Abstract
Background
Glioblastoma is the most common and aggressive type of primary brain tumor, as most patients succumb to the disease less than two years after diagnosis. Critically, studies demonstrate that glioma recruits surrounding blood vessels, while some work suggests that tumor stem cells themselves directly differentiate into endothelial cells, yet the molecular and cellular dynamics of the endothelium in glioma are poorly characterized. The goal of this study was to establish molecular and morphological benchmarks for tumor associated vessels (TAVs) and tumor derived endothelial cells (TDECs) during glioblastoma progression.
Methods
Using In-Utero Electroporation and CRISPR/Cas9 genome engineering to generate a native, immunocompetent mouse model of glioma, we characterized vascular-tumor dynamics in three dimensions during tumor progression. We employed bulk and single-cell RNA-Sequencing to elucidate the relationship between TAVs and TDECs. We confirmed our findings in a patient derived orthotopic xenograft (PDOX) model.
Results
Using a mouse model of glioma, we identified progressive alteration of vessel function and morphogenesis over time. We also showed in our mouse model that TDECs are a rare subpopulation that contributes to vessels within the tumor, albeit to a limited degree. Furthermore, transcriptional profiling demonstrates that both TAVs and TDECs are molecularly distinct, and both populations feature extensive molecular heterogeneity. Finally, the distinct molecular signatures of these heterogeneous populations are also present in human glioma.
Conclusions
Our findings show extensive endothelial heterogeneity within the tumor and tumor microenvironment and provide insights into the diverse cellular and molecular mechanisms that drive glioma vascularization and angiogenesis during tumorigenesis.
Keywords: angiogenesis, glioma, lymphangiogenesis, tumor-associated vessels (TAVs), tumor-derived endothelial cells (TDECs)
Key Points.
Tumor-associated vessels are molecularly distinct compared to tumor derived endothelial cells.
Tumor-associated vessels show exceptional heterogeneity and are molecularly distinct.
Tumors generate their own rare endothelium, and these cells are also heterogeneous.
Importance of the Study.
Excessive angiogenesis and altered vessel function, both surrounding and within a tumor, are key diagnostic features in glioma. However, to date anti-angiogenic therapies have largely failed to improve patient outcomes in glioma, suggesting a deeper understanding of vessel dynamics during tumorigenesis is warranted. Here, using a combination of 3D imaging and bulk and single-cell transcriptional profiling, we defined the structural, cellular, and molecular characteristics of tumor-associated vessels (TAVs) and tumor-derived vessels (TDECs) within two animal models of glioma. Our findings reveal that TAVs and TDECs are molecularly distinct and that both cell types are highly heterogeneous. These findings create a shift in our understanding of vascular dynamics in glioma pathogenesis and suggest that a one-size-fits all approach to targeting the vasculature in glioma may not be efficacious due to the inherent molecular diversity in tumor-associated vessels, as well as tumor, derived vessels.
Glioblastoma (GBM) and high-grade glioma (HGG) represent a lethal classification of brain tumors characterized by a highly invasive and angiogenic nature. The extensive vascularization of GBM presents a significant hurdle to treating these tumors, as poorly remodeled and angiogenic vessels are considered refractory to chemotherapy and likely contribute to increased patient mortality.1–3 To date, therapies targeting this “angiogenic switch” (eg, Avastin/bevacizumab) have failed to provide substantial clinical benefit in terms of overall patient survival.4–6 Factors which lead to this adaptive resistance to anti-angiogenic therapy include utilizing alternative pathways to sustain tumor growth,3,7–9 vasculogenic mimicry, tumor stem cell transdifferentiation into endothelium,10–12 alterations in blood brain barrier (BBB) integrity, and the underlying intratumor cellular heterogeneity of GBM.13,14 While many of these factors have been studied previously, whether morphological and cellular diversity exists in tumor-associated or tumor-derived endothelial populations, and if so what the contribution of this diversity is to tumor progression, remains unclear.10–12 Using an endogenous mouse model of glioma,15 we characterized tumor-associated- vessels (TAVs)and tumor-derived endothelial-cell (TDEC) populations, finding that these two endothelial cell populations are molecularly distinct. Further examination of the underlying diversity of TAVs and TDECs revealed extensive cellular heterogeneity within each of these subpopulations.
Materials and Methods
Generation of Endogenous Glioma in a Mouse Model Using In Utero Electroporation
All mouse CRISPR-IUE GBM gliomas were generated in the CD-1 IGS mouse background. In utero electroporations (IUEs) were performed as previously described.15–17 Briefly, a plasmid containing guide RNAs targeting the tumor suppressor genes Nf1, Tp53, and Pten was co-electroporated along with a fluorescent reporter to label tumor cells (Supplementary Figure 1). See Supplementary Materials and Methods for more details. All mouse experiments were approved by the Baylor College of Medicine Institutional Animal Care and Use Committee.
CLARITY and Lightsheet Confocal Imaging
Adult mice were perfused via tail vein injection of 100 μL of fluorescently labeled lectin (Lycopersicon Esculentum) (VECTOR Laboratories, #DL-1178-1). Specimens were perfused with 1× PBS, followed by 5–7 mL of hydrogel (Logos Biosciences). Brains were drop-fixed in hydrogel solution overnight at 4 °C and cleared in electrophoretic clearing solution (Logos Biosciences, C13001) using the X-CLARITY platform (Logos Biosciences). Cleared brains were equilibrated in sRIMS at 4 °C before mounting and imaging. For more details, see Supplementary Materials and Methods.
In Vivo Miles Assay (Evans Blue Extravasation)
Mice were anesthetized and injected intravenously with 100 μL 1% Evan’s blue solution (Sigma). Thirty minutes after injection, mice were euthanized and perfused with 2% PFA. Extravasation of Evans blue into the underlying brain parenchyma was determined by calculating the ratio of absorbance in nanometers per milligram of tissue. See Supplementary Materials and Methods for details.
Fluorescent Activated Cell Sorting of TAVs and TDECs
See Supplemental Materials and Methods for FACS gating conditions. Briefly, tumor tissue from dissected mouse brains was sorted using anti-mouse CD31 (Biolegend-102423), endogenous GFP signal, anti-mouse podoplanin (Biolegend-127417), anti-mouse Vegfr3/Flt4 (R&D Systems FAB743P); human: anti-human HLA-ABC (BD Bioscience-557348), anti-human CD31 (BD Bioscience-340297), anti-human podoplanin (BD Bioscience-566456), and anti-Human Vegfr3/Flt4 (R&D System-FAB3492A).
Total RNA Extraction, Library Preparation, Sequencing, and Bioinformatic Analysis
For details on mRNA isolation, library construction, and sequencing see Supplemental Materials and Methods. Briefly, total RNA was isolated from cells captured by FACS. 8 bp single index cDNA libraries were quality controlled using the Standard Sensitivity NGS Fragment Analysis Kit (DNF-473–0500, Agilent formerly AATI). Equal concentrations (2 nM) of cDNA libraries were pooled and subjected to paired-end (2 × 75) sequencing of approximately 40 million reads per sample on a NextSeq.
Single Cell RNA-seq Analysis
Please see Supplementary Materials and Methods for expanded details regarding cell isolation and informatic analysis. Briefly, enriched CD31+ cells isolated by Magnetic Activated Cell Sorting (MACS) were processed using the 10× Genomics Chromium single-cell gene expression platform and 9561 total cells were analyzed.
Culture of Human Glioma Cell Lines and Patient-Derived Xenografts in SCID Mice
Human glioblastoma cell lines were procured from the University of Texas MD Anderson Cancer Center, with the informed consent approved by Institutional Review Board (IRB) protocol LAB04-0001. Primary human glioma cell lines were injected into the brains of SCID mice15 as approved by Baylor College of Medicine Institutional Animal Care and Use Committee. In some cases, neurospheres were differentiated into TDECs using described methodologies11 and tube formation was analyzed to verify the endothelial properties of TDECs.
Results
Progressive Morphological and Functional Changes in Tumor-Associated Vessels
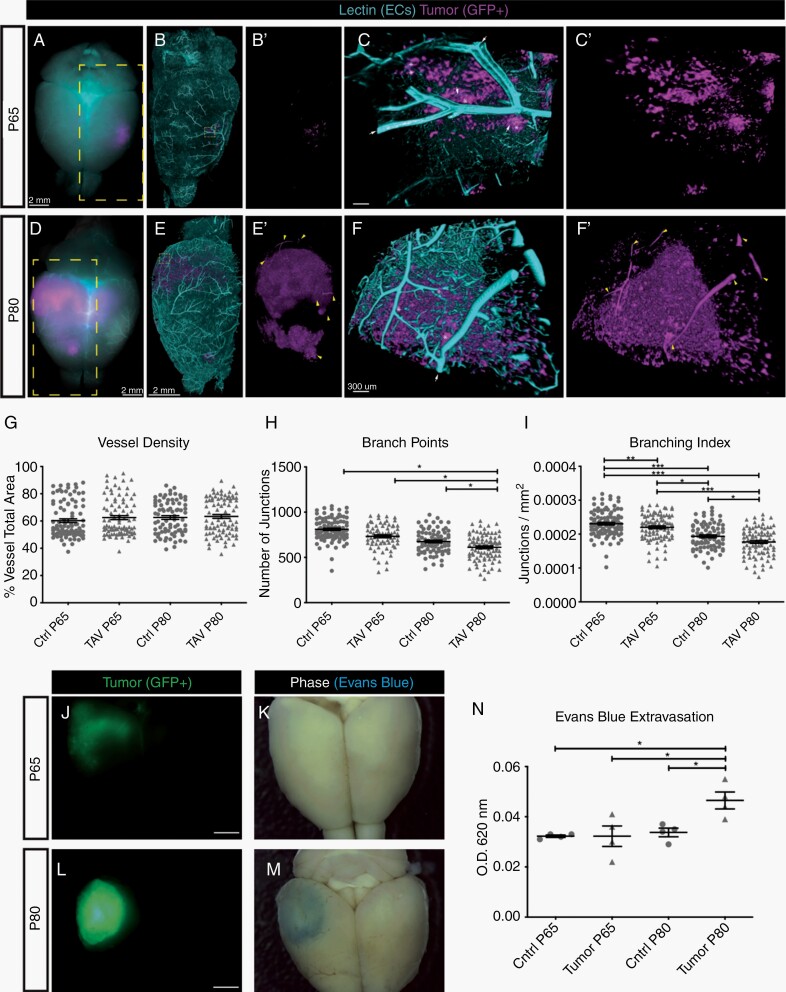
Previous studies described vascular development in xenografts of malignant glioma,18 however this process has not been characterized in a mouse model that features de novo tumor growth and an intact immune system. We utilized a native mouse model of glioma that features CRISPR/Cas9-mediated disruption of Nf1, p53, and Pten, tumor suppressor genes commonly mutated in glioma, combined with piggyBac driven expression of GFP for tumor labeling17 (Supplementary Figure 1A, B). This well-validated model15–17 recapitulates several hallmark features of malignant glioma including histopathology, diffuse and infiltrative growth, molecular signatures, and common genetic lesions. Using this system, we examined morphological changes in TAVs via CLARITY-based tissue clearing19 of fluorescent lectin perfused tumor-bearing brains followed by light-sheet microscopy at postnatal day 65 (P65) and P80 (Fig. 1A–F′) (Supp. Videos 1 and 2).
Fig. 1.
Progressive morphological and functional changes in tumor-associated vessels. (A,D) Whole-mount images of an intact mouse brain at P65 (A) or P80 (D), with tumor-derived cells labeled by GFP (magenta), and vessels labeled by fluorescent lectin (teal). The yellow boxed area in (A,D), after CLARITY-based tissue clearing and lightsheet confocal imaging, is magnified and shows (B,E) vessels and tumor together or (B′,E′) tumor alone. The yellow boxed area in (B,E) is magnified and shows (C,F) vessels and tumor and (C′,F′) tumor alone. White arrows denote vessels (teal) that are associated with tumors (magenta), denoted by asterisks. Yellow carets in (E′) denote GFP+ vessel-like tubes. Scale bar in A, B, B′, D, E, and E′ = 2 mM. Scale bar in C, C′, F, and F′ = 300 µm. (G-I) Quantification of vessel morphology at P65 and P80 for tumor-associated vasculature (TAV) and the contralateral non-tumor region. (J-M) Whole-mount and phase images show brain tumor progression from P65 (J, K) to P80 (L, M). Matching images of each brain following intravenous injection of Evans blue dye. (N) Quantification of Evans blue dye leak, as determined by the ratio of absorbance at OD620-405 nm per milligram of tissue from the tumor area of a contralateral region from the same brain.
Analyses of vessel morphometry revealed that TAV vessel density is unchanged between P65 and P80 compared to the contralateral side of the brain (Fig. 1G). Conversely, both the branching index and the number of branch point junctions are significantly reduced in TAVs, suggesting a failure to create productive vessel networks over time (Fig. 1H, I). In addition, we examined the functional properties of tumor brain vasculature by monitoring blood brain barrier (BBB) integrity in mice bearing tumors at P65 and P80. Increased extravasation of Evans blue dye in the brain after intravenous injection was evident at P80 in the tumor tissue compared to the contralateral side (Fig. 1J-N). Collectively, these morphological and functional analyses reveal progressive changes in both tumor-associated vessel morphology and function during glioma progression, consistent with previous observations in PDOX models.20
Tumor Derived Endothelial Cells are Present in a Native Glioblastoma Mouse Model
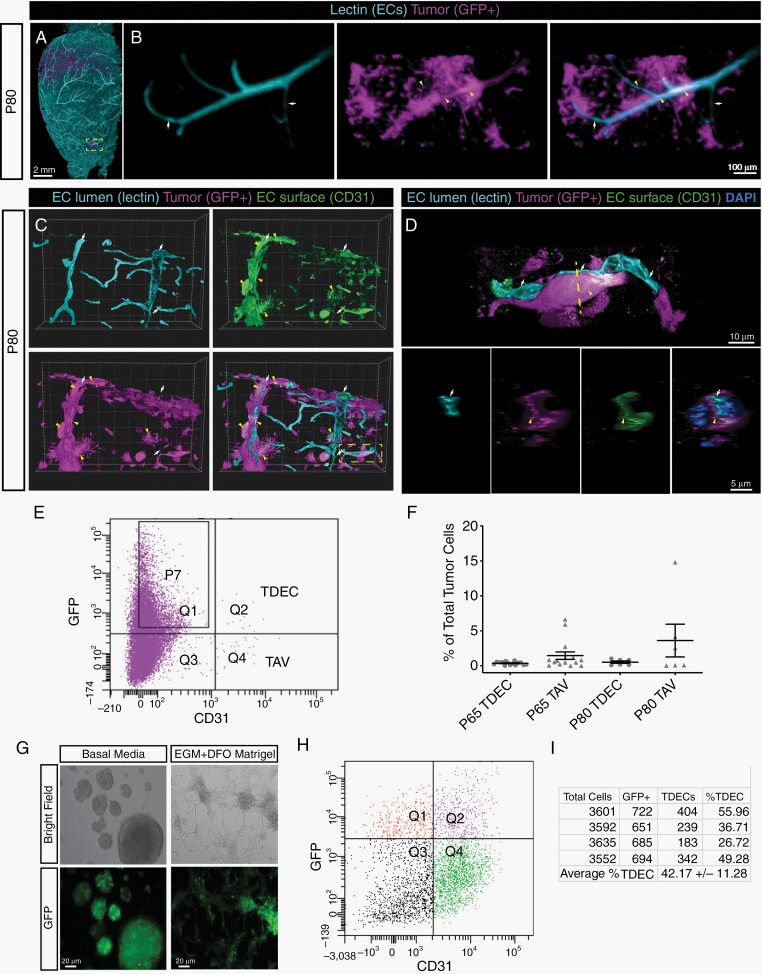
In the course of our analysis, we observed that a subset of tumor-derived cells (GFP+) contributed to vessels (lectin+) (Fig. 1E-F′). Prior reports suggest that glioma stem cells (GSCs) contribute to the glioma vascular endothelium,10–12 therefore we investigated whether glioma cells give rise to vessels in our model. We labeled vessels via perfusion of fluorescent lectin in P80 tumor-bearing mice and examined 3D reconstructed images for lectin-positive vessels that were labeled by the fluorescent GFP+ reporter transgene. Examination of 3D reconstructed brains showed GFP+ cells contributing to vessels (Fig. 2A, B). Immunohistochemistry on cryosections from P80 brains, followed by confocal imaging, identified lectin-labeled vessels that are GFP+ and express the endothelial-specific marker, CD31 (Fig. 2C-D). Together, these data suggest that tumor derived cells can adopt an endothelial lineage. To quantify the extent of TDECs, we dissociated P65 and P80 tumor-bearing brains and performed FACS analysis to isolate tumor-derived endothelial cells (GFP+, CD31+ aka TDECs) and normal endothelial cells (GFP−, CD31+ aka TAVs) (Fig. 2E). Quantification revealed an average TDEC contribution of 0.36% and 0.52% of the total tumor population at P65 and P80, respectively (Fig. 2F).
Fig. 2.
Tumor-derived endothelial cells are present in the native mouse model of glioma. (A) Whole mount reconstruction of a lightsheet confocal image from a P80 tumor-containing brain. The boxed area in (A) is magnified and shown in (B) with vessels (left), tumor cells (middle), and merged (right). White arrows denote vessels, yellow carets denote GFP+-positive vessels. (C) Reconstructions of 40 µm thick sections from fluorescent lectin (turquoise) perfused P80 tumors (magenta) stained with CD31 (green) and DAPI (pseudo yellow) in individual and merged channels. Red arrows indicate lumen of perfused vessels, white arrows indicates tumor-derived cells (magenta) positive for CD31 (green). Boxed area is magnified in (D), showing tumor-derived cells (white arrow, magenta) enwrapping a perfused vessel (red arrow, teal). The upper panel shows a longitudinal view and the lower panel is rotated 90 degrees to show cross section view of the vessel, X-Y-Z axis is indicated. (E) Representative FACS plot from a P65 brain shows GFP+ tumor-derived cells co-express the endothelial marker CD31 (TDECs). (F) Quantification of murine TAVs (CD31+, GFP−) and TDECs (GFP+, CD31+) at P65 and P80. (G) Glioma neurospheres produce tube-like structures when cultured in endothelial growth media (EGM), DFO, and grown in Matrigel. Scale bars = 20 µm. (H) Representative FACS plots of tumor neurospheres sorted for GFP+ and CD31+ shows co-expression of these two markers; quantification in (I).
To verify that FACS-isolated TDECs are tumor-derived, we performed sequencing of CD31+,GFP+ cells, identifying a substantial number of insertions and/or deletions (eg, in/dels) in the Pten, Nf1, and Tp53 loci, confirming their tumor origins (Supplementary Figure 1C). To determine whether tumor cells from our mouse glioma model can generate endothelium, we generated glioma neurospheres11 and cultured them in endothelial cell growth medium and Matrigel, which induced morphological changes that are reminiscent of tube-like structures (Fig. 2G). FACS confirmed the presence of CD31+, GFP+ TDECs, demonstrating that cultured gliomaspheres generate endothelial cells (Fig. 2H-I). These observations, in conjunction with our imaging data, demonstrate that our native model of glioma generates TDEC populations.
TAVs and TDECs Feature an Endothelial Signature
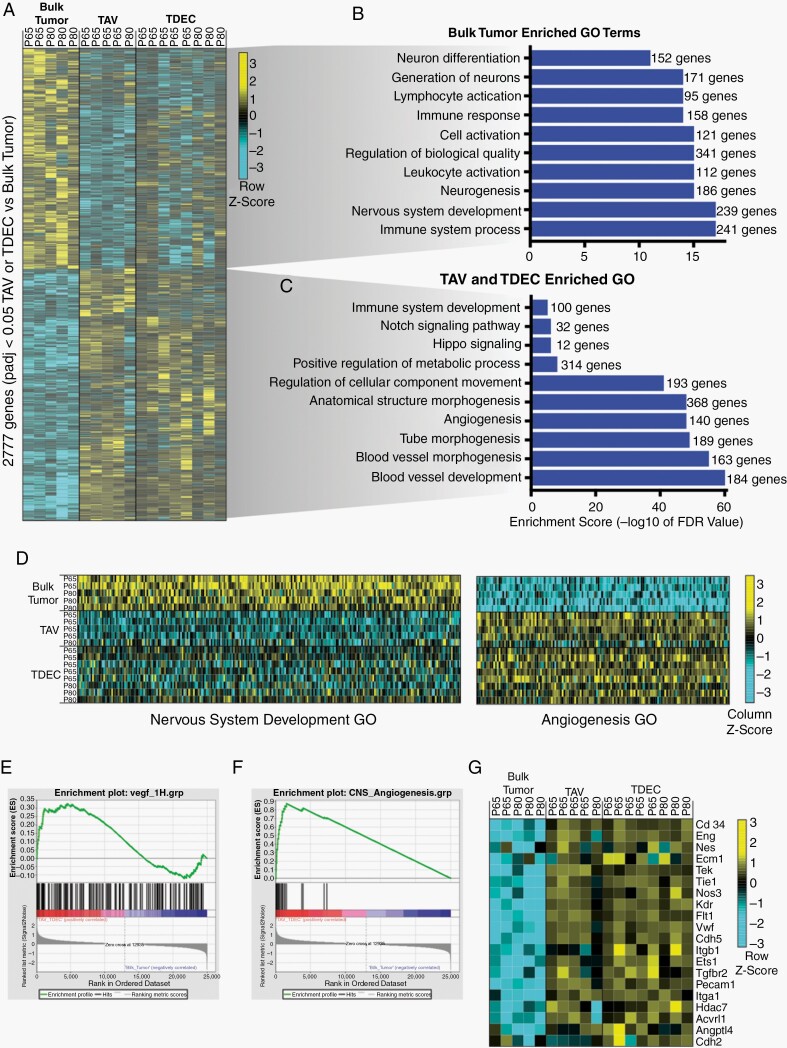
Having established that our mouse model contains both TAV (CD31+, GFP−) and TDEC (CD31+, GFP+) endothelial cell populations, we sought to further distinguish TAVs and TDECs from the bulk tumor. To this end, we performed RNA-sequencing of FACS isolated TAVs and TDECs from P65 and P80 tumors, along with CD31+ populations from the cortex of non-tumor bearing, age-matched mice. Comparative bioinformatics identified more than 2700 significantly differentially expressed genes between bulk tumor and the combined TAV and TDEC populations (Fig. 3A, Supplementary Table 1).
Fig. 3.
Tumor-associated vasculature and tumor-derived endothelium are molecularly distinct. (A) Heatmap of differentially expressed genes detected by RNA-seq. Genes enriched are yellow, genes depleted are turquoise. Associated select gene ontology (GO) terms for genes up-regulated in either (B) bulk tumor or (C) TAV and TDEC populations. (D) Heatmap showing the individual genes for the GO categories Nervous System Development and Angiogenesis. (E) Gene Set Expression Analysis (GSEA) shows enrichment of VEGF-induced transcripts in the combined TAV and TDEC populations. (F) GSEA shows enrichment of CNS angiogenic genes in TAV and TDECs. (G) Heatmap of individual genes used for the angiogenic analysis (panel E) depicts their enrichment in the TAV and TDEC populations and depletion from bulk tumor.
Gene Ontology (GO) analysis confirmed that TAVs and TDECs were enriched in blood vessel- and angiogenesis-related processes, while the bulk tumor displayed features of neural development (Fig. 3B, C). Analysis of individual genes within two representative GO categories, Nervous System Development and Angiogenesis, confirmed the endothelial identity of the TDEC and TAV populations (Fig. 3D). To verify that TAVs and TDECs exhibit signatures indicative of endothelial cells, we compared our results with endothelial gene expression datasets. TDEC/TAV enriched transcripts showed a strong enrichment for a VEGF-induced and angiogenic gene signature21 compared to the bulk tumor (Fig. 3E). Additionally, we found that the signatures of TDECs and TAVs bore a striking resemblance to an established CNS dataset taken from the E14.5 (angiogenic) developing mouse brain22 (Fig. 3F). Visualizing the angiogenic target genes (from panel “E”) showed upregulation in TAVs and TDECs (Fig. 3G), reinforcing their endothelial identity.
Single-Cell Sequencing Identifies Extensive Heterogeneity Among TAVs in Glioma
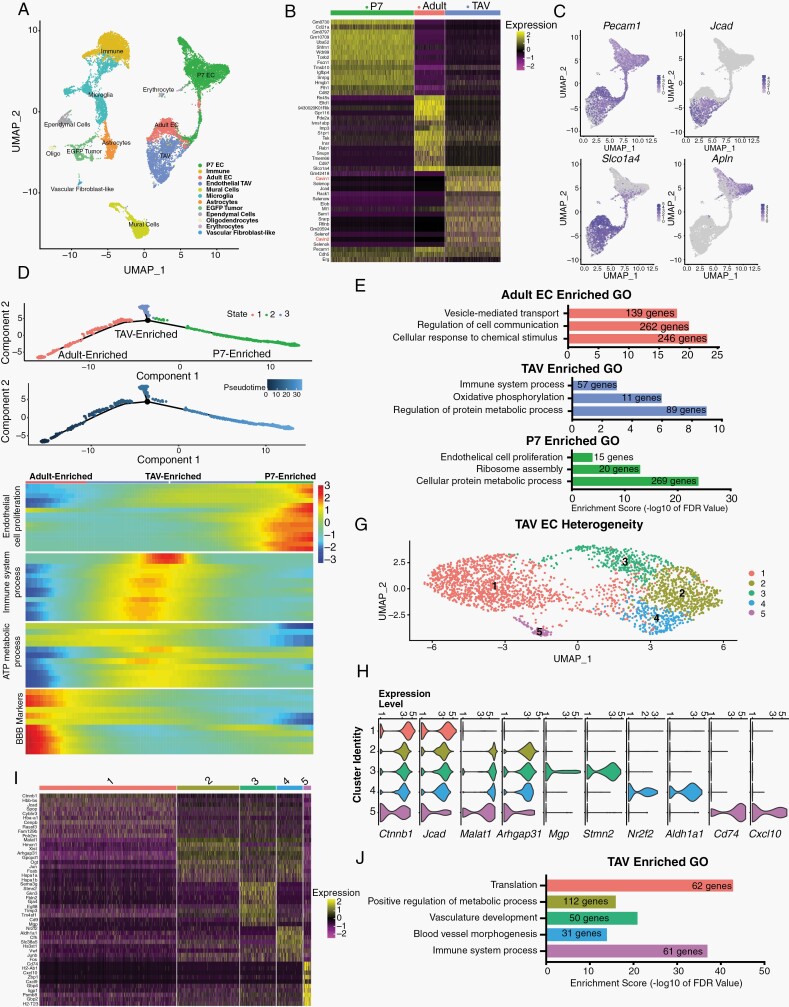
To further interrogate the heterogeneity of glioma vasculature we performed single-cell RNA-sequencing on endothelial cells isolated from glioma-bearing brains. P80 brains from tumor-bearing mice were dissociated, enriched for endothelial cells, and single cell transcriptomes were generated.23 Analysis of the cells’ transcriptomes using Seurat24 and established endothelial data sets25,26 identified 10 distinct cell types, ranging from mural cells to EGFP+ lineage tumor cells (identified by expression of GFP reporter) (Fig. 4A, Supplementary Figure 2A). Differential gene expression analysis revealed unique molecular signatures between these different clusters, as established markers of endothelium (Glut1/Slc2a1, Flt1, Cldn5), microglia (Ccl12), astrocytes (Cspg5, Apoe, Aldoc, Slc1a) and EGFP+ tumor cells expressing glial, oligodendrocyte, and astrocyte markers (eg, Rtn1, Olig1, and Fabp7, respectively) (see Supplementary Figure 2).
Fig. 4.
TAVs display extensive cellular heterogeneity and are not simply angiogenic adult endothelium. (A) UMAP representation of scRNA-seq individual transcriptomes from wildtype P7, adult brain endothelial cells, and P80 tumor-bearing brains; each population is color-coded according to their identity (determined by cell-type specific markers). The corresponding number of cells per cluster is shown in parenthesis. (B) Heatmap showing the top 15 gene markers for each unique endothelial cell cluster. (C) Population marker expression superimposed on only the three endothelial datasets (P7, adult, and TAV). (D) Monocle trajectory analysis of P7 brain ECs, adult brain ECs, and P80 TAVs. (E) GO terms associated with enriched transcripts detected in each cell state, from panel (D). (F) Trajectory heatmap showing dynamic gene expression changes between the endothelial clusters (TAV, Adult, and P7 ECs) plotted along pseudotime as determined by Monocle trajectory analysis from panel (E). (G) UMAP representation of scRNA-seq individual transcriptomes from P80 TAVs identifies 5 unique populations, each color-coded according to their unique identity. (H) Violin plots show gene expression between the 5 different TAV clusters. (I) Heatmap of the top 10 differentially expressed genes between each population. (J) GO terms for enriched transcripts specific to each TAV population.
Reanalysis of only the brain endothelium identified 3 separate clusters (Fig. 4C), each with a distinct molecular signature (Fig. 4B, C). While all P80 endothelial cells (ECs) were isolated by CD31+ FACS, scRNA-seq failed to identity any GFP+ ECs, suggesting these cells are TAVs and not TDECs. This likely reflects the rare nature of TDECs (Fig. 2E, F) and limited sequencing depth. While some pan-endothelial genes, such as Pecam1 (encoding CD31) were expressed across P7, adult, and TAV ECs, other transcripts like the angiogenic marker Apln was restricted to angiogenic P7 ECs, the BBB marker Slco1a4 (Oatp1a4)27 was enriched in adult and TAV ECs, and the cell-cell junction encoding gene Jcad was enriched in TAVs (Fig. 4B, C and Supplementary Table 5). To understand the relationships between ECs from these three clusters, we used trajectory analysis to order the cells in pseudotime, finding that TAVs branch away from angiogenic ECs (P7 ECs) transitioning to a quiescent, homeostatic ECs (adult ECs), suggesting they represent a unique cell state (Fig. 4D). GO analysis of enriched transcripts in each of these three states showed that genes involved in cell proliferation and metabolism are enriched in P7 ECs, vesicle transport and cell to cell communication are present in adult ECs, and immune system and protein metabolism genes are enriched in TAVs (Fig. 4E). Trajectory heatmaps identified dynamic expression of genes between these states, as transcripts enriched in cell proliferation skewed toward P7 ECs, mature BBB markers are enriched in adult ECs, while ATP metabolic processes and immune genes are centered in TAVs (Fig. 4F). These data reinforce our findings that TAVs are molecularly distinct (Fig. 3) and endowed with unique metabolic and motility signatures.
Reanalysis of the P80 TAV clusters showed that the tumor endothelium features extensive heterogeneity, identifying five distinct populations (Fig. 4G) that are defined by their enrichment of distinct endothelial markers. While Jcad, Spop, and Ctnnb1 (β-catenin, a regulator of canonical Wnt signaling and BBB integrity)28 are expressed across all 5 clusters, clusters 2–5 are marked by the long non-coding RNA Malat1,29 Jun, and Arhgap31, a critical GTPase that mediates VEGFR2 signaling.30 Cluster 3 expressed Matrix Gla Protein (Mgp, a regulator of arteriovenous patterning),31Stmn2, and Sema3g, as well as the arterial marker Gja4 (Connexin37). Cluster 4 was enriched for the venous marker Nr2f2,32 the endothelial marker Vwf, Aldh1a1, and Junb, while cluster 5 was marked by expression of the chemokine receptor CD74 and the proinflammatory chemokine Cxcl10 (Fig. 4H, I and Supplementary Figure 2). Gene ontology analysis between these clusters demonstrates that each population is enriched for unique cellular processes (Fig. 4J).
TDEC Populations are Molecularly and Cellularly Heterogeneous
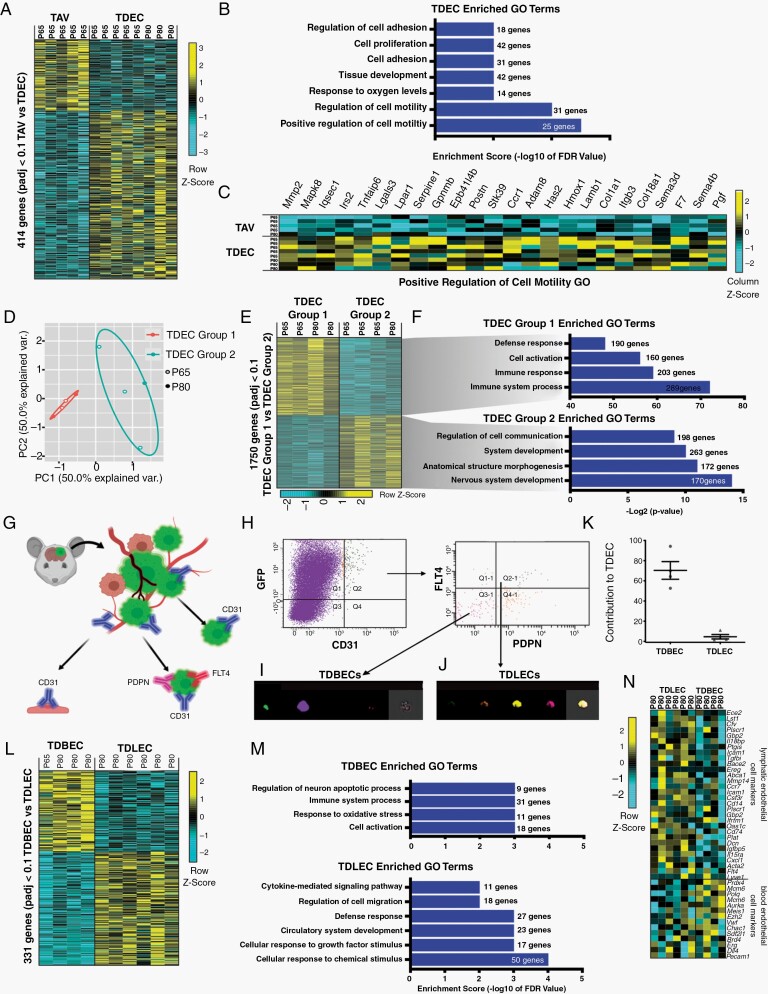
Given the extensive molecular heterogeneity within TAVs, we next sought to molecularly distinguish TAVs and TDECs. Towards this, we directly compared the TAV and TDEC transcriptomes, which revealed over 400 differentially expressed genes between the two populations (Fig. 5A). GO term analysis of these differential gene signatures revealed that genes linked to cell proliferation, cell adhesion, and cell motility, three hallmarks of angiogenesis33 are significantly enriched in TDECs (Supplementary Table 2) (Fig. 5B). Examination of the GO category Positive Regulation of Cell Motility revealed clear upregulation of migratory genes in TDECs compared to TAVs, suggesting they are more infiltrative and angiogenic than their TAV counterparts (Fig. 5C).
Fig. 5.
Murine glioma TDECs are comprised of molecularly and cellularly unique subpopulations. (A) Heatmap of differentially expressed genes in TAVs and TDECs. Genes enriched are indicated in yellow, genes depleted are turquoise. (B) Select gene ontology (GO) terms for genes up-regulated in TDEC populations are shown. (C) Heatmap showing individual genes for the GO category Positive Regulation of Cell Migration. (D) Principal component analysis of bulk RNA-Seq shows that TDEC transcriptomes segregate into 2 groups. (E) Heatmap of differentially expressed genes detected by RNA-seq shows Group 1 (n = 4) and Group 2 (n = 4) TDECs feature distinct signatures. (F) GO terms for select genes up-regulated in TDEC Group1 and Group 2 populations. (G) Schematic illustrating the strategy for isolating TDBECs from mouse glioma. (H) Representative FACS plots demonstrating the isolation of TDBEC and TDLEC populations. (I) Representative image stream analysis demonstrating expression of GFP and CD31 in TDBECs and (J) GFP, CD31, PDNP, and FLT4 in TDLECs. (K) Quantification of TDBECs and TDLECs to the total TDEC population. (L) A heatmap of differentially expressed genes in TDBECs and TDLECs. (M) Associated select GO terms for genes up-regulated in bulk tumor, TDBEC, or TDLEC populations. (N) Heatmap of select genes upregulated in murine blood lymphatic (TDLECs) versus blood endothelial cells (TDBECs).
To further probe the prospective TDEC heterogeneity, we segregated our existing bulk RNA-Sequencing TDEC transcriptome datasets using principle component analysis, finding that bulk TDEC datasets segregated into two populations: TDEC group 1 and group 2 (Fig. 5D). GO analysis of 1750 differentially expressed genes between the two populations revealed immune responses were significantly enriched in group 1, whereas cell-cell communication and nervous system development were upregulated in group 2 (Fig. 5E, F and Supplementary Table 2). Among the immune-related genes identified in group 1 were lymphatic cell surface markers, Podoplanin (Pdpn)34 and Flt4,35 suggesting that TDECs segregate into lymphatic endothelial cells (TDLECs-Group1) and blood endothelial cells (TDBECs-Group2). Next, we performed FACS on P80 native tumors, isolating GFP+, CD31+ endothelial cells, followed by isolation of PDPN+ and FLT44+ cells (Fig. 5G). This strategy revealed that TDECs can be fractionated into two subpopulations, with prospective TBECs marked by GFP+, CD31+, PDPN−, FLT4− and prospective TDLECs marked by GFP+, CD31+, PDPN+, FLT4+ (Fig. 5G, H). Image stream analysis of verified the existence of these subpopulations, while quantification revealed TDLECs constituted a minority of TDECs (Fig. 5I-K).
RNA-Sequencing of these two TDEC subpopulations, TDBECs and TDLECs, identified more than 330 differentially expressed genes (Fig. 5L). GO analysis revealed that circulatory system, cell migration, cytokine signaling, immune and inflammatory, and interferon-alpha response genes are enriched in TDLECs (Fig. 5M).36 Next, we used markers for blood endothelial cells (CD31+, FLT4−, PDPN−) and lymphatic endothelium (CD31+, FLT4+, PDPN+),34 identifying a unique signature for TDLECs that is enriched for immune and inflammatory genes found in lymphatics, including immune cell activation marker Icam1, the secreted proteoglycan Dcn, Tgfbi, and MHC class II-associated invariant chain and chemokine receptor CD74 (Fig. 5N). Collectively, these data show that glioma TDEC populations are comprised of distinct subpopulations exhibiting lymphatic and blood endothelial properties.
TDBECs and TDLECs are Present in PDOX-derived glioblastoma Tumors
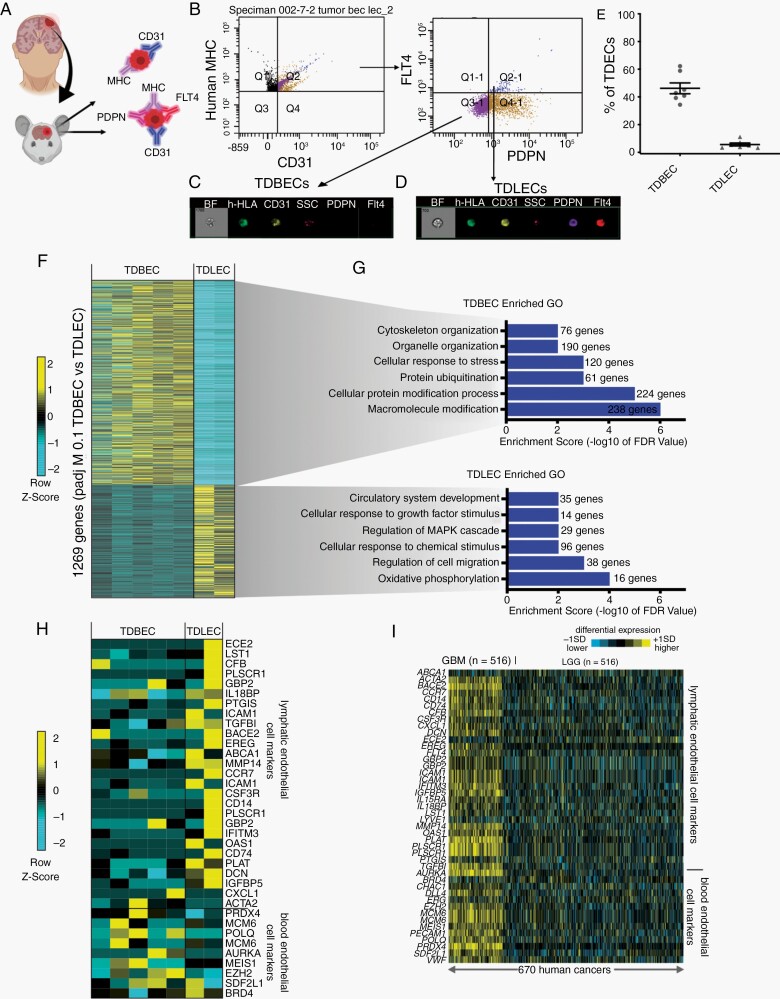
To determine whether TDBEC and TDLEC populations exist in human glioma models, we intracranially injected patient-derived glioma cell lines into SCID mice (ie, PDOX model) and upon tumor formation TDBECs and TDLECs were isolated based on a combination of cell surface expression of human leukocyte antigen, CD31, PDPN, and FLT4 by FACS (Fig. 6A). FACS and image stream analysis confirmed that each of these TDEC subpopulations is present in our PDOX model of glioma (Fig. 6B-E). Subsequent RNA-sequencing on these populations yielded over 1200 differentially expressed genes (Fig. 6F). Analysis of human PDOX TDBEC and TDLEC gene ontologies revealed significant enrichment in processes that paralleled our native mouse models, such as circulatory system development and cell migration (Fig. 6G). Further analysis revealed that PDOX-derived TDBEC and TDLEC populations express a repertoire of established markers of blood endothelial cells and lymphatic endothelium, similar to their mouse counterparts (Fig. 6H; see Fig. 5N). Finally, we evaluated the expression of key genes enriched in both our murine and human TDBEC and TDLEC datasets in low- and high-grade glioma, finding significant enrichment of both signatures in high grade, but not low grade, glioma (Fig. 6I). These PDOX data, combined with observations from our native mouse model of glioma and the human TCGA data, indicate that TDLECs and TDBECs represent two unique endothelial cell populations in glioma, and suggest that while TAVs and TDECs are molecularly distinct from normal brain vasculature, as well as one another, they each feature extensive heterogeneity at a population level.
Fig. 6.
TDEC heterogeneity is conserved in a human xenograft model of glioma. (A) Schematic of TDBECs and TDLECs FACS isolation from PDOX model. (B) FACS plots demonstrating isolation of human TDBEC and TDLEC cell populations from PDOX. (C) Image stream analysis demonstrating protein expression of GFP and CD31 TDBECs. (D) Image stream analysis demonstrating expression of GFP, CD31, PDNP, and FLT4 TDLECs. (E) Quantification of TDBECs and TDLECs relative to the entire TDEC population. (F) Heatmap of differentially expressed genes in PDOX TDBECs and PDOX TDLECs. (G) Select GO terms up-regulated in either human TDLEC or human TDBEC populations. (H) Heatmap depicting conserved upregulation of lymphatic endothelial transcripts in PDOX TDLECs and blood endothelial transcripts in PDOX TDBECs. (I) Heatmap showing enrichment of blood endothelial cell and lymphatic transcripts in high-grade human glioma (n = 154) compared to low-grade glioma (LGC) (n = 516) as determined by RNA-seq data from TCGA.
Discussion
Despite the importance of angiogenesis in cancer pathophysiology, defined functional and molecular benchmarks for vessels during glioma tumorigenesis are lacking. Utilizing both an endogenous glioma model in mice and a PDOX mouse model we characterized the morphological and functional properties of the vasculature during de novo, glioma progression. RNA-seq and scRNA-seq analysis revealed that TAVs are molecularly distinct from normal brain endothelium and that they exhibit extensive heterogeneity. Insights into the biology of TAVs has the potential to reveal new facets of the diverse microenvironment in which glioma progresses and may suggest novel therapeutic targets. For example, genes linked to the production of caveolae in the plasma membrane (eg, Cavin-1 and Cavin-2) are significantly upregulated in TAVs (Supplementary Figure 3). Caveolae regulate cellular senescence and play critical roles in cell signaling. Cavin-2 (also known as serum deprivation response, Sdpr) downregulates endothelial nitric-oxide synthase (eNOS) production and pathologic angiogenesis.37,38 Interestingly, Cavin-1 (Polymerase I and Transcript Release Factor) is elevated in glioma and increased expression is associated with decreased survival time in patients.39 Caveolae may control interstitial fluid pressure, which is dramatically elevated in GBM and impedes drug delivery, and these targets could represent a rationale target for future therapeutic interventions.40
In addition to TAVs, we also characterized the cellular and molecular properties of TDECs. While the existence of TDECs is controversial,10–12,41,42 prior studies on this population did not define how TDECs differ from TAVs. Our analysis represents the first molecular comparison between tumor-associated and tumor-derived vascular populations in an immune-competent animal model, where tumors arise in a de novo manner, in the native brain microenvironment. Through a combination of lightsheet imaging, FACS, and transcriptomics we show that (1) that TDECs are present in a native murine model of glioma and (2) that TAV and TDEC populations are molecularly distinct cell types. Finally, it’s important to note that GSC populations in PDOX models can also produce pericyte-like cells43 and it’s possible to misconstrue tumor-derived pericytes for TDECs. To resolve this, we performed immunostaining with CD13, a pericyte marker, and did not identify any GFP+, CD13+ cells (Supplementary Figure 4C-E). Moreover, comparative bioinformatics between pericyte gene signatures26,44 and our TDEC transcriptional profiling data failed to reveal a significant correlation between these datasets (Supplementary Figure 4F-G). Together, these data lend additional credence to our identification of TDEC populations.
Consistent with prior studies, we found that TDECs are a rare population, raising the question of whether such a rare population is functionally significant. Our investigations into TDEC heterogeneity provides some insight into their possible roles in tumorigenesis. Using FACS approaches with the lymphatic cell surface markers PDPN and FLT4 we found that TDECs are composed of two distinct endothelial subpopulations: TDBECs and TDLECs. Analysis of their gene expression profiles revealed that TDBECs exhibit profiles enriched for vascular development, while TDLECs possess pro-inflammatory and lymphatic signatures. Previous studies suggest that tumor-derived lymphatic cells facilitate metastasis, as primary tumor cells migrate along the lymphatic endothelium.45 However, glioma rarely metastasizes, therefore, TDLECs may potentially play a unique role in tumor inflammation and immune cell recruitment, as they exhibit interferon-alpha and interferon-beta response signatures. Future studies on TDLECs could provide insights to the sources of inflammation in the context of a complex tumor landscape.
The unique inflammatory signatures described in TAVs, TDLECs, and TDBECs are intriguing as their potential interactions with tumor-associated macrophages and other immune populations could explain a variety of emerging immune phenotypes, including immunosuppression.46 These data suggest that targeting specific molecular pathways controlling endothelial cell recruitment within the glioma microenvironment may be a strategy for inhibiting glioma progression or enhancing the efficacy of immunotherapy or other interventions. Moreover, we show that both TAVs and TDECs are molecularly distinct from healthy CNS endothelium, suggesting that strategies differentially targeting tumor-derived vessels, while sparing normal endothelium, may be possible.
Supplementary Material
Funding
This work was supported by grants from the Brockman Foundation (B.D. and C.J.C.), the National Cancer Institute-Cancer Therapeutic Discovery (U01-CA217842 to B.D.), the National Institutes of Health (T32-HL902332 to J.C., F31-CA243382 to E.H.H., 5T32GM088129-10 to W.D.T. 5T32HL007676-27/28 to A.M.H., NS094615 to G.R., and CA223388 to B.D.), American Heart Association (19POST34430008 to A.M.H., 19PRE34410104 to M.C.G.), Cancer Prevention Research Institute of Texas (RP150334 to B.D. and C.J.C., RP200402 to J.D.W., B.D., and C.J.C.), and a pilot award from the Baylor College of Medicine Cardiovascular Research Institute (J.D.W. and B.D.).
Authorship statement. J.C.C., J.D.W., and B.D. conceptualized the study; J.C.C. and M.C.G. designed experiments; J.C.C., M.C.G., B.L., W.D.T., E.H.H., and A.M.H. performed experiments; J.C.C., M.C.G., B.L., B.T., Y.Z., C.J.C., and J.D.W. analyzed data; J.C.C., M.C.G., J.D.W., and B.D. wrote the manuscript; M.C.G., W.D.T., A.M.H., C.J.C., J.D.W., and B.D. secured funding; All authors edited and approved the manuscript.
Conflict of interest statement. None.
References
- 1. Semrad TJ, O’Donnell R, Wun T, et al. Epidemiology of venous thromboembolism in 9489 patients with malignant glioma. J Neurosurg. 2007;106(4):601–608. [DOI] [PubMed] [Google Scholar]
- 2. Jain RK, di Tomaso E, Duda DG, Loeffler JS, Sorensen AG, Batchelor TT. Angiogenesis in brain tumours. Nat Rev Neurosci. 2007;8(8):610–622. [DOI] [PubMed] [Google Scholar]
- 3. Bergers G, Hanahan D. Modes of resistance to anti-angiogenic therapy. Nat Rev Cancer. 2008;8(8):592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kalpathy-Cramer J, Chandra V, Da X, et al. Phase II study of tivozanib, an oral VEGFR inhibitor, in patients with recurrent glioblastoma. J Neurooncol. 2017;131(3):603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xue W, Du X, Wu H, et al. Aberrant glioblastoma neovascularization patterns and their correlation with DCE-MRI-derived parameters following temozolomide and bevacizumab treatment. Sci Rep. 2017;7(1):13894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cha Y, Kim YJ, Lee SH, et al. Post-bevacizumab clinical outcomes and the impact of early discontinuation of bevacizumab in patients with recurrent malignant glioma. Cancer Res Treat. 2017;49(1):129–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lu KV, Bergers G. Mechanisms of evasive resistance to anti-VEGF therapy in glioblastoma. CNS Oncol. 2013;2(1):49–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rose SD, Aghi MK. Mechanisms of evasion to antiangiogenic therapy in glioblastoma. Clin Neurosurg. 2010;57:123–128. [PubMed] [Google Scholar]
- 9. Kerbel RS, Yu J, Tran J, et al. Possible mechanisms of acquired resistance to anti-angiogenic drugs: implications for the use of combination therapy approaches. Cancer Metastasis Rev. 2001;20(1–2):79–86. [DOI] [PubMed] [Google Scholar]
- 10. Ricci-Vitiani L, Pallini R, Biffoni M, et al. Tumour vascularization via endothelial differentiation of glioblastoma stem-like cells. Nature. 2010;468(7325):824–828. [DOI] [PubMed] [Google Scholar]
- 11. Soda Y, Marumoto T, Friedmann-Morvinski D, et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci U S A. 2011;108(11):4274–4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang R, Chadalavada K, Wilshire J, et al. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010;468(7325):829–833. [DOI] [PubMed] [Google Scholar]
- 13. Snuderl M, Fazlollahi L, Le LP, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. [DOI] [PubMed] [Google Scholar]
- 14. Eyler CE, Matsunaga H, Hovestadt V, Vantine SJ, van Galen P, Bernstein BE. Single-cell lineage analysis reveals genetic and epigenetic interplay in glioblastoma drug resistance. Genome Biol. 2020;21(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Glasgow SM, Zhu W, Stolt CC, et al. Mutual antagonism between Sox10 and NFIA regulates diversification of glial lineages and glioma subtypes. Nat Neurosci. 2014;17(10):1322–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu K, Lin CJ, Hatcher A, et al. PIK3CA variants selectively initiate brain hyperactivity during gliomagenesis. Nature. 2020;578(7793):166–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. John Lin CC, Yu K, Hatcher A, et al. Identification of diverse astrocyte populations and their malignant analogs. Nat Neurosci. 2017;20(3):396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lagerweij T, Dusoswa SA, Negrean A, et al. Optical clearing and fluorescence deep-tissue imaging for 3D quantitative analysis of the brain tumor microenvironment. Angiogenesis. 2017;20(4):533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chung K, Wallace J, Kim SY, et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497(7449):332–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mathivet T, Bouleti C, Van Woensel M, et al. Dynamic stroma reorganization drives blood vessel dysmorphia during glioma growth. EMBO Mol Med. 2017;9(12):1629–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fish JE, Cantu Gutierrez M, Dang LT, et al. Dynamic regulation of VEGF-inducible genes by an ERK/ERG/p300 transcriptional network. Development. 2017;144(13):2428–2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hupe M, Li MX, Kneitz S, et al. Gene expression profiles of brain endothelial cells during embryonic development at bulk and single-cell levels. Sci Signal. 2017;10(487). [DOI] [PubMed] [Google Scholar]
- 23. Macosko EZ, Basu A, Satija R, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161(5):1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36(5):411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sabbagh MF, Heng JS, Luo C, et al. Transcriptional and epigenomic landscapes of CNS and non-CNS vascular endothelial cells. eLife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vanlandewijck M, He L, Mäe MA, et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554(7693):475–480. [DOI] [PubMed] [Google Scholar]
- 27. Daneman R, Zhou L, Agalliu D, Cahoy JD, Kaushal A, Barres BA. The mouse blood-brain barrier transcriptome: a new resource for understanding the development and function of brain endothelial cells. PLoS One. 2010;5(10):e13741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zhou Y, Wang Y, Tischfield M, et al. Canonical WNT signaling components in vascular development and barrier formation. J Clin Invest. 2014;124(9):3825–3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Du B, Wang J, Zang S, Mao X, Du Y. Long non-coding RNA MALAT1 suppresses the proliferation and migration of endothelial progenitor cells in deep vein thrombosis by regulating the Wnt/β-catenin pathway. Exp Ther Med. 2020;20(4):3138–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Caron C, DeGeer J, Fournier P, et al. CdGAP/ARHGAP31, a Cdc42/Rac1 GTPase regulator, is critical for vascular development and VEGF-mediated angiogenesis. Sci Rep. 2016;6:27485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yao Y, Jumabay M, Wang A, Boström KI. Matrix Gla protein deficiency causes arteriovenous malformations in mice. J Clin Invest. 2011;121(8):2993–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, Tsai SY. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature. 2005;435(7038):98–104. [DOI] [PubMed] [Google Scholar]
- 33. Potente M, Gerhardt H, Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146(6):873–887. [DOI] [PubMed] [Google Scholar]
- 34. Kriehuber E, Breiteneder-Geleff S, Groeger M, et al. Isolation and characterization of dermal lymphatic and blood endothelial cells reveal stable and functionally specialized cell lineages. J Exp Med. 2001;194(6):797–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mäkinen T, Veikkola T, Mustjoki S, et al. Isolated lymphatic endothelial cells transduce growth, survival and migratory signals via the VEGF-C/D receptor VEGFR-3. EMBO J. 2001;20(17):4762–4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hu X, Deng Q, Ma L, et al. Meningeal lymphatic vessels regulate brain tumor drainage and immunity. Cell Res. 2020;30(3):229–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Boopathy GTK, Kulkarni M, Ho SY, et al. Cavin-2 regulates the activity and stability of endothelial nitric-oxide synthase (eNOS) in angiogenesis. J Biol Chem. 2017;292(43):17760–17776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Powter EE, Coleman PR, Tran MH, et al. Caveolae control the anti-inflammatory phenotype of senescent endothelial cells. Aging Cell. 2015;14(1):102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pu W, Nassar ZD, Khabbazi S, et al. Correlation of the invasive potential of glioblastoma and expression of caveola-forming proteins caveolin-1 and CAVIN1. J Neurooncol. 2019;143(2):207–220. [DOI] [PubMed] [Google Scholar]
- 40. Pu W, Qiu J, Nassar ZD, et al. A role for caveola-forming proteins caveolin-1 and CAVIN1 in the pro-invasive response of glioblastoma to osmotic and hydrostatic pressure. J Cell Mol Med. 2020;24(6):3724–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rodriguez FJ, Orr BA, Ligon KL, Eberhart CG. Neoplastic cells are a rare component in human glioblastoma microvasculature. Oncotarget. 2012;3(1):98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bougnaud S, Golebiewska A, Oudin A, et al. Molecular crosstalk between tumour and brain parenchyma instructs histopathological features in glioblastoma. Oncotarget. 2016;7(22):31955–31971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cheng L, Huang Z, Zhou W, et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153(1):139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. He L, Vanlandewijck M, Raschperger E, et al. Analysis of the brain mural cell transcriptome. Sci Rep. 2016;6:35108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alitalo K. The lymphatic vasculature in disease. Nat Med. 2011;17(11):1371–1380. [DOI] [PubMed] [Google Scholar]
- 46. Takenaka MC, Gabriely G, Rothhammer V, et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat Neurosci. 2019;22(5):729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.