of the 144 patients with mutated IGHV, 20 (13.9%) had a translocation, 124 (86.1%) did not have a translocation, 12 (8.3%) had complex karyotypes, and 132 (91.7%) had non-complex karyotypes.
Table 1.
The patients’ baseline characteristics and associations with time to first treatment.
Outcome
The median follow-up for censored patients was 30 months (range, 0.03-102 months). The median TFT for the entire cohort was 47 months (95% confidence interval [95% CI]: 39-61 months). Higher karyotype complexity was significantly associated with TFT (hazard ratio [HR]=1.16, 95% CI: 1.11-1.22; P<0.001), as was complex karyotype defined using ≥3 aberrations (HR=2.92, 95% CI: 1.98-4.31; P<0.001) and ≥5 aberrations (HR=2.93, 95% CI: 1.79-4.79; P<0.001). Since there was no difference in the ability to discriminate TFT when defining complexity as ≥3 or ≥5 aberrations (Online Supplementary Table S1, Online Supplementary Figure S1), all further analyses used the definition of ≥3 abnormalities.
In univariable models, the following variables were significant for a shorter TFT: presence of a translocation (HR=2.64, 95% CI: 1.87-3.71; P<0.001), Rai stage 3-4 (HR=3.73, 95% CI: 2.32-5.99; P<0.001), karyotype complexity (HR=2.92, 95% CI: 1.98-4.31, P<0.001), unmutated IGHV (HR=3.48, 95% CI: 2.38-5.08; P<0.001), three copies of MYC (HR=2.53, 95% CI: 1.36-4.71; P=0.003), del(17p) (HR=2.10, 95% CI: 1.31-3.37; P=0.002), del(11q) (HR=2.92, 95% CI: 1.98-4.31; P<0.001), 2-microglobulin >3.5 mg/L (HR=2.33, 95% CI: 1.11-4.88; P=0.025), and log-transformed WBC count (HR=1.72, 95% CI: 1.35-2.20; P<0.001) (Table 1).
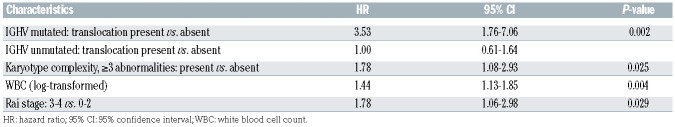
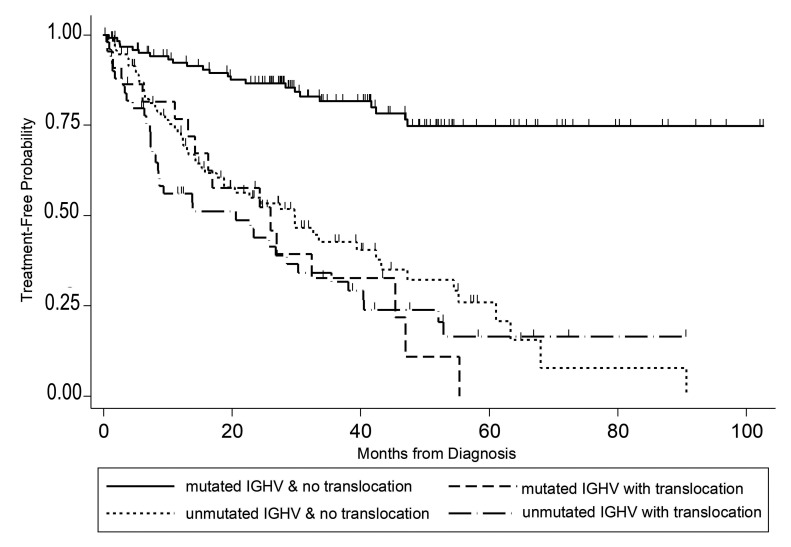
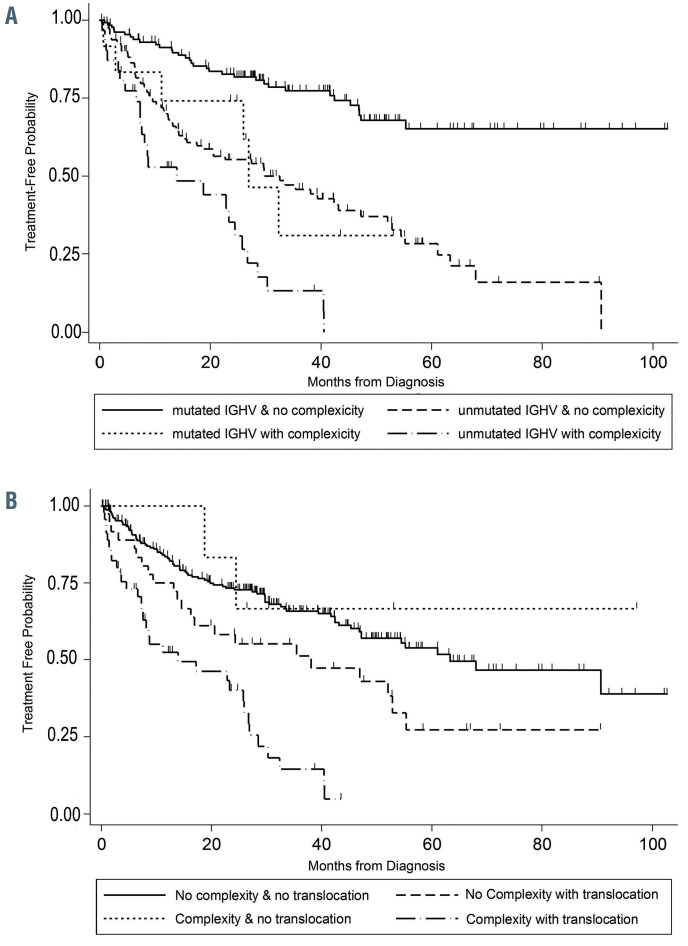
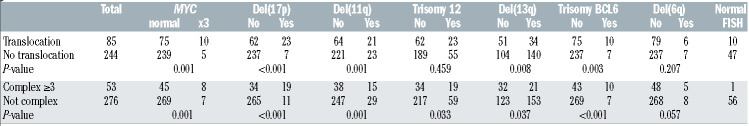
In the multivariable regression model, there was significant effect modification of IGHV status on the relationship between translocations and TFT (P=0.002) (Table 4). In IGHV-mutated patients, those with a translocation had over 3.5 times the risk of starting treatment relative to those without a translocation (HR=3.53, 95% CI: 1.76-7.06; P<0.001); however, in IGHV-unmutated patients, a translocation did not significantly increase the risk of starting treatment (HR=1.00, 95% CI: 0.61-1.64; P=0.99) (Figure 1). This effect modification appears consistent across groups defined by complex karyotype (Online Supplementary Figure S2). We did not detect a significant interaction between IGHV and karyotype complexity (Figure 2A), nor between translocations and complexity (Figure 2B). Examination of both the presence of a translocation and karyotype complexity in the analysis of TFT showed that among the 85 patients with a translocation, karyotype complexity added prognostic information (HR=2.31, 95% CI: 1.25-4.26; P=0.007). Independently of IGHV status and translocations, Rai stage 3-4 (HR=1.78, 95% CI: 1.06-2.98; P=0.029), log-transformed WBC count (HR=1.44, 95% CI: 1.13-1.85; P=0.004) and karyotype complexity (HR=1.78, 95% CI: 1.08-2.93; P=0.025) remained statistically significant in the multivariable model (Table 4). Notably, once these variables were accounted for in the model, del(17p) did not add significant information (P=0.51). Using a higher frequency of del(17p) with a 20% cut off, del(17p) still was not significant in the multivariable analysis (P=0.51). Similarly, del(11q) was no longer significant in the multivariable model.
Among 139 patients who started CLL treatment, 54 received chemoimmunotherapy. With a median follow-up of 43.9 (range, 3.4-94.0) months from the start of CLL treatment, 13 deaths occurred. The overall survival estimates are 91% (95% CI: 79%-96%) at 1 year, 84% (95% CI: 70%-92%) at 3 years, and 77% (95% CI 61%-87%) at 5 years after starting CLL treatment. In these treated patients, Rai stage 3-4 and del(17p) were associated with shorter survival in univariable analyses with a hazard ratio of 5.36 (95% CI: 1.62-17.76), and 6.88 (95% CI: 2.20-21.50), respectively.
Table 2.
Association between karyotype complexity (≥3 aberrations) and translocations. Unbalanced translocations are more frequent in patients with complex karyotypes, but balanced translocations are more frequent in those with non-complex karyotypes.
Table 3.
Associations of IGHV with karyotype complexity (≥3 aberrations) and translocations. Translocations and complex karyotypes occur more frequently in unmutated IGVH chronic lymphocytic leukemia.
Table 4.
Multivariable model of factors significantly associated with time to first treatment.
Table 5 shows correlations of interphase FISH abnormalities with both the presence of a translocation and with karyotype complexity. Del(11q), del(17p), three copies of MYC, and three copies of BCL6 were all highly associated both with the presence of a translocation and with karyotypic complexity. An opposite trend was seen for cases with del(13q); del(13q) was more frequent both in cases without a translocation and in non-complex cases. Trisomy 12 was frequent in complex cases, but was not associated with the presence of a translocation. Deletion of 6q was not associated with either karyotypic complexity or the presence of a translocation. Deletion of 13q as the sole FISH-identified abnormality, typically indicative of a good prognosis, occurred in 119 cases. Twelve of these cases had a translocation, and three had a complex karyotype. In addition, ten cases with a translocation had no abnormalities detected by FISH, emphasizing the need for metaphase cytogenetics.
Discussion
Previous studies have found a significant impact of translocations on the outcome of patients with CLL.5,9,17-19 We show that when detected within 1 year of diagnosis, the presence of a translocation in cases with mutated IGHV appears to negate the positive impact of mutated IGHV. However, in cases of unmutated IGHV, the presence of a translocation did not appear to influence prognosis. In this study, patients with mutated IGHV who did not have a translocation had a significantly longer TFT than those with mutated IGHV and a translocation or those with unmutated IGHV regardless of the presence of a translocation (Figure 1).
Figure 1.
Time to first treatment of patients with chronic lymphocytic leukemia. Data are plotted for patients according to whether they had mutated or unmutated IGHV and whether they did or did not have a translocation.
Baliakas et al.9 found that translocations were strongly associated with CD38+ expression, karyotypic complexity, abnormal 17p and abnormal 11q. However, the presence of a translocation did not influence TFT, although those cases with an unbalanced translocation and those with a complex karyotype had a significantly shorter TFT. Translocations were also associated with complexity, del(17p), and del(11q) in our series. However, when considering their impact on outcome, those cases with a mutated IGHV and a translocation had a poor TFT compared with cases with mutated IGHV and no translocation. Rigolin et al.19 showed that unbalanced translocations were associated with shorter TFT and overall survival. In our studies, both balanced and unbalanced translocations were considered together and overall had an impact on TFT.
These results add to the factors that may influence the outcome of CLL patients with mutated IGHV. IGHVmutated patients with high expression of CD49d and those with high expression of CXCR4 have an inferior outcome compared to patients with low expression of these variables.30-32 As neither CD49d nor CXCR4 was evaluated in our patients, we are unable to compare their effects with those of the presence of a translocation. Whether these factors influence chromosome instability, a possible factor in generation of a translocation, was not investigated.
Complex karyotypes have also been associated with an adverse prognosis in CLL.4-16,19 While some studies15,16 found that the association of karyotypic complexity was more significant when complexity was defined as ≥5 aberrations than when defined as ≥3 aberrations, we found that complexity defined as ≥3 abnormalities was as significant as complexity defined as ≥5 abnormalities. Although karyotypic complexity detected within 1 year of diagnosis of CLL remained significant in a multivariable analysis, the hazard ratio for the presence of a translocation in patients with mutated IGHV was much higher than that for karyotypic complexity (HR for complexity=1.78, HR for presence of a translocation=3.53). Among patients with a translocation, karyotypic complexity added prognostic information.
Figure 2.
Time to first treatment of patients with chronic lymphocytic leukemia. (A) Data are plotted for patients according to whether they had mutated or unmutated IGHV and a complex (≥3 aberrations) or non-complex karyotype. (B) Data are plotted for patients according to whether they did or did not have a translocation and whether they had a complex (≥3 aberrations) or non-complex karyotype.
This study also showed that CLL patients with a higher WBC count were likely to require treatment earlier than those with a lower WBC count. This is consistent with the findings of the study by Del Giudice et al.,33 who showed that in patients with Binet stage A CLL, a higher WBC count independently predicted a shorter time to treatment. Since a higher WBC count is consistent with a greater disease burden, this is compatible with the hypothesis that patients with a greater disease burden, regardless of other factors, have a shorter TFT.
A del(17p) has also been associated with a poor outcome. 1,5,34-38 However, when detected within 1 year of diagnosis, it was no longer significant in a multivariable analysis, independently of other variables. It is known that del(17p) frequently occurs with disease progression and that a higher frequency of del(17p) may be significant.39,40 We, therefore examined whether a higher frequency of this abnormality was significant. Using a 20% cutoff, higher frequency of del(17p) was also not significant in the multivariable analysis (P=0.51). Furthermore, our patients were within 1 year of diagnosis, which is consistent with the findings of Tam et al.,39 who showed that a del(17p) in asymptomatic CLL patients does not necessarily convey a poor prognosis. It also has been shown that del(17p) in association with a complex karyotype predicts a particularly poor outcome.15,20,41 Although our patients with del(17p) were statistically associated with a complex karyotype, 11/30 of our del(17p) cases did not have a complex karyotype. The loss of significance of del(17p) in a multivariable analysis may in part reflect this.
In these studies, FISH abnormalities typically associated with a poor outcome (three copies of MYC, del(17p), del(11q) and three copies of BCL6) were more frequent in cases with a translocation and in those with a complex karyotype. Trisomy 12 showed no association with the presence of a translocation, but was more frequent in complex cases than in non-complex cases (35.9% and 21.4%, respectively; P=0.033). Deletions of 13q, typically associated with a good outcome, were more frequent in cases without a translocation and in non-complex cases, consistent with its association with a good outcome. However, del(13q) as a sole FISH abnormality was present in 12 cases with a translocation and in three cases with a complex karyotype, thus 15 of 119 cases (12.6%) with del(13q) as the only FISH-detected abnormality had poor metaphase cytogenetics, identifying these patients as possibly not having the good prognosis typically associated with sole del(13q). Fifty-seven cases had no FISH abnormality (Table 5); ten of these had a translocation, and one had a complex karyotype. These results indicate the importance of metaphase cytogenetics in order to riskstratify patients accurately.
Table 5.
Table 5. Associations between translocations or karyotype complexity with abnormalities detected by interphase fluorescence in situ hybridization.
The reason a translocation may negate the good prognosis associated with mutated IGHV is not known. Both the presence of a translocation and a complex karyotype may reflect an underlying genetic instability in CLL. The strong correlation of translocations and complex karyotypes with del(17p), and therefore loss of TP53, which is known to be associated with genetic instability,42 supports this. However, the majority of patients with translocations or complex karyotypes did not have del(17p); thus, other factors must contribute to genetic instability in CLL. Other factors that have been associated with genetic instability include telomere shortening, abnormal ATM, lack of correct kinetochore-microtubule attachment, activationinduced cytidine deaminase, defective mitosis and replication stress. Further investigations are needed to identify whether these or other factors contribute to genetic instability in CLL.
These studies have some significant limitations. These include the relatively low number of patients with translocations (85/329), the retrospective nature of the study, and the younger age of patients (median, 60 years). Because of the retrospective nature of this study, some factors, significantly TP53 mutational status and CD49d expression, could not be included as they were not determined routinely. Additionally, both WBC count and β 2-microglobulin level had a 90-day window, as they were not collected routinely at diagnosis. Also, although all cytogenetic studies were performed within 1 year of diagnosis, they were done at different time-points within this 1-year timeframe.
These results support performing metaphase cytogenetic analysis on cultures stimulated with a CpG oligodeoxynucleotide prospectively at diagnosis on all patients with CLL to verify these findings and to determine whether these findings apply with current targeted therapies. The use of CpG oligodeoxynucleotide stimulation is required to adequately define abnormal clones in CLL,5,17,21-26 thus enabling the detection of translocations that are prognostic early in the course of the disease. This additional prognostic factor may help to provide a more precise prognosis for CLL patients, and may more accurately define specific treatment options that will be beneficial to these patients. While the presence of a translocation has previously been shown to correlate with a poor prognosis, this study indicates that in patients with unmutated IGHV, translocations may not affect prognosis. However, detection of a translocation in patients with mutated IGHV may affect prognosis as early as the time of diagnosis.
Supplementary Material
Funding Statement
Funding This work was supported by Leukemia and Lymphoma Society SCOR grant #7004-11, NCI-7 P50 CA140158-02, and NCI-P30 CA016058, which supports the OSU Comprehensive Cancer Center’s Shared Resources, and the D Warren Brown Foundation.
References
- 1.Byrd JC, Gribben JG, Peterson BL, et al. Select high-risk genetic features predict earlier progression following chemoimmunotherapy with fludarabine and rituximab in chronic lymphocytic leukemia: justification for risk-adapted therapy. J Clin Oncol. 2006;24(3):437-443. [DOI] [PubMed] [Google Scholar]
- 2.Oscier DG, Gardiner AC, Mould SJ, et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood. 2002;100(4):1177-1184. [PubMed] [Google Scholar]
- 3.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94(6):1848-1854. [PubMed] [Google Scholar]
- 4.Haferlach C, Dicker F, Weiss T, et al. Toward a comprehensive prognostic scoring system in chronic lymphocytic leukemia based on a combination of genetic parameters. Genes Chromosomes Cancer. 2010;49(9):851-859. [DOI] [PubMed] [Google Scholar]
- 5.Haferlach C, Dicker F, Schnittger S, Kern W, Haferlach T. Comprehensive genetic characterization of CLL: a study on 506 cases analysed with chromosome banding analysis, interphase FISH, IgV(H) status and immunophenotyping. Leukemia. 2007;21(12):2442-2451. [DOI] [PubMed] [Google Scholar]
- 6.Woyach JA, Ruppert AS, Guinn D, et al. BTK(C481S)-mediated resistance to ibrutinib in chronic lymphocytic leukemia. J Clin Oncol. 2017;35(13):1437-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woyach JA, Lozanski G, Ruppert AS, et al. Outcome of patients with relapsed or refractory chronic lymphocytic leukemia treated with flavopiridol: impact of genetic features. Leukemia. 2012;26(6):1442-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaglowski SM, Ruppert AS, Heerema NA, et al. Complex karyotype predicts for inferior outcomes following reduced-intensity conditioning allogeneic transplant for chronic lymphocytic leukaemia. Br J Haematol. 2012;159(1):82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baliakas P, Iskas M, Gardiner A, et al. Chromosomal translocations and karyotype complexity in chronic lymphocytic leukemia: a systematic reappraisal of classic cytogenetic data. Am J Hematol. 2014;89(3):249-255. [DOI] [PubMed] [Google Scholar]
- 10.Rogers KA, Huang Y, Ruppert AS, et al. A single-institution retrospective cohort study of first-line R-EPOCH chemoimmunotherapy for Richter syndrome demonstrating complex chronic lymphocytic leukaemia karyotype as an adverse prognostic factor. Br J Haematol. 2018;180(2):259-266. [DOI] [PubMed] [Google Scholar]
- 11.Rigolin GM, Formigaro L, Cavallari M, et al. An extensive molecular cytogenetic characterization in high-risk chronic lymphocytic leukemia identifies karyotype aberrations and TP53 disruption as predictors of outcome and chemorefractoriness. Oncotarget. 2017;8(17):28008-28020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigolin GM, Cavallari M, Quaglia FM, et al. In CLL, comorbidities and the complex karyotype are associated with an inferior outcome independently of CLL-IPI. Blood. 2017;129(26):3495-3498. [DOI] [PubMed] [Google Scholar]
- 13.Le Bris Y, Struski S, Guièze R, et al. Major prognostic value of complex karyotype in addition to TP53 and IGHV mutational status in first-line chronic lymphocytic leukemia. Hematol Oncol. 2017;35(4):664-670. [DOI] [PubMed] [Google Scholar]
- 14.Cavallari M, Cavazzini F, Bardi A, et al. Biological significance and prognostic/predictive impact of complex karyotype in chronic lymphocytic leukemia. Oncotarget. 2018;9(76):34398-34412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baliakas P, Jeromin S, Iskas M, et al. Cytogenetic complexity in chronic lymphocytic leukemia: definitions, associations, and clinical impact. Blood. 2019;133(11):1205-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigolin GM, del Giudice I, Formigaro L, et al. Chromosome aberrations detected by conventional karyotyping using novel mitogens in chronic lymphocytic leukemia: clinical and biologic correlations. Genes Chromosomes Cancer. 2015;54(12):818-826. [DOI] [PubMed] [Google Scholar]
- 17.Mayr C, Speicher MR, Kofler DM, et al. Chromosomal translocations are associated with poor prognosis in chronic lymphocytic leukemia. Blood. 2006;107(2):742-751. [DOI] [PubMed] [Google Scholar]
- 18.Van Den Neste E, Robin V, Francart J, et al. Chromosomal translocations independently predict treatment failure, treatment-free survival and overall survival in B-cell chronic lymphocytic leukemia patients treated with cladribine. Leukemia. 2007;21(8): 1715-1722. [DOI] [PubMed] [Google Scholar]
- 19.Rigolin GM, Saccenti E, Guardalben E, et al. In chronic lymphocytic leukaemia with complex karyotype, major structural abnormalities identify a subset of patients with inferior outcome and distinct biological characteristics. Br J Haematol. 2018;181(2): 229-233. [DOI] [PubMed] [Google Scholar]
- 20.Herling CD, Klaumünzer M, Rocha CK, et al. Complex karyotypes and KRAS and POT1 mutations impact outcome in CLL after chlorambucil-based chemotherapy or chemoimmunotherapy. Blood. 2016;128(3): 395-404. [DOI] [PubMed] [Google Scholar]
- 21.Decker T, Schneller F, Kronschnabl M, et al. Immunostimulatory CpG-oligonucleotides induce functional high affinity IL-2 receptors on B-CLL cells: costimulation with IL- 2 results in a highly immunogenic phenotype. Exp Hematol. 2000;28(5):558-568. [DOI] [PubMed] [Google Scholar]
- 22.Decker T, Schneller F, Sparwasser T, et al. Immunostimulatory CpG-oligonucleotides cause proliferation, cytokine production, and an immunogenic phenotype in chronic lymphocytic leukemia B cells. Blood. 2000;95(3):999-1006. [PubMed] [Google Scholar]
- 23.Dicker F, Schnittger S, Haferlach T, Kern W, Schoch C. Immunostimulatory oligonucleotide- induced metaphase cytogenetics detect chromosomal aberrations in 80% of CLL patients: a study of 132 CLL cases with correlation to FISH, IgVH status, and CD38 expression. Blood. 2006;108(9):3152-3160. [DOI] [PubMed] [Google Scholar]
- 24.Muthusamy N, Breidenbach H, Andritsos L, et al. Enhanced detection of chromosomal abnormalities in chronic lymphocytic leukemia by conventional cytogenetics using CpG oligonucleotide in combination with pokeweed mitogen and phorbol myristate acetate. Cancer Genet. 2011;204(2):77-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Put N, Konings P, Rack K, et al. Improved detection of chromosomal abnormalities in chronic lymphocytic leukemia by conventional cytogenetics using CpG oligonucleotide and interleukin-2 stimulation: a Belgian multicentric study. Genes Chromosomes Cancer. 2009;48(10):843-853. [DOI] [PubMed] [Google Scholar]
- 26.Struski S, Gervais C, Helias C, et al. Stimulation of B-cell lymphoproliferations with CpG-oligonucleotide DSP30 plus IL-2 is more effective than with TPA to detect clonal abnormalities. Leukemia. 2009;23(3): 617-619. [DOI] [PubMed] [Google Scholar]
- 27.Haferlach C, Jeromin S, Nadarajah N, et al. Cytogenetic and molecular genetic clonal evolution in CLL is associated with an unmutated IGHV status and frequently leads to a combination of loss of 17p and TP53 mutation. Blood. 2016;128(22):3213-3213. [DOI] [PubMed] [Google Scholar]
- 28.Stilgenbauer S, Sander S, Bullinger L, et al. Clonal evolution in chronic lymphocytic leukemia: acquisition of high-risk genomic aberrations associated with unmutated VH, resistance to therapy, and short survival. Haematologica. 2007;92(9):1242-1245. [DOI] [PubMed] [Google Scholar]
- 29.Hallek M, Cheson BD, Catovsky D, et al. iwCLL guidelines for diagnosis, indications for treatment, response assessment, and supportive management of CLL. Blood. 2018;131(25):2745-2760. [DOI] [PubMed] [Google Scholar]
- 30.Pepper C, Buggins AG, Jones CH, et al. Phenotypic heterogeneity in IGHV-mutated CLL patients has prognostic impact and identifies a subset with increased sensitivity to BTK and PI3Kinhibition. Leukemia. 2015;29(3):744-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumann T, Delgado J, Santacruz R, et al. CD49d (ITGA4) expression is a predictor of time to first treatment in patients with chronic lymphocytic leukaemia and mutated IGHV status. Br J Haematol. 2016;172(1):48-55. [DOI] [PubMed] [Google Scholar]
- 32.Gattei V, Bulian P, Del Principe MI, et al. Relevance of CD49d protein expression as overall survival and progressive disease prognosticator in chronic lymphocytic leukemia. Blood. 2008;111(2):865-873. [DOI] [PubMed] [Google Scholar]
- 33.Del Giudice I, Mauro FR, De Propris MS, et al. White blood cell count at diagnosis and immunoglobulin variable region gene mutations are independent predictors of treatment- free survival in young patients with stage A chronic lymphocytic leukemia. Haematologica. 2011;96(4):626-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dohner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910-1916. [DOI] [PubMed] [Google Scholar]
- 35.Foa R, Del Giudice I, Guarini A, Rossi D, Gaidano G. Clinical implications of the molecular genetics of chronic lymphocytic leukemia. Haematologica. 2013;98(5):675-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin TS, Ruppert AS, Johnson AJ, et al. Phase II study of flavopiridol in relapsed chronic lymphocytic leukemia demonstrating high response rates in genetically high-risk disease. J Clin Oncol. 2009;27(35):6012-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zenz T, Eichhorst B, Busch R, et al. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28(29): 4473-4479. [DOI] [PubMed] [Google Scholar]
- 38.Stephens DM, Ruppert AS, Weirda WG, et al. Externally validated predictive clinical model for untreated del(17p13.1) chronic lymphocytic leukemia patients. Am J Hematol. 2015;90(11):967-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tam CS, Shanafelt TD, Wierda WG, et al. De novo deletion 17p13.1 chronic lymphocytic leukemia shows significant clinical heterogeneity: the M. D. Anderson and Mayo Clinic experience. Blood. 2009;114(5):957-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Dyke DL, Werner L, Rassenti LZ, et al. The Dohner fluorescence in situ hybridization prognostic classification of chronic lymphocytic leukaemia (CLL): the CLL Research Consortium experience. Br J Haematol 2016;173(1):105-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu L, Kim HT, Kasar S, et al. Survival of del17p CLL depends on genomic complexity and somatic mutation. Clin Cancer Res 2017;23(3):735-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu G, Parant JM, Lang G, et al. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet. 2004;36(1):63-68. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.