Abstract
Since February-2020, the world has been battling a tragic public-health crisis with the emergence and spread of 2019-nCoV. Due to the lack of information about the pathogenesis-specific treatment of Covid-19, early diagnosis and timely treatment are important. However, there is still a lack of information about routine-blood-parameteres (RBP) findings and effects in the disease process. Although the literature includes various interventions, existing studies need to be generalized and their reliability improved. In this study, the efficacy of routine blood values used in the diagnosis and prognosis of Covid-19 and independent biomarkers obtained from them were evaluated retrospectively in a large patient group. Low lymphocyte (LYM) and white-blood-cell (WBC), high CRP and Ferritin were effective in the diagnosis of the disease. The (d-CWL) = and (d-CFL) = biomarkers derived from them were the most important risk factors in diagnosing the disease and were more successful than direct RBP values. High d-CWL and d-CFL values largely confirmed the Covid-19 diagnosis. The most effective RBP in the prognosis of the disease was CRP. (d-CIT) = CRP*INR*Troponin; (d-CT) = CRP*Troponin; (d-PPT) = PT*Troponin*Procalcitonin biomarkers were found to be more successful than direct RBP values and biomarkers used in previous studies in the prognosis of the disease. In this study, biomarkers derived from RBP were found to be more successful in both diagnosis and prognosis of Covid-19 than previously used direct RBP and biomarkers.
Keywords: Covid-19; Biochemical, hematological and inflammatory biomarkers; Blood routine parameters; Covid-19′s ındependent biomarkers
1. Introduction
2019-nCoV, which is a member of the Coronavirial and Nidovirale family, is an enveloped, positively oriented, single-stranded RNA beta coronavirus [1]. The virus originated in bats and was transmitted to humans through animals in Wuhan, China in December 2019 and spread from there all over the world. The Covid-19 outbreak (caused by severe acquired respiratory syndrome-coronavirus-2 [SARS-CoV-2]) posed and continues to pose a major threat to public health globally [2], [3]. Symptoms were generally seen as fever, cough, sore throat, shortness of breath, fatigue as well as weakness [1]. Moderate and general patients have milder symptoms and a good prognosis, but severe and critically ill patients are difficult to treat and have a high mortality rate [4], [5]. Unfortunately, information on early prediction factors for disease progression is relatively limited. Due to the lack of constructive information about the pathogenesis and specific treatment of Covid-19, early diagnosis and timely treatment are of clinical importance [6], [7]. The most common laboratory findings, including hematological and biochemical parameters, play an important role in the initial screening for Covid-19 [6], [8].
Routine laboratory parameters have great clinical importance in predicting the diagnosis and progression of the disease [5]. In addition, while the number of Covid-19 cases is increasing day by day, there is limited information about the hematological and laboratory findings of the disease and their effects on the disease [4], [5], [9]. For this reason, many academic studies are conducted to determine the change of blood routine laboratory parameters in Covid-19, prognosis, morbidity, mortality and their relationship with other comorbidities. Although the literature includes various attempts to address such issues, the place and importance of routine blood laboratory parameters in Covid-19 disease is not fully understood [4], [5].
Although the clinical features of Covid-19 have been identified, large sample size studies representing laboratory abnormalities of these patients are still needed [7], [10]. Therefore, the relationship between Covid-19 disease and laboratory parameters should be supported by large data sets. In addition, more successful biomarkers continue to be used as alternatives to current clinical approaches in the diagnosis and prognosis of the disease.
Within the scope of these important goals, this study compared the effect of predictive routine blood parameters frequently used in the literature in the prediction of the diagnosis and prognosis (in the distinction of subjects in all services unit and intensive care unit) and the success of independent biomarkers derived from these blood values in this sense. In addition, the success of the derived biomarkers and the biomarkers previously used for this purpose in the literature were compared. With the results to be obtained, it is thought that this study will guide useful strategies for clinicians in predicting the diagnosis and prognosis of the disease.
2. Material and methods
This retrospective single-center study was conducted in accordance with the 1989 Helsinki Declaration. This study was approved by the Ministry of Health and the Ethics Committee. Data matching our criteria were collected from Erzincan Binali Yıldırım University Mengücek Gazi Training and Research Hospital information system between March 2020 and December 2020 and were included in the study. The research only covered people over 18 years old. Laboratory data of the patients were the first blood values measured at the time of first admission to the hospital.
2.1. Study design and participants criteria
With the diagnosis of “Covid-19 identified virus”, patient information was scanned and the information of a total of 2648 patients was reported. The diagnosis of Covid-19 was defined only in cases detected as SARS-CoV-2 by rRT-PCR in nasopharyngeal or oropharyngeal swabs at the dates covered by this study in our hospital. Routine laboratory parameters generally used in the prognosis of Covid-19 were obtained from all patients. Biomarkers frequently used in the diagnosis, prognosis and mortality estimation of the disease were calculated for all patients. When the data were being scanned, they were reported in two groups, those treated in the intensive care unit (ICU) and subjects in all services unit (non-ICU). In addition, an equal number of control groups (2648 individuals) with the patient group were determined.
2.2. Blood routine laboratory parameters used in the study
The Sysmex XN-1000 Hematology System (Sysmex Corporation, Kobe, Japan) was used to perform cell blood count of individuals in the experimental and control groups. Biochemical tests were analyzed from serum by spectrophotometric method using Beckman Coulter Olympus AU2700 Plus Chemistry Analyzer (Beckman Coulter, Tokyo, Japan). Ferritin was evaluated by a chemiluminescence immunoassay (Centaur XP, Siemens Healthcare, Germany). Prothrombin time (PT), activated partial prothrombin time (aPTT), and fibrinogen were determined with a fully digital coagulation device of Ceveron-Alpha (Diapharma Group Inc., West Chester, Canada). C-reactive protein (CRP) was measured by the nephelometric method on the BN ™ II System (Siemens, Munich, Germany). Procalcitonin (PCT), D-dimer and Troponin were analyzed from whole blood on the AQT90 flex Radiometer® (Bronshoj, Denmark).
2.3. Biomarkers used in previous studies
Biomarkers from previous studies4-5 used in predicting the prognosis and mortality of Covid-19 disease were identified and summarized below. The effect of these biomarkers in the diagnosis of Covid-19 disease and in the separation of inpatients into intensive care/service (all non-intensive care units) units (ie the prognosis of the disease) was investigated.
NEU: Neutrophil count, LYM: Lymphocyte count, WBC: White blood cell count, PLT: Platelet count, CRP: C-reactive protein count.
2.4. Biomarkers derived for this study
Biomarkers have been derived for the diagnosis of Covid-19 disease and estimation of the separation of inpatients into units (intensive care/all non-intensive care units) (i.e., for the prognosis of the disease). In the conclusion part, the performances of the biomarkers were compared. The new biomarkers that have been derived are summarized below.
(d-TI = Troponin*INR); (d-PPT = PT*Procalcitonin*Troponin); (d-CIT: CRP*INR*Troponin)
LYM: Lymphocyte count, WBC: White blood cell count, PLT: Platelet count, CRP: C-reactive protein count, INR: international normalized ratio.
2.5. Design of independent biomarkers derived in this study and study workflow
Independent biomarkers derived from our approach in this study will provide an alternative to clinical approaches in predicting the diagnosis and prognosis of Covid-19. While deriving biomarkers, our research question was: Will the biomarkers we derive be more successful in predicting the diagnosis or prognosis of the disease than existing biomarkers in the literature or direct blood values?
In order to determine the predictors that are effective in the diagnosis of the disease, the difference of the predictors between the ICU-Control and nonICU-Control groups was examined. In addition, in determining the predictors that are effective in the prognosis of the disease, the difference of predictors between non-ICU and ICU was evaluated.
Direct values of blood values were used when designing biomarkers to predict Covid-19 or to separate patients into non-ICU and ICU units (ie, predict the prognosis of the disease). For this purpose, the following criteria were applied in the reporting of many different mathematical ratios (independent biomarkers) tested on the direct data of blood values: As a result of the Multivariate Logistic Regression, which was run to estimate the individual effect of the predictors for the diagnosis and prognosis of the disease, the predictor was 1) significant odds-ratio (OR) (ie, have significant influence); 2) significant AUC (area under the receiver operation characteristic curve (ROC)) value and 3) the multicollinearity problem between newly derived variables and direct blood values was controlled by the VIF (Variance Inflation Factor) criterion. As a result of all trials, new biomarkers found in models with VIF < 10 were taken into account. The dependent variable for the diagnosis of the disease was patient-control (binary), the dependent variable for prognosis was non-ICU and ICU (binary).
In addition, if the predictors were evaluated together, would there be an increase in success in predicting the diagnosis and prognosis according to their individual evaluations?
Therefore, decision trees suitable for the use of readers-clinicians were drawn for the collective evaluation of predictors that were found to be effective and successful in the diagnosis and prognosis of the disease. CHAID algorithm was used in drawing decision trees.
2.6. Evaluation of the predictors
The values of the predictors in terms of accuracy, sensitivity, specificity, area under the ROC curve (AUC) were obtained. Since false negatives for diagnosis (i.e., Covid-19 positive patients who were classified as negative and possibly allowed to go home) were more harmful than false positives in this screening task, accuracy (success in identifying patients and healthy individuals in total) and AUC (how positive were those determined as positive actually were)) as the main quality criteria.
Similarly, in differentiating patients between non-ICU and ICU units (i.e., in determining the prognosis of the disease) false negatives (i.e., covidites in non-ICU while they should be in the ICU) are more harmful than false positives in this screening task, was taken accuracy and AUC quality criteria evaluated as.
2.7. Statistical analysis of the study
Categorical variables were represented as frequency and percentage, while average, median, minimum, maximum, and standard deviation values were given for continuous variables. Shapiro-Wilk test was used to verify the normality of distributions of quantitative variables. Mann-WhitneyU test was used to compare continuous variables. Categorical variables were analyzed using the test. Multivariate logistic regression was used to evaluate the effect (oddsi-ratio) of age, gender and other predictors on admission to the intensive care unit / all non-intensive care units and disease detection [4]. The ROC curve was performed to determine the optimum breakpoints of laboratory parameters and biomarkers in the diagnosis and prognosis of the disease. CHAID analysis, one of the Decision Tree methods, was used to collectively evaluate the predictor variables in predicting the diagnosis and prognosis of the disease. SPSS (version 20.0, SPSS Inc, Chicago) program was used for statistical analysis of the data. p < 0.05 was considered statistically significantly.
2.8. CHAID analysis
CHAID analysis is a nonparametric method that can model categorical and continuous variables together, providing reliable estimates in large samples. In addition to these advantages, it can detail the relationships between independent variables and provide easy-to-understand outputs in the form of trees, even in the most complex models. CHAID analysis with these advantages has a wide range of uses.
3. Results
3.1. Comparison of blood routine parameters and various biomarkers of non-ICU group and contol group on admission
Table 1 shows the blood routine parameters and various biomarker values of the non-ICU patient group and the control group. 2458 of 2648 Covid-19 patients who had blood routine examinations on admission were treated in the non-ICU group. Gender variable was not correlated with the individual's presence in the non-ICU or Control group (p > 0.585). There were many differences between the non-ICU and control groups in terms of blood routine parameters and biomarker values. non-ICU patients had higher C-reactive protein count (CRP) (22.2 vs 9.4 × mg/L), higher D-Dimer value (545.1 vs 404.0 × μg/L), higher Ferritin value (183.7 vs 55.0 × μg/L), higher Fibrinogen value (320.5 vs 308.7 × mg/L), higher Procalcitonin value (0.2 vs 0.1 × μg/L), higher activated partial prothrombin time (aPPT) (31.2 vs 30.3 × sec). In addition, there was no significant difference in both groups in terms of international normalized ratio (INR), prothrombin time (PT) and Troponin levels (p > 0.05). Significant difference was observed between the non-ICU and ICU groups in all biomarkers (derived and available biomarkers) except d-TI.
Table 1.
Blood routine parameter and biomarker values of non-ICU group and Control group on admission.
| Non-ICU | Control | p-value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex Male (%) Female (%) |
1219 (49.6) 1239 (50.4) |
1293 (48.8) 1355 (51.2) |
0.585 | |||||||||
| Mean | Median | Min | Max | St D | Mean | Median | Min | Max | St D | |||
| Age | 53.6 | 55.0 | 18.0 | 102.0 | 18.6 | 49.0 | 48.0 | 18.0 | 102.0 | 19.5 | 0.000*** | |
| Blood routine parameters | CRP (mg/L) | 22.2 | 6.1 | 3.0 | 406.0 | 36.5 | 9.4 | 4.1 | 1.0 | 330.0 | 24.7 | 0.000*** |
| D-Dimer (μg/L) | 545.1 | 391.0 | 1.06 | 9240.0 | 684.44 | 404.0 | 391.0 | 104.0 | 9610.0 | 249.01 | 0.000*** | |
| Ferritin (μg/L) | 183.7 | 49.2 | 2.2 | 1650.0 | 281.8 | 55.0 | 49.2 | 0.2 | 1650.0 | 65.2 | 0.000*** | |
| Fibrinogen (mg/L) | 320.5 | 308.6 | 90.2 | 668.1 | 55.3 | 308.7 | 308.6 | 169.9 | 437.8 | 7.2 | 0.000*** | |
| INR | 1.11 | 1.10 | 0.8 | 17.8 | 0.4 | 1.2 | 1.15 | 0.8 | 7.0 | 0.4 | 0.620 | |
| PT (Sec) | 13.3 | 13.0 | 10.1 | 181.0 | 4.0 | 13.6 | 13.0 | 9.1 | 74.9 | 3.8 | 0.698 | |
| Procalcitonin (μg/L) | 0.2 | 0.1 | 0.1 | 24.0 | 0.6 | 0.1 | 0.1 | 0.1 | 27.0 | 0.8 | 0.000*** | |
| ESR (mm/h) | 17.8 | 11.0 | 2.0 | 124.0 | 18.0 | 13.9 | 11.0 | 2.0 | 120.0 | 10.8 | 0.000*** | |
| Troponin (ng/L) | 18.3 | 10.0 | 10.0 | 420.0 | 13.9 | 33.3 | 10.0 | 10.0 | 2500.0 | 541.9 | 0.889 | |
| aPPT (Sec) | 31.2 | 29.8 | 12.0 | 101.0 | 5.5 | 30.3 | 29.8 | 12.0 | 101.0 | 4.1 | 0.000*** | |
| PLT. ×109/L | 241.6 | 229.0 | 11.0 | 745.0 | 89.1 | 261.6 | 253.0 | 9.0 | 768.0 | 74.2 | 0.000*** | |
| LYM. ×109/L | 1.7 | 1.4 | 0.1 | 90.3 | 3.0 | 2.3 | 2.1 | 0.2 | 60.5 | 1.8 | 0.000*** | |
| NEU. ×109/L | 5.3 | 4.3 | 0.5 | 66.4 | 3.8 | 5.1 | 4.5 | 0.8 | 22.2 | 2.6 | 0.001** | |
| WBC. ×109/L | 7.7 | 6.7 | 0.4 | 127.0 | 5.2 | 8.2 | 7.6 | 1.5 | 68.3 | 3.3 | 0.000*** | |
| Biomarkers in the literature | LCR | 0.3 | 0.2 | 0.01 | 22.5 | 0.7 | 0.4 | 0.2 | 0.01 | 6.3 | 0.3 | 0.000*** |
| PLR | 214.5 | 155.2 | 1.6 | 3054.6 | 185.0 | 138.0 | 118.8 | 3.1 | 2527.3 | 93.2 | 0.000*** | |
| NLR | 5.8 | 2.7 | 0.1 | 255.7 | 10.9 | 2.8 | 2.1 | 0.1 | 50.9 | 2.9 | 0.000*** | |
| d-NLR | 3.3 | 1.9 | 0.1 | 84.0 | 4.2 | 1.9 | 1.5 | 0.1 | 25.5 | 1.5 | 0.000*** | |
| Biomarkers derived in this study | d-CL | 21.2 | 5.5 | 0.04 | 1923.1 | 59.3 | 5.19 | 1.25 | 0.1 | 357.1 | 16.5 | 0.000*** |
| d-CWL | 3.57 | 0.83 | 0.0 | 209.8 | 10.6 | 0.77 | 0.26 | 0.0 | 53.1 | 2.7 | 0.000*** | |
| d-CFL | 6663.4 | 450.0 | 2.18 | 621117.5 | 27224.5 | 341.1 | 95.2 | 0.16 | 56713.3 | 1887.6 | 0.000*** | |
| d-CI | 23.3 | 7.3 | 3.9 | 407.1 | 36.5 | 10.5 | 5.2 | 2.1 | 331.1 | 24.7 | 0.000*** | |
| d-CT | 414.7 | 64.1 | 30.0 | 195670.0 | 4916.1 | 326.3 | 40.7 | 10.0 | 335800.0 | 7118.9 | 0.000*** | |
| d-TI | 20.2 | 10.9 | 8.3 | 4578.0 | 146.9 | 41.1 | 10.9 | 7.5 | 37000.0 | 769.5 | 1.00 | |
| d-PPT | 42.15 | 15.6 | 12.1 | 8977.8 | 320.9 | 62.1 | 15.6 | 10.9 | 52200.0 | 1091.9 | 0.000*** | |
| d-CIT | 460.4 | 70.3 | 25.1 | 213280.3 | 5475.6 | 384.8 | 44.3 | 10.9 | 366022.0 | 8217.7 | 0.000*** | |
aPPT: activated partial prothrombin time; CRP: C-reactive protein; ESR: erythrocyte sedimentation rate; INR: international normalized ratio PT: prothrombin time; d-NLR: neutrophil count divided by the result of WBC count minus neutrophil count; LCR: lymphocyte to C-reactive protein ratio; NLR: neutrophil to lymphocyte ratio; PLR: platelet to lymphocyte ratio; INR: international normalized ratio; PLT: platelet count; LYM: lymphocyte count; WBC: white blood cell count; NEU: neutrophil count;; d-CL: CRP to lymphocyte ratio; d-CWL: CRP divided by WBC times LYM; d-CFL: CRP times Ferritin result divided by LYM; d-CI: CRP times INR; d-CT: CRP times Troponin; d-TI: Troponin times INR; d-PPT: PT times Procalcitonin times Troponin; d-CIT: CRP times INR times Troponin (see method section). Bold p values were found to be significant. *p < 0.05, **p < 0.01, ***p < 0.001 was considered significant. St D: Standart Devision.
3.2. Comparison of blood routine parameters and various biomarkers of ICU group and contol group on admission
Table 2 shows the blood routine parameters and biomarker values of the ICU patient group and the control group on admission. 190 of the 2648 Covid-19 patients who had blood routine examinations on admission were treated in the ICU group. The majority of this group consists of males and their mean age is significantly higher than the control group (p < 0.03). There were many differences between the ICU and control groups in terms of blood routine parameters and biomarker values. ICU patients had higher CRP (78.0 vs 9.4 × mg/L), higher D-Dimer value (1049.8 vs 404.0 × μg/L), higher Ferritin value (345.9 vs 55.0 × μg/L), higher Fibrinogen value (322.2 vs 308.7 × mg/L), higher INR (1.19 vs 1.15), higher PT (14.1 vs 13.6 × SEC), higher Procalcitonin value (1.1 vs 0.1 × μg/L), higher erythrocyte sedimentation rate (ESR) (38.0 vs 13.9 × (mm/h), higher Troponin value (46.2 vs 33.3 × ng/L), higher aPPT (32.6 vs 30.3 × SEC). However, the control group had higher platelet (PLT) count (261.6 vs 243.4 × 109/L, higher lymphocyte (LYM) count (2.3 vs 1.5 × 109/L), higher white blood cell (WBC) count (8.2 vs 7.2 × 109/L). Also, neutrophil (NEU) counts were not significantly different between control and ICU groups (p > 0.05). With the exception of LCR, all biomarkers (derived and available biomarkers) were higher in ICU patients.
Table 2.
Blood routine parameter and biomarker values of ICU group and Control group on admission.
| ICU | Control | p-value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex Male (%) Female (%) |
114 (60.0) 76 (40.0) |
1293 (48.8) 1355 (51.2) |
0.003** | |||||||||
| Mean | Median | Min | Max | St. D | Mean | Median | Min | Max | St D | |||
| Age | 72.9 | 75.0 | 20.0 | 100.0 | 11.9 | 49.0 | 48.0 | 18.0 | 102.0 | 19.5 | 0.000*** | |
| Blood routine parameters | CRP (mg/L) | 78.0 | 54.5 | 3.0 | 325.0 | 77.2 | 9.4 | 4.1 | 1.0 | 330.0 | 24.7 | 0.000*** |
| D-Dimer (μg/L) | 1049.8 | 391.0 | 353.0 | 9610.0 | 1755.1 | 404.0 | 391.0 | 104.0 | 9610.0 | 249.0 | 0.000*** | |
| Ferritin (μg/L) | 345.9 | 49.2 | 49.2 | 1650. | 493.2 | 55.0 | 49.2 | 0.2 | 1650.0 | 65.2 | 0.000*** | |
| Fibrinogen (mg/L) | 322.2 | 308.6 | 213.0 | 501.4 | 45.3 | 308.7 | 308.6 | 169.9 | 437.8 | 7.2 | 0.000*** | |
| INR | 1.19 | 1.1 | 0.9 | 2.4 | 0.2 | 1.15 | 1.1 | 0.8 | 7.0 | 0.4 | 0.000*** | |
| PT (Sec) | 14.1 | 13.1 | 10.9 | 27.7 | 2.3 | 13.6 | 13.0 | 9.1 | 74.9 | 3.8 | 0.000*** | |
| Procalcitonin (μg/L) | 1.1 | 0.1 | 0.12 | 88.0 | 6.7 | 0.1 | 0.1 | 0.1 | 27.0 | 0.8 | 0.000*** | |
| ESR (mm/h) | 38.0 | 11.0 | 2.0 | 140.0 | 41.1 | 13.9 | 11.0 | 2.0 | 120.0 | 10.8 | 0.000*** | |
| Troponin (ng/L) | 46.2 | 10.0 | 8.9 | 1500.0 | 150.0 | 33.3 | 10.0 | 10.0 | 2500.0 | 541.9 | 0.000*** | |
| aPPT (Sec) | 32.6 | 29.8 | 19.9 | 94.6 | 8.5 | 30.3 | 29.8 | 12.0 | 101.0 | 4.1 | 0.002 | |
| PLT, ×109/L | 243.4 | 232.5 | 17.0 | 715.0 | 87.2 | 261.6 | 253.0 | 9.0 | 768.0 | 74.2 | 0.000*** | |
| LYM, ×109/L | 1.5 | 1.4 | 0.17 | 3.6 | 0.75 | 2.3 | 2.1 | 0.2 | 60.5 | 1.8 | 0.000*** | |
| NEU, ×109/L | 5.04 | 4.27 | 1.02 | 20.0 | 3.0 | 5.1 | 4.5 | 0.8 | 22.2 | 2.6 | 0.453 | |
| WBC, ×109/L | 7.2 | 6.7 | 2.3 | 24.0 | 3.14 | 8.2 | 7.6 | 1.5 | 68.3 | 3.3 | 0.000*** | |
| Biomarkers in the literature | LCR | 0.1 | 0.04 | 0.01 | 0.75 | 0.12 | 0.4 | 0.2 | 0.01 | 6.3 | 0.3 | 0.000*** |
| PLR | 212.3 | 161.7 | 30.9 | 1380.7 | 180.5 | 138.0 | 118.8 | 3.1 | 2527.3 | 93.2 | 0.000*** | |
| NLR | 5.0 | 2.8 | 0.6 | 55.7 | 6.76 | 2.8 | 2.1 | 0.1 | 50.9 | 2.9 | 0.000*** | |
| d-NLR | 2.9 | 1.9 | 0.5 | 18.6 | 2.7 | 1.9 | 1.5 | 0.1 | 25.5 | 1.5 | 0.000*** | |
| Biomarkers derived in this study | d-CL | 76.9 | 34.0 | 1.2 | 1147.1 | 127.1 | 5.19 | 1.25 | 0.1 | 357.1 | 16.5 | 0.000*** |
| d-CWL | 12.4 | 4.6 | 0.11 | 154.2 | 21.4 | 0.77 | 0.26 | 0.0 | 53.1 | 2.7 | 0.000*** | |
| d-CFL | 34497.5 | 3285.8 | 65.4 | 720000.0 | 95114.0 | 341.1 | 95.2 | 0.16 | 56713.3 | 1887.6 | 0.000*** | |
| d-CI | 93.5 | 61.2 | 3.3 | 484.3 | 96.8 | 10.5 | 5.2 | 2.1 | 331.1 | 24.7 | 0.000*** | |
| d-CT | 5302.6 | 609.0 | 30.2 | 340500.0 | 27969.3 | 326.3 | 40.7 | 10.0 | 335800.0 | 7118.9 | 0.000*** | |
| d-TI | 55.5 | 11.85 | 9.0 | 1800.0 | 177.6 | 41.1 | 10.9 | 7.5 | 37000.0 | 769.5 | 0.000*** | |
| d-PPT | 2486.0 | 19.9 | 12.9 | 340800.0 | 25223.9 | 62.1 | 15.6 | 10.9 | 52200.0 | 1091.9 | 0.000*** | |
| d-CIT | 6674.7 | 770.8 | 32.6 | 408600.0 | 34496.3 | 384.8 | 44.3 | 10.9 | 366022.0 | 8217.7 | 0.000*** | |
aPPT: activated partial prothrombin time; CRP: C-reactive protein; ESR: erythrocyte sedimentation rate; INR: international normalized ratio PT: prothrombin time; d-NLR: neutrophil count divided by the result of WBC count minus neutrophil count; LCR: lymphocyte to C-reactive protein ratio; NLR: neutrophil to lymphocyte ratio; PLR: platelet to lymphocyte ratio; INR: international normalized ratio; PLT: platelet count; LYM: lymphocyte count; WBC: white blood cell count; NEU: neutrophil count; d-CL: CRP to lymphocyte ratio; d-CWL: CRP divided by WBC times LYM; d-CFL: CRP times Ferritin result divided by LYM; d-CL: CRP to lymphocyte ratio; d-CWL: CRP divided by WBC times LYM; d-CFL: CRP times Ferritin result divided by LYM; d-CI: CRP times INR; d-CT: CRP times Troponin; d-TI: Troponin times INR; d-PPT: PT times Procalcitonin times Troponin; d-CIT: CRP times INR times Troponin (see method section). Bold p values were found to be significant. * p < 0.05, ** p < 0.01, *** p < 0.001 was considered significant. St D: Standart Devision.
3.3. Comparison of blood routine parameters and various biomarkers of non-ICU group and ICU group on admission
Table 3, presents the blood routine parameters and biomarker values of the ICU patient group and the non-ICU patient group on admission of the 2648 Covid-19 patients who underwent blood routine examinations, 190 were treated in the ICU group, and 2458 in the non-ICU group. While the majority of the non-ICU group is women, the majority of the ICU group is men (p < 0.06). The mean age of the ICU group was significantly higher than the non-ICU group (p < 0.05). Some differences were seen in blood routine parameters and biomarker values between ICU and non-ICU groups. ICU patients had higher CRP (78.0 vs 22.2 × mg/L), higher D-Dimer value (1049.8 vs 545.1 × μg/L), higher Ferritin value (345.9 × 183.7 × μg/L), normalized ratio of higher INR (1.19 vs 1.11), higher PT (14.1 vs 13.3 × SEC), higher Procalcitonin value (1.1 vs 0.2 × μg/L), higher ESR (38.0 vs 17.8 × mm/h), higher Troponin value (46.2 vs 18.3 ng/L). Fibrinogen, aPPT, PLT, LYM, NEU, WBC count and other biomarkers did not differ significantly between non-ICU and ICU patients. However, non-ICU group had higher LCR biomarker value (0.3 vs 0.1). All biomarkers derived in this study were higher in the ICU group.
Table 3.
Blood routine parameter and biomarker values of ICU group and non-ICU group on admission.
| Non-ICU |
ICU |
p-value | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sex Male (%) Female (%) |
1219 (49.6) 1239 (50.4) |
114 (60.0) 76 (40.0) |
0.006** | |||||||||
| Mean | Median | Min | Max | St D | Mean | Median | Min | Max | St D | |||
| Age | 53.6 | 55.0 | 18.0 | 102.0 | 18.6 | 72.9 | 75.0 | 20.0 | 100.0 | 72.9 | 0.000*** | |
| Blood routine parameters | CRP (mg/L) | 22.2 | 6.1 | 3.0 | 406.0 | 36.5 | 78.0 | 54.5 | 3.0 | 325.0 | 78.0 | 0.000*** |
| D-Dimer (μg/L) | 545.1 | 391.0 | 1.06 | 9240.0 | 684.44 | 1049.8 | 391.0 | 353.0 | 9610.0 | 1049.8 | 0.000*** | |
| Ferritin (μg/L) | 183.7 | 49.2 | 2.2 | 1650.0 | 281.8 | 345.9 | 49.2 | 49.2 | 1650. | 345.9 | 0.028* | |
| Fibrinogen (mg/L) | 320.5 | 308.6 | 90.2 | 668.1 | 55.3 | 322.2 | 308.6 | 213.0 | 501.4 | 322.2 | 0.150 | |
| INR | 1.11 | 1.1 | 0.8 | 17.8 | 0.4 | 1.19 | 1.1 | 0.9 | 2.4 | 1.2 | 0.000*** | |
| PT (Sec) | 13.3 | 13.0 | 10.1 | 181.0 | 4.0 | 14.1 | 13.1 | 10.9 | 27.7 | 14.1 | 0.000*** | |
| Procalcitonin (μg/L) | 0.2 | 0.1 | 0.1 | 24.0 | 0.6 | 1.1 | 0.1 | 0.12 | 88.0 | 1.1 | 0.000*** | |
| ESR (mm/h) | 17.8 | 11.0 | 2.0 | 124.0 | 18.0 | 38.0 | 11.0 | 2.0 | 140.0 | 38.0 | 0.000*** | |
| Troponin (ng/L) | 18.3 | 10.0 | 10.0 | 420.0 | 13.9 | 46.2 | 10.0 | 8.9 | 1500.0 | 46.2 | 0.000*** | |
| aPPT (Sec) | 31.2 | 29.8 | 12.0 | 101.0 | 5.5 | 32.6 | 29.8 | 19.9 | 94.6 | 32.6 | 0.547 | |
| PLT, ×109/L | 241.6 | 229.0 | 11.0 | 745.0 | 89.1 | 243.4 | 232.5 | 17.0 | 715.0 | 243.4 | 0.841 | |
| LYM, ×109/L | 1.7 | 1.4 | 0.1 | 90.3 | 3.0 | 1.5 | 1.4 | 0.17 | 3.6 | 1.5 | 0.554 | |
| NEU, ×109/L | 5.3 | 4.3 | 0.5 | 66.4 | 3.8 | 5.04 | 4.27 | 1.02 | 20.0 | 5.04 | 0.115 | |
| WBC, ×109/L | 7.7 | 6.7 | 0.4 | 127.0 | 5.2 | 7.2 | 6.7 | 2.3 | 24.0 | 7.2 | 0.088 | |
| Biomarkers in the literature | LCR | 0.3 | 0.2 | 0.01 | 22.5 | 0.7 | 0.1 | 0.04 | 0.01 | 0.75 | 0.1 | 0.000*** |
| PLR | 214.5 | 155.2 | 1.6 | 3054.6 | 185.0 | 212.3 | 161.7 | 30.9 | 1380.7 | 212.3 | 0.090 | |
| NLR | 5.8 | 2.7 | 0.1 | 255.7 | 10.9 | 5.0 | 2.8 | 0.6 | 55.7 | 5.0 | 0.055 | |
| d-NLR | 3.3 | 1.9 | 0.1 | 84.0 | 4.2 | 2.9 | 1.9 | 0.5 | 18.6 | 2.9 | 0.268 | |
| Biomarkers derived in this study | d-CL | 21.2 | 5.5 | 0.04 | 1923.1 | 59.3 | 76.9 | 34.0 | 1.2 | 1147.1 | 127.1 | 0.000*** |
| d-CWL | 3.57 | 0.83 | 0.0 | 209.8 | 10.6 | 12.4 | 4.6 | 0.11 | 154.2 | 21.4 | 0.000*** | |
| d-CFL | 6663.4 | 450.0 | 2.18 | 621117.5 | 27224.5 | 34497.5 | 3285.8 | 65.4 | 720000.0 | 95114.0 | 0.000*** | |
| d-CI | 23.3 | 7.3 | 3.9 | 407.1 | 36.5 | 93.5 | 61.2 | 3.3 | 484.3 | 96.8 | 0.000*** | |
| d-CT | 414.7 | 64.1 | 30.0 | 195670.0 | 4916.1 | 5302.6 | 609.0 | 30.2 | 340500.0 | 27969.3 | 0.000*** | |
| d-TI | 20.2 | 10.9 | 8.3 | 4578.0 | 146.9 | 55.5 | 11.85 | 9.0 | 1800.0 | 177.6 | 0.000*** | |
| d-PPT | 42.15 | 15.6 | 12.1 | 8977.8 | 320.9 | 2486.0 | 19.9 | 12.9 | 340800.0 | 25223.9 | 0.000*** | |
| d-CIT | 460.4 | 70.3 | 25.1 | 213280.3 | 5475.6 | 6674.7 | 770.8 | 32.6 | 408600.0 | 34496.3 | 0.000*** | |
aPPT: activated partial prothrombin time; CRP: C-reactive protein; ESR: erythrocyte sedimentation rate; INR: international normalized ratio PT: prothrombin time; d-NLR: neutrophil count divided by the result of WBC count minus neutrophil count; LCR: lymphocyte to C-reactive protein ratio; NLR: neutrophil to lymphocyte ratio; PLR: platelet to lymphocyte ratio; INR: international normalized ratio; PLT: platelet count; LYM: lymphocyte count; WBC: white blood cell count; NEU: neutrophil count; d-CL: CRP to lymphocyte ratio; d-CWL: CRP divided by WBC times LYM; d-CFL: CRP times Ferritin result divided by LYM; d-CI: CRP times INR; d-CT: CRP times Troponin; d-TI: Troponin times INR; d-PPT: PT times Procalcitonin times Troponin; d-CIT: CRP times INR times Troponin (see method section). Bold p values were found to be significant. * p < 0.05, ** p < 0.01, *** p < 0.001 was considered significant. St D: Standart Devision.
3.4. Analysis of the efficiency of blood routine parameters and biomarkers in the diagnosis of Covid-19 on admission
Those who were treated in the hospital with the definitive diagnosis of Covid-19 (non-ICU and ICU total 2648 people) were determined as a positive group. No Covid-19 patients (2648 people who applied to the hospital for other complaints) were determined as the control group (negative group). We obtained the crude odds-ratio (OR) after performing multivariate logistic regression analysis to identify predictors that may affect the diagnosis of Covid-19 (Table 4 ). Given that the blood test results were influenced by age and gender, we excluded the possible effects of age and gender and obtained the adjusted OR after the adjustment of gender and age [4], [20].
Table 4.
Risk coefficients of indicators and biomarkers in the diagnosis of COVID-19 disease.
| Predictors Biomarkers | Crude odds ratio (OR) (95% CI) | p-value | Adjusted odds ratio (ORa)a (95% CI) | p-value |
|---|---|---|---|---|
| CRP (mg/L) | 2.02 (1.01–3.03) | 0.000*** | 1.07 (1.00–1.14) | 0.038* |
| Ferritin (μg/L) | 3.01 (1.11–3.25) | 0.000*** | 1.09 (1.08–1.11) | 0.000*** |
| Procalcitonin (μg/L) | 1.61 (1.21–1.70) | 0.000*** | 1.12 (0.96–1.09) | 0.000*** |
| LYM, x109/L | 0.79 (0.70–0.90) | 0.000*** | 1.28 (1.21–1.37) | 0.000*** |
| WBC, x109/L | 0.87 (0.83–0.91) | 0.000*** | 0.80 (0.76–0.83) | 0.124 |
| NLR | 1.44 (1.02–1.45) | 0.000*** | 1.14 (1.06–1.23) | 0.065 |
| PLR | 2.29 (1.09–3.99) | 0.000*** | 1.01 (1.00–1.02) | 0.046* |
| d-NLR | 1.37 (1.02–1.45) | 0.022* | 1.27 (1.11–1.46) | 0.000*** |
| d-CL | 1.11 (1.01–1.27) | 0.000*** | 1.045 (1.04–1.05) | 0.000*** |
| d-CWL | 1.33 (1.23–1.37) | 0.000*** | 1.81 (1.17–1.94) | 0.000*** |
| d-CFL | 1.10 (1.00–1.60) | 0.000*** | 1.17 (1.14–1.20) | 0.000*** |
CRP: C-reactive protein; d-NLR: neutrophil count divided by the result of WBC count minus neutrophil count; NLR: neutrophil to lymphocyte ratio; PLR: platelet to lymphocyte ratio; WBC: white blood cell count; d-CL: CRP to lymphocyte ratio; d-CWL: CRP divided by WBC times LYM; d-CFL: CRP times Ferritin result divided by LYM.
Adjustment for age and gender (* p < 0.05, **p < 0.01, ***p < 0.001 was considered significant).
By multivariate logistic regression analysis, CRP, Ferritin, Procalcitonin, LYM, WBC, NLR, PLR, d-NLR, d-CL, d-CWL and d-CFL values of ORs were found to be significant the diagnosis of the disease (p < 0.05). However, WBC and NLR, was not found to be significant when these variables were modeled according to age and gender. (p > 0.05) (see Table 4).
3.5. Evaluating the diagnostic performance of routine blood parameters and biomarkers in the diagnosis of Covid-19
Those who were treated with the definitive diagnosis of Covid-19 were determined as a positive group. The control group that was not diagnosed with Covid-19 was determined as a negative group. The performances of the predictors with significant odds-ratio in the diagnosis of the disease (Table 4) were obtained (Table 5 ) and cut off values were determined by drawing the ROC curves (Fig. 1 ).
Table 5.
Diagnostic values of parameters and biomarkers in the detection of Covid-19 on admission.
| Blood routine parameters and Biomarkers | AUC (95% CI) |
Cutt of value | Sensitivity (95% CI) |
Specificity (95% CI) |
Accuracy (%) | p-value |
|---|---|---|---|---|---|---|
| CRP (mg/L) | 0.618 (0.601–0.634) |
> 4.06 | 61.6 (60.4–66.8) |
81.0 (75.4–78.3) |
63.77 | 0.000*** |
| Ferritin (μg/L) | 0.671 (0.655–0.686) |
> 49.15 | 47.3 (55.4–59.2) |
92.97 (91.9–93.9) |
67.44 | 0.000*** |
| Procalcitonin (μg/L) | 0.613 (0.597–0.628) |
> 0.12 | 23.6 (15.7–53.4) |
98.7 (79.5–99.3) |
62.18 | 0.000*** |
| NLR | 0.621 (0.606–0.637) |
> 1.84 | 71.7 (70.0–73.3) |
49.8 (37.9–51.8) |
60.80 | 0.000*** |
| PLR | 0.662 (0.647–0.676) |
> 103.8 | 81.3 (79.8–82.7) |
54.6 (52.7–66.4) |
68.60 | 0.000*** |
| d-.NLR | 0.611 (0.595–0.628) |
> 1.41 | 68.7 (67.0–70.5) |
51.9 (50.0–43.8) |
60.70 | 0.000*** |
| LYM, x109/L | 0.730 (0.72–0.75) |
< 1.41 | 85.6 (76.0–90.2) |
50.8 (34.7–70.9) |
71.20 | 0.000*** |
| WBC, x109/L | 0.60 (0.58–0.61) |
< 6.25 | 54.9 (53.0–56.7) |
73.6 (71.9–75.3) |
64.30 | 0.000*** |
| d -CL | 0.766 (0.75–0.78) |
> 2.57 | 72.4 (59.1–83.3) |
70.0 (58.4–80.6) |
74.20 | 0.000*** |
| d-CWL | 0.778 (0.77–0.79) |
> 0.52 | 68.5 (51.3–76.2) |
80.6 (69.2–88.4) |
77.60 | 0.000*** |
| d-CFL | 0.783 (0.77–0.80) |
> 137.8 | 78.9 (64.6–83.6) |
72.2 (56.1–85.4) |
79.54 | 0.000*** |
CRP: C-reactive protein; d-NLR: neutrophil count divided by the result of WBC count minus neutrophil count; NLR: neutrophil to lymphocyte ratio; PLR: platelet to lymphocyte ratio; LYM: lymphocyte count; WBC: white blood cell count; d-CL: CRP to lymphocyte ratio; d-CWL: CRP divided by WBC times LYM; d-CFL: CRP times Ferritin result divided by LYM (see Section 2) (***p < 0.001 was considered significant).
Fig. 1.
The left figure and the middle figure are the ROC curves for the diagnosis of Covid-19 of biomarkers derived with blood parameters. The figure on the right is the ROC curves of the biomarkers used in the literature for the diagnosis of this study. CRP: C-reactive protein; d-NLR: derived neutrophil to lymphocyte ratio; NLR: neutrophil to lymphocyte ratio; PLR: platelet to lymphocyte ratio; LYM: lymphocyte count; WBC: white blood cell count; d-CL: CRP to lymphocyte ratio; d-CWL: CRP divided by WBC times LYM; d-CFL: CRP times Ferritin result divided by LYM (see Section 2).
When looking at the diagnostic values of blood parameters and independent biomarkers in Table 5, it is seen that the specificty value in favor of sensitivity sometimes decreases. Since false negatives for diagnosis were more harmful than false positives in this screening task, was taken accuracy (ACC) and AUC as the main quality criteria (see Section 2.6). When looking at the area under the curve (AUC) and the value of accuracy (ACC), the CRP is 0.62 (%) – 0.64 (%), the Ferritin is 0.67 (%) – 0.67 (%), the LYM is 0.73 (%) – 0.71 (%) and the WBC is 0.60 (%) – 0.64 (%), respectively. However, the biomarkers derived in this study were found to be more successful in detecting true positives (AUC) and patient-healthy discrimination (ACC) than older biomarkers and routine blood values. When looking at the AUC and the ACC, the d-CL is 0.76 (%) – 0.74 (%), the d-CWL is 0.78 (%) – 0.77 (%),the d-CFL is 0.78 (%) – 0.79 (%) (see Table 5) (the all p values < 0.001).
3.6. Analysis of the efficiency of blood routine parameters and biomarkers in the diagnosis of
3.6.1. ICU (intensive care unit) on admission
190 patients who were treated in the ICU unit with the diagnosis of Covid-19 in the hospital were determined as positive groups. 2458 patients who were treated in non-ICU units with the diagnosis of Covid-19 were determined as negative group. We obtained the crude odds-ratio (OR) after performing multivariate logistic regression analysis to determine the predictors that may affect the determination of patients to non-ICU and ICU (Table 6 ). Given that the blood test results were influenced by age and gender, we excluded the possible effects of age and gender and obtained the adjusted OR after the adjustment of gender and age [4], [5].
Table 6.
Risk coefficients of indicators and biomarkers in the diagnosis of ICU (intensive care unit) on admission.
| Predictors Biomarkers | Crude odds ratio (OR) (95% CI) |
p-value | Adjusted odds ratio (ORa)a (95% CI) | p-value |
|---|---|---|---|---|
| CRP (mg/L) | 1.34 (1.13–1.47) | 0.000*** | 0.97 (0.95–0.99) | 0.006** |
| INR | 1.28 (1.11–1.37) | 0.014* | 1.63 (1.01–2.60) | 0.046* |
| PT (Sec) | 1.23 (1.05–1.39) | 0.006** | 1.01 (0.96–1.05) | 0.836 |
| Procalcitonin (μg/L) | 1.15 (1.01–1.31) | 0.043* | 1.10 (0.93–1.28) | 0.250 |
| ESR (mm/h) | 1.12 (1.02–1.14) | 0.000*** | 1.02 (1.01–1.02) | 0.600 |
| Troponin (ng/L) | 1.10 (1.03–1.15) | 0.011* | 1.00 (0.94–1.06) | 0.874 |
| LCR | 0.30 (0.24–0.37) | 0.000*** | 0.28 (0.08–0.90) | 0.031* |
| d-CL | 1.71 (1.10–1.90) | 0.007** | 1.00 (1.00–1.01) | 0.186 |
| d-CI | 1.24 (1.10–1.71) | 0.000*** | 1.04 (1.02–1.05) | 0.000*** |
| d-CT | 1.02 (1.00–1.21) | 0.001** | 1.00 (0.99–1.00) | 0.780 |
| d-TI | 1.10 (1.00–1.12) | 0.014* | 1.04 (0.95–1.06) | 0.872 |
| d-PPT | 1.02 (1.00–1.11) | 0.030* | 1.00 (1.00–1.11) | 0.030* |
| d-CIT | 1.08 (1.04–1.31) | 0.016* | 1.06 (1.00–1.30) | 0.016* |
CRP: C-reactive protein; ESR: erythrocyte sedimentation rate; INR: international normalized ratio PT: prothrombin time; LCR: lymphocyte to C-reactive protein ratio; d- CL: CRP to lymphocyte ratio; d-CI: CRP times INR; d-CT: CRP times Troponin; d-TI: Troponin times INR; d-PPT: PT times Procalcitonin times Troponin; d-CIT: CRP times INR times Troponin (see Section 2).
Adjustment for age and gender. (* p < 0.05, ** p < 0.01, *** p < 0.001 was considered significant).
By multivariate logistic regression analysis, CRP, INR, PT, Procalcitonin, ESR, Troponin, LCR, d-CL, d-CI, d-CT, d-TI, d-PPT and d-CIT values of odds-ratio’s were found to be significant the diagnosis of the disease (p < 0.05). However, PT, Procalcitonin, ESR, Troponin, d-CL, d-CT and d-TI was not found to be significant when these variables were modeled according to age and gender. (p > 0.05) (see Table 6).
3.7. Evaluation of the diagnostic performance of routine blood parameters and biomarkers in differentiating Covid-19 patients into non-ICU and ICU (in the prognosis of the disease)
Covid-19 patients treated in the ICU unit were determined as a positive group, non-ICU were identified as negative group. The receiver operating characteristic (ROC) curve was created to analyze the diagnostic performance of various routine blood parameters and biomarkers (parameters in Table 6) whose effectiveness in determining the diagnosis of ICU (significant OR) on admission (Fig. 2 ).
Fig. 2.
In the distinction between non-ICU and ICU (ie, prognosis of the disease): the left figure is the blood parameters; the middle figure is of newly derived biomarkers; The figure on the right is the ROC curves of biomarkers in the literature. CRP: C-reactive protein; INR: international normalized ratio; ESR: erythrocyte sedimentation rate; PT: prothrombin time; LCR: lymphocyte to C-reactive protein ratio; d-CL: CRP to lymphocyte ratio; d-CI: CRP times INR; d-CT: CRP times Troponin; d-TI: Troponin times INR; d-PPT: PT times Procalcitonin times Troponin; d-CIT: CRP times INR times Troponin (see Section 2).
When we look at the diagnostic performance values of blood parameters and independent biomarkers in determining ICU patients in Table 7 , it is seen that the specificity value in favor of sensitivity sometimes decreases. Since false negatives (false non-ICU) are more harmful than false positives (false ICU) in determining the ICU group (in determining the prognosis of the disease), accuracy (ACC) and AUC were taken as the main quality criteria (see Section 2.6). When looking at the area under the curve (AUC) and the value of accuracy (ACC), the CRP is 0.77 (%) – 0.71 (%), the INR is 0.67 (%) – 0.66 (%), the Procalcitonin is 0.64 (%) – 0.63 (%), the PT is 0.67 (%) – 0.70 (%) and the Troponin is 0.61 (%) – 0.65 (%), respectively. However, biomarkers derived in this study were found to be more successful in identifying ICU patients (true positives-AUC) and identifying non-ICU and ICU patients (ACC) than older biomarkers and routine blood values (see Table 7). When looking at the AUC and the ACC, the d-CL is 0.74 (%) – 0.71 (%),the d-CI is 0.78 (%) – 0.76 (%), the d-CT is 0.79 (%) – 0.80 (%), the d-TI is 0.73 (%) – 0.72 (%), the d-PPT is 0.78 (%) – 0.79 (%),the d-CIT is 0.80 (%) – 0.83 (%). (the all p values < 0.001) (see Table 7).
Table 7.
Diagnostic values of parameters and biomarkers in the detection of ICU on admission.
| Blood routine parameters and Biomarkers | AUC (95% CI) |
Cutt of value | Sensitivity (95% CI) |
Specificity (95% CI) |
Accuracy (%) | p-value |
|---|---|---|---|---|---|---|
| CRP (mg/L) | 0.77 (0.74–0.81) |
> 25.95 | 90.73 (65.3–99.5) |
41.3 (34.5–77.9) |
71.5 | 0.000*** |
| INR | 0.67 (0.62–0.72) |
> 1.09 | 51.6 (54.2–68.9) |
81.8 (84.4–87.2) |
66.7 | 0.000*** |
| Procalcitonin (μg/L) | 0.64 (0.59–0.69) |
> 0.14 | 45.3 (40.7–54.3) |
80.7 (74.5–88.3) |
63.0 | 0.000*** |
| PT(Sec) | 0.67 (0.62–0.72) |
< 12.9 | 53.7 (49.0–59.3) |
87.2 (85.9–88.4) |
70.2 | 0.000*** |
| ESR (mm/h) | 0.63 (0.59–0.68) |
> 11.5 | 44.3 (56.1–70.7) |
81.7 (69.9–83.5) |
63.0 | 0.000*** |
| Troponin (ng/L) | 0.61 (0.56–0.66) |
> 23.6 | 60.8 (56.1–74.2) |
61.2 (51.2–71.8) |
64.7 | 0.000*** |
| LCR | 0.72 (0.56–0.66) |
< 0.24 | 45.2 (29.3–61.5) |
90.1 (90.3–97.0) |
70.1 | 0.000*** |
| d-CL | 0.74 (77.3–77.9) |
> 5.87 | 79.7 (64.2–83.4) |
55.0 (43.6–78.9) |
71.4 |
0.000*** |
| d-CI | 0.78 (0.75–0.82) |
> 4.44 | 91.1 (86.3–94.3) |
41.4 (22.11–63.6) |
76.7 |
0.000*** |
| d-CT | 0.79 (0.76–0.83) |
> 40.6 | 97.4 (95.3–98.6) |
44.1 (27.2–62.1) |
80.7 | 0.000*** |
| d-TI | 0.73 (0.68–0.78) |
> 10.96 | 63.2 (52.2–73.3) |
83.0 (72.1–91.4) |
71.9 | 0.000*** |
| d-PPT | 0.78 (0.75–0.82) |
> 15.7 | 84.7 (74.3–92.1) |
67.4 (49.4–82.6) |
79.2 |
0.000*** |
| d-CIT | 0.80 (0.77–0.84) |
>52.0 | 91.5 (82.1–94.7) |
43.2 (20.7–70.1) |
83.6 |
0.000*** |
CRP: C-reactive protein; ESR: erythrocyte sedimentation rate; INR: international normalized ratio PT: prothrombin time; LCR: lymphocyte to C-reactive protein ratio; d-CL: CRP to lymphocyte ratio; d-CI: CRP times INR; d-CT: CRP times Troponin; d-TI: Troponin times INR; d-PPT: PT times Procalcitonin times Troponin; d-CIT: CRP times INR times Troponin (see Section 2) (see method section) (***p < 0.001 was considered significant).
4. Discussion
Covid-19 is a systemic multi-organ damage disease causing severe acute respiratory syndrome (SARS-CoV-2), which still causes significant death and disease in the world and continues to spread rapidly [3], [11], [12]. Regarding immunity to Covid-19, it is still not entirely clear what responses occur in Covid-19 and whether people recovering from Covid-19 infection are protected from a second infection [13], [14], [15]. With these clinical features of Covid-19, there is no specific treatment approved yet [13].
Therefore, predicting the diagnosis and prognosis of the disease is important for early intervention [4]. In a sense, it is possible to determine both the diagnosis and the prognosis of the disease with routine blood values (biomarkers) that are easier to access, more economical and faster. Similarly, one study said that dynamic surveillance of the Peripheral blood system, particularly eosinophils, helped predict severe cases of Covid-19 [5]. Indeed, this study found abnormal peripheral routine blood results in Covid-19 patients. These advantages have further increased the importance of routine blood values and will be a predictive aid in the current crisis.
In the study conducted for this purpose, changes were found in most of the routine laboratory tests and biomarkers. It was observed that these changes were more in intensive care patients. This study showed that older age and male sex are seriously threatened in terms of both Covid-19 and disease progression. Similarly, previous studies recorded advanced age and male gender as severe prognosis in Covid-19 [4], [16], [17], [18], [19].
In this study, the efficacy of routine blood values and biomarkers in the diagnosis of the disease (patient-healthy) and classification of patients into non-ICU or ICU (prognosis of the disease) were evaluated.
In this study, a significant increase was observed in routine blood parameters of CRP, D-Dimer, Ferritin, Fibrinogen, Procalcitonin, NEU, ESR, aPPT and old-new derived biomarkers in patients. Only the d-TI in mild covid patients was not different from healthy. However, in all patient groups, a significant decrease was observed in platelet, white blood cell count and lymphocyte counts and independent biomarker values of LCR. Similarly, in another study, it was emphasized that an increase in many laboratory parameters such as ferritin, D-Dimer and Fibrinogen is expected in Covid-19 [20], [21], [22]. This change in routine blood values is thought to be caused by viral infection, autoimmune and inflammatory conditions, and mineral deficiency in patients. Similarly, Guan et al showed abnormal lymphocyte and platelet parameters in the peripheral blood of 1099 patients with Covid-19 [5], [23].
Similarly, previous studies reported that lymphopenia is common in Covid-19 cases and is an important and reliable indicator of disease severity [5], [24], [25]. It was stated that lymphopenia can be caused by the Covid-19 virus, either directly (for example, being the target of the virus with the ACE2 receptors on them) or indirectly, which suppresses lymphocyte production or shortens the half-life of lymphocytes [26], [27], [28], [29].
In this study, biomarkers (derived) that are effective in the diagnosis of the disease were found to be more successful than the diagnostic performance of direct blood values (Table 5). Specifically, d-CWL and d-CFL were the highest in the diagnosis of the disease, AUC: 77.8%, ACC: 77.6% and AUC: 78.3%, ACC: 79.54%, respectively (Fig. 1). LYM, NLR, LCR, PLR, d-NLR, which were used as important biomarkers in the diagnosis of the disease in previous studies [4], [5], [19], [20], [29], [30], [31], performed lower than the d-CFL and d-CWL produced in this study (Table 5). In addition, it was observed that biomarkers derived to determine the diagnosis of the disease were not affected by gender and age, while NLR, which is frequently used in the diagnosis of the disease in the literature, was found to be affected (adjusted odds-ratio, see Table 4). Accordingly, the d-CWL and d-CFL biomarkers that we have derived instead of direct blood values and existing biomarkers can be used in the diagnosis of the disease.
However, these diagnostic achievements of blood values and biomarkers are individual. For a user-friendly guide that can be used in suspicious cases in the diagnosis of the disease, a decision tree that evaluates the predictors together was drawn (Fig. 3 ). The total classification accuracy rate of the decision tree, which was created in a statistically significant way with eight predictors in the diagnosis of the disease, was 81.6%, the accuracy rate for classifying patients was 85.2%, and the accuracy rate for classifying the healthy was 77.9%, and it was promising (Table 8 ). The presence of CRP and its derived biomarkers at the root of the tree was an interpretable confirmation of the accuracy of our Decision Tree (Fig. 3) in the approach to diagnosing the disease. Similarly, many studies [4], [18], [30] have stated that CRP and LYM are important biomarkers in determining the presence and severity of the disease.
Fig 3.
Decision tree evaluating the effects of predictive variables together in the diagnosis of Covid-19.
Table 8.
Estimated accuracy of blood parameters and independent biomarkers in the diagnosis of Covid-19 using CHAID analysis.
| Observed | Predicted |
||
|---|---|---|---|
| non-Covid-19 | Covid-19 | Percent Correct | |
| non-Covid-19 | 2064 | 584 | 77,9% |
| Covid-19 | 391 | 2257 | 85,2% |
| Overall Percentage | 46,4% | 53,6% | 81,6% |
It also increased the advantage of this decision tree (Fig. 3), in identifying patients (Table 8) with higher accuracy than direct blood values and individual achievements of all biomarkers (Table 5).
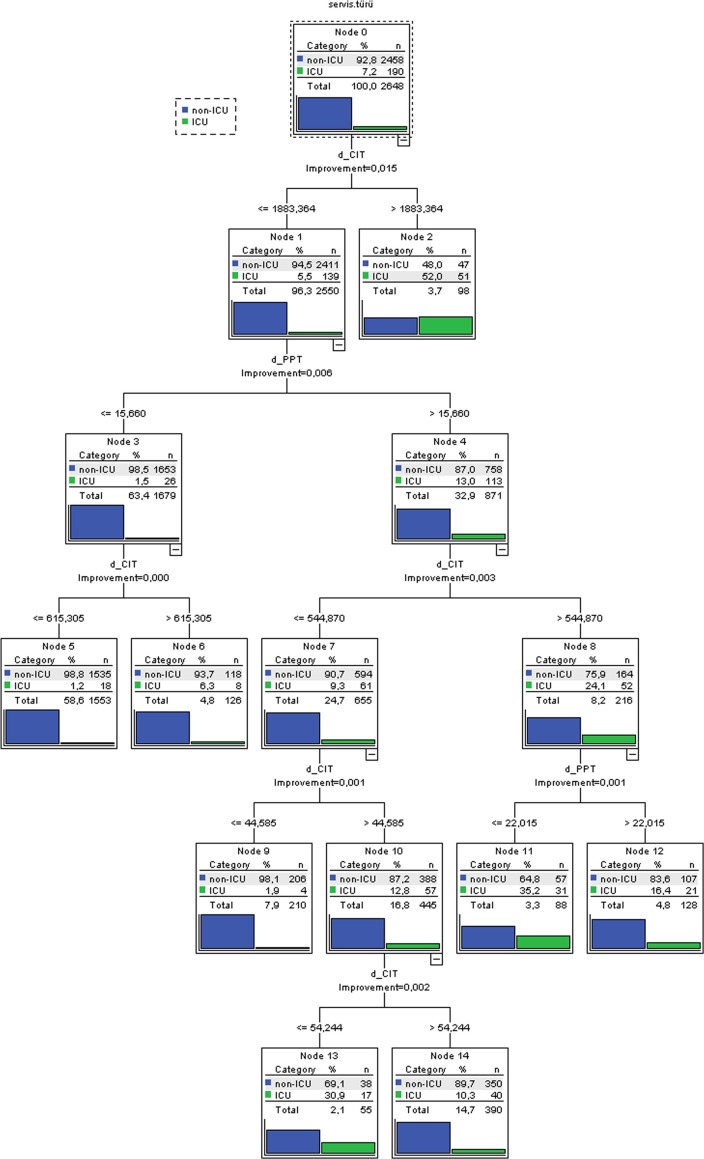
In this study, biomarkers (derived) that were effective in distinguishing ICU patients from non-ICU patients to determine disease progression were found to be more successful than the diagnostic performance of direct blood values (Table 7). Specifically, the d-CT, d-PPT, and d-CIT biomarkers reached AUC: 79.0%, ACC: 80.7% and AUC: 78.0%, ACC: 79.2%, AUC: 0.80%, ACC: 83.6 in identifying patients with ICU, respectively. Accordingly, instead of the direct values of CRP, INR, PT, Procalcitonin and Troponin, which are effective in the severity of the disease, d-CT, d-PPT and d-CIT biomarkers can be used more successfully in classifying patients into ICU.
A decision tree was drawn that interprets blood values and biomarkers that are effective in classifying patients into ICU and non-ICU together with a simple decision (Fig. 4 ). In determining patients to ICU or non-ICU, the total classification accuracy rate of the decision tree, which was created with d-CIT and d-PPT biomarkers in a statistically significant way, was 93.5%, the accuracy rate of classifying ICU patients was 26.8%, and the accuracy rate for classifying non-ICU patients was 98.1% (Table 9 ). It was not desirable that the general accuracy rate of the decision tree obtained in Fig. 4 was high in favor of non-ICU and low accuracy rate in determining ICU patients. Accordingly, our Covid-19 patient population could not be classified in a single decision tree with high accuracy for both non-ICU and ICU with current predictors. This situation showed that there are different factors that affect individuals' treatment in ICU and non-ICU (affecting the prognosis of the disease) than the changes in the current blood values.
Fig 4.
Decision tree evaluating the combined effect of predictive variables in determining patients to non-ICU and ICU in Covid-19.
Table 9.
Estimated accuracy of blood parameters and independent biomarkers in the differentiation of non-ICU and ICU (ie, prognosis of the disease) using CHAID analysis.
| Observed | Predicted |
||
|---|---|---|---|
| non-ICU | ICU | Percent Correct | |
| non-ICU | 2411 | 47 | 98,1% |
| ICU | 139 | 51 | 26,8% |
| Overall Percentage | 97,6% | 2,4% | 93,5% |
However, the success of this decision tree (Fig. 4) in identifying non-ICU patients was much higher than the individual success of the predictors presented in Table 7 for classifying non-ICUs. In addition, the d-CIT and d-PPT biomarkers, which are statistically significant in Fig. 4, perfectly classify non-ICU patients and identified a high percentage of ICU (Table 7). Therefore, if identification of ICU patients is more important and false positives (non-ICUs) are less important, the cut-off values of d-CIT and d-PPT in Table 7 should be considered. However, if identification of non-ICU patients is more important and false positives (ICUs) are less important, Fig. 4 can be examined.
The presence of CRP-derived biomarkers at the root of the tree provides a confirmation of our interpretable Decision Tree (Fig. 4) of the robustness of the approach to determining disease severity. Similarly, [31], [32] reported that CRP and ESR increased with disease severity.
The biomarkers previously (LCR, NLR, d-NLR, PLR) used to determine the severity of the disease were lower than the biomarkers derived from the study in determining ICU (Table 7) [17], [18], [30], [33].
Similarly, one study [41] identified age, D-Dimer, and CRP as the strongest early predictors of mortality using the CHAID classification tree structure. Similar to this study, patients in ICU were observed to have higher Fibrinogen, D-Dimer, CRP, PT, aPPT, ESR and Ferritin values than non-ICU [34], [35], [36].
In another study, it was noted that the platelet count decreased in the ICU group and was associated with the severity of the disease [34], [37], [38]. Similarly, in this study, the platelet count was similar between non-ICU and ICU and was greatly reduced compared to the control group. D-Dimer was found to be high in most of both ICU and non-ICU patients. Increased D-Dimer level with decreased platelet level in a significant proportion of patients was a sign of hypercoagulation for cases of Covid-19 promoting microthrombosis in the vascular system [4].
Changes in routine laboratory data in the study were similarly seen in other studies [19], [23], [26], [34], [39], [40]. In some studies, the difference in prametre values is mostly in the same direction in this study [9], [27]. Among the reasons for the difference, the severity stage of the disease, sampling time, additional diagnosis of the patients and the number of patients included in the studies can be said.
5. Conclusion
After discussing the clinical features of Covid-19, we analyzed the haematological and immunological characteristics of blood routine parameters in patients. It showed that advanced age and male gender are seriously threatened in terms of both Covid-19 and disease progression. Low LYM and WBC, high CRP and Ferritin were effective in the diagnosis of the disease. The d-CWL and d-CFL biomarkers derived from them were the most important risk factors in the diagnosis of the disease. In addition, these biomarkers were more successful in determining the diagnosis of the disease than the diagnostic success of direct blood values and previously used biomarkers (NLR, d-NLR, PLR, LCR). High d-CWL and d-CFL values can greatly support the diagnosis of Covid-19. Also, using the decision tree in Fig. 1, patients with Covid-19 can be identified with a high degree of accuracy in a simple way. d-CIT, d-CT and d-PPT obtained from these blood values were found to be more successful in determining patients to ICU (in the prognosis of the disease) compared to direct blood values and previously used biomarkers. High d-CT, d-CIT and d-PPT values can help identify patients with Covid-19 to ICU. However, if identification of non-ICU patients is more important, the decision tree in Fig. 4 can be used. It was understood that the biomarkers we obtained from routine laboratory tests determined the diagnosis and prognosis of Covid-19 more successfully than direct blood values and previously used biomarkers.
6. Limitations of the study
Various independent biomarkers used in the study need to be tested in the diagnosis and prognosis of many other infectious diseases. The low number of ICU patient groups compared to the non-ICU group was one of the limitations of this study. In addition, since it is a retrospective study from the records, inability to access comorbidity data and being single-centered are other limitations of this study. Retrospective studies naturally lack control of variables; Therefore, prospective cohorts are also needed to validate our study data. In addition, the data used in this study were obtained from the Covid-19 patient population in the spring, summer and autumn seasons. Therefore, some parameter values in this study showed seasonal or periodic differences from similar studies. Multicenter studies in larger patient groups will further clarify the importance of routine laboratory parameters in Covid-19.
Ethical approval
The study protocol was approved by the Institutional Ethics Review Board of Erzincan Binali Yıldırım University after being approved by the Ministry of Health of the Republic of Turkey in accordance with the Declaration of the Helsinki World Medical Association (Ethics Committee Decision number: 2021/02-07).
Author contributions
Huyut MT: He designed the study, edited and created the material and methodology, applied the analyses, interpreted, discussed, wrote the study. İlkbahar F: Organized the material, created the open access dataset.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Singhal T.A. Review of Coronavirus Disease-2019 (COVID-19) Indian J. Pediatrics. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bedford J., Enria D., Giesecke J., et al. COVID-19: towards controlling of a pandemic. Lancet. 2020;395:1015–1018. doi: 10.1016/S0140-6736(20)30673-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sogut O., Can M.M., Guven R., Kaplan O., et al. Safety and efficacy of hydroxychloroquine in 152 outpatients with confirmed COVID-19: a pilot observational study. Am. J. Emergency Med. 2021;40:41–46. doi: 10.1016/j.ajem.2020.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mertoglu C., Huyut M.T., Arslan Y., Ceylan Y., Çoban T.A. How do routine laboratory tests change in coronavirus disease 2019? Scand. J. Clin. Lab. Invest. 2021;81:24–33. doi: 10.1080/00365513.2020.1855470. [DOI] [PubMed] [Google Scholar]
- 5.Sun S., Cai X., Wang H., et al. Abnormalities of peripheral blood system in patients with COVID-19 in Wenzhou, China. Clin. Chim. Acta. 2020;507:174–180. doi: 10.1016/j.cca.2020.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.J. Wu, P. Zhang, L. Zhang, W. Meng, et al., Rapid and accurate identification of COVID-19 infection through machine learning based on clinical available blood test results, 2020, medRxiv preprint https://doi.org/10.1101/2020.04.02.20051136.
- 7.Gao Y., Li T., Han M., et al. Diagnostic utility of clinical laboratory data determinations for patients with the severe COVID-19. J. Med. Virol. 2020;92 doi: 10.1002/jmv.25770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu Y., Yang Y., Zhang C., et al. Clinical and biochemical indexes from 2019-nCoV infected patients linked to viral loads and lung injury. Sci. China Life Sci. 2020;63:364–374. doi: 10.1007/s11427-020-1643-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ponti G., Maccaferri M., Ruini C., et al. Biomarkers associated with COVID-19 disease progression. Crit. Rev. Clin. Lab Sci. 2020;57:389–399. doi: 10.1080/10408363.2020.1770685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mousavi S.A., Rad S., Rostami T., Rostami M., et al. Hematologic predictors of mortality in hospitalized patients with COVID-19: a comparative study. Hematology. 2020;25:383–388. doi: 10.1080/16078454.2020.1833435. [DOI] [PubMed] [Google Scholar]
- 11.Mattiuzzi C., Lippi G. Which lessons shall we learn from the 2019 novel coronavirus outbreak? Ann. Transl. Med. 2020;8:48. doi: 10.21037/atm.2020.02.06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wan Y., Shang J., Graham R., et al. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94 doi: 10.1128/jvi.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhavana V., Thakor P., Singh S.B., Mehra N.K. COVID-19: Pathophysiology, treatment options, nanotechnology approaches, and research agenda to combating the SARS-CoV2 pandemic. Life Sci. 2020;261:1–17. doi: 10.1016/j.lfs.2020.118336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vellingiri B., Jayaramayya K., Iyer M., Narayanasamy A., et al. COVID-19: a promising cure for the global panic. Sci. Total Environ. 2020;725 doi: 10.1016/j.scitotenv.2020.138277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Translation: Management of Coronavirus Disease 2019 (COVID-19): experience in Zhejiang Province, China, (n.d). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7227201/ (accessed June 15, 2020).
- 16.Mo P., Xing Y., Xiao Y., et al. Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin. Infect Dis. 2020 doi: 10.1093/cid/ciaa270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., Du X., Chen J., et al. Neutrophil-to-lymphocyte ratio as an independent risk factor for mortality in hospitalized patients with COVID-19. J. Infect. 2020;81:e6–e12. doi: 10.1016/j.jinf.2020.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang A.P., Liu Jp., Tao Wq., et al. The diagnostic and predictive role of NLR, d-NLR and PLR in COVID-19 patients. Int. Immunopharmacol. 2020;84 doi: 10.1016/j.intimp.2020.106504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonetti G., Manelli F., Patroni A., et al. Laboratory predictors of death from coronavirus disease 2019 (COVID-19) in the area of Valcamonica, Italy. Clin. Chem Lab Med. 2020;58:1100–1105. doi: 10.1515/cclm-2020-0459. [DOI] [PubMed] [Google Scholar]
- 20.Onur S.T., Altın S., Sokucu S.N., Fikri B.İ., Barça T., Bolat E., Toptaş M. Could ferritin level be an indicator of COVID-19 disease mortality? J. Med. Virol. 2021;93:1672–1677. doi: 10.1002/jmv.26543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.G. Lippi, M. Plebani, B.M. Henry, Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID‐19) infections: a metaanalysis, Clin. Chim. Acta 506 (2020) 145–148. https://doi.org/10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed]
- 22.Rosário C., Zandman-Goddard G., Meyron-Holtz E.G., D'Cruz D.P., Shoenfeld Y. The hyperferritinemic syndrome: macrophage activation syndrome, Still's disease, septic shock and catastrophic antiphospholipid syndrome. BMC Med. 2013;185:1–11. doi: 10.1186/1741-7015-11-185. http://www.biomedcentral.com/1741-7015/11/185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guan Wj., Hu Y., Ni Zy., et al. Clinical characteristics of coronavirus disease 2019 in China. New Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan B.E., Chong V.C.L., Chan S.S.W., et al. Hematologic parametersin patients with COVID-19 infection. Am. J. Hematol. 2020;95:E131–E134. doi: 10.1002/ajh.25774. [DOI] [PubMed] [Google Scholar]
- 25.Tan L., Wang Q., Zhang D., et al. Lymphopenia predicts disease severity of COVID-19: a descriptive and predictive study. Signal Transduct Target Ther. 2020;27:33. doi: 10.1038/s41392-020-0148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bohn M.K., Lippi G., Horvath A., et al. Molecular, serological, and biochemical diagnosis and monitoring of COVID-19: IFCC taskforce evaluation of the latest evidence. Clin. Chem. Lab. Med. 2020;58:1037–1052. doi: 10.1515/cclm-2020-0722. [DOI] [PubMed] [Google Scholar]
- 27.Wang F., Hou H., Luo Y., et al. The laboratory tests and host immunity of COVID-19 patients with different severity of illness. JCI Insight. 2020;5:1037–1052. doi: 10.1515/cclm-2020-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu H., Zhong L., Deng J., et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12 doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Y.C., Liang W.G., Chen F.W., et al. IL-19 induces production of IL-6 and TNF-alpha and results in cell apoptosis through TNF-alpha. J. Immunol. 2002;169:4288–4297. doi: 10.4049/jimmunol.169.8.4288. [DOI] [PubMed] [Google Scholar]
- 30.Demirdal T., Sen P. The significance of neutrophil-lymphocyte ratio, plateletlymphocyte ratio and lymphocyte-monocyte ratio in predicting peripheral arterial disease, peripheral neuropathy, osteomyelitis and amputation in diabetic foot infection. Diabetes Res. Clin. Pract. 2018;144:118–125. doi: 10.1016/j.diabres.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Zeng F., Huang Y., Guo Y., et al. Association of inflammatory markers with the severity of COVID-19: a meta-analysis. Int. J. Infect Dis. 2020;96:467–474. doi: 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Jj., Dong X., Cao Yy., et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China. Eur. J. Allergy Clin. Immunol. 2020;75:1730–1741. doi: 10.1111/all.14238. [DOI] [PubMed] [Google Scholar]
- 33.Wang S., Zhang S., Song S., et al. Fasting blood glucose at admission is an independent predictor for 28-day mortality in patients with COVID-19 without previous diagnosis of diabetes: a multi-centre retrospective study. Diabetologia. 2020;63:2102–2110. doi: 10.1007/s00125-020-05209-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amgalan A., Othman M. Hemostatic laboratory derangements in COVID-19 with a focus on platelet count. Platelets. 2020;31:740–746. doi: 10.1080/09537104.2020.1768523. [DOI] [PubMed] [Google Scholar]
- 35.Tang N., Li D., Wang X., et al. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thachil J., Agarwal S. Understanding the COVID-19 coagulopathy spectrum. Anaesthesia. 2020;75:1432–1436. doi: 10.1111/anae.15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bi X., Su Z., Yan H., et al. Prediction of severe illness due to COVID-19 based on an analysis of initial fibrinogen to albumin ratio and platelet count. Platelets. 2020;31:674–679. doi: 10.1080/09537104.2020.1760230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang S.Q., Huang Q.F., Xie W.M., et al. The association between severe COVID-19 and low platelet count: evidence from 31 observational studies involving 7613 participants. Br. J. Haematol. 2020;190:e29–33. doi: 10.1111/bjh.16817. [DOI] [PubMed] [Google Scholar]
- 39.Henry B.M., de Oliveira M.H.S., Benoit S., et al. Hematologic biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin. Chem. Lab Med. 2020;58:1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 40.Henry B.M., Benoit S.W., de Oliveira M.H.S., et al. Laboratory abnormalities in children with mild and severe coronavirus disease 2019 (COVID-19): a pooled analysis and review. Clin. Biochem. 2020;81:1–8. doi: 10.1016/j.clinbiochem.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]