Abstract
Microglia are the primary immune cells of the central nervous system and crucial to proper development and maintenance of the brain. Microglia have been recognized to be associated with neurodegenerative diseases and neuroinflammatory disorders. CX3C chemokine receptor 1 (CX3CR1), which is specifically expressed in microglia, regulates microglia homeostatic functions such as microglial activation and is downregulated in aged brain and disease-associated microglia in rodents, yet its role in human microglia is not fully understood. In this study, we investigated the function of CX3CR1 in human microglia using human induced pluripotent stem (iPS) cell-derived microglia-like cells. Human iPS cell-derived microglia-like cells expressed microglial markers and showed an activated state and phagocytic activity. Using CRISPR/Cas9 genome editing, we deleted CX3CR1 in human iPS cells and found increased inflammatory responses and phagocytic activity in mutant as compared to wild-type microglia-like cells. In addition, the CX3C chemokine ligand 1 (CX3CL1, a ligand for CX3CR1) significantly decreased the upregulation of IL-6 by lipopolysaccharide stimulation in human iPS cell-derived microglia-like cells. These results suggest that CX3CR1 in human microglia may contribute to microglial homeostasis by regulating inflammatory response and phagocytosis.
1. INTRODUCTION
Microglia are the resident immune cells of the central nervous system (CNS) (Tremblay et al., 2011). Microglia guide neural development by responding to local changes in the brain microenvironment such as phagocytosing apoptotic cells, pruning synapses, modulating neurogenesis and regulating synapse plasticity and myelin formation and provide key function of the homoeostasis in the CNS (Schafer & Stevens, 2015). As immune cells, microglia respond to disruptions caused by injury, pathology or ageing (Salter & Stevens, 2017). These responses, often termed “activation,” are defined as any physical or biochemical changes of the microglial homeostatic state and include proliferation, migration to the site of pathology, phagocytosis of debris and cells, and secretion of the cytokines and chemokines necessary to stimulate microglia and other brain cells (Hammond et al., 2019). In brain disease, microglia undergo substantial morphological, molecular and functional changes, which establish new biological states relevant to disease pathogenesis and progression (Wright-Jin & Gutmann, 2019). Microglia have been recognized to be associated with neurodegenerative diseases and neuroinflammatory disorders. For example, recent genome-wide association studies of neurodegenerative diseases have highlighted some genes which are specifically expressed in microglia (Chan et al., 2015; Korvatska et al., 2015). In addition, neuroinflammation including microglial activation is present in patients with neurodegenerative diseases such as Alzheimer’s disease, Parkinson’s disease, multiple system atrophy, depression, fibromyalgia, chronic fatigue syndrome (Airas, Nylund, & Rissanen, 2018; Albrecht et al., 2019; Gerhard, 2016; Nakatomi et al., 2014; Setiawan et al., 2018) and is associated with the severity of neuropsychologic symptoms (Albrecht et al., 2019; Nakatomi et al., 2014). Although microglial function and dysfunction are likely to be involved in numerous neurodegenerative conditions, their precise contributions to disease pathogenesis and progression are not fully understood.
Most insights into the role of microglia come from research using animal models of disease. Species-specific genetic variations indicate that the rodent microglia fail to fully recapitulate the biology of human microglia (Hasselmann et al., 2019) emphasizing the importance of studying human microglia. The availability of human induced pluripotent stem (iPS) cells allows the derivation of many differentiated cell types (Takahashi et al., 2007). Recently, several protocols to differentiate microglia from human iPS cells have been developed (Abud et al., 2017; Brownjohn et al., 2018; Douvaras et al., 2017; Haenseler et al., 2017; Muffat et al., 2016; Pandya et al., 2017). Primitive yolk sac macrophages were efficiently generated from human iPS cells and matured into microglia-like cells under fully defined conditions with a transcriptome closely mimicking that of foetal and adult microglia rather than blood macrophages (Muffat et al., 2016). The derivation of human iPS cell-derived microglia-like cells provides a platform to investigate their physiological function in human nervous system under highly defined conditions. In addition, combined with a genome editing and regulation technology such as CRISPR/Cas9, it is now possible to study human neurological disease in the relevant molecular and cellular context (Soldner & Jaenisch, 2018).
CX3CR1 is a G protein-coupled C-X3-C motif chemokine receptor 1. CX3CL1 (also known as fractalkine) is a C-X3-C motif chemokine ligand 1 and a sole ligand for CX3CR1. In the brain, the expression of CX3CR1 is microglia-specific (Cardona et al., 2006) and the expression of its ligand CX3CL1 is neuron-specific (Harrison et al., 1998). CX3CR1 regulates microglial functions such as cytokine production and motility as response to inflammatory signals. In addition, CX3CL1-CX3CR1 signalling represents an important signalling pathway between microglia and neurons (Mecca, Giambanco, Donato, & Arcuri, 2018). Under pathological conditions, CX3CR1 and/or CX3CL1 is downregulated and reduced in the brain of aged rodents (Bachstetter et al., 2011; Keren-Shaul et al., 2017; Krasemann et al., 2017; Mecca et al., 2018). Furthermore, CX3CR1 is one of the top suppressed genes in specific neurodegenerative microglial phenotypes such as DAM (disease-associated microglia) and MGnD (microglial neurodegenerative phenotype) microglia, which are recently defined as a unique population of microglia (Butovsky & Weiner, 2018; Keren-Shaul et al., 2017; Krasemann et al., 2017). However, the role of CX3CR1 in human microglia has not been defined.
In this study, we investigated the function of CX3CR1 in human iPS cell-derived microglia-like cells. We generated CX3CR1 KO human iPS cells using CRISPR/Cas9 genome editing and examined inflammatory responses and phagocytic activity in CX3CR1 KO human microglia-like cells derived from iPS cells.
2. MATERIALS AND METHODS
2.1. Human iPS cell culture
Human iPS cell line 657 was generated from female AG07657 fibroblasts obtained from Coriell using a single STEMCCA-LoxP polycistronic vector (gift from Dr. Gustavo Mostoslavsky, Boston University) encoding 4 reprogramming factors flanked by IoxP sites as previously described (Svoboda et al., 2019). Human iPS cells were cultured at 37°C in 5% O2 on mitomycin C-inactivated mouse embryonic fibroblasts in human embryonic stem (hES) cell medium, composed of Dulbecco’s modified Eagle’s medium(DMEM)/F12 (Invitrogen), 15% foetal bovine serum (FBS) (HyClone), 5% knockout serum replacement (Invitrogen), 1% nonessential amino acids (Invitrogen), 1 mM glutamine (Invitrogen), 1% penicillin/streptomycin, 0.1 mM β-mercaptoethanol (Sigma-Aldrich) and 4 ng/ml basic fibroblast growth factor (bFGF) (Life Technologies). Cells were passaged manually or with 1 mg/ml collagenase type IV (Invitrogen) every 5–7 days.
2.2. Differentiation towards microglia
Human iPS cells were differentiated into microglia-like cells as previously described (Muffat et al., 2016). Human iPS cell colonies were treated with 1 mg/ml collagenase type IV, mildly triturated to form a suspension of uniform clumps and transferred directly to hES cell medium for microglial differentiation initiation, composed of DMEM/F12, 15% FBS, 5% knockout serum replacement, 1% nonessential amino acids, 1 mM l-glutamine, 1% penicillin/streptomycin and 0.1 mM β-mercaptoethanol in ultra-low attachment six-well plates (Corning). Medium was changed to microglial medium (MGM), composed of neurobasal, neuroglial differentiation medium (1% Gem21 NeuroPlex supplement w/o vitamin A, 0.2% Albumax I, 0.5% N2 NeuroPlex supplement, 50 mM NaCl, 1mM sodium pyruvate, 1% penicillin/streptomycin, 1× Glutamax, 3.5 ng/ml biotin, 0.3% l-ascorbic acid, 0.01% sodium DL-lactate), 40 ng/ml Interleukin(IL)-34 and 40 ng/ml colony-stimulating factor 1 (CSF1), at 24 hr after starting microglial differentiation and every 5–7 days to remove dead cells and cell debris. From 2 weeks after starting microglial differentiation, yolk sac embryoid bodies were formed, and the supernatant with single cells was placed in a Primaria six-well plate (Corning) every 5–7 days. After 0.5 hr, attached cells were washed with MGM to remove unattached cells and small embryoid bodies. Further maintenance was performed with MGM for more 2–4 weeks for microglia maturation, and medium was changed every 5–7 days. Microglia-like cells were passaged with Accutase (Invitrogen). Microglial activation was stimulated for 24 hr by 100 ng/ml lipopolysaccharide (LPS) and/or 20 ng/ml interferonγ (IFNγ) for M1 activation, and 20 ng/ml IL-4 and 20 ng/ml IL-13 for M2 activation. CX3CL1 (CX3CL1 chemokine domain, R&D Systems) was added to microglia-like cells to examine the effect on microglial activation.
2.3. Differentiation towards neural progenitor cells (NPCs) and neurons
Human iPS cells were dissociated into single cells and plated at 3–5 × 106 density onto one well of matrigel-coated 6-well plate in mTeSR serum-free media (StemCell Technologies) supplemented with Rho-associated protein kinase inhibitor Y27632 (ROCK inhibitor, 10 μM, Stemgent). When cells reached full monolayer, medium was switched to DMEM/F12 supplemented with 20% knockout serum replacement, 2 mM l-glutamine, 1% penicillin/streptomycin, 1% nonessential amino acids, 0.1 mM β-mercaptoethanol and 4 ng/ml bFGF. Cells were cultured in this medium with 25% addition of NPC differentiation medium, composed of 50% DMEM/F12 with HEPES, 50% neurobasal, 0.5% of N2 NeuroPlex and 1% of Gem21 NeuroPlex supplements (Gembio), 1% nonessential amino acids, 2 mM l-glutamine, 1% penicillin/streptomycin and 0.1 mM β-mercaptoethanol, every 48 hr until completely replaced within 10 days. ROCK inhibitor (10 μM) was added 1 hr before and for 24 hr after passage in the medium during the first 3 passages. Cells were passaged after emerging rosette-like neuroepithelium structures (10–14 days after start of differentiation) 1:1 every 3 days with Accutase. After 2 weeks from starting differentiation, 10 ng/ml bFGF was added. With every passage, NPCs stabilized in culture and could be expanded by passaging to 1:2 and 1:3 and frozen as a stock at passage 3 to passage 6.
NPCs were differentiated into neurons in neuron differentiation medium, composed of neurobasal medium, 1% B27, 0.5% nonessential amino acids, 1 × Glutamax, 1% penicillin/streptomycin, 0.07% d-glucose, 10 ng/ml GDNF, 10 ng/ml BDNF. NPCs were plated at 1–2 × 106 cells/well on matrigel-coated 6-well plate with neuron differentiation medium. Medium was changed every 3 days to avoid yellow medium. After 2 days from starting neuron differentiation, immature neurons appeared in the culture. After 10–14 days from starting neuron differentiation, cells were passaged with Accutase on Poly-d-Lysine and laminin-coated plate at density of 1 × 106 cells/well of 6-well plate. Neurons continued to differentiate to more mature neuronal phenotype for another 4 weeks. Neuron-conditioned medium was collected from the medium of differentiated neurons at medium change after 48 hr culture of 2 ml of neuron differentiation medium on 1 × 106 neurons/well. Neuron-conditioned medium was spun by centrifugation to remove cells, and supernatants of neuron-conditioned medium were used for phagocytosis assay.
2.4. Immunostaining
Cells were fixed with 4% paraformaldehyde in PBS+ for 1 hr at room temperature. After fixation, cells were blocked with 3% normal donkey serum (NDS) with 0.3% Triton in PBS+ for 1 hr at room temperature. Primary antibodies were against Iba1 (AB5076, 1:500), P2 purinergic receptor Y12 (P2RY12) (Sigma, HPA014518, 1:500), transmembrane protein 119 (TMEM119) (Abcam, AB185333, 1:100), CX3CR1 (Santa Cruz, sc377227, 1:50) with 0.1% Triton and 3% NDS in PBS+ for O/N at 4°C and visualized by secondary antibodies conjugated with Alexa 488, 568, 594, 647 (Invitrogen, 1:500) with 0.1% Triton and 3% NDS in PBS+ for 2 hr at room temperature, followed by counterstaining with DAPI.
2.5. Quantitative polymerase chain reaction (qPCR)
Total RNA was extracted from cells using the RNeasy Plus Micro Kit (Qiagen) according to the manufacturer’s protocol. RNA was reverse transcribed into cDNA using SuperScript III First-Strand Synthesis SuperMix (Invitrogen). Transcript abundance was determined by quantitative PCR using SYBR Green PCR mix (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. The following primer sequences were used: P2RY12 (F: TTTGTGTGTCAAGTTACCTCCG, R: CTGGTGGTCTTCTGGTAGCG), TMEM119 (F: CTGGCCTTTCTGCTGATGTTC, R: TCACTCTGGTCCACGTACTTC), Iba1 (F: GGGAGACGTTCAGCTACCC, R: TGGCTTTTCCTTTTCTCTCG), CD11b (F: GCCTTGACCTTATGTCATGGG, R: CCTGTGCTGTAGTCGCACT), triggering receptor expressed on myeloid cells 2 (TREM2) (F: GGAGCACAGCCATCACAGAC, R: CACATGGGCATCCTCGAAGC), PU.1 (F: CCACTGGAGGTGTCTGACG, R: CAGGTCCAACAGGAACTGGT), tumour necrosis factor α (TNFα) (F: GAGGCCAAGCCCTGGTATG, R: CGGGCCGATTGATCTCAGC), IL-6 (F: GGTACATCCTCGACGGCATCT, R: GTGCCTCTTTGCTGCTTTCAC), IL-1β (F: CTGTCCTGCGTGTTGAAAGA, R: TGAAGACAAATCGCTTTTCCA), CD206 (F: AGCTGACACAAGGAAGATGGA, R: GCACCCGTTAAAATCAGGAG), IL-4 (F: TCTTTGCTGCCTCCAAGAAC, R: GTCCTTCTCATGGTGGCTGT), arginase 1 (ARG1) (F: CGTGGGAGGTCTGACATACA, R: GGGATGGGTTCACTTCCATT), CX3CR1 (F: ACCACCTGTATGGGAAATGC, R: AGAACACTTCCATGCCTGCT) and GAPDH (F: AGGGGTCTACATGGCAACTG, R: CGACCACTTTGTCAAGCTCA).
2.6. Western blotting
After PBS wash, cells were lysed with RIPA buffer containing protease and phosphatase inhibitors. Protein concentrations of cell lysates were measured with Qubit Protein Assay kits (Molecular probes). Cell lysates were mixed with NuPAGE LDS sample buffer, heated at 95°C for 10 min and subjected to SDS-PAGE and immunoblotting. Primary antibodies were directed towards Iba1 (Abcam, ab5076, 1:100), TMEM119 (Abcam, AB185333, 1:100), CX3CR1 (Santa Cruz, sc-377227, 1:100) and GAPDH (Abcam, ab125247, 1:4,000). Immunoblot detection was performed with ECL Western Blotting Detection Kit (Amersham).
2.7. Cytokine profiler
For analysis of the secreted cytokine profile of cells, the human cytokine array kit (R&D Systems) was used according to the manufacturer’s instructions. The supernatants of cell cultures were added to a prepared cytokine antibody panel membrane. After incubation, the membranes were revealed with ECL reagent.
2.8. Phagocytosis assay
Microglia-like cells were plated in Primaria 6-well plate (1–5 × 105 cells/well). Fluoresbrite Polychromatic Red Microspheres 1.0 μm (Polysciences) were added to cells at 1:100 ratio (cells:beads) and incubated for 1 hr at 37°C. Cells were washed with PBS, collected with Accutase and resuspended in fluorescence-activated cell sorting (FACS) buffer, composed of 1% FBS heat-inactivated and 1 mM EDTA in PBS, in FACS tube with cell strainer. Cells were examined on a flow cytometer (BD LSRFortessa).
2.9. Generation of CX3CR1 KO human iPS cell
The guide RNA targeting Cx3cr1 exon 2 (target sequence: CTGTTATATTGGGGACATCG) was designed according to the previous method (Ran et al., 2013). A pair of oligos (F: CACCGCTGTTATATTGGGGACATCG, R; AAACCGATGTCCCCAATATAACAGC) were annealed and ligated to vector PX458 (pSpCas9(BB)-2A-GFP, Addgene plasmid ID:48138). Constructs were sequenced to verify correct sequences with the following primer (LKO.1 5′ [hU6 promoter]: GACTATCATATGCTTACCGT). To test the efficiency of sgRNA-mediated cleavage, human embryonic kidney (HEK)293T cells were transfected with constructed vector in suspension with X-treameGENE 9 DNA Transfection Reagent (Roche) and Survey assay with the following primers (F: GCCTAGAGCCAAATGCTCAC, R: GACACTCTTGGGCTTCTTGC) were conducted. The constructed vector was transfected into 657 human iPS cells with electroporation. Transfected cells with green fluorescent protein (GFP) were isolated by FACS sorting. Cell clones were isolated, expanded and did DNA sequencing to assess target modifications. In addition, iPS cells were differentiated into microglia-like cells and the expression levels of CX3CR1 in microglia-like cells were examined by Western blotting.
2.10. Statistical analysis
Significance of differences between 2 groups was analysed using Student’s t test, while those among more than 2 groups was analysed using Dunnett’s multiple comparison test. p < .05 was considered statistically significant.
3. RESULTS
3.1. Expression of microglial markers in human iPS cell-derived microglia-like cells
We examined the expression of microglial markers in human iPS cell-derived microglia-like cells at 4 weeks after starting microglial differentiation. Human iPS cell-derived microglia-like cells expressed microglial markers such as P2RY12 and TMEM119, and also expressed CX3CR1 by immunostaining (Figure 1a). We verified that human iPS cell-derived microglia-like cells specifically expressed microglial markers such as P2RY12, TMEM119, Iba1, CD11b, TREM2 and PU.1 at the mRNA level by qPCR compared with human iPS cell-derived NPCs and neurons (Figure 1b). CX3CR1 was expressed at a higher level in microglia-like cells compared with NPCs, neurons and astrocytes derived from human iPS cells (Figure 1b). In addition, we confirmed the expressions of Iba1, TMEM119 and CX3CR1 at the protein level by Western blotting (Figure 1c).
Figure 1.
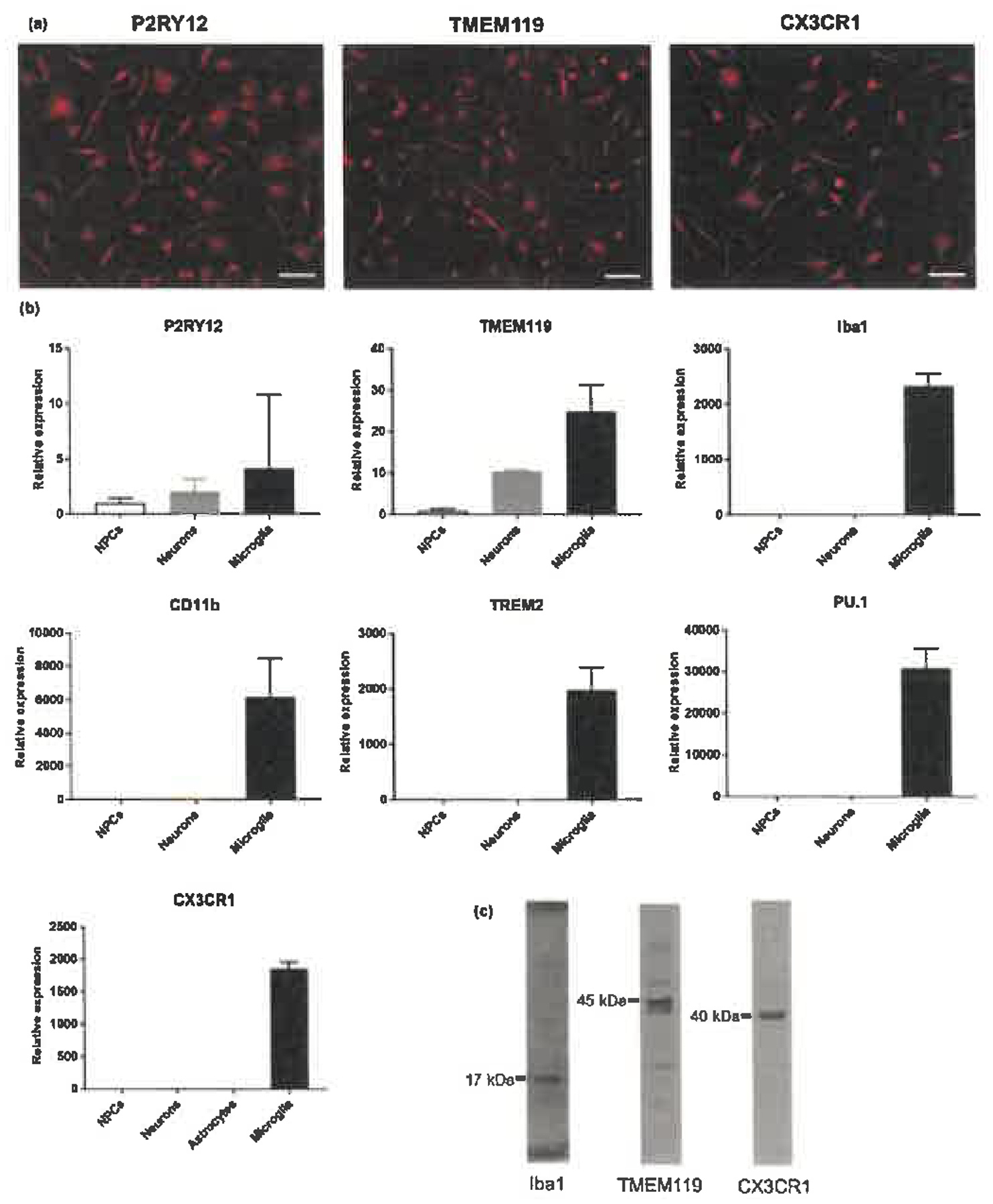
Expression of microglial markers in human iPS cell-derived microglia-like cells. (a) Immunostaining for P2RY12, TMEM119 and CX3CR1. Scale bars, 50 μm. (b) mRNA expression by qPCR in human iPS cell-derived neural progenitor cells (NPCs), neurons, astrocytes and microglia-like cells. Each bar represents the geometric mean ± SD. (c) Western blotting by Iba1, TMEM119 and CX3CR1
3.2. Expression of microglial activation markers and cytokines/chemokines
We examined the mRNA levels of activation markers in human iPS cell-derived microglia-like cells. Treatment of microglia-like cells with 100 ng/ml LPS and 20 ng/ml IFNγ (M1 activation inducer) or with 20 ng/ml IL-4 and 20 ng/ml IL-13 (M2 activation inducer) stimulation, respectively, significantly upregulated TNFα, IL-6 and IL-1β (M1 activation markers) or CD206, IL-4 and ARG1 (M2 activation markers) (Figure 2a). We also assessed cytokine and chemokine profiles released into the supernatant of human iPS cell-derived microglia-like cells. At the unstimulated condition, human iPS cell-derived microglia-like cells secreted detectable levels of cytokines and chemokines such as CC chemokine ligand 2 (CCL2), macrophage inflammatory protein (MIP)-1α and IL-8. In addition, stimulation by 100 ng/ml LPS and 20 ng/ml IFNγ increased CCL2, MIP-1α, IL-8, C-X-C motif chemokine ligand (CXCL)1, CXCL10 and IL-6 secretion (Figure 2b).
Figure 2.
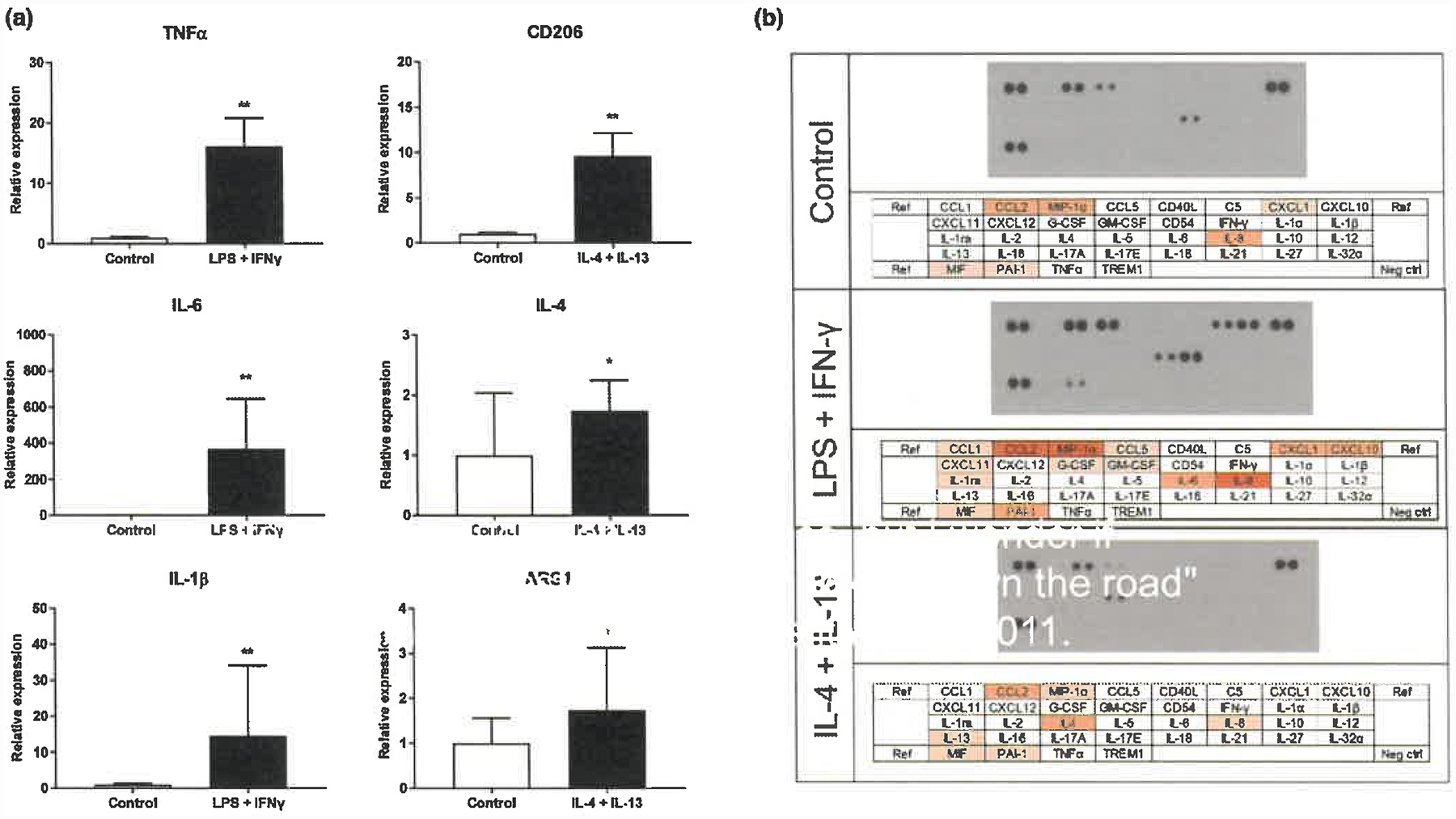
Expression of microglial activation markers and cytokines/chemokines in human iPS cell-derived microglia-like cells. (a) mRNA levels of microglia activation marker after microglial activation by qPCR in human iPS cell-derived microglia-like cells. Each bar represents the geometric mean ± SD. *p < .05, **p < .01 Statistical differences were assessed for ΔΔCt values by Student’s t test compared with control group. (b) Cytokine profiles of human iPS cell-derived microglia-like cells
We also assessed phagocytic activity of human iPS cell-derived microglia-like cells using a FACS-based phagocytosis assay with fluorescent beads. Figure 3 shows that human iPS cell-derived microglia-like cells exhibited phagocytic activity after incubating with fluorescent beads for 1 hr.
Figure 3.
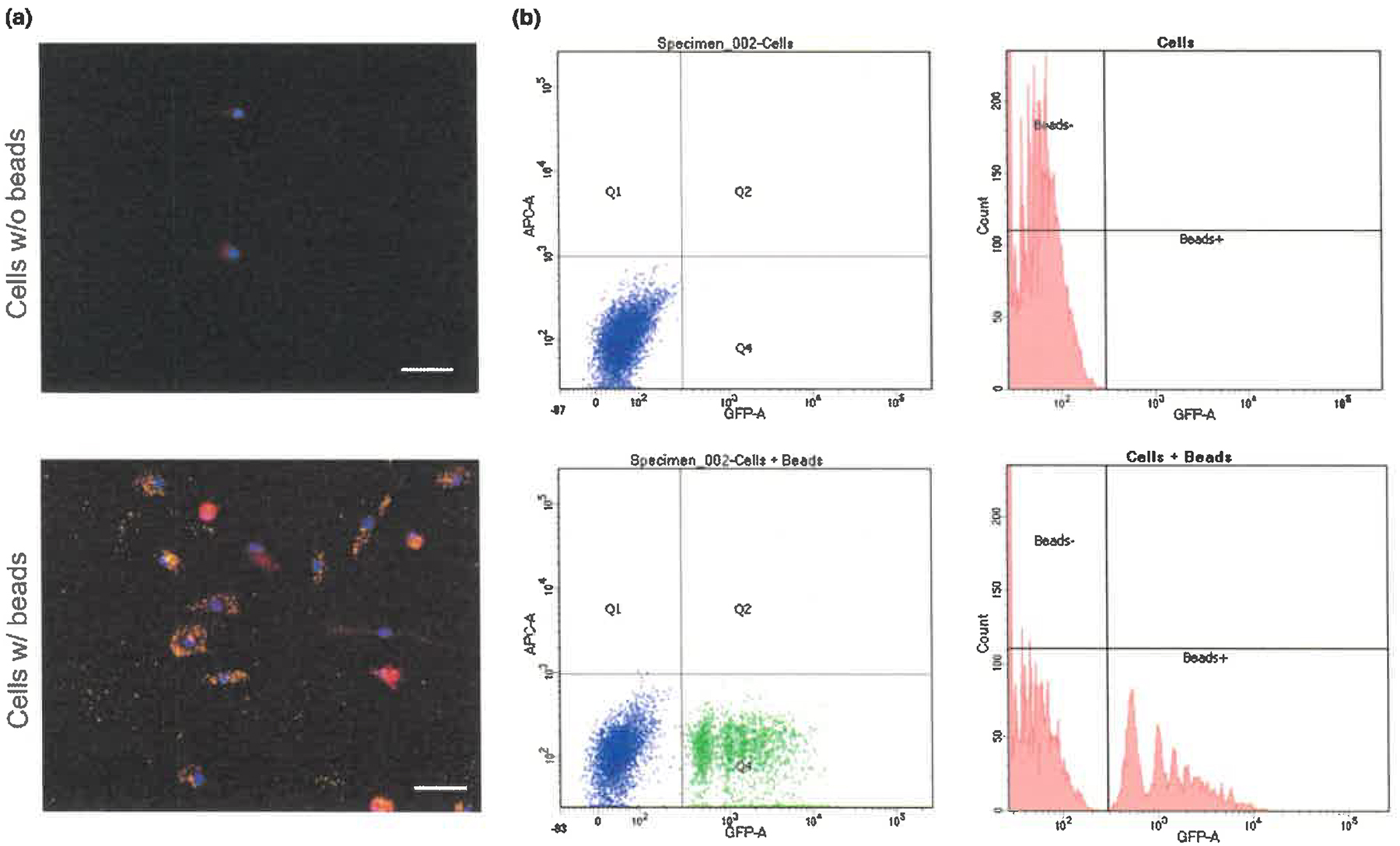
Phagocytic activity in human iPS cell-derived microglia-like cells. (a) Example images of microglia-like cells stained for beads (green), Iba1 (red) and DAPI (blue). Scale bars, 50 μm. (b) FACS-based phagocytosis assay with fluorescent beads. FACS analysis of human iPS cell-derived microglia-like cells showed cells are 38% fluorescent bead-positive (green dot) in a representative experiment
3.3. Functional analysis of CX3CR1
CX3CR1 is expressed at a higher level in microglia-like cells as compared to NPCs, neurons and astrocytes (Figure 1b). To investigate the role of CX3CR1 in microglia biology, we deleted the gene using CRISPR-Cas9 gene-editing. Optimal guide RNA expression constructs were selected after transfection into HEK293T and assaying for indels by the SURVEYOR nuclease assay (Figure 4a) and were transfected into human iPSC cells. We isolated GFP-positive cells by FACS sorting (Figure 4b) and identified clones homozygous for deletion of CX3CR1. Based on the sequencing results, the clone shown in Figure 4 has a homozygous frameshift mutation leading to a premature stop codon for CX3CR1 protein (Figure 4a). We differentiated microglia-like cells from wild-type and CX3CR1 KO human iPS cells and examined the expression level of CX3CR1 by Western blotting. Figure 4c confirms that microglia-like cells derived from CX3CR1 KO human iPS cells did not express CX3CR1 protein. The results are based on a single KO clone, with one extra repetition (data not shown).
Figure 4.
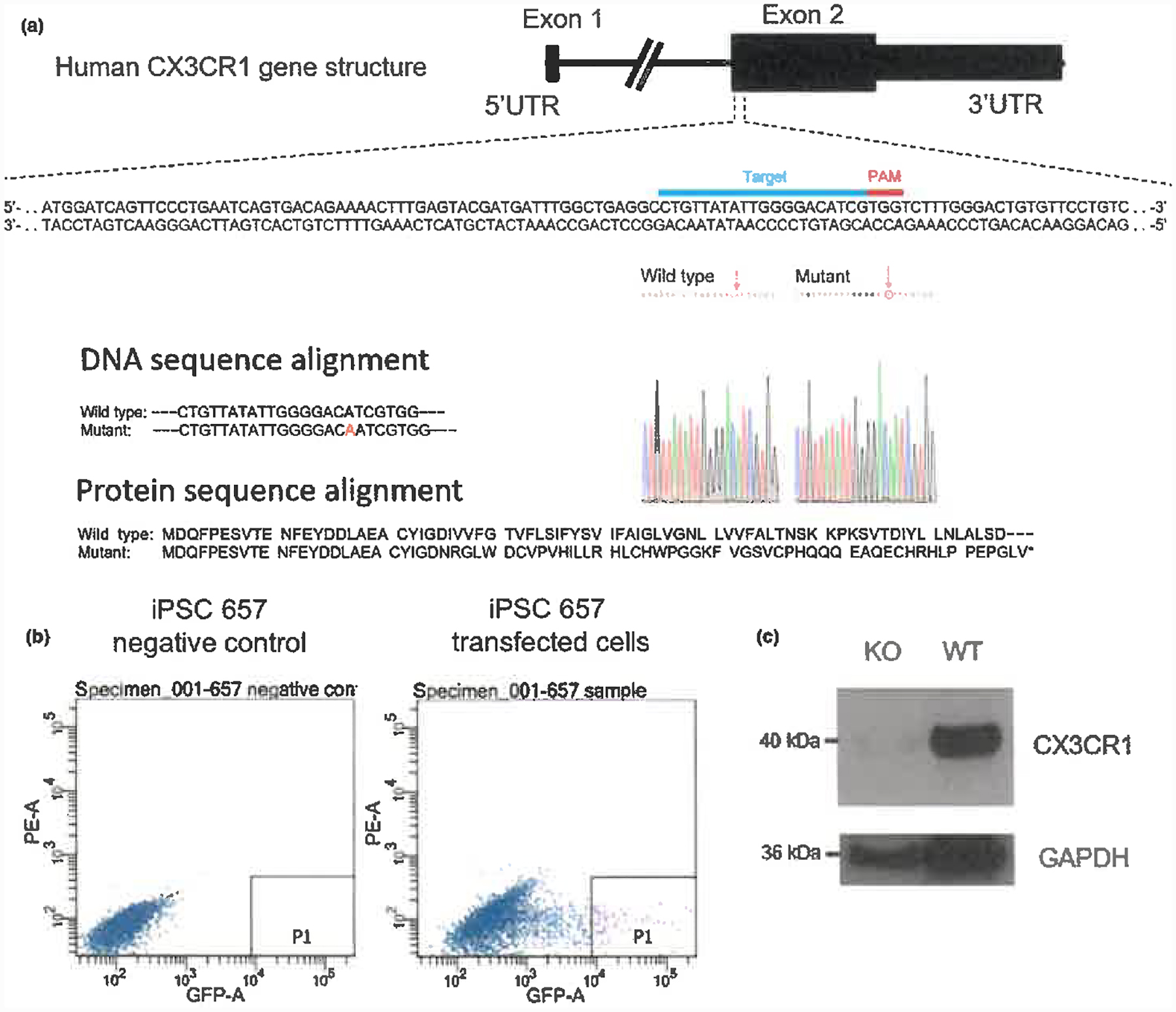
Generation of CX3CR1 KO human iPS cells. (a) Deletion of human CX3CR1 using CRISPR/Cas9 genome editing with the following truncated peptide sequence MDQFPESVTENFEYDDLAEACYIGDNRGLWDCVPVHILLRHLCHWPGGKFVGSVCPHQQQEAQECHRHLPPEPGLV*. (b) Isolation of GFP-positive iPS cells by FACS sorting. (c) Protein expression level of CX3CR1 in CX3CR1 KO and wild-type (WT) human iPS cell-derived microglia-like cells by Western blotting
To functionally assess the effect of deleting CX3CR1, we stimulated mutant and wild-type cells in MGM with 100 ng/ml LPS and determined mRNA expression levels of M1 activation markers. Figure 5a shows that the mRNA expression level of M1 activation markers such as TNFα, IL-6 and IL-1β in CX3CR1 KO microglia-like cells was significantly increased by stimulation with 100 ng/ml LPS as compared to wild-type microglia-like cells (Figure 5a). In addition, IL-18, CCL2 and MIP-1α secretion into the medium increased in mutant as compared to wild-type microglia-like cells in MGM (Figure 5b). The effects seen in the assays described in Figure 5a,b might include ligand-dependent effects because microglia also express CX3CL1 (Zujovic, Benavides, Vigé, Carter, & Taupin, 2000). Indeed, we have confirmed the expression of CX3CL1 in human iPSC-derived microglia-like cells by qPCR although microglial CX3CL1 expression remains controversial. Finally, we examined the effect of CX3CL1 (CX3CR1 ligand, chemokine domain) on microglial activation in wild-type microglia-like cells. Treatment of microglia-like cells with 100 ng/ml LPS stimulation significantly upregulated IL-6 (M1 activation marker). CX3CL1 significantly inhibited the upregulation of IL-6 upon LPS stimulation in wild-type human iPS cell-derived microglia-like cells (Figure 5d).
Figure 5.
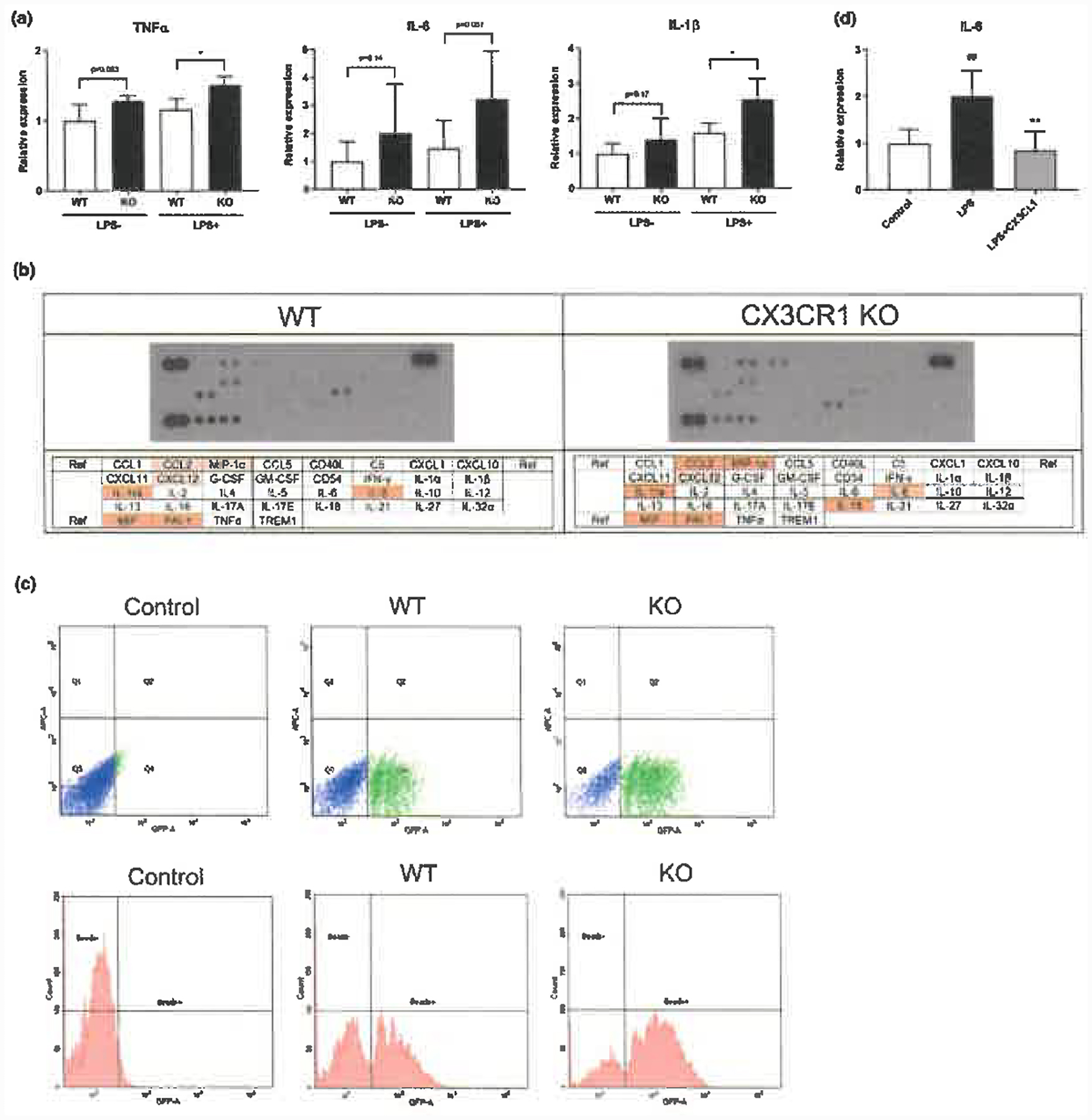
Functional analysis of CX3CR1 in human iPS cell-derived microglia-like cells. (a) mRNA levels of microglial activation markers (TNFα, IL-6, IL-1β) by qPCR in CX3CR1 KO and wild-type (WT) human iPS cell-derived microglia-like cells. Each bar represents the geometric mean ± SD. *p < .05 Statistical differences were assessed for ΔΔCt values by Student’s t test compared with WT group. (b) Cytokine profiles in CX3CR1 KO and WT human iPS cell-derived microglia-like cells. (c) Phagocytic activity in CX3CR1 KO and WT human iPS cell-derived microglia-like cells. Each bar represents the mean ± SD. **p < .01 Statistical differences were assessed by Student’s t test compared with WT group. (d) Effect of CX3CL1 on the upregulation of IL-6 (microglial activation marker) upon 100 ng/ml LPS stimulation by qPCR in wild-type human iPS cell-derived microglia-like cells. Each bar represents the geometric mean ± SD. ##p < .01 and **p < .01 Statistical differences were assessed for ΔΔCt values by Student’s t test compared with control group and LPS stimulation group, respectively
We also examined the phagocytic activity of wild-type and CX3CR1 KO microglia-like cells by a phagocytosis assay using fluorescent microspheres. The cells were cultured in neuron-conditioned medium for 24 hr; fluorescent beads were added, and cells were FACS sorted after 1 hr. Figure 5c shows that mutant microglia-like cells exhibited a significantly increased phagocytic activity in both % of phagocytosed cells (percentage of fluorescent-positive cells) and relative phagocytosis (ratio of the mean microsphere fluorescence intensity [nonstimulated control microglia-like cells = 1]). The effects in Figure 5c assay include ligand-dependent effects because neuron-conditioned medium contain endogenous CX3CL1 that neurons secreted.
4. DISCUSSION
We generated microglia-like cells from human iPS cells and showed that the cells expressed microglial markers, released inflammatory and neurotrophic cytokine/chemokines after stimulation of LPS/IFNγ or IL-4/IL-13 and exhibited phagocytic activity. Using CRISPR/Cas9-mediated genome editing, we deleted CX3CR1 in iPS cells and found that mutant microglia-like cells displayed an increased inflammatory response and phagocytic activity as compared to control microglia-like cells. These findings demonstrate that CX3CR1 may contribute to microglial homeostasis by regulating inflammatory response and phagocytosis.
In this study, we differentiated human microglia-like cells from primitive yolk-sac macrophages which were derived from human iPS cells (Muffat et al., 2016). Microglia are highly sensitive to their environment and exhibit deficiency of transcriptional signature upon isolation from the brain (Gosselin et al., 2017; Svoboda et al., 2019; Hasselman et al., 2019). For instance, human iPS cell-derived microglia-like cells differentiated by our protocol poorly expressed SALL1, which is a transcriptional regulator defining in vivo microglia identity and function (Buttgereit et al., 2016) but is one of the genes most affected by the in vitro culture conditions (Gosselin et al., 2017). Nevertheless, microglia-like cells differentiated from human iPS cells expressed signature microglial markers and induced microglial activation markers as inflammatory responses after LPS exposure and phagocytic activity. In addition, microglia-like cells differentiated from iPS cells did not express neuron markers such as MAP2 by immunostaining, suggesting that the cultures did not contain neurons. Thus, this system allows to study microglial functions in a defined culture system.
We show that microglial activation markers such as TNF-α, IL-6 and IL-1β in CX3CR1 KO human microglia-like cells are increased following LPS stimulation as compared to wild-type cells. This result is consistent with CX3CL1, the ligand of CX3CR1, inhibiting the upregulation of IL-6 upon LPS stimulation in wild-type microglia-like cells. Moreover, secretion of IL-18, CCL2 and MIP-1α was increased in CX3CR1 mutant microglia-like cells as compared with wild-type human microglia-like cells consistent with CX3CR1 deletion increasing the inflammatory responses. These results agree with results obtained in rodent microglia demonstrating that CX3CL1 reduces LPS-induced pro-inflammatory cytokine secretions (Lyons et al., 2009; Mizuno, Kawanokuchi, Numata, & Suzumura, 2003). Furthermore, CX3CL1 suppressed microglial activation in the striatum of a rat model of Parkinson’s disease (Pabon, Bachstetter, Hudson, Gemma, & Bickford, 2011) and in primary mouse microglia (Lee et al., 2010) consistent with CX3CL1 acting to reduce microglial activation. In summary, our findings suggest that CX3CL1-CX3CR1 signalling in human microglia may be involved in maintaining microglia in a deactivated state.
CX3CL1-CX3CR1 sensing may represent an immunological checkpoint which dampens microglia activation in response to external stimuli and thus functions in controlling the inflammatory response. Dysregulation of the CX3CL1-CX3CR1 pathway might induce an initiation or exacerbation of neurodegeneration (Hickman, Izzy, Sen, Morsett, & El Khoury, 2018) as suggested by the genetic ablation of CX3CR1 resulting in neurotoxicity (Cardona et al., 2006). Similarly, our study shows that CX3CR1 mutant microglia-like cells derived from human iPS cells display increased inflammatory responses and phagocytic activity compared with CX3CR1 human wild-type microglia-like cells. Thus, our data are consistent with CX3CR1 being involved in the homeostasis and maintenance of the deactivated state and inhibition of phagocytosis in microglia.
ACKNOWLEDGEMENTS
We thank Malkiel Cohen, Emile Wogram, Haiting Ma, Julien Muffat and Yuya Kunisada for advice, Dongdong Fu, Raaji Alagappan and Tenzin Lungiangwa for experimental assistance, and Patti Wisniewski, Hanna Aharonov and Eleanor Kincaid from the Whitehead FACS facility for FACS. This work was supported by grants from Novo Nordisk, by NIH grants R37HD045022, 1R01-NS088538 and 5R01-MH104610, and by a grant from the Cure Alzheimer Foundation.
Abbreviations
- bFGF
basic fibroblast growth factor
- CCL
CC chemokine ligand
- CNS
central nervous system
- CRISPR
clustered regularly interspaced short palindromic repeat
- CSF
colony-stimulating factor
- CX3CL1
C-X3-C motif chemokine ligand 1
- CX3CR1
C-X3-C motif chemokine receptor 1
- CXCL
C-X-C motif chemokine ligand
- DAM
disease-associated microglia
- DMEM
Dulbecco’s modified Eagle’s medium
- FACS
fluorescence-activated cell sorting
- FBS
foetal bovine serum
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- HEK
human embryonic kidney
- hES
human embryonic stem
- IFN
interferon
- IL
interleukin
- iPS
induced pluripotent stem
- KO
knockout
- LPS
lipopolysaccharide
- MAP2
microtubule-associated protein 2
- MGM
microglial medium
- MGnD
microglial neurodegenerative phenotype
- MIP
macrophage inflammatory protein
- NDS
normal donkey serum
- NPC
neural progenitor cells
- P2RY12
P2 purinergic receptor Y12
- qPCR
quantitative polymerase chain reaction
- TMEM119
transmembrane protein 119
- TNF
tumour necrosis factor
- TREM2
triggering receptor expressed on myeloid cells 2
- WT
wild type
Footnotes
CONFLICT OF INTERESTS
N.M. is an employee of Astellas Pharma Inc. R.J. is a cofounder of Fate Therapeutics, Fulcrum Therapeutics and Omega Therapeutics and an advisor to Dewpoint and Camp4 Therapeutics.
DATA AVAILABILITY STATEMENT
Data related to this study are available from the authors.
Contributor Information
Murai Nobohito, Astellas Pharma Inc..
Mitalipova Maisam, Whitehead Institute for Biomedical Research.
Jaenisch Rudolf, Whitehead Institute for Biomedical Research and the Massachusetts Institute of Technology.
REFERENCES
- Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, … Blurton-Jones M (2017). iPSC-derived human microglia-like cells to study neurological diseases. Neuron, 94, 278–293. 10.1016/j.neuron.2017.03.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Airas L, Nylund M, & Rissanen E (2018). Evaluation of microglial activation in multiple sclerosis patients using positron emission tomography. Frontiers in Neurology, 9, 181. 10.3389/fneur.2018.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albrecht DS, Forsberg A, Sandström A, Bergan C, Kadetoff D, Protsenko E, … Loggia ML (2019). Brain glial activation in fibromyalgia – A multi-site positron emission tomography investigation. Brain, Behavior, and Immunity, 75, 72–83. 10.1016/j.bbi.2018.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachstetter AD, Morganti JM, Jernberg J, Schlunk A, Mitchell SH, Brewster KW, … Gemma C (2011). Fractalkine and CX 3 CR1 regulate hippocampal neurogenesis in adult and aged rats. Neurobiology of Aging, 32, 2030–2044. 10.1016/j.neurobiolaging.2009.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownjohn PW, Smith J, Solanki R, Lohmann E, Houlden H, Hardy J, … Livesey FJ (2018). Functional studies of missense TREM2 mutations in human stem cell-derived microglia. Stem Cell Reports, 10, 1294–1307. 10.1016/j.stemcr.2018.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O, & Weiner HL (2018). Microglial signatures and their role in health and disease. Nature Reviews Neuroscience, 19, 622–635. 10.1038/s41583-018-0057-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit A, Lelios I, Yu X, Vrohlings M, Krakoski NR, Gautier EL, … Greter M (2016). Sall1 is a transcriptional regulator defining microglia identity and function. Nature Immunology, 17, 1397–1406. 10.1038/ni.3585 [DOI] [PubMed] [Google Scholar]
- Cardona AE, Pioro EP, Sasse ME, Kostenko V, Cardona SM, Dijkstra IM, … Ransohoff RM (2006). Control of microglial neurotoxicity by the fractalkine receptor. Nature Neuroscience, 9, 917–924. 10.1038/nn1715 [DOI] [PubMed] [Google Scholar]
- Chan G, White CC, Winn PA, Cimpean M, Replogle JM, Glick LR, … De Jager PL (2015). CD33 modulates TREM2: Convergence of Alzheimer loci. Nature Neuroscience, 18, 1556–1558. 10.1038/nn.4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douvaras P, Sun B, Wang M, Kruglikov I, Lallos G, Zimmer M, … Fossati V (2017). Directed differentiation of human pluripotent stem cells to microglia. Stem Cell Reports, 8, 1516–1524. 10.1016/j.stemcr.2017.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhard A (2016). TSPO imaging in parkinsonian disorders. Clinical and Translational Imaging, 4, 183–190. 10.1007/s40336-016-0171-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, … Glass CK (2017). An environment-dependent transcriptional network specifies human microglia identity. Science, 23, 356. 10.1126/science.aal3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haenseler W, Sansom SN, Buchrieser J, Newey SE, Moore CS, Nicholls FJ, … Cowley SA (2017). A highly efficient human pluripotent stem cell microglia model displays a neuronal-co-culture-specific expression profile and inflammatory response. Stem Cell Reports, 8, 1727–1742. 10.1016/j.stemcr.2017.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, … Stevens B (2019). Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity, 50, 253–271. 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JK, Jiang Y, Chen S, Xia Y, Maciejewski D, McNamara RK, … Feng L (1998). Role for neuronally derived fractalkine in mediating interactions between neurons and CX3CR1-expressing microglia. Proceedings of the National Academy of Sciences, USA, 95, 10896–10901. 10.1073/pnas.95.18.10896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann J, Coburn MA, England W, Figueroa Velez DX, Kiani Shabestari S, Tu CH, … Blurton-Jones M (2019). Development of a chimeric model to study and manipulate human microglia in vivo. Neuron, 103, 1016–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman S, Izzy S, Sen P, Morsett L, & El Khoury J (2018). Microglia in neurodegeneration. Nature Neuroscience, 21, 1359–1369. 10.1038/s41593-018-0242-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, … Amit I (2017). A unique microglia type associated with Restricting development of Alzheimer’s disease. Cell, 169, 1276–1290. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Korvatska O, Leverenz JB, Jayadev S, McMillan P, Kurtz I, Guo X, … Bird TD (2015). R47H variant of TREM2 associated with Alzheimer disease in a large late-onset family: Clinical, genetic, and neuropathological study. JAMA Neurology, 72, 920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, … Butovsky O (2017). The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity, 47, 566–581. 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Varvel NH, Konerth ME, Xu G, Cardona AE, Ransohoff RM, & Lamb BT (2010). CX3CR1 deficiency alters microglial activation and reduces beta-amyloid deposition in two Alzheimer’s disease mouse models. American Journal of Pathology, 177, 2549–2562. 10.2353/ajpath.2010.100265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons A, Lynch AM, Downer EJ, Hanley R, O’Sullivan JB, Smith A, & Lynch MA (2009). Fractalkine-induced activation of the phosphatidylinositol-3 kinase pathway attenuates microglial activation in vivo and in vitro. Journal of Neurochemistry, 110, 1547–1556. [DOI] [PubMed] [Google Scholar]
- Mecca C, Giambanco I, Donato R, & Arcuri C (2018). Microglia and aging: The role of the TREM2-DAP12 and CX3CL1-CX3CR1 axes. International Journal of Molecular Sciences, 19, 318. 10.3390/ijms19010318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno T, Kawanokuchi J, Numata K, & Suzumura A (2003). Production and neuroprotective functions of fractalkine in the central nervous system. Brain Research, 979, 65–70. 10.1016/S0006-8993(03)02867-1 [DOI] [PubMed] [Google Scholar]
- Muffat J, Li Y, Yuan B, Mitalipova M, Omer A, Corcoran S, … Jaenisch R (2016). Efficient derivation of microglia-like cells from human pluripotent stem cells. Nature Medicine, 22, 1358–1367. 10.1038/nm.4189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatomi Y, Mizuno K, Ishii A, Wada Y, Tanaka M, Tazawa S, … Watanabe Y (2014). Neuroinflammation in patients with chronic fatigue syndrome/myalgic encephalomyelitis: An 11C-(R)-PK11195 PET study. Journal of Nuclear Medicine, 55, 945–950. 10.2967/jnumed.113.131045 [DOI] [PubMed] [Google Scholar]
- Pabon MM, Bachstetter AD, Hudson CE, Gemma C, & Bickford PC (2011). CX3CL1 reduces neurotoxicity and microglial activation in a rat model of Parkinson’s disease. Journal of Neuroinflammation, 8, 9. 10.1186/1742-2094-8-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya H, Shen MJ, Ichikawa DM, Sedlock AB, Choi Y, Johnson KR, … Park JK (2017). Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nature Neuroscience, 20, 753–759. 10.1038/nn.4534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, & Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nature Protocols, 8, 2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter MW, & Stevens B (2017). Microglia emerge as central players in brain disease. Nature Medicine, 23, 1018–1027. 10.1038/nm.4397 [DOI] [PubMed] [Google Scholar]
- Schafer DP, & Stevens B (2015). Microglia function in central nervous system development and plasticity. Cold Spring Harbor Perspectives in Biology, 7, a020545. 10.1101/cshperspect.a020545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiawan E, Attwells S, Wilson AA, Mizrahi R, Rusjan PM, Miler L, … Meyer JH (2018). Association of translocator protein total distribution volume with duration of untreated major depressive disorder: A cross-sectional study. Lancet Psychiatry, 6, 339–347. 10.1016/S2215-0366(18)30048-8 [DOI] [PubMed] [Google Scholar]
- Soldner F, & Jaenisch R (2018). Stem cells, genome editing, and the path to translational medicine. Cell, 175, 615–632. 10.1016/j.cell.2018.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda DS, Barrasa MI, Shu J, Rietjens R, Zhang S, Mitalipova M, … Jaenisch R (2019). Human iPSC-derived microglia assume a primary microglia-like state after transplantation into the neonatal mouse brain. Proceedings of the National Academy of Sciences, USA, 116, 25293–25303. 10.1073/pnas.1913541116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, & Yamanaka S (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131, 861–872. 10.1016/j.cell.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Tremblay MÈ, Stevens B, Sierra A, Wake H, Bessis A, & Nimmerjahn A (2011). The role of microglia in the healthy brain. Journal of Neuroscience, 31, 16064–16069. 10.1523/JNEUROSCI.4158-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright-Jin EC, & Gutmann DH (2019). Microglia as dynamic cellular mediators of brain function. Trends in Molecular Medicine, 25, 967–979. 10.1016/j.molmed.2019.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zujovic V, Benavides J, Vigé X, Carter C, & Taupin V (2000). Fractalkine modulates TNF-alpha secretion and neurotoxicity induced by microglial activation. Glia, 29, 305–315. [PubMed] [Google Scholar]


