Abstract
Background and Aims:
Alterations in microRNA (miRNA) and in the intestinal barrier are putative risk factors for irritable bowel syndrome (IBS). We aimed to identify differentially expressed colonic mucosal miRNAs, their targets in IBS compared to healthy controls (HCs), and putative downstream pathways.
Methods:
Twenty-nine IBS patients (15 IBS with constipation [IBS-C], 14 IBS with diarrhea [IBS-D]), and 15 age-matched HCs underwent sigmoidoscopy with biopsies. A nCounter array was used to assess biopsy associated miRNA levels. A false discovery rate (FDR) <10% was considered significant. Real-time PCR (RT-PCR) was used to validate differentially expressed genes. To assess barrier function, trans-epithelial electrical resistance (TEER) and dextran flux assays were performed on Caco-2 intestinal epithelial cells that were transfected with miRNA-inhibitors or control inhibitors. Protein expression of barrier function associated genes was confirmed using western blots.
Results:
Four out of 247 miRNAs tested were differentially expressed in IBS compared to HCs (FDR<10%). RT-PCR validation suggested decreased levels of miR-219a-5p and miR-338-3p in IBS (0.026 and p=0.004), and IBS-C (p=0.020 and 0.06) vs. HCs as the strongest associations. Inhibition of miR-219a-5p resulted in altered expression of proteasome/barrier function genes. Functionally, miR-219a-5p inhibition enhanced the permeability of intestinal epithelial cells as TEER was reduced (25-50%, p<0.05) and dextran flux was increased (p<0.01). Additionally, inhibition of miR-338-3p in cells caused alterations in the mitogen-activated protein kinase (MAPK) signaling pathway genes.
Conclusion:
Two microRNAs that potentially affect permeability and visceral nociception were identified to be altered in IBS patients. MiR-219a-5p and miR-338-3p potentially alter barrier function and visceral hypersensitivity via neuronal and MAPK signaling and could be therapeutic targets in IBS.
Keywords: miRNA, miR-338-3p, miR-219a-5p, MAPK signaling, barrier function
Graphical Abstract
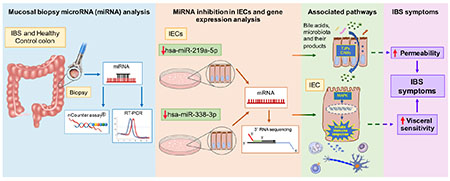
IBS, irritable bowel syndrome; miRNA, microRNA; RT-PCR, real time PCR; IECs, Intestinal epithelial cells; TJPs, tight junction proteins; CAMs, cell adhesion molecules; MAPK, mitogen activated protein kinase; dotted lines represent proposed mechanisms
Introduction
Irritable bowel syndrome (IBS) is a prevalent chronic gastrointestinal (GI) condition characterized by abdominal pain and altered bowel habits (BH) including diarrhea (IBS-D), constipation (IBS-C), or a mixture of both diarrhea and constipation (IBS-M). IBS is a disorder of altered gut-brain interactions1 and is associated with significant morbidity2. Reported findings in IBS include alterations in central sensory processing, neurohormonal regulation, motility and secretion, bile acid metabolism, gut microbiome, immune activation, and epithelial barrier function, and some of these alterations may contribute to IBS symptoms. Intestinal barrier dysfunction associated with altered BH and abdominal pain has been reported in some patients with IBS3,4. Some studies have reported the presence of immune activation via mast cells and T-lymphocytes5, which may mediate intestinal barrier dysfunction, motor abnormalities, and visceral pain in IBS6,7. Although the etiology of IBS is incompletely understood, there is evidence that genetic, environmental, and epigenetic8 factors play a role.
Expression of protein-coding genes (mRNAs) has been previously investigated in IBS9,10, however, a majority of transcripts are non-coding11. MicroRNAs (miRNAs) are small (21-23 bp) non-coding RNAs that regulate gene expression either by base-pairing to target mRNAs or via endonucleolytic mRNA cleavage12. MiRNAs have been implicated in several GI physiologic and pathophysiologic mechanisms and studied widely in intestinal immune and inflammatory diseases, however, studies in IBS are highly heterogeneous13–20. Most IBS-related miRNA studies were limited to IBS-D women. Some of the miRNAs studied were suggested to play a role in visceral hypersensitivity and barrier dysfunction, which are important pathophysiological mechanisms in IBS21. For example, miR-29a targets the glutamine synthetase gene (GLUL) and increases intestinal permeability20, and miR-199a/b targets transient receptor potential cation channel subfamily V member 1 (TRPV1), and a decreased expression of this miRNA correlates with visceral hypersensitivity15. However, there is a lack of a global overview of validated miRNA changes, differences in target gene expression, and associated pathways in IBS, specifically IBS-C. We hypothesize that 1) IBS and BH subtypes are associated with changes in expression of mucosal miRNA and their target genes 2) IBS-associated miRNAs regulate functions/pathways associated with IBS pathophysiology.
We addressed these hypotheses by aiming to identify: 1) differentially expressed miRNAs between IBS and BH subtypes vs. healthy controls (HCs), 2) targets of differentially regulated miRNA and associated pathways by silencing or overexpressing them in intestinal epithelial cell lines, 3) differentially regulated miRNA target genes in the colonic mucosa of IBS patients, and 4) testing potential functional roles for the miRNAs identified.
Methods
Study Population
IBS patients and HCs ages 18-55 were recruited primarily by community advertisement. The diagnosis of IBS and BH subtypes was based on Rome III criteria22 and confirmed by a clinician with expertise in IBS. HCs had no personal or family history of IBS or other chronic pain conditions. Additional exclusion criteria for all subjects included: infectious or inflammatory disorders, active psychiatric illness over the past six months assessed by structured clinical interview for the DSM-IV (MINI)23, use of corticosteroids or narcotics, or current tobacco or alcohol abuse. Participants were compensated. The study was approved by the UCLA Institutional Review Board, and subjects signed a written informed consent prior to the study. Overall IBS symptoms, abdominal pain, and bloating severity over the prior week were assessed with numeric rating scales (0-20)24. Current anxiety and depression symptoms were measured with the Hospital Anxiety and Depression (HAD) scale25. Scores were classified as non-case (0-7), doubtful case (8-10), or definite case (>11).
Colonic mucosal tissue collection
Sigmoid biopsies (30 cm from the anal verge) were obtained by flexible sigmoidoscopy following tap water enemas. Specimens were flash-frozen in liquid nitrogen. Multiple biopsies were obtained from each patient and one biopsy each was used for miRNA and mRNA sequencing. All participants were instructed to hold aspirin and non-steroidal anti-inflammatory drugs (NSAIDs) for 72 hours before the procedure.
NanoString nCounter assay for miRNA expression
Human v2 miRNA CodeSet was used to measure miRNA expression. RNA was extracted from tissues using TRIzol reagent (Invitrongen, Carlsbad, CA) and quantified using NanoDrop (Thermo Scientific, Waltham, MA). The nCounter assay was performed using Nanostring® Analysis Technologies Systems (Nanostring Technologies, Seattle, USA) as described in the Supplementary Methods. All the miRNAs referred to as miRs are human miRNAs (Hsa-miRs).
Real-time PCR (RT-PCR)
Semi-quantitative RT-PCR was used to determine the expression levels of selected miRNAs in biopsies and NCM460 cell line. A miRCURY LNA Universal cDNA synthesis Kit was used to reverse transcribe the RNA (Exiqon). ExiLENT SYBR® Green master mix (203421; Exiqon) and a CFX384 RT-PCR detection system (Bio-Rad, Hercules, CA) were used to perform RT-PCR. LNA miRNA PCR assays were used to measure miRNA expression as listed in Supplementary Table 1.
In vitro target identification
The human immortalized colonic epithelial cell line NCM460 (INCELL, San Antonio, TX; referred to as intestinal epithelial cell-lines, IECs herein) were cultured as described in the Supplementary Methods. At 48 hours post-transfection, the cells were harvested and TRizol was used to extract RNA. The expression of selected miRNAs in the cell lines was confirmed by RT-PCR.
Assessment of epithelial barrier function
For barrier function experiments, Caco-2 intestinal epithelial cells were used (American Type Culture Collection, Manassas, VA). Trans-epithelial electrical resistance (TEER) was measured every 24 hours over three days and dextran flux was measured every two hours over a 6 hour period as described in the Supplementary Methods.
3′ RNA sequencing of miRNA targets
Total RNA was extracted from biopsies and from IECs transfected with miRNA inhibitors or mimics using RNeasy Plus Mini Kit (Qiagen, 74134, Hilden, Germany). RNA was quantified using QuantiT™ RiboGreen™ RNA Assay Kit (Fisher Scientific, Waltham, MA) and samples were normalized to a concentration of 20 ng/μL. RNA quality (RNA integrity number, RIN) was assessed by Agilent TapeStation capillary electrophoresis (most samples had a RIN>7). Gene expression was measured using QuantSeq 3′ mRNA sequencing26. QuantSeq generates highly strand-specific next-generation sequencing libraries close to the 3′ end of polyadenylated RNA. RNA sequencing (RNA-seq) was performed in the UCLA Neuroscience Genomics Core Laboratory using Lexogen QuantSeq 3′ FWD cDNA library synthesis26 and multiplex DNA sequencing on an Illumina HiSeq 4000 instrument with single-strand 65 nucleotide sequence reads following standard manufacturer protocols.
Statistical and bioinformatic analysis:
nCounter
Data preprocessing and normalization: nCounter expression data was analyzed using the NanoStringNorm27 and limma28 packages in R. Sample content normalization was performed using the geometric mean of the top 75 expressed genes. The background was calculated using the geometric mean of negative controls and the data were log-transformed. Hierarchical clustering was used to identify clusters of similar miRNA expression patterns and to identify outliers. One outlier was detected and was excluded from the analysis. Differential expression analysis: Differential expression was determined using a linear model for arrays implemented in ‘limma’ package in R. The adjusted p-values were generated using the function ‘p.adjust’ in R. A false discovery rate (FDR)< 0.1 was considered significant, which is acceptable when additional validation experiments are planned29. MiRNA clusters and individual miRNAs were correlated with the clinical symptoms using logistic regression.
RT-PCR
Bioinformatic methods used for the selection of miRNAs for RT-PCR validation are described in Supplementary Methods. Experiments were performed in triplicates for each condition. MiRNA expression levels were normalized to the average level of 5S rRNA and U6 snRNA. Normalized expression levels were quantified to the plate control. Comparative Ct [DeltaDeltaC(T)] method30 was used to analyze the data. A p-value<0.05 was considered significant.
QuantSeq RNA sequencing
Samples yielded ~3 million reads. De-multiplexed raw reads (FASTQs) were then subjected to the QuantSeq 3’ mRNA-Seq Integrated Data Analysis Pipeline on the Bluebee© Genomics platform (https://www.bluebee.com/lexogen/), which uses standard tools but with parameter settings optimized for processing QuantSeq data. Additional analysis steps are described in the Supplementary Methods. A FDR<5% was considered significant. For gene ontology and pathway analyses, a FDR<0.05 and a fold change >1.2 was used.
Western Blot Analysis
Antibodies for tight junction protein-1 (TJP1/ZO1 cat# 61-7300) and E-cadherin (CDH1, clone 4A2C7) antibodies were from Invitrogen. The GAPDH antibody was from Cell Signaling Technologies (Danvers, MA; clone 14C10). The experimental methods are outlined in Supplementary Methods.
GO pathways and network analysis of miRNA targets
TarBase v.8 and Diana miRpath V3 were used to make in-silico predictions of miRNA targets and associated pathways31. The network of Gene Ontology (GO) terms associated with differentially expressed genes for selected GO terms in IEC models was visualized using ClueGO 2.5.4 in Cytoscape 3.5 (Cytoscape Consortium, http://www.cytoscapeconsortium.org/). The CluePedia plugin was used to enrich the genes with publicly available information from databases, including STRING (https://string-db.org/), IntAct (https://www.ebi.ac.uk/intact/), MiMI (http://www.ncibi.org/mimi.html), miRbase (http://www.mirbase.org/), and miRecords (http://c1.accurascience.com/miRecords/)32.
Identification of miRNA targets for drug development
For the identification of genes that are targets of known prescribed or experimental drugs, we used The International Union of Basic and Clinical Pharmacology (IUPHAR) as described in the Supplementary Methods.
Results
To identify miRNAs altered in IBS patients, we used a total of 44 subjects including 29 IBS patients (52% IBS-C and 48% IBS-D) and 15 HCs. Baseline characteristics are displayed in Table 1. IBS patients and HCs were similar in sex, age, BMI, ethnicity, and race (p>0.05). Overall symptom severity of IBS patients was moderate. Mean anxiety and depression scores were low (non-case) in both groups. Twenty-eight participants (IBS=22, HC=6) reported medication use; most used laxatives or antidiarrheals and NSAIDs only on an as-needed basis. One IBS patient used a benzodiazepine. None of the subjects were taking probiotics or antibiotics.
Table 1:
Demographic characteristics of IBS patients and healthy controls
| Group | IBS | IBS-C | IBS-D | HCs |
|---|---|---|---|---|
| N | 29 | 15 | 14 | 15 |
| Women (%) | 55 | 67 | 43 | 47 |
| Age [mean(sd)] | 33.9 (12.2) | 32 (13.7) | 35.93 (10.5) | 34.2 (9.2) |
| BMI [mean(sd)] | 24.69 (5.0) | 23.98 (3.47) | 25.45 (6.29) | 25.75 (3.1) |
| Ethnicity | ||||
| Hispanic N (%) | 4 (14) | 3 (20) | 1 (7) | 2 (13) |
| Race | ||||
| Asian (%) | 8 (28) | 3 (20) | 5 (36) | 3 (20) |
| Black (%) | 4 (13) | 4 (27) | 0 (0) | 1 (7) |
| White (%) | 12 (41) | 7 (47) | 5 (36) | 8 (53) |
| Other/mixed (%) | 5 (18) | 1 (7) | 4 (28) | 3 (20) |
| Bowel habit subtype | ||||
| IBS-C (%) | 15 (52) | NA | NA | NA |
| IBS-D (%) | 14 (48) | NA | NA | NA |
| HAD anxiety score (0-21) [mean(sd)] | 6.38 (5.9) | 4.27 (4.53) | 8.64 (6.57) | 4.0 (3.1) |
| HAD depression score (0-21) [mean(sd)] | 3.03 (2.9) | 2.07 (2.43) | 4.07 (3.08) | 1.73 (1.94) |
| Overall IBS severity (0-20) [mean(sd)] | 10.4 (4.3) | 9.43 (4.47) | 11.29 (4.05) | NA |
| Abdominal pain (0-20) [mean(sd)] | 10.4 (4.2) | 9.14 (4.4) | 11.57 (3.88) | NA |
| Bloating [mean(sd)] | 12.5 (4.1) | 12.07 (4.4) | 13.07 (3.97) | NA |
Table 1 shows the demographic characteristics of IBS subjects and healthy controls (HCs). N, number of subjects; sd, standard deviation; HAD, Hospital anxiety depression scale; IBS-C constipation-predominant IBS; IBS-D, diarrhea-predominant IBS, NA, not applicable.
MiRNAs associated with IBS and BH subtypes
nCounter platform was used to assess 800 miRNAs simultaneously. Of these, 247 miRNAs were expressed above the background and were tested for differential expression between IBS and BH subtypes, and HCs. Four out of 247 miRNAs were differentially expressed between IBS and HCs, and two were deregulated between IBS-C and HCs (FDR<0.1). MiR-363-3p and miR-338-3p were downregulated, whereas miR-106b-5p and miR-532-5p were upregulated in IBS vs. HCs (FDR=0.06, all miRNAs). When comparing IBS-C vs. HCs, the levels of miR-338-3p and miR-100-5p were decreased (FC=−1.82 and −1.72, FDR=0.04) and the levels of miR-106b-5p were increased (FC=1.31, FDR=0.04, Supplementary Table 2). In IBS-D vs. HCs, a marginal association of eight miRNAs was observed (p<0.05), with miR-219-5p being 3-fold decreased in IBS-D compared to HCs (p<0.05).
To validate the high-throughput miRNA data, we performed RT-PCR on 12 differentially expressed miRNAs shown in Figure 1, that were selected from significantly (FDR<0.1) and differentially expressed miRNAs at p<0.05, in IBS and BH subtypes vs. HCs. We prioritized the miRNAs to be included in the validation set using the ‘random forest’ classification algorithm (Supplementary Figure 1). The miRNAs were sorted based on their ability to discriminate between IBS and HCs as detailed in the Supplementary Results. Additionally, we included bowel habit subtype associated miRNAs for validation. Hierarchical clustering of the 12 miRNAs identified subtypes within IBS, however, they were not associated with IBS symptom severity. Of the 12 miRNAs validated, the strongest associations were decreased levels of miR-338-3p and miR-219-5p in IBS vs. HCs (p = 0.004 and 0.026 respectively, Supplementary Figure 2), and in IBS-C vs. HCs (p = 0.03 and 0.06, respectively). When comparing nCounter and RT-PCR results of the genes that were altered between IBS and HCs, 83% were in congruence (Table 2).
Figure 1.
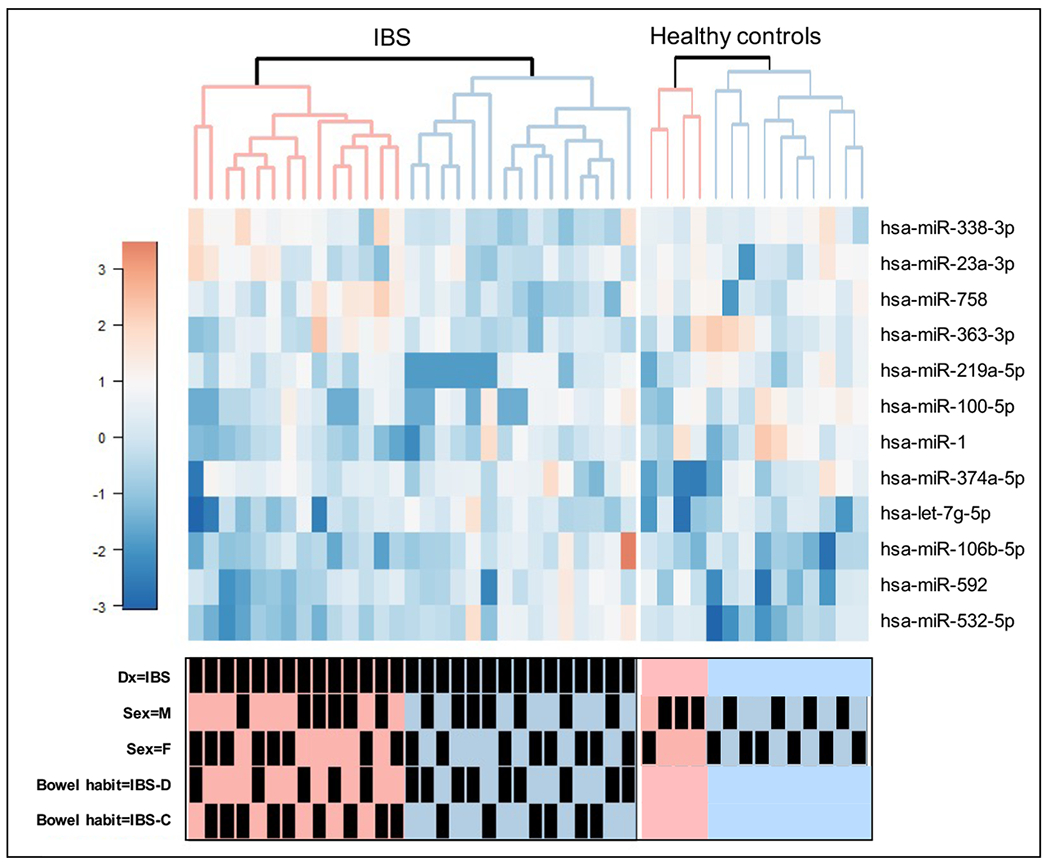
shows the heatmap of the normalized expression of 12 differentially expressed miRNAs. Heatmap colors range from dark blue, white, and dark red, representing low, average, and high expression, respectively. Dx, diagnosis; M, Male; F, Female; IBS-C constipation-predominant IBS; IBS-D, diarrhea-predominant IBS. The bottom-panel shows variables associated with the samples.
Table 2:
Differentially expressed microRNAs in IBS and IBS bowel habit subtypes compared to healthy controls
| nCounter P Values | FDR | nCounter Fold Change | RT-PCR p values | RT-PCR Fold Change | |
|---|---|---|---|---|---|
| IBS vs. HCs | N=29,15 | N=24,14 | |||
| hsa-miR-363-3p | 0.0004 | 0.063 | −1.750 | 0.056 | −1.138* |
| hsa-miR-338-3p | 0.001 | 0.063 | −1.602 | 0.004 | −1.342* |
| hsa-miR-106b-5p | 0.001 | 0.063 | 1.249 | 0.056 | −2.177 |
| hsa-miR-532-5p | 0.001 | 0.063 | 1.352 | 0.396 | 1.043* |
| hsa-miR-100-5p | 0.006 | 0.301 | −1.448 | 0.223 | −1.186* |
| hsa-miR-23a-3p | 0.020 | 0.598 | −1.198 | 0.348 | −1.193* |
| hsa-miR-758 | 0.022 | 0.598 | −2.355 | 0.936 | −1.038* |
| hsa-miR-592 | 0.032 | 0.626 | 1.343 | 0.525 | −1.582 |
| hsa-miR-1 | 0.038 | 0.626 | −1.367 | 0.120 | −2.318* |
| hsa-miR-219-5p | 0.09 | 0.911 | −2.280 | 0.026 | −1.441* |
| IBS-C vs. HCs | N=15, 15 | N=13, 14 | |||
| hsa-miR-338-3p | 0.0002 | 0.039 | −1.820 | 0.020 | −1.325* |
| hsa-miR-100-5p | 0.0004 | 0.039 | −1.720 | 0.230 | −1.221* |
| hsa-miR-106b-5p | 0.0005 | 0.039 | 1.310 | 0.025 | −1.782 |
| hsa-miR-532-5p | 0.002 | 0.128 | 1.380 | 0.699 | −1.008 |
| hsa-miR-363-3p | 0.003 | 0.166 | −1.690 | 0.100 | −1.172* |
| hsa-miR-23a-3p | 0.010 | 0.342 | −1.260 | 0.379 | −1.173* |
| hsa-miR-1 | 0.016 | 0.430 | −1.520 | 0.008 | −1.814* |
| hsa-miR-592 | 0.037 | 0.537 | 1.390 | 0.131 | −1.404 |
| hsa-miR-374a-5p | 0.048 | 0.537 | 1.120 | 0.049 | −1.600 |
| hsa-miR-219-5p | 0.5 | 0.235 | −1.610 | 0.060 | −1.246* |
| IBS-D vs. HCs | N=14, 15 | N=11, 14 | |||
| hsa-miR-338-3p | 0.034 | 0.999 | −1.390 | 0.119 | −1.364* |
| hsa-miR-106b-5p | 0.024 | 0.869 | 1.190 | 0.048 | −2.823 |
| hsa-miR-532-5p | 0.009 | 0.869 | 1.320 | 0.720 | 1.113* |
| hsa-miR-363-3p | 0.001 | 0.262 | −1.820 | 0.078 | −1.089* |
| hsa-miR-758 | 0.021 | 0.869 | −2.750 | 0.199 | −1.116* |
| hsa-miR-219-5p | 0.016 | 0.172 | −3.100 | 0.129 | −1.315* |
Table 2 shows differentially expressed microRNAs in IBS and IBS bowel habit subtypes compared to healthy controls (HCs).
direction of differential expression in nCounter was in agreement with real-time PCR (RT-PCR);
FDR, false discovery rate; IBS-C constipation-predominant IBS; IBS-D, diarrhea-predominant IBS; N= number of subjects.
Identification of change in mRNA expression associated with miR-219a-5p and miR-338-3p
For both nCounter miRNA and RT-PCR data, decreased levels of both miR-219a-5p and miR-338-3p were observed in IBS (and IBS bowel habit subtypes) compared to HCs. Additionally, computationally predicted targets of miR-219a-5p were associated with barrier function, which is important in IBS pathogenesis. To identify the targets of miR-219a-5p and miR-338-3p, we inhibited miRNAs in IECs and measured their expression (Supplementary Figure 3).
Inhibition of miR-219a-5p in NCM460 cells alters the expression of permeability associated genes
To study the role of miR-219a-5p downregulation in the pathophysiology of IBS, we inhibited miR-219a-5p in normal human IECs and performed 3′ mRNA sequencing. 1066 genes were upregulated, and 1187 genes were downregulated between miR-219a-5p-inhibited cells and control cells (FDR<0.05, absolute fold change > 1.2 fold). GO terms associated with the genes upregulated in miR-219a-inhibited cells vs. controls included, “cell-cell adherence junction” (count=55 genes, Benjamini_p= 2.1E-11) and “Neuropathy” (count=14 genes, Benjamini_p= 7.4E-3) among others.
GO network analysis of the differentially expressed genes resulted in a network of terms such as “cell-cell adherence junction”, “Wnt signaling”, “Parkinson’s disease” and “proteasome”, with miR-219-5p (Figure 2). Go term ‘cell-cell adherence junction’ was associated with genes including, Tight junction (TJ) protein 1 (TJP1 or ZO-1, FDR=0.03), E-Cadherin (CDH1), Carcinoembryonic antigen-related cell adhesion molecule 5 (CEACAM5) and, Catenin delta 1 (CTNND1, FDR=0.040), and ‘Wnt signaling’ was associated with Kruppel-like factor 5 (KLF5, FDR=0.02) and Zinc finger 148 (ZNF148, p=0.046), which were upregulated in miR-219-inhibited cells, and are suggested to play a role in barrier function33. Increased proteasome-mediated degradation of tight TJ has been associated with altered barrier function, increased permeability, and low-grade inflammation in IBS34.
Figure 2.

shows a network of deregulated genes and select gene ontology (GO) terms associated with the inhibition of miR-219a-5p. Nodes and edges are described in the top right panel of the figure.
Expression of barrier function-related miR-219a-5p associated genes in NCM460 cells.
The mRNA sequencing data showed an upregulation of barrier function associated genes including ZO1 and CDH1, which play an important role in barrier function. To test the differential expression at the protein level, we measured the protein expression of ZO1 and CDH1 in miR-219a-inhibited NCM460 cells compared to control cells by Western blot (Figure 3). ZO1 protein was significantly upregulated in miR-219a-inhibited cells compared to control cells. However, E-cadherin levels were decreased in miR-219a-inhibited cells compared to controls, which is in agreement with the previous studies35. A decreased E-cadherin expression has been associated with inflamed epithelium in Crohn’s disease and ulcerative colitis patients36.
Figure 3.
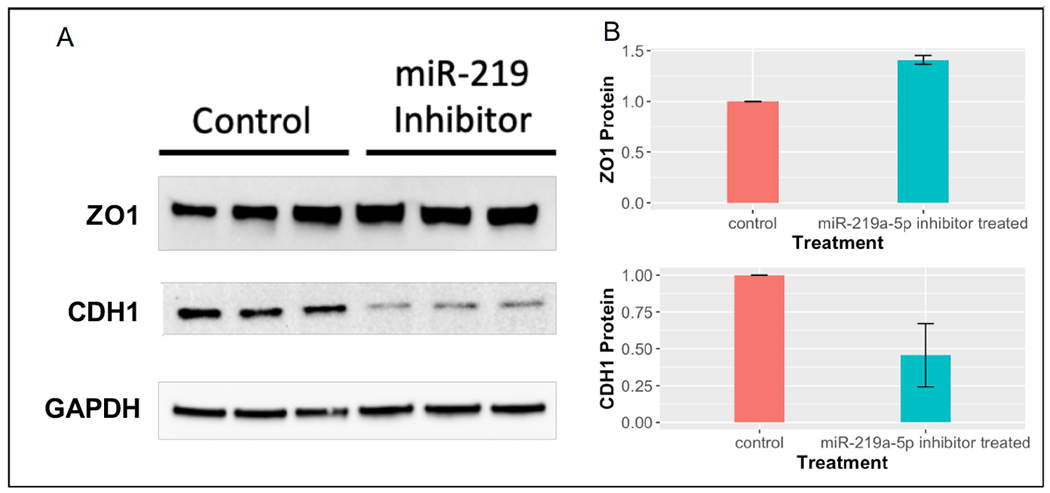
shows A) western blots of tight junction protein 1 (TJP1 or ZO1) and E-cadherin (CDH1) proteins performed in NCM460 cells, B) Protein levels of ZO1 and CDH1 by densitometry. The error bars represent mean ± standard deviation, and the data represent fold changes relative to control. The experiments were performed in duplicate.
Inhibition of miR-219a-5p disrupts intestinal barrier function
Given the observed association of miR-219a-5p inhibition and expression of cell-cell adherence junctions genes, we hypothesized that a decreased miR-219a-5p expression would result in perturbed barrier function and consequently increased paracellular permeability. Since NCM460 cells formed only weak barriers, we inhibited miR-219a-5p expression in a barrier-forming, intestinal epithelial cell line, Caco-237, and measured permeability. After one day, a reduced TEER was observed in miR-219a-5p inhibitor-treated monolayers compared to control-treated monolayers and this difference increased over time, with some experiments exhibiting a significant 50% reduction in TEER on the final day (p<0.05, Figure 4A). As another measure of barrier function, dextran flux was then measured at the end of each experiment. It was observed that 2, 4, and 6 hours after the addition of dextran, almost no dextran crossed the control-treated monolayers, however, both 4 and 10 kDa dextran were able to significantly cross miR-219a-5p treated monolayers (p<0.01, Figure 4B).
Figure 4.
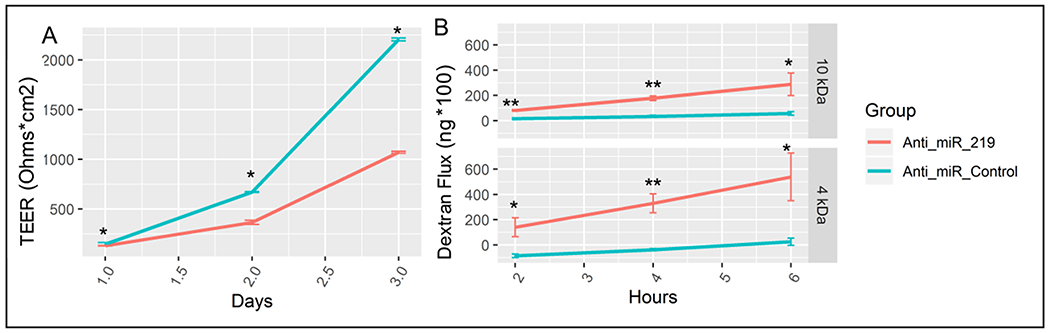
shows, A) a progressive decrease in trans-epithelial electrical resistance (TEER) over 3 days in Caco-2 cells treated with miR-219a-5p inhibitor (Anti_miR_219, red line) compared to cells treated with control miRNA (Anti_miR_Control, blue line), and B) an increased dextran flux over 6 hours (measured every 2 hours) in cells treated with miR-219a-5p inhibitor (Anti_miR_219, red line) compared to cells treated with control miRNA (Anti_miR_Control, blue line). *, p<0.05; **, p<0.01.
MiR-219a-5p is associated with neuronal genes
In addition to changes in barrier function associated genes, miR-219a-5p inhibition resulted in deregulation of mitochondrial genes along with the proteasome complex genes. The ubiquitin-proteasome system plays an important role in mitochondrial biogenesis and mitochondrial function38,39, which modulates neuronal function and participates in neuronal signaling40.
Additionally, Growth differentiation factor-15 (GDF15), which is a hormone conveying somatic distress to the brain41 was among the top upregulated expressed genes. Also, we observed an upregulation of Neuferricin (CYB5D2) which promotes neurogenesis, in addition to several physiologic functions including cholesterol/steroid biosynthesis, drug metabolism, and response, among others42.
Targets of miR-219a-5p are deregulated in the colonic mucosa of IBS patients
To identify the overlapping genes differentially regulated in the cellular model and colonic mucosa of IBS vs. HCs, we performed 3′ mRNA sequencing on the colonic mucosa of subjects associated with this study. We identified 134 genes, which included genes associated with the mitochondrial function such as oxidation-reduction process, including cytochrome b561(CYB561), cell-cell adhesion function, including (integrin subunit beta 1 binding protein 1(ITGB1BP1), channel proteins including, TRPM8 channel-associated factor 1 (TCAF1), ABC transporter genes, ABCC1 and ABCA5, and calcium/calmodulin dependent protein kinase ID (CAMK1D), that were deregulated in both the colonic mucosa of IBS patients compared to HCs as well as in the miR-219a-5p-inhibited cells (p<0.05 for colon and FDR<0.1 for cells, congruent fold changes, Supplementary Table 3).
MiR-338-3p inhibition is associated with MAPK signaling and immune pathway alterations
To uncover the role of miR-338-3p in the pathophysiology of IBS, we inhibited miR-338-3p in NCM460 cells and performed 3′ mRNA sequencing. Inhibition of miR-338-3p was associated with deregulation of 1368 genes (FDR<0.05, 737 up, and 631 downregulated). The deregulated genes were significantly associated with GO terms including “kinase activity” (Enrichment p-value = 8.5E-3, # of genes = 58). A network of kinase and MAPK (mitogen-activated protein kinase or protein serine/threonine kinase) pathway genes, some of which are the predicted targets of has-miR-338-3p included kinase-related and immune response genes (Figure 5). MAPK pathways are implicated in inflammation pain and hypersensitivity in animal models43,44. While the MAPK genes including MAPK1, MAPK8IP3, and MAPK9 were upregulated, genes associated with the ‘kinase inhibitor’ term, including tribbles related protein 3 (TRIB3), were downregulated in miR-338-3p-inhibited cells, suggesting activation of the MAPK pathway. TRIB3 inhibits key inflammatory signaling pathways, including the MAPK and phosphatidylinositol 3 kinase (PI3K) networks43. Additionally, innate immune response related and Wnt signaling related genes were upregulated in miR-338-inhibited cells compared to control cells. Additional MAPK associated genes included Stratifin (SFN, Keratinocyte-releasable 14-3-3-sigma) and Fatty acid amide hydrolase (FAAH) which were upregulated in miR-338 inhibited cells compared to controls. SFN is a proinflammatory cytokine that binds to CD13 (also known as aminopeptidase N, APN) which plays a role in pain sensation via MAPK pathway45. Fatty acid amide hydrolase (FAAH), which catalyzes the degradation of endocannabinoids (anandamide, 2-arachidonoylglycerol) and mediates promigratory effects via MAPK pathway46, was increased in miR-338-inhibited cells. FAAH inhibitors ameliorate signs of acute, inflammatory, visceral, and neuropathic pain in animal models47.
Figure 5.
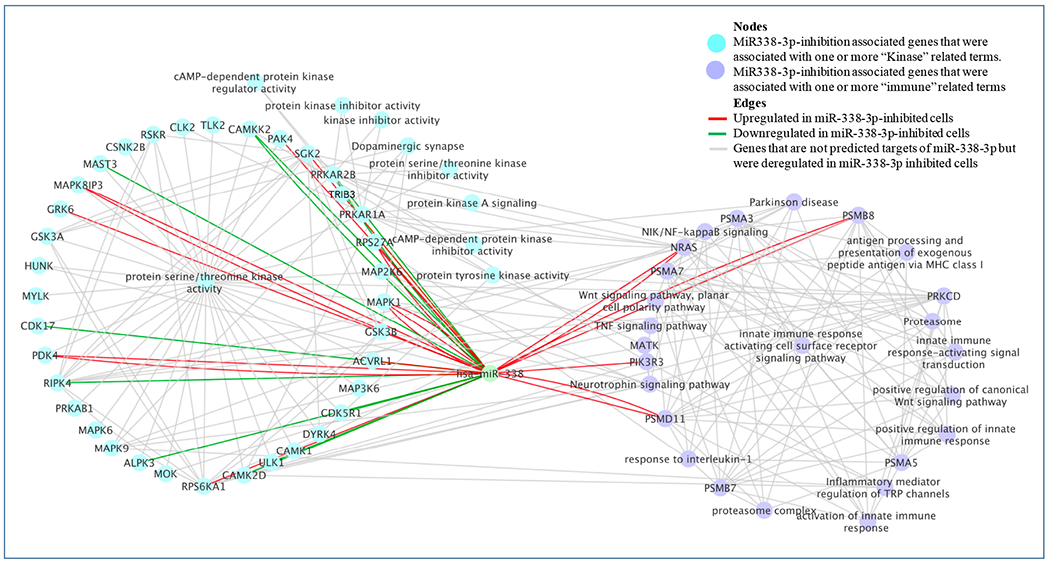
shows a network of deregulated genes and select gene ontology (GO) terms associated with the inhibition of hsa-miR-338-3p. Nodes and edges are described in the top right panel of the Figure.
MiR-338-3p associated genes are deregulated in the colonic mucosa of IBS patients
When comparing protein-coding genes that were differentially expressed in the colonic mucosa of IBS patients to HCs as well as miR-338-3p-inhibited IECs, 47 were downregulated and 45 were upregulated in both (p<0.05 for colon and FDR<0.1 for cells), Supplementary Table 4). These included upregulation of kinase domain-containing genes such as Serine/Threonine kinase (AKT2), and LIM domain kinase 1 (LIMK1) and downregulation of ATM serine/threonine kinase (ATM) and inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta (IKBKB). This suggests an involvement of the MAPK pathway, which has been associated with pronociception in a rat visceral pain model48. Additionally, solute carrier (SLC) family 2 member A3 (SLC2A3, GLUT3, facilitated glucose transporter) was downregulated in both IBS patients and the IEC miR-338-3p inhibition model. Mice deficient in GLUT3 show increased startle response, which is a functional pain paradigm in IBS women49.
Genes associated with miR-219a-5p and miR-338-3p deregulation are targets of known drugs
Next, we investigated whether the differentially expressed genes in colonic mucosa (IBS vs. HCs) and IEC miRNA inhibition models were targets of known drugs. Using the pharmacology database as described in the Methods, we identified genes including CAMK1D and TRPM8 Channel Associated Factor 1 (TCAF1), which were upregulated, and interleukin-7 receptor subunit alpha (IL-7R), which was downregulated in IBS vs. HCs and miR-219a-5p inhibited cells (Supplementary Table 5).
Additionally, we identified inhibitors for LIM Domain Kinase 1 (LIMK1) and Serine/Threonine Kinase 2 (AKT2) protein kinases, which were upregulated miR-338-3p inhibited cells as well as between IBS and HCs (Supplementary Table 6).
Discussion
The key findings of this study are 1) identification of two miRNAs (miR-219a-5p, miR-338-3p) in the colonic mucosa which are downregulated in IBS 2) miR-219a-5p dysregulation results in impaired barrier function in IECs, and 3) identification of pathways and networks through combined bioinformatic analysis of gene expression profiling in colonic mucosal biopsies from IBS and IEC models. Additionally, we identified a miRNA that was altered in IBS, which plays a potential role in visceral hypersensitivity via the MAPK pathway.
Our study identified novel colonic mucosal miRNAs associated with IBS. There is ample evidence that alterations in miRNAs may be important contributing factors in IBS pathogenesis. For example, downregulation of miR-16 and miR-125b have been associated with a dysregulation of barrier function in IBS-D patients via modulation of TJ signaling50. Changes in colonic mucosal expression of miR-29a20 are associated with altered permeability via regulation of glutamine synthase gene in IBS51. Similarly, miR-199 has been associated with visceral hypersensitivity via regulation of TRPV1 in IBS-D, further supporting the importance of miRNAs in IBS pathophysiology. These miRNAs were not associated with IBS or within either BH subset of IBS patients in this study, which may be due to differences in the IBS patient populations (race/ethnicity), tissue sampling (e.g., sigmoid colon vs. jejunum), sample size differences, and/or variable analysis platforms. The potential sources of differences between this and the previous miRNA studies on IBS and animal models of IBS are listed in Supplementary Table 7 and 8, respectively. While many studies focused on IBS-D women, a subset of patients with increased permeability, or miRNA in the small intestine, our study included colonic mucosal biopsies from men and women with IBS-C or IBS-D. Similar to our previous findings, we found most differences in IBS-C vs. HCs52.
While increased permeability is most often associated with IBS-D, it has also been observed in IBS-C53. We observed downregulation of miR-219a-5p in both IBS-C and IBS-D compared to HCs. Our findings and previous studies on miR-219a-5p suggest that its downregulation may impact many pathways with potential relevance to IBS. For example, miR-219a-5p inhibition was associated with TJP1/ZO1 upregulation and impaired barrier function. ZO1 is a TJ protein that interacts with claudins and occludins on epithelial cell membranes54. Although some studies have reported an association of decreased ZO1 expression in the colonic mucosa with increased permeability in IBS53,55, our results are consistent with studies demonstrating decreased permeability in cells depleted for ZO156. Another explanation for the discordance of phenotype and gene expression changes is that the mechanism may not involve direct alterations in TJ protein expression or that they may be compensatory. Increased Wnt-signaling has been associated with altered barrier function in epithelial cells33. Additionally, we found E-cadherin to be downregulated at the protein level. Mice models that lack E-cadherin highlighted its importance in maintaining intestinal epithelial integrity and in the defense against enteropathogenic bacteria36. Furthermore, an increase in expression of barrier function genes may be related to increased ion transport and increased fluid or electrolyte secretion as suggested in the previous studies57.
Functional analysis of miR-219-inhibited cells showed a significant difference in the TEER and dextran flux between controls and miR-219a-5p inhibited cells suggesting a role for mir-219a-5p in regulating barrier function. The differences in the baseline levels of rate of dextran flux between controls and the 219a-5p-inhibited cells may be due to the role of this miRNA in regulating cortical actomyosin contraction.
Additionally, we identified an association of miR-219a-5p with the proteasome and mitochondria-associated genes, linked to neurological disorders including Parkinson’s disease, which is associated with altered GI motility58. The proteasome system modulates neuronal function and plays an important role in neuronal signaling40. A study in a mouse model of spinal cord injury found downregulation of miR-219a-5p associated with an increase in inflammatory cytokines and reactive oxygen species (ROS), which were normalized by transfection with miR-219a mimics59. Similarly, reduced expression of miR-219 in spinal neurons in mouse models of chronic inflammation was significantly associated with inflammatory pain, while overexpression was associated with a reversal of thermal hyperalgesia, mechanical allodynia and, spinal neuronal sensitization60,61. These data suggest a role for miR-219a in reducing mitochondrial oxidative stress, which has been previously associated with IBS62,63. Colonic mucosal gene expression analysis corroborated the gene expression findings in IECs suggesting a deregulation of neuro-motor and neuronal cell adhesion functions associated with downregulation of miR-219a-5p in IBS. This is supported by our previous finding that colonic mucosal expression in IBS-C is involved in pathways mediating neuronal signaling10. Further studies are needed to determine if inhibition of miR-219a-5p is associated with visceral hypersensitivity or mucosal immune activation in IBS.
Similarly, changes in permeability with altered Wnt signaling may also result in alterations in homeostatic mechanisms associated with a proliferative vs. differentiated fate, which may include metabolism and apoptosis in addition to alteration in cellular junctions64. Both increased apoptosis and oxidative stress can increase permeability65. Upregulation of KLF5 and CTNND1 in IECs with miR-219-5p depletion is also supportive of a role of miR-219-5p in Wnt signaling66,67. Additionally, there is bioinformatic evidence for miR-219-5p regulating ZNF148, which was upregulated in our miR-219-5p depletion model and is a positive regulator of Wnt signaling68. Another cadherin-binding protein, cortactin (CTTN), was downregulated, a change that was associated with increased permeability in mice69. Oxidative stress-related barrier dysfunction could also be due to other signaling mechanisms as discussed above.
Our study identified differentially expressed genes common to both IBS colon and miR-219-inhibited cells that can be potential drug targets. TCAF1, which was increased in the colon and miR-219-inhibited cells codes for an ion binding protein that regulates TRPM8 trafficking and activity and plays a role in temperature sensing70. TRPM8 antagonists have been investigated to treat chronic pain and migraine and can be a potential therapeutic agent in IBS71. Additionally, CAMK1D has been associated with epigenetic changes associated with the transition from acute to chronic pain in mouse prefrontal cortex following nerve injury72 and was identified as a potential drug target (Supplementary Table 5).
Another interesting finding from this study was that miR-338-3p targets the MAPK pathway and its downregulation, as observed in IBS vs. HCs, results in downregulation of MAPK inhibitors including TRIB3. TRIB3 is regulated by cannabidiol (CBD), a non-psychotropic phytocannabinoid that modulates allodynia73 via TRPV4 signaling74. Additionally, miR-338-3p depletion resulted in deregulation of several MAPK pathway genes including MAPK1 and MAPK9, activated in response to stressful stimuli75. Animal studies showed that activation of MAPKs and PI3K pathways in dorsal horn neurons involved in the production of proinflammatory cytokines mediate inflammatory pain and visceral hypersensitivity43,44. Additionally, inhibitors of MAPKs have been shown to effectively alleviate inflammatory and neuropathic pain in animal models76. Colonic gene expression analysis corroborated the involvement of genes associated with MAPK and cell adhesion pathways in IBS. The role of the MAPK pathway in IBS, which is not a primarily inflammatory disorder, is unclear. However, there is evidence of immune activation and microscopic inflammation in some patients, particularly post-infection IBS (PI-IBS). In patients with PI-IBS, alterations in the innate and adaptive immune systems were associated with increased intestinal permeability which persisted after the enteric infection77,78.
Genes congruently deregulated in the colon of IBS and miR-338-inhibited cells included targets of known drugs. For example, we identified channel blockers for KCNJ14 and ligands for SLC2A3 that can be investigated as potential therapeutic agents.
This study has limitations. Although our study’s sample size was comparable to most other miRNA studies and included relatively well-characterized IBS and HC populations, some of the findings warrant replication in larger cohorts. We tried to overcome the sample size limitation in part by validating the findings independently using RT-PCR. Interestingly, some of the miR-219a-5p targets were neuronal even though we studied epithelial cell lines. Since the NCM460 cells have a multilineage capability for in vitro differentiation, they express neuroendocrine markers including chromogranin79, which may explain the expression of neuronal genes. Additionally, there are limitations associated with drawing conclusions regarding neuronal physiology based on findings from mucosal biopsies. While the mucosa is innervated by sensory nerve fibers, and biopsies frequently include intestinal submucosal elements, an alternative explanation is that the expression changes may reflect alterations in glial cells or enteroendocrine cells, both of which have neuronal properties. Both miR-219a-5p and miR-338-3p have well-defined roles in the maturation of oligodendrocytes which have some functional overlap with enteric glial cells80. Additionally, the drug targets predicted here are based on the assumption that increased mRNA translates into increased protein expression, which is not always true. Nevertheless, our study provides evidence for multiple new drug targets that can be potentially explored for IBS.
In conclusion, using integrative analysis on high throughput miRNA and 3′quantseq data, followed by validation of individual targets on a well-characterized, age and sex balanced IBS and HC groups, our study showed several altered miRNAs and miRNA-associated pathways that may play a role in intestinal permeability and visceral hypersensitivity, which are characteristics of IBS. Based on our observations, future studies investigating some of the proposed drug targets and focusing on pathways that lead to neuro-immune dysfunction in IBS may be warranted.
Supplementary Material
Funding acknowledgements
Grants: NIH P50 DK64539, R21 DK104078, UL1TR000124, UCLA/IMT-core CURE/P30 DK041301.
Abbreviations used in this paper:
- AKT2
serine/threonine kinase
- ATM
ATM serine/threonine kinase
- BH
bowel habits
- CAMK1D
calcium/calmodulin dependent protein kinase ID
- FAAH
fatty acid amide hydrolase
- FDR
false discovery rate
- GI
gastrointestinal
- GO
Gene Ontology
- GPCR
G protein-coupled receptors HC, healthy control
- IBS
irritable bowel syndrome
- IBS-C
irritable bowel syndrome with constipation
- IBS-D
irritable bowel syndrome with diarrhea
- IEC
intestinal epithelial cell
- IKBKB
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta
- LIMK1
LIM domain kinase 1
- MAPK
mitogen-activated protein kinase
- miRNA
microRNA
- mRNA
messenger RNA
- PCR
polymerase chain reaction
- TCAF1
TRPM8 channel-associated factor 1
- TEER
transepithelial electrical resistance
- TJ
tight junction
- TJP1
tight junction protein 1
- TRIB3
tribbles related protein 3
- TRPV1
transient receptor potential cation channel subfamily V member 1
- ZO1
zonula occludens-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
No conflict of interest to declare.
References
- 1.Drossman DA, Hasler WL. Rome IV-Functional GI Disorders: Disorders of Gut-Brain Interaction. Gastroenterology 2016;150:1257–1261. [DOI] [PubMed] [Google Scholar]
- 2.Lovell RM, Ford AC. Global prevalence of and risk factors for irritable bowel syndrome: a meta-analysis. Clin Gastroenterol Hepatol 2012;10:712–721.e4. [DOI] [PubMed] [Google Scholar]
- 3.Camilleri M, Lasch K, Zhou W. Irritable bowel syndrome: methods, mechanisms, and pathophysiology. The confluence of increased permeability, inflammation, and pain in irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol 2012;303:G775–785. [DOI] [PubMed] [Google Scholar]
- 4.Barbara G, Stanghellini V, De Giorgio R, et al. Activated mast cells in proximity to colonic nerves correlate with abdominal pain in irritable bowel syndrome. Gastroenterology 2004;126:693–702. [DOI] [PubMed] [Google Scholar]
- 5.Chadwick VS, Chen W, Shu D, et al. Activation of the mucosal immune system in irritable bowel syndrome. Gastroenterology 2002;122:1778–1783. [DOI] [PubMed] [Google Scholar]
- 6.Buhner S, Li Q, Vignali S, et al. Activation of human enteric neurons by supernatants of colonic biopsy specimens from patients with irritable bowel syndrome. Gastroenterology 2009;137:1425–1434. [DOI] [PubMed] [Google Scholar]
- 7.Barbara G, Wang B, Stanghellini V, et al. Mast cell-dependent excitation of visceral-nociceptive sensory neurons in irritable bowel syndrome. Gastroenterology 2007;132:26–37. [DOI] [PubMed] [Google Scholar]
- 8.Mahurkar-Joshi S, Chang L. Epigenetic Mechanisms in Irritable Bowel Syndrome. Front Psychiatry 2020;11:805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camilleri M, Carlson P, Acosta A, et al. Colonic mucosal gene expression and genotype in irritable bowel syndrome patients with normal or elevated fecal bile acid excretion. Am J Physiol Gastrointest Liver Physiol 2015;309:G10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Videlock Elizabeth J, Mahurkar-Joshi Swapna, Iliopoulos Dimitrios, et al. Dysregulation of the long-noncoding RNA, GHRLOS, in irritable bowel syndrome. Gastroenterology 2017;152:S722. [Google Scholar]
- 11.Taft RJ, Pheasant M, Mattick JS. The relationship between non-protein-coding DNA and eukaryotic complexity. Bioessays 2007;29:288–299. [DOI] [PubMed] [Google Scholar]
- 12.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010;79:351–379. [DOI] [PubMed] [Google Scholar]
- 13.Wohlfarth C, Schmitteckert S, Härtle JD, et al. miR-16 and miR-103 impact 5-HT 4 receptor signalling and correlate with symptom profile in irritable bowel syndrome. Sci Rep 2017;7:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martinez C, Rodino-Janeiro BK, Lobo B, et al. miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut 2017;66:1537–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Q, Yang L, Larson S, et al. Decreased miR-199 augments visceral pain in patients with IBS through translational upregulation of TRPV1. Gut 2016;65:797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou Q, Huang Y, Zhu S, et al. MiR-144 Increases Intestinal Permeability in IBS-D Rats by Targeting OCLN and ZO1. Cell Physiol Biochem 2017;44:2256–2268. [DOI] [PubMed] [Google Scholar]
- 17.Zhu H, Xiao X, Chai Y, et al. MiRNA-29a modulates visceral hyperalgesia in irritable bowel syndrome by targeting HTR7. Biochem Biophys Res Commun 2019;511:671–678. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y, Li Y, Hao Z, et al. Association of the Serotonin Receptor 3E Gene as a Functional Variant in the MicroRNA-510 Target Site with Diarrhea Predominant Irritable Bowel Syndrome in Chinese Women. J Neurogastroenterol Motil 2016;22:272–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hou Q, Huang Y, Zhang C, et al. MicroRNA-200a Targets Cannabinoid Receptor 1 and Serotonin Transporter to Increase Visceral Hyperalgesia in Diarrhea-predominant Irritable Bowel Syndrome Rats. J Neurogastroenterol Motil 2018;24:656–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chao G, Wang Y, Zhang S, et al. MicroRNA-29a increased the intestinal membrane permeability of colonic epithelial cells in irritable bowel syndrome rats. Oncotarget 2017;8:85828–85837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stanisor OI, Diest SA van, Yu Z, et al. Stress-Induced Visceral Hypersensitivity in Maternally Separated Rats Can Be Reversed by Peripherally Restricted Histamine-1-Receptor Antagonists. PLOS ONE 2013;8:e66884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longstreth GF, Thompson WG, Chey WD, et al. Functional bowel disorders. Gastroenterology 2006;130:1480–1491. [DOI] [PubMed] [Google Scholar]
- 23.Sheehan DV, Lecrubier Y, Sheehan KH, et al. The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry 1998;59 Suppl 20:22–33;quiz 34–57. [PubMed] [Google Scholar]
- 24.Munakata J, Naliboff B, Harraf F, et al. Repetitive sigmoid stimulation induces rectal hyperalgesia in patients with irritable bowel syndrome. Gastroenterology 1997;112:55–63. [DOI] [PubMed] [Google Scholar]
- 25.Zigmond AS, Snaith RP. The hospital anxiety and depression scale. Acta Psychiatr Scand 1983;67:361–370. [DOI] [PubMed] [Google Scholar]
- 26.Moll P, Ante M, Seitz A, et al. QuantSeq 3′ mRNA sequencing for RNA quantification. Nature Methods 2014. Available at: https://www.nature.com/articles/nmeth.f.376 [Accessed April 9, 2018].
- 27.Waggott D, Chu K, Yin S, et al. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics 2012;28:1546–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDonald JH In: Handbook of Biological Statistics. 3rd ed. Baltimore, Maryland: Sparky House Publishing; 2014:254–260. Available at: http://www.biostathandbook.com/multiplecomparisons.html. [Google Scholar]
- 30.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 31.Vlachos IS, Zagganas K, Paraskevopoulou MD, et al. DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 2015;43:W460–W466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics 2013;29:661–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cattin A-L, Le Beyec J, Barreau F, et al. Hepatocyte nuclear factor 4alpha, a key factor for homeostasis, cell architecture, and barrier function of the adult intestinal epithelium. Mol Cell Biol 2009;29:6294–6308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coëffier M, Gloro R, Boukhettala N, et al. Increased Proteasome-Mediated Degradation of Occludin in Irritable Bowel Syndrome. American Journal of Gastroenterology 2010;105:1181–1188. [DOI] [PubMed] [Google Scholar]
- 35.Smalley KSM, Brafford P, Haass NK, et al. Up-Regulated Expression of Zonula Occludens Protein-1 in Human Melanoma Associates with N-Cadherin and Contributes to Invasion and Adhesion. Am J Pathol 2005;166:1541–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schneider MR, Dahlhoff M, Horst D, et al. A Key Role for E-cadherin in Intestinal Homeostasis and Paneth Cell Maturation. PLoS One 2010;5. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3001873/ [Accessed November 25, 2020]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lea T Caco-2 Cell Line. In: Verhoeckx K, Cotter P, López-Expósito I, et al. , eds. The Impact of Food Bioactives on Health: in vitro and ex vivo models. Cham (CH): Springer; 2015. Available at: http://www.ncbi.nlm.nih.gov/books/NBK500149/ [Accessed November 13, 2020]. [PubMed] [Google Scholar]
- 38.Tian T, Wang Z, Zhang J. Pathomechanisms of Oxidative Stress in Inflammatory Bowel Disease and Potential Antioxidant Therapies. Oxidative Medicine and Cellular Longevity 2017. Available at: https://www.hindawi.com/journals/omcl/2017/4535194/ [Accessed August 14, 2019]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Poyton RO, Castello PR, Ball KA, et al. Mitochondria and Hypoxic Signaling. Annals of the New York Academy of Sciences 2009;1177:48–56. [DOI] [PubMed] [Google Scholar]
- 40.Ramachandran KV, Margolis SS. A mammalian nervous system-specific plasma membrane proteasome complex that modulates neuronal function. Nat Struct Mol Biol 2017;24:419–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lockhart SM, Saudek V, O’Rahilly S. GDF15: A Hormone Conveying Somatic Distress to the Brain. Endocr Rev 2020;41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryu CS, Klein K, Zanger UM. Membrane Associated Progesterone Receptors: Promiscuous Proteins with Pleiotropic Functions – Focus on Interactions with Cytochromes P450. Front Pharmacol 2017;8. Available at: https://www.frontiersin.org/articles/10.3389/fphar.2017.00159/full [Accessed December 1, 2020] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kiss-Toth E, Bagstaff SM, Sung HY, et al. Human tribbles, a protein family controlling mitogen-activated protein kinase cascades. J Biol Chem 2004;279:42703–42708. [DOI] [PubMed] [Google Scholar]
- 44.Ji R-R, Befort K, Brenner GJ, et al. ERK MAP Kinase Activation in Superficial Spinal Cord Neurons Induces Prodynorphin and NK-1 Upregulation and Contributes to Persistent Inflammatory Pain Hypersensitivity. J Neurosci 2002;22:478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nefla M, Sudre L, Denat G, et al. The pro-inflammatory cytokine 14–3-3ε is a ligand of CD13 in cartilage. J Cell Sci 2015;128:3250–3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wollank Y, Ramer R, Ivanov I, et al. Inhibition of FAAH confers increased stem cell migration via PPARα. J Lipid Res 2015;56:1947–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Booker L, Kinsey SG, Abdullah RA, et al. The fatty acid amide hydrolase (FAAH) inhibitor PF-3845 acts in the nervous system to reverse LPS-induced tactile allodynia in mice. Br J Pharmacol 2012;165:2485–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ji Y, Tang B, Traub RJ. Spinal estrogen receptor alpha mediates estradiol-induced pronociception in a visceral pain model in the rat. Pain 2011;152:1182–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naliboff BD, Waters AM, Labus JS, et al. Increased Acoustic Startle Responses in IBS Patients During Abdominal and Nonabdominal Threat. Psychosom Med 2008;70:920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martínez C, Rodiño-Janeiro BK, Lobo B, et al. miR-16 and miR-125b are involved in barrier function dysregulation through the modulation of claudin-2 and cingulin expression in the jejunum in IBS with diarrhoea. Gut. 2017. September;66(9):1537–1538. doi: 10.1136/gutjnl-2016-311477. Epub 2017 Jan 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou Q, Souba WW, Croce CM, et al. MicroRNA-29a regulates intestinal membrane permeability in patients with irritable bowel syndrome. Gut 2010;59:775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Videlock EJ, Mahurkar-Joshi S, Hoffman JM, et al. Sigmoid colon mucosal gene expression supports alterations of neuronal signaling in irritable bowel syndrome with constipation. Am J Physiol Gastrointest Liver Physiol 2018;315:G140–G157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piche T, Barbara G, Aubert P, et al. Impaired intestinal barrier integrity in the colon of patients with irritable bowel syndrome: involvement of soluble mediators. Gut 2009;58:196–201. [DOI] [PubMed] [Google Scholar]
- 54.Furuse M, Itoh M, Hirase T, et al. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. J Cell Biol 1994;127:1617–1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bertiaux-Vandaële N, Youmba SB, Belmonte L, et al. The expression and the cellular distribution of the tight junction proteins are altered in irritable bowel syndrome patients with differences according to the disease subtype. Am J Gastroenterol 2011;106:2165–2173. [DOI] [PubMed] [Google Scholar]
- 56.Kim S, Kim G-H. Roles of claudin-2, ZO-1 and occludin in leaky HK-2 cells. PLOS ONE 2017;12:e0189221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Camilleri M, Carlson P, Valentin N, et al. Pilot study of small bowel mucosal gene expression in patients with irritable bowel syndrome with diarrhea. Am J Physiol Gastrointest Liver Physiol 2016;311 :G365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fasano A, Visanji NP, Liu LWC, et al. Gastrointestinal dysfunction in Parkinson’s disease. Lancet Neurol 2015;14:625–639. [DOI] [PubMed] [Google Scholar]
- 59.Zhu Y, Xu Q, Sha W-P, et al. MiR-219-5p promotes spinal cord injury recovery by inhibiting NEUROD2-regulated inflammation and oxidative stress. Eur Rev Med Pharmacol Sci 2019;23:37–43. [DOI] [PubMed] [Google Scholar]
- 60.Pan Z, Zhu L-J, Li Y-Q, et al. Epigenetic modification of spinal miR-219 expression regulates chronic inflammation pain by targeting CaMKIlY. J Neurosci 2014;34:9476–9483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu X-M, Cao S-B, Zhang H-L, et al. Downregulation of miR-219 enhances brain-derived neurotrophic factor production in mouse dorsal root ganglia to mediate morphine analgesic tolerance by upregulating CaMKIIγ. Mol Pain 2016;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mete R, Tulubas F, Oran M, et al. The role of oxidants and reactive nitrogen species in irritable bowel syndrome: A potential etiological explanation. Med Sci Monit 2013;19:762–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mahurkar S, Polytarchou C, Iliopoulos D, et al. Genome-wide DNA methylation profiling of peripheral blood mononuclear cells in irritable bowel syndrome. Neurogastroenterol Motil 2016;28:410–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu Y, Elble RC. Homeostatic Signaling by Cell-Cell Junctions and Its Dysregulation during Cancer Progression. J Clin Med 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bhattacharyya A, Chattopadhyay R, Mitra S, et al. Oxidative stress: an essential factor in the pathogenesis of gastrointestinal mucosal diseases. Physiol Rev 2014;94:329–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H, Dai S-D, Zhang D, et al. Delta-catenin promotes the proliferation and invasion of colorectal cancer cells by binding to E-cadherin in a competitive manner with p120 catenin. Target Oncol 2014;9:53–61. [DOI] [PubMed] [Google Scholar]
- 67.Kim CK, Saxena M, Maharjan K, et al. Krüppel-like factor 5 regulates stemness, lineage specification, and regeneration of intestinal epithelial stem cells. Cell Mol Gastroenterol Hepatol 2020;9:587–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ocadiz-Ruiz R, Photenhauer AL, Hayes MM, et al. ZBP-89 function in colonic stem cells and during butyrate-induced senescence. Oncotarget 2017;8:94330–94344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Citlán-Madrid AF, Vargas-Robles H, García-Ponce A, et al. Cortactin deficiency causes increased RhoA/ROCK1-dependent actomyosin contractility, intestinal epithelial barrier dysfunction, and disproportionately severe DSS-induced colitis. Mucosal Immunol 2017;10:1237–1247. [DOI] [PubMed] [Google Scholar]
- 70.Gkika D, Lemonnier L, Shapovalov G, et al. TRP channel-associated factors are a novel protein family that regulates TRPM8 trafficking and activity. J Cell Biol 2015;208:89–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weyer AD, Lehto SG. Development of TRPM8 Antagonists to Treat Chronic Pain and Migraine. Pharmaceuticals (Basel) 2017;10. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5490394/ [Accessed December 11, 2020]. [DOI] [PMC free article] [PubMed]
- 72.Topham L, Gregoire S, Kang H, et al. The Transition from Acute to Chronic Pain: Dynamic Epigenetic Reprogramming of the Mouse Prefrontal Cortex up to One Year Following Nerve Injury. bioRxiv 2020:2020.02.22.956128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.De Gregorio D, McLaughlin RJ, Posa L, et al. Cannabidiol modulates serotonergic transmission and reverses both allodynia and anxiety-like behavior in a model of neuropathic pain. Pain 2019;160:136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oláh A, Tóth BI, Borbiró I, et al. Cannabidiol exerts sebostatic and antiinflammatory effects on human sebocytes. J Clin Invest 2014;124:3713–3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Baldari S, Ubertini V, Garufi A, et al. Targeting MKK3 as a novel anticancer strategy: molecular mechanisms and therapeutical implications. Cell Death Dis 2015;6:e1621–e1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Qu Y-J, Jia L, Zhang X, et al. MAPK Pathways Are Involved in Neuropathic Pain in Rats with Chronic Compression of the Dorsal Root Ganglion. Evid Based Complement Alternat Med 2016;2016. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4967678/ [Accessed November 12, 2019]. [DOI] [PMC free article] [PubMed]
- 77.Spiller RC, Jenkins D, Thornley JP, et al. Increased rectal mucosal enteroendocrine cells, T lymphocytes, and increased gut permeability following acute Campylobacter enteritis and in post-dysenteric irritable bowel syndrome. Gut 2000;47:804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barbara G, Grover M, Bercik P, et al. Rome Foundation Working Team Report on Post-Infection Irritable Bowel Syndrome. Gastroenterology 2019;156:46–58.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moyer MP, Manzano LA, Merriman RL, et al. NCM460, a Normal Human Colon Mucosal Epithelial Cell Line. In Vitro Cellular & Developmental Biology Animal 1996;32:315–317. [DOI] [PubMed] [Google Scholar]
- 80.Nguyen LH, Ong W, Wang K, et al. Effects of miR-219/miR-338 on microglia and astrocyte behaviors and astrocyte-oligodendrocyte precursor cell interactions. Neural Regen Res 2020;15:739–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


