Abstract
Background
The resistance mechanism of the third generation of epidermal growth factor (EGFR) tyrosine kinase inhibitor (TKI) osimertinib is complex. Epithelial mesenchymal transition (EMT) is a common mechanism of EGFR‐TKI acquired resistance. Snail is an important transcription factor related to EMT. Whether targeting Snail can reverse the resistance of osimertinib by downregulating Snail is unknown.
Methods
The presence of EMT in H1975/OR (osimertinib resistance) cells was confirmed by transwell assay. To explore the EMT role in resistance, the expression levels of EMT markers were detected in both parental cells H1975 and resistant cells H1975OR. We used RNA interference technology to knockdown the key regulator Snail in resistant cells. After the interference efficiency was confirmed, changes in EMT‐related molecules of Snail were explicitly downregulated, and changes in sensitivity and migration and invasion ability were also examined. We used CDK4/6 inhibitor to test the ability of reversing drug resistance by downregulating Snail.
Results
Compared with the H1975 cell line, the H1975/OR resistant cell line showed increased invasiveness, upregulated expression of vimentin and downregulation of E‐cadherin. EMT occurred in the H1975/OR resistant cell line. The expression of Snail was upregulated in the osimertinib‐resistant cell line H1975/OR. Knockdown of Snail increased the sensitivity of H1975/OR cells to osimertinib. CDK4/6 inhibitor palbociclib could downregulate the expression of Snail. CDK 4/6 inhibitor palbociclib combined with osimertinib could reverse the resistance of osimertinib in H1975/OR.
Conclusions
Snail plays an important role in the third generation of EGFR‐TKI osimertinib resistance, which may be reversed by downregulating Snail.
Keywords: acquired resistance, CDK4/6 inhibitor, epithelial‐mesenchymal transition (EMT), Snail, tyrosine kinase inhibitor (TKI)
In the osimertinib‐resistant cell line H1975/OR, the resistance to Osimertinib and EMT was successfully reversed by downregulating the Snail.

INTRODUCTION
Lung cancer is the leading cause of cancer‐related death in China and worldwide. Non‐small cell lung cancer (NSCLC) accounts for about 85% of all lung cancer diagnoses. 1 , 2 In Asian patients with lung cancer, about 30%–51.4% of lung adenocarcinoma patients have EGFR sensitive mutations3, 4, 5, 6 sensitive to epidermal growth factor (EGFR) tyrosine kinase inhibitor (TKI) therapy. 7 , 8 , 9 However, acquired resistance to EGFR‐TKIs is inevitable. The resistance mechanism of EGFR‐TKIs is complex. The most common resistance mechanism of first generation EGFR‐TKIs is T790M mutation. 10 , 11 , 12 , 13 In general, third generation EGFR TKI osimertinib has been approved globally for the treatment of T790M‐positive NSCLC patients who have disease progression after therapy with first‐ or second‐generation EGFR‐TKIs. 11 , 14 However, it is unfortunate that osimertinib is resistant in the process of continuous use. The mechanism of osimertinib‐resistance is more complex, including C797S, c‐Met amplification, bypass activation, pathological type changes and so on. 13 , 15 , 16 Epithelial mesenchymal transition (EMT) is one of the important mechanisms of EGFR‐TKI resistance. 17 , 18 , 19 , 20 Previous studies have shown that EMT is one of the important mechanisms of resistance to erlotinib, gefitinib in the first generation and osimertinib third generation of EGFR‐TKIs. 17 , 19 , 20 , 21 Epithelial‐mesenchymal transition (EMT), manifesting as a loss of E‐cadherin, and an increase in vimentin expression, is a process in which epithelial cells lose their polarity and adhesion to gain migratory ability and adopt a mesenchymal phenotype. 22 E‐cadherin expression is transcriptionally repressed by Zeb, Snail, Slug, and Twist. 23 These EMT transcriptional regulators have been suggested as potential therapeutic targets for human cancers. 24 For example, the expression of Twist has been shown to be increased in an EGFR‐TKI resistant cell line, involved in the regulation of EMT and genetic and pharmacological inhibition of TWIST resulted in growth inhibition and apoptosis in EGFR‐mutant NSCLC cell lines. 21 Knockdown of Hakai has also been reported to elevate E‐cadherin expression, attenuate stemness, and resensitize cells to gefitinib, and dual HDAC and HMGR inhibitor JMF3086 inhibited the Src/Hakai and Hakai/E‐cadherin interaction to reverse E‐cadherin expression, and attenuated vimentin and stemness to restore gefitinib sensitivity. 19 , 21
However, Snail is a key transcription factor of EMT, which has been found to promote the transformation of tumor epithelial cells into stromal cells, enhance the movement and invasion ability of tumor cells, promote the dryness of tumor cells and cell migration. 25 , 26 In our study, we demonstrated that genetic silencing of Snail or the downregulation of the expression of Snail by CDK4/6 inhibitor palbociclib in EGFR TKI resistant EGFR‐mutated cells increased sensitivity to EGFR‐TKI. At present, there is no specific inhibitor of Snail which has been approved by the FDA and CFDA. A previous study reported that CDK4/6 inhibitor palbociclib could regulate Snail expression, inhibit EMT and reduce the occurrence of distant metastasis in triple negative breast cancer. 27 The main mechanism might be that CDK4/6‐mediated activation of DUB3 is essential to deubiquitinate and stabilize Snail. DUB3 may also promote the proliferation of NSCLC by inhibiting Cyclin A degradation. 28 Therefore, in our study we used palbociclib to explore its effect on Snail expression and to ascertain whether it could reverse drug resistance.
METHODS
Cell lines and cell culture
Human lung adenocarcinoma cell lines H1975 (sensitivity to osimertinib, with a compound EGFR exon 21 L858R and T790M mutation) and H1975OR (osimertinib resistance) were used in this study. The NCI‐H1975 human lung adenocarcinoma cell line was obtained from the American Type Culture Collection (ATCC). The osimertinib‐resistant H1975 (H1975OR) cell line was established by our laboratory as previously described. 29 Cells were cultured in RPMI‐1640 medium supplemented with 10% fetal bovine serum (Gibco) at 37°C in a humidified incubator with 5% CO2.
Cell proliferation assay
The cells H1975 and H1975OR were seeded at a density of 1.0 × 104 cells/well in a 96‐well plate for 48, and 72 hours. Cell counting kit‐8 (CCK‐8) solution (10 μl, B34302, Selleck Chemicals) was added to each well prior to the endpoint of incubation. The absorbance, which represents the cell count, was determined with a microculture plate reader at 450 nm.
Cell migration assays
Transwell assays were performed using polyethylene terephthalate transwell filters (Millipore; 8.0 μm pore size) placed over a bottom chamber containing RPMI medium with 10% FBS. Cells suspended in serum‐free RPMI medium were added to the upper chamber at the indicated density. After 6–8 h of incubation at 37°C, cells that had migrated to the lower side of the filter were stained with crystal violet. Five fields per transwell were photographed using a microscope at 200× magnification. For wound‐healing assays, cells were seeded in a six‐well plate, and a wound area was generated by scraping with a 1–2 ml pipette tip. After 42 h of incubation, the wounded monolayer was photographed.
Flow cytometry analysis of apoptosis
A PE‐Annexin V/7‐AAD apoptosis detection kit was used to evaluate apoptosis. Briefly, for apoptosis analysis, cells were cultured 48 h and then collected and stained with PEAnnexin V and 7‐AAD for 15 min in the dark. The cells were analyzed by flow cytometry (BD Biosciences) within 1 h. The data were analyzed with FlowJo software.
Reverse transcription and quantitative real‐time PCR
Total RNA was isolated using Trizol (ThermoFisher Scientific) and an RNeasy Mini Kit (Qiagen), following the manufacturer's instructions. cDNA was synthesized using the M‐MLV Reverse Transcriptase Kit (Promega) according to the manufacturer's protocol. Quantitative real‐time PCR analysis was performed with ABI SYBR Green Master Mix (Thermofisher Scientific) in an ABI7500 Real‐time PCR System according to the manufacturer's protocol. Each sample was run in triplicate for each gene. Transcript levels were normalized to the housekeeping gene GAPDH and analyzed by the relative quantification 2‐ΔΔCt method. All gene primers were obtained from SBS. We used Affymetrix GeneChip probe to test the gene changes in the RNA of H1975 and H1975/OR cells. All the detected gene primers are shown in Table 1.
TABLE 1.
The detected gene primers
| Gene name | Forward prime(5′ to 3′) | Reverse prime (5′ to 3′) |
|---|---|---|
| GAPDH | GGAGTCAACGGATTTGGTCG | CTTGATTTTGGAGGGATCTCG |
| E‐cadherin | TGA AAAGAGAGTGGAAGTGTCCGAG | GATTAGGGCTGTGTACGTGCTGTTC |
| N‐cadherin | CAATCCTCCAGA GTTTACTGCCATG | GATTGGTTTGACCACGGTGACTAAC |
| Vimentin | GCA AAGCAGGAGTCCACTGAGTACC | TGTCAAGGGCCATCTTAACATTGA G |
| Snail | TCGGAAGCCTAACTACAGCGA | AGATGAGCATTGGCAGCGAG |
| DUB3 | CTATCATTGCGGTCTTTGTCTCC | AAGTGATGCTACAGGCAGTGA |
Western blotting
Protein samples were resolved by sodium dodecyl sulfatepolyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes (Merck Millipore). Membranes were blocked with fat‐free milk combined with tris‐buffered saline plus tween 20 for 1 h at room temperature and then incubated with the appropriate primary antibody and horseradish peroxidaseconjugated secondary antibodies. Imager was used to visualize the blots. The primary antibodies used in this study are as follows: anti‐phospho‐EGFR(Try 1068) (#3777, 1:1000, cell signaling), E‐cadherin (#3195, 1:1000, cell signaling), anti‐vimentin (#571, 1:1000, cell signaling), anti‐Snail (#3879, 1:1000, cell signaling), anti Twist1 (#46702, 1:1000, cell signaling), MMP2 (#40994, 1:1000, cell signaling); MMP9 (#13667, 1:1000, cell signaling); and β‐actin (ab92552, 1:1000).
Generation of Snail‐knockdown cells by transfecting plasmid
Development of transfection vectors and transfection of H1975/OR cells were performed as standard protocols, with some modifications. 30 In brief, H1975/OR cells were transduced with plasmid that expressed scrambled siRNA (control, forward: GCACAACAA GCCGAATACA, reverse: UGUAUUCGGCUUUGUUGUGC), Snail siRNA#1 (Forward:: GGACUUUGAUGAAGACCAU, reverse: AUGGUCUUCAUCAAAGUCC), Snail siRNA #2. (forward: GCGAGCUGCAGGACUCUAA, reverse: UUAGAGUCCUGCAGCUCGC). Cells were then washed.
with PBS and cultured before screening for gene expression. Once decreased expression of Snail was confirmed, the cells were used for experiments.
Statistical analysis
All data were analyzed using GraphPad Prism 7.0 (GraphPad Software). Independent samples t‐test were used to identify the statistical significance between groups. ANOVAs were used for analysis of more than two groups of data in all experiments. Significant differences between the means were considered. p < 0.05 indicated statistical significance.
RESULTS
Invasion of H1975OR cell line increased and apoptosis decreased indicating EMT
The inhibitory dose (IC50) of H1975OR cells was significantly higher than H1975. The IC50 of the two groups of cell lines was 0.1024 and 8.391 μM, respectively (Figure 1(a), p < 0.01). The drug resistance index was 81.94, which was much higher than 10, indicating that the cell line was resistant. The proportion of apoptotic cells in parent cell line H1975 was about 10%, and 6.1% in the H1975/OR cell line by flow cytometry (Figure 1(b)), indicating no statistically significant difference (p = 0.170, Figure 1(c)). Compared with H1975, the proapoptotic proteins Bax and cleaved PARP were decreased, and the expression of antiapoptotic protein Bcl‐2 was increased in H1975/OR (Figure 1(d)). Transwell migration test was used to compare the invasive ability of H1975 and H1975/OR cell lines. In 24 and 48 h, more H1975/OR cells crossed the transwell, indicating that the H1975/OR cell line was more invasive (Figure 1(e)). Similarly, the transwell invasion test confirmed that the number of H1975OR crossing matrix gel and transwell was more invasive (Figure 1(f)).
FIGURE 1.
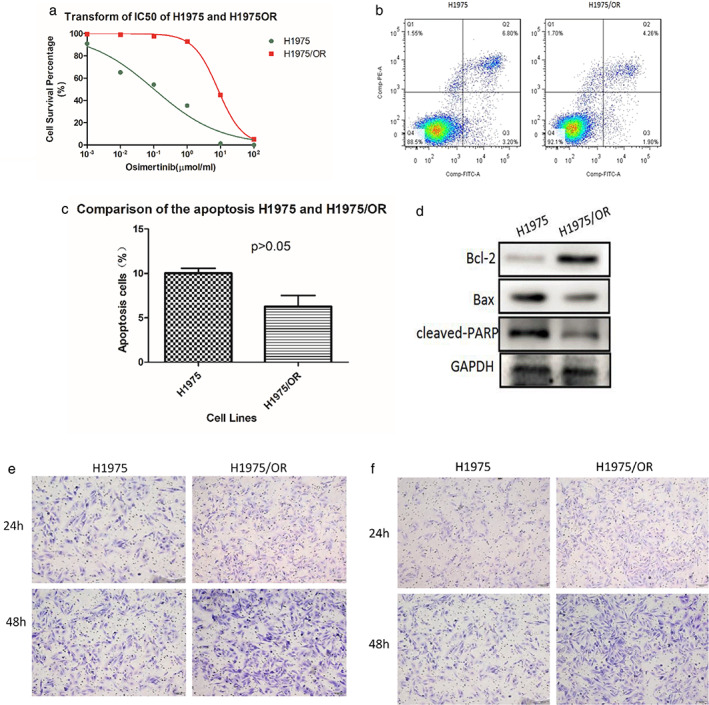
(a) IC50 of H1975 and H1975/OR. (b, c, d) Difference of apoptotic ability between H1975 and H1975OR. (e, f) The invasive ability of H1975 and H1975/OR by transwell assay
EMT related mRNA and protein in H1975OR
Compared with the H1975 cell line, N‐cadherin and vimentin mRNA were upregulated and E‐cadherin was downregulated in H1975/OR (Figure 2(a)). Western blot further confirmed that E‐cadherin expression was downregulated in H1975OR, while the expression of N‐cadherin and vimentin was upregulated. MMP‐2, which was related to the invasion ability of tumor cells, was downregulated. The expression of MMP‐9, Snail and Twist were upregulated (Figure 2(b)). In the sensitive cell line H1975, the expression of Snail gradually increased with the continuous application of osimertinib, suggesting that Snail might play an important role in the process of osimertinib resistance (Figure 2(c)).
FIGURE 2.
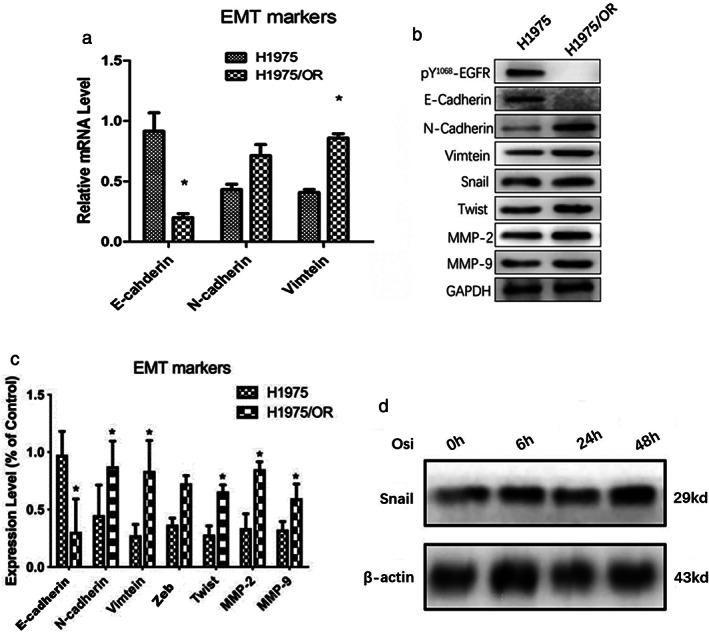
EMT related gene and protein in H1975/OR and H1975 ((a) The level of mRNA of N‐cadherin and vimentin was upregulated and E‐cadherin downregulated. (b) The protein of N‐cadherin, vimentin, Snail and Twist were upregulated and E‐cadherin downregulated. (c) The mRNA of E‐cadherin, N‐cadherin, vimtein, Zeb, Twist, MMP‐2 and MMP‐9. (d) The expression of Snail in H1975 in osimertinib 100 nM)
Knockdown of Snail reversed osimertinib‐resistance in H1975/OR
The expression of Snail was interfered by transfecting plasmid siRNA in H1975/OR. The knockdown of Snail was verified by Western blot (Figure 3(a)). In H1975/OR si‐snail#1, the expression of E‐cadherin was upregulated, while N‐cadherin and vimentin were downregulated (Figure 3(b)). In H1975/OR si‐snail#1, the number of cells penetrating the transwell was reduced (Figure 3(c)), and the wound healing was slow (Figure 3(d)), which indicated the decline of migration and invasion ability. Compared with H1975/OR, H1975/OR si‐snail#1 was more sensitive to osimertinib (Figure 3(e)) and was consistent with the change in EMT (Figure 3(f)).
FIGURE 3.
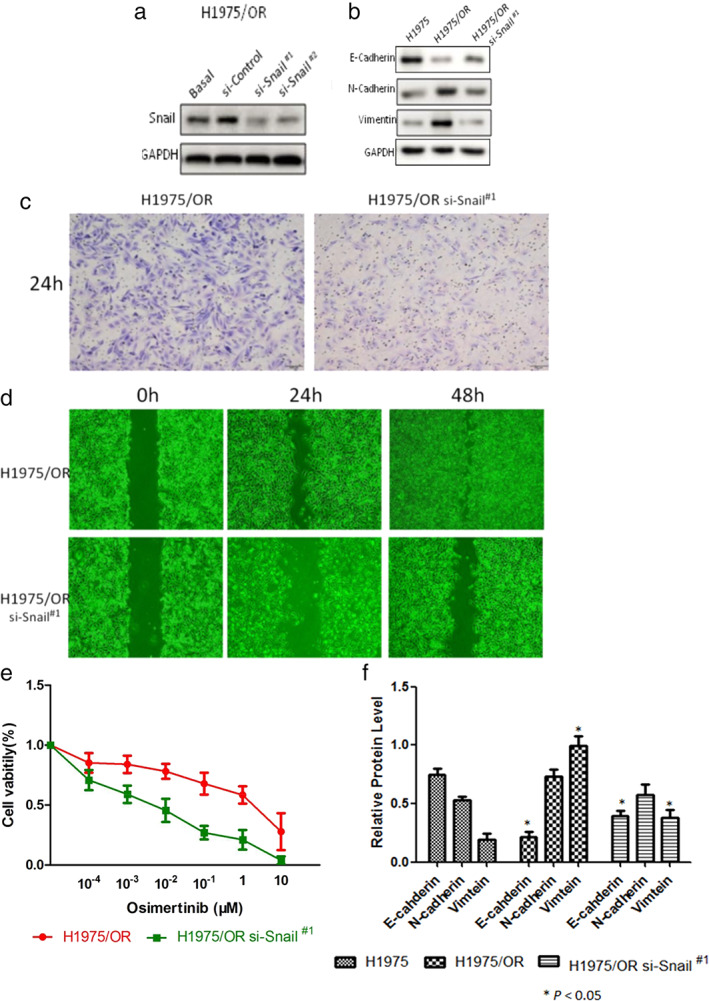
Knockdown of Snail in H1975/OR. (a) The knockdown of snail was verified by Western blot in H1975OR si‐snail#1 and si‐snail#2. (b) E‐cadherin was upregulated, while N‐cadherin and vimentin were downregulated in H1975OR si‐snail#1. (c) The number of cells penetrating the transwell was reduced in H1975OR si‐snail#1. (d) Wound healing was slow in H1975/OR si‐snail#1. (e) IC 50 of osimertinib was 11.5 nM in H1975OR si‐snail#1 (f) mRNA of EMT gene in H1975, H1975/OR and H1975/OR si‐snail#1
CDK4/6 inhibitor palbociclib regulated Snail and reversed osimertinib‐resistance
We then determined whether cell cycle arrest with the CDK4/6 inhibitor palbociclib indeed restored the sensitivity of osimertinib. CDK4/6 inhibitor palbociclib at the tested concentration ranges did not or only weakly suppressed the growth of H1975/OR. As expected, H1975/OR was resistant to osimertinib. However, the combination of palbociclib and osimertinib very effectively inhibited the growth of H1975/OR (Figure 4(a),(b)). The difference was statistically significant (p < 0.05). In the resistant cell line H1975OR, palbociclib combined with osimertinib could significantly downregulate Snail expression (Figure 4(c)). Furthermore, there were no statistically significant difference in the mRNA level of Snail in H1975 and H1975OR. However, the mRNA of DUB3 in H1975OR was higher than H1975 (Figure 4(d)).
FIGURE 4.
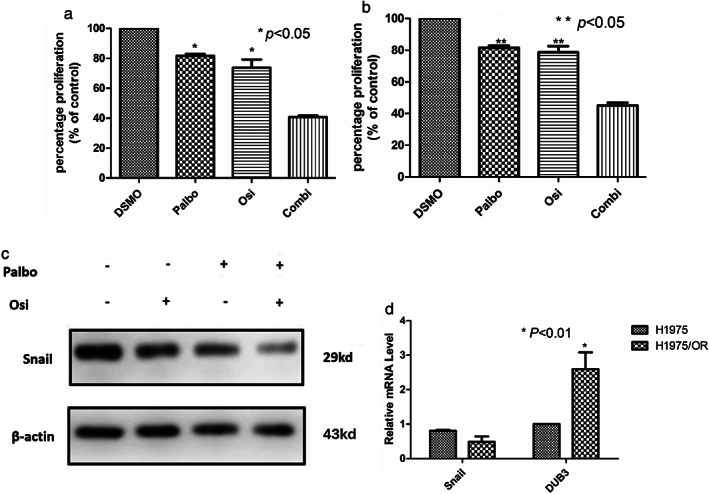
CDK4/6 inhibitor palbociclib combined with osimertinib restrained H1975OR and downregulated Snail. (a, b) Osimertinib (2.5 and 1.25 μM) combined CDK4/6 inhibitor palbociclib 1.25 μM increased inhibition of H1975OR. (c) Osimertinib (2.5 μM) and combined CDK4/6 inhibitor palbociclib (1.25 μM) downregulated Snail. (d) The mRNA of Snail and DUB3 in H1975 and H1975OR
DISCUSSION
EGFR‐TKI resistance is the main cause of failure in the treatment of patients with EGFR sensitive mutations. With the wide application of the third generation EGFR‐TKI osimertinib, more and more patients develop osimertinib‐resistance, which are not T790M. EMT might be one of the important mechanisms of the third generation EGFR‐TKI osimertinib‐resistance. In our study, we confirmed that, CDK4/6 inhibitor palbociclib combined with osimertinib could reverse osimertinib resistance by regulating Snail in the H1975OR cell line.
The expression of Snail in lung cancer is related to DFS and OS in NSCLC. High Snail expression has been reported to be a significant prognostic predictor in patients with pathological N0 NSCLC. 31 In lung cancer, UGDH promotes tumor metastasis by increasing the stability of Snail mRNA. 32 In our study, knockdown of Snail reversed the EMT phenotype, downregulated N‐cadherin and vimentin, upregulated E‐cadherin, and reduced the invasion and migration ability of H1975OR. Snail may therefore be a therapeutic target for reversion of EGFR‐TKI resistance.
Traditionally, Snail is considered to be difficult to become a drug target. The drugs which regulate the Snail upstream pathway and indirectly downregulate Snail expression have become the focus of research in the field. This study found that in H1975OR, CDK4/6 inhibitor might downregulate DUB3 by inhibiting the function of CDK4, and Snail may be ubiquitinated and degraded due to the loss of DUB3 protection, thus reducing tumor invasion and metastasis. Further studies are needed to clarify the possible mechanism.
In addition, Li et al. found that CYD19, a new synthetic compound, directly combined with the conserved 174 arginine (r174) in Snail evolution could significantly inhibit Snail protein expression in a variety of tumor cell lines at the level of nM. It mainly interfered with the binding of acetyltransferase CBP / P300 with Snail through the binding of CYD19 with Snail protein, which resulted in the loss of acetylation protection of the latter ubiquitination degradation. 33
Unfortunately, these studies were only performed in H1975OR, and not in more EGFR‐TKI resistant cell lines. In future, it is hoped that we can further verify these results in more EGFR‐TKI cell lines and a PDX model.
In conclusion, we believe that targeting EMT related transcription factor Snail is an important mechanism for reversing the resistance of EGFR‐TKI to osimertinib in the future. It is expected that clinical trials in more new drugs, such as CYD19, will soon be completed confirming its effectiveness in the human body. At the same time, it is also hoped that CDK4/6 inhibitors could be explored more in EGFR‐TKI‐resistant cell lines in order to provide more support for the combined application of CDK4/6 inhibitors palbociclib and osimertinib.
Qin Q, Li X, Liang X, et al. Targeting the EMT transcription factor Snail overcomes resistance to osimertinib in EGFR‐mutant non‐small cell lung cancer. Thorac Cancer. 2021;12:1708–1715. 10.1111/1759-7714.13906
Funding information National Natural Science Foundation of China, Grant/Award Numbers: 31301160, 81572268, 81972186; Natural Science Foundation of Tianjin, Grant/Award Numbers: 16JCYBJC24400, 17JCYBJC25500
Contributor Information
Linlin Sun, Email: lsun1@tmu.edu.cn.
Diansheng Zhong, Email: zhongdsh@hotmail.com.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. [DOI] [PubMed] [Google Scholar]
- 3. Kawaguchi T, Matsumura A, Fukai S, Tamura A, Saito R, Zell JA, et al. Japanese ethnicity compared with Caucasian ethnicity and never‐smoking status are independent favorable prognostic factors for overall survival in non‐small cell lung cancer. J Thorac Oncol. 2010;5(7):1001–10. [DOI] [PubMed] [Google Scholar]
- 4. Shi Y, Au JS, Thongprasert S, et al. A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non–small‐cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol. 2014;9(2):154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shi Y, Li J, Zhang S, et al. Molecular epidemiology of EGFR mutations in Asian patients with advanced non‐small‐cell lung cancer of adenocarcinoma histology – mainland China subset analysis of the PIONEER study. PLoS One. 2015;10(11):e143515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pi C, Xu CR, Zhang MF, Peng XX, Wei XW, Gao X, et al. EGFR mutations in early‐stage and advanced‐stage lung adenocarcinoma: analysis based on large‐scale data from China. Thorac Cancer. 2018;9(7):814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou C, Wu YL, Chen G, Feng J, Liu XQ, Wang C, et al. Erlotinib versus chemotherapy as first‐line treatment for patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (OPTIMAL, CTONG‐0802): a multicentre, open‐label, randomised, phase 3 study. Lancet Oncol. 2011;12(8):735–42. [DOI] [PubMed] [Google Scholar]
- 8. Sugawara S, Oizumi S, Minato K, Harada T, Inoue A, Fujita Y, et al. Randomized phase II study of concurrent versus sequential alternating gefitinib and chemotherapy in previously untreated non‐small cell lung cancer with sensitive EGFR mutations: NEJ005/TCOG0902. Ann Oncol. 2015;26(5):888–94. [DOI] [PubMed] [Google Scholar]
- 9. Seto T, Kato T, Nishio M, Goto K, Atagi S, Hosomi Y, et al. Erlotinib alone or with bevacizumab as first‐line therapy in patients with advanced non‐squamous non‐small‐cell lung cancer harbouring EGFR mutations (JO25567): an open‐label, randomised, multicentre, phase 2 study. Lancet Oncol. 2014;15(11):1236–44. [DOI] [PubMed] [Google Scholar]
- 10. Ke EE, Zhou Q, Zhang QY, Su J, Chen ZH, Zhang XC, et al. A higher proportion of the EGFR T790M mutation may contribute to the better survival of patients with exon 19 deletions compared with those with L858R. J Thorac Oncol. 2017;12(9):1368–75. [DOI] [PubMed] [Google Scholar]
- 11. Mok TS, Wu Y, Ahn M, et al. Osimertinib or platinum‐pemetrexed in EGFR T790M‐positive lung cancer. N Engl J Med. 2017;376(7):629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kobayashi S, Boggon TJ, Dayaram T, Jänne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non‐small‐cell lung cancer to gefitinib. N Engl J Med. 2005;352(8):786–92. [DOI] [PubMed] [Google Scholar]
- 13. Leonetti A, Sharma S, Minari R, Perego P, Giovannetti E, Tiseo M. Resistance mechanisms to osimertinib in EGFR‐mutated non‐small cell lung cancer. Br J Cancer. 2019;121(9):725–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Murtuza A, Bulbul A, Shen JP, Keshavarzian P, Woodward BD, Lopez‐Diaz FJ, et al. Novel third‐generation EGFR tyrosine kinase inhibitors and strategies to overcome therapeutic resistance in lung cancer. Cancer Res. 2019;79(4):689–98. [DOI] [PubMed] [Google Scholar]
- 15. John T, Akamatsu H, Delmonte A, Su WC, Lee JS, Chang GC, et al. EGFR mutation analysis for prospective patient selection in AURA3 phase III trial of osimertinib versus platinum‐pemetrexed in patients with EGFR T790M‐positive advanced non‐small‐cell lung cancer. Lung Cancer. 2018;126:133–8. [DOI] [PubMed] [Google Scholar]
- 16. Santoni‐Rugiu E, Melchior LC, Urbanska EM, et al. Intrinsic resistance to EGFR‐tyrosine kinase inhibitors in EGFR‐mutant non‐small cell lung cancer: differences and similarities with acquired resistance. Cancers. 2019;11(7):923–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu X, Chen L, Liu L, Niu X. EMT‐mediated acquired EGFR‐TKI resistance in NSCLC: mechanisms and strategies. Front Oncol. 2019;9:1044–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li L, Gu X, Yue J, Zhao Q, Lv D, Chen H, et al. Acquisition of EGFR TKI resistance and EMT phenotype is linked with activation of IGF1R/NF‐kappaB pathway in EGFR‐mutant NSCLC. Oncotarget. 2017;8(54):92240–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Weng C, Chen L, Lin Y, et al. Epithelial‐mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene. 2019;38(4):455–68. [DOI] [PubMed] [Google Scholar]
- 20. Yoshida T, Song L, Bai Y, et al. ZEB1 mediates acquired resistance to the epidermal growth factor receptor‐tyrosine kinase inhibitors in non‐small cell lung cancer. PLoS One. 2016;11(1):e147344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yochum ZA, Cades J, Wang H, Chatterjee S, Simons B, O'Brien JP, et al. Targeting the EMT transcription factor TWIST1 overcomes resistance to EGFR inhibitors in EGFR‐mutant non‐small‐cell lung cancer. Oncogene. 2019;38(5):656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial‐mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7(2):131–42. [DOI] [PubMed] [Google Scholar]
- 23. Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT‐inducing transcription factors. Nat Cell Biol. 2014;16(6):488–94. [DOI] [PubMed] [Google Scholar]
- 24. Shih JY, Tsai MF, Chang TH, Chang YL, Yuan A, Yu CJ, et al. Transcription repressor slug promotes carcinoma invasion and predicts outcome of patients with lung adenocarcinoma. Clin Cancer Res. 2005;11(22):8070–8. [DOI] [PubMed] [Google Scholar]
- 25. Wu Y, Zhou BP. Snail: more than EMT. Cell Adhes Migr. 2010;4(2):199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang Y, Shi J, Chai K, Ying X, Zhou B. The role of snail in EMT and tumorigenesis. Curr Cancer Drug Targets. 2013;13(9):963–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu T, Yu J, Deng M, Yin Y, Zhang H, Luo K, et al. CDK4/6‐dependent activation of DUB3 regulates cancer metastasis through SNAIL1. Nat Commun. 2017;8:13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu B, Deng T, Ma H, et al. Deubiquitinase DUB3 regulates cell cycle progression via stabilizing Cyclin a for proliferation of non‐small cell lung cancer cells. Cells. 2019;8(4):297–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin Q, Li X, Liang X, Zeng L, Wang J, Sun L, et al. CDK4/6 inhibitor palbociclib overcomes acquired resistance to third‐generation EGFR inhibitor osimertinib in non‐small cell lung cancer (NSCLC). Thorac Cancer. 2020;11(9):2389–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector‐mediated drug‐inducible RNA interference. J Virol. 2003;77(16):8957–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang G, Ma W, Li Y, Jiang Y, Ma G, Zhang X, et al. Prognostic value of Twist, Snail and E‐cadherin expression in pathological N0 non‐small‐cell lung cancer: a retrospective cohort study. Eur J Cardiothorac Surg. 2018;54(2):237–45. [DOI] [PubMed] [Google Scholar]
- 32. Wang X, Liu R, Zhu W, Chu H, Yu H, Wei P, et al. UDP‐glucose accelerates SNAI1 mRNA decay and impairs lung cancer metastasis. Nature. 2019;571(7763):127–31. [DOI] [PubMed] [Google Scholar]
- 33. Li HM, Bi YR, Li Y, Fu R, Lv WC, Jiang N, et al. A potent CBP/p300‐snail interaction inhibitor suppresses tumor growth and metastasis in wild‐type p53‐expressing cancer. Sci Adv. 2020;6(17):w8500. [DOI] [PMC free article] [PubMed] [Google Scholar]


