Abstract
The gut microbiome has important effects on human health, yet its importance in human aging remains unclear. Here we demonstrate that, starting in mid-to-late adulthood, gut microbiomes become increasingly unique to individuals with age. We leverage three independent cohorts comprising over 9000 individuals and find that compositional uniqueness is strongly associated with microbially produced amino acid derivatives circulating in the bloodstream. In older age (over ~80 years), healthy individuals show continued microbial drift toward a unique compositional state, whereas this drift is absent in less healthy individuals. The identified microbiome pattern of healthy aging is characterized by a depletion of core genera found across most humans, primarily Bacteroides. Retaining a high Bacteroides dominance into older age, or having a low gut microbiome uniqueness measure, predicts decreased survival in a four-year follow-up. Our analysis identifies increasing compositional uniqueness of the gut microbiome as a component of healthy aging, which is characterized by distinct microbial metabolic outputs in the blood.
Introduction
The ecological dynamics of the human gut microbiome have been characterized by rapid change in early life (0–3 years), followed by a long period of relative stability, ending with gradual changes associated with advanced age1,2. Particularly in older populations (65+ years), studies over the past several years have revealed a number of associations between gut microbiome composition and measures of physical fitness 3, frailty 4, and diet 5, highlighting the importance of proper gut microbiome function into the latter decades of human life. Despite substantial progress in our understanding of the human gut microbiome, very little is still known about when age-associated changes in the gut microbiome begin, how these changes influence host physiology, and whether aging patterns within the gut microbiome simply reflect, or contribute to, long-term health and survival outcomes. Importantly, identifying aging patterns within the gut microbiome could have major clinical implications for both monitoring and modifying gut microbiome health throughout the human lifespan.
Several studies conducted on centenarian populations provided potential insight into gut microbial trajectories associated with aging. Biagi et al.6 demonstrated that gut microbiomes of centenarians (≤104 years of age) and supercentenarians (104+ years) show a depletion in core abundant taxa (Bacteroides, Roseburia and Faecalibacterium, among others), complemented by an increase in the prevalence of rare taxa. Similar findings have since been reported in other centenarian populations across the world, such as in Sardinian, Chinese and Korean centenarians, relative to healthy, younger controls7–9. Some studies have also reported higher levels of gut α-diversity in centenarians compared to younger individuals8–10, indicating that gut microbiomes continue to develop within their hosts, even in the latest decades of human life. The loss of core taxa, the exact identities of which may vary across different human populations (Bacteroides vs. Prevotella)11, and the increase in α-diversity reported in long-lived individuals suggest that gut microbiomes may become increasingly divergent, or unique, to each individual as they age. This phenomenon of community compositional divergence seen in centenarians may be key to understanding how the gut microbiome contributes to the changing physiological landscape accompanying human aging.
Gut microbial associations reported in centenarians are often inconsistent with studies of elderly populations. In particular, studies on the ELDERMET cohort (i.e. the most extensively studied cohort of older persons with gut microbiome data to date) have reported an increased dominance of the core genera Bacteroides, Alistipes and Parabacteroides in those 65+ years old compared to healthy, younger controls 12. Measures of α-diversity have also been shown to negatively correlate with frailty 4, indicating changes in the gut microbiome correspond to age-associated decline. Studies on older long-term care residents further characterized a gradual shift in gut microbiome composition associated with the duration of stay in the care facility2,13, while a more recent study demonstrated that older individuals (60+) exhibited higher variability in gut microbiome composition relative to younger individuals (20–60 years old), which has been attributed to a putative increase in abundance of pathobionts at the expense of beneficial gut bacteria14. Collectively, these and other studies1 provide a view of the human gut microbiome across the adult lifespan as relatively stable up until old age, at which point gradual compositional shifts occur that reflect, and potentially contribute to, declining health.
The heterogeneity of findings in aging studies indicates there may exist multiple gut microbiome patterns of aging, some of which reflect better health and life expectancy outcomes than others. Although recent analyses have demonstrated a link between gut microbiome composition and long-term health outcomes 15,16, the scarcity of cohorts with longitudinal follow-up data, the lack of detailed molecular phenotyping and health metrics, and the relatively small sample sizes of existing studies on aging limit our understanding of gut microbial changes seen across the human lifespan. In the present study, we overcome these limitations and present an analysis of the gut microbiome and phenotypic data from over 9000 individuals from three independent cohorts spanning 18 to 101 years of age, with longitudinal follow-up data in an older cohort of predominantly community-dwelling individuals that allowed us to track survival outcomes.
Results
Study design and cohort descriptions
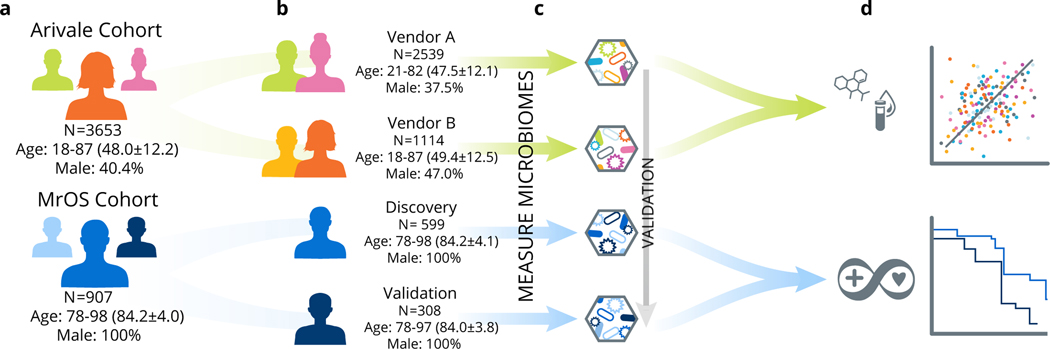
We primarily studied two distinct cohorts: a deeply phenotyped population of individuals who were enrolled in a scientific wellness company (the ‘Arivale cohort’, ages 18–87) (Extended Data 1) and the Osteoporotic Fractures in Men (MrOS) cohort (ages 78–98)17–19 (Extended Data 2) (Fig. 1). These cohorts further subdivide into two groups each. The Arivale cohort separates into Group A (N=2539) and Group B (N=1114), where the distinguishing factor is the use of different vendors for the collection and processing of stool samples (see Methods). The MrOS cohort separates into a discovery cohort (N=599) and a validation cohort (N=308), because although stool samples from this population were all collected at the same timepoint, they were processed in two separate batches approximately 3 years apart. We further confirmed the identified aging pattern in an additional external dataset from the American Gut Project (AGP) (N=4575)20. We began by analyzing baseline data from the Arivale cohort to identify gut microbial aging patterns across most of the adult human lifespan, and investigate how these patterns correspond to host physiology. We then extended our analysis into the MrOS cohort, where we had detailed health metrics and follow-up data on mortality, to evaluate how the patterns identified within the Arivale cohort correspond to health and survival in the latter decades of human life.
Fig. 1. Conceptual outline of study and analysis workflow.
(a) Two different study populations were used: the Arivale cohort and the Osteoporotic Fractures in Men (MrOS) cohort. (b) Each of these two study populations were further subdivided into two groups; the Arivale cohort was split based on the microbiome vendor used to collect and process samples while the MrOS cohort separated into Discovery and Validation groups based on the batch in which the samples were run (discovery samples were processed in the initial batch, validation samples were processed several years later). (c) We profiled the microbiomes from these four study populations beginning with the Arivale cohort and validating our findings across the additional populations. (d) Our analysis pipeline further explored associations between the identified gut microbial aging pattern, lifestyle factors, and host physiology in the combined Arivale cohort, as well as health metrics and mortality in the combined MrOS cohort.
A gut microbiome aging pattern spans much of the adult lifespan
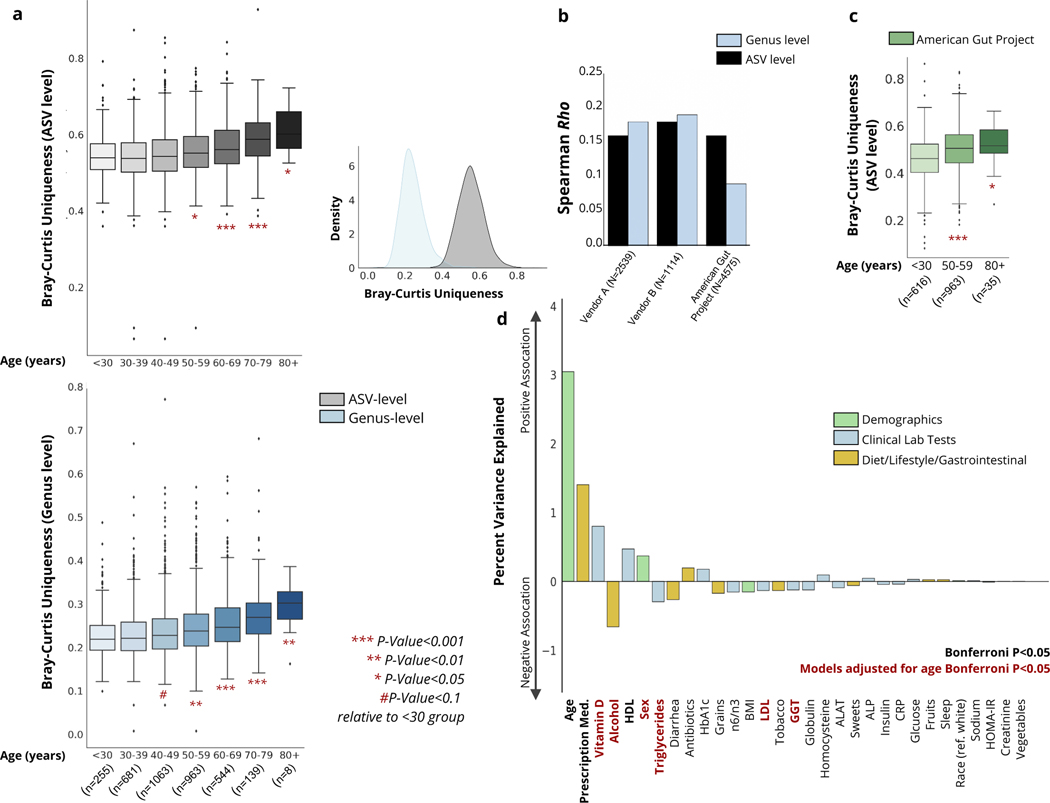
To characterize gut microbial patterns associated with aging, we initially performed a β-diversity analysis comparing all available baseline microbiome samples from a heterogenous, and relatively healthy Arivale population (Fig.1 and Extended Data 1). To capture the compositional divergence indicative of healthy aging observed in centenarians, our analysis involved extracting the minimum value for each individual from a calculated Bray-Curtis dissimilarity matrix. This value reflects how dissimilar an individual is from their nearest neighbor, given all other gut microbiome samples in the cohort. We refer to this as a measure of ‘uniqueness’: the higher the value, the more distinct the gut microbiome is from everyone else’s in the studied population. Arivale participants showed initial drift toward an increasingly unique gut microbiome composition starting between 40–50 years of age at the genus level, and 50–60 years at the amplicon sequence variant (ASV) level, and this continued to increase with every passing decade (Fig. 2a). The correlation between uniqueness and age was consistent across two different microbiome vendors used for gut microbiome processing, at both the ASV and genus level, independent of sex, BMI and Shannon diversity (Fig. 2b and Extended Data 3). We replicated our analysis using additional β-diversity metrics. Uniqueness based on Weighted UniFrac demonstrated a similar positive association with age across both vendors at both the ASV and genus levels, while the unweighted Jaccard metric resulted in comparable associations with age at the ASV level, but considerably weaker associations at the genus level (Extended Data 3). Similar results were obtained when using Unweighted UniFrac, though the association was weaker in Vendor A compared to Vendor B. Genus-level analysis once again showed weak to no association with age when using Unweighted UniFrac (Extended Data 3). We expected unweighted (i.e. presence-absence) β-diversity metrics to show a weaker association with aging, given our initial hypothesis that the shifting relative dominance between core taxa and accessory taxa would drive rising uniqueness (i.e. this pattern should only be apparent when using weighted metrics that incorporated taxon abundances). From this point forward, we focus primarily on the Bray-Curtis uniqueness measure, because it is a weighted measure not influenced by phylogeny and is well suited to capture changes in gut bacterial dominance previously observed in smaller cohorts of extremely long-lived individuals (decline in core taxa) 9.
Fig. 2. Associations between gut microbial uniqueness and age across the Arivale cohort.
(a) Boxplots showing gut microbiome uniqueness measures calculated using the ASV-level (grey) and genus-level (blue) Bray-Curtis dissimilarity metric across the adult lifespan across the Arivale cohort, adjusted for vendor. Asterisks indicate significant differences relative to the youngest <30 group, from a linear regression model adjusted for vendor, sex, BMI, and Shannon diversity (ASV-level: (50–59) P=3.52e-02, (60–69) P=1.88e-05, (70–79) P=1.47e-09, (80+) P=1.12e-02, genus-level: (40–49) P=7.15e-02, (50–59) P=3.57e-03, (60–69) P=4.33e-07, (70–79) P=8.16e-09, (80+) P=7.90e-03, two-sided). Also shown is the distribution of uniqueness calculated using the Bray-Curtis metric on both the ASV and genus level. (b) Spearman correlation coefficients for measures of Bray-Curtis uniqueness with age in individuals whose stool samples were processed by vendor A or B, as well as an additional external dataset (The American Gut Project). (c) Boxplots showing gut microbiome uniqueness scores calculated using the ASV-level Bray-Curtis across early, mid and late adulthood in the American Gut Project dataset. Asterisks indicate significant differences relative to the youngest <30 group, from a linear regression model adjusted for sex and Shannon diversity ((50–59) P=2.77e-09, (80+) P=2.95e-02). In both (A) and (C), box plots represent the interquartile range (25th to 75th percentile, IQR), with the middle line demarking the median; whiskers span 1.5 × IQR, points beyond this range are shown individually. (d) Percent of variance explained in genus-level Bray-Curtis uniqueness by a diverse number of demographic and lifestyle factors, as well as a subset of clinical laboratory tests.
We next validated the identified uniqueness-aging signal in the AGP data set20, a self-selected citizen-scientist cohort of thousands of individuals spanning a wide age range (18–101), which demonstrated a consistent pattern (Fig.2b–c, Extended Data 3). While the association was significant across ASV and genus level analysis across all three cohorts, we conservatively focused on the genus-level Bray-Curtis measure in our downstream analysis. As we have described previously, genus-level data are less sensitive to batch effects and more amenable to cross-cohort comparisons21.
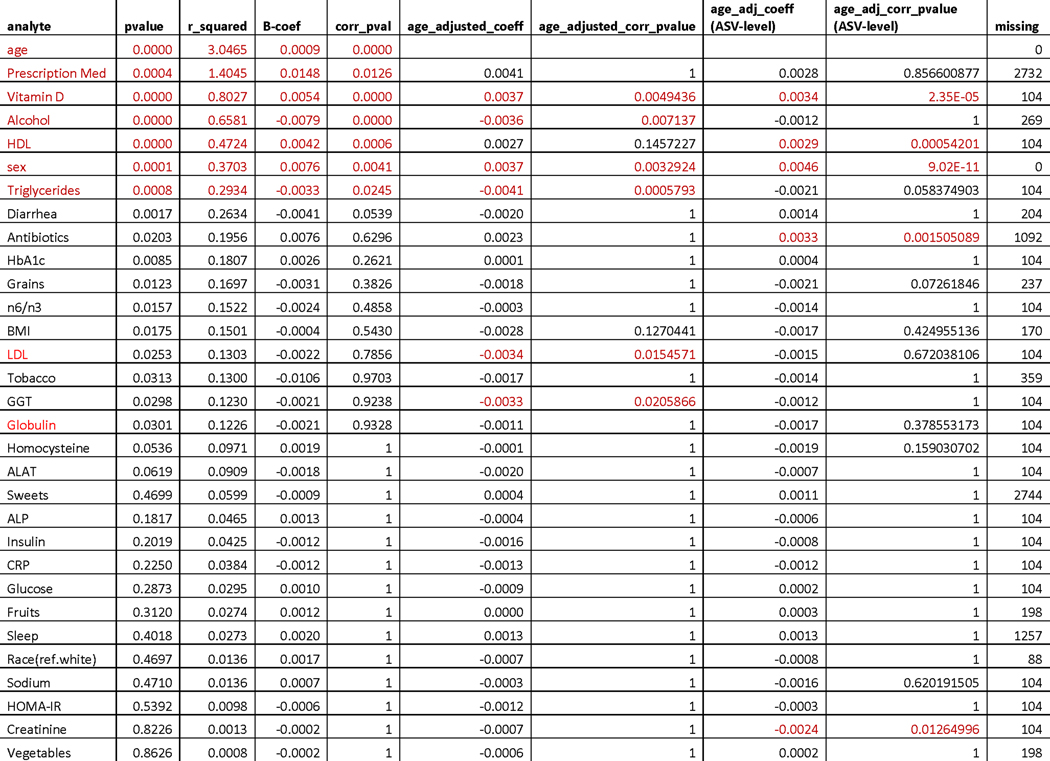
To further characterize the observed gut microbiome aging pattern, and understand how it is reflected in host physiology and health, we tested the correspondence between genus-level Bray-Curtis uniqueness and a wide variety of clinical laboratory tests, demographic information, and self-reported lifestyle/health measures in the Arivale cohort, adjusting for microbiome vendor (Fig. 2d, Extended Data 4). Of all the factors tested, age demonstrated the strongest association with gut microbiome uniqueness. Several other factors were significantly associated with uniqueness, including prescription medication use and alcohol consumption (Extended Data 4). However, after adjusting for age, mainly lipid markers remained significantly associated with gut microbiome uniqueness, with the direction of association indicating healthier metabolic and lipid profiles in individuals with more unique gut microbiomes: e.g. lower low-density lipoprotein (LDL) cholesterol, higher vitamin D, and lower triglycerides in individuals with more unique microbiomes (Fig. 2d). Interestingly, self-reported dietary measures showed little to no association with our gut microbiome uniqueness metric. However, the depth of information on dietary habits captured by our questionnaires was limited, hence these results need to be interpreted with caution. More detailed dietary questionnaires are likely required to capture the broad impact of diet on gut microbial composition reported previously 5. The same age-adjusted analysis was replicated using the ASV-level Bray-Curtis uniqueness measure, showing similar results in terms of lipid profiles (high-density lipoprotein (HDL), vitamin D) and sex, as well as additional significant associations with antibiotics use and creatinine levels (Extended Data 4).
Reflection of gut microbiome uniqueness in the host metabolome
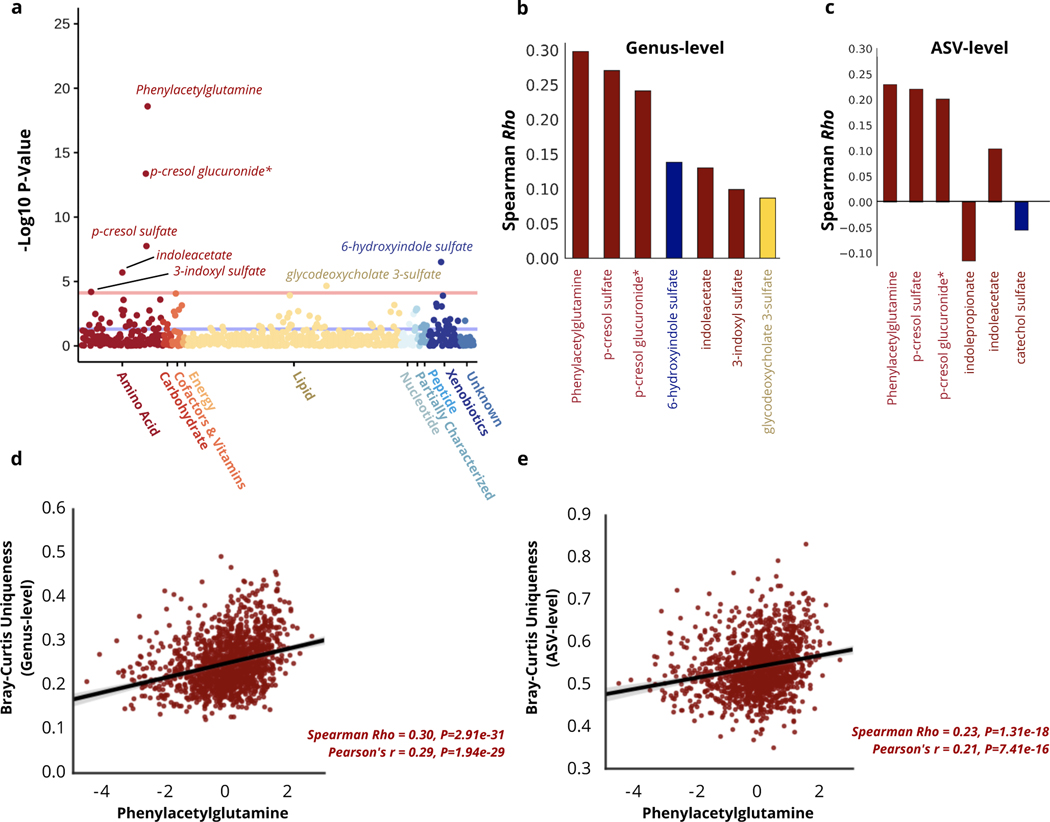
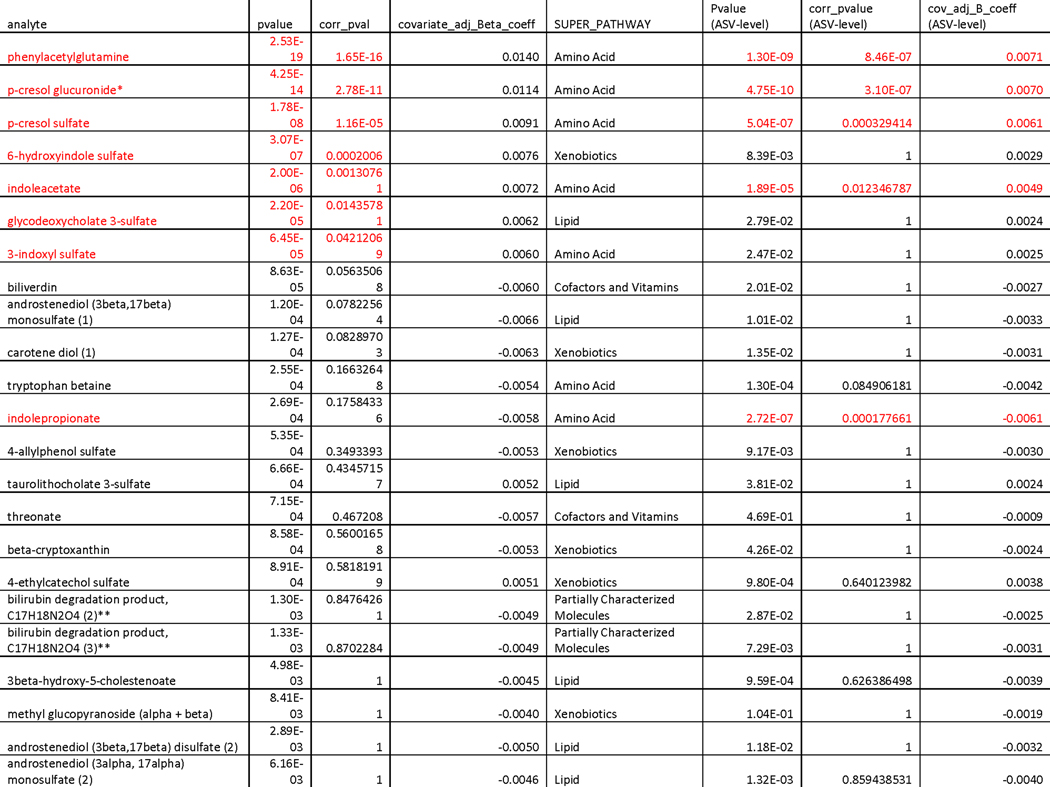
Our research group has previously demonstrated a strong reflection of gut microbiome community structure in the human plasma metabolome22. In order to better understand how host physiology reflects the increasingly unique composition of the gut microbiome seen with aging, and to gain potential mechanistic insight into the functional changes that take place in the microbiota, we regressed our uniqueness measure against each of the 653 plasma metabolites measured in the Arivale cohort, adjusting for covariates (see Methods). A total of seven metabolites, all microbial in origin, remained significantly associated with genus-level uniqueness after multiple hypothesis correction (Bonferroni P-Value<0.05) (Fig. 3a&b). Repeating the analysis at the ASV-level independently across all 653 metabolites identified six analytes significantly associated with uniqueness, 4/6 overlapping with the genus-level analysis, as well as an additional significant association with indolepropionate and catechol sulfate not captured by the genus-level measure (Fig.3c, Extended Data 5). The identified metabolites fell primarily into one of two classes: phenylalanine/tyrosine metabolites (phenylacetylglutamine, p-cresol glucuronide, p-cresol sulfate) and tryptophan metabolites (3-indoxyl sulfate, 6-hydroxyindole sulfate, indoleacetate, indolepropionate). Interestingly, significant changes in both tryptophan and phenylalanine pathways have been previously reported in centenarians relative to younger controls, with centenarians showing greater activation of these pathways in the gut microbiome 23,24. A metabolite previously enriched in centenarians relative to younger controls, phenylacetylglutamine 24, demonstrated the strongest correspondence with gut microbiome uniqueness in our analysis, explaining 7.7% of the variance at the genus level (adj. β (95%CI) = 0.014 (0.011,0.017), P-Value= 2.53e-19) and 3.6% at the ASV level (β (95%CI) = 0.007 (0.005,0.009), P-Value= 1.29e-09) (Fig. 3c&d, Extended Data 5). These findings indicate that the observed gut microbial drift towards a more unique compositional state seen with age is characterized by a distinct shift in gut microbial amino acid metabolism, which may serve as a useful biomarker for gut microbiome changes across the human lifespan.
Fig. 3. Reflection of gut microbiome uniqueness in plasma metabolites.
(a) A plot of -log10 p-values for each of the 653 plasma metabolites measured in the Arivale cohort, from OLS regression models predicting genus-level Bray-Curtis uniqueness adjusted for microbiome vendor, sex, age, age2, a sex*age interaction term, BMI, and Shannon diversity. Metabolites are color-coded by their super-family. All metabolites above the light red line are significant after multiple-hypothesis correction (Bonferroni P<0.05, two-sided), while the blue line indicates the unadjusted P-value threshold. Asterisks (*) indicate metabolites that were confidently identified on the basis of mass spectrometry data, but for which no reference standards are available to verify the identity. (b) Spearman correlation coefficients for each of the metabolites significantly associated with genus-level Bray-Curtis uniqueness after adjusting for covariates and multiple-hypothesis correction (Bonferroni P<0.05 two-sided). (c) Spearman correlation coefficients for each of the metabolites significantly associated with the ASV-level Bray-Curtis uniqueness measure after adjusting for covariates and multiple-hypothesis correction (Bonferroni P<0.05 two-sided). For both subfigures b) and c), bars are color-coded as in a). (d) Scatter plot of genus-level Bray-Curtis Uniqueness and the strongest metabolite predictor, phenylacetylglutamine, adjusted for vendor. (e) Scatter plot of ASV-level Bray-Curtis uniqueness and the strongest metabolite predictor, phenylacetylglutamine, adjusted for vendor. The lines shown are the y∼x regression lines, and the shaded regions are 95% confidence intervals for the slope of the line. The p-values reported in (d) and (e) are a result of two-sided statistical tests.
Gut microbial pattern of healthy aging in latest decades of human life
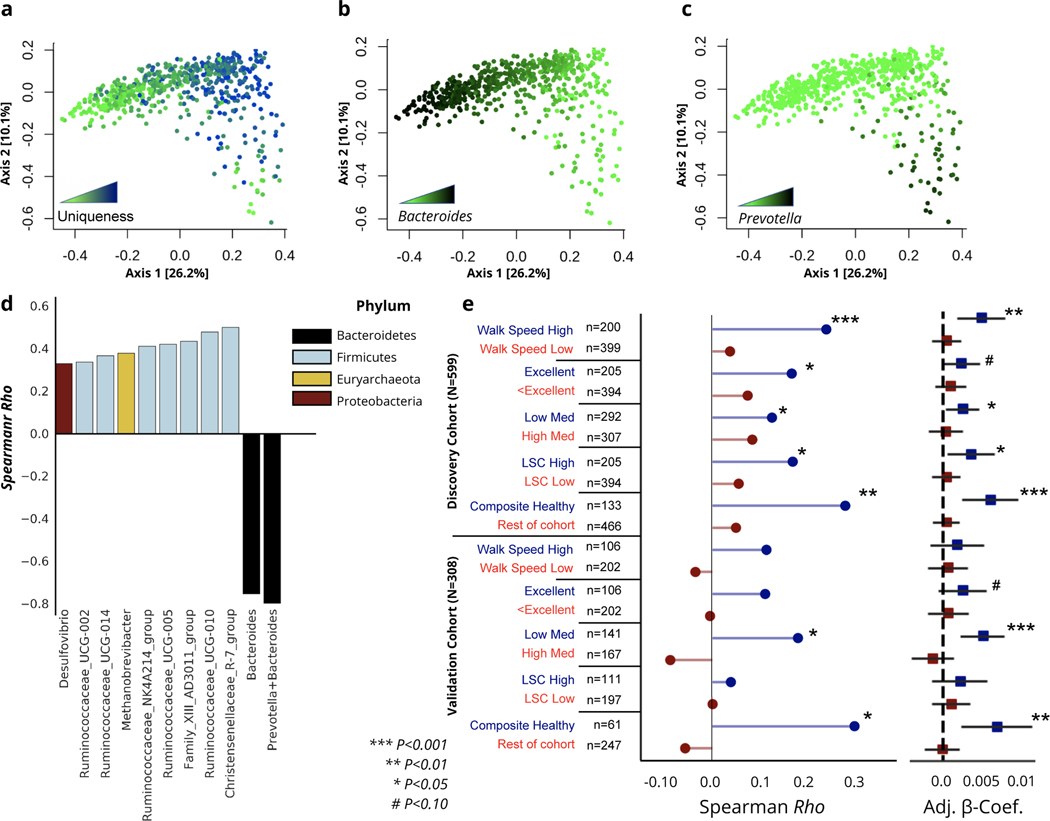
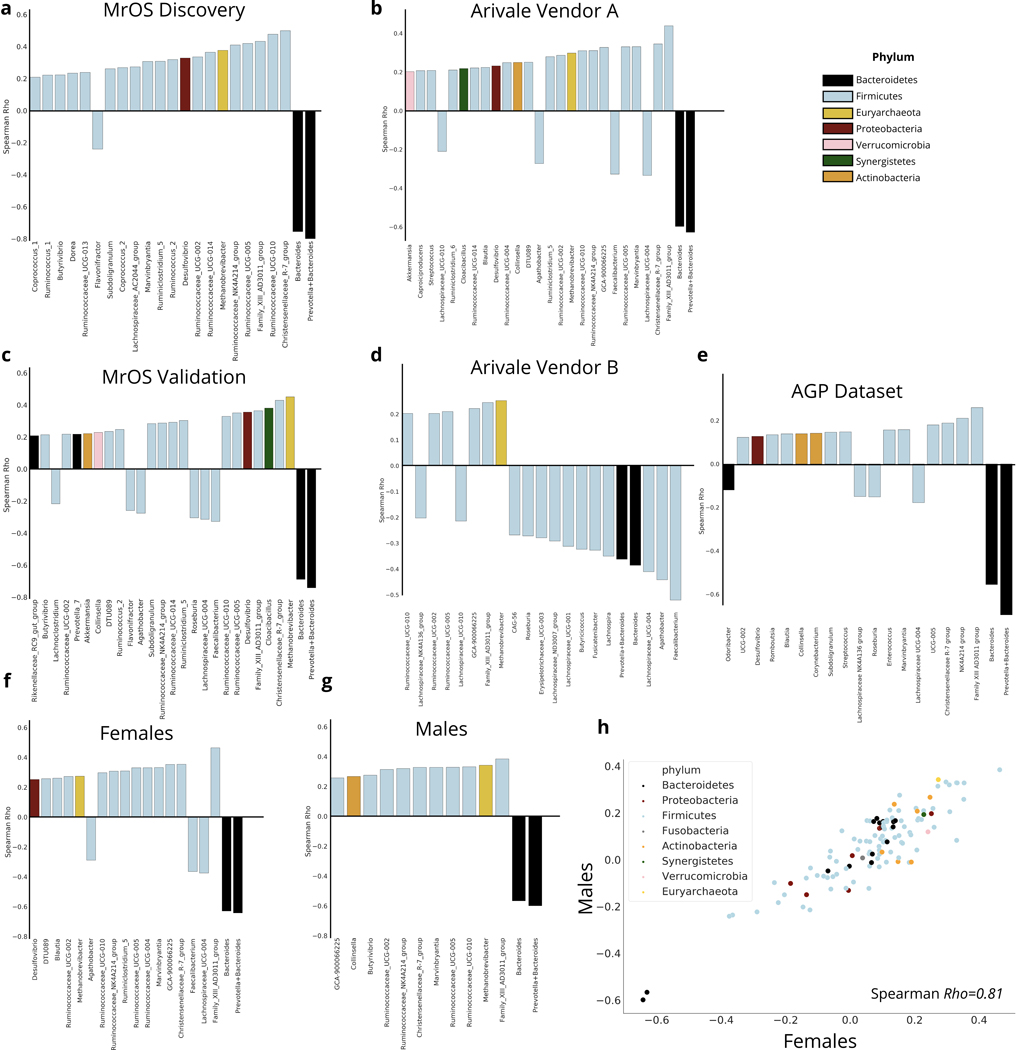
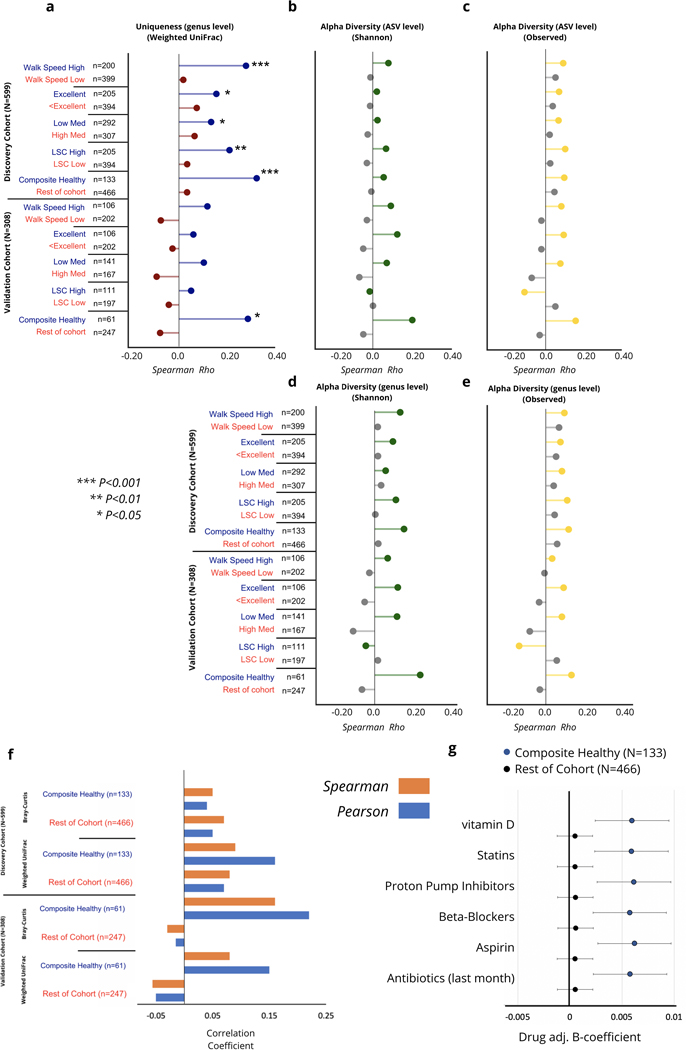
To better understand the long-term health implications of the identified aging dynamics of the gut microbiome, we extended our analysis into a separate cohort of older men with paired health and longitudinal follow-up data (the MrOS cohort, see Methods). The MrOS cohort is a prospective cohort study that recruited older male participants across the United States. At the fourth follow-up visit, a subset of the participants provided stool samples for 16S rRNA sequencing of their gut microbiome (discovery cohort N=599, validation cohort N=308)19. All participants who provided a stool sample exceeded 78 years of age at the time of sampling, allowing us to gain insight into the relationship between the gut microbiome and host health at the latest decades of human life (Fig.1 & Extended Data 2). Once again, we calculated a uniqueness score for each individual using the genus-level Bray-Curtis dissimilarity metric. Projecting MrOS microbiome data onto the first two Principal Coordinates revealed that samples with the highest Bray-Curtis uniqueness tended to fall away from common microbiome profiles, i.e. Bacteroides or Prevotella dominated ecosystems (Fig. 4a–c). We further correlated individual taxa with gut microbiome uniqueness. While the negative association of genus-level gut microbiome uniqueness with Bacteroides and the sum of Bacteroides/Prevotella was the strongest, a number of genera belonging to the Firmicutes phylum showed positive associations with the same measure of compositional divergence (Fig. 4D). Interestingly, a number of the identified taxa positively associated with uniqueness, both beneficial (Christensenellaceae) and potentially pathogenic (Methanobrevibacter, Desulfibrio) have been previously implicated in human longevity, enriched in long-lived individuals 8,9. We further replicated the same analysis in the MrOS validation cohort, the Arivale cohort across vendors and stratified by sex, as well as the AGP dataset. In each case, we observed a high level of congruence in the major taxa that correlate with compositional uniqueness, indicating that rising uniqueness with age is characterized by the rise and decline of the same sets of taxa across considerably diverse populations. (Extended Data 6).
Fig. 4. Increased dissimilarity of the gut microbiome as a function of healthy aging in the MrOS cohort.
(a-c) PCoA of the MrOS discovery cohort color-coded by (a) genus-level Bray-Curtis uniqueness, (b) relative Bacteroides abundance, and (c) relative Prevotella abundance. (d) Barplot demonstrating the correlation of strongest taxa associated with genus-level gut microbiome uniqueness in the MrOS discovery cohort, color-coded by phylum. (e) Correlation of genus-level Bray-Curtis uniqueness scores with age across the MrOS discovery and validation cohorts under different health stratifications. Also shown are age β-coefficients (slopes) with 95% confidence intervals from (OLS) linear regression models predicting genus-level Bray-Curtis uniqueness adjusted for BMI across the same stratifications. ‘Excellent’ corresponds to individuals who self-reported their health to be excellent, while ‘<Excellent’ incorporates all individuals who self-reported their health being anything less than excellent (good, fair, poor, or very poor).’Composite Healthy’ refers to individuals who fell into the healthy sub-group in at least 3 of the 4 stratifications performed. LSC: Life-Space Score. Significance of association was tested using a two-sided hypothesis, and p-values have not been corrected for multiple hypothesis testing. Exact correlation coefficients, β-coefficients, and corresponding p-values can be found in Extended Data 7.
Consistent with our initial analysis, age showed a positive association with our uniqueness score in the MrOS cohort, although to a weaker extent than in the larger Arivale population (genus-level Bray-Curtis Spearman Rho=0.11, P-Value= 0.0097, ASV-level Bray-Curtis Spearman Rho=0.07, P-Value= 0.072). Unlike the Arivale cohort, MrOS participants were considerably more health heterogenous at time of sampling, with a large proportion of participants reporting chronic conditions (Extended Data 2). The health heterogeneity of MrOS participants provided an opportunity to better understand whether the observed increase in microbiome dissimilarity with age depends on host health, and thus may be indicative of healthy aging. Hence, we re-ran the above analysis under four different stratifications based on: medication use, self-perceived health, life-space score (LSC), and walking speed. We chose these four health metrics because collectively they encompass a diverse repertoire of health in older populations (Table 1).
Table 1:
Description of health metrics used for stratification in the MrOS cohort.
| Health Metric | Description | Stratification |
|---|---|---|
| Medication use | Medication use is associated with chronic diseases and comorbidities, and is an important modulator of the gut microbiome 53. High medication use is particularly prevalent in older populations, with nearly 40% of individuals 65+ years old reporting taking ≥5 medications 54. | High: >8, Low: ≤8 medications. This allowed us to generate two groups of participants with similar age distribution but very different pharmacological profiles. |
| Self-perceived health | Self-perceived health has been previously shown to be an independent predictor of earlier mortality in older populations 55–57. | In the MrOS cohort, individuals chose one out of five possible responses (excellent, good, fair, poor, very poor). We stratified the cohort into individuals who reported excellent health and those who reported anything less than excellent. |
| Life-space Score (LSC) | LSC is an indicator of mobility, i.e. how often an individual leaves their room, house, or neighborhood and has been previously associated with risk of mortality in MrOS participants 52. Its strength as a measure lies in that it not only provides insight into whether an individual is physically capable of performing activities, but also whether that individual actually performs these activities 58. | For both the LSC and walking speed, we stratified the cohort into tertiles and defined the top tertile as the healthy group (High), while the bottom two tertiles were combined into the less healthy group (low). |
| Walking Speed | Walking speed is a validated measure used to assess functional status and overall health 59, and had been previously shown to be associated with executive function, and predictive of cognitive decline 60. | |
| Composite | A composite of all 4 of the above measures | Healthy - individuals who met 3+ of the above criteria (Extended Data 2) |
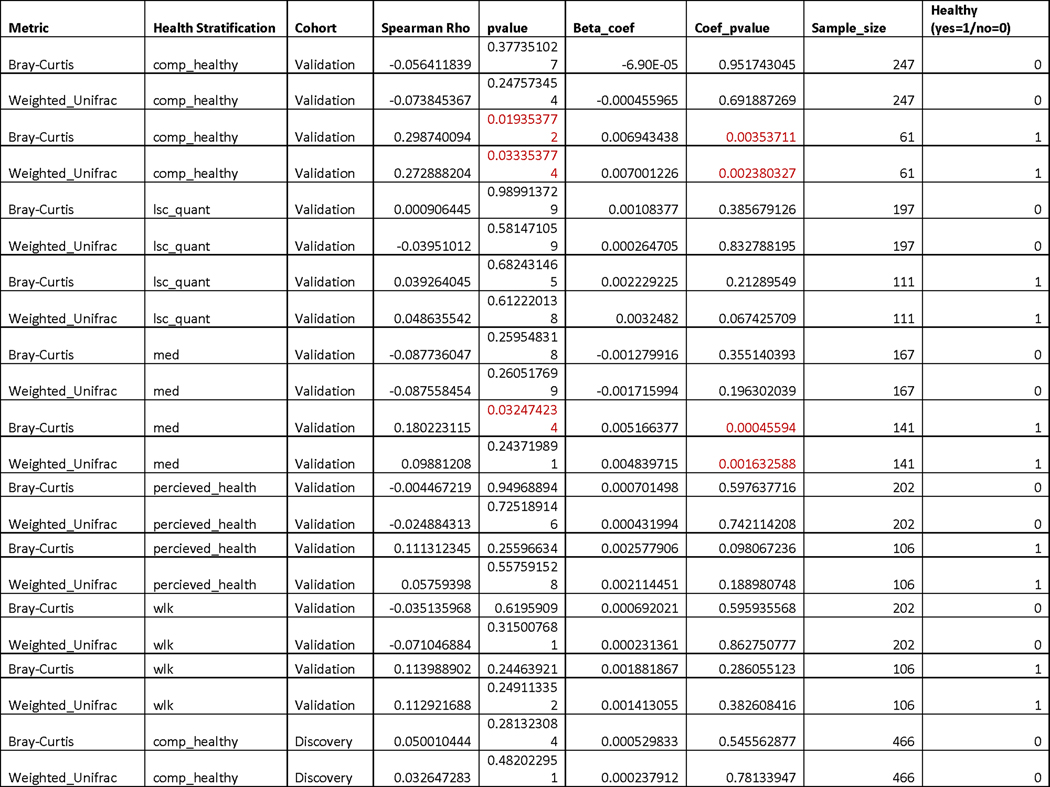
Under all stratifications considered, we observed a stronger positive association between age and genus-level microbiome uniqueness in healthier individuals, while the association was lower or absent altogether in individuals demonstrating worse health (Fig. 4e & Extended Data 7). We further generated a composite stratification (composite healthy), where MrOS participants had to meet at least three of the four criteria outlined above to be classified as healthy (Table 1 & Extended Data 2). In this limited group of 133 individuals we observed an even stronger association between gut microbiome uniqueness and age than under any individual stratification. We replicated the analysis on the second batch of MrOS gut microbiome samples (validation cohort, N=308), demonstrating very similar results (Fig. 4e & Extended Data 7). We also ran the same analysis using genus-level Weighted UniFrac dissimilarity, and observed high level of congruence between results (Extended Data 7 & 8). We further replicated the analysis on the ASV level. Although the strength of association between healthy aging and uniqueness at the ASV level was weaker, it still showed the same directional trends with healthier individuals showing a stronger positive correlation, particularly in the validation cohort (Extended Data 8). In contrast, measures of α-diversity at the genus and ASV levels were not significantly associated with age under any stratification considered (Extended Data 8).
Drugs are known to significantly impact the composition and function of the gut microbiome 25. Given the high number of medications taken by MrOS participants, we explored whether the identified healthy aging pattern is confounded by drug use. To this end, we performed additional analysis using the genus-level Bray-Curtis uniqueness measure in the MrOS discovery cohort. First, we focused specifically on the highly medicated individuals (>8 reported meds). In this high med. group, we identified a subset of individuals on multiple medications who are nevertheless healthy using the criteria for walking speed described in Table 1. The correlation between uniqueness and age was still significant in healthy, highly medicated individuals (Pearson’s r=0.26, P-Value =0.047, Spearman Rho=0.27, P-Value =0.01, n=81). Once again, less healthy individuals did not show the same pattern (Pearson’s r= −0.05, P-Value=0.50, Spearman Rho=0.02, P-Value =0.76, n=223).
Next we explored the impact of adjusting age-uniqueness linear regression models for individual medication use. The five most prevalent supplements and drugs reported by MrOS participants, as well as antibiotic use in the last month, did not impact the association between age and Bray-Curtis uniqueness in healthy composite individuals, and did not change the lack of the same relationship in the remainder of the cohort (Extended Data 8). Although drug-microbiome interactions are more complex than what can be captured by our approach, we conclude that medication use is not the main driver behind the observed variability in gut microbiome aging patterns across health states.
Gut microbiome predicts mortality in extreme aging
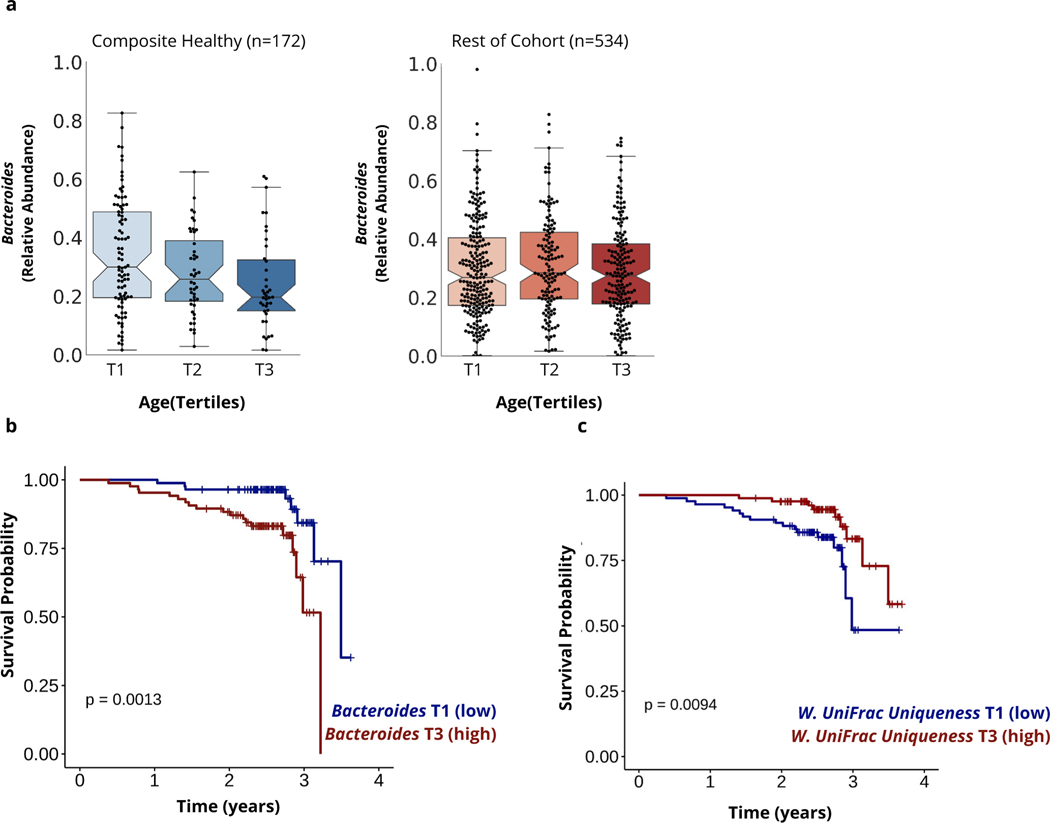
Next, we focused exclusively on community-dwelling individuals (i.e. excluding participants in assisted living, nursing homes, and/or who have been hospitalized in the past 12 months) from the two MrOS data sets, combined together for increased power (N=706) (Fig. 1). We performed genus-level differential abundance analysis to identify genera associated with age in healthy composite individuals (N=173) and the remainder of the cohort (N=533), separately, adjusting for batch (discovery/validation) and BMI. In healthy composite individuals, only the genus Bacteroides (adj. β (s.e.): -0.063 (0.017), P-Value=0.0004) demonstrated a significant negative association with age after multiple hypothesis correction (Fig. 5a). These findings are consistent with our gut dissimilarity analysis, where the uniting feature of unique microbiomes is the depletion of the most common and dominant genera. Consistently, there was no significant association between age and relative Bacteroides abundance in participants who did not meet our health criteria (adj. β (s.e.): −0.0085 (0.009), P-Value =0.35) (Fig. 5a). In contrast, individuals in worse health demonstrated a distinct gut microbiome aging pattern characterized by a decline in the genera Lachnoclostridium (adj. β (s.e.): −0.037 (0.0095), P-Value =1.59e-04) and the Rumminococace family genus UBA1819 (adj. β (s.e.): −0.075 (0.016), P-Value =3.72e-06). These results indicate there may be multiple gut microbiome aging patterns in the later stages of human life that are associated with distinct health outcomes.
Figure 5. Associations between identified gut microbial aging patterns and survival in older adults.
(a) Boxplots demonstrating the relative abundance of the genus Bacteroides across tertiles of age in community-dwelling individuals identified as healthy on 3+ criteria specified (composite healthy) and the remainder of the cohort. (b) Kaplan Meier Curve demonstrating the association between overall survival and relative Bacteroides abundance grouped into tertiles in community-dwelling MrOS participants 85+ years at time of sampling. (c) Kaplan Meier Curve demonstrating the association between overall survival and ASV-level Weighted UniFrac uniqueness grouped into tertiles in community-dwelling MrOS participants who were 85+ years at the time of sampling. P-values shown in (b) and (c) are a result of Log-rank tests (two-sided) comparing the two survival curves, and have not been corrected for multiple hypothesis testing.
Our findings from both β-diversity and differential abundance analysis in healthy elderly MrOS participants indicate that the identified gradual aging pattern may, in its end stages, resemble gut microbiome changes previously observed in extremely long-lived individuals (i.e. loss of core taxa) 8,9. Therefore, we utilized longitudinal data from the MrOS cohort to investigate whether the characterized gut microbiome pattern is not only reflective of healthy aging, but also predictive of survival. We performed the analysis in two steps: 1) on all community-dwelling participants (N=706) and 2) only on community-dwelling participants in the top age tertile (85+ years of age, N=256) at time of gut microbiome sampling, because these participants were the closest to achieving extreme age in the course of the study’s follow-up period (~4 years). When focusing on all individuals in the cohort, we identified a significant positive association between relative Bacteroides abundance and increased risk of all-cause mortality, independent of age, BMI, clinical site, self-perceived health, diagnosis of congestive heart failure, and batch in which stool samples were processed. Replicating the analysis in the oldest individuals (85+ years old) revealed a stronger association and higher Hazard Ratios compared to the whole cohort (Table 2 & Fig. 5b). Using the participants’ calculated Bray-Curtis and Weighted UniFrac uniqueness scores yielded comparable results in 85+ year olds, where mortality risk decreased in individuals with more unique gut microbiomes independent of the same covariates. In contrast, the same associations between Bray-Curtis and Weighted UniFrac uniqueness measures and mortality were not present when younger participants were included in the analysis (Table 2). Once again, we replicated the same analysis on the ASV level. Across both dissimilarity metrics, the trends were consistent with the genus level measures, however only the Weighted UniFrac metric passed the significance threshold across all multivariable models (Table 2 and Fig.5c).
Table 2:
Associations between gut microbiome measures and mortality in the MrOS cohort.Unadjusted, age, clinical site and batch adjusted and multivariable adjusted Hazard Ratios (HR) and the 95% confidence intervals (95%CI) of relative Bacteroides abundance, genus level Bray-Curtis and Weighted UniFrac uniqueness scores, as well as the same metrics calculated at the ASV level, from Cox Proportional Hazard Regression models evaluating mortality risk in all community-dwelling MrOS participants (n=706) and exclusively community-dwelling MrOS participants 85+ years old (n=256). Multivariable models were adjusted for age, clinical site, BMI, self-perceived health, diagnosis of congestive heart failure, and batch in which stool samples were processed. Both relative Bacteroides abundance and The Bray-Curtis uniqueness measures were scaled and centered prior to mortality analysis. Significant HRs are bolded and colored in red (P≤0.05, two-sided). HR and 95%CI reported have not been corrected for multiple hypothesis testing.
| Unadj. HR (95%CI) | Age, Clinical Site, & batch Adj. HR (95%CI) | Multivariable Adj. | |||
|---|---|---|---|---|---|
| HR (95% CI)* | |||||
| Genus level analysis | Rel. Bacteroides Abundance | 1.21 | 1.31 | 1.28 |
Community dwelling participants
all ages (N=706) |
| (0.95–1.54) | (1.02–1.68) | (1.00–1.65) | |||
| Uniqueness (Bray-Curtis) | 1.15 | 1.08 | 1.09 | ||
| (0.91–1.46) | (0.85–1.38) | (0.85–1.39) | |||
| Uniqueness (Weighted UniFrac) | 1.03 | 0.94 | 0.94 | ||
| (0.81–1.31) | (0.74–1.19) | (0.75–1.19) | |||
| ASV level analysis | Uniqueness (Bray-Curtis) | 1.26 | 1.19 | 1.13 | |
| (0.98–1.62) | (0.92–1.54) | (0.87–1.47) | |||
| Uniqueness (Weighted UniFrac) | 0.88 | 0.82 | 0.82 | ||
| (0.64–1.21) | (0.59–1.14) | (0.58–1.16) | |||
| Genus level analysis | Relative Bacteroides Abundance | 1.74 | 1.89 | 1.91 |
Community dwelling Participants
85+ years old only (N=256) |
| (1.27–2.37) | (1.36–2.63) | (1.38–2.66) | |||
| Uniqueness (Bray-Curtis) | 0.76 | 0.70 | 0.70 | ||
| (0.54–1.07) | (0.49–0.99) | (0.50–0.99) | |||
| Uniqueness (Weighted UniFrac) | 0.65 | 0.60 | 0.59 | ||
| (0.43–0.98) | (0.40–0.90) | (0.40–0.87) | |||
| ASV level analysis | Uniqueness (Bray-Curtis) | 0.89 | 0.86 | 0.77 | |
| (0.65–1.23) | (0.62–1.21) | (0.54–1.11) | |||
| Uniqueness (Weighted UniFrac) | 0.40 | 0.32 | 0.26 | ||
| (0.19–0.86) | (0.14–0.71) | (0.12–0.59) |
Discussion
There is limited understanding of how the human gut microbiome changes throughout adulthood and how these changes influence host physiology. Here, we evaluated gut microbiome patterns associated with aging across over 9000 individuals from three distinct study populations spanning 18–101 years of age. The major findings of our analysis were: 1) individual gut microbiomes became increasingly more unique to each individual with age, starting in mid-to-late adulthood, and this uniqueness was positively associated with known microbial metabolic markers previously implicated in immune regulation, inflammation, aging and longevity; 2) in the latter decades of human lifespan, healthy individuals continued to show an increasingly unique gut microbial compositional state (associated with a decline in core taxa) with age, while this pattern was absent in those in worse health; 3) in individuals approaching extreme age (85+ years old), retaining high relative Bacteroides abundance and having a low gut microbiome uniqueness measure were both associated with significantly decreased survival in the course of 4 year follow-up. These observations are strengthened by the presence of similar age-related trends across two independent microbiome vendors, and three demographically distinct cohorts (Arivale, MrOS, AGP).
While the ecological composition of the gut microbiome became increasingly divergent with aging, we found a corresponding convergence in plasma concentrations of microbial metabolites, indicating that aging is characterized by gradual shifts in gut metabolic capacity. A number of the identified microbial metabolites in our analysis have been previously characterized as mildly toxic phenylalanine/tyrosine microbial fermentation products (p-cresol sulfate, phenylacetylglutamine, p-cresol glucuronide), indicating a rising burden of gut xenobiotic metabolites in an aging host 26,27. This is consistent with a recent study in the ELDERMET cohort demonstrating that fecal concentrations of p-cresol correlated with increased frailty, particularly in long-term care residents 14. The reported association between p-cresol and frailty indicates that p-cresol may reflect, or potentially contribute to, age-associated decline in this population. However, the association between microbial protein fermentation products and aging in community dwelling individuals, as well as certain animal models of longevity, show conflicting results. For example, urine levels of phenylacetylglutamine and p-cresol sulfate have both been previously shown to be enriched in centenarians relative to elderly and young controls 24. Similarly, metabolomic profiles of naked mole rats, a model organism characterized by extreme lifespan that exhibits negligible age-associated decline, demonstrated increased levels of phenylalanine and tryptophan degradation products (phenylacetylglutamine, indoleacetate) in the blood relative to a group of control mice 28. Therefore, it’s likely that protein microbial metabolites reflect different aging gut ecosystem dynamics in community dwelling individuals versus severely frail long-term care residents. Alternatively, resilient individuals may be able to effectively neutralize and excrete these microbial compounds, while others succumb to the negative effects of these same metabolites on host physiology throughout the course of aging.
Additional metabolites associated with our observed gut microbial pattern were dominated by indoles (3-indoxyl sulfate, 6-hydroxyindole sulfate, indoleacetate and indolepropionate), which are gut bacterial degradation products of tryptophan. Bacterial indole metabolism is emerging as an important mediator of the gut microbiome-host immune homeostasis 29. Indole (the precursor to the human microbial co-metabolite indoxyl sulfate identified in our analysis) has been shown to increase healthspan and extend survival in a number of animal models 30. One of the most recognized mechanisms of action of indole metabolites is mediating inflammation through binding the aryl hydrocarbon receptor 31. Studies in mice demonstrated that indole metabolites can alleviate lipopolysaccharide induced liver inflammation 31,32 and protect from chemically induced colitis33, highlighting their role in mediating host inflammatory responses. Consistent with their positive effect on healthspan and inflammation in animal models, concentrations of certain plasma indole metabolites (indoxyl-sulfate and indolelactate) were positively associated with improved physical function in older adults34. A number of microbial indole metabolites (indoleacetate, indolepropionate, indoxyl-sulfate and indolelactate) have further been recently shown to be depleted in plasma of obese individuals relative to normal weight controls35. While the majority of indole metabolites identified in our study were positively associated with gut microbiome uniqueness, ASV-level analysis revealed a negative association with indolepropionate, indicating there is likely a more complex reprogramming of tryptophan metabolism with healthy aging than our present analysis is able to capture.
The immune regulatory function of tryptophan microbial metabolites implicates the gut microbiome in adapting to, and possibly modulating, the host immune system throughout human aging. This is supported by recent studies demonstrating that that first signs of immunological aging in adults occur at approximately 40 years of age36, consistent with when the first signs of increasing gut microbiome uniqueness with aging were observed in the Arivale cohort. Collectively, our metabolomic analysis suggests that microbial amino acid metabolism may be an important new component in how the host immune system changes with age. However, further studies in animal models and humans are needed to better understand these complex interactions across the human lifespan.
A striking finding in our analysis was that increasing compositional uniqueness was both reflective of healthy aging and predictive of survival in older adults. This suggests that the gut microbiome may not only reflect, but potentially contribute to, a longer host lifespan, which is consistent with some recent non-human animal studies 37,38. Concomitant with increasing uniqueness in healthy older individuals, a decline in the core genus Bacteroides emerged as a major characteristic of healthy aging. Bacteroides and other major taxa showed consistent associations with uniqueness across both men and women, and across the demographically distinct MrOS, Arivale and AGP cohorts. This indicates that the identified uniqueness trajectory can be traced back to consistent taxonomic changes, providing better interpretability to the identified pattern. The role of Bacteroides in aging has been inconsistent in the literature. Early investigations of the gut microbiome and aging reported an increased dominance of Bacteroides in older persons (65+ years) relative to healthy younger controls 12,39. Conversely, several studies have shown a decline in core taxa, like Bacteroides, in extremely long-lived individuals. These latter studies often focused on highly co-abundant groups of taxa (CAGs) and their association with age. Bacteroides was grouped into the CAG of core microbes that declines with age, along with the primary taxa that drove this signal, such as Faecalibacterium and Coprococus 8,9. There are several possible explanations for these conflicting results throughout the literature. For example, there may exist multiple patterns associated with aging in healthy and less-healthy individuals. In the MrOS cohort we were able to define healthy aging through a relatively strict cut-off. Thus, we were able to show that a gradual decline in Bacteroides with age was only observed in the healthiest individuals, which was not the case in less-healthy, yet still functionally independent, older adults of similar age. The importance of declining Bacteroides and other core taxa in aging requires further investigation. However, we propose that the optimal trajectory for the adaptation of the gut microbiome to an aging human host depends on an increase of rare taxa capable of synthesizing bioactive microbial metabolites (i.e. indoles) at the expense of Bacteroides species and other core taxa. This makes particular sense in the context of the changing immunological landscape with age, and the potential anti-inflammatory and immune-modulatory effects of several of the microbial metabolites associated with rising uniqueness described above.
Previously, studies on the gut microbiome in older cohorts have highlighted patterns separating community dwelling individuals from frailer older persons residing in long-term care facilities. These studies revealed significant associations between gut microbial composition and health markers, inflammation, and diet 4,5, and characterized gut microbial patterns indicative of age-associated decline 2. Coupling these findings with results from studies investigating gut microbiome development in children and adults1 has led some to hypothesize that the gut microbiome remains relatively stable throughout adulthood and into old age, at which point gradual compositional changes reflect, and potentially contribute to, declining health. In the current analyses, we have expanded the investigation of the gut microbiome in aging to cover majority of the adult lifespan in multiple, large deeply-phenotyped cohorts. While it is evident from previous research that gut microbiomes of older individuals (65+ years) change with deteriorating health, we propose that gut microbiomes of healthy individuals continue to develop along a distinct trajectory. This trajectory originates in adulthood, is accompanied by a rise in specific plasma microbial metabolites, reflects a healthy aging phenotype, and is predictive of extended survival in the latest decades of human life. As our understanding of the aging microbiome increases, monitoring and identifying modifiable features that may promote healthy aging and longevity will have important clinical implications for the world’s growing older population.
Methods
IRB Approval
Arivale Cohort:
Procedures for the current study were run under the Western Institutional Review Board (WIRB) with Institutional Review Board (IRB) Study Number 20170658 at the Institute for Systems Biology and 1178906 at Arivale (both in Seattle, WA).
MrOS Cohort:
The institutional review board at each of the 6 MrOS study sites approved the study protocol, and written informed consent was obtained from all participants.
Cohorts
The Arivale cohort consists of individuals over 18 years of age who between 2015 and 2019 self-enrolled in a now closed scientific wellness company. Briefly, majority of Arivale participants (~80%) were residents of Washington or California when in the program. While the cohort tends to be healthier than the general US population, it is more reflective of populations in these two states 40. The cohort is also predominantly female (~60%), which may be due to women being more likely to join these types of programs. For this study, only baseline measurements were considered for each participant, and individuals who provided a stool sample were included in the analysis. Demographic information on the cohort is provided in Extended Data 1.
The MrOS study is an ongoing prospective study of close to 6000 men recruited across six clinical U.S. sites. The cohort, recruitment criteria, and stool sample collections have been previously described in detail 18,19. Briefly, the recruitment for the MrOS cohort was designed to obtain a population sample representative of older (65+) U.S. men in the community. Since this was a volunteer population, it is likely enriched for healthier individuals. However, comparison between MrOS participants and NHANES data (representative of the US population), showed the two cohorts are quite similar 41. The cohort was not chosen to enrich for individuals with frailty, fractures, or osteoporosis. During the fourth follow-up visit of the original study, a subset of participants across all six clinics was asked if they would consent to have their stool sampled for microbiome analysis. Participants who agreed were given the OMNIgene-GUT stool/feces collection kit (OMR-200, DNA Genotek, Ottawa, Canada) and collected the fecal sample at their homes. Demographic information on MrOS participants is provided in Extended Data 2. In the initial uniqueness analysis, all participants with available high-quality microbiome data were used (N=907). Subsequent differential abundance analysis focused exclusively on community-dwelling individuals (N=706) (excluding individuals in assisted living, nursing homes and who have been hospitalized in the past 12 months). Finally, survival analysis was conducted on all community dwelling individuals as well as specifically on community dwelling individuals in the latest stages of aging (85+ years old, N=256). The number of deaths in the whole community dwelling group and in 85+ year old community dwelling group was 66 and 41, respectively.
Microbiome Analysis
Arivale cohort
Independent of the vendor used, stool samples were collected at the participants’ homes using DNA collection kits with a proprietary chemical DNA stabilizer provided by the microbiome vendor to maintain DNA integrity at ambient temperatures following collection. Gut microbiome sequencing data in the form of FASTQ files were provided on the basis of either the 300-bp paired-end MiSeq profiling of the 16S V3 + V4 region (DNAgenotek, vendor A) or 250-bp paired-end MiSeq profiling of the 16S V4 region (Second Genome, vendor B). Further analysis was performed using the denoise workflow from mbtools (https://github.com/gibbons-lab/mbtools) that wraps DADA2. In summary, we first trained DADA2 42 error models separately for each sequencing run and used those to obtain sequence variants for each sample. This was followed by de novo chimera removal which removed ~17% of all reads as chimeric and resulted in about 89,000 final sequence variants across all samples. Taxonomy assignment was performed using the RDP classifier with the SILVA database (version 132). Here 99% of the reads could be classified on the family level, 89% on the genus level and 32% on the species level. Species level taxonomy was identified by exact alignment to the SILVA reference sequences. Sequence variants were aligned to each other using DECIPHER 43 and the multiple sequence alignment was trimmed by removing each position that consisted of more than 50% gaps. The resulting core alignment had a length of 420 base pairs and was used to reconstruct a phylogenetic tree using FastTree 44. Downstream gut microbiome data analysis was conducted using the Phyloseq Package 45. In two separate analyses, gut microbiome samples were rarefied to 21123 (vendor A, DNAGenotek) and 25596 (Vendor B, Second Genome) reads, the minimum number of reads per sample for each vendor. For uniqueness analysis, the Bray-Curtis 46, Unweighted and Weighted UniFrac 47, and Jaccard matrices were calculated for all samples within each vendor using the rarefied genus and ASV tables. The minimum value for each row, corresponding to the dissimilarity of each sample to their nearest neighbor, was then extracted from the matrix and used for downstream analysis.
MrOS cohort
Stool samples were processed at the Alkek Center for Metagenomics and Microbiome Research (CMMR) at Baylor College of Medicine using their custom analytic pipeline in two separate batches (Discovery N=599, Validation N=320). Samples for both batches were collected during the same follow-up visit (visit 4), at which point all health-related data analyzed in this study was also collected. Sequencing data in the form of FASTQ files was then processed using the same pipeline as described above for the Arivale cohort. Preliminary microbiome data analysis was conducted using the Phyloseq Package. For α-diversity and uniqueness analysis, reads were rarefied to an even depth of 10000 reads. A total of 12 samples in the validation cohort had less reads than the specified cut-off, and hence were excluded from the analysis (Validation N=308). α-diversity measures were calculated at both the ASV and genus levels using the Phyloseq package 45. Both the rarefied ASV and genus tables were used for ß-diversity analysis comparing samples across the whole cohort. Uniqueness was calculated as described for the Arivale cohort. The calculated uniqueness measure for each participant was then used for downstream analysis. As part of our analytical pipeline, we also performed differential abundance analysis assessing the relationship of individual genera with age in individuals defined as healthy and unhealthy, separately. Analysis was performed in R (version 3.44) using beta-binomial regression through the Corncob package (version 1.0) 48. Models were adjusted for BMI, and batch (discovery/validation). Type 1 error was controlled using the Bonferroni method (P<0.1).
ASV abundances from the American Gut Project were obtained from the 125bp BIOM table deposited in figshare [6137192] 49 provided in the original manuscript by McDonald et. al.20. Metadata for study participants was obtained from the table provided in the same publication and also deposited in figshare [6137315] 50. Taxonomy was again assigned using the “assignTaxonomy” function contained in DADA2 on the raw ASVs returned by deblur and using the SILVA database (version 138). The combined data was then converted to Phyloseq objects for further downstream processing. Only samples with corresponding age in the metadata and a minimum of 10000 reads were used for analysis. Individuals <18 years old were further removed from the dataset. This resulted in a sample size of 4575. Additional information on the cohorts, data and software can be found in the Reporting Summary document.
Plasma Metabolomics & Clinical Laboratory Tests
Blood draws for all assays were performed by trained phlebotomists at LabCorp or Quest service centers. For the 24-hour period leading up to the blood draw, Arivale participants were required to avoid alcohol, vigorous exercise, aspartame and monosodium glutamate, and to begin fasting 12 hours in advance. Metabolomics analysis was conducted on baseline plasma samples from the Arivale cohort by Metabolon Inc. (North Carolina, USA) 51. Each EDTA-plasma Arivale sample was thawed on ice, after which a recovery standard was added to each sample for quality control purposes. As part of sample preparation, aqueous methanol extraction was performed to remove the protein fraction while retaining the maximum amount of small molecular weight compounds in the sample. The sample extract was then aliquoted into five separate fractions, one for each of the four methods used for subsequent metabolite quantification, and one aliquot as a potential backup. Excess organic solvent was removed from the aliquoted samples by placing the samples on a TurboVap® (Zymark). Aliquoted sample extracts were stored overnight under nitrogen before analysis. Regardless of which method for metabolite detection was used, all samples were run on the Waters ACQUITY ultra-performance liquid chromatography (UPLC) and a Thermo Scientific Q-Exactive high resolution/accurate mass spectrometer interfaced with a heated electrospray ionization (HESI-II) source and Orbitrap mass analyzer operated at 35,000 mass resolution. The four aliquoted sample extracts were dried then reconstituted in solvents compatible with each of the four methods used for downstream metabolite quantification. To ensure injection and chromatographic consistency, each solvent further contained a series of standards at fixed concentrations. Two of the four aliquots were analyzed using acidic positive ion conditions chromatographically optimized for either more hydrophobic (solvent consisting of water, methanol, acetonitrile, 0.05% perfluoropentanoic acid (PFPA) and 0.01% formic acid (FA)) or hydrophilic compounds (water and methanol, containing 0.05% PFPA and 0.1% FA). Both of these aliquots were eluted using a C18 column (Waters UPLC BEH C18-2.1×100 mm, 1.7 μm). Aliquot 3 was analyzed under basic negative ion optimized conditions with elution performed using a dedicated C18 column in solvent containing methanol and water, with 6.5mM Ammonium Bicarbonate at pH 8. The fourth and final aliquot was analyzed via negative ionization following elution from a HILIC column (Waters UPLC BEH Amide 2.1×150 mm, 1.7 μm) using a gradient consisting of water and acetonitrile with 10mM Ammonium Formate, pH 10.8. Mass spectrometry (MS) analysis was performed using dynamic exclusion and alternating between MS and data-dependent MSn scans. The scan range varied slightly between the four methods used, and covered 70–1000 m/z. To further account for potential run and day variability, process blanks and EDTA-plasma technical replicates were run intermittently throughout the study run-days. A biochemical library of over 3300 purified standards based on chromatographic properties and mass spectra was used for identification of known chemical entities. For statistical analysis, the raw metabolomics data were median scaled within each batch, such that the median value for each metabolite was one. To adjust for possible batch effects, further normalization across batches was performed by dividing the median-scaled value of each metabolite by the corresponding average value for the same metabolite in quality control samples of the same batch. In this study, we analyzed participants’ baseline plasma metabolomics data. Further filtering was performed to retain only individuals who provided blood and stool samples within 21 days of each other (median=0, interquartile range =−2 to 4 days). Individuals with Bray-Curtis uniqueness values greater or less than 3 standard deviations from the mean were removed prior to statistical analysis. A 10% missing value threshold was set, which was passed by 653 metabolites. Missing values for metabolites were imputed to be the median observed value for that metabolite. A total of 1459 Arivale participants had paired gut microbiome-plasma metabolome data and met the inclusion criteria. Values for each metabolite were log transformed prior to analysis. Clinical laboratory tests were conducted by either Quest or LabCorp. A 10% missing value threshold was set for each clinical laboratory test used in the analysis. All but 104 participants (N=3549) had paired clinical laboratory-gut microbiome data. Both metabolomics and clinical laboratory tests were scaled and centered prior to analysis and only baseline measures for each individual were used.
Lifestyle/Health Questionnaires in the Arivale Cohort
Data on lifestyle, diet and health were obtained through self-administered questionnaires completed by Arivale participants during their initial assessment. For reporting antibiotic use, participants chose from three possible responses (‘not in the past year’, ‘in the past year’ and ‘in the past three months’) which were recoded into ordinal variables 0, 0 and 1 respectively. Participants chose one of several possible frequencies in response to how often they experience diarrhea, that were recoded as follows: ‘infrequently/never’ = 0, ‘once a week or less’ = 1, ‘more than once a week’ = 2 and ‘daily’ = 2. Similarly, alcohol use (no. of drinks per day) was reported on the following scale which was recoded into corresponding ordinal variables: (0) ‘I do not drink’, (1) ‘1–2 drinks’: (2) ‘3–4 drinks’: (3) ‘5–6 drinks’: (4) ‘More than 6 drinks’. Current tobacco use and prescription medication were both modelled as binary variables (yes/no). Finally, for dietary variables (fruit, vegetables, grains, and sweets intake), participants chose one of the following responses, which were then recoded to the corresponding ordinal variables: (grains): (0) ‘Zero/less than 1 per day’: (1) ‘1–2’: (2) ‘3–4’: (3) ‘5–6’: and (4) ‘7 or more’. (fruits, vegetables): (0) ‘Zero/less than 1 per day’:’(1) ‘1’: (2) ‘2–3’: (3) ‘4–5’: (4) ‘6 or more’. (chocolates/sweets): (0) ‘Less than once per month’: (1) ‘1–3 times per month’: (2) ‘Once per week’: ‘(3) ‘2–4 times per week’: (4) 5–6 times per week’: (5) Once per day’: (6) 2–3 times per day’: (6) ‘4–5 times per day’: (6) ‘6+ times per day’. Sleep was reported as the average amount of sleep you get a day on a three-point scale: (0) ‘Less than 6 hours’: (1) ‘7 to 9 hours’: (2) ‘More than 9 hours’. As the Arivale cohort consists of self-enrolled participants, the response rates for different questionnaires vary. The number of missing values for each response is reported in Extended Data 4.
Health Measures in the MrOS Cohort
We utilized four different health measures that were collected on MrOS participants during their fourth follow-up visit, when stool samples were collected for microbiome analysis. Medication use, self-perceived health, and the Life-Space score (LSC) were all self-reported. Self-perceived health captured each individual’s rating of their own health compared to other individuals their own age. The implementation of the LSC in the MrOS cohort has been described in detail previously and is summarized below 52. The LSC consisted of five levels pertaining to the question: “During the past 4 weeks, have you been to: (1)-“other rooms of your home besides the room where you sleep?”; (2)-“an area outside your home such as your porch, deck, or patio, hallway (of an apartment building), or garage, in your own yard or driveway?”; (3)-“places in your neighborhood, other than your own yard or apartment building?”; (4)-“places outside your neighborhood, but within your town?”; (5)-“places outside your town?”. Within each of these levels, participants answered how often they traveled to that area and whether assistance in the form of equipment or another person was required. The LSC was then calculated by assigning a score to each of the 5 levels outlined above and summing them. Level scores were obtained by multiplying the level number (1–5) by an independence factor (2=no assistance; 1.5=use of equipment only; 1=use of another person with/without equipment) and a frequency factor (1=less than once/week; 2=1–3 times/week; 3=4–6 times/week; and 4=daily). The final LSC measure could range from 0 (restricted to one’s bedroom) to 120 (traveled outside one’s town daily without assistance). We defined healthy individuals as those in the top tertile of the LSC cohort distribution. This corresponded to an LSC value of ≥96. Walking speed was calculated based on the time it took each participant to walk 6 meters (m/s). Like with the LSC, we defined healthy individuals based on walking speed if their speed was in the top tertile (≥1.17). A total of 7 MrOS participants did not have available walking speed data. This is due to either the participants not coming to the clinic, or not being able to attempt the test. These individuals were classified in the walking speed low group in our analysis.
Statistical Analysis
Depending on the statistical approach, analysis was conducted using either R (v 3.6) or Python (v 3.7). The relationship between the calculated uniqueness measure and age in the Arivale cohort was modeled using Ordinary Least Square (OLS) linear regression (Python) where square root transformed Bray-Curtis uniqueness was modeled as the dependent variable and each age decade was compared to the youngest reference group (<30 years), adjusting for sex, BMI, and either genus or ASV-level Shannon diversity, depending on what level the uniqueness measure was calculated. We chose to adjust for Shannon diversity because, in the Arivale cohort, it was associated with both age and microbiome uniqueness (higher α-diversity makes you more likely to be unique). We wanted to assess the significance of our dissimilarity pattern independent of changes in α-diversity seen with age and previously reported in literature. The same adjustment was not made for MrOS participants, since α-diversity measures showed no association with age in that cohort. When assessing the relationship between clinical, lifestyle, and demographic variables with gut microbial uniqueness, Bray-Curtis uniqueness values greater or less than 3 standard deviations from the mean were removed. OLS linear regression was then used to assess the individual relationship between each factor and square root transformed Bray-Curtis gut microbial uniqueness, with microbiome vendor included as a covariate. Percent variance explained by each factor was calculated by taking the percent variance explained by the complete OLS model (variable of interest and microbiome vendor) and subtracting the percent variance explained by microbiome vendor alone. The same analysis was then repeated with age included as a covariate (age-adjusted models). When investigating the relationship between plasma metabolite concentrations and gut microbial uniqueness, each metabolite was log transformed and subsequently scaled and centered. The square root transformed Bray-Curtis uniqueness measure was then regressed against each metabolite individually, adjusting for microbiome vendor, sex, age, age2, a sex*age interaction term, BMI, and Shannon diversity using OLS regression. In each instance where multiple hypotheses were tested, type I error was controlled for using the Bonferroni method (P<0.05). In the MrOS Cohort, correlation between Bray-Curtis Uniqueness and age was calculated using the Python statistical functions package (scipy.stats) using the square root transformed uniqueness measure. Mortality analysis was conducted in R using the package survival (v 2.44–1.1). Relative Bacteroides abundance (after rarefaction) and uniqueness scores were scaled and centered prior to survival analysis. Cox-proportional hazard regression models were generated assessing the relationship between survival and Relative Bacteroides abundance, Bray-Curtis uniqueness or Weighted UniFrac uniqueness independently, adjusting for clinical site, batch (discovery/validation) and age, and adjusting for clinical site, age, BMI, self-perceived health (excellent, good, <good), diagnosis of congestive heart failure, and batch in which stool samples were processed (discovery/validation).
Extended Data
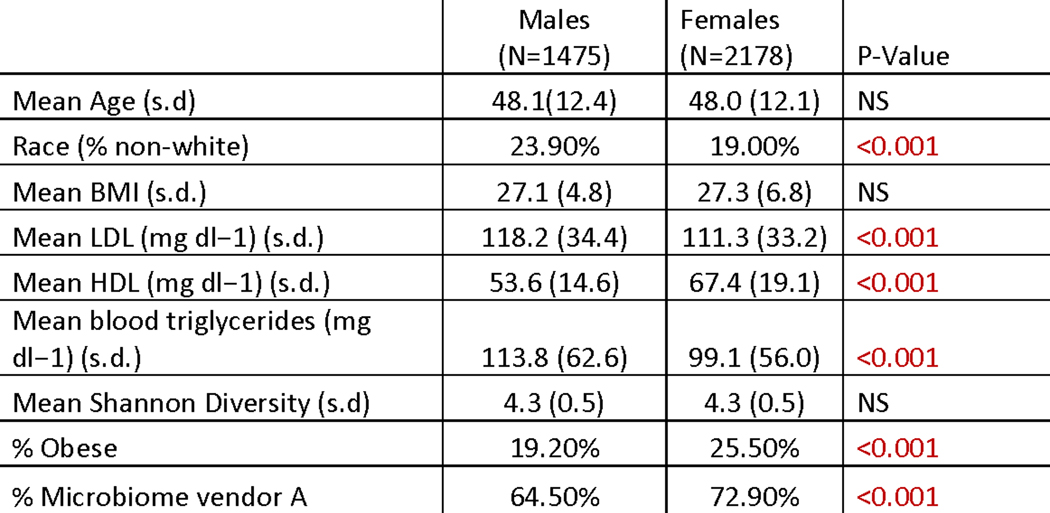
Extended Data Fig. 1. Arivale cohort Demographics table.
For comparisons between males and females, χ2 tests were run for categorical variables and two-sided t-tests for continuous variables. Obese was defined as BMI ≥30. Abbreviations: BMI- body mass index; LDL-low-density lipoprotein cholesterol; HDL-high-density lipoprotein cholesterol, s.d.-standard deviation. P-values <0.05 (two-sided) are colored in red.
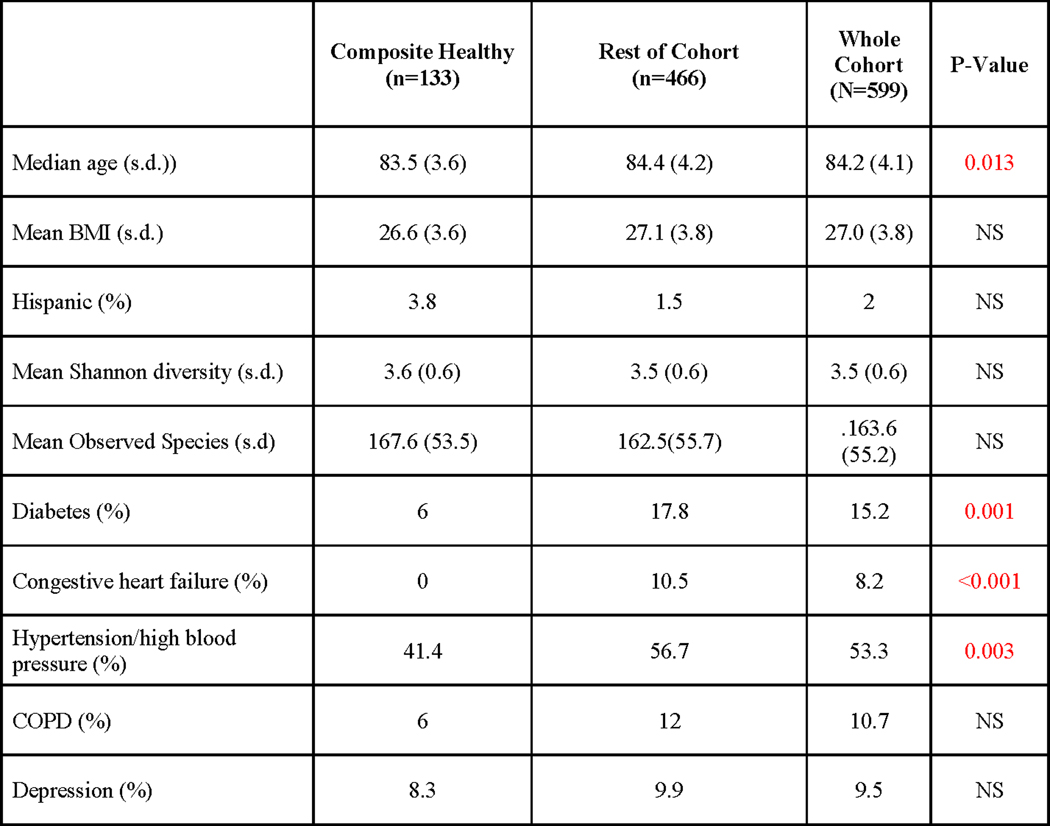
Extended Data Fig. 2. MrOS discovery cohort characteristics table stratified into composite healthy and remainder of cohort.
Statistical tests used to compare groups are as follows: independent samples t-tests were used for comparing age, body mass index (BMI), Shannon diversity and Observed Species; χ2 or Fisher’s exact (if assumptions of χ2 were not met) tests were used to compare ethnicity (percentage Hispanic), and prevalence of each of the specified diseases. P-values <0.05, two-sided are colored in red.
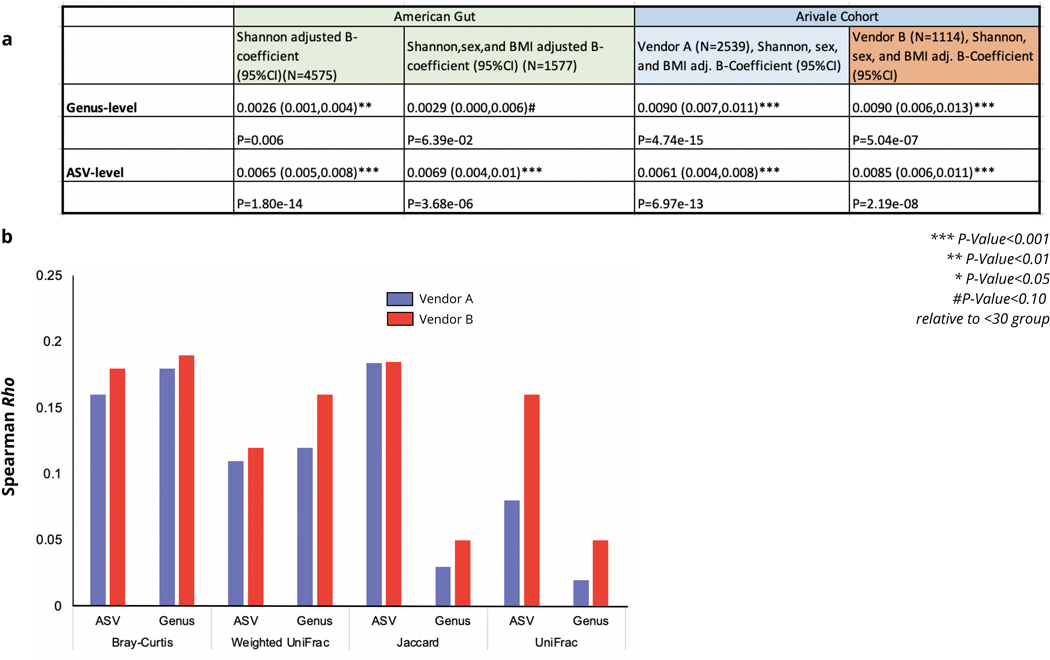
Extended Data Fig. 3. Associations between age and gut microbiome uniqueness across cohorts using different distance metrics.
(a) Age ß-coefficients and corresponding P-values from OLS models predicting Bray-Curtis uniqueness at the ASV- and genus-level in the American Gut Project (AGP) and two vendors in the Arivale cohort. In the AGP dataset, the analysis was performed on all samples, and then repeated on the subset of samples who had available sex and BMI data for covariate adjustment. P-values reported are derived from OLS linear regression models and result from a two-sided hypothesis. (b) Spearman correlations of different ß-diversity metrics with age on both the ASV- and genus-level independently in each vendor used for gut microbiome processing in the Arivale cohort.
Extended Data Fig. 4. Table with associations between Bray-Curtis gut microbiome uniqueness and clinical, demographic, and diet/lifestyle/health measures in the Arivale Cohort.
‘pvalue’ corresponds to the unadjusted P-Value of the ß-coefficient (B-coef column) for each analyte from an OLS model adjusted for gut microbiome vendor. ‘r_squared’ reflects the percent of variance explained beyond microbiome vendor for each analyte independently for the Genus-level Bray-Curtis measure. ‘age_adjusted_coeff’ and ‘age_adjusted_corr_pvalue’ correspond to the ß-coefficient and the Bonferroni corrected P-Value (two-sided) for each analyte predicting Genus-level Bray-Curtis Uniqueness, adjusting for gut microbiome vendor and age. The ‘age_adj_coeff (ASV-level)’ and the ‘age_adj_corr_pvalue (ASV-level)’ correspond to analysis done on the ASV-level Bray-Curtis Uniqueness measure, where models were adjusted for vendor and age. ‘Missing’ shows the number of missing observations for each analyte. Values highlighted in red are statistically significant after multiple-hypothesis correction (Bonferroni P-Value<0.05, two-sided).
Extended Data Fig. 5. Table of associations between Bray-Curtis gut microbiome uniqueness and plasma metabolites in the Arivale cohort.
‘pvalue’ corresponds to the unadjusted P-Value of the ß-coefficient (covariate_adj. Beta_coeff column) for each analyte from an OLS model adjusted for age, age2, sex, a sex*age interaction term, BMI, Shannon diversity, and vendor with Genus-level Bray-Curtis uniqueness as the dependent variable. ‘corr_pval’ corresponds to the Bonferoni corrected P-value. ‘SUPER_PATHWAY’ indicates what pathway the metabolite belongs to. The last three columns are the same as the first three, but for Bray-Curtis uniqueness calculated on the ASV level. All metabolites with an unadjusted P-Value<0.01 are shown. Values highlighted in red are statistically significant after multiple-hypothesis correction (Bonferroni P-Value<0.05, two-sided).
Extended Data Fig. 6. Associations between taxa and gut microbiome uniqueness across cohorts and sex.
(a-d) Plots demonstrating the correlation coefficients between genus-level Bray-Curtis gut microbiome uniqueness and individual taxa in the (a) Discovery MrOS cohort, (b) Vendor A in the Arivale Cohort, (c) Validation MrOS cohort, (d) and vendor B of the Arivale cohort. Only correlations > |0.20|) are shown. (e) Plots demonstrating the strength of correlation between genus-level Bray-Curtis microbiome uniqueness and individual taxa in the in the AGP dataset. The strongest 20 associations are shown. (d-e) Plots demonstrating the strength of correlation between gut microbiome uniqueness and individual taxa in Vendor A of the Arivale cohort in (f) females and (g) males. (h) Scatter plot of correlation coefficients for each genus tested between males and females. The correlation of the coefficients for each genus between sexes is shown. Only genera that had less than 5% zero values and a mean greater than five counts were tested.
Extended Data Fig. 7. Table of Spearman correlation coefficients and Beta-coefficients testing associations between age and uniqueness in the MrOS cohort.
Uniqueness measures reported in this table were calculated at the genus level. ‘Health Stratification’ corresponds to the metric used to define healthy individuals. ‘ Spearman Rho’ reports the Spearman correlation coefficient between age and microbiome uniqueness for the specified group of participants, while the ‘pvalue’ column provides the corresponding p-value. ‘Beta_coeff’ is the BMI adjusted age beta-coefficient predicting uniqueness across the same stratifications as the ‘Spearman Rho’ column. ‘Coef_pvalue’ provides the p-value corresponding to the age Beta-coefficient from linear regression models. ‘Sample_size’ is the number of participants in each stratification while the last column “Healthy (yes=1/no=0)” specifies whether the group of participants is the healthy subgroup (yes(1)), or the remainder of the cohort (No(0)). Significant p-values (P<0.05, two-sided) are highlighted in red. No multiple hypothesis correction was performed.
Extended Data Fig. 8. Associations between age and gut microbiome measures across health stratifications in the MrOS cohort.
(a-e) Plots demonstrating the strength of Spearman correlation between age and gut microbiome measures at different taxonomic resolutions. (a) The blue/red panel corresponds to the calculated Weighted UniFrac (ß-diversity) uniqueness score at the genus level, while (b) the grey/green and (c) grey/yellow panels correspond to Shannon diversity and Observed species (α-diversity measures) at the ASV level, respectively. Significant correlations (two-sided) are indicated with asterisks. Exact correlation coefficients and corresponding p-values for (a) are provided in Extended Data 7. (d-e) The same plots as in (b-c), with α-diversity calculated at the genus level. (f) Comparison of ASV level and genus-level analysis in healthy aging in the MrOS cohort. Barplots represent correlation coefficients comparing age and uniqueness at the ASV level across composite healthy MrOS individuals, and the remainder of the cohort in both the discovery and validation groups. (g) ß -coefficients for age from OLS regression models predicting genus-level Bray-Curtis uniqueness in healthy composite individuals and remainder of the cohort, adjusted individually for the most commonly reported supplements and medications in the MrOS cohort.
Acknowledgements
We thank C. Funk for helpful discussions throughout the course of this project. We also thank J. Dougherty and M. Brunkow for their coordination efforts.
Funding: This work was supported by the M.J. Murdock Charitable Trust (L.H. and N.D.P.), Arivale and a generous gift from C. Ellison. S.M.G., C.D. and S.P. were supported by a Washington Research Foundation Distinguished Investigator Award and by start-up funds from the Institute for Systems Biology. Further support comes from the National Academy of Medicine Catalyst Award (N.D.P., S.M.G., L.H., E.S.O.) and NIH grant U19AG023122 awarded by the National Institute on Aging (NIA). The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health funding. The following institutes provide support: the NIA, the National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS), the National Center for Advancing Translational Sciences (NCATS), and NIH Roadmap for Medical Research under the following grant numbers: U01 AG027810, U01 AG042124, U01 AG042139, U01 AG042140, U01 AG042143, U01 AG042145, U01 AG042168, U01 AR066160, and UL1 TR000128. NIAMS grant R01AR061445 provided support for the generation of 16S genotyping data.
Footnotes
Competing interests: The authors declare no competing interests.
Code Availability: Code used to process gut microbiome samples is available on the Gibbons lab GitHub page (https://github.com/gibbons-lab/mbtools) and code used for statistical analysis is available through the Hood-Price lab GitHub (https://github.com/PriceLab/AgingMicrobiome).
Data availability: Qualified researchers can access the full Arivale deidentified dataset supporting the findings in this study for research purposes. Requests should be sent to andrew.magis@isbscience.org. The MrOS data set is available to researchers through the following website: https://mrosdata.sfcc-cpmc.net. The data is available to qualified researchers upon submission and approval of a research plan. The American Gut Project biom table and the accompanying metadata is publicly available through figshare, reference numbers [6137192]49 and [6137192]50, respectively.
References:
- 1.Yatsunenko T. et al. Human gut microbiome viewed across age and geography. Nature 486, 222–227 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Toole PW & Jeffery IB Gut microbiota and aging. Science (80-. ). 350, 1214–1215 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Castro-Mejía JL et al. Physical fitness in community-dwelling older adults is linked to dietary intake, gut microbiota, and metabolomic signatures. Aging Cell (2020). doi: 10.1111/acel.13105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson M. et al. Signatures of early frailty in the gut microbiota. Genome Med. (2016). doi: 10.1186/s13073-016-0262-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Claesson MJ et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 488, 178–184 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Biagi E. et al. Gut Microbiota and Extreme Longevity. Curr Biol 26, 1480–1485 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Kim BS et al. Comparison of the Gut Microbiota of Centenarians in Longevity Villages of South Korea with Those of Other Age Groups. J. Microbiol. Biotechnol (2019). doi: 10.4014/jmb.1811.11023 [DOI] [PubMed] [Google Scholar]
- 8.Wu L. et al. A Cross-Sectional Study of Compositional and Functional Profiles of Gut Microbiota in Sardinian Centenarians. mSystems 4, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong F. et al. Gut microbiota signatures of longevity. Current Biology (2016). doi: 10.1016/j.cub.2016.08.015 [DOI] [PubMed] [Google Scholar]
- 10.Kong F, Deng F, Li Y. & Zhao J. Identification of gut microbiome signatures associated with longevity provides a promising modulation target for healthy aging. Gut Microbes (2019). doi: 10.1080/19490976.2018.1494102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vangay P. et al. US Immigration Westernizes the Human Gut Microbiome. Cell (2018). doi: 10.1016/j.cell.2018.10.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Claesson MJ et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci U S A 108 Suppl, 4586–4591 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffery IB, Lynch DB & O’Toole PW Composition and temporal stability of the gut microbiota in older persons. ISME J. (2016). doi: 10.1038/ismej.2015.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh TS, Das M, Jeffery IB & O’Toole PW Adjusting for age improves identification of gut microbiome alterations in multiple diseases. Elife (2020). doi: 10.7554/eLife.50240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tierney BT et al. The predictive power of the microbiome exceeds that of genome-wide association studies in the discrimination of complex human disease. bioRxiv 2019.12.31.891978 (2020). doi: 10.1101/2019.12.31.891978 [DOI] [Google Scholar]
- 16.Salosensaari A. et al. Taxonomic Signatures of Long-Term Mortality Risk in Human Gut Microbiota. medRxiv 2019.12.30.19015842 (2020). doi: 10.1101/2019.12.30.19015842 [DOI] [Google Scholar]
- 17.Zubair N. et al. Genetic Predisposition Impacts Clinical Changes in a Lifestyle Coaching Program. Sci. Rep (2019). doi: 10.1038/s41598-019-43058-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blank JB et al. Overview of recruitment for the osteoporotic fractures in men study (MrOS). Contemp. Clin. Trials (2005). doi: 10.1016/j.cct.2005.05.005 [DOI] [PubMed] [Google Scholar]
- 19.Abrahamson M, Hooker E, Ajami NJ, Petrosino JF & Orwoll ES Successful collection of stool samples for microbiome analyses from a large community-based population of elderly men. Contemp. Clin. Trials Commun (2017). doi: 10.1016/j.conctc.2017.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDonald D. et al. American Gut: an Open Platform for Citizen Science Microbiome Research. mSystems (2018). doi: 10.1128/msystems.00031-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gibbons SM, Duvallet C. & Alm EJ Correcting for batch effects in case-control microbiome studies. PLoS Comput. Biol (2018). doi: 10.1371/journal.pcbi.1006102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wilmanski T. et al. Blood metabolome predicts gut microbiome α-diversity in humans. Nat. Biotechnol (2019). doi: 10.1038/s41587-019-0233-9 [DOI] [PubMed] [Google Scholar]
- 23.Rampelli S. et al. Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany. NY) (2013). doi: 10.18632/aging.100623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Collino S. et al. Metabolic Signatures of Extreme Longevity in Northern Italian Centenarians Reveal a Complex Remodeling of Lipids, Amino Acids, and Gut Microbiota Metabolism. PLoS One (2013). doi: 10.1371/journal.pone.0056564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vich Vila A. et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun (2020). doi: 10.1038/s41467-019-14177-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nemet I. et al. A Cardiovascular Disease-Linked Gut Microbial Metabolite Acts via Adrenergic Receptors. Cell (2020). doi: 10.1016/j.cell.2020.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han H. et al. p-Cresyl sulfate aggravates cardiac dysfunction associated with chronic kidney disease by enhancing apoptosis of cardiomyocytes. J. Am. Heart Assoc (2015). doi: 10.1161/JAHA.115.001852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis KN, Rubinstein ND & Buffenstein R. A window into extreme longevity; the circulating metabolomic signature of the naked mole-rat, a mammal that shows negligible senescence. GeroScience (2018). doi: 10.1007/s11357-018-0014-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roager HM & Licht TR Microbial tryptophan catabolites in health and disease. Nature Communications (2018). doi: 10.1038/s41467-018-05470-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonowal R. et al. Indoles from commensal bacteria extend healthspan. Proc. Natl. Acad. Sci. U. S. A (2017). doi: 10.1073/pnas.1706464114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krishnan S. et al. Gut Microbiota-Derived Tryptophan Metabolites Modulate Inflammatory Response in Hepatocytes and Macrophages. Cell Rep. (2018). doi: 10.1016/j.celrep.2018.03.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beaumont M. et al. The gut microbiota metabolite indole alleviates liver inflammation in mice. FASEB J. (2018). doi: 10.1096/fj.201800544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexeev EE et al. Microbiota-Derived Indole Metabolites Promote Human and Murine Intestinal Homeostasis through Regulation of Interleukin-10 Receptor. Am. J. Pathol (2018). doi: 10.1016/j.ajpath.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lustgarten MS, Price LL, Chalé A. & Fielding RA Metabolites related to gut bacterial metabolism, peroxisome proliferator-activated receptor-alpha activation, and insulin sensitivity are associated with physical function in functionally-limited older adults. Aging Cell (2014). doi: 10.1111/acel.12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cussotto S. et al. Tryptophan Metabolic Pathways Are Altered in Obesity and Are Associated With Systemic Inflammation. Front. Immunol (2020). doi: 10.3389/fimmu.2020.00557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Márquez EJ et al. Sexual-dimorphism in human immune system aging. Nat. Commun (2020). doi: 10.1038/s41467-020-14396-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bárcena C. et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nature Medicine (2019). doi: 10.1038/s41591-019-0504-5 [DOI] [PubMed] [Google Scholar]
- 38.Kundu P. et al. Neurogenesis and prolongevity signaling in young germ-free mice transplanted with the gut microbiota of old mice. Sci. Transl. Med (2019). doi: 10.1126/scitranslmed.aau4760 [DOI] [PubMed] [Google Scholar]
- 39.Zwielehner J. et al. Combined PCR-DGGE fingerprinting and quantitative-PCR indicates shifts in fecal population sizes and diversity of Bacteroides, bifidobacteria and Clostridium cluster IV in institutionalized elderly. Exp. Gerontol (2009). doi: 10.1016/j.exger.2009.04.002 [DOI] [PubMed] [Google Scholar]
References:
- 40.Earls JC et al. Multi-Omic Biological Age Estimation and Its Correlation With Wellness and Disease Phenotypes: A Longitudinal Study of 3,558 Individuals. J. Gerontol. A. Biol. Sci. Med. Sci 74, S52–S60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cawthon PM, Shahnazari M, Orwoll ES & Lane NE Osteoporosis in men: Findings from the Osteoporotic Fractures in Men Study (MrOS). Therapeutic Advances in Musculoskeletal Disease (2016). doi: 10.1177/1759720X15621227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Callahan BJ et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13, 581–583 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright ES DECIPHER: Harnessing local sequence context to improve protein multiple sequence alignment. BMC Bioinformatics (2015). doi: 10.1186/s12859-015-0749-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price MN, Dehal PS & Arkin AP FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS One (2010). doi: 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMurdie PJ & Holmes S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One (2013). doi: 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bray JR & Curtis JT An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecol. Monogr (1957). doi: 10.2307/1942268 [DOI] [Google Scholar]
- 47.Lozupone C. & Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol (2005). doi: 10.1128/AEM.71.12.8228-8235.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin BD, Witten D. & Willis AD Modeling microbial abundances and dysbiosis with beta-binomial regression. Ann. Appl. Stat (2020). doi: 10.1214/19-AOAS1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDonald D. et al. American Gut Project fecal sOTU counts table. (2018). doi: 10.6084/m9.figshare.6137192.v1 [DOI] [Google Scholar]
- 50.McDonald D. et al. Full American Gut Project mapping file. (2018). doi: 10.6084/m9.figshare.6137315.v1 [DOI] [Google Scholar]
- 51.Manor O. et al. A Multi-omic Association Study of Trimethylamine N-Oxide. Cell Rep. (2018). doi: 10.1016/j.celrep.2018.06.096 [DOI] [PubMed] [Google Scholar]
- 52.Mackey DC et al. Life-space mobility and mortality in older men: A prospective cohort study. J. Am. Geriatr. Soc (2014). doi: 10.1111/jgs.12892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Maier L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature (2018). doi: 10.1038/nature25979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Charlesworth CJ, Smit E, Lee DSH, Alramadhan F. & Odden MC Polypharmacy among adults aged 65 years and older in the United States: 1988—2010. Journals of Gerontology - Series A Biological Sciences and Medical Sciences (2015). doi: 10.1093/gerona/glv013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Machón M, Vergara I, Dorronsoro M, Vrotsou K. & Larrañaga I. Self-perceived health in functionally independent older people: Associated factors. BMC Geriatr. (2016). doi: 10.1186/s12877-016-0239-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Idler EL & Benyamini Y. Self-Rated Health and Mortality: A Review of Twenty-Seven Community Studies. Journal of Health and Social Behavior (1997). doi: 10.2307/2955359 [DOI] [PubMed] [Google Scholar]
- 57.Mossey JM & Shapiro E. Self-rated health: a predictor of mortality among the elderly. Am. J. Public Health (1982). doi: 10.2105/AJPH.72.8.800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peel C. et al. Assessing Mobility in Older Adults: The UAB Study of Aging Life-Space Assessment. Phys. Ther (2005). doi: 10.1093/ptj/85.10.1008 [DOI] [PubMed] [Google Scholar]
- 59.Middleton A, Fritz SL & Lusardi M. Walking speed: The functional vital sign. Journal of Aging and Physical Activity (2015). doi: 10.1123/japa.2013-0236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mielke MM et al. Assessing the temporal relationship between cognition and gait: Slow gait predicts cognitive decline in the mayo clinic study of aging. Journals Gerontol. - Ser. A Biol. Sci. Med. Sci (2013). doi: 10.1093/gerona/gls256 [DOI] [PMC free article] [PubMed] [Google Scholar]