Abstract
Next-generation sequencing (NGS) is rapidly expanding into routine oncology practice. Genetic variations in both the cancer and inherited genomes are informative for hereditary cancer risk, prognosis, and treatment strategies. Herein, we focus on the clinical perspective of integrating NGS results into patient care to assist with therapeutic decision making. Five key considerations are addressed for operationalization of NGS testing and application of results to patient care as follows: (1) NGS test ordering and workflow design; (2) result reporting, curation, and storage; (3) clinical consultation services that provide test interpretations and identify opportunities for molecularly guided therapy; (4) presentation of genetic information within the electronic health record; and (5) education of providers and patients. Several of these key considerations center on informatics tools that support NGS test ordering and referencing back to the results for therapeutic purposes. Clinical decision support tools embedded within the electronic health record can assist with NGS test utilization and identifying opportunities for targeted therapy including clinical trial eligibility. Challenges for project and change management in operationalizing NGS-supported, evidence-based patient care in the context of current information technology systems with appropriate clinical data standards are discussed, and solutions for overcoming barriers are provided.
INTRODUCTION
The application of genetic information to assist with the clinical management of patients with cancer is rapidly expanding into routine care. Both cancer (ie, somatic) and inherited (ie, germline) genomes are clinically important, as variations in either genome can be informative for treatment strategies and drug response.1,2 More than 200 commercially available drugs contain genetic information in their US Food and Drug Administration–approved labeling for indications and usage or for potential impact on drug safety or response.3,4 The majority of these medications are either anticancer agents or drugs used in the oncology supportive care setting (eg, pain medications, antidepressants, and antifungal prophylaxis). Molecularly guided therapy may be applicable to front-line treatment for certain cancer diagnoses, if pertinent genetic variants are present, or applicable to later-line therapies including opportunities for off-label use.5,6 Molecularly focused clinical trials, such as basket trials, that require specific genetic alterations for enrollment eligibility are also expanding treatment options for patients with cancer.7,8
CONTEXT
Key Objective
How can next-generation sequencing (NGS) be seamlessly integrated into routine patient care and coupled with solutions for data storage that are linked to informatic systems supporting identification of therapeutic opportunities and clinical research?
Knowledge Generated
Operationalizing NGS is complex with institutional workflows and infrastructure needed to efficiently order, receive, and store NGS data. Clinical services are essential to guide systematic clinical implementation with informatic systems needed to organize and visualize complex NGS data.
Relevance
Somatic and germline NGS data can assist with clinical management of patients with cancer by identifying opportunities for targeted therapy and mitigating gene-drug interactions. The electronic health record can be leveraged for efficient NGS test ordering, with curation of discrete genetic information supporting decision support tools that alert clinicians in real time of important results. Precision medicine clinical services can assist with interpretation of the NGS results by bridging gaps between guideline limitations and application to patient care.
There are numerous genetic testing platforms that can be used for guiding cancer care. Genetic testing can range from single gene testing (eg, EGFR pyrosequencing) to comprehensive next-generation sequencing (NGS) of the whole genome or exome, although whole genome sequencing has not yet emerged as a customary test offering in oncology. Targeted NGS of the somatic genome that encompasses hundreds of genes is more commonly performed in the oncologic setting to identify opportunities for molecularly guided therapy. Germline testing for therapeutic purposes has not been as greatly adopted, but is likely to increase due in part to clinical trials such as Pancreas Cancer Olaparib Ongoing and Trial of PARP Inhibition in Prostate Cancer (TOPARP) showing clinical benefit of PARP (poly [ADP-ribose] polymerase) inhibitors when certain germline variants (eg, BRCA1 or BRCA2 alterations) are present.9-13 Furthermore, there is strong clinical evidence demonstrating that the germline pharmacogenetic results can assist with mitigating chemotherapy toxicity risks along with optimizing supportive care drug selection and dosage.2,14-21 Comprehensive NGS platforms are emerging that provide both somatic and germline genetic information, with efforts underway to extract pharmacogenetic information from sequencing results.22 Taken together, clinicians are increasingly being exposed to vast amounts of clinically important genetic data that can affect the medical management of patients with cancer.
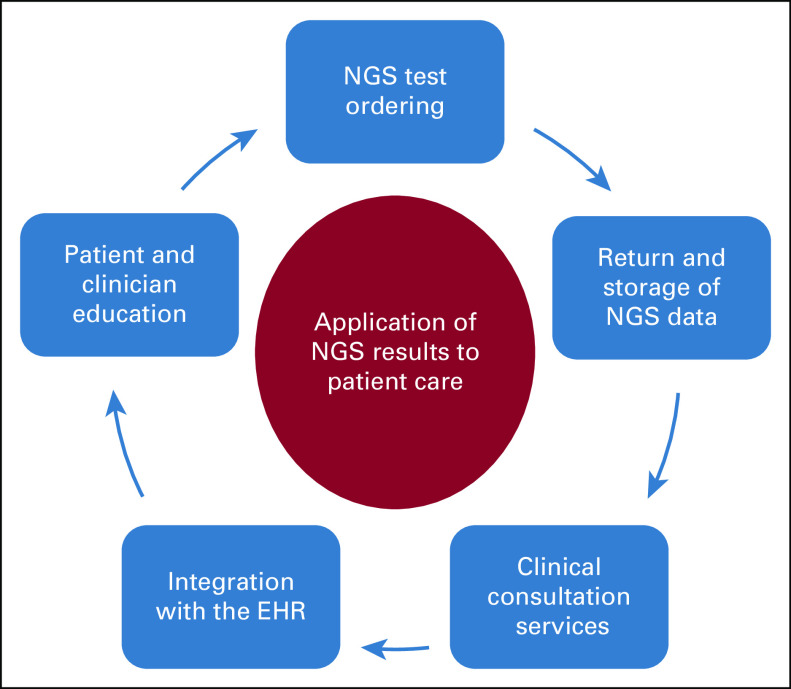
Strategies are needed to fully harness the enormous amounts of genetic information generated from NGS and to integrate clinically relevant results into patient care. Focusing on therapeutics, we addressed five key considerations for application of the NGS results to patient care as follows: (1) NGS test ordering and workflow design; (2) result reporting, curation, and storage; (3) clinical consultation services that provide test interpretations and identify opportunities for molecularly guided therapy; (4) presentation of important results and optimizing the human computer interface within the electronic health record (EHR); and (5) education of providers and patients (Fig 1). Several of these key considerations center on informatics tools that support test ordering algorithms that incorporate appropriate test utilization and support referencing back to the NGS results. Potential challenges for operationalizing these key considerations and solutions for overcoming barriers are discussed. Specific examples of integrating NGS into clinical care are provided on the basis of our experiences that are inclusive of precision medicine specialists (J.K.H.), oncologists (K.K.F., J.E.G., B.M., and J.M.), and clinical informaticists (R.M.P. and M.F.O.) along with population health and data scientists (J.K.T., E.R., and D.E.R.).23-25 Our review is intended to provide guidance for incorporating NGS into patient care, specifically applicable to health systems, executive administrators, precision medicine programs, health informaticists, and providers.
FIG 1.
Key considerations for integrating somatic and germline NGS results into patient care. EHR, electronic health record; NGS, next-generation sequencing.
NGS TEST ORDERING—WORKFLOW DESIGN
The selection of an NGS platform can depend on the cancer type, availability of a particular tissue specimen, genes of interest, whether information on somatic and/or germline is needed, and additional desired features including transcriptome sequencing, microsatellite instability status, homologous recombination deficiency status, and tumor mutation burden.26 Administrative and workflow factors can also influence the selection of an NGS assay, including test costs, patient billing, and methods for returning results (eg, physician and patient portals, portable document formats [PDFs], and bioinformatic pipelines). Genetic testing oversight committees, with key stakeholders inclusive of clinicians, molecular pathologists, bioinformaticists, patients, finance, and legal, can help identify their institution's optimal options for NGS testing. Larger medical centers often choose to develop in-house NGS assays that cover most testing requirements, supplemented with reference laboratories used as needed for certain clinical scenarios.26 After devising optimal NGS testing strategies, identifying which patients who should undergo NGS testing must be carefully considered on the basis of the clinical characteristics of each unique patient. Additionally, NGS testing may be influenced by the technical challenges of ordering the test, associated costs, and patient consenting.
A survey of oncologists found that most have used NGS testing in clinical practice, with guiding treatment for advanced refractory disease as the most common reason for ordering an NGS assay.27 However, there are limited consensus guidelines for which patients should be considered for NGS testing to guide treatment strategies, with the National Comprehensive Cancer Network guidelines and the European Society for Medical Oncology providing guidance for certain cancer diagnoses.5,28,29 Identifying patients who may benefit from NGS testing versus focused-indication molecular tests (eg, BRAFV600E testing for patients with melanoma), or no molecular testing at all, can be challenging. Institution-specific clinical pathways, which have been increasingly used to integrate evidence-based treatment standards into clinical workflows and decision support, can be a tool to help identify patients who may benefit from NGS testing on the basis of diagnosis, prognostic indicators, and prior therapy.30 As an example, Moffitt Cancer Center has developed almost 80 evidence-base, multidisciplinary clinical pathways covering 57 disease states with approximately half of these clinical pathways fully integrated into the EHR.25,31,32 Considerations for molecular testing, including NGS assays, are embedded into clinical pathway algorithms inclusive of recommendations for targeted therapies (Fig 2). As opportunities for molecularly guided therapy continue to grow, oncology practice may eventually move toward obtaining NGS tests on all patients with cancer or at least for those with advanced disease that failed first-line therapy.29,33 Furthermore, comprehensive NGS testing may be more efficient, and less costly, than ordering numerous single-gene or focused-indication molecular tests.27
FIG 2.
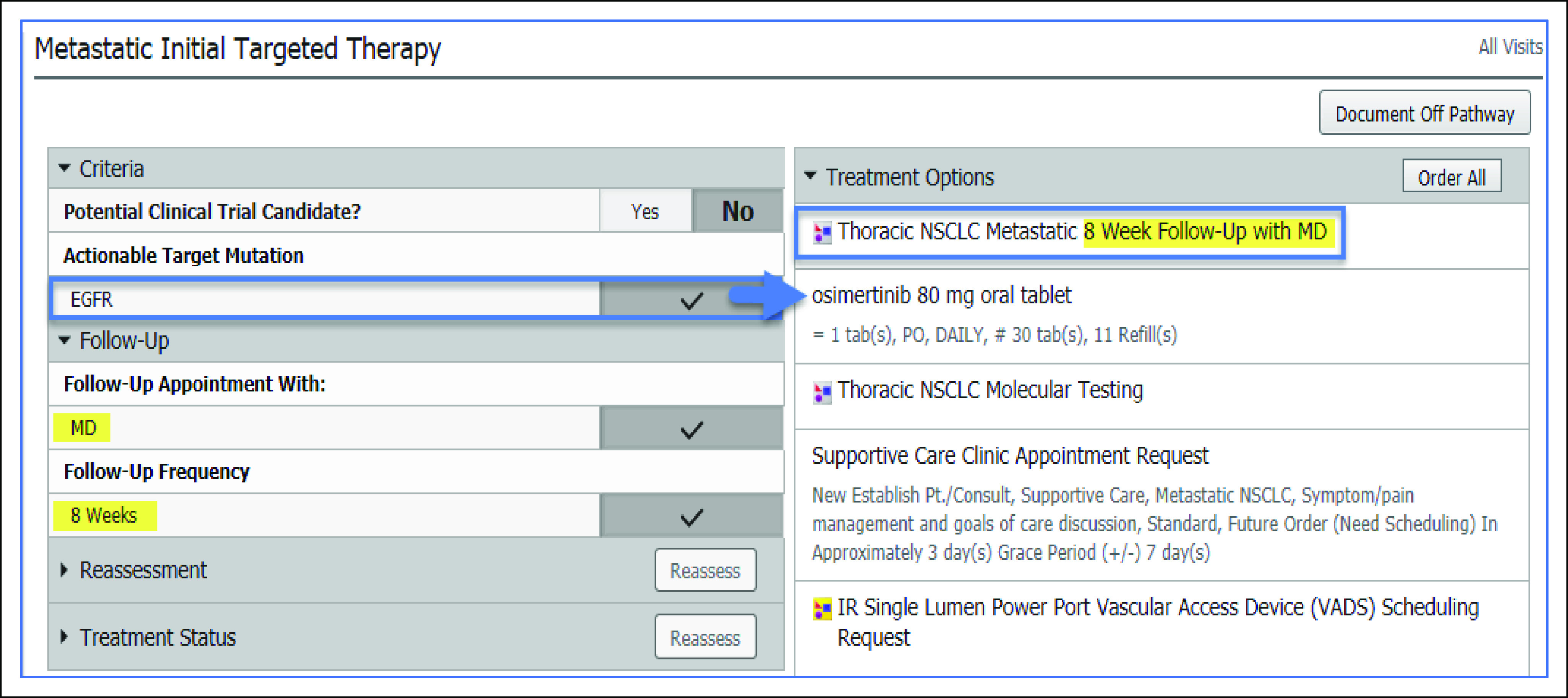
Example of a clinical pathway for metastatic NSCLC integrated into the EHR. Genetic testing considerations (ie, Thoracic NSCLC Molecular Testing) that are inclusive of NGS assays are embedded within the metastatic NSCLC pathway. Molecularly guided therapeutic recommendations are provided if an actionable genetic finding is present. For example, osimertinib is recommended as a treatment consideration for a tumor harboring an EGFR mutation that is amendable to tyrosine kinase inhibitor therapy. EHR, electronic health record; NGS, next-generation sequencing; NSCLC, non–small-cell lung cancer. © Cerner Corporation. Reproduced with permission.
For those patients identified for NGS testing, the actual assay order placement can be challenging and time consuming. Lack of standardization across reference laboratories for order input causes administrative burden for providers and facilities to accommodate various methodologies for order submission. Depending on the laboratory performing the test and that laboratory's technical abilities, the actual ordering may vary from electronic formats (eg, laboratory portal orderable) to paper forms.24 The required information may also vary by laboratory and/or order type and may require manual annotation of the diagnosis and other clinical information such as International Classification of Diseases-10 code, patient demographics, and insurance information. Completion of such forms, either in paper or electronic format, can be demanding for busy clinics; inadvertently missing information can delay the receipt of clinically important NGS results and therefore affect patient care. To assist with enabling efficient NGS test ordering practices, building discrete test orders instead of miscellaneous test ordering in the EHR can support institution-specific clinical pathway algorithms and be integrated into pre-existing order sets for easier physician order entry.20 Moreover, when paired with laboratory interfaces to enable sharing of diagnosis and insurance information, the burden of order entry for ancillary staff, and possible error entry, is further decreased. If a tissue-based NGS assay is ordered, linking a test order to a patient's biopsy specimen is vital.
Patient consent for NGS testing is also an important consideration, especially given the risk of secondary findings associated with NGS testing, even among somatic-focused tests.34 In oncology, there are a growing number of genes typically included on targeted-NGS assays that have implications for both therapeutic decision making and inherited disease.9,10,35,36 As NGS testing expands to the whole exome or genome, the exposure to secondary findings could potentially increase.37 Patient consenting procedures inclusive of secondary findings disclosure have been established for germline testing,38,39 but consenting processes for oncology-focused NGS tests are still evolving. Obtaining consent for NGS testing empowers patients to decide what genetic information they are comfortable receiving, as some patients may not want to know familial disease risks with preference for single-gene or focused-indication molecular tests.40 NGS secondary findings can also result in ethical quandaries, especially in those instances where a provider is exposed to likely pathogenic germline variants and the patient's preference is not to receive such information.41 There have been limited examples to date of liability cases regarding the return of secondary findings that were discovered from somatic NGS.42 If consenting for genetic testing was not performed and secondary findings are reported, then legal ramifications may exist, although liability risks have mostly focused on negligent acts where secondary findings are not disclosed to patients that could potentially cause future harm.42,43 The value of secondary findings that have uncertain prognostic value or have no currently viable clinical interventions has been debated, including whether such results should be reported.44,45 Although the focus of this review is on clinical applications of NGS, it should be noted that the return of NGS results to research participants continues to be debated including the reinterpretation of research-grade NGS information.46,47 Ultimately, patients should have the option of electing to receive or not receive secondary findings, and institutions in collaboration with legal experts can mitigate liability risks by developing opt-out policies and procedures that are applicable to scenarios where NGS results are returned but not yet disclosed to unconsented patients. If a patient's preference is to not receive secondary findings, certain reference laboratories may offer somatic NGS assays that filter out known germline variants that are associated with secondary findings. However, filtering of germline variants may decrease opportunities for individualized treatment strategies.
Clinical decision support (CDS) tools embedded within the EHR could assist with appropriate test utilization. For example, CDS tools could alert providers to which patients have not yet been consented for NGS testing. This approach could prevent the ordering of particular NGS assays for those patients who do not want to be exposed to secondary findings and mitigate risks of ethical dilemmas. In most clinical scenarios, comprehensive germline testing assays should only be performed once during a patient's lifetime. Decision support tools could also prevent duplicate ordering of germline tests that could be costly to the health system and/or patients.
RESULT REPORTING, CURATION, AND STORAGE
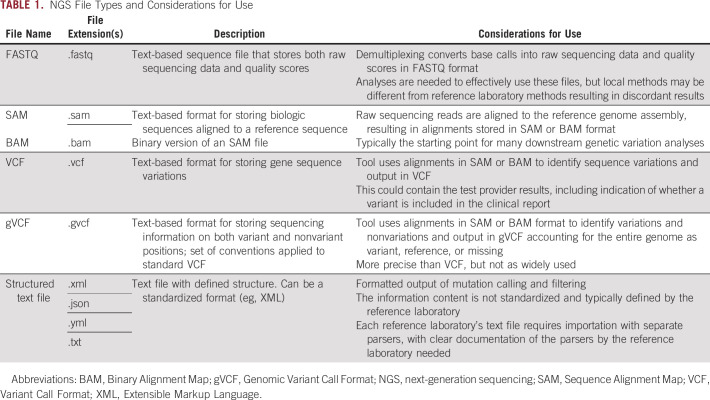
Numerous methods are used for return of NGS results including laboratory portals, electronic notifications, application programming interfaces, and EHRs. The results are typically presented to oncologists as a static PDF report that summarizes important findings and, depending on the laboratory, recommendations for molecularly guided therapy along with clinical trial eligibility. A challenge with PDF documents is that the results are not in a discrete, easily searchable format, which precludes the ability to search for specific molecular data months or potentially years later for clinical or research purposes. Natural language processing pipelines are being developed to extract and discretely curate genetic information from PDF clinical reports,48 but this may capture only a fraction of the data generated from NGS, thus not ultimately meeting all research needs. Data files (eg, raw FASTQ sequence results) that often supplement the clinical reports typically provide more granular NGS data.
There are inherent differences in how data files capture NGS results, with each file type having unique considerations for use (Table 1). A commonly encountered example is the distinction between the initial raw NGS data and the pathology-approved clinical report. Raw NGS data files may be of utility for research purposes, but if the NGS results are referred back to for clinical applications, then clinical reports, Clinical Laboratory Improvement Amendments (CLIA) compliant data files or CLIA compliant data pipelines, that translate the raw data are necessary. However, methodologies for analyzing NGS data can differ among reference laboratories. Thus, even if CLIA compliant data pipelines that accommodate numerous file types are built in-house, there can still be discordance with reference laboratories (eg, liquid versus tissue NGS assays) including which variants are reported, the genome reference used to annotate results (eg, Genome Reference Consortium version h37 versus h38), and how variants are interpreted.49-51 Database systems that are scalable to accept massive amounts of NGS data from differing file types can enable curation and storage. A Negative Storage Model database solution that can accommodate various data file format inputs was developed at Moffitt Cancer Center where each position in the genome is accounted for as reference, variant, or missing, and a single file output converter generates a gVCF file for each NGS assay performed.52
TABLE 1.
NGS File Types and Considerations for Use
After identifying the data file formats that are desired for clinical use along with curation and storage of NGS results, there remains complexity with developing a consistent route of transmission and ingestion of laboratory data. For instance, data files may be received as data dumps often at arbitrary or inconsistent time intervals. Application programming interfaces for seamless integration of clinical reports into the EHR and integration of supplemental data files with local storage solutions are arguably the optimal method for facilitating transmission of real-time results. Creating efficiencies for the transmission of data is particularly critical for clinical care, as some patients may urgently need the NGS results to help guide the next line of therapy.
Another potential barrier to curation and storage of NGS data is linkage of the returned results to the specimen used for molecular interrogation. Numerous specimens can be used for NGS including blood, tissue from the primary site at time of diagnosis or progression, and tissue from sites of metastatic disease across a continuum of time. The specimen used and the timing of NGS testing could influence future therapeutic considerations, such as recognizing acquired mutations over the course of disease treatment that may be predictive of drug resistance. Facilitating the linkage between NGS results, the patient, biologic specimen, test order, reference laboratory, and annotating results as germline or somatic remains a challenge, as each of these variables can affect the corresponding clinical actionability. A first step to addressing this barrier is recognizing unique identifiers that are associated with each variable, mapping how each unique identifier is interconnected, and incorporating data workflows into the overall architecture.
From the clinical perspective, an ultimate goal for curation and storage of NGS results is to enable the harmonization of discrete genetic information with other clinical data. Customized data marts have the potential to link patient demographics, performance status, renal and hepatic function, drug allergies, prior treatment history, molecular information, and other relevant clinical data. Incorporating genotype-phenotype associations from publicly available curated databases such as ClinVar, Precision Oncology Knowledge Base (OncoKB),53 and Clinical Pharmacogenetics Implementation Consortium (CPIC)54 into data marts could assist with quickly interpreting genetic results. Visualization tools embedded within customized data marts, such as a timeline of prior treatment and NGS testing, would enable clinicians to rapidly analyze clinical data and plan a next line of therapy. Data mart visualization tools could be used as part of tumor boards to summarize clinical cases.55 Curation of discrete data can also facilitate the querying of prior patients with similar disease and molecular characteristics to better understand treatment outcomes and guide future treatment decision making.
As molecularly focused clinical trials continue to grow in number, identifying those eligible for a trial can be difficult, especially assessing prior NGS results every time a new trial is opened to enrollment. Integrating molecularly focused clinical trial inclusion and exclusion criteria into NGS data marts has the potential to provide a real-time list of trial opportunities for each patient and enable clinicians along with clinical trial coordinators to retrospectively search for eligible patients. Generation of operational reports that summarize curated NGS results could assist with identifying gaps in clinical trial portfolios and for investigators considering opening a molecularly focused clinical trial help determine if there are likely to be a sufficient number of eligible patients. Beyond clinical trials, data marts could also support other research opportunities, such as observational studies. Institution-specific, de-identified molecular data could be linked to other sources of clinical and patient-generated data and visualized in open-source tools such as cBioPortal for hypothesis generation and cohort identification.56,57 Data marts could also support efficiently populating publicly accessible, shared resources inclusive of the American Association for Cancer Research's Project GENIE initiative to allow for discovery and outcomes research among large and diverse curated data sets.58
CLINICAL CONSULTATION SERVICES TO SUPPORT INTEGRATION OF NGS INTO PATIENT CARE
The NGS results can be complex and time-consuming to interpret, especially when rare or novel genetic variants are reported, numerous resistance mutations are present, or several options for targeted therapy are identified. Level of evidence classifications has been developed to assist clinicians with clinical decision making, which are based on strength of evidence supporting the use of a particular therapy to target a reported genetic variant.53,59-62 Although these classifications are valuable for application of NGS to patient care, there remain inherent limitations. Level of evidence classifications may provide limited assistance for how to interpret rare or novel genetic alterations. Patient characteristics and preferences, which are not accounted for by level of evidence classifications, can influence decision making. Furthermore, level of evidence classifications, along with recommendations provided by reference laboratories, often focuses on single gene-drug pairs, which may limit recommendations for targeting both a tumor driver and acquired resistance mutation.63 Perhaps more importantly, reference laboratory interpretations can be highly variable. The same genetic variant can be interpreted as pathogenic, a variant of uncertain significance or benign dependent on the reference laboratory used for NGS testing.49
Clinical consultation services can assist with interpretation of NGS results by bridging gaps between guideline limitations and application to patient care. As an example, a Precision Medicine Clinical Service was established at Moffitt Cancer Center dedicated to review and interpret all NGS results obtained as part of clinical care. Motivations for establishing the precision medicine service included the following: (1) the majority of reported genetic variants do not have well-defined clinical applicability, thus requiring in-depth review processes to determine actionability; (2) the need to harmonize variability that can be observed across reference laboratory NGS reports including how the genetic results are presented and strength of front page recommendations; and (3) the need to support busy clinics where there may be limited time for NGS result interpretation.64 Patients are increasingly undergoing multiple NGS tests inclusive of both liquid biopsies and tissue-based assays, in part due to serial NGS testing to identify acquired resistance mutations.65 Thus, reconciliation of discrepancies that may be observed when using multiple types of NGS assays is emerging as vital clinical services.49,65
As part of Moffitt's Precision Medicine Clinical Service, the NGS results are reviewed at a weekly multidisciplinary case conference to obtain consensus on the actionability of genetic findings. Members of this multidisciplinary team include precision medicine specialists, pharmacists, molecular pathologists, oncologists, and genetic counselors. If applicable, therapeutic recommendations are provided that consider patient characteristics, prior lines of therapy, and patient preferences for clinical trials. For those cases where consensus is not reached, a referral is made to a molecular tumor board. The molecular tumor board includes oncologists across all oncologic and hematologic diseases, pathologists, medical geneticists, pharmacists, and translational research scientists.64 Similar to the Precision Medicine Clinical Service, the molecular tumor board reviews NGS findings and incorporates the patient's medical history to provide evidence-based recommendations for patient care.
The rapid growth of clinical NGS is likely to continue for the foreseeable future. Both scalability and sustainability are barriers to precision medicine consultation services.66 There will also be a need for multidisciplinary approaches, such as precision medicine and genetic counselor clinical services collaborating on risk mitigation strategies inclusive of therapeutics and disease risk.21,39 Information technology tools inclusive of data marts and visualization tools could support scalability efforts by allowing for efficient summary and analysis of NGS findings. Automated approaches for genetic interpretations and consultation notes that flow directly into the EHR could expand clinical service capacity by minimizing the time spent in documenting recommendations.67 EHR message systems could be leveraged to directly communicate between consultation services and oncologists, with a consult orderable in the EHR providing a streamlined approach for oncologists to request NGS review. Challenges remain with reimbursement models for precision medicine clinical services.68 The potential for improved treatment outcomes, mitigation of toxicities, and freeing up time for oncologists to see patients instead of spending time in interpreting large NGS panels may outweigh the institutional support needed to provide precision medicine services.69
PRESENTATION OF THE NGS RESULTS WITHIN THE ELECTRONIC HEALTH RECORD
The EHR has become a focal point for patient care delivery within health systems, due in part to the Centers for Medicare and Medicaid Promoting Interoperability Programs. Less than a decade ago, there were only a few highly impactful genes rising to the level of being incorporated into EHRs for guidance of therapeutic decision making. In a relatively short period of time, NGS has emerged as a routine test offering in oncology practice with numerous genetic variants associated with drug response. Because of the rapid adoption of NGS, EHRs have not yet evolved to fully incorporate large amounts of clinically important genetic information.70,71 There are several considerations for operationalizing NGS results within EHRs including how to organize genetic information for quick access, annotation of discrete results to support CDS, and alerting providers in real time of targeted therapy options, clinical trial opportunities, and life-threatening gene-drug interactions.
Similar to how discrete test names built into the EHR can support test ordering, discrete test names can also facilitate organization with the EHR to support the rapid identification of desired test results.70,71 However, difficulties may still arise with verifying which specimen was used for a particular NGS assay, especially for tissue specimens that might have been collected several years before test ordering. For health systems with precision medicine or other genetic focused clinical services, additional efforts may be needed to locate consultation notes in the EHR. Organizing data in a patient-centric, time-independent manner supports quick access to NGS results and associated specimens used for testing, along with access to passive CDS tools consisting of interpretations and other comments.72,73 Customized views of genetic information within the EHR can streamline access to genetic results, pathology information, and associated interpretations (Fig 3).
FIG 3.
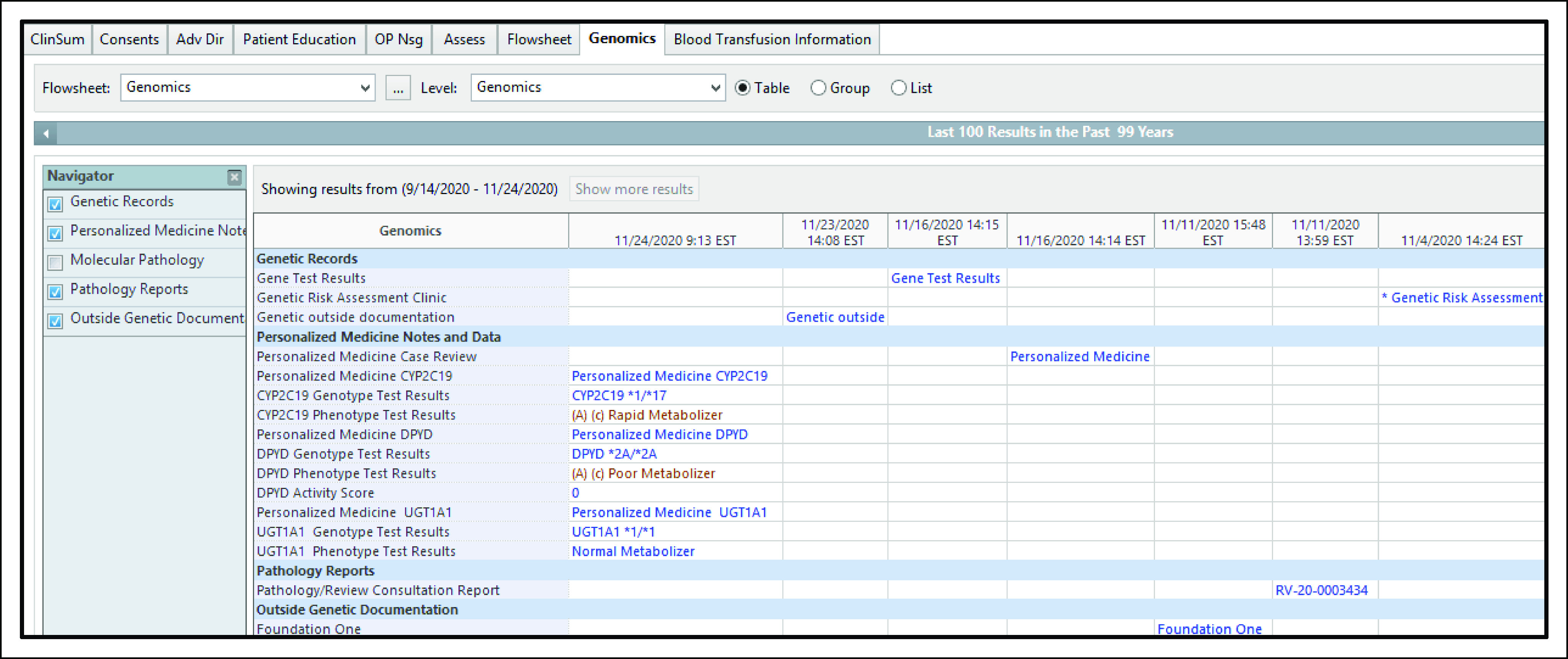
Example of a customized view of genetic information within an EHR. Pertinent information associated with germline and somatic genetic testing is summarized in a Genomics Tab. Genetic information includes pathology review and specimen reference number (ie, RV-20-0003434), genetic test results, and consultation notes (eg, Personalized Medicine Case Review). EHR, electronic health record. © Cerner Corporation. Reproduced with permission.
Active CDS inclusive of interruptive pop-up alerts that provide clinicians with meaningful information at the point of care can supplement EHR tools such as a Genomics Tab.74,75 A barrier to implementing active CDS is the need for discrete genetic data within the EHR, for example, using DPYD genotype (eg, DPYD*2A/*2A) or phenotype (eg, DPYD poor metabolizer) to trigger an alert warning of a high-risk gene-drug interaction when a fluoropyrimidine is ordered (Fig 4).15 Alert fatigue resulting in nonadherence can also be a barrier to active CDS,76 with consideration needed for what genetic information warrants an interruptive alert. Perhaps the field that has been at the forefront of leveraging EHR infrastructure to drive genetics-focused interruptive alerts is germline pharmacogenetics.77-79 EHR terminologies and standards (eg, LOINC, SNOMED, and HL7) have been developed to support the transfer of discrete pharmacogenetic results from laboratories to EHRs or other clinical databases.80 Comprehensive EHR terminologies and standards to support discrete annotation of somatic NGS results, though, have not been fully developed. Efforts from initiatives such as the Electronic Medical Records and Genomics network may illuminate innovative methods for discrete annotation of both somatic and germline NGS results in the EHR, although it is currently difficult to envision that in the near future EHRs will store the entirety of NGS results.78,79 A likely solution is database systems, which are separate from the EHR, will securely store massive amounts of NGS results with interconnectivity between data marts and EHRs to highlight clinically important results.
FIG 4.
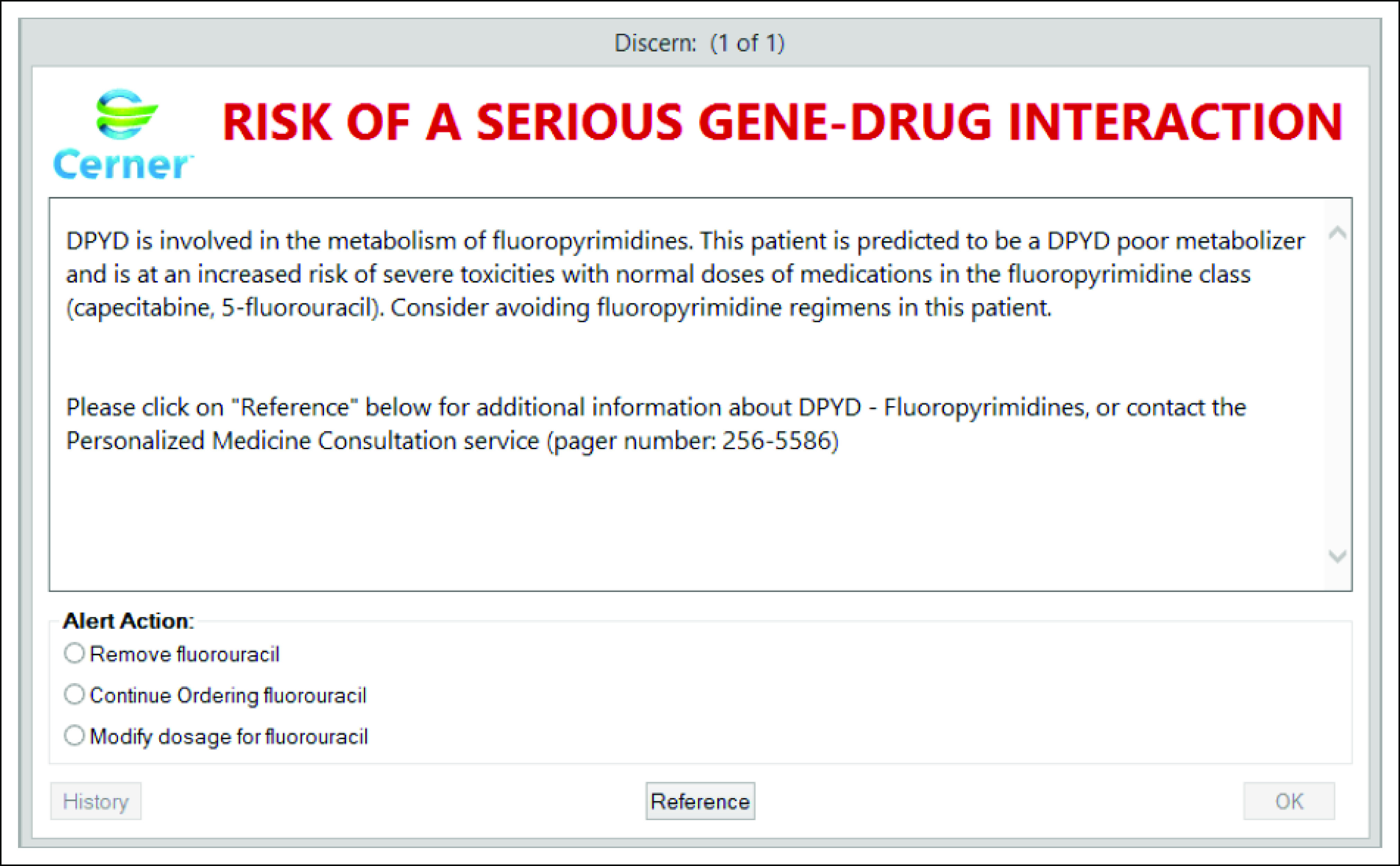
Example of an interruptive gene-drug interaction clinical decision support alert within an EHR. DYPD poor metabolizer is discretely documented in the EHR. Ordering of a fluoropyrimidine triggers a gene-drug interaction interruptive alert. DYPD, dihydropyrimidine dehydrogenase; EHR, electronic health record. © Cerner Corporation. Reproduced with permission.
CLINICIAN AND PATIENT EDUCATION
Clinician and patient education has consistently been identified as barriers to integrating genetic information into patient care.66,81 Specifically, clinicians have identified a lack of knowledge on how to interpret genetic test results and modify treatment plans as impeding clinical applications.66 As NGS testing moves toward standard of care for certain cancer diagnoses, there will be a need to develop educational programs for clinicians that are inclusive of physicians, pharmacists, and nurses. It has been proposed to incorporate clinical genetics into training programs.73,82 As an example, all hematology-oncology fellows at Moffitt Cancer Center must complete a one-month rotation on the Precision Medicine Clinical Service. For clinicians no longer in training, locally available continuing education seminars inclusive of where to find NGS results in the medical record, resources for interpretation, and application to patient care are potential solutions for increasing genetic knowledge. CDS can also be employed as a tool for clinical education.
Educating patients on the role of NGS in oncology practice will be of importance for genetic test consenting and subsequent application to patient care. A combination of education tools may be needed that range from printable materials to videos and other interactive media.83 Patient portals, mobile or voice applications, online resources, or virtual consultations are other routes for patient-focused genetic education. Consideration should be given to patient preferences for content and delivery. For example, patient portals may be a preferred route to receive education for some, whereas others may prefer in person education. Education content and delivery needs may also differ across age ranges inclusive of pediatrics, young adults, and senior adults. Additionally, education tools should also take into consideration cultural differences along with any preferences among racial and ethnic groups for content and delivery.84,85 Collaboration with patients and patient advocacy groups can help identify education needs and effective delivery methods.86,87
In conclusion, NGS is becoming part of routine oncology practice, driven in part by an exponential increase of molecularly guided therapies. NGS results can also assist with mitigating chemotherapy toxicity risks along with optimizing supportive care drug selection and dosage. We presented key considerations for application of NGS results to patient care ranging from test ordering and return of the results to clinical consultation services and leveraging the EHR to disseminate genetic information. As NGS is further integrated into patient care, education of providers and patients will be vital. Successfully integrating NGS into patient care will assist with identifying opportunities for targeted therapy and mitigating drug-induced adverse effects. Combining genetic information with informatics tools and specialized clinical services can create a powerful arsenal to identify opportunities for targeted therapy including clinical trial eligibility identification, assistance in guiding appropriate selection and dosage of chemotherapy drugs along with supportive care medicines, and mitigation of drug-induced adverse effects.
ACKNOWLEDGMENT
We would like to thank members of the Clinical Informatics, Health Informatics, and Pathology Departments for supporting efforts to integrate NGS results into patient care. We would like to thank Kalie Craven and Rhonda Whiteside for assistance with EHR screenshots and Kerry Kelly for development of interruptive CDS.
J. Kevin Hicks
Consulting or Advisory Role: Quest Diagnostics, 23andMe
Research Funding: OneOme
Karen K. Fields
Stock and Other Ownership Interests: Pfizer, AbbVie
Honoraria: NCCN, NACCME, Pacific Group on Business, HMP, MJH Healthcare Holdings LLC
Consulting or Advisory Role: United Health Group
Speakers' Bureau: Genentech/Roche
Travel, Accommodations, Expenses: Genentech/Roche, CBI, Discern Health, Tapestry, NACCME, Cigna Health Care, FLASCO
Jhanelle E. Gray
Consulting or Advisory Role: Bristol Myers Squibb, EMD Serono, Inivata, Merck Sharp & Dohme, Axiom Healthcare Strategies, Novartis, AstraZeneca, Blueprint Medicines, Lilly, Sanofi, Janssen Scientific Affairs
Research Funding: Array BioPharma, Merck, AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim, Genentech/Roche, G1 Therapeutics, Novartis, Pfizer, Ludwig Institute for Cancer Research
Travel, Accommodations, Expenses: Bristol Myers Squibb, Merck Sharp & Dohme, Inivata, Merck, EMD Serono, Novartis
Bryan McIver
Honoraria: Eisai, Loxo/Lilly, Blueprint Medicines, Exelixis
Consulting or Advisory Role: Eisai, Loxo/Lilly, Exelixis
Speakers' Bureau: Loxo/Lilly
Mandy F. O'Leary
Stock and Other Ownership Interests: Bristol Myers Squibb
Randa M. Perkins
Stock and Other Ownership Interests: Progyny
Other Relationship: Centene
Open Payments Link: https://openpaymentsdata.cms.gov/physician/263299
Jamie K. Teer
Patents, Royalties, Other Intellectual Property: Patent application: Large Data Set Negative Information Storage Model
Joseph Markowitz
Stock and Other Ownership Interests: Intel, Aflac, CVS, Amdocs, Consolidated Edison
Honoraria: Springer
Consulting or Advisory Role: Idera, Newlink Genetics, Array Biopharma
Research Funding: Reata Pharmaceuticals, Macrogenics, Morphogenesis, Genoptix, Idera, Jackson Laboratory for Genomic Medicine, Merck
Other Relationship: Springer
Dana E. Rollison
Leadership: NanoString Technologies
Stock and Other Ownership Interests: NanoString Technologies
Patents, Royalties, Other Intellectual Property: I am a co-inventor on a provisional patent application filed in July of 2020. The patent pertains to the use of spectrophotometer-measured UV radiation exposure in combination with regulatory T-cells measured in circulation to predict risk of subsequent skin cancer
Travel, Accommodations, Expenses: Caserta Analytics Inc (not a healthcare company specifically, but they do business with healthcare companies), NanoString Technologies
No other potential conflicts of interest were reported.
SUPPORT
Supported in part by H. Lee Moffitt Cancer Center and Research Institute NIH P30-CA076292 (all authors) and ASHP Foundation (J.K.H.).
AUTHOR CONTRIBUTIONS
Conception and design: J. Kevin Hicks, Rachel Howard, Phillip Reisman, Jacob J. Adashek, Karen K. Fields, Bryan McIver, Randa M. Perkins, Edmondo Robinson, Ankita Tandon, Jamie K. Teer, Joseph Markowitz, Dana E. Rollison
Administrative support: Dana E. Rollison
Financial support: Dana E. Rollison
Collection and assembly of data: J. Kevin Hicks, Jacob J. Adashek, Kelly McKee, Mandy F. O'Leary, Edmondo Robinson, Jamie K. Teer, Dana E. Rollison
Data analysis and interpretation: J. Kevin Hicks, Jacob J. Adashek, Jhanelle E. Gray, Mandy F. O'Leary, Edmondo Robinson, Jamie K. Teer, Joseph Markowitz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by the authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
J. Kevin Hicks
Consulting or Advisory Role: Quest Diagnostics, 23andMe
Research Funding: OneOme
Karen K. Fields
Stock and Other Ownership Interests: Pfizer, AbbVie
Honoraria: NCCN, NACCME, Pacific Group on Business, HMP, MJH Healthcare Holdings LLC
Consulting or Advisory Role: United Health Group
Speakers' Bureau: Genentech/Roche
Travel, Accommodations, Expenses: Genentech/Roche, CBI, Discern Health, Tapestry, NACCME, Cigna Health Care, FLASCO
Jhanelle E. Gray
Consulting or Advisory Role: Bristol Myers Squibb, EMD Serono, Inivata, Merck Sharp & Dohme, Axiom Healthcare Strategies, Novartis, AstraZeneca, Blueprint Medicines, Lilly, Sanofi, Janssen Scientific Affairs
Research Funding: Array BioPharma, Merck, AstraZeneca, Bristol Myers Squibb, Boehringer Ingelheim, Genentech/Roche, G1 Therapeutics, Novartis, Pfizer, Ludwig Institute for Cancer Research
Travel, Accommodations, Expenses: Bristol Myers Squibb, Merck Sharp & Dohme, Inivata, Merck, EMD Serono, Novartis
Bryan McIver
Honoraria: Eisai, Loxo/Lilly, Blueprint Medicines, Exelixis
Consulting or Advisory Role: Eisai, Loxo/Lilly, Exelixis
Speakers' Bureau: Loxo/Lilly
Mandy F. O'Leary
Stock and Other Ownership Interests: Bristol Myers Squibb
Randa M. Perkins
Stock and Other Ownership Interests: Progyny
Other Relationship: Centene
Open Payments Link: https://openpaymentsdata.cms.gov/physician/263299
Jamie K. Teer
Patents, Royalties, Other Intellectual Property: Patent application: Large Data Set Negative Information Storage Model
Joseph Markowitz
Stock and Other Ownership Interests: Intel, Aflac, CVS, Amdocs, Consolidated Edison
Honoraria: Springer
Consulting or Advisory Role: Idera, Newlink Genetics, Array Biopharma
Research Funding: Reata Pharmaceuticals, Macrogenics, Morphogenesis, Genoptix, Idera, Jackson Laboratory for Genomic Medicine, Merck
Other Relationship: Springer
Dana E. Rollison
Leadership: NanoString Technologies
Stock and Other Ownership Interests: NanoString Technologies
Patents, Royalties, Other Intellectual Property: I am a co-inventor on a provisional patent application filed in July of 2020. The patent pertains to the use of spectrophotometer-measured UV radiation exposure in combination with regulatory T-cells measured in circulation to predict risk of subsequent skin cancer
Travel, Accommodations, Expenses: Caserta Analytics Inc (not a healthcare company specifically, but they do business with healthcare companies), NanoString Technologies
No other potential conflicts of interest were reported.
REFERENCES
- 1.Wang L, McLeod HL, Weinshilboum RM: Genomics and drug response. N Engl J Med 364:1144-11532011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Relling MV, Evans WE: Pharmacogenomics in the clinic. Nature 526:343-3502015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.US Food and Drug Administration : Table of Pharmacogenomic Biomarkers in Drug Labeling. Silver Spring, MD, US Food and Drug Administration; 2020 [Google Scholar]
- 4.US Food and Drug Administration : Table of Pharmacogenetic Associations. Silver Spring, MD, US Food and Drug Administration; 2020 [Google Scholar]
- 5.Ettinger DS, Aisner DL, Wood DE, et al. : NCCN guidelines insights: Non-small cell lung cancer, version 5.2018. J Natl Compr Canc Netw 16:807-8212018 [DOI] [PubMed] [Google Scholar]
- 6.Vela CM, Knepper TC, Gillis NK, et al. : Quantitation of targetable somatic mutations among patients evaluated by a personalized medicine clinical service: Considerations for off-label drug use. Pharmacotherapy 37:1043-10512017 [DOI] [PubMed] [Google Scholar]
- 7.Redig AJ, Janne PA: Basket trials and the evolution of clinical trial design in an era of genomic medicine. J Clin Oncol 33:975-9772015 [DOI] [PubMed] [Google Scholar]
- 8.Beckman RA, Antonijevic Z, Kalamegham R, et al. : Adaptive design for a confirmatory basket trial in multiple tumor types based on a putative predictive biomarker. Clin Pharmacol Ther 100:617-6252016 [DOI] [PubMed] [Google Scholar]
- 9.Mateo J, Carreira S, Sandhu S, et al. : DNA-repair defects and Olaparib in metastatic prostate cancer. N Engl J Med 373:1697-17082015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golan T, Hammel P, Reni M, et al. : Maintenance Olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 381:317-3272019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giri VN, Knudsen KE, Kelly WK, et al. : Implementation of germline testing for prostate cancer: Philadelphia Prostate Cancer Consensus Conference 2019. J Clin Oncol 38:2798-28112020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mateo J, Porta N, Bianchini D, et al. : Olaparib in patients with metastatic castration-resistant prostate cancer with DNA repair gene aberrations (TOPARP-B): A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol 21:162-1742020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Bono J, Mateo J, Fizazi K, et al. : Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med 382:2091-21022020 [DOI] [PubMed] [Google Scholar]
- 14.Relling MV, Schwab M, Whirl-Carrillo M, et al. : Clinical Pharmacogenetics Implementation Consortium guideline for thiopurine dosing based on TPMT and NUDT15 genotypes: 2018 update. Clin Pharmacol Ther 105:1095-11052019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amstutz U, Henricks LM, Offer SM, et al. : Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for dihydropyrimidine dehydrogenase genotype and fluoropyrimidine dosing: 2017 update. Clin Pharmacol Ther 103:210-2162018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell GC, Caudle KE, Whirl-Carrillo M, et al. : Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther 102:213-2182017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crews KR, Gaedigk A, Dunnenberger HM, et al. : Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther 95:376-3822014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hicks JK, Bishop JR, Sangkuhl K, et al. : Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 and CYP2C19 genotypes and dosing of selective serotonin reuptake inhibitors. Clin Pharmacol Ther 98:127-1342015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hicks JK, Sangkuhl K, Swen JJ, et al. : Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther 102:37-442017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hicks JK, Quilitz RE, Komrokji RS, et al. : Prospective CYP2C19-guided voriconazole prophylaxis in patients with neutropenic acute myeloid leukemia reduces the incidence of subtherapeutic antifungal plasma concentrations. Clin Pharmacol Ther 107:563-5702020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hicks JK, McLeod HL: Probabilistic medicine: A pre-emptive approach is needed for cancer therapeutic risk mitigation. Biomark Med 13:987-9902019 [DOI] [PubMed] [Google Scholar]
- 22.Sangkuhl K, Whirl-Carrillo M, Whaley RM, et al. : Pharmacogenomics clinical annotation tool (PharmCAT). Clin Pharmacol Ther 107:203-2102020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perkins RM, Markowitz J: Development and optimization of clinical informatics infrastructure to support bioinformatics at an oncology center. Methods Mol Biol 2194:1-192021 [DOI] [PubMed] [Google Scholar]
- 24.O'Leary MF: Leveraging pathology informatics concepts to achieve discrete lab data for clinical use and translational research. Methods Mol Biol 2194:21-332021 [DOI] [PubMed] [Google Scholar]
- 25.Hooda SM, Fields KK: Transitioning clinical practice guidelines into the electronic health record through clinical pathways. Methods Mol Biol 2194:45-592021 [DOI] [PubMed] [Google Scholar]
- 26.Zeng J, Johnson A, Shufean MA, et al. : Operationalization of next-generation sequencing and decision support for precision oncology. JCO Clin Cancer Inform 3:1-122019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freedman AN, Klabunde CN, Wiant K, et al. : Use of next-generation sequencing tests to guide cancer treatment: Results from a nationally representative survey of oncologists in the United States. JCO Precis Oncol 2018. doi: 10.1200/PO.18.00169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson AB, Venook AP, Al-Hawary MM, et al. : NCCN guidelines insights: Colon cancer, version 2.2018. J Natl Compr Canc Netw 16:359-3692018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mosele F, Remon J, Mateo J, et al. : Recommendations for the use of next-generation sequencing (NGS) for patients with metastatic cancers: A report from the ESMO Precision Medicine Working Group. Ann Oncol 31:1491-15052020 [DOI] [PubMed] [Google Scholar]
- 30.Zon RT, Frame JN, Neuss MN, et al. : American Society of Clinical Oncology policy statement on clinical pathways in oncology. J Oncol Pract 12:261-2662016 [DOI] [PubMed] [Google Scholar]
- 31.Wong W: Optimizing resource utilization and the need for enhanced value frameworks. J Clin Pathways 5:7.2019. https://www.journalofclinicalpathways.com/depth-look-moffitt-cancer-centers-clinical-pathways-program-and-use-payer-strategies-part-i [Google Scholar]
- 32.An in-depth look at Moffitt Cancer Center's clinical pathways program and use in payer strategies—Part I. J Clin Pathways, 2019 [Google Scholar]
- 33.McKenzie AJ, Dilks HH, Jones SF, et al. : Should next-generation sequencing tests be performed on all cancer patients? Expert Rev Mol Diagn 19:89-932019 [DOI] [PubMed] [Google Scholar]
- 34.Bijlsma RM, Bredenoord AL, Gadellaa-Hooijdonk CG, et al. : Unsolicited findings of next-generation sequencing for tumor analysis within a Dutch consortium: Clinical daily practice reconsidered. Eur J Hum Genet 24:1496-15002016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry DK, Wang X, Michalski ST, et al. : Clinical cohort analysis of germline EGFR T790M demonstrates penetrance across ethnicities and races, sexes, and ages. JCO Precis Oncol 4:170-1752020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dumbrava EI, Brusco L, Daniels M, et al. : Expanded analysis of secondary germline findings from matched tumor/normal sequencing identifies additional clinically significant mutations. JCO Precis Oncol 2019. doi: 10.1200/PO.18.00143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schneider BP, Stout LA, Philips S, et al. : Implications of incidental germline findings identified in the context of clinical whole exome sequencing for guiding cancer therapy. JCO Precis Oncol 4:1109-11212020 [DOI] [PubMed] [Google Scholar]
- 38.ACMG Board of Directors : ACMG policy statement: Updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med 17:68-692015 [DOI] [PubMed] [Google Scholar]
- 39.Hicks JK, Shealy A, Schreiber A, et al. : Patient decisions to receive secondary pharmacogenomic findings and development of a multidisciplinary practice model to integrate results into patient care. Clin Transl Sci 11:71-762018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hamilton JG, Shuk E, Garzon MG, et al. : Decision-making preferences about secondary germline findings that arise from tumor genomic profiling among patients with advanced cancers. JCO Precis Oncol 1:PO.17.001822017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Johnson KJ, Gehlert S: Return of results from genomic sequencing: A policy discussion of secondary findings for cancer predisposition. J Cancer Policy 2:75-802014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marchant G, Barnes M, Evans JP, et al. : From genetics to genomics: Facing the liability implications in clinical care. J Law Med Ethics 48:11-432020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans BJ: Minimizing liability risks under the ACMG recommendations for reporting incidental findings in clinical exome and genome sequencing. Genet Med 15:915-9202013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pray LA: Questionable prognostic value of genetic testing. Nat Educ 1:74.2008 [Google Scholar]
- 45.McPherson E: Genetic diagnosis and testing in clinical practice. Clin Med Res 4:123-1292006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorogood A, Dalpe G, Knoppers BM: Return of individual genomic research results: Are laws and policies keeping step? Eur J Hum Genet 27:535-5462019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bombard Y, Brothers KB, Fitzgerald-Butt S, et al. : The responsibility to recontact research participants after reinterpretation of genetic and genomic research results. Am J Hum Genet 104:578-5952019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, Yang P, Xue S, et al. : Translating cancer genomics into precision medicine with artificial intelligence: Applications, challenges and future perspectives. Hum Genet 138:109-1242019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balmana J, Digiovanni L, Gaddam P, et al. : Conflicting interpretation of genetic variants and cancer risk by commercial laboratories as assessed by the prospective registry of multiplex testing. J Clin Oncol 34:4071-40782016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuderer NM, Burton KA, Blau S, et al. : Comparison of 2 commercially available next-generation sequencing platforms in oncology. JAMA Oncol 3:996-9982017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guo Y, Dai Y, Yu H, et al. : Improvements and impacts of GRCh38 human reference on high throughput sequencing data analysis. Genomics 109:83-902017 [DOI] [PubMed] [Google Scholar]
- 52.Gonzalez-Calderon G, Liu R, Carvajal R, et al. : A negative storage model for precise but compact storage of genetic variation data. Database 20202020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chakravarty D, Gao J, Phillips SM, et al. : OncoKB: A precision oncology knowledge base. JCO Precis Oncol 20172017. doi: 10.1200/PO.17.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Relling MV, Klein TE: CPIC: Clinical Pharmacogenetics Implementation Consortium of the pharmacogenomics research network. Clin Pharmacol Ther 89:464-4672011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rao S, Pitel B, Wagner AH, et al. : Collaborative, multidisciplinary evaluation of cancer variants through virtual molecular tumor boards informs local clinical practices. JCO Clin Cancer Inform 4:602-6132020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cerami E, Gao J, Dogrusoz U, et al. : The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401-4042012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao J, Aksoy BA, Dogrusoz U, et al. : Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Project GENIE goes public. Cancer Discov 7:118.2017 [DOI] [PubMed] [Google Scholar]
- 59.Andre F, Mardis E, Salm M, et al. : Prioritizing targets for precision cancer medicine. Ann Oncol 25:2295-23032014 [DOI] [PubMed] [Google Scholar]
- 60.Van Allen EM, Wagle N, Stojanov P, et al. : Whole-exome sequencing and clinical interpretation of formalin-fixed, paraffin-embedded tumor samples to guide precision cancer medicine. Nat Med 20:682-6882014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meric-Bernstam F, Johnson A, Holla V, et al. : A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst 107:djv098.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mateo J, Chakravarty D, Dienstmann R, et al. : A framework to rank genomic alterations as targets for cancer precision medicine: The ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT). Ann Oncol 29:1895-19022018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haura EB, Hicks JK, Boyle TA: Erdafitinib overcomes FGFR3-TACC3-mediated resistance to osimertinib. J Thorac Oncol 15:e154-e1562020 [DOI] [PubMed] [Google Scholar]
- 64.Knepper TC, Bell GC, Hicks JK, et al. : Key lessons learned from Moffitt's molecular tumor board: The clinical genomics action committee experience. Oncologist 22:144-1512017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hicks JK, Saller J, Wang E, et al. : Cell-free circulating tumor DNA supplementing tissue biopsies for identification of targetable mutations: Implications for precision medicine and considerations for reconciling results. Lung Cancer 111:135-1382017 [DOI] [PubMed] [Google Scholar]
- 66.Levy KD, Blake K, Fletcher-Hoppe C, et al. : Opportunities to implement a sustainable genomic medicine program: Lessons learned from the IGNITE network. Genet Med 21:743-7472019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hicks JK, Crews KR, Hoffman JM, et al. : A clinician-driven automated system for integration of pharmacogenetic interpretations into an electronic medical record. Clin Pharmacol Ther 92:563-5662012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frueh FW: Regulation, reimbursement, and the long road of implementation of personalized medicine—A perspective from the United States. Value Health 16:S27-S312013 [DOI] [PubMed] [Google Scholar]
- 69.Faulkner E, Holtorf AP, Walton S, et al. : Being precise about precision medicine: What should value frameworks incorporate to address precision medicine? A report of the personalized precision medicine special interest group. Value Health 23:529-5392020 [DOI] [PubMed] [Google Scholar]
- 70.Ohno-Machado L, Kim J, Gabriel RA, et al. : Genomics and electronic health record systems. Hum Mol Genet 27:R48-R552018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Warner JL, Jain SK, Levy MA: Integrating cancer genomic data into electronic health records. Genome Med 8:113.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hicks JK, Dunnenberger HM, Gumpper KF, et al. : Integrating pharmacogenomics into electronic health records with clinical decision support. Am J Health Syst Pharm 73:1967-19762016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hicks JK, Aquilante CL, Dunnenberger HM, et al. : Precision pharmacotherapy: Integrating pharmacogenomics into clinical pharmacy practice. J Am Coll Clin Pharm 2:303-3132019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bell GC, Crews KR, Wilkinson MR, et al. : Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc 21:e93-92014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hicks JK, Stowe D, Willner MA, et al. : Implementation of clinical pharmacogenomics within a large health system: From electronic health record decision support to consultation services. Pharmacotherapy 36:940-9482016 [DOI] [PubMed] [Google Scholar]
- 76.van der Sijs H, Aarts J, Vulto A, et al. : Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc 13:138-1472006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson JA, Caudle KE, Gong L, et al. : Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for pharmacogenetics-guided warfarin dosing: 2017 update. Clin Pharmacol Ther 102:397-4042017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gottesman O, Kuivaniemi H, Tromp G, et al. : The electronic medical records and genomics (eMERGE) network: Past, present, and future. Genet Med 15:761-7712013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gottesman O, Scott SA, Ellis SB, et al. : The CLIPMERGE PGx program: Clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther 94:214-2172013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Caudle KE, Dunnenberger HM, Freimuth RR, et al. : Standardizing terms for clinical pharmacogenetic test results: Consensus terms from the clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med 19:215-2232017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sperber NR, Carpenter JS, Cavallari LH, et al. : Challenges and strategies for implementing genomic services in diverse settings: Experiences from the implementing GeNomics in pracTicE (IGNITE) network. BMC Med Genomics 10:35.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Korf BR: Genetics and genomics education: The next generation. Genet Med 13:201-2022011 [DOI] [PubMed] [Google Scholar]
- 83.Mills R, Ensinger M, Callanan N, et al. : Development and initial assessment of a patient education video about pharmacogenetics. J Pers Med 7:4.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Permuth-Wey J, Vadaparampil S, Rumphs A, et al. : Development of a culturally tailored genetic counseling booklet about hereditary breast and ovarian cancer for Black women. Am J Med Genet A 152A:836-8452010 [DOI] [PubMed] [Google Scholar]
- 85.San Miguel-Majors SL, Whitaker DE, Davis BC, et al. : Education on cancer risk assessment and genetic counseling to address cancer health disparities among racial/ethnic groups and rural populations: Implementing culturally tailored outreach through community health educators. J Genet Couns 29:243-2462020 [DOI] [PubMed] [Google Scholar]
- 86.Batist G, Michaud S, Richards DP, et al. : Developing a model of a patient-group pathway to accessing cancer clinical trials in Canada. Curr Oncol 25:e597-e6092018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Koay PP, Sharp RR: The role of patient advocacy organizations in shaping genomic science. Annu Rev Genomics Hum Genet 14:579-5952013 [DOI] [PubMed] [Google Scholar]