Abstract
Multicellular organisms are composed of tissues with diverse cell sizes. Whether a tissue primarily consists of numerous, small cells as opposed to fewer, large cells can impact tissue development and function. The addition of nuclear genome copies within a common cytoplasm is a recurring strategy to manipulate cellular size within a tissue. Cells with more than two genomes can exist transiently, such as in developing germlines or embryos, or can be part of mature somatic tissues. Such nuclear collectives span multiple levels of organization, from mononuclear or binuclear polyploid cells to highly multinucleate structures known as syncytia. Here, we review the diversity of polyploid and syncytial tissues found throughout nature. We summarize current literature concerning tissue construction through syncytia and/or polyploidy and speculate why one or both strategies are advantageous.
Keywords : Polyploidy, Syncytia, Endocycle, Endomitosis, Fusion, Cyst, Multinucleate
Overview
In at least 9 different mammalian organ systems (Table 1), cells with greater than two genomes can be found. Focusing primarily on metazoans, this review focuses on such highly conserved increases in nuclear genome number and its impact on tissue biology. We discuss how nuclear genome number is increased and survey where cells with such increased genome content are found. We also broadly review our limited knowledge regarding how adding genomes can alter tissue function and speculate how future studies might tackle the largely unknown question of how extra nuclei impact tissue function. As part of this discussion, we compare and contrast the properties of large cells with extra genomes packed into a single nucleus versus those cells with multiple genomes in multiple nuclei.
Table 1.
Nuclear number and ploidy and mechanism of various cell types
| Cell type (species) | Organ system | Predominant nuclear number and ploidy per nucleus (C) | Mechanism | References |
|---|---|---|---|---|
| Germline cyst (female mouse) | Reproductive | Up to 25 × 1C | Incomplete cytokinesis | (Lei and Spradling 2016) |
| Germline cyst (female fruit fly) | Reproductive | 16 × 1C | Incomplete cytokinesis | (Lei and Spradling 2016) |
| Cardiomyocyte (pig) | Cardiovascular | 8 × 2C | Incomplete cytokinesis | (Velayutham et al. 2020) |
| Cardiomyocyte (mouse) | Cardiovascular | 2 × 2C | Incomplete cytokinesis | (Soonpaa et al. 1996) |
| Cardiomyocyte (human) | Cardiovascular | 1 × 4C and 2 × 2C | Incomplete cytokinesis or endocycle | (Brodsky et al. 1992; Hesse et al. 2012; Mollova et al. 2013; Bergmann et al. 2015) |
| Osteoclast (chicken) | Skeletal | 4 × 2C | Cell–cell fusion | (Piper et al. 1992) |
| Macrophage granuloma (human) | Lymphatic | 2 × 2C and 1 × 4C | Incomplete cytokinesis or endocycle | (Herrtwich et al. 2016) |
| Myocyte (mouse) | Muscular | ~ 220 × 2C (extensor digitorum longus) | Cell–cell fusion | (Hansson et al. 2020) |
| Myocyte (fruit fly) | Muscular | 10–15 × 32C (ventral longitudinal muscles 3 and 4) | Cell–cell fusion, endocycle | (Windner et al. 2019) |
| Megakaryocyte (human, rat) | Cardiovascular | 1 × 8C, 1 × 16C, 1 × 32C | Incomplete cytokinesis and incomplete karyokinesis | (Odell et al. 1976; Brown et al. 1997; Lordier et al. 2008) |
| Hepatocyte (human) | Digestive | 1 × 2C, 2 × 2C, 2 × 4C, 1 × 4C, 1 × 8C | Incomplete cytokinesis or endocycle | (Kudryavtsev et al. 1993) |
| Rectal papillar cells (fruit fly) | Digestive | 100 × 8C | Endocycle, cytoplasm-sharing | (Fox et al. 2010; Schoenfelder et al. 2014; Peterson et al. 2020) |
| Syncytiotrophoblast (human) | Extraembryonic | up to 6 × 10^10 × 2C | Cell–cell fusion | (Simpson et al. 1992) |
| Trophoblast giant cells (mouse) | Extraembryonic | Up to 1 × 512–1024C | Endocycle | (Barlow and Sherman 1972; MacAuley et al. 1998) |
| Salivary gland (fruit fly) | Digestive | 1 × 512–1024C | Endocycle | (Hammond and Laird 1985) |
|
Subperineurial glia (fruit fly) |
Nervous | 1–4 × 4–32C | Incomplete cytokinesis or endocycle | (Unhavaithaya and Orr-Weaver 2012; Von Stetina et al. 2018) |
Under construction: adding genome copies to cells during tissue growth
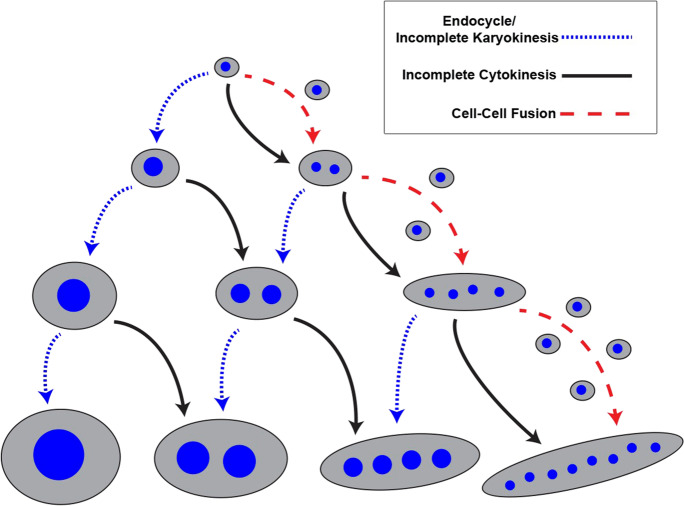
Diploid, sexually reproducing organisms pass on one set of chromosomes from each parent to their offspring. Cells in these organisms then grow and form tissues through mitotic cell cycles, whereby the diploid genome is fully replicated and the daughter cells wholly separate during mitosis and cytokinesis. But in specific tissues (for examples see Table1), the diploid genome can undergo further duplications to increase the total DNA content (C value) per cell (Fox and Duronio 2013; Øvrebø and Edgar 2018). Such cell cycle-dependent genome copy increases occur through two main processes. First, cells can switch from a mitotic cycle to a cycle commonly referred to as either the endocycle or the endoreduplication cycle. Endocycles consist of just one gap phase per cycle (known as G-phase) and an intervening genome duplication phase (S-phase), but no entry into mitosis. Endocycles lead to increased nuclear size and content within a single nucleus (Fig. 1). Examples of cells that undergo endocycles include the trophoblast giant cells in mammals and salivary gland and rectal papillar cells in insects (Table1). Second, mitotic cycling cells can switch to a cycle commonly known as endomitosis. Endomitosis involves truncating the cell cycle either in mitosis or cytokinesis and thus primarily differs from endocycles in that cells continue to at least enter mitosis. All endocycling cells, as well as any endomitotic cells that truncate mitosis prior to anaphase (incomplete karyokinesis), will yield mononucleated polyploid cells (Fig. 1). In contrast, endomitotic cells that progress to anaphase yet undergo incomplete cytokinesis will produce binucleate polyploid cells (Fig. 1), as occurs in many cardiomyocytes and hepatocytes (Table 1). Binucleate cells can also undergo subsequent endomitosis with incomplete cytokinesis to yield highly multinucleate syncytia (Fig. 1), as occurs in pig cardiomyocytes (Table1). For more information on regulation of the variant cell cycles that generate polyploid cells, we refer the reader to several recent reviews (Øvrebø and Edgar 2018; Shu et al. 2018; Lazzeri et al 2019; Lang and Schnittger 2020).
Fig. 1.
Multiple mechanisms lead to the same ploidy and nuclear number outcomes. Cycles with absent or incomplete karyokinesis can both yield increased nuclear size and ploidy. Cycles with full nuclear division but incomplete cytokinesis, as well as cell–cell fusion, increase the number of nuclei. Each blue arrow represents a doubling in genome content. Each black arrow represents an incomplete cytokinesis. Each red arrow represents the addition of the number of single nuclei adjacent to the arrow. This illustration does not depict reductive division, which can reverse increased cellular ploidy
Independent of the cell cycle, fusion of plasma membranes of adjacent cells provides an additional mechanism to generate multinucleated cells and syncytia (Fig. 1). Examples of multinucleated syncytial tissues formed by plasma membrane fusion are skeletal myocytes, osteoclasts, and the syncytiotrophoblast (Table 1). Diverse cell fusion mechanisms include establishing competence for adjacent cells to recognize each other and fuse and the action of fusogen molecules and the actin cytoskeleton to remodel neighboring cell membranes. We refer the reader to these recent reviews (Hernandez and Podbilewicz 2017; Lee and Chen 2019; Petrany and Millay 2019) for a comprehensive discussion of cell fusion mechanisms and molecules. As highlighted in the sub-section “skeletal muscle,” it is also possible for a tissue to increase genome copy numbers through a combination of both cell fusion and ploidy-increasing cell cycles.
The terminology surrounding cells with increased genome copy number can be confusing. Technically speaking, the product of endocycles, endomitosis, and cell fusion all produce polyploid cells. Polyploidy thus applies to multinucleated cells where each individual nucleus is diploid. However, as discussed in the sub-section “progenitor syncytia,” some instances of increased nuclear content (e.g. germline cysts) are transient, and are therefore not typically referred to as polyploid. Similarly, binucleate cells are not commonly referred to as syncytia. To further add to the complexity, polyploid cells can continue to divide (Fox et al. 2010; Miyaoka et al. 2012), and polyploid cells can fuse/share cytoplasm (Peterson et al. 2020) and ploidy may also reduce in some cases (Duncan et al. 2010; Lucchetta and Ohlstein 2017; Matsumoto et al. 2021). For the purposes of this review, we use the term polyploid to refer to an increase in cellular DNA content to at least three or more genome copies, and similarly we use the term syncytia to refer to cells containing three or more nuclei. In the next two sections, we focus on the diversity and differing biology regarding ploidy states and nuclear number in animal tissues.
Distinct architecture: a survey of tissues with extra genomes across nature
In this section, we present a survey of distinct examples of tissues with extra genomes, from mononucleate polyploid to highly multinucleated syncytia. We also discuss the potential benefits and tradeoffs of extra genomes in a number of tissues. A separate discussion of why tissues with extra genomes use different organizational strategies (e.g., mononucleate vs. multinucleated) is presented later, in the section “Form and Function.”
Progenitor syncytia
During progenitor stages of development, nuclei of germ cells and early embryos frequently share cytoplasm. This sub-section highlights the conservation of syncytia in metazoan germ cell lineages and extra-embryonic cells.
Germline syncytia
Nuclear division followed by incomplete cytokinesis (akin to endomitosis) is a widely conserved route to germline cyst formation in many animal species (Fig. 1; Table1; Fawcett et al. 1959; Hime et al. 1996; de Cuevas et al. 1997; Kloc et al. 2004; Maddox et al. 2005; Kosaka et al. 2007; Marlow and Mullins 2008; Lei and Spradling 2013; Amini et al. 2014). This process creates syncytial cyst structures in the female germline of fruit flies (Drosophila melanogaster), clawed frogs (Xenopus laevis), zebrafish (Danio rerio), and mice (Mus musculus). Within these cysts, organelles and other cytoplasmic materials can be shared (Zamboni and Gonndos 1968; Ruby et al. 1969; Gutzeit 1986; Bolívar et al. 2001; Pepling and Spradling 2001; Cox and Spradling 2003; Kloc et al. 2004; Kosaka et al. 2007; Marlow and Mullins 2008; Lei and Spradling 2013, 2016). Cyst cell number is invariantly 16 germ cells in female (and male) Drosophila germ cysts and can reach up to 25 cells in the mouse ovary (Lei and Spradling 2016, reviewed in Greenspan et al. 2015). Frequently, the oocyte is the only cell to survive through oogenesis, a phenomenon known as a meroistic ovary (McCall and Steller 1998; Foley and Cooley 1998 and reviewed in Lu et al. 2017). In such cases, syncytial organization can serve to nourish the growing oocyte.
However, nourishing of a future gamete cannot be the only function of germline syncytia.
For example, death of cyst cells supporting the future oocyte does not always occur in female syncytial germlines, such as in organisms with panoistic ovaries (reviewed in Lu et al. 2017). Similarly, while incomplete cytokinesis leads to syncytial cysts during male germ cell development in several species including in flies and mice, most nuclei of the cyst survive and eventually individualize into mature sperm (reviewed in Yoshida 2016; Yamashita 2018). As in the female syncytial germline, male syncytial germ nuclei can also share gene products (Braun et al. 1989; Caldwell and Handel 1991; Ventelä et al. 2003; Kaufman et al. 2020). Several models for why cytoplasm is shared in developing germ cysts where each germ cell goes on to become a gamete are outlined nicely in previous reviews (Greenbaum et al. 2011; Lu et al. 2017). Briefly, these include (1) the need to synchronize critical events in gamete production such as the onset of meiosis (which is highly synchronized in males), (2) the ability to neutralize a deleterious mutation in a single germ cell of the cyst that might otherwise outcompete neighboring gametes during fertilization, (3) the sharing of X and Y gene products to make early male gametes phenotypically diploid, and (4) increased sensitivity to DNA damage. More recently, study in C. elegans suggests that a syncytium can also compensate for the mechanical stress of oogenesis (Amini et al. 2014; Priti et al. 2018). Future study can determine the extent to which each of these mechanisms contributes to productive function of germline syncytia.
Extra-embryonic syncytia
A common theme across metazoan evolution is the syncytial nature of cells that support the growth of the embryo. Many insects have a syncytial yolk sac as part of the extra-embryonic tissue during embryogenesis (reviewed in Schmidt-Ott and Kwan 2016). As in germline syncytia, these extra-embryonic syncytia are formed by repeated rounds of nuclear division and incomplete cytokinesis of the yolk sac cytoplasm during blastoderm stages. In some insect species including in Drosophila melanogaster, most of these nuclei then migrate out of the future yolk sac to the embryo surface and contribute to the animal body plan (Campos-Ortega et al. 1997, reviewed in Zissler 1992; Davis and Patel 2002). The syncytial yolk nuclei that remain play important roles in embryonic morphogenesis, in part through maintaining integrin-based adhesion with the surrounding embryo (Reed et al. 2004; Benton et al. 2013). In all teleost fishes, the yolk syncytial layer is an extra-embryonic tissue. This tissue is derived from nuclear divisions with incomplete cytokinesis (Lentz and Trinkaus 1967; Kimmel and Law 1985a, b; Chu et al. 2012). The zebrafish yolk syncytial layer, which consists of several hundred nuclei (Carvalho et al. 2009), has numerous roles in early fish development, including nourishing the early embryo (Walzer and Schönenberger 1979a, b) and in signaling events that pattern the embryo (reviewed in Carvalho and Heisenberg 2010; Webb and Miller 2013). Relative to other syncytia, the role of yolk syncytia remains poorly understood. Future studies aimed at specifically disrupting the syncytial nature of the yolk in insect and fish species are needed to understand potential roles of yolk syncytia in embryos.
In many mammals, the extra-embryonic placenta that supports the embryo contains a syncytium. Unlike the examples discussed above in the germline and yolk, cell fusion is the mechanism of syncytium formation in the placenta (reviewed in Gerbaud and Pidoux 2015; Soygur and Sati 2016). In humans, there are three distinctly localized syncytial layers of trophoblast cells in the placenta, each thought to play an endocrine and immune barrier function (reviewed in Turco and Moffett 2019; Roberts et al. 2021). Trophoblast layers may be some of the most extreme examples of syncytia in nature, with up to 6 × 10^10 nuclei estimated in a human syncytial trophoblast (Simpson et al. 1992). In mice, genetic ablation of syncytin molecules, which are endogenous retroviral proteins required for syncytiotrophoblast layer formation, leads to a variety of growth phenotypes, including fetal death (Dupressoir et al. 2009, 2011). Further, altered syncytin expression is reported in human placental pathologies (Chen et al. 2006; Vargas et al. 2011; Soygur et al. 2016). A recent study (Buchrieser et al. 2019) revealed an acute sensitivity of trophoblast syncytium formation to immune signaling through interferons. Upon interferon stimulation in pregnant mice, trophoblast syncytium formation is blocked, leading to halted pregnancy. It is interesting to speculate that the formation of an extraembryonic syncytium provides a developmental checkpoint related to immune sensing. Future work in insect, fish, and mammalian models can further reveal the roles of extraembryonic syncytia, such as whether they allow for more rapid transmission of signals that determine whether embryogenesis should proceed.
Somatic polyploidy
Following germ cell and embryo stages, extra genomes are seen again as specific somatic lineages differentiate. One general theme of somatic cells with extra genomes is that they can enable the growth of much larger cells (Edgar and Orr-Weaver 2001). In general, cell size growth occurs according to nutrient availability and metabolic capacity (as reviewed in Melaragno et al. 1993; Lloyd 2013; Schoenfelder and Fox 2015). It follows then that adding additional genome copies can support larger cells due to increased metabolism and secretion. This sub-section presents a survey of mononuclear or multinuclear somatic polyploidy, focusing on tissue examples that are conserved in animals.
Cardiomyocytes
One of the most conserved examples of polyploidy in a somatic tissue is in the cardiac muscle. Cardiomyocytes in the simple heart tube of Drosophila are polyploid (Yu et al. 2013, 2015). In mammals, cardiomyocyte polyploidy appears to have co-evolved with endothermy (Hirose et al. 2019). Fish (including zebrafish), amphibians, and reptiles have little cardiomyocyte polyploidy (Wills et al. 2008; Hirose et al. 2019). Monotremes such as echidna and platypus have moderate levels of cardiomyocyte polyploidy (40–50% polyploid), while rodents, humans, and other mammals have largely (90% +) polyploid cardiomyocytes (Soonpaa et al. 1996; Bergmann et al. 2015; Hirose et al. 2019).
One potential tradeoff to polyploidy in mammals is a loss of regenerative capacity. The diploid zebrafish heart responds to tissue injury using cell division (hyperplasia) to replace dead or missing cells (Poss et al. 2002), and experimentally increasing cardiomyocyte ploidy in this organism decreases regeneration (Gonzalez-Rosa et al. 2018). Instead, polyploid cardiomyocytes in many mammals respond to tissue damage through scarring and cellular and tissue enlargement through further nuclear content addition (hypertrophy, Meckert et al. 2005; Patterson et al. 2017; Hirose et al. 2019; Derks and Bergmann 2020; Han et al. 2020). The low rate of cell division in mammals accompanies scarring, and the hypertrophy that results instead of cell division is insufficient to replace lost cardiomyocytes (Senyo et al. 2013).
Cardiomyocytes demonstrate a great range of mononucleate and multinucleate polyploidy throughout the animal kingdom. Within the category of the mammalian heart with 90% + polyploid adult cardiomyocytes, the number of nuclei per cardiomyocyte varies greatly. Mouse and rat cardiomyocytes are almost entirely binucleate (2 nuclei each with 2C DNA content, or 2 × 2C) and are formed by endomitosis with incomplete cytokinesis (Soonpaa et al. 1996; Li et al. 1996, 1997a, b; Engel et al. 2006; Liu et al. 2010, 2019). Adult human cardiomyocytes contain both mononucleated and binucleated cardiomyocytes with typical ploidies of 4–8C, with a range of diploid, tetraploid, and octoploid DNA content (Brodsky et al. 1992; Bergmann et al. 2009, 2015; Mollova et al. 2013). Pig cardiomyocytes typically contain anywhere between 4 and 16 diploid nuclei arranged in a line (Velayutham et al. 2020). Giraffe cardiomyocytes contain 4–8 diploid nuclei also organized linearly, although 4 nuclei are more common (Ostergaard et al. 2013). Most other mammals studied, including sheep, rat, dog, cat, and cow, have predominantly binucleate cardiomyocytes, with each nucleus containing diploid DNA content (Bensley et al. 2016; Velayutham et al. 2019). Future study is needed on non-humate primates and other large mammals to determine the evolution of mononucleate versus multinucleate cardiomyocyte polyploidy. Why mammalian cardiomyocytes have variable nuclear numbers is unknown (see section “Form and Function”).
In addition to developmental polyploidization, nuclear content is added in response to heart injury and disease. Human cardiomyocytes reach ploidies up to 32C in hearts hypertrophied in response to infarction, congenital heart defects, rheumatic heart diseases, and significant hypertension (Brodsky et al. 1994; Herget et al. 1997; Steinhauser and Lee 2011; Senyo et al. 2013). Interestingly, roughly equal populations of mononucleated and binucleated cardiomyocytes are retained after hypertrophy (Brodsky et al. 1994). This suggests that hypertrophic growth largely occurs through ploidy increase in individual nuclei and not the addition of new cells in humans.
Hepatocytes
Hepatocytes are the major cell type in the vertebrate liver. Like cardiomyocytes, hepatocytes also exhibit species-specific differences in ploidy and nuclear number. Several reviews explore hepatocyte polyploidy in depth (Gentric and Desdouets 2014; Zhang et al. 2019). Hepatocyte polyploidy and multinucleation varies between species. Rat and mouse hepatocytes are ~ 75–90% polyploid, human hepatocytes are ~ 30–40% polyploid, and guinea pig and woodchucks have very low levels of hepatocyte polyploidy and binucleation (Kreutz et al. 2017). Mammalian livers also exhibit mixed populations of binucleate and mononucleate polyploid cells, which makes the liver an intriguing system to study their differences. Within the ~ 30% polyploid segment of human hepatocytes, binucleate (2 × 2C and 2 × 4C) and mononucleate (1 × 4C, 1 × 8C) polyploid cells are found (Kudryavtsev et al. 1993). The mouse liver contains roughly equal fractions of mononucleate diploid, mononucleate polyploid, binucleate, and binucleate polyploid cells, whereas nuclear number beyond 2 is uncommon (Kreutz et al. 2017). Insects such as Drosophila also have hepatocyte-like cells, known as oenocytes, and these cells also are polyploid (Gutierrez et al. 2006; Cinnamon et al. 2016). The vertebrate liver is highly regenerative, and both diploid hepatocyte division and polyploidy-promoting endocycles or endomitosis are capable of full restoration of liver mass (Davoli et al. 2010; Diril et al. 2012; Miyaoka et al. 2012). In contrast, mice with livers that are incapable of diploid hepatocyte division are prone to a diabetic-like phenotype, inflammation, and fibrosis (Dewhurst et al. 2020; Ow et al. 2020). Further, polyploidy can both promote or suppress liver cancer, depending on the context (Zhang et al. 2018; Lin et al. 2021). Therefore, while multiple hepatocyte ploidies and nuclear numbers can compensate for liver injury, the absence of diploidy can impact hepatocyte metabolism and liver health.
Skeletal muscle
Skeletal muscle provides an interesting example of a syncytial tissue that uses often high levels of multinucleation and, in some cases, nuclear content increase to grow and adjust to changing demands. Skeletal muscle consists of long, multinucleated muscle fibers formed through cell–cell fusion events (reviewed in Deng et al. 2017; Petrany and Millay 2019). In the mouse extensor digitorum longus muscle, there are approximately 220 nuclei per myofiber (Hansson et al. 2020). In some organisms, both cell fusion and ploidy increase within individual nuclei contribute to muscle fiber size growth and function (Windner et al. 2019). One potential hypothesis for why skeletal muscle contains numerous nuclei is the myonuclear domain hypothesis (reviewed in Schwartz 2018), which proposes that each nucleus is responsible for supplying its own territory in the syncytial muscle fiber. The usage of many small nuclei minimizes transport distance of nuclear products to the cytoplasmic domain especially when forming a long cylinder as opposed to a sphere.
Drosophila skeletal muscle employs both multinucleation through cell fusion and nuclear scaling (through regulation of ploidal level) to reach a fixed muscle fiber size (Windner et al. 2019). This suggests that there is coordination between nuclei and sensing of global nuclear content within the muscle fiber. Windner et al. propose a model in which nuclear number, ploidy content, and nucleolus size inform and optimize cell metabolism for the desired muscle fiber size (Windner et al. 2019). A potential advantage of nuclear scaling over cell fusion is the lack of reliance on extrinsic myoblast production. Nuclei already incorporated into the fiber can increase in ploidy in response to signaling. Therefore, the relative contributions of cell fusion and polyploidization may be due to myoblast production potential. The use of both cell fusion and nuclear ploidy increase allows for precise tuning of muscle fiber size and metabolism in Drosophila.
In contrast to fly skeletal muscle, mammalian skeletal muscle appears to only grow through multinucleation. A recent study found that while myonuclear number impacts muscle fiber size within a lower range of nuclear number, the impact of additional myonuclei on fiber size diminishes at a higher range of nuclei. Further, fibers with fewer nuclei can adapt by increasing transcriptional output (Cramer et al. 2020). Skeletal myofibers also experience a limited amount of cellular hypertrophy without DNA replication. However, most adaptive growth after exercise or injury occurs via myonuclear accretion (the addition of nuclei, Goh et al. 2019). The diversity of ploidy organization in tissues such as cardiac and skeletal muscle brings to mind the question—when is mononucleate or multinucleate polyploidy advantageous?
Form and function: when is mononucleate or multinucleate polyploidy advantageous?
In the preceding sections, we discussed how extra genome copies arise within cells, and what is known about how these extra genomes impact tissue biology. Much remains to be known regarding the function of such whole genome duplication, and integrative studies across diverse models can provide answers (Fox et al. 2020). In this section, we discuss a related question: how does tissue function differ when extra genomes are in one versus many nuclei? In this section we highlight some of the similarities and differences between mononucleate and multinucleate/syncytial polyploidy and suggest avenues for future study.
Mononucleate and multinucleate polyploid cells are similar
It is not well understood why some tissues can be built or repaired using mononucleate polyploid cells, multinucleate cells, or mixture of the two. There is some evidence that mononucleate and multinucleate cells of the same ploidy do not functionally differ. In the injured Drosophila abdominal epithelium, cell fusion and endocycles collaborate to repair the tissue, yet either mode of genome addition can suffice (Losick et al. 2013, 2016). In the mouse liver, Kreutz et al. and others found no significant difference in cell size and shape between binucleate and mononucleate hepatocytes nor a difference in liver zone location (Martin et al. 2002; Kreutz et al. 2017). However, binucleate and mononucleate polyploid cells do differ in gene expression, with 32% of differentially expressed genes between these ploidal states (Kreutz et al. 2017). Kreutz et al. speculate that binucleate and mononucleate polyploid cells contribute biological heterogeneity to the liver, although how nuclear number affects hepatic gene expression is unknown (Kreutz et al. 2017).
In the heart, mononucleate and multinucleate murine cardiomyocytes of varying ploidy have similar transcriptional profiles. Single-cell sequencing revealed no substantial differences between mononucleate and multinucleate rod-shaped cardiomyocytes isolated from adult mouse ventricle (Yekelchyk et al. 2019). However, transcripts do differ between cardiomyocytes from wild type and hypertrophic hearts (Nomura et al. 2018; Yekelchyk et al. 2019). Megakaryocytes are large, polyploid cells that produce platelets. Mammalian megakaryocytes typically use endomitosis to form polyploid, lobed nuclei (Lordier et al. 2008). The mechanism of ploidy increase does not matter as long as the desired ploidy is achieved. Switching to endocycles for ploidy increase does not impact platelet-forming by megakaryocytes (Trakala et al. 2015). It would be interesting to generate fully multinucleated megakaryocytes to test whether nuclear number similarly does not affect cell function. These findings support the common idea that multinucleate and mononucleate cells of the same ploidy are functionally equivalent.
How mononucleate and multinucleate polyploid cells might differ
We propose here possible reasons that multiple or single nuclei might be beneficial in tissue organization. Some potential differentiating factors could include cell shape, nuclear specialization, and cell division potential. We propose that future studies should consider the existence of a gradient from mononucleated diploid to highly multinucleated cell types (Fig. 2) with distinctive advantages and constraints of each nuclear organization strategy.
Fig. 2.
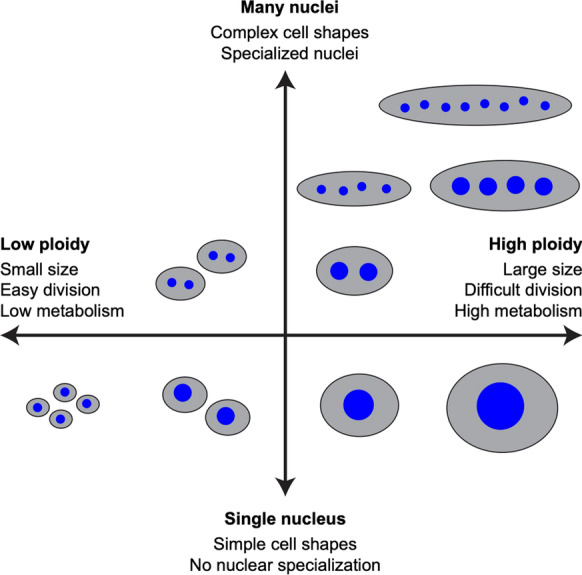
Ploidy and multinucleation. Axes showing changing cellular properties based on a spectrum from low to high ploidy and nuclear number. “High” and “low” ploidy are kept intentionally vague in this figure, as cell-type-specific biology may impact the properties listed
Diploid mononucleate cells have unique properties unmatched by polyploid or multinucleated cells such as conventional cell division (Storchova 2014; Schoenfelder and Fox 2015). On the other side of the gradient, highly multinucleated, polyploid cells have unique properties, such as large, complex shapes, that diploid and/or mononucleate cells cannot achieve (Melaragno et al. 1993; Schoenfelder and Fox 2015). In the middle, ploidy and nucleation states may be similar enough to one another to achieve some degree of functional redundance.
Multinucleation may facilitate cell shapes that are not practical or even possible for mononucleated polyploid cells. Cell size is limited by the distance that nuclear products must travel to reach the plasma membrane (Marshall et al. 2012; Schoenfelder and Fox 2015). Human skeletal muscle fibers are giant cells, sometimes containing thousands of nuclei. A giant cell with a singular, 1000C nucleus could not distribute its genetic material and transcriptional activity in the same way that a highly multinucleated cell can. Similarly, the human placental syncytiotrophoblast layer contains hundreds of thousands of nuclei and forms a long, thin, single-celled barrier between the mother and child. The seamless barrier formed by a multinucleated cell does not allow cells or materials to pass. Again, mononucleated polyploidy would be less efficient for a slender barrier. It is less obvious why pig cardiomyocytes grow longitudinally and form elongated cells with 4, 8, or 16 diploid nuclei arranged in a line (Velayutham et al. 2020). The benefit, if any, of building a heart from long cardiomyocytes instead of the shorter, squatter cardiomyocytes seen in mice or humans remains to be seen and should be tested experimentally. The diploid, mononucleated neuron seems to be an exemption to the challenge of long range cellular transport challenges. However, neurons have evolved unique long-distance transport methods (Grafstein and Forman 1980; Steward and Banker 1992). Multinucleation and long-distance transport both facilitate elongated cell shape.
Another potential advantage of multinucleated cells is the ability to specialize or even remove nuclei. Nuclei in skeletal muscle and syncytial placenta have been shown to have differential transcription within shared cytoplasm (Bursztajn et al. 1989; Fogarty et al. 2011; Petrany et al. 2020). Multinucleate cells formed through cell–cell fusion as opposed to endomitosis, such as skeletal myocyte and syncytiotrophoblast, have an additional advantage as they incorporate newly divided cells. This separation of genetic material allows for transcriptional tuning, such as favoring younger nuclei or nuclei without aneuploidies (imbalanced chromosome number) or DNA damage. In some cases, multinucleated cells can also favor certain nuclei for division. Only the undamaged nucleus divides in binucleate budding yeast with one damaged and one undamaged nucleus (Demeter et al. 2000). Skeletal myocytes gain nuclei after repeated exercise and may lose nuclei upon muscle atrophy although loss of myonuclei through apoptosis was recently disputed (Allen et al. 1999; Schwartz 2018).
Adding extra genomes creates both challenges and opportunities regarding gene dosage regulation (as reviewed in Schoenfelder and Fox 2015). Some degree of specialization is possible in mononucleated polyploid cells through monoallelic gene expression (Huang et al. 2018). Mononucleate and multinucleate are both able to target their gene expression to certain alleles (Bursztajn et al. 1989; Demeter et al. 2000; Fogarty et al. 2011; Huang et al. 2018). However, multinucleate cells have more options to add, remove, and spatially specialize nuclei.
Mononucleated and multinucleated polyploid cells may also differ in their (albeit limited) capacity for cell division. Polyploidy, regardless of nuclear number, is associated with terminal differentiation and a post-mitotic cell state (Lee et al. 2009). However, polyploid cell division has been observed in development and disease. The polyploid mononucleate fruit fly rectal papillar cells undergo programmed, though error-prone cell divisions (Fox et al. 2010; Schoenfelder et al. 2014). Nuclear division also occurs within some syncytial cells such as the synchronous divisions in the Drosophila embryo (reviewed in Foe et al. 1993) and the asynchronous divisions in Ashbya gossypii (Gladfelter et al. 2006). At equivalent ploidy, mononucleate and multinucleate cells face quite different challenges to successfully complete mitosis and cytokinesis. Mononucleate polyploid cells must organize and separate their many additional sister chromatids, which may have altered chromosome structure (reviewed in Stormo and Fox 2017). Multinucleate cells must separate additional nuclei, which can create a challenge for maintaining ploidy (Duncan et al. 2010; Matsumoto et al. 2020). Binucleate cardiomyocytes are capable of forming pseudo-bipolar spindles in order to divide in vitro, although these divisions are error-prone (Leone and Engel 2019). Both multinuclear and mononuclear polyploid cells face difficulties to complete cell division, although these challenges are specific to the nuclear arrangement. Control of the mitotic spindle assembly checkpoint was found to be a specific vulnerability of polyploid Drosophila and cancer cells, highlighting the heightened challenge of segregating a polyploid genome during mitosis (Stormo and Fox 2016, 2019; Quinton et al. 2021).
Multinucleate polyploid cells may allow slightly larger cell size than equivalently polyploid mononucleate cells. The Drosophila subperineurial glia (SPG) grow and form a blood–brain barrier through both mononucleate and multinucleate polyploidy (Unhavaithaya and Orr-Weaver 2012; Von Stetina et al. 2018). The SPG exhibit regional differences in multinucleation. Seventy percent of the brain lobe SPG are multinucleate and each nucleus is polyploid, while SPG in the peripheral nervous system and ventral nerve cord are solely mononucleate polyploid (Unhavaithaya and Orr-Weaver 2012). The maintenance of a barrier surrounding the proliferating brain requires the presence of both mononucleate and multinucleate polyploid SPG (Von Stetina et al. 2018). Multinucleate SPG are larger than mononucleate SPG of equivalent ploidy but do not differ in cell shape (Von Stetina et al. 2018). However, Von Stetina et al. (Von Stetina et al. 2018) note that size alone may not totally account for the requirement of multinucleate SPG for barrier function. Even when cell shape is comparable, multinucleated cells may be slightly larger than similar mononucleated cells, which could be advantageous for biological barrier maintenance. These findings could be relevant to the recurring role of extra embryonic syncytia as a barrier as discussed in the earlier sub-section “Extraembryonic syncytia.” In addition to the placenta, we note that extra nuclei are frequently found to play a barrier protective role in diverse cell types, where they may permit tissue growth/regrowth without compromising cell junctions (Unhavaithaya and Orr-Weaver 2012; Losick et al. 2013, 2016; Cohen et al. 2018; Wang et al. 2018).
Finally, cell types that increase in both nuclear ploidy and in nuclear number represent a unique, understudied class of tissues with increased genome content. If multinucleation and mononucleated polyploidy are functionally equivalent, why are both strategies used in the same cell? For example, Drosophila muscle fiber size and metabolism are tuned by both cell fusion and endoreplication as discussed above (Windner et al. 2019). The order of events may also be important. Drosophila muscle fiber fuses to form an elongated cell shape before undergoing endocycles (Windner et al. 2019). We can also imagine the reverse, in which mononucleated cells endocycle to increase in ploidy and then fuse or undergo polyploid mitosis to form a multinucleated cell with polyploid nuclei. For example, mononucleated Drosophila rectal papillar cells undergo endocycles to increase in ploidy and subsequently begin to share cytoplasm. (Peterson et al. 2020). Furthermore, rectal papillar endocycles are required for cytoplasm sharing to occur. We also predict that cells with both nuclear polyploidy and multinucleation would face increased barriers to faithful cell division than cells with only one of the two. Perhaps the combination of the two strategies totally blocks the possibility of division. The intersection of mononucleate and multinucleate polyploidy should be a point of future study.
The mechanisms and roles for polyploidy in development, disease, stress, and injury responses are better understood every year. Mononucleate and multinucleate polyploid cells were once thought to be functionally equivalent. However, we maintain that these two states have unique and intersecting properties that affect tissue architecture and function.
We suggest that additional study aimed at the distinct functions and regulation of mononucleated and multinucleated polyploid cells can reveal benefits of each tissue organization strategy. This should be tested in the in vivo systems described above. Hearts from various species built using different nuclear organization strategies (Table1) should be directly compared given the diversity in cardiomyocyte strategies. In addition, pharmacological and genetic techniques blocking either nuclear division or cytokinesis could be used to convert mononucleate or multinucleate polyploid cells within a certain cell type. For example, cyclin-dependent kinase 1 (Cdk1) ablation can convert endomitoses into endocycles in megakaryocytes, and additional ablation of Cdk2 prevents aberrant re-replication in endocycled megakaryocytes (Trakala et al. 2015). Knock down of fizzy-related (fzr), an anaphase-promoting complex/cyclosome regulator, may convert endocycles into endomitoses or mitoses (Sigrist and Lehner 1997; Schoenfelder et al. 2014; Cohen et al. 2018), depending on whether the cell type has a cytokinetic block such as the partial Rho/Rock defect observed in megakaryocytes (Lordier et al. 2008). In the mouse heart, manipulation of the LaminB2 gene can also impact the degree of mononucleate vs. multinucleate cardiomyocyte ploidy (Han et al. 2020). Between-species comparisons and within-species manipulations will help to determine why tissues use multinucleate and/or mononucleate polyploidy. In addition to the study of naturally occurring cells with extra genomes, future study should include the study of extra genomes in disease, including pathogenic syncytia, such as those produced by SARS-CoV-2 (Buchrieser et al. 2020) and the highly prevalent whole genome duplications found in cancer (Zack et al. 2013; Bielski et al. 2018).
Conclusion
In this review, we have emphasized the wide conservation of cells with greater than diploid genome content in animal tissues. Tissues with extra genomes are frequently required across evolution for gamete formation, embryonic development, and for proper cardiac, liver, and skeletal muscle function. While we have focused on animals, this phenomenon is found throughout nature (e.g., as reviewed in Bomblies 2020). Perhaps due to nomenclature, mononucleate and multinucleate polyploid cells are not often discussed alongside multinucleate syncytia in tissues such as the germline. We argue here that future efforts should compare findings between all examples of extra genomes. Doing so can reveal commonalities in function among diverse tissues with extra genome content, as well as specialized roles for, e.g., large cells with few, highly polyploid nuclei vs. many, lower ploidy nuclei. Much remains to be learned regarding the need for cellular collectives in tissue biology.
Acknowledgements
We thank those whose work contributed to the knowledge base discussed in this review but whom we were unable to cite due to space limitations. We thank Rebeccah Stewart and Yarui Diao for comments on the manuscript. This project was supported by NHLBI grant HL140811 to NP and NIGMS grant GM118447 to DF.
Abbreviations
- SPG
Subperineurial glia
- Fzr
Fizzy-related
- Cdk
Cyclin-dependent kinase
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Allen DL, Roy RR, Edgerton VR. Myonuclear domains in muscle adaptation and disease. Muscle Nerve. 1999;22:1350–1360. doi: 10.1002/(sici)1097-4598(199910)22:10[1350::aid-mus3]3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Amini R, Goupil E, Labella S, et al. C. Elegans Anillin proteins regulate intercellular bridge stability and germline syncytial organization. J Cell Biol. 2014;206:129–143. doi: 10.1083/jcb.201310117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow PW, Sherman MI. The biochemistry of differentiation of mouse trophoblast: studies on polyploidy. J Embryol Exp Morphol. 1972;27:447–465. [PubMed] [Google Scholar]
- Bensley JG, De Matteo R, Harding R, Black MJ. Three-dimensional direct measurement of cardiomyocyte volume, nuclearity, and ploidy in thick histological sections. Sci Rep. 2016;6:23756. doi: 10.1038/srep23756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton MA, Akam M, Pavlopoulos A. Cell and tissue dynamics during Tribolium embryogenesis revealed by versatile fluorescence labeling approaches. Development. 2013;140:3210–3220. doi: 10.1242/dev.096271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Bhardwaj RD, Bernard S, et al. Evidence for cardiomyocyte renewal in humans. Science. 2009;324:98–102. doi: 10.1126/science.1164680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann O, Zdunek S, Felker A, et al. Dynamics of cell generation and turnover in the human heart. Cell. 2015;161:1566–1575. doi: 10.1016/j.cell.2015.05.026. [DOI] [PubMed] [Google Scholar]
- Bielski CM, Zehir A, Penson AV, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet. 2018;50:1189–1195. doi: 10.1038/s41588-018-0165-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolívar J, Huynh JR, López-Schier H, et al. Centrosome migration into the Drosophila oocyte is independent of BicD and egl, and of the organisation of the microtubule cytoskeleton. Development. 2001;128:1889–1897. doi: 10.1242/dev.128.10.1889. [DOI] [PubMed] [Google Scholar]
- Bomblies K. When everything changes at once: finding a new normal after genome duplication. Proc R Soc B Biol Sci. 2020;287:20202154. doi: 10.1098/rspb.2020.2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun RE, Behringer RR, Peschon JJ, et al. Genetically haploid spermatids are phenotypically diploid. Nature. 1989;337:373–376. doi: 10.1038/337373a0. [DOI] [PubMed] [Google Scholar]
- Brodsky VY, Chernyaev AL, Vasilyeva IA. Variability of the cardiomyocyte ploidy in normal human hearts. Virchows Arch B Cell Pathol Incl Mol Pathol. 1992;61:289–294. doi: 10.1007/BF02890430. [DOI] [PubMed] [Google Scholar]
- Vy B, Sarkisov DS, Arefyeva AM, et al. Polyploidy in cardiac myocytes of normal and hypertrophic human hearts; range of values. Virchows Arch. 1994;424:429–435. doi: 10.1007/BF00190566. [DOI] [PubMed] [Google Scholar]
- Brown AS, Hong Y, De Belder A, et al. Megakaryocyte ploidy and platelet changes in human diabetes and atherosclerosis. Arterioscler Thromb Vasc Biol. 1997;17:802–807. doi: 10.1161/01.ATV.17.4.802. [DOI] [PubMed] [Google Scholar]
- Buchrieser J, Degrelle SA, Couderc T, et al. IFITM proteins inhibit placental syncytiotrophoblast formation and promote fetal demise. Science. 2019;365:176–180. doi: 10.1126/science.aaw7733. [DOI] [PubMed] [Google Scholar]
- Buchrieser J, Dufloo J, Hubert M, et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020;39:e106267. doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursztajn S, Berman SA, Gilbert W. Differential expression of acetylcholine receptor mRNA in nuclei of cultured muscle cells. Proc Natl Acad Sci U S A. 1989;86:2928–2932. doi: 10.1073/pnas.86.8.2928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell KA, Handel MA. Protamine transcript sharing among postmeiotic spermatids. Proc Natl Acad Sci U S A. 1991;88:2407–2411. doi: 10.1073/pnas.88.6.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V, Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. Berlin Heidelberg: Springer; 1997. [Google Scholar]
- Carvalho L, Heisenberg CP. The yolk syncytial layer in early zebrafish development. Trends Cell Biol. 2010;20:586–592. doi: 10.1016/j.tcb.2010.06.009. [DOI] [PubMed] [Google Scholar]
- Carvalho L, Stühmer J, Bois JS, et al. Control of convergent yolk syncytial layer nuclear movement in zebrafish. Development. 2009;136:1305–1315. doi: 10.1242/dev.026922. [DOI] [PubMed] [Google Scholar]
- Chen C-P, Wang K-G, Chen C-Y, et al. Altered placental syncytin and its receptor ASCT2 expression in placental development and pre-eclampsia. BJOG an Int J Obstet Gynaecol. 2006;113:152–158. doi: 10.1111/j.1471-0528.2005.00843.x. [DOI] [PubMed] [Google Scholar]
- Chu LT, Fong SH, Kondrychyn I, et al. Yolk syncytial layer formation is a failure of cytokinesis mediated by Rock1 function in the early zebrafish embryo. Biol Open. 2012;1:747–753. doi: 10.1242/bio.20121636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cinnamon E, Makki R, Sawala A, et al. Drosophila Spidey/Kar Regulates Oenocyte Growth via PI3-Kinase Signaling. PLOS Genet. 2016;12:e1006154. doi: 10.1371/journal.pgen.1006154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen E, Allen SR, Sawyer JK, Fox DT. Fizzy-related dictates a cell cycle switch during organ repair and tissue growth responses in the drosophila hindgut. Elife. 2018;7:e38327. doi: 10.7554/eLife.38327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox RT, Spradling AC. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development. 2003;130:1579–1590. doi: 10.1242/dev.00365. [DOI] [PubMed] [Google Scholar]
- Cramer AAW, Prasad V, Eftestol E, et al. Nuclear numbers in syncytial muscle fibers promote size but limit the development of larger myonuclear domains. Nat Commun. 2020;11:6287. doi: 10.1038/s41467-020-20058-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis GK, Patel NH. Short, long, and beyond: Molecular and embryological approaches to insect segmentation. Annu Rev Entomol. 2002;47:669–699. doi: 10.1146/annurev.ento.47.091201.145251. [DOI] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Cuevas M, Lilly M, Spradling A. Germline cyst formation in drosophila. Annu Rev Genet. 1997;31:405–428. doi: 10.1146/annurev.genet.31.1.405. [DOI] [PubMed] [Google Scholar]
- Demeter J, Lee SE, Haber JE, Stearns T. The DNA damage checkpoint signal in budding yeast is nuclear limited. Mol Cell. 2000;6:487–492. doi: 10.1016/s1097-2765(00)00047-2. [DOI] [PubMed] [Google Scholar]
- Deng S, Azevedo M, Baylies M. Acting on identity: myoblast fusion and the formation of the syncytial muscle fiber. Semin Cell Dev Biol. 2017;72:45–55. doi: 10.1016/j.semcdb.2017.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derks W, Bergmann O. Polyploidy in cardiomyocytes: roadblock to heart regeneration? Circ Res. 2020;126:552–565. doi: 10.1161/CIRCRESAHA.119.315408. [DOI] [PubMed] [Google Scholar]
- Dewhurst MR, Ow JR, Zafer G, et al (2020) Loss of hepatocyte cell division leads to liver inflammation and fibrosis. PLoS Genet 16: 10.1371/journal.pgen.1009084 [DOI] [PMC free article] [PubMed]
- Diril MK, Ratnacaram CK, Padmakumar VC, et al. Cyclin-dependent kinase 1 (Cdk1) is essential for cell division and suppression of DNA re-replication but not for liver regeneration. Proc Natl Acad Sci U S A. 2012;109:3826–3831. doi: 10.1073/pnas.1115201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AW, Taylor MH, Hickey RD, et al. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Bawa O, et al. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci U S A. 2009;106:12127–12132. doi: 10.1073/pnas.0902925106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Harper F, et al. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci U S A. 2011;108:E1164–E1173. doi: 10.1073/pnas.1112304108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar BA, Orr-Weaver TL. Endoreplication cell cycles: more for less. Cell. 2001;105:297–306. doi: 10.1016/S0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- Engel FB, Schebesta M, Keating MT. Anillin localization defect in cardiomyocyte binucleation. J Mol Cell Cardiol. 2006;41:601–612. doi: 10.1016/j.yjmcc.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Fawcett DW, Ito S, Slautterback D. The occurrence of intercellular bridges in groups of cells exhibiting. J Biophys Biochem Cytol. 1959;5:453–460. doi: 10.1083/jcb.5.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foe VE, Odell GM, Edgar BA (1993) Mitosis and morphogenesis in the Drosophila embryo. In: Bate M, Martinez-Arias A (eds) The Development of Drosophila melanogaster. Cold Spring Harbor Press, pp 149–300
- Fogarty NM, Mayhew TM, Ferguson-Smith AC, Burton GJ. A quantitative analysis of transcriptionally active syncytiotrophoblast nuclei across human gestation. J Anat. 2011;219:601–610. doi: 10.1111/j.1469-7580.2011.01417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley K, Cooley L. Apoptosis in late stage Drosophila nurse cells does not require genes within the H99 deficiency. Development. 1998;125:1075–1082. doi: 10.1242/dev.125.6.1075. [DOI] [PubMed] [Google Scholar]
- Fox DT, Duronio RJ. Endoreplication and polyploidy: new insights into development and disease. Development. 2013;140:3–12. doi: 10.1242/dev.080531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Gall JG, Spradling AC. Error-prone polyploid mitosis during normal Drosophila development. Genes Dev. 2010;24:2294–2302. doi: 10.1101/gad.1952710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox DT, Soltis DE, Soltis PS, et al. Polyploidy: a biological force from cells to ecosystems. Trends Cell Biol. 2020;30:688–694. doi: 10.1016/j.tcb.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentric G, Desdouets C. Polyploidization in liver tissue. Am J Pathol. 2014;184:322–331. doi: 10.1016/j.ajpath.2013.06.035. [DOI] [PubMed] [Google Scholar]
- Gerbaud P, Pidoux G. Review: An overview of molecular events occurring in human trophoblast fusion. Placenta. 2015;36:S35–S42. doi: 10.1016/j.placenta.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Gladfelter AS, Hungerbuehler AK, Philippsen P. Asynchronous nuclear division cycles in multinucleated cells. J Cell Biol. 2006;172:347–362. doi: 10.1083/jcb.200507003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh Q, Song T, Petrany MJ, et al (2019) Myonuclear accretion is a determinant of exercise-induced remodeling in skeletal muscle. Elife 8: 10.7554/eLife.44876 [DOI] [PMC free article] [PubMed]
- Gonzalez-Rosa JM, Sharpe M, Field D, et al. Myocardial Polyploidization Creates a Barrier to Heart Regeneration in Zebrafish. Dev Cell. 2018;44(433–446):e7. doi: 10.1016/j.devcel.2018.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grafstein B, Forman DS. Intracellular transport in neurons. Physiol Rev. 1980;60:1167–1283. doi: 10.1152/physrev.1980.60.4.1167. [DOI] [PubMed] [Google Scholar]
- Greenbaum MP, Iwamori T, Buchold GM, Matzuk MM. Germ cell intercellular bridges. Cold Spring Harb Perspect Biol. 2011;3:1–18. doi: 10.1101/cshperspect.a005850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan LJ, De Cuevas M, Matunis E. Genetics of Gonadal Stem Cell Renewal. Annu Rev Cell Dev Biol. 2015;31:291–315. doi: 10.1146/annurev-cellbio-100913-013344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E, Wiggins D, Fielding B, Gould AP. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2006;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- Gutzeit HO. The role of microfilaments in cytoplasmic streaming in Drosophila follicles. J Cell Sci. 1986;80:159–169. doi: 10.1242/jcs.80.1.159. [DOI] [PubMed] [Google Scholar]
- Hammond MP, Laird CD. Control of DNA replication and spatial distribution of defined DNA sequences in salivary gland cells of Drosophila melanogaster. Chromosoma. 1985;91:279–286. doi: 10.1007/BF00328223. [DOI] [PubMed] [Google Scholar]
- Han L, Choudhury S, Mich-Basso JD, et al. Lamin B2 levels regulate polyploidization of cardiomyocyte nuclei and myocardial regeneration. Dev Cell. 2020;53:42–59.e11. doi: 10.1016/j.devcel.2020.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson KA, Eftestøl E, Bruusgaard JC, et al. Myonuclear content regulates cell size with similar scaling properties in mice and humans. Nat Commun. 2020;11:1–14. doi: 10.1038/s41467-020-20057-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herget GW, Neuburger M, Plagwitz R, Adler CP. DNA content, ploidy level and number of nuclei in the human heart after myocardial infarction. Cardiovasc Res. 1997;36:45–51. doi: 10.1016/S0008-6363(97)00140-5. [DOI] [PubMed] [Google Scholar]
- Hernandez JM, Podbilewicz B. The Hallmarks of Cell-Cell Fusion. Development. 2017;144:4481–4495. doi: 10.1242/dev.155523. [DOI] [PubMed] [Google Scholar]
- Herrtwich L, Nanda I, Evangelou K, et al. DNA damage signaling instructs polyploid macrophage fate in granulomas. Cell. 2016;167:1264–1280.e18. doi: 10.1016/j.cell.2016.09.054. [DOI] [PubMed] [Google Scholar]
- Hesse M, Raulf A, Pilz G-A, et al. Direct visualization of cell division using high-resolution imaging of M-phase of the cell cycle. Nat Commun. 2012;3:1076. doi: 10.1038/ncomms2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J Cell Sci. 1996;109:2779–2788. doi: 10.1242/jcs.109.12.2779. [DOI] [PubMed] [Google Scholar]
- Hirose K, Payumo AY, Cutie S, et al. Evidence for hormonal control of heart regenerative capacity during endothermy acquisition. Science. 2019;364:184–188. doi: 10.1126/science.aar2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang WC, Bennett K, Gregg C. Epigenetic and cellular diversity in the brain through allele-specific effects. Trends Neurosci. 2018;41:925–937. doi: 10.1016/j.tins.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman RS, Price KL, Mannix KM, et al. Drosophila sperm development and intercellular cytoplasm sharing through ring canals do not require an intact fusome. Development. 2020;147:dev190140. doi: 10.1242/dev.190140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Law RD. Cell lineage of zebrafish blastomeres. I. Cleavage pattern and cytoplasmic bridges between cells. Dev Biol. 1985;108:78–85. doi: 10.1016/0012-1606(85)90010-7. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Law RD. Cell lineage of zebrafish blastomeres. II. Formation of the yolk syncytial layer. Dev Biol. 1985;108:86–93. doi: 10.1016/0012-1606(85)90011-9. [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Dougherty MT, et al. Formation, architecture and polarity of female germline cyst in Xenopus. Dev Biol. 2004;266:43–61. doi: 10.1016/j.ydbio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Kosaka K, Kawakami K, Sakamoto H, Inoue K. Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech Dev. 2007;124:279–289. doi: 10.1016/j.mod.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Kreutz C, MacNelly S, Follo M, et al (2017) Hepatocyte ploidy is a diversity factor for liver homeostasis. Front Physiol 8: 10.3389/fphys.2017.00862 [DOI] [PMC free article] [PubMed]
- Kudryavtsev BN, Kudryavtseva MV, Sakuta GA, Stein GI. Human hepatocyte polyploidization kinetics in the course of life cycle. Virchows Arch B Cell Pathol Incl Mol Pathol. 1993;64:387–393. doi: 10.1007/BF02915139. [DOI] [PubMed] [Google Scholar]
- Lang L, Schnittger A. Endoreplication - a means to an end in cell growth and stress response. Curr Opin Plant Biol. 2020;54:85–92. doi: 10.1016/j.pbi.2020.02.006. [DOI] [PubMed] [Google Scholar]
- Lazzeri E, Angelotti ML, Conte C, Anders HJ RP (2019) Surviving acute organ failure: cell polyploidization and progenitor proliferation. Trends Mol Med Epub ahead of print:30041–3. 10.1016/j.molmed.2019.02.006 [DOI] [PubMed]
- Lee DM, Chen EH. Drosophila myoblast fusion: invasion and resistance for the ultimate union. Annu Rev Genet. 2019;53:67–91. doi: 10.1146/annurev-genet-120116-024603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HO, Davidson JM, Duronio RJ. Endoreplication: polyploidy with purpose. Genes Dev. 2009;23:2461–2477. doi: 10.1101/gad.1829209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Spradling AC. Mouse primordial germ cells produce cysts that partially fragment prior to meiosis. Dev. 2013;140:2075–2081. doi: 10.1242/dev.093864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Spradling AC. Mouse oocytes differentiate through organelle enrichment from sister cyst germ cells. Science. 2016;352:95–99. doi: 10.1126/science.aad2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentz TL, Trinkaus JP. A fine structural study of cytodifferentiation during cleavage, blastula, and gastrula stages of Fundulus heteroclitus. J Cell Biol. 1967;32:121–138. doi: 10.1083/jcb.32.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M, Engel FB. Pseudo-bipolar spindle formation and cell division in postnatal binucleated cardiomyocytes. J Mol Cell Cardiol. 2019;134:69–73. doi: 10.1016/j.yjmcc.2019.07.005. [DOI] [PubMed] [Google Scholar]
- Li F, McNelis MR, Lustig K, Gerdes AM (1997a) Hyperplasia and hypertrophy of chicken cardiac myocytes during gposthatching development. Am J Physiol - Regul Integr Comp Physiol 273: 10.1152/ajpregu.1997.273.2.r518 [DOI] [PubMed]
- Li F, Wang X, Bunger PC, Gerdes AM. Formation of binucleated cardiac myocytes in rat heart: I. Role of actin-myosin contractile ring. J Mol Cell Cardiol. 1997;29:1541–1551. doi: 10.1006/jmcc.1997.0381. [DOI] [PubMed] [Google Scholar]
- Li F, Wang X, Capasso JM, Gerdes AM. Rapid transition of cardiac myocytes from hyperplasia to hypertrophy during postnatal development. J Mol Cell Cardiol. 1996;28:1737–1746. doi: 10.1006/jmcc.1996.0163. [DOI] [PubMed] [Google Scholar]
- Lin H, Huang YS, Fustin JM, et al. Hyperpolyploidization of hepatocyte initiates preneoplastic lesion formation in the liver. Nat Commun. 2021;12:1–18. doi: 10.1038/s41467-020-20572-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang CH, Ammanamanchi N, et al (2019) Control of cytokinesis by β-adrenergic receptors indicates an approach for regulating cardiomyocyte endowment. Sci Transl Med 11: 10.1126/scitranslmed.aaw6419 [DOI] [PMC free article] [PubMed]
- Liu Z, Yue S, Chen X, et al. Regulation of cardiomyocyte polyploidy and multinucleation by CyclinG1. Circ Res. 2010;106:1498–1506. doi: 10.1161/CIRCRESAHA.109.211888. [DOI] [PubMed] [Google Scholar]
- Lloyd AC. The Regulation of Cell Size. Cell. 2013;154:1194–1205. doi: 10.1016/j.cell.2013.08.053. [DOI] [PubMed] [Google Scholar]
- Lordier L, Jalil A, Aurade F, et al. Megakaryocyte endomitosis is a failure of late cytokinesis related to defects in the contractile ring and Rho/Rock signaling. Blood. 2008;112:3164–3174. doi: 10.1182/blood-2008-03-144956. [DOI] [PubMed] [Google Scholar]
- Losick VP, Fox DT, Spradling AC. Polyploidization and cell fusion contribute to wound healing in the adult Drosophila epithelium. Curr Biol. 2013;23:2224–2232. doi: 10.1016/j.cub.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losick VP, Jun AS, Spradling AC. Wound-induced polyploidization: Regulation by hippo and JNK signaling and conservation in Mammals. PLoS One. 2016;11:e0151251-. doi: 10.1371/journal.pone.0151251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu K, Jensen L, Lei L, Yamashita YM. Stay connected: a germ cell strategy. Trends Genet. 2017;33:971–978. doi: 10.1016/j.tig.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchetta EM, Ohlstein B. Amitosis of polyploid cells regenerates functional stem cells in the Drosophila intestine. Cell Stem Cell. 2017;20:609–620.e6. doi: 10.1016/j.stem.2017.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAuley A, Cross JC, Werb Z. Reprogramming the cell cycle for endoreduplication in rodent trophoblast cells. Mol Biol Cell. 1998;9:795–807. doi: 10.1091/mbc.9.4.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox AS, Habermann B, Desai A, Oegema K. Distinct roles for two C. elegans anillins in the gonad and early embryo. Development. 2005;132:2837–2848. doi: 10.1242/dev.01828. [DOI] [PubMed] [Google Scholar]
- Marlow FL, Mullins MC. Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev Biol. 2008;321:40–50. doi: 10.1016/j.ydbio.2008.05.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Young KD, Swaffer M, et al. What determines cell size? BMC Biol. 2012;10:101. doi: 10.1186/1741-7007-10-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin NC, McCullough CT, Bush PG, et al. Functional analysis of mouse hepatocytes differing in DNA content: Volume, receptor expression, and effect of IFNγ. J Cell Physiol. 2002;191:138–144. doi: 10.1002/jcp.10057. [DOI] [PubMed] [Google Scholar]
- Matsumoto T, Wakefield L, Peters A, et al (2021) Proliferative polyploid cells give rise to tumors via ploidy reduction. Nat Commun 12:. 10.1038/s41467-021-20916-y [DOI] [PMC free article] [PubMed]
- Matsumoto T, Wakefield L, Tarlow BD, Grompe M. In vivo lineage tracing of polyploid hepatocytes reveals extensive proliferation during liver regeneration. Cell Stem Cell. 2020;26:34–47.e3. doi: 10.1016/j.stem.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall K, Steller H. Requirement for DCP-1 caspase during Drosophila oogenesis. Science. 1998;279:230–234. doi: 10.1126/science.279.5348.230. [DOI] [PubMed] [Google Scholar]
- Meckert PC, Rivello H a n G, Vigliano C, et al (2005) Endomitosis and polyploidization of myocardial cells in the periphery of human acute myocardial infarction. Cardiovasc Res 67:116–123 [DOI] [PubMed]
- Melaragno JE, Mehrotra B, Coleman AW. Relationship between endopolyploidy and cell size in epidermal tissue of arabidopsis. Plant Cell. 1993;5:1661–1668. doi: 10.2307/3869747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyaoka Y, Ebato K, Kato H, et al. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Curr Biol. 2012;22:1166–1175. doi: 10.1016/j.cub.2012.05.016. [DOI] [PubMed] [Google Scholar]
- Mollova M, Bersell K, Walsh S, et al. Cardiomyocyte proliferation contributes to heart growth in young humans. Proc Natl Acad Sci U S A. 2013;110:1446–1451. doi: 10.1073/pnas.1214608110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura S, Satoh M, Fujita T, et al. Cardiomyocyte gene programs encoding morphological and functional signatures in cardiac hypertrophy and failure. Nat Commun. 2018;9:4435. doi: 10.1038/s41467-018-06639-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell TT, Murphy JR, Jackson CW. Stimulation of megakaryocytopoiesis by acute thrombocytopenia in rats. Blood. 1976;48:765–775. doi: 10.1182/blood.v48.5.765.765. [DOI] [PubMed] [Google Scholar]
- Ostergaard KH, Baandrup UT, Wang T, et al. Left ventricular morphology of the giraffe heart examined by stereological methods. Anat Rec. 2013;296:611–621. doi: 10.1002/ar.22672. [DOI] [PubMed] [Google Scholar]
- Øvrebø JI, Edgar BA (2018) Polyploidy in tissue homeostasis and regeneration. Dev 145: 10.1242/dev.156034 [DOI] [PMC free article] [PubMed]
- Ow JR, Cadez MJ, Zafer G, et al. Remodeling of whole-body lipid metabolism and a diabetic-like phenotype caused by loss of CDK1 and hepatocyte division. Elife. 2020;9:1–34. doi: 10.7554/eLife.63835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson M, Barske L, Van Handel B, et al. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nat Genet. 2017;49:1346–1353. doi: 10.1038/ng.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol. 2001;234:339–351. doi: 10.1006/dbio.2001.0269. [DOI] [PubMed] [Google Scholar]
- Peterson NG, Stormo BM, Schoenfelder KP, et al (2020) Cytoplasmic sharing through apical membrane remodeling. Elife 9:. 10.7554/eLife.58107 [DOI] [PMC free article] [PubMed]
- Petrany MJ, Millay DP. Cell fusion: merging membranes and making muscle. Trends Cell Biol. 2019;29:964–973. doi: 10.1016/j.tcb.2019.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrany MJ, Swoboda CO, Sun C, et al. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat Commun. 2020;11:1–12. doi: 10.1038/s41467-020-20063-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper K, Boyde A, Jones SJ. The relationship between the number of nuclei of an osteoclast and its resorptive capability in vitro. Anat Embryol. 1992;186:291–299. doi: 10.1007/BF00185977. [DOI] [PubMed] [Google Scholar]
- Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Sci (new York, NY) 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- Priti A, Ong HT, Toyama Y, et al. Syncytial germline architecture is actively maintained by contraction of an internal actomyosin corset. Nat Commun. 2018;9:1–15. doi: 10.1038/s41467-018-07149-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinton RJ, DiDomizio A, Vittoria MA, et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature. 2021;590:492–497. doi: 10.1038/s41586-020-03133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed BH, Wilk R, Schöck F, Lipshitz HD. Integrin-dependent apposition of Drosophila extraembryonic membranes promotes morphogenesis and prevents anoikis. Curr Biol. 2004;14:372–380. doi: 10.1016/j.cub.2004.02.029. [DOI] [PubMed] [Google Scholar]
- Roberts RM, Ezashi T, Schulz LC, et al. Syncytins expressed in human placental trophoblast. Placenta. 2021 doi: 10.1016/j.placenta.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby JR, Dyer RF, Skalko RG. The occurrence of intercellular bridges during oogenesis in the mouse. J Morphol. 1969;127:307–339. doi: 10.1002/jmor.1051270304. [DOI] [PubMed] [Google Scholar]
- Schmidt-Ott U, Kwan CW. Morphogenetic functions of extraembryonic membranes in insects. Curr Opin Insect Sci. 2016;13:86–92. doi: 10.1016/j.cois.2016.01.009. [DOI] [PubMed] [Google Scholar]
- Schoenfelder KP, Fox DT. The expanding implications of polyploidy. J Cell Biol. 2015;209:485–491. doi: 10.1083/jcb.201502016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenfelder KP, Montague RA, Paramore SV, et al. Indispensable pre-mitotic endocycles promote aneuploidy in the Drosophila rectum. Development. 2014;141:3551–3560. doi: 10.1242/dev.109850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz LM. Skeletal muscles do not undergo apoptosis during either atrophy or programmed cell death-revisiting the myonuclear domain hypothesis. Front Physiol. 2018;9:1887. doi: 10.3389/fphys.2018.01887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senyo SE, Steinhauser ML, Pizzimenti CL, et al. Mammalian heart renewal by pre-existing cardiomyocytes. Nature. 2013;493:433–436. doi: 10.1038/nature11682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Z, Row S, Deng WM. Endoreplication: The Good, the Bad, and the Ugly. Trends Cell Biol. 2018;28:465–474. doi: 10.1016/j.tcb.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigrist SJ, Lehner CF. Drosophila fizzy-related down-regulates mitotic cyclins and is required for cell proliferation arrest and entry into endocycles. Cell. 1997;90:671–681. doi: 10.1016/S0092-8674(00)80528-0. [DOI] [PubMed] [Google Scholar]
- Simpson RA, Mayhew TM, Barnes PR. From 13 weeks to term, the trophoblast of human placenta grows by the continuous recruitment of new proliferative units: A study of nuclear number using the disector. Placenta. 1992;13:501–512. doi: 10.1016/0143-4004(92)90055-X. [DOI] [PubMed] [Google Scholar]
- Soonpaa MH, Kim KK, Pajak L, et al. Cardiomyocyte DNA synthesis and binucleation during murine development. Am J Physiol. 1996;271:H2183–H2189. doi: 10.1152/ajpheart.1996.271.5.H2183. [DOI] [PubMed] [Google Scholar]
- Soygur B, Sati L. The role of syncytins in human reproduction and reproductive organ cancers. Reproduction. 2016;152:R167–R178. doi: 10.1530/REP-16-0031. [DOI] [PubMed] [Google Scholar]
- Soygur B, Sati L, Demir R. Altered expression of human endogenous retroviruses syncytin-1, syncytin-2 and their receptors in human normal and gestational diabetic placenta. Histol Histopathol. 2016;31:1037–1047. doi: 10.14670/HH-11-735. [DOI] [PubMed] [Google Scholar]
- Steinhauser ML, Lee RT. Regeneration of the heart. EMBO Mol Med. 2011;3:701–712. doi: 10.1002/emmm.201100175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O, Banker GA. Getting the message from the gene to the synapse: sorting and intracellular transport of RNA in neurons. Trends Neurosci. 1992;15:180–186. doi: 10.1016/0166-2236(92)90170-d. [DOI] [PubMed] [Google Scholar]
- Storchova Z. Ploidy changes and genome stability in yeast. Yeast. 2014;31:421–430. doi: 10.1002/yea.3037. [DOI] [PubMed] [Google Scholar]
- Stormo BM, Fox DT (2019) Interphase cohesin regulation ensures mitotic fidelity after genome reduplication. Mol Biol Cell 30: 10.1091/mbc.E17-10-0582 [DOI] [PMC free article] [PubMed]
- Stormo BM, Fox DT. Polyteny: still a giant player in chromosome research. Chromosom Res. 2017;25:201–214. doi: 10.1007/s10577-017-9562-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo BM, Fox DT. Distinct responses to reduplicated chromosomes require distinct Mad2 responses. Elife. 2016;5:2275. doi: 10.7554/eLife.15204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakala M, Rodríguez-Acebes S, Maroto M, et al. Functional Reprogramming of Polyploidization in Megakaryocytes. Dev Cell. 2015;32:155–167. doi: 10.1016/j.devcel.2014.12.015. [DOI] [PubMed] [Google Scholar]
- Turco MY, Moffett A (2019) Development of the human placenta. Dev. 146 [DOI] [PubMed]
- Unhavaithaya Y, Orr-Weaver TL. Polyploidization of glia in neural development links tissue growth to blood-brain barrier integrity. Genes Dev. 2012;26:31–36. doi: 10.1101/gad.177436.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas A, Toufaily C, Lebellego F, et al. Reduced expression of both syncytin 1 and syncytin 2 correlates with severity of preeclampsia. Reprod Sci. 2011;18:1085–1091. doi: 10.1177/1933719111404608. [DOI] [PubMed] [Google Scholar]
- Velayutham N, Agnew EJ, Yutzey KE. Postnatal cardiac development and regenerative potential in large mammals. Pediatr Cardiol. 2019;40:1345–1358. doi: 10.1007/s00246-019-02163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velayutham N, Alfieri CM, Agnew EJ, et al. Cardiomyocyte cell cycling, maturation, and growth by multinucleation in postnatal swine. J Mol Cell Cardiol. 2020;146:95–108. doi: 10.1016/j.yjmcc.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventelä S, Toppari J, Parvinen M. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: Mechanisms of haploid gene product sharing. Mol Biol Cell. 2003;14:2768–2780. doi: 10.1091/mbc.E02-10-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Stetina JR, Frawley LE, Unhavaithaya Y, Orr-Weaver TL. Variant cell cycles regulated by Notch signaling control cell size and ensure a functional blood-brain barrier. Development. 2018;145:dev157115. doi: 10.1242/dev.157115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer C, Schönenberger N. Ultrastructure and cytochemistry of the yolk syncytial layer in the alevin of trout (Salmo fario trutta L. and Salmo gairdneri R.) after hatching - II. The Cytoplasmic Zone Cell Tissue Res. 1979;196:75–93. doi: 10.1007/BF00236349. [DOI] [PubMed] [Google Scholar]
- Walzer C, Schönenberger N. Ultrastructure and cytochemistry study of the yolk syncytial layer in the alevin of trout (Salmo fario trutta L.) after hatching - I. The Vitellolysis Zone Cell Tissue Res. 1979;196:59–73. doi: 10.1007/BF00236348. [DOI] [PubMed] [Google Scholar]
- Wang J, Batourina E, Schneider K, et al. Polyploid superficial cells that maintain the urothelial barrier are produced via incomplete cytokinesis and endoreplication. Cell Rep. 2018;25:464–477.e4. doi: 10.1016/j.celrep.2018.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb SE, Miller AL (2013) Calcium signaling in extraembryonic domains during early teleost development. In: International Review of Cell and Molecular Biology. Elsevier Inc., pp 369–418 [DOI] [PubMed]
- Wills AA, Holdway JE, Major RJ, Poss KD. Regulated addition of new myocardial and epicardial cells fosters homeostatic cardiac growth and maintenance in adult zebrafish. Development. 2008;135:183–192. doi: 10.1242/dev.010363. [DOI] [PubMed] [Google Scholar]
- Windner SE, Manhart A, Brown A, et al. Nuclear Scaling Is Coordinated among Individual Nuclei in Multinucleated Muscle Fibers. Dev Cell. 2019;49(48–62):e3. doi: 10.1016/j.devcel.2019.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM. Subcellular specialization and organelle behavior in germ cells. Genetics. 2018;208:19–51. doi: 10.1534/genetics.117.300184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekelchyk M, Guenther S, Preussner J, Braun T. Mono- and multi-nucleated ventricular cardiomyocytes constitute a transcriptionally homogenous cell population. Basic Res Cardiol. 2019;114:36. doi: 10.1007/s00395-019-0744-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S. From cyst to tubule: Innovations in vertebrate spermatogenesis. Wiley Interdiscip Rev Dev Biol. 2016;5:119–131. doi: 10.1002/wdev.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Daniels J, Glaser AE, Wolf MJ. Raf-mediated cardiac hypertrophy in adult Drosophila. Dis Model Mech. 2013;6:964–976. doi: 10.1242/dmm.011361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Daniels JP, Wu H, Wolf MJ. Cardiac hypertrophy induced by active Raf depends on Yorkie-mediated transcription. Sci Signal. 2015;8:ra13. doi: 10.1126/scisignal.2005719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni L, Gonndos B. Intercellular bridges and synchronization of germ cell differentiation during oogenesis in the rabbit. J Cell Biol. 1968;36:276–282. doi: 10.1083/jcb.36.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Lin YH, Tarlow B, Zhu H. The origins and functions of hepatic polyploidy. Cell Cycle. 2019;18:1302–1315. doi: 10.1080/15384101.2019.1618123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhou K, Luo X, et al. The polyploid state plays a tumor-suppressive role in the liver. Dev Cell. 2018;44:447–459.e5. doi: 10.1016/j.devcel.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zissler D. From egg to pole cells: ultrastructural aspects of early cleavage and germ cell determination in insects. Microsc Res Tech. 1992;22:49–74. doi: 10.1002/jemt.1070220106. [DOI] [PubMed] [Google Scholar]