Abstract
The cell surface receptor Neuropilin-1 (Nrp1) was recently identified as a host factor for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) entry. The Spike protein of SARS-CoV-2 is cleaved into two segments, the S1 (residues (res.) 1–685) and the S2 (res. 686–1273) domains by furin protease. Nrp1 predominantly binds to the C-terminal RRAR amino acid motif (res. 682–685) of the S1 domain. In this study, we firstly modeled the association of an Nrp1 protein (consisting of domains a2-b1-b2) with the Spike protein. Next, we studied the separation of S2 from the S1 domain, with and without Nrp1 bound, by utilizing molecular dynamics pulling simulations. During the separation, Nrp1 stabilizes the S1 C-terminal region (res. 640–685) and thereby assists the detachment of S2 N-terminal region (res. 686–700). Without Nrp1 bound, S1 tends to become stretched, whereas the bound Nrp1 stimulates an earlier separation of S2 from the S1 domain. The liberated S2 domain is known to mediate the fusion of virus and host membranes; thus, Nrp1 likely increases virus infectivity by facilitating the S1 and S2 separation. We further analyzed the possible topological structure of the SARS-CoV-2 Spike protein when bound with Nrp1 and angiotensin-converting enzyme 2 (ACE2). Understanding of such an Nrp1-assisted viral infection opens the gate for the generation of protein-protein inhibitors, such as antibodies, which could attenuate the infection mechanism and protect certain cells in a future Nrp1-ACE2 targeted combination therapy.
Significance
Here, we explore the separation mechanism for the Spike protein by computational simulation, utilizing pulling molecular dynamics. The study provides insight into the first steps in the molecular mechanism of virus infection, especially because it suggests how the newly discovered virus co-receptor Neuropilin-1 facilitates separation of two subunits of the virus Spike protein prior to virus-host cell membrane fusion.
Introduction
The virus severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is responsible for the world-wide 2020 and 2021 coronavirus disease 2019 (COVID-19) pandemic. It is increasingly appreciated for its deleterious effects on the human nervous and cardiovascular systems, including longer-term symptoms in some of the patients. SARS-CoV-2 is known to associate with the host cell surface receptor angiotensin-converting enzyme 2 (ACE2) via the viral surface glycoprotein: the Spike or S protein (1). Upon the initial contact of Spike protein and ACE2, the viral Spike protein can be further cleaved via a host cell protease furin, which appears to be a crucial step of SARS-CoV-2 for cell entry (2,3). This type of cleavage generates two Spike protein fragments, S1 and S2 (4,5). S2 displays a membrane fusion domain, which is capped by S1 in the structure of the viral Spike protein. The separation of S2 from S1 is needed to expose the membrane fusion domain, which then triggers fusion between virus and host cell, allowing the flow of virus RNA into the host cell for replication, protein synthesis, and new virus assembly (3). S1 displays a C-terminal polypeptide motif with the sequence RRAR (4,5). This amino acid motif was recently found to be recognized by another human cell surface receptor Neuropilin-1 (Nrp1) (6, 7, 8). Following this observation, Nrp1 was classified as a novel host factor that may work together with ACE2 in SARS-CoV-2 entry.
Neuropilin (Nrp,with two forms of Neuropilin-1 and Neuropilin-2) is a transmembrane protein receptor active in neurons and other cells. It can interact with several proteins that have a C-end rule (CendR) sequence—a motif of the R/KXXR/K sequence exposed at the protein’s C-terminus, allowing them to bind to neuropilin. One of the most well-known binding partners of neuropilin is the vascular endothelial growth factor (VEGF) (9, 10, 11). Nrps, together with the VEGF receptor, VEGFR, are involved in the development and function of axons and synapses. VEGF binding to VEGFR:Nrp complexes are crucial events in the growth and leakiness of blood vessels. Thus, the Spike protein binding with Nrp1 may directly or indirectly interfere with the formation of the endogenous receptor complex, with its signaling and functions in the host cell (12, 13, 14). This connection between the SARS-CoV-2 Spike protein and neuropilin may relate the longer-term effects of COVID-19 on the biological processes in neurons and blood vessels (15, 16, 17). Specifically, a significant number of critically ill COVID-19 patients experience impairments such as brain inflammation, an increased permeability of the blood-brain barrier, as well as an elevated occurrence in the number of strokes (18, 19, 20, 21). These observations make the Nrp1:SARS-CoV-2 interaction an urgent focus for investigation.
The S1 RRAR amino acid motif is the predominant recognition site between Nrp1 and the S1 subunit. However, a construct of the Spike protein with the deletion of the S1 RRAR motif still interacts with Nrp1 (6). Therefore, the binding between Nrp1 and the Spike protein also appears to involve other regions on both proteins. In addition, the underlying molecular mechanism by which Nrp1 facilitates SARS-CoV-2 viral infection is not yet understood. In this study, we predict possible binding models between the Nrp1 a2-b1-b2 domain-containing protein with the ACE2-bound Spike protein (Fig. 1 a). By considering topological constraints, our docking calculations identify probable binding models of Nrp1 with the Spike protein trimer. Importantly, molecular dynamics pulling simulations suggest that the separation of S2 from the S1 domain is assisted by the interaction with Nrp1, proposing a molecular mechanism by which Nrp1 helps a viral attack by stimulating Spike protein mediated infection.
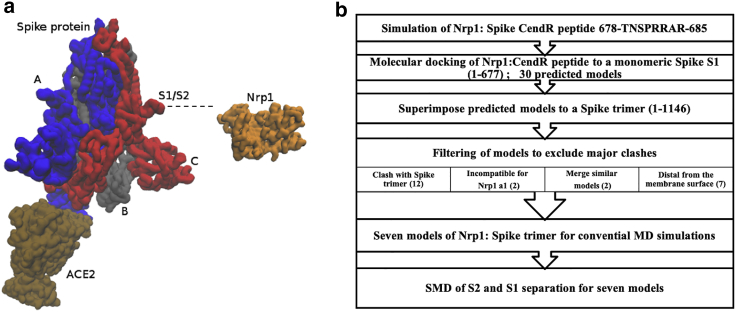
Figure 1.
Overview of a Spike protein trimer with ACE2 and Nrp1 and the workflow of the study. (a) ACE2 binds to Spike trimer RBD (chain A). Nrp1 associates with Spike trimer at the S1-S2 cleavage site (chain C). (b) Flow chart of the computational modeling and simulation approach utilized in this study. To see this figure in color, go online.
Materials and methods
The overall flow chart of the study is shown in Fig. 1 b. All simulated systems mentioned earlier are listed in Table S1.
Complex of Nrp1-a2-b1-b2-domain-containing protein with Spike protein segment 678-TNSPRRAR-685
The polybasic sequence 678-TNSPRRAR-685 of the Spike protein fragment S1 associates with the b1 domain of Nrp1 (6). We firstly modeled a complex of an Nrp1 (a2-b1-b2-domain-containing) protein bound to this polybasic peptide. The structure of this human Nrp-1 fragment was built, based on available crystal structure (Protein Data Bank, PDB: 2qqm). Missing loops in the crystal structure were added using MODELLER (22). The VEGF-A:Nrp1 b1 domain complex (PDB: 4deq) was used as a template to construct the model of the 678-TNSPRRAR-685 peptide bound to Nrp1 (11). The modeled complex was set up and then relaxed for 20 ns at 310 K by an all-atom molecular dynamics simulation. In the simulation, the association between the 678-TNSPRRAR-685 segment and the Nrp1 b1 domain was largely maintained by electrostatic attraction of Spike S1 residues res. R683 and R685 with Nrp1 ress. D320 and D348, as described in the study of Daly et al. (6). The final simulation structure of the complex of Nrp1:678-TNSPRRAR-685 motif was extracted and used for further molecular docking.
Molecular docking of Nrp1-678-TNSPRRAR-685 motif to a monomeric Spike S1 (res.1–677)
To identify binding sites (between Nrp1 and the Spike protein) other than the RRAR motif, the relaxed structure of the Nrp1 C-terminal octamer peptide obtained from the earlier simulation was treated as a ligand and was then docked to the receptor—here, a monomeric Spike S1 (res. 1–677) with ClusPro 2.0 (23). The receptor structure of Spike protein (res. 1–1146) was based on the available crystal structure (PDB: 6vsb) and models (adding missing loops and residues) developed by Woo et al. (24,25). The ligand in this case consisted of Nrp1 (a2-b1-b2) and the Spike protein 678-TNSPRRAS-685 motif. The -N-terminus of the peptide (res. T678) needs to fuse with residue Q677 of the receptor Spike S1. So, a distance restraint of 3 Å was applied between residues 677 and 678 in the docking. For simplicity, glycosylation of the Spike protein was not considered in the docking. Finally, ClusPro 2.0 gave 30 predicted models.
Filtration of seven highly probable binding models of Nrp1:monomeric S1
Next, we excluded some models based on several criteria (Fig. 1 b). First, structures that clash with the Spike protein trimer were excluded. Because the docking was done between Nrp1 with a monomeric S1 (chain C); when the predicted models are superimposed to a trimeric Spike protein, some of them clash with either S2 of chain C or chains A and B. In total, 12 models were excluded in this step (see representative instances in Fig. S1 a). In two models, the N-terminal of the Nrp1 domain a2 was buried inside the space between chain C N-terminal domain (NTD), chain B, and chain C receptor-binding domain (RBD), leaving no space to place Nrp1's a1 domain(see Fig. S1 b). In the other two cases, the steric position of Nrp1 relative to S1 was similar (root mean-square deviation of 1.2 or 1.5 nm, respectively; see Fig. S1 c), we only kept one of two models with the better docking score. Sixteen models were excluded so far. Finally, we also excluded seven models that position the C-terminus of the Nrp1 b2 domain (res. 586) too away from the membrane (see representative models in Fig. S1 d). We will further discuss the topological constraint in the Results and discussion. Altogether, after the filtering, we focused on seven models of Nrp1-a2-b1-b2: Spike-protein binding, which fulfilled these criteria (see Fig. 2 a).
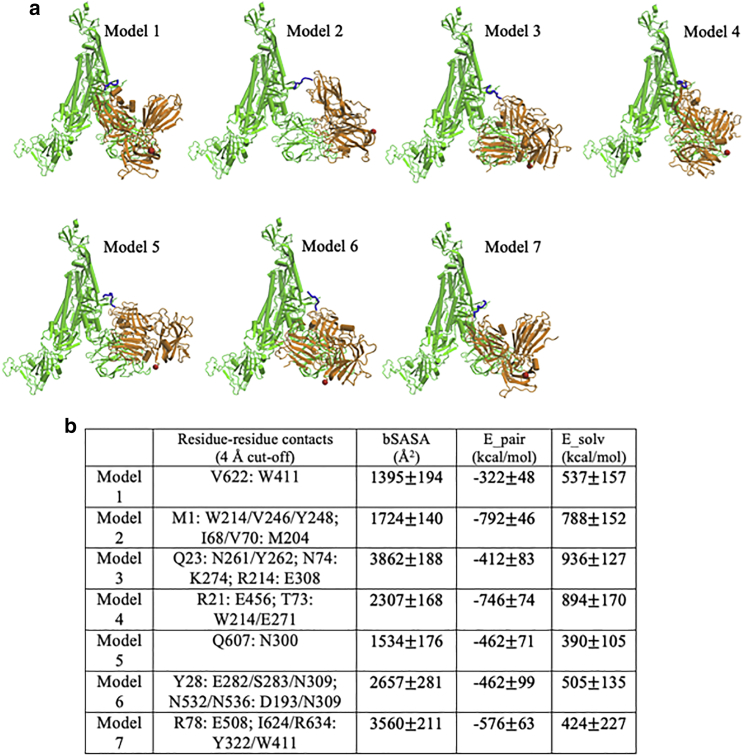
Figure 2.
Binding modes for seven predicted models. (a) Models, shown for Spike trimer (chain-C): Nrp1 (a2-b1-b2) binding. The TNSPRRAR motif is marked in blue. The last residue of the Nrp1 a2-b1-b2 domain segment is marked in red. (b) Noticeable residue-residue contacts (<0.4 nm) between Spike protein and Nrp1 in different models (except for the RRAR binding region). Together with additional residue-residue contacts (<0.6 nm), the proteins are seen to interact via domains a2, b1, and b2 on Nrp1 and a limited number of regions N1–N4, RBDN, RBDC, C1–C3, the very C-terminus of S1 and beginning of S2 (see Fig. S3 for details). Listed in Table b are the buried solvent-accessible surface area (SASA), pair-interaction energy, and changes of solvation energy for the different models. The interaction energies (based upon pairwise vdW and electrostatic forces) are calculated between Nrp1 (domains a2-b1-b2) with a full Spike trimer (chains A, B, and C). The models shown are listed in order of their population near their cluster centers (most to least). Standard errors were calculated over different frames of the simulation trajectories (saved at every 100 ps). To see this figure in color, go online.
Molecular simulation of Nrp1-Spike-protein trimer
The seven models of Nrp1:monomeric-S1 (res. 1–677) were considered for further analysis. Firstly, the modeled complex was superimposed with the full-length Spike protein trimer (res. 1–1146; chain C was used for superimposition). To relax and better accommodate the initial docked structures in the context of the full-length (1–1146) Spike protein trimer, we performed all-atom molecular dynamics simulation with one Nrp1 bound to the Spike protein trimer for each of the seven models for 20 ns. Standard simulation procedures were used with the NAMD version 2.12 package (26). Specifically, all proteins were solvated by TIP3P water with 150 mM NaCl. Simulation parameters were set as 2 fs for time step, thermostat at 310 K, and barostat of 1 bar, using the CHARMM36m force field (27). The system contained ∼530,000 atoms in a cubic periodic boundary, including water and ions. The root mean-square deviation of the Nrp1: Spike complex is plotted in Fig. S2, showingthat all the simulations are equilibrated (perhaps with exception of model 3). The pair interaction, solvation energy, and surface-accessible surface area between Nrp1 and Spike were calculated and listed in Fig. 2 b.
Steered molecular dynamics simulation of the separation of S2 domain from S1 subunit
We applied steered molecular dynamics with Generalized Born Implicit Solvent method to investigate the separation of S2 from the S1 unit (28). In the implicit solvent simulation, the cutoff for calculating the Born radius was set as 15 Å. A cutoff of 14 Å was used for calculating the interactions between atoms. Ion concentration was set as 150 mM. For the steered molecular dynamics simulations, a time step of 1 fs was used. The S2 domain was pulled out of S1 domain with a constant velocity of 0.00001 Å per time step over 20 ns of simulations, run in duplicate with different velocity starting seeds. A force constant of 25 kcal/(mol.Å2) was used for the harmonic spring that generates the force to pull S2 away from S1. During the pulling, the ACE2 protein as well as Nrp1 were restrained. To resist the drift of the S1 domain, the RBDs (res. 318–541) of three Spike protein units were also restrained. The principal axis of the Spike protein trimer was aligned to the Z direction. The pulling direction was set in the Z direction. Because of the application of biasing force, the association between Nrp1 and S1 678-TNSPRRAR-685 motif could be quickly broken during the pulling. This happened for models 2, 3, and 6 and especially for model 5. To study the effect of Nrp1 binding on the S1:S2 dissociation, we excluded simulations in which Nrp1: S1-TNSPRRAR associations were not maintained during the pulling.
The pulling force was plotted versus the displacement of the S2 domain. The total work was calculated as the cumulative work over all steps . It should be noted that the pulling work can be significantly overestimated because the pulling was done over a very limited timescale (20 ns) (29, 30, 31). For an estimate of the free energy, methods such as umbrella sampling simulations could be utilized (32). Here, the main purpose of pulling is to qualitatively but not quantitatively estimate the energy barrier for separating the S2 and S1 subunits, especially to observe the separation details at the Nrp1:Spike S and, the S1:S2 interface, including around the S1-S2 cleavage site. We chose an implicit solvent model for the steered molecular dynamics for the following reasons: to pull S2 out of S1, the system needs to be long enough in the Z direction (estimated >45 nm). For an explicit solvent simulation, we estimated the system would have >1,325,000 atoms. In addition to the huge size of the simulation system, the greater challenge is that water molecules would find it hard to fit in the space between S1 and S2 subunits during a pulling over a limited simulation time. The S2 subunit is big in size. During the pulling, an excessive pressure would be generated by pushing away the many water molecules in a size-limited simulation box because the water molecules have not enough time to relax. Therefore, although generally the explicit solvent model would probably give a better characterization of the detailed molecular interactions, here, for the pulling simulation of a sizeable protein subunit (S2), an explicit solvent simulation would introduce a high level of noise. In the implicit solvent model, the water interaction is modeled as a dielectric continuum. Importantly, the direct protein-protein interaction and protein entropy components are less affected by the choice of the implicit versus the explicit solvent model. So, we chose an implicit solvent model in this study for the pulling simulation.
Model construction of Spike-Nrp1-ACE2 complex with viral and host membrane
We used the membrane-bound ACE2 (PDB: 6m17) built with CHARMM GUI as a template. Next, the Spike protein is docked to ACE2 based on the template structure of the ACE2:RBD complex. Specifically, the “up” RBD of chain A was bound to ACE2. The Spike protein (1–1174) was tilted relative to the membrane because it was bound to an ACE2. Next, we fused the Spike protein to the transmembrane helix segment (Carbon Alpha, CA distance ∼0.1 nm), again built with CHARMM GUI. The distance between the viral and host membrane was estimated around 30 nm. Then, Nrp1 (a2-b1-b2) is docked to the Spike protein with templates of model 1–7.
Results and discussion
Binding modes of between Nrp1 and Spike protein
The simulated binding models between Spike protein with an Nrp1 are shown in Fig. 2. Sterically, for all the seven models, the Nrp1 are located in between the NTD (res. 13–305) and the bottom of the RBD (res. 509–541) of the Spike protein. Within this region, Nrp1 has different domain orientations relative to the S1 subunit (Fig. 2 a). Beyond the major interaction site between the Nrp1 and the S1 CendR (681-RRAR-685) motif, in all seven systems, there are several additional interactions between the Nrp1 and the Spike protein. We summarized major interaction residues/regions in Fig. 2 b and Fig. S3. On the side of the S1 subunit, S1 NTD (res. 13–305), the N- and C-termini of the RBD (res. 318–437; res. 509–541), and the C-terminal S1 regions are major binding sites. On the side of the Nrp1, each one of the three domains (a1, b1, and b2) more or less make contact with the Spike protein. The Nrp1 domains as well as the Spike protein binding regions are relatively extended, with only three pairs of the 12 defined regions in Fig. S3 being in close proximity (N2–N4, RBDN-RBCC, and C2–C3). Several models overlap with respect to at least some of the regions, which suggests that a certain type of interface is involved in forming a complex. Intriguingly, there is only one model that has all three Nrp1 domains involved, and one with only one domain (b1) involved (models 4 and 5, respectively). Models involving Nrp1 domains a2, b1, or b2 are considerably varied in the regions that are being contacted on the side of the Spike protein.
Looking at models 1, 2, and 7 at the residue level, noticeable hydrophobic contacts such as V622:W411, I68/V70:M204, or I624/R634:Y322/W411 are seen. In models 3, 4, and 7, electrostatic pairs such as R21:E456, R214:E308, or R78:E507 are apparent. The remaining contacts are mostly established between polar amino acids with other polar, charged, and aromatic amino acid side chains, which have the possibility to form hydrogen bonds. Experimentally, a relatively weak binding affinity (13.0 or 20.3 μM) was measured between the Nrp1 b1 domain and the Spike S1 CendR peptide (679–685) (6). Additional contacts beyond the CendR peptide may help to strengthen the Nrp1:Spike protein complex. The calculated buried surface area, paired interaction as well as solvation interaction energy between Nrp1 and Spike protein are listed for reference in Fig. 2 b. At this point, however, we have no strong evidence that any one of the seven models may be more strongly bound than the remaining ones. Thus, currently, we suggest that rather than forming a single or very few well-defined complex structures with specific domain-domain interactions, several binding interfaces may coexist and can interconvert with each other, similar to other dynamic protein complexes (e.g., K-Ras at membrane) (33,34). It is worth mentioning that Nrp1 does not interact with the RBD’s motif for binding ACE2 (res. 437–538); thus, the ACE2 association with the Spike protein should be unperturbed unless there are allosteric effects (35, 36, 37). Moreover, for simplicity, glycosylation of the Spike protein is not considered in the docking and simulation. The Spike protein has multiple glycosylation sites in the S1 domain. The great majority of these sites (N122, N149, N165, N234, N282, N331, N343, N603, and N657) are distant from the binding interface between Nrp1 and S1. Only N17, N74, N323, and N616 are within or close to the interacting regions between Nrp1 and S1 (Fig. S4). These four glycosylation sites may have an influence on Nrp1 binding, but this issue is beyond the scope of this report and is deferred to a future article that will also include experimental data on the system.
The association of Nrp1 with the Spike protein relies on the cleavage of Spike protein into the S1 and S2 domain segments by furin protease. The purpose of the cleavage is to free the S2 subunit for subsequent viral-host membrane fusion. We inferred that the Nrp1 may have a role in the separation process of S1 and S2. Next, instead of investigating further details on the Nrp1: Spike protein binding modes, we explore a possible role of the Nrp1 in modulating the separation of the Spike protein S2 and S1 segments.
The separation of S2 from the S1 domain
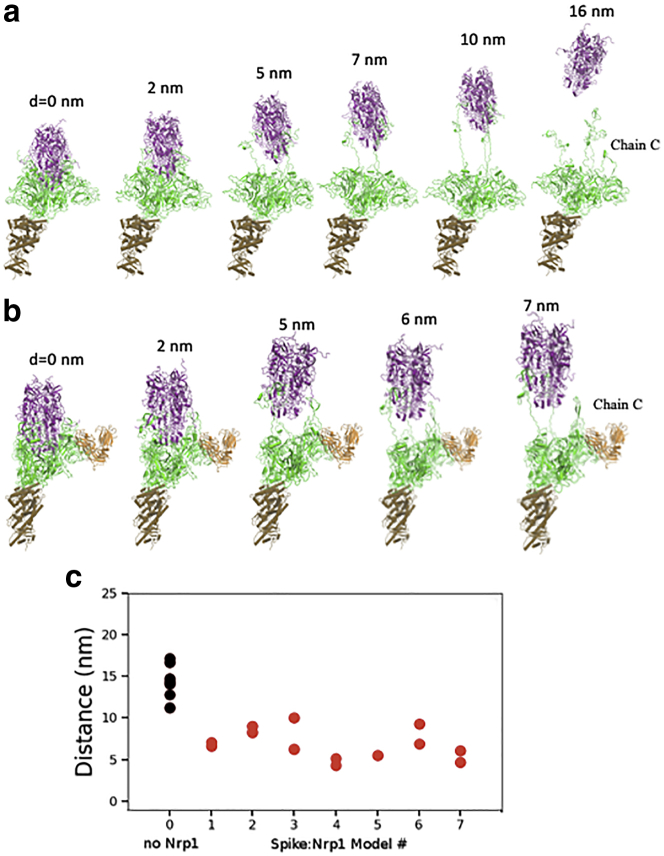
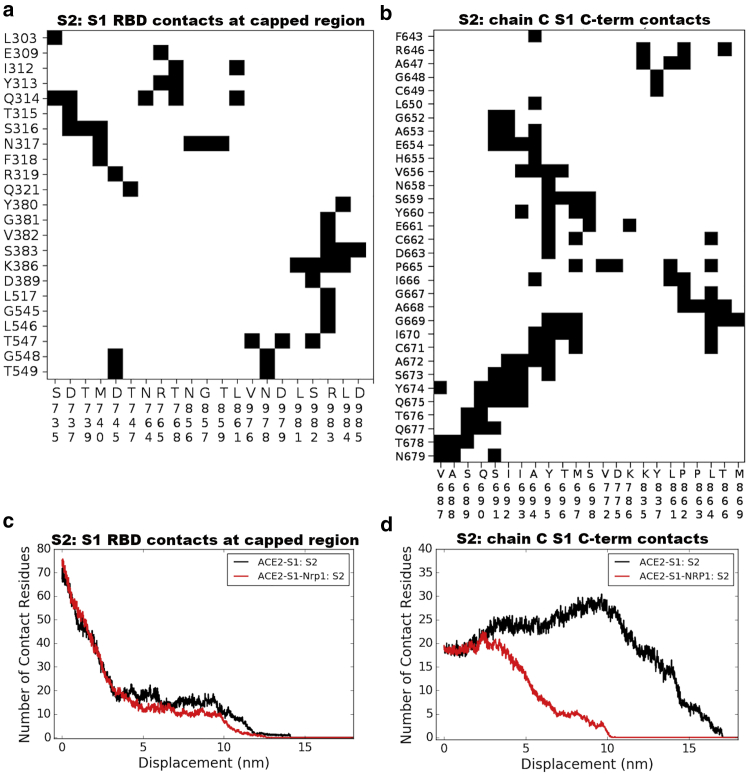
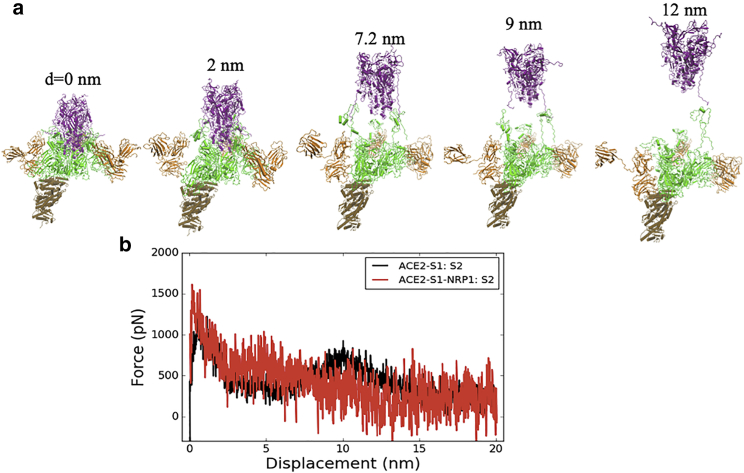
We characterized the separation process of the Spike protein by pulling the S2 out of the S1 subunit using a steered molecular dynamics simulation of 20 ns, with a constant pulling velocity of 1 nm/ns. The peptide bond between residues 685 (S1) and 686 (S2) were cut for all three units of the Spike protein trimer. We pulled S2 out of S1 in two system setups, with and without Nrp1, i.e., the ACE2-S1:S2 complex and ACE2-S1-Nrp1:S2 complex (a “-” indicates that those units are moved together, whereas “:” indicates a non-covalent interaction, whose dissociation is being monitored). Fig. 3, a and b shows snapshots at the stated times of the S2 separation from the S1 domain with and without Nrp1 bound. In each case, the S2 subdomain (res. ∼730–770 and 960–1000) is gradually pulled out from the S1 subunit, which capped it. Interactions are completely lost at an S2 displacement ∼5–6 nm. The detachment of S2 from S1 at the S1-S2 cleavage region (res. ∼640–700), however, takes longer time and distance. Remarkably, the S2:S1 dissociation, observed at the S1/S2 cleavage region for chain C of the Spike protein happens at a shorter distance of 7 nm in the presence of Nrp1 bound there (Fig. 3 b), in comparison with a distance of 16 nm in the absence of Nrp1 (Fig. 3 a). Fig. 3 c shows the separation position of S2 from S1 for chain C of the Spike protein for all simulated complex systems. The separation position was measured by the displacement of center of mass (c.o.m) of S2 from its initial position. The displacement of the S2 domain is typically <10 nm, mostly ∼5–7 nm with the assistance of Nrp1 binding. By contrast, in the absence of Nrp1 binding, the displacement of the S2 domain is around 14.42.0 nm. Thus, for all seven simulations, the separation of S2 and S1 for the chain C of Spike protein happens with a shorter displacement of the S2 domain.
Figure 3.
Separation process of the ACE2-S1 and S2 domain of chain C with and without Nrp1, here shown (a) in absence of Nrp1 (first of seven simulations shown) and (b) for ACE2-S1-Nrp1:S2 model 7, bottom. (S1 is in green, S2 in purple, and Nrp1 in orange.) The displacement of the S2 domain c.o.m relative to initial position is given as distance d. The time interval in the pulling simulations (1 nm/ns) between the structures shown is 2, 5, 7, 10, and 16 ns without Nrp1 bound and 2, 5, 6, and 7 ns with Nrp1 bound, respectively. (c) Plot of the distance between the S1 and the S2 c.o.ms of chain C at the time of separation (when nearest distance S1:S2 distance becomes >5 Å). Two simulations were run and reported for each model except for model 5, in which Nrp1:S1 binding is easily disconnected during the pulling of the S2 domain. To see this figure in color, go online.
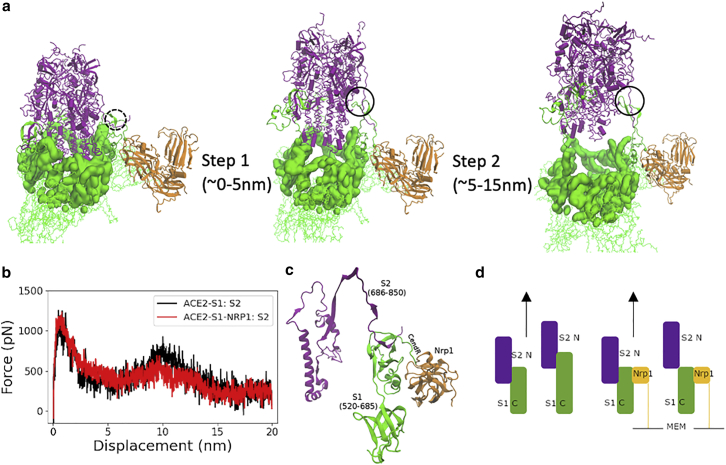
In Fig. 4, we looked into the separation process in greater detail. Overall, the separation of S2 from S1 involves two steps (Fig. 4 a): the separation at the cap region (S1 res. 303–549 and S2 res. 735–768 and 856–985) and the separation at the S1/S2 cleavage region (S1 res. 640–685 and S2 res. 686–700). For the first step, the S2 region escapes from a cavern surrounded by S1 residues (Fig. 4 a). In this cavern, several noticeable interactions between S1 (mainly RBD residues) and S2 were seen as E309-R765, I312-L861, Y313-R765, F318-M740, R319-D745, and Y380-L984 (see Fig. 5 a for full contact maps). Actually, ACE2 binding appears to require at least one of RBD to be in “up” conformation, which reduces the interaction between that one RBD unit and the S2 segment (38, 39, 40). The S2-S1 dissociation in this cap region involves the breaking of the majority of contact interactions between S1 and S2. This constitutes the first and highest energy barrier for the S2-S1 dissociation (Fig. 4 b; Table 1). After step 1 separation (∼5 nm displacement of S2), the number of contact residues at the capped region were largely reduced (Fig. 5 c); however, the connection between S2 and S1 at the S1-S2 cleavage region is still maintained at this point in time (Fig. 5 d). As shown in Fig. 5 c, the Nrp1 does not have a noticeable influence on the detachment process of S2 and S1 at the cap region. At around 5-nm displacement of S2, most of the initial contact interactions at the cap region are broken in both cases.
Figure 4.
Two-step separation between S2 and S1. (a) Two steps of S2 exit from the S1 domain: S1 regions that cap S2 domain (purple) are highlighted in a surface representation in green. (b) Pulling force versus displacement of S2 domain (with and without Nrp1). Results shown were averaged over duplicate simulations of the seven models without and with Nrp1. (c) S2-S1 contacts at the cleavage site at the start of the simulation (here model 7 at 0 ns). (d) Shows a schematic picture of the “holding effect” of Nrp1—essentially, Nrp1 binding stabilizes the region it associates with and does not allow the stretching of this region. (S2 is in violet, S1 in green, and Nrp1's primary binding domain b1 in yellow.)
Figure 5.
Contacts between S2 and S1 at capped region and at the S1-S2 cleavage region. The cap region comprises S1 res. 303–549 (mostly RBD region) and S2 res. 735–768/856–985. The S1-S2 cleavage region consists of S1 res. 640–685 and S2 res. 686–700. (a) Contact map between S1 RBD and S2 subunit. (b) Contact map between S1 C-terminal region and S2. (c) Number of residues of S2 (chains A, B, and C) that interact with S1 RBD (chains A, B, and C) at a different position of S2 displacement with and without Nrp1 bound. (d) Number of residues of S2 (chains A, B, and C) that interact with S1 C-terminal region (res. 640–685 only of chain C), again with and without Nrp1 bound. Note that the number of contacts is counted to all three S protein units and temporarily increases as the S1-S2 cleavage region becomes distorted. To see this figure in color, go online.
Table 1.
The pulling force averaged over every 1-nm interval with Nrp1 bound and without Nrp1
| d (nm) | Force (pN) w Nrp1 | Force (pN) w/o Nrp1 | Δ (pN) | p |
|---|---|---|---|---|
| 0–1 | 935 | 833 | 102 | ∗∗∗ |
| 1–2 | 881 | 814 | 66 | ∗∗∗ |
| 2–3 | 614 | 500 | 113 | ∗∗∗ |
| 3–4 | 511 | 380 | 131 | ∗∗∗ |
| 4–5 | 441 | 377 | 63 | ∗∗∗ |
| 5–6 | 393 | 341 | 51 | ∗∗∗ |
| 6–7 | 384 | 383 | 0 | 0.98 |
| 7–8 | 434 | 460 | −26 | ∗ |
| 8–9 | 443 | 512 | −69 | ∗∗∗ |
| 9–10 | 518 | 663 | −144 | ∗∗∗ |
| 10–11 | 433 | 680 | −246 | ∗∗∗ |
| 11–12 | 412 | 582 | −169 | ∗∗∗ |
| 12–13 | 392 | 431 | −39 | ∗ |
| 13–14 | 331 | 357 | −26 | 0.03 |
| 14–15 | 239 | 313 | −73 | ∗∗∗ |
| 15–16 | 218 | 270 | −52 | ∗∗∗ |
| 16–17 | 206 | 235 | −28 | 0.03 |
| 17–18 | 245 | 236 | 9 | 0.41 |
| 18–19 | 246 | 242 | 3 | 0.77 |
| 19–20 | 236 | 251 | −15 | 0.22 |
The difference and significance (p-values) are calculated over simulation trajectories every 1 ns interval (saved at every 100 fs or 100 steps)
p < 0.01;
p < 0.0001; w, with; w/o, without.
At the S1-S2 cleavage region, noticeably, S2 residues 692–697 form a β-sheet with S1 residues 669–674 (Figs. 4 c and 5 b). This interaction contributes to the second barrier during the S2-S1 dissociation (Fig. 4 b; Table 1). When Nrp1 is bound to the S1 CendR peptide, it has a critical role in this S1-S2 cleavage region. In the absence of Nrp1, the S1 C-terminal region tends to move together with the S2 subunit when it is pulled. Thus, it takes longer time and distance for the detachment of S2 and S1 at the S1-S2 cleavage region (Figs. 4 d and 5 d). In the presence of Nrp1, however, the Nrp1 binding at the cleavage site provides a mechanical support for the adjacent regions (S1 res. 640–685) by stabilizing them. This interaction largely restricts the stretching and local unfolding of the S1 C-terminal region (Figs. 4 d and 5 d) and, in turn, appears to destabilize S1-S2 contacts in this region. This leads to the earlier separation of the N-terminal segment of S2 (res. ∼686–700) from the C-terminal of S1 (res. ∼640–685).
The separation of S2 and S1 for chain A and chain B is not influenced by Nrp1 binding with chain C. Typically, the full separation of S2 from S1 for chain A and chain B still needs a big displacement of the S2 domain ∼13–17 nm. Comparing the work needed to separate S2 and S1, the value with and without Nrp1 is 111385 and 117891 kcal/mol, respectively. Thus, there is little difference in the total work that we estimate is needed to separate the S2 and S1 domains. However, in all cases (Fig. S5), Nrp1 binding reduces the second energy barrier for separation. This is likely a reflection of a reduced need of pulling force at 7–10 nm displacement of S2, as in the presence of Nrp1, S2 already detaches from S1 at the cleavage region for one unit of Spike protein (chain C). There is a caveat here in that the fast pulling of S2 from S1 could significantly overestimate the need of force/work for the S2-S1 separation. Thus, we cannot conclude whether or not the bound Nrp1 may actually reduce or alter the dissociation free energy of S1 from S1. Nevertheless, our analysis clearly demonstrates that the Nrp1 can accelerate the S2-S1 separation by allowing it to occur at a shorter displacement of S2. This is crucial when considering that there is a limited distance for S2 movement that is possible spatially between the viral and host membranes as discussed later.
Global topological constraints in an Nrp1-Spike-ACE2 protein complex
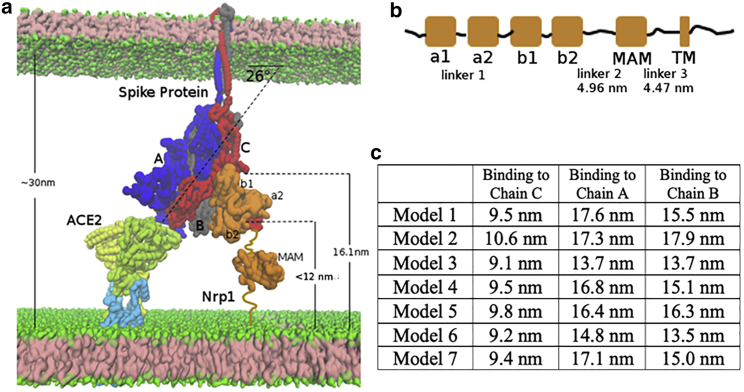
The Spike protein must be close enough to the host membrane to access the Nrp1 extracellular region; however, the steric position of Spike protein is influenced by the host ACE2 receptor. ACE2 forms dimer at the membrane surface in complex with the neutral amino acid transporter B0AT1 dimer (1). Overall, the configuration of the ACE2 dimer lacks flexibility and is perpendicular to the cell membrane, i.e., facing away from it. The Spike protein domains are tilted relative to the membrane by binding to the ACE2 molecules with its RBD (from chain A; Fig. 6 a). We measured the tilting angle as ∼26° (orientation of the vector connecting c.o.ms of S1 (1–685) and c.o.m. of S2 (686–1146)). Spike chain C is the closest subunit to the host membrane, and thus, the easiest one for Nrp1 association. The distance of the S1-S2 cleavage site (chain C) is ∼16.1 nm to the cell membrane surface. So, Nrp1 roughly needs to extend ∼16.1 nm above the membrane. We next estimate the probable length of the Nrp1 extracellular region in this context.
Figure 6.
Topological constraints of Nrp1 on binding with Spike protein trimer, bound to an ACE2 dimer. (a) ACE2 (yellow and lime, which also go into and through the membrane) and B0AT1 (light blue) 2:2 dimer complex at the bottom left. Shown is the Spike protein trimer (chains A, B, and C in blue, gray, and red, respectively) in the middle. Shown is Nrp1 (orange) at the bottom right of the figure. (b) Linear domain arrangement in the sequence of the Nrp1 receptor. (c) Estimated distance of C-terminus of the Nrp1 b2 domain to the membrane bilayer lipid headgroups in the different models with Nrp1 bound to chains A, B, and C. To see this figure in color, go online.
The Nrp1 extracellular region (res. 1–856) consists of five domains: a1 (27–141), a2 (147–265), b1 (275–424), b2 (431–586) and an MAM domain (640–813) (Fig. 6 b). The fold of the protein fragment containing the a2-b1-b2 domains is relatively compact with short linkers of five to nine amino acids between these domains. By contrast, the linkers between the b2 and MAM domains (linker 2, 587–639), as well as the connection of the MAM domain to the transmembrane helix (linker 3, 814–856) are long. We estimated the end-to-end distance of linker 2 and linker 3 by applying the free rotation model of a polymer chain:, with n as 53 and 43 residue equivalents (n), respectively (41). The length () of a segment is set as 0.34 nm, and the rotation angle () is set as 134°, as typically used in the coarse-grained model for a random coil polypeptide chain (42). The average end-to-end distance is estimated as 4.96 and 4.47 nm, respectively (Fig. 6 b). Based on the available structure (43), the distance between N- and C-terminus of the MAM domain is 2.6 nm. Supposing linker 2, linker 3, and MAM all align in a way to maximize the distance in membrane normal direction (Z direction), we estimated the probable length of linker-2-MAM-linker-3 region is 12.0 nm. The orientation of the Nrp1 (a2-b1-b2 domain) protein fragment relative to the Spike protein determines whether the C-terminus of the Nrp1 b2 domain is close enough to fuse with N-terminus of its linker 2. Generally, the Nrp1 a1-b1-b2 fragment should be placed in a position in which the distance of the C-terminus of its b2 domain (residue 586) to the membrane surface is <12.0 nm. With this criterion, we excluded seven predicted docking models as mentioned in the Materials and methods (Fig. 1 b), which have distances that are too far, between 13.5 and 17.3 nm. For the remaining seven models we selected, the distances of the b2 C-terminus to the membrane surface are estimated to be between 9.1 and 10.6 nm (Fig. 6 c).
Nrp1 is able to access the Spike protein chain C as analyzed earlier, but we predict that Nrp1 would need to be stretched to bind with Spike chain A and chain B because the estimated distances (b2 C-terminus to membrane) are large for all seven models (Fig. 6 c). However, we cannot exclude the possibility that linker 2 and 3 of Nrp1 may largely straighten themselves so that Nrp1 can still access the CendR peptide of chains A and B spatially. Furthermore, in the case that Nrp1 could interact with Spike protein in absence of ACE2, the spatial constraint can be released. Thus, next we further explore the separation process when Spike protein is bound to three Nrp1. Fig. 7 shows the separation process of S2 from S1 with the assistance of three Nrp1s (for model 1). The complete separation of S2 and S1 for chains A, B, and C occurs with a displacement of the S2 domain of 6.8, 9, and 12 nm and 13, 2.3, and 5.8 nm, respectively, for duplicate simulations. Again, these displacements are much shorter than the displacement of the S2 domain (∼13–17 nm for all three units) in the absence of Nrp1. Similar to Fig. 4, Nrp1 binding lowers the second activation barrier (Fig. 7 b). However, the pulling work needed to separate S2 and S1 with the support of Nrp1 is also only slightly smaller than that without the support of Nrp1: 1026123 vs. 117891 kcal/mol. Therefore, Nrp1 does not significantly alter the overall work needed for S2-S1 separation; however, it makes the separation happen earlier by stabilizing the S1 C-terminal region during the dissociation process. Again, as in the case of the singly bound Nrp1 to one Spike protein unit, this would accelerate the separation of S2 from the S1 domain also for the three Nrp1/three Spike protein units (i.e., a trimer).
Figure 7.
Separation process of S2 and S1 with three Nrp1s, i.e., one binding to each unit of the Spike protein trimer. (a) Snapshots and the displacement of the S2 domain relative to initial position are shown. (b) Pulling force versus displacement of the S2 domain with three Nrp1 bound. Data are shown as averaged over two simulations for model 1 (red), that is the separation of two times three S1:S2 contacts, whereas the ACE2 data are taken from Fig. 4b. To see this figure in color, go online.
Conclusions
In this study, we modeled the structures of Nrp1 (domains a2-b1-b2) binding to the ACE2 complexed SARS-CoV-2 Spike protein. We analyzed the topological constraints that are generated in such a complex. Importantly, we studied the separation process when the S2 subunit of the Spike protein moves out of the S1 subunit in the presence and absence of S1 bound Nrp1. We found that the Nrp1 association with the Spike protein could facilitate an earlier or more probable separation of S2 from the S1 domain by stabilizing S1 at the C-terminal region and by destabilizing S1-S2 contacts, allowing earlier dissociation of the S2 N-terminal segment from this region. Lastly, we suggest that the tilting of the Spike protein with respect to the membrane is also helpful for the separation of S2 and S1 subunits. Even without ACE2 binding, the recent cryo-electron tomography (cryo-ET) structure of an isolated SARS-CoV-2 virus shows a prevalent tilting of the entire Spike protein by 40° relative to the virus surface/membrane (44). The lateral diffusion of the Spike protein relative to the ACE2 and Nrp1 receptors may provide a source of kinetic energy that drives the separation of the S2 and S1 domains. Future computational and experimental work are needed to unveil the details of the underlying molecular mechanism.
Author contributions
M.B. and Z.L. conceived the project and wrote the manuscript.
Acknowledgments
We thank Prof. Tracy Tran (Rutgers University) for insightful discussion. This work is supported by a National Institutes of Health R01 grant from the National Eye Institute, R01EY029169, and previous grants from National Institute of General Medical Sciences (R01GM073071 and R01GM092851) to the Buck lab.
Editor: Alan Grossfield.
Footnotes
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.05.026.
Supporting material
References
- 1.Yan R., Zhang Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., Kleine-Weber H., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shang J., Wan Y., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vankadari N. Structure of furin protease binding to SARS-CoV-2 spike glycoprotein and implications for potential targets and virulence. J. Phys. Chem. Lett. 2020;11:6655–6663. doi: 10.1021/acs.jpclett.0c01698. [DOI] [PubMed] [Google Scholar]
- 5.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daly J.L., Simonetti B., Yamauchi Y. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantuti-Castelvetri L., Ojha R., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moutal A., Martin L.F., Khanna R. SARS-CoV-2 spike protein co-opts VEGF-A/neuropilin-1 receptor signaling to induce analgesia. Pain. 2021;162:243–252. doi: 10.1097/j.pain.0000000000002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guo H.-F., Vander Kooi C.W. Neuropilin functions as an essential cell surface receptor. J. Biol. Chem. 2015;290:29120–29126. doi: 10.1074/jbc.R115.687327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teesalu T., Sugahara K.N., Ruoslahti E. C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. USA. 2009;106:16157–16162. doi: 10.1073/pnas.0908201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker M.W., Xu P., Vander Kooi C.W. Structural basis for selective vascular endothelial growth factor-A (VEGF-A) binding to neuropilin-1. J. Biol. Chem. 2012;287:11082–11089. doi: 10.1074/jbc.M111.331140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin X.-X., Zheng X.-R., Mao X.-Y. Vascular endothelial growth factor (VEGF) as a vital target for brain inflammation during the COVID-19 outbreak. ACS Chem. Neurosci. 2020;11:1704–1705. doi: 10.1021/acschemneuro.0c00294. [DOI] [PubMed] [Google Scholar]
- 13.Kong Y., Han J., Zhang H. VEGF-D: a novel biomarker for detection of COVID-19 progression. Crit. Care. 2020;24:373. doi: 10.1186/s13054-020-03079-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu C., Rodriguez E.R., Ginty D.D. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Dev. Cell. 2003;5:45–57. doi: 10.1016/s1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mey L., Hörmann M., Al-Fakhri N. Neuropilin-1 modulates vascular endothelial growth factor-induced poly(ADP-ribose)-polymerase leading to reduced cerebrovascular apoptosis. Neurobiol. Dis. 2013;59:111–125. doi: 10.1016/j.nbd.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 16.Hota P.K., Buck M. Plexin structures are coming: opportunities for multilevel investigations of semaphorin guidance receptors, their cell signaling mechanisms, and functions. Cell. Mol. Life Sci. 2012;69:3765–3805. doi: 10.1007/s00018-012-1019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu M.-H., Ying N.-W., Chen Y.-L. Galectin-1 induces vascular permeability through the neuropilin-1/vascular endothelial growth factor receptor-1 complex. Angiogenesis. 2014;17:839–849. doi: 10.1007/s10456-014-9431-8. [DOI] [PubMed] [Google Scholar]
- 18.Najjar S., Najjar A., Azhar S. Central nervous system complications associated with SARS-CoV-2 infection: integrative concepts of pathophysiology and case reports. J. Neuroinflammation. 2020;17:231. doi: 10.1186/s12974-020-01896-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khan S., Gomes J. Neuropathogenesis of SARS-CoV-2 infection. eLife. 2020;9:e59136. doi: 10.7554/eLife.59136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellul M.A., Benjamin L., Solomon T. Neurological associations of COVID-19. Lancet Neurol. 2020;19:767–783. doi: 10.1016/S1474-4422(20)30221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ackermann M., Verleden S.E., Jonigk D. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in covid-19. N. Engl. J. Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Webb B., Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2017;1654:39–54. doi: 10.1007/978-1-4939-7231-9_4. [DOI] [PubMed] [Google Scholar]
- 23.Alekseenko A., Ignatov M., Kozakov D. Protein-protein and protein-peptide docking with ClusPro server. Methods Mol. Biol. 2020;2165:157–174. doi: 10.1007/978-1-0716-0708-4_9. [DOI] [PubMed] [Google Scholar]
- 24.Alekseenko A., Ignatov M., Kozakov D. Protein-protein and protein-peptide docking with ClusPro server. Methods Mol. Biol. 2020;367:1260–1263. doi: 10.1007/978-1-0716-0708-4_9. [DOI] [PubMed] [Google Scholar]
- 25.Woo H., Park S.-J., Im W. Developing a fully glycosylated full-length SARS-CoV-2 spike protein model in a viral membrane. J. Phys. Chem. B. 2020;124:7128–7137. doi: 10.1021/acs.jpcb.0c04553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phillips J.C., Braun R., Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J., Rauscher S., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onufriev A.V., Case D.A. Generalized Born implicit solvent models for biomolecules. Annu. Rev. Biophys. 2019;48:275–296. doi: 10.1146/annurev-biophys-052118-115325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao M., Wilmanns M., Schulten K. Steered molecular dynamics studies of titin I1 domain unfolding. Biophys. J. 2002;83:3435–3445. doi: 10.1016/S0006-3495(02)75343-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Izrailev S., Stepaniants S., Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Batchelor M., Gowdy J., Paci E. Effect of external pulling forces on the length distribution of peptides. Biochim. Biophys. Acta. 2015;1850:903–910. doi: 10.1016/j.bbagen.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 32.Gumbart J.C., Roux B., Chipot C. Efficient determination of protein-protein standard binding free energies from first principles. J. Chem. Theory Comput. 2013;9:3789–3798. doi: 10.1021/ct400273t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z.-L., Buck M. Computational modeling reveals that signaling lipids modulate the orientation of K-Ras4A at the membrane reflecting protein topology. Structure. 2017;25:679–689.e2. doi: 10.1016/j.str.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z.-L., Prakash P., Buck M. A “Tug of War” maintains a dynamic protein-membrane complex: molecular dynamics simulations of C-raf RBD-CRD bound to K-Ras4B at an anionic membrane. ACS Cent. Sci. 2018;4:298–305. doi: 10.1021/acscentsci.7b00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghorbani M., Brooks B.R., Klauda J.B. Exploring dynamics and network analysis of spike glycoprotein of SARS-COV-2. Biophys. J. 2021;120:2902–2913. doi: 10.1016/j.bpj.2021.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhattacharjee S., Bhattacharyya R., Sengupta J. Dynamics and electrostatics define an allosteric druggable site within the receptor-binding domain of SARS-CoV-2 spike protein. FEBS Lett. 2021;595:442–451. doi: 10.1002/1873-3468.14038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casalino L., Dommer A., Amaro R.E. AI-driven multiscale simulations illuminate mechanisms of SARS-CoV-2 spike dynamics. bioRxiv. 2020 doi: 10.1101/2020.11.19.390187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benton D.J., Wrobel A.G., Gamblin S.J. Receptor binding and priming of the spike protein of SARS-CoV-2 for membrane fusion. Nature. 2020;588:327–330. doi: 10.1038/s41586-020-2772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreira R.A., Guzman H.V., Poma A.B. Characterization of structural and energetic differences between conformations of the SARS-CoV-2 spike protein. Materials (Basel) 2020;13:5362. doi: 10.3390/ma13235362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cai Y., Zhang J., Chen B. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020;369:1586–1592. doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rubinstein M., Colby R.H. Oxford University Press; Oxford, UK: 2003. Polymer Physics. [Google Scholar]
- 42.Monticelli L., Kandasamy S.K., Marrink S.-J. The MARTINI coarse-grained force field: extension to proteins. J. Chem. Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- 43.Yelland T., Djordjevic S. Crystal structure of the neuropilin-1 MAM domain: completing the neuropilin-1 ectodomain picture. Structure. 2016;24:2008–2015. doi: 10.1016/j.str.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yao H., Song Y., Li S. Molecular architecture of the SARS-CoV-2 virus. Cell. 2020;183:730–738.e13. doi: 10.1016/j.cell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.