Conspectus
RNA lies upstream of nearly all biology and functions as the central conduit of information exchange in all cells. RNA molecules encode information both in their primary sequences and in complex structures that form when an RNA folds back on itself. From the time of discovery of messenger RNA in the late 1950s until quite recently, we had only a rudimentary understanding of RNA structure across vast regions of most messenger and non-coding RNAs. This deficit is now rapidly being addressed, especially by SHAPE chemistry, mutational profiling (MaP), and closely related platform technologies that, collectively, create chemical microscopes for RNA. These technologies make it possible to interrogate RNA structure, quantitatively and at nucleotide resolution and at large scales, for entire mRNAs, non-coding RNAs, and viral RNA genomes. By applying comprehensive structure probing to diverse problems, we and others are showing that control of biological function mediated by RNA structure is ubiquitous across prokaryotic and eukaryotic organisms. Work over the past decade using SHAPE-based analyses has clarified key principles. First, the method of RNA structure probing matters. SHAPE-MaP, with its direct and one-step readout that probes nearly every nucleotide by reaction at the 2’-hydroxyl, gives a more detailed and accurate readout than alternatives. Second, comprehensive chemical probing is essential. Focusing on fragments of large RNAs or using meta-gene or statistical analyses to compensate for sparse datasets misses critical features and often yields structure models with poor predictive power. Finally, every RNA has its own internal structural personality. There are myriad ways in which RNA structure modulates sequence accessibility, protein binding, translation, splice-site choice, phase separation, and other fundamental biological processes. In essentially every instance where we have applied rigorous and quantitative SHAPE technologies to study RNA structure-function interrelationships, new insights regarding biological regulatory mechanisms have emerged. RNA elements with more complex higher-order structures appear more likely to contain high-information-content clefts and pockets that bind small molecules, broadly informing a vigorous field of RNA-targeted drug discovery. The broad implications of this collective work are two-fold. First, it is long past time to abandon depiction of large RNAs as simple noodle-like or gently flowing molecules. Instead, we need to emphasize that nearly all RNAs are punctuated with distinctive internal structures, a subset of which modulate function in profound ways. Second, structure probing should be an integral component of any effort that seeks to understand the functional nexuses and biological roles of large RNAs.
Key References
Siegfried, N. A.; Busan, S.; Rice, G. M.; Nelson, J. A. E.; Weeks, K. M. RNA Motif Discovery by SHAPE and Mutational Profiling (SHAPE-MaP). Nature Methods 2014, 11, 959–965.1
Introduces mutational profiling (MaP) for one-step detection of chemical adducts in RNA, the low SHAPE-low Shannon entropy metric for discovery of potential regulatory motifs, and emphasizes the now-ubiquitous result that large RNAs are punctuated by extensive internal structure.
Mustoe, A. M.; Busan, S.; Rice, G. M.; Hajdin, C. E.; Peterson, B. K.; Ruda, V. M.; Kubica, N.; Nutiu, R.; Baryza, J. L.; Weeks, K. M. Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing. Cell 2018, 173, 181–195.2
The first large-scale RNA structure probing study to focus specifically on transcripts with comprehensive, high-quality, per-nucleotide probing data. RNA structure is shown to be a pervasive regulator of gene expression, effectively comprising a distinct level of the genetic code.
Dethoff, E. A.; Boerneke, M. A.; Gokhale, N. S.; Muhire, B. M.; Martin, D. P.; Sacco, M. T.; McFadden, M. J.; Weinstein, J. B.; Messer, W. B.; Horner, S. M.; Weeks, K. M. Pervasive Tertiary Structure in the Dengue Virus RNA Genome. Proc Natl Acad Sci U S A 2018, 115, 11513– 11518.3
Provides a glimpse into the future emphasizing that large RNAs form (of course!) extensive internal base paired structures. But, distinctively, a subset of these structures also fold to create more complex higher-order structures that govern overall RNA compaction and function.
The Ubiquity and Fundamentalness of RNA Structure
As soon as they are synthesized, cellular and viral RNAs fold back on themselves to form (typically extensive) internal base pairs and (rarer) higher-order interactions. These structures may have intrinsic function, bind proteins, or occlude functional structures and thereby be switch-like (Figure 1). mRNAs, long non-coding RNAs (lncRNA), and viral RNAs therefore encode biological information both in sequence and in structure4–10.
Figure 1.
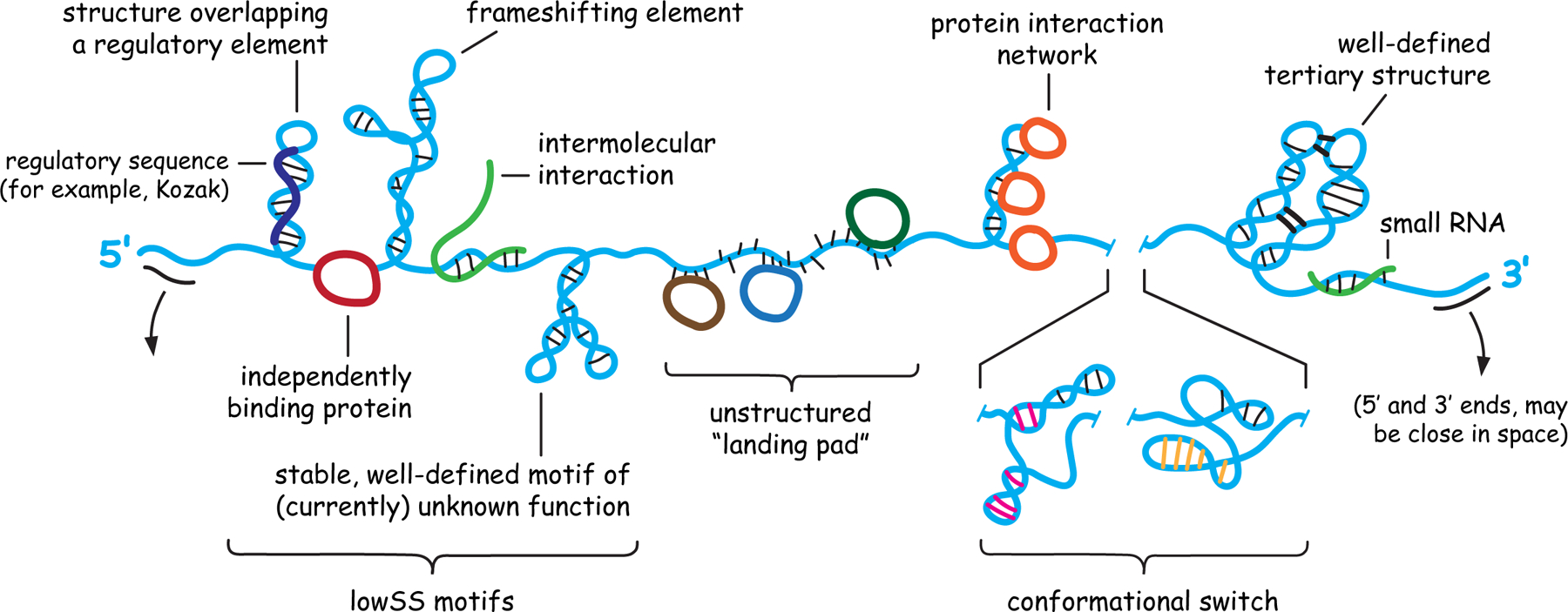
Architecture and potential internal structures of RNA molecules. Important classes of motifs are shown, a subset of which might occur in any given RNA.
Until recently, rigorous methods for comprehensive examination of the long-range, in-solution structure of RNA molecules did not exist. First, it was difficult to measure RNA structure with nucleotide resolution and in a biophysically quantitative way. Second, it was difficult to model RNA structure accurately, especially for long RNAs. Third, many data-directed RNA structure models yield so many plausible structures that it was unclear where to look to identify the most impactful biology.
Advances in chemical probing have yielded powerful approaches for identifying, modeling, and characterizing the functional roles of RNA structure. Chemical probing is a venerable method11 that has been revolutionized by melding new chemistries, readouts by massively parallel sequencing, and innovative data analysis strategies. Especially impactful have been selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE)12–14, mutational profiling (MaP) (including both SHAPE-MaP1,15 and DMS-MaP16–19 approaches), and pseudo-free energy change strategies for data-driven secondary structure modeling20,21. Secondary structure landscapes can be now profiled for large RNAs efficiently and for many RNAs simultaneously in living cells. SHAPE and closely related technologies are now used in innovative ways by research labs and biotechnology companies worldwide22–31.
In this communication, we will accept that SHAPE-based methods work as advertised and focus on examples from our lab emphasizing how comprehensive, per-nucleotide examination of RNA structure can identify functional elements in complex RNAs and define mechanisms by which RNA structure governs biological function. This overview emphasizes the importance of incorporating structural information into all efforts to understand RNA biology and provides a jumping off point for further investigations.
SHAPE Chemistry and Mechanism
SHAPE chemistry interrogates free RNA 2’-hydroxyl groups, which nearly all nucleotides contain. SHAPE probing involves two basic steps: (1) treatment of an RNA with a hydroxyl-selective reagent and (2) read-out of the pattern of chemical modification in the RNA. From the earliest experiments12,13, it was clear that SHAPE measures local nucleotide flexibility (Figure 2A). Analyses of the geometric conformations of nucleotides with high reactivities, as visualized crystallographically, subsequently revealed that SHAPE reactivity at 2’-hydroxyl groups is facilitated by intramolecular general base catalysis and by specific conformations adopted by the proximal 3’-phosphodiester group32 (Figure 2B). SHAPE thus measures local nucleotide flexibility because conformationally dynamic nucleotides transiently adopt states that facilitate reactivity.
Figure 2.
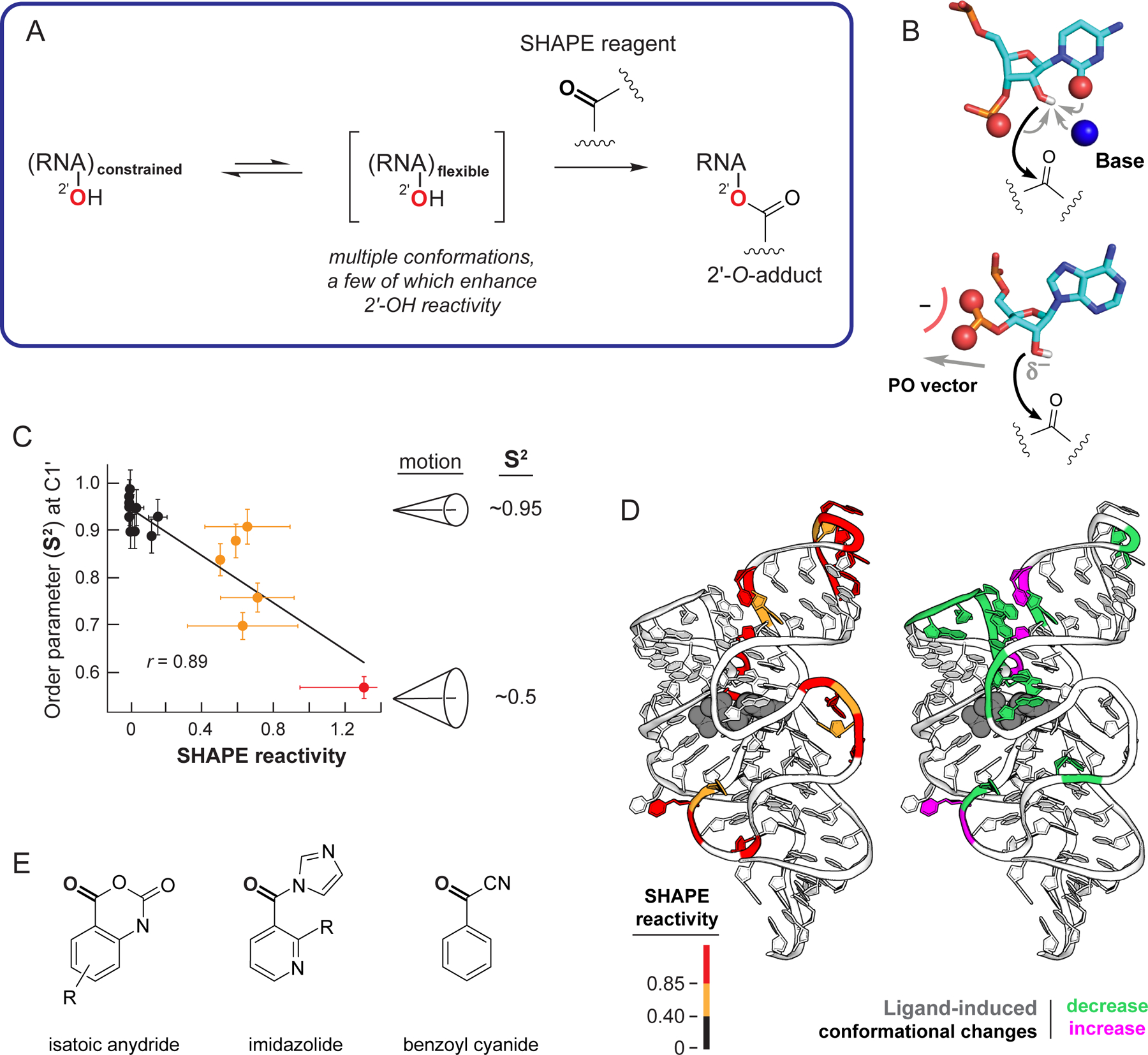
Mechanism of RNA SHAPE chemistry. (A) Reaction scheme. (B) Nucleotide conformations that enhance SHAPE reactivity involve (top) general base catalysis by the pyrimidine O2 (or purine N3), pro-S oxygen, or through-space groups and (bottom) stabilization of the 2’-oxyanion via conformations that direct nonbridging oxygen groups away from the 2’-OH group. (C) Correlation between the model-free generalized order parameter, S2, and SHAPE reactivity, exemplified by the U1A-binding RNA element33. (D) SHAPE reactivities superimposed on the TPP riboswitch aptamer domain showing (left) absolute SHAPE reactivities and (right) changes in SHAPE reactivities (and thus local nucleotide dynamics) upon ligand binding (gray). (E) Classes of SHAPE reagents.
Local nucleotide flexibility, as measured by SHAPE, correlates well with a fundamental biophysical measurement, the model-free generalized order parameter33, S2, which varies from 0.0 (disordered) to 1.0 (fully ordered). The correlation between S2 and SHAPE emphasizes that SHAPE is a biophysical measurement of spatial disorder at single-nucleotide resolution (Figure 2C). SHAPE distinguishes well-structured from conformationally dynamic regions of an RNA and detects changes in RNA structure as a function of perturbants, including binding by small molecules29,34–36 (Figure 2D). The biophysically rigorous relationship between SHAPE and local RNA structure becomes impressively meaningful because SHAPE can interrogate thousands of nucleotides at a time, including in living cells, for a significant fraction of a cellular transcriptome.
Three broad classes of SHAPE reagents have been developed based on isatoic anhydride12,14,37, imidazolide38,39, and benzoyl cyanide40 scaffolds (Figure 2E). The most important considerations for selecting a SHAPE reagent are half-life and solubility. Among the most useful reagents are 1M7 for fast, quantitative probing of RNAs under simplified conditions, in viruses, and in bacterial cells14; 5NIA, NAI, and 2A3 for probing in eukaryotic cells37–39, and benzoyl cyanide for fast, time-resolved experiments40–42. Differential reactivities between reagents sharing the isatoic anhydride scaffold can be used to “fingerprint” nucleotides that show distinct one-sided stacking patterns or that form the rarer C2’-endo ribose pucker and undergo slow conformational changes35,41,43.
The Mutational Profiling (MaP) Readout
Massively parallel sequencing44 has transformed characterization of the genome, RNA and DNA modifications and protein interactions, and (of course!) RNA structure15,25. Sequencing-detected genomics technologies involve cleavage, covalent modification, or enrichment of an RNA or DNA and then acquiring thousands to millions of “measurements” by sequencing. There are significant challenges to sequencing-based experiments, notably their complexity, the inability to validate but a small fraction of the results, and, usually, the lack of pre-genomics ground-truth reference experiments.
Within this context, RNA structure probing occupies a special niche because RNA chemical probing experiments conducted over the past 40 years have been read out by well-validated methods11,20,45,46 and RNAs with known structures can be used to evaluate new methods. In our work, it has proven invaluable to create a continuous trail of validation from simple biochemical to whole-transcript experiments.
SHAPE chemical adducts have been read out by several generations of technology. SHAPE 2’-O-adducts induce stops to primer extension by conventional reverse transcription, which converts an RNA to a complementary DNA (cDNA) such that the 3’-end of the cDNA indicates the site of a chemical adduct. Such “STOP-RT” SHAPE adducts were initially detected by gel electrophoresis and then by capillary electrophoresis20,47,48, which ultimately resolved RNA structures over a few thousand nucleotides20,47 including in cells47,49,50.
To read out SHAPE data by massively parallel sequencing, we initially (like many other laboratories), used the STOP-RT approach and prepared libraries for sequencing by the now-standard approach of ligating DNA adapters on to the initial cDNAs. However, our lab immediately noticed that these data correlated poorly with prior well-validated capillary electrophoresis methods, a feature now widely recognized51–54. The multiple complex biochemical steps required for library preparation, after the STOP-RT step, bias the final reactivity profiles.
To avoid throwing away the biophysical rigor of SHAPE technologies (Figure 2) upon integration with massively parallel sequencing, we invented the MaP strategy1,15. MaP leverages relaxed fidelity reverse transcription, whereby the polymerase reads through chemical adducts in an RNA, resulting in a mutation or short deletion. cDNAs are aligned and sites of mutation are counted to yield per-nucleotide reactivity profiles (Figure 3A). The MaP strategy has several impactful features: (1) Chemical adducts are recorded in a single, direct step1. (2) Sites of chemical adducts are recorded internally in the cDNA and thus can be amplified without biasing internal structural information. (3) Almost any sequencing library preparation approach can be used, without biasing the MaP detection step. (4) MaP readouts correlate strongly with validated capillary electrophoresis readouts15,55. (5) Each per-nucleotide measurement originates from multiple discrete mutation events, thus defining a standard error for each measurement. And (6) multiple chemical adducts can be detected in single RNA molecules, which laid the foundation for a new category of technologies involving single-molecule correlated chemical probing3,16,17,19,56–59 (Figure 3A, bottom). The term MaP emphasizes the independence of the strategy from library sequencing (-seq) steps; in this view, names that link MaP and seq are oxymorons. This focus on maintaining a quantitative relationship between probing and readout has motivated diverse research groups to adopt MaP-based strategies. For example, most work investigating SARS-CoV-2 RNA genome structure at nucleotide resolution has used SHAPE-MaP or related methods60–65.
Figure 3.
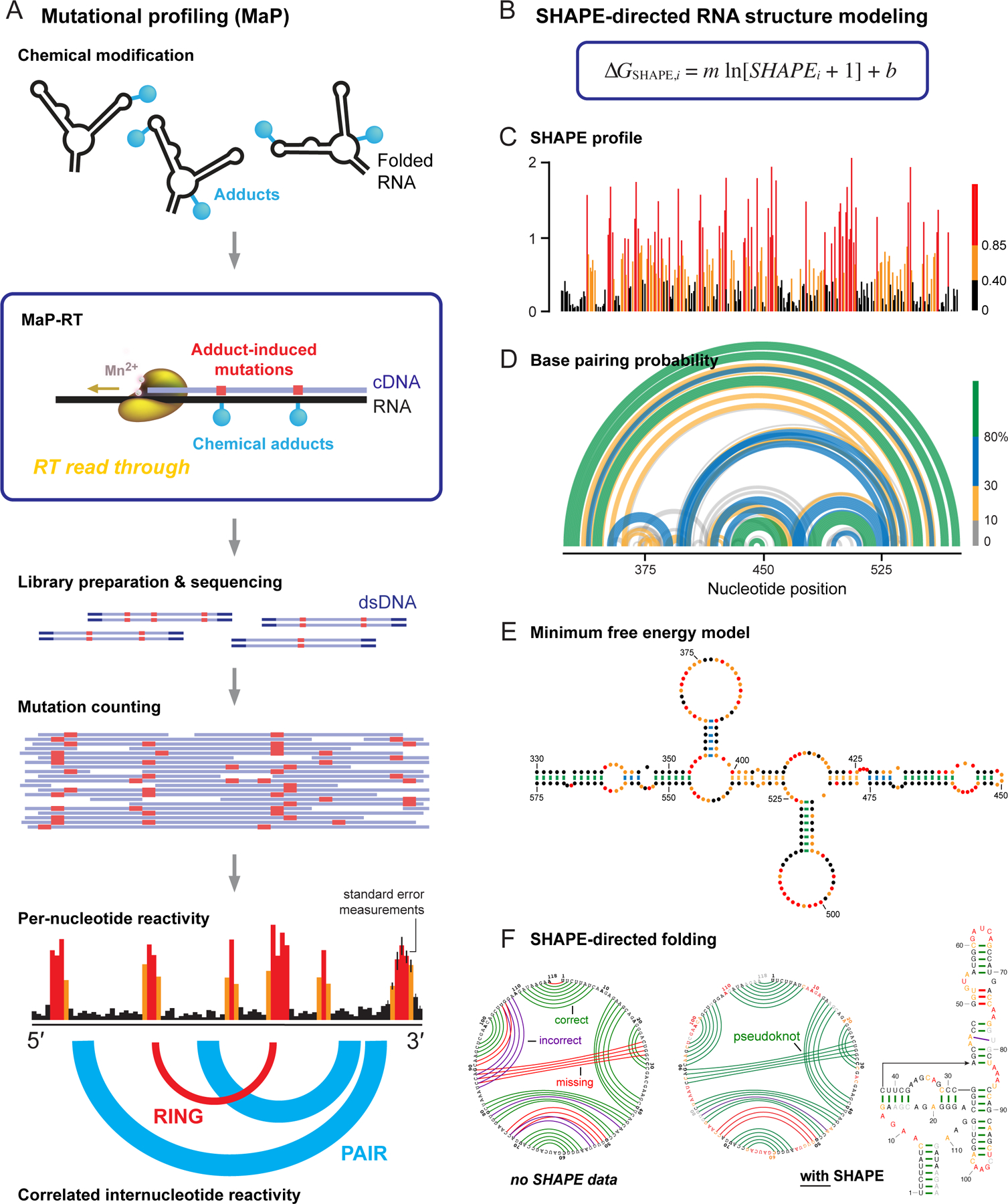
Overview of SHAPE, MaP, and RNA structure modeling. (A) Mutational profiling. RNA is treated with a reagent that reports secondary or tertiary structure; relaxed fidelity reverse transcription records chemical adducts as mutations relative to the original sequence (in red) internally in the cDNA; cDNAs are sequenced; and reads are aligned and used to create reactivity profiles. Data may be interpreted on a per-nucleotide basis or as through-space internucleotide correlations. (B) The ∆GSHAPE pseudo-free energy change relationship that enables SHAPE-directed structure modeling. (C) Per-nucleotide reactivity profile for a domain of the STMV RNA genome56,91. (D, E) Secondary structure models (based on data in panel C) shown as (D) probability arc plots and (E) minimum free energy secondary structure diagram. In probability arcs and lines connecting base pairs, colors indicate the likelihood of unique pairing for a given nucleotide. (F) Representative secondary structure modeling, showing the SAM-I riboswitch21 (left) without and (right) with SHAPE data. SHAPE-directed modeling often yields dramatic improvements, including for RNAs containing pseudoknots. Arcs indicate correct (green), incorrect (purple), and missing (red) pairs, relative to accepted structure.
SHAPE-Directed RNA Structure Modeling
SHAPE data are approximately inversely correlated with the probability that a nucleotide forms a base pair. As the logarithm of a probability corresponds to an energy, SHAPE data can be converted into a pseudo-free-energy change term, ∆GSHAPE, and used to modify the free energy terms in nearest-neighbor RNA folding algorithms20,21 (Figure 3B). There are two parameters in ∆GSHAPE, the slope, m, is the penalty for base pairing that increases with SHAPE reactivity; the b term is negative and reflects a favorable term for pairing at nucleotides with low SHAPE reactivities. SHAPE information (Figure 3C) is used to create data-directed structure models that can be visualized in several ways. The most comprehensive is as probability arcs15,66–68 (Figure 3D), which illustrate the structural complexity of an RNA ensemble. Alternatively, the most probable, minimum free energy structure can be shown in a base-pairing diagram, which simplifies the underlying complexity of an RNA (Figure 3E). SHAPE-directed secondary structure modeling achieves good-to-excellent agreement with accepted structures for diverse RNAs1,20 including RNAs containing pseudoknots21 (Figure 3F).
Finding Functional Motifs: The Low SHAPE-Low Shannon Entropy Metric
Not all structures adopted by long RNAs are well-defined or functionally important. We have found that regions with both low SHAPE reactivities and low Shannon entropy (lowSS) – the former reflecting constrained nucleotides and a highly structured RNA region and the latter indicative of a well-defined structure – are much more likely to be functional (Figure 4, center box). Shannon entropy is calculated from the probability of formation of each base pair across all possible structures in the ensemble compatible with the SHAPE data1,69.
Figure 4.
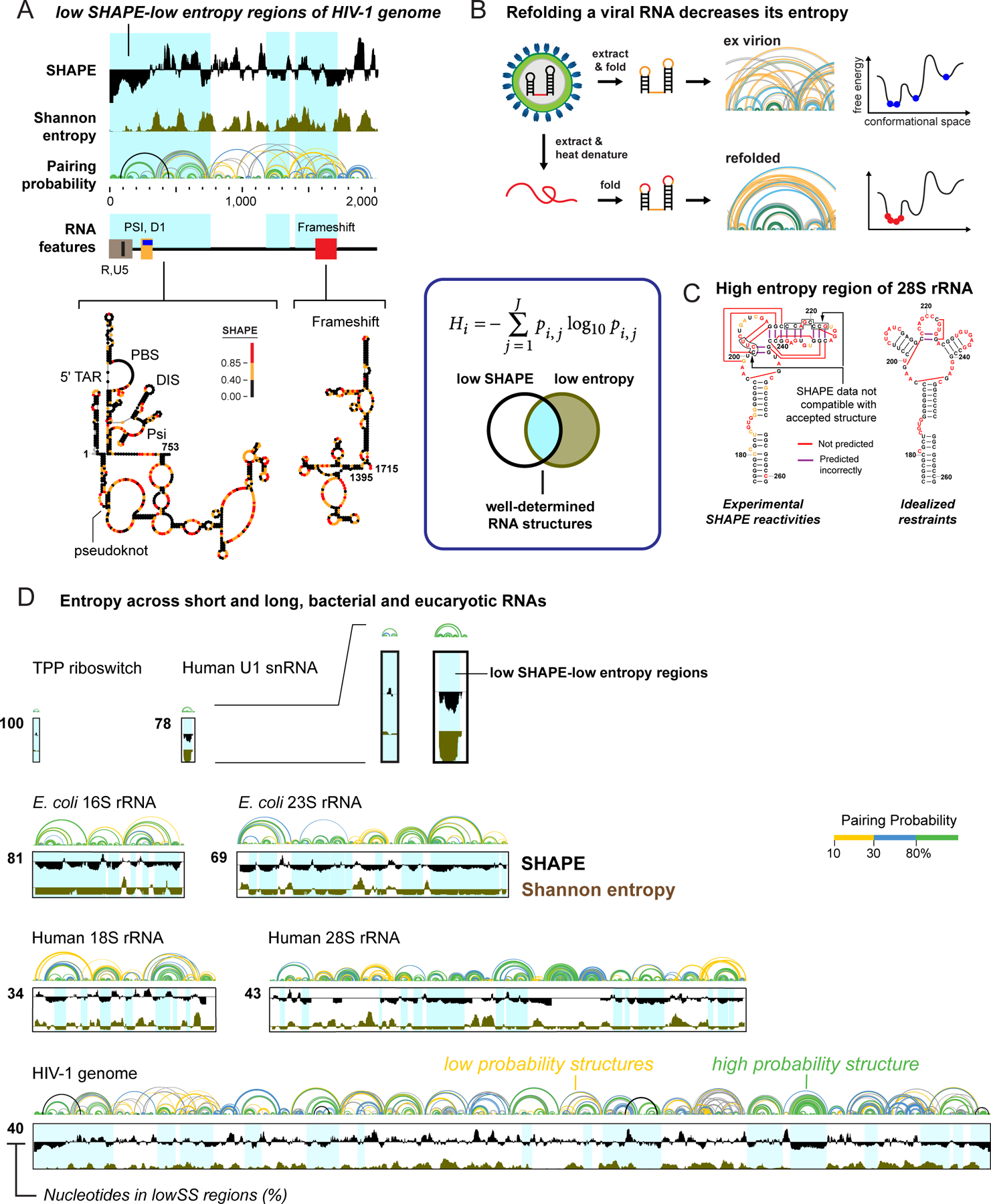
Identification of well-determined RNA structures. (box) Equation for Shannon entropy (H) and illustration of overlap of low SHAPE and low Shannon entropy (lowSS) regions. (A) Representative analysis illustrating that functional RNA elements tend to overlap with lowSS regions (blue shading), here corresponding to the 5’-UTR and frameshift elements of HIV-1. (B and C) lowSS analysis of (B) native-like and refolded dengue RNA genomes72 and (C) human rRNAs74. (D) Illustration of the enormous diversity in lowSS regions, extending from the TPP riboswitch (79 nts, 100% lowSS) to the HIV-1 RNA genome (9,173 nts, 40% lowSS), and including bacterial and human rRNAs15,74. All RNAs shown on the same length scale.
The usefulness of the lowSS metric first became clear in studies of the HIV-1 genome (Figure 4A). Most known functional motifs were found in lowSS regions, and the metric was predictive of novel, and validatable, pseudoknot-containing elements1. Functional motifs have since been shown to be overrepresented among lowSS regions in RNA viruses1,3,61,70, lncRNAs71, and mRNAs2. The lowSS metric also allows de novo identification of well-folded and recapitulatable structural elements in synthetic or refolded RNAs studied under simplified conditions. For example, refolding the dengue virus RNA genome (~10 kb nucleotides) yielded an RNA that was more highly structured and sampled fewer conformations than the more native-like RNA extracted from virions72. The lowSS metric identified the subset of elements that folded similarly in both states (Figure 4B).
Bacterial1,20,21 and archaeal73 ribosomal RNAs (rRNAs) have been widely used to validate SHAPE-directed RNA structure modeling. In strong contrast, when we obtained SHAPE profiles for the human 18S and 28S rRNAs, as gently extracted from cells, numerous regions were incompatible with accepted structures74. This deficit was not a limitation of SHAPE-directed modeling itself, as many motifs could be modelled accurately using idealized reactivities (Figure 4C). Instead, only about 40% of human rRNAs formed well-determined secondary structures. Prior studies using STOP-RT readouts had also obtained structure probing data for eukaryotic rRNAs, but the discrepancy between modeled and accepted structures was not noted, likely due to the non-quantitative nature of STOP-RT methods. Eukaryotic rRNAs thus have less well-determined structures than prokaryotic rRNAs and are extensively unfolded when gently extracted from cells (Figure 4C, 4D).
The lowSS criterion reveals extensive differences between and across RNAs. The base-paired secondary structures of the bacterial TPP riboswitch and human U1 snRNA are extremely well determined15,74 (Figure 4D). Bacterial rRNAs similarly contain large expanses of lowSS regions, whereas human rRNAs contain large regions of high-entropy regions15,74 (Figure 4D). Large RNAs like the HIV-1 RNA genome1 and the Xist noncoding RNA71 have heterogeneous profiles, with high-entropy regions punctuated by regions of well-determined structure (Figure 4D). The extent of lowSS regions is one way every cellular RNA has its own “structural personality”.
The Importance of Comprehensiveness
A dominant lesson from large-scale RNA structure analysis is that RNA molecules should be studied comprehensively. For example, early models for the CAG repeat sequence in the HTT mRNA, implicated in Huntington’s disease, and for the translational frameshift element in the HIV-1 genome emphasized simple helices that form when short RNAs spanning these regions are studied75,76 (Figure 5A, B). In the context of native flanking sequences, however, much more complex structures are observed: For the HTT mRNA, short CAG repeats primarily pair with flanking poly-CCN sequences; whereas, only long, disease-associated repeats form CAG stem-loop structures77 (Figure 5A). For the HIV-1 frameshift element, SHAPE-based analyses of full-length genome transcripts identified a second frameshifting sequence and revealed the functional importance of RNA helices that only form in long transcripts78,79 (Figure 5B). These complex structures may support selective binding by small-molecule ligands80. These results emphasize the importance of analyzing RNA structure in the context of full-length RNAs.
Figure 5.
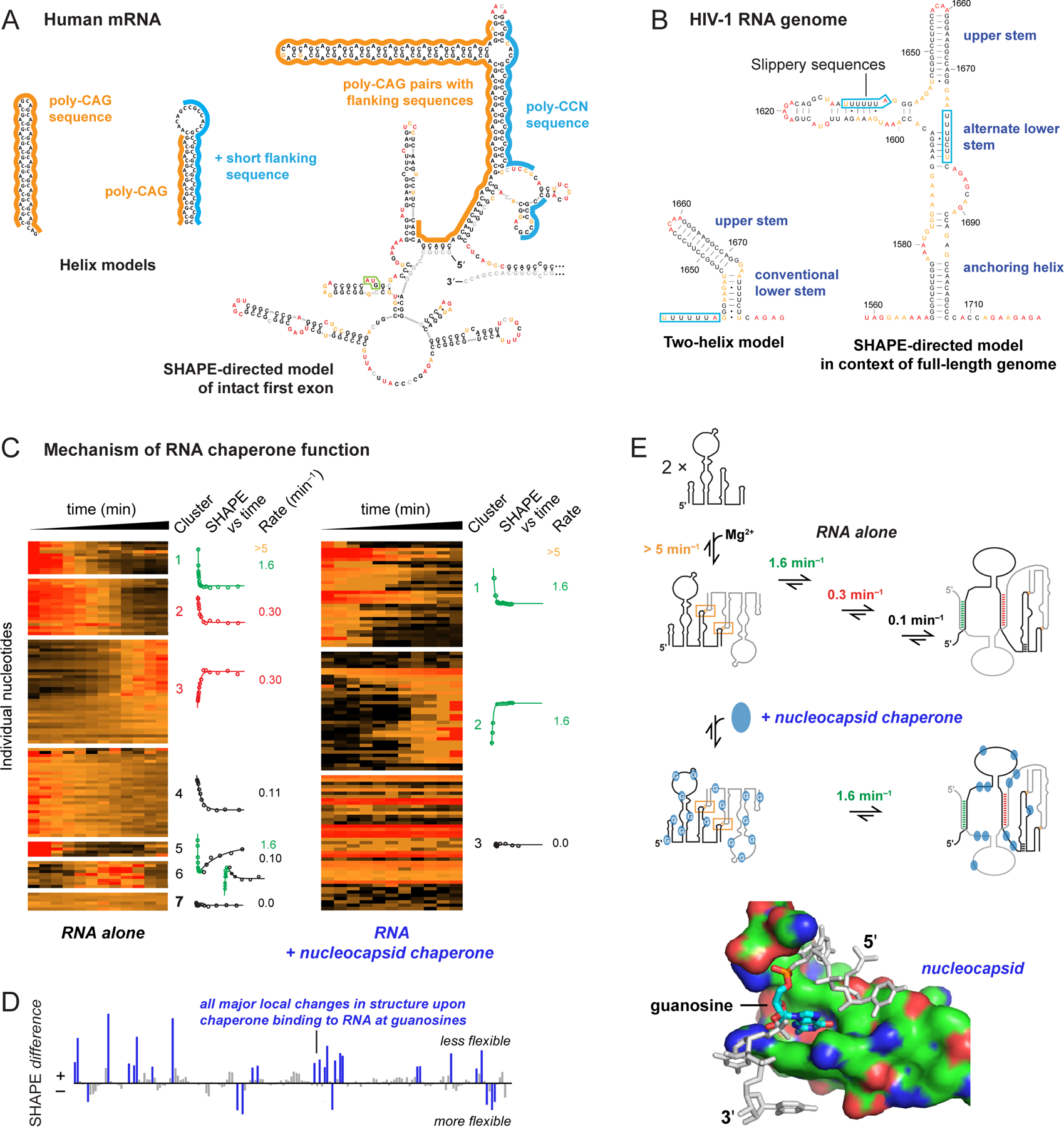
Comprehensiveness in understanding RNA structure and RNA-protein interactions. (A) Secondary structure models for CAG repeat-containing sequences from human HTT mRNA based on analysis of (left) local structures and (right) the full-length first exon (360–500 nts, depending on number of CAG repeats)77. Start codon is boxed in green. (B) Secondary structure models of the HIV-1 RNA frameshift element based on analysis of (left) local structure and (right) the entire HIV-1 genome78. (C) Time-resolved folding of a retroviral RNA packaging domain in the absence and presence of nucleocapsid chaperone83. Per-nucleotide reactivities are shown on a scale from red (high) to black (low). Nucleotides are grouped by k-means clustering; kinetic profiles are shown for each cluster. (D) SHAPE-detected protein-RNA contacts 7 seconds after protein addition reveals interactions at guanosine (blue). (E) Model for chaperone-mediated facilitation of RNA folding by destabilizing interactions involving guanosine83.
Another class of comprehensiveness is the ability to interrogate all four ribonucleotides. Folding of many large RNAs is facilitated by proteins called RNA chaperones that accelerate formation of RNA-RNA interactions, catalyze conformational changes and acquisition of unique functional structures, and act over long sequence distances81,82. We used time-resolved SHAPE – employing the fast reacting benzoyl cyanide reagent40 (Figure 2E; half-life 0.25 seconds) – to interrogate the structure of a retroviral RNA element that undergoes a complex dimerization reaction essential for correct packaging of two RNA genomes in a virion83. The RNA-alone dimerization reaction is slow, showing seven distinct kinetic behaviors in four rate processes. In the presence of the viral nucleocapsid chaperone protein, dimerization occurs rapidly, in a single step (Figure 5C). The chaperone specifically destabilizes RNA interactions involving guanosine (Figure 5D). Guanosine forms highly stable base pairs with cytidine and promiscuously pairs with cytidine and uridine, features that likely create a complex RNA folding landscape. Our SHAPE-directed model emphasizes that the nucleocapsid RNA chaperone acts over large distances via a simple mechanism: weakening interactions involving guanosine83 (Figure 5E).
Effects of the Cellular Environment on RNA Structure
The cellular environment is vastly different from conditions in test-tube experiments. Cells are crowded and contain numerous classes of RNA binding proteins. Diverse processes including translation, splicing, transport, and macromolecular assembly are likely to affect RNA structure. When we compared the structure of the aptamer domain of the adenine riboswitch under simplified test-tube conditions versus in Escherichia coli cells, our SHAPE analysis revealed that the riboswitch RNA is much more highly structured in cells than under simplified conditions50 (Figure 6A). Even addition of Mg2+ to 30 mM in test tube experiments, a highly stabilizing condition for RNA, does not yield an RNA that is as structured as in cells. Molecular crowding agents, like PEG, stabilized long-range loop-loop interactions but, overall, failed to stabilize the structure of the riboswitch as well as the in-cell state84. In cells, ligand binding only altered SHAPE reactivities of nucleotides close to the ligand pocket, indicating that the RNA is essentially fully folded in cells (Figure 6A). The intracellular environment thus has a large effect on RNA structure that is difficult to replicate in test-tube experiments.
Figure 6.
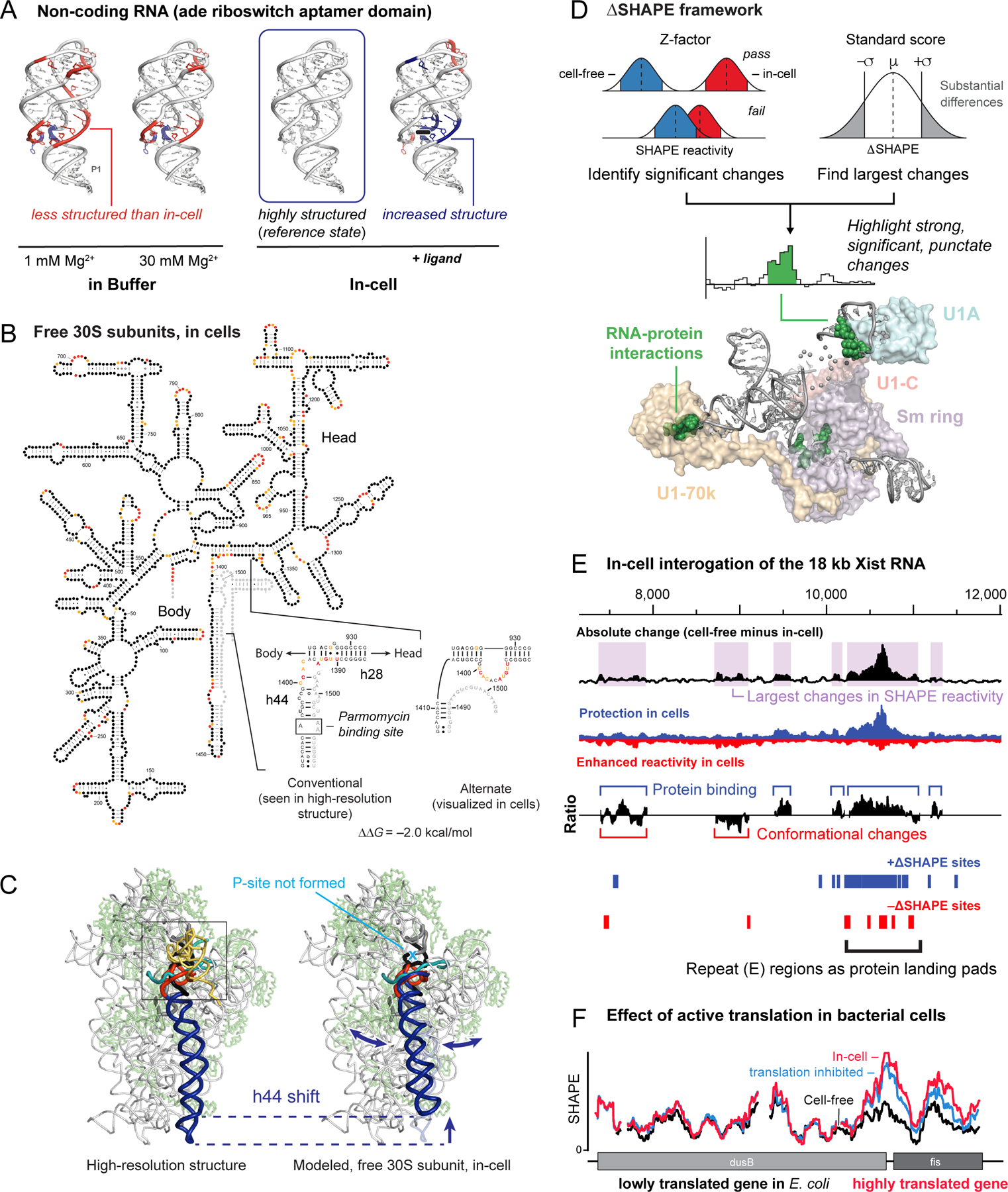
Cellular environment and RNA structure. (A) SHAPE analysis of the adenine riboswitch aptamer domain under simplified conditions, in cells (the reference state, box), and in the presence of ligand50. Higher and lower SHAPE reactivities relative to in-cell RNA are red and blue, respectively. (B) Model for the structure of the 16S rRNA in free 30S subunits in cells49. SHAPE reactivity pattern in helices 28 and 44 is incompatible with the structure visualized in high-resolution structures. (C) Movement of helix 44 in the in-cell state, emphasizing a large-scale conformational switch. (D) The ∆SHAPE framework for identifying significant changes between two states. Structure shows ∆SHAPE sites in the human U1 snRNP complex (green spheres) and their proximal proteins. (E) Protein interactions across the mouse Xist lncRNA mapped using large-scale difference analysis. (top) Effects of the in-cell environment categorized by absolute differences in SHAPE reactivity (50-nt sliding window). (middle) Ratio of positive to negative differences is suggestive of protein binding and RNA structural rearrangement. (bottom) Positive and negative ∆SHAPE sites are blue and red, respectively. (F) Effect of translation on RNA structure in E. coli cells. SHAPE reactivities increase, relative to the cell-free state, specifically in highly translated genes; kasugamycin treatment partially abrogates this increase.
It had been known for decades that the small subunit of the ribosome can interconvert between “active” and “inactive” states85,86. We found that in cells, 16S rRNA in 30S subunits exists primarily in an inactive conformation that has a SHAPE profile in helices 28 and 44 incompatible with high-resolution data49 (Figure 6B). Binding by the antibiotic paromomycin switches the 16S rRNA into the active conformation, implying that the energy barrier between active and inactive states is low. Mutants that inhibit interconversion between states compromise translation in cells. Modeling revealed that the inactive and active structures differ in the position of helix 44, which likely has large effects on interactions between the 30S subunit and mRNA and for assembly with the large subunit49 (Figure 6C). Thus, SHAPE revealed that the classic 30S ribosomal subunit inactive state is an abundant in-cell structure that regulates ribosome function.
SHAPE-MaP is unique among structure probing strategies because counting mutation events enables both measurement of local nucleotide flexibility and quantification of the uncertainty in each measurement (Figure 3A, bottom)1,53. These features enable a statistical framework, ∆SHAPE53,87, that tests for significant local structural differences between two states (Figure 6D). SHAPE reactivity differences can be summed over larger windows and used to identify positive and negative differences reflecting transitions between structured and unstructured as a result of cellular interactions, especially protein binding.
We used the ∆SHAPE framework to examine the effects of the cellular environment on the 18-kb Xist lncRNA71. Roughly one-half of all Xist nucleotides are changed by the cellular environment (Figure 6E). Xist contains several partial tandem repeat sequences that show large differences in SHAPE reactivity under simplified conditions as compared to in cells. Tandem repeats are among the few motifs that are consistently unstructured over large regions, and we showed that repeat regions function as landing pads for protein binding (Figure 6E). Other regions appear to either modulate protein binding or undergo conformational changes upon forming protein interactions. Our studies of Xist demonstrated that in-cell SHAPE allows efficient identification of important functional regions of large RNAs71 that can then be the focus of mechanism-based studies.
Ongoing cellular processes also affect RNA structure. SHAPE probing revealed that translation disrupts RNA structure in coding regions in E. coli cells and that this effect is larger specifically in highly translated coding regions2. This in-cell RNA destabilization effect is reduced when translation is inhibited, and disappears for cell-free RNA (Figure 6F). In sum, RNA structure is heavily modulated by the cellular environment and can increase, decrease, or rearrange dramatically due to crowding, structural rearrangements, or interaction with proteins, including in surprising ways. SHAPE enables direct discovery and modeling of these effects.
mRNAs Have Distinct Structural Personalities
Data from most “transcription-wide” studies are sparse, noisy, and often analyzed in bulk statistical or meta-gene frameworks. As emphasized above, focusing analysis on such data is often misleading. For the E. coli transcriptome, meta-gene analysis would suggest that mRNAs are featureless (Figure 7A), but comprehensive, high-quality data reveal a strikingly different picture: E. coli transcripts show enormous structural diversity2 (Figure 7B). Highly structured lncRNAs, like the RNase P RNA, have low SHAPE reactivities. By comparison, SHAPE reactivities for coding regions vary dramatically (Figure 7B). Each transcript, and indeed individual regions within transcripts, have distinct “structural personalities”.
Figure 7.
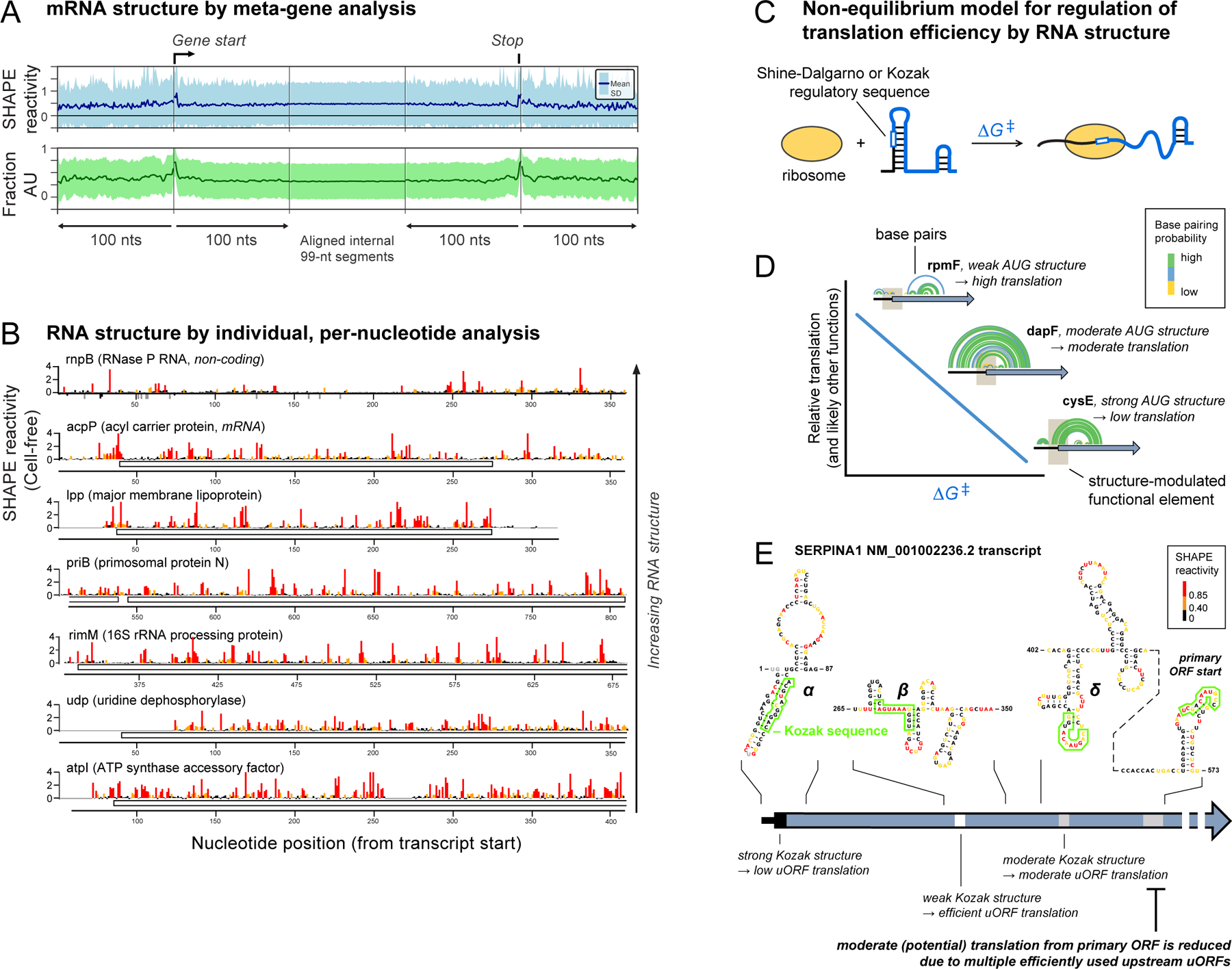
“Structural personalities” of bacterial and human RNAs. (A) Meta-gene representation of averaged mRNA structure in E. coli based on SHAPE reactivity and A/U nucleotide content. (B) Per-nucleotide SHAPE reactivities for one lncRNA and six mRNAs from E. coli2. (C) Model for how RNA structure modulates accessibility to regulatory sequences. Unfolding an RNA motif (∆G‡) imposes an energetic penalty on translation initiation (and likely many other processes). (D) RNA structures that tune translation initiation in E. coli. Brown box indicates AUG translation start site; arcs illustrate RNA base pairs. (E) Example of SERPINA1 mRNA structure containing a primary ORF and three (α, β, γ) competing upstream open reading frames (uORFs). Minimum free energy structures are shown, nucleotides are colored by SHAPE reactivity, and Kozak sequences are boxed.
In one example, mRNAs vary substantially in translation efficiency (TE), the amount of protein made from a given mRNA. Classical studies had shown that mRNA structure can enhance or impede the accessibility of regulatory sequences in an mRNA, influencing TE88. We used high-quality SHAPE data from both bacterial and eukaryotic mRNAs to develop a general model for how RNA structure regulates TE89. For E. coli mRNAs, a non-equilibrium model in which translation requires local unfolding of 30 nucleotides of RNA structure at the site of the Shine-Dalgarno sequence and start codon explains TE well2 (Figure 7C). The best relationship between RNA structure and translation was obtained when we calculated the cost of disrupting pre-existing structure without considering refolding, suggesting that TE is governed by the cost of disrupting pre-existing structure, with unfolding rapidly followed by ribosome binding89 (Figure 7D).
We extended these results to the 11 spliced isoforms of the human SERPINA1 mRNA. Each isoform encodes exactly the same protein in a primary open reading frame (ORF) downstream of different combinations of up to three upstream open reading frames (uORFs), each with their own Kozak sequence start site sequence of differing strengths90. Use of any of the uORFs reduces translation efficiency at the primary ORF. When we used SHAPE data to model the structures of the 11 isoforms, we found that the same non-equilibrium model (Figure 7C), developed for bacterial RNAs, explained the relative accessibility of the uORFs and the primary ORF. Our model yielded a dramatic improvement over the so-called leaky-scanning model for translation of SERPINA1 isoforms90 (Figure 7E). In sum, these results emphasize the importance of obtaining high-quality, per-nucleotide data across entire RNA transcripts and reveal that apparently unremarkable RNA structures can tune RNA functions by modulating the interactive accessibility of RNA to diverse ligands and proteins.
Structure-based Discovery of RNA Regulatory Elements in E. coli
In high-quality E. coli transcriptome data (Figure 8A), we identified 58 lowSS motifs, situated in 51 of 147 (35%) non-coding RNA regions2 (Figure 8B). Of these, 49 motifs were uncharacterized, 80% showed evolutionary conservation, and roughly half overlapped regions with literature evidence for function (Figure 8C). Several were validated by our lab. For example, we identified a three-helix junction in the 5’-UTR of the operon that encodes the ribosomal proteins L28 and L33. Remarkably, the SHAPE-directed structure of this motif is similar to that of the large subunit rRNA in the region that binds L28 (Figure 8A). We showed that L28 and L33, plus L9 encoded in another operon, bind this three-helix junction element: L9 and L28 bind together, and L9 binding competes with L33. This motif, identified de novo by SHAPE-informed discovery, exhibits remarkable regulatory complexity2. Every E. coli transcript we studied, for which we could obtain high-quality SHAPE data, appears to have some element of its expression regulated by RNA structure2 (Figure 8D).
Figure 8.
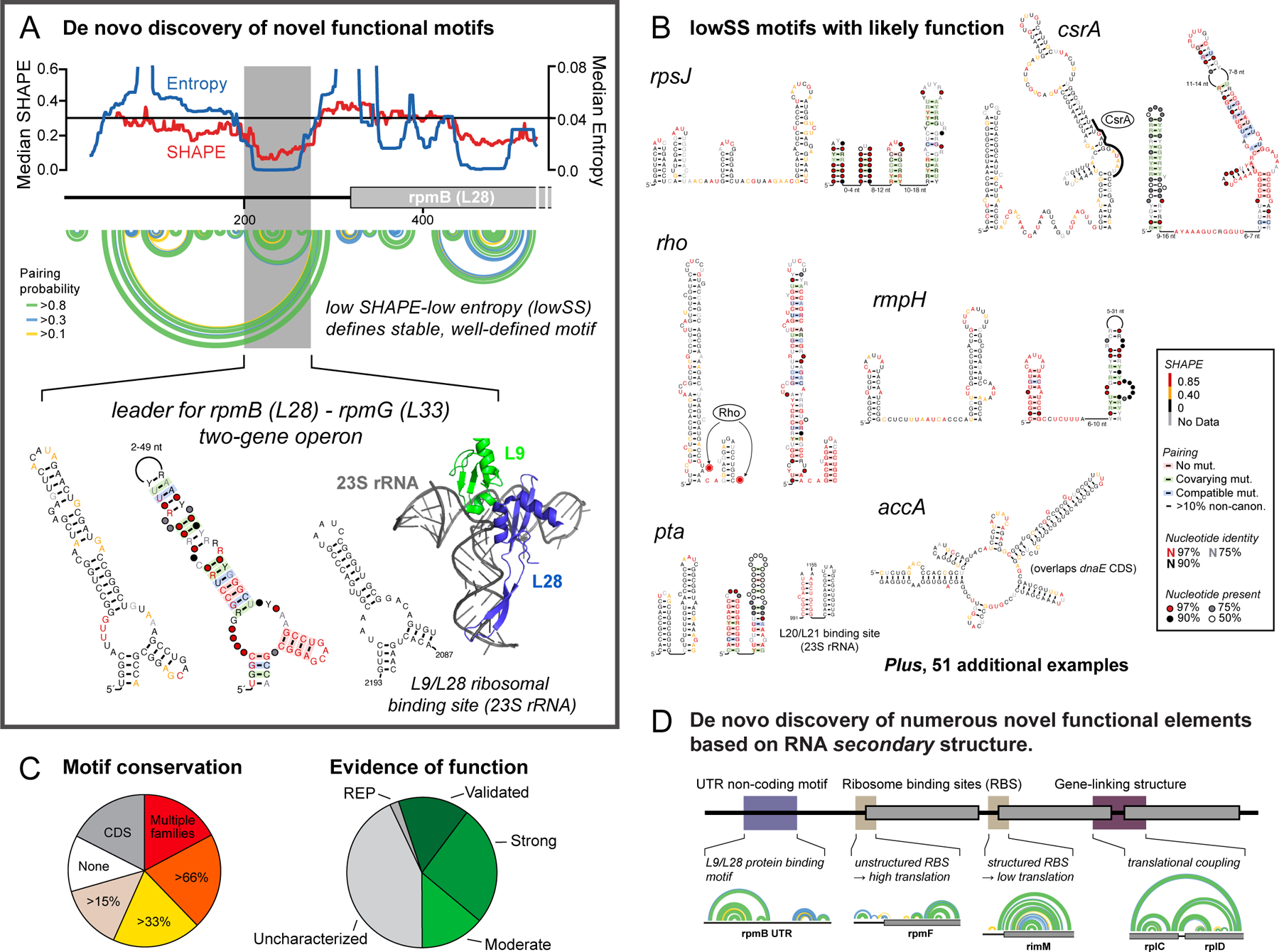
De novo discovery of functional RNA elements as lowSS regions. (A) A lowSS region (gray box) in the rpmB mRNA forms an autoregulatory element that mimics L9/L28 binding sites in the 28S rRNA. (B) Novel RNA regulatory elements identified in E. coli. Structures are annotated by SHAPE reactivity and evidence for conservation. (C) Conservation of lowSS structures identified in enterobacteria and evidence of function based on literature. (D) Mechanisms by which RNA structure regulates gene expression across the E. coli transcriptome based on identification of well-determined secondary structures2. Arcs indicate base pairs.
The Frontier in Motif Discovery: Higher-order and Tertiary RNA Structure
The next frontier opportunity is to extend large-scale RNA structure analysis to more complex structures. We recently probed the structure of the entire dengue serotype 2 virus (DENV2) RNA genome by SHAPE and identified 24 lowSS elements3 (half are shown in Figure 9A). These elements overlap with previously identified functional elements or show evidence of evolutionary pressure to maintain the modeled secondary structure, consistent with functional roles in viral replication. The existence of numerous higher-order structures in a large RNA is now an utterly unremarkable observation. A critical goal now is to determine which of these elements form higher-order functional structures.
Figure 9.
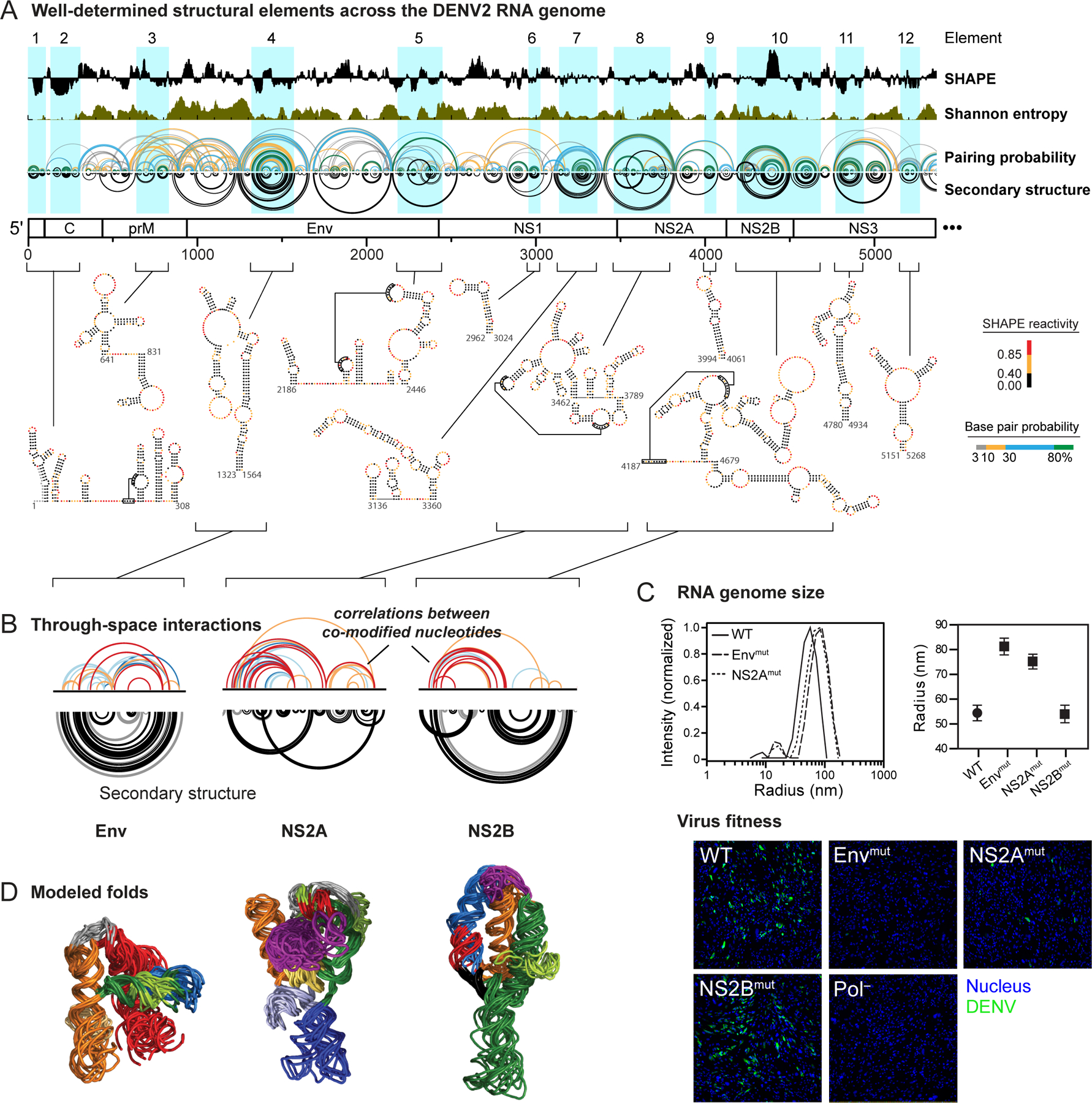
Discovery of higher-order RNA structure. (A) Well-determined secondary structure elements across the first half of the DENV2 genome based on the lowSS metric. (B) RING correlated chemical probing analysis of three RNA elements with well-determined structures and higher-order folds. Elements are named by the gene in which they occur. (C) Effect of mutations in regions with significant RING correlations on (top) global compaction of the DENV2 RNA genome (measured by dynamic light scattering) and on (bottom) replication fitness (visualized by immunostaining of DENV2 envelope protein and nuclei). (D) Modeled three-dimensional folds of three elements in the DENV2 RNA based on RING correlations used to restrain discrete molecular dynamics simulations.
We used a new chemical probing strategy called RING-MaP16,17 to assess the likelihood that individual elements in the DENV2 RNA genome form higher-order structures. RING-MaP exploits the ability of MaP reverse transcription to measure multiple chemical adducts in a single RNA strand. Nucleotides that experience structural communication, because they are in the same helix19,57 or because they are linked via through-space interactions16,17,59, show correlated internucleotide reactivities more often than expected by chance (Figure 3A, bottom). At present, RING uses the conventional structural probing reagent dimethyl sulfate16,19. RINGs can be approximately separated into those corresponding to secondary versus tertiary interactions by the number of nucleotides between sites after excluding internal stem loops3; longer contact distances reflect tertiary structure. Eight regions in the DENV2 RNA showed RING correlations supportive of higher-order tertiary structure (three are shown in Figure 9B).
Mutating these RNA elements with higher-order structure, located in the Env and NS2A genes (although not the NS2B gene), increased the hydrodynamic radius of RNA genome transcripts and compromised viral fitness3 (Figure 9C). The mutant viruses replicated poorly even after 60 days of passage in cell culture, suggesting that disrupting RNA tertiary structure is not easily revertible by the virus. We used RING measurements as restraints in molecular dynamics simulations to model the overall folds of regions with dense RINGs. Most elements fold into compact, well-determined structures (Figure 9D). The precise mechanisms by which these higher-order RNA structures affect viral fitness are not fully known.
In sum, RING experiments revealed that RNA motifs with complex, higher-order architectures are pervasive across the DENV2 RNA, are functionally integrated with viral replication, and – we posit – are harbingers of complex, functional structures likely to be found in the future in numerous viral, messenger, and non-coding RNAs.
Long-term Opportunities
RNAs fold back on themselves to form substantial internal structures, many of which play direct roles in function. With the advent of experimentally concise, comprehensive, quantitative, and high-throughput RNA structure probing technologies, we are now poised to identify and focus on those structures most likely to impact function and regulatory mechanisms. In essentially every instance where my laboratory, and many other research groups, have applied SHAPE to interrogate biological systems, new insights regarding RNA structure-function interrelationships have emerged. RNA structure likely regulates expression and biological roles of every class of RNA, is sufficiently ubiquitous to comprise a distinct level of the genetic code, and likely creates higher-order folds broadly targetable with small molecules.
Although additional innovations will certainly be developed, per-nucleotide RNA structure probing by SHAPE is now a mature, well validated, and robust technology. SHAPE, mutational profiling (MaP), and related strategies should be applied to essentially all projects involving RNA. The RNA structure probing and analysis field is poised to undergo a transition. The next frontier is to devise experimentally concise, information-rich, and accurate strategies for measuring higher-order and tertiary interactions at large scales.
Acknowledgements
K.M.W. is indebted to the many creative student, postdoctoral, and faculty colleagues who co-created projects described here. Our laboratory is supported by the National Institutes of Health (R35 GM122532, R01 AI068462 and R01 HL111527), the National Science Foundation (MCB-2027701), and private foundations and biotechnology companies.
Biography
Kevin Weeks is Kenan Distinguished Professor of Chemistry at the University of North Carolina. His laboratory leverages chemical principles to devise definitive and quantitative technologies for understanding how RNA structure governs biological function and uses these technologies to define new principles of RNA-mediated regulation, spanning virus replication, mRNA and lncRNA biology, and RNA-targeted drug discovery. The overarching vision of the Weeks laboratory is to prepare team members for long-term leadership at the interfaces of chemistry, biology, technology, and genomics.
Footnotes
Disclosure
K.M.W. is an advisor to and holds equity in Ribometrix.
References
- (1).Siegfried NA; Busan S; Rice GM; Nelson JAE; Weeks KM RNA Motif Discovery by SHAPE and Mutational Profiling (SHAPE-MaP). Nature Methods 2014, 11, 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Mustoe AM; Busan S; Rice GM; Hajdin CE; Peterson BK; Ruda VM; Kubica N; Nutiu R; Baryza JL; Weeks KM Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing. Cell 2018, 173, 181–195.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Dethoff EA; Boerneke MA; Gokhale NS; Muhire BM; Martin DP; Sacco MT; McFadden MJ; Weinstein JB; Messer WB; Horner SM; Weeks KM Pervasive Tertiary Structure in the Dengue Virus RNA Genome. Proc Natl Acad Sci U S A 2018, 115, 11513–11518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Sharp PA The Centrality of RNA. Cell 2009, 136, 577–580. [DOI] [PubMed] [Google Scholar]
- (5).Atkins JF; Gesteland RF; Cech T RNA Worlds: From Life’s Origins to Diversity in Gene Regulation; 2011.
- (6).Mauger DM; Siegfried NA; Weeks KM The Genetic Code as Expressed Through Relationships Between mRNA Structure and Protein Function. FEBS Lett 2013, 587, 1180–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Cech TR; Steitz JA The Noncoding RNA Revolution-Trashing Old Rules to Forge New Ones. Cell 2014, 157, 77–94. [DOI] [PubMed] [Google Scholar]
- (8).Herbert A; Rich A RNA Processing and the Evolution of Eukaryotes. Nat Genet 1999, 21, 265–269. [DOI] [PubMed] [Google Scholar]
- (9).Cruz JA; Westhof E The Dynamic Landscapes of RNA Architecture. Cell 2009, 136, 604–609. [DOI] [PubMed] [Google Scholar]
- (10).Lieberman J Tapping the RNA World for Therapeutics. Nat. Struct. Mol. Biol 2018, 25, 357–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Peattie DA; Gilbert W Chemical Probes for Higher-Order Structure in RNA. Proc Natl Acad Sci U S A 1980, 77, 4679–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Merino EJ; Wilkinson KA; Coughlan JL; Weeks KM RNA Structure Analysis at Single Nucleotide Resolution by Selective 2’-Hydroxyl Acylation and Primer Extension (SHAPE). J Am Chem Soc 2005, 127, 4223–4231. [DOI] [PubMed] [Google Scholar]
- (13).Wilkinson KA; Merino EJ; Weeks KM RNA SHAPE Chemistry Reveals Nonhierarchical Interactions Dominate Equilibrium Structural Transitions in tRNA(Asp) Transcripts. J Am Chem Soc 2005, 127, 4659–4667. [DOI] [PubMed] [Google Scholar]
- (14).Mortimer SA; Weeks KM A Fast-Acting Reagent for Accurate Analysis of RNA Secondary and Tertiary Structure by SHAPE Chemistry. J Am Chem Soc 2007, 129, 4144–4145. [DOI] [PubMed] [Google Scholar]
- (15).Smola MJ; Rice GM; Busan S; Siegfried NA; Weeks KM Selective 2’-Hydroxyl Acylation Analyzed by Primer Extension and Mutational Profiling (SHAPE-MaP) for Direct, Versatile and Accurate RNA Structure Analysis. Nat Protoc 2015, 10, 1643–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Homan PJ; Favorov OV; Lavender CA; Kursun O; Ge X; Busan S; Dokholyan NV; Weeks KM Single-Molecule Correlated Chemical Probing of RNA. Proc Natl Acad Sci U S A 2014, 111, 13858–13863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Sengupta A; Rice GM; Weeks KM Single-Molecule Correlated Chemical Probing Reveals Large-Scale Structural Communication in the Ribosome and the Mechanism of the Antibiotic Spectinomycin in Living Cells. PLoS Biol 2019, 17, e3000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Zubradt M; Gupta P; Persad S; Lambowitz AM; Weissman JS; Rouskin S DMS-MaPseq for Genome-Wide or Targeted RNA Structure Probing in Vivo. Nature Methods 2017, 14, 75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Mustoe AM; Lama NN; Irving PS; Olson SW; Weeks KM RNA Base-Pairing Complexity in Living Cells Visualized by Correlated Chemical Probing. Proc Natl Acad Sci U S A 2019, 116, 24574–24582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Deigan KE; Li TW; Mathews DH; Weeks KM Accurate SHAPE-Directed RNA Structure Determination. Proc Natl Acad Sci U S A 2009, 106, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hajdin CE; Bellaousov S; Huggins W; Leonard CW; Mathews DH; Weeks KM Accurate SHAPE-Directed RNA Secondary Structure Modeling, Including Pseudoknots. Proc Natl Acad Sci U S A 2013, 110, 5498–5503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Weeks KM; Mauger DM Exploring RNA Structural Codes with SHAPE Chemistry. Acc Chem Res 2011, 44, 1280–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Lu Z; Chang HY Decoding the RNA Structurome. Curr Opin Struct Biol 2016, 36, 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Rausch JW; Sztuba-Solinska J; Le Grice SFJ Probing the Structures of Viral RNA Regulatory Elements with SHAPE and Related Methodologies. Front Microbiol 2017, 8, 2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Strobel EJ; Yu AM; Lucks JB High-Throughput Determination of RNA Structures. Nat Rev Genet 2018, 19, 615–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Mitchell D; Assmann SM; Bevilacqua PC Probing RNA Structure in Vivo. Curr Opin Struct Biol 2019, 59, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Mauger DM; Cabral BJ; Presnyak V; Su SV; Reid DW; Goodman B; Link K; Khatwani N; Reynders J; Moore MJ; McFadyen IJ mRNA Structure Regulates Protein Expression Through Changes in Functional Half-Life. Proc Natl Acad Sci U S A 2019, 116, 24075–24083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Boerneke MA; Ehrhardt JE; Weeks KM Physical and Functional Analysis of Viral RNA Genomes by SHAPE. Annu Rev Virol 2019, 6, 93–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Martin S; Blankenship C; Rausch JW; Sztuba-Solinska J Using SHAPE-MaP to Probe Small Molecule-RNA Interactions. Methods 2019, 167, 105–116. [DOI] [PubMed] [Google Scholar]
- (30).Li B; Cao Y; Westhof E; Miao Z Advances in RNA 3D Structure Modeling Using Experimental Data. Front Genet 2020, 11, 574485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Manfredonia I; Incarnato D Structure and Regulation of Coronavirus Genomes: State-of-the-Art and Novel Insights From SARS-CoV-2 Studies. Biochem Soc Trans 2020, 58, 343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).McGinnis JL; Dunkle JA; Cate JHD; Weeks KM The Mechanisms of RNA SHAPE Chemistry. J Am Chem Soc 2012, 134, 6617–6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Gherghe CM; Shajani Z; Wilkinson KA; Varani G; Weeks KM Strong Correlation Between SHAPE Chemistry and the Generalized NMR Order Parameter (S2) in RNA. J Am Chem Soc 2008, 130, 12244–12245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Wang B; Wilkinson KA; Weeks KM Complex Ligand-Induced Conformational Changes in tRNA(Asp) Revealed by Single-Nucleotide Resolution SHAPE Chemistry. Biochemistry 2008, 47, 3454–3461. [DOI] [PubMed] [Google Scholar]
- (35).Steen K-A; Rice GM; Weeks KM Fingerprinting Noncanonical and Tertiary RNA Structures by Differential SHAPE Reactivity. J Am Chem Soc 2012, 134, 13160–13163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Warner KD; Homan P; Weeks KM; Smith AG; Abell C; Ferre-D’Amare AR Validating Fragment-Based Drug Discovery for Biological RNAs: Lead Fragments Bind and Remodel the TPP Riboswitch Specifically. Chem Biol 2014, 21, 591–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Busan S; Weidmann CA; Sengupta A; Weeks KM Guidelines for SHAPE Reagent Choice and Detection Strategy for RNA Structure Probing Studies. Biochemistry 2019, 58, 2655–2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Spitale RC; Crisalli P; Flynn RA; Torre EA; Kool ET; Chang HY RNA SHAPE Analysis in Living Cells. Nat. Chem. Biol 2013, 9, 18–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Marinus T; Fessler AB; Ogle CA; Incarnato D A Novel SHAPE Reagent Enables the Analysis of RNA Structure in Living Cells with Unprecedented Accuracy. Nucleic Acids Res 2021, 15, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Mortimer SA; Weeks KM Time-Resolved RNA SHAPE Chemistry. J Am Chem Soc 2008, 130, 16178–16180. [DOI] [PubMed] [Google Scholar]
- (41).Mortimer SA; Weeks KM C2’-Endo Nucleotides as Molecular Timers Suggested by the Folding of an RNA Domain. Proc Natl Acad Sci U S A 2009, 106, 15622–15627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Watters KE; Strobel EJ; Yu AM; Lis JT; Lucks JB Cotranscriptional Folding of a Riboswitch at Nucleotide Resolution. Nat. Struct. Mol. Biol 2016, 23, 1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Gherghe CM; Mortimer SA; Krahn JM; Thompson NL; Weeks KM Slow Conformational Dynamics at C2’-Endo Nucleotides in RNA. J Am Chem Soc 2008, 130, 8884–8885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Goodwin S; McPherson JD; McCombie WR Coming of Age: Ten Years of Next-Generation Sequencing Technologies. Nat Rev Genet 2016, 17, 333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Ehresmann C; Baudin F; Mougel M; Romby P; Ebel JP; Ehresmann B Probing the Structure of RNAs in Solution. Nucleic Acids Res 1987, 15, 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Moazed D; Stern S; Noller HF Rapid Chemical Probing of Conformation in 16 S Ribosomal RNA and 30 S Ribosomal Subunits Using Primer Extension. J. Mol. Biol 1986, 187, 399–416. [DOI] [PubMed] [Google Scholar]
- (47).McGinnis JL; Weeks KM Ribosome RNA Assembly Intermediates Visualized in Living Cells. Biochemistry 2014, 53, 3237–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Karabiber F; McGinnis JL; Favorov OV; Weeks KM QuShape: Rapid, Accurate, and Best-Practices Quantification of Nucleic Acid Probing Information, Resolved by Capillary Electrophoresis. RNA 2013, 19, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).McGinnis JL; Liu Q; Lavender CA; Devaraj A; McClory SP; Fredrick K; Weeks KM In-Cell SHAPE Reveals That Free 30S Ribosome Subunits Are in the Inactive State. Proc Natl Acad Sci U S A 2015, 112, 2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Tyrrell J; McGinnis JL; Weeks KM; Pielak GJ The Cellular Environment Stabilizes Adenine Riboswitch RNA Structure. Biochemistry 2013, 52, 8777–8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Weeks KM RNA Structure Probing Dash Seq. Proc Natl Acad Sci U S A 2011, 108, 10933–10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Weeks KM Toward All RNA Structures, Concisely. Biopolymers 2015, 103, 438–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Smola MJ; Calabrese JM; Weeks KM Detection of RNA-Protein Interactions in Living Cells with SHAPE. Biochemistry 2015, 54, 6867–6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Sexton AN; Wang PY; Rutenberg-Schoenberg M; Simon MD Interpreting Reverse Transcriptase Termination and Mutation Events for Greater Insight Into the Chemical Probing of RNA. Biochemistry 2017, 56, 4713–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Busan S; Weeks KM Accurate Detection of Chemical Modifications in RNA by Mutational Profiling (MaP) with ShapeMapper 2. RNA 2018, 24, 143–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Larman BC; Dethoff EA; Weeks KM Packaged and Free Satellite Tobacco Mosaic Virus (STMV) RNA Genomes Adopt Distinct Conformational States. Biochemistry 2017, 56, 2175–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Krokhotin A; Mustoe AM; Weeks KM; Dokholyan NV Direct Identification of Base-Paired RNA Nucleotides by Correlated Chemical Probing. RNA 2017, 23, 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Tomezsko PJ; Corbin VDA; Gupta P; Swaminathan H; Glasgow M; Persad S; Edwards MD; Mcintosh L; Papenfuss AT; Emery A; Swanstrom R; Zang T; Lan TCT; Bieniasz P; Kuritzkes DR; Tsibris A; Rouskin S Determination of RNA Structural Diversity and Its Role in HIV-1 RNA Splicing. Nature 2020, 582, 438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Ehrhardt JE; Weeks KM Time-Resolved, Single-Molecule, Correlated Chemical Probing of RNA. J Am Chem Soc 2020, 142, 18735–18740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Manfredonia I; Nithin C; Ponce-Salvatierra A; Ghosh P; Wirecki TK; Marinus T; Ogando NS; Snijder EJ; van Hemert MJ; Bujnicki JM; Incarnato D Genome-Wide Mapping of SARS-CoV-2 RNA Structures Identifies Therapeutically-Relevant Elements. Nucleic Acids Res 2020, 48, 12436–12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Huston NC; Wan H; Strine MS; de Cesaris Araujo Tavares R; Wilen CB; Pyle AM Comprehensive in Vivo Secondary Structure of the SARS-CoV-2 Genome Reveals Novel Regulatory Motifs and Mechanisms. Mol. Cell 2021, 81, 584–598.e585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Zhao J; Qiu J; Aryal S; Hackett JL; Wang J The RNA Architecture of the SARS-CoV-2 3’-Untranslated Region. Viruses 2020, 12, 1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Iserman C; Roden CA; Boerneke MA; Sealfon RSG; McLaughlin GA; Jungreis I; Fritch EJ; Hou YJ; Ekena J; Weidmann CA; Theesfeld CL; Kellis M; Troyanskaya OG; Baric RS; Sheahan TP; Weeks KM; Gladfelter AS Genomic RNA Elements Drive Phase Separation of the SARS-CoV-2 Nucleocapsid. Mol. Cell 2020, 80, 1078–1091.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Lan TCT;, Allan MF; Malsick LE; Khandwala S; Nyeo SSY; Bathe M; Griffiths A; Rouskin S Structure of the Full SARS-CoV-2 RNA Genome in Infected Cells 10.1101/2020.06.29.178343v1 2020. [DOI]
- (65).Sanders W; Fritch EJ; Madden EA; Graham RL; Vincent HA; Heise MT; Baric RS; Moorman NJ Comparative Analysis of Coronavirus Genomic RNA Structure Reveals Conservation in SARS-Like Coronaviruses. bioRxiv 10.1101/2020.06.15.153197 2020. [DOI]
- (66).Lu ZJ; Gloor JW; Mathews DH Improved RNA Secondary Structure Prediction by Maximizing Expected Pair Accuracy. RNA 2009, 15, 1805–1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Busan S; Weeks KM Visualization of RNA Structure Models Within the Integrative Genomics Viewer. RNA 2017, 23, 1012–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Busan S; Weeks KM Visualization of lncRNA and mRNA Structure Models Within the Integrative Genomics Viewer. Methods Mol. Biol 2021, 2254, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Reuter JS; Mathews DH RNAstructure: Software for RNA Secondary Structure Prediction and Analysis. BMC Bioinformatics 2010, 11, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Mauger DM; Golden M; Yamane D; Williford S; Lemon SM; Martin DP; Weeks KM Functionally Conserved Architecture of Hepatitis C Virus RNA Genomes. Proc Natl Acad Sci U S A 2015, 112, 3692–3697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Smola MJ; Christy TW; Inoue K; Nicholson CO; Friedersdorf M; Keene JD; Lee DM; Calabrese JM; Weeks KM SHAPE Reveals Transcript-Wide Interactions, Complex Structural Domains, and Protein Interactions Across the Xist lncRNA in Living Cells. Proc Natl Acad Sci U S A 2016, 113, 10322–10327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Dethoff EA; Weeks KM Effects of Refolding on Large-Scale RNA Structure. Biochemistry 2019, 58, 3069–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Lavender CA; Lorenz R; Zhang G; Tamayo R; Hofacker IL; Weeks KM Model-Free RNA Sequence and Structure Alignment Informed by SHAPE Probing Reveals a Conserved Alternate Secondary Structure for 16S rRNA. PLoS Comput. Biol 2015, 11, e1004126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Giannetti CA; Busan S; Weidmann CA; Weeks KM SHAPE Probing Reveals Human rRNAs Are Largely Unfolded in Solution. Biochemistry 2019, 58, 3377–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Krzyzosiak WJ; Sobczak K; Wojciechowska M; Fiszer A; Mykowska A; Kozlowski P Triplet Repeat RNA Structure and Its Role as Pathogenic Agent and Therapeutic Target. Nucleic Acids Res 2012, 40, 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (76).Brakier-Gingras L; Charbonneau J; Butcher SE Targeting Frameshifting in the Human Immunodeficiency Virus. Expert Opin. Ther. Targets 2012, 16, 249–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Busan S; Weeks KM Role of Context in RNA Structure: Flanking Sequences Reconfigure CAG Motif Folding in Huntingtin Exon 1 Transcripts. Biochemistry 2013, 52, 8219–8225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Low JT; Garcia-Miranda P; Mouzakis KD; Gorelick RJ; Butcher SE; Weeks KM Structure and Dynamics of the HIV-1 Frameshift Element RNA. Biochemistry 2014, 53, 4282–4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Garcia-Miranda P; Becker JT; Benner BE; Blume A; Sherer NM; Butcher SE Stability of HIV Frameshift Site RNA Correlates with Frameshift Efficiency and Decreased Virus Infectivity. J Virol 2016, 90, 6906–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Warner KD; Hajdin CE; Weeks KM Principles for Targeting RNA with Drug-Like Small Molecules. Nat Rev Drug Discov 2018, 17, 547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Herschlag D RNA Chaperones and the RNA Folding Problem. J. Biol. Chem 1995, 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- (82).Rajkowitsch L; Chen D; Stampfl S; Semrad K; Waldsich C; Mayer O; Jantsch MF; Konrat R; Bläsi U; Schroeder R RNA Chaperones, RNA Annealers and RNA Helicases. RNA Biol 2007, 4, 118–130. [DOI] [PubMed] [Google Scholar]
- (83).Grohman JK; Gorelick RJ; Lickwar CR; Lieb JD; Bower BD; Znosko BM; Weeks KM A Guanosine-Centric Mechanism for RNA Chaperone Function. Science 2013, 340, 190–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Tyrrell J; Weeks KM; Pielak GJ Challenge of Mimicking the Influences of the Cellular Environment on RNA Structure by PEG-Induced Macromolecular Crowding. Biochemistry 2015, 54, 6447–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (85).Zamir A; Miskin R; Elson D Interconversions Between Inactive and Active Forms of Ribosomal Subunits. FEBS Lett 1969, 3, 85–88. [DOI] [PubMed] [Google Scholar]
- (86).Moazed D; Van Stolk BJ; Douthwaite S; Noller HF Interconversion of Active and Inactive 30 S Ribosomal Subunits Is Accompanied by a Conformational Change in the Decoding Region of 16 S rRNA. J. Mol. Biol 1986, 191, 483–493. [DOI] [PubMed] [Google Scholar]
- (87).Smola MJ; Weeks KM In-Cell RNA Structure Probing with SHAPE-MaP. Nat Protoc 2018, 13, 1181–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Kozak M Regulation of Translation via mRNA Structure in Prokaryotes and Eukaryotes. Gene 2005, 361, 13–37. [DOI] [PubMed] [Google Scholar]
- (89).Mustoe AM; Corley M; Laederach A; Weeks KM Messenger RNA Structure Regulates Translation Initiation: A Mechanism Exploited From Bacteria to Humans. Biochemistry 2018, 57, 3537–3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Corley M; Solem A; Phillips G; Lackey L; Ziehr B; Vincent HA; Mustoe AM; Ramos SBV; Weeks KM; Moorman NJ; Laederach A An RNA Structure-Mediated, Posttranscriptional Model of Human Α−1-Antitrypsin Expression. Proc Natl Acad Sci U S A 2017, 114, E10244–E10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Archer EJ; Simpson MA; Watts NJ; O’Kane R; Wang B; Erie DA; McPherson A; Weeks KM Long-Range Architecture in a Viral RNA Genome. Biochemistry 2013, 52, 3182–3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Lavender CA; Ding F; Dokholyan NV; Weeks KM Robust and Generic RNA Modeling Using Inferred Constraints: a Structure for the Hepatitis C Virus IRES Pseudoknot Domain. Biochemistry 2010, 49, 4931–4933. [DOI] [PMC free article] [PubMed] [Google Scholar]


