In this review, Hanna and Kelsey discuss what is known about the underlying mechanisms of establishing and maintaining allelic epigenetic marks, the role of endogenous retroviruses, and the potential conservation of canonical and noncanonical imprinting in mice and humans.
Keywords: chromatin, DNA methylation, endogenous retroviruses, epigenetics, genomic imprinting
Abstract
Genomic imprinting is the monoallelic expression of a gene based on parent of origin and is a consequence of differential epigenetic marking between the male and female germlines. Canonically, genomic imprinting is mediated by allelic DNA methylation. However, recently it has been shown that maternal H3K27me3 can result in DNA methylation-independent imprinting, termed “noncanonical imprinting.” In this review, we compare and contrast what is currently known about the underlying mechanisms, the role of endogenous retroviral elements, and the conservation of canonical and noncanonical genomic imprinting.
Introduction to genomic imprinting
Genomic imprinting is the monoallelic expression of a gene based on parent of origin. Imprinted genes are essential for fetal and placental growth and development. It is hypothesized that imprinting arose in placental mammals due to the conflict between maternal and paternal genomes in the fetus to regulate maternal resources during and immediately after pregnancy, with maternal imprints repressing fetal growth while paternal imprints promote it (Moore and Haig 1991). To date, there are several examples of imprinted genes that fit this model, including key regulators of fetal growth such as the insulin growth factor 2 (IGF2) and its receptor IGF2R, which are reciprocally imprinted (DeChiara et al. 1990; Barlow et al. 1991; DeChiara et al. 1991; Weksberg et al. 1993).
Shortly after the discovery of the first imprinted genes, it was shown that imprinted gene expression was regulated by allelic epigenetic marks, in particular repressive DNA methylation, inherited from the parental germline (Bartolomei et al. 1993; Brandeis et al. 1993; Ferguson-Smith et al. 1993; Li et al. 1993). This canonical form of imprinting has since been characterized across mammals and is highly conserved at a number of imprinted gene clusters. Imprinted genes and their regulatory features have been most extensively characterized in the mouse and human genomes, and genome-wide screens have identified not only species-specific but also tissue-specific imprinting. It was recently demonstrated that several placental-specific imprinted genes in mice are in fact regulated by an alternative epigenetic mechanism, histone 3 lysine 27 trimethylation (H3K27me3) inherited from the maternal germline. This form of DNA methylation-independent imprinting has been termed “noncanonical” imprinting. Despite its recent discovery, noncanonical imprinting has already been shown to be distinct in its genomic characteristics and underlying mechanisms from canonical imprinting, opening up a new field of study. In this review, we discuss what is known about the underlying mechanisms of establishing and maintaining allelic epigenetic marks, the role of endogenous retroviruses, and the potential conservation of canonical and noncanonical imprinting in mice and humans (summarized in Table 1).
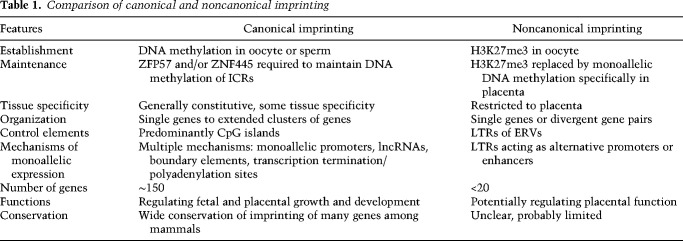
Table 1.
Comparison of canonical and noncanonical imprinting
Mechanisms of canonical imprinting
Gametic DNA methylation differences
The major driver of genomic imprinting has long been recognized as DNA methylation, specifically differences in methylation between the oocyte and sperm at imprinting control regions (ICRs). An ICR is the discrete genomic element that is necessary to orchestrate in cis the monoallelic expression of single or multiple imprinted genes within a domain (Spahn and Barlow 2003). ICRs coincide with germline differentially methylated regions (gDMRs), and most ICRs correspond to promoter CpG islands that acquire methylation in the female germline (Schulz et al. 2010).
De novo methylation in germ cells requires the methyltransferase DNMT3A together with the catalytically inactive cofactor DNMT3L (Bourc'his et al. 2001; Hata et al. 2002; Bourc'his and Bestor 2004; Kaneda et al. 2004; Smallwood et al. 2011), which are recruited to an appropriate underlying histone modification landscape. In the oocyte, DNA methylation is almost exclusively restricted to transcribed gene bodies (Kobayashi et al. 2012). The widespread use of oocyte-specific alternative transcription start sites means that the majority of maternal ICRs are spanned by transcription (Fig. 1; Chotalia et al. 2009; Veselovska et al. 2015; Singh et al. 2017). The establishment of DNA methylation at maternal ICRs is a consequence of acquiring a permissive chromatin state for the recruitment of DNMTs. Loss of histone 3 lysine 4 dimethylation (H3K4me2) at intragenic CpG islands is catalyzed by the transcription-coupled lysine demethylase KDM1B (Ciccone et al. 2009; Stewart et al. 2015; Veselovska et al. 2015), and deposition of H3K36me2 and H3K36me3 over transcribed regions by the histone lysine methyltransferase SETD2 (Xu et al. 2019; Shirane et al. 2020). Conversely, sperm is highly methylated throughout much of the genome, a pattern that is conferred by DNMT3A and DNMT3L (Bourc'his and Bestor 2004; Kaneda et al. 2004), with the addition of DNMT3C in rodents (Barau et al. 2016). Unlike the oocyte, the deposition of DNA methylation in spermatogenesis is not dependent on H3K36me3, but rather H3K36me2, which shows broad genomic distribution through the activity of methyltransferase NSD1 (Shirane et al. 2020).
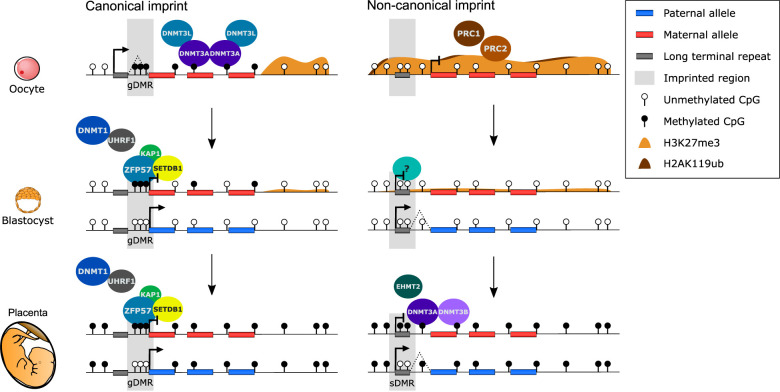
Figure 1.
Mechanisms of establishment and maintenance of maternal canonical and noncanonical imprinting. (Left) Canonical imprinting: DNA methylation is targeted to transcribed gene bodies, including canonical imprinted gDMRs, in oogenesis by tetramers of DNMT3A and DNMT3L. There is widespread usage of long terminal repeats (LTRs) as alternative upstream promoters in the oocyte. In the preimplantation and postimplantation embryo, a complex of ZFP57 (or ZNF445), TRIM28 (KAP1), and H3K9 methyltransferase SETDB1 localizes to gDMRs recruiting DNMT1 to maintain DNA methylation on the maternal allele. In the postimplantation embryo, the gDMR is present in the fetus and placenta, enabling imprinted gene expression of a single gene or cluster of genes. (Right) Noncanonical imprinting: H3K27me3 is established by PRC2, which is in part dependent on PRC1 ubiquitination of H2AK119, across untranscribed regions in oogenesis, including noncanonically imprinted LTRs. In the preimplantation embryo, maternal H3K27me3 is progressively lost genome-wide, and whether an unknown factor is required to mark the maternal allele of noncanonically imprinted LTRs remains unclear. In the postimplantation embryo, noncanonical imprints become placental specific, acquiring DNA methylation on the maternal allele, creating secondary DMRs (sDMRs), in placental and extraembryonic cell types. In the fetus (not shown), the noncanonically imprinted LTRs become biallelically methylated. The acquisition of DNA methylation in postimplantation development at these domains is dependent on EHMT2 activity, through either the deposition of H3K9me2 or post-translational modification of proteins integral for de novo DNMT activity.
Thus, it appears that there is no mechanism of de novo methylation in the germline specifically targeted to imprinted loci per se. Rather, the distinctive dependence of the de novo DNMTs on H3K36 methylation in the oocyte and sperm results in dimorphic DNA methylation landscapes, providing the opportunity for imprinting to emerge at gDMRs. Consequently, locus-specific differences in gamete methylation between species is one mechanism that enables species-specific imprinting to arise (Brind'Amour et al. 2018).
Postfertilization maintenance mechanisms
While distinct patterns of DNA methylation in the egg and sperm are the prerequisite for imprinting, gametic methylation differences are far more extensive than the number of imprinted loci; for example, there are ∼2000 CpG islands highly methylated in oocytes but not sperm (Kobayashi et al. 2012). Therefore, the maintenance of gamete-derived methylation in the embryo is critical in specifying the number of persistent gDMRs and, consequently, the number of imprinted loci. The discovery of the involvement of the zinc finger protein ZFP57 demonstrated that imprint maintenance relies on sequence-specific factors (Li et al. 2008; Mackay et al. 2008). ZFP57 is a member of the large family of Krüppel-associated box (KRAB)-containing zinc finger proteins (ZFPs) that provide DNA sequence binding specificity to the KRAB repressor complex. ZFP57 binds a CpG-containing hexanucleotide motif present in multiple copies in most ICRs (Quenneville et al. 2011; Strogantsev et al. 2015; Anvar et al. 2016) and, critically, binds the motif when the central CpG is methylated. ZFP57 recruits cofactors TRIM28 (KAP1) and SETDB1, targeting H3K9me3 to the locus, which in turn can be bound by the UHRF1 and DNMT1 complex following DNA replication, enabling maintenance methylation of the newly replicated DNA and the reinstatement of symmetric CpG methylation (Fig. 1; Sharif et al. 2007; Quenneville et al. 2011; Liu et al. 2013; Ming et al. 2020).
A second KRAB-ZFP, ZNF445, was recently identified as an alternative imprinting maintenance factor on the basis of its genomic binding profile in human and mouse embryonic stem cells, DNA methylation-dependent binding, and expression in human oocytes (Monteagudo-Sánchez et al. 2019; Takahashi et al. 2019). In mice, the two ZFPs both bind all imprinted DMRs and exhibit some functional redundancy, except for the Peg10 DMR, raising the possibility that additional ZFPs may play a role in protecting imprints. Notably, based on differences in the severity of the phenotypes and gDMR methylation defects in zygotic mutants, ZFP57 appears to be the predominant methylation protective factor in mice (Takahashi et al. 2019). On the other hand, its greater evolutionary conservation and expression profile give more prominence to ZNF445 in maintenance of imprints in humans.
Although most germline differentially methylated CpG islands lose methylation during the phase of reprogramming in the preimplantation embryo (Smallwood et al. 2011; Kobayashi et al. 2012), some enjoy a transitory imprinted status before their methylation is erased or overwritten, a phenomenon referred to as transient imprinting (Proudhon et al. 2012). The significance of transient imprints is not fully understood; strikingly, however, their influence can outlast their very limited differential methylation status. In the case of the Gpr1/Zdbf2 locus, monoallelic transcription of the long noncoding RNA (lncRNA) Liz from the Gpr1 gDMR sets up persistent allelic differences in the embryo postimplantation (a secondary DMR [sDMR] and allele-specific enrichment of H3K27me3 upstream of the Zdbf2 promoter), while the gDMR becomes biallelically methylated (Duffie et al. 2014). This early epigenetic programming event results in a long-term phenotypic effect: Mice deficient in Liz expression develop normally but do not activate Zdbf2 in the brain postnatally and exhibit growth retardation (Greenberg et al. 2017).
Imprinted long noncoding RNAs and cis-regulation of clusters
ICRs are incredibly potent genomic elements because they can specify the imprinted monoallelic expression of genes over tens of kilobases, or even megabases, of DNA. In the case of the Airn/Igf2r domain in the mouse placenta, imprinted expression extends to over a dozen genes spanning a genomic interval of >10 Mb (Andergassen et al. 2017). Much of the domain is also monoallelically enriched in H3K27me3 in extraembryonic tissues (Andergassen et al. 2019; Hanna et al. 2019). Deletion of the ICR (which is the promoter of the Airn lncRNA), or prevention of de novo methylation of the ICR in the oocyte, abolishes the imprinted status of the entire domain (Andergassen et al. 2017; Hanna et al. 2019). Just how an ICR enforces monoallelic transcription over such extended domains has been the focus of attention for many years. It is likely that a combination of mechanisms emanating from the ICR applies over such large domains, especially if there is sufficient evolutionary pressure to select imprinting of multiple genes; moreover, different mechanisms may have evolved at different imprinted clusters. For example, monoallelic silencing of Igf2r depends on interference of the promoter by Airn transcription, rather than the lncRNA itself (Latos et al. 2012). On the other hand, for genes in the domain not overlapped by Airn transcription, other mechanisms must operate. By extension of the model of promoter interference, one possibility is that monoallelic Airn transcription through essential placenta-specific enhancers represses the distal genes that depend on these enhancers. However, this model has been discounted by genetic experiments deleting the entire Airn transcribed region (Andergassen et al. 2019). This finding returns to the frame a long-established model that lncRNAs bind and recruit repressive chromatin modifiers, such as G9a (EHMT2) or polycomb repressor complexes (PRCs), to imprinted domains (Nagano et al. 2008; Terranova et al. 2008). For the megabase-scale imprinted domains, parallels with the lncRNA Xist and X-chromosome inactivation re-emerge (Khamlichi and Feil 2018). Molecular investigations in trophoblast and embryonic stem cells have demonstrated that 3D folding is essential to bring CpG islands within close proximity to ICRs at the Airn and Kcnq1ot1 loci in cis, enabling PRCs to facilitate allelic silencing (Schertzer et al. 2019). Nevertheless, what features are critical for determining the extent of these imprinted domains and how exactly imprinted lncRNAs function remain to be fully elucidated.
Noncanonical imprinting
Discovery and properties
For many years, we have understood that DNA methylation is central to regulating imprinting; however, there have been examples of imprinted loci that appeared not to be controlled by DNA methylation, which compelled us to entertain alternative mechanisms of imprinting. For example, there were no detectable promoter DMRs at the placenta-specifically imprinted genes Gab1 and Sfmbt2; in addition, their imprinting is retained even in conceptuses lacking oocyte-derived DNA methylation (Okae et al. 2012). An explanation for these anomalies has emerged with the discovery of a parallel mechanism of imprinting, which has been termed “noncanonical” imprinting.
Work that profiled DNase I-hypersensitive sites (DHSs) separately in isolated maternal and paternal pronuclei of mouse zygotes found that a subset of paternal-specific DHSs was not associated with known imprinted genes but with genes with paternal allele-biased expression (Inoue et al. 2017a). This was further evidenced from analysis of gynogenetic or androgenetic preimplantation embryos, as well as reciprocal hybrids. These genes do not map into regions of DNA methylation in oocytes, and their imprinting is maintained when oocytes are deprived of DNA methylation (Chen et al. 2019; Hanna et al. 2019). Critically, forced expression of the H3K27me3 demethylase KDM6B in zygotes abrogates their imprinted status (Inoue et al. 2017a). Genetic confirmation of the role of H3K27me3 has subsequently been obtained by conditional deletion of Eed, which encodes an essential component of the PRC2, in oocytes (Inoue et al. 2018).
An intriguing property of noncanonical imprinting is its tissue specificity. Although multiple genes with paternal allele bias in preimplantation embryos were identified, all (with the exception of Slc38a4) became biallelically expressed or repressed in the postimplantation epiblast (which gives rise to the embryonic lineages), while a subset retained imprinted expression in extraembryonic lineages and the placenta (Inoue et al. 2017a). This subset included Gab1 and Sfmbt2 previously identified as being independent of oocyte-derived methylation for their imprinting (Okae et al. 2012).
Functions
Canonical imprinted genes participate in a range of developmental and physiological adaptations in mammals, notably in placental and fetal growth but also the central control of metabolism and some cognitive behaviors. It is too soon to conclude the functional domains of noncanonical imprinting, but the restriction of this form of imprinting to the placenta would implicate involvement in fetal growth control or placental endocrine adaptations to pregnancy. As a measure of its relative importance, global elimination of canonical imprinting by prevention of de novo DNA methylation in the female germline is incompatible with embryo development beyond mid gestation (Bourc'his et al. 2001). In contrast, abrogation of noncanonical imprinting by oocyte-specific ablation of Eed does allow survival to term but with reduced litter size, indicating embryonic losses (Inoue et al. 2018; Prokopuk et al. 2018).
A major role for oocyte-derived H3K27me3 has been identified in X-chromosome regulation. In rodents, the paternal X chromosome in female preimplantation embryos is silenced as a means of dosage compensation. This imprinted inactivation state persists in the extraembryonic lineages postimplantation, while in the embryo proper, both X chromosomes are reactivated before undergoing random X-chromosome inactivation. The epigenetic mark in the oocyte that suppresses the Xist locus to ensure activity of the maternal X chromosome in cleavage embryos has remained elusive, after reports that maternal DNA methylation was dispensable (Chiba et al. 2008). Recently, it was demonstrated that imprinted X-chromosome inactivation depended on oocyte H3K27me3 (Inoue et al. 2017b). Accordingly, both male and female embryos display aberrant inactivation of the maternal X chromosome upon oocyte-specific deletion of Eed, which could explain the excess of male embryo losses (Inoue et al. 2018). Because of this effect on X-chromosome dynamics, the global impact of perturbed noncanonical imprinting of autosomal genes is difficult to infer; in addition, there are discrepancies between studies. Oocyte-specific ablation of Eed is reported to result in reduced litter size and overgrowth of offspring but no sex ratio distortion (Prokopuk et al. 2018), or reduced litter size, deficit of males but ostensibly normal offspring (Inoue et al. 2018), while ablation of Ezh2 in oocytes, which encodes the H3K27 methyltransferase, causes substantial growth retardation (Erhardt et al. 2003).
The importance of noncanonical imprinting has been demonstrated in somatic cell nuclear transfer (SCNT). In this procedure, nuclei from somatic cells are reprogrammed in the egg cytoplasm, but the process is inefficient and many cloned embryos fail owing, in part, to placental abnormalities. Whereas donor cells of embryonic or adult origin should have normal canonical imprints, they lack imprinting of noncanonical imprinted genes (Okae et al. 2014; Matoba et al. 2018). In an attempt to mitigate against this lack of noncanonical imprinting, SCNT was performed with donor cells carrying heterozygous deletions for each of the Gab1, Sfmbt2, and Slc38a4 protein-coding genes to restore normal expression levels; however, none of the deletions prevented placental hyperplasia in clones. Strikingly, however, use of an allele that deleted the large miRNA cluster within an intron of Sfmbt2 did substantially ameliorate the placental phenotype (Inoue et al. 2020).
Apart from Sfmbt2, noncanonical imprinted genes with significant phenotypic effects when ablated include Slc38a4 (placental and fetal growth) (Matoba et al. 2019) and Gab1 (placental labyrinth hypoplasia) (Itoh et al. 2000). However, the expression of either gene is not limited to extraembryonic tissues, and they have significant domains of imprinted or nonimprinted expression in other tissues, so it is difficult to conclude how much of the respective knockout phenotypes relate to their noncanonical imprinting. Thus, further work is needed to definitively demonstrate the importance of noncanonically imprinted genes in placental development.
Establishment mechanism: targeting of H3K27me3 in the oocyte
In the oocyte, H3K27me3 is distributed in atypically broad domains and is anticorrelated with DNA methylation (Fig. 1; Zheng et al. 2016). Hence, H3K27me3 and any potential noncanonically imprinted domains are restricted to the untranscribed fraction of the genome. The mutual exclusivity of H3K27me3 and DNA methylation appears, at least in part, to be dependent on the establishment of H3K36me3 and DNA methylation across transcribed regions. Ablation of H3K36me3, and consequently DNA methylation, through the deletion of the H3K36me3 methyltransferase Setd2 causes the patterning of H3K27me3 to become widespread (Xu et al. 2019). H3K27me3 and the self-interacting polycomb-associated domains (PADs)—a unique 3D organization of the oocyte genome—are established during oogenesis through the action of PRC2 and PRC1, respectively (Inoue et al. 2018; Du et al. 2020). Recent evidence further suggests that PRC2 activity is, at least in part, dependent on PRC1 ubiquitination of H2AK119, which is colocalized with the broad domains of H3K27me3 (Fig. 1; Mei et al. 2021). PRC2 enzymatic activity is tightly regulated throughout oogenesis through the action of the germline-specific PRC2 cofactor EZHIP, which represses deposition of H3K27me3 in the late stages of oogenesis (Ragazzini et al. 2019). In the absence of EZHIP, H3K27me3 is dramatically increased in mature oocytes, and female fertility is impaired.
Notably, PADs are lost in MII oocytes and reset postfertilization in the two-cell embryo, and it has been suggested that maternal H3K27me3 may be critical for this resetting (Du et al. 2020). However, as with H3K27me3, PADs are progressively lost during preimplantation development (Zheng et al. 2016; Du et al. 2020). At typical polycomb targets (e.g., Hox gene clusters), it has become evident that PRC1 activity at the two-cell stage is critical for later targeting of PRC2, but depletion of PRC1 in the zygote had no impact on noncanonical imprinting, suggesting it does not play a role in propagating maternal allelic silencing (Chen et al. 2021). These data support the notion that polycomb-independent mechanisms may be required for the maintenance of noncanonically imprinted domains past the embryonic cleavage stages.
Maintenance mechanism: transition for H3K27me3 to DNA methylation control
What do we know about the mechanism of maintaining noncanonical imprints, and how does it differ from conventional DNA methylation-dependent imprinting? H3K27me3 is the mark inherited from oocytes that initially provides parental allele asymmetry by preventing establishment of DHSs and H3K4me3 on maternal alleles (Inoue et al. 2017a; Chen et al. 2019). However, H3K27me3 is reprogrammed in preimplantation embryos, resulting in very little genomic occupancy by the blastocyst stage (Zheng et al. 2016). DNA methylation is also reprogrammed globally in preimplantation embryos, but ICRs crucially are exempt from this loss. In contrast, noncanonical imprinted genes do not retain allelic H3K27me3 (Fig. 1), including in the extraembryonic tissues in which their imprinting persists (Chen et al. 2019; Hanna et al. 2019). Instead, almost all of these genes acquire sDMRs selectively in extraembryonic tissues (Fig. 1), such that their persistent imprinting is maintained by DNA methylation (Chen et al. 2019; Hanna et al. 2019). Recent evidence suggests that EHMT2 is critical in the establishment of sDMRs at noncanonically imprinted domains (Zeng et al. 2021), but it remains to be determined whether EHMT2-mediated H3K9me2 or histone-independent activities are required for DNA methylation establishment at these regions. Notably, the Sfmbt2 locus is an exception in retaining maternal allele enrichment for H3K27me3 and lacking an sDMR (Andergassen et al. 2019; Chen et al. 2019). An important clue in understanding noncanonical imprinting regulation has been the identification of the long terminal repeats (LTRs) of endogenous retroviruses (ERVs) as the candidate regulatory elements (Hanna et al. 2019), as discussed in more detail below. Potentially, these LTRs could mediate both the activity of these elements at preimplantation stages and the placenta-specific expression, possibly via different transcriptional regulators, but their precise roles need to be experimentally determined.
Endogenous retroviruses and genomic imprinting
ERVs are repetitive elements derived from retroviruses and comprise ∼10% of the mouse genome (Friedli and Trono 2015). Intact ERVs contain the retroviral genes necessary for viral replication, flanked by LTRs. However, the vast majority of ERVs in the genome have lost their viral genes through recombination between the complementary flanking LTRs and thus remain as “solo-LTRs” (Belshaw et al. 2007). LTRs have been frequently commandeered as cis-regulatory elements and significantly contribute to the gene regulatory landscape (Faulkner et al. 2009). LTR sequences can contain, or acquire through mutagenesis, sites for transcription factor binding, transcription initiation, splicing, and/or polyadenylation and thus can impact gene regulation in a multitude of ways (Thompson et al. 2016). Because of the mutagenic nature of retrotransposition events, intact ERVs are targeted for silencing by a rapidly evolving network of KRAB-ZFP proteins, which recruit H3K9me3 and DNA methylation (Matsui et al. 2010; Rowe et al. 2013; Ecco et al. 2016). However, solo LTRs often evade these silencing mechanisms and therefore have an increased likelihood of being co-opted into the gene regulatory landscape (Leung and Lorincz 2012). In particular, subclasses of solo LTRs appear to have benefitted from epigenetic reprogramming events in gametogenesis and early embryogenesis, acquiring essential roles in gene regulation during these developmental windows (Macfarlan et al. 2012; Chuong et al. 2013; Xie et al. 2013; Veselovska et al. 2015).
Shortly after the discovery of the first imprinted genes, Barlow (1993) proposed that genomic imprinting may have evolved as a host defense mechanism to silence viral genomic elements using DNA methylation. This hypothesis was based, in part, on work demonstrating that a transgene carrying an LTR acquired imprinting in gametogenesis, which persisted into the embryo (Chaillet et al. 1991). Since these early findings, there has been accumulating evidence that some aspects of genomic imprinting are linked to the silencing of LTRs and to transcriptional activity of solo-LTRs in the germline and placental trophoblast.
The setting of imprinted DNA methylation in the male and female germlines has been linked to ERVs. There are three imprinted gDMRs established in spermatogenesis; however, only the Rasgrf1 gDMR is attributable to an ERV. Expression of a solo-LTR (RMER4B) upstream of the Rasgrf1 gene generates small RNAs that enable targeting of repressive PIWI-piRNA to the locus, resulting in its de novo methylation in sperm (Fig. 2A; Watanabe et al. 2011). Conversely, there is a widespread role for ERVs in the establishment of maternal imprinted gDMRs in oogenesis. As previously discussed, maternal gDMRs are predominantly located at CpG island promoters. In oogenesis, de novo DNA methylation is almost exclusively targeted to actively transcribed gene bodies (Kobayashi et al. 2012; Veselovska et al. 2015); thus, for imprinted maternal gDMRs to become methylated, they must be intragenic in the oocyte. This is achieved, in part, through the occurrence of transcriptionally active LTRs upstream of canonical promoters (Fig. 2B; Veselovska et al. 2015; Brind'Amour et al. 2018). Ablation of transcription across maternal gDMRs in the oocyte results in a failure of these regions to gain DNA methylation (Chotalia et al. 2009; Veselovska et al. 2015; Bogutz et al. 2019). As the majority of intergenic regions, and consequently LTRs, are unmethylated in the oocyte genome, it has yet to be explored why this specific subset of LTRs, mostly malRs and ERVKs, are active in oogenesis.
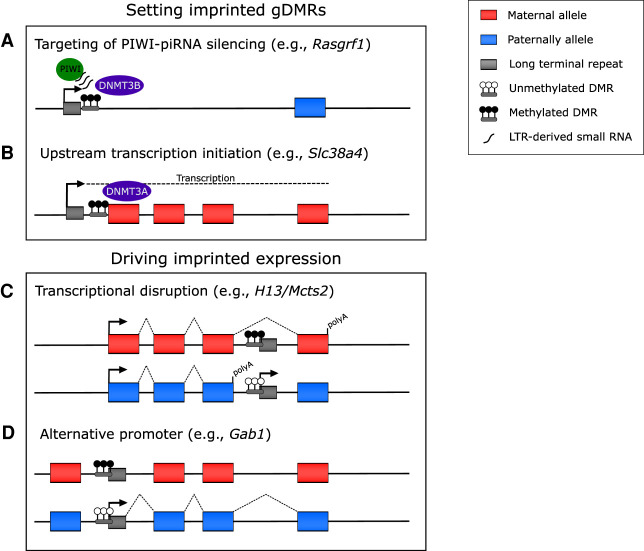
Figure 2.
Role for LTRs in genomic imprinting. Endogenous retroviral LTRs have been demonstrated to be essential in both the setting of imprinted differentially methylated regions (DMRs) in the germline (A,B) and in driving imprinting gene expression at a number of loci (C,D). (A) In spermatogenesis, expression of LTR-derived small RNAs upstream of Rasgrf1 is targeted by the piRNA-PIWI silencing pathway, which in turn recruits de novo methyltransferase DNMT3B to methylate the locus. (B) In oogenesis, the widespread occurrence of LTR-derived transcripts traversing canonical promoters results in their methylation via the recruitment of DNMT3A to sites of elongating transcription. (C) Within the intron of imprinted gene H13, an LTR-derived gene (Mcts2) harbors a maternal gDMR, resulting in imprinted expression of Mcts2 from the paternal allele. The allelic transcriptional activity of Mcts2 disrupts the transcriptional elongation of H13, causing its premature polyadenylation. (D) The allelic expression of several noncanonically imprinted genes is a consequence of LTRs acting as alternative promoters, forming chimeric transcripts with nearby genes.
Several imprinted loci contain ERVs that facilitate their imprinting in the embryo. For example, the imprinted gene H13 has an imprinted gDMR within its intron, which is a transcriptional start site for the retroviral-derived gene Mcts2. Mcts2 is expressed from the paternal allele, and this transcription is proposed to interfere with the transcriptional elongation of paternal H13, resulting in its premature truncation and polyadenylation (Fig. 2C; Wood et al. 2008). At a number of noncanonically imprinted loci, LTR-initiated transcripts are spliced on to nearby protein-coding or noncoding RNA genes, resulting in imprinted chimeric transcripts (Fig. 2D; Hanna et al. 2019). Consistent with LTRs demonstrating tissue-specific activity, noncanonically imprinted LTRs are exclusively expressed in extraembryonic tissues, including the placenta and visceral endoderm (Hanna et al. 2019).
Imprinted gDMRs have also co-opted the ERV silencing machinery, KRAB-ZFPs, to enable the protection and maintenance of monoallelic DNA methylation during developmental reprogramming (Li et al. 2008; Pathak and Feil 2018), as previously discussed. While the vast majority of gDMRs do not contain an identifiable ERV, each (with the exception of one) contains motifs that are recognized by ZFP57 and/or ZNF445 (Quenneville et al. 2011; Takahashi et al. 2019). Notably, ZFP57 binds not only imprinted gDMRs but also a number of ERVs throughout the genome (Shi et al. 2019). Despite using a common KRAB-ZFP, the underlying mechanisms silencing imprinted gDMRs and ERVs appear to be distinct; in the absence of ZFP57, imprinted gDMRs become derepressed, while ERVs remain silenced (Shi et al. 2019). Beyond ZFP57 and ZNF445, there are also KRAB-ZFPs that act in a locus-specific manner. ZFP568 is essential for the establishment of DNA methylation at a sDMR at the placental-specific promoter of Igf2 (Yang et al. 2017). Deletion of Zfp568 results in up-regulation of Igf2 and embryonic lethality, a phenotype that was partially rescued by deletion of Igf2 (Yang et al. 2017). As the mechanisms regulating the establishment of sDMRs are investigated further, we may discover additional roles for KRAB-ZFPs in targeting allelic de novo DNA methylation in the postimplantation embryo.
Overall, ERVs have contributed to the evolution of genomic imprinting in mammals by several distinct mechanisms. The ability of LTRs to direct tissue-specific transcription, taking advantage of epigenetic programming events, has permitted (1) the expression of LTR-derived small RNAs that allow targeting of silencing piRNA-PIWI in spermatogenesis, (2) the targeting of DNA methylation to LTR-initiated transcription units in oocytes, and (3) the expression of noncanonical imprinted chimeric transcripts in the preimplantation embryo and placenta. Furthermore, the exploitation of the KRAB-ZFPs by gDMRs has enabled protection of their epigenetic state through early embryonic reprogramming events. With the advent of in vivo CRISPR technologies and ultra-low-input sequencing approaches, it is now possible to study and functionally test epigenetic reprogramming events in detail; these investigations may offer exciting new insights into the roles for ERVs in regulating genomic imprinting.
Extent and conservation of imprinting
Genome-wide surveys of imprinting
Since the early days of imprinting, there have been continual efforts to identify imprinted genes in a systematic manner, using the best current methods in species with useful available genetic resources or for which embryo manipulations were feasible. In the last decade, these efforts have been dominated by next-generation sequencing approaches, such as applying RNA-seq to reciprocal hybrid mouse crosses to quantify parental biases in expression using allelic variants. Early application of RNA-seq to the mouse brain resulted in the identification of >1000 genes exhibiting parent of origin allelic expression (Gregg et al. 2010), far higher than prevailing estimates of the number of imprinted genes. Such findings led to intense discussion about technical and bioinformatic errors in allelic RNA-seq data and the need for validation by independent methods, as well as debate about what threshold should be applied for calling an imprinted state from a parental allele expression bias (DeVeale et al. 2012; Kelsey and Bartolomei 2012). Subsequent studies have revealed an apparently complex repertoire of “nongenetic” allele-specific expression in the developing brain beyond conventional imprinting (Huang et al. 2017).
The issue of when parental-biased expression qualifies as imprinting is complicated by tissue complexity where cells with imprinted and nonimprinted expression for a given gene may exist side by side. Single-cell RNA-seq should help resolve this issue but may be limited by the problem of allele dropout. In recent single-cell analysis of the mouse cortex, consistent monoallelic/biased expression was found in all cells of the major cell types, at least for known imprinted genes (Laukoter et al. 2020); but this may differ for uncharacterized imprinted genes or genes at the extremities of imprinted domains. In the placenta, there is the additional issue of maternal tissue contamination, which has been a recurrent problem that probably led to an overestimate of the number of maternally expressed imprinted loci and dictates additional measures for verification (Okae et al. 2012; Andergassen et al. 2017). Nevertheless, it is possible that we are approaching a final listing of imprinted genes in mice: A recent RNA-seq survey of 34 tissues and developmental stages concluded with 93 high-confidence imprinted genes, and although this included 17 novel imprinted loci, they all belonged to characterized imprinted domains (Andergassen et al. 2017).
In human studies, an inevitable limitation is the inability to engineer crosses, necessitating other strategies to distinguish bona fide imprinted effects from allele-specific expression caused by cis-acting genetic variants. Recent surveys have mined large RNA-seq data sets from human tissues, supported by genotyped pedigrees or trios, to provide parent of origin information. Such studies have generally found known imprinted genes to be monoallelically expressed in multiple tissues, and provide a resource of novel candidates, many of which tend to exhibit tissue-restricted monoallelic expression (Babak et al. 2015; Baran et al. 2015). However, RNA-seq analysis based on short-read sequencing can have difficulties in reporting allelic biases of isoforms from complex transcription units, and tissue heterogeneity may still limit the ability to detect cell type-specific imprinting.
Genome-wide DNA methylation approaches to imprinted gene identification in humans have exploited abnormalities in imprinting, such as hydatidiform mole, uniparental disomies, or triploid placenta samples (Hanna et al. 2016; Sanchez-Delgado et al. 2016) or allelic variants (Hamada et al. 2016). A recent study took advantage of the unique genetic resource of the Icelandic population by conducting whole-genome bisulfite sequencing on peripheral blood DNA from 285 individuals for which parent of origin phased haplotypes, as well as RNA-seq data, were available (Zink et al. 2018). Such screens provide the basis for a comprehensive account of imprinting in humans, reveal the imprinting landscape in the human genome with a resolution previously only obtained in mice, and identify cases of polymorphic imprinting.
LTRs and imprinting in humans
Many canonically imprinted gene clusters are conserved between mice and humans in both epigenetic regulation and synteny. This conservation extends to the role of LTR-derived transcripts in targeting DNA methylation in the oocyte and the requirement for ZFP57/ZNF445 in maintaining imprinted DNA methylation throughout early embryonic development. Nevertheless, some key differences have already been observed in the mechanisms of LTR-associated imprinting in humans, and the extent to which noncanonical imprinting may act on LTRs expressed in the human placenta has yet to be explored.
The genome of the human oocyte has more than twice as many methylated regions as the mouse oocyte, yet both exhibit DNA methylation largely restricted to transcribed domains (Okae et al. 2014; Hanna et al. 2018). LTRs contribute significantly in forming this transcriptional landscape, with >15% of all transcripts in the mouse oocyte initiating from an LTR (Veselovska et al. 2015). While LTRs contribute to a smaller proportion of the human oocyte transcriptome (Brind'Amour et al. 2018), LTR-initiated transcription mediates at least 15% of human-specific maternal gDMRs (Bogutz et al. 2019), demonstrating that LTR transcription in oocytes is a key player in the evolution of genomic imprinting.
A notable difference between mice and humans is the timing and necessity for KRAB-ZFPs in protecting imprinted gDMRs. Unlike in mice, ZFP57 is not expressed in the human oocyte, meaning that the necessity for ZFP57 to protect imprinted DNA methylation must be later in development (Okae et al. 2014). Notably, ZFP57 is also not expressed in bovine or porcine oocytes (Ivanova et al. 2020). Furthermore, mutations in ZFP57 result in incomplete loss of imprinting (Bak et al. 2016), which may in part be explained by a predominant role for other ZFPs, such as ZNF445. ZNF445 appears to be complementary to ZFP57 in protecting human imprinted gDMRs and is highly expressed in human oocytes (Takahashi et al. 2019). However, it should also be noted that in the preimplantation human embryo, maternally inherited DNA methylation is not passively lost nearly to the extent it is in mice (Okae et al. 2014). Intriguingly, this persistence of inherited maternal DNA methylation appears particularly to affect the placenta, which exhibits hundreds of genomic loci with a maternal bias in DNA methylation (Hamada et al. 2016; Hanna et al. 2016; Sanchez-Delgado et al. 2016). Whether these placental-specific imprinted DMRs contain ZFP57/ZNF445 binding sites is currently unknown, but their polymorphic imprinting in the human population (Hanna et al. 2016; Sanchez-Delgado et al. 2016) suggests that, if present, there may be sequence variants in associated motifs.
Recurrent evolution of noncanonical imprinted domains?
Given the evidence that noncanonical imprinting may be regulated by LTRs, and especially because the elements implicated in mice are rodent-specific, there is little expectation that the same genes will be imprinted in different mammalian lineages, although the process may be conserved across placental mammals. Consistent with this prediction, GAB1 and SFMBT2 are not imprinted in the human placenta (Okae et al. 2012); moreover, GAB1 is not within a domain of H3K27me3 in human oocytes (Xia et al. 2019). Slc38a4 is an interesting case because, in mice, the gene seems to combine canonical and noncanonical modes of imprinting. The promoter of Slc38a4 contains a conventional maternal gDMR that depends on transcription from upstream MaLR elements in oocytes (Bogutz et al. 2019). Notably, the maintenance of allelic DNA methylation at the Slc38a4 gDMR is uniquely dependent on H3K9 dimethyltransferase EHMT2 in the embryo (Auclair et al. 2016). Furthermore, Slc38a4 is subject to noncanonical imprinted regulation in the placenta (Okae et al. 2012; Inoue et al. 2017a), possibly from upstream LTRs acting as enhancers that are within an H3K27me3-marked domain in oocytes (Hanna et al. 2019). SLC38A4 is not imprinted in the human placenta (G Kelsey, unpubl.) but is reported to be imprinted in porcine placenta and polymorphically so in bovine placenta (Bischoff et al. 2009; Xu et al. 2018). The mechanism of imprinting in these species is not known. These observations suggest a selective pressure to imprint this gene and possibly independent evolutionary events. Slc38a4 encodes the system A neutral amino acid transporter SNAT4, and mice lacking Slc38a4 expression exhibit placental and fetal growth restriction (Matoba et al. 2019). The situation for the gene Smoc1 is ambiguous: It is noncanonically imprinted in mice, and although imprinting of SMOC1 has been described in human fibroblasts, the imprinted allele is opposite to that of mice (Santoni et al. 2017).
Noncanonical imprinting: conserved or not?
An approach to evaluate the existence of H3K27me3-determined noncanonical imprinting in humans has been to examine the genomic distribution of H3K27me3 in human gametes or preimplantation embryos. Two studies have done this, but come to different conclusions. By performing CUT&RUN for H3K27me3 in human morulae, combined with whole-genome or exome sequencing of donor material to identify allelic variants, Zhang et al. (2019) identified regions of preferential maternal allele enrichment for H3K27me3. Furthermore, RNA-seq analysis identified genes with paternal allele-biased expression unlinked to sites of oocyte DNA methylation and therefore unlikely to be conventional imprinted genes. For one of these candidates, evidence of allelically enriched H3K27me3 was obtained. This could represent the type of transient imprinting observed in mouse preimplantation embryos, and it will be important to determine whether allelically biased expression persists in the placenta, as in noncanonical imprinting in mice. In contrast, in their survey of histone modifications in human oocytes and cleavage embryos, Xia et al. (2019) report that domains of H3K27me3 in oocytes are rapidly lost after fertilization and absent in the eight-cell stage, including at most of the above candidate genes, leading them to conclude that noncanonical imprinting is unlikely to exist in humans. Consistent with the absence of imprinted X-inactivation in humans (Migeon and Do 1979; Looijenga et al. 1999; Moreira de Mello et al. 2010), the XIST locus is devoid of H3K27me3 in preimplantation embryos (Xia et al. 2019).
Because noncanonical imprinted regions in mice transition from H3K27me3 in the oocyte to become imprinted sDMRs in placenta, we sought an alternative approach to identify potential noncanonically imprinted DMRs in humans. Taking advantage of candidate maternal placental DMRs identified previously using the Illumina HumanMethylation450 (450K) array (Hanna et al. 2016), we selected those that were unmethylated (<25%) and enriched for H3K27me3 (more than −0.75 log2RPKM corrected for DMR length) in human oocytes using publicly available data (Okae et al. 2014; Xia et al. 2019), resulting in 65 putative maternal “noncanonical” imprinted DMRs (Fig. 3A,B; Supplemental Table S1). We evaluated allelic DNA methylation by allelically mapping whole-genome bisulfite sequencing data from first trimester placental trophoblasts (Hamada et al. 2016), using the dbSNP common SNP annotation (http://www.ncbi.nlm.nih.gov/SNP) and the SNPsplit mapping program (Krueger and Andrews 2016). Allelic analyses confirmed a >10% allelic difference in DNA methylation at 16/26 of informative DMRs (Fig. 3C), although parent of origin was not assessed (Supplemental Table S1). Together, these data provide preliminary evidence that noncanonical imprinting may also exist in the human genome, while further work will be needed to validate candidate loci and demonstrate whether these DMRs regulate allelic expression. Notably, the putative noncanonical imprinted DMRs, compared with the Illumina 450K array probes, were significantly enriched for CpG islands and SINEs (Fig. 3D), rather than LTRs as in mice. This suggests that while the mechanism may be conserved between species, the underlying regulatory features are likely not. This difference may reflect the dissimilarities in the prevalence of repetitive elements between the mouse and human genomes (Thomas et al. 2003). However, it is important to highlight that repetitive elements in general are underrepresented on the Illumina 450K array (Fig. 3D); therefore, it is likely that loci have been missed by this approach.
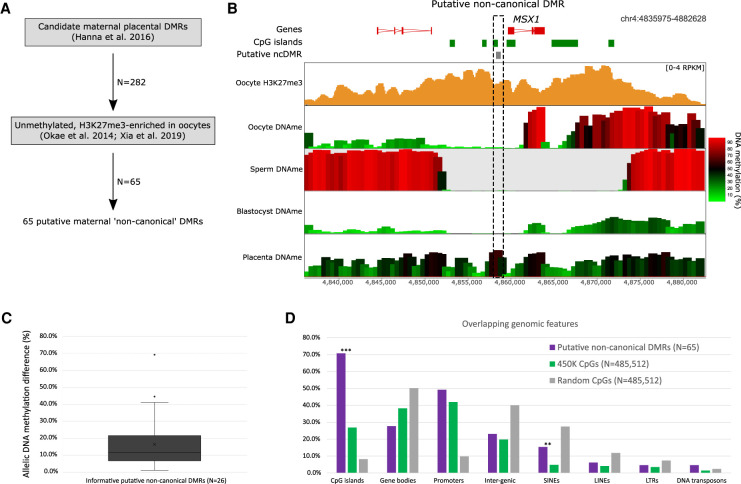
Figure 3.
Identification of putative noncanonical imprinted DMRs in the human placenta. Using the epigenetic patterning characteristic of noncanonical DMRs in mice, we investigated publicly available data to identify putative maternal “noncanonical” DMRs in humans. (A) Previously reported candidate maternal DMRs from placenta were evaluated for oocyte DNA methylation and H3K27me3, selecting those domains that were unmethylated and enriched for H3K27me3 in oocytes (N = 65). (B) Screenshot of putative noncanonically imprinted DMR upstream of the MSX1 gene. Enrichment for H3K27me3 in human oocytes is shown using running 500-bp windows, with a 100-bp step, quantitated as RPKM. DNA methylation in human oocytes, sperm, blastocyst, and first trimester placental trophoblast is shown using 1-kb running windows, with a 500-bp step. (C) The box plot shows the allelic difference in DNA methylation at informative putative noncanonical imprinted genes (N = 26). Informative DMRs were defined as those with at least three CpGs covered by at least two reads on each allele. (D) Overlapping genomic features were compared between putative noncanonical DMRs (N = 65) and CpGs on the Illumina 450K array (N = 485,512) using the χ2 statistic. P-value significance was adjusted for multiple comparisons using Bonferroni correction. (**) P < 0.001, (***) P < 0.0001. A random subset of genomic CpGs is shown in gray for context (N = 485,512).
The pursuit of comprehensively identifying human imprinted domains continues to present challenges, including the necessity for deep sequencing of genomics data sets, the scarcity of informative SNPs, obtaining parent of origin information for relevant SNPs, and the cellular heterogeneity of human samples, as previously discussed. The initial identification and characterization of noncanonical imprinting in mice emphasizes the value in using animal models to direct our approaches for investigating molecular and epigenetic phenomena in human development.
Supplementary Material
Acknowledgments
We thank Felix Krueger in the Bioinformatics group at the Babraham Institute for processing and allelically mapping the publicly available data sets used in this review, sourced from either Gene Expression Omnibus (GEO) or the DNA Data Bank of Japan (DDBJ) (Hanna et al. 2016 [GSE74738], Okae et al. 2014 [DRP002710], Xia et al. 2019 [GSE124718], Hamada et al. 2016 [JGAS00000000038]). Work in G.K.’s laboratory is supported by grants from the UK Biotechnology and Biological Sciences Research Council (BBS/E/B/000C0423) and Medical Research Council (MR/S000437/1); C.W.H. is supported by a Next-Generation Fellowship from the Centre for Trophoblast Research.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.348422.121.
Freely available online through the Genes & Development Open Access option.
Competing interest statement
The authors declare no competing interests.
References
- Andergassen D, Dotter CP, Wenzel D, Sigl V, Bammer PC, Muckenhuber M, Mayer D, Kulinski TM, Theussl HC, Penninger JM, et al. 2017. Mapping the mouse allelome reveals tissue-specific regulation of allelic expression. Elife 6: e25125. 10.7554/eLife.25125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andergassen D, Muckenhuber M, Bammer PC, Kulinski TM, Theussl HC, Shimizu T, Penninger JM, Pauler FM, Hudson QJ. 2019. The Airn lncRNA does not require any DNA elements within its locus to silence distant imprinted genes. PLoS Genet 15: e1008268. 10.1371/journal.pgen.1008268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anvar Z, Cammisa M, Riso V, Baglivo I, Kukreja H, Sparago A, Girardot M, Lad S, De Feis I, Cerrato F, et al. 2016. ZFP57 recognizes multiple and closely spaced sequence motif variants to maintain repressive epigenetic marks in mouse embryonic stem cells. Nucleic Acids Res 44: 1118–1132. 10.1093/nar/gkv1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclair G, Borgel J, Sanz LA, Vallet J, Guibert S, Dumas M, Cavelier P, Girardot M, Forné T, Feil R, et al. 2016. EHMT2 directs DNA methylation for efficient gene silencing in mouse embryos. Genome Res 26: 192–202. 10.1101/gr.198291.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babak T, DeVeale B, Tsang EK, Zhou Y, Li X, Smith KS, Kukurba KR, Zhang R, Li JB, van der Kooy D, et al. 2015. Genetic conflict reflected in tissue-specific maps of genomic imprinting in human and mouse. Nat Genet 47: 544–549. 10.1038/ng.3274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak M, Boonen SE, Dahl C, Hahnemann JM, Mackay DJ, Tümer Z, Grønskov K, Temple IK, Guldberg P, Tommerup N. 2016. Genome-wide DNA methylation analysis of transient neonatal diabetes type 1 patients with mutations in ZFP57. BMC Med Genet 17: 29. 10.1186/s12881-016-0292-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baran Y, Subramaniam M, Biton A, Tukiainen T, Tsang EK, Rivas MA, Pirinen M, Gutierrez-Arcelus M, Smith KS, Kukurba KR, et al. 2015. The landscape of genomic imprinting across diverse adult human tissues. Genome Res 25: 927–936. 10.1101/gr.192278.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Hérault Y, Guillou F, Bourc'his D. 2016. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354: 909–912. 10.1126/science.aah5143 [DOI] [PubMed] [Google Scholar]
- Barlow DP. 1993. Methylation and imprinting: from host defense to gene regulation? Science 260: 309–310. 10.1126/science.8469984 [DOI] [PubMed] [Google Scholar]
- Barlow DP, Stöger R, Herrmann BG, Saito K, Schweifer N. 1991. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature 349: 84–87. 10.1038/349084a0 [DOI] [PubMed] [Google Scholar]
- Bartolomei MS, Webber AL, Brunkow ME, Tilghman SM. 1993. Epigenetic mechanisms underlying the imprinting of the mouse H19 gene. Genes Dev 7: 1663–1673. 10.1101/gad.7.9.1663 [DOI] [PubMed] [Google Scholar]
- Belshaw R, Watson J, Katzourakis A, Howe A, Woolven-Allen J, Burt A, Tristem M. 2007. Rate of recombinational deletion among human endogenous retroviruses. J Virol 81: 9437–9442. 10.1128/JVI.02216-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff SR, Tsai S, Hardison N, Motsinger-Reif AA, Freking BA, Nonneman D, Rohrer G, Piedrahita JA. 2009. Characterization of conserved and nonconserved imprinted genes in swine. Biol Reprod 81: 906–920. 10.1095/biolreprod.109.078139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogutz AB, Brind'Amour J, Kobayashi H, Jensen KN, Nakabayashi K, Imai H, Lorincz MC, Lefebvre L. 2019. Evolution of imprinting via lineage-specific insertion of retroviral promoters. Nat Commun 10: 5674. 10.1038/s41467-019-13662-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99. 10.1038/nature02886 [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294: 2536–2539. 10.1126/science.1065848 [DOI] [PubMed] [Google Scholar]
- Brandeis M, Kafri T, Ariel M, Chaillet JR, McCarrey J, Razin A, Cedar H. 1993. The ontogeny of allele-specific methylation associated with imprinted genes in the mouse. EMBO J 12: 3669–3677. 10.1002/j.1460-2075.1993.tb06041.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brind'Amour J, Kobayashi H, Richard Albert J, Shirane K, Sakashita A, Kamio A, Bogutz A, Koike T, Karimi MM, Lefebvre L, et al. 2018. LTR retrotransposons transcribed in oocytes drive species-specific and heritable changes in DNA methylation. Nat Commun 9: 3331. 10.1038/s41467-018-05841-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaillet JR, Vogt TF, Beier DR, Leder P. 1991. Parental-specific methylation of an imprinted transgene is established during gametogenesis and progressively changes during embryogenesis. Cell 66: 77–83. 10.1016/0092-8674(91)90140-T [DOI] [PubMed] [Google Scholar]
- Chen Z, Yin Q, Inoue A, Zhang C, Zhang Y. 2019. Allelic H3K27me3 to allelic DNA methylation switch maintains noncanonical imprinting in extraembryonic cells. Sci Adv 5: eaay7246. 10.1126/sciadv.aay7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Djekidel MN, Zhang Y. 2021. Distinct dynamics and functions of H2AK119ub1 and H3K27me3 in mouse preimplantation embryos. Nat Genet 53: 551–563. 10.1038/s41588-021-00821-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba H, Hirasawa R, Kaneda M, Amakawa Y, Li E, Sado T, Sasaki H. 2008. De novo DNA methylation independent establishment of maternal imprint on X chromosome in mouse oocytes. Genesis 46: 768–774. 10.1002/dvg.20438 [DOI] [PubMed] [Google Scholar]
- Chotalia M, Smallwood SA, Ruf N, Dawson C, Lucifero D, Frontera M, James K, Dean W, Kelsey G. 2009. Transcription is required for establishment of germline methylation marks at imprinted genes. Genes Dev 23: 105–117. 10.1101/gad.495809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MA, Soares MJ, Baker JC. 2013. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet 45: 325–329. 10.1038/ng.2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccone DN, Su H, Hevi S, Gay F, Lei H, Bajko J, Xu G, Li E, Chen T. 2009. KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints. Nature 461: 415–418. 10.1038/nature08315 [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Efstratiadis A, Robertsen EJ. 1990. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature 345: 78–80. 10.1038/345078a0 [DOI] [PubMed] [Google Scholar]
- DeChiara TM, Robertson EJ, Efstratiadis A. 1991. Parental imprinting of the mouse insulin-like growth factor II gene. Cell 64: 849–859. 10.1016/0092-8674(91)90513-X [DOI] [PubMed] [Google Scholar]
- DeVeale B, van der Kooy D, Babak T. 2012. Critical evaluation of imprinted gene expression by RNA-Seq: a new perspective. PLoS Genet 8: e1002600. 10.1371/journal.pgen.1002600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zheng H, Kawamura YK, Zhang K, Gassler J, Powell S, Xu Q, Lin Z, Xu K, Zhou Q, et al. 2020. Polycomb group proteins regulate chromatin architecture in mouse oocytes and early embryos. Mol Cell 77: 825–839.e7. 10.1016/j.molcel.2019.11.011 [DOI] [PubMed] [Google Scholar]
- Duffie R, Ajjan S, Greenberg MV, Zamudio N, Escamilla del Arenal M, Iranzo J, Okamoto I, Barbaux S, Fauque P, Bourc'his D. 2014. The Gpr1/Zdbf2 locus provides new paradigms for transient and dynamic genomic imprinting in mammals. Genes Dev 28: 463–478. 10.1101/gad.232058.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecco G, Cassano M, Kauzlaric A, Duc J, Coluccio A, Offner S, Imbeault M, Rowe HM, Turelli P, Trono D. 2016. Transposable elements and their KRAB-ZFP controllers regulate gene expression in adult tissues. Dev Cell 36: 611–623. 10.1016/j.devcel.2016.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. 2003. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development 130: 4235–4248. 10.1242/dev.00625 [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, et al. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571. 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- Ferguson-Smith AC, Sasaki H, Cattanach BM, Surani MA. 1993. Parental-origin-specific epigenetic modification of the mouse H19 gene. Nature 362: 751–755. 10.1038/362751a0 [DOI] [PubMed] [Google Scholar]
- Friedli M, Trono D. 2015. The developmental control of transposable elements and the evolution of higher species. Annu Rev Cell Dev Biol 31: 429–451. 10.1146/annurev-cellbio-100814-125514 [DOI] [PubMed] [Google Scholar]
- Greenberg MV, Glaser J, Borsos M, Marjou FE, Walter M, Teissandier A, Bourc'his D. 2017. Transient transcription in the early embryo sets an epigenetic state that programs postnatal growth. Nat Genet 49: 110–118. 10.1038/ng.3718 [DOI] [PubMed] [Google Scholar]
- Gregg C, Zhang J, Butler JE, Haig D, Dulac C. 2010. Sex-specific parent-of-origin allelic expression in the mouse brain. Science 329: 682–685. 10.1126/science.1190831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H, Okae H, Toh H, Chiba H, Hiura H, Shirane K, Sato T, Suyama M, Yaegashi N, Sasaki H, et al. 2016. Allele-specific methylome and transcriptome analysis reveals widespread imprinting in the human placenta. Am J Hum Genet 99: 1045–1058. 10.1016/j.ajhg.2016.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, Peñaherrera MS, Saadeh H, Andrews S, McFadden DE, Kelsey G, Robinson WP. 2016. Pervasive polymorphic imprinted methylation in the human placenta. Genome Res 26: 756–767. 10.1101/gr.196139.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, Demond H, Kelsey G. 2018. Epigenetic regulation in development: is the mouse a good model for the human? Hum Reprod Update 24: 556–576. 10.1093/humupd/dmy021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, Pérez-Palacios R, Gahurova L, Schubert M, Krueger F, Biggins L, Andrews S, Colomé-Tatché M, Bourc'his D, Dean W, et al. 2019. Endogenous retroviral insertions drive non-canonical imprinting in extra-embryonic tissues. Genome Biol 20: 225. 10.1186/s13059-019-1833-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata K, Okano M, Lei H, Li E. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129: 1983–1993. 10.1242/dev.129.8.1983 [DOI] [PubMed] [Google Scholar]
- Huang WC, Ferris E, Cheng T, Hörndli CS, Gleason K, Tamminga C, Wagner JD, Boucher KM, Christian JL, Gregg C. 2017. Diverse non-genetic, allele-specific expression effects shape genetic architecture at the cellular level in the mammalian brain. Neuron 93: 1094–1109.e7. 10.1016/j.neuron.2017.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Suzuki T, Zhang Y. 2017a. Maternal H3K27me3 controls DNA methylation-independent imprinting. Nature 547: 419–424. 10.1038/nature23262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Jiang L, Lu F, Zhang Y. 2017b. Genomic imprinting of Xist by maternal H3K27me3. Genes Dev 31: 1927–1932. 10.1101/gad.304113.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Chen Z, Yin Q, Zhang Y. 2018. Maternal Eed knockout causes loss of H3K27me3 imprinting and random X inactivation in the extraembryonic cells. Genes Dev 32: 1525–1536. 10.1101/gad.318675.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Ogonuki N, Kamimura S, Inoue H, Matoba S, Hirose M, Honda A, Miura K, Hada M, Hasegawa A, et al. 2020. Loss of H3K27me3 imprinting in the Sfmbt2 miRNA cluster causes enlargement of cloned mouse placentas. Nat Commun 11: 2150. 10.1038/s41467-020-16044-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Yoshida Y, Nishida K, Narimatsu M, Hibi M, Hirano T. 2000. Role of Gab1 in heart, placenta, and skin development and growth factor- and cytokine-induced extracellular signal-regulated kinase mitogen-activated protein kinase activation. Mol Cell Biol 20: 3695–3704. 10.1128/MCB.20.10.3695-3704.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanova E, Canovas S, Garcia-Martínez S, Romar R, Lopes JS, Rizos D, Sanchez-Calabuig MJ, Krueger F, Andrews S, Perez-Sanz F, et al. 2020. DNA methylation changes during preimplantation development reveal inter-species differences and reprogramming events at imprinted genes. Clin Epigenetics 12: 64. 10.1186/s13148-020-00857-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda M, Okano M, Hata K, Sado T, Tsujimoto N, Li E, Sasaki H. 2004. Essential role for de novo DNA methyltransferase Dnmt3a in paternal and maternal imprinting. Nature 429: 900–903. 10.1038/nature02633 [DOI] [PubMed] [Google Scholar]
- Kelsey G, Bartolomei MS. 2012. Imprinted genes … and the number is? PLoS Genet 8: e1002601. 10.1371/journal.pgen.1002601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khamlichi AA, Feil R. 2018. Parallels between mammalian mechanisms of monoallelic gene expression. Trends Genet 34: 954–971. 10.1016/j.tig.2018.08.005 [DOI] [PubMed] [Google Scholar]
- Kobayashi H, Sakurai T, Imai M, Takahashi N, Fukuda A, Yayoi O, Sato S, Nakabayashi K, Hata K, Sotomaru Y, et al. 2012. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet 8: e1002440. 10.1371/journal.pgen.1002440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR. 2016. SNPsplit: allele-specific splitting of alignments between genomes with known SNP genotypes. F1000Res 5: 1479. 10.12688/f1000research.9037.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latos PA, Pauler FM, Koerner MV, Senergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE, et al. 2012. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338: 1469–1472. 10.1126/science.1228110 [DOI] [PubMed] [Google Scholar]
- Laukoter S, Pauler FM, Beattie R, Amberg N, Hansen AH, Streicher C, Penz T, Bock C, Hippenmeyer S. 2020. Cell-type specificity of genomic imprinting in cerebral cortex. Neuron 107: 1160–1179.e9. 10.1016/j.neuron.2020.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DC, Lorincz MC. 2012. Silencing of endogenous retroviruses: when and why do histone marks predominate? Trends Biochem Sci 37: 127–133. 10.1016/j.tibs.2011.11.006 [DOI] [PubMed] [Google Scholar]
- Li E, Beard C, Jaenisch R. 1993. Role for DNA methylation in genomic imprinting. Nature 366: 362–365. 10.1038/366362a0 [DOI] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC. 2008. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15: 547–557. 10.1016/j.devcel.2008.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao Q, Li P, Zhao Q, Zhang J, Li J, Koseki H, Wong J. 2013. UHRF1 targets DNMT1 for DNA methylation through cooperative binding of hemi-methylated DNA and methylated H3K9. Nat Commun 4: 1563. 10.1038/ncomms2562 [DOI] [PubMed] [Google Scholar]
- Looijenga LH, Gillis AJ, Verkerk AJ, van Putten WL, Oosterhuis JW. 1999. Heterogeneous X inactivation in trophoblastic cells of human full-term female placentas. Am J Hum Genet 64: 1445–1452. 10.1086/302382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, et al. 2008. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet 40: 949–951. 10.1038/ng.187 [DOI] [PubMed] [Google Scholar]
- Matoba S, Wang H, Jiang L, Lu F, Iwabuchi KA, Wu X, Inoue K, Yang L, Press W, Lee JT, et al. 2018. Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell 23: 343–354.e5. 10.1016/j.stem.2018.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoba S, Nakamuta S, Miura K, Hirose M, Shiura H, Kohda T, Nakamuta N, Ogura A. 2019. Paternal knockout of Slc38a4/SNAT4 causes placental hypoplasia associated with intrauterine growth restriction in mice. Proc Natl Acad Sci 116: 21047–21053. 10.1073/pnas.1907884116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931. 10.1038/nature08858 [DOI] [PubMed] [Google Scholar]
- Mei H, Kozuka C, Hayashi R, Kumon M, Koseki H, Inoue A. 2021. H2AK119ub1 guides maternal inheritance and zygotic deposition of H3K27me3 in mouse embryos. Nat Genet 53: 539–550. 10.1038/s41588-021-00820-3 [DOI] [PubMed] [Google Scholar]
- Migeon BR, Do TT. 1979. In search of non-random X inactivation: studies of fetal membranes heterozygous for glucose-6-phosphate dehydrogenase. Am J Hum Genet 31: 581–585. [PMC free article] [PubMed] [Google Scholar]
- Ming X, Zhang Z, Zou Z, Lv C, Dong Q, He Q, Yi Y, Li Y, Wang H, Zhu B. 2020. Kinetics and mechanisms of mitotic inheritance of DNA methylation and their roles in aging-associated methylome deterioration. Cell Res 30: 980–996. 10.1038/s41422-020-0359-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteagudo-Sánchez A, Sánchez-Delgado M, Mora JRH, Santamaría NT, Gratacós E, Esteller M, de Heredia ML, Nunes V, Choux C, Fauque P, et al. 2019. Differences in expression rather than methylation at placenta-specific imprinted loci is associated with intrauterine growth restriction. Clin Epigenetics 11: 35. 10.1186/s13148-019-0630-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Haig D. 1991. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet 7: 45–49. 10.1016/0168-9525(91)90040-W [DOI] [PubMed] [Google Scholar]
- Moreira de Mello JC, de Araújo ES, Stabellini R, Fraga AM, de Souza JE, Sumita DR, Camargo AA, Pereira LV. 2010. Random X inactivation and extensive mosaicism in human placenta revealed by analysis of allele-specific gene expression along the X chromosome. PLoS One 5: e10947. 10.1371/journal.pone.0010947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. 2008. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 322: 1717–1720. 10.1126/science.1163802 [DOI] [PubMed] [Google Scholar]
- Okae H, Hiura H, Nishida Y, Funayama R, Tanaka S, Chiba H, Yaegashi N, Nakayama K, Sasaki H, Arima T. 2012. Re-investigation and RNA sequencing-based identification of genes with placenta-specific imprinted expression. Hum Mol Genet 21: 548–558. 10.1093/hmg/ddr488 [DOI] [PubMed] [Google Scholar]
- Okae H, Chiba H, Hiura H, Hamada H, Sato A, Utsunomiya T, Kikuchi H, Yoshida H, Tanaka A, Suyama M, et al. 2014. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet 10: e1004868. 10.1371/journal.pgen.1004868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak R, Feil R. 2018. Environmental effects on chromatin repression at imprinted genes and endogenous retroviruses. Curr Opin Chem Biol 45: 139–147. 10.1016/j.cbpa.2018.04.015 [DOI] [PubMed] [Google Scholar]
- Prokopuk L, Stringer JM, White CR, Vossen RHAM, White SJ, Cohen ASA, Gibson WT, Western PS. 2018. Loss of maternal EED results in postnatal overgrowth. Clin Epigenetics 10: 95. 10.1186/s13148-018-0526-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudhon C, Duffié R, Ajjan S, Cowley M, Iranzo J, Carbajosa G, Saadeh H, Holland ML, Oakey RJ, Rakyan VK, et al. 2012. Protection against de novo methylation is instrumental in maintaining parent-of-origin methylation inherited from the gametes. Mol Cell 47: 909–920. 10.1016/j.molcel.2012.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quenneville S, Verde G, Corsinotti A, Kapopoulou A, Jakobsson J, Offner S, Baglivo I, Pedone PV, Grimaldi G, Riccio A, et al. 2011. In embryonic stem cells, ZFP57/KAP1 recognize a methylated hexanucleotide to affect chromatin and DNA methylation of imprinting control regions. Mol Cell 44: 361–372. 10.1016/j.molcel.2011.08.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragazzini R, Pérez-Palacios R, Baymaz IH, Diop S, Ancelin K, Zielinski D, Michaud A, Givelet M, Borsos M, Aflaki S, et al. 2019. EZHIP constrains polycomb repressive complex 2 activity in germ cells. Nat Commun 10: 3858. 10.1038/s41467-019-11800-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Friedli M, Offner S, Verp S, Mesnard D, Marquis J, Aktas T, Trono D. 2013. De novo DNA methylation of endogenous retroviruses is shaped by KRAB-ZFPs/KAP1 and ESET. Development 140: 519–529. 10.1242/dev.087585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Delgado M, Riccio A, Eggermann T, Maher ER, Lapunzina P, Mackay D, Monk D. 2016. Causes and consequences of multi-locus imprinting disturbances in humans. Trends Genet 32: 444–455. 10.1016/j.tig.2016.05.001 [DOI] [PubMed] [Google Scholar]
- Santoni FA, Stamoulis G, Garieri M, Falconnet E, Ribaux P, Borel C, Antonarakis SE. 2017. Detection of imprinted genes by single-cell allele-specific gene expression. Am J Hum Genet 100: 444–453. 10.1016/j.ajhg.2017.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schertzer MD, Braceros KCA, Starmer J, Cherney RE, Lee DM, Salazar G, Justice M, Bischoff SR, Cowley DO, Ariel P, et al. 2019. lncRNA-induced spread of polycomb controlled by genome architecture, RNA abundance, and CpG island DNA. Mol Cell 75: 523–537.e10. 10.1016/j.molcel.2019.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz R, Proudhon C, Bestor TH, Woodfine K, Lin CS, Lin SP, Prissette M, Oakey RJ, Bourc'his D. 2010. The parental non-equivalence of imprinting control regions during mammalian development and evolution. PLoS Genet 6: e1001214. 10.1371/journal.pgen.1001214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. 2007. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450: 908–912. 10.1038/nature06397 [DOI] [PubMed] [Google Scholar]
- Shi H, Strogantsev R, Takahashi N, Kazachenka A, Lorincz MC, Hemberger M, Ferguson-Smith AC. 2019. ZFP57 regulation of transposable elements and gene expression within and beyond imprinted domains. Epigenetics Chromatin 12: 49. 10.1186/s13072-019-0295-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirane K, Miura F, Ito T, Lorincz MC. 2020. NSD1-deposited h3k36me2 directs de novo methylation in the mouse male germline and counteracts Polycomb-associated silencing. Nat Genet 52: 1088–1098. 10.1038/s41588-020-0689-z [DOI] [PubMed] [Google Scholar]
- Singh VB, Sribenja S, Wilson KE, Attwood KM, Hillman JC, Pathak S, Higgins MJ. 2017. Blocked transcription through KvDMR1 results in absence of methylation and gene silencing resembling Beckwith-Wiedemann syndrome. Development 144: 1820–1830. 10.1242/dev.145136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smallwood SA, Tomizawa S, Krueger F, Ruf N, Carli N, Segonds-Pichon A, Sato S, Hata K, Andrews SR, Kelsey G. 2011. Dynamic CpG island methylation landscape in oocytes and preimplantation embryos. Nat Genet 43: 811–814. 10.1038/ng.864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spahn L, Barlow DP. 2003. An ICE pattern crystallizes. Nat Genet 35: 11–12. 10.1038/ng0903-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart KR, Veselovska L, Kim J, Huang J, Saadeh H, Tomizawa S, Smallwood SA, Chen T, Kelsey G. 2015. Dynamic changes in histone modifications precede de novo DNA methylation in oocytes. Genes Dev 29: 2449–2462. 10.1101/gad.271353.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogantsev R, Krueger F, Yamazawa K, Shi H, Gould P, Goldman-Roberts M, McEwen K, Sun B, Pedersen R, Ferguson-Smith AC. 2015. Allele-specific binding of ZFP57 in the epigenetic regulation of imprinted and non-imprinted monoallelic expression. Genome Biol 16: 112. 10.1186/s13059-015-0672-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N, Coluccio A, Thorball CW, Planet E, Shi H, Offner S, Turelli P, Imbeault M, Ferguson-Smith AC, Trono D. 2019. ZNF445 is a primary regulator of genomic imprinting. Genes Dev 33: 49–54. 10.1101/gad.320069.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. 2008. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell 15: 668–679. 10.1016/j.devcel.2008.08.015 [DOI] [PubMed] [Google Scholar]
- Thomas JW, Touchman JW, Blakesley RW, Bouffard GG, Beckstrom-Sternberg SM, Margulies EH, Blanchette M, Siepel AC, Thomas PJ, McDowell JC, et al. 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788–793. 10.1038/nature01858 [DOI] [PubMed] [Google Scholar]
- Thompson PJ, Macfarlan TS, Lorincz MC. 2016. Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol Cell 62: 766–776. 10.1016/j.molcel.2016.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veselovska L, Smallwood SA, Saadeh H, Stewart KR, Krueger F, Maupetit-Mehouas S, Arnaud P, Tomizawa S, Andrews S, Kelsey G. 2015. Deep sequencing and de novo assembly of the mouse oocyte transcriptome define the contribution of transcription to the DNA methylation landscape. Genome Biol 16: 209. 10.1186/s13059-015-0769-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Tomizawa S, Mitsuya K, Totoki Y, Yamamoto Y, Kuramochi-Miyagawa S, Iida N, Hoki Y, Murphy PJ, Toyoda A, et al. 2011. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 332: 848–852. 10.1126/science.1203919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weksberg R, Shen DR, Fei YL, Song QL, Squire J. 1993. Disruption of insulin-like growth factor 2 imprinting in Beckwith-Wiedemann syndrome. Nat Genet 5: 143–150. 10.1038/ng1093-143 [DOI] [PubMed] [Google Scholar]
- Wood AJ, Schulz R, Woodfine K, Koltowska K, Beechey CV, Peters J, Bourc'his D, Oakey RJ. 2008. Regulation of alternative polyadenylation by genomic imprinting. Genes Dev 22: 1141–1146. 10.1101/gad.473408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Xu J, Yu G, Yao G, Xu K, Ma X, Zhang N, Liu B, Li T, Lin Z, et al. 2019. Resetting histone modifications during human parental-to-zygotic transition. Science 365: 353–360. 10.1126/science.aaw5118 [DOI] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153: 1134–1148. 10.1016/j.cell.2013.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Zhang C, Li J, Wang G, Chen W, Li D, Li S. 2018. Polymorphic imprinting of SLC38A4 gene in bovine placenta. Biochem Genet 56: 639–649. 10.1007/s10528-018-9866-5 [DOI] [PubMed] [Google Scholar]
- Xu Q, Xiang Y, Wang Q, Wang L, Brind'Amour J, Bogutz AB, Zhang Y, Zhang B, Yu G, Xia W, et al. 2019. SETD2 regulates the maternal epigenome, genomic imprinting and embryonic development. Nat Genet 51: 844–856. 10.1038/s41588-019-0398-7 [DOI] [PubMed] [Google Scholar]
- Yang P, Wang Y, Hoang D, Tinkham M, Patel A, Sun MA, Wolf G, Baker M, Chien HC, Lai KN, et al. 2017. A placental growth factor is silenced in mouse embryos by the zinc finger protein ZFP568. Science 356: 757–759. 10.1126/science.aah6895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng T, Pierce N, Szabó PE. 2021. H3k9 methyltransferase EHMT2/G9a controls ERVK-driven non-canonical imprinted genes. bioRxiv 2021.03.29.437617. [DOI] [PubMed] [Google Scholar]
- Zhang W, Chen Z, Yin Q, Zhang D, Racowsky C, Zhang Y. 2019. Maternal-biased H3K27me3 correlates with paternal-specific gene expression in the human morula. Genes Dev 33: 382–387. 10.1101/gad.323105.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Huang B, Zhang B, Xiang Y, Du Z, Xu Q, Li Y, Wang Q, Ma J, Peng X, et al. 2016. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol Cell 63: 1066–1079. 10.1016/j.molcel.2016.08.032 [DOI] [PubMed] [Google Scholar]
- Zink F, Magnusdottir DN, Magnusson OT, Walker NJ, Morris TJ, Sigurdsson A, Halldorsson GH, Gudjonsson SA, Melsted P, Ingimundardottir H, et al. 2018. Insights into imprinting from parent-of-origin phased methylomes and transcriptomes. Nat Genet 50: 1542–1552. 10.1038/s41588-018-0232-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.