Abstract
Background:
Pacing artifacts must be excluded from the analysis of paced ECG waveform. This study aimed to develop and validate an algorithm to identify and remove the pacing artifacts on ECG and vectorcardiogram (VCG).
Methods:
We developed a semi-automatic algorithm that identifies the onset and offset of a pacing artifact based on the VCG signal slope steepness and designed a graphical user interface that permits quality control and fine-tuning the constraining threshold values. We used 1,054 ECGs from the retrospective, multicenter cohort study “Global Electrical Heterogeneity and Clinical Outcomes,” including 3,825 atrial and 10,031 ventricular pacing artifacts for the algorithm development and 22 ECGs including 108 atrial and 241 ventricular pacing artifacts for validation. Validation was performed per digital sample. We used the kappa-statistic of interrater agreement between manually labeled sample (ground-truth) and automated detection.
Results:
The constraining parameter values were for onset threshold 13.06±6.21 μV/ms, offset threshold 34.77±17.80 μV/ms, and maximum window size 27.23 ± 3.53 ms. The automated algorithm detected a digital sample belonging to pacing artifact with a sensitivity of 74.5% and specificity of 99.6% and classified correctly 98.8% of digital samples (ROC AUC 0.871; 95%CI 0.853–0.878). The kappa-statistic was 0.785, indicating substantial agreement. The agreement was on 98.81% digital samples, significantly (P<0.00001) larger than the random agreement on 94.43% of digital samples.
Conclusions:
The semi-automated algorithm can detect and remove ECG pacing artifacts with high accuracy and provide a user-friendly interface for quality control.
Keywords: pacemaker, pacing artifact, ECG, signal processing
Introduction
Since the 1950s, cardiac pacing became a vital treatment modality for a growing number of cardiovascular patients. Innovations in cardiac pacing expanded indications for implantable devices capable of delivering cardiac pacing.[1] Pacemaker implantation rates increased from 467 per million in 1993 to 616 per million in 2009.[2] In 2014, an estimated 351,000 pacemaker inpatient procedures were performed in the US.[2] The number of patients living with an implanted cardiac pacemaker is steadily growing.
An electrocardiogram (ECG) is widely used to determine the heart rhythm and to evaluate the performance of pacemaker functioning, especially in emergency settings.[3] Atrial-paced and ventricular-paced rhythm and atrioventricular (AV) dual-paced rhythm are included in the list of core primary ECG diagnostic statements, endorsed by the American Heart Association (AHA), the American College of Cardiology (ACC), the Heart Rhythm Society (HRS), and the International Society for Computerized Electrocardiography (ISCE).[4]
Notably, the ECG diagnostic standards enforced the rule that no secondary statements can accompany the primary diagnostic statement of paced rhythm or paced complexes.[4] The rule[4] stemmed from the dogma about secondary repolarization abnormalities, stating that paced ventricular complexes are examples of secondary repolarization abnormalities.[5] Therefore, the consensus is that paced ECG could only be used to diagnose paced rhythm, but, otherwise, it cannot be clinically useful.[5]
Nevertheless, we, and others, showed that cardiac memory could be detected during continued altered activation.[6, 7] Cardiac memory is neither pure primary nor pure secondary repolarization abnormality.[8, 9] Spatial ventricular gradient (SVG) is independent of the activation sequence.[7, 10, 11] Measurement of SVG on paced ECG furnished clinically useful information.[12, 13] Thus, paced ECG carries important data for meaningful analysis, which needs to be further studied.
The first step in the automated analysis of paced ECG is the removal of pacing artifacts. There have been many proprietary patented algorithms designed to detect the pacing artifacts at the front end. Several algorithms act before the signal passed an analog-to-digital converter.[14–16] Other approaches combine analog and digital detection [17, 18] or use fully digital detection.[19, 20] Because of the proprietary nature of such algorithms, the exact characteristics and performance of pacing artifact detection algorithms implemented in clinically-used ECG recording equipment are not publicly available. There was no open-code algorithm for pacing artifact detection and removal. This paper presents the development and validation of a new semi-automated algorithm to detect and remove pacing artifacts. We tested the hypothesis that the semi-automated algorithm can accurately detect and remove pacing artifacts from a vectorcardiogram (VCG) obtained from 12-lead ECG.
Methods
Study population
We analyzed data from the retrospective, multicenter cohort study “Global Electrical Heterogeneity and Clinical Outcomes” (GEHCO).[13, 21] The study was approved by the Institutional Review Boards at the Oregon Health & Science University and each participating institution. The study collected digital 12-lead ECG signal recorded before implantation of implantable cardioverter-defibrillator (ICD) or cardiac resynchronization therapy defibrillator (CRT-D), or upgrade from the previous pacemaker, as well as before each subsequent generator change or upgrade. The present study included only patients with atrial-paced (AP), ventricular- (including bi-ventricular-) paced (VP), and AV dual- (including atrial and bi-ventricular) paced (AVP) rhythm on available digital 12-lead ECG. Only one ECG per patient was included in this study.
Algorithm description
The algorithm and open-source software code written in MATLAB (MathWorks, Natick, MA, USA) are provided at https://github.com/Tereshchenkolab/Pacing_spike_removal. The algorithm was developed using a 10-second digital ECG signal with a sampling rate 500Hz. The amplitude resolution was either 1 μV or 5 μV.
Before automated analysis, every ECG was reviewed by at least two physicians as previously described, [13] and each cardiac beat was manually labeled as AP, VP, AVP, or fusion beat, as appropriate.
Figure 1 shows the flowchart of the developed algorithm. First, baseline wander was removed from the 12-lead ECG, using inverse continuous 1-D wavelet transform (icwt Matlab function). First, the continuous wavelet transform of the raw ECG signal was obtained from the “CWT” Matlab module. CWT returns the continuous wavelet transform of the raw ECG signal. The input raw ECG signal was passed as a real vector of regularly sampled timetable. The analytic Morse wavelet with the symmetry parameter (gamma) equal to 3 and the time-bandwidth product equal to 60 is used to obtain CWT. Based on the energy spread of the wavelet in frequency and time the minimum and maximum scales are determined automatically. Matlab CWT module uses L1 normalization.
Figure 1.
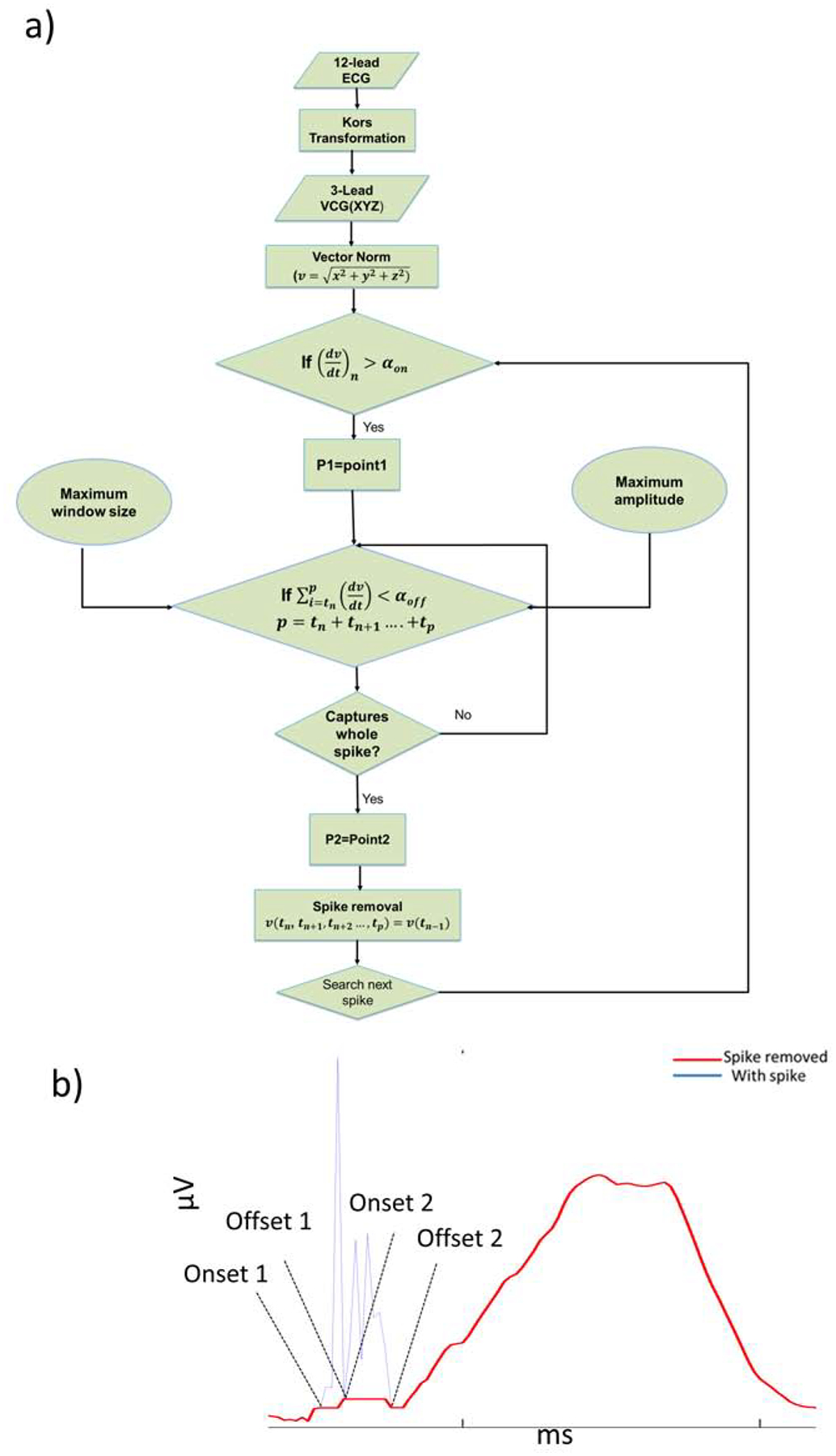
A. Flowchart representation of the algorithm to detect and remove a pacing artifact. B. Representative example of a pacing artifact on vector magnitude ECG signal.
Then XYZ orthogonal ECG was obtained from a 12-lead ECG signal by using the Kors transformation matrix.[22] Once the XYZ leads were obtained, the vector magnitude was calculated from Eq.1.
| Eq.1 |
where is the vector norm, and x, y, and z are the XYZ orthogonal vectors.
Step 1. Pacing artifact onset detection
Slope was calculated for each pair of the consecutive samples on the 10-second ECG recording. The algorithm automatically selected the pacing artifact’s onset (P1) if the following condition was satisfied.
| Eq 2. |
where αon = pacing artifact onset threshold.
Step 2. Pacing artifact offset detection
Next, the algorithm searched for the pacing artifact’s offset. Since the vector norm is an absolute number, the sum of the slopes measured for each pair of the consecutive samples within the pacing artifact tends to zero, assuming that the pacing artifact’s offset approaches the baseline, which has a value close to zero. However, in practice, the pacing artifact’s offset has a non-zero value. Therefore, we assumed that the offset was an arbitrary positive value, which we referred to as pacing artifact offset threshold, αoff. Thus, the offset point (P2) was obtained when the following condition was satisfied.
| Eq. 3. |
where p = tn + tn+1 … … … … … … . +tp, tn= spike onset time point, tp=spike offset time point, and αoff = pacing artifact’s offset threshold.
Step 3. Pacing artifact removal
Once the onset (tn) and offset (tp) time points of a pacing artifact were determined, the spike was removed by making the signal values within the detected pacing artifact time window (tn < t < tp) equal to the value tn. This was given by,
| Eq. 4. |
where v = signal amplitude.
Since the onset and offset points are not replaced and they may not have the same value, the connecting line may have non-zero slope.
Graphic user interface
A graphic user interface was developed that allows users to choose the threshold values from a given range (Figure 2). Additional user-defined parameters were included. Maximum spike amplitude (Max_amp) defines the range of the upper limit value of the pacing artifact. Maximum window size (Max_window) defines the range of the sample points considered as the time window of the pacing spike.
Figure 2.
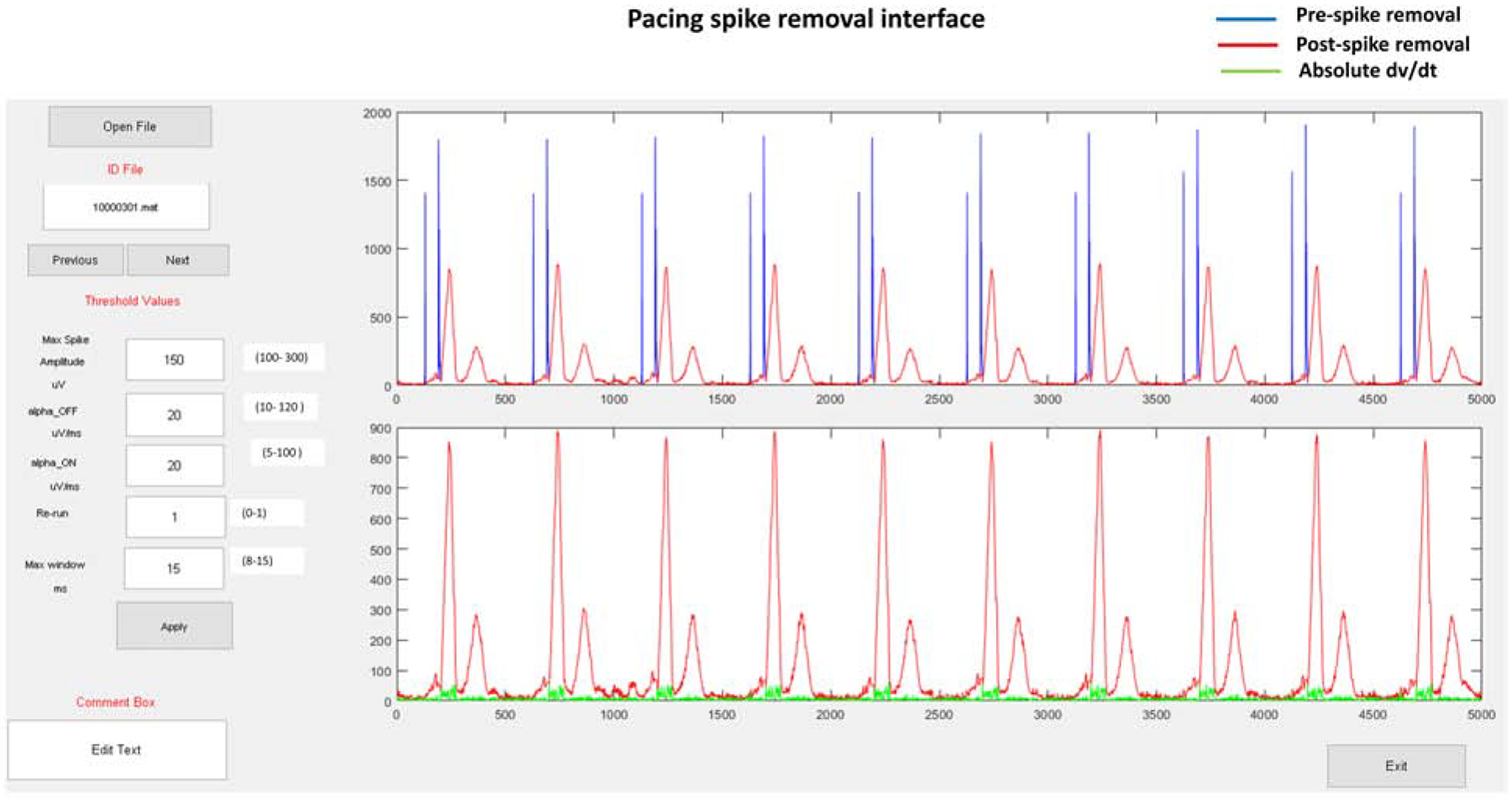
Graphic user interface for pacing artifact removal. The interface allows users to input threshold parameters and review the output for each set of chosen parameters.
Re-run button allows running the algorithm recursively if a given set of parameters remove the artifact partially only. “Re-run” runs the algorithm with another set of parameters to remove the remaining of a spike (after partial removal with the first set of parameters). Generally, satisfactory results can be obtained by a couple of recursive re-run applications. The condition of satisfactory spike removal is to check whether absolute maximum dv/dt falls within the QRS complex time window (Figure 2). As shown in the figure, absolute dv/dt curve (green) is superimposed on the vector magnitude plot, which allows the user to check whether this condition is satisfied.
Validation of the automated pacing artifacts detection
Considering the broad differences in pacing artifacts morphology and duration (because of differences in ECG recording equipment and implanted device manufacturer), we used 98% of the data (1,054 ECGs including 1,399 atrial, 7,605 ventricular, and 2,426 AV pacing artifacts) for the algorithm development and 2% of the data (22 ECGs including 108 atrial and 241 ventricular pacing artifacts) for validation.
For validation, to obtain the ground truth, one investigator (NJ) manually labeled each sample on digital ECG signal as either belonging to pacing artifact (Yes) or not (No). Another investigator (KTH) ran the algorithm and similarly reported whether each data sample is a part of a pacing artifact (Yes or No). Before starting validation, NJ and KTH carried out a training step with a different training subset of the ECG data to set the criteria for manual labeling of a pacing spike, which they both agreed on. The onset point was defined as the point just before the maximum upward dv/dt. The offset point was defined as the point where the spike-tail becomes equal to the onset value or where it approaches the nearest of the onset value. All the sample points between the onset and offset were considered pacing artifact data points. During the validation, the investigators (NJ and KTH) were blinded to each other results. The third investigator (LGT) conducted a statistical analysis.
Statistical analysis
Statistical analysis was performed using STATA MP 16.1 (StataCorp LLC, College Station, TX, USA). Continuous variables were reported as mean ± standard deviation (SD). We used the kappa-statistic measure of interrater agreement for two independent raters. Nonparametric receiver operating characteristic (ROC) analysis with a rating and discrete classification data was performed to calculate the area under the ROC curve (ROC AUC) and measure the automated pacing artifact detection’s sensitivity and specificity.
Results
Study population
Clinical and demographic characteristics of the study population are reported in Table 1. This study included heart failure patients, mostly white men. Approximately half of the patients had nonischemic cardiomyopathy, and one-third had diabetes. Most of the patients (70%) had CRT-D implanted, and 30% had ICD implanted. Implanted devices were manufactured by four companies (Table 1), which allowed us to develop the algorithm that considered various features of pacing pulses by different manufacturers. The vast majority of patients (86%) had a VP or AVP rhythm on analyzed ECG. An average heart rate was 70 beats per minute.
Table 1.
Study population characteristics
| All patients (n=454) | |
|---|---|
| Age(mean±SD), yrs | 67.6±12.3 |
| Female, % | 23.8 |
| White, % | 81.1 |
| Nonischemic cardiomyopathy, % | 42.4 |
| Hypertension, % | 73.1 |
| Diabetes, % | 33.4 |
| Use class I or III antiarrhythmic drugs, % | 16.7 |
| Use beta-blockers, % | 84.5 |
| Left ventricular ejection fraction(mean±SD), % | 29.3±12.2 |
| New York Heart Association heart failure class I-II, % | 49.3 |
| New York Heart Association heart failure class III-IV, % | 50.7 |
| Single-chamber implantable cardioverter-defibrillator, % | 7.5 |
| Dual-chamber implantable cardioverter-defibrillator, % | 22.8 |
| Cardiac resynchronization therapy defibrillator, % | 69.7 |
| Device manufacturer Medtronic, % | 67.7 |
| Device manufacturer Guidant/Boston Scientific, % | 17.5 |
| Device manufacturer St. Jude/Abbott, % | 13.5 |
| Device manufacturer Biotronic, % | 1.3 |
| Atrial-paced rhythm, n(%) | 62(13.7) |
| Ventricular-paced rhythm, n(%) | 282(62.1) |
| Atrio-ventricular paced rhythm, n(%) | 110(24.2) |
| Heart rate (mean±SD), beats per minute | 70.1±12.3 |
| QRS duration (mean±SD), ms | 130.7±35.3 |
| Bazett-corrected QT interval (mean±SD), ms | 484.0±51.3 |
Algorithm development and validation
In the algorithm development phase, in the process of detecting and removing 1,399 AP, 7,605 VP, and 2,426 AVP artifacts, we obtained a range of constraining values for the pacing spike artifacts (∝on = 5–100 μV/ms, ∝off =10–120 μV/ms, Max_amp=100–300 μV , Max_window=8–15 ms). Initial values of the constraining parameters were obtained from manual measurement of each of the parameters from a few beats in the first ECG recording. The same set parameter values were applied for the next recording to see pacing spike artifacts were removed satisfactorily. If the pacing spikes were not removed satisfactorily using the parameter values used for the previous ECG recording, each of the parameter values were gradually increased or decreased by the following increment: ∝on = 5 μV/ms, ∝off =5 μV/ms, Max spike amplitude=20 μV , Max window=2 ms. We recorded the minimum and maximum values of each of the parameters used to remove pacing spikes from 1,054 ECGs in the development phase, which produced the range of parameter values for an user. Figure 3 shows a representative example of pacing artifacts removal from a VP and AVP VCG vector magnitude signal, and Figure 4 illustrates their removal from 12-lead ECG.
Figure 3.
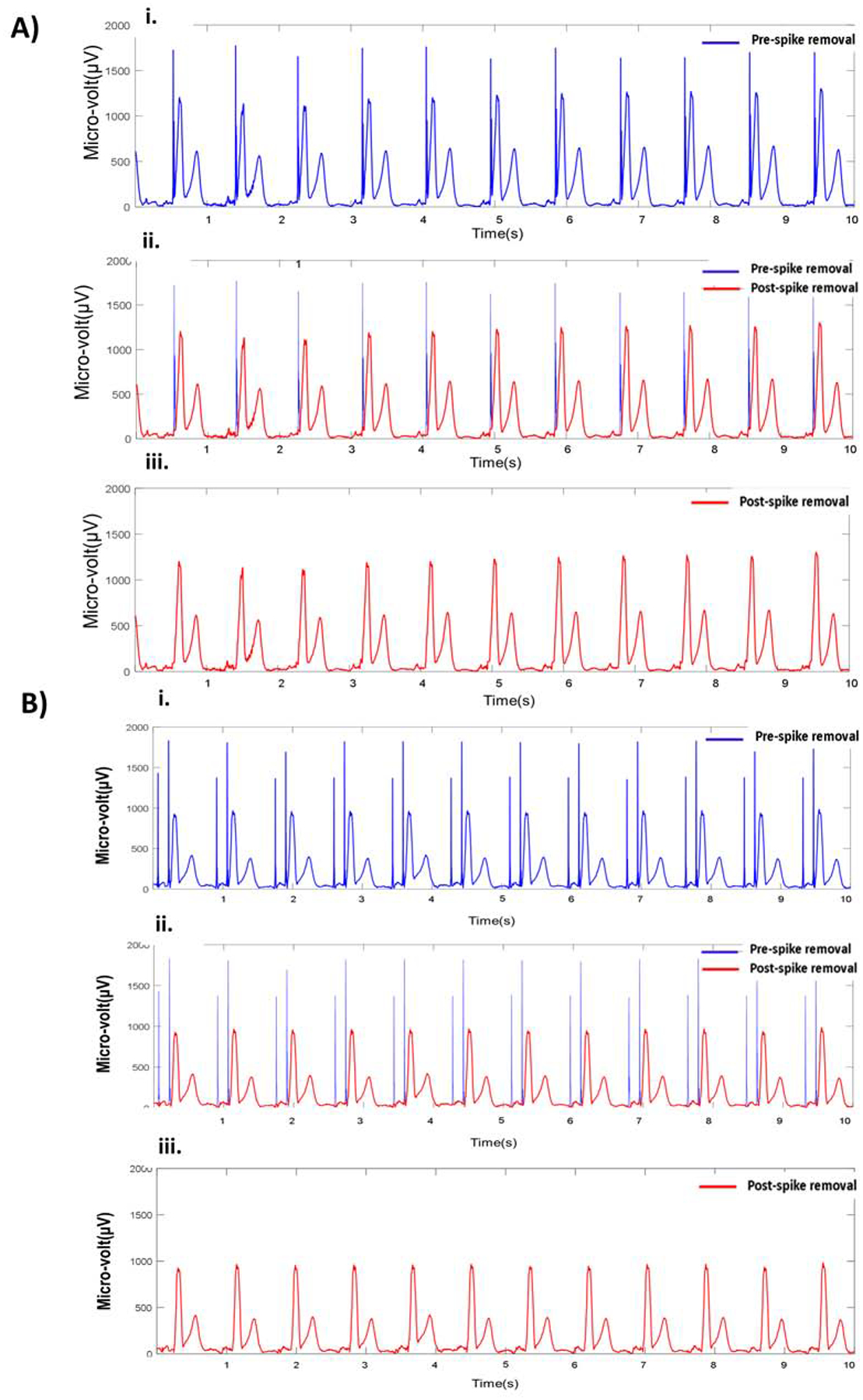
Example of pacing artifact removal from a ventricular-paced (A) and AV-paced (B) VCG. i) Original ventricular-paced VCG vector magnitude signal. i) Detected pacing artifacts (blue) and VCG signal (red).i) VCG vector magnitude signal after removal of pacing artifacts. (A) The pacing spike onset (αon) and offset (αoff) threshold values were 5 and 10 μV/ms, respectively. (B) The corresponding value of αon and αoff were 7.5 and 15.5 μV/ms, respectively.
Figure 4.
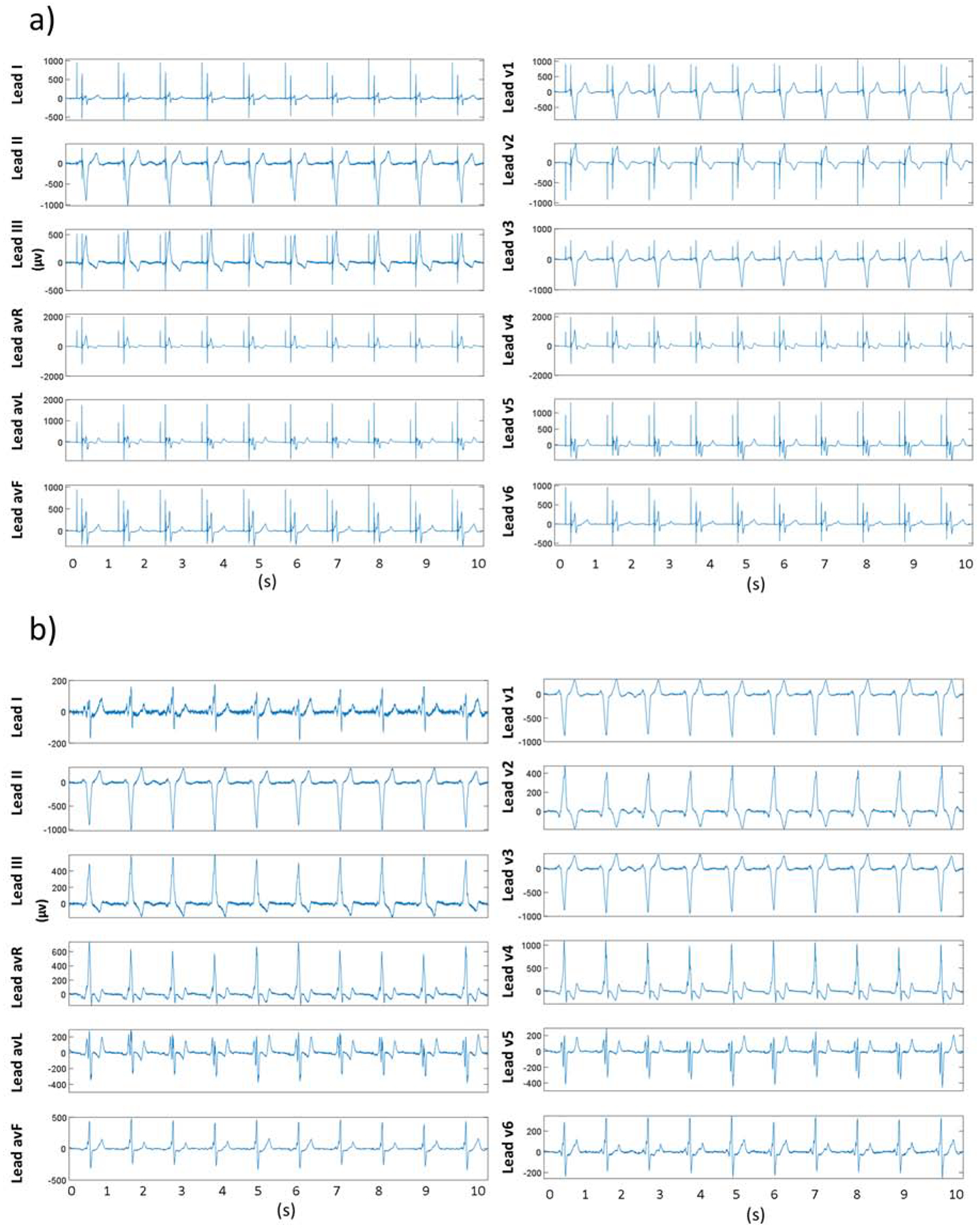
Example of pacing artifacts removal from 12-lead ECG a) 12-lead ECG with pacing artifacts. b) 12-lead ECG trace after pacing artifacts removal.
From the validation dataset (n= 22 ECGs) the mean of 4 constraining parameter values was found: mean maximum amplitude 146.36±76.36 μV, mean onset threshold (αon) 13.06±6.21 μV/ms, mean offset threshold (αoff) 34.77±17.80 μV/ms, and mean maximum window size 27.23 ± 3.53 ms. Notably, the automated algorithm detected the presence of pacing artifact with 100% accuracy, 100% sensitivity, and 100% specificity.
For per-sample validation of the algorithm, we analyzed 110,000 digital signal samples. Each ECG had 5000 samples. On average, a pacing artifact occupied 9.2±3.2 samples or 18.4±6.4 ms. A two-by-two table (Table 2) reports an agreement between the ground truth and automated detection of pacing artifacts for each sample of the digital ECG data. If the automated algorithm had made the determination of whether a given data sample belongs to pacing artifact or not randomly (but with probabilities equal to the overall proportions), we would expect the agreement on 94.43% of digital samples. In fact, they agreed on 98.81%. The amount of agreement indicated that we could reject the null hypothesis that they were making their determination randomly (P<0.00001). The kappa-statistic was 0.785, indicating substantial agreement.
Table 2.
Two by two table of a per-sample agreement between the ground trith and automated algorithm detection of pacing artifacts
| Automated algorithm pacing artifact digital sample assessment | Ground truth digital sample of the pacing artifact | ||
|---|---|---|---|
| YES | NO | Total | |
| YES | 2,494 | 462 | 2,956 |
| NO | 853 | 106,192 | 107,044 |
| Total | 3,346 | 106,654 | 110,000 |
In a per-sample validation, the automated algorithm assigned a digital sample to pacing artifact with ROC AUC 0.871 (95% confidence interval 0.853–0.878), a sensitivity of 74.5%, and specificity of 99.6% and classified correctly 98.8% of digital samples. While the middle of the pacing artifact was always accurately detected, the onset and especially the pacing artifact’s offset can be detected with an error. For the subsequent ECG morphology analysis, it is important to ensure that the pacing artifact was removed completely, but the physiological ECG waveforms were preserved.
Discussion
In this work, we developed and validated the semi-automated algorithm to detect and remove pacing artifacts from a routinely clinically available (diagnostic bandwidth 0.5–150 Hz and sampling rate 500Hz) digital ECG signal. The fully automated algorithm was perfectly 100% accurate in the detection of a pacing artifact’s presence and demonstrated 75% sensitivity and 100% specificity for the per-digital-sample automated detection of pacing artifacts, whereas a user-friendly interface allowed additional fine-tuning and quality control of the artifact removal.
Current ECG machines convert the analog ECG signal to digital at the front end.[23] Modern pacemaker’s stimulus output is frequently ≤ 0.25 ms in duration. Therefore, front-end sampling has to be ≥ 10,000 samples per second in order to detect and represent the pacemaker’s stimulus output. Furthermore, contemporary bipolar pacemaker’s stimulus output is usually small (2–4 Volt). The AHA/ACC/HRS/ISCE-endorsed recommendations for the standardization and interpretation of the ECG[23] emphasized that ECG manufacturers should maintain the required front-end sampling rate for reliable and accurate detection of narrow pacemaker pulses but should not artificially increase pulses’ amplitude to avoid ECG’s morphology distortion. However, in clinical settings, the information about the front-end sampling rate and an approach to handling the presentation of pacemaker stimulus outputs by a specific ECG recording machine is not readily available. In a nonselective CRT patient population, cardiologists observed visible ventricular pacing artifacts in 90% and visible atrial pacing artifacts – in 70% of patients with paced rhythm, whereas the automated ECG reading algorithm detected only 20% of patients with VP rhythm, and none with AP rhythm.[24]
In accordance with the growing population of patients with implanted pacemakers, ICD, and CRT devices, the list of meaningful interpretations of paced ECG morphology is expanding. Acute myocardial infarction can manifest on ECG in patients with VP rhythm, although the sensitivity of acute myocardial infarction diagnosis on paced ECG is low.[25, 26] Left ventricular paced QRS width and the difference between biventricular-paced and pre-implant QRS width predict CRT response.[27] Our previous study showed that the addition of global electrical heterogeneity (GEH) ECG metrics to clinical risk factors of sudden cardiac death (SCD) is especially rewarding in the presence of paced rhythms.[12] In a subgroup of participants with VP ECG (Supplemental Table 13 in [12]), with the addition of GEH parameters, 33% of SCD victims were appropriately reclassified into a higher-risk category (from low to high risk). In contrast, only 10% were similarly appropriately reclassified amongst participants without a paced rhythm on ECG. Furthermore, in the VP rhythm subgroup, no SCD victims were inappropriately reclassified from high to low risk. The addition of GEH also improved SCD-specific risk prediction. The proportion of SCD decreased from 12% to 6% in the intermediate-risk group and increased from 15% to 18% in the high-risk group. The large prospective study of more than 20,000 participants with a median of 14 years of follow-up showed the clinical usefulness of GEH measurements on VP ECG.[12]
Therefore, there is a growing need for reliable detection of pacing artifacts and accurate analysis of paced ECG morphology. Particularly, while the pacing artifact detection is important for the diagnosis of a paced rhythm, the pacing artifact itself should be removed from the analysis of the paced ECG waveform. As discussed above, an ideal solution endorsed by the AHA/ACC/HRS/ISCE involves the oversampling at the ECG front end at ≥10,000 Hz, saving pacing artifacts data (beat labels) on a unique marker-channel, removing pacing artifact, and subsequent downsampling to 500–1000Hz for conventional ECG signal analysis and storage.[23] Such front-end solution provides high accuracy of pacing artifact detection, and removes only a tiny fraction of the ECG signal (0.25–0.5 ms). For example, the algorithm described by Polpetta and Banelli[28] follows the recommended approach and reports promising results. Unfrtunately, currently, workflow carries pacing artifacts all the way to the diagnostic bandwidth (0.05–150Hz), where they become wide (up to 30–40 ms), and thus their removal dramatically affects ECG morphology and negatively impacts clinically important ECG measurements. Our algorithm provides the solution for academic electrocardiology research, allowing accurate removal of pacing artifacts, and calls industry attention to this problem asking for implementing contemporary front-end solutions.
Many proprietary algorithms have been proposed for the detection of pacing artifacts.[14–20, 28–33] Many of the previously developed algorithms proposed more sophisticated and elegant solutions to the problem than our algorithm. However, none of them reported their performance in a large-scale real-life clinical study. Nearly all previously developed algorithms were proprietary, and none provided open-source software code. Therefore, it was impossible to compare the performance of our algorithm to those that have been developed previously. In response to the unmet need of academic electrocardiology research, we provided simple, easy-to-implement algorithm, open-source code and user-friendly GUI, which facilitates the implementation of our tool in clinical research by investigators without an access to advanced computing resourses.
Unfortunately, in clinical settings, it is usually unknown which front-end algorithm is utilized by a given ECG machine manufacturer. If an ECG has been recorded using unknown front-end characteristics, signal processing is the only option. A few previous fully automated algorithms addressed pacing artifacts detection. Helfenbein et al.[30] developed an algorithm for pacing artifacts detection on a commonly used ECG signal (sampling rate 500Hz, bandwidth 0.05–150 Hz), capitalizing on the fact that low-pass filtering broadens the pacing artifact width. The authors reported a sensitivity of 97.2% for detecting a paced rhythm [30] which is lower than our results. Notably, the authors did not report an accuracy and algorithm performance per each sample of the digital ECG signal. Furthermore, widely used algorithms[30] are based on filtering the ECG signal, which distorts the beginning of the QRS and widens the QRS complex. The distortion of the QRS complex challenges the clinical use of paced QRS morphology measurements.[27]
In the past, ECG manufacturers offered an option to turn ON or OFF a feature that enhances an automatically detected pacing artifact, by drawing a big spike. The feature was designed to make it easier for a clinician to see the spike. The AHA/ACC/HRS/ISCE-endorsed recommendations ECG[23] discouraged the use a such “confabulator” to avoid ECG’s morphology distortion.
At the end of the pacing artifact, two phenomena should be considered. The first phenomenon is caused by ECG filtering (0.5–150 Hz or even 1–40 Hz passband), resulting in aliasing, distortion, and ringing, which manifests as widening of pacing artifact and ECG signal.[34] In this retrospective multicenter study, we analyzed ECGs that were stored for more than 20 years, and the details of the used filter specifications were unknown. The second phenomenon is an interaction of pacing stimulus with the myocardium, which is complex.[35] The QRS latency (time from the pacing stimulus to QRS onset) depends on (1) location of pacing lead, (2) proximity, and distribution of post-infarction myocardial scar or diffuse fibrosis, (3) scarring of myocardium around the lead that is growing over time and in extreme cases causing failure to capture, (4) pacing stimulus strength.[35] Therefore, theoretically, both artificial and physiological events and their combination could be responsible for the voltage difference at the onset and offset of pacing artifact. As we were not able to rule out the physiological mechanism, we permitted the larger αoff value. Ultimately, we provided the user with an opportunity to select the thresholds based on their data and goals of the analysis.
Limitations
This paper presented a semi-automated algorithm to detect and remove pacing spikes artifacts from ECG. However, the algorithm itself is not fully automatic. The semi-automated approach is time-consuming. However, the pacing spikes from diverse patient populations and pacemaker settings always come with a high degree of variability, making automated dynamic threshold estimation challenging, though not impossible. We have to emphasize that the ideal approach for the analysis of paced ECG waveform morphology has to include ECG front-end oversampling[23], recording the presence of pacing artifacts on a separate marker-channel, and immediate its removal at the front end. Removal of a pacing artifact at the front end preserves paced ECG morphology for its subsequent analysis. The growing number of patients with implanted pacemaker devices and clinical needs for a meaningful analysis of paced ECG waveform [13, 27] calls for ECG manufacturers’ attention to handling ECG front-end manufacturing.
Conclusions
We developed a semi-automated algorithm to detect and remove pacing spike artifacts from digital ECG signal. The algorithm demonstrated its ability to detect and remove pacing spike artifacts with high sensitivity and specificity.
Highlights.
We developed and validated a semi-automated algorithm to detect and remove pacing spike artifacts from a digital ECG signal.
The semi-automated algorithm can detect and remove pacing spike artifacts with high accuracy and provide a user-friendly interface for quality control.
Acknowledgment
The authors thank all GEHCO investigators.
Funding:
This work was partially supported by the National Institutes of Health (HL118277), the American Heart Association (17GRNT33670428), Medical Research Foundation of Oregon, and OHSU President Bridge funding to Tereshchenko.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
None declared.
References
- [1].Cheng A, Tereshchenko LG, Evolutionary innovations in cardiac pacing, Journal of Electrocardiology, 44 (2011) 611–615. [DOI] [PubMed] [Google Scholar]
- [2].Virani SS, Alonso A, Benjamin EJ, Bittencourt MS, Callaway CW, Carson AP, Chamberlain AM, Chang AR, Cheng S, Delling FN, Djousse L, Elkind MSV, Ferguson JF, Fornage M, Khan SS, Kissela BM, Knutson KL, Kwan TW, Lackland DT, Lewis TT, Lichtman JH, Longenecker CT, Loop MS, Lutsey PL, Martin SS, Matsushita K, Moran AE, Mussolino ME, Perak AM, Rosamond WD, Roth GA, Sampson UKA, Satou GM, Schroeder EB, Shah SH, Shay CM, Spartano NL, Stokes A, Tirschwell DL, VanWagner LB, Tsao CW, American Heart Association Council on Epidemiology, Prevention Statistics Committee, Stroke Statistics Subcommittee, Heart Disease and Stroke Statistics-2020 Update: A Report From the American Heart Association, Circulation, 141 (2020) e139–e596. [DOI] [PubMed] [Google Scholar]
- [3].Greenhut SE, Jenkins JM, Dicarlo LA, Computerized interpretation of the paced ECG, Journal of Electrocardiology, 24 (1991) 146–152. [DOI] [PubMed] [Google Scholar]
- [4].Mason JW, Hancock EW, Gettes LS, Recommendations for the standardization and interpretation of the electrocardiogram, Heart Rhythm, 4 (2007) 413–419. [DOI] [PubMed] [Google Scholar]
- [5].Rautaharju PM, Surawicz B, Gettes LS, AHA/ACCF/HRS Recommendations for the Standardization and Interpretation of the Electrocardiogram, Journal of the American College of Cardiology, 53 (2009) 982–991. [DOI] [PubMed] [Google Scholar]
- [6].Shvilkin A, Bojovic B, Vajdic B, Gussak I, Ho KK, Zimetbaum P, Josephson ME, Vectorcardiographic and electrocardiographic criteria to distinguish new and old left bundle branch block, Heart Rhythm, 7 (2010) 1085–1092. [DOI] [PubMed] [Google Scholar]
- [7].Haq KT, Cao J, Tereshchenko LG, Characteristics of Cardiac Memory in Patients with Implanted Cardioverter-defibrillators: The Cardiac Memory with Implantable Cardioverter-defibrillator (CAMI) Study, J Innov Card Rhythm Manag, 12 (2021) 4395–4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Rosenbaum MB, Blanco HH, Elizari MV, Lázzari JO, Davidenko JM, Electrotonic modulation of the T wave and cardiac memory, American Journal of Cardiology, 50 (1982) 213–222. [DOI] [PubMed] [Google Scholar]
- [9].Ozgen N, Rosen MR, Cardiac memory: a work in progress, Heart Rhythm., 6 (2009) 564–570. [DOI] [PubMed] [Google Scholar]
- [10].Tereshchenko LG, Ghanem RN, Abeyratne A, Swerdlow CD, Intracardiac QT integral on far-field ICD electrogram predicts sustained ventricular tachyarrhythmias in ICD patients, Heart Rhythm, 8 (2011) 1889–1894. [DOI] [PubMed] [Google Scholar]
- [11].Burger HC, A theoretical elucidation of the notion ventricular gradient, American Heart Journal, 53 (1957) 240–246. [DOI] [PubMed] [Google Scholar]
- [12].Waks JW, Sitlani CM, Soliman EZ, Kabir M, Ghafoori E, Biggs ML, Henrikson CA, Sotoodehnia N, Biering-Sorensen T, Agarwal SK, Siscovick DS, Post WS, Solomon SD, Buxton AE, Josephson ME, Tereshchenko LG, Global Electric Heterogeneity Risk Score for Prediction of Sudden Cardiac Death in the General Population: The Atherosclerosis Risk in Communities (ARIC) and Cardiovascular Health (CHS) Studies, Circulation, 133 (2016) 2222–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Waks JW, Haq KT, Tompkins C, Rogers AJ, Ehdaie A, Bender A, Minnier J, Dalouk K, Howell S, Peiris A, Raitt M, Narayan SM, Chugh SS, Tereshchenko LG, Competing risks in patients with primary prevention implantable cardioverter-defibrillators: Global Electrical Heterogeneity and Clinical Outcomes (GEHCO) study, Heart Rhythm, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Regan RJ, Pace pulse signal conditioning circuit, US Patent 4,574,813, Hewlett-Packard Company, 1986. [Google Scholar]
- [15].Shaya MN, Wyshogrod BL, Pace pulse identification apparatus for heart pacemaker, US Patent 4,664,116, Hewlett-Packard Company, 1987. [Google Scholar]
- [16].Jin Y, Yu Y, Jin J, Huang Y, Development on pacing ECG monitoring system, Proceedings of the 20th Annual International Conference of the IEEE Engineering in Medicine and Biology Society. Vol.20 1998, pp. 230–232. [Google Scholar]
- [17].Wang JY, Shay MN, Shaya MN, Wang J, Pace pulse elimination apparatus for ECG, US Patent 4,832,041, Hewlett-Packard Company, 1989. [Google Scholar]
- [18].Yonce DJ, Ormsby PJ, System and method for detection of pacing pulses within ECG signals, US Patent 6,477,404 B1, Cardiac Pacemakers Inc, 2002. [Google Scholar]
- [19].Herleikson EC, ECG pace pulse detection and processing, US Patent 5,682,902, Hewlett-Packard Company, 1997. [Google Scholar]
- [20].Kruse JM, Kaszas CJ, Nelson CG, Heart pacing pulse detection system, US Patent 5,448,997, Medtronic Inc, 1995. [Google Scholar]
- [21].Waks JW, Hamilton C, Das S, Ehdaie A, Minnier J, Narayan S, Niebauer M, Raitt M, Tompkins C, Varma N, Chugh S, Tereshchenko LG, Improving sudden cardiac death risk stratification by evaluating electrocardiographic measures of global electrical heterogeneity and clinical outcomes among patients with implantable cardioverter-defibrillators: rationale and design for a retrospective, multicenter, cohort study, J Interv Card Electrophysiol, 52 (2018) 77–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kors JA, van HG, Sittig AC, van Bemmel JH, Reconstruction of the Frank vectorcardiogram from standard electrocardiographic leads: diagnostic comparison of different methods, Eur.Heart J, 11 (1990) 1083–1092. [DOI] [PubMed] [Google Scholar]
- [23].Kligfield P, Gettes LS, Bailey JJ, Childers R, Deal BJ, Hancock EW, van HG, Kors JA, Macfarlane P, Mirvis DM, Pahlm O, Rautaharju P, Wagner GS, Josephson M, Mason JW, Okin P, Surawicz B, Wellens H, Recommendations for the standardization and interpretation of the electrocardiogram: part I: the electrocardiogram and its technology a scientific statement from the American Heart Association Electrocardiography and Arrhythmias Committee, Council on Clinical Cardiology; the American College of Cardiology Foundation; and the Heart Rhythm Society endorsed by the International Society for Computerized Electrocardiology, J Am Coll.Cardiol, 49 (2007) 1109–1127. [DOI] [PubMed] [Google Scholar]
- [24].Andersson HB, Hansen MB, Thorsberger M, Biering-Sørensen T, Nielsen JB, Graff C, Pehrson S, Svendsen JH, Diagnostic accuracy of pace spikes in the electrocardiogram to diagnose paced rhythm, Journal of electrocardiology, 48 (2015) 834–839. [DOI] [PubMed] [Google Scholar]
- [25].Sgarbossa EB, Pinski SL, Gates KB, Wagner GS, Early electrocardiographic diagnosis of acute myocardial infarction in the presence of ventricular paced rhythm. GUSTO-I investigators, Am J Cardiol, 77 (1996) 423–424. [DOI] [PubMed] [Google Scholar]
- [26].Brady WJ, Lentz B, Barlotta K, Harrigan RA, Chan T, ECG Patterns Confounding the ECG Diagnosis of Acute Coronary Syndrome: Left Bundle Branch Block, Right Ventricular Paced Rhythms, and Left Ventricular Hypertrophy, Emergency Medicine Clinics of North America, 23 (2005) 999–1025. [DOI] [PubMed] [Google Scholar]
- [27].Hsing JM, Selzman KA, Leclercq C, Pires LA, McLaughlin MG, McRae SE, Peterson BJ, Zimetbaum PJ, Paced left ventricular QRS width and ECG parameters predict outcomes after cardiac resynchronization therapy: PROSPECT-ECG substudy, Circ Arrhythm Electrophysiol, 4 (2011) 851–857. [DOI] [PubMed] [Google Scholar]
- [28].Polpetta A, Banelli P, Fully digital pacemaker detection in ECG signals using a non-linear filtering approach, Annu Int Conf IEEE Eng Med Biol Soc, 2008 (2008) 5406–5410. [DOI] [PubMed] [Google Scholar]
- [29].Donehoo RF, Browne DW, Pacemaker pulse detection and artifact rejection, US Patent 5,660,184, Johnson & Johnson Medical Inc, 1997. [Google Scholar]
- [30].Helfenbein ED, Lindauer JM, Zhou SH, Gregg RE, Herleikson EC, A software-based pacemaker pulse detection and paced rhythm classification algorithm, Journal of Electrocardiology, 35 (2002) 95–103. [DOI] [PubMed] [Google Scholar]
- [31].Luo S, Johnston P, Hong W, Performance study of digital pacer spike detection as sampling rate changes, 2008 Computers in Cardiology, 2008, pp. 349–352. [Google Scholar]
- [32].Lall C, Zhang Z, Chen Y, Performance challenges in ECG pacemaker pulse detection systems, 2012 Computing in Cardiology, 2012, pp. 765–768. [Google Scholar]
- [33].Andrla P, Plesinger F, Halamek J, Leinveber P, Viscor I, Jurak P, A Method For Removing Pacing Artifacts From Ultra-High-Frequency Electrocardiograms, 2018 Computing in Cardiology Conference (CinC), 2018, pp. 1–4. [Google Scholar]
- [34].Luo S, Johnston P, A review of electrocardiogram filtering, Journal of Electrocardiology, 43 (2010) 486–496. [DOI] [PubMed] [Google Scholar]
- [35].Tedrow U, Maisel WH, Epstein LM, Soejima K, Stevenson WG, Feasibility of adjusting paced left ventricular activation by manipulating stimulus strength, J.Am.Coll.Cardiol, 44 (2004) 2249–2252. [DOI] [PubMed] [Google Scholar]


