Here, Norton et al. applied genome-scale CRISPR/Cas9 inactivation screens to cell lines derived from a murine model of small cell lung cancer (SCLC) and found SCLC cells were particularly sensitive to the deletion of NEDD8 and other neddylation pathway genes. Genetic suppression or pharmacological inhibition of this pathway caused cell death not only in mouse SCLC cell lines but also in patient-derived xenograft (PDX) models of pulmonary and extrapulmonary small cell carcinoma treated ex vivo or in vivo, and these findings identify neddylation as a regulator of neuroendocrine cell state and potential therapeutic target for small cell carcinomas.
Keywords: cancer, neuroendocrine, small cell lung cancer
Abstract
Small cell lung carcinoma (SCLC) is among the most lethal of all solid tumor malignancies. In an effort to identify novel therapeutic approaches for this recalcitrant cancer type, we applied genome-scale CRISPR/Cas9 inactivation screens to cell lines that we derived from a murine model of SCLC. SCLC cells were particularly sensitive to the deletion of NEDD8 and other neddylation pathway genes. Genetic suppression or pharmacological inhibition of this pathway using MLN4924 caused cell death not only in mouse SCLC cell lines but also in patient-derived xenograft (PDX) models of pulmonary and extrapulmonary small cell carcinoma treated ex vivo or in vivo. A subset of PDX models were exceptionally sensitive to neddylation inhibition. Neddylation inhibition suppressed expression of major regulators of neuroendocrine cell state such as INSM1 and ASCL1, which a subset of SCLC rely upon for cell proliferation and survival. To identify potential mechanisms of resistance to neddylation inhibition, we performed a genome-scale CRISPR/Cas9 suppressor screen. Deletion of components of the COP9 signalosome strongly mitigated the effects of neddylation inhibition in small cell carcinoma, including the ability of MLN4924 to suppress neuroendocrine transcriptional program expression. This work identifies neddylation as a regulator of neuroendocrine cell state and potential therapeutic target for small cell carcinomas.
Small cell lung cancer (SCLC) is a highly metastatic neuroendocrine tumor type that accounts for roughly 15% of all lung cancers (Rudin et al. 2021). Typically responsive to a chemotherapy doublet of a platinum and etoposide, rapid emergence of chemoresistance results in a 5-yr survival rate of only 5%–6%, and addition of immune checkpoint inhibition to chemotherapy has only modestly improved responses (Horn et al. 2018; Paz-Ares et al. 2019; Rudin et al. 2020). SCLC is driven by the inactivation of the tumor suppressors TP53 and RB1 in almost all cases and does not exhibit frequent mutations in “druggable” oncogenic targets common in lung adenocarcinoma (Peifer et al. 2012; Rudin et al. 2012; George et al. 2015). SCLC also exhibits inactivating mutations in chromatin regulators such as KMT2D and CREBBP (Augert et al. 2017) as well as amplifications of MYC family members (Peifer et al. 2012; George et al. 2015), but targeted therapies directed toward SCLC harboring any specific genetic alteration have not demonstrated clinical efficacy.
SCLC exhibits neuroendocrine cell characteristics, and, consistent with neuroendocrine cells representing a major cell-of-origin, targeting Rb1/Trp53 deletion to mouse pulmonary neuroendocrine cells efficiently gives rise to SCLC (Sutherland et al. 2011). Transcriptional analyses can stratify SCLC into subtypes based on activity of key transcription factors that regulate neuroendocrine and neural cell state, including ASCL1 (SCLC-A), NEUROD1 (SCLC-N), and POU2F3 (SCLC-P), with the ASCL1-driven subset of SCLC reflecting the majority of SCLC (Rudin et al. 2019; Wooten et al. 2019). ASCL1 is a master regulator of pulmonary neuroendocrine cell fate, and deletion of ASCL1 prevents this cell population from forming in mouse models (Borges et al. 1997; Ito et al. 2000). ASCL1 is also necessary for SCLC, as its deletion completely prevented SCLC emergence in mouse models (Borromeo et al. 2016). Another transcription factor important for development of the pulmonary neuroendocrine cell lineage is INSM1, expressed in both SCLC-A and SCLC-N subtypes (Rudin et al. 2019). INSM1 promotes pulmonary neuroendocrine cell differentiation, with lungs of embryonic mice lacking Insm1 failing to develop terminally differentiated neuroendocrine cells (Fujino et al. 2015; Jia et al. 2015). Furthermore, INSM1 positively regulates the expression of ASCL1 and suppression of INSM1 slows growth in SCLC cell lines (Fujino et al. 2015). These studies suggest that, like ASCL1, INSM1 might also constitute a therapeutic target for SCLC. However, transcription factors are challenging to target directly using small molecules. As maintenance of neuroendocrine cell differentiation appears essential for SCLC viability, increasing effort is being placed on finding druggable targets to suppress neuroendocrine signaling in SCLC. For example, pharmacologic inhibition of the LSD1 demethylase (Augert et al. 2019) indirectly leads to suppression of neuroendocrine differentiation and of SCLC growth.
Extrapulmonary small cell carcinomas are high-grade neuroendocrine tumors that can arise in almost any organ of the body and resemble SCLC in both appearance and clinical behavior (Dasari et al. 2018). Many key genetic alterations in SCLC are also found in high-grade extrapulmonary small cell carcinomas, including TP53 and RB1 inactivation, ASCL1 induction, and NOTCH pathway suppression (Meder et al. 2016). Extrapulmonary small cell carcinomas may arise from transformation of resident neuroendocrine cells found throughout the body or can represent tumor evolution under therapeutic drug selection from either adenocarcinomas or squamous cell carcinomas (Quintanal-Villalonga et al. 2020).
In this study, we used CRISPR/Cas9 inactivation screens to identify new druggable targets essential for SCLC cell proliferation and viability. We identify protein neddylation as a druggable target in both SCLC and extrapulmonary small cell carcinoma. In these high-grade neuroendocrine carcinomas, we link neddylation inhibition to regulation of INSM1 expression and control of neuroendocrine cell state.
Results
Genome-scale CRISPR/Cas9 inactivation screens of murine SCLC cell lines
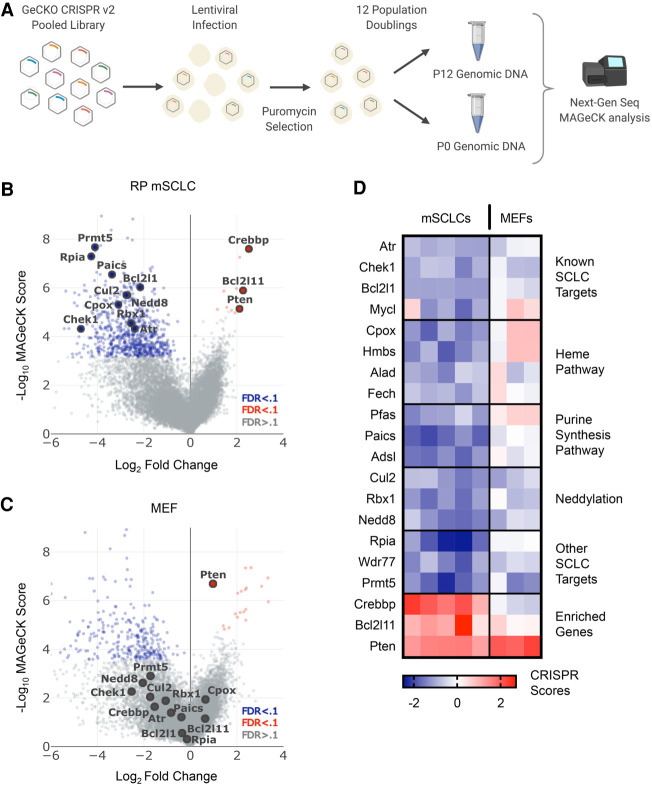
We conducted a series of genome-scale CRISPR/Cas9 screens employing five cell lines derived from a genetically engineered autochthonous Rb1/Trp53-deleted SCLC mouse model (Fig. 1A; Meuwissen et al. 2003). SCLC arising in this mouse model exhibits molecular and histological features of human SCLC. For comparison, we also performed similar screens on immortalized mouse embryonic fibroblasts (MEFs) derived from Trp53lox/lox embryos (Augert et al. 2020). As detailed previously (Augert et al. 2020), we infected cells with the GeCKO v2 pooled lentiviral sgRNA library (Sanjana et al. 2014) containing 130,209 sgRNAs targeting 20,611 genes, with most genes harboring six distinct sgRNAs per gene. Each Trp53-null MEF isolate and SCLC cell line was infected at a MOI of <1 and maintained at a population to ensure 500-fold coverage of transduced cells. Immediately after selection, we collected a P0 sample for next-generation sequencing to assess baseline sgRNA representation. Cell lines were passaged until reaching 12 population doublings (P12), and genomic DNA was then isolated followed by library construction and sequencing. We employed MAGeCK (Li et al. 2014, 2015) to identify genes targeted by sgRNA sets that are significantly enriched or depleted in the P12 samples when compared with the P0 set. Volcano plots for the SCLC (Fig. 1B) and Trp53-null MEF (Fig. 1C) data sets show many common essential genes targeted by sgRNAs depleted in the screens (also see Venn diagrams in Supplemental Fig. S1A). Of the genes targeted by sgRNAs enriched in both Trp53-null MEFs and mSCLC cells, and therefore acting as functional candidate tumor suppressors, only Pten was significant across both data sets (Fig. 1B,C). We also calculated CRISPR scores (Wang et al. 2015), that is, the log2 fold change in sgRNA frequency relative to P0 averaged, across all sgRNAs targeting a given gene. A heat map of CRISPR scores (Fig. 1D) also reveals different dependencies between SCLC cells and Trp53-null MEFs. sgRNAs enriched in CRISPR screening of SCLC cells targeted genes with known tumor suppressor activity in SCLC, including Pten, Bcl2l11 (BIM), and the histone acetyltransferase Crebbp (Fig. 1D). We and others have used mouse models to validate both Pten and Crebbp as SCLC tumor suppressors (Cui et al. 2014; McFadden et al. 2014; Jia et al. 2018); thus, these data support the effectiveness of the screen and provide information on tumor suppressors in SCLC. Our main goal, however, was to identify therapeutically targetable genes necessary for SCLC growth and survival. sgRNAs targeting orthologs of known SCLC dependencies (BCL2L1, ATR, CHEK1, and MYCL) were significantly depleted in mSCLC cells but not in Trp53-null MEFs (Fig. 1B,D). BCL2 family members (Gandhi et al. 2011; Rudin 2012), ATR (Doerr et al. 2017), CHEK1 (Sen et al. 2017b), and MYCL (Kim et al. 2016) are all genes for which pharmacologic inhibition or genetic deletion impairs SCLC cell growth and/or viability. In addition to identifying known SCLC dependencies, we also identified novel hits. sgRNAs targeting genes in the heme synthesis pathway, including Cpox, Hmbs, and Fech, significantly dropped out in SCLC cells (Fig. 1B,D; Supplemental Table S1). Also, members of the de novo purine synthesis pathway, including Pfas, Paics, Adsl, and Impdh2, were essential in SCLC. Previous work using a focused CRISPR/Cas9 library also identified Paics and Pfas as essential genes in murine SCLC cells (Li et al. 2019), and IMPDH proteins were shown as potential therapeutic targets in a subset of SCLC (Huang et al. 2018). Pyrimidine biosynthesis pathway genes, including Cad and Ctps, were also SCLC-essential (Supplemental Table S1), consistent with recent findings (Li et al. 2019). We were particularly interested in novel druggable dependencies identified in our screen for which clinical grade pathway inhibitors are available. For example, we found that sgRNAs targeting the methyltransferase Prmt5 and its binding partner Wdr77 dropped out in the SCLC cells. sgRNAs targeting numerous regulators of protein neddylation including Nedd8, Rbx1, and Cul2 also significantly dropped out in mSCLCs. Neddylation is a post-translational modification involving the covalent addition of the ubiquitin-like NEDD8 (neuronal precursor cell-expressed developmentally down-regulated protein 8) molecule to lysine residues (Kamitani et al. 1997; Xirodimas 2008). Cullin proteins are the major target of protein neddylation, and they require neddylation to function; thus, a critical function of the neddylation pathway is control of protein ubiquitination mediated by cullin-RING ligase (CRL) complexes (Chiba and Tanaka 2004; Pan et al. 2004; Petroski and Deshaies 2005). We did not observe statistically significant dropout of sgRNAs targeting additional cullin family members or other neddylation-related genes (Uba3, Nae1, and Ube2m) in mSCLC cells (Supplemental Fig. S1B,C). However, lack of dropout for any given gene is challenging to interpret owing to heterogeneity in sgRNA effectiveness.
Figure 1.
Whole genome CRISPR/Cas9 screen identifies potential therapeutic targets and pathways in SCLC (A) Schematic of the CRISPR/Cas9 dropout screen conducted using the GeCKO v2 lentiviral mouse library. (B,C) Volcano plots of MAGeCK analysis results from five screened mSCLC cell lines (B) and three screened Trp53-null MEF isolates (C). (D) CRISPR score heat map of the top depleted and enriched genes for each of the five mSCLC cell lines and three Trp53-null MEF isolates screened.
To validate a subset of hits identified in our screen, including neddylation pathway genes, we applied an inducible lentiviral two-vector CRISPR/Cas9 sgRNA expression system to an mSCLC cell line (G6263) used in our screen. This cell line was transduced with a vector conferring constitutive U6 promoter-driven sgRNA expression, and a second vector that expresses Cas9 in a doxycycline-inducible manner. Ten-day proliferation assays revealed reduced viability of cells infected with sgRNAs targeting Nedd8, Rbx1, Cul2, Insm1, Ascl1, Rpia, Bcl2l1, and Cpox, in comparison with control neutral guides (Supplemental Fig. S1D). Thus, our genome scale functional screens revealed essential genes in SCLC, including novel targets for potential therapeutics.
Murine SCLC lines and a subset of PDX models of small cell carcinoma display sensitivity to pharmacological neddylation inhibition
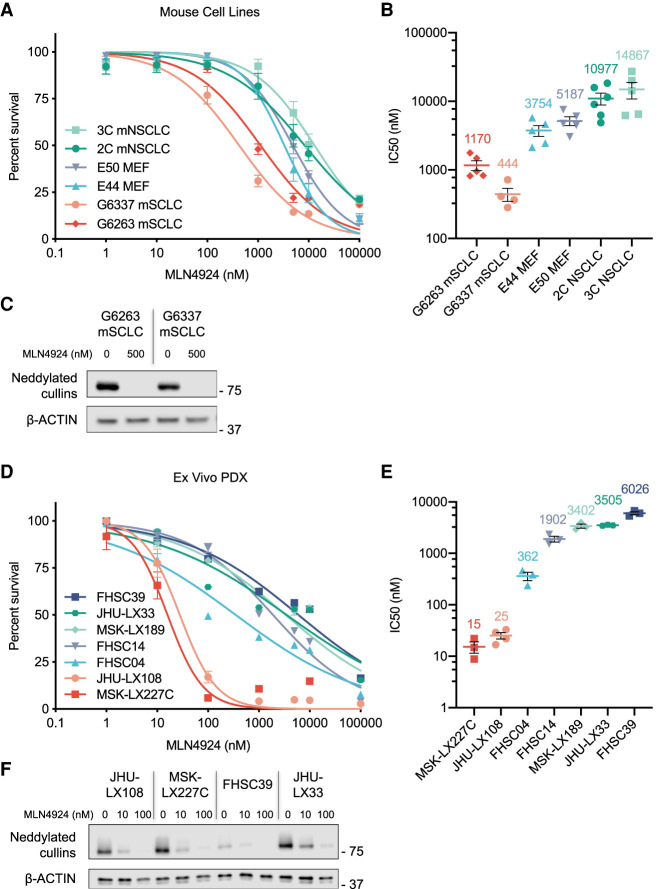
The neddylation pathway was of particular interest in part due to the availability of MLN4924, an ATP-competitive small molecule inhibitor of the neddylation activation enzyme (NAE1) that is being tested in advanced clinical trials for other indications (Soucy et al. 2009). NAE1 inhibition prevents the neddylation of cullin proteins (among other targets) and subsequent ubiquitylation and degradation of the cullin protein substrate (Soucy et al. 2009; Brownell et al. 2010; Liao et al. 2011). To determine whether pharmacologic neddylation inhibition recapitulates effects of neddylation pathway gene deletion, we treated a subset of mSCLC cell lines and Trp53-null MEF isolates employed in our screens with MLN4924. For comparison, we also included two murine non-small cell lung cancer lines (NSCLC) that we derived from a KrasG12D, Trp53-deleted mouse model (Fig. 2A). In line with our screen results, mSCLC lines were indeed more sensitive to neddylation inhibition than MEFs or mouse NSCLC cells, with an IC50 range between 0.4 and 1.2 μM for the mSCLC lines screened compared with 3–5 μM for the MEFs and 10–15 μM for the mNSCLC lines (Fig. 2B). The vast majority of cellular neddylation occurs on cullin proteins and Western blotting analysis revealed loss of neddylated cullins following 48 h of MLN4924 treatment (Fig. 2C), confirming target engagement.
Figure 2.
Treatment of murine SCLC cell lines and human PDX models with the neddylation inhibitor MLN4924 (A) Combined dose response curve for MLN4924-treated Rb1;Trp53–null mSCLC cell lines, Trp53–null MEFs, and Trp53-null; KRASG12D mNSCLC cell lines. Cell viability measured at 72 h by CellTiter-Glo. Data are means ± SEM. (B) Graph depicting experimental MLN4924 IC50 value replicates for mSCLC cell lines, MEF isolates, and mNSCLC cell lines. Mean IC50 value is shown above each sample set. Data are means ± SEM. The same biological replicates used for A are shown. (C) Immunoblot analysis for NEDD8 showing the levels of neddylated cullins for two mSCLC cell lines treated with MLN4924. Cells were collected after 48-h treatment of MLN4924 at two concentrations (0 and 500 nM). β-ACTIN was used as loading control. (D) Combined dose response curve for MLN4924-treated ex vivo PDX models of SCLC and extrapulmonary small cell carcinoma. Cell viability measured at 72 h by CellTiter-Glo. Data are means ± SEM. (E) Graph depicting experimental MLN4924 IC50 value replicates for PDX lines. Mean IC50 value is shown above each sample set. Data are means ± SEM. The same biological replicates used for D are shown. (F) Immunoblot analysis for NEDD8, showing the levels of neddylated cullins for MLN4924-sensitive PDX models JHU-LX108 and MSK-LX227C and the more resistant PDX models FHSC39 and JHU-LX33. Cells were collected after 48-h treatment of MLN4924 at three concentrations (0, 10, and 100 nM). β-ACTIN was used as loading control.
To better assess therapeutic potential of the neddylation inhibition in human SCLC, we tested MLN4924 sensitivity across a panel of SCLC PDX models (including the major SCLC-A and SCLC-N subtypes) that we briefly cultured ex vivo (Fig. 2D; Augert et al. 2019). To extend this analysis to extrapulmonary small cell carcinoma, we included a PDX (MSK-LX227C) derived from a de novo small cell cancer of the cervix, histologically indistinguishable from SCLC, with expression of synaptophysin, chromogranin, and CD56 and with high-level MYC amplification; in this tumor, TP53 and RB1 are WT, being presumably inactivated by oncogenic HPV E6 and E7 rather than by mutation. Remarkably, two models, a SCLC (JHU-LX108, subtype SCLC-A) and the extrapulmonary small cell carcinoma MSK-LX227C (also ASCL1-positive), exhibited IC50 values of 25 and 15 nM, which were orders of magnitude lower than other ex vivo SCLC PDX models and human cell lines tested (Fig. 2D,E), as well as our sensitive mouse SCLC cell lines (Fig. 2A,B). Western blotting analysis of four of the PDX models cultured ex vivo show a reduction of neddylated cullins 48 h after MLN4924 (10 nM) treatment, again confirming target engagement (Fig. 2F). These data suggest that mouse and human SCLC cells are generally sensitive to neddylation inhibition but that a subset of human high-grade neuroendocrine cancers may be exquisitely sensitive to this therapeutic approach.
MLN4924 treatment in SCLC and extrapulmonary small cell PDX models dramatically slows growth
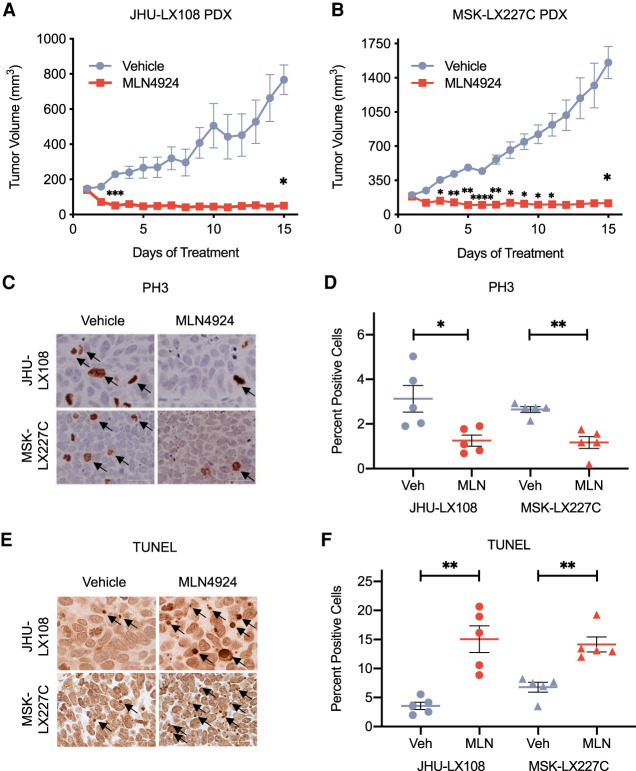
We next determined whether exquisite sensitivity to MLN4924 observed in MSK-LX227C and JHU-LX108 PDX models cultured ex vivo translated in vivo. Approximately 1 million disassociated cells were injected into the flanks of NOD-SCID-γ (NSG) mice and allowed to grow to a tumor volume of 150–200 mm3. Mice were then randomized to receive either 60 mg/kg MLN4924 or the (2-hydroxypropyl)-B-cyclodextrin (HPBCD) vehicle control, given once daily by subcutaneous injection. Treatments continued for 15 d with daily measurements for body weight and tumor volume before resecting the tumors for histological and molecular analysis. The treatment regimen was well tolerated based on assessments of body weight (Supplemental Fig. S2A). We found that both the JHU-LX108 (Fig. 3A; Supplemental Fig. S2B) and MSK-LX227C (Fig. 3B; Supplemental Fig. S2C) PDX models exhibit lack of tumor growth when treated with MLN4924. In contrast, the FHSC39 PDX model with modest sensitivity ex vivo (Fig. 2D,E) was comparatively less sensitive to in vivo treatment but nonetheless exhibited a clear initial response that was not sustained over the 15 d of treatment (Supplemental Fig. S2D,E). To elucidate effects of neddylation inhibition on proliferation and apoptosis, we performed immunohistochemical staining for phospho-histone-H3 (Ser10) (PH3) and TUNEL analyses on fixed tumor samples. JHU-LX108 and MSK-LX227C (Fig. 3C) showed a significant decrease in the number of proliferating PH3-positive mitotic cells with MLN4924 treatment (quantified in Fig. 3D). TUNEL staining (Fig. 3E) revealed a significant increase in apoptosis with MLN4924 in both JHU-LX108 and MSK-LX227C (quantified in Fig. 3F). Thus, monotherapy with a neddylation inhibitor in a subset of PDX models led to lack of tumor growth that was associated with reduced proliferation and increased apoptosis.
Figure 3.
MLN4924 treatment of PDX models in vivo (A,B) Tumor growth inhibition curves of vehicle (HPBCD) and MLN4924-treated (60 mg/kg, 7 dpw) JHU-LX108 PDX (A) and MSK-LX227C PDX (B) flank tumors over 15 d. Data are means ± SEM (n = 5 animals per treatment group per PDX model). (*) P < 0.05, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001, by two-way ANOVA with Sidak's post-test. (C) Representative IHC images of PH3 staining of JHU-LX108 and MSK-LX227C tumors treated by vehicle or MLN4924. Scale bar, 100 μm. (D) Quantification of five representative PH3 IHC field images for each of the MLN4924 and vehicle-treated JHU-LX108 or MSK-LX227C tumors. (E) Representative TUNEL staining of JHU-LX108 and MSK-LX227C tumors treated by vehicle or MLN4924. Scale bar, 100 μm. (F) Quantification of five representative TUNEL field images for each of the MLN4924 and vehicle-treated JHU-LX108 or MSK-LX227C tumors. For D and F, data are means ± SEM (n = 5 in all groups). (*) P < 0.05, (**) P < 0.01, by unpaired Student's t-test.
MLN4924 treatment suppresses neuroendocrine cell state
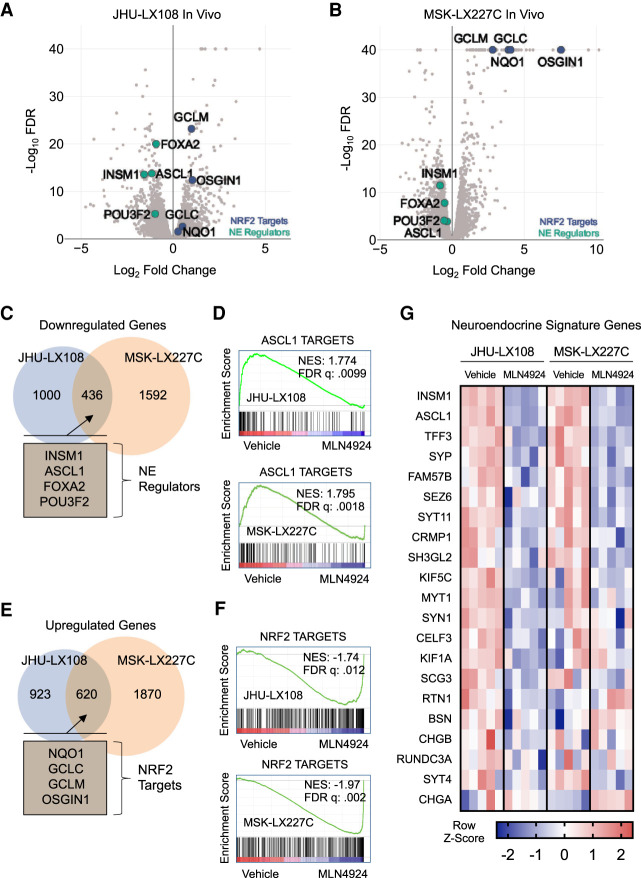
Both SCLC and extrapulmonary small cell carcinoma characteristically express multiple neuroendocrine markers. To understand the molecular pathways that underlie strong responsiveness to MLN4924 in high-grade neuroendocrine cancers, we employed RNA-seq analyses. We used edgeR (Robinson et al. 2010) to identify differentially expressed genes in PDX tumors treated for 15 d with vehicle or MLN4924. Volcano plots showing MLN4924-dependent genes in JHU-LX108 and MSK-LX227C models revealed striking down-regulation of neuroendocrine master regulators including INSM1, ASCL1, FOXA2, and POU3F2 upon inhibition of neddylation (Fig. 4A,B). We also observed up-regulation of NRF2 transcription factor targets, including NQO1, GCLC, GCLM, and OSGIN1. NRF2, which is encoded by NFE2L2, controls response to oxidative stress and its protein stability is tightly regulated by cullin-dependent ubiquitylation. Thus, stabilization of NRF2 is expected upon neddylation inhibition (Kobayashi et al. 2004; Soucy et al. 2009). Analysis of the less sensitive PDX model, FHSC39, did not show a decrease in neuroendocrine gene expression upon MLN4924 treatment but did reveal up-regulation of NRF2 targets upon neddylation inhibition (Supplemental Fig. S3A). Of note, FHSC39 is a SCLC-N model that expresses NEUROD1. Overlapping significant down-regulated and up-regulated genes in the two MLN4924-sensitive PDX model tumors (JHU-LX108 and MSK-LX227C) revealed 436 commonly down-regulated and 620 up-regulated transcripts. Included in the genes down-regulated upon MLN4924 treatment (Fig. 4C) are key regulators of neuroendocrine differentiation: INSM1, ASCL1, FOXA2, and POU3F2. We were particularly interested in ASCL1 suppression upon neddylation inhibition given the requirement of mouse mSCLC for ASCL1 expression (Borromeo et al. 2016). Using a curated list of ASCL1 transcription factor target genes (Borromeo et al. 2016), we performed gene set enrichment analysis (GSEA) (Subramanian et al. 2005). GSEA showed that both JHU-LX108 and MSK-LX227C exhibit significant down-regulation of ASCL1 targets when treated with MLN4924 (Fig. 4D). Conversely, treatment of FHSC39 did not result in ASCL1 target down-regulation (Supplemental Fig. S3B) but did result in up-regulation of NQO1 and other NRF2 targets according to the NFE2L2.V2 gene set (M2870) (Supplemental Fig. S3C). The significant up-regulation of NRF2 targets was also seen in the JHU-LX108 and MSK-LX227C models (Fig. 4E,F). Since ASCL1 is a master regulator of neuroendocrine differentiation, we next determined whether a neuroendocrine signature gene set (Zhang et al. 2018) was also suppressed with neddylation inhibition. Upon MLN4924 treatment, the JHU-LX108 and MSK-LX227C PDX models indeed exhibit transcriptional down-regulation of neuroendocrine signature genes (Fig. 4G) while FHSC39 model lacked a similar transcriptional trend (Supplemental Fig. S3D). Pathway analysis of the common significantly down-regulated genes in JHU-LX108 and MSK-LX227C showed significant enrichment in pathways related to dopaminergic neurogenesis (Supplemental Fig. S3E), while genes commonly up-regulated upon MLN4924 treatment are enriched in pathways for benzo(a)pyrene metabolism, glutathione metabolism, proteasome degradation, and NRF2 activation. Taken together, these data demonstrate that, although neddylation inhibition activates NRF2 signaling across all models tested, high sensitivity toward MLN4924 inhibition is instead correlated with suppression of a neuroendocrine gene expression program. Remarkably, suppression of an overlapping set of critical neuroendocrine regulators including INSM1, ASCL1, FOXA2, and POU3F2 was consistently observed in both the highly responsive SCLC and extrapulmonary small cell models. To determine whether the transcriptional suppression of neuroendocrine regulators is also observed outside of the in vivo treated tumors with high associated cell death, we performed transcriptional analysis on ex vivo PDX models following a 48-h MLN4924 treatment (Supplemental Fig. S4A–D). We again observed decreases in INSM1, ASCL1, FOXA2, and POU3F2 mRNA transcripts, similar to our in vivo results.
Figure 4.
Transcriptional analysis of MLN4924-treated tumors identify changes in neuroendocrine signature in high-grade neuroendocrine cancers (A,B) Volcano plots of edgeR analysis results from RNA-seq performed on five MLN4924-treated and five vehicle-treated JHU-LX108 (A) or MSK-LX227C (B) PDX tumors. Green markers indicate select neuroendocrine genes and blue markers indicate select NRF2 targets. (C) Venn diagram showing common significant down-regulated genes (FDR < .05) determined by edgeR analysis of MLN4924-treated JHU-LX108 and MSK-LX227C PDX tumors. Notable common genes highlighted in inset. (D) Gene set enrichment analysis plots for treated JHU-LX108 (top) and MSK-LX227C (bottom) tumors using list of identified ASCL1 direct targets in SCLC. (E) Venn diagram showing common significant up-regulated genes (FDR < .05) determined by edgeR analysis of MLN4924-treated JHU-LX108 and MSK-LX227C PDX tumors. Notable common genes highlighted in the inset. (F) Gene set enrichment analysis plots for treated JHU-LX108 (top) and MSK-LX227C (bottom) tumors using list of NRF2 targets. (G) Heat map of select differentially expressed neuroendocrine signature genes for MLN4924-treated JHU-LX108 or MSK-LX227C PDX tumors compared with vehicle treatment. Color indicates Z-score for each comparison (five vehicle vs. five MLN4924).
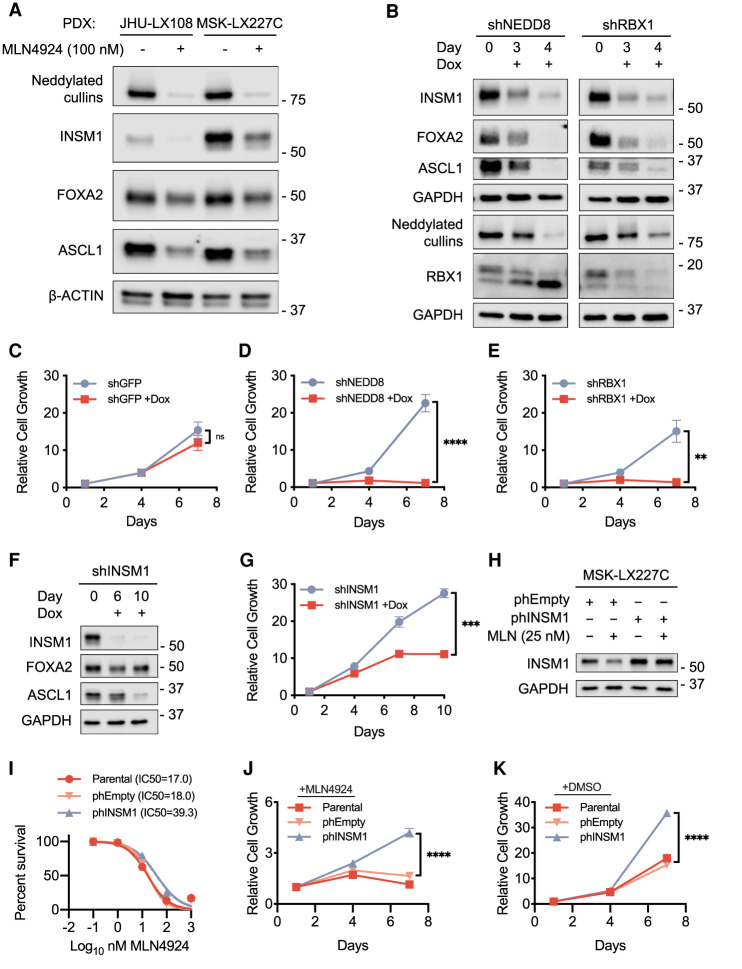
After identifying reduced expression of neuroendocrine-associated transcripts upon MLN4924 treatment in the highly sensitive PDX models, we next examined protein levels. Treatment of both JHU-LX108 and MSK-LX227C for 48 h with MLN4924 (100 nM) resulted in decreased protein levels of neuroendocrine regulators INSM1, ASCL1, FOXA2, and POU3F2 (Fig. 5A; Supplemental Fig. S5A). Conversely, MLN4924 treatment consistently led to increases in NRF2 and P21, targets of ubiquitin-mediated degradation that are increased upon MLN4924 treatment in other systems (Soucy et al. 2009). The NRF2 and P21 protein increases were observed across a panel of PDX models ex vivo but without direct correlation to MLN4924 sensitivity (Supplemental Fig. S5A). Western blot analysis of MSK-LX227C cells treated with MLN4924 again confirmed reduction of neddylated cullins and increased free NEDD8 (Supplemental Fig. S5B). To assess whether neddylation inhibition generally inhibits neuroendocrine regulators in both human and mouse SCLC, we returned to the mSCLC lines used in our CRISPR screens. Western blot analysis of the mouse G6263 and G6337 mSCLC cell lines reveal decreased levels of neuroendocrine lineage proteins upon MLN4924 treatment (Supplemental Fig. S5C) similar to the observations made in the human MSK-LX227C and JHU-LX108 PDX ex vivo cells. Additionally, Western blot analysis of the MLN4924-treated tumors from the in vivo JHU-LX108 PDX model revealed similar down-regulation of ASCL1 and INSM1 at the protein level (Supplemental Fig. S5D), consistent with the RNA-seq results. In the NEUROD1-expressing model, FHSC39 protein levels of INSM1, ASCL1, and NEUROD1 were not reduced (Supplemental Fig. S5E), consistent with the transcriptional data of the treated in vivo tumors.
Figure 5.
Perturbation of the neddylation pathway disrupts the neuroendocrine signature in high-grade neuroendocrine cancers. (A) Immunoblot analysis for NEDD8, showing the levels of neddylated cullins and for neuroendocrine regulators in the MLN4924-sensitive ex vivo PDX models JHU-LX108 and MSK-LX227C. Cells were treated for 48 h with DMSO or MLN4924 (100 nM). β-ACTIN was used as loading control. (B) Immunoblot analysis for the levels of neuroendocrine regulators and neddylation pathway members upon inducible knockdown of NEDD8 or RBX1 in the ex vivo PDX model MSK-LX227C. Samples were supplemented with 1 μg/mL doxycycline and collected at days 0, 3, and 4. GAPDH was used as loading control. (C–E) Proliferation assay results comparing on-dox and off-dox MSK-LX227C shGFP (C), shNEDD8 (D), or shRBX1 (E) knockdown cell lines. Samples were assayed at days 1, 4, and 7 with readout determined by CellTiter-Glo. (F) Immunoblot analysis for the levels of INSM1 upon the inducible knockdown of INSM1 in MSK-LX227C ex vivo cells. Samples were supplemented with 1 μg/mL doxycycline and collected at day 0, 6, and 10. GAPDH was used as loading control. (G) Proliferation assay results comparing on-dox and off-dox MSK-LX227C shINSM1 knockdown cells. Samples were assayed at days 1, 4, 7, and 10 with readout determined by CellTiter-Glo. (H) Immunoblot analysis for the levels of INSM1 in MSK-LX227C ex vivo cells constitutively overexpressing INSM1 (phINSM1) or an empty vector control (phEmpty). Cells were treated for 96 h with DMSO or MLN4924 (25 nM). GAPDH was used as loading control. (I) Dose response curve for MLN4924-treated ex vivo MSK-LX227C cells overexpressing INSM1 or empty vector control. Percent survivability determined at 72 h by CellTiter-Glo. Data are means ± SEM (n = 4 biological replicates). (J) Proliferation of ex vivo MSK-LX227C cells overexpressing INSM1 or empty vector control cultured with MLN4924 (25 nM). Samples were assayed at days 1, 4, and 7. Readout was determined at each time point by CellTiter-Glo. (K) Proliferation of ex vivo MSK-LX227C cells overexpressing INSM1 or empty vector control cultured in standard growth media. Samples were assayed at days 1, 4, and 7. Readout was determined at each time point by CellTiter-Glo. For C–E,G, J, and K, data are means ± SEM (n ≥ 3 biological replicates in all groups). Relative cell growth determined by normalizing average luminescence (RLU) values for the initial time point to 1. (ns) Not significant, (**) P < 0.01, (***) P < 0.001, (****) P < 0.0001, by unpaired Student's t-test.
To our knowledge, neddylation inhibition has not previously been linked to control of neuroendocrine cell state and therapeutic efficacy in tumor types that require the maintenance of neuroendocrine state for viability. To complement our pharmacological observation with genetic perturbations, we next used a dox-inducible system with shRNA-expressing lentiviral vectors to knock down the expression of neddylation pathway members NEDD8 and RBX1. Doxycycline was supplemented to MSK-LX227C cells every 48 h, and cells were collected at time points of 72 and 96 h to validate knockdown by Western blot analysis (Fig. 5B). Knockdown of NEDD8 and RBX1 phenocopied MLN4924 treatment in leading to striking down-regulation of the neuroendocrine regulators INSM1, ASCL1, and FOXA2. These molecular observations were coupled with a strong reduction in proliferation upon NEDD8 and RBX1 knockdown compared with the shGFP controls (Fig. 5C–E). Thus, suppression of proliferation and neuroendocrine cell master regulator gene expression was not an off-target effect of MLN4924 but reflects an unappreciated role for neddylation pathway control of neuroendocrine cell state.
Pharmacologic and genetic suppression of neddylation led to particularly strong suppression of the expression of INSM1, a regulator of ASCL1 (Figs. 4A–C, 5A). To examine the functional importance of reduced INSM1, we again used the inducible hairpin system (Fig. 5F). Knockdown of INSM1 in MSK-LX227C cells, over a period of 10 d, reduced proliferation compared with the off-dox condition (Fig. 5G). Thus, despite MLN4924 treatment resulting in altered expression of hundreds of transcripts, reduction in INSM1 alone can suppress proliferation/viability of MSK-LX227C cells. We found that ASCL1 levels are reduced with the knockdown of INSM1, consistent with previous observations in a developmental context in which INSM1 loss abrogated ASCL1 expression (Fujino et al. 2015). Next, we overexpressed INSM1 in the MLN4924-sensitive MSK-LX227C ex vivo model to determine whether this would confer resistance to neddylation inhibition. Indeed, overexpression of INSM1, as validated by Western blot (Fig. 5H), resulted in a twofold increase in IC50 upon MLN4924 treatment compared with an empty vector control (Fig. 5I). MSK-LX227C cells overexpressing INSM1 also exhibited a significant increase in cell viability when grown in the presence of MLN4924 (25 nM) over 7 d (Fig. 5J). Interestingly, INSM1 overexpression also resulted in increased proliferation even in the absence of neddylation inhibition (Fig. 5K). As INSM1 regulates ASCL1 (Fig 5F), we next overexpressed ASCL1 using the same system. Overexpression of ASCL1 also promoted MLN4924 resistance (Supplemental Fig. S6A–D). This suggests that the strong growth effect of neddylation inhibition occurs through an axis involving INSM1 and ASCL1. Our data support neddylation as a critical mechanism for the maintenance of neuroendocrine cell state, confirming neddylation inhibition as a potential treatment strategy for high-grade neuroendocrine cancers.
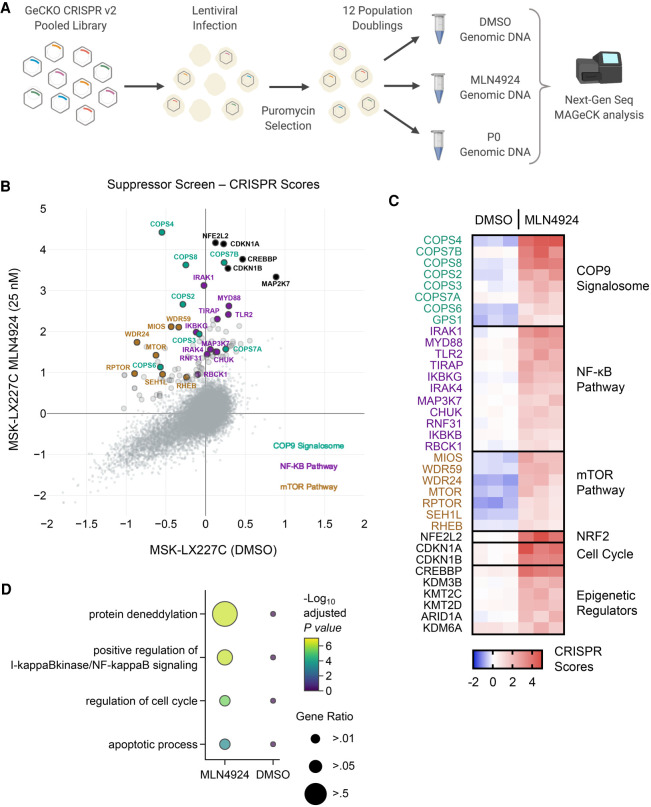
CRISPR/Cas9 suppressor screen to identify mechanisms of resistance to MLN4924
Targeted therapies for cancer treatments are typically effective for a transient period before acquired resistance emerges. Previous candidate gene approaches revealed point mutations in the UBA3 protein as one potential mechanism of resistance to MLN4924 treatment in colorectal carcinoma, leukemic, and NSCLC cells, as these mutations reduced NAE heterodimer binding to the NEDD8-MLN4924 adduct (Milhollen et al. 2012; Toth et al. 2012; Xu et al. 2014). To gain broader insights into mechanisms through which resistance to MLN4924 may develop in highly responsive tumors, we performed a genome-scale suppressor screen. We transduced MSK-LX227C cells grown ex vivo with the GeCKO v2 human CRISPR/Cas9 inactivation library and passaged cells under DMSO or MLN4924 (25 nM) (Fig. 6A). After infection at an MOI of <1 and puromycin selection, 65 million cells were collected for each initial time point (P0), and 65 million cells were retained for each of the DMSO and MLN4924 (25 nM) treatment arms. DMSO-treated cells were passaged for 12 population doublings while MLN4924-treated cells were passaged until the DMSO arm reached completion. We quantified the average CRISPR Score for each gene across three replicates for both our vehicle and MLN4924-treated samples (Fig. 6B,C). MAGeCK analyses revealed that neddylation pathway members NEDD8, RBX1, UBE2M, and NAE1 were essential in DMSO control MSK-LX227C cells (Supplemental Fig. S7A), confirming that human small cell carcinomas are sensitive to genetic suppression of neddylation similar to their mouse model counterparts. To identify gene deletions that confer resistance to MLN4924, MAGeCK analyses revealed number of genes/pathways as strongly enriched in the MLN4924-treated cells (FDR < 0.1) (Supplemental Fig. S7B). Pathway analysis performed using Enrichr (Chen et al. 2013; Kuleshov et al. 2016) identified deneddylation, NF-κB activation, cell cycle regulation, and apoptosis as enriched in the screen (Fig. 6D). Most strikingly, sgRNAs targeting COPS1 (GPS1), COPS2, COPS3, COPS4, COPS6, COPS7A, and COPS8, all part of the COP9 signalosome, were highly enriched in the MLN4924-treated group. The COP9 complex deneddylates and deactivates cullin-RING E3 ligase (CRL) complexes (Wei and Deng 2003), suggesting that impairment of deneddylation may be a means to suppress effects of neddylation inhibition. We also found that sgRNAs targeting the NF-κB pathway, including MYD88, TIRAP, MAP3K7, and the three IKK complex members (CHUK, IKBKB, and IKBKG), were highly enriched in the MLN4924-treated replicates. While deletion of mTOR pathway genes was deleterious in the DMSO control, sgRNAs targeting MTOR, RHEB, RPTOR, and members of the GATOR2 complex (MIOS, WDR59, WDR24, and SEH1L) were enriched in the MLN4924-treated samples. In addition, NRF2, and CDK inhibitors CDKN1A (P21) and CDKN1B (P27) and several epigenetic regulators including CREBBP and KDM3B scored highly in our suppressor screen (Fig. 6C).
Figure 6.
Whole-genome CRISPR suppressor screen of MLN4924-treated MSK-LX227C cells identifies the COP9 signalosome, NF-κB, and mTOR pathways as potential mechanisms of sensitivity and resistance. (A) Schematic of the CRISPR/Cas9 dropout screen conducted using the GeCKO v2 lentiviral human library. (B) X,Y plot of average CRISPR scores for the MSK-LX227C MLN4924 (25 nM) and MSK-LX227C DMSO data sets. Enlarged markers indicate genes falling above the y = x + 1 line. Notable genes are labeled and colorized according to pathway identity (n = 3 biological replicates for each screen data set). (C) Heat map of top enrichment CRISPR scores organized by select pathways for the three MSK-LX227C DMSO replicates and three MSK-LX227C MLN4924 (25 nM) replicates. (D) Pathway dot plot for significant enriched genes filtered by MAGeCK analysis for both the MSK-LX227C MLN4924-treated and DMSO data sets (FDR < 0.1). Pathway analysis performed with Enrichr and GO biological process gene ontology. Dot color indicates significance and dot size represents the proportion of genes identified in the pathway.
Inhibition of the COP9 signalosome abrogates the effects of neddylation pathway perturbation
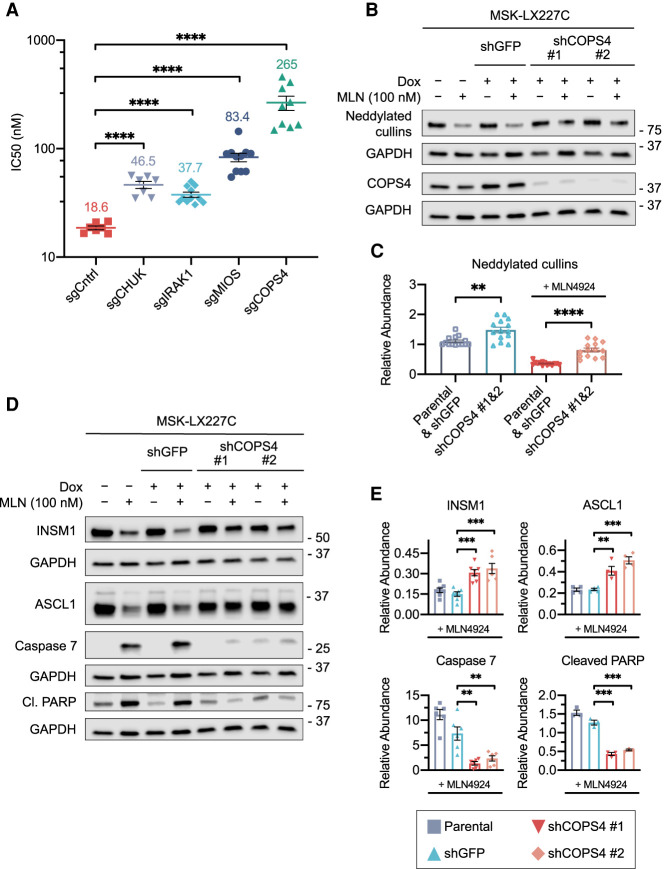
For suppressor screen hit validation, we next tested sgRNAs targeting components of the COP9 signalosome (COPS4), the NF-κB pathway (CHUK, IRAK1), and mTORC1 signaling (MIOS) in MSK-LX227C cells (Fig. 7A). For each gene targeted, we validated sgRNA knockout efficiency by Western blot (Supplemental Fig. S8A,B). Most strikingly, knockout of the COP9 signalosome subunit COPS4 led to dramatic resistance toward MLN4924, increasing the IC50 > 10-fold. Knockouts of the NF-κB pathway members and of the mTORC1-activating GATOR2 complex member MIOS also led to resistance to MLN4924 treatment, albeit to a lesser extent than COPS4 (Fig. 7A).
Figure 7.
Abrogation of the COP9 signalosome and other pathways provides resistance to MLN4924. (A) Response to MLNL4924 treatment (72-h) upon CHUK, IRAK1, MIOS, or COPS4 invalidation in MSK-LX227C ex vivo cells. IC50s are shown. Data are means ± SEM, from at least n = 7 biological replicates. (****) P < 0.0001, unpaired Student's t-test. (B) Representative immunoblot analysis for NEDD8, showing the levels of neddylated cullins and for COPS4 in MSK-LX227C cells treated with DMSO or MLN4924 (100 nM) for 96 h. Parental MSK-LX227C ex vivo cells were compared with cells infected with an inducible shRNA targeting GFP or COPS4. On-dox cells were supplemented with 1 μg/mL doxycycline. GAPDH was used as loading control. (C) Quantification of relative levels of neddylated cullin protein abundance from multiple replicate immunoblot experiments described in B, showing combined parental MSK-LX227C and shGFP data compared with the combined data of two independent COPS4 shRNAs. Data are means ± SEM (n = 7 biological replicates). (**) P < 0.01, (****) P < 0.0001, unpaired Student's t-test. (D) Representative immunoblot analysis for the levels of neuroendocrine regulators and pro-apoptotic proteins for samples described in B. GAPDH was used as loading control. (E) Quantification of the relative levels of cleaved Caspase 7, cleaved PARP, INSM1, and ASCL1 protein abundance for multiple replicate immunoblot experiments described in D. Data are means ± SEM (n ≥ 3 biological replicates). (**) P < 0.01, (***) P < 0.001, unpaired Student's t-test. For C and E, relative abundance levels calculated by normalization to individual GAPDH loading controls with the untreated parental MSK-LX227C set to a relative abundance of 1.
We next generated a MSK-LX227C cell line with inducible shRNA-mediated suppression of COPS4 to further validate our screen results. Western blot analysis of MSK-LX227C cells infected with shRNAs targeting COPS4 and treated with doxycycline confirmed reduction in COPS4 protein levels (Fig. 7B). We found that protein levels of neddylated cullins, identified via Western blotting for NEDD8, increased upon COPS4 knockdown in the MLN4924-treated condition (Fig. 7B, quantified C). These data suggest that loss of COP9 signalosome function may provide resistance to MLN4924 by increasing neddylation.
We next examined functional effects of COPS4 depletion. CellTiter-Glo assays showed that COPS4 knockdown allowed MSK-LX227C cells to proliferate in the presence of MLN4924 (Supplemental Fig. S8C). Western blot analyses showed that COPS4 knockdown leads to marked reduction in apoptotic markers such as cleaved Caspase 7 and cleaved PARP following MLN4924 treatment (Fig. 7D,E). Furthermore, COPS4 knockdown partially restored protein levels of neuroendocrine master regulators INSM1 and ASCL1 under MLN4924 treatment (Fig. 7D,E). Thus, inhibiting deneddylation via COP9 subunit deletion provides a mechanism of resistance to neddylation inhibition (see model, Supplemental Fig. S9), which correlates with impaired ability of MLN4924 to suppress the expression of master regulators of neuroendocrine cell state.
Discussion
To identify new therapeutic targets for refractory high-grade neuroendocrine cancers, we applied whole-genome CRISPR/Cas9 inactivation screens to murine Rb1/Trp53-deleted mSCLC cell lines. We were encouraged to see sgRNAs targeting known SCLC dependencies, such as orthologs of MYCL, ATR, CHEK1, and BCL2L1, emerge in our screen data. Inactivation of Mycl in the Rb1/Trp53 SCLC mouse model decreases tumor burden and progression (Kim et al. 2016), and inhibitors of ATR and CHEK1 have shown anti-tumor activity in preclinical studies (Byers et al. 2012; Cardnell et al. 2013; Doerr et al. 2017; Sen et al. 2017a,b). We were most interested in identifying novel dependencies for which pharmacologic inhibitors could rapidly be applied in the clinic. Highly significant in our list of SCLC-essential genes were NEDD8, RBX1, and CUL2, leading us to investigate neddylation inhibition as a therapeutic strategy. The selective inhibitor of the neddylation pathway, MLN4924, precludes the initial step of NEDD8-mediated enzymatic cascade (Soucy et al. 2009). MLN4924 has been tested in numerous tumor types, slowing proliferation in both cell lines and in vivo PDX models (Lin et al. 2010; Milhollen et al. 2010, 2011; Swords et al. 2010; Wei et al. 2012; Tong et al. 2017). In our study, mSCLC cell lines were more sensitive to MLN4924 treatment than either mNSCLC cell lines or Trp53-null MEFs, and additionally we identified two PDX models, representing SCLC and extrapulmonary small cell carcinoma, that responded exceptionally well to MLN4924 both ex vivo and in vivo. Thus, our studies reveal that targeting the neddylation pathway should be investigated as a potential therapeutic option for a subset of high-grade neuroendocrine cancers including SCLC.
Extrapulmonary small cell carcinomas have been less extensively studied than SCLC but share many histologic and clinical characteristics, including aggressive growth, metastatic capacity, and a marked but transient response to platinum-based chemotherapy (Zheng et al. 2015). Here, we show that an HPV-positive high-grade cervical small cell carcinoma, positive for the critical SCLC transcription factor ASCL1, demonstrates remarkable similarities to a SCLC-A subtype SCLC in response to inhibition of neddylation. These similarities include concomitant down-regulation of a set of known neuroendocrine transcriptional regulators and striking single agent response to MLN4924 in vivo. These data are encouraging, suggesting that inhibition of neddylation as a therapeutic strategy, initially identified in murine SCLC lines, may have broad application to a subset of human SCLC and extrapulmonary small cell carcinomas.
Maintenance of neuroendocrine differentiation can be essential for small cell carcinomas (Borromeo et al. 2016; Augert et al. 2019), and we found that MLN4924 treatment resulted in the suppression of expression of master regulators of neuroendocrine cell differentiation including INSM1, ASCL1, FOXA2, and POU3F2 in sensitive PDX models. Additionally, genetic suppression of the neddylation pathway genes NEDD8 and RBX1 phenocopies the MLN4924 response by causing a similar suppression of INSM1 and ASCL1, which accompanies strong antiproliferative and apoptotic effects. While knockdown of INSM1 in MSK-LX227C cells decreases proliferation, we also note that MLN4924 induces a far broader suppression of neuroendocrine-related transcripts and leads to a stronger antiproliferative effect than loss of INSM1. While we did not delineate the molecular pathway through which MLN4924 results in suppression of INSM1, we hypothesize the existence of a transcriptional repressor of INSM1 regulated by ubiquitin-mediated proteolysis and stabilized upon MLN4924 treatment. It is also possible that it is an upstream regulator of such a repressor that is neddylation-controlled or that neddylation of a non-cullin target could underlie MLN4924 control of INSM1 transcription and neuroendocrine cell state.
Targeted therapies, even when effective, typically lead to acquired resistance, and previous studies identified point mutations in UBA3 as one possible mechanism of resistance to MLN4924 (Milhollen et al. 2012; Toth et al. 2012; Xu et al. 2014). To ascertain potential mechanisms of resistance to MLN4924 using an unbiased approach, we performed a genome scale CRISPR/Cas9 suppressor screen. We identified genes for which deletions promote cell viability in the presence of MLN4924. As top hits, sgRNAs targeting the entire COP9 signalosome were enriched in our screen. The primary function of this multiunit complex is to deneddylate substrates, including cullin-RING ubiquitin ligases (Lyapina et al. 2001; Petroski and Deshaies 2005). Our data support knockout of COP9 signalosome genes as impairing deneddylation, essentially freezing proteins in the neddylated state. Studies have suggested that deneddylation precedes and promotes degradation of cullin substrates, and thus proper COP9 signalosome function is a necessary step in preventing accumulation of cullin substrates (Miyauchi et al. 2008). While COP9 complex suppression conferred particularly strong resistance to MLN4924, deletion of genes in the NF-κB and mTOR pathways as well as NRF2, CDKN1A (P21), and CDKN1B (P27) also contributed to resistance to MLN4924. Control of P27 stability has previously been implicated in mediating effects of MLN4924 in SCLC (Zhao et al. 2020), and these data overall suggest that drug effects on multiple signaling pathways contribute to the impaired viability upon MLN4924 treatment in responsive models. Deletion of mTORC1 complex genes promoting MLN4924 resistance was particularly surprising, as kinome shRNA screening recently revealed mTOR as essential in a subset of SCLC (Kern et al. 2020) and our mouse SCLC screen data showed that deletion of the same genes was deleterious in the absence of MLN4924. MLN4924 is being advanced in clinical trials across various cancer types, and we hypothesize that some of the mechanisms of resistance coming through our screen in small cell carcinoma cells, such as impairment of deneddylation, may emerge as mechanisms of resistance to MLN4924 relevant to multiple cancer types.
SCLC remains an intractable tumor type with new therapeutic strategies greatly needed. Our study shows the utility of genome-wide CRISPR/Cas9 inactivation screens to identify genes and pathways as therapeutic targets for SCLC. We focused on neddylation pathway inhibition, which suppressed neuroendocrine signaling in human models of both SCLC and extrapulmonary small cell carcinoma. Whether the sensitivity toward neddylation inhibition observed in the small cell carcinoma of the cervix extends to other sites of origin for high-grade neuroendocrine tumors remains to be determined. While we focused on MLN4924 as monotherapy, this agent can enhance the effects of chemotherapy (Jazaeri et al. 2013; Vanderdys et al. 2018; Ferris et al. 2020) and is currently being tested in clinical trials as combination therapy with various chemotherapeutics across solid and liquid cancer types (Swords et al. 2018; Lockhart et al. 2019). Moreover, first-line therapy for SCLC now includes both chemotherapeutic agents as well as PD1-axis inhibition, and MLN4924 has recently been shown to augment responses to PD1 inhibition (McGrail et al. 2020). Thus, while we hypothesize, based on our PDX data, that there may be a subset of SCLC with potential to respond exceptionally well to MLN4924 monotherapy, future work should also test the potential for MLN4924 to augment responses to chemotherapy and/or immune checkpoint inhibition more broadly in SCLC. Pharmacological agents that can suppress neuroendocrine cell state in SCLC may provide a new inroad toward improving the dismal prognosis of patients with SCLC and high-grade neuroendocrine carcinomas.
Materials and methods
Genetically engineered mouse models of SCLC and cell line generation
We thank Anton Berns for the Trp53lox/lox and Tyler Jacks for the Rb1lox/lox strains. We obtained Adenovirus containing a CMV-promoted Cre (Ad5-CMV-Cre) from the University of Iowa Vector Core. Intratracheal infection of Ad-CMV-Cre was performed on adult Trp53lox/lox; Rb1lox/lox mice at a titer of 1.25 × 109 pfu (3 × 108 pfu for G6337 and G6263) per mouse. Monitoring of mice continued each week after infection until mice showed noticeable signs of labored breathing. After euthanasia, tumor fragments were extracted to generate cell lines, snap-frozen for molecular analysis, or fixed in NBF for histology and immunochemistry. All animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) at the Fred Hutchinson Cancer Research Center.
Trp53lox/lox MEF generation
At 13.5 d postcoitum, embryos from a Trp53lox/lox mouse were collected and moved to a sterile 10-cm dish in a tissue culture hood. Individual embryos were cleaned with PBS before all maternal tissues, innards, and embryo heads were removed. After shifting the embryo body to a clean dish, 1 mL of .05% trypsin in EDTA was added and the embryo body chopped with a sterilized razor blade. The resulting embryo mince was incubated for 1 h at 37°C. Fresh DMEM was added to the digested MEF cells and then moved to a fresh sterile 10-cm culture dish. Cells were cultured for 72–96 h, splitting before reaching confluence, and frozen for liquid nitrogen stock. To inactivate p53 expression, Ad5-CMV-Cre was added to the media of cultured MEFs at a MOI of 100. The virus addition was repeated to ensure complete p53 loss.
Cell culture
MEF and 293TN cells were both cultured in DMEM supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin (Pen/Strep;Thermo Fisher Scientific 15140122) and 10% FBS (Omega Scientific FB-01). Mouse SCLC and PDX ex vivo cell lines were grown in RPMI Complete media. The recipe was as follows: RPMI 1640 (Thermo Fisher Scientific 11875-093), 100 U/mL penicillin, 100 μg/mL streptomycin, 15% FBS, 1 mM sodium pyruvate (Thermo Fisher Scientific 11360070), 100 μM β-mercaptoethanol (Thermo Fisher Scientific 31350010), and 10 μg/mL insulin, human recombinant zinc solution (Thermo Fisher Scientific 12585014). Doxycycline (Sigma 44577) was used for inducible lentiviral systems at noted concentrations. All cells were incubated at 5% CO2 and 95% air.
Pooled library DNA and virus creation
The DNA and viral productions of both the GeCKO v2 mouse (Addgene 1000000052 and 1000000053; a gift from Feng Zhang) and human (Addgene 1000000048 and 1000000049; a gift from Feng Zhang) libraries were performed as described previously (Sanjana et al. 2014).
CRISPR library titering
Titer of half libraries (for both mouse and human GeCKO v2) for SCLC cells was determined by spinfection of 3 million cells of individual cell lines in a well of a 12-well plate. Cells were supplemented with virus and 8 μg/mL of polybrene at a volume of 600 μL. Plates were centrifuged at 2000 rpm for 2 h at 37°C. After spinfection, cell wells were mixed with 900 μL of media and incubated overnight. Virus was removed the day after transduction. At 48 h after transduction, lines were treated with puromycin for 72 h. Calculation of virus for the desired 0.3 MOI was determined by comparing number of puromycin-surviving cells to the unselected control.
CRISPR/Cas9 screening
For the Trp53-null MEF screen (Augert et al. 2020), 2.5 × 108 cells were transduced with the GeCKO v2 mouse pooled library at a MOI of <1 to ensure a 30% successful transduction rate. After overnight virus addition, cells were selected with puromycin for 3 d. For P0, 6.6 × 107 cells were collected, and 6.6 × 107 cells were passaged until 12 population doublings were reached, maintaining 500-fold coverage of all guides. A P12 pellet was collected. Three independent MEF isolates were screened. For screening of mouse SCLC cell lines, 4 × 108 cells were transduced with both halves of the GeCKO v2 mouse pooled library at a MOI of <1 to ensure a 30% successful transduction rate. Three million cells in a 600-μL volume were spinfected in the presence of polybrene and virus at 2000 rpm for 2 h at 37°C. After completion of the spin, each well was resuspended with 900 μL of fresh media and the cell-virus mixture left overnight. Virus was removed the next day and the wells pooled. The following day, transduced cells were selected with puromycin for 3 d. For a P0 pellet sample, 6.6 × 107 cells were collected, and 6.6 × 107 cells were passaged until 12 population doublings were reached, maintaining 500-fold coverage of all guides. A P12 pellet was collected. Five mouse SCLC lines were screened. For the MSK-LX227C PDX ex vivo resistance screen, transduction of MSK-LX227C cells was performed similar to the mouse SCLC screen. We used the GeCKO v2 human pooled library for this screen. After selection with puromycin, 6.6 × 107 cells were collected for a P0 pellet, 6.6 × 107 cells were cultured with DMSO as an untreated control, and 6.6 × 107 cells were cultured with a final MLN4924 concentration of 25 nM. DMSO control cells were passaged until 12 population doublings were achieved, maintaining 500-fold coverage of guides. MLN4924-treated cells were split every 3–4 d with fresh MLN4924-supplemented media until the DMSO control cells reached endpoint. Pellets for the DMSO and MLN4924-treated cells were collected for each of the three replicates.
Library generation and CRISPR/Cas9 screen analysis
To prepare libraries for sequencing, 430 μg of extracted genomic DNA per sample were amplified and prepared for sequencing using Phusion Flash high-fidelity master mix (Thermo Fisher F548L) according to a previously described protocol (Chen et al. 2015). A two-step PCR reaction was employed to first amplify the sgRNA cassette and maintain library complexity (12 cycles), and a second PCR reaction was performed to add barcodes and Illumina adapters (16 cycles). Reactions were pooled after each PCR. Following the second PCR, reaction products were run on a 2.5% agarose gel and the DNA purified using a gel extraction kit (PureLink gel extraction kit; Invitrogen K210012). Once purified, the concentration of individual libraries was quantified using the Kapa Biosystems library quantification kit (Roche KK4824) according to kit instructions. Libraries were sequenced using an Illumina HiSeq 2500. The sequencing results were analyzed using the MAGeCK-VISPR pipeline (Li et al. 2015). For CRISPR score analyses of aligned and normalized read counts, the following formula was used: CRISPR score = log2 (final sgRNA abundance/initial sgRNA abundance) (Wang et al. 2015).
Xenograft models and drug treatment
Patient-derived xenografts from SCLC circulating tumor cells or tumor biopsies were developed as previously described (Augert et al. 2019). Cells were reinjected as 100-μL aliquots of a 1:1 mixture of matrigel and dissociated cells into the flanks of NSG mice. Treatments of either MLN4924 or vehicle control were started at a flank tumor volume size of ∼150–200 mm3. Mice were administered MLN4924 or vehicle control at 60 mg/kg once daily for 15 d and individual flank tumors measured using calipers. At 15 d, tumors were resected after euthanasia of the mice.
Plasmids
We cloned individual guide sgRNA vectors for screen validation using the lentiGuide-Puro (Addgene 52963; a gift from Feng Zhang) vector in conjunction with Lenti-iCas9-neo (Addgene 85400; a gift from Qin Yan). For knockouts of human CHUK, MIOS, IRAK1, COPS4, and other validations of the MSK-LX227C suppressor screen, we used the lentiCRISPR v2 vector (Addgene 52961; a gift from the Zhang laboratory). To induce knockdown of GFP, NEDD8, RBX1, INSM1, and COPS4, we used the EZ-Tet-pLKO series of plasmids (EZ-Tet-pLKO-Puro [Addgene 85966; a gift from Cindy Miranti], EZ-Tet-pLKO-Blast [Addgene 85973; a gift from Cindy Miranti], and EZ-Tet-pLKO-Hygro [Addgene 85972; a gift from Cindy Miranti]). shRNA and sgRNA sequences are listed in Supplemental Table S2. The INSM1 cDNA was a gift from the Peter Nelson laboratory (Origene RG215007). The ASCL1 lentiviral overexpression plasmid (phASCL1-N106; Addgene 31781; a gift from Gerald Crabtree) was used as the base vector for the constitutive neuroendocrine overexpression construct. The INSM1 cDNA was cloned into the phASCL1 vector backbone. To create the empty control (phEmpty), the ASCL1 cDNA was removed and the vector religated.
Lentiviral infections
LentiCRISPR v2 sgRNAs, lentiGuide-Puro, lenti-iCas9-neo, EZ-Tet-pLKO shRNAs, and ORF overexpression plasmids were infected using the spinfection protocol described above.
Drug sensitivity, cell viability, and proliferation assays
MLN4924 IC50 determination assays were conducted by seeding 17,000–20,000 cells (depending on basal proliferation rate) into an opaque-walled, clear-bottom 96-well culture plate in combination with increasing concentration of MLN4924. The volume of cells, diluted MLN4924, and media totaled 100 μL per well. Each assay was done in triplicate with at least three independent replicates. At 72 h, viability was assessed using the CellTiter-Glo luminescent cell viability assay (Promega) following the manufacturer's protocol. The plate was mixed on a plate shaker and the luminescent signal read using a BioTek plate reader. Proliferation assays were conducted by seeding 1750–2000 cells into an opaque-walled, clear-bottom 96-well tissue culture plate at a total volume of 100 μL. Cells were seeded in replicate plates to account for each day of cell viability readings needed (day 1, day 4, day 7, and, if noted, day 10). For inducible lentiviral constructs, on-doxycycline (on-dox) wells included 1 μg/mL doxycycline upon seeding and were readministered to appropriate wells on even days of the assay. The collection of cell pellets for accompanying immunoblotting and RNA analysis followed similar setups to those described above except seeded in cell culture dishes with scaled up cell numbers and volumes.
Immunohistochemistry
Fragments of dissected PDX tumors were fixed in neutral-buffer formalin for 48 h and then stored in 70% ethanol. Tumors were embedded in paraffin blocks and sectioned at 4 μm. Hematoxylin and eosin staining was completed using standard procedures. The primary antibody anti-phospho-histone-H3 (Ser10; 1:500; EMD Millipore 06-570) was used for the immunohistochemical staining of slides. Briefly, slides were dewaxed in xylene and rehydrated through a decreasing gradient of ethanol in TBST. Antigen unmasking was performed in a microwave and slides were allowed to cool. Prior to overnight antibody incubation, sections were blocked with a 5% goat serum solution (Jackson ImmunoReseaerch 005-000-121) followed by blocking of endogenous peroxidases with a 3.5% H2O2 solution. A biotin-conjugated secondary antibody was used (Vector Laboratories BA-1000) followed by the VectaStain ABC kit (Vector Laboratories PK-4000). Finally, detection was performed (Vector Laboratories SK-4100) and imaged using a Nikon E800 microscope. The POD in situ cell death detection kit (Roche 11684817910) was used for the TUNEL assays according to the manufacturer's protocol.
RNA extraction, RNA-seq libraries, and differential expression analysis
Extraction of RNA from tumor tissues was performed with TRIzol according to the manufacturer's specifications. To generate RNA-seq libraries from 500 ng of total RNA, the Ultra RNA library preparation kit for Illumina (New England BioLabs E7530L) was used according to the manufacturer's instructions. Sequencing of 50-bp single ends was performed with an Illumina HiSeq 2500. Reads of low quality were omitted prior to independent alignments to both human (UCSC hg19) and mouse (UCSC mm9) genomes using TopHat (Trapnell and Schatz 2009). Employing a custom Python script, human and mouse alignments were evaluated for each read, where reads with fewer mismatches against the human reference than mouse were retained for downstream processing. The FPKM values were quantified using Cuffdiff (Trapnell et al. 2013). The HTSeq Python package (Anders et al. 2015) was used to generate counts from the TopHat alignments. Differentially expressed genes upon MLN4924 treatment of PDX tumors were identified using the edgeR software package (Robinson et al. 2010). The significance of differentially expressed genes was set at a false discovery rate (FDR) of .05. Genomic data sets were submitted to the Gene Expression Omnibus repository under the accession number GSE171262.
Gene set enrichment and pathway analyses
Common significantly down-regulated or up-regulated genes from the JHU-LX108 and MSK-LX227C data sets were analyzed using the Enrichr website application (http://amp.pharm.mssm.edu/Enrichr) (Chen et al. 2013; Kuleshov et al. 2016). Gene set enrichment analysis (GSEA) of differentially expressed genes from the JHU-LX108, MSK-LX227C, and FHSC39 data sets was performed using the GenePattern site (http://www.genepattern.org) (Subramanian et al. 2005).
Protein extraction and immunoblotting
Lysis from whole-cell extracts of cultured murine and human cell lines was prepared in diluted RIPA buffer (10× concentration; Cell Signaling 9806). Phosphatase and protease inhibitors were added to the RIPA buffer prior to extraction. Cells were dissociated by mechanical homogenization followed by repeated passaging through successive small gauge needles. Protein concentration was determined using Pierce BCA protein assay kit (Thermo Fisher Scientific 23227). Immunoblotting was performed using standard protocols. The following antibodies were used in this study: anti-β-ACTIN (1:2000; Cell Signaling 4970), anti-GAPDH (1:2000; Santa Cruz Biotechnology sc-32233), anti-NEDD8 (1:1000; Cell Signaling 2745), anti-RBX1 (1:1000; Cell Signaling 11922), anti-INSM1 (1:1000; Santa Cruz Biotechnology sc-271408), anti-MASH1 (ASCL1; 1:1000; BD 24B72D11.1), anti-FOXA2 (1:1000; Cell Signaling 8186), anti-BRN2 (POU3F2; 1:1000; Proteintech 18998-1-AP), anti-NRF2 (1:1000; Cell Signaling 12721), anti-P21 (1:1000; Santa Cruz Biotechnology sc-6246), anti-CSN4 (COPS4; 1:1000; Bethyl Laboratories A300-013A), anti-PARP (1:1000; Cell Signaling 9542), anti-Caspase 7 (1:1000; Cell Signaling 12827), anti-IKKα (1:1000; Cell Signaling 2682), anti-MIOS (1:1000; Bethyl Laboratories A304-699A), and anti-IRAK1 (1:1000; Santa Cruz Biotechnology sc-5288).
Graphics and illustrations
Screen diagram and model illustrations were created using the BioRender.com website. Volcano plots, pathway dot plots, and X,Y plots were created using the Plotly Python graphing library. All other plots were generated with GraphPad Prism 8.
Statistical analysis
Statistical significance between two groups was calculated with a two-tailed unpaired Student's t-test. For the tumor growth curves, a mixed model two-way ANOVA with Sidak's multiple comparison test was carried out to compare vehicle with MLN4924 treatment groups. Statistics were calculated using GraphPad Prism 8.
Supplementary Material
Acknowledgments
We thank the Fred Hutchinson Genomics and Bioinformatics core for their aid in the generation and analysis of next-generation sequencing data. We also recognize the support from the Fred Hutchinson Comparative Resources and Experimental Histopathology staff. We also thank Eli Grunblatt for help with the immunohistochemistry analysis. J.P.N. was supported by funding from National Institutes of Health (NIH) training grant T32 GM007270, and E.E. was supported by funding from NIH grant T32CA009657. A.A. acknowledges support from the Lung Cancer Research Foundation/Uniting Against Lung Cancer. NIH grants U01CA235625 to D.M. and U24CA213274 and R01CA197936 to C.M.R. provided funding support. This research was funded in part through the NIH/National Cancer Institute Cancer Center support grant P30 CA015704, the Specialized Program of Research Excellence (SPORE) in lung cancer award P50CA228944, and the Evergreen Fund Pilot Award.
Author contributions: J.P.N. and D.M. conceived the project and analyzed and interpreted data. J.P.N. executed methodology and collected data. A.A. performed the Trp53-null MEF screen. E.E. performed PDX model generation and propagation. R.B. provided bioinformatics analysis. C.M.R. provided PDX model reagents and contributed to writing the manuscript. J.P.N. and D.M. wrote the manuscript. All authors aided in reviewing and editing.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348316.121.
Competing interest statement
C.M.R. has consulted regarding oncology drug development with AbbVie, Amgen, Astra Zeneca, Epizyme, Genentech/Roche, Ipsen, Jazz, Lilly, and Syros, and serves on the scientific advisory boards of Bridge Medicines, Earli, and Harpoon Therapeutics. D.M. has previously received research funding from Roche and from Janssen Pharmaceuticals.
References
- Anders S, Pyl PT, Huber W. 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augert A, Zhang Q, Bates B, Cui M, Wang X, Wildey G, Dowlati A, MacPherson D. 2017. Small cell lung cancer exhibits frequent inactivating mutations in the histone methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance). J Thorac Oncol 12: 704–713. 10.1016/j.jtho.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augert A, Eastwood E, Ibrahim AH, Wu N, Grunblatt E, Basom R, Liggitt D, Eaton KD, Martins R, Poirier JT, et al. 2019. Targeting NOTCH activation in small cell lung cancer through LSD1 inhibition. Sci Signal 12: eaau2922. 10.1126/scisignal.aau2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augert A, Mathsyaraja H, Ibrahim AH, Freie B, Geuenich MJ, Cheng PF, Alibeckoff SP, Wu N, Hiatt JB, Basom R, et al. 2020. MAX functions as a tumor suppressor and rewires metabolism in small cell lung cancer. Cancer Cell 38: 97–114.e7. 10.1016/j.ccell.2020.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges M, Linnoila RI, van de Velde HJ, Chen H, Nelkin BD, Mabry M, Baylin SB, Ball DW. 1997. An achaete-scute homologue essential for neuroendocrine differentiation in the lung. Nature 386: 852–855. 10.1038/386852a0 [DOI] [PubMed] [Google Scholar]
- Borromeo MD, Savage TK, Kollipara RK, He M, Augustyn A, Osborne JK, Girard L, Minna JD, Gazdar AF, Cobb MH, et al. 2016. ASCL1 and NEUROD1 reveal heterogeneity in pulmonary neuroendocrine tumors and regulate distinct genetic programs. Cell Rep 16: 1259–1272. 10.1016/j.celrep.2016.06.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, et al. 2010. Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ. Mol Cell 37: 102–111. 10.1016/j.molcel.2009.12.024 [DOI] [PubMed] [Google Scholar]
- Byers LA, Wang J, Nilsson MB, Fujimoto J, Saintigny P, Yordy J, Giri U, Peyton M, Fan YH, Diao L, et al. 2012. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov 2: 798–811. 10.1158/2159-8290.CD-12-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardnell RJ, Feng Y, Diao L, Fan YH, Masrorpour F, Wang J, Shen Y, Mills GB, Minna JD, Heymach JV, et al. 2013. Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN 673 in small cell lung cancer. Clin Cancer Res 19: 6322–6328. 10.1158/1078-0432.CCR-13-1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma'ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics 14: 128. 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Sanjana NE, Zheng K, Shalem O, Lee K, Shi X, Scott DA, Song J, Pan JQ, Weissleder R, et al. 2015. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160: 1246–1260. 10.1016/j.cell.2015.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Tanaka K. 2004. Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Curr Protein Pept Sci 5: 177–184. 10.2174/1389203043379783 [DOI] [PubMed] [Google Scholar]
- Cui M, Augert A, Rongione M, Conkrite K, Parazzoli S, Nikitin AY, Ingolia N, MacPherson D. 2014. PTEN is a potent suppressor of small cell lung cancer. Mol Cancer Res 12: 654–659. 10.1158/1541-7786.MCR-13-0554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari A, Mehta K, Byers LA, Sorbye H, Yao JC. 2018. Comparative study of lung and extrapulmonary poorly differentiated neuroendocrine carcinomas: a SEER database analysis of 162,983 cases. Cancer 124: 807–815. 10.1002/cncr.31124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerr F, George J, Schmitt A, Beleggia F, Rehkamper T, Hermann S, Walter V, Weber JP, Thomas RK, Wittersheim M, et al. 2017. Targeting a non-oncogene addiction to the ATR/CHK1 axis for the treatment of small cell lung cancer. Sci Rep 7: 15511. 10.1038/s41598-017-15840-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris J, Espona-Fiedler M, Hamilton C, Holohan C, Crawford N, McIntyre AJ, Roberts JZ, Wappett M, McDade SS, Longley DB, et al. 2020. Pevonedistat (MLN4924): mechanism of cell death induction and therapeutic potential in colorectal cancer. Cell Death Discov 6: 61. 10.1038/s41420-020-00296-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino K, Motooka Y, Hassan WA, Ali Abdalla MO, Sato Y, Kudoh S, Hasegawa K, Niimori-Kita K, Kobayashi H, Kubota I, et al. 2015. Insulinoma-associated protein 1 is a crucial regulator of neuroendocrine differentiation in lung cancer. Am J Pathol 185: 3164–3177. 10.1016/j.ajpath.2015.08.018 [DOI] [PubMed] [Google Scholar]
- Gandhi L, Camidge DR, Ribeiro de Oliveira M, Bonomi P, Gandara D, Khaira D, Hann CL, McKeegan EM, Litvinovich E, Hemken PM, et al. 2011. Phase I study of Navitoclax (ABT-263), a novel Bcl-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors. J Clin Oncol 29: 909–916. 10.1200/JCO.2010.31.6208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, Leenders F, Lu X, Fernandez-Cuesta L, Bosco G, et al. 2015. Comprehensive genomic profiles of small cell lung cancer. Nature 524: 47–53. 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn L, Mansfield AS, Szczęsna A, Havel L, Krzakowski M, Hochmair MJ, Huemer F, Losonczy G, Johnson ML, Nishio M, et al. 2018. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med 379: 2220–2229. 10.1056/NEJMoa1809064 [DOI] [PubMed] [Google Scholar]
- Huang F, Ni M, Chalishazar MD, Huffman KE, Kim J, Cai L, Shi X, Cai F, Zacharias LG, Ireland AS, et al. 2018. Inosine monophosphate dehydrogenase dependence in a subset of small cell lung cancers. Cell Metab 28: 369–382.e5. 10.1016/j.cmet.2018.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Udaka N, Yazawa T, Okudela K, Hayashi H, Sudo T, Guillemot F, Kageyama R, Kitamura H. 2000. Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 127: 3913–3921. [DOI] [PubMed] [Google Scholar]
- Jazaeri AA, Shibata E, Park J, Bryant JL, Conaway MR, Modesitt SC, Smith PG, Milhollen MA, Berger AJ, Dutta A. 2013. Overcoming platinum resistance in preclinical models of ovarian cancer using the neddylation inhibitor MLN4924. Mol Cancer Ther 12: 1958–1967. 10.1158/1535-7163.MCT-12-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia S, Wildner H, Birchmeier C. 2015. Insm1 controls the differentiation of pulmonary neuroendocrine cells by repressing Hes1. Dev Biol 408: 90–98. 10.1016/j.ydbio.2015.10.009 [DOI] [PubMed] [Google Scholar]
- Jia D, Augert A, Kim DW, Eastwood E, Wu N, Ibrahim AH, Kim KB, Dunn CT, Pillai SPS, Gazdar AF, et al. 2018. Crebbp loss drives small cell lung cancer and increases sensitivity to HDAC inhibition. Cancer Discov 8: 1422–1437. 10.1158/2159-8290.CD-18-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamitani T, Kito K, Nguyen HP, Yeh ET. 1997. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem 272: 28557–28562. 10.1074/jbc.272.45.28557 [DOI] [PubMed] [Google Scholar]
- Kern JA, Kim J, Foster DG, Mishra R, Gardner EE, Poirier JT, Rivard C, Yu H, Finigan JH, Dowlati A, et al. 2020. Role of mTOR as an essential kinase in SCLC. J Thorac Oncol 15: 1522–1534. 10.1016/j.jtho.2020.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Wu N, Kim YC, Cheng PF, Basom R, Kim D, Dunn CT, Lee AY, Kim K, Lee CS, et al. 2016. Genetic requirement for Mycl and efficacy of RNA Pol I inhibition in mouse models of small cell lung cancer. Genes Dev 30: 1289–1299. 10.1101/gad.279307.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. 2004. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol 24: 7130–7139. 10.1128/MCB.24.16.7130-7139.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, Liu XS. 2014. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol 15: 554. 10.1186/s13059-014-0554-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Köster J, Xu H, Chen CH, Xiao T, Liu JS, Brown M, Liu XS. 2015. Quality control, modeling, and visualization of CRISPR screens with MAGeCK-VISPR. Genome Biol 16: 281. 10.1186/s13059-015-0843-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ng SR, Colon CI, Drapkin BJ, Hsu PP, Li Z, Nabel CS, Lewis CA, Romero R, Mercer KL, et al. 2019. Identification of DHODH as a therapeutic target in small cell lung cancer. Sci Transl Med 11: eaaw7852. 10.1126/scitranslmed.aaw7852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao H, Liu XJ, Blank JL, Bouck DC, Bernard H, Garcia K, Lightcap ES. 2011. Quantitative proteomic analysis of cellular protein modulation upon inhibition of the NEDD8-activating enzyme by MLN4924. Molecular & Cellular Proteomics 10: M111.009183. 10.1074/mcp.M111.009183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Milhollen MA, Smith PG, Narayanan U, Dutta A. 2010. NEDD8-targeting drug MLN4924 elicits DNA rereplication by stabilizing Cdt1 in S phase, triggering checkpoint activation, apoptosis, and senescence in cancer cells. Cancer Res 70: 10310–10320. 10.1158/0008-5472.CAN-10-2062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockhart AC, Bauer TM, Aggarwal C, Lee CB, Harvey RD, Cohen RB, Sedarati F, Nip TK, Faessel H, Dash AB, et al. 2019. Phase Ib study of pevonedistat, a NEDD8-activating enzyme inhibitor, in combination with docetaxel, carboplatin and paclitaxel, or gemcitabine, in patients with advanced solid tumors. Invest New Drugs 37: 87–97. 10.1007/s10637-018-0610-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Shevchenko A, Deshaies RJ. 2001. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science 292: 1382–1385. 10.1126/science.1059780 [DOI] [PubMed] [Google Scholar]
- McFadden DG, Papagiannakopoulos T, Taylor-Weiner A, Stewart C, Carter SL, Cibulskis K, Bhutkar A, McKenna A, Dooley A, Vernon A, et al. 2014. Genetic and clonal dissection of murine small cell lung carcinoma progression by genome sequencing. Cell 156: 1298–1311. 10.1016/j.cell.2014.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail DJ, Garnett J, Yin J, Dai H, Shih DJH, Lam TNA, Li Y, Sun C, Li Y, Schmandt R, et al. 2020. Proteome instability is a therapeutic vulnerability in mismatch repair-deficient cancer. Cancer Cell 37: 371–386.e12. 10.1016/j.ccell.2020.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meder L, König K, Ozretić L, Schultheis AM, Ueckeroth F, Ade CP, Albus K, Boehm D, Rommerscheidt-Fuss U, Florin A, et al. 2016. NOTCH, ASCL1, p53 and RB alterations define an alternative pathway driving neuroendocrine and small cell lung carcinomas. Int J Cancer 138: 927–938. 10.1002/ijc.29835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. 2003. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell 4: 181–189. 10.1016/S1535-6108(03)00220-4 [DOI] [PubMed] [Google Scholar]
- Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, et al. 2010. MLN4924, a NEDD8-activating enzyme inhibitor, is active in diffuse large B-cell lymphoma models: rationale for treatment of NF-κB-dependent lymphoma. Blood 116: 1515–1523. 10.1182/blood-2010-03-272567 [DOI] [PubMed] [Google Scholar]
- Milhollen MA, Narayanan U, Soucy TA, Veiby PO, Smith PG, Amidon B. 2011. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res 71: 3042–3051. 10.1158/0008-5472.CAN-10-2122 [DOI] [PubMed] [Google Scholar]
- Milhollen MA, Thomas MP, Narayanan U, Traore T, Riceberg J, Amidon BS, Bence NF, Bolen JB, Brownell J, Dick LR, et al. 2012. Treatment-emergent mutations in NAEβ confer resistance to the NEDD8-activating enzyme inhibitor MLN4924. Cancer Cell 21: 388–401. 10.1016/j.ccr.2012.02.009 [DOI] [PubMed] [Google Scholar]
- Miyauchi Y, Kato M, Tokunaga F, Iwai K. 2008. The COP9/signalosome increases the efficiency of von Hippel-Lindau protein ubiquitin ligase-mediated hypoxia-inducible factor-α ubiquitination. J Biol Chem 283: 16622–16631. 10.1074/jbc.M710599200 [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Kentsis A, Dias DC, Yamoah K, Wu K. 2004. Nedd8 on cullin: building an expressway to protein destruction. Oncogene 23: 1985–1997. 10.1038/sj.onc.1207414 [DOI] [PubMed] [Google Scholar]
- Paz-Ares L, Dvorkin M, Chen Y, Reinmuth N, Hotta K, Trukhin D, Statsenko G, Hochmair MJ, Özgüroğlu M, Ji JH, et al. 2019. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 394: 1929–1939. 10.1016/S0140-6736(19)32222-6 [DOI] [PubMed] [Google Scholar]
- Peifer M, Fernández-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, Plenker D, Leenders F, Sun R, Zander T, et al. 2012. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 44: 1104–1110. 10.1038/ng.2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petroski MD, Deshaies RJ. 2005. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6: 9–20. 10.1038/nrm1547 [DOI] [PubMed] [Google Scholar]
- Quintanal-Villalonga A, Chan JM, Yu HA, Pe'er D, Sawyers CL, Sen T, Rudin CM. 2020. Lineage plasticity in cancer: a shared pathway of therapeutic resistance. Nat Rev Clin Oncol 17: 360–371. 10.1038/s41571-020-0340-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM. 2012. Vismodegib. Clin Cancer Res 18: 3218–3222. 10.1158/1078-0432.CCR-12-0568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Durinck S, Stawiski EW, Poirier JT, Modrusan Z, Shames DS, Bergbower EA, Guan Y, Shin J, Guillory J, et al. 2012. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet 44: 1111–1116. 10.1038/ng.2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Poirier JT, Byers LA, Dive C, Dowlati A, George J, Heymach JV, Johnson JE, Lehman JM, MacPherson D, et al. 2019. Molecular subtypes of small cell lung cancer: a synthesis of human and mouse model data. Nat Rev Cancer 19: 289–297. 10.1038/s41568-019-0133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Awad MM, Navarro A, Gottfried M, Peters S, Csőszi T, Cheema PK, Rodriguez-Abreu D, Wollner M, Yang JC, et al. 2020. Pembrolizumab or placebo plus etoposide and platinum as first-line therapy for extensive-stage small-cell lung cancer: randomized, double-blind, phase III Keynote-604 study. J Clin Oncol 38: 2369–2379. 10.1200/JCO.20.00793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudin CM, Brambilla E, Faivre-Finn C, Sage J. 2021. Small-cell lung cancer. Nat Rev Dis Primers 7: 3. 10.1038/s41572-020-00235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11: 783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen T, Tong P, Diao L, Li L, Fan Y, Hoff J, Heymach JV, Wang J, Byers LA. 2017a. Targeting AXL and mTOR pathway overcomes primary and acquired resistance to WEE1 inhibition in small-cell lung cancer. Clin Cancer Res 23: 6239–6253. 10.1158/1078-0432.CCR-17-1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen T, Tong P, Stewart CA, Cristea S, Valliani A, Shames DS, Redwood AB, Fan YH, Li L, Glisson BS, et al. 2017b. CHK1 inhibition in small-cell lung cancer produces single-agent activity in biomarker-defined disease subsets and combination activity with cisplatin or olaparib. Cancer Res 77: 3870–3884. 10.1158/0008-5472.CAN-16-3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458: 732–736. 10.1038/nature07884 [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland KD, Proost N, Brouns I, Adriaensen D, Song JY, Berns A. 2011. Cell of origin of small cell lung cancer: inactivation of Trp53 and Rb1 in distinct cell types of adult mouse lung. Cancer Cell 19: 754–764. 10.1016/j.ccr.2011.04.019 [DOI] [PubMed] [Google Scholar]
- Swords RT, Kelly KR, Smith PG, Garnsey JJ, Mahalingam D, Medina E, Oberheu K, Padmanabhan S, O'Dwyer M, Nawrocki ST, et al. 2010. Inhibition of NEDD8-activating enzyme: a novel approach for the treatment of acute myeloid leukemia. Blood 115: 3796–3800. 10.1182/blood-2009-11-254862 [DOI] [PubMed] [Google Scholar]
- Swords RT, Coutre S, Maris MB, Zeidner JF, Foran JM, Cruz J, Erba HP, Berdeja JG, Tam W, Vardhanabhuti S, et al. 2018. Pevonedistat, a first-in-class NEDD8-activating enzyme inhibitor, combined with azacitidine in patients with AML. Blood 131: 1415–1424. 10.1182/blood-2017-09-805895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Si Y, Yu H, Zhang L, Xie P, Jiang W. 2017. MLN4924 (Pevonedistat), a protein neddylation inhibitor, suppresses proliferation and migration of human clear cell renal cell carcinoma. Sci Rep 7: 5599. 10.1038/s41598-017-06098-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth JI, Yang L, Dahl R, Petroski MD. 2012. A gatekeeper residue for NEDD8-activating enzyme inhibition by MLN4924. Cell Rep 1: 309–316. 10.1016/j.celrep.2012.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Schatz MC. 2009. Optimizing data intensive GPGPU computations for DNA sequence alignment. Parallel Comput 35: 429–440. 10.1016/j.parco.2009.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. 2013. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31: 46–53. 10.1038/nbt.2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderdys V, Allak A, Guessous F, Benamar M, Read PW, Jameson MJ, Abbas T. 2018. The neddylation inhibitor pevonedistat (MLN4924) suppresses and radiosensitizes head and neck squamous carcinoma cells and tumors. Mol Cancer Ther 17: 368–380. 10.1158/1535-7163.MCT-17-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM. 2015. Identification and characterization of essential genes in the human genome. Science 350: 1096–1101. 10.1126/science.aac7041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW. 2003. The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286. 10.1146/annurev.cellbio.19.111301.112449 [DOI] [PubMed] [Google Scholar]
- Wei D, Li H, Yu J, Sebolt JT, Zhao L, Lawrence TS, Smith PG, Morgan MA, Sun Y. 2012. Radiosensitization of human pancreatic cancer cells by MLN4924, an investigational NEDD8-activating enzyme inhibitor. Cancer Res 72: 282–293. 10.1158/0008-5472.CAN-11-2866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooten DJ, Groves SM, Tyson DR, Liu Q, Lim JS, Albert R, Lopez CF, Sage J, Quaranta V. 2019. Systems-level network modeling of small cell lung cancer subtypes identifies master regulators and destabilizers. PLoS Comput Biol 15: e1007343. 10.1371/journal.pcbi.1007343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xirodimas DP. 2008. Novel substrates and functions for the ubiquitin-like molecule NEDD8. Biochem Soc Trans 36: 802–806. 10.1042/BST0360802 [DOI] [PubMed] [Google Scholar]
- Xu GW, Toth JI, da Silva SR, Paiva SL, Lukkarila JL, Hurren R, Maclean N, Sukhai MA, Bhattacharjee RN, Goard CA, et al. 2014. Mutations in UBA3 confer resistance to the NEDD8-activating enzyme inhibitor MLN4924 in human leukemic cells. PLoS One 9: e93530. 10.1371/journal.pone.0093530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Girard L, Zhang YA, Haruki T, Papari-Zareei M, Stastny V, Ghayee HK, Pacak K, Oliver TG, Minna JD, et al. 2018. Small cell lung cancer tumors and preclinical models display heterogeneity of neuroendocrine phenotypes. Transl Lung Cancer Res 7: 32–49. 10.21037/tlcr.2018.02.02 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Iqbal NJ, Sukrithan V, Nicholas C, Xue Y, Yu C, Locker J, Zou J, Schwartz EL, Zhu L. 2020. Targeted inhibition of the E3 ligase SCF(Skp2/Cks1) has antitumor activity in RB1-deficient human and mouse small-cell lung cancer. Cancer Res 80: 2355–2367. 10.1158/0008-5472.CAN-19-2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Liu D, Fallon JT, Zhong M. 2015. Distinct genetic alterations in small cell carcinoma from different anatomic sites. Exp Hematol Oncol 4: 2. 10.1186/2162-3619-4-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.