Abstract
The precise mechanism of transcription termination of the eukaryotic RNA polymerase III (Pol III) has been a subject of considerable debate. Although previous studies have clearly shown that multiple uracils at the end of RNA transcripts are required for Pol III termination, the effects of upstream RNA secondary structure in the nascent transcript on transcriptional termination is still unclear. To address this, we developed an in cellulo Pol III transcription termination assay using the recently developed Tornado-Corn RNA aptamer system to create a Pol III-transcribed RNA that produces a detectable fluorescent signal when transcribed in human cells. To study the effects of RNA sequence and structure on Pol III termination, we systematically varied the sequence context upstream of the aptamer and identified sequence characteristics that enhance or diminish termination. For transcription from Pol III type 3 promoters, we found that only poly-U tracts longer than the average length found in the human genome efficiently terminate Pol III transcription without RNA secondary structure elements. We observed that RNA secondary structure elements placed in proximity to shorter poly-U tracts induced termination, and RNA secondary structure by itself was not sufficient to induce termination. For Pol III type 2 promoters, we found that the shorter poly-U tract lengths of 4 uracils were sufficient to induce termination. These findings demonstrate a key role for sequence and structural elements within Pol III-transcribed nascent RNA for efficient transcription termination, and demonstrate a generalizable assay for characterizing Pol III transcription in human cells.
Keywords: transcription, termination, RNA polymerase III, aptamer, synthetic biology, RNA
Introduction
Cells require mechanisms for precisely terminating transcribed RNAs at the appropriate genetic loci in order to maintain proper genetic regulation and minimize undesired expression of downstream genomic regions [1–3]. The process of transcription termination in prokaryotes is well understood [4], but there remain questions regarding the mechanisms of termination in eukaryotes [1, 5–8]. This knowledge gap is particularly substantial for transcription mediated by eukaryotic RNA polymerase III (Pol III), which transcribes non-coding RNAs such as the 5S ribosomal RNA, tRNAs, snRNA, and a variety of miRNA [9, 10]. Given the important roles played by these classes of RNA in health and disease processes [11, 12], elucidating the mechanisms of transcription termination could provide insights into these important components of cellular regulation.
Pol III transcriptional termination occurs when the transcribing polymerase reaches a stretch of adenosines which is encoded into the nascent RNA as a poly-uracil (poly-U) tract [4]. The average lengths of these genomic tracts vary across eukaryotic species, with an average of 5–7 uracil nucleotides (nt) within the genome of Schizosaccharomyces pombe (S. pombe), 6–9 nt in Saccharomyces cerevisiae (S. cerevisiae) and 4–5 nt in humans [13]. In this regard, eukaryotic Pol III termination signals are similar to those employed in the bacterial intrinsic termination mechanism, which also occurs at a poly-U stretch [14]. In bacterial transcriptional termination, weak interactions between the A-U bases within the nascent RNA-DNA hybrid signal the elongating RNA polymerase (RNAP) to transition into a pause conformation, and the resulting structural rearrangement contributes to the dissociation of the transcription complex from the template.
In addition to the poly-U tract, RNA secondary structure has been shown to play a role in transcription termination mechanisms [14]. In particular, secondary structural elements such as hairpins, are known to be a critical component of the intrinsic termination mechanism for prokaryotic RNAPs [14]. For example, bacterial intrinsic transcriptional terminators require an intramolecular base pairing interaction to form a hairpin structure immediately adjacent to the poly-U tract, with more stable secondary structures contributing to greater termination efficiency [15, 16]. However, the necessity of RNA secondary structure for eukaryotic Pol III termination is currently debated [6, 7, 17–20]. Some reports indicate that Pol III termination is enhanced when RNA structural elements are proximal to a poly-U tract [6, 17, 20], while other reports indicate that secondary structure is dispensable and Pol III termination is not enhanced in its presence [5, 18, 21]. Most studies of Pol III termination have utilized in vitro transcription assays using reassembled purified components of the S. cerevisiae Pol III transcription complex on DNA templates designed to contain various RNA sequence and structure contexts. In these assays, transcription components were first assembled into the full complex by loading on DNA templates, and then tested for their ability to read through poly-U tracts in the presence or absence of upstream RNA secondary structure [18]. While there is general agreement that poly-U tract length and the context of the sequence flanking this region impact Pol III transcription [5, 20, 22–26], there were conflicting results to whether RNA secondary structural elements adjacent to the poly-U tract enhance termination efficiency [5–7, 17, 18]. This could be due to differences in enzyme preparations used, differences in mechanisms of transcription termination across eukaryotic species, or the type of Pol III promoters employed in these assays which are known to recruit different transcription factor subunits making up the pre-initiation complex (PIC) [6, 7, 17–19, 27–29].
To provide additional insight into the RNA sequence and structural determinants of Pol III termination, we decided to address a gap in these observations by studying this phenomenon in human cell lines. Specifically, we adapted an in cellulo Pol III transcriptional reporter system based on fluorescent RNA aptamers that are active in human cell lines [30]. Specifically, we utilized the Tornado system which contains the Corn aptamer embedded within RNA transcripts for direct reporting of Pol III transcriptional activity in cells (Fig. 1A). Corn was specifically chosen over other fluorescent aptamers such as Broccoli due to its particularly bright in cellulo fluorescence and smaller sequence space that enhances its capacity for manipulation [30]. Placing specific RNA sequences upstream of the Tornado reporter system allowed us to assess the impact of these sequences on transcription efficiency. This system enabled us to determine how various transcript characteristics, including poly-U tract length and the presence and location of predicted RNA structural elements, influence Pol III termination efficiency in human cells. This setup additionally allowed us to test these phenomena in the context of Pol III type 2 and 3 promoters. We anticipate that this work will further clarify the mechanism of Pol III transcription termination and enable the forward design of synthetic variants for precise control of Pol III expression in human cells.
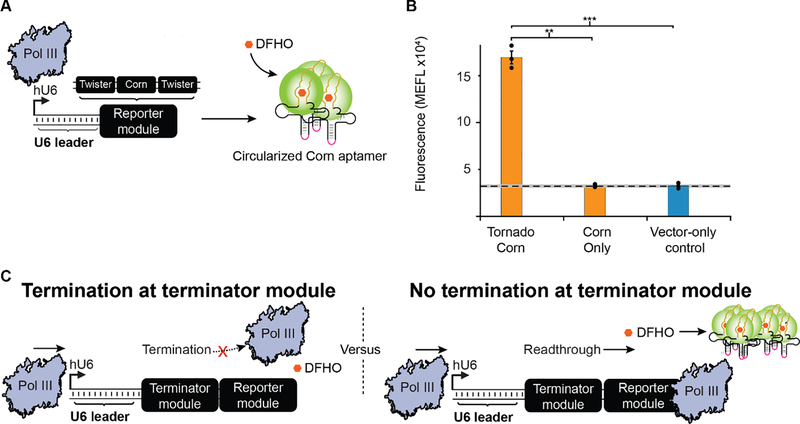
Figure 1: Development of an assay for measuring Pol III termination in human cells.
A) Schematic overview of the construct design of the Tornado system. A human U6 promoter drives Pol III transcription of a ‘Reporter module’, comprising a Corn aptamer (yellow line) which is embedded within a tRNA scaffold and flanked by two twister self-cleaving ribozymes. The ribozymes’ self-cleavage results in a 5’ hydroxyl and a 3’ end consisting of a 2’,3’-cyclic phosphate which is recognized and ligated (pink line) by the endogenous protein RtcB, increasing its stability. Ribozyme self-cleavage and ligation leaves a circularized Corn aptamer that is stabilized by the tRNA scaffold. When DFHO is bound to the Corn aptamer, this dye becomes fluorescent. B) The addition of Tornado to the Reporter module enables quantification of Pol III transcription in cellulo. Colored bars represent the average of 3 biological replicates (circles). The dashed line indicates the average signal from the vector-only (negative) control cells, while the grey horizontal bar represents the standard error of the mean (S.E.M) of the signal from these cells; this convention is applied in subsequent figures. Statistical significance was measured using a one-tailed heteroscedastic Welch’s t-test (***p < 0.001). Error bars represent the S.E.M. C) Termination modules were introduced in this study to investigate the effect of different RNA sequences and structures on Pol III termination. Shown here are expected assay outcomes as a function of termination (left) or readthrough (right) at the Terminator module.
An Assay for Quantifying Pol III Transcription Termination in Human Cells
To investigate the Pol III termination mechanism in cellulo, we first sought to develop a method that could quantitatively characterize the abundance of Pol III-generated transcripts within a human cell line. We started with previous work which used the fluorescent RNA aptamer, Corn, to study the subcellular localization of Pol III transcripts [30]. When transcribed, Corn forms a secondary structure that binds the ligand 3,5-difluoro-4-hydroxybenzylidene-imidazolinone-2-oxime (DFHO) with nanomolar affinity. This binding event then activates fluorescence of DFHO, which when excited with light at a wavelength of 505 nm emits fluorescence at 545 nm. To enhance its stability to enable detection, the Corn aptamer is included within the middle of a tRNA scaffold sequence, which folds in such a way as to reduce RNA degradation [30]. Importantly for our purposes, the Corn aptamer system is both sufficiently photostable and transcribed at sufficient levels from the human U6 (hU6) Pol III type 3 promoter to enable the transcripts to be detected in cells using flow cytometry [30–32].
To employ these parts for the current study, we refined methods for quantifying RNA transcript levels in human cells [30]. We first sought to detect Pol III-driven transcription by expressing Corn aptamer-containing transcripts in the human embryonic kidney (HEK293FT) cell line (Supplementary Fig. 1A) [30]. Specifically, we transfected HEK293FT cells with the plasmid pAV-U6+27-tCORN, a plasmid construct containing in order, a human hU6 promoter, a 27 bp U6 leader sequence commonly included for optimal expression [32], the Corn aptamer fused to a tRNA scaffold, and a SV40 termination site [30] (Supplementary Fig. 2) (Supplementary Table 1). In our experiments this initial design did not yield a signal that was significantly different than the background signal (Fig. 1B).
We hypothesized that this lack of signal may arise from inadequate transcript stability. Therefore, to boost the observable signal, we adapted the recently developed Tornado system which was designed to enhance the detectable signal from the Corn aptamer [31]. In the Tornado system, the Corn aptamer-tRNA scaffold is flanked by two twister ribozyme sequences (Fig. 1A). Following transcription, these self-cleaving ribozymes cleave the RNA in two locations, which allows the nuclear protein RtcB to ligate the free ends together, producing a circularized RNA containing the Corn aptamer [31]. This circular RNA is protected from endogenous exonucleases, allowing Corn aptamer transcripts to accumulate to higher concentrations and thus conferring an enhancement in fluorescence (Fig. 1A) [31]. To utilize the Tornado system, we introduced a Reporter module into our constructs, consisting of an hU6 promoter, the same U6 leader sequence, a twister ribozyme, the Corn aptamer fused to a tRNA scaffold, a second twister ribozyme, and SV40 termination site (which is commonly used for Pol II-driven expression) due to its internal poly-U tract. When transfected into HEK293FT cells, this Tornado construct enabled robust detection of Pol III-driven transcription (Fig. 1B). We concluded that this Tornado-based system is well-suited for quantifying Pol III-driven expression and termination in cellulo.
Poly-U Sequence Length Modulates Pol III Termination From Type 3 Promoters
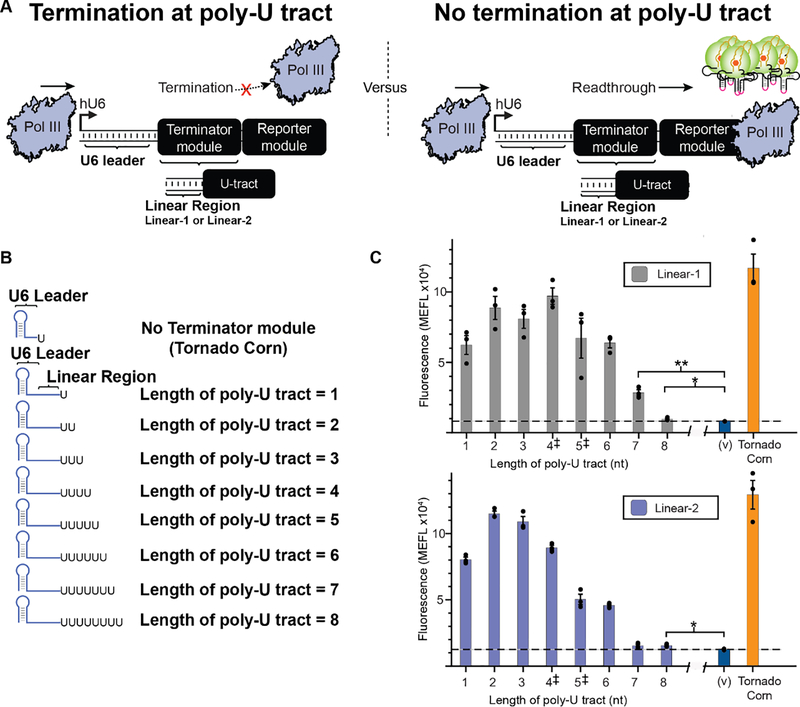
We next sought to investigate how Pol III termination efficiency varies with the length of the poly-U sequence tract. To study this, we modified the Tornado reporter construct to include an additional Terminator module downstream of the U6 leader sequence and upstream of the Reporter module sequence elements (Fig. 1C). In this manner, RNA elements that impact termination can be identified due to decreases in the amount of generated fluorescent signal. Using this approach, we assayed the impact of Terminator modules incorporating varying numbers of U nucleotides in the transcribed RNA (Fig. 2A). To study the effect of poly-U sequence length on termination independent of RNA structure, we included a computationally designed ‘linear’ sequence within the Terminator module, upstream of the poly-U sequence, which is predicted to lack any intramolecular RNA structures. Candidate linear sequences were designed using the Nucleic Acids PACKage (NUPACK) [33], and this algorithm was utilized to predict the absence of secondary structure throughout this region when included in a transcript alongside the entire Tornado Reporter module (Fig. 2AB, right; Methods; Supplementary Fig. 3). Two different linear sequences were employed—Linear-1 and Linear-2—in order to evaluate whether any one specific choice of linear sequence contributes to termination.
Figure 2: Varying poly-U tract length modulates termination of Pol III-driven transcription in cellulo.
A) Assay design for quantifying relative termination efficiency as a function of poly-U tract sequence length. A ‘Terminator module’ was constructed by placing varying lengths of poly-U tract sequence downstream of RNA sequences designed to be completely linear—i.e., lacking secondary structure (Linear-1 or Linear-2). This combined Terminator module was placed immediately downstream of the U6 promoter and upstream of the Reporter module. In this system, the efficiency of the Terminator module is inversely related to the magnitude of the fluorescent output. B) Illustration of predicted RNA structures and designs. The U6 leader sequence is predicted to fold into a 5’ hairpin structure. C) Termination efficiency was experimentally quantified for constructs varying in poly-U length for two different linear sequences Linear-1 (top) and Linear-2 (bottom). The outputs were compared to the fluorescence observed from cells transfected with a vector-only negative control (v = no Reporter module), and a construct lacking a Terminator module (Tornado corn). For both choices of linear sequence, poly-U tract lengths of 1–6 nt were found to be significantly different (p < 0.05) from the vector-only control using a one-tailed heteroscedastic Welch’s t-test followed by the Benjamini-Hochberg procedure with a false discovery rate cutoff of 0.05 (Supplementary Table 2). By this test, Linear-1 constructs with poly-U tracts of 7–8 nt also differed from the vector only control (p < 0.05). The Linear-2 construct with a poly-U tract of 8 nt differed from the vector only control (p < 0.05), but the construct with the poly-U tract length of 7 nt did not. Colored bars represent the average of 3 biological replicates with individual points plotted as circles. Error bars represent the S.E.M. The dashed line represents the average of three replicates for the vector-only control (v), and the grey horizontal bar represents the S.E.M. of the vector-only control. ‡ denotes the two most commonly found poly-U tract lengths within the human genome.
Using this Terminator module approach, we evaluated the effect of poly-U tract lengths ranging from 1 to 8 nt on Pol III termination in cellulo (Fig. 2B). For both linear sequence contexts, we observed that past a certain length, increasing the poly-U tract length decreased reporter signal, indicating more efficient termination (Fig. 2C). Interestingly, within this assay, we actually observed a small increase in signal when comparing constructs containing poly-U tracts of length 4 nt to a length of 1 nt (Supplementary Table 2). This was surprising, as tract length of 4 uracils is the average length of all poly-U tracts in human Pol III-expressed genes [13]. Although most Pol III-responsive promoters are type 2, our analysis of a subset of transcripts expressed from Pol III type 3 promoters demonstrates similar poly-U lengths to this average calculated across all Pol III-responsive promoters (Supplementary Table 3) [28]. We observed a consistent trend of decreasing signal output only after poly-U tracts reached a size of 7 nt or greater in both contexts. When comparing against the background signal from our vector-only control, only poly-U tracts of 7 uracils demonstrated no significant difference in observed signal for the Linear-2 construct, indicating the difficulty of achieving complete termination with only poly-U sequences in these contexts (Supplementary Table 2). We speculated that if our model transcripts require longer poly-U tracts to achieve efficient termination than do endogenous Pol III-driven transcripts [13], perhaps other transcript features could confer efficient termination with shorter poly-U tracts.
RNA Structure Adjacent to the Poly-U Tract Enhances Pol III Termination From Type 3 Promoters
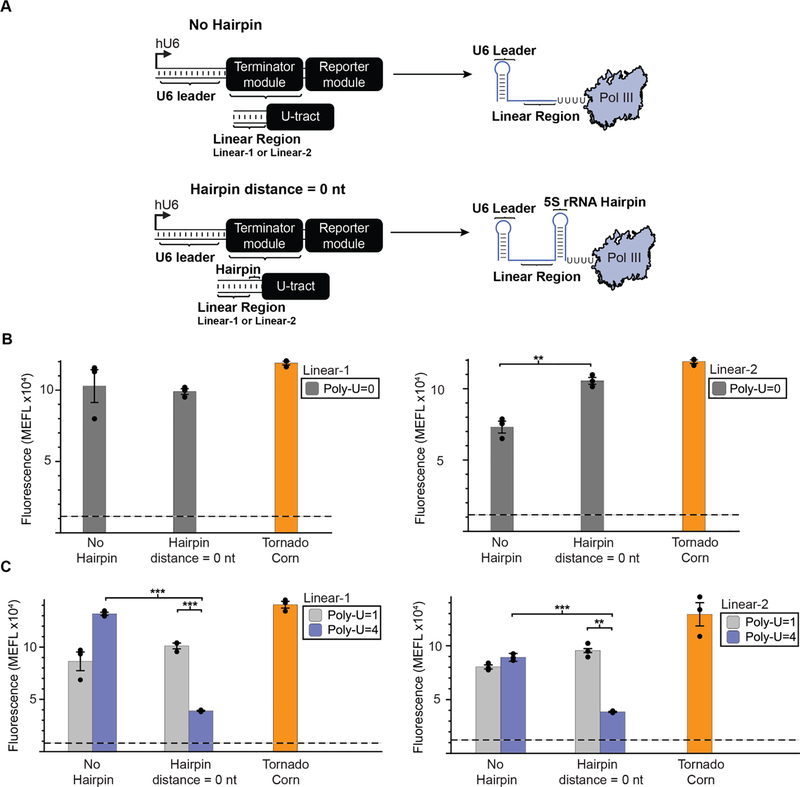
We next sought to investigate how upstream RNA structure might influence Pol III termination at poly-U tracts. We started by adapting our expression constructs to include a sequence that introduces a well-known secondary structural element by encoding a portion of the 5S ribosomal RNA (rRNA) hairpin. This 5S rRNA hairpin is predicted to fold into a 9 bp RNA hairpin structure with a 5 nt loop, and it was previously employed to investigate the impact of RNA secondary structure on Pol III termination using in vitro transcription assays [6]. In our investigation, we placed this sequence immediately upstream of the poly-U tract (Fig. 3A). NUPACK analysis was then used to confirm that (i) the upstream linear region was still predicted to assume a single-stranded conformation, and (ii) no other competing RNA structures were predicted as a consequence of introducing the 5S rRNA hairpin (Supplementary Fig. 3).
Figure 3: Upstream RNA secondary structure enhances termination as a function of poly-U tract length.
A) A schematic depicting the positioning of an RNA secondary structure element immediately upstream of the poly-U tract within the Terminator module. The secondary structure utilized is a 23 nt portion of the 5S ribosomal RNA (rRNA) predicted to fold into a 9 bp hairpin following previous studies of Pol III termination in vitro. This structure was either omitted (No Hairpin) or positioned after the linear sequence and immediately upstream of the poly-U tract (Hairpin distance = 0 nt). B) Impacts of upstream RNA structure to termination was evaluated for both the Linear-1 (left) and Linear-2 (right) sequence contexts when the poly-U tract is completely removed. C ) Contribution of upstream RNA structure to termination was evaluated for both the Linear-1 (left) and Linear-2 (right) sequence contexts, when the poly-U tract is changed from 1 to 4 uracils. Colored bars represent the average of 3 biological replicates with individual points plotted as circles. Error bars represent the S.E.M. The dashed line represents the average of three replicates for the vector-only control, and the grey horizontal bar represents the S.E.M. of the vector-only control, as reported in Fig. 2C. Statistical significance of the indicated comparisons (brackets) was measured using one-tailed heteroscedastic Welch’s t-tests followed by the Benjamini-Hochberg procedure with a false discovery rate cutoff of 0.05 (Supplementary Table 2). * = p < 0.05, ** = p< 0.01, *** = p < 0.001.
We first evaluated how adding this hairpin influences termination in the absence of a poly-U tract. In particular we found that without the poly-U tract, adding a hairpin had no effect on termination for the Linear-1 context and actually resulted in an increase in fluorescence for the Linear-2 context (Fig. 3B). We next investigated adding the RNA hairpin to poly-U tracts of length 1 and 4 nt (Fig. 3C). For poly-U tract lengths of 1 nt, we did not observe a decrease in fluorescence when the hairpin was added indicating that the RNA structure did not influence transcription termination efficiency. In contrast, for poly-U tract lengths of 4 nt, we observed a significant decrease in fluorescence when the hairpin was added for both the Linear-1 and Linear-2 contexts. This suggests that both RNA sequence and structure elements are needed to enhance transcription termination within our assay.
The Distance of the RNA Secondary Structure Impacts its Ability to Enhance Pol III Termination From Type 3 Promoters
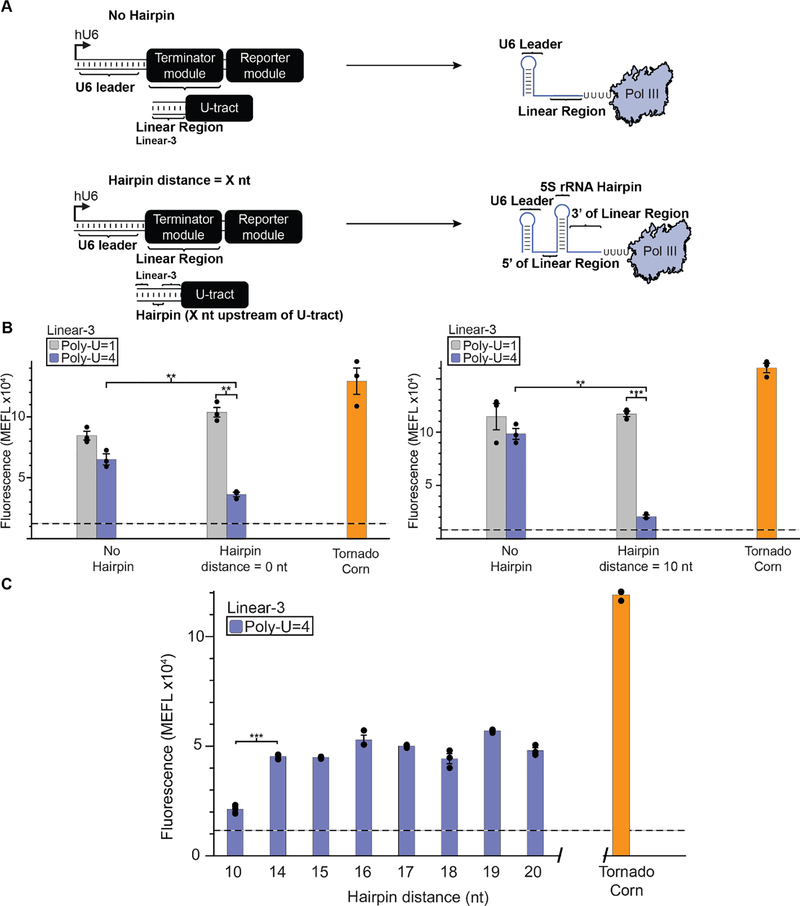
We next investigated whether the position of the secondary structural element within the RNA transcript impacts its enhancement of termination efficiency. In the constructs analyzed in Figure 3, the hairpin was placed immediately adjacent to the poly-U track (i.e., a distance of 0 nt upstream). To test how the placement of this hairpin affects termination efficiency, we generated constructs in which the hairpin was instead placed on the other side of the linear sequence—at a distance of 10 nt upstream of the poly-U tract (Supplementary Fig. 4). We again employed NUPACK analysis to determine that new transcripts were predicted to assume the expected conformations (Supplementary Fig. 3). Similar to the above results, the placement of the hairpin 10 nt upstream of the poly-U tract of length 1 nt had no influence on termination efficiency, and instead the addition of a hairpin led to an increase in fluorescence (Supplementary Fig. 4B, Supplementary Table 4). In contrast, inserting a hairpin 10 nt upstream of poly-U tract of length 4 nt led to a significant decrease in fluorescence and thus an enhancement of termination for both Linear-1 and Linear-2 contexts (Supplementary Fig. 4B). These data demonstrated that inclusion of a secondary structural element 10 nt upstream from the poly-U tract enhances Pol III termination in a similar fashion to that which occurs when the hairpin is directly adjacent to the poly-U tract.
Interestingly, the above observation contrasts with the prokaryotic intrinsic termination mechanism, where the RNA hairpin must be placed immediately adjacent to the poly-U tract in order to confer effective termination [14]. We therefore sought to investigate how far upstream RNA structure can be placed and still enhance Pol III termination. To do so, we used NUPACK to design a new transcript with a longer linear sequence (Linear-3) that enables insertion of the RNA hairpin up to 20 nt upstream from the poly-U tract. First, we confirmed that Linear-3-based constructs exhibited the same patterns observed for Linear-1- and Linear-2-based constructs when the hairpin was placed 0 nt or 10 nt upstream from the poly-U tract (Fig. 4B). We then created a series of constructs by incrementally increasing the distance between the hairpin and the poly-U tract. Overall, we observed increasing fluorescence (decreasing termination efficiency) when the hairpin was moved to a distance of 14 nt, and we saw no meaningful increase in fluorescence when moving the hairpin further away. (Fig. 4C). Overall, these data suggest that within the context of our assay, for transcription from Pol III type 3 promoters the location of RNA secondary structure and the length of the poly-U tract interact to modulate Pol III transcription termination efficiency in human cells.
Figure 4: Distance of RNA structure from the poly-U tract impacts termination efficiency.
A) A schematic depicting the positioning of predicted secondary structure upstream of the poly-U tract. This structure was either omitted (No Hairpin) or positioned X nt upstream of the poly-U tract (where Hairpin distance = X nt). The secondary structure utilized is a 23 nt portion of the 5S ribosomal RNA (rRNA) predicted to fold into a 9 bp hairpin. In these constructs, the Linear-3 sequence was used to enable X to be up to 20 nt. B) Constructs based upon Linear-3 exhibit a pattern which is similar to those based upon Linear-1 and −2 for hairpin distances of 0 or 10 nt. Significance was measured using a one-tailed heteroscedastic Welch’s t-test followed by the Benjamini-Hochberg procedure with a false discovery rate cutoff of 0.05 (Supplementary Table 2). ** = p < 0.01, *** = p < 0.001. C) Moving the hairpin further upstream from the poly-U tract reduces termination efficiency. Observed fluorescence for hairpin distances 10, differed significantly from those with a hairpin distance of 14, 15, 16, 17, 18, 19 and 20 (Supplementary Table 4). Colored bars represent the average of 3 biological replicates with individual points plotted as circles. Error bars represent the S.E.M. The dashed line represents the average of three replicates for the vector-only control (v) and the grey horizontal bar represents the S.E.M. of the vector-only control.
Poly-U Tract Affects Pol III Termination More Than RNA Secondary Structure Elements for Some Type 2 Promoters
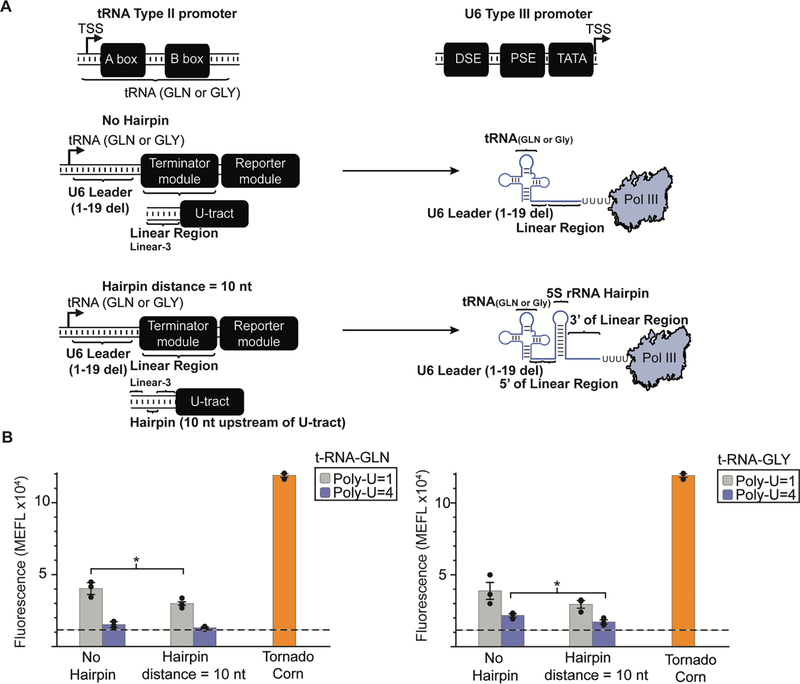
We next sought to evaluate the impact of sequence and structure on transcription termination originating from a Pol III type 2 promoter. All tRNA, which represent the vast majority of Pol III transcripts, originate from type 2 promoters [28]. The type 2 promoters differ from their type 3 counterparts as they possess an internal transcription start site (TSS) flanked by A box and B box elements, whereas the U6 type 3 promoter contains a proximal sequence element (PSE) along with a distal sequence element (DSE) upstream of the TSS (Fig. 5A) [27]. These different elements confer promoter-specific recruitment of various transcription factor subunits, ultimately recruiting Pol III for transcription initiation [27, 34]. To interrogate the impact of transcript structure and sequence on transcription from this promoter type in our system, we completely replaced the human U6 promoter in our constructs with two different tRNA sequences bearing an internal Pol III type 2 promotor (Fig. 5A). For this purpose, we employed the tRNA GLN and GLY as these tRNA are known to provide the greatest Pol III transcription rates when fused to artificial constructs [35, 36]. Compared to poly-U tracts of length 1 nt, we observed that poly-U tracts of length 4 nt enhanced termination efficiency in both the absence and presence of an included hairpin, respectively, for both tRNA sequence contexts. The addition of the RNA hairpin alone did not meaningfully enhance termination (for either poly-U tract). These data indicate that for the type 2 promoters evaluated here, the length of the poly-U tract affects Pol III transcription termination to a greater extent than does the presence of RNA secondary structure elements.
Figure 5: The impacts of sequence and structure on termination of transcription from type 2 Pol III promoters.
A) A schematic depicting the construction of our reporters with type 2 promoters versus the previously utilized type 3 promoters. Our type 2 promoter encompassed the transcription start site (TSS) as well as the complete tRNA (GLN or GLY) sequence. This diagram illustrates the positioning of predicted RNA secondary structure upstream of the poly-U tract for type 2 Pol III promoters. Constructs were made by fusing each of two type 2 tRNA promoters with a portion of the U6 leader sequence (minus the first 19 nt) to increase the length of the predicted linear region before the poly-U tract. An additional 23 nt portion of the 5S ribosomal RNA (rRNA), predicted to fold into a 9 bp hairpin structure, was either omitted (No Hairpin) or positioned 10 nt upstream of the poly-U tract. In these constructs, the Linear-3 sequence was utilized. B) Constructs using type 2 Pol III promoters show different patterns of termination from those using a type 3 promoter. Significant termination is seen whenever a poly-U tract of 4 uracils is present, independent of the presence of a predicted hairpin structure. Significance was measured using a one-tailed heteroscedastic Welch’s t-test (Supplementary Table 4). ** = p < 0.01, *** = p < 0.001. Colored bars represent the average of 3 biological replicates with individual points plotted as circles. Error bars represent the S.E.M. The dashed line represents the average of three replicates for the vector-only control (v) and the grey horizontal bar represents the S.E.M. of the vector-only control.
Discussion:
In this study, we developed a means for quantitatively interrogating Pol III termination in human cells. We found that for transcription from Pol III type 3 promoters, poly-U sequence length, and the presence and position of an RNA hairpin structure can both influence Pol III termination. Specifically, we found that poly-U tracts alone can enhance transcription termination if they are at least 7 or 8 nt in length (Fig. 2). In addition, an RNA hairpin structure can enhance the termination when used in conjunction with shorter poly-U lengths (Fig. 3), although this effect is diminished the further away this hairpin structure is from the poly-U tract (Fig. 4). Notably, RNA structure by itself did not appear to cause termination (Fig. 3B). In contrast, we observed that termination for transcription from Pol III type 2 promoters can occur due to the presence of the poly-U tract alone (Fig. 5), similar to previous in vitro observations [18, 19]. This is an important advance in our understanding of Pol III termination, since previous studies evaluating the impact of multiple RNA sequence and structure elements on Pol III termination offered conflicting findings about the importance of these features [5–7, 17, 18, 20]. This study thus offers a potential resolution in supporting the interpretation that poly-U sequence and RNA structure are important for Pol III termination.
It is important to note the differences between model systems used in this study and previous work. Notably, previous studies utilized either in vitro transcription assays with purified polymerase components, or assays in yeast that are perhaps closest to our study [6, 18, 37]. For the latter, Arimbasseri et al. employed an in vivo yeast reporter system where transcription readthrough resulted in the coloration of plated yeast colonies [18]; thus, this readout is more indirect compared to the fluorescent RNA approach we utilized. In Arimbasseri et al.’s yeast assay using a type 2 promoter system, Pol III termination was observed to be unimpacted by the presences of upstream structural elements, which was similar to our observation for type 2 promoters in human cells. In contrast, we observed a meaningful impact of structure on enhancing termination of transcription driven by type 3 promoters in human cells. More work will be needed to elucidate whether these differences are organism-specific or how they might reflect nuances about Pol III termination.
Interestingly, our finding that both poly-U tract length and RNA secondary structure can enhance Pol III transcription from type 3 promoters is similar to the case of prokaryotic intrinsic termination [4]. Prokaryotic intrinsic termination occurs when the RNAP encounters a poly-U tract and changes from an elongation to paused state. During this pause, secondary structure encoded within the nascent RNA forms and acts to further destabilize the transcription complex resulting in transcription termination [16, 38]. In addition, the RNA secondary structure is often immediately adjacent to the poly-U tract in prokaryotes [14]. Notably, our findings for eukaryotic Pol III termination differ from the prokaryotic case in that the RNA structure still has an influence when not placed immediately adjacent to the poly-U tract. Potentially, this lack of spatial requirements may be due to the ability of Pol III to undergo extensive backtracking following interaction with the poly-U tract [6]. This backtracking of the polymerase may result in the repositioning of RNA secondary structure to be adjacent to the transcription complex, enabling termination. It is also possible that the weaker hybridization forces between average length poly-U tracts and template result in a confirmational shift of the polymerase in a similar manner to what is seen during prokaryotic transcription termination [39, 40]. This shift may make the polymerase more sensitive to further destabilization forces, resulting in termination either from an upstream secondary structure or larger poly-U tracts. Further work will be needed to uncover the exact biomolecular interactions that are occurring during Pol III termination.
We also sought to address the possibility that some of our observed phenomena may be influenced by the nucleotide composition flanking the poly-U tract. Previous studies have shown that higher GC content with respect to the content of the dinucleotides flanking the poly-U tract increases termination, although the magnitude of this effect appears to dramatically change with even slight variations in overall sequence context [5, 20, 22–26]. We therefore designed the sequences used in this study to control for these effects. Three different dinucleotide sequences consisting of GC, TG, and AA were employed 5’ of the poly-U while the region 3’ of the poly-U tract was held constant across all constructs (Supplementary Fig. 2). The fact that we observed identical trends with respect to sequence and structural determinants of Pol III termination in all three 5’ dinucleotide contexts suggests that flanking dinucleotide composition does not influence the conclusions drawn in this study (Fig. 3B & Fig. 4B). However, it is likely that sequence context may help to explain the small variations seen among comparable constructs which differ only in the choice of linear regions.
We also note that our synthetic biology approach for studying transcription termination using fluorescent RNA aptamers should be able to be used to study other features of Pol III termination. It would be of interest to assay the impacts of some notable factors such as the degree of upstream inter-nucleotide base stacking and the minimum free energy (MFE) of secondary structure [16]. Perhaps this work could also be utilized to perform functional enzymatic assays in cellulo to study how changes to the Pol III subunits, involved in termination, coordinate termination events with various nascent RNA sequence and structural elements [40]. We can envision testing an orthogonal Pol III mutant within our system following depletion of wild type Pol III that has been tagged with an inducible degradation systems [41]. As we currently do not expect any issues with adapting this assay for other culturable eukaryotic species, further testing may provide a more comprehensive model for Pol III termination across the eukaryotic domain.
This study adds to the growing body of knowledge of the RNA sequence and structural determinants of Pol III termination. Altogether, our system enables one to characterize termination within a context that may be most relevant for understanding the natural regulation of cellular processes which are known to impact both human development and disease [11, 42]. This could also be important from a biotechnology standpoint, as an increased understanding of Pol III termination may lead to forward-design of novel termination sequences that possess desired levels of termination in different genetic contexts, which could be useful for defining expression of genes useful in a range of biotechnology applications [15].
Methods:
Design of RNA sequences:
RNA sequences were designed, and structure prediction analysis performed using the Nucleic Acids PACKage [33]. RNA secondary structures were predicted from sequence utilizing the NUPACK online web portal in analysis mode. All folding queries were run under the RNA setting at 37oC using default parameters. Novel linear regions (i.e. RNA sequences predicted to not fold into secondary structures) were designed using the NUPACK web server in design mode by utilizing dot bracket notation of the desired length with the design feature. For example, to produce a hairpin with 5 base pairs and a loop of 4 nt, we input the notation (((((….))))), which NUPACK used to generate an RNA sequence predicted to fold into that structure. RNA sequence outputs were then inserted into the complete sequence construct to ascertain whether they were predicted to fold as designed in that context. Only sequences that were predicted to fold as designed were used in the study.
Plasmid construction:
All plasmid construction was performed utilizing the Gibson assembly protocol or inverse PCR [43, 44]. All gBlocks® and oligo primers were ordered from Integrated DNA Technologies. Assembled plasmids were transformed into and stored within NEB® Turbo Competent Cells (Supplementary Table 1). All constructs were sequenced verified using Quintara Biosciences. The type 3 promoter construct variants utilized in the main text were based off the construct pAV-U6+27-Tornado-Corn. The construct pAV-U6+27-tCORN, also from the Jaffrey lab, was obtained from Addgene (Addgene plasmid #106233) (Supplementary Fig. 1). The type 2 promoter constructs were developed by starting with the tRNA sequences, including the transcription start site, from Mefferd et al. [36]. Nine nucleotides of the unstructured region of the U6 leader sequence were then appended to this to keep the distance between the termination module and the tRNA’s structure as long as possible to facilitate our studies. A table of Addgene accession numbers for constructs utilized in this study, except for pAV-U6+27-Tornado-Corn, can be found in Supplementary Table 5. The construct pAV-U6+27-Tornado-Corn was received as a gift from Dr. Samie Jaffrey.
Plasmid preparation for transfection:
Plasmids were transformed into NEB® Turbo Competent Cells. Single colony forming units of each construct were then resuspended into 100 mL LB media with appropriate antibiotics and shook overnight at 37oC. Plasmids were harvested using Qiagen Midiprep Kits. Neutralized cell lysate was both spun down at 16xg for 30 min and the supernatant was then run through a Qiagen tip-100. Plasmids were resuspended in TE buffer at 200 ng/μL using a Thermo Scientific Nanodrop for quantification.
Plasmid Purity Quality Control:
In preliminary work, next-generation sequencing demonstrated that some of our plasmid stocks possessed a small amount of recombined construct, a fact that was not detected using traditional sanger sequencing (not shown). To rectify this, plasmid stocks were retransformed into Invitrogen™’s recombinase limited DH10B competent cells. These transformations were harvested using the Qiagen Miniprep Kit and a small percentage of product was linearized and run on a 1% agarose gel. Only those preparations demonstrating a single band at the expected length were again retransformed into new Invitrogen™ DH10B cells and were then harvested using the Qiagen Midiprep Kit, for final reagent material. A sample from each new plasmid stock was again linearized and verified to be homogeneous on a 1% agarose gel prior to use in transfection.
Cell culturing:
HEK293FT cells (Life Technologies/Thermo) were maintained at 37°C and 5% CO2. Cells were cultured in DMEM (Gibco 3F1600–091) supplemented with 10% FBS, 6mM L-glutamine (2 mM from Gibco 31600–091 and 4 mM from additional Gibco 25030–081), penicillin (100 U/μL), and streptomycin (100 μg/mL) (Gibco 15140122).
Transfection:
HEK293FT cells were seeded at 1.5 × 105 cells in 0.5 mL of supplemented DMEM media in 24 well plates. At 6–8 h post seeding, cells were transfected using calcium phosphate precipitation: DNA—200ng of construct (unless otherwise stated) (Supplementary Fig. 5) and 200 ng of plasmid encoding blue fluorescent protein (BFP) as a transfection control—was mixed in H2O, and 2M CaCl2 was added to a final concentration of 0.3 M CaCl2. The vector-only control was generated by transfecting with only 200 ng of the plasmid encoding BFP. For all samples, transfected DNA mass was brought to a total of 800 ng/well though the addition of empty pcDNA™3.1(+) Mammalian Expression Vector (pcDNA) (ThermoFisher Scientific). This mixture was added dropwise to an equal-volume solution of 2x HEPES-Buffered Saline (280 mM NaCl, 0.5 M HEPES, 1.5 mM Na2HPO4) and gently pipetted up and down four times. After 2.5 min, the solution was mixed vigorously by pipetting ten times. 100 μL of this mixture was added dropwise to each well in a 24-well plate of cells. The next morning, the medium was aspirated and replaced with fresh medium.
Sample harvest:
At 24–30 h post media change, cells were harvested for flow cytometry with 0.05% Trypsin EDTA (Thermo Fisher Scientific #25300120), incubating for 3 min at 37oC followed by quenching with phenol red-free DMEM. The resulting cell solution was added to 500 μL of flow buffer consisting of 1X phosphate-buffered saline (PBS), 4% FBS, 5 mM MgSO4 [30]. Cells were spun at 150xg for 5 minutes, supernatant was aspirated, and 200 μL of flow buffer supplemented to a final concentration of 5 μM DFHO dye (TOCRIS Bioscience) was added prior to analysis by flow cytometry.
Analytical flow cytometry:
Flow cytometry was performed using on a BD LSR Fortessa Special Order Research Product. Approximately 3,000–6,000 single transfected cells were analyzed per sample, using BFP as the transfection control. BD LSR Fortessa settings used were as follows: BFP was collected in the Pacific Blue channel (405 nm excitation, 450/50 nm filter) and DFHO dye signal was collected in the FITC channel (488 nm excitation, 505 LP and 530/30 nm filter). Samples were analyzed using FlowJo v10 software (FlowJo, LLC). Fluorescence data were compensated for spectral bleed-through, the HEK293FT cell population was identified by SSC-A vs. FSC-A gating, and single cells were identified by FSC-A vs. FSC-H gating (Supplementary Fig. 6). To distinguish between transfected and non-transfected cells, a control sample of cells was transfected with empty vector only (pcDNA). This empty vector control was used to identify cells that were positive for the constitutive fluorescent protein (BFP) used as a transfection control in all other samples. The gate was drawn to include no more than 1% of cells in the empty vector control. Constructs with reporters for respective fluorescence channels were analyzed and compensation was applied to account for spectral overlap (Supplementary Fig. 6).
After gating for transfection, the intensity of the DFHO dye signal was quantified as Mean Fluorescence Intensity (MFI) by taking the geometric mean of fluorescence intensity in the FITC channel within each transfected cell population. MFI was then converted to Mean Equivalent of Fluorescien (MEFL) using UltraRainbow Calibration Particles (Spherotech URCP-100–2H), which were incorporated as part of each individual experiment (Supplementary Fig. 7). The bead population was identified by FSC-A vs. SSC-A gating, and 9 bead subpopulations were identified through two fluorescent channels. MEFL values corresponding to each subpopulation were supplied by the manufacturer and a calibration curve was generated for the experimentally determined MFI vs. the manufacturer specified MEFLs. A linear regression was performed with the constraint that 0 MFI equals 0 MEFL, and the slope from the regression was used to convert MFI to MEFL for each cellular population.
We also evaluated an alternative data analysis approach where MEFL values are normalized to the magnitude of BFP fluorescence in case there is a correlation between increased observed MEFL and transfection efficiency. To test this approach, a subset of experimental data was analyzed using both MEFL and MEFL normalized to the BFP signal output from our transfection control (Supplementary Fig. 8.) Since similar trends were observed in both approaches, we did not employ normalization by BFP in our data analysis pipeline; by minimizing data processing, we also avoid potential artifacts due to differential saturation of output between DFHO and BFP signals.
Finally, we confirmed that reporter output did not vary significantly across the course of a representative 1.5 h flow cytometry data collection experiment (Supplementary Fig. 9), ruling out potential artifacts due to the order in which samples were analyzed.
Data Analysis:
The mean fluorescence intensity (MFI) described above of the single-cell, transfected population was calculated and exported for further analysis. To calculate signal, MEFLs were averaged across three biological replicates. A vector-only control sample transfected with the BFP transfection control and empty vector (pcDNA) was treated with 5 μM DFHO dye, averaged across three biological replicates, and used to measure background signal. Statistical significance of measured fluorescence differences between specified cell populations was measured by utilizing a one-tailed heteroscedastic Welch’s t-test either alone, or followed by the Benjamini-Hochberg procedure [45]. Data and statistical analysis were performed using Excel (Microsoft). (Supplementary Table 2 & Supplementary Table 4).
Supplementary Material
The role of RNA sequence and structure on Pol III termination is uncertain.
We developed an in cellulo assay for interrogating human Pol III termination.
Type 3 promoters show robust termination only at longer than average poly-U tracts.
For type 3 promoters, RNA structure enhances termination, but only near poly-U tracts.
Type 2 promoters demonstrate efficient termination without additional RNA structure.
Acknowledgment:
We wish to thank Dr. Samie Jaffrey for sharing a stock of the original Tornado construct bearing the Corn aptamer. We wish to acknowledge Michael Sumner for initial conversations around the ideas that led to this work. We would also like to acknowledge Adam Silverman for critical reading of this manuscript. We wish to acknowledge Katherine Berman for conversations leading to the methodology used within this paper. M.S.V and W.K.C. were supported in part by Northwestern University’s Graduate School Cluster in Biotechnology, System, and Synthetic Biology, which is affiliated with the Biotechnology Training Program. W.K.C. was supported in part by the National Institutes of Health Training Grant (T32GM008449) through Northwestern University’s Biotechnology Training Program. D.Z.B. was supported by the National Institutes of Health Training Grant (T32GM008382) through Northwestern University’s Molecular Biophysics Training Program. This work was also supported by the Northwestern University – Flow Cytometry Core Facility supported by Cancer Center Support Grant (NCI CA060553). This work was supported in part by the National Institute of Biomedical Imaging and Bioengineering (grant no. 1R01EB026510 to J.N.L.), an NSF CAREER (grant no. 1452441 to J.B.L.), and through the National Institute for General Medicine Sciences (grant no. R01GM130901 to J.B.L).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability:
All processed analytical flow cytometry data, as well as calculations of averages, S.E.M. and statistical comparisons, was deposited within Northwestern’s open access arch database (https://arch.library.northwestern.edu/). The data can be accessed at https://doi.org/10.21985/n2-9je1-6c04
References
- [1].Roberts J Mechanisms of Bacterial Transcription Termination. Journal of Molecular Biology. 2019;431:4030–9. [DOI] [PubMed] [Google Scholar]
- [2].Maier K, Marchfelder A. It’s all about the T: transcription termination in archaea. Biochem Soc Trans. 2019;47:461–8. [DOI] [PubMed] [Google Scholar]
- [3].Porrua O, Libri D. Transcription termination and the control of the transcriptome: why, where and how to stop. Nat Rev Mol Cell Biol. 2015;16:190–202. [DOI] [PubMed] [Google Scholar]
- [4].Ray-Soni A, Bellecourt M, Landick R. Mechanisms of Bacterial Transcription Termination: All Good Things Must End. Annual Reviews of Biochem. 2016;85:319–47. [DOI] [PubMed] [Google Scholar]
- [5].Bogenhagen D, Brown D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–70. [DOI] [PubMed] [Google Scholar]
- [6].Nielsen S, Yuzenkova Y, & Zenkin N Mechanism of Eukaryotic RNA polymerase III transcription termination. Science. 2013;340:1577–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Arimbasseri A, Rijal K, Maraia R. Transcription termination by the eukaryotic RNA Polymerase III. Biochim Biophys Acta. 2013;1829:318–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Proudfoot N Transcription termination in mammals: Stopping the RNA polymerase II juggernaut. Science. 2016;352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Borchert G, Lanier W, Davidson B. RNA polymerase III transcribes human microRNAs. Nat Structure & Mol Biol. 2006;13:1097–101 [DOI] [PubMed] [Google Scholar]
- [10].Turowski T, Tollervey D Transcription by RNA polymerase III: insights into mechanism and regulation. Biochem Soc Trans. 2016;44:1367–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Li Y, Kowdley K. MicroRNAs in Common Human Diseases. Genomics, Proteomics & Bioinformatics. 2012;10:246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Padgett R New connections between splicing and human disease. Trends Genet. 2012;28:147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Braglia P, Percudani R, Dieci G. Sequence Context Effects on Oligo(dT) Termination Signal Recognition by Saccharomyces cerevisiae RNA Polymerase III*. journal of biological chemistry. 2005. [DOI] [PubMed] [Google Scholar]
- [14].Wilson K, Hippel Pv. Transcription termination at intrinsic terminators: the role of the RNA hairpin. PNAS. 1995;92:8793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen Y-J, Liu P, Nielsen A, Brophy J, Clancy K, Peterson T, et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat Methods. 2013;10:659–64. [DOI] [PubMed] [Google Scholar]
- [16].Chappell J, Westbrook A, Verosloff M, & Lucks J Computational design of small transcription activating RNAs for versatile and dynamic gene regulation. Nat Comm. 2017;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nielsen S, & Zenkin N Response to Comment on “Mechanism of eukaryotic RNA polymerase III transcription termination”. Science. 2014;345:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Arimbasseri A, Kassavetis G, & Maraia R Comment on “Mechanism of eukaryotic RNA polymerase III transcription termination”. Science. 2014;345:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Arimbasseri A, Maraia R. Mechanism of Transcription Termination by RNA Polymerase III Utilizes a Non-template Strand Sequence-Specific Signal Element. Mol Cell. 2015;58:1124–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Chu W, Ballard R, Schmid C. Palindromic sequences preceding the terminator increase polymerase III template activity. Nucleic Acids Res. 1997;25:2077–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chedin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998:3857–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Maraia R, Chang D-y, Wolffe A, Vorce R, Hsu K. The RNA Polymerase III Terminator Used by a B1-Alu Element Can Modulate 3’ Processing of the Intermediate RNA Product. American Society for Microbiology. 1992;12:1500–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Goodier J, Maraia R. Terminator-specific recycling of a B1-Alu transcription complex by RNA polymerase III is mediated by the RNA terminus-binding protein La. J Biol Chem. 1998;273:26110–6. [DOI] [PubMed] [Google Scholar]
- [24].Wang X, Folk W. Termination of transcription by RNA polymerase III from wheat germ. The Journal of Biological Chemistry. 1994;269:4993–5004. [PubMed] [Google Scholar]
- [25].Mazabraud A, Scherly D, Müller F, Rungger D, Clarkson S. Structure and transcription termination of a lysine tRNA gene from Xenopus laevis. J Mol Biol. 1987;195:834–45. [DOI] [PubMed] [Google Scholar]
- [26].Allison D, Hall B. Effects of alterations in the 3′ flanking sequence on in vivo and in vitro expression of the yeast SUP4-o tRNATyr gene. EMBO J. 1985;4:2657–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Schramm L, Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes & Dev. 2002;16:2593–620. [DOI] [PubMed] [Google Scholar]
- [28].Arimbasseri AG, Rijal K, Maraia RJ. Comparative overview of RNA polymerase II and III transcription cycles, with focus on RNA polymerase III termination and reinitiation. Transcription. 2014;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lata E, Teichmann M. Activation and repression at the heart of human RNA polymerase III. Nature Structural & Molecular Biology volume 2021;28:124–6. [DOI] [PubMed] [Google Scholar]
- [30].Song W, Filonov G, Kim H, Hirsch M, Li X, Moon J, et al. Imaging RNA polymerase III transcription using a photobleach stable RNA-fluorophore complex. Nat Chem Biol. 2017;13:1187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Litke J, Jaffrey S. Highly Efficient Expression of Circular RNA Aptamers in Cells Using Autocatalytic Transcripts. Nat Biotech. 2019;37:667–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Good PD, Krikos AJ, Li SXL, Bertrand E, Lee NS, Giver L, et al. Expression of small, therapeutic RNAs in human cell nuclei. Gene Therapy. 1997;4:45–54. [DOI] [PubMed] [Google Scholar]
- [33].Zadeh JN, Steenberg CD, Bois JS, Wolfe BR, Pierce MB, Khan AR, et al. NUPACK: analysis and design of nucleic acid systems. J Comput Chem. 2011;32:170–3. [DOI] [PubMed] [Google Scholar]
- [34].Wang Q, Li S, Wan F, Xu Y, Wu Z, Cao M, et al. Structural insights into transcriptional regulation of human RNA polymerase III. Nature Structural & Molecular Biology. 2021;28:220–7. [DOI] [PubMed] [Google Scholar]
- [35].Knapp D, Michaels Y, Jamilly M, Ferry Q, Barbosa H, Milne T, et al. Decoupling tRNA promoter and processing activities enables specific Pol-II Cas9 guide RNA expression. Nature Communications. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mefferd A, Kornepati A, Bogerd H, Kennedy E, Cullen B. Expression of CRISPR/Cas single guide RNAs using small tRNA promoters. RNA. 2015;21:1683–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Arimbasseri AG, Maraia RJ. Biochemical analysis of transcription termination by RNA polymerase III from yeast Saccharomyces cerevisiae. Methods in Molecular Biology. New York, NY: Humana Press; 2015. p. 185–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol Cell. 1999;3:495–504. [DOI] [PubMed] [Google Scholar]
- [39].Peters J, Vangeloff A, Landick R. Bacterial Transcription Terminators: The RNA 3′-End Chronicles. J Mol Biol. 2011;412:793–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25:118–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].McGinness K, Baker T, Sauer R. Engineering Controllable Protein Degradation. Molecular Cell. 2006;22:701–7. [DOI] [PubMed] [Google Scholar]
- [42].Xing’er-Koo C, Kobiyama K, Shen Y, LeBert N, Ahmad S, Khatoo M, et al. RNA Polymerase III Regulates Cytosolic RNA:DNA Hybrids and Intracellular MicroRNA Expression. Journal of Biological Chemistry. 2015;290:7463–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ochman H, Gerber A, Hartl D. Genetic applications of an inverse polymerase chain reaction. Genetics. 1988;120:621–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gibson D, Young L, Chuang R, Venter J, Hutchinson C, Smith H. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–5. [DOI] [PubMed] [Google Scholar]
- [45].Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All processed analytical flow cytometry data, as well as calculations of averages, S.E.M. and statistical comparisons, was deposited within Northwestern’s open access arch database (https://arch.library.northwestern.edu/). The data can be accessed at https://doi.org/10.21985/n2-9je1-6c04