Abstract
Late gadolinium enhanced (LGE) cardiac magnetic resonance (CMR) imaging, the current benchmark for assessment of myocardium viability, enables the identification and quantification of the compromised myocardial tissue regions, as they appear hyper-enhanced compared to the surrounding, healthy myocardium. However, in LGE CMR images, the reduced contrast between the left ventricle (LV) myocardium and LV blood-pool hampers the accurate delineation of the LV myocardium. On the other hand, the balanced-Steady State Free Precession (bSSFP) cine CMR imaging provides high resolution images ideal for accurate segmentation of the cardiac chambers. In the interest of generating patient-specific hybrid 3D and 4D anatomical models of the heart, to identify and quantify the compromised myocardial tissue regions for revascularization therapy planning, in our previous work, we presented a spatial transformer network (STN) based convolutional neural network (CNN) architecture for registration of LGE and bSSFP cine CMR image datasets made available through the 2019 Multi-Sequence Cardiac Magnetic Resonance Segmentation Challenge (MS-CMRSeg). We performed a supervised registration by leveraging the region of interest (RoI) information using the manual annotations of the LV blood-pool, LV myocardium and right ventricle (RV) blood-pool provided for both the LGE and the bSSFP cine CMR images. In order to reduce the reliance on the number of manual annotations for training such network, we propose a joint deep learning framework consisting of three branches: a STN based RoI guided CNN for registration of LGE and bSSFP cine CMR images, an U-Net model for segmentation of bSSFP cine CMR images, and an U-Net model for segmentation of LGE CMR images. This results in learning of a joint multi-scale feature encoder by optimizing all three branches of the network architecture simultaneously. Our experiments show that the registration results obtained by training 25 of the available 45 image datasets in a joint deep learning framework is comparable to the registration results obtained by stand-alone STN based CNN model by training 35 of the available 45 image datasets and also shows significant improvement in registration performance when compared to the results achieved by the stand-alone STN based CNN model by training 25 of the available 45 image datasets.
Keywords: Image Registration, Late Gadolinium Enhanced MRI, Cine Cardiac MRI, Deep Learning, Multi-task Learning
1. INTRODUCTION
Myocardial viability assessment is an essential step for diagnosis and optimal therapy planning for patients diagnosed with cardiovascular diseases such as myocardial infarction, cardiomyopathy or myocarditis. Hence, it is important to accurately detect, localize and quantify the diseased myocardium regions, also referred as infarct/scar, in order to determine the part of the heart that may benefit from revascularization therapy.1 In LGE CMR imaging, the scars exhibit hyper-enhanced intensities, enabling the assessment of their transmural extent.2 This feature renders LGE CMR as the current gold standard for viability assessment of the myocardium.
Nevertheless, in LGE CMR imaging, the gadolinium-based contrast agent that is responsible for the brighter scar tissue reduces the contrast between the LV myocardium and the LV blood-pool, making it difficult to accurately delineate the myocardium. On the contrary, the bSSFP cine CMR images provide high resolution, dynamic 3D images of the cardiac chambers with very good contrast between myocardium and blood-pool; however, they do not show the scarred myocardial regions (Fig 1). To this end, it is crucial to segment and co-register the LGE and bSSFP cine CMR images to generate hybrid 3D and 4D anatomical models of the heart in the effort to help localize, quantify and clearly visualize the compromised myocardial regions, for use in patient-specific revascularization therapy planning and guidance. On account of this, a number of researchers proposed extracting the myocardium and blood-pool contours from the cine CMR images and superimposing them on the LGE CMR images, thus, formulating it as a multimodal image registration problem.
Figure 1:
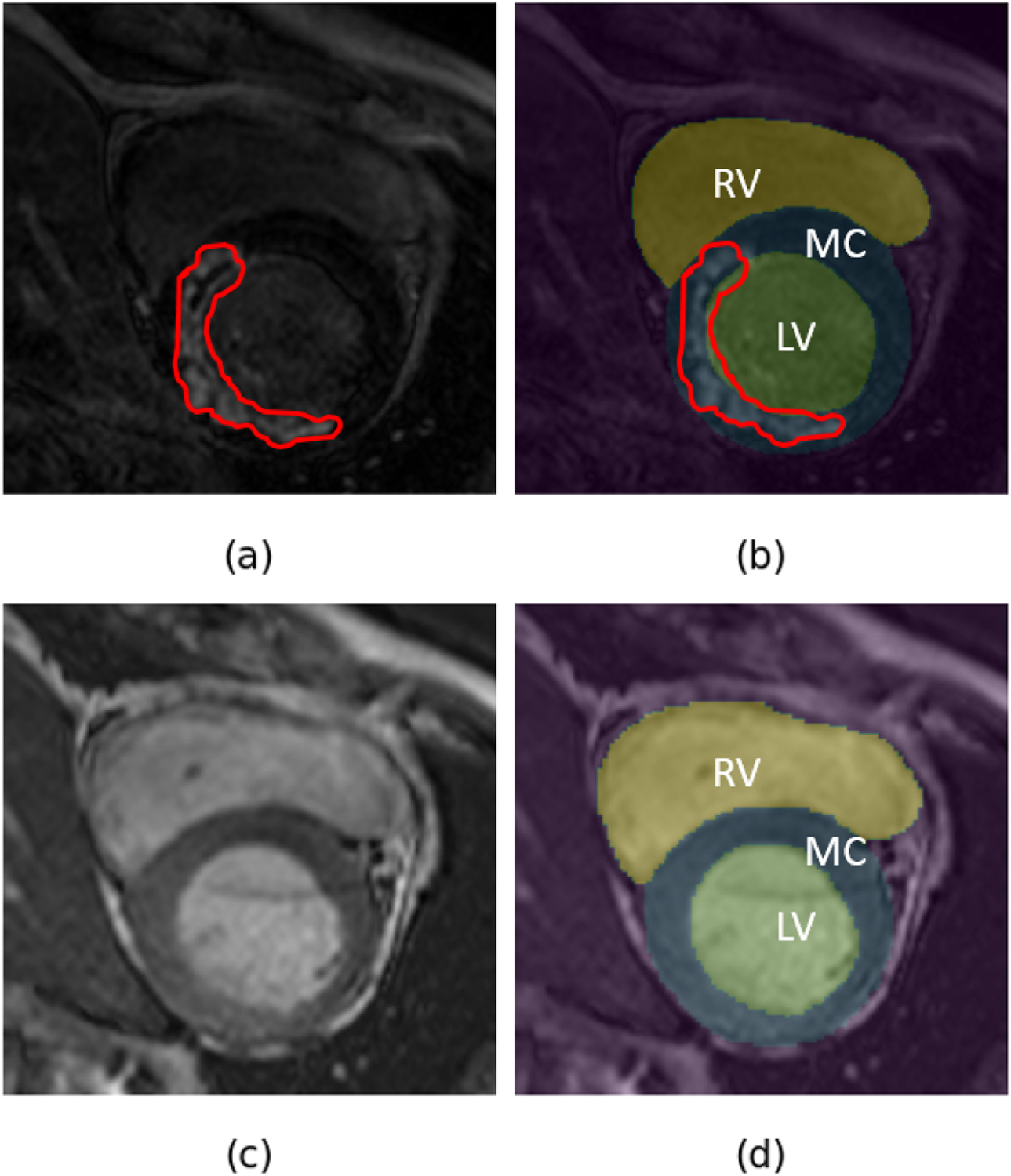
Example of (a) LGE CMR image and associated hyper-enhanced regions marked by red contour with (b) overlaid manual annotations - LV blood-pool (LV), LV myocardium (MC) and RV blood-pool (RV), and (c) bSSFP cine CMR image with its (d) manual annotations overlaid on it.
Prior to deep learning, a number of algorithms like rigid registration using normalized mutual information as similarity measure,3 affine registration using pattern intensity as similarity measure,1 as well as multi-step registration approaches (rigid followed by deformable registration4) have been proposed for registering the cine images with contrast delay-enhanced images. These traditional approaches iteratively optimize the registration cost function for a given pair of images.
In recent years, several researchers proposed utilizing deep learning for optimization of these registration cost functions.5 These unsupervised deep learning registration algorithms help speed up the registration process compared to the traditional unsupervised algorithms, however, they do not significantly improve the registration accuracy, as the similarity measures used are the same.
In the recent 2019 MS-CMRSeg challenge,6, 7 researchers proposed an alternative approach to learning features from one image type and using them to segment the other image type - training adversarial networks to generate synthetic LGE CMR images from bSSFP cine CMR images, followed by training U-Net architectures on these synthetic LGE images to segment cardiac chambers from the LGE images.8–11 Although these methods result in good segmentation performance, they are time consuming, as they involve training adversarial networks followed by U-Net models.
In our previous work, we showed that a STN based RoI-guided CNN can be used to accurately register bSSFP cine CMR images with its corresponding LGE CMR images in a time-efficient manner.12 Here, we propose a joint deep learning framework that consists of three branches - a STN-inspired CNN for supervised registration of bSSFP cine CMR images and LGE CMR images, an U-Net model13 for segmentation of bSSFP cine CMR images and an U-Net model for segmentation of LGE CMR images. Inspired by Qin et al.,14 we optimize a composite loss function by training all three networks simultaneously. The aim of the proposed joint deep learning model is to further improve registration accuracy by sharing the weights learned from the segmentation models. We train the proposed joint network on the 2019 MS-CMRSeg challenge dataset using the provided manual annotations of the LV blood-pool, LV myocardium, and RV blood-pool.
2. METHODOLOGY
2.1. Dataset
The dataset used in our experiments was made available through the 2019 MS-CMRSeg challenge.6, 7 The dataset consists of LGE and bSSFP cine CMR images acquired at end-diastole for 45 patients diagnosed with cardiomyopathy.
The bSSFP cine CMR images consisted of 8–12 slices with an in-plane resolution of 1.25 mm × 1.25 mm and a slice thickness of 8–13 mm, while the LGE CMR images consisted of 10–18 slices featuring an in-plane resolution of 0.75 mm × 0.75 mm and a 5 mm slice thickness. To deal with the heterogeneity in slice thickness, image sizes and in-plane image resolution between LGE and bSSFP cine CMR images, all the images are resampled to a slice thickness of 5 mm, in-plane image resolution of 0.75 mm × 0.75 mm and resized to 224 × 224 pixels.
2.2. STN-based Registration
The conventional STN consists of three parts - a localisation network, a grid generator and a differentiable image sampler. In our experiments, the input to the localisation network is a concatenated bSSFP cine CMR image and a LGE CMR image. The localization network outputs a six-dimensional vector θ, that results in the transformation matrix Tθ,
| (1) |
containing the parameters for affine registration. The grid generator uses these predicted transformation parameters to generate a sampling grid i.e., a set of points where the input map should be sampled to produce the transformed output. In the training phase, this sampling grid and the ground truth (GT) map of the LGE CMR image are the inputs to the differentiable image sampler, resulting in a transformed GT map of the LGE CMR image (Fig. 2). In the testing phase, the sampling grid generated by the grid generator is input to the differentiable image sampler along with the LGE image slice to produce a transformed LGE image slice.
Figure 2:
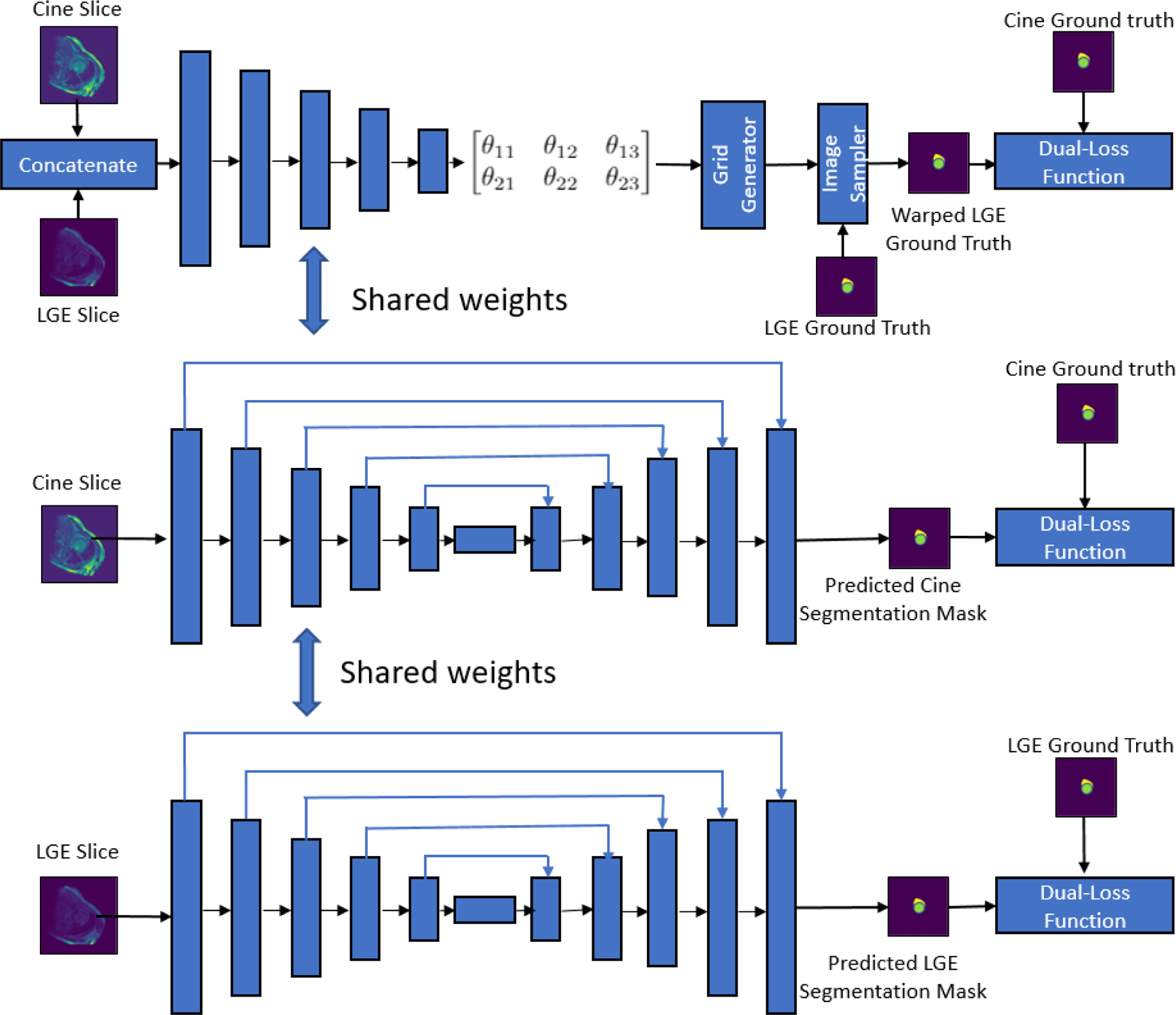
Schematic architecture of the proposed joint deep learning framework consisting of three branches - a STN based CNN for registration of bSSFP cine CMR and LGE CMR images, an U-Net model for segmentation of bSSFP cine CMR images and an U-Net model for segmentation of LGE CMR images.
2.3. Joint Deep Learning Model for Registration and Segmentation
As shown in earlier works,14–16 image registration and segmentation tasks are closely related and it has been shown that learning features from one task can benefit the other task. In this work, we explore a joint deep learning model for registration of bSSFP cine CMR and LGE CMR images, and segmentation of cardiac chambers (LV blood-pool, LV myocardium and RV blood-pool) from the bSSFP cine CMR and LGE CMR images (Fig. 2). The coupling of these registration and segmentation tasks result in sharing of the weights learnt from the segmentation task with the registration branch of the network, improving the registration accuracy.
2.4. Experiments and Implementation Details
In this study, our experiments are focused on comparing the registration results of the proposed joint deep learning model with the stand-alone STN based model.12 We also compare the results obtained by training our networks by splitting the available 45 bSSFP cine and LGE MRI datasets to 35 for training, 5 for validation and 5 for testing, and the results obtained by training our networks by splitting to 25 for training, 15 for validation and 5 for testing.
The three branches of our joint deep learning model are trained using the following dual-loss function:
| (2) |
where and are cross-entropy loss and Dice loss, respectively. The loss function is calculated using the predicted segmentation maps and their corresponding GT maps for the bSSFP cine and LGE CMR segmentation networks ( and , respectively). For the supervised RoI-guided registration network, the dual-loss function is computed using the transformed GT map of the LGE images and the GT of cine bSSFP images (). Therefore, the resulting composite loss function is given by
| (3) |
where λ1, λ2 and λ3 are the trade-off parameters for the three branches of the joint deep learning model.
We train our networks by randomly augmenting both the bSSFP cine CMR and the LGE CMR images on-the-fly using a series of translation, rotation and gamma correction operations. In all our experiments, the networks are trained using the Adam optimizer with a learning rate of 10−4 and a gamma decay of 0.99 every alternate epoch for fine-tuning for 100 epochs on a machine equipped with NVIDIA RTX 2080 Ti GPU with 11GB of memory.
2.5. Evaluation metrics
To quantify our registration accuracy, we calculate the Euclidean distance between the LV and RV blood-pool centers i.e. center distance (CD) of the LGE CMR image and bSSFP cine CMR image, and compare it with the CD of the transformed LGE CMR image and the bSSFP cine CMR image. These blood-pool centers are identified as the centroid of the LV and RV blood-pool segmentation masks. We also evaluate our registration using the average surface distance (ASD) between the LV blood-pool, LV myocardium and RV blood-pool segmentation masks of the LGE and bSSFP cine CMR images, before and after registration.
3. RESULTS AND DISCUSSION
In Table 1, we summarize the registration performance of the stand-alone STN-based RoI-guided CNN and the proposed joint deep learning model. We compare the mean CD and mean ASD achieved by both the stand-alone STN based CNN and the joint deep learning model by training 25 of the 45 available image datasets and by training 35 of the 45 available image datasets. We can observe that the registration performance of the joint deep learning model achieved by training only 25 image datasets is comparable to that of the stand-alone STN based registration when trained using 35 image datasets and significantly better than the stand-alone STN based registration when trained using 25 image datasets (p-value < 0.1 for RV blood-pool CD, and LV blood-pool CD and ASD). We can also observe that when the joint deep learning model is trained using 35 image datasets, the LV blood-pool CD and LV myocardium ASD is significantly lower than the rest of the models (Fig. 3). In Fig. 4, we show an example of the manual annotations of the cardiac chambers of a bSSFP cine CMR image overlaid on its corresponding LGE CMR image before registration and after registration using both stand-alone STN-based RoI-guided CNN model and the joint deep learning model.
Table 1:
Summary of registration evaluation.
| LV CD (mm) |
LV ASD (mm) |
MC ASD (mm) |
RV CD (mm) |
RV ASD (mm) |
|
|---|---|---|---|---|---|
| Before Registration | 3.28 (1.83) |
2.53 (1.23) |
1.78 (0.78) |
4.36 (3.79) |
2.42 (1.20) |
| Stand-alone STN Model Training: 25 patients |
2.46 (1.31)** |
2.20 (1.23)* |
1.52 (0.84)* |
2.92 (2.18)** |
1.95 (1.02)* |
| Stand-alone STN Model Training: 35 patients |
2.27 (1.38)** |
2.09 (1.14)** |
1.40 (1.12)* |
2.52 (2.66)** |
1.73 (1.02)** |
| Joint Model Training: 25 patients |
2.26 (1.34)** |
1.96 (0.93)** |
1.41 (0.71)* |
2.60 (2.02)** |
1.77 (0.84)** |
| Joint Model Training: 35 patients |
2.18 (1.46)** |
1.94 (0.93)** |
1.33 (0.73)** |
2.53 (2.14)** |
1.72 (0.97)** |
Mean (std-dev) center-to-center distance (CD) and average surface distance (ASD) for LV blood-pool (LV), LV myocardium (MC) and RV blood-pool (RV). The best evaluation metrics achieved are labeled in bold.
Statistically significant differences between the registration metrics before and after registration were evaluated using the Student t-test and are reported using * for p < 0.05 and ** for p < 0.005.
Figure 3:
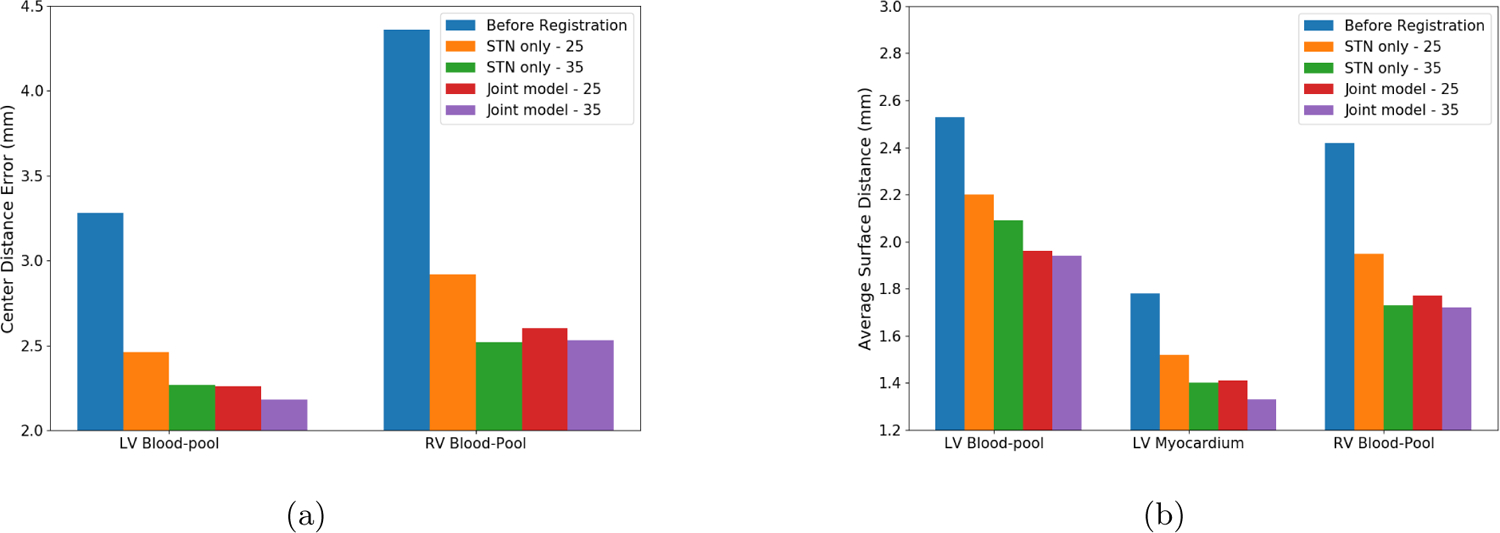
Comparison of (a) mean CD and (b) mean ASD values before registration, stand-alone STN based supervised registration (training data: 25 patients), stand-alone STN based supervised registration (training data: 35 patients), joint deep learning model (training data: 25 patients) and joint deep learning model (training data: 35 patients)
Figure 4:
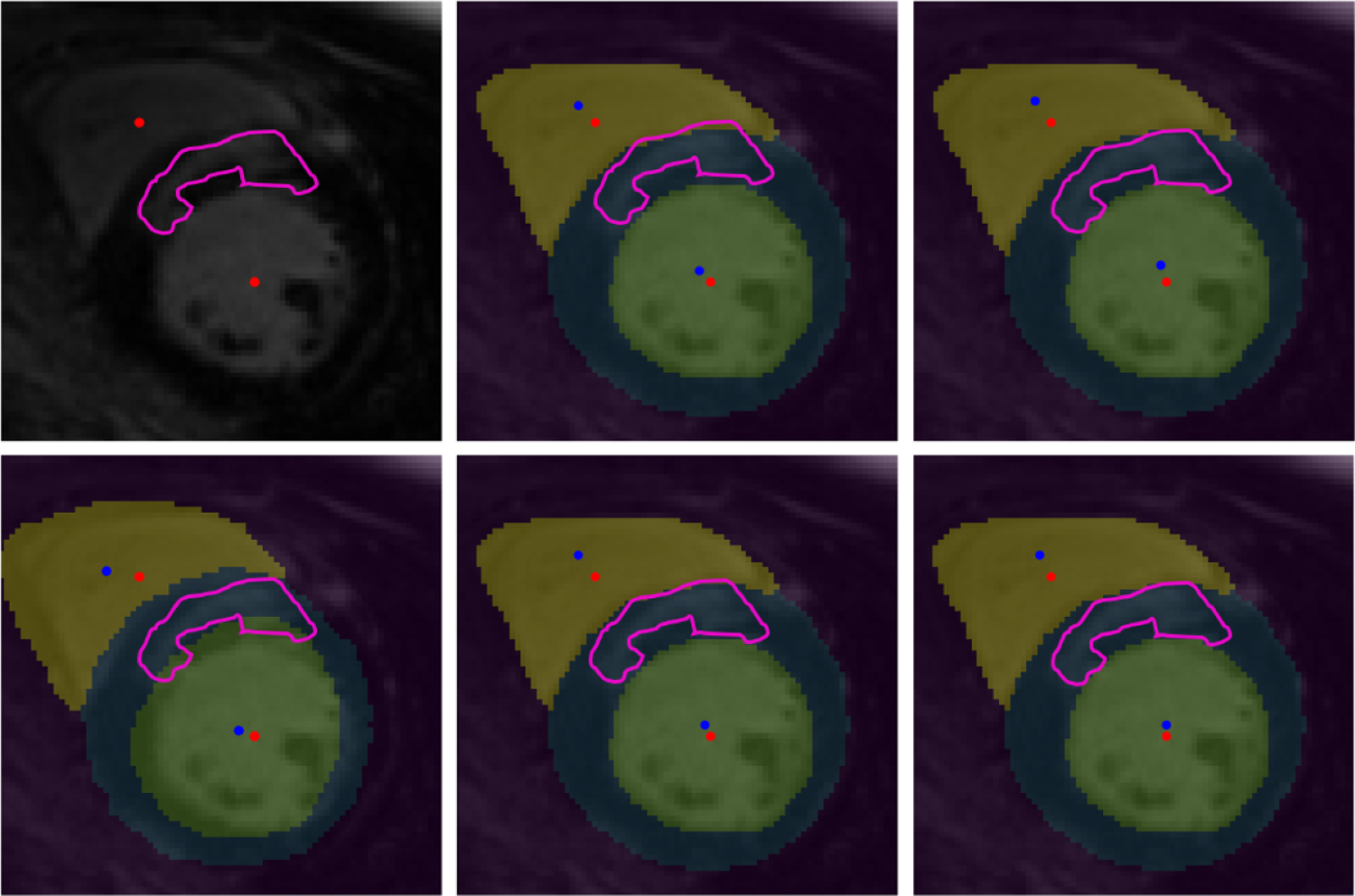
Panel 1–1: LGE CMR image and associated hyper-enhanced regions marked by pink contour and LV and RV blood-pool centers marked by red dots; Panel 2–1: before registration (CD: 3.46 mm, ASD: 1.87 mm); overlaid unregistered LGE CMR image and features (from Panel 1–1) onto the bSFFP image showing the RV blood-pool (yellow) and LV blood-pool (green) and their centers (marked by blue dots) and the LV myocardium (blue) marked on the bSSFP image; Panel 1–2: overlaid LGE CMR image onto the bSSFP image following stand-alone STN model registration using 25 patients for training (CD: 2.77 mm, ASD: 1.62 mm); Panel 2–2: stand-alone STN model registration using 35 patients for training (CD: 2.72 mm, ASD: 1.45 mm); Panel 1–3: joint deep learning model registration using 25 patients for training (CD: 2.77 mm, ASD: 1.51 mm); and Panel 2–3: joint deep learning model registration using 35 patients for training (CD: 2.65 mm, ASD: 1.47 mm).
In our previous work,12 we showed that a STN inspired RoI-guided CNN architecture can be reliably used to register the bSSFP cine CMR and LGE CMR images. The major drawback of the method is the need for annotations of cardiac structures for large number of training data. In this paper, we investigate whether the joint deep learning framework is a viable option for registration of LGE and bSSFP cine CMR images. Our results reveal that the proposed joint deep learning model leverages the weights learnt from the segmentation task to improve the registration accuracy and produces reliable registration results using lesser number of training data and manual annotations.
The mean Dice scores achieved by the segmentation branches of the bSSFP cine CMR images and LGE CMR images are 84.73% and 71.49%, respectively. The poor results of the segmentation branches are due to the limited number of the training data, however, the weights learnt from these segmentation branches improve the registration accuracy in the joint deep learning model. The computational time required for each epoch for a stand-alone STN model is around 63 seconds and 67 seconds to train 25 and 35 of the 45 available image datasets, respectively, whereas the joint deep learning model requires around 110 seconds and 155 seconds to train 25 and 35 of the 45 available image datasets, respectively. It is worth to be noted that although the stand-alone STN model takes less training time for the registration, it nevertheless requires manual annotations, while the joint model requires relatively more time for training, the number of manual annotations needed are fewer.
4. CONCLUSION AND FUTURE WORK
In this paper, we present a joint deep learning model for registration of LGE and bSSFP cine CMR images, and the segmentation of cardiac chambers from the LGE CMR and the bSSFP CMR images. The coupling of the segmentation and the registration tasks enables a multi-task training and results in obtaining reliable registration results using a lower number of training datasets, reducing the need for a large number of manual annotations.
As part of our future work, we will be investigating other variants of U-Net architecture to improve the segmentation performance of the joint deep learning model and these obtained segmentation masks can be used to further fine-tune the registration in case of sparsely annotated datasets, resulting in a weakly-supervised method for registration. Ultimately, we intend to build 3D models that help quantify and visualize the compromised myocardial regions.
ACKNOWLEDGMENTS
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award No. R35GM128877 and by the Office of Advanced Cyber-infrastructure of the National Science Foundation under Award No. 1808530.
REFERENCES
- [1].Wei D, Sun Y, Chai P, Low A, and Ong SH, “Myocardial segmentation of late gadolinium enhanced MR images by propagation of contours from cine MR images,” in [International Conference on Medical Image Computing and Computer-Assisted Intervention], 428–435, Springer; (2011). [DOI] [PubMed] [Google Scholar]
- [2].Juan LJ, Crean AM, and Wintersperger BJ, “Late gadolinium enhancement imaging in assessment of myocardial viability: techniques and clinical applications,” Radiologic Clinics 53(2), 397–411 (2015). [DOI] [PubMed] [Google Scholar]
- [3].Chenoune Y, Constantinides C, El Berbari R, Roullot E, Frouin F, Herment A, and Mousseaux E, “Rigid registration of delayed-enhancement and cine cardiac MR images using 3D normalized mutual information,” in [2010 Computing in Cardiology], 161–164, IEEE; (2010). [Google Scholar]
- [4].Guo F, Li M, Ng M, Wright G, and Pop M, “Cine and multicontrast late enhanced MRI registration for 3D heart model construction,” in [International Workshop on Statistical Atlases and Computational Models of the Heart], 49–57, Springer; (2018). [Google Scholar]
- [5].Khalil A, Ng S-C, Liew YM, and Lai KW, “An overview on image registration techniques for cardiac diagnosis and treatment,” Cardiology research and practice 2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhuang X, “Multivariate mixture model for cardiac segmentation from multi-sequence MRI,” in [International Conference on Medical Image Computing and Computer-Assisted Intervention], 581–588, Springer; (2016). [Google Scholar]
- [7].Zhuang X, “Multivariate mixture model for myocardial segmentation combining multi-source images,” IEEE transactions on pattern analysis and machine intelligence 41(12), 2933–2946 (2018). [DOI] [PubMed] [Google Scholar]
- [8].Chen C, Ouyang C, Tarroni G, Schlemper J, Qiu H, Bai W, and Rueckert D, “Unsupervised multi-modal style transfer for cardiac MR segmentation,” arXiv preprint arXiv:1908.07344 (2019).
- [9].Liu Y, Wang W, Wang K, Ye C, and Luo G, “An automatic cardiac segmentation framework based on multi-sequence MR image,” arXiv preprint arXiv:1909.05488 (2019).
- [10].Petersen SE, Ballester MAG, and Lekadir K, “Combining multi-sequence and synthetic images for improved segmentation of late gadolinium enhancement cardiac MRI,” in [Statistical Atlases and Computational Models of the Heart. Multi-Sequence CMR Segmentation, CRT-EPiggy and LV Full Quantification Challenges: 10th International Workshop, STACOM 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, October 13, 2019, Revised Selected Papers], 290, Springer Nature. [Google Scholar]
- [11].Tao X, Wei H, Xue W, and Ni D, “Segmentation of multimodal myocardial images using shape-transfer gan,” arXiv preprint arXiv:1908.05094 (2019).
- [12].Upendra RR, Simon R, and Linte CA, “A supervised image registration approach for late gadolinium enhanced MRI and cine cardiac MRI using convolutional neural networks,” in [Annual Conference on Medical Image Understanding and Analysis], 208–220, Springer; (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ronneberger O, Fischer P, and Brox T, “U-net: Convolutional networks for biomedical image segmentation,” in [International Conference on Medical image computing and computer-assisted intervention. Lecture Notes in Computer Science], 9351, 234–241, Springer; (2015). [Google Scholar]
- [14].Qin C, Bai W, Schlemper J, Petersen SE, Piechnik SK, Neubauer S, and Rueckert D, “Joint learning of motion estimation and segmentation for cardiac MR image sequences,” in [International Conference on Medical Image Computing and Computer-Assisted Intervention], 472–480, Springer; (2018). [Google Scholar]
- [15].Tsai Y-H, Yang M-H, and Black MJ, “Video segmentation via object flow,” in [Proceedings of the IEEE conference on computer vision and pattern recognition], 3899–3908 (2016). [Google Scholar]
- [16].Cheng J, Tsai Y-H, Wang S, and Yang M-H, “Segflow: Joint learning for video object segmentation and optical flow,” in [Proceedings of the IEEE international conference on computer vision], 686–695 (2017). [Google Scholar]


