In this study, Zhao et al. set out to characterize how plants respond to cold through regulation of FLC expression. Using genetics and genomics approaches, the authors reveal how natural temperature fluctuations promote COOLAIR regulation of FLC, with the first autumn frost acting as a key indicator of autumn/winter arrival.
Keywords: FLC, COOLAIR, noncoding RNA, vernalization, temperature-sensing
Abstract
Plants monitor many aspects of their fluctuating environments to help align their development with seasons. Molecular understanding of how noisy temperature cues are registered has emerged from dissection of vernalization in Arabidopsis, which involves a multiphase cold-dependent silencing of the floral repressor locus FLOWERING LOCUS C (FLC). Cold-induced transcriptional silencing precedes a low probability PRC2 epigenetic switching mechanism. The epigenetic switch requires the absence of warm temperatures as well as long-term cold exposure. However, the natural temperature inputs into the earlier transcriptional silencing phase are less well understood. Here, through investigation of Arabidopsis accessions in natural and climatically distinct field sites, we show that the first seasonal frost strongly induces expression of COOLAIR, the antisense transcripts at FLC. Chamber experiments delivering a constant mean temperature with different fluctuations showed the freezing induction of COOLAIR correlates with stronger repression of FLC mRNA. Identification of a mutant that ectopically activates COOLAIR revealed how COOLAIR up-regulation can directly reduce FLC expression. Consistent with this, transgenes designed to knockout COOLAIR perturbed the early phase of FLC silencing. However, all transgenes designed to remove COOLAIR resulted in increased production of novel convergent FLC antisense transcripts. Our study reveals how natural temperature fluctuations promote COOLAIR regulation of FLC, with the first autumn frost acting as a key indicator of autumn/winter arrival.
As sessile organisms, plants have to extract specific temperature cues from fluctuating environments to time their developmental transitions. Knowledge of these mechanisms will be key to understanding the consequences of climate change. In Arabidopsis, a major determinant of seasonal flowering is the floral repressor FLC, which is epigenetically silenced by prolonged cold during vernalization (Michaels and Amasino 1999; Sheldon et al. 2000). Vernalization involves a multiphase cold-dependent silencing, where cold-induced transcriptional silencing precedes a low probability PRC2 epigenetic switching mechanism. The epigenetic switch is promoted by the PRC2-accessory protein VIN3, which is induced slowly, taking weeks to reach maximal levels (Sung and Amasino 2004). VIN3 accumulation requires absence of warm temperature spikes, in addition to long-term cold exposure (Hepworth et al. 2018; Zhao et al. 2020). The transcriptional silencing involves cold-induced FLC antisense transcripts, COOLAIR, whose induction correlates with FLC transcriptional shutdown and switching of the chromatin states at FLC (Swiezewski et al. 2009; Csorba et al. 2014; Rosa et al. 2016). COOLAIR also functions to promote rapid cycling in plants in warm conditions in a cotranscriptional chromatin silencing mechanism that links promotion of proximal polyadenylation to FLC histone H3K4me1 demethylation (Costa and Dean 2019; Fang et al. 2020; Wu et al. 2020).
COOLAIR is highly conserved across different species (Castaings et al. 2014; Hawkes et al. 2016; Jiao et al. 2019) and has been shown to contribute to the variation in FLC regulation across natural Arabidopsis accessions (Shindo et al. 2006; Coustham et al. 2012; Li et al. 2014). Notably, a noncoding single-nucleotide polymorphism (SNP) alters COOLAIR splicing, increasing FLC transcription levels (Li et al. 2015). This mechanism involves the activator FRIGIDA protein and distally polyadenylated COOLAIR (Johanson et al. 2000; Geraldo et al. 2009; Li et al. 2018). The natural variation studies implicating a functional role for COOLAIR in FLC regulation have been followed up by transgene experiments designed to further explore the mechanism of COOLAIR action (Csorba et al. 2014; Wang et al. 2014; Rosa et al. 2016). However, in some cases, studies of transgenes aimed at attenuating antisense expression have concluded that COOLAIR expression is not required for vernalization (Helliwell et al. 2011; Li et al. 2018; Luo et al. 2019; Luo and He 2020). This led to the suggestion that COOLAIR functions in FLC regulation at warm temperatures but potentially not in the cold. In order to test this, we investigated the role of COOLAIR in vernalization, in both natural and laboratory conditions and in different accessions. Our study demonstrates the importance of COOLAIR-mediated FLC silencing in natural conditions, with the first seasonal freezing temperatures leading to COOLAIR-mediated FLC transcriptional silencing.
Result
COOLAIR shows variable temperature sensitivity and is highly up-regulated by freezing temperatures
Through the analysis of >1000 worldwide Arabidopsis natural accessions, we previously identified five predominant FLC haplotypes defined by noncoding SNPs (Li et al. 2014). According to the varied vernalization responses measured by flowering time after growth at a constant 5°C, these haplotypes were classified into “rapid vernalizing” (RV) and “slow vernalizing” (SV) types (Li et al. 2014). To further investigate their vernalization response in field conditions, we selected representative accessions of these five major haplotypes (RV: Edi-0 and Col FRISF2; SV: Var2-6, Ull2-5, and Bro1-6), as well as an extra SV accession, Löv-1, collected from the North Swedish field site (Duncan et al. 2015; Qüesta et al. 2020). To allow the comparison among these haplotypes, we generated new near-isogenic lines (NILs) by repeatedly backcrossing each FLC haplotype to Col FRISF2, our reference genetic background (“Col FRI”) (Duncan et al. 2015; Li et al. 2015). Recent work has shown that these accessions and their associated NILs show variation in response to autumn cold in the field (Hepworth et al. 2020).
Here, we analyzed COOLAIR induction in field conditions to determine whether COOLAIR plays a role in natural conditions in different Arabidopsis accessions and their associated NILs. Three climatically different field sites were chosen: Norwich, United Kingdom (52° 62.2191′ N, 1°22.1695´ E; temperate oceanic); Ullstorp, Sweden (56°06.6721′ N, 13°94.4655′ E; “South Sweden”; a warm-summer continental); and Ramsta, Sweden (62°50.988′ N, 18°11.570′ E; “North Sweden”; subarctic) (Supplemental Figs. S1, S2; Antoniou-Kourounioti et al. 2018; Hepworth et al. 2018, 2020). Norwich generally has mild winters with few frosts, while in Ullstorp temperatures often fall below freezing, whereas the Ramsta site is usually snow-covered during part of the winter. The experiments ran from late summer/early autumn 2014 until spring 2015 and were repeated in North Sweden from late summer 2016 to the spring of 2017. Sowing times were adjusted to each site (earlier further north), and in the first year, two plantings were performed in North Sweden, a fortnight apart.
Three SV accessions (Löv-1, Ull2-5, Bro1-6) showed higher COOLAIR levels than the RV Col FRI across all the plantings (Supplemental Fig. S2). In contrast, Col-0 (which is a rapid cycler with no vernalization requirement for flowering) had very low levels of total COOLAIR in Norwich and at most time points in Sweden. COOLAIR levels in the RV accession Edi-0 and its NIL were very similar to Col FRI in Norwich, but in Sweden, especially North Sweden, COOLAIR was higher in Edi-0 than Col FRI. This was also the case for Var2-6, with early differences only in Sweden. These data suggest COOLAIR induction in the different accessions shows different temperature sensitivity.
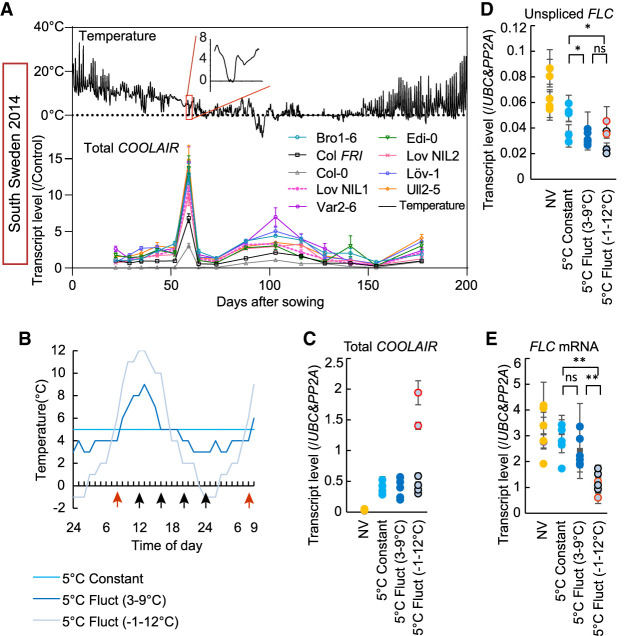
We noticed a strong peak of expression of COOLAIR in all genotypes, even Col-0, in the second week of measurements (the 26th day after sowing) in the first North Sweden planting and the sixth week (the 59th day after sowing) in the South (Fig. 1A; Supplemental Figs. S1, S2). These unusual peaks occurred when temperatures dipped below 0°C on the morning of the day of measurement (e.g., for South Sweden 2014, see Fig. 1A; for North Sweden 2016, see Supplemental Fig. S3A,B).
Figure 1.
COOLAIR expression is highly induced by freezing temperature. (A) COOLAIR expression in all genotypes in a field in South Sweden over winter 2014–2015. Plots in the top panel show the temperature profile in the field with the first appearance of freezing temperature highlighted and expanded in the red box. Plots in the bottom panel show the relative transcript level of COOLAIR as analyzed by RT-qPCR. (B) The temperature profiles of chambers set up to analyze the interrelationship of freezing, COOLAIR expression, and FLC expression. Plants grown in these chambers were used to generate data shown in C–E. (C–E) Relative transcript level of COOLAIR (C), unspliced FLC (D), and FLC mRNA (E) measured throughout the day (NV [nonvernalized]) and after 2 wk of different cold exposure. Results were presented by combining the data of the six sampling points from each treatment. The sampling times are indicated by arrows in B. More details are described in the Materials and Methods. (C) COOLAIR data points in red (indicated by the red arrow in B) were those taken ∼8 h after freezing. Vernalized treatments compared by ANOVA with Tukey's post hoc test. (*) P < 0.05, (**) P < 0.01, (ns) no significance. Expression data were normalized as indicated in each panel. Error bars show SEM.
Recapitulation of the COOLAIR expression spike in temperature-controlled chambers
We confirmed the up-regulation of COOLAIR by freezing by reproducing the temperature profile of the week before the first peak in South Sweden in a growth cabinet. COOLAIR expression rose within an hour of experiencing freezing and peaked ∼8 h after freezing but returned to cool-temperature levels within 24 h (Supplemental Fig. S3C). There was a small reduction in the level of FLC transcripts immediately after freezing exposure (Supplemental Fig. S3D,E), reflecting the coordination of antisense–sense transcriptional circuitry. To further analyze the interrelationship of freezing, COOLAIR expression, and FLC expression, we grew seedlings for 2 wk in matched chambers at an average temperature of 5°C, but given either as a constant temperature or two different fluctuating temperature regimes repeated every 24 h (Fig. 1B). We saw the expected induction of proximal polyadenylation of COOLAIR by constant cold; however, this was significantly enhanced in the chamber where the fluctuations included a below-freezing period (Fig. 1C; Supplemental Fig. S3F–I). Of note, this hyperinduction of COOLAIR is not just a consequence of a steep temperature drop, as COOLAIR induction was lower after transfer from nonvernalized (NV) 20°C to constant 5°C (Supplemental Fig. S3J). All the temperature regimes led to a reduction in FLC expression, including the FLC mRNA and the unspliced transcript level (nascent transcript containing intron 2 and intron 3) used as a proxy for transcription (Fig. 1D,E). However, the enhanced COOLAIR induction in the freezing regime caused significantly lower FLC mRNA levels (Fig. 1E). The mechanism behind the different behavior of the unspliced transcripts and mRNA after exposure to freezing is currently unclear.
Genetic up-regulation of COOLAIR is associated with FLC transcriptional shutdown
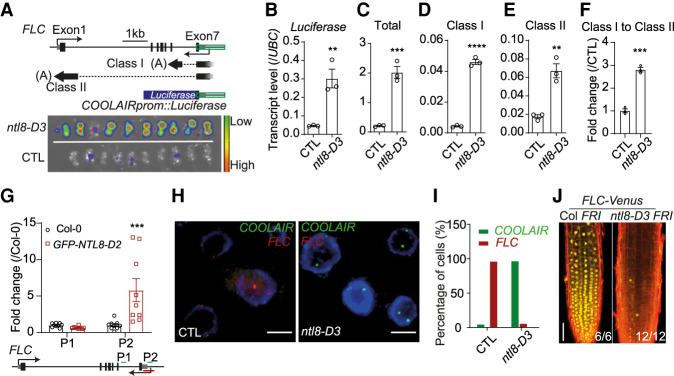
The complex relationship of the FLC down-regulation and the freezing-induced COOLAIR spike was intriguing with respect to the registration of external temperature and FLC regulation. In order to better understand the relationship between COOLAIR up-regulation and FLC repression, we sought to identify a mutant constitutively expressing high levels of COOLAIR. A forward genetic screen using a COOLAIRprom::luciferase reporter (Swiezewski et al. 2009; Sun et al. 2013), identified a dominant mutant, ntl8-D3, that had a high luciferase signal in warm conditions (Fig. 2A,B). All endogenous COOLAIR transcripts, including both proximal (class I) and distal (class II) polyadenylated forms, showed ectopic up-regulation in ntl8-D3 (Fig. 2C–E), with a relative increase in proximal polyadenylation as seen in wild-type plants after cold (Fig. 2F; Supplemental Fig. S3I). Two separate mutant alleles, ntl8-D1 and ntl8-D2, and three overexpression transgenics, ntl8-OE1, ntl8-OE2, and ntl8-OE3, showed the same COOLAIR up-regulation (Supplemental Fig. S4A–F; Zhao et al. 2020). ntl8-D alleles cause a truncation in the NTL8 protein that deletes the C-terminal transmembrane domain and results in enhanced nuclear localization (Supplemental Fig. S4G; Zhao et al. 2020). NTL8 encodes a NAC domain transcription factor that directly binds VIN3 and the COOLAIR promoter (Fig. 2G; Supplemental Fig. S4H; O'Malley et al. 2016; Xi et al. 2020; Zhao et al. 2020). Interestingly, the ntl8-D mutant alleles are also defective in long-term temperature regulation of VIN3 (Zhao et al. 2020), and the same logic that we have described for VIN3 accumulation—namely, reduced dilution of wild-type NTL8 protein during cold exposure—may be important in promoting long-term COOLAIR expression in autumn conditions. Previous analysis of ntl8-D2 vin3-6 double mutants revealed that the VIN3 overexpression, which also occurs in the ntl8-D mutant, was not necessary for the lower FLC expression before vernalization (Zhao et al. 2020). In order to better understand whether the higher COOLAIR expression was a reflection of more expression from the same cells or a higher fraction of cells expressing COOLAIR, we performed single-molecule RNA fluorescence in situ hybridization (smRNA FISH) on the ntl8-D3 mutant in warm conditions (Rosa et al. 2016). COOLAIR expression was found at high levels in all cells of the root (Fig. 2H,I); thus, ntl8-D3 had expanded the expression zone normally seen for COOLAIR, from just prevasculature cells to all cell types (Rosa et al. 2016). This is consistent with the reduced FLC-Venus signals in the ntl8-D3 mutant in warm conditions (Fig. 2J). This mutually exclusive transcription of COOLAIR and FLC supports the repressive effect of COOLAIR on FLC transcription from the same gene copy, agreeing with the previous study (Rosa et al. 2016).
Figure 2.
Genetic up-regulation of COOLAIR expression is associated with FLC transcriptional reduction. (A) Luminescence assay for warm-grown ntl8-D3 plants (20°C). (CTL) Progenitor line carrying transgenic COOLAIRprom::luciferase reporter. A diagram of the COOLAIRprom::luciferase reporter is shown in the top panel. Untranslated region (UTR) of FLC is indicated by a gray box, and exons are represented by black boxes. A green box indicates the COOLAIR promoter. Simplified diagrams are included to show the alternative polyadenylation of COOLAIR: proximal polyadenylated class I and distal polyadenylated class II. (Black solid line) Exons, (black dashed line) introns, (gray lines) transcriptional start region. (B–E) qPCR analyzing luciferase (B), total COOLAIR (C), class I COOLAIR (D), and class II COOLAIR (E) transcript levels in warm conditions (20°C) for CTL and ntl8-D3. Levels normalized to UBC. Error bars show SEM of three biological replicates. (F) Ratio of proximal class I to distal class II COOLAIR with the data from C–E, normalized to CTL. Unpaired two-tailed t-test was performed, and significances for each comparison are shown. (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001. (G) ChIP analysis of NTL8 binding at COOLAIR region. (Control) Col-0. Error bars show SEM of eight replicates. Two-way analysis of variance (ANOVA) with Turkey's multiple comparisons test was performed, and significances for individual comparison of interest are shown. (***) P < 0.001. Positions of amplicon P1 and P2 are indicated on the diagram. (H) Representative images of nuclei (indicated by DAPI staining; blue) hybridized with intronic smRNA FISH probes for COOLAIR (green) and FLC (red) showing mutually exclusive transcription in CTL and ntl8-D3 mutants. Plants were grown at 20°C. Scale bars, 5 μm. (I) Percentage of cells with FLC or COOLAIR signal in nonprevascular cells in CTL (n = 125 for FLC, 207 for COOLAIR) and ntl8-D3 (n = 171 for FLC, 269 for COOLAIR). (J) Imaging of FLC-Venus in Col FRI and ntl8-D3 FRI grown at 20°C. Numbers of independent roots assayed for each genotype are indicated in the bottom right corner. Scale bar, 50 μm.
COOLAIR is required for FLC transcriptional shutdown in natural and laboratory conditions
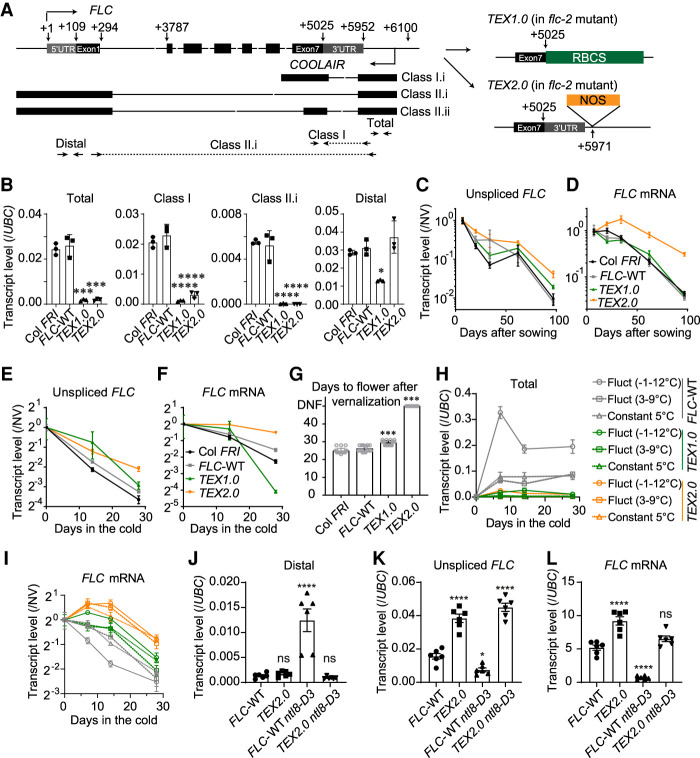
We then investigated how COOLAIR may mediate FLC shutdown by interrogating transgenic lines carrying FLC disrupted in production of COOLAIR (Terminator Exchange 1.0 and 2.0, FLC::FLC-TEX1.0 and FLC-TEX2.0). The TEX1.0 line was generated in a previous study (Csorba et al. 2014), and the TEX2.0 line was newly generated by inserting a NOS terminator to terminate COOLAIR transcription without disturbing the 3′ UTR of FLC (Fig. 3A). COOLAIR transcripts (detected by primers for total, class I, and class II.i) (Csorba et al. 2014) were greatly reduced in both the TEX1.0 and TEX2.0 lines (Fig. 3B). In natural conditions, the unspliced FLC transcript levels in both TEX1.0 and TEX2.0 reduced more slowly than in wild-type controls (transgenic FLC-WT and nontransgenic Col FRI) during vernalization (Fig. 3C), supporting a role for COOLAIR in mediating FLC transcriptional shutdown. Moreover, the FLC mRNA level was accordingly higher in TEX2.0 but not in TEX1.0 (Fig. 3D), likely due to the change in the 3′ UTR causing instability of the FLC mRNA in TEX1.0 (Csorba et al. 2014). Consistently, the same FLC expression in TEX1.0 and TEX2.0 and late-flowering phenotypes were also observed in laboratory conditions (Fig. 3E–G), reinforcing the view that COOLAIR plays a role in FLC transcriptional shutdown.
Figure 3.
COOLAIR is required for FLC regulation in both field and laboratory conditions. (A) Schematic illustration of wild-type FLC, TEX 1.0, and TEX 2.0. Untranslated region (UTR) of FLC is indicated by gray box, and exons are represented by black boxes. Black arrows indicate positions of primers for detecting COOLAIR (the detailed positions are distal, 40–135; class I, 5634–5953; total, 5792–6034). (B) Antisense transcript levels in nonvernalized (NV) wild-type reference Col FRI, wild-type transgenic control (FLC-WT), and two TEX lines. Levels normalized to UBC. Error bars show SEM of three biological replicates. (C,D) Unspliced FLC transcripts (C) and FLC mRNA (D) level in wild-type and TEX lines in the Norwich field in 2016–2017. Expression normalized first to an internal control (see the Materials and Methods) and then to the initial level (time point 1) of each genotype. Error bars show SEM. Data of the Col FRI were previously reported (Antoniou-Kourounioti et al. 2018). (E,F) Unspliced FLC transcripts (E) and FLC mRNA (F) for the wild-type and two TEX lines under constant laboratory vernalization conditions (5°C). Transcript level was normalized first to an internal control and then to nonvernalized (NV; 20°C). Error bars show SEM of three biological replicates. (G) Flowering time phenotype of wild-type and two TEX lines after 4 wk of cold in laboratory conditions. (UNF) Not flowering after 50 d, n = 20 plants for each genotype. (H,I) Plants were grown in conditions shown in Figure 1B. Relative transcript level of total COOLAIR (H) and FLC mRNA (I) was measured ∼8 h after freezing and after 7, 14, and 28 d of cold exposure. Levels were normalized first to UBC and then to the NV of each genotype. Error bars show SEM of two biological replicates for the 7-d time point, and three biological replicates for all other time points. (J–L) qPCR analysis of distal COOLAIR (J), unspliced FLC (K), and FLC mRNA (L) transcript levels in warm conditions in FLC-WT, TEX2.0, FLC-WT ntl8-D3, and TEX2.0 ntl8-D3 mutants. Levels were normalized to UBC. Errors show SEM of six biological replicates. One-way analysis of variance (ANOVA) with ’Dunnett's multiple comparisons test was performed, and significances for individual comparisons of interest are shown. (*) P < 0.05, (***) P < 0.01, (****) P < 0.001, (ns) no significance.
Given that the freezing-enhanced COOLAIR induction led to significantly lower FLC mRNA levels (Fig. 1E), these two TEX lines were also tested in the temperature regimes of Figure 1B. Similar to the observation in Figure 1, COOLAIR was more induced in wild-type plants in the freezing regime compared with the other two temperature regimes (Fig. 3H), and FLC mRNA levels in the freezing regimes were accordingly lower (Fig. 3I). In contrast, such lower FLC mRNA levels associated with freezing temperature were not observed in either of the TEX lines, where COOLAIR induction is greatly reduced (Fig. 3H,I). This supports the view that COOLAIR is responsible for the enhanced decrease in FLC mRNA levels in the freezing regimes. To further determine the causality of COOLAIR up-regulation and FLC down-regulation, we generated a double mutant of ntl8-D3 with TEX2.0, as well as a double mutant of ntl8-OE3 with a COOLAIR promoter deletion line, FLCΔCOOLAIR (Luo et al. 2019). In both double mutants, there is no longer COOLAIR up-regulation by NTL8 (Fig. 3J; Supplemental Fig. S4I–L). We found that FLC down-regulation in ntl8-D3 and ntl8-OE3 is substantially suppressed by the COOLAIR knockout (Fig. 3K,L; Supplemental Fig. S4M). Therefore, COOLAIR up-regulation is a major causal factor for the FLC down-regulation in the ntl8-D or ntl8-OE mutants, supporting a direct role of COOLAIR or COOLAIR transcription in FLC transcriptional shutdown.
Transgenes aimed at knocking out COOLAIR promote production of novel convergent antisense transcripts
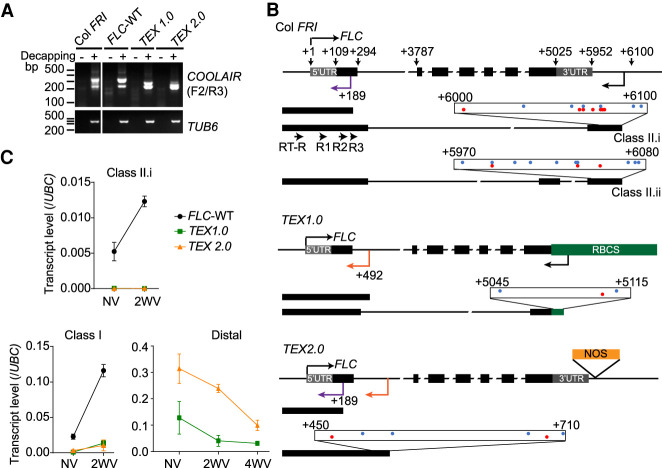
During the analysis of the TEX lines, it became clear that distally polyadenylated COOLAIR was still produced (Fig. 3B). This was unexpected given the presence of the upstream terminator sequence. To understand the origin of the COOLAIR distal amplicon in the TEX1.0 and TEX2.0 lines, 5′ RACE (rapid amplification of cDNA ends) was performed (Supplemental Fig. S5A). In wild-type plants, grown without (NV) or with 2 wk of cold treatment (2WV), two major spliced distal COOLAIR forms with a range of transcriptional start sites were identified (Fig. 4A,B), consistent with the previous study (Swiezewski et al. 2009). In addition, a low abundance convergent antisense transcript (hereafter referred to as CAS as previously defined by Kindgren et al. [2020]) possessing a 5′ cap was identified with a transcriptional start site within FLC exon 1 (Fig. 4A,B; Supplemental Table S1). In the TEX1.0 line, antisense transcription was found to initiate within the RBCS terminator fragment, producing a distal COOLAIR with the same splice sites as on the endogenous locus (Fig. 4A,B; Supplemental Table S1). In the TEX 2.0 line, no COOLAIR transcripts were detected except for the CAS transcript initiating within FLC exon 1 (Fig. 4A,B). Interestingly, additional CASs containing a 5′ cap were identified in both TEX lines originating from different positions inside FLC intron 1 (Fig. 4A,B; Supplemental Table S1). The multiple transcriptional start sites of CAS (Fig. 4B) are likely to contribute to the higher antisense expression in TEX2.0 (Figs. 3B, 4C). The proximally polyadenylated COOLAIR (class I) present at very low levels in both TEX lines is cold induced (Figs. 3B, 4C) and originates from the residual FLC fragment in the flc-2 background, rather than the TEX transgene (Supplemental Fig. S5B). Thus, the major COOLAIR-CAS transcripts in the TEX lines are novel CAS and are not cold induced (Fig. 4C). Given the low frequency of CAS in the wild type, their transcription is likely to be suppressed by COOLAIR transcription from the upstream native promoter.
Figure 4.
Removal of COOLAIR promotes intragenic convergent antisense transcripts in TEX lines. (A) Agarose gel showing the antisense transcripts detected by 5′ RACE in wild-type reference Col FRI, wild-type transgenic control (FLC-WT), and two TEX lines after 2 wk of cold (5°C). TUB6 was used as a control. Samples without (−) or with (+) decapping treatment are indicated. Primers used are mapped in B. (B) Schematic illustrations showing the antisense transcriptional start sites (TSSs) mapped by 5′ RACE in the wild-type and TEX lines. The untranslated region (UTR) of FLC is indicated by a gray box, and exons are represented by black boxes. Black arrows show the positions of COOLAIR TSS in the wild type and TEX 1.0. Purple arrows show the positions of CAS TSS common in the wild type and TEX 2.0, while orange arrows show that of CAS TSSs only in TEX lines. Antisense TSSs were mapped in the scaled boxes with red dots representing those in NV samples and blue dots representing those in 2WV samples. Numbers indicate distance (in base pairs) from the FLC TSS. Primers (RT-R, R1, R2, and R3) used for 5′ RACE are indicted in the Col FRI schematic. (C) Expression level of antisense transcripts in NV and cold-treated wild-type and TEX lines. (2WV) Two weeks of cold treatment; (4WV) 4 wk of cold treatment. Error bars show SEM for three biological replicates. Primers used are illustrated in Figure 3A.
We further investigated three other transgenic lines (FLC + MAF2-T, FLC + NOS-T, and FLCΔCOOLAIR) that had been designed to remove COOLAIR from FLC (Li et al. 2018; Luo et al. 2019). The FLC + MAF2-T and the FLC + NOS-T lines were generated by replacing the COOLAIR promoter with the MAF2 terminator and NOS terminator, respectively (Li et al. 2018), while the FLCΔCOOLAIR line was generated by deleting a 324-bp region in the COOLAIR promoter, downstream from FLC 3′ UTR, using the CRISPR method (Luo et al. 2019). The novel CAS transcripts originating from FLC intron 1 described above were detected in all three lines, with an even higher level in FLC + MAF2-T and FLC + NOS-T (Supplemental Fig. S5B–D). In addition, similar to TEX1.0, antisense transcript start sites were detected in the MAF2-T terminator in the FLC + MAF2-T line (Supplemental Fig. S5D). Similar findings of cryptic promoter usage after disruption of upstream promoters, and alternative transcripts arising when antisense transcription is perturbed, have been reported in S. cerevisiae (Mayer et al. 2015; Kim et al. 2016). The generation of novel COOLAIR-CAS transcripts from intragenic regions in FLC at each attempt to remove COOLAIR, including the CRISPR deletion of the endogenous COOLAIR promoter (FLCΔCOOLAIR), suggests a tight interconnection between antisense transcription and chromatin state at the locus.
Discussion
Plants effectively use fluctuating temperature cues to judge seasonal progression, but the molecular mechanisms underlying this are poorly understood. Through field studies of different Arabidopsis genotypes, we have shown how a temperature dip below freezing hyperinduces transcription of antisense transcripts at the floral repressor locus FLC to facilitate transcriptional silencing (Fig. 5). Since in natural field conditions, colder weather generally follows the first autumn frost, the hyperinduction of COOLAIR by freezing may be one of the many cues used by plants to monitor seasonal progression. The evolutionary significance of this remains to be explored. The ability to respond to acute or ambient cold temperature may provide the plasticity important for adaptation to different climates. This would then have parallels to environmentally regulated gene regulation in yeast, where antisense transcription has been shown to induce faster and higher amplitude changes in the associated gene in response to environmental cues (Xu et al. 2011; Beck et al. 2016; Cloutier et al. 2016). Subsequent frosts show weaker and variable effects on COOLAIR expression (Fig. 1; Supplemental Figs. S1, S2), likely due to the epigenetic silencing of the whole locus by the VIN3-dependent Polycomb switching mechanism that occurs after the cold-induced transcriptional silencing (Antoniou-Kourounioti et al. 2018; Hepworth et al. 2018).
Figure 5.
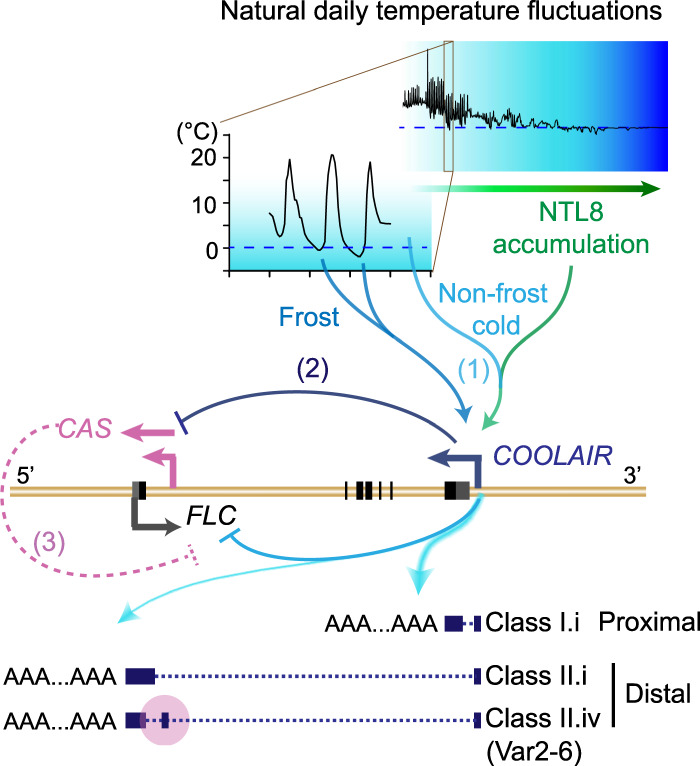
A schematic illustration of COOLAIR regulation of FLC expression. (1) Natural low temperatures (indicated by graded blue) promote NTL8 slow accumulation (graded green arrow), providing the long-term cold information for COOLAIR up-regulation and FLC regulation. Frosts function as strong cues to enhance COOLAIR up-regulation, conferring transcriptional plasticity to FLC. Furthermore, proximal polyadenylation of COOLAIR is enhanced by natural low temperatures, including freezing, supporting the mechanism tightly linking the altered 3′ processing/polyadenylation of COOLAIR to the transcriptional state at the FLC locus (Liu et al. 2010; Marquardt et al. 2014; Fang et al. 2020). (2) CAS transcription is inhibited by transcription from the upstream COOLAIR promoter, possibly influenced by the cold-promoted proximal polyadenylation of COOLAIR. (3) CAS originates from the region (shaded in pink) where the class II.iv exon is alternatively spliced (Li et al. 2015), highlighting its importance in the COOLAIR regulation of FLC transcription.
How such small changes in temperature—from just above to below freezing—cause such a strong up-regulation of COOLAIR expression remains to be determined. An R loop generated by the invasion of COOLAIR into the DNA duplex at the COOLAIR promoter limits further rounds of antisense transcription (Sun et al. 2013). Whether freezing temperature alters the biophysical behavior of this structure to enable strong up-regulation of expression is an interesting possibility (Sun et al. 2007). NTL8 may also interface with the R-loop to influence COOLAIR expression. We have recently found that NTL8 is a direct regulator of VIN3, providing long-term cold information to VIN3 through the mechanism of reduced dilution from slower growth (Zhao et al. 2020). It would seem likely that NTL8 provides long-term cold information to COOLAIR (Fig. 5), but whether NTL8 or its homologs are also involved in the response of COOLAIR to acute freezing temperature remains to be addressed.
How COOLAIR transcription influences the transcriptional output of FLC remains to be resolved, but the ectopic transcription of COOLAIR in the ntl8-D mutant demonstrated causality between up-regulation of COOLAIR and FLC down-regulation at the same gene copy (Fig. 2). Antisense transcription has been shown to alter sense transcription dynamics in a chromatin-dependent manner in yeast and human cells (Murray et al. 2015; Brown et al. 2018). This appears to also be the case for FLC as removal of COOLAIR disrupted the synchronized replacement of H3K36me3 with H3K27me3 at the intragenic FLC nucleation site during the cold (Csorba et al. 2014). It is also possible that COOLAIR transcription reduces functionality of trans factors or gene/intronic loops that promote FLC transcription (Crevillén et al. 2013; Li et al. 2018).
The failed attempts with a range of transgenes to completely remove COOLAIR helps explain the confusion on COOLAIR function. All the independent transgenes analyzed produce new antisense transcripts initiating in the first intron. Consistent with this, recent genome-wide measurements of TSSs showed that extensive alternative intragenic transcriptional initiation occurs in Arabidopsis, and this can be affected by cold (Kindgren et al. 2018), cotranscriptional RNA degradation (Thieffry et al. 2020; Thomas et al. 2020), or mutants disrupted in chromatin signaling (Nielsen et al. 2019; Le et al. 2020). The activation of alternative TSSs significantly influences transcription from nearby TSSs, and thus is important for plant development and adaptation to environmental changes (Kindgren et al. 2018; Nielsen et al. 2019; Le et al. 2020; Thieffry et al. 2020; Thomas et al. 2020). Such CAS transcripts have been found to initiate globally from promoter-proximal exon–intron boundaries across the Arabidopsis genome and are correlated with promoter-proximal RNA Pol II stalling, a checkpoint for transcriptional regulation (Adelman and Lis 2012; Core and Adelman 2019; Kindgren et al. 2020). Interestingly, the major COOLAIR CAS transcripts originate in a region encompassing the alternative splice site of the distal polyadenylated COOLAIR class II.iv. This isoform has higher abundance in the natural accession Var2-6 and is associated with increased FLC expression through a cotranscriptional mechanism involving capping of the FLC nascent transcript (Fig. 5; Li et al. 2015). Moreover, the slower FLC shutdown rate in Var2-6 in the field supports the importance of this checkpoint in FLC transcription regulation (Hepworth et al. 2020). Similar to other systems (Lenstra et al. 2015), these CAS transcripts are likely suppressed at the endogenous locus by COOLAIR transcription from the upstream native promoter. This suppression may be influenced by the cold promotion of proximal 3′ processing/polyadenylation of COOLAIR. This would parallel FLC silencing in warm conditions, promoted by the alternative 3′ processing of COOLAIR (Liu et al. 2010; Marquardt et al. 2014; Fang et al. 2020). We envision that these CAS transcripts might be differentially expressed when COOLAIR transcription is altered, for example, during the initial freezing-dependent COOLAIR spike, possibly contributing to the switch of the local chromatin/transcription states. It will be informative in the future to capture the dynamics of these low abundant CAS transcripts during vernalization. Our current understanding of the many ways COOLAIR regulates FLC is shown in Figure 5.
Overall, our work reveals how first frost acts as a seasonal cue to up-regulate COOLAIR and transcriptionally repress FLC. Such a temperature-regulated antisense–sense circuitry endows a transcriptional plasticity to the FLC locus to effectively respond to the natural fluctuating temperatures of autumn. On a different timescale, but also cold-induced, the Polycomb nucleation mechanism then locks in the FLC silenced transcriptional state to maintain the epigenetic memory of cold exposure. Dissection of the vernalization response has thus elucidated the mechanisms used by plants to translate natural temperature fluctuations into long-term seasonal information.
Materials and methods
No statistical methods were used to predetermine sample size. Field experiments were randomized in a complete-block design as described (Hepworth et al. 2020), with sample size chosen on the basis of feasibility and to buffer against sample loss. Laboratory experiments were not randomized and investigators were not blinded to allocation during experiments and outcome assessment. Sampling in all cases was performed by collecting material from new plants (not repeated sampling) for replicates and also between time points.
Plant materials
All near-isogenic lines in this study were previously described in Hepworth et al. (2020). FLC::FLC-TEX1.0 and FLC::FLC15 WT707 were previously described in Csorba et al. (2014). FLC + MAF2-T, FLC + NOS-T, and FLCΔCOOLAIR were generated as previously described (Li et al. 2018; Luo et al. 2019). The other two ntl8-D alleles (ntl8-D1 and ntl8-D2), GFP-NTL8-D2 transgenic line, and NTL8 overexpression lines (ntl8-OE1 [Salk_866741], ntl8-OE2 [Salk_587226], and ntl8-OE3 [35S::HA-NTL8 transgenic line]) were previously described (Zhao et al. 2020).
The COOLAIRprom::luciferase reporter line was generated by transforming the COOLAIRprom-luciferase construct, as previously described (Sun et al. 2013), into Col FRI plants. A transgenic line containing a single-copy transgene was selected as the progenitor line (named as CTL) for the forward genetic screen.
FLC::FLC-TEX2.0 was generated by inserting a NOS terminator fragment within the first exon of COOLAIR within a ∼12-kb genomic fragment, using adjacent EcoRI and SalI restriction sites immediately downstream from the FLC sense 3′ UTR and the primers “fragment-1-F-1653” (5′-GCTTAACGAGCTTGCACACA-3′) with “fragment-1-R-EcoRI-1653” (5′-GAGGAATTcaagatctcgatgcaattctcac-3′) and “fragment-2-F-SalI” (5′-AGAGTCGACagtgtatgtgttcttcacttctgtcaa-3′) with “fragment-2-R” (5′-TATGGAAGAGGTCGGTCACG-3′). The assembled fragment was cloned into pCambia1300, which was transformed into the Arabidopsis flc-2 FRI genotype with a floral dipping method. Transformants were selected on medium supplemented with hygromycin (Sigma-Aldrich H0654), and single-copy transformants were selfed to generate homozygous T3 lines. Both FLC and COOLAIR expressions were screened before vernalization and after a 2-wk vernalization treatment to identify a single representative line, TEX2.0-472.
Plant growth conditions
Plant were generally grown in growth conditions as described previously (Berry et al. 2015; Rosa et al. 2016; Antoniou-Kourounioti et al. 2018; Hepworth et al. 2018, 2020; Zhao et al. 2020).
Field experiments
Details for field experiments have been described previously (for 2016–2017 winter, see Antoniou-Kourounioti et al. 2018; for 2014–2015 winter, see Hepworth et al. 2018; for both 2016–2017 and 2014–2015 winters, see Hepworth et al. 2020). Briefly, the experiment site in the north was at Ramsta (62°50.988′ N, 18°11.570′ E), and in the south was at Ullstorp (56°06.6721′ N, 13°94.4655′ E). Plants were sown and moved to the field site as follows: Norwich, sown into position on September 29, 2014; South Sweden, sown on September 24, 2014, and moved on October 8, 2014; North Sweden—first year, early planting sown on August 26, 2014, and moved on September 11, 2014, and late planting sown on September 8, 2014, and moved on September 24, 2014; and North Sweden—second year, sown on August 12, 2016, and moved on August 24, 2016.
Laboratory experiments
In general, seeds were stratified after sowing for 3 d at ∼4°C–5°C. Briefly, for RNA analysis and ChIP experiments, plants were grown on Murashige and Skoog (MS) agar plates without glucose. For microscopy, plants were grown on MS plates with 1% agar placed vertically. For nonvernalized (NV) conditions, plants were grown for 10–12 d at long photoperiod conditions (16-h light, 8-h dark with constant 20°C), while for vernalization treatment, plants were moved to 5°C cold treatment at short photoperiod conditions (8-h light, 16-h dark with constant 5°C) after growing at long photoperiod conditions for 7 d if not specified.
Response of TEX1.0 and TEX2.0 lines in laboratory conditions (Fig. 3E–G)
Seeds were sown on plates with selective antibiotic and MS agar media without glucose. Plants were grown for 10 d postgermination in long days at 20°C and then grown for an additional 3 d before NV sampling or moved to a growth room for vernalization with 8-h light and grown for 2 or 4 wk (2WV or 4WV) at 5°C, before transferring seedlings to soil and growing for 20 d at 22°C/20°C with 16-h light/8-h darkness (4WT20). Three replicates of >15 seedlings (NV, 2WV, and 4WV) or three replicates of three pooled leaf/meristem tissue (4WT20) were screened for expression.
Flowering time was analyzed as previously described (Liu et al. 2010). Briefly, plants were vernalized for 4 wk at 5°C with 8-h light before being transferred to soil at 22°C/20°C with 16-h light/8-h darkness. The days plants took to bolting when flower buds were visible at the shoot apical meristem were counted as a measurement of flowering time. Counting was stopped after 50 d as all plants had flowered for controls and TEX1.0 and nothing for TEX2.0.
Recreation of field freezing conditions in the laboratory (Supplemental Fig. S3C–E)
Plants were grown in a nonvernalizing growth chamber in long days for 1 wk at 22°C/20°C 16-h light/8-h darkness and then transferred to a growth chamber (Conviron) with the temperatures and light period matching the week before the first frost (to allow acclimation) and the day following the morning frost. Temperatures and light times are shown in Supplemental Table S2. Three biological replicates of more than five seedlings were sampled at 17:00 and 20:00 on the seventh day, the night before freezing; at 07:00, 09:00, 11:00, 13:00, 15:00, 17:00, and 20:00 on the eighth day, when freezing occurred at 10:00 and 13:00–14:00; and at 20:00 on the following day.
Fluctuating freezing experiments (Figs. 1B–E, 3H,I; Supplemental Fig. S3F–J)
Col FRI seeds were sown onto soil. Nonvernalized plants were grown for 10 d in the nonvernalizing growth chamber and sampled at six time points over 24 h before plants were then moved to 8-h light/16-h darkness growth chambers for vernalization Light was on from 09:00 to 17:00. For vernalization, plants were treated with three different temperature regimes: constant 5°C, daily fluctuating 5°C (3°C–9°C) and daily fluctuating 5°C (−1°C to 12°C) (Fig. 1B). Three biological replicate samples were taken at six time points over 24 h after 2-wk exposure to vernalization. For the NV samples, plants were sampled at 12:00, 16:00, 20:00, and 24:00 and at 08:00 twice 24 h apart. For the vernalization samples, plants were sampled at 12:00, 16:00, 20:00, and 24:00 and at 09:00 twice 24 h apart. Lights came on at midnight for NV and at 09:00 in vernalization treatments. For Figure 3, H and I, experiments were performed as described above, except sampling was performed at the time point (09:00). For Supplemental Figure S3, F–J, experiments were performed following the above procedures, except that Col FRI seeds were sown onto medium in Petri dishes and were sampled at the time point (09:00).
Mutagenesis, genetic screening, and gene cloning of ntl8-D3 mutation
Mutagenesis was carried out following the procedures previously described (Liu et al. 2010). Around 20 M2 (mutagenesis generation 2) seeds from each single M1 plant were screened by being sown on MS medium and stratified for 3 d in the cold (5°C). After growing in a growth cabinet for 10 d, the M2 seedlings were assayed for the bioluminescence with 1 μM luciferin (Promega E1603) under a CCD camera (NightOwl). A mutant was identified to also show high VIN3 expression in warm conditions (Supplemental Fig. S4N), similar to what we found in ntl8-D1 and ntl8-D2 mutants (Zhao et al. 2020). Further Sanger sequencing showed that the mutant carries a mutation in AT2G27300 and so was named ntl8-D3 (Supplemental Fig. S4G,O).
RNA extraction and QPCR
Total RNA was extracted as previously described (Box et al. 2011). Genomic DNA was digested with TURBO DNA-free (Ambion Turbo DNase kit AM1907) according to the manufacturer's guidelines, before reverse transcription was performed. The reverse transcription was performed with the SuperScript III reverse transcriptase (ThermoFisher 18080093) following the manufacturer's protocol using gene-specific primers. Relevant primers are listed in Supplemental Tables S3 and S4. For field experiments, RNA extraction and QPCR were performed as described (Antoniou-Kourounioti et al. 2018; Hepworth et al. 2018, 2020). In brief, analysis of qPCR results was performed with LinReg with normalization to the geometric means of the At5g25760 (“PP2A”) and At1g13320 (“UBC”) control genes. The same analysis was also used in Figure 1, C–E, and Supplemental Figure S3, C–E. The rest results were normalized to single reference gene, UBC. Primers used are described in Supplemental Table S3.
Microscopy
For detecting the fluorescence of FLC-Venus, confocal imaging was performed using a 20×/0.7 NA multi-immersion lens, with water as the immersion fluid on a Leica TCS SP8 X confocal microscope following the procedures in Berry et al. (2015). Roots were immersed in 2 μg/mL propidium iodide (Sigma-Aldrich P4864) to label the cell wall. FLC-Venus was excited with illumination at 514 nm (Argon ion laser). Emissions from Venus were detected between 518 nm and 555 nm using a cooled Leica HyD SMD detector in photon-counting mode. Propidium iodide was detected simultaneously with FLC-Venus by collecting emissions between wavelengths 610 nm and 680 nm. To allow comparison between treatments, the same settings were used for images in both the wild type and ntl8-D3 mutant.
The smRNA FISH experiment was performed following the procedures described previously (Rosa et al. 2016).
Chromatin immunoprecipitation (ChIP)
NTL8 protein ChIP experiments were carried out following the methods described in Zhao et al. (2020). Briefly, nuclei were extracted from 3 g of materials with 30 mL of Honda buffer (0.4 M sucrose, 2.5% Ficoll, 5% dextran T40, 25 mM Tris-HCl at pH 7.4, 10 mM MgCl2, 0.5% Triton X-100, 0.5 mM PMSF, proteinase inhibitor cocktail [Roche 04693159001], 5 mM DTT). After nuclei extraction, purified nuclei were lysed by RIPA buffer (1× PBS, 1% Igepal CA-630 [Sigma I8896], 0.5% sodium deoxycholate, 0.1% SDS, proteinase inhibitor cocktail) and then fragmented by sonication (Diagenode Bioruptor). After sonication, the fragmented chromatin extract was cleared by centrifugation at 13,000 rpm for 15 min at 4°C before immunoprecipitation. The immunoprecipitations were performed with GFP-trap beads (Chromotek GTMA-20) for GFP-NTL8-D2 (Fig. 2G) and anti-HA magnetic beads (Pierce 88836) for HA-NTL8 (Supplemental Fig. S4H) in the ntl8-OE3 line. Relevant primers are listed in Supplemental Table S3.
5′ RACE
Five micrograms of total RNA was first treated with calf intestine alkaline phosphatase (CIAP; Merk Sigma P4978) for 1 h at 37°C and purified with phenol:chloroform. After 5′ cap removing for 1 h at 37°C with cap-clip acid pyrophosphatase (CCAP; Cambio C-CC15011H), 1 μL of 5′ RACE adapter 0.3 μg/μL; (5′-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA-3′) was ligated to a half volume of the CIAP/CCAP-treated RNA (2.5 μL) by T4 RNA ligase (NEB M0204S) for 2 h at 37°C. The CIAP-treated RNA without following CCAP treatment was used as control. All the 5′ RACE adapter-ligated RNA was then reverse transcribed by SuperScript III reverse transcriptase (ThermoFisher 18080093) with a distal COOLAIR-specific primer, RT-R, with TUB6 as a control (Fig. 4B). The cDNA was submitted to nested PCR, and the subsequently purified PCR products were ligated to a T-vector (Supplemental Fig. S5A). At least 48 colonies from each RNA sample were examined by PCR before at least two colonies from each different sized PCR product were sent to Sanger sequencing. Oligos for 5′ RACE are listed in Supplemental Table S4.
Supplementary Material
Acknowledgments
For genetic materials, we are indebted to Dr. Zhe Wu (Southern University of Science and Technology, China) for designing the plasmid construct for the TEX2.0 line, Huamei Wang for generating the TEX2.0 construct, and Shuqin Chen for preparing seed stocks. We also appreciate the gift of FLCΔCOOLAIR, FLC + MAF2-T, and FLC + NOS-T seeds from Dr. Yuehui He (Shanghai Center for Plant Stress Biology). For the field work, we thank Kristina Berggren, Catja Selga, Deborah Cox, Barley R. Collier Harris, Torbjörn Säll, Svante Holm, and the family of Öhman and Nils Jönsson. This work was funded by the European Research Council grant “MEXTIM” (339462), a Royal Society Professorship (RP\R1\180002), and the BBSRC Institute Strategic Programmes GRO (BB/J004588/1) and GEN (BB/P013511/1). Finally, we reiterate our appreciation of all of the members of the Dean laboratory for their discussions.
Author contributions: C.D., Y.Z., and P.Z. conceived the study. P.Z. performed the experiments in Figure 4 and Supplemental Figure S5 and the model in Figure 5. Y.Z. performed the experiments in Figures 2, B–G and J, and 3, H–L, and Supplemental Figures S3, F–I, and S4. J.H. performed the experiments in Figure 1A and in Supplemental Figures S1, S2, and S3, A and B. R.B. performed the experiments in Figure 3, A–G. J.D. performed the experiments in Figure 1, B–E. A.H. performed the experiment in Supplemental Figure S3, C–E. C.X. performed the smRNA FISH in Figure 2, H and I. H.Y. screened the ntl8-D3 mutant in Figure 2A. C.D., Y.Z., P.Z., R.L.A.-K., and J.H. interpretated the data. C.D. acquired the funding. C.D. administered the project. C.D. supervised the study. C.D., Y.Z., P.Z., and J.H. wrote the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348362.121.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Adelman K, Lis JT. 2012. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13: 720–731. 10.1038/nrg3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou-Kourounioti RL, Hepworth J, Heckmann A, Duncan S, Qüesta J, Rosa S, Säll T, Holm S, Dean C, Howard M. 2018. Temperature sensing is distributed throughout the regulatory network that controls FLC epigenetic silencing in vernalization. Cell Syst 7: 643–655.e9. 10.1016/j.cels.2018.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck ZT, Xing Z, Tran EJ. 2016. lncRNAs: bridging environmental sensing and gene expression. RNA Biol 13: 1189–1196. 10.1080/15476286.2016.1240139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry S, Hartley M, Olsson TSG, Dean C, Howard M. 2015. Local chromatin environment of a Polycomb target gene instructs its own epigenetic inheritance. Elife 4: e07205. 10.7554/eLife.07205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box MS, Coustham V, Dean C, Mylne JS. 2011. Protocol: a simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods 7: 7. 10.1186/1746-4811-7-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T, Howe FS, Murray SC, Wouters M, Lorenz P, Seward E, Rata S, Angel A, Mellor J. 2018. Antisense transcription-dependent chromatin signature modulates sense transcript dynamics. Mol Syst Biol 14: e8007. 10.15252/msb.20178007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castaings L, Bergonzi S, Albani MC, Kemi U, Savolainen O, Coupland G. 2014. Evolutionary conservation of cold-induced antisense RNAs of FLOWERING LOCUS C in Arabidopsis thaliana perennial relatives. Nat Commun 5: 4457. 10.1038/ncomms5457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier SC, Wang S, Ma WK, Al Husini N, Dhoondia Z, Ansari A, Pascuzzi PE, Tran EJ. 2016. Regulated formation of lncRNA-DNA hybrids enables faster transcriptional induction and environmental adaptation. Mol Cell 62: 148. 10.1016/j.molcel.2016.03.012 [DOI] [PubMed] [Google Scholar]
- Core L, Adelman K. 2019. Promoter-proximal pausing of RNA polymerase II: a nexus of gene regulation. Genes Dev 33: 960–982. 10.1101/gad.325142.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa S, Dean C. 2019. Storing memories: the distinct phases of Polycomb-mediated silencing of Arabidopsis FLC. Biochem Soc Trans 47: 1187–1196. 10.1042/BST20190255 [DOI] [PubMed] [Google Scholar]
- Coustham V, Li P, Strange A, Lister C, Song J, Dean C. 2012. Quantitative modulation of polycomb silencing underlies natural variation in vernalization. Science 337: 584–587. 10.1126/science.1221881 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Sonmez C, Wu Z, Dean C. 2013. A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J 32: 140–148. 10.1038/emboj.2012.324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C. 2014. Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci 111: 16160–16165. 10.1073/pnas.1419030111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S, Holm S, Questa J, Irwin J, Grant A, Dean C. 2015. Seasonal shift in timing of vernalization as an adaptation to extreme winter. Elife 4: e6620. 10.7554/eLife.06620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Wu Z, Raitskin O, Webb K, Voigt P, Lu T, Howard M, Dean C. 2020. The 3′ processing of antisense RNAs physically links to chromatin-based transcriptional control. Proc Natl Acad Sci 117: 15316–15321. 10.1073/pnas.2007268117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geraldo N, Bäurle I, Kidou S, Hu X, Dean C. 2009. FRIGIDA delays flowering in Arabidopsis via a cotranscriptional mechanism involving direct interaction with the nuclear cap-binding complex. Plant Physiol 150: 1611–1618. 10.1104/pp.109.137448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes EJ, Hennelly SP, Novikova IV, Irwin JA, Dean C, Sanbonmatsu KY. 2016. COOLAIR antisense RNAs form evolutionarily conserved elaborate secondary structures. Cell Rep 16: 3087–3096. 10.1016/j.celrep.2016.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Robertson M, Finnegan EJ, Buzas DM, Dennis ES. 2011. Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS One 6: e21513. 10.1371/journal.pone.0021513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth J, Antoniou-Kourounioti RL, Bloomer RH, Selga C, Berggren K, Cox D, Collier Harris BR, Irwin JA, Holm S, Säll T, et al. 2018. Absence of warmth permits epigenetic memory of winter in Arabidopsis. Nat Commun 9: 639. 10.1038/s41467-018-03065-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hepworth J, Antoniou-Kourounioti RL, Berggren K, Selga C, Tudor EH, Yates B, Cox D, Collier Harris BR, Irwin JA, Howard M, et al. 2020. Natural variation in autumn expression is the major adaptive determinant distinguishing Arabidopsis FLC haplotypes. Elife 9: e57671. 10.7554/eLife.57671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao F, Pahwa K, Manning M, Dochy N, Geuten K. 2019. Cold induced antisense transcription of FLOWERING LOCUS C in distant grasses. Front Plant Sci 10: 72. 10.3389/fpls.2019.00072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C. 2000. Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347. 10.1126/science.290.5490.344 [DOI] [PubMed] [Google Scholar]
- Kim JH, Lee BB, Oh YM, Zhu C, Steinmetz LM, Lee Y, Kim WK, Lee SB, Buratowski S, Kim T. 2016. Modulation of mRNA and lncRNA expression dynamics by the Set2–Rpd3S pathway. Nat Commun 7: 13534. 10.1038/ncomms13534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Ard R, Ivanov M, Marquardt S. 2018. Transcriptional read-through of the long non-coding RNA SVALKA governs plant cold acclimation. Nat Commun 9: 4561. 10.1038/s41467-018-07010-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindgren P, Ivanov M, Marquardt S. 2020. Native elongation transcript sequencing reveals temperature dependent dynamics of nascent RNAPII transcription in Arabidopsis. Nucleic Acids Res 48: 2332–2347. 10.1093/nar/gkz1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le NT, Harukawa Y, Miura S, Boer D, Kawabe A, Saze H. 2020. Epigenetic regulation of spurious transcription initiation in Arabidopsis. Nat Commun 11: 3224. 10.1038/s41467-020-16951-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, Coulon A, Chow CC, Larson DR. 2015. Single-molecule imaging reveals a switch between spurious and functional ncRNA transcription. Mol Cell 60: 597–610. 10.1016/j.molcel.2015.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Filiault D, Box MS, Kerdaffrec E, van Oosterhout C, Wilczek AM, Schmitt J, McMullan M, Bergelson J, Nordborg M, et al. 2014. Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev 28: 1635–1640. 10.1101/gad.245993.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Tao Z, Dean C. 2015. Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev 29: 696–701. 10.1101/gad.258814.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Jiang D, He Y. 2018. FRIGIDA establishes a local chromosomal environment for FLOWERING LOCUS C mRNA production. Nat Plants 4: 836–846. 10.1038/s41477-018-0250-6 [DOI] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. 2010. Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327: 94–97. 10.1126/science.1180278 [DOI] [PubMed] [Google Scholar]
- Luo X, He Y. 2020. Experiencing winter for spring flowering: a molecular epigenetic perspective on vernalization. J Integr Plant Biol 62: 104–117. 10.1111/jipb.12896 [DOI] [PubMed] [Google Scholar]
- Luo X, Chen T, Zeng X, He D, He Y. 2019. Feedback regulation of FLC by FLOWERING LOCUS T (FT) and FD through a 5′ FLC promoter region in Arabidopsis. Mol Plant 12: 285–288. 10.1016/j.molp.2019.01.013 [DOI] [PubMed] [Google Scholar]
- Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C. 2014. Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell 54: 156–165. 10.1016/j.molcel.2014.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. 2015. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161: 541–554. 10.1016/j.cell.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM. 1999. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. 10.1105/tpc.11.5.949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray SC, Haenni S, Howe FS, Fischl H, Chocian K, Nair A, Mellor J. 2015. Sense and antisense transcription are associated with distinct chromatin architectures across genes. Nucleic Acids Res 43: 7823–7837. 10.1093/nar/gkv666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, Ard R, Leng X, Ivanov M, Kindgren P, Pelechano V, Marquardt S. 2019. Transcription-driven chromatin repression of intragenic transcription start sites. PLoS Genet 15: e1007969. 10.1371/journal.pgen.1007969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR. 2016. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 166: 1598. 10.1016/j.cell.2016.08.063 [DOI] [PubMed] [Google Scholar]
- Qüesta JI, Antoniou-Kourounioti RL, Rosa S, Li P, Duncan S, Whittaker C, Howard M, Dean C. 2020. Noncoding SNPs influence a distinct phase of Polycomb silencing to destabilize long-term epigenetic memory at Arabidopsis FLC. Genes Dev 34: 446–461. 10.1101/gad.333245.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S, Duncan S, Dean C. 2016. Mutually exclusive sense–antisense transcription at FLC facilitates environmentally induced gene repression. Nat Commun 7: 13031. 10.1038/ncomms13031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. 2000. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC). Proc Natl Acad Sci 97: 3753–3758. 10.1073/pnas.97.7.3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo C, Lister C, Crevillen P, Nordborg M, Dean C. 2006. Variation in the epigenetic silencing of FLC contributes to natural variation in Arabidopsis vernalization response. Genes Dev 20: 3079–3083. 10.1101/gad.405306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Li JM, Wartell RM. 2007. Conversion of stable RNA hairpin to a metastable dimer in frozen solution. RNA 13: 2277–2286. 10.1261/rna.433307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C. 2013. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340: 619–621. 10.1126/science.1234848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM. 2004. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164. 10.1038/nature02195 [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C. 2009. Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462: 799–802. 10.1038/nature08618 [DOI] [PubMed] [Google Scholar]
- Thieffry A, Vigh ML, Bornholdt J, Ivanov M, Brodersen P, Sandelin A. 2020. Characterization of Arabidopsis thaliana promoter bidirectionality and antisense RNAs by inactivation of nuclear RNA decay pathways. Plant Cell 32: 1845–1867. 10.1105/tpc.19.00815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas QA, Ard R, Liu J, Li B, Wang J, Pelechano V, Marquardt S. 2020. Transcript isoform sequencing reveals widespread promoter-proximal transcriptional termination in Arabidopsis. Nat Commun 11: 2589. 10.1038/s41467-020-16390-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZW, Wu Z, Raitskin O, Sun Q, Dean C. 2014. Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc Natl Acad Sci 111: 7468–7473. 10.1073/pnas.1406635111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Fang X, Zhu D, Dean C. 2020. Autonomous pathway: FLOWERING LOCUS C repression through an antisense-mediated chromatin-silencing mechanism. Plant Physiol 182: 27–37. 10.1104/pp.19.01009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Park S-R, Kim D-H, Kim E-D, Sung S. 2020. Transcriptome and epigenome analyses of vernalization in Arabidopsis thaliana. Plant J 103: 1490–1502. 10.1111/tpj.14817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Clauder-Münster S, Smolik M, Huber W, Steinmetz LM. 2011. Antisense expression increases gene expression variability and locus interdependency. Mol Syst Biol 7: 468. 10.1038/msb.2011.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Antoniou-Kourounioti RL, Calder G, Dean C, Howard M. 2020. Temperature-dependent growth contributes to long-term cold sensing. Nature 583: 825–829. 10.1038/s41586-020-2485-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.