In this review, Li et al. detail research advances pertaining to the genetics and biology of colorectal cancer, emerging concepts gleaned from immune and single-cell profiling, and critical advances and remaining knowledge gaps influencing the development of effective therapies for this cancer.
Keywords: CRC, colorectal cancer, cancer genetics, cancer therapy development, tumor microenviornment
Abstract
Colorectal cancer has served as a genetic and biological paradigm for the evolution of solid tumors, and these insights have illuminated early detection, risk stratification, prevention, and treatment principles. Employing the hallmarks of cancer framework, we provide a conceptual framework to understand how genetic alterations in colorectal cancer drive cancer cell biology properties and shape the heterotypic interactions across cells in the tumor microenvironment. This review details research advances pertaining to the genetics and biology of colorectal cancer, emerging concepts gleaned from immune and single-cell profiling, and critical advances and remaining knowledge gaps influencing the development of effective therapies for this cancer that remains a major public health burden.
Colorectal cancer (CRC) is the second most common adult cancer in women and the third most common in men, and it is the fourth leading cause of cancer death, accounting for 9.2% of deaths worldwide (Bray et al. 2018; Dekker et al. 2019). The 5-yr and 10-yr survival rates are 65% and 58%, respectively (Siegel et al. 2017), and incidence and mortality rates are 25% higher in men than in women (Dekker et al. 2019). CRC incidence and mortality rates also vary by race and ethnicity, being highest in non-Hispanic blacks and lowest in Asian Americans/Pacific Islanders (Siegel et al. 2017). In recent years, the overall incidence of CRC, particularly rectal and distal colon cancers, has declined in individuals older than 50 yr but increased in those younger than 50 yr (Siegel et al. 2017). Extensive CRC screening has substantially reduced incidence and mortality by enabling early detection and removal of precancerous adenomas (Wolf et al. 2018). Although increased nonsystematic screening of young adults may be contributing to increased incidence (Siegel et al. 2017), multiple pervasive instigators may drive early-onset CRC (in patients <50 yr old), including global adoption of a westernized diet, chronic stress, and widespread use of antibiotics with alteration of the gut microbiota (Hofseth et al. 2020). Moreover, 10%–20% of all patients with CRC possess a positive family history (Dekker et al. 2019), and ∼5% of all cases of CRC are linked to a known hereditary CRC syndrome detectable by germline testing (Dekker et al. 2019). Finally, increased sporadic CRC incidence is associated with longstanding inflammatory bowel disease (Dekker et al. 2019) and variable lifestyle factors such as physical inactivity, unhealthy diet, smoking, obesity, and heavy alcohol consumption (Chan and Giovannucci 2010; Doubeni et al. 2012), which can be reduced dramatically with improved disease management and implementation of wellness programs.
Intriguingly, differences in clinical outcomes and drug responsiveness are dependent on the location of cancer along the colon and rectum. Relevant contributing factors include their distinct physiological functions, gut microbiome (Tropini et al. 2017), regionally resident immune cell types (Stenstad et al. 2007; Lee et al. 2015; Agace and McCoy 2017), dietary carcinogens (McMichael and Potter 1985; West et al. 1989), timing of disease detection, and ontogeny factors (Iacopetta 2002). With respect to intestinal development, the proximal (right) large intestine (cecum, ascending colon, and the transverse colon) derives from the embryonic midgut, whereas the distal (left) large intestine (splenic flexure, descending colon, sigmoid colon, and rectum) derives from the embryonic hindgut (LaPointe et al. 2008). These ontogenetic differences are associated with differential gene expression patterns along the proximal–distal axis (LaPointe et al. 2008). Proximal sporadic colon tumors, more frequently diagnosed in women (51%–62% of cases) (Hansen and Jess 2012) and in African-Americans (Augustus and Ellis 2018), show a higher TNM stage at first diagnosis, display a pattern with high levels of genome-wide promoter hypermethylation referred to as CpG island methylator phenotype (CIMP), exhibit microsatellite instability (MSI) due to deficient DNA mismatch repair mechanisms (dMMR), are more frequently mutated in KRAS and BRAF and have a worse prognosis in terms of survival (Dienstmann et al. 2017; Venook et al. 2017; Dekker et al. 2019). In contrast, distal colorectal tumors are more likely to present with chromosomal instability (CIN) (Iacopetta 2002) and show a more favorable prognosis (Loupakis et al. 2015). A few studies have suggested that proximal or distal colorectum favors different levels of constitutive β-catenin signaling controlled by retained β-catenin binding affinity in truncated APC (Albuquerque et al. 2011). An increased number of retained 20-amino-acid β-catenin binding repeats in truncated APC is correlated with dMMR, suggesting the potential mechanisms for these regional differences in the occurrence of MSI (Albuquerque et al. 2011; Leedham et al. 2013), while additional studies are needed to understand the mechanisms of regional differences in the occurrence of CIN and CIMP.
The normal colon epithelium contains crypts that are comprised of a variety of cells. At the crypt bottoms, there are rapidly cycling colonic stem cells. The colonic stem cells express a variety of markers such as LGR5 (Barker et al. 2007; Zeki et al. 2011) and OLFM4 (Zeki et al. 2011). The colonic stem cells reside at the base of the colonic crypts, “stem cell niche,” and are supported by pericryptal myofibroblasts producing signaling factors that maintain the stemness of the colonic stem cells (Zeki et al. 2011). The colonic stem cells generate precursor cells that differentiate into cells with specialized physiological functions: enterocytes for nutrient uptake, goblet cells for mucus production, and enteroendocrine cells for hormone production (Vermeulen and Snippert 2014). Some characteristics of colonic stem cells are shared by CRC stem cells (Dalerba et al. 2011; Li et al. 2017a). The concept that CRC may originate from CRC stem cells arises from the longevity and self-renewal of stem cells enabling accumulation and propagation of oncogenic mutations (Zeki et al. 2011). Limiting dilution transplantation experiments show that only a subset of CRC cancer cells possesses tumor-initiating potential (O'Brien et al. 2007); moreover, a single CRC cell can produce various differentiated colon epithelial lineage types (Kirkland 1988), further supporting the existence of cancer cells with stem cell-like properties in CRC tumors. These CRC cancer stem cells show increased chemotherapy resistance (Dallas et al. 2009), underscoring the importance of better defining markers, signaling circuits, and therapeutic targets that differentiate normal versus neoplastic stem cells for improved CRC therapy (Nakanishi et al. 2013).
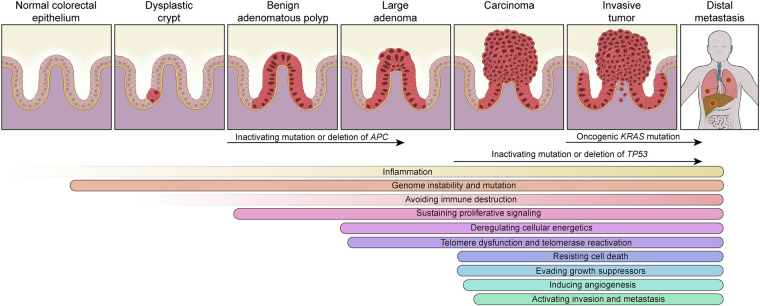
The highly chronicled evolution of CRC reveals its origin first as an aberrant crypt that evolves into a benign adenomatous polyp, which ultimately transforms into sporadic CRC over a long period of time of ∼10–15 yr (Dekker et al. 2019). This conventional adenoma-carcinoma-metastasis model occurs in most cases of CRC (70%–90%) (Fig. 1). These phenotypic transitions are associated with the accumulation of specific signature genetic events of “APC-KRAS-TP53,” known as the Vogelstein model (Fearon and Vogelstein 1990). Emerging data from the TCGA, mouse genetic models, and clinic-pathological correlations have revised the sequence of gene events as “APC-TP53-KRAS” (see “CRC Immunity” and “Activating Invasion and Metastasis”) (Boutin et al. 2017; Liao et al. 2019). As illustrated in Figure 1, adenoma emergence coincides with inactivating mutation or deletion of APC (Fearon and Vogelstein 1990), adenocarcinoma sustains inactivating mutations or deletion of TP53 (which may be dispensable [Janssen et al. 2006]) with telomere dysfunction and double-stranded DNA breakage driving CIN (Rudolph et al. 2001), and invasive/metastatic disease often shows activating mutations in KRAS (Boutin et al. 2017). Alternatively, ∼10% of CRCs can evolve along a so-called serrated neoplasia pathway featuring one of two progression presentations: (1) a sessile serrated pathway, in which a microvesicular hyperplastic polyp progresses to a sessile serrated adenoma and then to either MSI or microsatellite stable (MSS) carcinoma, or (2) a traditional serrated pathway, in which a goblet cell-rich hyperplastic polyp progresses to a traditional serrated adenoma and then to MSS carcinoma (Snover 2011; Rashtak et al. 2017). These serrated neoplasms exhibit a higher frequency of activating mutations in BRAF and KRAS, increased CIMP (Dekker et al. 2019), hypermutation rates, but rarely APC mutations (Rashtak et al. 2017; Chang et al. 2018). Another specific form of CRC, the colitis-associated cancer (CAC), most frequently appears in patients with inflammatory bowel disease (IBD). CAC accounts for about 2% of CRC. Compared with the sporadic/familial CRC, it shares similarities but also presents distinct features in terms of etiology, genetic alterations, and treatment interventions, as comprehensively reviewed elsewhere (Shalapour and Karin 2020).
Figure 1.
Colorectal cancer “conventional adenoma–carcinoma–metastasis” model and corresponding cancer hallmarks.
In addition to the CRC signature mutations of APC, TP53, and KRAS, in-depth genomic and transcriptomic profiling has revealed disease heterogeneity reflected by numerous lower-frequency mutations and transcriptional profiles, classified into four consensus molecular subtypes (CMS). CMS1 features hypermutation, MSI, and an active immune response. CMS2 presents with epithelial features and significant activation of Wnt and Myc signaling. CMS3 exhibits epithelial features and metabolic dysregulation. CMS4 possesses strong TGF-β activation, stromal invasion, and angiogenesis (Guinney et al. 2015). The CMS classification provides a framework for prognostication and improved assignment of targeted therapies in precision trials (Dekker et al. 2019).
The versatile features of CRC subtypes are captured by a diverse array of models, including chemically induced and genetically engineered mouse models, as comprehensively discussed elsewhere (Heyer et al. 1999; Mouradov et al. 2014; McIntyre et al. 2015; Medico et al. 2015; Brown et al. 2016; Bürtin et al. 2020), as well as patient-derived cell lines, organoids, and xenografts. Each model system offers unique experimental merits. With respect to genetically engineered mouse models, the first CRC model, ApcMin/+, was developed in 1990 (Moser et al. 1990; Su et al. 1992). This model expresses truncated APC protein and is largely used to model human familial adenomatous polyposis and CRC. Of note, tumors developed in the ApcMin/+ mouse are restricted to the small intestine, which is rare in humans. This caveat of the ApcMin/+ model has been overcome by the generation of mouse models with gene recombination driven by CDX2-Cre, or particularly Villin-Cre induced by tamoxifen enema, which are more specific for colonic epithelium and develop mostly CRC (el Marjou et al. 2004; Hinoi et al. 2007). One such example is the recently generated iKAP model that harbors the three major signature mutations (Apc/Trp53/Kras) induced by Villin-Cre, recapitulates CRC progression, and serves as a platform to understand the biological functions of specific genetic mutations as well as dissect the cellular interactions operating in the tumor microenvironment (TME) (Boutin et al. 2017; Liao et al. 2019). Moreover, one of the most popular mouse models of CRC is based on a combination of dextran sulfate sodium-induced experimental colitis with carcinogen (azoxymethane) exposure, which researchers have used for studying CAC.
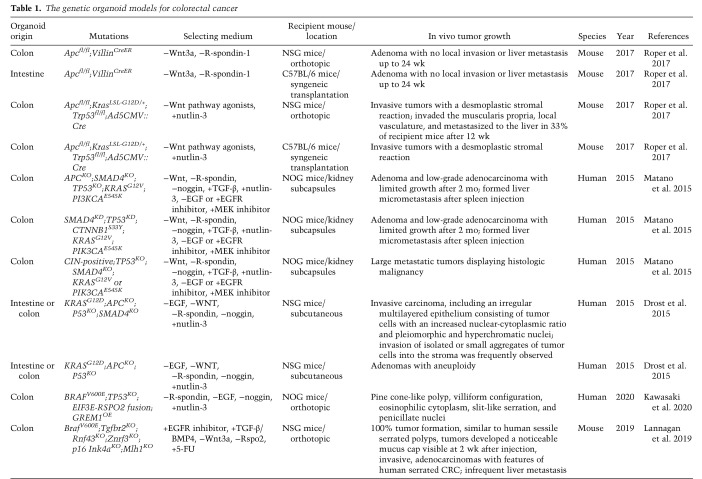
More recently, three-dimensional “organoid” model systems have been developed via genetic editing of normal intestinal epithelium, summarized in Table 1, and appear to mirror well the complex genetic and transcriptomic features of various CRC subtypes. CRC tumor organoids derived from human or mouse tumors are also being established and used increasingly to understand the genetic bases of response to therapies (Calon et al. 2015; Vlachogiannis et al. 2018; Ganesh et al. 2019; Ooft et al. 2019; Arena et al. 2020; Yao et al. 2020), profile genomic alterations (Bolhaqueiro et al. 2019) and intratumoral heterogeneity (Roerink et al. 2018; Bruun et al. 2020), and create organoid-xenografts for realistic disease modeling (Roper et al. 2017). In addition, patient-derived tumor organoids may also serve as avatar systems for ex vivo testing of drug regimens with the goal of guiding personalized clinical treatment (Narasimhan et al. 2020). An important enhancement in organoid modeling has been the tumor organoid–T-cell coculture systems using tumor tissues from CRC patients and immune cells, particularly T cells derived from autologous peripheral blood lymphocytes (Cattaneo et al. 2020). In these models, the autologous T cells react to tumor organoids but not healthy organoids or tissue (Dijkstra et al. 2018). As such, these models may facilitate the investigation of immune therapies in defined genetic contexts as well as determine adaptive and resistance mechanisms to immune and targeted therapies. Moreover, the tumor-reactive T cells derived from these coculture models may illuminate potential adoptive T-cell transfer therapies.
Table 1.
The genetic organoid models for colorectal cancer
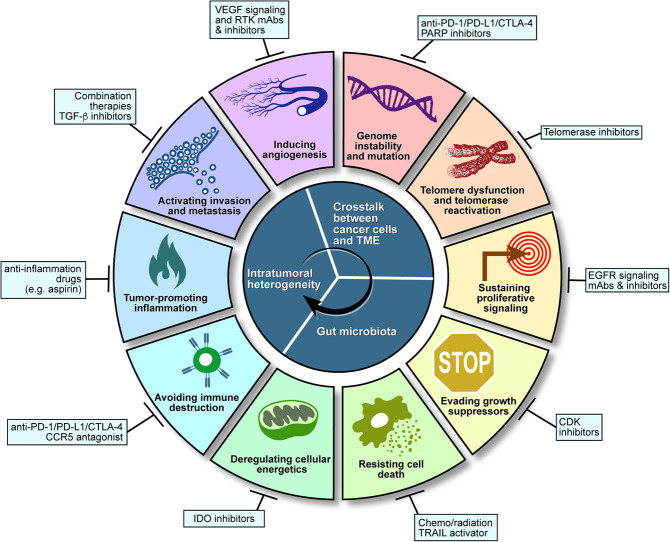
In summary, major progress across multiple fronts has positioned the field to make a decisive assault on one of the most prevalent and lethal cancers. This review provides an extensive summary of signature genetic alterations and epigenetic alterations (such as DNA methylation, histone modification, and co-/post-transcriptional regulation of RNA expression), refined molecular classification of disease subsets, and a maturing pipeline of immune and targeted therapies coupled with immune and molecular profiles of treated human tumors. The next subsection details some of the prominent genetic and biological features of CRC in the framework of hallmarks of the cancer paradigm (Hanahan and Weinberg 2011).
The hallmarks of CRC
Genome instability and mutation
Comprehensive genomic profiling of CRC has revealed significant intratumoral and intertumoral heterogeneity resulting from the accumulation of genetic mutations and chromosomal aberrations during disease initiation and progression. Genomic instability in CRC presents as one of two major forms: CIN and MSI (Lengauer et al. 1998). The CRCs lacking CIN or MSI are classified as genome stable (GS) (Liu et al. 2018b). In GS CRC, the DNA repair genes and tumor suppressors are likely to be transcriptionally silenced through CIMP, though a large proportion of MSI CRCs and a small population of CIN CRCs are also CIMP-positive, and about 10% of CRCs are negative for CIN, MSI, or CIMP (Simons et al. 2013).
Chromosome instability (CIN)
CIN is characterized by chromosomal numerical alterations (aneuploidy) and structural alterations (somatic copy number alterations, deletions, insertions, amplifications, or loss of heterozygosity) (Nguyen et al. 2020), occurring in 65%–70% of sporadic CRCs. Nearly all CIN tumors show activated Wnt signaling, and 80% harbor mutational inactivation of APC, a negative regulator of the Wnt pathway (Nguyen et al. 2020). Mutational inactivation/deletion of TP53 occurs in 60% of CIN tumors (Nguyen et al. 2020), and p53 loss of function (1) directly drives CIN and (2) provides a permissive context for genome instability mechanisms. With respect to genome instability, combined telomere dysfunction and p53 deficiency is a major CIN mechanism, as revealed by the occurrence of anaphase bridges in early-stage carcinomas of human CRC (Rudolph et al. 2001) and spontaneous CRC occurrence in telomerase-deficient p53 mutant mice (Artandi et al. 2000). Aneuploidy may also result from defects in mitotic checkpoints, microtubule attachment, centrosomes, mitotic spindles, and chromosome cohesion, resulting in errors in chromosome partitioning during cell division. For example, microdeletions in the MACROD2 gene, present in one-third of sporadic CRCs, interrupts the transferase catalytic activity of PARP1, leading to DNA repair deficiency during mitotic entry, spindle checkpoint relaxation, and aneuploidy (Sakthianandeswaren et al. 2018). Tolerance of aneuploidy-induced cell death in CRC has been attributed to BCL9L deficiency and subsequent reduced basal caspase-2 levels and insufficient cleavage of MDM2 and BID death agonist (López-García et al. 2017).
Microsatellite instability (MSI)
Microsatellites are DNA sequences containing repetitive motifs that tend to accumulate higher mutation rates than other genomic regions. MSI is the phenotypic manifestation of dMMR resulting from mutational inactivation of MMR genes, including MLH1, MSH2, MSH3, MSH6, and PMS2. MSH2 forms heterodimers with MSH6 or MSH3 and regulates mismatch recognition and repair initiation (Li 2008). Loss of the MMR gene function results in genetic hypermutation, fueling CRC development. In sporadic CRC, MLH1 promoter hypermethylation leading to gene silencing is the most common cause of MSI (Cunningham et al. 1998). In some Lynch syndrome patients, hypermethylation of the MSH2 promoter is caused by a transcriptional read-through due to the germline deletion of its direct upstream gene, TACSTD1 (encoding EPCAM) (Ligtenberg et al. 2009). MSH3, responsible for the repair of mismatches in dinucleotides and tetranucleotides, is repressed or mislocalized by hypoxia, inflammation (Tseng-Rogenski et al. 2015), and cellular stress (Søreide et al. 2016) in CRC. MSI-high (MSI-H) tumors possess many point mutations that create abundant neoantigens that can engender an inflammatory phenotype characterized by dense infiltration of lymphocytes, and this phenotype shows more frequent and robust responses to immune checkpoint inhibitor therapies approved by the US Food and Drug Administration (FDA) (see “CRC Immunity”; Willis et al. 2020).
In summary, CRC genome instability mechanisms, presenting as CIN or MSI, contribute to tumor heterogeneity but with distinct genetic and biological features. These designations have important diagnostic and therapeutic implications for patients with CRC.
Telomere dysfunction and telomerase reactivation
Telomeres, along with the shelterin complex, protect and maintain chromosomal integrity (van Steensel et al. 1998; de Lange 2005). The pathogenetic relevance of telomere dysfunction in CRC was reinforced further by studies in the telomerase-deficient ApcMin/+ mouse model (Rudolph et al. 2001). Correspondingly, in humans, progressive telomere erosion occurs in the intestinal epithelium during aging (O'Sullivan et al. 2006), and telomere dysfunction (anaphase bridging) has been documented in the adenoma-to-carcinoma transition, indicating that a telomere-based crisis plays a role in driving CIN in the early stages of human CRC (Fig. 1; Rudolph et al. 2001). Finally, cancer progression is associated with telomerase reactivation, which is present in 85%–90% of all cancer types, including CRC (Tang et al. 1998).
Beyond CIN, telomere biology may contribute to key processes governing carcinogenesis, such as inflammation, which is a well-known instigator of colon cancer. Patients with IBD, particularly ulcerative colitis (UC), are at high risk for CRC development (Eaden et al. 2001; O'Sullivan et al. 2002; Jess et al. 2005; Risques et al. 2008). In UC, the cumulative risk of CRC is 2%, 8%, and 18% after 10, 20, and 30 yr, respectively, of disease duration (Eaden et al. 2001). Accelerated telomere attrition has been documented in the colon of UC patients, supporting the hypothesis that resultant CIN contributes to increased CRC occurrence (O'Sullivan et al. 2002; Risques et al. 2008). Interestingly, age-dependent telomere attrition in the intestinal epithelium could also be a primary cause of late-onset IBD and may also contribute to disease recurrence. Specifically, telomere dysfunction can activate ATM/cABL, which phosphorylates and activates YAP1, resulting in up-regulation of prointerleukin (IL)-18 (Chakravarti et al. 2020). In the colon, the gut microbiome-activated inflammasome-mediated caspase-1 cleaves pro-IL-18 into mature IL-18, resulting in inflammation. As a result, increased intracellular ROS levels accelerate telomere damage and attrition, generating a feed-forward loop of telomere dysfunction-inflammation, which could fuel genomic instability and ultimately cancer (Jurk et al. 2014). In addition to the IL-18 mechanism, extrachromosomal telomere fragments present in cells undergoing crisis can induce the cGAS/STING pathway (Nassour et al. 2019), which induces chronic inflammation through a type I interferon-mediated pathway. Thus, the telomere–cGAS/STING connection could potentially be another driver of cancers associated with inflammation and form the basis of novel therapeutic interventions.
Another important intersection of telomeres and other CRC hallmarks relates to mitochondrial biology and oxidative defense. Specifically, telomere dysfunction activates p53, which in turn represses PGC1-α and PGC1-β, the master regulators of genes for mitochondrial biogenesis and oxidative defense. Telomere dysfunction-induced ROS and reactive nitrogen species (Valko et al. 2007) damage lipids, proteins, and DNA. ROS can oxidize lipids known as polyunsaturated fatty acids and produce other free radicals and substances such as malondialdehyde, conjugated dienes, hydroperoxides, lipoperoxides, and toxic aldehydes (Marnett 1999; Cejas et al. 2004). Lipid peroxidation in turn can change membrane fluidity and increase membrane permeability, leading to defects in signaling as well as inflammation. Protein oxidation can impair protein function, which could impact the fidelity of polymerases and DNA repair enzymes (Shringarpure and Davies 2002) as well as the proteasome system responsible for clearing misfolded and damaged proteins (Shringarpure and Davies 2002; Grune et al. 2003). This accumulation of damaged proteins can drive several diseases, including CRC (Marnett 2000). Finally, high ROS levels can increase the oxidization of DNA nucleotides (adenine, guanine, cytosine, and thymine), which is mutagenic by itself and is present in several types of cancers, including CRC (Bjelland and Seeberg 2003). Another interesting aspect is that high ROS states, brought about by telomere dysfunction driving inflammation, elevate expression of cyclooxygenase enzyme-2 (COX2) (Uchida 2017), which catalyzes the conversion of arachidonic acid to prostaglandins. The role of this axis in CRC was validated in prevention trials for patients with familial adenomatous polyposis, as well as multiple coclinical trials with different murine models, which demonstrated that COX2 inhibitors were efficacious in reducing tumor multiplicity and size, leading to the initial approval by the FDA. However, cardiovascular safety concerns prompted the withdrawal of the indication (Koehne and Dubois 2004; Ricciardiello et al. 2016).
The relevance of telomerase reactivation in CRC pathogenesis is evident on several levels. First, telomerase reverse transcriptase (TERT) levels and telomerase activity increase during adenoma-carcinoma progression (Tatsumoto et al. 2000; Terrin et al. 2008), and high levels of telomerase correlate positively with poor prognosis (Bertorelle et al. 2013). This correlation may relate directly to procarcinogenic activities of TERT and/or reflect the level of MYC or WNT, which can up-regulate TERT gene transcription. Although TERT gene promoter mutations are common in many cancer types (up to 80% in some cancers [Killela et al. 2013]), such mutations occur in only 10% of cases of CRC (Siraj et al. 2020). Because these somatic mutations augment TERT transcription via enhanced ETS transcription factor binding (GAPB) to the newly formed ETS binding motifs (Bell et al. 2015; Stern et al. 2015), it is tempting to speculate that this low frequency reflects the high c-MYC levels in CRC, which induces TERT transcription via conserved E-boxes in the TERT gene promoter (Wu et al. 1999). Similarly, in high Wnt signaling, β-catenin interacts with KLF4 to engage four conserved TCF4 binding sites in the TERT gene promotor (Hoffmeyer et al. 2012).
In summary, telomere dysfunction and telomerase reactivation play pivotal roles in adenoma-to-carcinoma progression through a variety of mechanisms, including CIN. Moreover, telomere dysfunction itself can drive inflammation and decrease ROS defense, thereby promoting carcinogenesis on multiple levels.
Sustaining proliferative signaling
Somatic genetic alterations activate key signal transduction pathways that endow cancer cells with a proproliferative state. In CRC, the major proproliferative signaling pathways are the EGFR-RAS and WNT–β-catenin pathways.
EGFR signaling
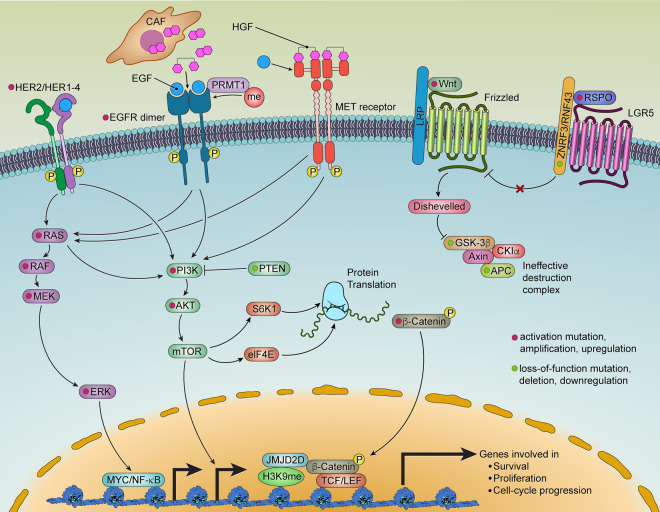
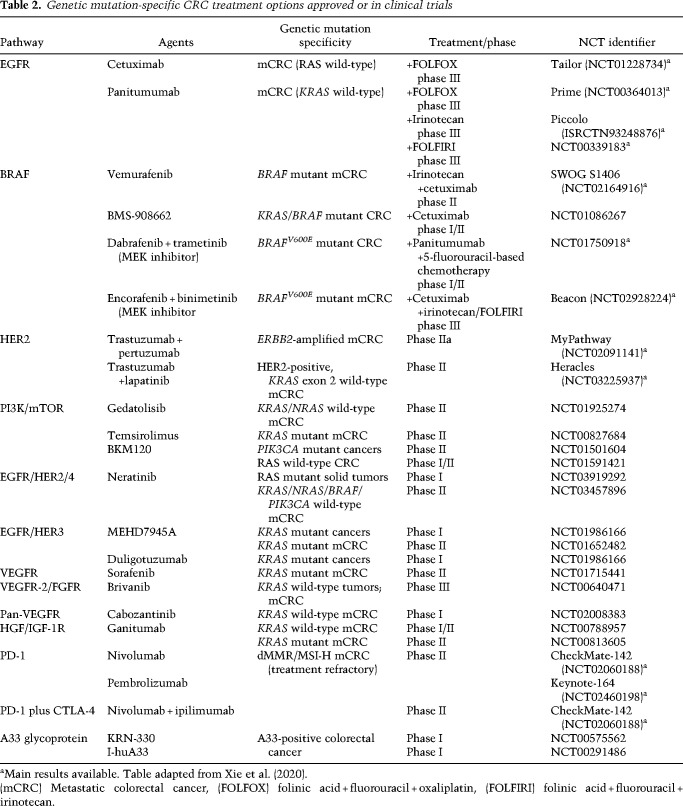
EGFR activation triggers downstream RAS/RAF/MEK/ERK and PI3K/AKT signaling cascades. These signaling components are frequently mutated in CRC (Fig. 2). The exception is EGFR itself, which is rarely mutated (1% of cases of CRC [Barber et al. 2004]) and instead shows overexpression in ∼80% of CRCs (Spano et al. 2005). Enhanced EGFR activation can occur via post-translational modifications involving methylation of R198 and R200 by protein arginine methyltransferase 1 (PRMT1), which enhances its binding to EGF and consequent signaling activation, even in the presence of the EGFR inhibitor (Liao et al. 2015). Moreover, consistent with the receptor tyrosine kinase (RTK) coactivation hypothesis (Stommel et al. 2007), the MET receptor could substitute for EGFR signaling. Specifically, HGF, the MET receptor ligand secreted by cancer-associated fibroblasts (Woolston et al. 2019), can fully replace EGF in driving normal intestinal stem cells into intestinal organoids and promoting the expansion of Apc mutant mouse organoids, findings consistent with the ability of MET to bypass EGFR inhibition in CRC (Joosten et al. 2017, 2019). HER2/ERBB2 activation can also activate EGFR downstream signaling. Notably, HER2 somatic mutations and gene amplifications occur in 7% of cases of CRC and activating mutations of HER2 endow anchorage-independent growth in colon epithelial cells (Fig. 2; Kavuri et al. 2015).
Figure 2.
Growth signaling and their interconnection in CRC.
The clinical effectiveness of EGFR blockade is dictated by the mutational status of its downstream signaling components (Di Nicolantonio et al. 2008); specifically, gain-of-function mutations in RAS, RAF, MEK, or ERK can maintain cancer cell proliferation and survival upon EGFR inhibition. Activating mutations in KRAS, NRAS, or HRAS are collectively present in ∼50% of cases of CRC; these mutations involve codons 12 or 13 (Vaughn et al. 2011) and, less frequently, codons 61, 117, or 146 (Edkins et al. 2006). Such mutations increase the stability of the KRAS-GTP complex resulting in constitutive activation of KRAS (Boguski and McCormick 1993). Recent studies have shown that oncogenic KRAS, long considered undruggable, can be inhibited by small molecules that covalently bind to the less common G12C variant (KRASG12C) (Canon et al. 2019). Such inhibitors have shown antitumor activity in early-stage clinical trials, with most of the success in non-small cell lung cancers and less so in CRC (Hong et al. 2020). In these trials, the lack of responses or resistance may relate to several mechanisms, including (1) a high level of basal RTK activation and phospho-ERK signaling rebound, which suggests that combining EGFR and KRASG12C inhibitors could potentially overcome resistance (Amodio et al. 2020); (2) the coactivation of multiple RTKs, as observed in other cancers (Stommel et al. 2007); and (3) the possibility that oncogenic KRAS plays a role in sustaining metastases rather than primary tumor growth, as suggested by preclinical models (Boutin et al. 2017; Liao et al. 2019).
With respect to the RAS pathway in CRC, the RAS effector, BRAF, is mutated in 10%–15% (Davies et al. 2002; Rajagopalan et al. 2002; Vaughn et al. 2011) of early-stage CRC and around 5% in stage IV CRC, in the hotspot codon 600 (V600E) (Vaughn et al. 2011), and is mutually exclusive of RAS mutations (De Roock et al. 2011). BRAF mutation is a negative prognosis factor for CRC (Sanz-Garcia et al. 2017). BRAFV600E activates MEK-ERK, which in turn activates transcription factors such as MYC (Meyer and Penn 2008) and NF-κB (Suh et al. 2008). The inhibition of BRAF alone showed limited activity in metastatic BRAFV600E CRC owing to EGFR-mediated reactivation of MAPK signaling (Corcoran et al. 2012), leading to the approval of combinations of EGFR inhibitors with BRAF and MEK inhibitors that have now become the standard of care in patients with metastatic BRAF mutant tumors. In addition, the clinical application of specific RAS pathway inhibitors should be mindful of the impact of the drug on host components in the TME. Along these lines, it is worth noting that MEK inhibitors inhibit both cancer cells and T cells, diminishing antitumor immunity, whereas the KRASG12C inhibitor would have an advantage because it is specific to cancer cells and spares T cells (Canon et al. 2019). It is also worth noting that oncogenic KRAS up-regulates cytokines, which recruit immunosuppressive myeloid cells to impair responses to immunotherapy (see “CRC Immunity”).
With respect to the PI3K/AKT/mTOR pathway in CRC, a variety of activating mechanisms can target its signaling components, such as activating mutations in the PI3K regulatory p85 subunit (Cantley 2002) (8% of cases [Luo et al. 2003; Jaiswal et al. 2009]), amplification of AKT1 (6%) (Yaeger et al. 2018), and loss of heterozygosity (23%–35%) or epigenetic silencing (19%) (Zhang et al. 2011) of PTEN, the negative regulator of PI3K signaling (Fig. 2). PIK3CA mutations often coexist with KRAS mutations (Haigis 2017), and mutant KRAS can also activate PI3K signaling via direct interaction (Rodriguez-Viciana et al. 1994; Gupta et al. 2007). The PI3K/AKT/mTOR signaling pathway responds to both intracellular (stress and energy) and extracellular (growth factor and hormone) signals (Francipane and Lagasse 2014). Activation of mTOR leads to increased S6K1 and eIF4E activity to promote protein translation and cell growth in terms of both size and proliferation (Fingar et al. 2002). Accordingly, therapeutic inhibition of PIK3CA and mTOR individually and collectively has shown antitumor activity in preclinical models in CRC (Foley et al. 2017). However, this is not recapitulated in clinical trials, suggesting a highly complex signaling network in vivo that dampened the antitumor activity of single agents and the necessity of combinatorially targeting multiple components of the main pathway and the alternative feedback loops.
Wnt/β-catenin signaling
Wnt/β-catenin signaling maintains normal and cancer cell stemness and chronic Wnt activation promotes CRC. The level of β-catenin in the cytoplasm is controlled by the destruction complex, consisting of AXIN, APC, CK1, and GSK3. The accumulation of Wnt ligands causes β-catenin to dissociate from the destruction complex and migrate to the nucleus. Coupled with TCF or LEF, β-catenin activates many genes enabling tumor growth, including TERT, as mentioned earlier (Hoffmeyer et al. 2012). Wnt itself can be epigenetically activated by lysine demethylase 3 (KDM3) (Li et al. 2017b) or by DNA hypermethylation (Tao et al. 2019). Wnt signaling activating mutations can be divided into ligand-independent alterations of the intracellular signal transduction proteins, such as APC and β-catenin, and ligand-dependent mutations, which amplify endogenous Wnt signal transmembrane transduction, such as R-spondin (RSPO) fusions (Fig. 2). Significantly distinct transcriptional, epigenetic, morphological, and clinical characteristics have been identified in CRCs with ligand-dependent or ligand-independent Wnt activation (Kleeman et al. 2020). Tumors with ligand-dependent Wnt activation remain sensitive to Wnt ligand inhibition (Kleeman et al. 2020).
Ligand-independent Wnt signaling activation is most frequently driven by alterations in APC and CTNNB1 (the gene encoding β-catenin), occurring in 80% and 5% of cases of CRC, respectively (The Cancer Genome Atlas Network 2012). In mouse models of intestinal cancer engineered with loss of Apc and Trp53 and oncogenic Kras activation, genetic restoration of Apc resulted in cancer cell differentiation, tumor regression without relapse, and re-establishment of normal crypt-villus morphology, indicating that Wnt signaling serves a role in tumor maintenance and therefore represents a prime target for intervention (Dow et al. 2015). Besides mutational inactivation, loss of APC can result from degradation by lysosomes due to enhanced vesicular trafficking induced by β-catenin through positive feedback regulation (Jung et al. 2018a). Of note, APC also regulates other β-catenin-independent signaling, such as binding to actin to regulate cell migration and chromosomal fidelity (Jung and Park 2020) and binding to MINK1 to regulate CRC cell proliferation (Popow et al. 2019). Conversely, CRC can down-regulate Wnt signaling repressors that trap β-catenin in inactive complexes (Jung et al. 2018b) and promote β-catenin degradation (Zhang et al. 2016a; Yuan et al. 2017). In CRC, β-catenin's transcription transactivation potential can be enhanced by phosphorylation induced by RTK (van Veelen et al. 2011) and up-regulation of coactivator proteins interacting with the β-catenin/TCF4 complex (Takada et al. 2012; Hua et al. 2019). For example, increased H3K9me3 demethylase JMJD2D interacts with β-catenin to up-regulate target gene transcription (Fig. 2; Peng et al. 2019). Even though loss of function of APC and gain of function of β-catenin could activate Wnt signaling independent of the Wnt ligands, it has been shown that CRC cells with these mutations remain responsive to Wnt ligands blockade (He et al. 2005; Voloshanenko et al. 2013; Jung and Park 2020). Targeting β-catenin is challenging because of its intracellular location and absence of enzymatic activity. A recent study demonstrated the efficacy of direct pharmacological inhibition of β-catenin with RNA interference in a preclinical model, shedding light on further mechanistic study and clinical trials (Ganesh et al. 2016). In addition, targeting downstream genes of β-catenin through inhibition of the precursor mRNA splicing enzyme, CDC-like kinase (CLK), has shown potent antitumor effects in gastrointestinal tumors in xenograft mouse models and is currently being tested in solid tumor in clinical trials (Tam et al. 2020).
Ligand-dependent Wnt signaling is initiated by Wnt ligands binding to the seven-pass transmembrane frizzled (Fz) receptor and its coreceptors, LRP5 and LRP6 (MacDonald et al. 2009). Norrin and RSPO act through the Fz/LRP complex as potent Wnt agonists (Clevers and Nusse 2012). The Wnt ligands secretion is dependent on their palmitoylation by Porcupine (PORCN). PORCN is now spotlighted as a target in several clinical trials for CRC (Jung and Park 2020). RSPO, feeding into the canonical Wnt pathway, strongly promotes intestinal crypt proliferation. The receptor for RSPO, LGR5, is a marker for intestinal stem cells and is a Wnt target gene in CRC. LGR5 physically resides within the Fz/LRP complex and mediates RSPO engagement of the canonical Wnt pathway (MacDonald et al. 2009). LGR5+ stem cells appear to be the preferred cells of origin for intestinal cancer (Barker et al. 2009; Schepers et al. 2012) even though CRC is speculated to have multiple cell of origin lineages, such as LRIG1+ intestinal stem cells (Powell et al. 2012), an area that may benefit from in-depth single-cell omics studies. Besides their essentiality in primary CRC growth, LGR5+ stem cells also appear critical for the long-term growth of CRC liver metastasis (de Sousa e Melo et al. 2017; Fumagalli et al. 2020). In CRC, LGR5 could be degraded by E3 ligases NEDD4 and NEDD4L, the loss of which (occurring in 5% of CRC cases) enhances intestinal stem cell proliferation, induces high-grade dysplasia, and accelerates CRC progression (Novellasdemunt et al. 2020). In tumors without APC mutation or downstream Wnt pathway mutations, up-regulation of RSPO induced by EIF3E-RSPO2 and PTPRK-RSPO3 chromosome rearrangements (10% of cases of CRC) (Seshagiri et al. 2012) can initiate Wnt-dependent hyperplasia and tumor development and synergize with mutant KRAS (Han et al. 2017; Hilkens et al. 2017). RSPO fusion also sensitizes CRC cells to asparaginase therapy by inhibiting GSK3 and limiting protein degradation and free asparagine generation (Hinze et al. 2020). Of note, in a subset of stromal-rich CRC, desmoplastic stromal expression of RSPO ligands could compensate for the absence of epithelial mutation (Kleeman et al. 2020). Cell-surface transmembrane E3 ubiquitin ligase ZNRF3 (deleted in 0.5% of cases of CRC) and its functional homolog RNF43 (mutated in ∼18% of CRC) (Hao et al. 2016a) can negatively regulate Wnt signaling by promoting the turnover of cell membrane Fz and LRP6. Specifically, the two recurrent hotspot mutations in RNF43 in CRC and endometrial cancer are G659fs and R177fs. Both mutations are close to MSI loci, which may explain the high frequency (79.7%) of RNF43 mutations in MSI CRC (Hao et al. 2016a). The membrane clearance of ZNRF3/RNF43 induced by RSPO results in membrane accumulation of Wnt receptors and activation of Wnt signaling (Fig. 2; Hao et al. 2012).
In summary, the EGFR and Wnt/β-catenin pathways sustain genetic and epigenetic alterations in virtually all CRCs and play central roles in driving cancer cell proliferation, as well as other tumor biological hallmarks, as detailed in the following subsections. Moreover, an understanding of the circuitry of these pathways has yielded meaningful therapeutic advances, and the elucidation of resistance mechanisms continues to illuminate novel strategies for improved management via combination therapies.
Evading growth suppressors
Cancer cells overcome growth constraints operative in normal cells via deactivation of cell cycle checkpoints, tolerance of DNA damage, and override of senescence. The mechanisms underlying the genetics and biology of evasion of growth suppression in CRC are as follows.
Bypassing cell cycle checkpoints
Regulation of cell cycle phases—G0/G1, S, G2, and M—involves the orchestrated actions of cyclin-dependent kinases (CDKs), checkpoint kinases, aurora kinases, and Polo-like kinases (PLKs). In concert with activating cyclins, CDKs are a focal point for cell cycle control and receive activating signals originating from mitogenic pathways such as RAS and inhibitory signaling from DNA damage-sensing checkpoint molecules such as p53 (Otto and Sicinski 2017). The G1 cyclin/CDK complex phosphorylates and inactivates Rb to unleash E2F transcription factor activity to up-regulate genes promoting progression to later phases of the cell cycle. The progression through S phase and the transition from G2 to M phases are controlled by cyclin–CDK complex, PLK1, and aurora A/B (Otto and Sicinski 2017). The up-regulation of aurora A kinase in CRC increases microtubule assembly rates and causes transient abnormalities in mitotic spindle geometry, which promotes the generation of lagging chromosomes and aneuploidy (Ertych et al. 2014). Inhibiting aurora A kinase alone in CRC patients showed no significant response in a phase I clinical trial (Cervantes et al. 2012). However, a recent study showed that the tumor suppressor ARID1A, a component of the SWI/SNF chromatin remodeling complex, is synthetically lethal with up-regulated aurora A in CRC. Inhibiting aurora A activity in ARID1A-deficient cells significantly increases G2/M arrest and induces cellular multinucleation and apoptosis (Wu et al. 2018), suggesting an increased efficacy of aurora A kinase inhibition in stratified CRC patients with ARID1A-deficiency. PLK1 reactivates CDK1 after cells recover from DNA damage (Otto and Sicinski 2017). In CRC, inhibition of PLK1 causes prometaphase accumulation and subsequent death of KRAS mutant cells, indicating that a synthetic lethal relationship exists between PLK1 and mutant KRAS (Luo et al. 2009).
Tolerating DNA damage
As noted above, CRC cells incur genetic mutations driven by mutagenic mechanisms and unrepaired DNA damage. The conventional DNA damage response (DDR) initiates with “damage sensing” by DDR sensors specific to DNA double-strand breaks (DSBs), including MRN, and by sensors specific to single-strand DNA (ssDNA) damage, including RPA. The signals are then transduced by ATM for DSBs and ATR for ssDNA damage (Mirza-Aghazadeh-Attari et al. 2018) and amplified by phosphorylation of DDR mediators CHK2 and CHK1 (Liu et al. 2006), respectively. As the downstream effector of CHK1 and CHK2, p53 plays a central role in DDR, determining the fate of damaged cells: cell cycle arrest, senescence, or apoptosis, aligning with its frequent mutation in CRC. Both p53 deletion and increased p53 degradation by MDM2 contribute to unrepaired DNA damage and promotion of tumorigenesis. p53 degradation by MDM2 is inhibited by ARF encoded by the Ink4a/Arf tumor suppressor locus (Pomerantz et al. 1998; Zhang et al. 1998). Telomere-dysfunctional p53 mutant mice develop epithelial cancers, including CRC, whereas telomere-dysfunctional Ink4a/Arf-/- mice do not, underscoring the role of p53-dependent DNA damage signaling in dictating whether telomere dysfunction can serve as a mutational mechanism driving carcinomas (Khoo et al. 2007).
Beyond loss of p53, other DNA damage tolerance mechanisms have been detected in CRC. For example, the efficacy of irinotecan, the topoisomerase I (TOP1) inhibitor, which traps TOP1 on DNA and generates protein-linked DNA breaks that trigger cell death in CRC, is compromised by the fast repair of protein-linked DNA breaks mediated by faster recruitment or retention of 53BP1 at the DNA damage site due to a loss of H4K16 acetylation. Accordingly, histone deacetylase (HDAC) inhibitors can sensitize CRC cells to irinotecan (Meisenberg et al. 2017). Especially in MSI CRC, mutations in a microsatellite tract of MRE11, a component of the MRN complex, lead to a truncated MRE11 protein and deficient DSB repair. This deficiency sensitizes MSI CRC to PARP-1 inhibition in a synthetic lethality manner (Vilar et al. 2011). Along this line, MMR deficiency also showed a synthetically lethal relationship with PTEN-induced putative kinase 1 (PINK1) inhibition (Martin et al. 2011) or DNA polymerase POLG inhibition (Martin et al. 2010).
Overriding senescence
Cell senescence is induced by either a cell division counting mechanism termed “replicative senescence” or stress signaling brought about by DNA damage, oncogenic activation, or oxidative stress, termed “premature senescence” (Holliday 1983). Premature senescence protects cells from tumorigenesis. p53/p21 signaling-dependent senescence (Roninson 2003) is compromised by p53 degradation by MDM2, p53 inactivation via loss of acetylation (Wang et al. 2014), and mutation that impairs the formation of senescence-associated heterochromatin foci (de Barrios et al. 2017). In addition, p21 expression in CRC is inhibited by up-regulated TRIB2 (Hou et al. 2018) and LRH-1 (Kramer et al. 2016). CRC cell senescence, however, is also paradoxically associated with inflammation, which can promote tumorigenesis. CK1α down-regulation induces a senescence-associated inflammatory response, leading to loss of growth control capacity in the absence of p53 and acceleration of cancer cell growth and invasiveness (Pribluda et al. 2013).
In summary, multiple reinforcing genetic events neutralize diverse growth suppressor mechanisms linked to the cell cycle, genomic instability, and senescence. These mechanisms are governed by the orchestrated actions of components of cancer-relevant pathways of p53/MDM2 and Rb/CDK. As these pathways are universally deregulated in CRC, a deepening understanding of these circuitries and their interconnectedness could yield novel targets and synthetic lethal insights with therapeutic potential.
Resisting cell death
Cancer cells circumvent cell death mechanisms triggered by diverse stresses stemming from DNA damage, limited nutrients and oxygenation, and various anticancer treatments. Apoptosis resistance is the most prominent survival strategy adopted by cancer cells, and this strategy can also involve mechanisms of resistance to nonapoptotic forms of cell death such as necrosis and ferroptosis, as well as induction of prosurvival mechanisms such as autophagy.
Resisting intrinsic and extrinsic apoptosis
The major forms of apoptosis are broadly categorized as “intrinsic” or “extrinsic” pathways of apoptosis. The “intrinsic pathway” is regulated by Bcl-2 family proteins, which are either apoptosis promoters (e.g., BAX and BAK) or inhibitors (e.g., BCL-2 and BCL-XL) (Youle and Strasser 2008). Activation of the intrinsic apoptotic cascade initiates with antiapoptotic Bcl-2 proteins inhibition and BAX and BAK activation by BH3-only proteins such as PUMA and NOXA, followed by outer mitochondrial membrane permeabilization and subsequent release of cytochrome C. Cytochrome C activates the caspase-9–caspase-3 cascade, resulting in cleavage of a series of substrates, activation of DNase, and dismantling of dying cells (Youle and Strasser 2008). In CRC, hypoxia-induced YAP activation up-regulates its target gene BCL2L1 (encoding for BCL-XL) to protect cancer cells from apoptosis during oxygen deprivation (Greenhough et al. 2018). In the intestine, procarcinogenic signals can also emanate from dysregulated gut microbiota, which up-regulates IL-17C in intestinal epithelial cells, leading to induction of BCL-2 and BCL-XL to promote cell survival and tumorigenesis (Song et al. 2014). The “extrinsic pathway” is also operative in CRC pathogenesis. This pathway bypasses the mitochondrial step and instead uses cell surface death receptors, such as FAS and TNF receptor, which directly activate the central initiator of apoptosis, caspase-8, leading to caspase-3 activation and cell demolition (Youle and Strasser 2008). In CRC cells, FAS mutation impairs the formation of the death-inducing signal complex, which normally sensitizes cells to FAS ligand (Leon-Bollotte et al. 2011). Germline mutation in a FAS signaling component, FAF1, results in unstable FAF1 and is linked to hereditary CRC (Bonjoch et al. 2020).
p53 and cell death
p53 stands at the nexus of intrinsic and extrinsic pathways by activating transcription of components in both pathways, including PUMA, NOXA, FAS, and other death receptors (Aubrey et al. 2018). It is worth noting that p53 itself can directly evoke apoptosis via its translocation to mitochondria, the process of which is constrained by TRAF6-mediated ubiquitination of p53 in CRC (Zhang et al. 2016b). p53 degradation by MDM2 can be suppressed by mutant KRAS-induced LATS1 activation; however, wild-type KRAS counteracts this process (Matallanas et al. 2011). MYC is another context-specific regulator of p53-mediated apoptosis. In normal cells, MYC amplifies DNA damage-induced apoptosis by inhibiting BCL-XL and BCL-2 and activating BAK, BAX, and BH3-only proteins (Hoffman and Liebermann 2008), and MYC sensitizes cells to death ligands, including TNF, FAS ligands, and TRAIL (Hoffman and Liebermann 2008). In CRC cells, the loss of APC increases MYC (Schmidt et al. 2019) in the context of p53 loss of function, resulting in proliferation, survival, and malignant transformation (McMahon 2014). Moreover, p53 is also involved in other processes integral to cell survival such as autophagy, the process of removing damaged organelles, which prevents necrosis in apoptosis-deficient cells, thereby reducing inflammation (Mokarram et al. 2017). In CRC, the balance between apoptosis and autophagy is maintained by a complex composed of HMGB1 and p53, which regulates cytosolic localization of the reciprocal binding partner (Livesey et al. 2012b). p53 loss leads to cytosolic accumulation of HMGB1, which increases autophagy and decreases apoptosis, whereas HMGB1 loss increases cytosolic p53, resulting in increased apoptosis and decreased autophagy (Livesey et al. 2012a). Beyond its control of cancer cell survival, autophagy impacts other cancer hallmarks, including tumor immunity via degradation of MHC-I (Yamamoto et al. 2020) and cancer metabolism via its scavenging of nutrients.
Resisting nonapoptotic cell death
Other forms of cell death, such as necrosis and ferroptosis, are also deactivated in CRC cells. Necrosis is observed in 96% of cases of CRC and correlates positively with higher tumor stages (Pollheimer et al. 2010) and intestinal inflammation. CRC cells can resist necroptosis, a programmed form of necrosis, through elevated HGF-MET signaling, which reduces the level of necroptosis mediator RIPK1 (Seneviratne et al. 2015). In a caspase-8-deficient CRC mouse model, high levels of RIP3 sensitized cancer cells to second mitochondria-derived activator of caspase (SMAC) mimetic-induced necroptosis, pointing to a potential clinical strategy for patients with caspase-8 deficiency (He et al. 2017a). Finally, ferroptosis, the iron-dependent oxidative cell death, can be suppressed by phospholipid glutathione peroxidase GPX4 (Sui et al. 2018). Accordingly, a small molecule inhibitor of GPX4 (RSL3) induces ferroptosis in CRC (Sui et al. 2018).
In summary, CRC cells use various mechanisms to deactivate cell death programs via alterations in key regulators (e.g., p53 and MYC) and effectors (e.g., Bcl-2 family and FAS) of the apoptosis machinery. Activation of the death resistance strategies is critical for cancer cells to survive under various stresses. The universal activation of these mechanisms underscores their importance as therapeutic targets across diverse cancer types, genotypes, and etiologies.
Deregulating cellular energetics and metabolism
Normal cells primarily use oxidative phosphorylation (OXPHOS) to meet energy demands, and under low-oxygen conditions, normal cells undergo anaerobic glycolysis, which produces abundant lactate and limited ATP (Vander Heiden et al. 2009). Cancer cells have elevated requirements and needs for the nutrient to maintain their increased demand for energy. Therefore, extracellular and available nutrients regulate cancer cell proliferation and differentiation. However, unlike normal cells, cancer cells can adapt to nutrient conditions due to their greater metabolic plasticity. For example, cancer cells undergo metabolic reprogramming known as “aerobic glycolysis” or “the Warburg effect,” generating lactate even in the presence of oxygen. Glycolysis generates metabolic intermediates serving as the building blocks for biosynthesis (Vander Heiden et al. 2009; Liberti and Locasale 2016). In addition to aerobic glycolysis, CRC cell growth is also supported by enhanced glutamine utilization, lipid metabolism, one-carbon metabolism, and short-chain fatty acids metabolism. Of note, CMS3 epithelial CRCs show prominent activation of multiple metabolism signatures (Guinney et al. 2015). Therefore, manipulation of metabolic pathways will be targeting specifically this CRC subtype. Moreover, cancer cells can also regulate and reshape TME and its metabolic milieu, which also profoundly affects the function of diverse host cell types of the tumor.
Aerobic glycolysis
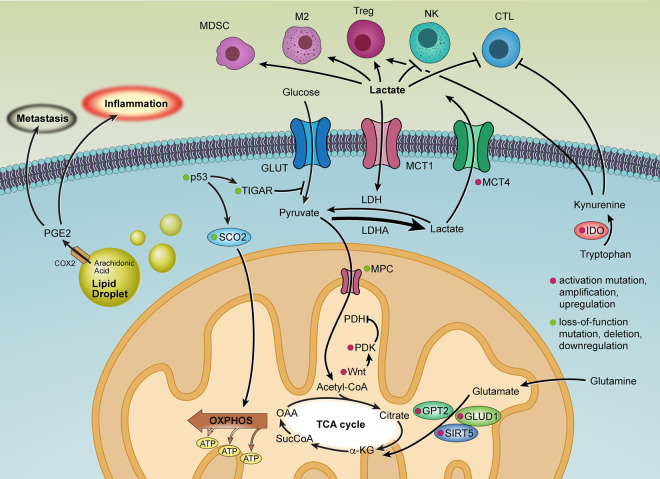
In CRC, cancer cell metabolic reprogramming reflects the impact of signature genetic alterations, which regulate key metabolic enzymes and mechanisms (Fig. 3). For example, Wnt signaling drives glycolysis via up-regulation of PDK1, which inhibits pyruvate flux to support mitochondrial respiration (Pate et al. 2014). Similarly, the frequent loss of p53 function in CRC reflects the neutralization of p53's induction of TIGAR, which normally restricts glycolytic flux (Bensaad et al. 2006), and loss of p53's up-regulation of SCO2, which normally drives OXPHOS and mitochondrial respiration (Wang et al. 2018a), thus collectively facilitating the shift to glycolysis (Matoba et al. 2006). Moreover, CRC stem cells have elevated LDHA levels, enhancing glycolysis (Ji et al. 2017). In early stages of CRC development, the Warburg effect can be reinforced by deletion or underexpression of mitochondrial pyruvate carrier (MPC) resulting from mitochondrial DNA truncating mutations (Bensard et al. 2020; Yuan et al. 2020). Accordingly, re-expressing MPC increases mitochondrial pyruvate oxidation and impairs anchorage-independent growth of CRC cells (Schell et al. 2014). Finally, CRC cells are known to display metabolic heterogeneity in the TME; i.e., some cancer cells show anaerobic glycolysis with low MPC while others use OXPHOS with high MPC to generate ATP (Brown et al. 2018). Such heterogeneity enables symbiotic metabolic exchange across cancer cells and host cells in the tumor ecosystem, as reviewed elsewhere (Dey et al. 2021).
Figure 3.
Aerobic glycolysis and metabolic remodeling of the tumor microenvironment in colorectal cancer.
In TME, nutrient deprivation dampens cytotoxic T lymphocyte (CTL) mTOR activity, glycolytic capacity, and IFN-γ production, which impairs antitumor activity (Chang et al. 2015); and, notably, checkpoint inhibitors can reverse the CTL metabolic profile and restore antitumor immunity. Conversely, in CRC cells, checkpoint blockade inhibits glycolysis via down-regulation of glycolysis enzymes, thereby increasing glucose availability in the TME and aiding CTL (Chang et al. 2015). Interestingly, immunosuppressive regulatory T cells (Tregs) rely on folic acid oxidation (Kinoshita et al. 2012) and OXPHOS and thus can thrive in the low-glucose conditions of the TME (He et al. 2017b; Wang et al. 2017). Moreover, the abundant lactate in the TME is profoundly immunosuppressive, that is, this oncometabolite stimulates immune suppressor cells (Tregs, myeloid-derived suppressor cells [MDSCs], and M2 macrophages) and inhibits antitumor immune cells (natural killer cells [NKs] and CTLs) via epigenetic mechanisms involving histone lactylation (Fischer et al. 2007; Husain et al. 2013; Hobson-Gutierrez and Carmona-Fontaine 2018; de la Cruz-López et al. 2019; Zhang et al. 2019). Along these lines, in CRC, the highly glycolytic cancer cells and fibroblasts up-regulate MCT4 to export high intracellular lactate into the TME (Brown et al. 2018; Fisel et al. 2018). Meanwhile, extracellular lactate is imported via up-regulated MCT1 in some cancer cells and LGR5+ intestinal stem cells to replenish the TCA cycle and sustain OXPHOS (Fig. 3; Fu et al. 2017; Rodríguez-Colman et al. 2017; Brown et al. 2018).
Glutamine, lipid, and one-carbon metabolism
In glutamine metabolism, the anaplerotic entry (to replenish intermediates of a metabolic event) of glutamine into the TCA cycle is supported by GLUD1, which is activated by overexpressed mitochondrial SIRT5 (Wang et al. 2018b), and reinforced by up-regulated GPT2, which is induced by PIK3CA mutations (Hao et al. 2016b). In lipid metabolism, CRC cells show increased uptake of extracellular lipids and up-regulation of enzymes driving de novo lipid biogenesis (Brown et al. 2018), supporting critical cell processes such as membrane formation, signal transduction, protein post-translational modifications, and energy storage (Brown et al. 2018). CRC cells also possess abundant lipid droplets, serving as reservoirs for COX2 and the site for PGE2 synthesis (Accioly et al. 2008; Brown et al. 2018), which in turn can increase inflammation, as specified earlier (Fig. 3). Finally, CRC cells use one-carbon metabolism (1CM), which provides nucleotides and fatty acids for cell proliferation and chromatin remodeling (Newman and Maddocks 2017; Brown et al. 2018). The levels of 1CM metabolites such as S-adenosylmethionine and 1CM enzymes such as PHGDH are elevated in CRC (Locasale 2013; Ryall et al. 2015; Brown et al. 2018).
Short-chain fatty acids (SCFAs)
SCFAs, like acetate, propionate, and butyrate, are (by)products of fiber fermentation in the gastrointestinal tract. They are produced via anaerobic bacterial fermentation within the colon and are thought to be protective of colon carcinogenesis (Ríos-Covián et al. 2016). Interestingly, while intestinal microbiota and their metabolites, like bile acids (Gadaleta et al. 2017), are known to fuel intestinal carcinogenesis through promotion of inflammation, recent data has shown that SCFAs can decrease CRC through suppression of inflammation (Louis et al. 2014; Singh et al. 2014). SCFAs are known to act as signaling molecules that can inhibit HDACs and as ligands for G protein-coupled receptors (Rooks and Garrett 2016). SCFA-driven HDAC inhibition tends to promote anti-inflammatory cell phenotypes (Rooks and Garrett 2016). SCFAs enhance barrier function and immune tolerance and promote gut homeostasis through increased mucus production by goblet cells (Gaudier et al. 2004; Burger-van Paassen et al. 2009; Rooks and Garrett 2016). Together, these findings provide a basis for a healthy and fiber-rich diet in inhibiting colon cancer development. Moreover, SCFAs are tryptophan metabolites that bind to the aryl hydrocarbon receptor (AhR), and retinoic acid (Levy et al. 2016; Rooks and Garrett 2016; Shibata et al. 2017). Commensal Lactobacillus uses tryptophan to produce AhR ligands, such as indole-3-aldehyde (Zelante et al. 2013). AhR activation is critical for the organogenesis of intestinal lymphoid follicles (ILFs) and AhR-expressing immune cells, including RORγt+ type 3 innate lymphoid cells (ILC3s) that are involved in ILF genesis (Kiss and Diefenbach 2012). In addition, AhR-induced IL-22 production by ILCs drives secretion of the antimicrobial peptides lipocalin-2, S100A8, and S100A9, which provide protection from translocating microbes (Kiss and Diefenbach 2012; Lee et al. 2012). Furthermore, IL-22 is a potent regenerative cytokine that restores barrier integrity (Sonnenberg et al. 2011). Tryptophan metabolism in CRC also contributes to an immunosuppressive TME. Specifically, CRC expresses indoleamine 2,3-dioxygenase (IDO), which metabolizes tryptophan to kynurenine, facilitating tumor immune escape and supporting tumor growth (Fig. 3; Thaker et al. 2013).
Together, these cancer cell-intrinsic and intercellular metabolic cross-talk mechanisms enable cancer cells to adapt to limited nutrient availability in the TME and serve to suppress immune surveillance via several mechanisms. Thus, inhibition of such metabolic processes may impact both cancer cell anabolic growth and tumor immunity.
Tumor-promoting inflammation
Inflammation is the manifestation of a host immune response toward exogenous or endogenous signals. The intestine is a unique environment in host defense in which inflammation can play a central role in resolving pathogenic infection, maintaining normal intestinal function, or promoting tumorigenesis (Balkwill and Mantovani 2001; Mantovani et al. 2008; Netea et al. 2017). The increased risk of colon cancer in patients with IBD historically established the role of inflammation in the development of CRC and the early interest of researchers in this particular malignancy (Rubin et al. 2012; Baker et al. 2019).
CRC inflammatory cytokine milieu
Multiple factors contribute to local and systemic inflammatory cytokine networks including cancer-driving mutations (Cooks et al. 2014; West et al. 2015). Extensive work shows that NF-κB is a crucial transcription regulator of inflammation, where constitutive activation of NF-κB in CRC mouse models promotes tumorigenesis through accelerated loss of Apc (Shaked et al. 2012). NF-κB is also known to induce DNA damage through production of ROS (Tilstra et al. 2012) and facilitate neoplasia and antiapoptotic capabilities of intestinal epithelial cells through IL-6 production (Greten et al. 2004) and TNF activation (Schwitalla et al. 2013a). The NF-κB pathway regulates the expression of most tumor-promoting cytokines both in cancer cells and immune cells. Aberrant NF-κB activation was detected in >50% of CRC and CAC (Kojima et al. 2004; Karin and Greten 2005). In CRC, the classical NF-κB activation usually occurs via pattern recognition receptors (PAMPs) or cytokines like IL-1β, TNF, and IL-17 (Greten et al. 2004). The alternative pathways, including p52/RelB, could be activated by RANKL and lymphotoxin-β (Terzić et al. 2010; Ramakrishnan et al. 2019), both of which further regulate the gut immune cells and microbiome homeostasis. Moreover, loss of p53 has been shown to increase intestinal permeability, initiating NF-κB-dependent inflammation and the induction of epithelial-mesenchymal transition (Schwitalla et al. 2013b).
Similarly, in IL-10-deficient mice, a spontaneous inflammation model, total mutation rates in colon are five times higher compared with those in wild-type mice, underscoring that inflammation can induce DNA damage and potentially MSI (Sato et al. 2006). A better understanding of cancer cell mutations modulating the cytokine network and inflammatory response can shed light on leveraging and targeting the TME to improve therapeutic responses.
Microbiota and inflammation in CRC
In CRC, tumor stages have been linked to microbiota landscape changes denoted “dysbiosis,” often responsible for initiating the inflammatory response and influencing tumor progression (Sears and Garrett 2014). A few microbial species have emerged as potentially important species enriched in CRC (Irrazábal et al. 2014). Fusobacterium nucleatum (Castellarin et al. 2012; Kostic et al. 2012; Nejman et al. 2020) is found to confer chemoresistance in a TLR4- and MyD88-dependent manner (Yu et al. 2017). Enterotoxigenic Bacteroides fragilis (ETBF) contributes to colonic barrier damage and promotes Wnt and NF-κB downstream signaling (Wu et al. 1998; Housseau and Sears 2010). Interestingly, if treated with the antibiotic cefoxitin, ETBF can be eliminated in murine models followed by reduced tumorigenesis and IL-17A expression, indicating that ETBF plays a role in Th17 response (DeStefano Shields et al. 2016; Housseau et al. 2016). Human CRC with Streptococcus gallolyticus showed increased production of proinflammatory cytokines such as COX2, IL-1, and IL-8 (Abdulamir et al. 2010). Reciprocally, intestinal inflammation can increase the proportion of highly genotoxic microbes and promote tumorigenesis (Arthur et al. 2012). Stool transplants from patients with CRC (but not from healthy individuals) into the colons of germ-free mice can increase the emergence of polyps, intestinal dysplasia, inflammation, and Th1 and Th17 cell accumulation (Wong et al. 2017). Application of next-generation sequencing allows sequencing of traditionally “unculturable” microbial species (Single-cell microbiology. [Editorial] 2016; Quince et al. 2017). Research on the complex interactions and cross-talk among diet, lifestyle, genetics, host immune response, and microbial activity is actively ongoing and may yield new insights into CRC pathogenesis and treatment strategies (Elinav et al. 2013; Brennan and Garrett 2016).
Innate lymphoid cells (iLC)
iLC is a critical immune cell population that regulates some of the host–commensal bacteria relationships that can impact immunity, inflammation, and tissue homeostasis in the intestine (Bouskra et al. 2008; Maloy and Powrie 2011; Sonnenberg and Artis 2012). They reside in mucosal surfaces to potentiate immune responses, sustain mucosal integrity, and maintain tissue homeostasis. ILC2 subset promotes CRC progression, mostly through the secretion of IL-22. IL-22 selectively acts on epithelial cells to induce STAT3 phosphorylation and proliferation (Kirchberger et al. 2013; Loyon et al. 2019; Wang et al. 2020). Besides the functions of supporting intestinal cell regeneration and inhibition of bacterial translocation, the IL-22 pathway has also recently been shown to cooperate with mutant KRAS and enhance cancer cell proliferation, in part through augmentation of the Myc pathway (McCuaig et al. 2020). iLCs regulate bacterial homeostasis and are regulated by the microbiome to produce a variety of cytokines, which can be both protumorigenic and antitumorigenic (Langowski et al. 2006; Geremia et al. 2011; Abt et al. 2012; Sonnenberg and Artis 2012).
In summary, inflammation is known to play important roles in tumorigenesis, the local cytokine milieu, and interaction between cancer-driving mutations and gut microbiome. Although many proinflammatory cytokines have been heavily studied, the net effect on local immune populations, host-microbiome interaction, and cancer progression remain to be fully elucidated in detail.
CRC immunity
Although CRC usually does not respond to immune checkpoint blockade (ICB) therapy, the correlation between certain immune cells and relapse and metastasis (Van den Eynde et al. 2018) points to the role of antitumor immunity in the development and progression of CRC. The FDA approval of anti-PD-1 therapy for MSI-H CRC represents a landmark advance. However, disease recurrence in patients with MSI-H CRC who received anti-PD-1 therapy and the complete lack of response in patients with MSS CRC underscore the need for a detailed analysis of immune composition and associated mechanisms operating in the CRC TME. For a full contextual understanding of tumor immunity to be possible, immune analyses must be integrated with information on specific CRC genetic alterations, the CMS subtypes, tumor mutational burden, and the gut microbiome, among other features as detailed in the following subsections.
Immunosuppressive TME
The abundance of several antitumor immune populations, including CTLs, NKs, and activated dendritic cells (DCs), is inversely correlated with TNM stage in CRC (Schwaab et al. 2001; Mlecnik et al. 2011; Jobin et al. 2017) and is significantly lower in CRC compared with normal mucosa, as revealed by single-cell RNA sequencing (scRNA-seq) (Zhang et al. 2018, 2020). CTLs and Th1 cells are critical contributors to IFN-γ-mediated antitumor immune response (Dunn et al. 2006; Mucida et al. 2013), and NKs have been shown to effectively target cancer cells that lose MHC-I antigens (Vivier et al. 2012). In addition to poor tumor infiltration, resident T and NK cells often exhibit exhaustion phenotypes with low cytotoxicity, increased PD-1, and decreased IFN-γ expression (Jobin et al. 2017). The exhaustion of T cells can be triggered by high PD-L1 expressed by regulatory B cells and immature DCs (Legitimo et al. 2014; Liu et al. 2018a). In addition, although Treg density correlates inversely with CRC PD-L1 levels (Masugi et al. 2017), apoptotic Tregs convert ATP to adenosine, which potently contributes to T-cell exhaustion by A2A-mediated IL-2 suppression (Maj et al. 2017).
A prominent feature of the immunosuppressive CRC TME is a preponderance of suppressive myeloid cells, such as tumor-associated macrophages (TAMs) and tumor-associated neutrophils (TANs), often collectively referred to as polymorphonuclear MDSCs (PMN MDSCs). It is important to appreciate the high degree of complexity in function and cell states of PMN MDSCs, especially in the context of TAM depletion therapies such as anti-CSF1R therapy (Zhang et al. 2020). scRNA-seq analyses have revealed anti-CSF1R resistant subpopulations such as Vegfa+ TAMs, purported to promote angiogenesis and tumorigenesis via secretion of VEGFA (see “Inducing Angiogenesis”). Besides, TAMs also secrete anti-inflammatory cytokines and growth factors that support tumor growth and proteolytic enzymes that accommodate tissue remodeling and tumor expansion (Erreni et al. 2011). Furthermore, TANs and TAMs are known to inhibit CTL activity via high production of ARG1 and ROS (Doedens et al. 2010; Movahedi et al. 2010; Wu et al. 2014) and promote CRC metastasis, given that inhibition of PMN MDSCs by blocking the key receptor CXCR2 leads to a reduction in metastatic disease (Jackstadt et al. 2019). Thus, the prominent role of PMN MDSCs in the hallmarks of CRC emphasizes the importance of defining various myeloid populations, elucidating their tumor biological roles, and devising and testing myeloid-targeted therapies.
Immune features of MSI-H and MSS subtypes
Among the CMSs, CMS1 includes most MSI-H CRCs whereas CMS2–4 are generally MSS CRCs. Correspondingly, MSI-H CRCs, comprising 15% of CRC cases and exhibiting dense immune-infiltrates, show high responsiveness to ICB therapy. In contrast, MSS CRCs, representing 85% of cases and characterized as “immune-cold” tumors, show no response to ICB therapy. Consistent with increased neoantigens, MSI-H tumors show a more than twofold increase in T-cell infiltration relative to MSS CRCs and increased CD8+/CD45RO+ memory T-cell populations, which portend better responses to ICB (Watanabe et al. 2001; Bauer et al. 2013; Sherwood et al. 2013; Kather and Halama 2019). Furthermore, scRNA-seq analyses of MSI-H CRC have shown increased proliferative Th1-like cells, which are known to enhance the effector activity of macrophages, B cells, and CD8+ T cells. In contrast, MSS tumors show increased proliferative Th17 cells, which antagonize Th1 cells, although the precise functions of Th1 and Th17 cells in CRC have yet to be fully dissected (Zhang et al. 2018). Finally, the distinct immune profiles of MSI-H and MSS CRCs are reinforced by multiple studies showing the presence and absence, respectively, of Crohn's-like lymphoid reaction (CLR) structures. These CLR structures are peritumoral lymphoid nodules, a type of tertiary lymphoid structure that is increasingly recognized as a critical modulator of local immunity (Schürch et al. 2020). A majority of MSI-H CRCs possess CLRs (Kim et al. 1994; Risio et al. 1996; Schürch et al. 2020), whereas most MSS CRCs do not. Such distinct infiltration patterns encourage in-depth analysis of these differentially infiltrated populations and corresponding recruiting mechanisms in CRC.
Although patients with MSI-H CRC present a robust response to ICB that is superior to the use of conventional chemotherapy in the setting of first-line treatment of metastatic CRC (Andre et al. 2020), it is important to emphasize that, in most of these patients, the disease rapidly develops immune-escape mechanisms (Asaoka et al. 2015). Such resistance mechanisms are a high-priority area of active investigation and may theoretically relate to (1) activation of alternative immune checkpoint molecules such as LAG3; (2) up-regulation of immunosuppressive factors, such as IDO and CXCL3 (see below), which recruit suppressive myeloid cells; (3) JAK1/2 loss-of-function mutations, which critically distort JAK1/2–STAT1 signaling and IFN-γ-mediated immune response (Dunn et al. 2006; Shin et al. 2017; Syn et al. 2017; Xu et al. 2018); and (4) mutations in key components of antigen presentation processes, such as biallelic losses of B2M and HLA genes and HLA class I master regulator NLRC5 (Grasso et al. 2018). Genome-wide CRISPR screening of multiple cancer cells including CRC revealed core cancer-intrinsic CTL evasion genes, particularly involved in the autophagy–NF-κB axis (Lawson et al. 2020).
CMS2-4/MSS CRCs, each with distinct genetic characteristics, display significant heterogeneity with immune cell infiltration and Immunoscore (Mlecnik et al. 2016; Pagès et al. 2018), suggesting the relevance of specific genetic mutations and potential variations of resistance mechanisms to ICB. For example, in CMS2, APC inactivation and β-catenin activation correlate with decreased T-cell infiltration, which would diminish responsiveness to ICB therapy (Grasso et al. 2018; Luke et al. 2019). In CMS3, the highly frequent KRAS mutation correlates with down-regulation of MHC-I, hence leading to low immunogenicity (Atkins et al. 2004; Koelzer et al. 2015). KRAS activation also drives increased GM-CSF production, which recruits immunosuppressive PMN MDSCs (Pylayeva-Gupta et al. 2012; Busch et al. 2016) into the CRC TME (Liao et al. 2019). In the iKAP model (Boutin et al. 2017), oncogenic KRAS also represses Irf2 expression, which in turn represses Cxcl3 expression. Thus, an oncogenic KRAS-driven increase in CXCL3 secretion results in the recruitment of immunosuppressive CXCR2+ PMN MDSCs. These murine observations appear to have clinical relevance on several levels including (1) KRAS mutation is mutually exclusive of IRF2 deletion in human CRC, (2) high IRF2 expression correlates positively with responsiveness to anti-PD-1 therapy in patients with MSI-H CRC, and (3) MSS iKAP CRCs can respond to anti-PD-1 therapy upon inhibition of the CXCL3–CXCR2 axis (Liao et al. 2019), suggesting targeting MDSCs combinatorially with ICB will synergistically increase treatment efficacy. Finally, CMS4 cancers are characterized by exuberant stromal infiltration and abundant fibroblast-derived TGF-β (Guinney et al. 2015). TGF-β is a powerful immunosuppressive factor that can block the recruitment of CD8+ and CD4+ T cells (Gorelik and Flavell 2000; Thomas and Massagué 2005) and antagonize the Th1-effector cell phenotype, both of which are reversed by TGF-β inhibitor treatment (Mariathasan et al. 2018; Tauriello et al. 2018; Batlle and Massagué 2019). TGF-β also polarizes TANs to a “protumor” N2 state (Fridlender and Albelda 2012; Masucci et al. 2019; Shaul and Fridlender 2019), which secretes extracellular matrix (ECM) remodeling enzymes and proangiogenesis factors to promote metastasis and angiogenesis (Gregory and Houghton 2011; Khanh et al. 2011; Mantovani et al. 2011; Piccard et al. 2012; Zhang et al. 2016c). In addition, scRNA-seq profiling (Lee et al. 2020) has shown that CMS4 is enriched with SPP1+ macrophages and myofibroblasts, which may contribute to ICB resistance by promoting inflammation (O'Regan et al. 2000; Irby et al. 2004) and survival of CRC stem cells (Vermeulen et al. 2010), respectively.
In summary, acquired resistance to ICB therapy, coupled with the varied features of the immune composition in CRC subtypes, underscores the need for systematic dissection of the contribution of specific signature mutations, immune cells, and their immune-modulatory factors in maintaining antitumor immunity in specific subsets of CRC. Such knowledge will guide the rational design of combination therapies targeting key oncogenic pathways with immunosuppressive functions and the major immunosuppressive cell populations.
Activating invasion and metastasis
Metastasis, predominantly to the liver, is the major cause of death in CRC. Although the conventional bottleneck model, where metastasis occurs at later stages of CRC development is more prevalent (Norton and Massagué 2006; Klein 2009; Leung et al. 2017), disseminated cells can also seed metastatic sites early when the primary carcinoma is still clinically undetectable (Hu et al. 2019; Ryser et al. 2020). Accumulating genetic alterations in the primary CRC cells, especially KRAS mutations and TGF-β signaling activation, enable CRC cell metastatic potential via promotion of epithelial–mesenchymal transition (EMT), increased intra-/extravasation, and colonization of secondary organs.
KRAS-driven TGF-β signaling activation
Although many signaling pathways contribute to CRC metastasis, TGF-β and oncogenic KRAS signaling play prominent roles. A ligand-bound TGF-β receptor initiates complex formation and nuclear translocation of SMAD2/3/4 to activate its target genes. In CRC, SMAD4 tumor suppressor is mutated in 12% of patients with metastatic or unresectable CRC (Mehrvarz Sarshekeh et al. 2017) and is among the most common additionally mutated genes in metastasis compared with primary CRC tumors (Goswami et al. 2015). Loss of SMAD4 switches BMP signaling from tumor-suppressive to prometastatic by activating RHO signaling via ROCK, promoting EMT (Voorneveld et al. 2014), and by recruiting CCR1+ myeloid cells that secrete MMP9 to facilitate cancer cell invasion and metastasis (Itatani et al. 2013). In the iKAP model, inhibition of TGF-β dramatically decreased tumor invasion, indicating that the TGF-β pathway is a key mediator of KRAS-driven invasiveness (Boutin et al. 2017). In another model, dual blockade of TGF-β and PD-L1 cured established CRC metastases (Tauriello et al. 2018), motivating clinical trials. Stromal TGF-β also stimulates the secretion of IL-11 from cancer-associated fibroblasts and triggers GP130/STAT3 signaling in cancer cells, increasing the efficiency of organ colonization and metastasis formation (Calon et al. 2012), the cross-talk of which is blocked by TGF-β inhibition (Calon et al. 2015).
Metastasis colonization
Many cancer cell- and liver-derived factors cooperate to promote liver colonization, consistent with the seed-and-soil concept (Fidler 2003). CRC cells form an inflammatory premetastatic niche via secretion of TIMP (Seubert et al. 2015) and ITGBL1-rich extracellular vesicles (Ji et al. 2020). In the liver, profibrotic hepatic stellate cells produce a supportive stromal matrix for CRC metastases (Badiola et al. 2012). In addition, hepatic ANGPTL6, a soluble factor enriched in hepatic blood vessels, complexes with E-cadherin and α(6) integrin on CRC cells to enhance their liver homing (Marchio et al. 2012). Following colonization, the metastatic cancer cells express PAD4, which induces citrullination of the ECM to promote greater adhesion and increase expression of characteristic epithelial markers (Yuzhalin et al. 2018). CRC cells also undergo adaptation to the new TME through metabolic reprogramming via up-regulation of ALDOB required for fructose metabolism, fueling cell proliferation (Bu et al. 2018). Moreover, liver metastatic CRC cells shift from a colon-specific to a liver-specific transcription profile through remodeling of the enhancer and superenhancer landscape (Teng et al. 2020).
In summary, driven by oncogenic KRAS mutation and the subsequent TGF-β signaling activation, CRC metastasis requires active cross-talk between primary cancer cells and the secondary organ, as well as the adaptation of primary cells by metabolic and transcriptional reprogramming. TGF-β signaling inhibition strategies in clinical trials would be expected to yield promising results through combination with other targeted therapies and immunotherapies.
Inducing angiogenesis
Angiogenesis plays a critical role in tumor growth by supplying nutrients and oxygen to meet energy demands (Carmeliet and Jain 2000) and is tightly regulated by the balance between pro- and antiangiogenetic ligands, such as VEGFA and TSP-1, respectively. Components of the VEGF pathway are often aberrantly activated in CRC. For example, up-regulated VEGFC and its receptor VEGFR3 in CRC result in increased lymphatic vessel density and permeability, as well as metastases in the lymph nodes, lungs, and livers (Tacconi et al. 2015). CRC angiogenesis is also promoted by tumor-infiltrating immune cells, such as neutrophils (Gordon-Weeks et al. 2017), CD11b+ myeloid cells (Lim et al. 2015), and macrophages (Zheng et al. 2013). Exosomes containing proangiogenic mRNA and microRNAs are secreted from CRC cells and regulate VEGFR2 and ZO-1 in endothelial cells and consequently promote vascular permeability and angiogenesis (Zeng et al. 2018).
Antiangiogenesis therapy is the standard of care for the treatment of metastatic CRC, although the survival benefit is limited due to acquired resistance linked to genetic mutations (Zhou et al. 2020). For example, amplification of POLR1D-induced up-regulation of VEGFA is associated with acquired resistance to the anti-VEGFA antibody therapy (Zhou et al. 2020). Another resistance mechanism involves vessel co-option, a process in which cancer cells use pre-existing normal tissue blood vessels to support growth (Kuczynski et al. 2019). This nonangiogenic mechanism of acquired resistance (Frentzas et al. 2016) is observed in nearly all CRC lung and liver metastases (Kuczynski et al. 2019), indicating that a combination of anti-VEGF and vessel co-option inhibition can confer better responses (Frentzas et al. 2016). In addition, the higher stiffness in CRC liver metastases than in primary tumors, acquired through the remodeling of ECM driven by hypoxia-induced elevation of hyaluronic acid and sulfated glycosaminoglycans (Rahbari et al. 2016), limits perfusion of drug delivery (Shen et al. 2020). Drugs normally prescribed for the treatment of hypertension can target the renin-angiotensin system, inhibit fibroblast contraction and ECM deposition, and reduce stiffness of liver metastatic tissue. Correspondingly, patients receiving combined anti-VEGF antibody and renin-angiotensin inhibitor therapy experience longer survival (Shen et al. 2020).
In summary, CRC angiogenesis driven by aberrant up-regulation of the VEGF pathway is targeted through antiangiogenesis targeted therapies, but these have limited benefit due to acquired resistance. A deeper understanding of the resistance mechanisms such as compensational signaling pathways (e.g., EGFR), vessel co-option, and ECM remodeling is expected to reveal novel targets and inform combination treatments to overcome therapy resistance.
Outlook of CRC therapeutics and future perspectives
A maturing CRC genomic atlas at the single-cell level, refined genetic model systems, and targeted therapies provide a foundation to advance our understanding of the biological hallmarks of CRC. As illustrated in Figure 4 and summarized in Table 2, this framework has led to approved and investigational therapeutics showing clinical benefit but has also highlighted critical knowledge gaps that must be addressed to make meaningful clinical advances.
Figure 4.
Current treatment strategies approved for or tested in clinical trials targeting the hallmarks of colorectal cancer and knowledge gaps remaining as obstacles to improve treatment efficacy.
Table 2.
Genetic mutation-specific CRC treatment options approved or in clinical trials
Therapies targeting the proliferative signaling in CRC include anti-EGFR monoclonal antibodies (e.g., cetuximab and panitumumab), and combinations with small-molecule inhibitors targeting the BRAF-MEK-ERK pathway are now standard of care. For example, in BRAFV600E mutant CRC, triplet therapy with BRAF (encorafenib), EGFR (cetuximab), and MEK (binimetinib) inhibitors increases overall survival (OS) compared with the standard treatment (Kopetz et al. 2019). In KRASG12C CRCs, the KRASG12C covalent inhibitor is showing promising antitumor activity in early-stage clinical trials (Canon et al. 2019). Similarly, HER2 activation, which sustains MAPK phosphorylation and confers resistance to EGFR inhibition (Kavuri et al. 2015), appears to be a viable target; two clinical trials of HER2 inhibitors (using a combination of trastuzumab plus lapatinib [Sartore-Bianchi et al. 2016] and trastuzumab and pertuzumab [Meric-Bernstam et al. 2019], respectively) have shown responses in KRAS wild-type, HER2-positive metastatic CRC. These trials, with overall response rates of >30%, are notable because metastatic CRC can be notoriously refractory to standard treatments such as cetuximab or panitumumab. The well-established HER2 diagnostics and active HER2 inhibitors set the stage for biomarker-driven trials for KRAS wild-type metastatic CRC with HER2 activation (Sartore-Bianchi et al. 2018).
Antiangiogenesis therapies, such as FDA-approved monoclonal antibodies against VEGF (e.g., bevacizumab [Kuipers et al. 2015], aflibercept [Holash et al. 2002], and ramucirumab [Krupitskaya and Wakelee 2009]), have shown activity in CRC. Several clinical trials have established that combined anti-VEGF and chemotherapy regimens can benefit patients with CRC (Chiorean et al. 2015; Tabernero et al. 2015; Van Cutsem et al. 2015; Cremolini et al. 2016; Kuboki et al. 2017). In a phase 1b/2 trial in KRAS mutant metastatic CRC, the inhibitor of PLK1, onvansertib, in combination with FOLFIRI + bevacizumab showed a 10-fold greater objective response rate (ORR) relative to the 4% ORR of FOLFIRI + bevacizumab (Lenz 2019). Finally, antiangiogenesis inhibitors may also target immunosuppressive mechanisms, suggesting potential combinations with immunotherapies. In addition to previously mentioned TAMs (see “CRC Immunity”), Tregs from mice bearing CRC tumors express VEGFR2 and proliferate in response to VEGF. Accordingly, anti-VEGF antibody (bevacizumab) therapy in patients with metastatic CRC has been shown to reduce the level of Tregs in the peripheral blood (Terme et al. 2013). These insights led to a phase 1b study combining anti-PD-L1 antibody (atezolizumab) and bevacizumab in patients with MSI-H metastatic CRC, yielding an encouraging ORR of 30% (Hochster et al. 2017).
Therapies that trigger cancer cell apoptosis also show promise in early-stage clinical trials with CRC. TRAIL selectively induces apoptosis in cancer cells by binding to proapoptotic death receptors DR4/5 and recruits FADD and procaspase-8 (Allen and El-Deiry 2011). ONC201 is a TRAIL inducer that selectively antagonizes DRD2, causing dampened AKT-ERK signaling, leading to up-regulation of TRAIL (Allen et al. 2013). Preclinical data showed that in CRC, ONC201 not only inhibits metastasis through cell death signaling but also has an immunostimulatory function, as suggested by increased intratumoral infiltration and activation of NKs (Wagner et al. 2018), leading to a phase 1b/2 trial combining ONC201 with the anti-PD-1 antibody nivolumab in patients with MSS metastatic CRC (https://clinicaltrials.gov/ct2/show/NCT03791398?term=BrUOG+379+Phase+Ib%2FII+Trial+ONC201%E2%80%89%2B%E2%80%89Nivolumab+in+MSS+mCRC+%28379%29&draw=2&rank=1).
ICB therapy has yielded meaningful responses in metastatic CRC with dMMR/MSI-H profiles. Patients who received anti-PD-1 antibody (nivolumab) exhibited an ORR of 31% (one-third of these are durable responses) and a 1-yr OS of 73% (Overman et al. 2017). Combining anti-PD-1 and anti-CTLA-4 antibodies in patients with dMMR/MSI-H metastatic CRC increased ORR to 55% and the 1-yr OS to 85% (Overman et al. 2018). At the same time, dual ICB therapy causes immune-related adverse events in 90% of patients, thus emphasizing the need for combinations with both increased efficacy and safety. Cetuximab increases CTL infiltration and expression of immune checkpoints such as PD-L1 and LAG3, suggesting a potential combination treatment of cetuximab with ICB therapies (Woolston et al. 2019). Similarly, trials combining nivolumab and epacadostat (IDO inhibitor) are ongoing in patients with advanced cancers, including CRC (https://clinicaltrials.gov/ct2/show/NCT02327078?term=A+Study+of+the+Safety%2C+Tolerability%2C+and+Efficacy+of+Epacadostat+Administered+in+Combination+With+Nivolumab+in+Select+Advanced+Cancers+%28ECHO-204%29&draw=2&rank=1). In addition, activation of CCR5 on macrophages, which leads to MMP production, promoting ECM destruction, tumor invasion, and metastasis, has prompted a phase 1 trial with a CCR5 antagonist in patients with CRC liver metastases. This trial showed objective clinical responses, as well as repolarization of TAMs to an antitumor phenotype (Halama et al. 2016). Finally, for tumors with lower inflammation and T-cell infiltration, which could be due to defects on priming or the absence of high-affinity T cells, vaccinations or more specific approaches like adoptive T-cell therapy (ATC), which are specific for a particular mutated antigen, may prove favorable options in combination with ICB. Therapeutic cancer vaccines can induce an immune response by delivering antigens to antigen-presenting cells, which prime and activate CD4+ and CD8+ T cells to initiate tumor destruction (Sahin and Türeci 2018). Moreover, ATC or CAR T cells provide tumor antigen-specific approaches and may be useful when the neoantigen load is lower, or if information regarding this neoantigen load is unavailable (Blankenstein et al. 2015). For example, CD8+ T cells targeting mutant KRAS (Tran et al. 2016) or p53 “hotspot” mutations (Malekzadeh et al. 2019) have been identified. Moreover, circulating PD-1+ lymphocytes have recently been shown to recognize human gastrointestinal cancer neoantigens (Gros et al. 2019), which can be isolated and retransferred.
For CRC early detection, besides the colonoscopy, a stool test can be used to look for occult (hidden) blood from the damaged blood vessels or mutant DNA in polyps or tumors (D'Souza et al. 2021). The positive results will need to be followed up and confirmed with colonoscopy. For the individuals not accessible or reticent to stool test or colonoscopy, detecting the circulating tumor cells (CTC) (Tsai et al. 2019) and circulating tumor DNA (ctDNA) (Dasari et al. 2020) have emerged as less invasive and more compliable alternatives for CRC detection, with high sensitivity and accuracy. In addition, ctDNA is also a useful parameter for monitoring patients’ response to therapies as well as tracking the clonal dynamics during the treatment (Dasari et al. 2020).
Although advances in science-based treatment and early detection for CRC are laudable, there remain significant knowledge gaps that must be addressed to better understand disease etiology, improve treatment responses, and prolong patient survival while preserving the quality of life. Although the range of research opportunities is broad, we consider five priority areas to be timely and potentially impactful.
The first area relates to systematically defining and validating the redundant signaling pathways that sustain CRC cell growth, such as interactive EGFR and HGF-MET coactivation, which impairs the efficacy of targeted therapies against these RTKs and their downstream effectors as a result of signaling compensation and rebound. A comprehensive analysis of adaptive signaling mechanisms in the setting of CRC targeted therapies in patients and cognate animal models is critical to identify combined inhibitors that may overcome signaling redundancy mechanisms while maintaining vigilance of additive side effects. Along these lines, CRC's immunosuppressive TME, comprising varied and versatile populations of immune cells, provides multiple axes of immune evasion mechanisms such as T-cell exhaustion and MDSC infiltration. Systematic cotargeting of these immune evasion mechanisms with combined ICB and myeloid inhibitors has shown great promise in preclinical models and should be pursued vigorously in the clinic (Liao et al. 2019).
The second area of opportunity looks beyond cancer and immune cells and encompasses the roles of other components of the CRC TME and the intestinal tissue environment, such as stromal fibroblasts and the gut microbiome. Cancer-associated fibroblasts secrete cancer-promoting factors such as FGF, HGF, TGF-β, and PGE2. In addition, fibroblast contraction and matrix deposition are known to increase CRC metastatic tumor stiffness, which may limit intratumoral drug delivery. The gut microbiome and its metabolites not only cause intestinal-epithelial cell DNA damage but also induce inflammation and modulate the immune response. Strikingly, the gut microbiome can also influence the function of p53 by switching mutant p53 from tumor-suppressive to oncogenic through a single microbiota-derived metabolite, gallic acid (Kadosh et al. 2020), further emphasizing the need to understand the role of the microbiome in tumor biology. Finally, scRNA-seq and high-throughput meta-omics profiling such as metaproteomics and metabolomics provide an unprecedented opportunity to better define novel TME components and their interactions. Genetic model systems manipulating these components are also critical in elucidating their biological roles in CRC tumorigenesis.
The third frontier relates to dissecting the underlying genetics and biology of CRC metastasis, which remains one of the greatest obstacles in the effective treatment of CRC and other solid tumors. Understanding the mechanisms that enable metastasis, determining and validating the key targets that maintain metastasis, and identifying how these targets may also influence primary tumor maintenance are all critical gaps in our knowledge. Some insights are beginning to surface from the study of mouse models, in which oncogenic KRAS and TGF-β have been shown to promote and maintain CRC metastasis via regulation of immunity and cell–cell contact mechanisms (Boutin et al. 2017). In addition, the fact that metastasis can occur at very early stages of CRC emphasizes the need for the identification of metastasis drivers in primary tumor specimens, which may also guide early detection strategies, precision prognostication, and neoadjuvant therapies for those with high metastatic risk profiles.
The fourth area relates to the emerging role of RNA (such as microRNAs and long intergenic noncoding RNAs) as well as RNA binding proteins (RBPs) in intestinal epithelial biology and CRC biology. Several RBPs have been shown to regulate intestinal epithelial homeostasis and contribute to malignant transformation in CRC. For example, LIN28A and LIN28B are overexpressed in 70% and 30%, respectively, of CRC. Their overexpression is correlated with tumor invasiveness, worse survival, and recurrence (Chatterji and Rustgi 2018). Musashi proteins, another group of RBPs, are linked to cancer therapeutic resistance due to their roles in EMT and stem cell functions (Chatterji and Rustgi 2018). In addition, many RBPs share conserved structural domains, raising the possibility that they may serve as targets of inhibitor development (Chatterji and Rustgi 2018).
The fifth and final area relates to deciphering the immeasurable and comprehensive source of publicly available databases on CRC and other cancers, such as TCGA, Oncomine, The Human Protein Atlas, and a variety of data on GEO. Analyzing such big data has led to the identification of frequent mutations in CRC, such as ARID1A and SOX9, the sidedness of CRC, as well as ERBB2 amplification as the new therapeutic target of CRC (Tomczak et al. 2015). Advanced bioinformatics tools and potential applications of artificial intelligence are imperatively needed to integrate the multidimensional data, increase the data resolutions, and provide readable information for clinicians and the public, ultimately improving CRC early detection, identifying novel drivers of CRC, accelerating drug discoveries, and aiding the design of clinical trials and personalized medicine.
In conclusion, the collective efforts of the CRC field have resulted in meaningful advances in our understanding and treatment of this disease. This foundation of improved understanding of the genetic and biological hallmarks of CRC has positioned the field for continued progress and clinical impact. Advances in molecular profiling and adoption of the precision trials paradigm should accelerate the detection, prevention, and treatment of many forms of CRC. As we further harness the power of profiling technologies, genetic model systems, and novel targeted therapies, we can expect to be continually humbled by the biological complexity of the CRC TME, the adaptive nature of interconnected signaling pathways, and the extreme plasticity of the genome and cellular states. Notwithstanding these challenges, the pace of meaningful advances predicts that this coming decade will be marked by major breakthroughs for this major cancer killer.
Acknowledgments
We thank Dr. Kevin Haigis, Dr. Winfried Edelmann, Dr. Anil Rustgi, Dr. Eduardo Vilar-Sanchez, Dr. Raju Kucherlapati, Dr. Yan Xia, Dr. Denise Spring, and Yiqiu (Mary) Zhang for insightful comments and edits on the manuscript. We thank Erica Goodoff, Senior Scientific Editor in the Research Medical Library at The University of Texas MD Anderson Cancer Center, for editorial assistance. We thank Dave Aten, MA, CMI in the Department of Strategic Communications at The University of Texas MD Anderson Cancer Center, for the preparation of the figures. J.L. was supported by the Cancer Prevention and Research Institute of Texas (CPRIT) Research Training Program (RP170067). D.C. was supported by the Triumph postdoctoral training program at MD Anderson supported by CPRIT (RP170067) and by pilot funding through the Digestive Disease Center-National Institute of Diabetes and Digestive and Kidney Diseases award (P30CA16672). S.S. was supported by the National Institutes of Health (NIH)-National Institute on Alcohol Abuse and Alcoholism (U01AA027681). The work in R.A.D.’s laboratory was supported by the NIH (R01CA231360-03), MD Anderson SPORE in Gastrointestinal Cancer-DRP Award, and the Harry Graves Burkhart III Distinguished University Chair in Cancer Research.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.348226.120.
Competing interest statement
R.A.D. is a founder, advisor, and/or director of Tvardi Therapeutics, Inc., which is developing a new class of medications for diverse cancers, chronic inflammatory diseases, and fibrotic diseases. R.A.D. is also cofounder and advisor of Asylia Therapeutics, Nirogy Therapeutics, Stellanova Therapeutics, and Sporos Bioventures.
References
- Abdulamir AS, Hafidh RR, Bakar FA. 2010. Molecular detection, quantification, and isolation of Streptococcus gallolyticus bacteria colonizing colorectal tumors: inflammation-driven potential of carcinogenesis via IL-1, COX-2, and IL-8. Mol Cancer 9: 249. 10.1186/1476-4598-9-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abt MC, Osborne LC, Monticelli LA, Doering TA, Alenghat T, Sonnenberg GF, Paley MA, Antenus M, Williams KL, Erikson J, et al. 2012. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37: 158–170. 10.1016/j.immuni.2012.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Accioly MT, Pacheco P, Maya-Monteiro CM, Carrossini N, Robbs BK, Oliveira SS, Kaufmann C, Morgado-Diaz JA, Bozza PT, Viola JP. 2008. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer Res 68: 1732–1740. 10.1158/0008-5472.CAN-07-1999 [DOI] [PubMed] [Google Scholar]
- Agace WW, McCoy KD. 2017. Regionalized development and maintenance of the intestinal adaptive immune landscape. Immunity 46: 532–548. 10.1016/j.immuni.2017.04.004 [DOI] [PubMed] [Google Scholar]
- Albuquerque C, Bakker ER, van Veelen W, Smits R. 2011. Colorectal cancers choosing sides. Biochim Biophys Acta 1816: 219–231. [DOI] [PubMed] [Google Scholar]
- Allen JE, El-Deiry WS. 2011. Oxaliplatin uses JNK to restore TRAIL sensitivity in cancer cells through Bcl-xL inactivation. Gastroenterology 141: 430–434. 10.1053/j.gastro.2011.06.026 [DOI] [PubMed] [Google Scholar]
- Allen JE, Krigsfeld G, Mayes PA, Patel L, Dicker DT, Patel AS, Dolloff NG, Messaris E, Scata KA, Wang W, et al. 2013. Dual inactivation of Akt and ERK by TIC10 signals Foxo3a nuclear translocation, TRAIL gene induction, and potent antitumor effects. Sci Transl Med 5: 171ra17. 10.1126/scitranslmed.3004828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodio V, Yaeger R, Arcella P, Cancelliere C, Lamba S, Lorenzato A, Arena S, Montone M, Mussolin B, Bian Y, et al. 2020. EGFR blockade reverts resistance to KRAS G12C inhibition in colorectal cancer. Cancer Discov 10: 1129–1139. 10.1158/2159-8290.CD-20-0187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre T, Shiu KK, Kim TW, Jensen BV, Jensen LH, Punt C, Smith D, Garcia-Carbonero R, Benavides M, Gibbs P, et al. 2020. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N Engl J Med 383: 2207–2218. 10.1056/NEJMoa2017699 [DOI] [PubMed] [Google Scholar]
- Arena S, Corti G, Durinikova E, Montone M, Reilly NM, Russo M, Lorenzato A, Arcella P, Lazzari L, Rospo G, et al. 2020. A subset of colorectal cancers with cross-sensitivity to olaparib and oxaliplatin. Clin Cancer Res 26: 1372–1384. 10.1158/1078-0432.CCR-19-2409 [DOI] [PubMed] [Google Scholar]
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. 2000. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406: 641–645. 10.1038/35020592 [DOI] [PubMed] [Google Scholar]
- Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan TJ, Campbell BJ, Abujamel T, Dogan B, Rogers AB, et al. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338: 120–123. 10.1126/science.1224820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaoka Y, Ijichi H, Koike K. 2015. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med 373: 1979. 10.1056/NEJMc1510353 [DOI] [PubMed] [Google Scholar]
- Atkins D, Breuckmann A, Schmahl GE, Binner P, Ferrone S, Krummenauer F, Störkel S, Seliger B. 2004. MHC class I antigen processing pathway defects, ras mutations and disease stage in colorectal carcinoma. Int J cancer 109: 265–273. 10.1002/ijc.11681 [DOI] [PubMed] [Google Scholar]
- Aubrey BJ, Kelly GL, Janic A, Herold MJ, Strasser A. 2018. How does p53 induce apoptosis and how does this relate to p53-mediated tumour suppression? Cell Death Differ 25: 104–113. 10.1038/cdd.2017.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augustus GJ, Ellis NA. 2018. Colorectal cancer disparity in African Americans: risk factors and carcinogenic mechanisms. Am J Pathol 188: 291–303. 10.1016/j.ajpath.2017.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiola I, Olaso E, Crende O, Friedman SL, Vidal-Vanaclocha F. 2012. Discoidin domain receptor 2 deficiency predisposes hepatic tissue to colon carcinoma metastasis. Gut 61: 1465–1472. 10.1136/gutjnl-2011-300810 [DOI] [PubMed] [Google Scholar]
- Baker AM, Cross W, Curtius K, Al Bakir I, Choi CR, Davis HL, Temko D, Biswas S, Martinez P, Williams MJ, et al. 2019. Evolutionary history of human colitis-associated colorectal cancer. Gut 68: 985–995. 10.1136/gutjnl-2018-316191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F, Mantovani A. 2001. Inflammation and cancer: back to Virchow? Lancet 357: 539–545. 10.1016/S0140-6736(00)04046-0 [DOI] [PubMed] [Google Scholar]
- Barber TD, Vogelstein B, Kinzler KW, Velculescu VE. 2004. Somatic mutations of EGFR in colorectal cancers and glioblastomas. N Engl J Med 351: 2883. 10.1056/NEJM200412303512724 [DOI] [PubMed] [Google Scholar]
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. 2007. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449: 1003–1007. 10.1038/nature06196 [DOI] [PubMed] [Google Scholar]
- Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, Danenberg E, Clarke AR, Sansom OJ, Clevers H. 2009. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature 457: 608–611. 10.1038/nature07602 [DOI] [PubMed] [Google Scholar]
- Batlle E, Massagué J. 2019. Transforming growth factor-β signaling in immunity and cancer. Immunity 50: 924–940. 10.1016/j.immuni.2019.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer K, Nelius N, Reuschenbach M, Koch M, Weitz J, Steinert G, Kopitz J, Beckhove P, Tariverdian M, von Knebel Doeberitz M, et al. 2013. T cell responses against microsatellite instability-induced frameshift peptides and influence of regulatory T cells in colorectal cancer. Cancer Immunol Immunother 62: 27–37. 10.1007/s00262-012-1303-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RJ, Rube HT, Kreig A, Mancini A, Fouse SD, Nagarajan RP, Choi S, Hong C, He D, Pekmezci M, et al. 2015. Cancer. The transcription factor GABP selectively binds and activates the mutant TERT promoter in cancer. Science 348: 1036–1039. 10.1126/science.aab0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, Gottlieb E, Vousden KH. 2006. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126: 107–120. 10.1016/j.cell.2006.05.036 [DOI] [PubMed] [Google Scholar]
- Bensard CL, Wisidagama DR, Olson KA, Berg JA, Krah NM, Schell JC, Nowinski SM, Fogarty S, Bott AJ, Wei P, et al. 2020. Regulation of tumor initiation by the mitochondrial pyruvate carrier. Cell Metab 31: 284–300.e7. 10.1016/j.cmet.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertorelle R, Briarava M, Rampazzo E, Biasini L, Agostini M, Maretto I, Lonardi S, Friso ML, Mescoli C, Zagonel V, et al. 2013. Telomerase is an independent prognostic marker of overall survival in patients with colorectal cancer. Br J Cancer 108: 278–284. 10.1038/bjc.2012.602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjelland S, Seeberg E. 2003. Mutagenicity, toxicity and repair of DNA base damage induced by oxidation. Mutat Res 531: 37–80. 10.1016/j.mrfmmm.2003.07.002 [DOI] [PubMed] [Google Scholar]
- Blankenstein T, Leisegang M, Uckert W, Schreiber H. 2015. Targeting cancer-specific mutations by T cell receptor gene therapy. Curr Opin Immunol 33: 112–119. 10.1016/j.coi.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, McCormick F. 1993. Proteins regulating Ras and its relatives. Nature 366: 643–654. 10.1038/366643a0 [DOI] [PubMed] [Google Scholar]
- Bolhaqueiro ACF, Ponsioen B, Bakker B, Klaasen SJ, Kucukkose E, van Jaarsveld RH, Vivie J, Verlaan-Klink I, Hami N, Spierings DCJ, et al. 2019. Ongoing chromosomal instability and karyotype evolution in human colorectal cancer organoids. Nat Genet 51: 824–834. 10.1038/s41588-019-0399-6 [DOI] [PubMed] [Google Scholar]
- Bonjoch L, Franch-Exposito S, Garre P, Belhadj S, Munoz J, Arnau-Collell C, Diaz-Gay M, Gratacos-Mulleras A, Raimondi G, Esteban-Jurado C, et al. 2020. Germline mutations in FAF1 are associated with hereditary colorectal cancer. Gastroenterology 159: 227–240.e7. 10.1053/j.gastro.2020.03.015 [DOI] [PubMed] [Google Scholar]
- Bouskra D, Brezillon C, Bérard M, Werts C, Varona R, Boneca IG, Eberl G. 2008. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature 456: 507–510. 10.1038/nature07450 [DOI] [PubMed] [Google Scholar]
- Boutin AT, Liao WT, Wang M, Hwang SS, Karpinets TV, Cheung H, Chu GC, Jiang S, Hu J, Chang K, et al. 2017. Oncogenic Kras drives invasion and maintains metastases in colorectal cancer. Genes Dev 31: 370–382. 10.1101/gad.293449.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. 2018. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68: 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Brennan CA, Garrett WS. 2016. Gut microbiota, inflammation, and colorectal cancer. Annu Rev Microbiol 70: 395–411. 10.1146/annurev-micro-102215-095513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KM, Xue A, Mittal A, Samra JS, Smith R, Hugh TJ. 2016. Patient-derived xenograft models of colorectal cancer in pre-clinical research: a systematic review. Oncotarget 7: 66212–66225. 10.18632/oncotarget.11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RE, Short SP, Williams CS. 2018. Colorectal cancer and metabolism. Curr Colorectal Cancer Rep 14: 226–241. 10.1007/s11888-018-0420-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruun J, Kryeziu K, Eide PW, Moosavi SH, Eilertsen IA, Langerud J, Rosok B, Totland MZ, Brunsell TH, Pellinen T, et al. 2020. Patient-derived organoids from multiple colorectal cancer liver metastases reveal moderate intra-patient pharmacotranscriptomic heterogeneity. Clin Cancer Res 26: 4107–4119. 10.1158/1078-0432.CCR-19-3637 [DOI] [PubMed] [Google Scholar]
- Bu P, Chen KY, Xiang K, Johnson C, Crown SB, Rakhilin N, Ai Y, Wang L, Xi R, Astapova I, et al. 2018. Aldolase B-mediated fructose metabolism drives metabolic reprogramming of colon cancer liver metastasis. Cell Metab 27: 1249–1262.e4. 10.1016/j.cmet.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger-van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G, van Goudoever JB, van Seuningen I, Renes IB. 2009. The regulation of intestinal mucin MUC2 expression by short-chain fatty acids: implications for epithelial protection. Biochem J 420: 211–219. 10.1042/BJ20082222 [DOI] [PubMed] [Google Scholar]
- Bürtin F, Mullins CS, Linnebacher M. 2020. Mouse models of colorectal cancer: past, present and future perspectives. World J Gastroenterol 26: 1394–1426. 10.3748/wjg.v26.i13.1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch SE, Hanke ML, Kargl J, Metz HE, MacPherson D, Houghton AM. 2016. Lung cancer subtypes generate unique immune responses. J Immunol 197: 4493–4503. 10.4049/jimmunol.1600576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calon A, Espinet E, Palomo-Ponce S, Tauriello DV, Iglesias M, Céspedes MV, Sevillano M, Nadal C, Jung P, Zhang XH, et al. 2012. Dependency of colorectal cancer on a TGF-β-driven program in stromal cells for metastasis initiation. Cancer Cell 22: 571–584. 10.1016/j.ccr.2012.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, Sevillano M, Palomo-Ponce S, Tauriello DV, Byrom D, et al. 2015. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 47: 320–329. 10.1038/ng.3225 [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Network. 2012. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, Gaida K, Holt T, Knutson CG, Koppada N, et al. 2019. The clinical KRAS(G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 575: 217–223. 10.1038/s41586-019-1694-1 [DOI] [PubMed] [Google Scholar]
- Cantley LC. 2002. The phosphoinositide 3-kinase pathway. Science 296: 1655–1657. 10.1126/science.296.5573.1655 [DOI] [PubMed] [Google Scholar]
- Carmeliet P, Jain RK. 2000. Angiogenesis in cancer and other diseases. Nature 407: 249–257. 10.1038/35025220 [DOI] [PubMed] [Google Scholar]
- Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. 2012. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res 22: 299–306. 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo CM, Dijkstra KK, Fanchi LF, Kelderman S, Kaing S, van Rooij N, van den Brink S, Schumacher TN, Voest EE. 2020. Tumor organoid–T-cell coculture systems. Nat Protoc 15: 15–39. 10.1038/s41596-019-0232-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejas P, Casado E, Belda-Iniesta C, De Castro J, Espinosa E, Redondo A, Sereno M, Garcia-Cabezas MA, Vara JA, Dominguez-Caceres A, et al. 2004. Implications of oxidative stress and cell membrane lipid peroxidation in human cancer (Spain). Cancer Causes Control 15: 707–719. 10.1023/B:CACO.0000036189.61607.52 [DOI] [PubMed] [Google Scholar]
- Cervantes A, Elez E, Roda D, Ecsedy J, Macarulla T, Venkatakrishnan K, Roselló S, Andreu J, Jung J, Sanchis-Garcia JM, et al. 2012. Phase I pharmacokinetic/pharmacodynamic study of MLN8237, an investigational, oral, selective aurora a kinase inhibitor, in patients with advanced solid tumors. Clin Cancer Res 18: 4764–4774. 10.1158/1078-0432.CCR-12-0571 [DOI] [PubMed] [Google Scholar]
- Chakravarti D, Hu B, Mao X, Rashid A, Li J, Li J, Liao WT, Whitley EM, Dey P, Hou P, et al. 2020. Telomere dysfunction activates YAP1 to drive tissue inflammation. Nat Commun 11: 4766. 10.1038/s41467-020-18420-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AT, Giovannucci EL. 2010. Primary prevention of colorectal cancer. Gastroenterology 138: 2029–2043.e10. 10.1053/j.gastro.2010.01.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Qiu J, O'Sullivan D, Buck MD, Noguchi T, Curtis JD, Chen Q, Gindin M, Gubin MM, van der Windt GJ, et al. 2015. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162: 1229–1241. 10.1016/j.cell.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Willis JA, Reumers J, Taggart MW, San Lucas FA, Thirumurthi S, Kanth P, Delker DA, Hagedorn CH, Lynch PM, et al. 2018. Colorectal premalignancy is associated with consensus molecular subtypes 1 and 2. Ann Oncol 29: 2061–2067. 10.1093/annonc/mdy337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji P, Rustgi AK. 2018. RNA binding proteins in intestinal epithelial biology and colorectal cancer. Trends Mol Med 24: 490–506. 10.1016/j.molmed.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiorean EG, Hurwitz HI, Cohen RB, Schwartz JD, Dalal RP, Fox FE, Gao L, Sweeney CJ. 2015. Phase I study of every 2- or 3-wk dosing of ramucirumab, a human immunoglobulin G1 monoclonal antibody targeting the vascular endothelial growth factor receptor-2 in patients with advanced solid tumors. Ann Oncol 26: 1230–1237. 10.1093/annonc/mdv144 [DOI] [PubMed] [Google Scholar]
- Clevers H, Nusse R. 2012. Wnt/β-catenin signaling and disease. Cell 149: 1192–1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Cooks T, Harris CC, Oren M. 2014. Caught in the cross fire: p53 in inflammation. Carcinogenesis 35: 1680–1690. 10.1093/carcin/bgu134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran RB, Ebi H, Turke AB, Coffee EM, Nishino M, Cogdill AP, Brown RD, Della Pelle P, Dias-Santagata D, Hung KE, et al. 2012. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2: 227–235. 10.1158/2159-8290.CD-11-0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cremolini C, Loupakis F, Masi G, Lonardi S, Granetto C, Mancini ML, Chiara S, Moretto R, Rossini D, Vitello S, et al. 2016. FOLFOXIRI or FOLFOXIRI plus bevacizumab as first-line treatment of metastatic colorectal cancer: a propensity score-adjusted analysis from two randomized clinical trials. Ann Oncol 27: 843–849. 10.1093/annonc/mdw052 [DOI] [PubMed] [Google Scholar]
- Cunningham JM, Christensen ER, Tester DJ, Kim CY, Roche PC, Burgart LJ, Thibodeau SN. 1998. Hypermethylation of the hMLH1 promoter in colon cancer with microsatellite instability. Cancer Res 58: 3455–3460. [PubMed] [Google Scholar]
- Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, et al. 2011. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 29: 1120–1127. 10.1038/nbt.2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas NA, Xia L, Fan F, Gray MJ, Gaur P, van Buren GII, Samuel S, Kim MP, Lim SJ, Ellis LM. 2009. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res 69: 1951–1957. 10.1158/0008-5472.CAN-08-2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari A, Morris VK, Allegra CJ, Atreya C, Benson AB III, Boland P, Chung K, Copur MS, Corcoran RB, Deming DA, et al. 2020. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper. Nat Rev Clin Oncol 17: 757–770. 10.1038/s41571-020-0392-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, et al. 2002. Mutations of the BRAF gene in human cancer. Nature 417: 949–954. 10.1038/nature00766 [DOI] [PubMed] [Google Scholar]
- de Barrios O, Győrffy B, Fernández-Aceñero MJ, Sánchez-Tillo E, Sánchez-Moral L, Siles L, Esteve-Arenys A, Roué G, Casal JI, Darling DS, et al. 2017. ZEB1-induced tumourigenesis requires senescence inhibition via activation of DKK1/mutant p53/Mdm2/CtBP and repression of macroH2A1. Gut 66: 666–682. 10.1136/gutjnl-2015-310838 [DOI] [PubMed] [Google Scholar]
- Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. 2019. Colorectal cancer. Lancet 394: 1467–1480. 10.1016/S0140-6736(19)32319-0 [DOI] [PubMed] [Google Scholar]
- de la Cruz-López KG, Castro-Muñoz LJ, Reyes-Hernández DO, García-Carrancá A, Manzo-Merino J. 2019. Lactate in the regulation of tumor microenvironment and therapeutic approaches. Front Oncol 9: 1143. 10.3389/fonc.2019.01143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev 19: 2100–2110. 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- De Roock W, De Vriendt V, Normanno N, Ciardiello F, Tejpar S. 2011. KRAS, BRAF, PIK3CA, and PTEN mutations: implications for targeted therapies in metastatic colorectal cancer. Lancet Oncol 12: 594–603. 10.1016/S1470-2045(10)70209-6 [DOI] [PubMed] [Google Scholar]
- de Sousa e Melo F, Kurtova AV, Harnoss JM, Kljavin N, Hoeck JD, Hung J, Anderson JE, Storm EE, Modrusan Z, Koeppen H, et al. 2017. A distinct role for Lgr5(+) stem cells in primary and metastatic colon cancer. Nature 543: 676–680. 10.1038/nature21713 [DOI] [PubMed] [Google Scholar]
- DeStefano Shields CE, Van Meerbeke SW, Housseau F, Wang H, Huso DL, Casero RA Jr, O'Hagan HM, Sears CL. 2016. Reduction of murine colon tumorigenesis driven by enterotoxigenic bacteroides fragilis using cefoxitin treatment. J Infect Dis 214: 122–129. 10.1093/infdis/jiw069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey P, Kimmelman AC, DePinho RA. 2021. Metabolic codependencies in the tumor microenvironment. Cancer Discov 11: 1067–1081. 10.1158/2159-8290.CD-20-1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dienstmann R, Mason MJ, Sinicrope FA, Phipps AI, Tejpar S, Nesbakken A, Danielsen SA, Sveen A, Buchanan DD, Clendenning M, et al. 2017. Prediction of overall survival in stage II and III colon cancer beyond TNM system: a retrospective, pooled biomarker study. Ann Oncol 28: 1023–1031. 10.1093/annonc/mdx052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijkstra KK, Cattaneo CM, Weeber F, Chalabi M, van de Haar J, Fanchi LF, Slagter M, van der Velden DL, Kaing S, Kelderman S, et al. 2018. Generation of tumor-reactive T cells by co-culture of peripheral blood lymphocytes and tumor organoids. Cell 174: 1586–1598.e12. 10.1016/j.cell.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, et al. 2008. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 26: 5705–5712. 10.1200/JCO.2008.18.0786 [DOI] [PubMed] [Google Scholar]
- Doedens AL, Stockmann C, Rubinstein MP, Liao D, Zhang N, DeNardo DG, Coussens LM, Karin M, Goldrath AW, Johnson RS. 2010. Macrophage expression of hypoxia-inducible factor-1α suppresses T-cell function and promotes tumor progression. Cancer Res 70: 7465–7475. 10.1158/0008-5472.CAN-10-1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doubeni CA, Major JM, Laiyemo AO, Schootman M, Zauber AG, Hollenbeck AR, Sinha R, Allison J. 2012. Contribution of behavioral risk factors and obesity to socioeconomic differences in colorectal cancer incidence. J Natl Cancer Inst 104: 1353–1362. 10.1093/jnci/djs346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow LE, O'Rourke KP, Simon J, Tschaharganeh DF, van Es JH, Clevers H, Lowe SW. 2015. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell 161: 1539–1552. 10.1016/j.cell.2015.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, van Jaarsveld RH, Ponsioen B, Zimberlin C, van Boxtel R, Buijs A, Sachs N, Overmeer RM, Offerhaus GJ, Begthel H, et al. 2015. Sequential cancer mutations in cultured human intestinal stem cells. Nature 521: 43–47. 10.1038/nature14415 [DOI] [PubMed] [Google Scholar]
- D'Souza N, Georgiou Delisle T, Chen M, Benton S, Abulafi M. 2021. Faecal immunochemical test is superior to symptoms in predicting pathology in patients with suspected colorectal cancer symptoms referred on a 2WW pathway: a diagnostic accuracy study. Gut 70: 1130–1138. 10.1136/gutjnl-2020-321956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GP, Koebel CM, Schreiber RD. 2006. Interferons, immunity and cancer immunoediting. Nat Rev Immunol 6: 836–848. 10.1038/nri1961 [DOI] [PubMed] [Google Scholar]
- Eaden JA, Abrams KR, Mayberry JF. 2001. The risk of colorectal cancer in ulcerative colitis: a meta-analysis. Gut 48: 526–535. 10.1136/gut.48.4.526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edkins S, O'Meara S, Parker A, Stevens C, Reis M, Jones S, Greenman C, Davies H, Dalgliesh G, Forbes S, et al. 2006. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther 5: 928–932. 10.4161/cbt.5.8.3251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. 2013. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer 13: 759–771. 10.1038/nrc3611 [DOI] [PubMed] [Google Scholar]
- el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. 2004. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis 39: 186–193. 10.1002/gene.20042 [DOI] [PubMed] [Google Scholar]
- Erreni M, Mantovani A, Allavena P. 2011. Tumor-associated macrophages (TAM) and inflammation in colorectal cancer. Cancer Microenviron 4: 141–154. 10.1007/s12307-010-0052-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ertych N, Stolz A, Stenzinger A, Weichert W, Kaulfuß S, Burfeind P, Aigner A, Wordeman L, Bastians H. 2014. Increased microtubule assembly rates influence chromosomal instability in colorectal cancer cells. Nat Cell Biol 16: 779–791. 10.1038/ncb2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. 1990. A genetic model for colorectal tumorigenesis. Cell 61: 759–767. 10.1016/0092-8674(90)90186-I [DOI] [PubMed] [Google Scholar]
- Fidler IJ. 2003. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer 3: 453–458. 10.1038/nrc1098 [DOI] [PubMed] [Google Scholar]
- Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev 16: 1472–1487. 10.1101/gad.995802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer K, Hoffmann P, Voelkl S, Meidenbauer N, Ammer J, Edinger M, Gottfried E, Schwarz S, Rothe G, Hoves S, et al. 2007. Inhibitory effect of tumor cell-derived lactic acid on human T cells. Blood 109: 3812–3819. 10.1182/blood-2006-07-035972 [DOI] [PubMed] [Google Scholar]
- Fisel P, Schaeffeler E, Schwab M. 2018. Clinical and functional relevance of the monocarboxylate transporter family in disease pathophysiology and drug therapy. Clin Transl Sci 11: 352–364. 10.1111/cts.12551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley TM, Payne SN, Pasch CA, Yueh AE, Van De Hey DR, Korkos DP, Clipson L, Maher ME, Matkowskyj KA, Newton MA, et al. 2017. Dual PI3K/mTOR inhibition in colorectal cancers with APC and PIK3CA mutations. Mol Cancer Res 15: 317–327. 10.1158/1541-7786.MCR-16-0256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francipane MG, Lagasse E. 2014. mTOR pathway in colorectal cancer: an update. Oncotarget 5: 49–66. 10.18632/oncotarget.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frentzas S, Simoneau E, Bridgeman VL, Vermeulen PB, Foo S, Kostaras E, Nathan M, Wotherspoon A, Gao ZH, Shi Y, et al. 2016. Vessel co-option mediates resistance to anti-angiogenic therapy in liver metastases. Nat Med 22: 1294–1302. 10.1038/nm.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridlender ZG, Albelda SM. 2012. Tumor-associated neutrophils: friend or foe? Carcinogenesis 33: 949–955. 10.1093/carcin/bgs123 [DOI] [PubMed] [Google Scholar]
- Fu Y, Liu S, Yin S, Niu W, Xiong W, Tan M, Li G, Zhou M. 2017. The reverse Warburg effect is likely to be an Achilles’ heel of cancer that can be exploited for cancer therapy. Oncotarget 8: 57813–57825. 10.18632/oncotarget.18175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli A, Oost KC, Kester L, Morgner J, Bornes L, Bruens L, Spaargaren L, Azkanaz M, Schelfhorst T, Beerling E, et al. 2020. Plasticity of Lgr5-negative cancer cells drives metastasis in colorectal cancer. Cell Stem Cell 26: 569–578.e7. 10.1016/j.stem.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta RM, Garcia-Irigoyen O, Moschetta A. 2017. Bile acids and colon cancer: is FXR the solution of the conundrum? Mol Aspects Med 56: 66–74. 10.1016/j.mam.2017.04.002 [DOI] [PubMed] [Google Scholar]
- Ganesh S, Koser ML, Cyr WA, Chopda GR, Tao J, Shui X, Ying B, Chen D, Pandya P, Chipumuro E, et al. 2016. Direct pharmacological inhibition of β-catenin by RNA interference in tumors of diverse origin. Mol Cancer Ther 15: 2143–2154. 10.1158/1535-7163.MCT-16-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesh K, Wu C, O'Rourke KP, Szeglin BC, Zheng Y, Sauvé CG, Adileh M, Wasserman I, Marco MR, Kim AS, et al. 2019. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat Med 25: 1607–1614. 10.1038/s41591-019-0584-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudier E, Jarry A, Blottière HM, de Coppet P, Buisine MP, Aubert JP, Laboisse C, Cherbut C, Hoebler C. 2004. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am J Physiol Gastrointest Liver Physiol 287: G1168–G1174. 10.1152/ajpgi.00219.2004 [DOI] [PubMed] [Google Scholar]
- Geremia A, Arancibia-Cárcamo CV, Fleming MP, Rust N, Singh B, Mortensen NJ, Travis SP, Powrie F. 2011. IL-23-responsive innate lymphoid cells are increased in inflammatory bowel disease. J Exp Med 208: 1127–1133. 10.1084/jem.20101712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon-Weeks AN, Lim SY, Yuzhalin AE, Jones K, Markelc B, Kim KJ, Buzzelli JN, Fokas E, Cao Y, Smart S, et al. 2017. Neutrophils promote hepatic metastasis growth through fibroblast growth factor 2-dependent angiogenesis in mice. Hepatology 65: 1920–1935. 10.1002/hep.29088 [DOI] [PubMed] [Google Scholar]
- Gorelik L, Flavell RA. 2000. Abrogation of TGFβ signaling in T cells leads to spontaneous T cell differentiation and autoimmune disease. Immunity 12: 171–181. 10.1016/S1074-7613(00)80170-3 [DOI] [PubMed] [Google Scholar]
- Goswami RS, Patel KP, Singh RR, Meric-Bernstam F, Kopetz ES, Subbiah V, Alvarez RH, Davies MA, Jabbar KJ, Roy-Chowdhuri S, et al. 2015. Hotspot mutation panel testing reveals clonal evolution in a study of 265 paired primary and metastatic tumors. Clin Cancer Res 21: 2644–2651. 10.1158/1078-0432.CCR-14-2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasso CS, Giannakis M, Wells DK, Hamada T, Mu XJ, Quist M, Nowak JA, Nishihara R, Qian ZR, Inamura K, et al. 2018. Genetic mechanisms of immune evasion in colorectal cancer. Cancer Discov 8: 730–749. 10.1158/2159-8290.CD-17-1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhough A, Bagley C, Heesom KJ, Gurevich DB, Gay D, Bond M, Collard TJ, Paraskeva C, Martin P, Sansom OJ, et al. 2018. Cancer cell adaptation to hypoxia involves a HIF-GPRC5A-YAP axis. EMBO Mol Med 10: e8699. 10.15252/emmm.201708699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory AD, Houghton AM. 2011. Tumor-associated neutrophils: new targets for cancer therapy. Cancer Res 71: 2411–2416. 10.1158/0008-5472.CAN-10-2583 [DOI] [PubMed] [Google Scholar]
- Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M. 2004. IKKβ links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 118: 285–296. 10.1016/j.cell.2004.07.013 [DOI] [PubMed] [Google Scholar]
- Gros A, Tran E, Parkhurst MR, Ilyas S, Pasetto A, Groh EM, Robbins PF, Yossef R, Garcia-Garijo A, Fajardo CA, et al. 2019. Recognition of human gastrointestinal cancer neoantigens by circulating PD-1+ lymphocytes. J Clin Invest 129: 4992–5004. 10.1172/JCI127967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Merker K, Sandig G, Davies KJ. 2003. Selective degradation of oxidatively modified protein substrates by the proteasome. Biochem Biophys Res Commun 305: 709–718. 10.1016/S0006-291X(03)00809-X [DOI] [PubMed] [Google Scholar]
- Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. 2015. The consensus molecular subtypes of colorectal cancer. Nat Med 21: 1350–1356. 10.1038/nm.3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Ramjaun AR, Haiko P, Wang Y, Warne PH, Nicke B, Nye E, Stamp G, Alitalo K, Downward J. 2007. Binding of ras to phosphoinositide 3-kinase p110α is required for ras-driven tumorigenesis in mice. Cell 129: 957–968. 10.1016/j.cell.2007.03.051 [DOI] [PubMed] [Google Scholar]
- Haigis KM. 2017. KRAS alleles: the devil is in the detail. Trends Cancer 3: 686–697. 10.1016/j.trecan.2017.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halama N, Zoernig I, Berthel A, Kahlert C, Klupp F, Suarez-Carmona M, Suetterlin T, Brand K, Krauss J, Lasitschka F, et al. 2016. Tumoral immune cell exploitation in colorectal cancer metastases can be targeted effectively by anti-CCR5 therapy in cancer patients. Cancer Cell 29: 587–601. 10.1016/j.ccell.2016.03.005 [DOI] [PubMed] [Google Scholar]
- Han T, Schatoff EM, Murphy C, Zafra MP, Wilkinson JE, Elemento O, Dow LE. 2017. R-spondin chromosome rearrangements drive Wnt-dependent tumour initiation and maintenance in the intestine. Nat Commun 8: 15945. 10.1038/ncomms15945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. 2011. Hallmarks of cancer: the next generation. Cell 144: 646–674. 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Hansen IO, Jess P. 2012. Possible better long-term survival in left versus right-sided colon cancer – a systematic review. Dan Med J 59: A4444. [PubMed] [Google Scholar]
- Hao HX, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, Lei H, Mickanin C, Liu D, Ruffner H, et al. 2012. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 485: 195–200. 10.1038/nature11019 [DOI] [PubMed] [Google Scholar]
- Hao HX, Jiang X, Cong F. 2016a. Control of Wnt receptor turnover by R-spondin-ZNRF3/RNF43 signaling module and its dysregulation in cancer. Cancers 8: 54. 10.3390/cancers8060054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Samuels Y, Li Q, Krokowski D, Guan BJ, Wang C, Jin Z, Dong B, Cao B, Feng X, et al. 2016b. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat Commun 7: 11971. 10.1038/ncomms11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Reguart N, You L, Mazieres J, Xu Z, Lee AY, Mikami I, McCormick F, Jablons DM. 2005. Blockade of Wnt-1 signaling induces apoptosis in human colorectal cancer cells containing downstream mutations. Oncogene 24: 3054–3058. 10.1038/sj.onc.1208511 [DOI] [PubMed] [Google Scholar]
- He GW, Gunther C, Thonn V, Yu YQ, Martini E, Buchen B, Neurath MF, Sturzl M, Becker C. 2017a. Regression of apoptosis-resistant colorectal tumors by induction of necroptosis in mice. Journal of Experimental Medicine 214: 1655–1662. 10.1084/jem.20160442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, Fan W, Henriquez B, Yu RT, Atkins AR, Liddle C, Zheng Y, Downes M, Evans RM. 2017b. Metabolic control of regulatory T cell (Treg) survival and function by Lkb1. Proc Natl Acad Sci 114: 12542–12547. 10.1073/pnas.1715363114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer J, Yang K, Lipkin M, Edelmann W, Kucherlapati R. 1999. Mouse models for colorectal cancer. Oncogene 18: 5325–5333. 10.1038/sj.onc.1203036 [DOI] [PubMed] [Google Scholar]
- Hilkens J, Timmer NC, Boer M, Ikink GJ, Schewe M, Sacchetti A, Koppens MAJ, Song JY, Bakker ERM. 2017. RSPO3 expands intestinal stem cell and niche compartments and drives tumorigenesis. Gut 66: 1095–1105. 10.1136/gutjnl-2016-311606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, Cho KR, Fearon ER. 2007. Mouse model of colonic adenoma–carcinoma progression based on somatic Apc inactivation. Cancer Res 67: 9721–9730. 10.1158/0008-5472.CAN-07-2735 [DOI] [PubMed] [Google Scholar]
- Hinze L, Labrosse R, Degar J, Han T, Schatoff EM, Schreek S, Karim S, McGuckin C, Sacher JR, Wagner F, et al. 2020. Exploiting the therapeutic interaction of WNT pathway activation and asparaginase for colorectal cancer therapy. Cancer Discov 10: 1690–1705. 10.1158/2159-8290.CD-19-1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson-Gutierrez SA, Carmona-Fontaine C. 2018. The metabolic axis of macrophage and immune cell polarization. Dis Model Mech 11: dmm034462. 10.1242/dmm.034462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochster HS, Bendell JC, Cleary JM, Foster P, Zhang W, He X, Hernandez G, Iizuka K, Eckhardt SG. 2017. Efficacy and safety of atezolizumab (atezo) and bevacizumab (bev) in a phase Ib study of microsatellite instability (MSI)-high metastatic colorectal cancer (mCRC). J Clin Oncol 35: 673–673. 10.1200/JCO.2017.35.4_suppl.673 [DOI] [Google Scholar]
- Hoffman B, Liebermann DA. 2008. Apoptotic signaling by c-MYC. Oncogene 27: 6462–6472. 10.1038/onc.2008.312 [DOI] [PubMed] [Google Scholar]
- Hoffmeyer K, Raggioli A, Rudloff S, Anton R, Hierholzer A, Del Valle I, Hein K, Vogt R, Kemler R. 2012. Wnt/β-catenin signaling regulates telomerase in stem cells and cancer cells. Science 336: 1549–1554. 10.1126/science.1218370 [DOI] [PubMed] [Google Scholar]
- Hofseth LJ, Hebert JR, Chanda A, Chen H, Love BL, Pena MM, Murphy EA, Sajish M, Sheth A, Buckhaults PJ, et al. 2020. Early-onset colorectal cancer: initial clues and current views. Nat Rev Gastroenterol Hepatol 17: 352–364. 10.1038/s41575-019-0253-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holash J, Davis S, Papadopoulos N, Croll SD, Ho L, Russell M, Boland P, Leidich R, Hylton D, Burova E, et al. 2002. VEGF-Trap: a VEGF blocker with potent antitumor effects. Proc Natl Acad Sci 99: 11393–11398. 10.1073/pnas.172398299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. 1983. Cancer and cell senescence. Nature 306: 742. 10.1038/306742a0 [DOI] [PubMed] [Google Scholar]
- Hong DS, Fakih MG, Strickler JH, Desai J, Durm GA, Shapiro GI, Falchook GS, Price TJ, Sacher A, Denlinger CS, et al. 2020. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N Engl J Med 383: 1207–1217. 10.1056/NEJMoa1917239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Z, Guo K, Sun X, Hu F, Chen Q, Luo X, Wang G, Hu J, Sun L. 2018. TRIB2 functions as novel oncogene in colorectal cancer by blocking cellular senescence through AP4/p21 signaling. Mol Cancer 17: 172. 10.1186/s12943-018-0922-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housseau F, Sears CL. 2010. Enterotoxigenic Bacteroides fragilis (ETBF)-mediated colitis in Min (Apc+/−) mice: a human commensal-based murine model of colon carcinogenesis. Cell Cycle 9: 3–5. 10.4161/cc.9.1.10352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Housseau F, Wu S, Wick EC, Fan H, Wu X, Llosa NJ, Smith KN, Tam A, Ganguly S, Wanyiri JW, et al. 2016. Redundant innate and adaptive sources of IL17 production drive colon tumorigenesis. Cancer Res 76: 2115–2124. 10.1158/0008-5472.CAN-15-0749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Ding J, Ma Z, Sun R, Seoane JA, Scott Shaffer J, Suarez CJ, Berghoff AS, Cremolini C, Falcone A, et al. 2019. Quantitative evidence for early metastatic seeding in colorectal cancer. Nat Genet 51: 1113–1122. 10.1038/s41588-019-0423-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua F, Shang S, Yang YW, Zhang HZ, Xu TL, Yu JJ, Zhou DD, Cui B, Li K, Lv XX, et al. 2019. TRIB3 interacts with β-catenin and TCF4 to increase stem cell features of colorectal cancer stem cells and tumorigenesis. Gastroenterology 156: 708–721.e15. 10.1053/j.gastro.2018.10.031 [DOI] [PubMed] [Google Scholar]
- Husain Z, Huang Y, Seth P, Sukhatme VP. 2013. Tumor-derived lactate modifies antitumor immune response: effect on myeloid-derived suppressor cells and NK cells. J Immunol 191: 1486–1495. 10.4049/jimmunol.1202702 [DOI] [PubMed] [Google Scholar]
- Iacopetta B. 2002. Are there two sides to colorectal cancer? Int J Cancer 101: 403–408. 10.1002/ijc.10635 [DOI] [PubMed] [Google Scholar]
- Irby RB, McCarthy SM, Yeatman TJ. 2004. Osteopontin regulates multiple functions contributing to human colon cancer development and progression. Clin Exp Metastasis 21: 515–523. 10.1007/s10585-004-2873-4 [DOI] [PubMed] [Google Scholar]
- Irrazábal T, Belcheva A, Girardin SE, Martin A, Philpott DJ. 2014. The multifaceted role of the intestinal microbiota in colon cancer. Mol Cell 54: 309–320. 10.1016/j.molcel.2014.03.039 [DOI] [PubMed] [Google Scholar]
- Itatani Y, Kawada K, Fujishita T, Kakizaki F, Hirai H, Matsumoto T, Iwamoto M, Inamoto S, Hatano E, Hasegawa S, et al. 2013. Loss of SMAD4 from colorectal cancer cells promotes CCL15 expression to recruit CCR1+ myeloid cells and facilitate liver metastasis. Gastroenterology 145: 1064–1075.e11. 10.1053/j.gastro.2013.07.033 [DOI] [PubMed] [Google Scholar]
- Jackstadt R, van Hooff SR, Leach JD, Cortes-Lavaud X, Lohuis JO, Ridgway RA, Wouters VM, Roper J, Kendall TJ, Roxburgh CS, et al. 2019. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell 36: 319–336.e7. 10.1016/j.ccell.2019.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal BS, Janakiraman V, Kljavin NM, Chaudhuri S, Stern HM, Wang W, Kan Z, Dbouk HA, Peters BA, Waring P, et al. 2009. Somatic mutations in p85α promote tumorigenesis through class IA PI3K activation. Cancer Cell 16: 463–474. 10.1016/j.ccr.2009.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen KP, Alberici P, Fsihi H, Gaspar C, Breukel C, Franken P, Rosty C, Abal M, El Marjou F, Smits R, et al. 2006. APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology 131: 1096–1109. 10.1053/j.gastro.2006.08.011 [DOI] [PubMed] [Google Scholar]
- Jess T, Gamborg M, Matzen P, Munkholm P, Sorensen TI. 2005. Increased risk of intestinal cancer in Crohn's disease: a meta-analysis of population-based cohort studies. Am J Gastroenterol 100: 2724–2729. 10.1111/j.1572-0241.2005.00287.x [DOI] [PubMed] [Google Scholar]
- Ji Y, Yang C, Tang Z, Yang Y, Tian Y, Yao H, Zhu X, Zhang Z, Ji J, Zheng X. 2017. Adenylate kinase hCINAP determines self-renewal of colorectal cancer stem cells by facilitating LDHA phosphorylation. Nat Commun 8: 15308. 10.1038/ncomms15308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Q, Zhou L, Sui H, Yang L, Wu X, Song Q, Jia R, Li R, Sun J, Wang Z, et al. 2020. Primary tumors release ITGBL1-rich extracellular vesicles to promote distal metastatic tumor growth through fibroblast-niche formation. Nat Commun 11: 1211. 10.1038/s41467-020-14869-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobin G, Rodriguez-Suarez R, Betito K. 2017. Association between natural killer cell activity and colorectal cancer in high-risk subjects undergoing colonoscopy. Gastroenterology 153: 980–987. 10.1053/j.gastro.2017.06.009 [DOI] [PubMed] [Google Scholar]
- Joosten SPJ, Zeilstra J, van Andel H, Mijnals RC, Zaunbrecher J, Duivenvoorden AAM, van de Wetering M, Clevers H, Spaargaren M, Pals ST. 2017. MET signaling mediates intestinal crypt-villus development, regeneration, and adenoma formation and is promoted by stem cell CD44 isoforms. Gastroenterology 153: 1040–1053.e4. 10.1053/j.gastro.2017.07.008 [DOI] [PubMed] [Google Scholar]
- Joosten SPJ, Mizutani T, Spaargaren M, Clevers H, Pals ST. 2019. MET signaling overcomes epidermal growth factor receptor inhibition in normal and colorectal cancer stem cells causing drug resistance. Gastroenterology 157: 1153–1155.e1. 10.1053/j.gastro.2019.06.029 [DOI] [PubMed] [Google Scholar]
- Jung YS, Park JI. 2020. Wnt signaling in cancer: therapeutic targeting of Wnt signaling beyond β-catenin and the destruction complex. Exp Mol Med 52: 183–191. 10.1038/s12276-020-0380-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YS, Jun S, Kim MJ, Lee SH, Suh HN, Lien EM, Jung HY, Lee S, Zhang J, Yang JI, et al. 2018a. TMEM9 promotes intestinal tumorigenesis through vacuolar-ATPase-activated Wnt/β-catenin signalling. Nat Cell Biol 20: 1421–1433. 10.1038/s41556-018-0219-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung YS, Wang W, Jun S, Zhang J, Srivastava M, Kim MJ, Lien EM, Shang J, Chen J, McCrea PD, et al. 2018b. Deregulation of CRAD-controlled cytoskeleton initiates mucinous colorectal cancer via β-catenin. Nat Cell Biol 20: 1303–1314. 10.1038/s41556-018-0215-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurk D, Wilson C, Passos JF, Oakley F, Correia-Melo C, Greaves L, Saretzki G, Fox C, Lawless C, Anderson R, et al. 2014. Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nat Commun 5: 4172. 10.1038/ncomms5172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadosh E, Snir-Alkalay I, Venkatachalam A, May S, Lasry A, Elyada E, Zinger A, Shaham M, Vaalani G, Mernberger M, et al. 2020. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 586: 133–138. 10.1038/s41586-020-2541-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M, Greten FR. 2005. NF-κB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol 5: 749–759. 10.1038/nri1703 [DOI] [PubMed] [Google Scholar]
- Kather JN, Halama N. 2019. Harnessing the innate immune system and local immunological microenvironment to treat colorectal cancer. Br J Cancer 120: 871–882. 10.1038/s41416-019-0441-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavuri SM, Jain N, Galimi F, Cottino F, Leto SM, Migliardi G, Searleman AC, Shen W, Monsey J, Trusolino L, et al. 2015. HER2 activating mutations are targets for colorectal cancer treatment. Cancer Discov 5: 832–841. 10.1158/2159-8290.CD-14-1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K, Fujii M, Sugimoto S, Ishikawa K, Matano M, Ohta Y, Toshimitsu K, Takahashi S, Hosoe N, Sekine S, et al. 2020. Chromosome engineering of human colon-derived organoids to develop a model of traditional serrated adenoma. Gastroenterology 158: 638–651.e8. 10.1053/j.gastro.2019.10.009 [DOI] [PubMed] [Google Scholar]
- Khanh DT, Mekata E, Mukaisho K, Sugihara H, Shimizu T, Shiomi H, Murata S, Naka S, Yamamoto H, Endo Y, et al. 2011. Prognostic role of CD10(+) myeloid cells in association with tumor budding at the invasion front of colorectal cancer. Cancer Sci 102: 1724–1733. 10.1111/j.1349-7006.2011.01987.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo CM, Carrasco DR, Bosenberg MW, Paik JH, Depinho RA. 2007. Ink4a/Arf tumor suppressor does not modulate the degenerative conditions or tumor spectrum of the telomerase-deficient mouse. Proc Natl Acad Sci 104: 3931–3936. 10.1073/pnas.0700093104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killela PJ, Reitman ZJ, Jiao Y, Bettegowda C, Agrawal N, Diaz LA Jr, Friedman AH, Friedman H, Gallia GL, Giovanella BC, et al. 2013. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci 110: 6021–6026. 10.1073/pnas.1303607110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Jen J, Vogelstein B, Hamilton SR. 1994. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol 145: 148–156. [PMC free article] [PubMed] [Google Scholar]
- Kinoshita M, Kayama H, Kusu T, Yamaguchi T, Kunisawa J, Kiyono H, Sakaguchi S, Takeda K. 2012. Dietary folic acid promotes survival of Foxp3+ regulatory T cells in the colon. J Immunol 189: 2869–2878. 10.4049/jimmunol.1200420 [DOI] [PubMed] [Google Scholar]
- Kirchberger S, Royston DJ, Boulard O, Thornton E, Franchini F, Szabady RL, Harrison O, Powrie F. 2013. Innate lymphoid cells sustain colon cancer through production of interleukin-22 in a mouse model. J Exp Med 210: 917–931. 10.1084/jem.20122308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland SC. 1988. Clonal origin of columnar, mucous, and endocrine cell lineages in human colorectal epithelium. Cancer 61: 1359–1363. [DOI] [PubMed] [Google Scholar]
- Kiss EA, Diefenbach A. 2012. Role of the aryl hydrocarbon receptor in controlling maintenance and functional programs of RORγt(+) innate lymphoid cells and intraepithelial lymphocytes. Front Immunol 3: 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleeman SO, Koelzer VH, Jones HJ, Vazquez EG, Davis H, East JE, Arnold R, Koppens MA, Blake A, Domingo E, et al. 2020. Exploiting differential Wnt target gene expression to generate a molecular biomarker for colorectal cancer stratification. Gut 69: 1092–1103. 10.1136/gutjnl-2019-319126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CA. 2009. Parallel progression of primary tumours and metastases. Nat Rev Cancer 9: 302–312. 10.1038/nrc2627 [DOI] [PubMed] [Google Scholar]
- Koehne CH, Dubois RN. 2004. COX-2 inhibition and colorectal cancer. Semin Oncol 31: 12–21. 10.1053/j.seminoncol.2004.03.041 [DOI] [PubMed] [Google Scholar]
- Koelzer VH, Dawson H, Andersson E, Karamitopoulou E, Masucci GV, Lugli A, Zlobec I. 2015. Active immunosurveillance in the tumor microenvironment of colorectal cancer is associated with low frequency tumor budding and improved outcome. Transl Res 166: 207–217. 10.1016/j.trsl.2015.02.008 [DOI] [PubMed] [Google Scholar]
- Kojima M, Morisaki T, Sasaki N, Nakano K, Mibu R, Tanaka M, Katano M. 2004. Increased nuclear factor-kB activation in human colorectal carcinoma and its correlation with tumor progression. Anticancer Res 24: 675–681. [PubMed] [Google Scholar]
- Kopetz S, Grothey A, Yaeger R, Van Cutsem E, Desai J, Yoshino T, Wasan H, Ciardiello F, Loupakis F, Hong YS, et al. 2019. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med 381: 1632–1643. 10.1056/NEJMoa1908075 [DOI] [PubMed] [Google Scholar]
- Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. 2012. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res 22: 292–298. 10.1101/gr.126573.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer HB, Lai CF, Patel H, Periyasamy M, Lin ML, Feller SM, Fuller-Pace FV, Meek DW, Ali S, Buluwela L. 2016. LRH-1 drives colon cancer cell growth by repressing the expression of the CDKN1A gene in a p53-dependent manner. Nucleic Acids Res 44: 582–594. 10.1093/nar/gkv948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupitskaya Y, Wakelee HA. 2009. Ramucirumab, a fully human mAb to the transmembrane signaling tyrosine kinase VEGFR-2 for the potential treatment of cancer. Curr Opin Investig Drugs 10: 597–605. [PubMed] [Google Scholar]
- Kuboki Y, Nishina T, Shinozaki E, Yamazaki K, Shitara K, Okamoto W, Kajiwara T, Matsumoto T, Tsushima T, Mochizuki N, et al. 2017. TAS-102 plus bevacizumab for patients with metastatic colorectal cancer refractory to standard therapies (C-TASK FORCE): an investigator-initiated, open-label, single-arm, multicentre, phase 1/2 study. Lancet Oncol 18: 1172–1181. 10.1016/S1470-2045(17)30425-4 [DOI] [PubMed] [Google Scholar]
- Kuczynski EA, Vermeulen PB, Pezzella F, Kerbel RS, Reynolds AR. 2019. Vessel co-option in cancer. Nat Rev Clin Oncol 16: 469–493. 10.1038/s41571-019-0181-9 [DOI] [PubMed] [Google Scholar]
- Kuipers EJ, Grady WM, Lieberman D, Seufferlein T, Sung JJ, Boelens PG, van de Velde CJ, Watanabe T. 2015. Colorectal cancer. Nat Rev Dis Primers 1: 15065. 10.1038/nrdp.2015.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langowski JL, Zhang X, Wu L, Mattson JD, Chen T, Smith K, Basham B, McClanahan T, Kastelein RA, Oft M. 2006. IL-23 promotes tumour incidence and growth. Nature 442: 461–465. 10.1038/nature04808 [DOI] [PubMed] [Google Scholar]
- Lannagan TRM, Lee YK, Wang T, Roper J, Bettington ML, Fennell L, Vrbanac L, Jonavicius L, Somashekar R, Gieniec K, et al. 2019. Genetic editing of colonic organoids provides a molecularly distinct and orthotopic preclinical model of serrated carcinogenesis. Gut 68: 684–692. 10.1136/gutjnl-2017-315920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPointe LC, Dunne R, Brown GS, Worthley DL, Molloy PL, Wattchow D, Young GP. 2008. Map of differential transcript expression in the normal human large intestine. Physiol Genomics 33: 50–64. 10.1152/physiolgenomics.00185.2006 [DOI] [PubMed] [Google Scholar]
- Lawson KA, Sousa CM, Zhang X, Kim E, Akthar R, Caumanns JJ, Yao Y, Mikolajewicz N, Ross C, Brown KR, et al. 2020. Functional genomic landscape of cancer-intrinsic evasion of killing by T cells. Nature 586: 120–126. 10.1038/s41586-020-2746-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Cella M, Colonna M. 2012. AHR and the transcriptional regulation of type-17/22 ILC. Front Immunol 3: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GH, Malietzis G, Askari A, Bernardo D, Al-Hassi HO, Clark SK. 2015. Is right-sided colon cancer different to left-sided colorectal cancer? - a systematic review. Eur J Surg Oncol) 41: 300–308. 10.1016/j.ejso.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Lee HO, Hong Y, Etlioglu HE, Cho YB, Pomella V, Van den Bosch B, Vanhecke J, Verbandt S, Hong H, Min JW, et al. 2020. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet 52: 594–603. 10.1038/s41588-020-0636-z [DOI] [PubMed] [Google Scholar]
- Leedham SJ, Rodenas-Cuadrado P, Howarth K, Lewis A, Mallappa S, Segditsas S, Davis H, Jeffery R, Rodriguez-Justo M, Keshav S, et al. 2013. A basal gradient of Wnt and stem-cell number influences regional tumour distribution in human and mouse intestinal tracts. Gut 62: 83–93. 10.1136/gutjnl-2011-301601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legitimo A, Consolini R, Failli A, Orsini G, Spisni R. 2014. Dendritic cell defects in the colorectal cancer. Hum Vaccin Immunother 10: 3224–3235. 10.4161/hv.29857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. 1998. Genetic instabilities in human cancers. Nature 396: 643–649. 10.1038/25292 [DOI] [PubMed] [Google Scholar]
- Lenz HJ, Ahn D, Erlander M, Barzi A. 2019. 667TiP—a phase Ib/II study of onvansertib (PCM-075) in combination with FOLFIRI and bevacizumab for second-line treatment of metastatic colorectal cancer in patients with a KRAS mutation. Ann Oncol 30: v250. 10.1093/annonc/mdz246.144 [DOI] [Google Scholar]
- Leon-Bollotte L, Subramaniam S, Cauvard O, Plenchette-Colas S, Paul C, Godard C, Martinez-Ruiz A, Legembre P, Jeannin JF, Bettaieb A. 2011. S-nitrosylation of the death receptor fas promotes fas ligand-mediated apoptosis in cancer cells. Gastroenterology 140: 2009–2018.e4. 10.1053/j.gastro.2011.02.053 [DOI] [PubMed] [Google Scholar]
- Leung ML, Davis A, Gao R, Casasent A, Wang Y, Sei E, Vilar E, Maru D, Kopetz S, Navin NE. 2017. Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res 27: 1287–1299. 10.1101/gr.209973.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy M, Thaiss CA, Elinav E. 2016. Metabolites: messengers between the microbiota and the immune system. Genes Dev 30: 1589–1597. 10.1101/gad.284091.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GM. 2008. Mechanisms and functions of DNA mismatch repair. Cell Res 18: 85–98. 10.1038/cr.2007.115 [DOI] [PubMed] [Google Scholar]
- Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL, Kong SL, Chua C, Hon LK, Tan WS, et al. 2017a. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 49: 708–718. 10.1038/ng.3818 [DOI] [PubMed] [Google Scholar]
- Li J, Yu B, Deng P, Cheng Y, Yu Y, Kevork K, Ramadoss S, Ding X, Li X, Wang CY. 2017b. KDM3 epigenetically controls tumorigenic potentials of human colorectal cancer stem cells through Wnt/β-catenin signalling. Nat Commun 8: 15146. 10.1038/ncomms15146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HW, Hsu JM, Xia W, Wang HL, Wang YN, Chang WC, Arold ST, Chou CK, Tsou PH, Yamaguchi H, et al. 2015. PRMT1-mediated methylation of the EGF receptor regulates signaling and cetuximab response. J Clin Invest 125: 4529–4543. 10.1172/JCI82826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W, Overman MJ, Boutin AT, Shang X, Zhao D, Dey P, Li J, Wang G, Lan Z, Li J, et al. 2019. KRAS–IRF2 axis drives immune suppression and immune therapy resistance in colorectal cancer. Cancer Cell 35: 559–572.e7. 10.1016/j.ccell.2019.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberti MV, Locasale JW. 2016. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci 41: 211–218. 10.1016/j.tibs.2015.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligtenberg MJ, Kuiper RP, Chan TL, Goossens M, Hebeda KM, Voorendt M, Lee TY, Bodmer D, Hoenselaar E, Hendriks-Cornelissen SJ, et al. 2009. Heritable somatic methylation and inactivation of MSH2 in families with Lynch syndrome due to deletion of the 3′ exons of TACSTD1. Nat Genet 41: 112–117. 10.1038/ng.283 [DOI] [PubMed] [Google Scholar]
- Lim SY, Gordon-Weeks A, Allen D, Kersemans V, Beech J, Smart S, Muschel RJ. 2015. Cd11b+ myeloid cells support hepatic metastasis through down-regulation of angiopoietin-like 7 in cancer cells. Hepatology 62: 521–533. 10.1002/hep.27838 [DOI] [PubMed] [Google Scholar]
- Liu S, Bekker-Jensen S, Mailand N, Lukas C, Bartek J, Lukas J. 2006. Claspin operates downstream of TopBP1 to direct ATR signaling towards Chk1 activation. Mol Cell Biol 26: 6056–6064. 10.1128/MCB.00492-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Lu Z, Gu J, Liu J, Huang E, Liu X, Wang L, Yang J, Deng Y, Qian J, et al. 2018a. MicroRNAs 15A and 16-1 activate signaling pathways that mediate chemotaxis of immune regulatory B cells to colorectal tumors. Gastroenterology 154: 637–651.e7. 10.1053/j.gastro.2017.09.045 [DOI] [PubMed] [Google Scholar]
- Liu Y, Sethi NS, Hinoue T, Schneider BG, Cherniack AD, Sanchez-Vega F, Seoane JA, Farshidfar F, Bowlby R, Islam M, et al. 2018b. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell 33: 721–735.e8. 10.1016/j.ccell.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey KM, Kang R, Vernon P, Buchser W, Loughran P, Watkins SC, Zhang L, Manfredi JJ, Zeh HJ III, Li L, et al. 2012a. P53/HMGB1 complexes regulate autophagy and apoptosis. Cancer Res 72: 1996–2005. 10.1158/0008-5472.CAN-11-2291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey KM, Kang R, Zeh HJ III, Lotze MT, Tang D. 2012b. Direct molecular interactions between HMGB1 and TP53 in colorectal cancer. Autophagy 8: 846–848. 10.4161/auto.19891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale JW. 2013. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat Rev Cancer 13: 572–583. 10.1038/nrc3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-García C, Sansregret L, Domingo E, McGranahan N, Hobor S, Birkbak NJ, Horswell S, Grönroos E, Favero F, Rowan AJ, et al. 2017. BCL9L dysfunction impairs caspase-2 expression permitting aneuploidy tolerance in colorectal cancer. Cancer Cell 31: 79–93. 10.1016/j.ccell.2016.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Hold GL, Flint HJ. 2014. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12: 661–672. 10.1038/nrmicro3344 [DOI] [PubMed] [Google Scholar]
- Loupakis F, Yang D, Yau L, Feng S, Cremolini C, Zhang W, Maus MK, Antoniotti C, Langer C, Scherer SJ, et al. 2015. Primary tumor location as a prognostic factor in metastatic colorectal cancer. J Natl Cancer Inst 107: dju427. 10.1093/jnci/dju427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loyon R, Jary M, Salomé B, Gomez-Cadena A, Galaine J, Kroemer M, Romero P, Trabanelli S, Adotevi O, Borg C, et al. 2019. Peripheral innate lymphoid cells are increased in first line metastatic colorectal carcinoma patients: a negative correlation with Th1 immune responses. Front Immunol 10: 2121. 10.3389/fimmu.2019.02121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke JJ, Bao R, Sweis RF, Spranger S, Gajewski TF. 2019. WNT/β-catenin pathway activation correlates with immune exclusion across human cancers. Clin Cancer Res 25: 3074–3083. 10.1158/1078-0432.CCR-18-1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Manning BD, Cantley LC. 2003. Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell 4: 257–262. 10.1016/S1535-6108(03)00248-4 [DOI] [PubMed] [Google Scholar]
- Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. 2009. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137: 835–848. 10.1016/j.cell.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald BT, Tamai K, He X. 2009. Wnt/β-catenin signaling: components, mechanisms, and diseases. Dev Cell 17: 9–26. 10.1016/j.devcel.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maj T, Wang W, Crespo J, Zhang H, Wang W, Wei S, Zhao L, Vatan L, Shao I, Szeliga W, et al. 2017. Oxidative stress controls regulatory T cell apoptosis and suppressor activity and PD-L1-blockade resistance in tumor. Nat Immunol 18: 1332–1341. 10.1038/ni.3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malekzadeh P, Pasetto A, Robbins PF, Parkhurst MR, Paria BC, Jia L, Gartner JJ, Hill V, Yu Z, Restifo NP, et al. 2019. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J Clin Invest 129: 1109–1114. 10.1172/JCI123791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy KJ, Powrie F. 2011. Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474: 298–306. 10.1038/nature10208 [DOI] [PubMed] [Google Scholar]
- Mantovani A, Allavena P, Sica A, Balkwill F. 2008. Cancer-related inflammation. Nature 454: 436–444. 10.1038/nature07205 [DOI] [PubMed] [Google Scholar]
- Mantovani A, Cassatella MA, Costantini C, Jaillon S. 2011. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat Rev Immunol 11: 519–531. 10.1038/nri3024 [DOI] [PubMed] [Google Scholar]
- Marchio S, Soster M, Cardaci S, Muratore A, Bartolini A, Barone V, Ribero D, Monti M, Bovino P, Sun J, et al. 2012. A complex of α6 integrin and E-cadherin drives liver metastasis of colorectal cancer cells through hepatic angiopoietin-like 6. EMBO Mol Med 4: 1156–1175. 10.1002/emmm.201101164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariathasan S, Turley SJ, Nickles D, Castiglioni A, Yuen K, Wang Y, Kadel EE III, Koeppen H, Astarita JL, Cubas R, et al. 2018. TGFβ attenuates tumour response to PD-L1 blockade by contributing to exclusion of T cells. Nature 554: 544–548. 10.1038/nature25501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnett LJ. 1999. Lipid peroxidation—DNA damage by malondialdehyde. Mutat Res 424: 83–95. 10.1016/S0027-5107(99)00010-X [DOI] [PubMed] [Google Scholar]
- Marnett LJ. 2000. Oxyradicals and DNA damage. Carcinogenesis 21: 361–370. 10.1093/carcin/21.3.361 [DOI] [PubMed] [Google Scholar]
- Martin SA, McCabe N, Mullarkey M, Cummins R, Burgess DJ, Nakabeppu Y, Oka S, Kay E, Lord CJ, Ashworth A. 2010. DNA polymerases as potential therapeutic targets for cancers deficient in the DNA mismatch repair proteins MSH2 or MLH1. Cancer Cell 17: 235–248. 10.1016/j.ccr.2009.12.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SA, Hewish M, Sims D, Lord CJ, Ashworth A. 2011. Parallel high-throughput RNA interference screens identify PINK1 as a potential therapeutic target for the treatment of DNA mismatch repair-deficient cancers. Cancer Res 71: 1836–1848. 10.1158/0008-5472.CAN-10-2836 [DOI] [PubMed] [Google Scholar]
- Masucci MT, Minopoli M, Carriero MV. 2019. Tumor associated neutrophils. Their role in tumorigenesis, metastasis, prognosis and therapy. Front Oncol 9: 1146. 10.3389/fonc.2019.01146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masugi Y, Nishihara R, Yang J, Mima K, da Silva A, Shi Y, Inamura K, Cao Y, Song M, Nowak JA, et al. 2017. Tumour CD274 (PD-L1) expression and T cells in colorectal cancer. Gut 66: 1463–1473. 10.1136/gutjnl-2016-311421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallanas D, Romano D, Al-Mulla F, O'Neill E, Al-Ali W, Crespo P, Doyle B, Nixon C, Sansom O, Drosten M, et al. 2011. Mutant K-Ras activation of the proapoptotic MST2 pathway is antagonized by wild-type K-Ras. Mol Cell 44: 893–906. 10.1016/j.molcel.2011.10.016 [DOI] [PubMed] [Google Scholar]
- Matano M, Date S, Shimokawa M, Takano A, Fujii M, Ohta Y, Watanabe T, Kanai T, Sato T. 2015. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med 21: 256–262. 10.1038/nm.3802 [DOI] [PubMed] [Google Scholar]
- Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. 2006. P53 regulates mitochondrial respiration. Science 312: 1650–1653. 10.1126/science.1126863 [DOI] [PubMed] [Google Scholar]
- McCuaig S, Barras D, Mann EH, Friedrich M, Bullers SJ, Janney A, Garner LC, Domingo E, Koelzer VH, Delorenzi M, et al. 2020. The interleukin 22 pathway interacts with mutant KRAS to promote poor prognosis in colon cancer. Clin Cancer Res 26: 4313–4325. 10.1158/1078-0432.CCR-19-1086 [DOI] [PubMed] [Google Scholar]
- McIntyre RE, Buczacki SJ, Arends MJ, Adams DJ. 2015. Mouse models of colorectal cancer as preclinical models. Bioessays 37: 909–920. 10.1002/bies.201500032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon SB. 2014. MYC and the control of apoptosis. Cold Spring Harb Perspect Med 4: a014407. 10.1101/cshperspect.a014407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMichael AJ, Potter JD. 1985. Diet and colon cancer: integration of the descriptive, analytic, and metabolic epidemiology. Natl Cancer Inst Monogr 69: 223–228. [PubMed] [Google Scholar]
- Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, Buscarino M, Isella C, Lamba S, Martinoglio B, et al. 2015. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun 6: 7002. 10.1038/ncomms8002 [DOI] [PubMed] [Google Scholar]
- Mehrvarz Sarshekeh A, Advani S, Overman MJ, Manyam G, Kee BK, Fogelman DR, Dasari A, Raghav K, Vilar E, Manuel S, et al. 2017. Association of SMAD4 mutation with patient demographics, tumor characteristics, and clinical outcomes in colorectal cancer. PLoS One 12: e0173345. 10.1371/journal.pone.0173345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisenberg C, Ashour ME, El-Shafie L, Liao C, Hodgson A, Pilborough A, Khurram SA, Downs JA, Ward SE, El-Khamisy SF. 2017. Epigenetic changes in histone acetylation underpin resistance to the topoisomerase I inhibitor irinotecan. Nucleic Acids Res 45: 1159–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric-Bernstam F, Hurwitz H, Raghav KPS, McWilliams RR, Fakih M, VanderWalde A, Swanton C, Kurzrock R, Burris H, Sweeney C, et al. 2019. Pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer (MyPathway): an updated report from a multicentre, open-label, phase 2a, multiple basket study. Lancet Oncol 20: 518–530. 10.1016/S1470-2045(18)30904-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Penn LZ. 2008. Reflecting on 25 years with MYC. Nature Reviews Cancer 8: 976–990. 10.1038/nrc2231 [DOI] [PubMed] [Google Scholar]
- Mirza-Aghazadeh-Attari M, Darband SG, Kaviani M, Mihanfar A, Aghazadeh Attari J, Yousefi B, Majidinia M. 2018. DNA damage response and repair in colorectal cancer: defects, regulation and therapeutic implications. DNA Repair 69: 34–52. 10.1016/j.dnarep.2018.07.005 [DOI] [PubMed] [Google Scholar]
- Mlecnik B, Tosolini M, Kirilovsky A, Berger A, Bindea G, Meatchi T, Bruneval P, Trajanoski Z, Fridman WH, Pagès F, et al. 2011. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J Clin Oncol 29: 610–618. 10.1200/JCO.2010.30.5425 [DOI] [PubMed] [Google Scholar]
- Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, Church SE, Lafontaine L, Fischer M, Fredriksen T, et al. 2016. Integrative analyses of colorectal cancer show Immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity 44: 698–711. 10.1016/j.immuni.2016.02.025 [DOI] [PubMed] [Google Scholar]
- Mokarram P, Albokashy M, Zarghooni M, Moosavi MA, Sepehri Z, Chen QM, Hudecki A, Sargazi A, Alizadeh J, Moghadam AR, et al. 2017. New frontiers in the treatment of colorectal cancer: autophagy and the unfolded protein response as promising targets. Autophagy 13: 781–819. 10.1080/15548627.2017.1290751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser AR, Pitot HC, Dove WF. 1990. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science 247: 322–324. 10.1126/science.2296722 [DOI] [PubMed] [Google Scholar]
- Mouradov D, Sloggett C, Jorissen RN, Love CG, Li S, Burgess AW, Arango D, Strausberg RL, Buchanan D, Wormald S, et al. 2014. Colorectal cancer cell lines are representative models of the main molecular subtypes of primary cancer. Cancer Res 74: 3238–3247. 10.1158/0008-5472.CAN-14-0013 [DOI] [PubMed] [Google Scholar]
- Movahedi K, Laoui D, Gysemans C, Baeten M, Stangé G, Van den Bossche J, Mack M, Pipeleers D, In't Veld P, De Baetselier P, et al. 2010. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res 70: 5728–5739. 10.1158/0008-5472.CAN-09-4672 [DOI] [PubMed] [Google Scholar]
- Mucida D, Husain MM, Muroi S, van Wijk F, Shinnakasu R, Naoe Y, Reis BS, Huang Y, Lambolez F, Docherty M, et al. 2013. Transcriptional reprogramming of mature CD4+ helper T cells generates distinct MHC class II-restricted cytotoxic T lymphocytes. Nat Immunol 14: 281–289. 10.1038/ni.2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi Y, Seno H, Fukuoka A, Ueo T, Yamaga Y, Maruno T, Nakanishi N, Kanda K, Komekado H, Kawada M, et al. 2013. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet 45: 98–103. 10.1038/ng.2481 [DOI] [PubMed] [Google Scholar]
- Narasimhan V, Wright JA, Churchill M, Wang T, Rosati R, Lannagan TRM, Vrbanac L, Richardson AB, Kobayashi H, Price T, et al. 2020. Medium-throughput drug screening of patient-derived organoids from colorectal peritoneal metastases to direct personalized therapy. Clin Cancer Res 26: 3662–3670. 10.1158/1078-0432.CCR-20-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassour J, Radford R, Correia A, Fusté JM, Schoell B, Jauch A, Shaw RJ, Karlseder J. 2019. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 565: 659–663. 10.1038/s41586-019-0885-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nejman D, Livyatan I, Fuks G, Gavert N, Zwang Y, Geller LT, Rotter-Maskowitz A, Weiser R, Mallel G, Gigi E, et al. 2020. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368: 973–980. 10.1126/science.aay9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Balkwill F, Chonchol M, Cominelli F, Donath MY, Giamarellos-Bourboulis EJ, Golenbock D, Gresnigt MS, Heneka MT, Hoffman HM, et al. 2017. A guiding map for inflammation. Nat Immunol 18: 826–831. 10.1038/ni.3790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AC, Maddocks ODK. 2017. One-carbon metabolism in cancer. Br J Cancer 116: 1499–1504. 10.1038/bjc.2017.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen LH, Goel A, Chung DC. 2020. Pathways of colorectal carcinogenesis. Gastroenterology 158: 291–302. 10.1053/j.gastro.2019.08.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton L, Massagué J. 2006. Is cancer a disease of self-seeding? Nat Med 12: 875–878. 10.1038/nm0806-875 [DOI] [PubMed] [Google Scholar]
- Novellasdemunt L, Kucharska A, Jamieson C, Prange-Barczynska M, Baulies A, Antas P, van der Vaart J, Gehart H, Maurice MM, Li VS. 2020. NEDD4 and NEDD4L regulate Wnt signalling and intestinal stem cell priming by degrading LGR5 receptor. EMBO J 39: e102771. 10.15252/embj.2019102771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien CA, Pollett A, Gallinger S, Dick JE. 2007. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature 445: 106–110. 10.1038/nature05372 [DOI] [PubMed] [Google Scholar]
- Ooft SN, Weeber F, Dijkstra KK, McLean CM, Kaing S, van Werkhoven E, Schipper L, Hoes L, Vis DJ, van de Haar J, et al. 2019. Patient-derived organoids can predict response to chemotherapy in metastatic colorectal cancer patients. Sci Transl Med 11: eaay2574. 10.1126/scitranslmed.aay2574 [DOI] [PubMed] [Google Scholar]
- O'Regan AW, Hayden JM, Berman JS. 2000. Osteopontin augments CD3-mediated interferon-γ and CD40 ligand expression by T cells, which results in IL-12 production from peripheral blood mononuclear cells. J Leukoc Biol 68: 495–502. [PubMed] [Google Scholar]
- O'Sullivan JN, Bronner MP, Brentnall TA, Finley JC, Shen WT, Emerson S, Emond MJ, Gollahon KA, Moskovitz AH, Crispin DA, et al. 2002. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat Genet 32: 280–284. 10.1038/ng989 [DOI] [PubMed] [Google Scholar]
- O'Sullivan J, Risques RA, Mandelson MT, Chen L, Brentnall TA, Bronner MP, Macmillan MP, Feng Z, Siebert JR, Potter JD, et al. 2006. Telomere length in the colon declines with age: a relation to colorectal cancer? Cancer Epidemiol Biomarkers Prev 15: 573–577. 10.1158/1055-9965.EPI-05-0542 [DOI] [PubMed] [Google Scholar]
- Otto T, Sicinski P. 2017. Cell cycle proteins as promising targets in cancer therapy. Nat Rev Cancer 17: 93–115. 10.1038/nrc.2016.138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al. 2017. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol 18: 1182–1191. 10.1016/S1470-2045(17)30422-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R, Hill A, et al. 2018. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol 36: 773–779. 10.1200/JCO.2017.76.9901 [DOI] [PubMed] [Google Scholar]
- Pagès F, Mlecnik B, Marliot F, Bindea G, Ou FS, Bifulco C, Lugli A, Zlobec I, Rau TT, Berger MD, et al. 2018. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 391: 2128–2139. 10.1016/S0140-6736(18)30789-X [DOI] [PubMed] [Google Scholar]
- Pate KT, Stringari C, Sprowl-Tanio S, Wang K, TeSlaa T, Hoverter NP, McQuade MM, Garner C, Digman MA, Teitell MA, et al. 2014. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J 33: 1454–1473. 10.15252/embj.201488598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K, Kou L, Yu L, Bai C, Li M, Mo P, Li W, Yu C. 2019. Histone Demethylase JMJD2D interacts with β-catenin to induce transcription and activate colorectal cancer cell proliferation and tumor growth in mice. Gastroenterology 156: 1112–1126. 10.1053/j.gastro.2018.11.036 [DOI] [PubMed] [Google Scholar]
- Piccard H, Muschel RJ, Opdenakker G. 2012. On the dual roles and polarized phenotypes of neutrophils in tumor development and progression. Crit Rev Oncol Hematol 82: 296–309. 10.1016/j.critrevonc.2011.06.004 [DOI] [PubMed] [Google Scholar]
- Pollheimer MJ, Kornprat P, Lindtner RA, Harbaum L, Schlemmer A, Rehak P, Langner C. 2010. Tumor necrosis is a new promising prognostic factor in colorectal cancer. Hum Pathol 41: 1749–1757. 10.1016/j.humpath.2010.04.018 [DOI] [PubMed] [Google Scholar]
- Pomerantz J, Schreiber-Agus N, Liegeois NJ, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee HW, et al. 1998. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2's inhibition of p53. Cell 92: 713–723. 10.1016/S0092-8674(00)81400-2 [DOI] [PubMed] [Google Scholar]
- Popow O, Paulo JA, Tatham MH, Volk MS, Rojas-Fernandez A, Loyer N, Newton IP, Januschke J, Haigis KM, Näthke I. 2019. Identification of endogenous adenomatous polyposis coli interaction partners and β-catenin-independent targets by proteomics. Mol Cancer Res 17: 1828–1841. 10.1158/1541-7786.MCR-18-1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell AE, Wang Y, Li Y, Poulin EJ, Means AL, Washington MK, Higginbotham JN, Juchheim A, Prasad N, Levy SE, et al. 2012. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell 149: 146–158. 10.1016/j.cell.2012.02.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribluda A, Elyada E, Wiener Z, Hamza H, Goldstein RE, Biton M, Burstain I, Morgenstern Y, Brachya G, Billauer H, et al. 2013. A senescence-inflammatory switch from cancer-inhibitory to cancer-promoting mechanism. Cancer Cell 24: 242–256. 10.1016/j.ccr.2013.06.005 [DOI] [PubMed] [Google Scholar]
- Pylayeva-Gupta Y, Lee KE, Hajdu CH, Miller G, Bar-Sagi D. 2012. Oncogenic Kras-induced GM-CSF production promotes the development of pancreatic neoplasia. Cancer Cell 21: 836–847. 10.1016/j.ccr.2012.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. 2017. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol 35: 833–844. 10.1038/nbt.3935 [DOI] [PubMed] [Google Scholar]
- Rahbari NN, Kedrin D, Incio J, Liu H, Ho WW, Nia HT, Edrich CM, Jung K, Daubriac J, Chen I, et al. 2016. Anti-VEGF therapy induces ECM remodeling and mechanical barriers to therapy in colorectal cancer liver metastases. Sci Transl Med 8: 360ra135. 10.1126/scitranslmed.aaf5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan H, Bardelli A, Lengauer C, Kinzler KW, Vogelstein B, Velculescu VE. 2002. Tumorigenesis: RAF/RAS oncogenes and mismatch-repair status. Nature 418: 934. 10.1038/418934a [DOI] [PubMed] [Google Scholar]
- Ramakrishnan SK, Zhang H, Ma X, Jung I, Schwartz AJ, Triner D, Devenport SN, Das NK, Xue X, Zeng MY, et al. 2019. Intestinal non-canonical NFκB signaling shapes the local and systemic immune response. Nat Commun 10: 660. 10.1038/s41467-019-08581-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashtak S, Rego R, Sweetser SR, Sinicrope FA. 2017. Sessile serrated polyps and colon cancer prevention. Cancer Prev Res (Phila) 10: 270–278. 10.1158/1940-6207.CAPR-16-0264 [DOI] [PubMed] [Google Scholar]
- Ricciardiello L, Ahnen DJ, Lynch PM. 2016. Chemoprevention of hereditary colon cancers: time for new strategies. Nat Rev Gastroenterol Hepatol 13: 352–361. 10.1038/nrgastro.2016.56 [DOI] [PubMed] [Google Scholar]
- Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. 2016. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 7: 185. 10.3389/fmicb.2016.00185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risio M, Reato G, di Celle PF, Fizzotti M, Rossini FP, Foa R. 1996. Microsatellite instability is associated with the histological features of the tumor in nonfamilial colorectal cancer. Cancer Res 56: 5470–5474. [PubMed] [Google Scholar]
- Risques RA, Lai LA, Brentnall TA, Li L, Feng Z, Gallaher J, Mandelson MT, Potter JD, Bronner MP, Rabinovitch PS. 2008. Ulcerative colitis is a disease of accelerated colon aging: evidence from telomere attrition and DNA damage. Gastroenterology 135: 410–418. 10.1053/j.gastro.2008.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Colman MJ, Schewe M, Meerlo M, Stigter E, Gerrits J, Pras-Raves M, Sacchetti A, Hornsveld M, Oost KC, Snippert HJ, et al. 2017. Interplay between metabolic identities in the intestinal crypt supports stem cell function. Nature 543: 424–427. 10.1038/nature21673 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Warne PH, Dhand R, Vanhaesebroeck B, Gout I, Fry MJ, Waterfield MD, Downward J. 1994. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature 370: 527–532. 10.1038/370527a0 [DOI] [PubMed] [Google Scholar]
- Roerink SF, Sasaki N, Lee-Six H, Young MD, Alexandrov LB, Behjati S, Mitchell TJ, Grossmann S, Lightfoot H, Egan DA, et al. 2018. Intra-tumour diversification in colorectal cancer at the single-cell level. Nature 556: 457–462. 10.1038/s41586-018-0024-3 [DOI] [PubMed] [Google Scholar]
- Roninson IB. 2003. Tumor cell senescence in cancer treatment. Cancer Res 63: 2705–2715. [PubMed] [Google Scholar]
- Rooks MG, Garrett WS. 2016. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 16: 341–352. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roper J, Tammela T, Cetinbas NM, Akkad A, Roghanian A, Rickelt S, Almeqdadi M, Wu K, Oberli MA, Sanchez-Rivera FJ, et al. 2017. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat Biotechnol 35: 569–576. 10.1038/nbt.3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin DC, Shaker A, Levin MS. 2012. Chronic intestinal inflammation: inflammatory bowel disease and colitis-associated colon cancer. Front Immunol 3: 107. 10.3389/fimmu.2012.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph KL, Millard M, Bosenberg MW, DePinho RA. 2001. Telomere dysfunction and evolution of intestinal carcinoma in mice and humans. Nat Genet 28: 155–159. 10.1038/88871 [DOI] [PubMed] [Google Scholar]
- Ryall JG, Cliff T, Dalton S, Sartorelli V. 2015. Metabolic reprogramming of stem cell epigenetics. Cell Stem Cell 17: 651–662. 10.1016/j.stem.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryser MD, Mallo D, Hall A, Hardman T, King LM, Tatishchev S, Sorribes IC, Maley CC, Marks JR, Hwang ES, et al. 2020. Minimal barriers to invasion during human colorectal tumor growth. Nat Commun 11: 1280. 10.1038/s41467-020-14908-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin U, Türeci O. 2018. Personalized vaccines for cancer immunotherapy. Science 359: 1355–1360. 10.1126/science.aar7112 [DOI] [PubMed] [Google Scholar]
- Sakthianandeswaren A, Parsons MJ, Mouradov D, MacKinnon RN, Catimel B, Liu S, Palmieri M, Love C, Jorissen RN, Li S, et al. 2018. MACROD2 Haploinsufficiency impairs catalytic activity of PARP1 and promotes chromosome instability and growth of intestinal tumors. Cancer Discov 8: 988–1005. 10.1158/2159-8290.CD-17-0909 [DOI] [PubMed] [Google Scholar]
- Sanz-Garcia E, Argiles G, Elez E, Tabernero J. 2017. BRAF mutant colorectal cancer: prognosis, treatment, and new perspectives. Ann Oncol 28: 2648–2657. 10.1093/annonc/mdx401 [DOI] [PubMed] [Google Scholar]
- Sartore-Bianchi A, Trusolino L, Martino C, Bencardino K, Lonardi S, Bergamo F, Zagonel V, Leone F, Depetris I, Martinelli E, et al. 2016. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): a proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol 17: 738–746. 10.1016/S1470-2045(16)00150-9 [DOI] [PubMed] [Google Scholar]
- Sartore-Bianchi A, Marsoni S, Siena S. 2018. Human epidermal growth factor receptor 2 as a molecular biomarker for metastatic colorectal cancer. JAMA Oncol 4: 19–20. 10.1001/jamaoncol.2017.3323 [DOI] [PubMed] [Google Scholar]
- Sato Y, Takahashi S, Kinouchi Y, Shiraki M, Endo K, Matsumura Y, Kakuta Y, Tosa M, Motida A, Abe H, et al. 2006. IL-10 deficiency leads to somatic mutations in a model of IBD. Carcinogenesis 27: 1068–1073. 10.1093/carcin/bgi327 [DOI] [PubMed] [Google Scholar]
- Schell JC, Olson KA, Jiang L, Hawkins AJ, Van Vranken JG, Xie J, Egnatchik RA, Earl EG, DeBerardinis RJ, Rutter J. 2014. A role for the mitochondrial pyruvate carrier as a repressor of the Warburg effect and colon cancer cell growth. Mol Cell 56: 400–413. 10.1016/j.molcel.2014.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers AG, Snippert HJ, Stange DE, van den Born M, van Es JH, van de Wetering M, Clevers H. 2012. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science 337: 730–735. 10.1126/science.1224676 [DOI] [PubMed] [Google Scholar]
- Schmidt S, Gay D, Uthe FW, Denk S, Paauwe M, Matthes N, Diefenbacher ME, Bryson S, Warrander FC, Erhard F, et al. 2019. A MYC–GCN2–eIF2α negative feedback loop limits protein synthesis to prevent MYC-dependent apoptosis in colorectal cancer. Nat Cell Biol 21: 1413–1424. 10.1038/s41556-019-0408-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürch CM, Bhate SS, Barlow GL, Phillips DJ, Noti L, Zlobec I, Chu P, Black S, Demeter J, McIlwain DR, et al. 2020. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell 182: 1341–1359.e19. 10.1016/j.cell.2020.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwaab T, Weiss JE, Schned AR, Barth RJ Jr. 2001. Dendritic cell infiltration in colon cancer. J Immunother 24: 130–137. 10.1097/00002371-200103000-00007 [DOI] [PubMed] [Google Scholar]
- Schwitalla S, Fingerle AA, Cammareri P, Nebelsiek T, Göktuna SI, Ziegler PK, Canli O, Heijmans J, Huels DJ, Moreaux G, et al. 2013a. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 152: 25–38. 10.1016/j.cell.2012.12.012 [DOI] [PubMed] [Google Scholar]
- Schwitalla S, Ziegler PK, Horst D, Becker V, Kerle I, Begus-Nahrmann Y, Lechel A, Rudolph KL, Langer R, Slotta-Huspenina J, et al. 2013b. Loss of p53 in enterocytes generates an inflammatory microenvironment enabling invasion and lymph node metastasis of carcinogen-induced colorectal tumors. Cancer Cell 23: 93–106. 10.1016/j.ccr.2012.11.014 [DOI] [PubMed] [Google Scholar]
- Sears CL, Garrett WS. 2014. Microbes, microbiota, and colon cancer. Cell Host Microbe 15: 317–328. 10.1016/j.chom.2014.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seneviratne D, Ma J, Tan X, Kwon YK, Muhammad E, Melhem M, DeFrances MC, Zarnegar R. 2015. Genomic instability causes HGF gene activation in colon cancer cells, promoting their resistance to necroptosis. Gastroenterology 148: 181–191.e17. 10.1053/j.gastro.2014.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, et al. 2012. Recurrent R-spondin fusions in colon cancer. Nature 488: 660–664. 10.1038/nature11282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seubert B, Grunwald B, Kobuch J, Cui H, Schelter F, Schaten S, Siveke JT, Lim NH, Nagase H, Simonavicius N, et al. 2015. Tissue inhibitor of metalloproteinases (TIMP)-1 creates a premetastatic niche in the liver through SDF-1/CXCR4-dependent neutrophil recruitment in mice. Hepatology 61: 238–248. 10.1002/hep.27378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaked H, Hofseth LJ, Chumanevich A, Chumanevich AA, Wang J, Wang Y, Taniguchi K, Guma M, Shenouda S, Clevers H, et al. 2012. Chronic epithelial NF-κB activation accelerates APC loss and intestinal tumor initiation through iNOS up-regulation. Proc Natl Acad Sci 109: 14007–14012. 10.1073/pnas.1211509109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalapour S, Karin M. 2020. Cruel to be kind: epithelial, microbial, and immune cell interactions in gastrointestinal cancers. Annu Rev Immunol 38: 649–671. 10.1146/annurev-immunol-082019-081656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul ME, Fridlender ZG. 2019. Tumour-associated neutrophils in patients with cancer. Nat Rev Clin Oncol 16: 601–620. 10.1038/s41571-019-0222-4 [DOI] [PubMed] [Google Scholar]
- Shen Y, Wang X, Lu J, Salfenmoser M, Wirsik NM, Schleussner N, Imle A, Freire Valls A, Radhakrishnan P, Liang J, et al. 2020. Reduction of liver metastasis stiffness improves response to bevacizumab in metastatic colorectal cancer. Cancer Cell 37: 800–817.e7. 10.1016/j.ccell.2020.05.005 [DOI] [PubMed] [Google Scholar]
- Sherwood AM, Emerson RO, Scherer D, Habermann N, Buck K, Staffa J, Desmarais C, Halama N, Jaeger D, Schirmacher P, et al. 2013. Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the T cells in adjacent mucosal tissue. Cancer Immunol Immunother 62: 1453–1461. 10.1007/s00262-013-1446-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Kunisawa J, Kiyono H. 2017. Dietary and microbial metabolites in the regulation of host immunity. Front Microbiol 8: 2171. 10.3389/fmicb.2017.02171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, Hugo W, Sandoval S, Torrejon DY, et al. 2017. Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov 7: 188–201. 10.1158/2159-8290.CD-16-1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shringarpure R, Davies KJ. 2002. Protein turnover by the proteasome in aging and disease. Free Radic Biol Med 32: 1084–1089. 10.1016/S0891-5849(02)00824-9 [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. 2017. Colorectal cancer statistics, 2017. CA Cancer J Clin 67: 177–193. 10.3322/caac.21395 [DOI] [PubMed] [Google Scholar]
- Simons CC, Hughes LA, Smits KM, Khalid-de Bakker CA, de Bruïne AP, Carvalho B, Meijer GA, Schouten LJ, van den Brandt PA, Weijenberg MP, et al. 2013. A novel classification of colorectal tumors based on microsatellite instability, the CpG island methylator phenotype and chromosomal instability: implications for prognosis. Ann Oncol 24: 2048–2056. 10.1093/annonc/mdt076 [DOI] [PubMed] [Google Scholar]
- Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H, Thangaraju M, Prasad PD, Manicassamy S, Munn DH, et al. 2014. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40: 128–139. 10.1016/j.immuni.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Single-cell microbiology. [Editorial] 2016. Nat Biotechnol 34: 1077. 10.1038/nbt.3728 [DOI] [PubMed] [Google Scholar]
- Siraj AK, Bu R, Iqbal K, Parvathareddy SK, Siraj N, Siraj S, Diaz MRF, Rala DR, Benito AD, Sabido MA, et al. 2020. Telomerase reverse transcriptase promoter mutations in cancers derived from multiple organ sites among Middle Eastern population. Genomics 112: 1746–1753. 10.1016/j.ygeno.2019.09.017 [DOI] [PubMed] [Google Scholar]
- Snover DC. 2011. Update on the serrated pathway to colorectal carcinoma. Hum Pathol 42: 1–10. 10.1016/j.humpath.2010.06.002 [DOI] [PubMed] [Google Scholar]
- Song X, Gao H, Lin Y, Yao Y, Zhu S, Wang J, Liu Y, Yao X, Meng G, Shen N, et al. 2014. Alterations in the microbiota drive interleukin-17C production from intestinal epithelial cells to promote tumorigenesis. Immunity 40: 140–152. 10.1016/j.immuni.2013.11.018 [DOI] [PubMed] [Google Scholar]
- Sonnenberg GF, Artis D. 2012. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity 37: 601–610. 10.1016/j.immuni.2012.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenberg GF, Fouser LA, Artis D. 2011. Border patrol: regulation of immunity, inflammation and tissue homeostasis at barrier surfaces by IL-22. Nat Immunol 12: 383–390. 10.1038/ni.2025 [DOI] [PubMed] [Google Scholar]
- Søreide K, Watson MM, Hagland HR. 2016. Deciphering the molecular code to colorectal liver metastasis biology through microsatellite alterations and allelic loss: the good, the bad, and the ugly. Gastroenterology 150: 811–814. 10.1053/j.gastro.2016.02.060 [DOI] [PubMed] [Google Scholar]
- Spano JP, Lagorce C, Atlan D, Milano G, Domont J, Benamouzig R, Attar A, Benichou J, Martin A, Morere JF, et al. 2005. Impact of EGFR expression on colorectal cancer patient prognosis and survival. Ann Oncol 16: 102–108. 10.1093/annonc/mdi006 [DOI] [PubMed] [Google Scholar]
- Stenstad H, Svensson M, Cucak H, Kotarsky K, Agace WW. 2007. Differential homing mechanisms regulate regionalized effector CD8αβ+ T cell accumulation within the small intestine. Proc Natl Acad Sci 104: 10122–10127. 10.1073/pnas.0700269104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern JL, Theodorescu D, Vogelstein B, Papadopoulos N, Cech TR. 2015. Mutation of the TERT promoter, switch to active chromatin, and monoallelic TERT expression in multiple cancers. Genes Dev 29: 2219–2224. 10.1101/gad.269498.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, et al. 2007. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 318: 287–290. 10.1126/science.1142946 [DOI] [PubMed] [Google Scholar]
- Su LK, Kinzler KW, Vogelstein B, Preisinger AC, Moser AR, Luongo C, Gould KA, Dove WF. 1992. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 256: 668–670. 10.1126/science.1350108 [DOI] [PubMed] [Google Scholar]
- Suh Y, Afaq F, Johnson JJ, Mukhtar H. 2008. A plant flavonoid fisetin induces apoptosis in colon cancer cells by inhibition of COX2 and Wnt/EGFR/NF-κB-signaling pathways. Carcinogenesis 30: 300–307. 10.1093/carcin/bgn269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui X, Zhang R, Liu S, Duan T, Zhai L, Zhang M, Han X, Xiang Y, Huang X, Lin H, et al. 2018. RSL3 drives ferroptosis through GPX4 inactivation and ROS production in colorectal cancer. Front Pharmacol 9: 1371. 10.3389/fphar.2018.01371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syn NL, Teng MWL, Mok TSK, Soo RA. 2017. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol 18: e731–e741. 10.1016/S1470-2045(17)30607-1 [DOI] [PubMed] [Google Scholar]
- Tabernero J, Yoshino T, Cohn AL, Obermannova R, Bodoky G, Garcia-Carbonero R, Ciuleanu TE, Portnoy DC, Van Cutsem E, Grothey A, et al. 2015. Ramucirumab versus placebo in combination with second-line FOLFIRI in patients with metastatic colorectal carcinoma that progressed during or after first-line therapy with bevacizumab, oxaliplatin, and a fluoropyrimidine (RAISE): a randomised, double-blind, multicentre, phase 3 study. Lancet Oncol 16: 499–508. 10.1016/S1470-2045(15)70127-0 [DOI] [PubMed] [Google Scholar]
- Tacconi C, Correale C, Gandelli A, Spinelli A, Dejana E, D'Alessio S, Danese S. 2015. Vascular endothelial growth factor C disrupts the endothelial lymphatic barrier to promote colorectal cancer invasion. Gastroenterology 148: 1438–1451.e8. 10.1053/j.gastro.2015.03.005 [DOI] [PubMed] [Google Scholar]
- Takada K, Zhu D, Bird GH, Sukhdeo K, Zhao JJ, Mani M, Lemieux M, Carrasco DE, Ryan J, Horst D, et al. 2012. Targeted disruption of the BCL9/β-catenin complex inhibits oncogenic Wnt signaling. Sci Transl Med 4: 148ra117. 10.1126/scitranslmed.3003808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam BY, Chiu K, Chung H, Bossard C, Nguyen JD, Creger E, Eastman BW, Mak CC, Ibanez M, Ghias A, et al. 2020. The CLK inhibitor SM08502 induces anti-tumor activity and reduces Wnt pathway gene expression in gastrointestinal cancer models. Cancer Lett 473: 186–197. 10.1016/j.canlet.2019.09.009 [DOI] [PubMed] [Google Scholar]
- Tang R, Cheng AJ, Wang JY, Wang TC. 1998. Close correlation between telomerase expression and adenomatous polyp progression in multistep colorectal carcinogenesis. Cancer Res 58: 4052–4054. [PubMed] [Google Scholar]
- Tao Y, Kang B, Petkovich DA, Bhandari YR, In J, Stein-O'Brien G, Kong X, Xie W, Zachos N, Maegawa S, et al. 2019. Aging-like spontaneous epigenetic silencing facilitates Wnt activation, stemness, and Braf(V600E)-induced tumorigenesis. Cancer Cell 35: 315–328.e6. 10.1016/j.ccell.2019.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumoto N, Hiyama E, Murakami Y, Imamura Y, Shay JW, Matsuura Y, Yokoyama T. 2000. High telomerase activity is an independent prognostic indicator of poor outcome in colorectal cancer. Clin Cancer Res 6: 2696–2701. [PubMed] [Google Scholar]
- Tauriello DVF, Palomo-Ponce S, Stork D, Berenguer-Llergo A, Badia-Ramentol J, Iglesias M, Sevillano M, Ibiza S, Canellas A, Hernando-Momblona X, et al. 2018. TGFβ drives immune evasion in genetically reconstituted colon cancer metastasis. Nature 554: 538–543. 10.1038/nature25492 [DOI] [PubMed] [Google Scholar]
- Teng S, Li YE, Yang M, Qi R, Huang Y, Wang Q, Zhang Y, Chen S, Li S, Lin K, et al. 2020. Tissue-specific transcription reprogramming promotes liver metastasis of colorectal cancer. Cell Res 30: 34–49. 10.1038/s41422-019-0259-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terme M, Pernot S, Marcheteau E, Sandoval F, Benhamouda N, Colussi O, Dubreuil O, Carpentier AF, Tartour E, Taieb J. 2013. VEGFA-VEGFR pathway blockade inhibits tumor-induced regulatory T-cell proliferation in colorectal cancer. Cancer Res 73: 539–549. 10.1158/0008-5472.CAN-12-2325 [DOI] [PubMed] [Google Scholar]
- Terrin L, Rampazzo E, Pucciarelli S, Agostini M, Bertorelle R, Esposito G, DelBianco P, Nitti D, De Rossi A. 2008. Relationship between tumor and plasma levels of hTERT mRNA in patients with colorectal cancer: implications for monitoring of neoplastic disease. Clin Cancer Res 14: 7444–7451. 10.1158/1078-0432.CCR-08-0478 [DOI] [PubMed] [Google Scholar]
- Terzić J, Grivennikov S, Karin E, Karin M. 2010. Inflammation and colon cancer. Gastroenterology 138: 2101–2114.e5. 10.1053/j.gastro.2010.01.058 [DOI] [PubMed] [Google Scholar]
- Thaker AI, Rao MS, Bishnupuri KS, Kerr TA, Foster L, Marinshaw JM, Newberry RD, Stenson WF, Ciorba MA. 2013. IDO1 metabolites activate β-catenin signaling to promote cancer cell proliferation and colon tumorigenesis in mice. Gastroenterology 145: 416–425.e4. 10.1053/j.gastro.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas DA, Massagué J. 2005. TGF-β directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell 8: 369–380. 10.1016/j.ccr.2005.10.012 [DOI] [PubMed] [Google Scholar]
- Tilstra JS, Robinson AR, Wang J, Gregg SQ, Clauson CL, Reay DP, Nasto LA, St Croix CM, Usas A, Vo N, et al. 2012. NF-κB inhibition delays DNA damage-induced senescence and aging in mice. J Clin Invest 122: 2601–2612. 10.1172/JCI45785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomczak K, Czerwinska P, Wiznerowicz M. 2015. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol (Pozn) 19: A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al. 2016. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 375: 2255–2262. 10.1056/NEJMoa1609279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tropini C, Earle KA, Huang KC, Sonnenburg JL. 2017. The gut microbiome: connecting spatial organization to function. Cell Host Microbe 21: 433–442. 10.1016/j.chom.2017.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WS, You JF, Hung HY, Hsieh PS, Hsieh B, Lenz HJ, Idos G, Friedland S, Yi-Jiun Pan J, Shao HJ, et al. 2019. Novel circulating tumor cell assay for detection of colorectal adenomas and cancer. Clin Transl Gastroenterol 10: e00088. 10.14309/ctg.0000000000000088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng-Rogenski SS, Hamaya Y, Choi DY, Carethers JM. 2015. Interleukin 6 alters localization of hMSH3, leading to DNA mismatch repair defects in colorectal cancer cells. Gastroenterology 148: 579–589. 10.1053/j.gastro.2014.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K. 2017. HNE as an inducer of COX-2. Free Radic Biol Med 111: 169–172. 10.1016/j.freeradbiomed.2017.02.004 [DOI] [PubMed] [Google Scholar]
- Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. 2007. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol 39: 44–84. 10.1016/j.biocel.2006.07.001 [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Prenen H, D'Haens G, Bennouna J, Carrato A, Ducreux M, Bouché O, Sobrero A, Latini L, Staines H, et al. 2015. A phase I/II, open-label, randomised study of nintedanib plus mFOLFOX6 versus bevacizumab plus mFOLFOX6 in first-line metastatic colorectal cancer patients. Ann Oncol 26: 2085–2091. 10.1093/annonc/mdv286 [DOI] [PubMed] [Google Scholar]
- Van den Eynde M, Mlecnik B, Bindea G, Fredriksen T, Church SE, Lafontaine L, Haicheur N, Marliot F, Angelova M, Vasaturo A, et al. 2018. The link between the multiverse of immune microenvironments in metastases and the survival of colorectal cancer patients. Cancer Cell 34: 1012–1026.e3. 10.1016/j.ccell.2018.11.003 [DOI] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. 2009. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324: 1029–1033. 10.1126/science.1160809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B, Smogorzewska A, de Lange T. 1998. TRF2 protects human telomeres from end-to-end fusions. Cell 92: 401–413. 10.1016/S0092-8674(00)80932-0 [DOI] [PubMed] [Google Scholar]
- van Veelen W, Le NH, Helvensteijn W, Blonden L, Theeuwes M, Bakker ER, Franken PF, van Gurp L, Meijlink F, van der Valk MA, et al. 2011. β-Catenin tyrosine 654 phosphorylation increases Wnt signalling and intestinal tumorigenesis. Gut 60: 1204–1212. 10.1136/gut.2010.233460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn CP, Zobell SD, Furtado LV, Baker CL, Samowitz WS. 2011. Frequency of KRAS, BRAF, and NRAS mutations in colorectal cancer. Genes Chromosomes Cancer 50: 307–312. 10.1002/gcc.20854 [DOI] [PubMed] [Google Scholar]
- Venook AP, Ou F-S, Lenz H-J, Kabbarah O, Qu X, Niedzwiecki D, Zemla T, Goldberg RM, Hochster HS, O'Neil BH, et al. 2017. Primary (1°) tumor location as an independent prognostic marker from molecular features for overall survival (OS) in patients (pts) with metastatic colorectal cancer (mCRC): analysis of CALGB/SWOG 80405 (Alliance). J Clin Oncol 35: 3503–3503. 10.1200/JCO.2017.35.15_suppl.3503 [DOI] [Google Scholar]
- Vermeulen L, Snippert HJ. 2014. Stem cell dynamics in homeostasis and cancer of the intestine. Nat Rev Cancer 14: 468–480. 10.1038/nrc3744 [DOI] [PubMed] [Google Scholar]
- Vermeulen L, De Sousa EMF, van der Heijden M, Cameron K, de Jong JH, Borovski T, Tuynman JB, Todaro M, Merz C, Rodermond H, et al. 2010. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol 12: 468–476. 10.1038/ncb2048 [DOI] [PubMed] [Google Scholar]
- Vilar E, Bartnik CM, Stenzel SL, Raskin L, Ahn J, Moreno V, Mukherjee B, Iniesta MD, Morgan MA, Rennert G, et al. 2011. MRE11 deficiency increases sensitivity to poly(ADP-ribose) polymerase inhibition in microsatellite unstable colorectal cancers. Cancer Res 71: 2632–2642. 10.1158/0008-5472.CAN-10-1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivier E, Ugolini S, Blaise D, Chabannon C, Brossay L. 2012. Targeting natural killer cells and natural killer T cells in cancer. Nat Rev Immunol 12: 239–252. 10.1038/nri3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernández-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R, et al. 2018. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359: 920–926. 10.1126/science.aao2774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voloshanenko O, Erdmann G, Dubash TD, Augustin I, Metzig M, Moffa G, Hundsrucker C, Kerr G, Sandmann T, Anchang B, et al. 2013. Wnt secretion is required to maintain high levels of Wnt activity in colon cancer cells. Nat Commun 4: 2610. 10.1038/ncomms3610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorneveld PW, Kodach LL, Jacobs RJ, Liv N, Zonnevylle AC, Hoogenboom JP, Biemond I, Verspaget HW, Hommes DW, de Rooij K, et al. 2014. Loss of SMAD4 alters BMP signaling to promote colorectal cancer cell metastasis via activation of Rho and ROCK. Gastroenterology 147: 196–208.e13. 10.1053/j.gastro.2014.03.052 [DOI] [PubMed] [Google Scholar]
- Wagner J, Kline CL, Zhou L, Campbell KS, MacFarlane AW, Olszanski AJ, Cai KQ, Hensley HH, Ross EA, Ralff MD, et al. 2018. Dose intensification of TRAIL-inducing ONC201 inhibits metastasis and promotes intratumoral NK cell recruitment. J Clin Invest 128: 2325–2338. 10.1172/JCI96711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Qian J, Hu Y, Kong X, Chen H, Shi Q, Jiang L, Wu C, Zou W, Chen Y, et al. 2014. ArhGAP30 promotes p53 acetylation and function in colorectal cancer. Nat Commun 5: 4735. 10.1038/ncomms5735 [DOI] [PubMed] [Google Scholar]
- Wang H, Franco F, Ho PC. 2017. Metabolic regulation of Tregs in cancer: opportunities for immunotherapy. Trends Cancer 3: 583–592. 10.1016/j.trecan.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Wang S, Peng Z, Wang S, Yang L, Chen Y, Kong X, Song S, Pei P, Tian C, Yan H, et al. 2018a. KRAB-type zinc-finger proteins PITA and PISA specifically regulate p53-dependent glycolysis and mitochondrial respiration. Cell Res 28: 572–592. 10.1038/s41422-018-0008-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang YQ, Wang HL, Xu J, Tan J, Fu LN, Wang JL, Zou TH, Sun DF, Gao QY, Chen YX, et al. 2018b. Sirtuin5 contributes to colorectal carcinogenesis by enhancing glutaminolysis in a deglutarylation-dependent manner. Nat Commun 9: 545. 10.1038/s41467-018-02951-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Qu Y, Xia P, Chen Y, Zhu X, Zhang J, Wang G, Tian Y, Ying J, Fan Z. 2020. Transdifferentiation of tumor infiltrating innate lymphoid cells during progression of colorectal cancer. Cell Res 30: 610–622. 10.1038/s41422-020-0312-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Wu TT, Catalano PJ, Ueki T, Satriano R, Haller DG, Benson AB III, Hamilton SR. 2001. Molecular predictors of survival after adjuvant chemotherapy for colon cancer. N Engl J Med 344: 1196–1206. 10.1056/NEJM200104193441603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West DW, Slattery ML, Robison LM, Schuman KL, Ford MH, Mahoney AW, Lyon JL, Sorensen AW. 1989. Dietary intake and colon cancer: sex- and anatomic site-specific associations. Am J Epidemiol 130: 883–894. 10.1093/oxfordjournals.aje.a115421 [DOI] [PubMed] [Google Scholar]
- West NR, McCuaig S, Franchini F, Powrie F. 2015. Emerging cytokine networks in colorectal cancer. Nat Rev Immunol 15: 615–629. 10.1038/nri3896 [DOI] [PubMed] [Google Scholar]
- Willis JA, Reyes-Uribe L, Chang K, Lipkin SM, Vilar E. 2020. Immune activation in mismatch repair-deficient carcinogenesis: more than just mutational rate. Clin Cancer Res 26: 11–17. 10.1158/1078-0432.CCR-18-0856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf AMD, Fontham ETH, Church TR, Flowers CR, Guerra CE, LaMonte SJ, Etzioni R, McKenna MT, Oeffinger KC, Shih YT, et al. 2018. Colorectal cancer screening for average-risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin 68: 250–281. 10.3322/caac.21457 [DOI] [PubMed] [Google Scholar]
- Wong SH, Zhao L, Zhang X, Nakatsu G, Han J, Xu W, Xiao X, Kwong TNY, Tsoi H, Wu WKK, et al. 2017. Gavage of fecal samples from patients with colorectal cancer promotes intestinal carcinogenesis in germ-free and conventional mice. Gastroenterology 153: 1621–1633.e6. 10.1053/j.gastro.2017.08.022 [DOI] [PubMed] [Google Scholar]
- Woolston A, Khan K, Spain G, Barber LJ, Griffiths B, Gonzalez-Exposito R, Hornsteiner L, Punta M, Patil Y, Newey A, et al. 2019. Genomic and transcriptomic determinants of therapy resistance and immune landscape evolution during anti-EGFR treatment in colorectal cancer. Cancer Cell 36: 35–50.e9. 10.1016/j.ccell.2019.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Lim KC, Huang J, Saidi RF, Sears CL. 1998. Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc Natl Acad Sci 95: 14979–14984. 10.1073/pnas.95.25.14979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, Dalla-Favera R. 1999. Direct activation of TERT transcription by c-MYC. Nat Genet 21: 220–224. 10.1038/6010 [DOI] [PubMed] [Google Scholar]
- Wu P, Wu D, Ni C, Ye J, Chen W, Hu G, Wang Z, Wang C, Zhang Z, Xia W, et al. 2014. γδT17 cells promote the accumulation and expansion of myeloid-derived suppressor cells in human colorectal cancer. Immunity 40: 785–800. 10.1016/j.immuni.2014.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Lyu J, Yang EJ, Liu Y, Zhang B, Shim JS. 2018. Targeting AURKA-CDC25C axis to induce synthetic lethality in ARID1A-deficient colorectal cancer cells. Nat Commun 9: 3212. 10.1038/s41467-018-05694-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie YH, Chen YX, Fang JY. 2020. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct Target Ther 5: 22. 10.1038/s41392-020-0116-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Xu J, Wu J, Hu Y, Han Y, Gu Y, Zhao K, Zhang Q, Liu X, Liu J, et al. 2018. Phosphorylation-mediated IFN-γR2 membrane translocation is required to activate macrophage innate response. Cell 175: 1336–1351.e17. 10.1016/j.cell.2018.09.011 [DOI] [PubMed] [Google Scholar]
- Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, Jayakumaran G, Middha S, Zehir A, Donoghue MTA, et al. 2018. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell 33: 125–136.e3. 10.1016/j.ccell.2017.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Venida A, Yano J, Biancur DE, Kakiuchi M, Gupta S, Sohn ASW, Mukhopadhyay S, Lin EY, Parker SJ, et al. 2020. Autophagy promotes immune evasion of pancreatic cancer by degrading MHC-I. Nature 581: 100–105. 10.1038/s41586-020-2229-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Xu X, Yang L, Zhu J, Wan J, Shen L, Xia F, Fu G, Deng Y, Pan M, et al. 2020. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell 26: 17–26.e6. 10.1016/j.stem.2019.10.010 [DOI] [PubMed] [Google Scholar]
- Youle RJ, Strasser A. 2008. The BCL-2 protein family: opposing activities that mediate cell death. Nature Reviews Molecular Cell Biology 9: 47–59. 10.1038/nrm2308 [DOI] [PubMed] [Google Scholar]
- Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, Qian Y, Kryczek I, Sun D, Nagarsheth N, et al. 2017. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170: 548–563.e16. 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Li N, Fu D, Ren J, Hui J, Peng J, Liu Y, Qiu T, Jiang M, Pan Q, et al. 2017. Histone methyltransferase SETD2 modulates alternative splicing to inhibit intestinal tumorigenesis. J Clin Invest 127: 3375–3391. 10.1172/JCI94292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Ju YS, Kim Y, Li J, Wang Y, Yoon CJ, Yang Y, Martincorena I, Creighton CJ, Weinstein JN, et al. 2020. Comprehensive molecular characterization of mitochondrial genomes in human cancers. Nat Genet 52: 342–352. 10.1038/s41588-019-0557-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuzhalin AE, Gordon-Weeks AN, Tognoli ML, Jones K, Markelc B, Konietzny R, Fischer R, Muth A, O'Neill E, Thompson PR, et al. 2018. Colorectal cancer liver metastatic growth depends on PAD4-driven citrullination of the extracellular matrix. Nat Commun 9: 4783. 10.1038/s41467-018-07306-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeki SS, Graham TA, Wright NA. 2011. Stem cells and their implications for colorectal cancer. Nat Rev Gastroenterol Hepatol 8: 90–100. 10.1038/nrgastro.2010.211 [DOI] [PubMed] [Google Scholar]
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D'Angelo C, Massi-Benedetti C, Fallarino F, et al. 2013. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity 39: 372–385. 10.1016/j.immuni.2013.08.003 [DOI] [PubMed] [Google Scholar]
- Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, Zhou K, Liu X, Ren X, Wang F, et al. 2018. Cancer-derived exosomal miR-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun 9: 5395. 10.1038/s41467-018-07810-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Xiong Y, Yarbrough WG. 1998. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell 92: 725–734. 10.1016/S0092-8674(00)81401-4 [DOI] [PubMed] [Google Scholar]
- Zhang J, Roberts TM, Shivdasani RA. 2011. Targeting PI3K signaling as a therapeutic approach for colorectal cancer. Gastroenterology 141: 50–61. 10.1053/j.gastro.2011.05.010 [DOI] [PubMed] [Google Scholar]
- Zhang J, Tsoi H, Li X, Wang H, Gao J, Wang K, Go MY, Ng SC, Chan FK, Sung JJ, et al. 2016a. Carbonic anhydrase IV inhibits colon cancer development by inhibiting the Wnt signalling pathway through targeting the WTAP-WT1-TBL1 axis. Gut 65: 1482–1493. 10.1136/gutjnl-2014-308614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Li CF, Zhang L, Wu CY, Han L, Jin G, Rezaeian AH, Han F, Liu C, Xu C, et al. 2016b. TRAF6 restricts p53 mitochondrial translocation, apoptosis, and tumor suppression. Mol Cell 64: 803–814. 10.1016/j.molcel.2016.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang W, Yuan X, Fu M, Qian H, Xu W. 2016c. Neutrophils in cancer development and progression: roles, mechanisms, and implications (review). Int J Oncol 49: 857–867. 10.3892/ijo.2016.3616 [DOI] [PubMed] [Google Scholar]
- Zhang L, Yu X, Zheng L, Zhang Y, Li Y, Fang Q, Gao R, Kang B, Zhang Q, Huang JY, et al. 2018. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature 564: 268–272. 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- Zhang D, Tang Z, Huang H, Zhou G, Cui C, Weng Y, Liu W, Kim S, Lee S, Perez-Neut M, et al. 2019. Metabolic regulation of gene expression by histone lactylation. Nature 574: 575–580. 10.1038/s41586-019-1678-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li Z, Skrzypczynska KM, Fang Q, Zhang W, O'Brien SA, He Y, Wang L, Zhang Q, Kim A, et al. 2020. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell 181: 442–459.e29. 10.1016/j.cell.2020.03.048 [DOI] [PubMed] [Google Scholar]
- Zheng J, Yang M, Shao J, Miao Y, Han J, Du J. 2013. Chemokine receptor CX3CR1 contributes to macrophage survival in tumor metastasis. Mol Cancer 12: 141. 10.1186/1476-4598-12-141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Perakis SO, Ulz P, Mohan S, Riedl JM, Talakic E, Lax S, Totsch M, Hoefler G, Bauernhofer T, et al. 2020. Cell-free DNA analysis reveals POLR1D-mediated resistance to bevacizumab in colorectal cancer. Genome Med 12: 20. 10.1186/s13073-020-0719-6 [DOI] [PMC free article] [PubMed] [Google Scholar]