Abstract
PAS domains are widespread, versatile domains found in proteins from all kingdoms of life. The PAS fold is composed of an antiparallel β-sheet with several flanking α-helices, and contains a conserved cleft for cofactor or ligand binding. The last few years have seen a prodigious increase in identified PAS domains and resolved PAS structures, including structures with effector and other domains. New bacterial PAS ligands have been discovered, and structure-function studies have improved our understanding of PAS signaling mechanisms. The list of bacterial PAS functions has now expanded to include roles in signal sensing, modulation, transduction, dimerization, protein interaction, and cellular localization.
Keywords: PAS domain, signal transduction, sensing, dimerization, protein interaction, cellular localization
Graphical abstract
PAS DOMAINS: WIDESPREAD, VERSATILE AND ADAPTABLE
Per-ARNT-Sim (PAS) domains are widespread sensing, signaling and dimerization domains found in proteins from all kingdoms of life [1]. The number of identified PAS domains has rapidly risen due to increased sequencing and algorithm refinements. As of May 2020, Pfam 33.1 includes 222,716 PAS sequences of which 84% derive from bacterial proteins, 12% are from eukaryotic proteins, and 4% are archaeal [2]. PAS domains are associated with 18,219 different protein architectures [2], although in bacteria, they most often reside at the N-terminus of histidine kinases, diguanylate cyclases/phosphodiesterases, and methyl-accepting chemotaxis proteins, where they regulate the functions of those proteins [3].
PAS domains are 100–120 amino acids in length and classically have divergent sequences (<20% average pairwise sequence identity), but conserved three-dimensional structures. The globular PAS core comprises an antiparallel β-sheet (with strands Aβ, Bβ, Gβ, Hβ, and Iβ) and several flanking α-helices (Cα, Dα, Eα, and Fα) (Figure 1a–d). The PAS core can also have adjoining N-terminal (Aα′) or C-terminal (Jα) helices that pack against the β-sheet. The β-sheet is the most conserved region of PAS domains, whereas the orientation, length, and number of α-helices, as well as loop lengths, vary between domains [4]. Commonly, PAS domains assemble into homodimers through a patch of hydrophobic residues on the outer surface of their β-sheet, often with contributions from the adjoining N-or C-terminal helices (Figure 1e, [4]). Less commonly, the β-sheet interfaces with another domain, e.g., PAS/HAMP interaction in the aerotaxis receptor Aer (Figure 1f, [5]).
Figure 1.
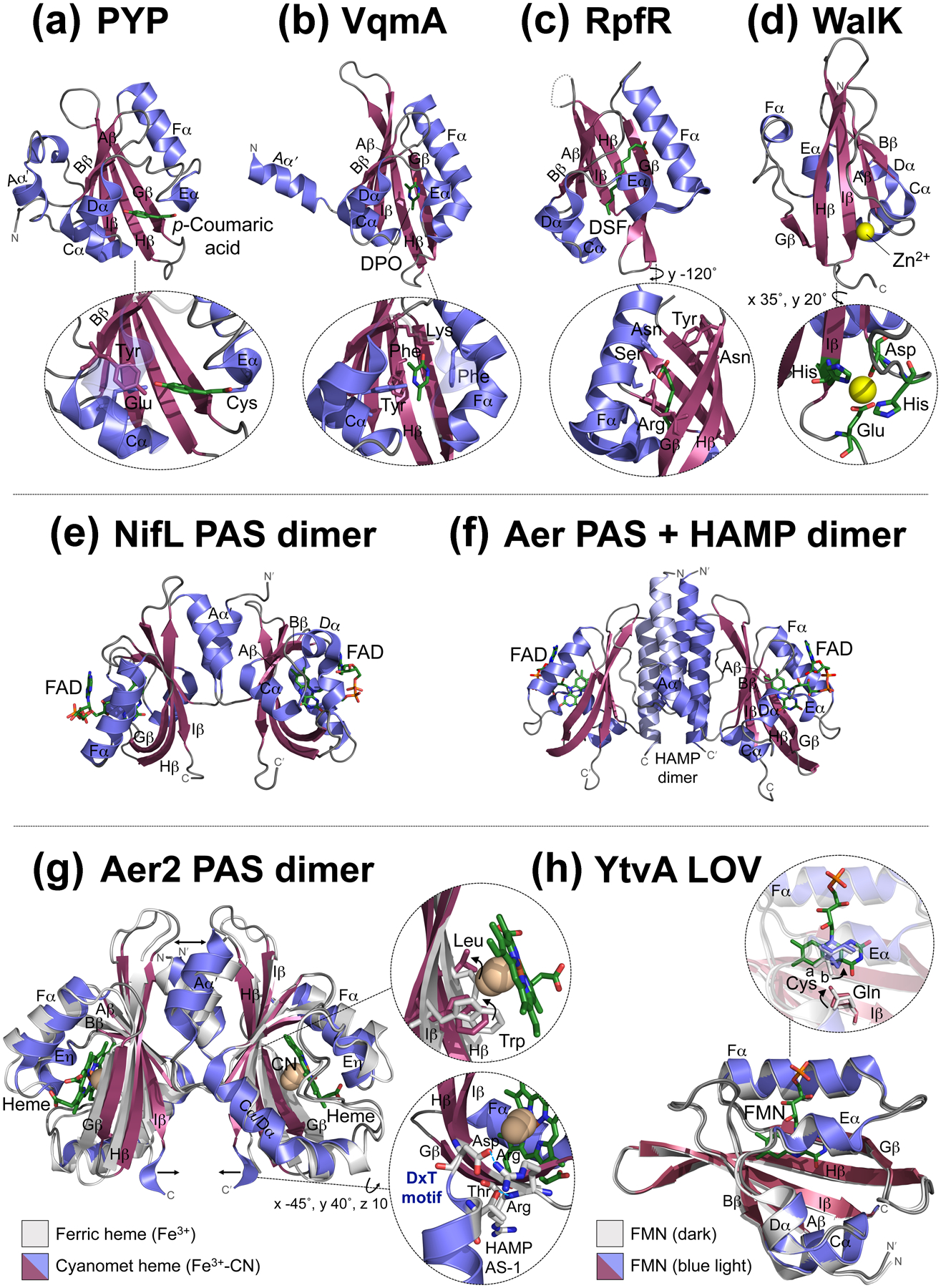
Examples of PAS domain structures and models with bound cofactors and/or ligands. In all examples, the PAS β-strands occur in the order B-A-I-H-G, and the β-sheet is surrounded by helices C-D-E-F. Adjoining N-terminal Aα′ helices are also shown. (a-d) PAS monomer structures: (a) The PAS structural prototype photoactive yellow protein (PYP) from Halorhodospira halophila with its cofactor p-coumaric acid (PDB: INWZ). The inset shows p-coumaric acid covalently bound to PAS via an Eα-Cys residue and stabilized by Bβ-Tyr and Cα-Glu residues [30]. (b) Vibrio cholerae VqmA PAS with its ligand 3,5-dimethyl-pyrazine-2-ol (DPO) (PDB: 6UGL). The inset shows DPO, which is stabilized by PAS Cα-Tyr, Fα-Phe, Hβ-Phe, and Hβ-Lys residues [7]. (c) Cronobacter turicensis RpfR PAS with its ligand (2Z)-dodec-2-eoic acid (DSF) (PDB: 6DGJ). The disordered HI loop in RpfR is represented by a dashed line. The inset shows DSF, which is stabilized by Fα-Ser, Fα-Asn, Gβ-Tyr, Gβ-Arg and Hβ-Asn residues [9]. (d) Staphylococcus aureus WalK PAS with Zn2+ (yellow sphere) bound to the surface (PDB: 4MN5). The inset shows the four metal-coordinating residues, Asp and His residues on a loop N-terminal to Aβ, an Iβ-His residue, and a Glu residue on a loop C-terminal to Iβ [11]. (e-f) Example of PAS dimerization (e) via the PAS β-sheet and N-terminal Aα′ helix [Azotobacter vinelandii NifL PAS1 dimer with FAD (PDB: 2GJ3)], compared with (f) the PAS β-sheet interfacing with a HAMP domain instead (Escherichia coli Aer). Aer PAS and HAMP models are based on the structures of NifL PAS1 (PDB: 2GJ3) and Af1503 HAMP (PDB: 2ASX), whereas the Aer PAS-HAMP interface is based on experimental data [5]. (g-h) PAS structures in different signaling states: (g) Overlay of two Aer2-PAS structures from Pseudomonas aeruginosa including an unliganded dimer bound to heme cofactor (Fe3+, PDB: 4HI4, [25]) and a liganded monomer with cyanomet heme (Fe3+-CN, PDB: 3VOL, [31]). For clarity, b-type heme is only shown bound to the liganded structure. Unlike canonical PAS structures, Aer2 PAS has a combined Cα/Dα helix and an Eη (instead of Eα) helix. Arrows depict ligand-induced movements of the Aα′ helices and the DxT motifs. The top inset shows how the heme-bound ligand displaces the Hβ-Leu residue from the ligand-binding site and the Iβ-Trp rotates ~90° to H-bond with the ligand [24,25]. Although this structure has CN as the ligand, O2 is the natural ligand for Aer2. The bottom inset shows the conserved DxT motif that couples Aer2-PAS to the C-terminal AS-1 (amphipathic sequence-1) helix of the adjoining HAMP domain. The Iβ-Asp of the DxT motif forms a putative salt-bridge with an Arg residue that begins PAS-Hβ, whereas the Thr residue (at the beginning of HAMP AS-1) H-bonds to the backbone amide of the subsequent Hβ-Arg. For clarity, only one liganded PAS monomer is shown, as is a portion of HAMP AS-1 C-terminal to the PAS domain. (h) Overlay of two YtvA-LOV monomers from Bacillus subtilis with bound FMN cofactor in dark (PDB: 2PR5), and blue-light illuminated (PDB: 2PR6), states. Both structures are bound to FMN, but for clarity, FMN is only shown bound to the light-illuminated structure. The inset shows how light induces the formation of a covalent bond between the Eα-Cys residue and FMN, which causes the side chain of the Iβ-Gln to flip 180° [32]. In the dark state, the side chain of the Eα-Cys assumes two distinct conformations, which are labelled “a” and “b” [32].
PAS domains were originally defined as intracellular sensors and it was recently argued that extracytoplasmic PAS domains should instead be classified as Cache domains [6]. For this review, we have only included PAS domains as defined by Pfam and have focused on recent findings on sensing and signaling in bacterial PAS, rather than Cache, domains.
PAS SENSORS: DIVERSE COFACTORS, LIGANDS AND SIGNALS
The best studied PAS domains have sensory functions derived from their ability to bind small molecules. The first small molecule identified was p-coumaric acid bound to PYP from Halorhodospira halophila (Figure 1a and Table 1), while the newest ligand is the pyrazine autoinducer 3,5-dimethyl-pyrazine-2-ol (DPO) bound to VqmA-PAS from Vibrio cholerae (Figure 1b and Table 1). PYP is a blue light sensor implicated in phototaxis, whereas VqmA is a member of the quorum-sensing LuxR-family that regulates gene expression (Figure 2a, [7,8]). Ligands can serve as either a primary PAS signal (like DPO binding to VqmA) or as a secondary signal after binding to an embedded cofactor. Examples include flavin cofactors, which are activated by electron reduction or photon absorption, and hemes, which are activated by gas binding (Table 1). Despite the diversity of PAS cofactors and ligands, most are bound within a conserved cleft formed by the inner surface of the PAS β-sheet and the Eα and Fα helices (Figure 1, [4]). The region around Eα-Fα is the least structurally conserved part of the PAS core [4]. In the case of VqmA, DPO recognition and binding requires four key PAS residues, including an Fα-Phe whose benzene ring provides π-π stacking interactions with the pyrazine ring of DPO (Figure 1b, [7]).
Table 1:
Cofactors and Ligands of Bacterial PAS Domains
| Example PAS Proteins | ||
|---|---|---|
| p-Coumaric acid | Blue light | Halorhodospira halophila PYP [30] |
| Riboflavin | Blue light | Erythrobacter litoralis EL346 [39] |
| Heme | O2, CO, NO |
b-type heme: Bradyrhizobium japonicum FixL [40] E. coli DOS [41] Pseudomonas aeruginosa Aer2 [42] and BdlA [43] c-type heme: Geobacter sulfurreducens GSU0935 and GSU0582 [44] |
| FeS cluster | O2 | Staphylococcus carnosus NreB [45] |
| - | Carbohydrates | Cellvibrio japonicus AbfS [46] |
| - | Fatty acids |
Chloroflexus aurantiacus MltR [47] Mycobacterium tuberculosis Rv1364 Cronobacter turicensis RpfR [9] |
| - | Pyrazine | Vibrio cholerae VqmA [7] |
| - | Aromatic hydrocarbons | Pseudomonas putida TodS [48] |
| - | Metal2+ |
E. coli PhoQ [10] Staphylococcus aureus WalK [11] |
Figure 2.
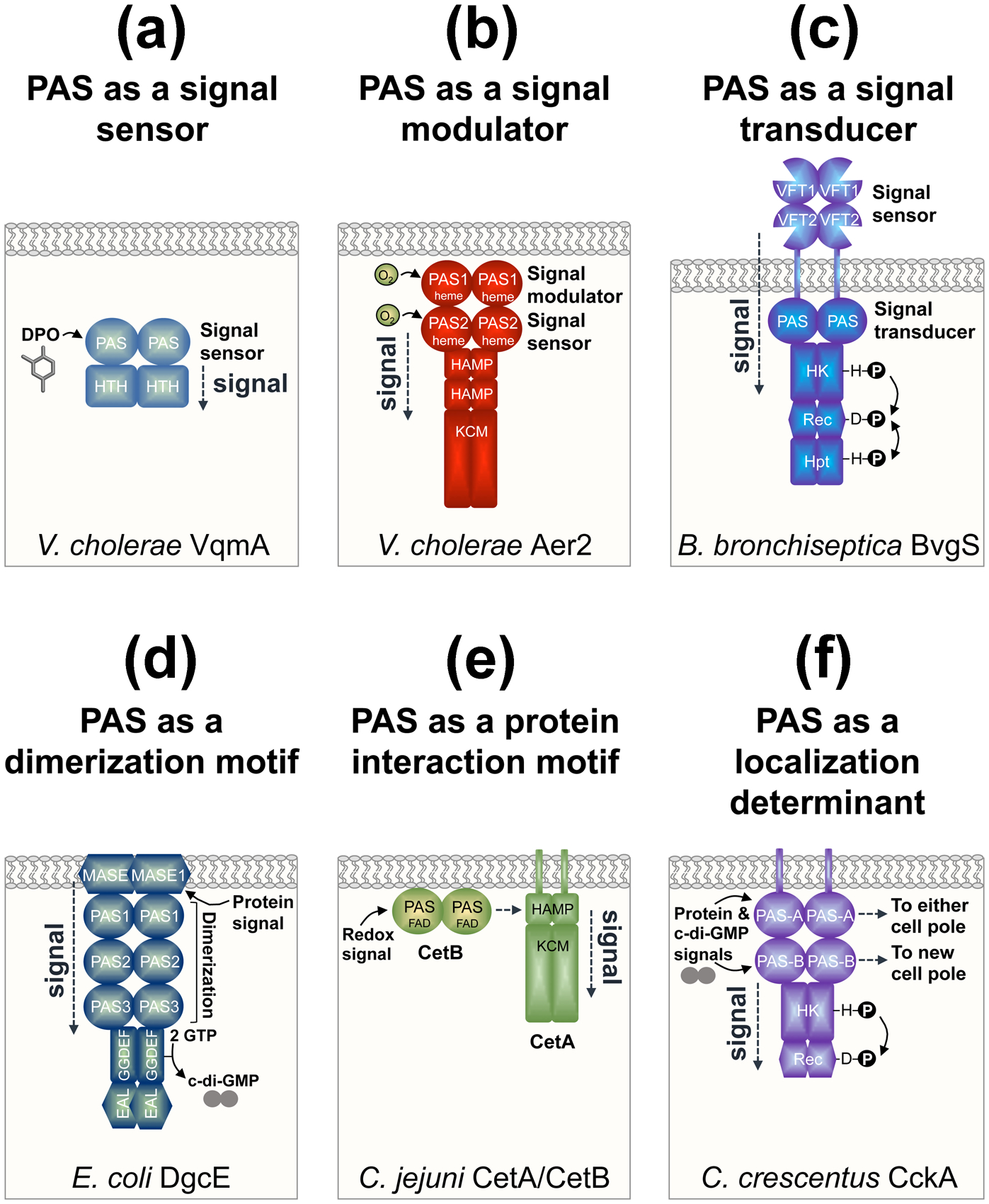
The diversity of PAS domain roles in bacterial proteins. All cartoons show the bacterial cytoplasmic membrane and the dimeric form of each protein. (a) PAS as a signal sensor as illustrated by VqmA from Vibrio cholerae. VqmA-PAS binds to the autoinducer 3,5-dimethyl-pyrazine-2-ol (DPO) and signals to a helix-turn-helix (HTH) DNA-binding domain to regulate transcription [7,8]. (b) PAS as a signal modulator as illustrated by Aer2 from V. cholerae. PAS1 modulates the extent of O2-mediated signaling from PAS2, which sends a conformational signal through two HAMP domains and a kinase control module (KCM) [15]. The KCM regulates a series of downstream chemosensory proteins. (c) PAS as a signal transducer as illustrated by BvgS from Bordetella bronchiseptica. BvgS transduces signals from a periplasmic Venus fly-trap (VFT) domain via a cytoplasmic PAS domain to C-terminal histidine kinase (HK; comprised of DHp and CA subdomains), response regulator (Rec), and histidine phosphotransfer (Hpt) domains, thus regulating kinase and phosphatase functions [16]. (d) PAS as a dimerization motif as illustrated by DgcE from Escherichia coli. The three DgcE PAS domains are essential for protein dimerization, which activates DgcE’s GGDEF (diguanylate cyclase) domain to produce c-di-GMP, and overcomes the anti-oligomerizing effect of the EAL (phosphodiesterase) domain [18]. PAS dimerization occurs after a protein complex binds to the membrane-associated sensor (MASE1) domain of DgcE. The EAL domain is degenerate and does not degrade c-di-GMP to guanosine monophosphate (GMP). (e) PAS as a protein interaction motif as illustrated by CetA and CetB from Camplyobacter jejuni. CetA and CetB constitute a bipartite energy-taxis system with the domains of the aerotaxis receptor, Aer, divided between two proteins. CetB is a PAS domain that is predicted to bind FAD, sense redox, and transmit signals through interaction with the HAMP domain of CetA [19]. (f) PAS as a cellular localization determinant as illustrated by CckA from Caulobacter crescentus. PAS-A directs CckA to either cell pole, whereas PAS-B targets CckA to new cell poles [20]. PAS-A and PAS-B also have sensory roles that regulate CckA’s kinase (HK; comprised of DHp and CA subdomains) and phosphatase (Rec) functions.
The most recently discovered bacterial PAS ligands are autoinducers and fatty acids, including fatty acid autoinducers (Table 1). The fatty acid autoinducer, diffusible signal factor (DSF) [(2Z)-dodec-2-eoic acid; an α,β-unsaturated carboxylic acid], was recently observed in the structure of the Cronobacter turicensis RpfR PAS domain (Figure 1c, [9]). The specificity of ligand binding was facilitated by the amide nitrogen of a PAS-Hβ Asn that interacts with the electron deficient Cβ of DSF (Figure 1c). DSF binding to RpfR activates its phosphodiesterase domain, resulting in c-di-GMP degradation and bacterial biofilm dispersal [9].
In addition to PAS domains that bind ligands within a conserved cleft, some bind metal ligands to the PAS surface (Table 1). This was previously demonstrated for PhoQ from E. coli [10], and more recently for WalK from Staphylococcus aureus. WalK tetrahedrally coordinates Zn2+ to a negatively-charged region on the PAS surface where it negatively regulates WalK’s autokinase activity (Figure 1d, [11]).
Lastly, PAS domains can act as adjuvants for cofactor binding to other domains. Bacteriophytochromes, for example, harness a PAS-Aα′ extension to assist with GAF domain-biliverdin interactions (Table 2). The PAS-Aα′ extension threads through an elongated loop of the GAF domain, and a PAS-Cys residue covalently binds to the tetrapyrrole chromophore via a thioether linkage (see [12] and [13] for the highest resolution crystal and NMR structures of a photosensory module, respectively).
Table 2:
Bacterial Multi-Domain Structures Containing PAS Domains
| Protein (with PDB Identifier and Year Deposited)3 | ||
|---|---|---|
| PAS | PAS-linker4-PAS PAS-linker-PAS-PAS GAF-PAS-PAS PAS-linker-PAS |
M. capsulatus MmoS (PDB: 3EWK; 2008) Rhodobacter sphaeroides PpsR (PDB: 4HH2; 2012) M. tuberculosis Rv3651 (PDB: 4Q6U; 2014) B. abortus LOV-HK (PDB: 6PPS; 2019) |
| ACT | ACT-linker-PAS | E. coli TyrR (PDB: 2JHE: 2007) |
| ANTAR | PAS-ANTAR-linker-PAS | Nakamurella multipartita PAL (PDB: 6HMJ; 2018) |
| DHp | PAS-DHp-CA HAMP-linker-PAS-DHp-CA PAS-DHpL-CA |
Thermatoga maritima ThkA (PDB: 3A0R; 2009) Streptococcus mutans VicK (PDB: 4I5S; 2012) E. litoralis EL346 (PDB: 4R3A; 2014) |
| GAF |
N5-PAS-GAF N-PAS-GAF N-PAS-GAF-PHY N-PAS-GAF-PHY N-PAS-GAF N-PAS-GAF-PHY-linker-PAS N-PAS-GAF N-PAS-GAF-PHY-linker-PAS N-PAS-GAF-PHY N-PAS-GAF-PHY-linker-GGDEF N-PAS-GAF-PHY GAF-linker-PAS GAF-PAS-PAS |
Bacteriophytochromes (Bph): Deinococcus radiodurans BphP (PDB: 1ZTU; 2005) Rhodopseudomonas palustris BphP3 (PDB: 2OOL; 2007) Synechocystis sp. Cph1 (PDB: 2VEA; 2007) P. aeruginosa BphP (PDB: 3C2W; 2008) R. palustris BphP2 (PDB: 4E04; 2012) R. palustris BphP1 (PDB: 4GW9; 2012) Stigmatella aurantiaca BphP (PDB: 4RQ9; 2014) Xanthomonas campestris BphP (PDB: 5AKP; 2015) Agrobacterium fabrum Agp1 (PDB: 5I5L; 2016) Idiomarina sp. PadC (PDB: 5LLX; 2016) A. fabrum Agp2 (PDB: 6G1Y; 2018) Others: E. coli DhaR (PDB: 4LRZ; 2013) M. tuberculosis Rv3651 (PDB: 4Q6U; 2014) |
| GGDEF | PAS-linker-GGDEF-linker-EAL | P. aeruginosa RbdA (PDB: 5XGB; 2017) |
| HAMP | HAMP-linker-PAS-DHp-CA | S. mutans VicK (PDB: 4I5S; 2012) |
| Hemerythrin | Hemerythrin-PAS | Pyrococcus furiosus PF0695 (PDB: 3CAX; 2008) |
| HTH | HTH-linker-PAS PAS-linker-HTH PAS-linker-HTH |
C. aurantiacus MltR (PDB: 3PXP; 2010) E. litoralis EL222 (PDB: 3P7N; 2010) V. cholerae VqmA (PDB: 6IDE; 2019) |
| STAS | PAS-linker-STAS | B. subtilis YtvA (PDB: 2MWG; 2014) |
PAS (Per-ARNT-Sim); ACT (Aspartate kinase, Chorismate mutase, and TyrA); ANTAR (AmiR and NasR Transcription Antitermination Regulators); DHp (Dimerization and phospho-accepting Histidine); GAF (cGMP-specific phosphodiesterases, Adenylate cyclases and FhlA); HAMP (Histidine kinases, Adenylate cyclases, Methyl-accepting chemotaxis proteins, and Phosphatases); HTH (Helix-Turn-Helix); STAS (Sulfate Transporter and AntiSigma factor antagonist).
CA (Catalytic); DHpL (DHp-Like); PHY (Phytochrome).
Where multiple structures exist for the same protein, the earliest structure is cited
α-Helical linkers (e.g., Jα- or S-helices)
N-terminal α-helix or unstructured coil containing the covalent Cys anchor for biliverdin
PAS DOMAINS AS SIGNAL MODULATORS AND TRANSDUCERS
A majority of PAS domains have not been shown to bind cofactors or ligands. For some of these domains, the ligand-binding cleft is too small to bind ligands; instead these domains function as protein interaction or dimerization domains and/or have roles in signal modulation or transduction. These functions differ from those of pure sensors. For example, the two PAS domains of the Xanthomonas campestris histidine kinase RavS regulate RavS function by inhibiting autokinase activity [14]. In RavS, the sensor role appears to be in the catalytic domain, which binds c-di-GMP. Complexity increases with V. cholerae Aer2, which has tandem PAS domains that both coordinate b-type heme and bind O2 [15]. However, PAS2 is an O2 sensor whereas PAS1 modulates the extent of O2-mediated signaling from PAS2 (Figure 2b, [15]). PAS domains are often located between input and output domains such that signal transduction is likely to be a major role. The PAS domain of Bordetella BvgS, for example, transduces signals from a periplasmic Venus fly-trap sensing domain to a cytoplasmic histidine kinase (Figure 2c). However, BvgS PAS might also sense cytoplasmic signals, and could potentially override Venus fly-trap signals, thus tuning BvgS responses to the bacterial environment [16]. Another example is the histidine kinase NifL from Azotobacter vinelandii, which has tandem PAS domains: dimeric PAS1 binds FAD and serves as a redox sensor (Figure 1e), whereas PAS2 has no cofactor and transduces PAS1-generated redox signals via changes in PAS oligomerization [17].
PAS DIMERIZATION OR PROTEIN INTERACTION AS A REQUIREMENT FOR FUNCTION
PAS domains commonly dimerize, but this can also be essential for protein function. The three consecutive PAS domains of the E. coli diguanylate cyclase DgcE were recently shown to be essential for dimerization and activation of DgcE’s GGDEF (diguanylate cyclase) domain (Figure 2d, [18]). It was suggested that PAS occurs proximal to GGDEF domains because GGDEF functions as a dimer and can’t dimerize on its own. In DgcE, the three PAS domains are sandwiched between the membrane-bound signal input domain (MASE1) and the cytoplasmic catalytic (GGDEF) domain, and are also required for signal transduction. Some bacterial PAS domains also promote protein-protein interaction. CetA and CetB from Campylobacter jejuni comprise a bipartite Aer-type receptor, whereby the PAS-CetB sensor interacts with the CetA-HAMP domain to control the activity of the kinase control module (Figure 2e, [19]).
PAS DOMAINS AS REGULATORS OF CELLULAR LOCALIZATION
In addition to the recognized roles of PAS in sensing, signaling and dimerization, a newly identified role for PAS domains is to regulate cellular localization for the spatiotemporal control of cell signaling. This has been elegantly demonstrated for the bifunctional histidine kinase/phosphatase CckA from the asymmetrically-dividing bacterium Caulobacter crescentus. CckA has two PAS domains, PAS-A and PAS-B, both of which direct CckA to cell poles: PAS-A directs CckA to either cell pole, whereas PAS-B targets CckA to new cell poles (Figure 2f, [20]). PAS-A is also required for density-dependent kinase activity through PAS-A mediated oligomerization of CckA [20,21]. In contrast, PAS-B inhibits kinase activity and stimulates CckA phosphatase activity through c-di-GMP-binding and interaction with a kinase called DivL [20,21]. CckA thus uses its two PAS domains to integrate multiple signals and modulate kinase versus phosphatase activity depending on its subcellular location and environment. Kinase activity is favored at new cell poles, with subsequent phosphotransfer to the transcription factor CtrA, which controls the expression of cell cycle-regulated promoters. In addition to CckA, the single domain PAS protein MopJ from C. crescentus also localizes to cell poles [22]. In Sinorhizobium meliloti, the PAS domain of RgsP similarly regulates RgsP localization [23]. RgsP is a phosphodiesterase involved in cell wall biogenesis, and its PAS domain helps target RgsP to new cell poles and to cell division sites where it connects c-di-GMP signaling to the control of peptidoglycan synthesis [23].
PAS SIGNALING MECHANISMS
Signals that originate within PAS cores most often propagate from the cofactor/ligand site to the PAS β-sheet as a conformational or dynamic signal. Aer2 receptors, for example, have a PAS-heme whose ligand-binding site is occupied by an Hβ-Leu. When O2 binds, the Hβ-Leu moves out of the ligand-binding site and an Iβ-Trp rotates ~90° to bond with and stabilize heme-bound O2 (Figure 1g, [15,23,24]). This initiates a conformational signal at the PAS β-sheet that propagates to C-terminal HAMP and kinase control domains. Perturbations in PAS β-sheets can also cause flanking N- or C-terminal helices to reposition or unfold, altering dimer interactions and inducing signaling. Ligand-binding to Aer2-PAS moves the Aα′ helices ~4 Å (Figure 1g), and a PAS signal-on mutant has an unfolded Aα′ helix that is dissociated from the PAS core [15]. Perhaps the best understood mechanism comes from a subset of PAS called LOV (Light-O2-Voltage) domains (Table 1). In LOV domains, light-induced protonation of the flavin cofactor (e.g., via formation of a thioadduct, Figure 1h) results in a PAS Iβ-Gln flipping its sidechain 180°; its oxygen-atom then bonds with the proton while the amide bonds with a residue on PAS-Aβ. This destabilizes interactions between the PAS β-sheet and flanking N- or C-terminal helices, promoting their detachment or rearrangement (reviewed by [26]). In a chimeric protein of the YtvA LOV domain (Figure 1h) fused to the effector module of FixL (YF1), weakening PAS β-sheet/Aα′ interactions rotates and tilts the PAS domains and separates the Jα helices, providing a mechanistic basis for signaling to the effector domain [26]. The tilting and rotating of short α-helices (many of which are amphipathic and form coiled-coils) appears to be a common theme in PAS-to-effector-domain signaling. PAS-Iβ often connects to this helix via a conserved “DxT” motif. The Asp residue of “DxT” forms a salt-bridge to a positively-charged PAS-GH loop residue, whereas the Thr actually begins the adjoining helix and H-bonds to a backbone amide in PAS-Hβ, thus providing a structural basis for coupling PAS to other domains (Figure 1g, [4]). The critical role of the “DxT” motif is evident for the Treponema denticola histidine kinase Hpk2, where an Asp to Lys mutant reduces PAS cofactor (hemin) binding, protein dimerization, and autophosphorylation [27].
The last decade has seen substantial increases in PAS structures containing effector and other domains (Table 2), but few structures have been resolved for the same protein in different signaling states. Generally, PAS signaling is thought to alter effector domain dynamics and quaternary structure. For example, DPO-binding to VqmA likely stabilizes the PAS β-sheet and modulates the PAS-effector domain interface, promoting enhanced DNA binding (Figure 2a, [7]). Similarly, zinc-binding to WalK stabilizes PAS and potentially modulates the angle between the PAS and C-terminal catalytic domains, regulating kinase activity [11]. Bacteriophytochrome PAS domains show minimal crystallographic changes between dark-adapted and photoactivated states (e.g., [28]), but recent NMR data suggests substantial light-induced remodeling at the PAS-GAF interface that could be involved in signaling [13]. Finally, protein interactions can also induce conformational shifts in PAS domains. When the DhaL kinase binds to the E. coli transcription factor DhaR, the DhaR coiled-coil linker region reorganizes and the connected PAS domain pivots, displacing some Cα atoms up to 15 Å; this triggers DhaR’s ability to activate gene expression [29].
CONCLUSIONS
The last few years has seen a surge in identified PAS domains, resolved PAS structures, and multidomain structures. New PAS ligands have been discovered, and structure-function studies have improved our understanding of PAS sensing and signaling mechanisms. The list of PAS functions is growing, highlighting their incredible versatility and utility. In addition to classic roles as sensors and dimerization domains, PAS domains modulate protein function and interactions, transduce signals, and determine cellular localization. Future progress will be facilitated by developing tools to identify non-covalently bound PAS ligands, and by exploiting PAS-effector domains in different signaling states to uncover structure-function relationships.
HIGHLIGHTS.
PAS domains are common sensing, signaling, dimerization, and localization domains
Sensing versatility derives from a conserved cleft that binds cofactors and ligands
PAS signaling involves repositioning of the PAS β-sheet and adjoining helices
PAS connects to diverse effector domains to regulate distinct cellular functions
PAS-effector domain structures have begun to clarify protein signaling mechanisms
ACKNOWLEDGEMENTS
This work was supported by the National Institute of General Medical Sciences (NIGMS) of the National Institutes of Health award number R01GM108655 to K. Watts. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Taylor BL, Zhulin IB: PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol Mol Biol Rev 1999, 63:479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. : The Pfam protein families database in 2019. Nucleic Acids Res 2019, 47:D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henry JT, Crosson S: Ligand-binding PAS domains in a genomic, cellular, and structural context. Annu Rev Microbiol 2011, 65:261–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moglich A, Ayers RA, Moffat K: Structure and signaling mechanism of Per-ARNT-Sim domains. Structure 2009, 17:1282–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garcia D, Watts KJ, Johnson MS, Taylor BL: Delineating PAS-HAMP interaction surfaces and signalling-associated changes in the aerotaxis receptor Aer. Mol Microbiol 2016, 100:156–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Upadhyay AA, Fleetwood AD, Adebali O, Finn RD, Zhulin IB: Cache domains that are homologous to, but different from PAS domains comprise the largest superfamily of extracellular sensors in prokaryotes. PLoS Comput Biol 2016, 12:e1004862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H, Li M, Guo H, Zhou H, Li B, Xu Q, Xu C, Yu F, He J: Crystal structure of the Vibrio cholerae VqmA-ligand-DNA complex provides insight into ligand-binding mechanisms relevant for drug design. J Biol Chem 2019, 294:2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Presents the crystal structure of the LuxR-family protein VqmA from V. cholerae with a new PAS ligand (DPO), in addition to structure-function studies providing details on transcriptional regulation and the impact of DPO binding.
- 8.Huang X, Duddy OP, Silpe JE, Paczkowski JE, Cong J, Henke BR, Bassler BL: Mechanism underlying autoinducer recognition in the Vibrio cholerae DPOVqmA quorum-sensing pathway. J Biol Chem 2020, 295:2916–2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Waldron EJ, Snyder D, Fernandez NL, Sileo E, Inoyama D, Freundlich JS, Waters CM, Cooper VS, Neiditch MB: Structural basis of DSF recognition by its receptor RpfR and its regulatory interaction with the DSF synthase RpfF. PLoS Biol 2019, 17:e3000123. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• A tour-de-force structure-function study of the C. turicensis RpfR protein bound to diffusible signal factor autoinducer and its involvement in quorum sensing, second messenger signaling, and regulation of autoinducer production.
- 10.Cheung J, Bingman CA, Reyngold M, Hendrickson WA, Waldburger CD: Crystal structure of a functional dimer of the PhoQ sensor domain. J Biol Chem 2008, 283:13762–13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monk IR, Shaikh N, Begg SL, Gajdiss M, Sharkey LKR, Lee JYH, Pidot SJ, Seemann T, Kuiper M, Winnen B, et al. : Zinc-binding to the cytoplasmic PAS domain regulates the essential WalK histidine kinase of Staphylococcus aureus. Nat Commun 2019, 10:3067. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Presents the crystal structure of the S. aureus WalK cytoplasmic PAS domain coordinated to zinc and demonstrates how zinc-binding regulates WalK autophosphorylation.
- 12.Sanchez JC, Carrillo M, Pandey S, Noda M, Aldama L, Feliz D, Claesson E, Wahlgren WY, Tracy G, Duong P, et al. : High-resolution crystal structures of a myxobacterial phytochrome at cryo and room temperatures. Struct Dyn 2019, 6:054701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isaksson L, Gustavsson E, Persson C, Brath U, Vrhovac L, Karlsson G, Orekhov V, Westenhoff S: Signaling mechanism of phytochromes in solution. Structure 2020. 10.1016/j.str.2020.08.009 [DOI] [PubMed] [Google Scholar]; • Evaluates the first NMR structure of a complete bacteriophytochrome photosensory module in two different states and indicates a different signaling pathway to that proposed from crystal structures.
- 14.Cheng ST, Wang FF, Qian W: Cyclic-di-GMP binds to histidine kinase RavS to control RavS-RavR phosphotransfer and regulates the bacterial lifestyle transition between virulence and swimming. PLoS Pathog 2019, 15:e1007952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greer-Phillips SE, Sukomon N, Chua TK, Johnson MS, Crane BR, Watts KJ: The Aer2 receptor from Vibrio cholerae is a dual PAS-heme oxygen sensor. Mol Microbiol 2018, 109:209–224. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Demonstrates distinct roles for two PAS-heme domains in the Vibrio cholerae Aer2 receptor in addition to the crystal structure of an Aer2 PAS domain in the signal-on conformation.
- 16.Sobran MA, Cotter PA: The BvgS PAS domain, an independent sensory perception module in the Bordetella bronchiseptica BvgAS phosphorelay. J Bacteriol 2019, 201:e00286–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slavny P, Little R, Salinas P, Clarke TA, Dixon R: Quaternary structure changes in a second Per-Arnt-Sim domain mediate intramolecular redox signal relay in the NifL regulatory protein. Mol Microbiol 2010, 75:61–75. [DOI] [PubMed] [Google Scholar]
- 18.Pfiffer V, Sarenko O, Possling A, Hengge R: Genetic dissection of Escherichia coli’s master diguanylate cyclase DgcE: Role of the N-terminal MASE1 domain and direct signal input from a GTPase partner system. PLoS Genet 2019, 15:e1008059. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Shows how three PAS domains of the E. coli diguanylate cyclase DgcE are essential for protein dimerization and activation of DgcE’s catalytic GGDEF domain.
- 19.Elliott KT, Dirita VJ: Characterization of CetA and CetB, a bipartite energy taxis system in Campylobacter jejuni. Mol Microbiol 2008, 69:1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mann TH, Seth Childers W, Blair JA, Eckart MR, Shapiro L: A cell cycle kinase with tandem sensory PAS domains integrates cell fate cues. Nat Commun 2016, 7:11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mann TH, Shapiro L: Integration of cell cycle signals by multi-PAS domain kinases. Proc Natl Acad Sci U S A 2018, 115:E7166–E7173. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Demonstrates the different roles of two PAS domains in the bifunctional histidine kinase/phosphatase CckA from C. crescentus and shows how multiple signals are integrated by CckA to spatiotemporally control cell signaling.
- 22.Sanselicio S, Berge M, Theraulaz L, Radhakrishnan SK, Viollier PH: Topological control of the Caulobacter cell cycle circuitry by a polarized single-domain PAS protein. Nat Commun 2015, 6:7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaper S, Yau HCL, Krol E, Skotnicka D, Heimerl T, Gray J, Kaever V, Sogaard-Andersen L, Vollmer W, Becker A: Seven-transmembrane receptor protein RgsP and cell wall-binding protein RgsM promote unipolar growth in Rhizobiales. PLoS Genet 2018, 14:e1007594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia D, Orillard E, Johnson MS, Watts KJ: Gas sensing and signaling in the PAS-heme domain of the Pseudomonas aeruginosa Aer2 receptor. J Bacteriol 2017, 199:e00003–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Airola MV, Huh D, Sukomon N, Widom J, Sircar R, Borbat PP, Freed JH, Watts KJ, Crane BR: Architecture of the soluble receptor Aer2 indicates an in-line mechanism for PAS and HAMP domain signaling. J Mol Biol 2013, 425:886–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moglich A: Signal transduction in photoreceptor histidine kinases. Protein Sci 2019, 28:1923–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Contains a detailed review of signal transduction in photoreceptors, including signaling mechanisms with LOV domains and from LOV to the effector domain.
- 27.Sarkar J, Miller DP, Oliver LD Jr., Marconi RT: The Treponema denticola PAS domain-containing histidine kinase Hpk2 is a heme binding sensor of oxygen levels. J Bacteriol 2018, 200:e00116–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burgie ES, Zhang J, Vierstra RD: Crystal structure of Deinococcus phytochrome in the photoactivated state reveals a cascade of structural rearrangements during photoconversion. Structure 2016, 24:448–457. [DOI] [PubMed] [Google Scholar]
- 29.Shi R, McDonald L, Cygler M, Ekiel I: Coiled-coil helix rotation selects repressing or activating state of transcriptional regulator DhaR. Structure 2014, 22:478–487. [DOI] [PubMed] [Google Scholar]
- 30.Borgstahl GE, Williams DR, Getzoff ED: 1.4 Å structure of photoactive yellow protein, a cytosolic photoreceptor: unusual fold, active site, and chromophore. Biochemistry 1995, 34:6278–6287. [DOI] [PubMed] [Google Scholar]
- 31.Sawai H, Sugimoto H, Shiro Y, Ishikawa H, Mizutani Y, Aono S: Structural basis for oxygen sensing and signal transduction of the heme-based sensor protein Aer2 from Pseudomonas aeruginosa. Chem Comm 2012, 48:6523–6525. [DOI] [PubMed] [Google Scholar]
- 32.Moglich A, Moffat K: Structural basis for light-dependent signaling in the dimeric LOV domain of the photosensor YtvA. J Mol Biol 2007, 373:112–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rebbapragada A, Johnson MS, Harding GP, Zuccarelli AJ, Fletcher HM, Zhulin IB, Taylor BL: The Aer protein and the serine chemoreceptor Tsr independently sense intracellular energy levels and transduce oxygen, redox, and energy signals for Escherichia coli behavior. Proc Natl Acad Sci U S A 1997, 94:10541–10546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bibikov SI, Biran R, Rudd KE, Parkinson JS: A signal transducer for aerotaxis in Escherichia coli. J Bacteriol 1997, 179:4075–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Key J, Hefti M, Purcell EB, Moffat K: Structure of the redox sensor domain of Azotobacter vinelandii NifL at atomic resolution: signaling, dimerization, and mechanism. Biochemistry 2007, 46:3614–3623. [DOI] [PubMed] [Google Scholar]
- 36.Ukaegbu UE, Rosenzweig AC: Structure of the redox sensor domain of Methylococcus capsulatus (Bath) MmoS. Biochemistry 2009, 48:2207–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Losi A, Polverini E, Quest B, Gartner W: First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys J 2002, 82:2627–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, et al. : Blue-light-activated histidine kinases: two-component sensors in bacteria. Science 2007, 317:1090–1093. [DOI] [PubMed] [Google Scholar]
- 39.Rivera-Cancel G, Ko WH, Tomchick DR, Correa F, Gardner KH: Full-length structure of a monomeric histidine kinase reveals basis for sensory regulation. Proc Natl Acad Sci U S A 2014, 111:17839–17844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gong W, Hao B, Mansy SS, Gonzalez G, Gilles-Gonzalez MA, Chan MK: Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction. Proc Natl Acad Sci U S A 1998, 95:15177–15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delgado-Nixon VM, Gonzalez G, Gilles-Gonzalez MA: Dos, a heme-binding PAS protein from Escherichia coli, is a direct oxygen sensor. Biochemistry 2000, 39:2685–2691. [DOI] [PubMed] [Google Scholar]
- 42.Watts KJ, Taylor BL, Johnson MS: PAS/poly-HAMP signalling in Aer-2, a soluble haem-based sensor. Mol Microbiol 2011, 79:686–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrova OE, Sauer K: PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 2012, 194:5817–5828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pokkuluri PR, Pessanha M, Londer YY, Wood SJ, Duke NE, Wilton R, Catarino T, Salgueiro CA, Schiffer M: Structures and solution properties of two novel periplasmic sensor domains with c-type heme from chemotaxis proteins of Geobacter sulfurreducens: implications for signal transduction. J Mol Biol 2008, 377:1498–1517. [DOI] [PubMed] [Google Scholar]
- 45.Mullner M, Hammel O, Mienert B, Schlag S, Bill E, Unden G: A PAS domain with an oxygen labile [4Fe-4S](2+) cluster in the oxygen sensor kinase NreB of Staphylococcus carnosus. Biochemistry 2008, 47:13921–13932. [DOI] [PubMed] [Google Scholar]
- 46.Emami K, Topakas E, Nagy T, Henshaw J, Jackson KA, Nelson KE, Mongodin EF, Murray JW, Lewis RJ, Gilbert HJ: Regulation of the xylan-degrading apparatus of Cellvibrio japonicus by a novel two-component system. J Biol Chem 2009, 284:1086–1096. [DOI] [PubMed] [Google Scholar]
- 47.Xu Q, van Wezel GP, Chiu HJ, Jaroszewski L, Klock HE, Knuth MW, Miller MD, Lesley SA, Godzik A, Elsliger MA, et al. : Structure of an MmyB-like regulator from C. aurantiacus, member of a new transcription factor family linked to antibiotic metabolism in actinomycetes. PLoS One 2012, 7:e41359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koh S, Hwang J, Guchhait K, Lee EG, Kim SY, Kim S, Lee S, Chung JM, Jung HS, Lee SJ, et al. : Molecular insights into toluene sensing in the TodS/TodT signal transduction system. J Biol Chem 2016, 291:8575–8590. [DOI] [PMC free article] [PubMed] [Google Scholar]


