In this study, Chen et al. analyzed expression of piRNAs and their targets during Drosophila spermatogenesis and compared it with the female counterpart. Using sequencing approaches and a novel bioinformatic pipeline, the authors define novel piRNA clusters and show adaptation of piRNAs that is dependent on sex-specific expression of transposons.
Keywords: Drosophila mauritiana, Drosophila melanogaster, Y chromosome, piRNA, satellite DNA, sexual dimorphism, spermatogenesis, transposable element
Abstract
Small noncoding piRNAs act as sequence-specific guides to repress complementary targets in Metazoa. Prior studies in Drosophila ovaries have demonstrated the function of the piRNA pathway in transposon silencing and therefore genome defense. However, the ability of the piRNA program to respond to different transposon landscapes and the role of piRNAs in regulating host gene expression remain poorly understood. Here, we comprehensively analyzed piRNA expression and defined the repertoire of their targets in Drosophila melanogaster testes. Comparison of piRNA programs between sexes revealed sexual dimorphism in piRNA programs that parallel sex-specific transposon expression. Using a novel bioinformatic pipeline, we identified new piRNA clusters and established complex satellites as dual-strand piRNA clusters. While sharing most piRNA clusters, the two sexes employ them differentially to combat the sex-specific transposon landscape. We found two piRNA clusters that produce piRNAs antisense to four host genes in testis, including CG12717/pirate, a SUMO protease gene. piRNAs encoded on the Y chromosome silence pirate, but not its paralog, to exert sex- and paralog-specific gene regulation. Interestingly, pirate is targeted by endogenous siRNAs in a sibling species, Drosophila mauritiana, suggesting distinct but related silencing strategies invented in recent evolution to regulate a conserved protein-coding gene.
PIWI-interacting (pi)RNA is a class of small noncoding RNAs named after their interaction with PIWI-clade Argonaute proteins. piRNAs guide PIWI proteins to complementary RNAs, thereby specifying the target of PIWI silencing. Unlike miRNAs and siRNAs that are ubiquitously expressed, the expression of piRNAs is restricted to gonads in many animals. As a result, perturbation of the piRNA program often compromises reproductive functions with no obvious defects in soma. Drosophila melanogaster is one of the most used model organisms to study piRNA biogenesis and function. In fact, piRNAs were first described in fly testes (Aravin et al. 2001; Vagin et al. 2006). However, most subsequent studies were performed using ovaries as a model system. Work on female gonads has shown that most piRNAs have homology to transposable elements (TEs), suggesting TEs as major targets of piRNAs (Brennecke et al. 2007). Studies on fly ovaries also identified large intergenic regions dubbed piRNA clusters that harbor nested TE fragments, which act as genomic source loci of piRNAs. A pericentromeric region on chr2R called 42AB was found to be the most active piRNA cluster in ovaries. It remains largely unexplored to what extent these findings from ovaries are applicable to the male counterpart. To date, we still know very little about how sexually dimorphic the Drosophila piRNA program is, besides that there is a single locus on the Y chromosome called Suppressor of Stellate [Su(Ste)] that produces piRNAs only in males.
Importantly, Drosophila as an animal model offers unique value for studying sexual dimorphism of the piRNA program in general. In zebrafish, piRNA pathway mutants are always phenotypically males (Houwing et al. 2007, 2008; Kamminga et al. 2010), rendering it nearly impossible to probe the impact of piRNA loss in females. In mice, an intact piRNA program is only required for male fertility, while murine females are insensitive to piRNA loss (Deng and Lin 2002; Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007). Contrary to fish and mouse, fly fertility is dependent on a functional piRNA pathway in both sexes (Lin and Spradling 1997; Aravin et al. 2001; Vagin et al. 2006; Brennecke et al. 2007). Therefore, Drosophila provides an unparalleled opportunity to study whether, and if so how, the piRNA program can be modified in each sex to safeguard reproductive functions.
In this study, we comprehensively analyzed the piRNA profile in Drosophila melanogaster testis and compared it with the female counterpart. Besides TEs, we found complex satellites as another class of selfish genetic elements targeted by the piRNA pathway in gonads of both sexes. Our analysis showed that the TE silencing piRNA program is sexually dimorphic, and it shows evidence of adaptation to the sex-specific TE landscape. To understand the genomic origins of differentially produced piRNAs, we sought to de novo define genome-wide piRNA clusters in testis. However, we noticed that the standard pipeline used for ovary piRNAs failed to detect known piRNA clusters in testis, so we developed a new bioinformatic algorithm to tackle this problem. Using the new algorithm, we were able to identify novel piRNA clusters and to quantify their activities more accurately in both sexes. Notably, piRNA source loci are employed differentially in males and females, and the sex bias of piRNA cluster expression appears to match that of their TE contents. We also found two loci producing piRNAs with the potential to repress host protein-coding genes, including a newly identified locus on Y that we named petrel, which produces piRNAs against CG12717/pirate. Expression of pirate, but not its close paralog verloren, is derepressed in multiple piRNA pathway mutants, indicating that piRNAs silence its expression and can distinguish paralogs with sequence similarities. Finally, we explored the evolutionary history of pirate and found it to be a young gene conserved in the melanogaster subgroup. Intriguingly, pirate is targeted by another class of small noncoding RNAs, endogenous siRNAs, in the sibling species Drosophila mauritiana, suggesting distinct small RNA-based silencing strategies invented in recent evolution to regulate a young, yet conserved, gene.
Results
Drosophila piRNA program is sexually dimorphic
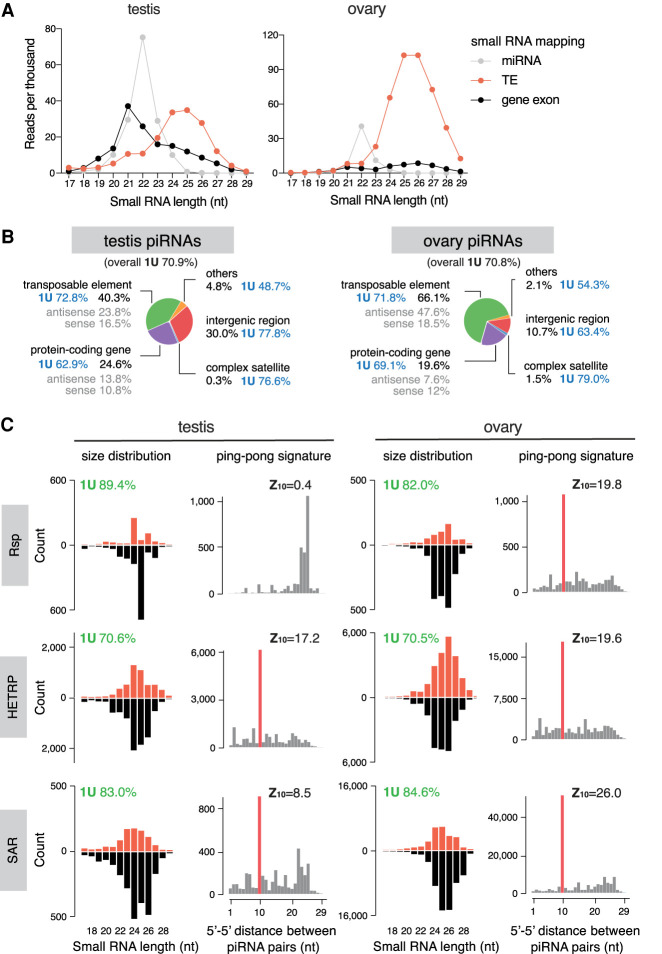
To characterize the piRNA profile in male gonads, we sequenced 18- to 30-nt small RNAs from testes and compared them with published ovary small RNA data sets (ElMaghraby et al. 2019). Mapping and annotation of small RNA reads using the pipeline shown in Supplemental Figure S1 revealed large differences in the expression of major classes of small RNAs between testes and ovaries. In agreement with previous findings (Czech et al. 2008), TE-mapping 23- to 29-nt piRNAs are the most abundant class of small RNAs in ovaries, while 21- to 23-nt microRNAs constitute a minor fraction and an even smaller one for 21-nt endogenous (endo-) siRNAs (Fig. 1A). In contrast, miRNAs constitute a larger fraction in testes, as do endo-siRNAs that map to protein-coding genes, consistent with a previous report (Wen et al. 2015). To define the piRNA population, we eliminated reads mapping to other types of noncoding RNA (rRNA, miRNA, snRNA, snoRNA, and tRNA) from 23- to 29-nt small RNAs. Remaining reads show a strong bias for U at the first nucleotide (“1U bias”: 70.9%), the feature of bona fide piRNAs (Fig. 1B). The piRNA-to-miRNA ratio is distinct between sexes: ∼10 in ovary and ∼2 in testis. In both sexes, piRNAs mapping to TEs take up the largest fraction of total piRNAs. However, whereas 66% of piRNAs mapped to TEs in ovaries, only 40% mapped to TEs in testes (Fig. 1B). Meanwhile, larger fractions of total piRNAs mapped to protein-coding genes (including introns) and intergenic regions in testes (24.6% and 30.0%, respectively) than ovaries (19.6% and 10.7%, respectively). These results suggest that distinct piRNA programs operate in male and female gonads.
Figure 1.
Analysis of small RNA profiles in testis and ovary. (A) Size distribution plots of microRNAs (gray), remaining small RNAs that map to TE consensus (red), and protein-coding gene exons (black), in the testis (left) and ovary (right). (B) Annotation of piRNA reads in the testis (left) and ovary (right). 1U nucleotide bias (percentage) for overall piRNA population and each category is shown next to labels. See also Supplemental Figure S1. (C) Characterization of piRNAs mapping to three known complex satellites in the two sexes. The left panels of each sex are size distribution of piRNAs mapping to consensus sequences of each complex satellite. The right panels are distributions of 5′-to-5′ distances of piRNA pairs, showing an enrichment for 10 nt (i.e., ping-pong signature), except for Rsp in testis (P < 0.05 for z > 1.96). 1U nucleotide bias (percentage) and ping-pong z-score are shown above the plots. See also Supplemental Figure S2.
Testis piRNAs also map to several known complex satellites: HETRP/TAS (a subtelomeric satellite repeat), Responder (Rsp), and SAR (related to 1.688 repeat family) (Fig. 1C; Supplemental Fig. S2A). Complex satellite-mapping small RNAs in testis exhibit 1U bias and size distribution that peaks around 24–26 nt, consistent with their piRNA identities. Both strands of complex satellites produce piRNAs, and their production depends on Rhino (Chen et al. 2020), a protein that marks dual-strand piRNA clusters and is required for their expression (Klattenhoff et al. 2009; Mohn et al. 2014; Zhang et al. 2014). Similarly, ovary small RNAs also map to complex satellites and show features of bona fide piRNAs, including 1U bias, size distribution that peaks around 24 to 26 nt, small RNA production from both strands, and dependency on Rhino. Moreover, piRNAs from complex satellites show a ping-pong signature, an enrichment for a 10-nt overlap between the 5′ ends of complementary piRNA pairs, except for Rsp in testis (Fig. 1C; Supplemental Fig. S2C). Finally, we examined the phasing pattern, the presence of piRNAs arranged tail to head one after another as a result of phased processing of piRNA precursors (Han et al. 2015; Mohn et al. 2015). We found such a phasing signature for two complex satellites in ovary, but not in testis (Supplemental Fig. S2B). These results show that complex satellites are sources of piRNAs in both sexes, pointing to a possible role of piRNAs in regulating satellite DNA and associated heterochromatin in the gonad.
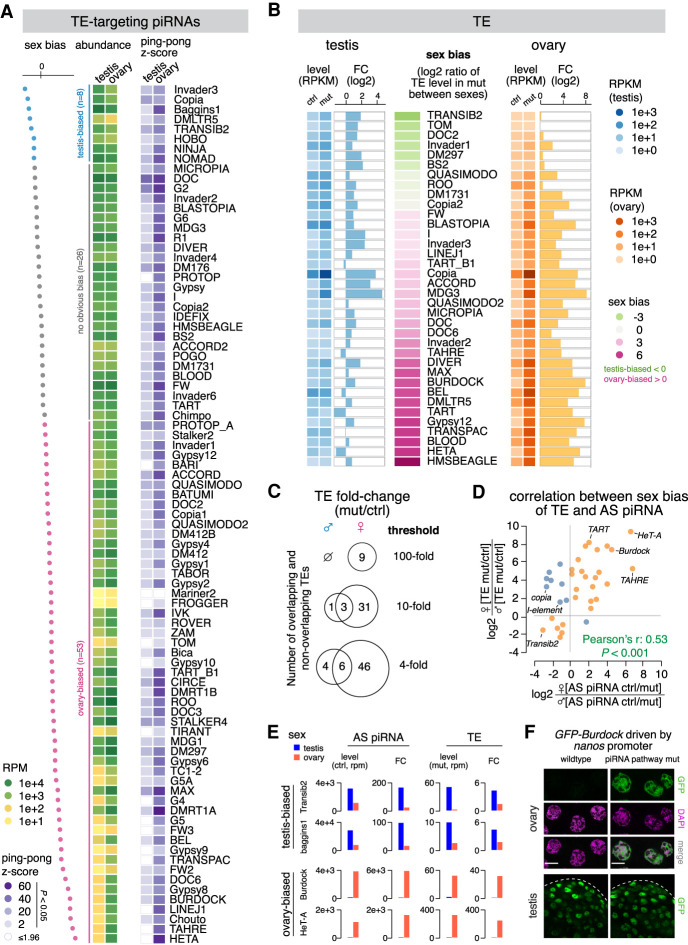
We next analyzed piRNAs targeting different TE families. Comparison of small RNA profiles in testis and ovary showed that piRNAs targeting different TEs are expressed at different levels in the two sexes (Fig. 2A). The top three TEs targeted by piRNA are all different in testis and ovary, and, among the top 10, only three are shared between the sexes (Supplemental Fig. S2D). The most differentially targeted TEs are two telomere-associated TEs, HeT-A and TAHRE, against which ovary makes 106 and 74 times more antisense piRNAs, respectively, than testis. In contrast, several elements such as baggins1, invader3, and copia are targeted by more piRNAs in testis. piRNAs targeting all but one (copia) TE families show a stronger ping-pong signature in ovary, as measured by the ping-pong z-score (Fig. 2A). In conclusion, different TE families are targeted by piRNAs differentially in the two sexes.
Figure 2.
Sexually dimorphic piRNA programs parallel sex-specific TE expression. (A) Heat maps showing the abundance of antisense piRNA (left) and ping-pong z-score (right) for each TE family in the two sexes. Statistically significant ping-pong z-scores (z > 1.96, equivalent to P < 0.05) are color-coded, while the remaining are marked as blank. TE families are sorted by sex bias of piRNA expression, defined as the log2 ratio of antisense piRNA abundance in testis over ovary. TEs with more than twofold differences in antisense piRNAs are colored as testis-biased (blue) and ovary-biased (pink), respectively, with the remaining having no obvious bias (gray). (B) Expression of 36 TE families that are regulated by rhi (see the Materials and Methods) in the testis (left) and ovary (right). TE families are sorted by sex bias of their expression in piRNA pathway mutant (rhi−/−), defined as the log2 ratio between sexes. Heat maps display TE levels in control and mutant, while bar graphs show the fold change of expression in mutant over control. (C) Venn diagrams of the number of TEs showing 100-fold, 10-fold, and fourfold derepression in rhi mutant over control of the two sexes. (D) Scatterplot displaying the correlation between sex biases of TE and TE antisense piRNA. For each TE family, the loss of antisense piRNAs in rhi mutants was calculated in each sex (ctrl over mut). The sex bias of piRNAs was defined as the log2 ratio of piRNA loss in female over male. Similarly, TE derepression in rhi mutants was calculated in each sex (mut over ctrl), and the sex bias was defined as the log2 ratio of TE derepression in female over male. TE families that show a correlation between the sex bias of antisense piRNA and that of TE derepression are colored as orange, with the rest as blue. (E) Histograms showing profiles of two sex-biased TEs for each sex. Antisense piRNA levels refer to those in control gonads, TE levels refer to those in piRNA pathway mutants (rhi−/−), and the fold change is calculated as mutant over control for TEs and the reverse for antisense piRNAs. (F) Confocal images of stage 7–8 nurse cells in ovary (top) and the apical tip of testis (bottom) that express a Burdock fused GFP reporter in wild-type and piRNA pathway mutant (rhi−/−) background, respectively. The reporter is expressed by nanos promoter that drives germline expression in both sexes, thus enabling the examination of piRNA silencing of Burdock sequences independent of natural expression patterns of Burdock transposon. Scale bars, 20 µm.
Distinct piRNA programs in two sexes parallel sex-specific TE expression
To explore whether sex differences in TE targeting piRNA programs are accompanied by differential expression of TEs themselves, we set out to compare expression levels of different TE families in the two sexes. Since the piRNA pathway efficiently represses TEs, their expression in wild-type animals does not reflect their full expression potential that can be achieved when piRNA silencing is removed. Hence, we analyzed TE expression in testes and ovaries of rhi mutants that lose piRNA production from dual-strand clusters in both sexes (Chen et al. 2020) and controls.
Profiling TE expression in the two sexes by polyA-selected (polyA+) RNA-seq demonstrated clear sexual dimorphism. Overall, TE expression in the piRNA pathway mutant testes and ovaries is weakly correlated (Spearman's ρ: 0.18) (Supplemental Fig. S3A). Among the 10 most expressed TE families in the two sexes, only four overlap, although the same element, copia, has the highest expression in both ovary and testis (Supplemental Fig. S3B). There are more TE families expressed above each of the three expression cutoffs (1000, 100, and 10 RPKM) in ovaries than testes (Supplemental Fig. S3A). The most ovary-biased TEs include Blood, gypsy12, and Burdock, and two telomere-associated TEs, HeT-A and TART (Fig. 2B). Only a few TE families are expressed higher in testis than ovary (Fig. 2B; Supplemental Fig. S3A). In this group, Transib2 and doc2 show the strongest bias for expression in testis (14-fold and 6.5-fold higher in testis than ovary, respectively). Overall, the majority of TE families demonstrate strong differences in their expression between sexes.
To quantify the effect of the piRNA pathway in suppressing TEs in the two sexes, we calculated the levels of TE derepression upon disruption of the piRNA pathway. Few TE families remained unaffected by rhi mutation, often accompanied by unperturbed antisense piRNA production (e.g., gypsy, gypsy10, and tabor). There are nine TE families up-regulated >100-fold in ovary. In contrast, no TE is up-regulated that strongly in testis (Fig. 2C). Overall, the vast majority of TEs show stronger derepression in ovaries, with gypsy12 (389-fold), Burdock (317-fold), HeT-A (239-fold), and TART (80-fold) being the most prominent examples, as all of them exhibited no or mild derepression (less than fourfold) in testes (Fig. 2B). We found only six TEs that show stronger (at least fourfold) derepression in testis than ovary (Transib2, BS2, baggins1, Dm297, invader3, and invader6). Altogether, our results show that piRNAs regulate the expression of different TE families to distinct extents in the two sexes, with many TEs silenced more in ovary and only a few silenced more in testis.
To explore the link between TE expression and piRNA programs in the two sexes, we identified a set of 36 TE families repressed by the piRNA pathway in at least one sex (see the Materials and Methods). For these TE families, there is a positive correlation between sex bias of piRNA production and sex bias of TE derepression (Pearson's ρ: 0.53, P <0.001) (Fig. 2D). For example, disruption of the piRNA pathway by rhi mutations dramatically increases expression of three telomere-associated TEs (HeT-A, TAHRE, and TART) in ovaries, where there are abundant piRNAs targeting these elements. On the contrary, much fewer piRNAs target these telomeric TEs in testes, and expression of these TEs remained very low in rhi mutant males (Fig. 2A,B,E). This result indicates that telomeric TEs have a strong, intrinsic bias in their expression toward the female germline and that the piRNA pathway appears to have adapted to this bias, generating respective antisense piRNAs in female, but not male, gonads. In contrast to ovary-biased TEs like telomeric elements, testis-biased TEs such as Transib2 and baggins1 are targeted by more antisense piRNAs in testis than ovary (Fig. 2A,B,E). Some TEs, such as copia, mdg3, and I-element are strongly repressed by piRNAs in both sexes. For such elements, the sex bias in piRNA production does not always match that of TE repression (Fig. 2D). Taken together, these findings suggest that, for most TEs, piRNA programs in males and females have adapted to differential TE activities between the sexes.
To further explore whether differential expression of piRNAs between the sexes has functional consequences, we studied Burdock, an LTR retrotransposon targeted by 53 times more piRNAs in ovary (3756 RPM) than testis (70 RPM) (Fig. 2A). We used a reporter composed of a fragment of Burdock expressed under the control of the heterologous nanos promoter that drives expression in the germline of both sexes (Handler et al. 2013). While the reporter was efficiently silenced in ovaries of wild-type flies, it was strongly derepressed in the piRNA pathway mutants (rhi−/−) (Fig. 2F), indicating that the piRNA program efficiently silences Burdock in the female germline. In contrast, we observed strong reporter expression in testes of wild-type males, and the disruption of the piRNA pathway in rhi mutants did not lead to an observable increase in its expression (Fig. 2F). This finding shows that Burdock is not silenced in testes, likely as a result of very few Burdock targeting piRNAs in males (Fig. 2A). Notably, expression of endogenous Burdock is high in ovary (when piRNA production is disrupted) but low in both wild-type and mutant testis (Fig. 2B,E). Thus, similar to telomeric TEs, the ability of the piRNA pathway to repress Burdock in the female but not the male germline correlates with an intrinsic bias for its expression in females. We conclude that differential expression of TE targeting piRNAs in male and female gonads can have functional consequences in their abilities to silence TEs.
Definition of piRNA clusters in testis with a new algorithm
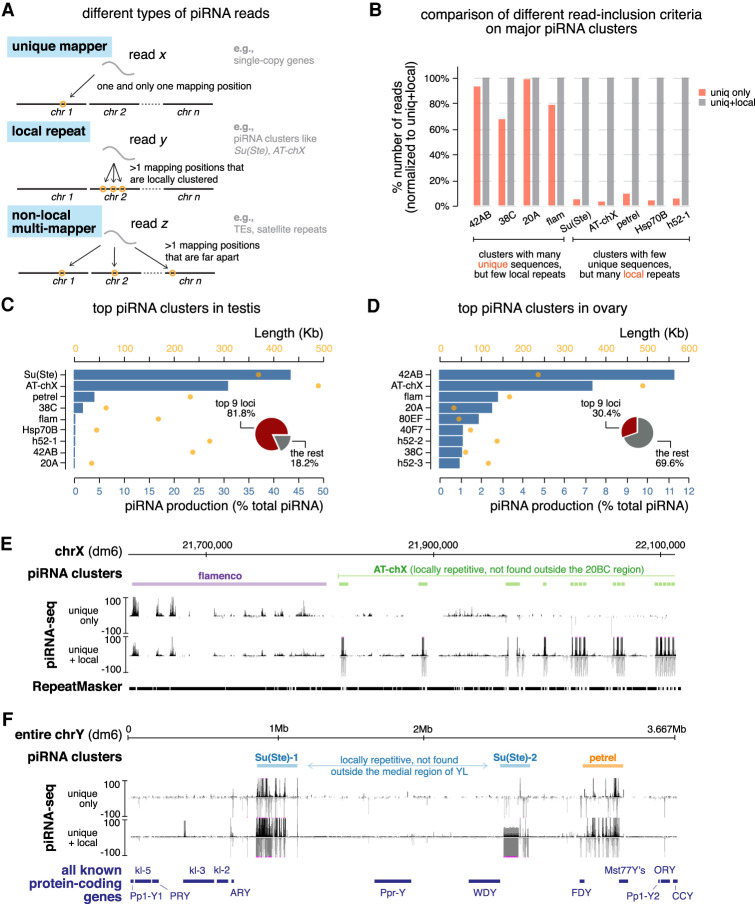
To get a deeper understanding of the piRNA program in male gonads, we sought to define the genomic origin of piRNAs and compare it between the two sexes. Since genome-wide identification of piRNA clusters has only been done in ovary, we decided to systematically search for genomic loci that generate piRNA in testis. We noticed that two major clusters in testis identified to date, Su(Ste) and AT-chX, both contain internal tandem repeats, that is, they are made of many copies of almost identical sequences (Aravin et al. 2001; Kotov et al. 2019). As a result, most piRNAs produced by these two loci mapped to the genome at multiple positions. However, the algorithm employed in previous studies to systematically define piRNA clusters in ovary only uses piRNAs that map to the genome at single unique positions (Brennecke et al. 2007; Mohn et al. 2014; George et al. 2015), raising the question of whether it is an appropriate approach to detect clusters like Su(Ste) composed primarily of internal tandem repeats. In fact, both Su(Ste) and AT-chX clusters were initially identified by different approaches (Aravin et al. 2001; Nishida et al. 2007).
Even though piRNAs produced from Su(Ste) and AT-chX cannot be mapped to single unique genomic loci, most of them mapped to several local repeats inside the respective clusters but nowhere else in the genome (Fig. 3A). Taking advantage of this property, we developed a new algorithm that takes into account local repeats to define piRNA clusters (Supplemental Fig. S4A,B). Briefly, in addition to uniquely mapped piRNAs, the algorithm searches for piRNA sequences that map to multiple positions within a single genomic region but nowhere else in the genome. This approach ensures that the identified region as a whole generates piRNAs, though the exact origin within the region remains unknown. Unlike the previous approach that uses exclusively uniquely mapped piRNAs, this algorithm successfully identified Su(Ste) and AT-chX, two major piRNA clusters in testis that contain local repeats (Fig. 3E,F).
Figure 3.
Definition of piRNA clusters in testis and ovary using a new algorithm. (A) Three types of piRNA reads, defined based on their mapping positions. Uniquely mapped reads can be mapped to only one position in the genome and their origin is unambiguous. Reads derived from local repeats can be mapped to several positions in the genome; however, all of these mapping positions are locally clustered in a single genomic region. On the other hand, nonlocal multimappers can be mapped to multiple positions that are not restricted to one genomic region (typically mapped to more than one chromosome). Previously, only uniquely mapped reads were used to define piRNA clusters and quantify their expression, as the genomic origin of multimappers is ambiguous. Inclusion of multimappers derived from local repeats, as shown in this study, allows identification of new piRNA clusters as well as a more accurate quantification of piRNA production from known clusters. At the same time, it preserves the certainty that reads are generated from genomic loci in question. See Supplemental Figure S4 for detailed pipeline. (B) Histogram comparing numbers of mapped reads for major piRNA clusters using different read inclusion criteria as defined in A. For each cluster, the number of mapped reads generated by different methods is normalized to the method that includes both unique and local repeat reads (the right column). See also Supplemental Figure S4 and the Materials and Methods. (C) Expression of the top nine most active piRNA clusters in testis. Blue bars depict the contribution of each cluster to total piRNAs (percentage) and orange dots show cluster lengths according to the dm6 genome assembly. Insert is a pie chart of the contribution of the top nine loci to total piRNAs in testis. (D) Same as in C but for ovary. (E) UCSC genome browser view of a pericentromeric region (chrX) encompassing the entire flamenco locus (purple) and the distal part of AT-chX piRNA cluster (green). Below the genomic coordinates (dm6) are piRNA coverage tracks using different read inclusion criteria. Note that, whereas flamenco produces piRNAs that can be mostly mapped to unique genomic positions, AT-chX generates piRNAs that map to local repeats in this cluster, but nowhere else in the genome. (F) UCSC genome browser view of the entire Y chromosome that harbors two Su(Ste) loci (blue) and the novel petrel piRNA cluster (orange). piRNA coverage tracks using different read inclusion criteria are shown below genomic coordinates (dm6). At the bottom, all known Y-linked protein-coding genes are drawn for reference (not to exact scale). Note that piRNA profiles of Su(Ste) and petrel clusters collapse if piRNAs derived from local repeats are excluded.
We applied this new algorithm to systematically identify piRNA clusters active in testes. We recovered piRNA clusters known to be active in testes as well as piRNA clusters previously defined in ovaries (e.g., 42AB, 38C, 20A, and flam) (Fig. 3C; Supplemental Table S1). Furthermore, our search identified several novel piRNA loci. One of the novel piRNA clusters is located on the Y chromosome flanked by FDY and Mst77Y genes (Fig. 3C,F) around heterochromatin band h17 (Gatti and Pimpinelli 1983). We named this locus petrel for “proximal to fertility regions on YL.” Another novel locus is h52-1, flanked by eIF4B and CG17514 genes on chr3L. h52-1 harbors tandem local repeats composed of nested TE fragments that cannot be found elsewhere in the genome. Similar to piRNA clusters identified in ovaries, only a few clusters active in testes produce piRNAs from one genomic strand (e.g., flam and 20A, so-called “unistrand clusters”), and the majority are dual-strand clusters that generate piRNAs from both genomic strands (Fig. 3E). In sum, our algorithm successfully found previously known piRNAs clusters and identified novel ones in Drosophila testes.
To compare the new algorithm with the approach that considers only uniquely mapped piRNAs, we applied both techniques to analyze the same testis piRNA data set. This comparison showed that major piRNA clusters in testis can be divided into two groups (Fig. 3B). The first group (42AB, 38C, 20A, and flam) contains piRNA clusters that harbor many unique sequences, so including local repeats does not substantially change their identification and quantification. On the other hand, the second group of genomic loci [Su(Ste), AT-chX, petrel, Hsp70B, and h52-1] is composed of piRNA clusters that contain few unique sequences but many local repeats, and, accordingly, our new algorithm identified >10-fold more piRNAs produced from these loci (Fig. 3B). Thus, this algorithm is not only useful for finding new piRNA source loci but also provides a more accurate quantification of piRNA production from previously known clusters.
Sex difference in piRNA cluster expression
To compare the expression of piRNA clusters between the sexes, we first applied our algorithm to published ovary piRNA data sets (Fig. 3D; Supplemental Table S1; ElMaghraby et al. 2019). Thus, piRNA clusters were defined and their activities were quantified in both sexes using the same algorithm, allowing for fair comparison. Surprisingly, our analysis revealed that AT-chX, originally described as a piRNA cluster in testes, is also highly active in ovaries. The AT-chX locus consists of local repeats (Kotov et al. 2019), so piRNAs produced from this locus were excluded in previous studies that analyzed only uniquely mapped reads. In fact, AT-chX is the second most active piRNA cluster in ovary, producing ∼7% of total piRNAs.
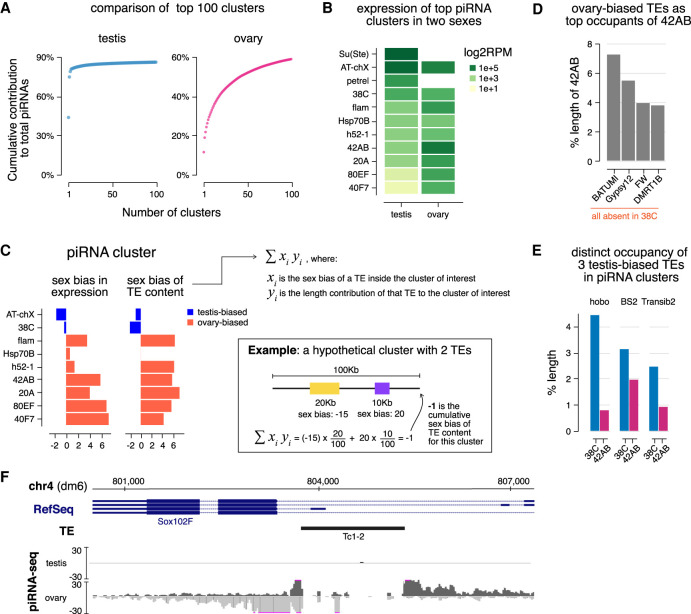
Comparison between piRNA clusters in males and females revealed a clear sex difference: A small number of loci produce the majority of piRNAs in testis, which is not the case for ovary (Fig. 4A). The two most active piRNA clusters in testes, Su(Ste) on the Y chromosome and AT-chX on the X chromosome, produce ∼43% and ∼31% of total piRNAs in testes, respectively (Fig. 3C). They are followed by the novel piRNA cluster on the Y chromosome, petrel, that produces ∼4% piRNAs. Along with another six loci, the top nine piRNA clusters in testis account for 81.8% of total piRNAs. In comparison, only 30.4% of total piRNAs are made from the top nine clusters in ovary, with the most active locus 42AB producing ∼11% of total piRNAs (Fig. 3D). Whereas a few loci dominate the global piRNA population in testis, the ovary piRNA profile is shaped by many loci producing piRNAs in comparable amounts.
Figure 4.
piRNA clusters are differentially employed to tame sex-specific TE expression. (A) Plot showing the cumulative contribution of top piRNA clusters to the total piRNA populations in the testis (left) and ovary (right), up to 100 clusters. (B) Heat maps showing piRNA production from major piRNA clusters. Note that Su(Ste) and petrel clusters are Y-linked so there are no piRNAs from these loci in females that lack the Y chromosome. (C) Bar graphs displaying the sex bias of piRNA cluster expression (left) and cumulative sex bias of the TE context for each cluster (right). Sex bias of piRNA cluster expression is defined as the log2 ratio of piRNA cluster expression in ovary over testis shown in B, so ovary-biased ones are positive in value. Cumulative sex bias of cluster TE content is calculated by summing the sex bias of TEs (as described for Fig. 2B) weighted by their length contributions to the cluster (equation shown at the right). An example is shown on the bottom right for a hypothetical cluster composed of two TEs with lengths and sex biases labeled accordingly for illustration. Only TEs showing strong sex biases were used in calculation. See also the Materials and Methods. (D) TE composition of ovary-biased 42AB cluster. Shown are fractions of 42AB cluster occupied by sequences from the top four TE families. These four TEs are completely absent in 38C, a testis-biased piRNA cluster. Expression of these four TEs is all ovary-biased (Supplemental Fig. S3A). (E) Contributions of three testis-biased TEs (Supplemental Fig. S3A) to the ovary-biased 42AB cluster and testis-biased 38C cluster. These TEs were selected as the most enriched by length in 38C compared with 42AB. (F) The Sox102F gene generates piRNAs in ovary, but not in testis. This locus harbors a single autonomous TE, Tc1-2, that has ovary-biased expression (Supplemental Fig. S3A). piRNA coverage tracks show both uniquely mapped and local repeat-derived reads.
Next, we compared expression levels of different piRNA clusters in male and female gonads. Females lack the Y chromosome, so they do not have piRNAs produced by Y-linked Su(Ste) and petrel clusters. For major clusters present in both male and female genomes, we observed pronounced sex differences (Spearman's ρ: 0.07) (Fig. 4B). For instance, 38C produces more piRNAs than 42AB, 80EF, and 40F7 in testes, but the opposite trend is found in ovaries. Some loci such as Sox102F on chr4 (Mohn et al. 2014; Zhang et al. 2014) appear to be active only in ovaries but not in testes (Fig. 4F). These differentially expressed piRNA clusters located on autosomes, of which both males and females have two copies, exemplify the sex-specific usage of piRNA loci. Moreover, we examined expression levels of major piRNA clusters on chrX (AT-chX, flam, and 20A), of which females have two copies (XX) and males have only one (XY). We found that a larger fraction of piRNAs originate from AT-chX in testes than ovaries, but the reverse was found for flam and 20A, suggesting that copy numbers of piRNA clusters do not correlate well with their expression. Altogether, these findings illustrate a sexually dimorphic employment of piRNA clusters, where different loci are engaged differentially in a sex-specific manner.
Different piRNA clusters have distinct TE contents, so their differential expression might sculpt sex-specific piRNA programs with distinct TE silencing capacities in males and females. To explore a link between the expression of a piRNA cluster and its TE content, we computed the cumulative sex bias of the TE content of each major piRNA cluster (Fig. 4C). This was done by summing sex biases of individual TEs in the piRNA cluster weighted by their length contributions to the cluster (see example in Fig. 4C). The sex bias of cluster TE content matches the sex bias in piRNA cluster expression, suggesting a link between the expression of piRNA clusters and the TEs they control. To substantiate this finding, we analyzed sequence compositions of three differentially expressed piRNA clusters: 42AB (ovary-biased), 38C (testis-biased), and Sox102F (ovary-specific). The top four TEs most enriched by length in ovary-biased 42AB (batumi, gypsy12, FW, and DMRT1b) are all ovary-biased in their expression (Fig. 4D; Supplemental Fig. S3A). Importantly, these four TEs are completely absent in the testis-biased 38C cluster. In contrast, three testis-biased TE families, hobo, BS2, and Transib2, are more enriched in 38C than in 42AB (Fig. 4E; Supplemental Fig. S3A). Moreover, the ovary-specific Sox102F cluster harbors a single autonomous transposon, Tc1-2, which has higher activity in ovary (Fig. 4F; Supplemental Fig. S3A). These examples show that differential expression of piRNA clusters in the two sexes often matches the differential activities of the TEs they control, supporting the notion that piRNA clusters are employed in a sex-specific fashion to cope with distinct TE landscape in male and female gonads.
piRNA clusters composed of local repeats produce piRNAs that target host genes
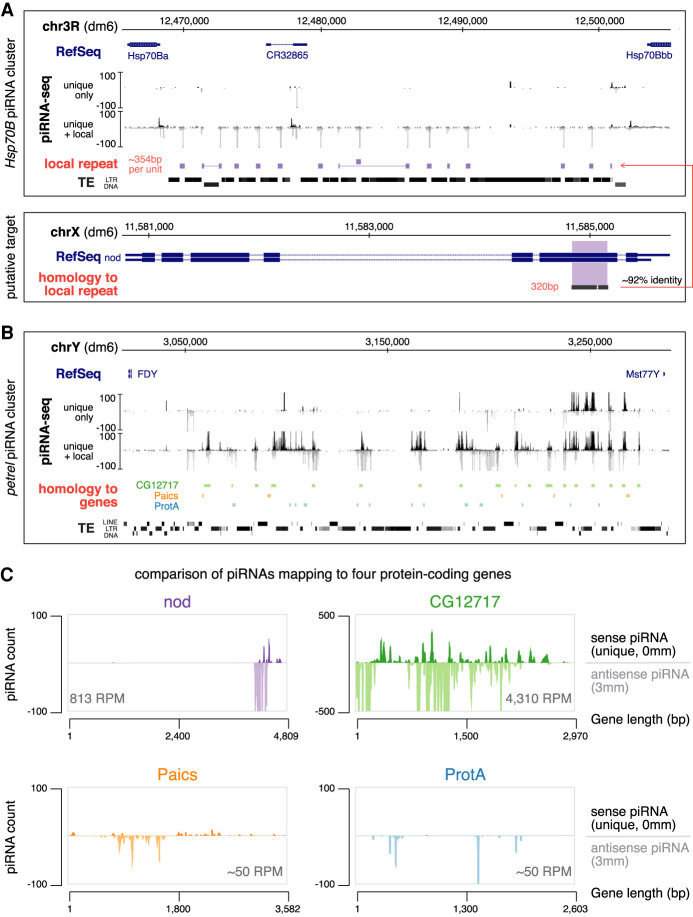
Our analysis indicated that 13.8% of testis piRNAs might potentially be involved in targeting host genes as they can be mapped to protein-coding genes in antisense orientation with a small number (zero to three) of mismatches between piRNA and gene sequences (Fig. 1B). To understand the genomic origin of these piRNAs, we further analyzed sequence compositions of piRNA clusters. We found that two clusters, Hsp70B and petrel, both of which contain local repeats, generate piRNAs that have the potential to target host genes.
The Hsp70B cluster spans ∼35 kb between two paralogous Hsp70B genes on chr3R, and it is active in both ovary and testis (Fig. 5A). The body of the Hsp70B cluster contains several TEs. Even though there are piRNAs mapping to these TEs, they can be mapped elsewhere in the genome as well, rendering it impossible to be certain that they originate from the Hsp70B locus. In fact, this cluster was previously identified through the presence of uniquely mapped piRNAs from flanking nonrepetitive genes (Mohn et al. 2014). However, our algorithm that takes into account local repeats revealed piRNAs generated from an ∼354-bp local repeat at the Hsp70B locus, which occupies nearly all intertransposon space within this cluster. Importantly, these piRNAs mapped exclusively to this local repeat at the Hsp70B cluster but nowhere else in the genome. Every copy of this local repeat is flanked by sequences of the copia2 retrotransposon and corresponds to a tandem repeat at Hsp70B described ∼40 yr ago (Lis et al. 1978; Hackett and LIs 1981). The entire repeat-rich region between two Hsp70B genes is present in D. melanogaster but not in either of its sibling species D. simulans or D. mauritiana (Livak et al. 1978; Leigh Brown and Ish-Horowicz 1981), suggesting a recent evolutionary origin. Intriguingly, these repeats have an ∼92% sequence identity to an exon of the nod gene, which encodes a kinesin-like protein necessary for chromosome segregation during meiosis (Carpenter 1973; Zhang et al. 1990; Hawley and Theurkauf 1993). The Hsp70B cluster generates piRNAs that are antisense to nod with a 91.3% averaged nucleotide identity to it. This level of sequence similarity is close to that between Suppressor of Stellate piRNAs and their Stellate targets, the first known case of piRNA repression (Aravin et al. 2001; Vagin et al. 2006), suggesting that piRNAs produced from the Hsp70B locus might be able to repress the nod gene.
Figure 5.
Hsp70B and petrel piRNA clusters encode piRNAs that target host genes. (A) Hsp70B piRNA cluster (top) and the putative target, nod (bottom). piRNA coverage tracks using different read inclusion criteria are shown below RefSeq and genomic coordinates (dm6) for the Hsp70B cluster. Approximately 354-bp local repeats homologous to a 320-bp exonic region of nod are depicted as solid blocks, which fill up most inter-TE space at this locus. Note that the “unique + local” piRNA track does not include TE-derived piRNAs that map outside this locus, but it picks up bona fide local repeats that are homologous, but not identical, to nod. (B) petrel piRNA cluster on the Y chromosome. piRNA coverage tracks using different read inclusion criteria are shown. Sequences with high levels of sequence similarity to protein-coding genes are depicted as colored blocks (not to exact scale). (Green) CG12717, (orange) Paics, (blue) ProtA. Note that gene homologous islands fill up most inter-TE space at this locus. Genomic coordinates are based on the dm6 genome assembly. (C) Coverage of sense (genome-unique, 0 mismatch) and antisense piRNAs (with up to three mismatches) over four putative, protein-coding gene targets of testis piRNAs. Antisense piRNA abundance is shown for each gene.
The second locus producing piRNAs that might target host genes is the novel piRNA cluster petrel on the Y chromosome, which is only present in XY males (Fig. 5B). This cluster spans >200 kb and includes two loci duplicated from chr2L and chrX, respectively, that contain almost the entire CG12717 gene (which encodes a SUMO protease) and small parts of Paics (which encodes an enzyme involved in purine biogenesis) and ProtA (which encodes protamine, a sperm chromatin protein) (Mendez-Lago et al. 2011). These gene homologous sequences are further duplicated locally on Y to >20 copies and take up nearly all the space in between TEs at petrel locus (Fig. 5B; Supplemental Fig. S5B). However, these gene-related sequences likely do not retain coding potentials as they are frequently interrupted by TE sequences. The petrel locus produces piRNAs antisense to CG12717, Paics, and ProtA genes, with averaged levels of nucleotide identity 92.5%, 93.9%, and 91.0%, respectively. Together, two piRNA clusters, Hsp70B and petrel, encode piRNAs with the potential to target both TEs and host genes.
We quantified expression of piRNAs antisense to nod, CG12717, Paics, and ProtA genes from these two clusters. Even though these piRNAs all possess over 90% identity to their putative targets, their abundances differ dramatically (Fig. 5C). The CG12717 gene is targeted by abundant piRNAs (4310 RPM), comparable with the 15th most targeted TE family in testis. piRNAs against nod are expressed at 813 RPM (approximately fivefold less compared with CG12717), while the levels of piRNA against Paics or ProtA are low (both ∼50 RPM). In addition, nearly the entire length of the CG12717 gene is targeted by piRNAs, whereas only small parts of nod, Paics, and ProtA are targeted. These findings suggest that CG12717 and nod might be regulated by piRNAs in testis.
piRNA-guided repression of SUMO protease CG12717/pirate during spermatogenesis
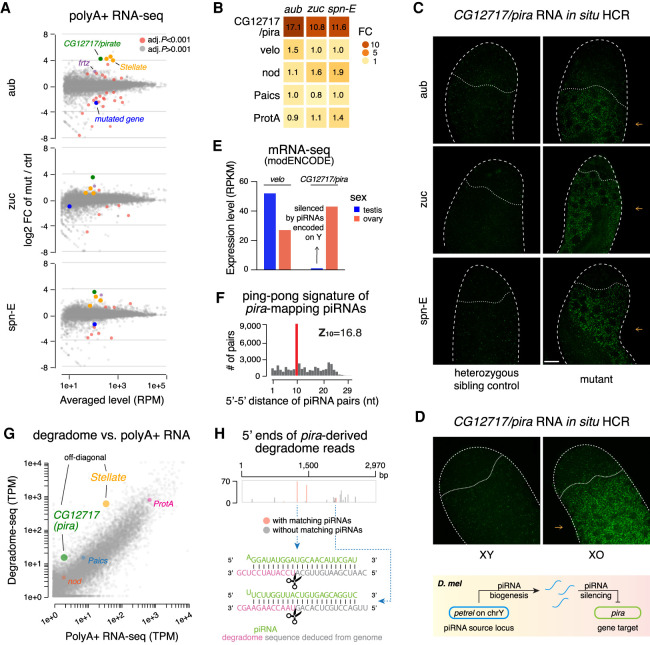
To examine the role of piRNAs in gene regulation, we employed RNA-seq to analyze expression of host genes in testes of three different piRNA pathway mutants: aub, zuc, and spn-E (Schmidt et al. 1999; Stapleton et al. 2001; Nishida et al. 2007; Pane et al. 2007). Transcriptome profiling revealed that only two genes, CG12717 and frtz, exhibited twofold or greater up-regulation in all three piRNA pathway mutants (Fig. 6A). Unlike CG12717, there are very few, if any, antisense piRNAs targeting frtz, so its up-regulation likely reflects a secondary phenotype following TE derepression. Strikingly, expression of CG12717 increased more than 10-fold in all three mutants (Fig. 6B), indicating that it is indeed strongly repressed by the piRNA pathway. Meanwhile, we observed no statistically significant up-regulation of nod, Paics, or ProtA in these three mutants (Fig. 6B), correlating with fewer piRNAs against these genes than CG12717 (Fig. 5C). Transcriptome profiling thus identifies CG12717 as a target of piRNA silencing and suggests that abundant antisense piRNAs with high target coverage might be required for efficient silencing.
Figure 6.
Regulation of CG12717/pira by the piRNA pathway. (A) MA plots showing gene expression changes from polyA+ RNA-seq of aub (top), zuc (middle), and spn-E (bottom) mutant testes versus heterozygous sibling controls. Genes are marked red when passing a stringent statistical cutoff (adjusted P < 0.001, from DESeq2). Additional coloring includes CG12717/pira (green), annotated Stellate transcripts (orange), frtz (purple), and the mutated gene in each mutant (blue). (B) Heat maps showing fold change of five protein-coding genes in three mutant testes according to the polyA+ RNA-seq shown in A. (C) Confocal images of pira mRNAs detected by in situ HCR in aub (top), zuc (middle), and spn-E (bottom) mutant testes along with respective heterozygous sibling controls. Probes were designed against an ∼400-bp sequence unique to pira and absent on Y (Supplemental Fig. S5B), so they do not target petrel piRNA precursors. Note that derepression of pira in piRNA pathway mutants is observed specifically in differentiating spermatocytes (pointed to by orange arrows). Scale bar, 20 µm. (D) Confocal images of pira transcripts detected by in situ HCR in XY and XO testes. Same scale as in C. A schematic of the Y chromosome- and piRNA-dependent silencing of pira is shown at the bottom. (E) Bar graphs displaying modENCODE data of pira and its paralog velo expression in D. melanogaster gonads of both sexes. (F) Analysis of ping-pong processing of pira-mapping piRNAs. Histogram shows distribution of 5′-to-5′ distances of complementary piRNA pairs with an enrichment for 10 nt (i.e., ping-pong signature). To select secondary piRNAs processed from pira transcripts, only reads that map perfectly to pira mRNAs in sense orientation and do not map perfectly to the petrel cluster were used in this analysis. Antisense piRNAs were selected allowing up to three mismatches. (G) Analysis of cellular transcripts enriched in degradome-seq library. Scatterplot shows the number of degradome-seq reads for each gene relative to its expression measured by polyA+ RNA-seq. Transcripts enriched in the degradome-seq library relative to their expression are located above the diagonal. These include Stellate, a known target of piRNA repression, and CG12717/pira, while nod, Paics, and ProtA transcripts are not enriched in degradome-seq. Different annotated copies of Stellate genes were merged. (H) Analysis of pira-derived degradome reads. The abundance of 5′ ends of pira-derived degradome reads is plotted, with the ones that have 10-nt 5′–5′ overlap with antisense piRNAs marked in red. Examples of two such degradome and piRNA pairings are shown at the bottom.
To further examine CG12717 expression, we performed RNA in situ hybridization chain reaction (in situ HCR). Expression of CG12717 is very low in control testis, but it was significantly increased in testes of aub, zuc, and spn-E mutants, establishing this gene as a bona fide gene target of the piRNA pathway (Fig. 6C). We also found strongly elevated CG12717 expression in testes of XO males that have an intact piRNA pathway but lack the Y chromosome (Fig. 6D), confirming that CG12717 silencing piRNAs are encoded on the Y chromosome. Consistent with the Y-linkage of piRNAs against CG12717, it is silenced in testes but expressed in ovaries (Fig. 6E). When derepressed, CG12717 is specifically expressed in differentiating spermatocytes but not in germline stem cells or mitotic spermatogonia. Interestingly, Stellate is expressed at the same stage when the silencing by Su(Ste) piRNAs is removed (Aravin et al. 2004).
piRNA-guided cleavage of target RNAs often triggers the production of secondary piRNAs from target RNAs in a process dubbed a ping-pong cycle (Brennecke et al. 2007). Examination of piRNA sequences revealed abundant piRNAs derived from the entire length of CG12717 mRNAs (Fig. 5C). In contrast, we found few piRNAs processed from transcripts of nod, Paics, or ProtA. Furthermore, sense piRNAs derived from CG12717 mRNAs and antisense piRNAs produced from the petrel locus demonstrated a strong ping-pong signature (Z10 = 16.8) (Fig. 6F), characteristic of an active ping-pong cycle. This finding suggests the direct cleavage of CG12717 transcripts guided by petrel piRNAs. Following the generation of secondary piRNAs by ping-pong, some target RNAs continue to be processed into tail-to-head strings of phased piRNAs dubbed trailing piRNAs (Han et al. 2015; Mohn et al. 2015). We observed a statistically significant phasing signature among CG12717-derived sense piRNAs (Z1 = 3.2) (Supplemental Fig. S5C), and for a quarter of the secondary piRNAs, we could identify trailing piRNAs following the ping-pong sites, consistent with CG12717 being a bona fide piRNA target.
To gain further confidence in piRNA-guided cleavage of CG12717 transcripts, we performed degradome-seq to profile cellular RNAs that bear 5′ monophosphate, which include 3′ products of piRNA-guided cleavage. Analysis of the testis degradome revealed that both Stellate and CG12717 are enriched among degradome fragments relative to their expression levels measured by polyA+ RNA-seq, consistent with both genes being subject to piRNA-guided cleavage (Fig. 6G). In contrast, fragments from nod, Paics, and ProtA were not enriched in the degradome library. In total, we obtained 354 degradome reads from two replicates that correspond to 19 unique 5′ ends that are derived from CG12717 mRNAs (Fig. 6H). Importantly, 39% (138 of 354) of these reads have corresponding antisense piRNAs that overlap 10 nt 5′ to 5′ and thus might be responsible for cleavage at these sites (Fig. 6H). Together, the presence of CG12717-derived sense piRNAs and identification of piRNA-guided cleavage products place CG12717 mRNA as a direct target of piRNAs in testis. As our results indicate that expression of CG12717, a SUMO protease gene related to Ulp2 in yeast (Berdnik et al. 2012), is strongly repressed by piRNAs in testis, we propose to name it pirate (pira; “piRNA target in testis”).
Evolution of pirate and pirate targeting small RNAs
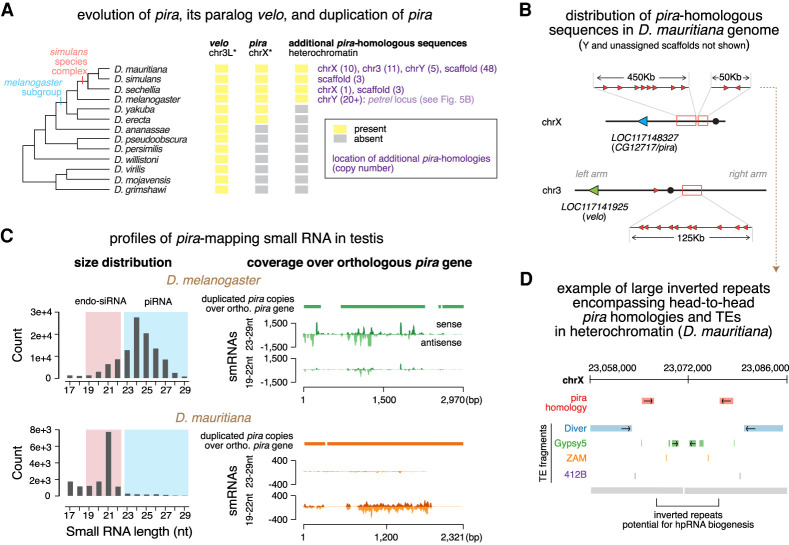
To understand how piRNA-mediated gene regulation of pira has evolved, we performed a tblastn search using the Drosophila melanogaster pira gene against genomes of other Drosophila species. We found multiple copies of pira-related sequences in genomes of the Drosophila simulans species complex (D. simulans, D. sechellia, and D. mauritiana) (Fig. 7A), but not in more distantly related species like D. erecta or D. yakuba. Similar to the petrel locus in D. melanogaster, these pira-related sequences reside in TE-rich regions (either pericentromeric heterochromatin or unassigned scaffolds) in the D. simulans species complex. While all pira-related sequences are exclusively located at petrel on the Y chromosome of D. melanogaster, pira homologous sequences can be found on different chromosomes in genomes of the D. simulans species complex. For instance, in D. mauritiana, pira homologous sequences can be found on at least chrY, chrX, chr3L, and chr3R (Fig. 7B). Therefore, duplications of pira-related sequences into heterochromatin have occurred in all four species.
Figure 7.
Evolution of pira and pira targeting small RNAs. (A) Cladogram of major species in the Drosophila genus (left) and the evolutionary history of velo, pira, and pira-related sequences in genomes of these species (right). Orthologs were identified based on sequence homology and synteny. Shown in purple are locations of additional pira copies in each species and copy numbers in parentheses. Asterisk marks the chromosome name in the melanogaster subgroup, as karyotype differs in more distantly related species. (B) Cartoon depicting distribution of pira homologous sequences in the D. mauritiana genome. Orthologous pira is marked in blue, orthologous velo is marked in green, and the duplicated, candidate sources of pira targeting endo-siRNAs are marked in red. Note that they scatter across pericentromeric heterochromatin of chrX and chr3, as well as chrY and scaffolds (not shown). (C) Profiles of pira-mapping small RNAs in testes of D. melanogaster (top) and D. mauritiana (bottom). Size distributions are shown at the left. Coverage plots over the orthologous pira gene in each species are shown on the right, including: cumulative alignment of heterochromatic, duplicated copies of pira over the syntenic, orthologous pira (top; solid bar); stranded coverage of 23- to 29-nt piRNAs (middle; histogram); and 19- to 22-nt endo-siRNAs (bottom; histogram) over the orthologous pira gene. (D) Illustration showing two representative head-to-head copies of pira homology (red) in the pericentromeric heterochromatin of the D. mauritiana X chromosome. pira-related sequences are flanked by TEs and are part of a large inverted repeat that could potentially permit hpRNA biogenesis.
To investigate whether heterochromatic pira homologous sequences produce small RNAs in testes of other species, we analyzed published small RNA data sets from testes of D. simulans and D. mauritiana (Lin et al. 2018; Kotov et al. 2019). We found no small RNAs mapping to the orthologous pira gene in D. simulans testes but abundant ones in D. mauritiana testes (Fig. 7C). Unexpectedly, unlike 23- to 29-nt pira-mapping piRNAs in D. melanogaster, pira-mapping small RNAs in D. mauritiana are mostly 21 nt long, indicating that they are endo-siRNAs. These endo-siRNAs have on average 93.5% identity with the D. mauritiana pira gene. Notably, similar to other dual-strand piRNA clusters described in D. melanogaster ovaries (Czech et al. 2008; Ghildiyal et al. 2008; Le Thomas et al. 2014), petrel in D. melanogaster testes also generates pira-mapping endo-siRNAs, though much less abundant than 23- to 29-nt piRNAs (Fig. 7C). Examination of heterochromatic, pira homologous sequences in the D. mauritiana genome revealed that most of them are arranged head-to-tail (Fig. 7B). However, there are four instances where pira homologous sequences are arranged head to head (Fig. 7B,D), which could potentially generate hairpin RNAs (hpRNAs), the preferred substrate for processing into endo-siRNAs by Dicer. Thus, targeting of pira by small RNAs in testis seems to be conserved in two Drosophila species. While pira is repressed mostly by piRNAs in D. melanogaster, it is targeted nearly exclusively by endo-siRNAs in D. mauritiana, suggesting two related but distinct regulation strategies employed in sibling species that diverged <3 million years ago.
In addition to pira, there is another Ulp2-like SUMO protease gene, verloren (velo) in the D. melanogaster genome. According to modENCODE data, both genes are expressed throughout the body across development, except that pira has a very low expression level in testis (Brown et al. 2014). pira and velo are paralogs whose homologous domains share 75% nucleotide identity (Supplemental Fig. S5A). In agreement with the sequence similarity, functions of Pira and Velo in the SUMO deconjugation pathway were shown to be partially redundant (Berdnik et al. 2012). Phylogenetic analysis showed that, while velo is found at syntenic locations throughout the Drosophila genus, pira is much younger and was only born after the split of D. melanogaster and ananassae species subgroups (Fig. 7A). These results indicate that pira and velo have evolved from a common ancestor gene, via interchromosomal duplication.
Considering the 75% nucleotide identity between the parts of pira and velo genes in D. melanogaster, pira targeting petrel piRNAs have a potential to target velo transcripts. However, we found that none of the pira antisense piRNAs can be mapped to velo transcript perfectly. Moreover, ∼200-fold fewer piRNAs have a potential to target velo with one to three mismatches. Transcriptome profiling in testes of aub, zuc, and spn-E mutants showed that, unlike pira, velo is not repressed by piRNAs (Fig. 6B). In addition, while pira is only expressed in ovaries, velo is expressed in both testes and ovaries and, in fact, has a higher expression level in testes (Fig. 6E). These results show that Y-linked petrel piRNAs repress specifically pira, but not its paralog, velo, suggesting that a high degree of complementarity is required for efficient piRNA silencing. Therefore, piRNAs distinguish closely related paralogs with high sequence similarity to achieve sex- and paralog- specific gene regulation.
Taken together, our results allowed us to reconstruct the evolutionary history of two paralogous, Ulp2-like SUMO protease genes. First, the pira gene was born via interchromosomal duplication after the split of D. melanogaster and ananassae species subgroups. This then permitted the differentiation of velo and pira functions, though these two genes remain in part functionally redundant in D. melanogaster (Berdnik et al. 2012). Next, divergence between pira and velo sequences created an opportunity for paralog-selective gene regulation by small RNA-guided mechanisms. This was achieved by duplications of pira sequences into heterochromatin in genomes of D. melanogaster and D. simulans species complex. It is plausible that, initially, heterochromatic, pira homologous sequences did not play a role in gene regulation, as illustrated by the absence of pira-mapping small RNAs in D. simulans. However, subsequent expansion and interaction with TE sequences might have enabled the evolution of two distinct repression mechanisms, via production of pira targeting piRNAs and endo-siRNAs, that dominated in D. melanogaster and D. mauritiana, respectively. Repression of pira by Y-linked piRNAs led to its specific repression in D. melanogaster testis, implicating the piRNA pathway in establishing distinct expression patterns of closely related paralogs after gene duplication.
Discussion
Previous studies systematically analyzed piRNA profiles in female gonads of D. melanogaster, revealing an essential role of piRNAs in regulation of many TEs (Brennecke et al. 2007; Li et al. 2009; Malone et al. 2009). However, these studies only provided a single snapshot of the relationship between TE and piRNA defense system, as they are insufficient to understand how the piRNA program might adapt to the changing TE repertoire and different levels of their expression. To this end, several studies explored the piRNA pathway in other species of Drosophila (Malone et al. 2009; Rozhkov et al. 2010; Saint-Leandre et al. 2020). These studies revealed that piRNA profiles are different across species, suggesting an adaptation of the defense mechanism to distinct challenges. However, drastic differences in both TE contents and piRNA cluster sequences even among closely related Drosophila species (Malone et al. 2009; Lerat et al. 2011; Kofler et al. 2015) make it difficult to disentangle different factors that sculpt species-specific piRNA programs. Here, we examined TE expression in males and females of the same species, revealing strong differences in TE activities between the sexes. This allowed us to compare piRNA programs in the two sexes with similar genomic contents (except the Y chromosome).
Another obstacle to understanding responses of the piRNA program to TEs is properly assessing TE expression. The D. melanogaster genome includes >100 different TE families whose expression levels can be measured by standard methods such as RNA-seq. However, TE expression in wild-type animals is greatly suppressed by the piRNA pathway (>100-fold for some families) (ElMaghraby et al. 2019). Therefore, in order to understand true expression potentials of TEs, it is necessary to study their expression upon removal of piRNA silencing, which is difficult to do in species other than model organisms like D. melanogaster. In this work, we examined the TE expression in piRNA pathway mutants, revealing genuine potentials of TE expression in both sexes. Combined analysis of TE and piRNA expression showed responses of the piRNA program to distinct TE expression profiles in the two sexes.
Analysis of the genomic origin of piRNAs represents an important but challenging task. As piRNA sequences are short (23–29 nt) and often derive from repetitive genomic regions, a large fraction of sequenced piRNA reads can be mapped to multiple genomic loci, preventing an unambiguous assignment of their origin. Accordingly, algorithms employed in previous studies only used the small fraction of piRNA reads that can be mapped to the genome at single unique positions to identify genomic regions that generate piRNAs. We took advantage of the fact that some genomic repeats are local (i.e., they reside within one genomic region and are absent in the rest of the genome) to develop a new algorithm for piRNA cluster definition and analysis (Fig. 3A; Supplemental Fig. S4). This approach was successful in identifying new piRNA clusters. Furthermore, it also provided a more accurate quantification of the piRNA cluster expression. We found that the Hsp70B cluster generates piRNAs against the nod gene. In addition, we discovered a novel cluster, petrel, on the Y chromosome that generates piRNAs against three host genes and ensures the strong silencing of the SUMO protease, CG12717/pira, during spermatocyte differentiation.
Our identification of the novel petrel locus on Y expanded known functions of the entirely heterochromatic Y chromosome (Figs. 3F, 5B). Three functionalities have been assigned to Y by the early 1980s (Gatti and Pimpinelli 1983). First, together with the X chromosome, Y encodes rDNA loci that express rRNAs and mediate homolog pairing. Second, Y encodes six protein-coding genes, so-called “fertility factors,” whose protein products are required for completion of spermatogenesis. Finally, the Y chromosome harbors the Su(Ste) locus that generates piRNAs to suppress Stellate genes to safeguard normal spermatogenesis (Aravin et al. 2001; Vagin et al. 2006). A handful of new protein-coding genes were discovered on Y in the past two decades (Bernardo Carvalho et al. 2009; Krsticevic et al. 2010); however, many of them appeared dispensable. Our finding that the Y chromosome encodes a novel piRNA cluster and produces piRNAs to regulate expression of the pira gene assigns a new function to the Y chromosome.
Sexual dimorphism of TE expression and TE silencing piRNA programs
D. melanogaster is an excellent model to study TE regulations and host-TE interactions, as its genome harbors many TE families that are transcriptionally and transpositionally active, generating new insertions in the population (Kofler et al. 2015). As ovaries and testes have complex and distinct tissue compositions, expression levels measured by RNA-seq and small RNA-seq cannot be used directly for comparison of cellular concentrations of transposon transcripts and piRNAs in male and female germlines. Therefore, we have compared rank orders of transposon and piRNA expression in the two sexes as well as fold changes in their levels upon disruption of the piRNA pathway between sexes. Our results indicate that expression of both TEs and piRNAs is sexually dimorphic. The majority of TE families are strongly expressed in ovaries, though some TEs are more active in testes. In line with this, our results indicate a stronger TE silencing piRNA program in female gonads (Fig. 2).
For TEs to be evolutionarily successful, they need to evolve strategies to maximize their chance to be inherited and expanded through generations. For example, TEs often hijack germline gene expression programs to be preferentially active in germ cells. Germline-biased expression leaves the choice of expression to either the female or male germline, or both. Importantly, the two sexes employ distinct evolutionary strategies and have different contributions toward the zygote. While the major contribution of sperm is its genome, the oocyte contributes large amounts of yolk, various protein factors, RNAs, and organelles such as mitochondria, in addition to its genome. This sexual asymmetry in their contributions to the next generation has important implications for reproduction strategies of TEs. TEs active in the male germline need to complete the entire life cycle from transcription to genomic insertion before sperm maturation, in order to propagate. In contrast, once transcribed, TEs active during oogenesis could finish their life cycle in the zygote after fertilization, as long as transcribed TE transcripts are deposited into the oocyte. The latter strategy is also used by the mammalian L1 retrotransposon that is expressed during gametogenesis, but genomic insertions might occur later during early embryogenesis (Kano et al. 2009). Thus, the expression bias toward ovaries observed for most TEs can be explained by an advantage for their proliferation, specifically, the extended window to finish their life cycle, in the female germline.
There are a few TEs that bias testis for expression, suggesting that there are likely male-specific vulnerabilities exploitable by these elements. For example, male germ cells use a testis-specific gene expression machinery (e.g., tTAF and tMAC) to transcribe meiotic and postmeiotic genes (Hiller et al. 2004; Beall et al. 2007). TEs might exploit this tissue-specific transcriptional machinery to enable their sex-biased expression. It will be important in the future to uncover molecular mechanisms underlying differentially expressed TEs between the sexes.
Analysis of piRNA profiles in testis and ovary indicates that piRNA programs have adapted to sex-biased TE expression (Fig. 2). The most striking example is the nearly exclusive expression of telomeric TEs and corresponding antisense piRNAs in the female germline. Our results suggest that differential expression of piRNA clusters in the two sexes together with the differential TE targeting capacity of each cluster contributes to the sex-specific, TE targeting piRNA program. We found that piRNA cluster expression is sexually dimorphic. Besides the Su(Ste) locus, we identified another major cluster on the Y chromosome that is only active in XY males. However, sex-biased expression is not restricted to Y-linked clusters, as many X-linked and autosomal clusters have differential activities between the sexes as well. Besides differential expression, genomic analysis showed differences in piRNA cluster TE contents, suggesting that different piRNA clusters are, to some extent, specialized to target different sets of TEs. Importantly, sex bias in cluster expression and their TE targeting potentials are linked: Clusters preferentially targeting ovary-biased TEs are more active in ovary, while testis-biased clusters tend to target testis-biased TEs (Fig. 4). Hence, piRNA clusters appear to be employed differentially by the two sexes to counteract specific TE threats they face. What determines the differential expression of piRNA clusters between the sexes awaits future studies. Previous work suggests that TE promoters embedded in piRNA clusters retain their activities (Mohn et al. 2014). Contribution of TE promoters to piRNA precursor transcription from piRNA clusters might explain the correlation between expression of clusters and their TE targets.
Satellite DNA as target of piRNA silencing
Satellite DNAs can be classified as either simple or complex satellites based on the length of repeating units, and they occupy large portions of the Drosophila genome, particularly at pericentromeric and subtelomeric regions (Hsieh and Brutlag 1979; Karpen and Spradling 1992; Lohe et al. 1993; Larracuente and Presgraves 2012). We found piRNAs expressed from three major families of complex satellites: subtelomeric HETRP/TAS, Responder (Rsp), and SAR/1.688 (including 359-bp). In fact, piRNAs can be mapped to both strands of complex satellites in gonads of both sexes, and they often possess ping-pong signature (Fig. 1C). Thus, our results expand the previous observation of piRNAs mapping to one strand of Rsp (Saito et al. 2006) and establish complex satellites as dual-strand piRNA clusters and potential targets of piRNA silencing in the Drosophila germline of both sexes. Our analysis was focused on complex satellites, as simple satellite repeats are still largely intractable to sequencing technologies today (Khost et al. 2017). However, a recent study reported that transcripts from AAGAG simple satellite repeats regulate heterochromatin in the male germline and are required for male fertility (Mills et al. 2019). It will be interesting to determine whether simple satellites produce piRNAs and, if so, whether their piRNA production is required for male fertility.
piRNAs loaded onto the nuclear Piwi protein guide heterochromatin assembly (Wang and Elgin 2011; Sienski et al. 2012; Le Thomas et al. 2013; Rozhkov et al. 2013). For this reason, satellite piRNAs might play a role in establishing germline heterochromatin, similar to heterochromatin formation guided by siRNAs in fission yeast (Hall et al. 2002; Volpe et al. 2002). While the function of complex satellites remains mostly elusive, Rsp has been implicated in a meiotic drive system called segregation distortion (Hartl 1973; Wu et al. 1988; Larracuente and Presgraves 2012). During male meiosis, the Segregation distorter (Sd) allele enhances its own transmission to haploid cells at the cost of the wild-type (Sd+) allele in Sd/Sd+ heterozygous males, violating the Mendelian law of inheritance. Importantly, segregation distortion requires the presence of a sufficient number of Rsp satellite repeats in trans. Although described >60 yr ago (Sandler et al. 1959), the molecular mechanism of segregation distortion remains unknown. Intriguingly, mutations of aubergine (aub), a PIWI protein, were found to be enhancers of segregation distortion (Gell and Reenan 2013). Together with our data, these data suggest that the piRNA pathway may play a role in segregation distortion during spermatogenesis.
Regulation of host genes by piRNAs
Though the central and conserved function of the piRNA pathway seems to be TE repression, other functions were also described in several organisms (for review, see Ozata et al. 2019). The role of piRNAs in regulating host gene expression is particularly intriguing and remains somewhat controversial. The first described piRNAs, Su(Ste) piRNAs, silence the expression of Stellate genes (Aravin et al. 2001; Vagin et al. 2006). However, Stellate genes and their piRNA suppressors appear to resemble selfish toxin–antitoxin systems rather than representing an example of host gene regulation (Aravin 2020). Since the discovery of piRNA pathway, there have been several studies reporting host protein-coding genes regulated by Drosophila piRNAs (for review, see Rojas-Ríos and Simonelig 2018). In this work, we analyzed the ability of Drosophila piRNAs to regulate host genes in testes, by examining gene targeting piRNAs and changes in host gene expression across three piRNA pathway mutants. We found piRNAs targeting four host genes: nod (a kinesin-like protein), CG12717/pira (a SUMO protease), Paics (a metabolic enzyme), and ProtA (a sperm chromatin protein). These four genes are targeted by antisense piRNAs generated from two piRNA clusters that contain sequence homology to them. However, only one of the four, CG12717/pira, is substantially repressed (>10-fold) by piRNAs (Fig. 6). As pira silencing piRNAs are encoded on the Y chromosome and thus only expressed in males, they are responsible for differential expression of pira in the two sexes. Indeed, in wild-type flies, pira is specifically silenced in male gonads while highly expressed in female counterparts. Thus, our results establish the ability of piRNAs to repress host protein-coding genes, and, at the same time, suggest that this role is likely restricted to a small number of genes.
Our results indicate that piRNA-guided repression of host genes requires a sufficient number of targeting piRNAs. While all four genes are targeted by piRNAs with similar levels of sequence identity (91%–94%; i.e., about two mismatches per piRNA), the abundance of piRNAs against each gene differs drastically. There are many more pira targeting piRNAs than the other three gene targets, at a level comparable with the 15th most targeted TE. Furthermore, while pira is targeted along almost the entire length, only small regions of other three genes are targeted by piRNAs. These differences in piRNA abundance and distribution of target sites could explain strong repression of pira, in contrast to the other three genes. It is possible that these genes are still regulated by piRNAs at specific stages, the question that remains to be further investigated. Importantly, abundant pira silencing piRNAs do not repress the pira paralog, velo, that has a 75% sequence identity with pira, indicating that a high complementarity between piRNA and target may be important for efficient silencing. In agreement with these, a previous report indicated that a similar level of sequence identity (∼76%) is insufficient for the silencing of vasa by AT-chX piRNAs (Kotov et al. 2019). Therefore, both high expression and high complementarity with targets might be required for efficient piRNA silencing in D. melanogaster.
This conclusion is important for analyzing the potential of piRNAs to repress host protein-coding genes. Unlike miRNAs, sequences of piRNAs are extremely diverse. Accordingly, if mismatches between piRNA and its target are well tolerated, a large number of cellular mRNAs should be targeted and repressed by piRNAs. Indeed, some host genes were proposed to be repressed by a few piRNA species that have multiple mismatches to mRNA sequences (Saito et al. 2009; Gonzalez et al. 2015; Klein et al. 2016; Rojas-Ríos et al. 2017). Our results suggest that such a spurious targeting by individual piRNAs is unlikely to cause repression. In fact, a high threshold for efficient target repression might permit production of diverse piRNA sequences against genuine targets such as TEs, without unintended interference with host gene expression.
The role of piRNA in evolution
Analysis of pira repression revealed a remarkable picture of evolutionary innovation (Fig. 7). piRNA-dependent repression of pira occurs in D. melanogaster but not in its sibling species, suggesting its rather recent origin. Efficient silencing of pira is linked to the presence of multiple copies of pira homologous sequences in a piRNA cluster inside heterochromatin. Interestingly, duplications of pira sequences into, and their expansion within, heterochromatin can be found in three closely related species of the D. simulans complex, in addition to D. melanogaster. However, the distribution and copy number of pira-related sequences differ among these four species. In fact, both the petrel locus that generates pira silencing piRNAs and its two flanking protein-coding genes, FDY and Mst77Y, evolved after the split of the D. melanogaster and D. simulans species complex (Krsticevic et al. 2010; Mendez-Lago et al. 2011; Carvalho et al. 2015), suggesting that the entire locus is unique to D. melanogaster. Furthermore, no small RNAs are generated from heterochromatic pira sequences in D. simulans, while endo-siRNAs are made against pira in D. mauritiana. The neutral theory of molecular evolution provides the most parsimonious interpretation of these results. This theory suggests that the initial duplication of pira sequences into heterochromatin might have been a random event that did not play a role in regulating the ancestral pira gene. However, subsequent evolution of pira-related sequences inside heterochromatin gave rise to two different modes of regulation, piRNA and endo-siRNA, in two different but closely related species. Emergence of small RNA-mediated repression was probably facilitated by the fact that pira itself was recently evolved and retains partially redundant functions with its paralog, velo (Berdnik et al. 2012), allowing independent regulation of two paralogs.
The evolutionarily innovative role of piRNAs in regulating host genes in Drosophila has interesting parallels in other organisms. The piRNA pathway was shown to regulate expression of the xol-1 gene involved in sex determination and dosage compensation in two different worm species, C. elegans and C. briggsae, that diverged >50 million years ago (Tang et al. 2018). On the other hand, pachytene piRNAs expressed during spermatogenesis in mammals evolved very fast and are generally poorly conserved (Özata et al. 2020). The function of pachytene piRNAs is under active debate as no obvious targets can be easily discerned by analysis of their sequences (Aravin et al. 2006; Girard et al. 2006; Vourekas et al. 2012). Recently, knockout of one pachytene piRNA cluster led to unexpected conclusion that a small fraction of piRNAs promote biogenesis from other piRNA clusters and regulate the expression of a few host genes, while the vast majority do not target any transcripts (Wu et al. 2020). Thus, mammalian pachytene piRNAs can be considered a selfish system that is occasionally involved in regulation of the host gene expression. Species-specific regulation of host genes by piRNAs in both Drosophila and mouse suggests that the piRNA pathway is used in evolution to create innovation in gene regulatory networks that might contribute to speciation. More generally, piRNAs might promote the evolvability of animal species. Though it is difficult to establish the function of any molecular mechanism in evolution, this proposal makes a testable prediction that host genes repressed by piRNAs differ even among closely related species. Future studies in nonmodel organisms will shed light on the role of piRNAs in evolution and speciation.
Materials and methods
Fly stocks
Stocks and crosses were raised at 25°C. The following stocks were used: aubQC42 (BDSC4968), aubHN2 (BDSC8517), zucDf (BSDC3079), spn-Ehls3987 (BDSC24853), and spn-E1 (BDSC3327) were obtained from the Bloomington Drosophila Stock Center; rhi2 and rhiKG were gifts of William Theurkauf; zucHM27 was a gift from Trudi Schüpbach; nosP-GFP-Burdock was a gift from Julius Brennecke. Heterozygous siblings were used as controls for all experiments, unless noted otherwise. The XO male was generated by crossing XY males to C(1)RM females (BDSC9460).
RNA in situ hybridization chain reaction (HCR)
A kit containing a DNA probe set, a DNA probe amplifier, and hybridization, amplification, and wash buffers was purchased from Molecular Instruments for CG12717 transcripts. To avoid targeting the petrel locus on Y, we designed probes against an ∼400-bp unique region present in CG12717 on X but absent on the Y chromosome. The CG12717 probe set (unique identifier: 3916/E064) initiated the B3 (Alexa546) amplifier. In situ HCR v3.0 (Choi et al. 2018) was performed according to the manufacturer's recommendations for generic samples in solution.
Image acquisition and analysis
Confocal images were acquired with a Zeiss LSM 800 using a 63× oil immersion objective (NA = 1.4) and processed using Fiji (Schindelin et al. 2012). Single focal planes were shown in all images, where dotted outlines were drawn for illustration purposes.
RNA-seq
RNA was extracted from 160 to 200 pairs of dissected testes of aubQC42/HN2, spn-E1/hls3987, zucHM27/Df, and respective heterozygous sibling controls in TRIzol (Invitrogen). PolyA+ selection was done using an NEBNext Poly(A) mRNA Magnetic Isolation Module (NEB E7490), followed by strand-specific library preparation with an NEBNext Ultra Directional RNA Library Prep kit for Illumina (NEB E7760) according to the manufacturer's instructions. Libraries were sequenced on an Illumina HiSeq 2500, yielding 11 million to 17 million 50-bp single-end reads. PolyA-selected RNA-seq of rhi mutants and controls were downloaded from NCBI SRA (two biological replicates per sex per genotype) (see Chen et al. 2020 for testis and GSE126578 for ovary).
RNA-seq analysis
To quantify expression levels of protein-coding genes across different piRNA pathway mutants (aub, zuc, and spn-E), we used kallisto 0.46.1 (Bray et al. 2016). Three heterozygous controls were pooled as triplicates of controls to be analyzed against duplicates of each of the three piRNA pathway mutants. Transcript-level quantification was pooled to obtain gene-level quantification. Differential gene expression was done with DESeq2 (Love et al. 2014). Expression of CG12717 and velo in ovary and testis from modENCODE (Brown et al. 2014) was extracted from FlyBase (Thurmond et al. 2019).
For analysis of TE expression and TE fold change in piRNA pathway mutants of both sexes, rhi mutants were used where piRNA production from germline-specific dual-strand clusters was abolished. Reads mapped to rRNA were discarded using bowtie 1.2.2 allowing three mismatches. Reads were then mapped to TE consensus from RepBase17.08 using bowtie 1.2.2 with -v 3 -k 1 and normalized to the total number of reads mapped to dm6 genome. For simplicity, reads mapped to LTR and internal sequences were merged for each LTR TE given their well correlative behaviors. Only TEs that have five or more RPM and ≥2.5 RPKM expression in piRNA pathway mutants of either sex were kept for the analysis (n = 87). A pseudocount of one was added before calculating TE fold change in piRNA pathway mutants.
Degradome-seq and analysis
Degradome-seq was done as previously described (Wang et al. 2014). Briefly, RNA was extracted from 50 pairs of wild-type testes (w1118) in TRIzol, followed by DNase treatment (Turbo DNase) and rRNA depletion (ribo-zero). Next, RNA bearing 5′ monophosphate was enriched by ligating a 5′ adaptor (5′-GUUCAGAGUUCUACAGUCCGACGAUC-3′) with T4 RNA ligase, followed by size selection for RNA >200 nt (RNA Clean & Concentrator-5). Reverse transcription was performed with SuperScript III and a primer containing a degenerate sequence at its 3′ end (5′-GCACCCGAGAATTCCANNNNNNNN-3′), which also introduced the 3′ adaptor. PCR was done to amplify cDNA and to introduce sequencing primer and index sequences. Two replicates were sequenced on an Illumina HiSeq 2500 for 150-bp single-end reads. To compare degradome with polyA+ RNA-seq, we used kallisto to assign reads to transcripts as described above. To identify degradome sequences that are 3′ piRNA-guided cleavage products of CG12717 transcripts, we mapped degradome reads (trimmed to 50 bp) to the dm6 genome with bowtie 1.2.2 -v 0 -m 1 and extracted those mapping to the coding strand of CG12717 (note that it is an intronless gene). CG12717-derived degradome reads were extended upstream of their 5′ ends based on dm6 genome sequence to examine their overlap with antisense piRNAs and extent of complementarity.
Identification of TEs regulated by rhi
To identify a set of TEs regulated by rhi in at least one sex, we looked for TEs that have at least 100 RPM in rhi mutant ovaries or at least 25 RPM in rhi mutant testes. Next, we filtered out TEs that showed less than threefold derepression in both sexes. From the initial 87 TEs defined above, these led to a total of 36 TEs regulated by rhi in at least one sex, shown in Figure 2B,D. See Supplemental Figure S3C for detailed profiles of these 36 TEs.
piRNA-seq
RNA extraction was done as above for RNA-seq. Small RNAs (18–30 nt) were purified by PAGE (15% polyacrylamide gel) from ∼1 µg of total RNA. Purified small RNA was subject to library prep using an NEBNext Multiplex Small RNA Sample Prep Set for Illumina (NEB E7330) according to the manufacturer's instructions. Adaptor-ligated, reverse-transcribed, PCR-amplified samples were purified again by PAGE (6% polyacrylamide gel). Two biological replicates per genotype were sequenced on an Illumina HiSeq 2500, yielding 15 million to 20 million 50-bp single-end reads.
piRNA-seq analysis of TEs, complex satellites and genes
To isolate piRNAs, adaptor-trimmed total small RNAs were size-selected for 23–29 nt (cutadapt 2.5), and those mapped to rRNA, miRNA, snRNA, snoRNA, and tRNA were discarded (bowtie 1.2.2 with -v 3). piRNAs were first mapped to RepBase17.08 to obtain the portion mapping to TEs and complex satellites; the rest was then mapped to gene sequences derived from the gtf file downloaded from Ensembl (BDGP6.28.99) (Yates et al. 2019); reads unmapped to repeats and genes were then mapped to dm6 to infer the portion mapping to intergenic regions, and the unmapped ones were listed under the “others” category. A pipeline is also drawn in Supplemental Figure S1. For TE antisense piRNA analysis, piRNA reads were mapped, normalized, and processed as done for polyA+ RNA-seq (see above). For complex satellite-mapping small RNAs, we plotted size distribution, analyzed nucleotide bias at position 1, and calculated coverage along consensus sequences using bedtools v2.28.0. Analysis of ping-pong signature (i.e., 5′-to-5′ distances between complementary piRNA pairs) and phasing signature (i.e., 3′-to-5′ distances on the same strand) were done with custom scripts. Ping-pong z-score was calculated using 1–9 nt and 11–23 nt as the background distribution for an enrichment of 10 nt, whereas phasing z-score, Z1 as defined in Han et al. (2015), was calculated using 2–50 nt as the background distribution for an enrichment of 1 nt. For piRNAs antisense to protein-coding genes of interest, we downloaded gene sequences from FlyBase (Thurmond et al. 2019) and mapped piRNAs to them using bowtie 1.2.2. For mRNA-derived sense piRNAs, we mapped piRNAs to the genome and kept ones with unique mapping and zero mismatches (bowtie 1.2.2 with -v 0 -m 1) to the gene regions and orientations of interest.
A pipeline tolerating local repeats for piRNA cluster analysis
We first separated rRNA-depleted 23- to 29-nt small RNA reads that map to one unique location in the genome and others that have multiple mapping positions (“multimappers”). For all multimappers, we filtered out those who map to more than one chromosome arm, retaining only ones with multimapping positions on a single chromosome arm (“intrachromosomal repeats”). Then, for each of the reads we kept as intrachromosomal repeats, we calculated the maximum distance (“max distance”) of all mapping positions. In order to enforce the local requirement, we hoped to identify a cutoff distance for max distances, which is large enough to contain known piRNA loci but small enough to allow certain resolution of neighboring loci. To this end, we analyzed a pool of 50-bp DNA fragments tiling the entire dm6 genome and plotted a histogram of max distances for all intrachromosomal repeats (Supplemental Fig. S4B). This revealed a density of intrachromosomal repeats having max distances smaller than ∼500 kb, as well as four pronounced peaks with larger max distances. Sequence analysis uncovered the identities of these peaks: The peak with an ∼600-kb max distance corresponds to AT-chX, the peak with an ∼1.8-Mb max distance represents Su(Ste), and the other two peaks mostly contain Y-specific simple repeats. We thus set a 2-Mb tolerance threshold of max distances to allow local repeats in piRNA cluster analysis. In other words, we defined local repeats as repeats that have all copies contained within a window <2 Mb and merged their normalized counts with unique sequences for piRNA cluster analysis. Alignment was done using bowtie2 to the dm6 genome. To compare this new pipeline with the convention that uses only unique mappers, we calculated the number of reads mapped to major piRNA clusters using both methods (Fig. 3B). A summary of this pipeline is shown in Supplemental Figure S4A.
Definition of piRNA clusters
Small RNAs (23–29 nt) were mapped to the dm6 genome using the above-mentioned pipeline tolerating local repeats and generated coverage profiles across 1-kb windows that tile the genome. One-kilobase windows including highly expressed miRNA, snRNA, snoRNA, hpRNA, or 7SL SRP RNA were excluded. One-kilobase windows with low read-coverage (≤100 bp) were also excluded. Then, 1-kb windows that produce at least certain amounts of piRNAs were extracted for cluster definition (≥10 RPM for testis, ≥50 RPM for ovary). Neighboring 1-kb widows within 3 kb were merged. If merged windows were ≥5 kb, they were merged again within 15 kb. This yields 844 piRNA clusters in testis and 525 piRNA clusters in ovary, after manual curation. Major piRNA clusters described before in ovaries (Brennecke et al. 2007; Mohn et al. 2014) were all recovered with similar resolution. To compare expression levels of major piRNA clusters between sexes, cluster boundaries were manually curated to guarantee identical regions being compared. piRNA clusters defined in this study for both sexes are listed in Supplemental Table S1.
TE content of piRNA clusters
TE annotation in the dm6 genome was downloaded from UCSC Table Browser (Karolchik et al. 2004). piRNA cluster boundaries were defined as described above. For a piRNA cluster of interest, the TE content is calculated as length contribution to the entire cluster length by individual TEs. TE contents add up to <100%, as TEs do not fill completely the cluster length.
Sex bias of piRNA cluster TE content
Sex bias of individual TEs was first computed as the log2 ratio of expression levels in piRNA pathway mutants (rhi) between sexes (ovary over testis). The sex bias of piRNA cluster TE content was then computed as the cumulative sex bias of individual TEs inside the cluster, weighted by their length contribution to the cluster. Using all expressed TEs or only ones that show pronounced sex bias generated comparable results. To eliminate noise, we only used TEs that exhibit strong, ≥10-fold sexual difference in expression (n = 24). An equation and an example are shown in Figure 4C.
BLAT and BLAST analysis
To characterize the unannotated sequence between annotated repeats in piRNA clusters, interrepeat sequences were analyzed using BLAT on UCSC genome browser (Kent 2002). For example, an inter-TE sequence at Hsp70B locus was used to BLAT against the dm6 genome, which revealed the homology with an exon of the nod gene (Fig. 5A). Homology between CG12717 and velo was done with both BLAT and BLAST, which yielded similar results. Characterization of CG12717 homologous sequences at the petrel locus (Supplemental Fig. S5B) was done by multiple sequence alignment with the Needle program (ebi.ac.uk/Tools/psa/emboss_needle).
Phylogenetic analysis
The longest transcripts of velo and CG12717 in the D. melanogaster genome were used to BLAST against the nucleotide collection with the tblastn program. Orthologs of these two genes in other Drosophila species were identified based on high nucleotide similarity and synteny. In all orthologs identified for both genes, we found the same flanking protein-coding genes, confirming their ortholog identities. Occasionally, BLAST with CG12717 revealed the velo ortholog in that species as well, but only in D. mauritiana, D. simulans, and D. sechellia genomes are there additional hits with high sequence homology to CG12717, other than the orthologous CG12717 and velo. These additional CG12717-related sequences are in some cases annotated as predicted genes but all buried in TE-rich heterochromatin (close to the centromere or in highly repetitive unassigned scaffolds). To examine the organization of CG12717-related sequences in the D. mauritiana genome in detail, we ran BLAST using the D. mauritiana CG12717 gene against its genome (assembly GCA_004382145.1), which revealed additional unannotated regions with high sequence similarity to CG12717. Those located on chrX and chr3 were drawn in Figure 7B. The instance where two adjacent CG12717-related sequences are arranged head-to-head on chrX is illustrated in Figure 7D, and the other three such instances are found in unassigned scaffolds. To uncover the identity of flanking unannotated sequences, we BLAST the 50-kb region encompassing CG12717-related sequences against the TE consensus (RepBase17.08). The cladogram was drawn for illustration (Drosophila 12 Genomes Consortium 2007).
Analysis of testis small RNAs in non-D. melanogaster species
The following testis small RNA libraries from non-D. melanogaster species were downloaded from NCBI SRA: D. simulans SRR7410589 (Lin et al. 2018) and D. mauritiana SRR7961897 (Kotov et al. 2019). Adaptor-trimmed reads were mapped to the orthologous CG12717 gene, D. simulans GD15918 and D. mauritiana LOC117148327, respectively (bowtie 1.2.2 with -v 3 -k 1). Coverage was plotted along the orthologous CG12717 gene.
Data visualization and statistical analysis
Most data visualization and statistical analysis were done in Python 3 via JupyterLab with the following software packages: numpy (Oliphant 2015), pandas (https://conference.scipy.org/proceedings/scipy2010/mckinney.html), and altair (VanderPlas et al. 2018). The UCSC genome brower (Kent et al. 2002) and IGV (Robinson et al. 2011; Thorvaldsdóttir et al. 2013) were used to explore sequencing data and to prepare the browser track panels shown.
Data availability
Sequencing data can be accessed via NCBI SRA with the following accession numbers: PRJNA646006 (rhi), PRJNA646216 (aub, zuc, and spn-E), and PRJNA719671 (degradome).
Supplementary Material
Acknowledgments
We are grateful to William Theurkauf, Trudi Schüpbach, Julius Brennecke, and the Bloomington Drosophila Stock Center for fly stocks. We thank Katalin Fejes Toth and members of the Aravin laboratory for discussion, and Silke Jensen and Emilie Brasset for valuable comments on the preprint. We appreciate the help of Maria Ninova and Fan Gao (Bioinformatics Resource Center, California Institute of Technology) with bioinformatics analysis, the help of Pei-Hsuan Wu and Ildar Gainetdinov (Phil Zamore's laboratory) with degradome-seq experiments, the help of Grace Shin and Maayan Schwarzkopf with HCR experiments, the help of Giada Spigolon and Andres Collazo (Biological Imaging Facility, California Institute of Technology) with microscopy, and the help of Igor Antoshechkin (Millard and Muriel Jacobs Genetics and Genomics Laboratory, California Institute of Technology) with sequencing. This work was supported by grants from the National Institutes of Health (R01 GM097363) and by the Howard Hughes Medical Institute Faculty Scholar Award to A.A.A.
Author contributions: P.C. conducted most experiments and most bioinformatic analysis. B.K.G. performed testis RNA-seq. All authors designed experiments and analyzed the data. P.C. and A.A.A. wrote the manuscript with input from all other coauthors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.345041.120.
Competing interest statement
The authors declare no competing interests.
References
- Aravin AA. 2020. Pachytene piRNAs as beneficial regulators or a defense system gone rogue. Nat Genet 52: 644–645. 10.1038/s41588-020-0656-8 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Naumova NM, Tulin AV, Vagin VV, Rozovsky YM, Gvozdev VA. 2001. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr Biol 11: 1017–1027. 10.1016/S0960-9822(01)00299-8 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Klenov MS, Vagin VV, Bantignies F, Cavalli G, Gvozdev VA. 2004. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. MCB 24: 6742–6750. 10.1128/MCB.24.15.6742-6750.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207. 10.1038/nature04916 [DOI] [PubMed] [Google Scholar]
- Beall EL, Lewis PW, Bell M, Rocha M, Jones DL, Botchan MR. 2007. Discovery of tMAC: a Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev 21: 904–919. 10.1101/gad.1516607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D, Favaloro V, Luo L. 2012. The SUMO protease Verloren regulates dendrite and axon targeting in olfactory projection neurons. J Neurosci 32: 8331–8340. 10.1523/JNEUROSCI.6574-10.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo Carvalho A, Koerich LB, Clark AG. 2009. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet 25: 270–277. 10.1016/j.tig.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103. 10.1016/j.cell.2007.01.043 [DOI] [PubMed] [Google Scholar]
- Brown JB, Boley N, Eisman R, May GE, Stoiber MH, Duff MO, Booth BW, Wen J, Park S, Suzuki AM, et al. 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512: 393–399. 10.1038/nature12962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJG, Bourc'his D, Bestor TH, de Rooij DG, Hannon GJ. 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514. 10.1016/j.devcel.2007.03.001 [DOI] [PubMed] [Google Scholar]
- Carpenter AT. 1973. A meiotic mutant defective in distributive disjunction in Drosophila melanogaster. Genetics 73: 393–428. 10.1093/genetics/73.3.393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Vicoso B, Russo CAM, Swenor B, Clark AG. 2015. Birth of a new gene on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci 112: 12450–12455. 10.1073/pnas.1516543112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Luo Y, Aravin AA. 2020. RDC complex executes a dynamic piRNA program during Drosophila spermatogenesis to safeguard male fertility. bioRxiv 10.1101/2020.08.25.266643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, Pierce NA. 2018. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development 145: dev165753. 10.1242/dev.165753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. 2008. An endogenous small interfering RNA pathway in Drosophila. Nature 453: 798–802. 10.1038/nature07007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H. 2002. Miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830. 10.1016/S1534-5807(02)00165-X [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218. 10.1038/nature06341 [DOI] [PubMed] [Google Scholar]
- ElMaghraby MF, Andersen PR, Pühringer F, Hohmann U, Meixner K, Lendl T, Tirian L, Brennecke J. 2019. A heterochromatin-specific RNA export pathway facilitates piRNA production. Cell 178: 964–979.e20. 10.1016/j.cell.2019.07.007 [DOI] [PubMed] [Google Scholar]
- Gatti M, Pimpinelli S. 1983. Cytological and genetic analysis of the Y chromosome of Drosophila melanogaster: I. Organization of the fertility factors. Chromosoma 88: 349–373. 10.1007/BF00285858 [DOI] [Google Scholar]
- Gell SL, Reenan RA. 2013. Mutations to the piRNA pathway component Aubergine enhance meiotic drive of segregation distorter in Drosophila melanogaster. Genetics 193: 771–784. 10.1534/genetics.112.147561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George P, Jensen S, Pogorelcnik R, Lee J, Xing Y, Brasset E, Vaury C, Sharakhov IV. 2015. Increased production of piRNAs from euchromatic clusters and genes in Anopheles gambiae compared with Drosophila melanogaster. Epigenetics Chromatin 8: 50. 10.1186/s13072-015-0041-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler ELW, Zapp ML, Weng Z, et al. 2008. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science 320: 1077–1081. 10.1126/science.1157396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202. 10.1038/nature04917 [DOI] [PubMed] [Google Scholar]
- Gonzalez J, Qi H, Liu N, Lin H. 2015. Piwi is a key regulator of both somatic and germline stem cells in the Drosophila testis. Cell Rep 12: 150–161. 10.1016/j.celrep.2015.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett RW, Lis JT. 1981. DNA sequence analysis reveals extensive homologies of regions preceding hsp70 and αβ heat shock genes in Drosophila melanogaster. Proc Natl Acad Sci 78: 6196–6200. 10.1073/pnas.78.10.6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K-I, Ayoub N, Cohen A, Grewal SIS. 2002. Establishment and maintenance of a heterochromatin domain. Science 297: 2232–2237. 10.1126/science.1076466 [DOI] [PubMed] [Google Scholar]
- Han BW, Wang W, Li C, Weng Z, Zamore PD. 2015. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 348: 817–821. 10.1126/science.aaa1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handler D, Meixner K, Pizka M, Lauss K, Schmied C, Gruber FS, Brennecke J. 2013. The genetic makeup of the Drosophila piRNA pathway. Mol Cell 50: 762–777. 10.1016/j.molcel.2013.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl DL. 1973. Complementation analysis of male fertility among the segregation distorter chromosomes of Drosophila melanogaster. Genetics 73: 613–629. 10.1093/genetics/73.4.613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley RS, Theurkauf WE. 1993. Requiem for distributive segregation: achiasmate segregation in Drosophila females. Trends Genet 9: 310–316. 10.1016/0168-9525(93)90249-H [DOI] [PubMed] [Google Scholar]
- Hiller M, Chen X, Pringle MJ, Suchorolski M, Sancak Y, Viswanathan S, Bolival B, Lin T-Y, Marino S, Fuller MT. 2004. Testis-specific TAF homologs collaborate to control a tissue-specific transcription program. Development 131: 5297–5308. 10.1242/dev.01314 [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in zebrafish. Cell 129: 69–82. 10.1016/j.cell.2007.03.026 [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF. 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 27: 2702–2711. 10.1038/emboj.2008.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh T, Brutlag D. 1979. Sequence and sequence variation within the 1.688 g/cm3 satellite DNA of Drosophila melanogaster. J Mol Biol 135: 465–481. 10.1016/0022-2836(79)90447-9 [DOI] [PubMed] [Google Scholar]
- Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJT, Roovers EF, Ladurner P, Berezikov E, Ketting RF. 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J 29: 3688–3700. 10.1038/emboj.2010.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH. 2009. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev 23: 1303–1312. 10.1101/gad.1803909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496. 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen GH, Spradling AC. 1992. Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics 132: 737–753. 10.1093/genetics/132.3.737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT–the BLAST-like alignment tool. Genome Res 12: 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12: 996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khost DE, Eickbush DG, Larracuente AM. 2017. Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Res 27: 709–721. 10.1101/gr.213512.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klattenhoff C, Xi H, Li C, Lee S, Xu J, Khurana JS, Zhang F, Schultz N, Koppetsch BS, Nowosielska A, et al. 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 138: 1137–1149. 10.1016/j.cell.2009.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein JD, Qu C, Yang X, Fan Y, Tang C, Peng JC. 2016. c-Fos repression by Piwi regulates Drosophila ovarian germline formation and tissue morphogenesis. PLoS Genet 12: e1006281. 10.1371/journal.pgen.1006281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Nolte V, Schlötterer C. 2015. Tempo and mode of transposable element activity in Drosophila. PLoS Genet 11: e1005406. 10.1371/journal.pgen.1005406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotov AA, Adashev VE, Godneeva BK, Ninova M, Shatskikh AS, Bazylev SS, Aravin AA, Olenina LV. 2019. piRNA silencing contributes to interspecies hybrid sterility and reproductive isolation in Drosophila melanogaster. Nucleic Acids Res 47: 4255–4271. 10.1093/nar/gkz130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krsticevic FJ, Santos HL, Januário S, Schrago CG, Carvalho AB. 2010. Functional copies of the Mst77F gene on the Y chromosome of Drosophila melanogaster. Genetics 184: 295–307. 10.1534/genetics.109.107516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849. 10.1242/dev.00973 [DOI] [PubMed] [Google Scholar]
- Larracuente AM, Presgraves DC. 2012. The selfish segregation distorter gene complex of Drosophila melanogaster. Genetics 192: 33–53. 10.1534/genetics.112.141390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh Brown AJ, Ish-Horowicz D. 1981. Evolution of the 87A and 87C heat-shock loci in Drosophila. Nature 290: 677–682. 10.1038/290677a0 [DOI] [PubMed] [Google Scholar]
- Lerat E, Burlet N, Biémont C, Vieira C. 2011. Comparative analysis of transposable elements in the melanogaster subgroup sequenced genomes. Gene 473: 100–109. 10.1016/j.gene.2010.11.009 [DOI] [PubMed] [Google Scholar]
- Le Thomas A, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, Hur JK, Aravin AA, Tóth KF. 2013. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev 27: 390–399. 10.1101/gad.209841.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Thomas A, Stuwe E, Li S, Du J, Marinov G, Rozhkov N, Chen Y-CA, Luo Y, Sachidanandam R, Toth KF, et al. 2014. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev 28: 1667–1680. 10.1101/gad.245514.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521. 10.1016/j.cell.2009.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC. 1997. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124: 2463–2476. [DOI] [PubMed] [Google Scholar]
- Lin C-J, Hu F, Dubruille R, Vedanayagam J, Wen J, Smibert P, Loppin B, Lai EC. 2018. The hpRNA/RNAi pathway is essential to resolve intragenomic conflict in the Drosophila male germline. Dev Cell 46: 316–326.e5. 10.1016/j.devcel.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis JT, Prestidge L, Hogness DS. 1978. A novel arrangement of tandemly repeated genes at a major heat shock site in D. melanogaster. Cell 14: 901–919. 10.1016/0092-8674(78)90345-8 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Freund R, Schweber M, Wensink PC, Meselson M. 1978. Sequence organization and transcription at two heat shock loci in Drosophila. Proc Natl Acad Sci 75: 5613–5617. 10.1073/pnas.75.11.5613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Hilliker AJ, Roberts PA. 1993. Mapping simple repeated DNA sequences in heterochromatin of Drosophila melanogaster. Trends Genet 9: 379. 10.1016/0168-9525(93)90135-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CD, Brennecke J, Dus M, Stark A, McCombie WR, Sachidanandam R, Hannon GJ. 2009. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 137: 522–535. 10.1016/j.cell.2009.03.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez-Lago M, Bergman CM, de Pablos B, Tracey A, Whitehead SL, Villasante A. 2011. A large palindrome with interchromosomal gene duplications in the pericentromeric region of the D. melanogaster Y chromosome. Mol Biol Evol 28: 1967–1971. 10.1093/molbev/msr034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills WK, Lee YCG, Kochendoerfer AM, Dunleavy EM, Karpen GH. 2019. RNA from a simple-tandem repeat is required for sperm maturation and male fertility in Drosophila melanogaster. Elife 8: e48940. 10.7554/eLife.48940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn F, Sienski G, Handler D, Brennecke J. 2014. The Rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell 157: 1364–1379. 10.1016/j.cell.2014.04.031 [DOI] [PubMed] [Google Scholar]
- Mohn F, Handler D, Brennecke J. 2015. piRNA-guided slicing specifies transcripts for Zucchini-dependent, phased piRNA biogenesis. Science 348: 812–817. 10.1126/science.aaa1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC. 2007. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13: 1911–1922. 10.1261/rna.744307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliphant TE. 2015. Guide to NumPy. Continuum Press, Austin, TX. [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O'Carroll D, Zamore PD. 2019. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet 20: 89–108. 10.1038/s41576-018-0073-3 [DOI] [PubMed] [Google Scholar]
- Özata DM, Yu T, Mou H, Gainetdinov I, Colpan C, Cecchini K, Kaymaz Y, Wu P-H, Fan K, Kucukural A, et al. 2020. Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat Ecol Evol 4: 156–168. 10.1038/s41559-019-1065-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A, Wehr K, Schüpbach T. 2007. Zucchini and squash encode two putative nucleases required for rasiRNA production in the Drosophila germline. Dev Cell 12: 851–862. 10.1016/j.devcel.2007.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative Genomics Viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Ríos P, Simonelig M. 2018. piRNAs and PIWI proteins: regulators of gene expression in development and stem cells. Development 145: dev161786. 10.1242/dev.161786 [DOI] [PubMed] [Google Scholar]
- Rojas-Ríos P, Chartier A, Pierson S, Simonelig M. 2017. Aubergine and piRNAs promote germline stem cell self-renewal by repressing the proto-oncogene Cbl. EMBO J 36: 3194–3211. 10.15252/embj.201797259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Aravin AA, Zelentsova ES, Schostak NG, Sachidanandam R, McCombie WR, Hannon GJ, Evgen'ev MB. 2010. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA 16: 1634–1645. 10.1261/rna.2217810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozhkov NV, Hammell M, Hannon GJ. 2013. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev 27: 400–412. 10.1101/gad.209767.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Leandre B, Capy P, Hua-Van A, Filée J. 2020. piRNA and transposon dynamics in Drosophila: a female story. Genome Biol Evol 12: 931–947. 10.1093/gbe/evaa094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Nishida KM, Mori T, Kawamura Y, Miyoshi K, Nagami T, Siomi H, Siomi MC. 2006. Specific association of Piwi with rasiRNAs derived from retrotransposon and heterochromatic regions in the Drosophila genome. Genes Dev 20: 2214–2222. 10.1101/gad.1454806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC. 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299. 10.1038/nature08501 [DOI] [PubMed] [Google Scholar]
- Sandler L, Hiraizumi Y, Sandler I. 1959. Meiotic drive in natural populations of Drosophila melanogaster. I. The cytogenetic basis of segregation-distortion. Genetics 44: 233–250. 10.1093/genetics/44.2.233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat Methods 9: 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Palumbo G, Bozzetti MP, Tritto P, Pimpinelli S, Schäfer U. 1999. Genetic and molecular characterization of sting, a gene involved in crystal formation and meiotic drive in the male germ line of Drosophila melanogaster. Genetics 151: 749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienski G, Dönertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell 151: 964–980. 10.1016/j.cell.2012.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton W, Das S, McKee BD. 2001. A role of the Drosophila homeless gene in repression of Stellate in male meiosis. Chromosoma 110: 228–240. 10.1007/s004120100136 [DOI] [PubMed] [Google Scholar]
- Tang W, Seth M, Tu S, Shen E-Z, Li Q, Shirayama M, Weng Z, Mello CC. 2018. A sex chromosome piRNA promotes robust dosage compensation and sex determination in C. elegans. Dev Cell 44: 762–770.e3. 10.1016/j.devcel.2018.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics 14: 178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurmond J, Goodman JL, Strelets VB, Attrill H, Gramates LS, Marygold SJ, Matthews BB, Millburn G, Antonazzo G, Trovisco V, et al. 2019. FlyBase 2.0: the next generation. Nucleic Acids Res 47: D759–D765. 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD. 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324. 10.1126/science.1129333 [DOI] [PubMed] [Google Scholar]
- VanderPlas J, Granger B, Heer J, Moritz D, Wongsuphasawat K, Satyanarayan A, Lees E, Timofeev I, Welsh B, Sievert S. 2018. Altair: interactive statistical visualizations for Python. JOSS 3: 1057. 10.21105/joss.01057 [DOI] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. 2002. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837. 10.1126/science.1074973 [DOI] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. 2012. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol 19: 773–781. 10.1038/nsmb.2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SH, Elgin SCR. 2011. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci 108: 21164–21169. 10.1073/pnas.1107892109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Yoshikawa M, Han BW, Izumi N, Tomari Y, Weng Z, Zamore PD. 2014. The initial uridine of primary piRNAs does not create the tenth adenine that is the hallmark of secondary piRNAs. Mol Cell 56: 708–716. 10.1016/j.molcel.2014.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen J, Duan H, Bejarano F, Okamura K, Fabian L, Brill JA, Bortolamiol-Becet D, Martin R, Ruby JG, Lai EC. 2015. Adaptive regulation of testis gene expression and control of male fertility by the Drosophila hairpin RNA pathway. Mol Cell 57: 165–178. 10.1016/j.molcel.2014.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Lyttle TW, Wu ML, Lin GF. 1988. Association between a satellite DNA sequence and the responder of segregation distorter in D. melanogaster. Cell 54: 179–189. 10.1016/0092-8674(88)90550-8 [DOI] [PubMed] [Google Scholar]
- Wu P-H, Fu Y, Cecchini K, Özata DM, Arif A, Yu T, Colpan C, Gainetdinov I, Weng Z, Zamore PD. 2020. The evolutionarily conserved piRNA-producing locus pi6 is required for male mouse fertility. Nat Genet 52: 728–739. 10.1038/s41588-020-0657-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, et al. 2019. Ensembl 2020. Nucleic Acids Res 48: D682–D688. 10.1093/nar/gkz966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Knowles BA, Goldstein LS, Hawley RS. 1990. A kinesin-like protein required for distributive chromosome segregation in Drosophila. Cell 62: 1053–1062. 10.1016/0092-8674(90)90383-P [DOI] [PubMed] [Google Scholar]
- Zhang Z, Wang J, Schultz N, Zhang F, Parhad SS, Tu S, Vreven T, Zamore PD, Weng Z, Theurkauf WE. 2014. The HP1 homolog Rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 157: 1353–1363. 10.1016/j.cell.2014.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data can be accessed via NCBI SRA with the following accession numbers: PRJNA646006 (rhi), PRJNA646216 (aub, zuc, and spn-E), and PRJNA719671 (degradome).