Abstract
Chromatin looping plays an important role in genome regulation. However, because ChIP-seq and loop-resolution Hi-C (DNA-DNA proximity ligation) are extremely challenging in mammalian early embryos, the developmental stage at which cohesin-mediated loops form remains unknown. Here, we study early development in medaka (the Japanese killifish, Oryzias latipes) at 12 time points before, during, and after gastrulation (the onset of cell differentiation) and characterize transcription, protein binding, and genome architecture. We find that gastrulation is associated with drastic changes in genome architecture, including the formation of the first loops between sites bound by the insulator protein CTCF and a large increase in the size of contact domains. In contrast, the binding of the CTCF is fixed throughout embryogenesis. Loops form long after genome-wide transcriptional activation, and long after domain formation seen in mouse embryos. These results suggest that, although loops may play a role in differentiation, they are not required for zygotic transcription. When we repeated our experiments in zebrafish, loops did not emerge until gastrulation, that is, well after zygotic genome activation. We observe that loop positions are highly conserved in synteny blocks of medaka and zebrafish, indicating that the 3D genome architecture has been maintained for >110–200 million years of evolution.
Genomes are partitioned into thousands of contact domains or topologically associated domains (TADs): intervals that exhibit enhanced contact frequency within themselves (Dixon et al. 2012; Nora et al. 2012; Rao et al. 2014). Loops often demarcate contact domains by physical tethering of a pair of loci, which are mediated by the insulator protein CTCF and the ring-shaped cohesin complex (Nora et al. 2017; Rao et al. 2017). Compartment domains are formed by cosegregation of chromatin intervals with similar histone marks, which is a phenomenon more generally known as compartmentalization (Lieberman-Aiden et al. 2009; Rao et al. 2014). Genome architecture plays a critical role in gene regulation, but how genome architecture is established during vertebrate development remains poorly understood. Embryogenesis itself consists of a highly orchestrated cascade of genetically encoded events, beginning with the fusion of the haploid male and female pronuclei. At first, the zygotic genome is transcriptionally silent, and rapid and synchronous cell cycles proceed under the control of maternally provided factors (Jukam et al. 2017). During this process, epigenetic modifications and chromatin accessibility are globally reprogrammed (Lee et al. 2014). Next, zygotic genome activation (ZGA), simultaneous transcription of thousands of zygotic genes, occurs, and development proceeds under the control of zygotic gene products (Tadros and Lipshitz 2009; Lee et al. 2014; Onichtchouk and Driever 2016; Wragg and Müller 2016). Subsequently, gastrulation begins, as the cells in the single-layered blastula differentiate into three germ layers, and rudimentary organs begin to form (Wolpert et al. 2006). Thus, by the end of gastrulation, the vertebrate body plan is established.
Although extensive progress has been made by studying nuclear architecture during embryogenesis in mammals (Du et al. 2017; Flyamer et al. 2017; Gassler et al. 2017; Ke et al. 2017; Chen et al. 2019; Zheng and Xie 2019; Collombet et al. 2020), studies of local folding features are greatly constrained by the fact that both ChIP-seq and loop-resolution Hi-C are extremely difficult to perform in mammals. This is because an adequate number of cells is difficult to obtain: mammals tend to have a small number of in vivo embryos, which must be extracted by sacrificing the mouse and because much of development takes place when an embryo contains only a handful of cells. This has prevented study of loop formation and protein binding during mammalian development. Fish, in particular, medaka (the Japanese killifish, Oryzias latipes), have many advantages for embryological and genome studies because they lay large numbers of eggs that are easy to collect and which develop ex vivo (Takeda and Shimada 2010). Medaka also benefit from a compact genome (roughly 800 Mb) with an excellent assembly, comparable in quality with that of mouse (Ichikawa et al. 2017). This greatly facilitates the generation of high-resolution contact maps, because the resolution varies inversely with the genome size. Here, we investigated genome architecture, protein binding, and transcriptional activation before, during, and after gastrulation in medaka. We also examined whether the late emergence of loops and loop positions are conserved across teleost fishes by repeating our experiments in zebrafish, which diverged from medaka 110–200 million years ago (Furutani-Seiki and Wittbrodt 2004; Takeda and Shimada 2010).
Results
The genome of mature medaka fibroblasts exhibits compartments and loop domains
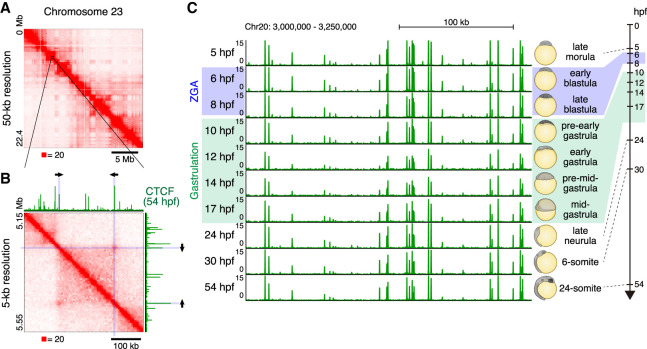
We began by examining the 3D genome structure of a medaka embryonic fibroblast cell line established from an embryo 4 day postfertilization (dpf; referred to as mature fibroblasts) using in situ Hi-C (Rao et al. 2014), generating 952 million read pairs. The Hi-C data was processed using Juicer (Durand et al. 2016), and the resulting matrices exhibited a typical plaid pattern associated with genome compartmentalization (Fig. 1A; Supplemental Fig. S1A). Specifically, [1] most the loci could be assigned to one of the two compartments, such that all loci in the same compartment exhibited an enhanced contact frequency with one another; [2] contiguous chromatin intervals in the same compartment manifest as bright squares along the diagonal of the contact map, that is, domains in which all pairs of loci exhibit an enhanced contact frequency with one another; and [3] comparison of the matrices with RNA-seq data confirmed that loci in one of the compartments are associated with active chromatin (A compartment), whereas loci in the other compartment are associated with inactive chromatin (B compartment) (Supplemental Fig. S1A). Such compartmentalization has been observed in many studies and species (Lieberman-Aiden et al. 2009; Dixon et al. 2012; Sexton et al. 2012; Kaaij et al. 2018).
Figure 1.
3D genome structure of medaka fibroblast cells and CTCF positioning throughout medaka development. (A) Hi-C contact map of medaka fibroblast cells. Normalized observed Hi-C contacts are shown as a heat map. Whole Chromosome 23 at 50-kb resolution shows plaid pattern. (B) Zoomed view of Chromosome 23 Hi-C contact map at 5-kb resolution and CTCF ChIP-seq track from 54-hpf embryos are shown. Arrows indicate the orientation of CTCF binding motifs within CTCF ChIP-seq peak at the loop anchor position. (C) CTCF positioning is stable throughout medaka development; representative view of CTCF ChIP-seq tracks across the developmental stages.
At higher resolutions, we observed numerous peaks in contact frequency indicating that a pair of anchor loci exhibit enhanced contact frequency with one another as compared to their neighbors along the contour of the chromosome (Fig. 1B; Supplemental Fig. S1B). These peaks are consistent with the presence of point-to-point loops between pairs of anchor loci and often demarcate contact domains, or topologically associated domains. We also found that these peaks were closely associated with pairs of loci bound by the insulator protein CTCF, with the corresponding CTCF-bound motifs in the convergent orientation (i.e., pointing at one another) (Fig. 1B; Supplemental Fig. S1C). The loop domains we observed in medaka closely resemble those seen in numerous studies in human and mouse. They are also consistent with a recent study in zebrafish which found that CTCF motifs in the convergent orientation were enriched in the vicinity of contact domains (Kaaij et al. 2018). Taken together, these observations demonstrate that all of the features seen in mammalian Hi-C maps are also seen in medaka.
CTCF positioning is largely fixed throughout medaka embryogenesis
Next, we performed CTCF ChIP-seq experiments over a longitudinal time-course spanning 10 stages of medaka development: four time points before gastrulation, three during gastrulation, and three thereafter. Specifically, we sampled at stage 9 (late morula [256–512 cells], 5 hours post fertilization [hpf]); stage 10 (early blastula [1 k cells], 6 hpf); stage 11 (late blastula [2–4 k cells], 8 hpf); stage 12 (pre-early gastrula, 10 hpf) stage 13 (early gastrula, 12 hpf); stage 14 (pre-mid-gastrula, 14 hpf); stage 15 (mid-gastrula, 17hpf); stage 18 (late neurula, 24 hpf); stage 21 (6-somite, 30 hpf); and stage 27 (24-somite, 54 hpf). Crucially, this time-course is difficult in mammals, where the collection of a sufficient number of cells is prohibitive. No such data are available in zebrafish so far. We found that CTCF peak positions were extremely stable across all developmental stages examined (Fig. 1C; Supplemental Fig. S1D). Indeed, most of the CTCF motifs in the convergent orientation observed at loop anchor loci in mature fibroblasts were bound by CTCF as early as 5 hpf (Supplemental Fig. S1E).
To further test the stable binding of CTCF during development, we performed quantitative ChIP-seq using spike-in chromatin (Bonhoure et al. 2014; Li et al. 2014; Orlando et al. 2014). We prepared chromatin from different medaka strains and mixed before ChIP; the two polymorphic medaka strains, d-rR and Hamochi, were used as experimental and spike-in chromatin, respectively. The spike-in normalized CTCF ChIP-seq signals of 6 and 30 hpf showed comparable levels (Supplemental Fig. S2A,B), suggesting that the CTCF is indeed binding to chromatin from the early stages. The immunofluorescence staining using CTCF antibody further supported the result (Supplemental Fig. S2C). These results imply that, although CTCF may play a role in early genome architecture and regulation, changes in the positioning of CTCF are not a principal driver of the transcriptional and architectural changes seen during medaka embryonic development (see below).
Generation of loop-resolution chromatin contact maps for 12 time points spanning medaka development
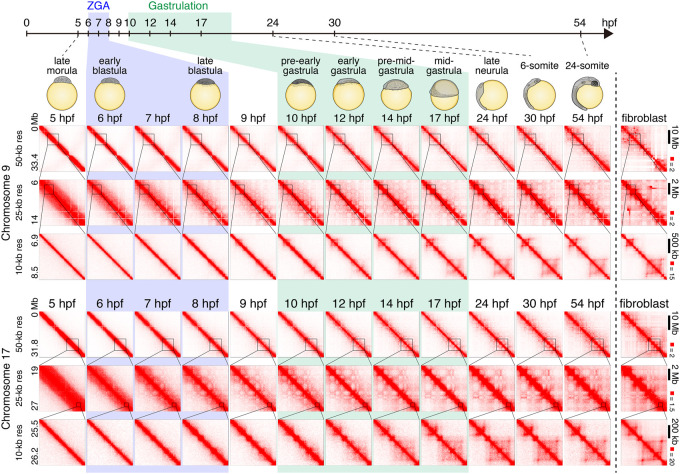
We then examined the dynamics of genome architecture. For this, we performed in situ Hi-C (Rao et al. 2014) at 12 distinct stages. These include the 10 stages examined for CTCF mapping, as well as two additional intermediate stages (stage 10.5, 7 hpf; and stage 11.5, 9 hpf). The proportion of interphase cells were also assessed at early stages by microscope observation (Supplemental Fig. S3A,B). We generated two replicates for all developmental stages, and the data showed high reproducibility between the replicates (Supplemental Fig. S4A). The replicates were combined for the further analyses to obtain loop-resolution (5–10 kb) in situ Hi-C maps for every time point studied (Fig. 2; Supplemental Fig. S4B). We found that neither compartments nor domains were present in the 5-hpf sample. Instead, the heat map was a bright, undifferentiated diagonal, indicating the absence of any local features of genome architecture (Fig. 2). Starting at 7 hpf—during ZGA—we observed the emergence of a plaid pattern, consistent with the presence of two compartments. This pattern became increasingly intense at each time point as development proceeded (Fig. 2; Supplemental Figs. S5A, S6). When we profiled open chromatin during ZGA and the surrounding time points (5 hpf–10 hpf) using ATAC-seq, as well as ChIP-seq using antibodies for H3K27 acetylation, we found that these features also appear around 7 hpf and that their positioning was strongly associated with one of the two long-range patterns (A compartments) seen in the Hi-C map (Supplemental Fig. S5A,B). Overall, our findings are consistent with previous studies showing that the emergence of compartmentalization occurs after fertilization in mice and Drosophila (Du et al. 2017; Hug et al. 2017; Ke et al. 2017), and this accompanies the formation of open chromatin and the activation of the zygotic genome in medaka.
Figure 2.
Chromatin contact dynamics across medaka development. Representative examples of Hi-C contact matrices from 12 time points across medaka development and fibroblast cells. Examples from Chromosomes 9 and 17 are shown for indicated resolutions. Normalized observed Hi-C contacts are shown as a heat map. Note that Chromosome 9 in fibroblast cells has chromosomal rearrangements.
Small contact domains emerge during zygotic genome activation and increase greatly in size during gastrulation
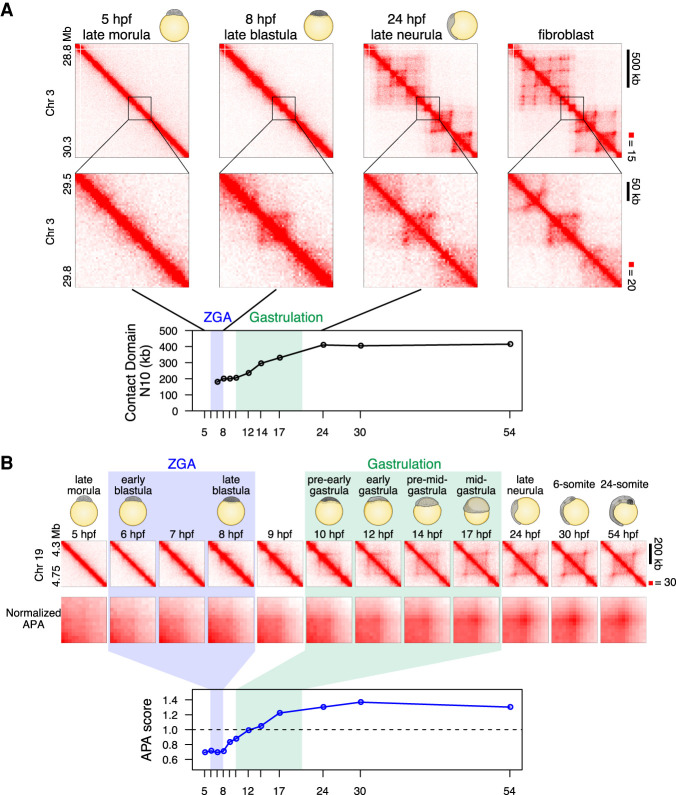
Next, we looked for contact domains (or TADs). The contact domains were identified using the Arrowhead algorithm of Juicer (Rao et al. 2014; Durand et al. 2016). We found that contact domains also began to be visible in our data at roughly 7–10 hpf (Supplemental Fig. S7A,B). These domains were mostly small and relatively ambiguous, as they were close to the diagonal, but a 3D-FISH experiment at the selected loci confirmed the increase in contact bias in interphase cells at this stage (Supplemental Fig. S7C). However, the size of domains increased greatly during gastrulation, and the domains at 24 hpf (late neurula) were much larger than those seen at 8 hpf (late blastula) (Fig. 3A; Supplemental Fig. S8A). These large domains were not detected at early stages, at the level of Hi-C contact enrichment. To quantify the genome-wide change in domain size, we calculated an N10 statistic for the domain size. This was calculated by examining all regions of the genome that are covered by a domain and determining the minimum domain size required to cover 10% of these regions; 10% of the entire genome covered by contact domains is covered by contact domains equal to or larger than this N10 value (thus, the remaining 90% are covered by domains that are smaller than the N10 value). Thus, the N10 statistic reflects the size of the largest contact domains observed. The domain N10 at 7 hpf, when we first observed domains, was 180 kb. This value did not change until the early gastrulation, and the domain N10 grew during gastrulation to 410 kb (24 hpf) (Fig. 3A). Notably, the number of contact domains shared with fibroblasts monotonically increased during development (Supplemental Fig. S8B), and the N10 in mature fibroblasts was 465 kb, suggesting that the large contact domains associated with mature cells emerge during, but not before, gastrulation.
Figure 3.
Establishment of loop domains during medaka development. (A) An example of establishment of large loop and small loop, and contact domain N10 statistics over stages are shown. Normalized observed Hi-C contacts are shown as a heat map at 10-kb (upper portion) and 5-kb (middle portion) resolution. Higher N10 values mean that larger contact domains cover longer intervals of genome (see Methods for detail). (B) Loops are established during gastrulation. Representative Hi-C contact map (normalized observed Hi-C contacts at 10-kb resolution) of a loop domain is shown in upper portion. Normalized APA plots in middle portion show that loops become evident during gastrulation. APA scores (loop signal enrichment compared to lower-right signal of APA plot) across the developmental stages are shown in lower portion. Values greater than 1 indicate the presence of loops. Hi-C contacts around loops identified in fibroblast cells were aggregated for each stage.
Loops emerge during gastrulation
We did not observe the majority of loops in the data until gastrulation (Fig. 3A,B; Supplemental Fig. S8A). Because automatic detection of loops suffered from many false positives due to higher noise near the diagonal, we filtered them out and counted the remaining reliable loops, that is, loops associated with contact domains (Rao et al. 2014) and 100 kb or larger in size (see Methods). Thus, short-range (<100 kb) loops, if any, were not subjected to further study. This method detected only two loops at 5 hpf (late morula) and 16 loops at 8 hpf (late blastula), whereas the number increased during gastrulation and to 405 at 24 hpf (Supplemental Fig. S8C).
To rule out the possibility that loops were present at earlier time points but were too weak to be detected individually, we performed aggregate peak analysis (APA) (Durand et al. 2016) to examine whether the 1869 loops (which are 100 kb or larger) observed in mature fibroblasts were reflected in maps from earlier time points. APA aggregates the Hi-C signal from a set of peaks: the center of the APA plot indicates the aggregate signal for the peak set, whereas adjacent values indicate the result of translating the peak set in various directions. Thus, an enhanced contact frequency in the central pixel of an APA plot indicates the presence of a signal from the loop list, in aggregate. We found no enrichment in the central pixel until 17 hpf (mid-gastrula) (Fig. 3B). These results indicate that, despite consistent patterns of CTCF binding throughout development, point-to-point loops between CTCF-bound sites do not form until 17 hpf.
We noticed that, even for the regions where large-size domains emerge, domain boundaries are already detectable near the diagonal before the loops emerge (6 hpf) (Fig. 3B). To examine this genome-wide, we aggregated Hi-C signals across all loop anchors identified in fibroblast cells. We confirmed that these loop domain boundaries are indeed detectable from the blastula stage and gradually became evident at later stages (Supplemental Fig. S8D), which is consistent with the analyses of insulation score (Supplemental Fig. S8E). Taken together, although loops are largely absent until gastrulation, local chromatin interaction is not uniform at blastula stage; that is, contact domain boundaries and small domains are beginning to form.
Blocking of ZGA has been shown to inhibit contact domain establishment in human embryos but not in mouse or Drosophila (Du et al. 2017; Hug et al. 2017; Ke et al. 2017; Chen et al. 2019). We examined whether the emergence of small contact domains and domain boundaries require ZGA in medaka. Injection of α-amanitin into medaka embryos almost completely blocked zygotic transcription (Supplemental Fig. S9A,B). We compared the Hi-C maps between uninjected and α-amanitin-injected embryos at 10 hpf and found that small contact domains and boundaries had been established in α-amanitin-injected embryos (Supplemental Fig. S9C–E). These results suggest that the formation of small contact domains or domain boundaries does not rely on zygotic transcription in medaka.
Extensive change in the genome architecture might be regulated by the activity of the cohesin complex. Cohesin proteins are essential for cell division, and thus they exist as maternal factors from the beginning of development, well before the emergence of domain structures (Supplemental Fig. S10A,B). However, cohesin and cohesin-regulators are dynamically regulated at the level of transcription during development (Supplemental Fig. S10A), and these proteins may have a role in the genome structure changes during development. Taken together, these observations give an insight into the mechanism of CTCF-mediated loop domain establishment during development based on a loop extrusion model (see Discussion).
Loop anchors colocalize with genes that are expressed late in development
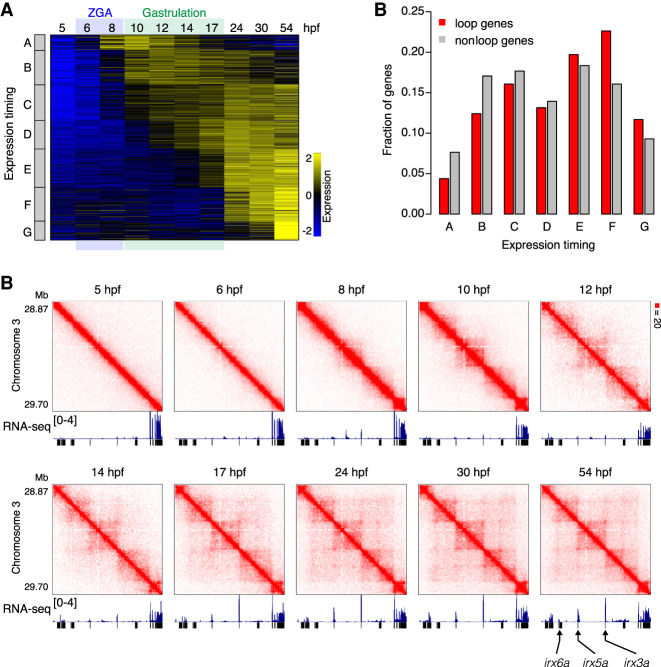
Our findings imply that zygotic genome activation and the onset of gastrulation do not require the formation of point-to-point loops between CTCF-bound sites. However, CTCF-mediated loops have been associated with gene transcription in numerous studies (Rao et al. 2014; Lupiáñez et al. 2015; Flavahan et al. 2016; Ji et al. 2016). To explore this relationship further, we performed RNA-seq for each of the 10 time points. Thus, we created a profile showing expression of every gene over time. To eliminate any confound from maternal transcripts that do not arise from zygotic transcription, we excluded all genes for which mRNA is detectable at 5 hpf. We then compared the expression timing of genes that were associated with loops (i.e., the TSS was in the same 5-kb bin as a CTCF loop anchor) in mature fibroblasts. We found that loop-associated genes (N = 137) tend to be expressed later in development than genes that are not associated with loops (N = 3101). We first classified genes into two categories, early-expressed genes and late-expressed genes (Supplemental Fig. S11A), and found that the loop genes are slightly more enriched in the class of late-expressed genes (P = 0.0145, Fisher's exact test) (Supplemental Fig. S11B). Furthermore, when we classified genes into seven classes depending on the timing of gene transcription, loop genes tended to be enriched in the classes of genes that are activated from mid-gastrula or later after gastrulation. (Fig. 4A,B). One example is the irx3a/irx5a/irx6a locus; the three genes are activated from mid-gastrula stage, and the timing coincides with the emergence of loops (Fig. 4C). It has been reported that knockout of CTCF in zebrafish embryos disrupts both the interaction between irx3a and irx5a and their expression pattern (Tena et al. 2011). These data, together with the observation that loops are generally established at mid-gastrula, suggest that, although loops are not required for transcription of the majority of genes, loops have a certain contribution to transcription of genes that is activated after or during gastrulation.
Figure 4.
CTCF-mediated loops are associated with late-activating genes. (A) Classification of zygotically expressed genes into seven (A to G) groups by expression dynamics. (B) Fraction of genes for each class is shown for loop genes and nonloop genes. Loop genes are enriched with genes activated from mid-gastrula (17 hpf) or later (gene class E, F, and G). (C) Examples of loop-associating late-activating genes. Normalized observed Hi-C contacts as a heat map at 10-kb resolution and RNA-seq tracks are shown. Loops emerge from 17 hpf (mid-gastrula) between irx genes, and transcription is activated at this stage.
Loop establishment during zebrafish development
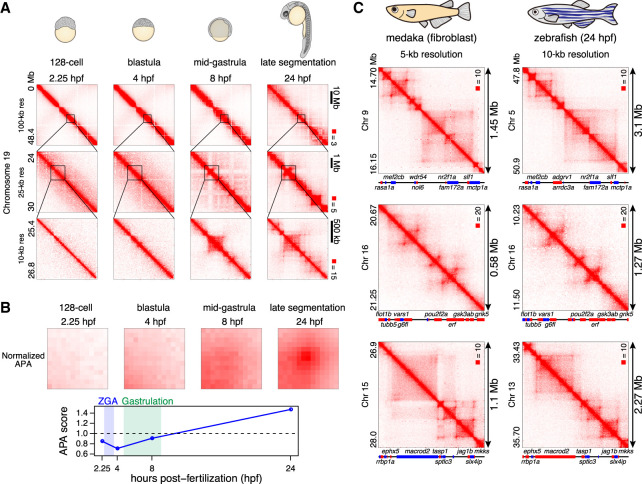
To examine whether the late establishment of loops during development is conserved among teleost species, we generated Hi-C maps of zebrafish embryos at selected four developmental stages; a stage before ZGA (128-cell stage, 2.25 hpf), stage just after ZGA (sphere stage [blastula], 4 hpf), mid-gastrula (75% epiboly stage, 8 hpf), and late segmentation stage (24 hpf). As a result, both compartmentalization and contact domains were not detectable before mid-gastrula (8 hpf), and strong compartmentalization and loops were observed at the late segmentation stage (24 hpf) (Fig. 5A; Supplemental Fig. S12). Similar to medaka embryos, the contact domains became detectable from gastrulation (8 hpf) (Fig. 5A). APA analyses confirmed that loops are established after mid-gastrula (8 hpf) (Fig. 5B); stable loops were not detectable at the mid-gastrula stage in zebrafish embryos (8 hpf), while they were formed by the late segmentation stage (24 hpf). Taken together, these results suggest that, in zebrafish also, loops and the majority of contact domains do not exist before gastrulation but are established during or after gastrulation. Thus, loops are not required for ZGA in zebrafish.
Figure 5.
Loop establishment in zebrafish embryos. (A) Representative examples of Hi-C contact matrices of zebrafish embryos. Normalized observed Hi-C contacts are shown as a heat map at indicated resolutions. (B) Normalized APA plots are shown in the upper portion, and APA scores are shown in the lower portion. Values greater than 1 indicate the presence of loops. Hi-C contacts around loops identified in 24-hpf embryos were aggregated for each stage. (C) Examples of conserved 3D structure in synteny blocks. Hi-C maps of medaka fibroblast (5-kb resolution), and 24-hpf zebrafish embryos (10-kb resolution) are shown. The orientations of genes is indicated by different colors (forward: red; reverse: blue). Gene names are indicated for selected genes because of the space limitation.
3D genome structure is conserved in synteny blocks of medaka and zebrafish
It has been reported that loops are frequently conserved between human and mouse (Rao et al. 2014) after separation of 80 myr (Mouse Genome Sequencing Consortium 2002). Medaka and zebrafish are two distantly related teleosts separated by an evolutionary distance of 115–200 myr, and unlike mouse and human which have the similar genome size, the genome size of zebrafish (1.7 Gb) is two times larger than medaka (800 Mb) with considerable chromosome rearrangements (Nakatani and Mclysaght 2017). With our loop-resolution Hi-C maps, we asked whether loops are still conserved between such a distant species. As synteny blocks between medaka and zebrafish tend to be small with limited sequence similarity, it was difficult to evaluate genome-wide conservation of 3D structures. Instead, we searched for relatively long synteny blocks and compared their Hi-C maps (Supplemental Fig. S14; see Methods). Among 11 long synteny blocks we examined, seven regions were found to exhibit quite similar Hi-C maps (Fig. 5C; Supplemental Fig. S13). Although orthologous regions are about twice the size of medaka in zebrafish, the size of their loops were in proportion to the genome size, and the relative boundary positions were conserved. This suggests that genome topology is conserved across the teleost species regardless of the genome size, in particular within synteny blocks.
Discussion
This study provides a detailed overview of the establishment and maturation of genome architecture during embryogenesis, profiling the distribution of CTCF and the emergence of loops and domains before, during, and after gastrulation. We find that gastrulation is a critical event associated with numerous changes in genome architecture of medaka. Although we find that small contact domains emerge during ZGA (consistent with earlier studies), much larger ones emerge during gastrulation, matching the sizes seen in mature cells. We also find that CTCF-CTCF loops first form during gastrulation. Furthermore, late emergence of mature loop structure was conserved among distantly related teleost species (medaka and zebrafish). The embryos we used in this study consist of multiple cell types especially after gastrulation, and some of the rare cell type–specific loops might have been missed in the present study. However, a large fraction of loops are known to be conserved among cell types (Rao et al. 2014), and thus our study describes the dynamics of most common loops.
Our study showed that, before ZGA, chromatin does not have any domain structures both in medaka and zebrafish. However, this appears to conflict with the main results of a recent study of nuclear architecture during zebrafish development (Kaaij et al. 2018) that the genome is highly structured at 2.25 h, followed by its loss and re-establishment throughout early embryogenesis. Our accompanying paper of Wike et al. (2021) confirmed our result of zebrafish and suggested that the discrepancy between ours and that of Kaaij et al. (2018) is due to the different timing of dechorionation of embryos in the experimental procedure. The teleost embryos are covered by chorion, which needs to be removed at some point before enzymatic reaction in the Hi-C experiment. A certain amount of somatic cells could be attached to the chorion, and this serves as a source of contamination of highly structured genomes. The effect of this is that somatic cells will become greater in early stage embryos when the cell number of an embryo is small, such as in the 128-cell stage (Wike et al. 2021). In this study, the chorion was removed completely at the one-cell stage, well before cross-linking of cells, to prevent the contamination. As seen in other previous studies using mice, human, and Drosophila, the gradual emergence of domain structure during early embryogenesis is a fundamental feature of animal chromatin.
We noticed that the timing of emergence of compartments and loops in medaka is a little earlier than in zebrafish. Medaka compartments begin to form during the blastula stage simultaneously with emergence of accessible chromatin and H3K27ac peaks (Supplemental Fig. S5A), but zebrafish compartmentalization starts after the blastula stage (Supplemental Fig. S12; Wike et al. 2021). In medaka embryos, loops are already formed at the mid-gastrula stage (Fig. 3B), but they form later in zebrafish, based on our APA analyses (Fig. 5B). The shorter cell-cycle length of zebrafish embryos (30 min in medaka and 15 min in zebrafish during cleavage stage) might partly account for the late emergence of these structures, as they require certain amount of time to be established after mitosis (Abramo et al. 2019; Zhang et al. 2019). However, higher temporal resolution investigation is required to examine the precise timing of loop emergence in zebrafish embryos.
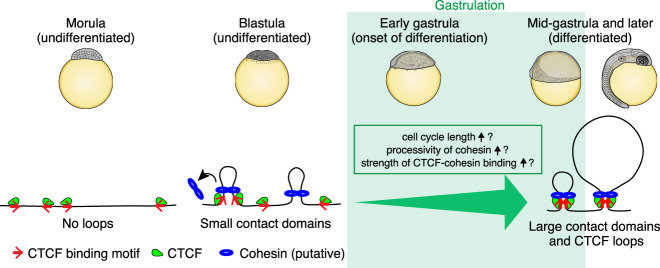
Why do loops and domains suddenly form during gastrulation in medaka? The simplest model is that the distribution of CTCF changes, because its binding sites demarcate both loops and domains. Indeed, a recent study has shown that, in human early embryos, CTCF is expressed at low levels before ZGA and that high expression of CTCF induced at ZGA is necessary for the establishment of TADs (Chen et al. 2019). Using medaka embryos that can routinely procure adequate numbers of cells across a developmental timeline, we directly investigated the distribution of CTCF on chromatin by ChIP-seq. We found that the distribution of CTCF is largely unchanged during development. Thus, in contrast to human embryos, CTCF is bound at early stages of medaka embryos, but loops do not form, suggesting that additional factors are involved in loop establishment.
It is interesting to consider these findings in light of the loop extrusion model (Sanborn et al. 2015; Fudenberg et al. 2016; Davidson et al. 2019; Kim et al. 2019). In this model, loop domains form when a cohesin-based extrusion complex comprising two physically tethered subunits lands on chromatin, and the two subunits slide in opposite directions until they arrive at an inward-pointing CTCF site. The model suggests that, when cohesin is present, loops between CTCF sites should form spontaneously. This correspondence has been seen in all prior studies in which loop-resolution Hi-C and CTCF ChIP-seq data were generated. Thus, one possibility that accounts for the increase in domain size during gastrulation is as a result of an increase in the processivity of the loop extrusion complex. Alternatively, an increase in the cell cycle length may allow the loop extrusion complex to process sufficiently. Indeed, cell division occurs every 30 min until the cleavage stage and the cycle becomes longer from the blastula stage in medaka, but loops require several hours to be reestablished after mitosis in mammalian cells (Abramo et al. 2019). Furthermore, the emergence of CTCF-CTCF loops during gastrulation might be driven by an increase in the strength of CTCF-cohesin binding (Fig. 6). The factors that might be responsible for such changes will be an important topic for future studies.
Figure 6.
A model of CTCF-mediated loop establishment during medaka development. At the morula stage, loops and contact domains do not exist, but CTCF is already bound to the future loop anchors. At the blastula stage, zygotic genome activation occurs, and small contact domains start to emerge by loop extrusion of cohesin. The boundaries of large contact domains are also detectable at this stage, but stable loops are not formed. During gastrulation, large contact domains and stable loops emerge, and this could be explained by an increase in the processivity of cohesin or the strength of CTCF-cohesin binding, or prolonged cell cycle length.
The late emergence of mature domain structures, including loop domains, suggests that they are not needed for initiating and regulating genome-wide transcription (i.e., ZGA). This finding is reminiscent of previous studies exploring the role of CTCF and cohesin disruption in mammalian cell lines, which have shown that the transcriptional consequences of such treatments are modest (Nora et al. 2017; Rao et al. 2017). The subsequent emergence of these features instead raises the possibility that loop domains play a role in the process of differentiation and lineage commitment or cell competency. Indeed, a significant increase in the number of long-distance CTCF-mediated loops is associated with differentiation of naive ES cells into neural stem cells (Pękowska et al. 2018).
Finally, our study revealed strong conservation of 3D genome structure at particular synteny blocks between medaka and zebrafish—distantly related species—further suggesting an irreplaceable role of loops in gene regulation.
Methods
Fish strains and sampling
We used the medaka d-rR wild-type strain. For RNA-seq data of hybrid embryos, we used a female HNI-II strain and male d-rR strain and obtained F1 hybrid medaka embryos. Medaka fish were maintained and raised under standard conditions. Embryos were dechorionated and incubated at 28°C. Developmental stages were determined based on a previous study (Iwamatsu 2004). A medaka embryonic fibroblast cell line was previously established in our laboratory. For zebrafish experiments, we used TL2E strain. All experimental procedures and animal care were carried out according to the animal ethics committee of the University of Tokyo (Approval No. 14-05).
In situ Hi-C
In situ Hi-C experiments were performed as previously described (Rao et al. 2014) with some modifications. For all developmental stages, two biological replicates were generated. Embryos were collected at the one-cell stage and dechorionated immediately. Dechorionated embryos were incubated at 28°C until a certain developmental stage, collected in L-15 medium with 15% FBS, and dissociated by pushing through a 21G needle using a syringe. Dissociated embryos or fibroblast cells were collected by centrifuge at 500g for 3 min and cross-linked with 1% formaldehyde in 1.5 mL L-15 medium (15% FBS) for 10 min at room temperature. Cross-linking was quenched by adding 2.5M glycine (125 mM final) and incubating for 5 min at room temperature, then 15 min on ice. Cells were pelleted at 500g for 5 min, and supernatant was removed and stored at −80°C.
Cells were thawed on ice, washed with PBS, resuspended in 250 μL of ice-cold Hi-C lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, 0.2% Igepal CA-630) with 50 μL of protease inhibitors (Sigma-Aldrich P8340), and incubated on ice for 20 min. The lysate was centrifuged at 2500g for 5 min, washed with ice-cold Hi-C lysis buffer, resuspended in 50 μL of 0.5% SDS, and incubated for 10 min at 62°C; 145 μL of water and 25 μL of 10% Triton X-100 were added and incubated for 15 min at 37°C. Twenty-five microliters of NEBuffer 2 and 100 U of MboI were added, and chromatin was digested overnight at 37°C with rotation. MboI was inactivated by incubating at 62°C for 20 min. DNA ends were labeled with biotin by adding 50 μL of fill-in master mix (37.5 μL of 0.4 mM biotin-14-dATP [Life Technologies], 1.5 μL of 10 mM dCTP, 1.5 μL of 10 mM dGTP, 1.5 μL of 10 mM dTTP, and 8 μL of 5 U/μL DNA Polymerase I Large Klenow Fragment [NEB]), and incubated at 37°C for 1.5 h with rotation. Proximal ligation was performed by adding 900 μL of ligation master mix (669 μL of water, 120 μL of 10× NEB T4 DNA ligase buffer, 100 μL of 10% Triton X-100, 6 μL of 10 mg/mL BSA, and 5 μL of 400 U/μL T4 DNA ligase [NEB]), and incubated at room temperature for 4 h. Proteins were degraded by adding 50 μL of 20 mg/mL Proteinase K and 120 μL of 10% SDS and incubated at 55°C for 2 h. Cross-linking was reversed by adding 130 μL of 5M NaCl and incubated at 68°C overnight.
The biotinylated DNA was collected by ethanol precipitation and resuspended in 130 μL of Tris buffer (10 mM Tris-HCl, pH 8.0). DNA was sheared using Covaris S220 with the following parameters: Peak Incident Power: 140, Duty Factor: 10, Cycle per Burst: 200, time: 80 sec. The sheared DNA was size-selected to a size of 100–500 bp and purified using AMPure XP beads (Beckman Coulter). DNA was eluted in 60 μL of Tris buffer.
The sheared DNA was end-repaired by adding 40 μL of master mix (10 μL of 10× NEB T4 DNA ligase buffer with 10 mM dATP, 20 μL of 2.5 mM dNTP, 5 μL of 10 U/μL NEB T4 PNK, 4 μL of 3 U/μL NEB T4 DNA polymerase I, and 1 μL of NEB DNA polymerase I Large Klenow Fragment) and incubated at room temperature for 30 min. End-repaired DNA was then purified using a MinElute PCR purification kit (Qiagen) and eluted in 40 μL of water. DNA was A-tailed by adding 20 μL of master mix (10 μL of 10× NEBuffer 2, 5 μL of 10 mM dATP, and 5 μL of 5 U/μL NEB Klenow exo minus) and incubated at 37°C for 30 min. The sample was then incubated at 65°C for 20 min, and 200 μL of water was added.
One hundred fifty microliters of 10 mg/mL Dynabeads MyOne Streptavidin T1 beads (Life Technologies) were washed with 400 μL of 1× Tween Washing Buffer (1× TWB: 5 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 M NaCl, and 0.05% Tween 20) and resuspended in 300 μL of 2× Binding Buffer (2× BB: 10 mM Tris-HCl, pH 7.5, 1 mM EDTA, and 2 M NaCl). The beads were added to the A-tailed DNA sample and incubated at room temperature for 15 min with rotation. The biotinylated DNA-bound beads were collected with a magnet, and the supernatant was discarded. The beads were washed twice by adding 600 μL of 1× TWB, transferred to a new tube, incubated at 55°C for 2 min on a Thermomixer, and the supernatant was discarded using a magnet. Then, the beads were washed with 100 μL of 1× Quick ligase buffer (NEB), resuspended in 50 μL of 1× Quick ligation buffer; 2 μL of NEB DNA Quick ligase and 1.5 μL of 30 μM indexed Illuminar adapter were added and incubated at room temperature for 15 min. The beads were washed twice by adding 600 μL of 1× TWB, transferred to a new tube, incubated at 55°C for 2 min on a Thermomixer, and the supernatant was discarded using a magnet. The beads were washed with 100 μL of Tris buffer, transferred to a new tube, and resuspended in 50 μL of Tris buffer. The libraries were amplified directly off of the beads with 6–7 cycles of PCR, using KAPA HiFi HotStart ReadyMix (KAPA Biosystems), and DNA was purified using AMPure XP beads. Paired-end sequencing of the Hi-C libraries was performed using the Illumina HiSeq 1500 platform.
ATAC-seq
ATAC-seq was performed as previously described (Buenrostro et al. 2013) with some modifications. Embryos were homogenized in PBS, and cells were harvested by centrifugation at 500g for 5 min. Approximately 5000 cells were used for each experiment. After washing with PBS, cells were resuspended in 500 μL of cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.1% Igepal CA-630), centrifuged for 10 min at 500g, and the supernatant was removed. A tagmentation reaction was performed as described previously (Buenrostro et al. 2013) with a Nextera Sample Preparation kit (Illumina). After tagmented DNA was purified using a MinElute kit (Qiagen), two sequential PCRs were performed to enrich small DNA fragments. First, nine-cycle PCR were performed using indexed primers from a Nextera Index kit (Illumina) and KAPA HiFi HotStart ReadyMix (KAPA Biosystems), and amplified DNA was size-selected to a size of <500 bp using AMPure XP beads (Beckman Coulter). Then, a second seven-cycle PCR was performed using the same primer as the first PCR and purified by AMPure XP beads. Libraries were sequenced using the Illumina HiSeq 1500 platform.
RNA-seq
Embryos were homogenized in 1 mL of Isogen (Nippongene), 200 μL of chloroform was added, and total RNA was isolated using an RNeasy MinElute Cleanup kit (Qiagen). Total RNA from fibroblast cells was extracted using an RNeasy Mini kit (Qiagen). Ribosomal RNA was removed using RiboMinus Eukaryote System v2 (Thermo Fisher Scientific), and RNA-seq libraries were generated using a KAPA Stranded RNA-seq kit (KAPA Biosystems). Libraries were sequenced using the Illumina HiSeq 1500 platform.
ChIP-seq
ChIP was performed as previously described with modifications (Nakamura et al. 2014). Dechorionated embryos were dissociated using a 21G needle in PBS containing 20 mM Na-butyrate, complete protease inhibitor (Roche), and 1 mM PMSF, and fixed with 1% formaldehyde for 8 min at room temperature, then quenched by adding glycine (200 mM final) and incubating on ice for 5 min. After washing with PBS, cell pellets were stored at −80°C. Cells were thawed on ice, suspended in lysis buffer (50 mM Tris-HCl, pH 8.0, 10 mM EDTA, 1% SDS, 20 mM Na-butyrate, complete protease inhibitors, and 1 mM PMSF), sonicated 10 times using a sonifier (Branson) at power seven, and centrifuged for collecting chromatin lysates. The chromatin lysates were diluted with RIPA ChIP buffer (10 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, 0.2% sodium deoxycholate, 20 mM Na-butyrate, complete protease inhibitors, and 1 mM PMSF) and rotated with an antibody/protein A Dynabeads complex overnight at 4°C. Immunoprecipitated samples were washed three times with RIPA buffer (10 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% SDS, and 0.2% sodium deoxycholate) and once with TE buffer, followed by elution with lysis buffer at 65°C overnight. Eluted samples were treated with RNase A for 2 h at 37°C, Proteinase K for 2 h at 55°C, and DNA was purified by phenol/chloroform extraction and ethanol precipitation. Input DNA was simultaneously treated from the elution step. ChIP-seq libraries were generated using a KAPA Hyper Prep kit (KAPA Biosystems). Antibodies used in this study are as follows; H3K27ac: ab4729 (abcam), CTCF: see below.
For spike-in ChIP-seq, we mixed the chromatin from 6 and 30 hpf d-rR strain embryos with a half of that amount of chromatin from 30 hpf Hamochi strain embryos, and used for ChIP.
Generation of medaka CTCF antibody
Four peptides derived from medaka CTCF (gene ID: ENSORLG00000008771) were synthesized by MAB Institute, Inc. (Nagano, Japan) as listed below:
1–36+C: METGQATALASDGKVLSEGGEALIQTGQGDEAGTMEC
50–70+C: MKTEVLEGGGTVTVTGGDEGQC
699–730: CTDETTEQVITGGGKPGAQSEELSQADAAAQE
C+734–753: CSAAPSNGDLTPEMILSMMDR
The mixture of these peptides was used as an antigen to generate antibodies against medaka CTCF in mice, and hybridoma clones were established by MAB Institute, Inc. We screened hybridoma clones by western blotting using the lysate of HEK293 cells transfected with pCS2-MT-olCtcf1. For the selected clones, the supernatant of the culture medium was concentrated by ammonium sulfate precipitation, and antibodies were purified by protein A sepharose and eluted with 100 mM Glycine, pH 4.0, then dialyzed with PBS + 0.05% NaN3.
3D-FISH
3D-FISH analyses were carried out following protocols described previously with some modifications (Kondo et al. 2014). Medaka fish embryos were fixed with 4% paraformaldehyde (PFA), embedded in paraffin, and sectioned into 5-μm thickness. After deparaffinization, HistoVT ONE (Nakalai Tesque 06380-05) treatment and a wash process by water, hybridizations with medaka BAC as probes proceeded. BACs, ola1-199A02, ola1-190N08, and ola1-182E06 were labeled with ChromaTide AlexaFluor 488-5-dUTP (Thermo Fisher Scientific C11397), Cy3-dCTP (GE Healthcare PA53021), or Cy5-dCTP (GE Healthcare PA55021) by nick-translation (Roche 976776), respectively.
The confocal images were captured by microscopy with Olympus UPlanSApo 100× NA 1.40 and PlanApo N 60× NA 1.42, images with 65-nm pixels in X-Y and 300-nm steps in Z axes. Those images were deconvoluted and distances of foci (centroid) were measured by using ImageJ.
Counting mitotic cell ratio in early embryos
Embryos were fixed with 4% PFA at room temperature for 2 h, then 4°C overnight. The fixed embryos were stained with DAPI, and mitotic cells were identified manually based on the morphology of condensed chromosomes. Cell number was counted using ImageJ.
Blocking zygotic genome activation by alpha-amanitin treatment
To block zygotic genome activation, embryos were collected at the one-cell stage and injected with 10 ng/µL of α-amanitin (Sigma-Aldrich A2263), incubated for 10 h, and used for RNA-seq and in situ Hi-C experiments.
Western blotting
Embryos were washed with PBS and lysed on ice for 10 min in a lysis buffer (25 mM Tris-HCl, pH 7.5, 250 mM NaCl, 5 mM MgCl2, 10% [v/v] glycerol, 0.2% [v/v] NP-40, and 1 mM NaF). Chromatin-enriched fractions were collected by low-speed centrifugation at 1500g for 5 min, washed with the lysis buffer, and further lysed with 1× SDS buffer. Proteins were separated using polyacrylamide gels (5.5% for SMC3 and 12.5% for histone H3) and transferred onto polyvinylidene difluoride membrane (MilliporeSigma). After blocking with 5% skim milk for 1 h at room temperature, membranes were incubated with primary antibodies (SMC3: abcam ab9263, histone H3: abcam ab1791) overnight at 4°C. After several washes, membranes were incubated with secondary antibody (anti-rabbit IgG Peroxidase, Sigma-Aldrich A0545) for 1 h at room temperature. Protein bands were visualized using the ECL Select Western Blotting Detection Reagent (GE Healthcare) and detected by ImageQuant (GE Healthcare).
Hi-C data processing
The sequenced reads were mapped, filtered, and normalized using Juicer (version 1.5) (Durand et al. 2016). Medaka reference genome version 2.2.4 or zebrafish danRer11 was used for mapping. In order to compare between different stages, the KR-normalized matrix was further normalized by average total contacts of each row, so that all samples will have the same total contact number. We used Juicer Tools (version 1.7.6) to calculate Pearson's correlation matrices, eigenvectors, and others in what follows.
The reproducibility between the replicates was calculated using HiCRep (Yang et al. 2017) using the default parameter. Normalized contact matrices at 100-kb resolution were used.
Loops were identified using Juicer Tools HiCCUPS at 5- and 10-kb resolution. As our fibroblast cells were maintained in a laboratory environment for a long period of time, chromosomes could have been rearranged, thereby yielding a number of peaks identified by HiCCUPS in the off-diagonal areas of the Hi-C contact maps. In particular, off-diagonal loops of >1 Mb in size were unexpectedly long, were likely to be false-positives, and were therefore removed. We also noticed that skewed sequence coverage bins sometimes caused abnormal enrichment on contact maps. To filter out these possibly false-positives loops, we selected loops on rows and columns of each contact matrix where the value of the KR normalization vector produced for matrix balancing was in the range of 0.5 to 2.0.
As we did not have CTCF ChIP-seq data for fibroblast cells, as substitutes we instead used CTCF ChIP-seq peaks identified from 54 hpf embryos. The CTCF motif orientation of loops was determined using MotifFinder of Juicer Tools. Specifically, we used motif MA0139.1_CTCF from Jasparcore 2014 as the CTCF motif, and the positions of the CTCF motif in the medaka genome were identified using FIMO (Grant et al. 2011) from the MEME Suite with the default parameters.
Aggregate peak analysis was performed using Juicer Tools using all filtered fibroblast loops that are 100 kb or larger at 5-kb resolution by using commands: apa -n 20 -w 6 -q 4 -r 5000. In the presence of strong background signals near the diagonal, we considered loops of size 100 kb or larger (see the rationale in Supplemental Fig. S8F). Normalized APA plots were used to depict loop signal enrichment across stages. The P2LL score (the ratio of the central pixel to the mean of the pixels in the lower left corner) from Juicer output was used for the quantification of loop strength.
Contact domains were annotated using the Juicer Tools Arrowhead algorithm at 5-kb resolution, changing the default filter size from 300 kb to 50 kb. To quantify the change in size of contact domains during development, we used the N10 statistics (Fig. 3A), where the N10 maximizes such that non-overlapping contact domains of size N10 or longer occupy at least 10% of the genome covered by domains. To calculate the N10 value, one can order non-overlapping contact domains in descending order and scan the ordered list from the top until the sum of domain sizes becomes 10% or larger. A higher N10 value means that larger contact domains occupy at least 10% of the genome covered by contact domains. The statistics show that contact domains with larger size are established later during gastrulation (Fig. 3A).
ATAC-seq and ChIP-seq data processing
The sequenced reads were preprocessed to remove low-quality bases and adapter-derived sequences using Trimmomatic v0.32 (Bolger et al. 2014) and then aligned to the medaka reference genome version 2.2.4 by BWA (Li and Durbin 2009). Reads with mapping quality (MAPQ) larger than or equal to 20 were used for further analyses. For ATAC-seq and ChIP-seq, MACS2 (version 2.1.1.20160309) (Zhang et al. 2008) was used to call peaks and generate signals per million reads tracks using the following commands; ATAC-seq: macs2 callpeak ‐‐nomodel ‐‐extsize 200 ‐‐shift -100 -g 600000000 -q 0.01 -B –SPMR, H3K27ac and CTCF ChIP-seq: macs2 callpeak -q 0.01 -B –SPMR. The option -g 600000000 is the effective genome size estimated by remapping 50-mer of the medaka reference genome version 2.2.4 to itself using GEM library tools; gem-indexer (build 1.423), gem-mappability (build 1.315), and counting the uniquely remapped reads denoted as “!” in the output (Marco-Sola et al. 2012). The number of peaks called by MACS2 depends on the total number of reads. Thus, to balance the total read number, 10 million reads were randomly sampled from each of the replicates for CTCF ChIP-seq data. For Figure 1C and Supplemental Figure S1D,E, replicates were combined for each stage after random sampling.
Normalization of spike-in ChIP-seq
The sequenced reads were preprocessed to remove low-quality bases and adapter-derived sequences using Trimmomatic v0.32 (Bolger et al. 2014) and then aligned to the medaka Hd-rR and HNI reference genome version 2.2.4 by BWA (Li and Durbin 2009). Reads with mapping quality larger than or equal to 20 were used for the further analyses. MACS2 (version 2.1.1.20160309) (Zhang et al. 2008) was used to call peaks and generate signals per million reads tracks. ChIP signals were then divided by Hamochi read (reads mapped on HNI genome) counts, and further divided by the ratio of d-rR reads to Hamochi reads in the input sample, which represents the relative amounts of the d-rR and Hamochi in the starting chromatin samples used for ChIP.
RNA-seq data processing for d-rR embryos (5 to 54 hpf) and fibroblast cells
The sequenced reads were preprocessed to remove low-quality bases and adapter-derived sequences using Trimmomatic v0.32 (Bolger et al. 2014) and were aligned to the medaka reference genome version 2.2.4 by STAR (Dobin et al. 2013), and reads with mapping quality larger than or equal 20 were used for further analyses.
Analysis of RNA-seq data from d-rR/HNI hybrid F1 embryos (5 to 10 hpf)
To investigate the transcriptional activity in early zygotic stages, we employed hybrid F1 of d-rR and HNI strains—the former was used as a paternal strain and the latter as a maternal strain—for RNA-seq analysis. We mapped RNA-seq reads of hybrid F1 embryos to both Hd-rR and HNI reference genomes (v2.2.4) (Ichikawa et al. 2017). For reliable identification of the source strain of expressed RNA, paternal Hd-rR, or maternal HNI, we utilized the genomic polymorphism between the two strains because the genetic divergence is sufficiently high and the single nucleotide variant (SNV) rate is ∼2.5% (Ichikawa et al. 2017). First, to call SNVs between the two genomes, we obtained publicly available whole-genome shotgun sequencing data of the HNI strain with accession numbers DRR002216 and DRR002217 from the DNA Data Bank of Japan (DDBJ; https://www.ddbj.nig.ac.jp/index-e.html). Raw Illumina reads were preprocessed to remove low-quality bases and adapter-derived sequences using Trimmomatic v0.32 (Bolger et al. 2014). Subsequently, a total of 109-fold coverage short-read data were aligned with the medaka Hd-rR reference genome (v2.2.4) by using BWA v0.7.8 (Li and Durbin 2009). After refining alignments around indel sites with GATK v3.6 (McKenna et al. 2010), SNVs and indels were called by SAMtools v1.2 (Li et al. 2009). High quality variants, SNVs, and indels, with read depth ≥20 and variant quality ≥20, were used for downstream analysis. To further filter erroneous regions where short reads from HNI matched to the Hd-rR genome, we aligned HNI blastula RNA-seq reads (NCBI Sequence Read Archive [SRA; https://www.ncbi.nlm.nih.gov/sra] accession number SRR3168578) with the two genomes using STAR v2.5.4 (Dobin et al. 2013) and discarded variant sites where HNI RNA-seq reads matched erroneously, retaining 17,516,984 sites for hybrid F1 embryo RNA-seq analyses. After mapping RNA-seq reads of hybrid F1 of each developmental stage the two genomes using STAR, we determined that reads were paternally expressed if they matched the Hd-rR sides of polymorphic SNVs. For Supplemental Figures S5 and S9, paternal read coverage was normalized by the sum of paternal and maternal read numbers to calculate the rpm.
Gene expression at loop anchors
The RPKM values were computed from RNA-seq read counts for the medaka gene model version 2.2.4 (http://utgenome.org/medaka_v2/#!Medaka_gene_model.md) in each time point. Out of the medaka gene model v2.2.4, genes associated with Ensembl protein ID were used. Then, we defined zygotically expressed genes (ZEGs) such that the read counts <30 and the RPKM <2 at 5 hpf, and read counts >30 and RPKM >4 in one of later stages than 5 hpf, which means that ZEGs were inactive before ZGA and began to be transcribed during or after that. Then, we got the vectors whose elements were the gene expression levels of ZEGs in developmental time course. To separate ZEGs by expression timing, the vectors were grouped into seven clusters according to a widely used method (Trapnell et al. 2014). Loop genes in ZEGs were defined so that their TSSs and loop anchors with loop size >100 kb (see the rationale in Supplemental Fig. S8F) were within the same 5-kb bin. Nonloop genes were the rest of ZEGs.
Association of large contact domains with mature cells
We compared contact domains between mature fibroblast cells and each time point during development. Two contact domains were treated identically when their boundaries were close, which was, however, a difficult task to do because the boundaries changed between samples. Indeed, the boundaries of 54 hpf and fibroblast (both were the most matured samples in our data) should be consistent; however, only 8% of boundaries at 54 hpf exactly matched those at fibroblast. To increase the ratio, we allowed the positional difference between pairs of matching domain boundaries to be at most 25 kb (5 bins), so that 71% domains at 54 hpf were associated with those at fibroblast. We counted the number of large contact domains which were shared with mature fibroblast cells and plotted for three different domain size ranges; ≥200 kb, ≥300 kb, and ≥ 400 kb (Supplemental Fig. S8B).
Detecting more reliable loops
Automatic detection of loops from Hi-C contact maps generally suffered from many false positives. To filter out false positive loops, we imposed two additional constraints on loops; namely, loops were associated with contact domains, and loops were 100 kb or larger in size by considering the resolution limitation of identifying loops (Supplemental Fig. S8F). The first condition was supported by the fact that a large fraction of peaks in mammals were associated with domains (Rao et al. 2014), which was also the case in the medaka genome. To meet the first condition, we extracted loops (called by HiCCUPS) associated with contact domains by selecting those located in boundaries of contact domains within a distance of 20 kb (4 bins).
Insulation score
The insulation scores were computed from the contact maps with 5-kb bins according to the method (Crane et al. 2015). We set the square size to 500 kb and smoothed the scores by averaging the neighboring five bins for each bin.
Detection of synteny blocks between medaka and zebrafish and comparison of Hi-C maps
We downloaded all protein-coding gene trees from Ensembl (release 75) and sorted the protein-coding genes with respect to the chromosomal position. The sorted genes were arranged along the x-axis (zebrafish) and the y-axis (medaka), and the Nth gene was represented by the Nth point. Then, a one-to-one ortholog pair between the Nth zebrafish gene and the Mth medaka gene was represented by a line from (N,M) to (N+1,M+1) if the two orthologs were encoded on the same strand, or a line from (N,M+1) to (N+1,M) if they were encoded on the reverse strand. Next, for each chromosome pair, we chained these orthologs into longer, approximately diagonal lines using a chaining algorithm with a geometric gap cost calculated as described previously (Abouelhoda and Ohlebusch 2003; Shibuya and Kurochkin 2003) (match score: 1, gap cost: 0.25). For example, a chain consisting of five medaka genes and five zebrafish orthologs has a chain score 5 if those genes are consecutive and non-inverted in both species; on the other hand, the chain score decreases as 5 − 0.25 × (i + j) if the chained regions include i medaka nonorthologous (or inverted) genes and j zebrafish nonorthologous (or inverted) genes. We chose these parameter values because the ortholog distribution between the medaka and zebrafish genomes showed that smaller gap costs (≤0.2) cause artifactual extension of chains and erroneous merger of distinct synteny blocks. During the chain extension procedure, we computed “increasing” chains and “decreasing” chains separately: specifically, “increasing” chains can extend to an orthologous pair of medaka and zebrafish genes only if the two genes are encoded on the same strand of the medaka and zebrafish chromosomes. On the other hand, “decreasing” chains can extend to an orthologous pair of medaka and zebrafish genes only if they are encoded on the opposite strands. In this way, locally inverted genes (by small-scale rearrangements or by misassembly) were treated as gaps and excluded from the chains. Finally, we defined high-scoring chains (score >9) as synteny blocks (i.e., regions with conserved gene order) between the medaka and zebrafish genomes (Supplemental Fig. S14) and presented the list of 101 synteny blocks in Supplemental Data 1. We assumed that orthologous regions are about twice the size of medaka in zebrafish according to their genome size. We therefore selected regions that are relatively long (medaka region size >0.7 Mb) and the size ratio of zebrafish to medaka is within the range of 1.33 to 3. Among 101 synteny blocks, 21 met these criteria, but 10 regions contained large unmapped regions in either of species. Consequently, 11 synteny blocks were examined for Hi-C map comparison. For Figure 5C and Supplemental Figure S13, we translated the genomic coordinates from the older genome assemblies (Ensembl release 75) to the more recent genome assemblies (i.e., Hd-rR v2.2.4 and zebrafish danRer11), and the most similar regions within the synteny blocks were shown.
Data access
All sequencing data generated in this study have been submitted to the NCBI BioProject database (https://www.ncbi.nlm.nih.gov/bioproject/) under accession number PRJDB7492. The data analysis pipeline is available as Supplemental Code and also at Bitbucket (https://bitbucket.org/tashiraka/medaka_hic/).
Supplementary Material
Acknowledgments
We thank H. Koseki for helpful discussion; M. Shamim for assistance with Juicer parameter setting; and Y. Yamagishi for assistance with animal care and embryo sampling. This work was supported by Core Research for Evolutional Science and Technology (CREST) grant numbers JPMJCR13W3 and JP17gm0510016, Japan Society for the Promotion of Science (JSPS) grant number JP16K20975, and Japan Agency for Medical Research and Development (AMED) grant number JP20gm1110007.
Author contributions: R.N. conceived the project. Y.M. found loops in fibroblast to explore analysis in gastrulation. R.N. and Y.M. analyzed the data. R.N. performed embryonic Hi-C and RNA-seq experiments. M.K. performed fibroblast Hi-C and RNA-seq experiments. R.N. and C.L.W. performed zebrafish Hi-C experiments. R.N. and H.N. performed ChIP-seq experiments. T.K., K.K., and A.S. performed 3D-FISH experiments. T.T. designed the CTCF antibody. M.K. analyzed the d-rR/HNI hybrid RNA-seq data. N.C.D. assisted in analyzing the Hi-C data. Y.N. searched for the synteny blocks between medaka and zebrafish. H.T., S.M., B.R.C., and E.L.A. supervised the project. R.N., Y.M., H.T., S.M., and E.L.A. prepared the manuscript with input from all authors.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at https://www.genome.org/cgi/doi/10.1101/gr.269951.120.
Competing interest statement
The authors declare no competing interests.
References
- Abouelhoda MI, Ohlebusch E. 2003. A local chaining algorithm and its applications in comparative genomics. In Algorithms in bioinformatics. WABI 2003. Lecture notes in computer science (ed. Benson G, Page RDM), Vol. 2812, pp. 1–16. Springer, Berlin, Heidelberg. 10.1007/978-3-540-39763-2_1 [DOI] [Google Scholar]
- Abramo K, Valton A, Venev SV, Ozadam H, Fox AN, Dekker J. 2019. A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat Cell Biol 21: 1393.–. 10.1038/s41556-019-0406-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhoure N, Bounova G, Bernasconi D, Praz V, Lammers F, Canella D, Willis IM, Herr W, Hernandez N, Delorenzi M, et al. 2014. Quantifying ChIP-seq data: a spiking method providing an internal reference for sample-to-sample normalization. Genome Res 24: 1157–1168. 10.1101/gr.168260.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10: 1213–1218. 10.1038/nmeth.2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ke Y, Wu K, Zhao H, Sun Y, Gao L, Liu Z, Zhang J, Tao W, Hou Z, et al. 2019. Key role for CTCF in establishing chromatin structure in human embryos. Nature 576: 306–310. 10.1038/s41586-019-1812-0 [DOI] [PubMed] [Google Scholar]
- Collombet S, Ranisavljevic N, Nagano T, Varnai C, Shisode T, Leung W, Piolot T, Galupa R, Borensztein M, Servant N, et al. 2020. Parental-to-embryo switch of chromosome organization in early embryogenesis. Nature 580: 142–146. 10.1038/s41586-020-2125-z [DOI] [PubMed] [Google Scholar]
- Crane E, Bian Q, McCord RP, Lajoie BR, Wheeler BS, Ralston EJ, Uzawa S, Dekker J, Meyer BJ. 2015. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature 523: 240–244. 10.1038/nature14450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters J-M. 2019. DNA loop extrusion by human cohesin. Science 366: 1338–1345. 10.1126/science.aaz3418 [DOI] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15.– . 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, He J, Xiang Y, Wang Q, Li Y, et al. 2017. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 547: 232–235. 10.1038/nature23263 [DOI] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL. 2016. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3: 95–98. 10.1016/j.cels.2016.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmer-Rachamimov AO, Suvà ML, Bernstein BE. 2016. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 529: 110–114. 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyamer IM, Gassler J, Imakaev M, Brandão HB, Ulianov S V, Abdennur N, Razin S V, Mirny LA, Tachibana-Konwalski K. 2017. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature 544: 110–114. 10.1038/nature21711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–2049. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani-Seiki M, Wittbrodt J. 2004. Medaka and zebrafish, an evolutionary twin study. Mech Dev 121: 629–637. 10.1016/j.mod.2004.05.010 [DOI] [PubMed] [Google Scholar]
- Gassler J, Brandão HB, Imakaev M, Flyamer IM, Ladstätter S, Bickmore WA, Peters J, Mirny LA, Tachibana K. 2017. A mechanism of cohesin-dependent loop extrusion organizes zygotic genome architecture. EMBO J 36: 3600–3618. 10.15252/embj.201798083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS. 2011. FIMO: scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018. 10.1093/bioinformatics/btr064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. 2017. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169: 216–228.e19. 10.1016/j.cell.2017.03.024 [DOI] [PubMed] [Google Scholar]
- Ichikawa K, Tomioka S, Suzuki Y, Nakamura R, Doi K, Yoshimura J, Kumagai M, Inoue Y, Uchida Y, Irie N, et al. 2017. Centromere evolution and CpG methylation during vertebrate speciation. Nat Commun 8: 1833. 10.1038/s41467-017-01982-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamatsu T. 2004. Stages of normal development in the medaka Oryzias latipes. Mech Dev 121: 605–618. 10.1016/j.mod.2004.03.012 [DOI] [PubMed] [Google Scholar]
- Ji X, Dadon DB, Powell BE, Fan ZP, Borges-Rivera D, Shachar S, Weintraub AS, Hnisz D, Pegoraro G, Lee TI, et al. 2016. 3D chromosome regulatory landscape of human pluripotent cells. Cell Stem Cell 18: 262–275. 10.1016/j.stem.2015.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, Shariati SAM, Skotheim JM. 2017. Zygotic genome activation in vertebrates. Dev Cell 42: 316–332. 10.1016/j.devcel.2017.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaij LJT, van der Weide RH, Ketting RF, de Wit E. 2018. Systemic loss and gain of chromatin architecture throughout zebrafish development. Cell Rep 24: 1–10.e4. 10.1016/j.celrep.2018.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke Y, Xu Y, Chen X, Feng S, Liu Z, Sun Y, Yao X, Li F, Zhu W, Gao L, et al. 2017. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 170: 367–381.e20. 10.1016/j.cell.2017.06.029 [DOI] [PubMed] [Google Scholar]
- Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. 2019. Human cohesin compacts DNA by loop extrusion. Science 366: 1345–1349. 10.1126/science.aaz4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Isono K, Kondo K, Endo TA, Itohara S, Vidal M, Koseki H. 2014. Polycomb potentiates Meis2 activation in midbrain by mediating interaction of the promoter with a tissue-specific enhancer. Dev Cell 28: 94–101. 10.1016/j.devcel.2013.11.021 [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Giraldez AJ. 2014. Zygotic genome activation during the maternal-to-zygotic transition. Annu Rev Cell Dev Biol 30: 581–613. 10.1146/annurev-cellbio-100913-013027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Harrison MM, Villalta JE, Kaplan T, Hughes H, Berkeley C. 2014. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. eLife 3: e03737. 10.7554/eLife.03737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupiáñez DG, Kraft K, Heinrich V, Krawitz P, Brancati F, Klopocki E, Horn D, Kayserili H, Opitz JM, Laxova R, et al. 2015. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell 161: 1012–1025. 10.1016/j.cell.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marco-Sola S, Sammeth M, Guigó R, Ribeca P. 2012. The GEM mapper : fast, accurate and versatile alignment by filtration. Nat Methods 9: 1185–1188. 10.1038/nmeth.2221 [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20: 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562. 10.1038/nature01262 [DOI] [PubMed] [Google Scholar]
- Nakamura R, Tsukahara T, Qu W, Ichikawa K, Otsuka T, Ogoshi K, Saito TL, Matsushima K, Sugano S, Hashimoto S, et al. 2014. Large hypomethylated domains serve as strong repressive machinery for key developmental genes in vertebrates. Development 141: 2568–2580. 10.1242/dev.108548 [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Mclysaght A. 2017. Genomes as documents of evolutionary history: a probabilistic macrosynteny model for the reconstruction of ancestral genomes. Bioinformatics 33: i369–i378. 10.1093/bioinformatics/btx259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, Van Berkum NL, Meisig J, Sedat J, et al. 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. 10.1038/nature11049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e22. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onichtchouk D, Driever W. 2016. Zygotic genome activators, developmental timing, and pluripotency. Curr Top Dev Biol 116: 273–297. 10.1016/bs.ctdb.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Orlando DA, Chen MW, Bradner JE, Guenther MG, Orlando DA, Chen MW, Brown VE, Solanki S, Choi YJ, Olson ER. 2014. Quantitative ChIP-Seq normalization reveals global modulation of the epigenome. Cell Rep 9: 1163–1170. 10.1016/j.celrep.2014.10.018 [DOI] [PubMed] [Google Scholar]
- Pękowska A, Klaus B, Xiang W, Severino J, Daigle N, Klein FA, Oleś M, Casellas R, Ellenberg J, Steinmetz LM, et al. 2018. Gain of CTCF-anchored chromatin loops marks the exit from naive pluripotency. Cell Syst 7: 482–495.e10. 10.1016/j.cels.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SSP, Huang SC, Glenn St Hilaire B, Engreitz JM, Perez EM, Kieffer-Kwon KR, Sanborn AL, Johnstone SE, Bascom GD, Bochkov ID, et al. 2017. Cohesin loss eliminates all loop domains. Cell 171: 305–320.e24. 10.1016/j.cell.2017.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanborn AL, Rao SSP, Huang S-C, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. 2015. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci 112: E6456–E6465. 10.1073/pnas.1518552112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. 2012. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. 10.1016/j.cell.2012.01.010 [DOI] [PubMed] [Google Scholar]
- Shibuya T, Kurochkin I. 2003. Match chaining algorithms for cDNA mapping. In Algorithms in bioinformatics. WABI 2003. Lecture notes in computer science (ed. Benson G, Page RDM), Vol. 2812, pp. 462–475. Springer, Berlin, Heidelberg. 10.1007/978-3-540-39763-2_33 [DOI] [Google Scholar]
- Tadros W, Lipshitz HD. 2009. The maternal-to-zygotic transition: a play in two acts. Development 136: 3033–3042. 10.1242/dev.033183 [DOI] [PubMed] [Google Scholar]
- Takeda H, Shimada A. 2010. The art of medaka genetics and genomics: What makes them so unique? Annu Rev Genet 44: 217–241. 10.1146/annurev-genet-051710-151001 [DOI] [PubMed] [Google Scholar]
- Tena JJ, Alonso ME, De La Calle-Mustienes E, Splinter E, De Laat W, Manzanares M, Gómez-Skarmeta JL. 2011. An evolutionarily conserved three-dimensional structure in the vertebrate Irx clusters facilitates enhancer sharing and coregulation. Nat Commun 2: 310. 10.1038/ncomms1301 [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol 32: 381–386. 10.1038/nbt.2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wike CL, Guo Y, Tan M, Nakamura R, Shaw DK, Díaz N, Whittaker-Tademy AF, Durand NC, Lieberman Aiden E, Vaquerizas JM, et al. 2021. Chromatin architecture transitions from zebrafish sperm through early embryogenesis. Genome Res (this issue) 31: 981–994. 10.1101/gr.269860.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolpert L, Jessel T, Lawrence P, Meyerowitz E, Robertson E, Smith J. 2006. Principles of development. Oxford University Press, Oxford. [Google Scholar]
- Wragg J, Müller F. 2016. Transcriptional regulation during zygotic genome activation in zebrafish and other anamniote embryos. Adv Genet 95: 161–194. 10.1016/bs.adgen.2016.05.001 [DOI] [PubMed] [Google Scholar]
- Yang T, Zhang F, Yardımcı GG, Song F, Hardison RC, Noble WS, Yue F, Li Q. 2017. HiCRep: assessing the reproducibility of Hi-C data using a stratum-adjusted correlation coefficient. Genome Res 27: 1939–1949. 10.1101/gr.220640.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based Analysis of ChIP-Seq (MACS). Genome Biol 9: R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Emerson DJ, Gilgenast TG, Titus KR, Lan Y, Huang P, Zhang D, Wang H, Keller CA, Giardine B, et al. 2019. Chromatin structure dynamics during the mitosis-to-G1 phase transition. Nature 576: 158–162. 10.1038/s41586-019-1778-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Xie W. 2019. The role of 3D genome organization in development and cell differentiation. Nat Rev Mol Cell Biol 20: 535–550. 10.1038/s41580-019-0132-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.