Abstract
Retinitis Pigmentosa (RP) is a mostly incurable inherited retinal degeneration affecting approximately 1 in 4000 individuals globally. The goal of this work was to identify drugs that can help patients suffering from the disease. To accomplish this, we screened drugs on a zebrafish autosomal dominant RP model. This model expresses a truncated human rhodopsin transgene (Q344X) causing significant rod degeneration by 7 days post-fertilization (dpf). Consequently, the larvae displayed a deficit in visual motor response (VMR) under scotopic condition. The diminished VMR was leveraged to screen an ENZO SCREEN-WELL REDOX library since oxidative stress is postulated to play a role in RP progression. Our screening identified a beta-blocker, carvedilol, that ameliorated the deficient VMR of the RP larvae and increased their rod number. Carvedilol may directly on rods as it affected the adrenergic pathway in the photoreceptor-like human Y79 cell line. Since carvedilol is an FDA-approved drug, our findings suggest that carvedilol can potentially be repurposed to treat autosomal dominant RP patients.
Subject terms: Neuroscience, Visual system, Retina, Neurodegeneration, Drug discovery, Drug screening, Phenotypic screening
Introduction
Retinitis Pigmentosa (RP) is a mostly incurable retinal-degenerative disease affecting approximately 1 in 4000 individuals globally1–3. Non-syndromic RP is caused by multiple mutations found in at least 65 causative genes with different modes of inheritance, while 271 causative genes have been identified in all RP subtypes. (RetNet database: https://sph.uth.edu/RetNet/)1,4–6. Patients suffering from RP have a cost burden of over $7000 per year on average higher than healthy individuals7. When patients lose their vision, they suffer from increased likelihood of injury, and increased anxiety and depression which decrease their quality of life8. Unfortunately, there are currently no effective treatment options available for the vast majority of patients suffering from the disease. Research into technologies including gene therapy, stem-cell therapy, and retinal prosthesis is being performed, however these options are still experimental and costly9. The only FDA-approved method for treating any form of RP is a recently developed gene therapy called Luxturna for the treatment of Lebar’s Congenital Amaurosis (LCA). Patients with biallelic RPE65 mutations preventing normal expression of the gene can be treated with Luxturna, which delivers functional RPE65 with an adeno-associated virus10. While Luxturna is very effective in restoring some vision to LCA patients, they represent only a small proportion of all RP patients. In addition, the Luxturna treatment strategy aims to replace a deficient enzyme in an autosomal recessive case of RP, however, this will not work in autosomal dominant cases of RP (adRP). This highlights an urgent need for RP therapeutics that are effective and inexpensive. To address this need, we utilized an adRP zebrafish model to perform phenotypic drug screening and identified the FDA-approved drug carvedilol as a positive hit.
The zebrafish can provide a powerful system to model RP, and they have been used to model a number of human retinal-degenerative diseases11–15. These models include transgenic zebrafish expressing human rhodopsin (RHO) with autosomal dominant mutations found in RP patients16. Up to 30% of RP cases are autosomal dominant, and of all autosomal dominant cases, and approximately 30% arise due to over 150 mutations in RHO5,17,18. These mutations include Q344X/Q344ter, a truncation mutation, which shortens RHO at the C-terminus by 5 amino acids19. Patients with this mutation suffer an early onset, severe form of autosomal-dominant RP20–22. Q344X RHO loses a VXPX ciliary trafficking motif on the C-terminus leading to its mislocalization to the inner segment and apoptotic cell death21,23,24. Despite the C-terminal truncation, Q344X RHO is a catalytically active protein that is still capable of G protein signaling. It is hypothesized that mislocalized RHO in the inner segment causes aberrant ADCY signaling which would ultimately trigger apoptosis through an increase of cAMP signaling15,25.
In zebrafish, a transgenic model was made to express a human Q344X RHO in rods under the zebrafish rho promoter15. This model exhibits significant rod degeneration as early as 5 days post-fertilization (dpf). The model also possesses a nose EGFP reporter in the transgenic cassette, which allows for an efficient mutant screening starting at 2 dpf. Previous work with the Q344X zebrafish has shown that adenylyl cyclase (ADCY) inhibition can lead to modest rod survival15. However, it has also been shown that the activation of mislocalized RHO is not necessary to induce cell death15,26. These findings indicate that Q344X can cause rod degeneration through more than one mechanism. In this study, we utilized this Q344X zebrafish model to develop an in vivo drug-screening platform for identifying drugs that may treat adRP.
The zebrafish is an ideal model for in vivo screening for drugs to treat RP9 due to its low cost of use, high fecundity, amenability to genetic manipulation27. It can also facilitate RP drug discovery because of the rapid development of its visual system28. In particular, zebrafish rod precursors begin to differentiate into rods as early as 36 h post-fertilization (hpf) in the ventral region of the retina by expressing rho29–31. The rod outer segments begin to form by 50 hpf, and fully formed outer segments have been found as early as 4 dpf32–34. These rods begin to form synapses by 62 hpf32,35. The earliest visually-evoked startle can be detected by 68 hpf36. After that, several visual behaviors gradually appear from 3 to 5 dpf, including the optokinetic response and the visual motor response (VMR)37–40. The VMR is a startle response triggered by a sudden light onset or offset, which results in increased locomotor behavior9,38,40–43. This behavior can be measured from multiple larvae simultaneously in 96-well plate format and is thus ideal for high-throughput, in vivo drug screening experiments9,44. The VMR has been utilized to identify oculotoxic drugs, and discover drugs that can benefit retinal degeneration44,45. Zebrafish have also been used to perform high-throughput drug screening based on fluorescent signals in the retina, but this approach does not provide direct functional insight46,47. On the contrary, utilizing the VMR as a drug-screening platform identifies compounds that improve visual function. To date, visual behavior including the VMR has not been used to screen drugs to treat RP. One reason is that due to the rods are not deemed functional until around 15 dpf30,48,49. However, recent works have detected rod ERG, and rod-mediated VMR and optokinetic response (OKR) in fish larvae as early as 5–6 dpf50,51. This indicates that the rod-driven, scotopic behavior of the larval zebrafish can potentially be utilized to screen drugs to treat RP.
In this study, we utilized a scotopic VMR assay utilizing the Q344X zebrafish model to screen for drugs that can treat RP. We found that this adRP model exhibited a diminished scotopic VMR behavior by 7 dpf. This response was driven by rods, as confirmed by specific rod ablation. Since it has been suggested that oxidative stress in the retina acts as one of the extrinsic factors to RP progression52, we leveraged this assay to screen a Redox library to determine if modulating oxidative stress could improve vision and increase rod survival in the Q344X zebrafish. The discovery of a drug that could alleviate oxidative stress could broadly treat RP regardless of the causative mutation, and zebrafish have proved to be an effective model for investigating the effects of oxidative stress in the eye53,54. Our screen uncovered carvedilol, a β-adrenergic receptor antagonist, enhanced the Q344X zebrafish VMR and increased rod number. We provided evidence that this drug acted on rods autonomously. Since carvedilol is already approved by the FDA to treat heart failure and high blood pressure, this drug can potentially be repurposed for the treatment of RP.
Results
VMR assay utilization for drug screening on Q344X zebrafish with scotopic illumination
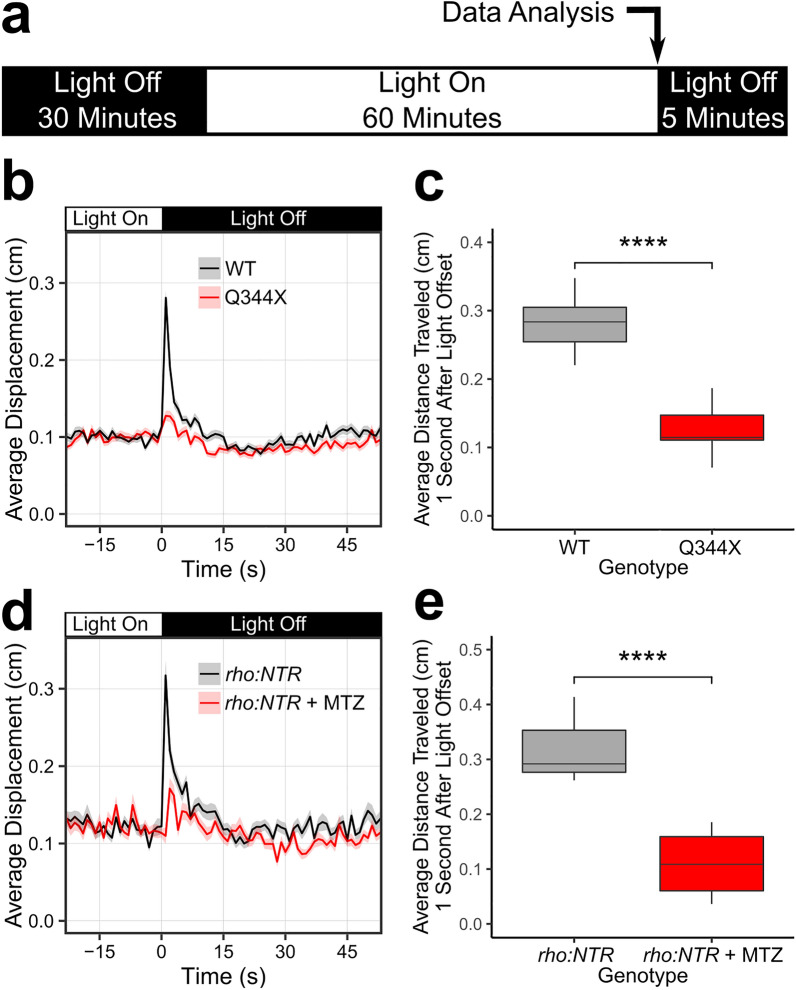
We utilized the VMR assay to screen drugs with the Q344X zebrafish model using a scotopic light stimulus. This fish model was selected for our drug screening as its rods begin to degenerate at 5 dpf, and the rod degeneration becomes severe by 7 dpf15. This rapid rod degeneration facilitates rapid evaluation of many compounds on many individual larvae. To determine the visual consequences of rod degeneration in the Q344X zebrafish, their VMR were measured under scotopic light illumination. An appropriate scotopic intensity was identified by systematically attenuating light intensity with neutral density filters until the light was 0.01 lx (Supplementary Fig. S1). To conduct the VMR assay, Q344X transgenic larvae were identified and sorted at 2 dpf by nose fluorescence. These larvae were dark adapted overnight at 6 dpf in a 96-well plate, and their VMR assessed at 7 dpf. To conduct a VMR experiment, these larvae were acclimated to the machine in darkness for 30 min, exposed to the scotopic light of 0.01 lx for 60 min, and then exposed to darkness again (Fig. 1a). The larval displacement was recorded per second for the duration of the experiment. When exposed to a light intensity of 0.01 lx, wild-type (WT) larvae displayed a robust startle response immediately after light offset (light-off VMR), while Q344X larvae displayed a significantly diminished light-off VMR (Fig. 1b). Specifically, WT larvae traveled significantly further on average than the Q344X larvae (µ ± standard error of the mean (s.e.m): 0.281 ± 0.036 cm vs. 0.127 ± 0.031 cm) one second after light offset (Fig. 1c). Both Q344X and WT larvae did not show a response to the light onset at 0.01 lx, and both groups displayed a similar VMR at higher photopic intensities (Supplementary Fig. S2). These results indicate that the expression of Q344X RHO diminished the light-off VMR of Q344X larvae at 0.01 lx.
Figure 1.
The Q344X larvae displayed a diminished scotopic light-off VMR driven by rods. (a) Schematic of the VMR protocol. On 7 dpf, larvae were habituated to the machine in darkness for 30 min. Then, the light stimulation was turned on and the plate was illuminated for 60 min. After that, the light was turned off. In this study, we mainly analyzed the VMR at light offset (light-off VMR) as indicated by the arrow. (b) The light-off VMR of wildtype (WT, black trace) and Q344X (red trace) larvae at 0.01 lx. The light was turned off at Time = 0. Each trace shows the average larval displacement of 18 biological replicates with 48 larvae per condition per replicate. The corresponding color ribbon indicates ± 1 standard error of the mean (s.e.m.). (c) Boxplot of the average larval displacement of WT and Q344X larvae one second after light offset. The average displacement of WT larvae (µ ± s.e.m.): 0.281 ± 0.036 cm, N = 18) was significantly larger than that of Q344X larvae (0.127 ± 0.031 cm, N = 18) (Welch’s Two Sample t-test, T = 13.2, df = 33.2, p value < 0.0001). To confirm this scotopic VMR was driven by rods, we chemically-ablated rods in larvae and subjected them to the same scotopic VMR assay (d, e). (d) The light-off VMR of larvae with nitroreductase-expressing rods treated with metronidazole (rho:NTR + MTZ, red trace) and without metronidazole (rho:NTR, black trace). Each trace shows the average displacement of 6 biological replicates with 24 larvae per condition per replicate. The corresponding color ribbon indicates ± 1 s.e.m. (e) Boxplot of the average displacement of rho:NTR and rho:NTR + MTZ larvae one second after light offset. The average displacement of untreated rho:NTR larvae (µ ± s.e.m.): 0.317 ± 0.061 cm, N = 6) was significantly larger than that of rho:NTR + MTZ larvae (0.110 ± 0.062 cm, N = 6) (Welch’s Two Sample t-test, T = 5.9, df = 10, p value < 0.0001).
The diminished VMR of Q344X larvae was likely caused by rod degeneration. We confirmed rods were responsible for the diminished scotopic VMR of Q344X larvae by rod ablation. To this end, we utilized a zebrafish line expressing nitroreductase (NTR) specifically in rods under the control of the rhodopsin promotor (rho:NTR)46. This enzyme would convert a prodrug metronidazole (MTZ) into a cytotoxic substance and specifically ablate rods. In this study, the NTR-expressing larvae were treated with 2.5 mM MTZ (rho:NTR + MTZ) from 5 to 7 dpf, and their scotopic light-off VMR was measured at 7 dpf. Like the Q344X line, the rod-ablated larvae showed a significantly diminished light-off VMR compared with the untreated larvae (Fig. 1d). The average displacement of rho:NTR group (0.317 ± 0.061 cm) was significantly further than that of rho:NTR + MTZ group (0.110 ± 0.062 cm) (Fig. 1e). The reduction of scotopic light-off VMR by rod ablation indicates that the response was substantially driven by rods. The rho:NTR line displayed a strong VMR to photopic stimuli with and without MTZ treatment indicating the cone pathway is not ablated and intact (Supplementary Fig. S3). This scotopic light-off VMR was then used to screen drugs that might improve rod response with the Q344X zebrafish model.
Drug screening revealed that carvedilol ameliorates the attenuated Q344X VMR
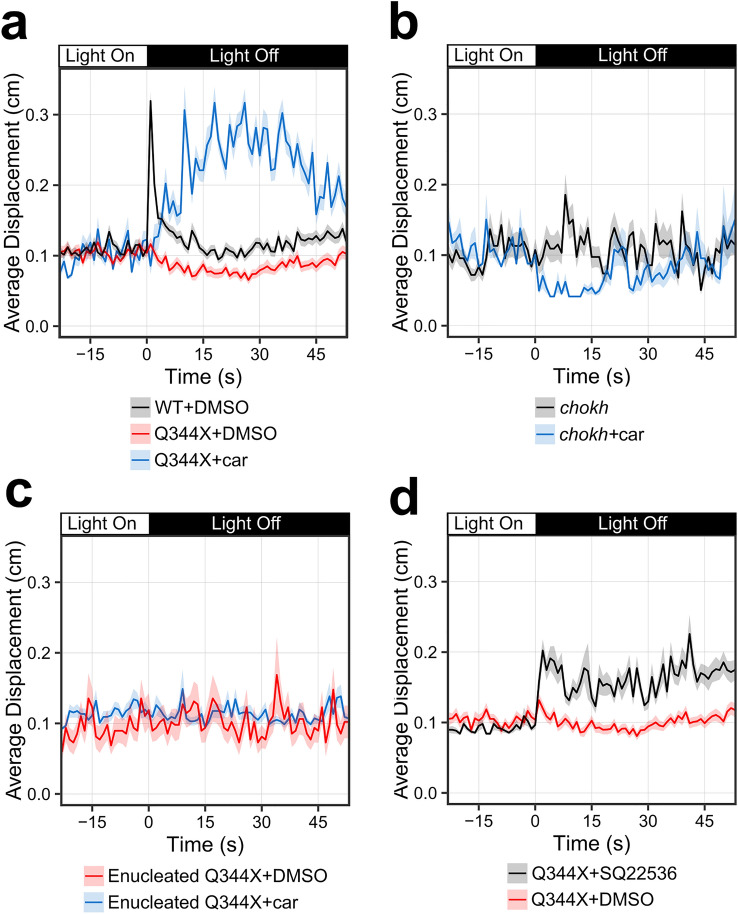
One of the prominent theories about RP pathogenesis is oxidative stress52. Since attenuating such stress might slow or prevent RP progression, we chose to screen and ENZO SCREEN-WELL REDOX library against the Q344X zebrafish model. We chose to begin drug treatment at 5 dpf to find drugs that can ameliorate the attenuated Q344X scotopic light-off VMR because rod degeneration in this model begins at this stage. 5 dpf larvae were exposed to compounds in this library dissolved in E3 media at a final concentration of 10 μM55, and their scotopic light-off VMR was tested at 7 dpf. The drugs of the library come dissolved in DMSO, thus all control larvae were treated with a matching concentration of 0.1% DMSO. All larvae were maintained in the same drug solution throughout the experiment. Each drug was tested twice using embryos collected on different dates. Of the 84 drugs tested, 16 were toxic to the zebrafish at 10 μM. The VMR of the remaining 68 drug-treated larval groups was normalized56 and then ranked based on the following selection criteria: Firstly, the two biological replicates must be consistent. The consistency was determined by a High-Dimensional Nonparametric Multivariate Test57 between the replicates. A small p value would indicate the replicates were dissimilar, whereas a high p value would indicate the replicates were similar. A cut off p value of 0.9 was chosen in this study to select those replicates that were highly similar to each other. Secondly, the drug-treated VMR must be significantly different from the DMSO-treated VMR, as determined by the Hotelling's T-squared test40. These criteria were applied to two timeframes: just 1 s after light offset to capture immediate response, and from 1 to 30 s after light offset to capture changes in any of the components of the VMR (Table 1). In the 1-s timeframe, 5 drug treatments gave rise to consistent larval behavior, but none of these drug treatments gave rise to a larval VMR that was significantly different from that displayed by DMSO-treated Q344X. However, in the 30-s timeframe, four drug treatments gave rise to a consistent larval behavior, and one drug treatment, carvedilol, provided both a consistent and significant change from the DMSO-treated Q344X VMR. Carvedilol-treated Q344X exhibited a sustained scotopic light-off VMR compared with DMSO-treated WT and Q344X controls (Fig. 2a). To determine if carvedilol was working through the retina, eyeless chokh/rx3 zebrafish58 were treated with the drug and their VMR was assessed. The chokh/rx3 larvae did not display a light-off VMR with or without carvedilol (Fig. 2b). Similarly, Q344X larvae were treated with carvedilol or DMSO at 5 dpf and were enucleated at 6 dpf to determine if carvedilol was exerting an effect on extraocular photoreceptors. Neither carvedilol-treated nor DMSO-treated enucleated Q344X larvae displayed a significant scotopic VMR (Fig. 2c). These results suggest that carvedilol is working at the level of the retina. Previous work with the Q344X line has shown that treatment with the ADCY inhibitor SQ 22,536 at a concentration of 100 μM improved rod survival15. To determine if this rod survival can translate into improved vision, the Q344X larvae were treated with 100 μM SQ 22,536 from 3 to 7 dpf at a concentration of 100 μM, and their scotopic light-off VMR was assessed at 7 dpf. The ADCY inhibitor was able to produce a significant Q344X VMR (Fig. 2d), however, this response was smaller than that produced by carvedilol treatment. Our screen therefore identified carvedilol, a drug that could functionally improve the vision of the Q344X adRP model.
Table 1.
Summary of drug-screening results.
| 1 s timeframe | 30 s timeframe | |
|---|---|---|
| Number of starting drugs in the library | 84 | 84 |
| Number of drugs not toxic | 68 | 68 |
| Number of drugs which induced consistent light-off scotopic VMR in both replicates | 5 | 4 |
| Number of drugs which induced consistent light-off scotopic VMR in both replicates, and significantly different from DMSO-treated controls (p value < 0.05) | 0 | 1 |
The 84 drugs in the ENZO Redox library were each applied to the Q344X larvae at 10 μM (N = 24 larvae) in two independent replicates. Of these 84 drugs, 16 were toxic. The two replicates were then compared to each other with a High-Dimensional Nonparametric Multivariate Test to determine similarity in either 1-s or 30-s timeframe. The drugs that induced consistent light-off scotopic VMR were compared to DMSO-treated Q344X controls to determine if they caused a significant change in behavior (High-Dimensional Nonparametric Multivariate Test, p value < 0.05). In the 1-s timeframe, no drugs met all criteria, but in the 30-s timeframe, one drug (carvedilol) was both consistent in the two replicates and caused a significant change in light-off scotopic VMR compared to controls.
Figure 2.
Drug screening on the Q344X zebrafish identified carvedilol as a beneficial drug. (a) Carvedilol treatment on Q344X larvae resulted in a sustained scotopic light-off VMR (blue trace, N = 2 replicates of 24 larvae) compared to that of both DMSO-treated WT larvae and DMSO-treated Q344X larvae (black and red trace respectively, N = 9 replicates of 48 larvae in each group). Each trace shows the average displacement of each replicate, and the color ribbons indicate µ ± s.e.m. The two carvedilol replicates were highly consistent and not different from each other (High-Dimensional Nonparametric Multivariate Test, N = 24, THD = 1.78, p value = 0.91). Each replicate demonstrated a significant change in behavior for the duration of 30 s after light offset above DMSO-treated Q344X larvae (Hotelling’s T-squared test, N = 24, T = 378.0 and 456.0, df = 30, p value < 0.0001 for each replicate). (b) To determine if carvedilol’s effects are elicited through the retina, eyeless chokh fish were treated with carvedilol (blue trace) and their VMR was compared with untreated control (black trace). Carvedilol treatment did not increase the chokh VMR (Hotelling’s T-squared test, N = 24 larvae, T = 37.8, df = 30, p value = 0.946). (c) Q344X larvae were enucleated to determine if extraocular expression of Q344X RHO was causing the VMR seen with carvedilol treatment. Larvae were treated with carvedilol (blue trace) or DMSO (red trace) at 5 dpf and enucleated on the morning of 6 dpf. VMR was assessed at 7 dpf. Carvedilol showed no effect on enucleated Q344X larvae. (Hotelling’s T-squared test, N = 24 larvae, T = 28.8, df = 30, p value = 0.948). (d) Q344X larvae were treated with 100 μM adenylyl cyclase (ADCY) inhibitor SQ 22,536 (black trace) at 3 dpf to determine if inhibiting ADCY would improve the VMR compared to DMSO treatment (red trace). Treatment with SQ 22,563 significantly improve the Q344X VMR over DMSO treatment (Hotelling’s T-squared test, N = 3 replicates 24 larvae, T = 118, df = 30, p value < 0.0001).
Carvedilol treatment increased rod number in the Q344X retina
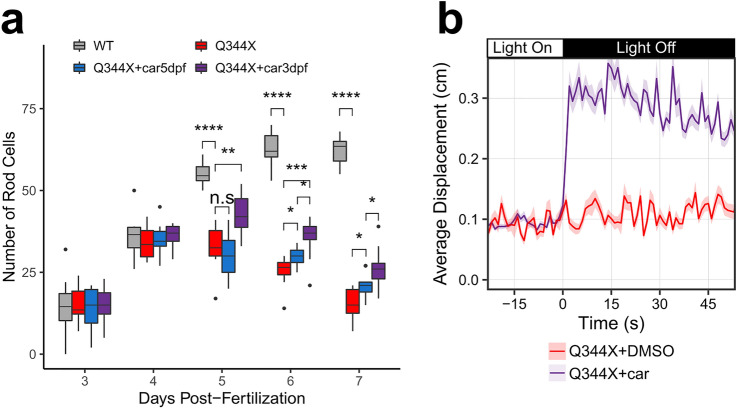
Since carvedilol enhanced the scotopic VMR of the Q344X larvae and acted through the retina, it likely exhibited benefits on the degenerating rods. The drug effect on rods was evaluated by quantification of rho:EGFP-positive cells on wholemount and sectioned retinae (Fig. 3). On cryosections, Q344X larvae exhibited significant rod degeneration on 5 dpf at which point they were treated with carvedilol. Carvedilol-treated Q344X larvae show increased rod number in the retina compared to DMSO-treated Q344X larvae on 6 dpf and 7 dpf (Fig. 3a–d). Next, to determine the anatomical distribution of the increased number of rods in the Q344X retina, whole-mount retinae were imaged to assess rod distribution. WT larvae had a high density of rods in the dorsal retina and ventral patch on 7 dpf while Q344X exhibited excessive rod degeneration in these areas (Fig. 3e). Carvedilol-treated Q344X showed an increased number of rods in both the dorsal retina and the ventral patch. To quantify these observations, WT, Q344X, and carvedilol-treated Q344X were binned into three classifications based on the distribution of EGFP signal: Strong, Intermediate, and Weak (Table 2). All WT larvae were classified as Strong. The carvedilol-treated Q344X larvae had significantly more Intermediate phenotypes in the lateral and ventral views compared to the DMSO-treated Q344X group. No larvae from the carvedilol or DMSO-treated Q344X groups was classified as Strong. The correlation between rod number increase and enhanced light-off VMR of Q344X larvae suggests that the increase in rod number with carvedilol treatment mediated the visual improvement.
Figure 3.
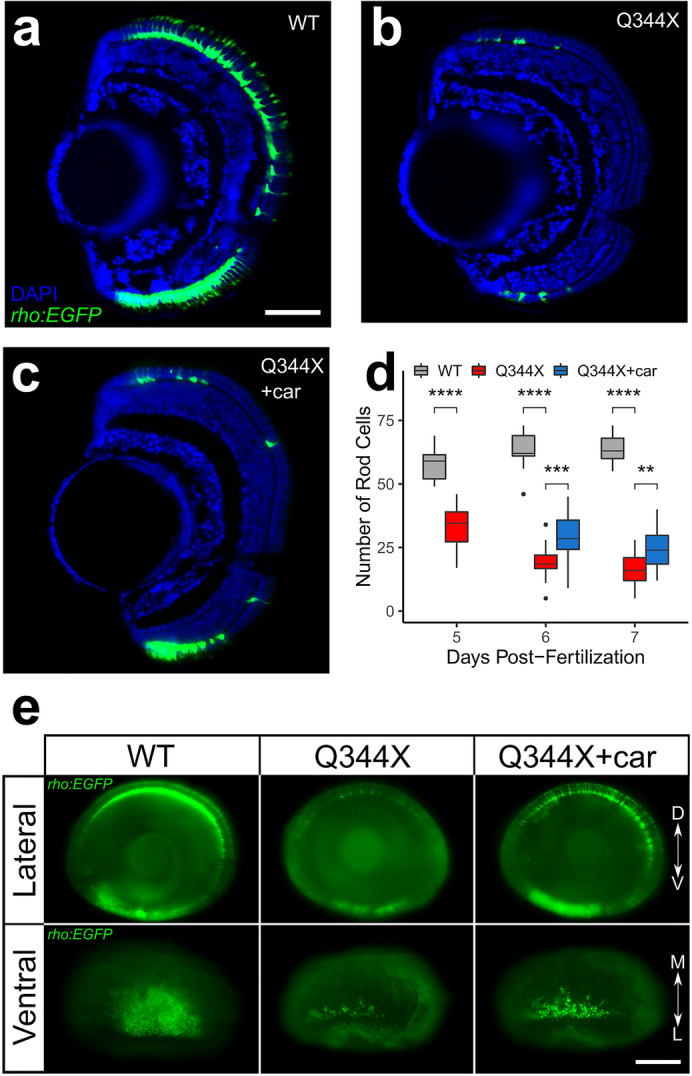
Carvedilol treatment increased rod numbers in the Q344X larvae. Representative retinal cryosection of (a) a wildtype larva (WT), (b) a DMSO-treated Q344X larva, and (c) a carvedilol-treated Q344X (car) larva at 7 dpf. Rods were labeled by EGFP expression driven by rho promoter, and the nuclei were counterstained with DAPI. Scale = 50 μm. (d) Quantification of rod number in WT, DMSO-treated Q344X, and carvedilol-treated Q344X retinal cryosections from 5 to 7 dpf. There was a statistically significant difference in rod number between groups at all stages determined by one-way ANOVA at 5 dpf (WT, N = 11; Q344X, N = 16; F(1,25) = 71.04, p value < 0.0001), at 6 dpf (WT, N = 9; Q344X, N = 20; Q344X + car, N = 21 ; F(2,44) = 96.9, p value < 0.0001), and at 7 dpf (WT, N = 9; Q344X, N = 17; Q344X + car, N = 11; F(2,41) = 167.9, p value < 0.0001). The effect of Q344X rod degeneration and carvedilol treatment on rod number was assessed post hoc by pairwise t-test with false discovery rate correction at 6 dpf (WT − Q344X, p value < 0.0001; Q344X − Q344X + car, p value < 0.001) and at 7 dpf (WT − Q344X, p value < 0.0001; Q344X − Q344X + car, p value < 0.001). (e) Representative whole-eye images of WT, Q344X, and carvedilol-treated Q344X larvae at 7 dpf. Rods were labeled by EGFP expression. Left column: WT rods were mainly found on dorsal and ventral retina (top). They were abundantly present in the ventral patch of the retina extending medially (bottom). Middle column: Q344X rods were mostly degenerated at the same stage (top). There were only a handful of rods remaining near the lateral edge of the ventral patch in the Q344X retina (bottom). Right column: carvedilol treatment increased the number of Q344X rods on both dorsal and ventral retina (top); however, gaps of missing rods were still apparently on dorsal retina. More rods were observed in the ventral patch of the carvedilol-treated retina (bottom). Statistical analysis of whole-mount data is shown in Table 2. Scale = 100 μm. D dorsal, V ventral, M medial, L lateral.
Table 2.
Rod analysis on whole-mount eyes.
| Strong | Intermediate | Weak | |
|---|---|---|---|
| WT lateral | 10 | 0 | 0 |
| Q344X lateral | 0 | 9 | 15 |
| Q344X + car lateral | 0 | 16 | 8 |
| WT ventral | 10 | 0 | 0 |
| Q344X ventral | 0 | 8 | 16 |
| Q344X + car ventral | 0 | 16 | 8 |
All larvae were bleached and examined from the lateral and ventral sides. The rho:EGFP signal were classified into 3 categories by the extent of its fluorescence. The Strong group contains the samples with high rod number/signal intensity in the dorsal retina and ventral patch; the Intermediate group contains the samples with distinct rods in the dorsal retina with noticeable gaps, and some rods in the ventral patch extending medially; and the Weak group contains the samples with sparse rods in the dorsal retina and the most lateral edge of the ventral patch. The WT image in Fig. 3e. is representative of the “Strong” group, the Q344X image in Fig. 3e. is representative of the “Weak” group, and the Q344X + car in Fig. 3e. is representative of the “Intermediate” group. Carvedilol treatment increased the number of Q344X larvae with Intermediate phenotypes and reduced the number of Weak phenotypes in both the lateral (Chi-square test, = 4.09, df = 1, p value < 0.05) and ventral views (Chi-square test, = 5.33, df = 1, p value < 0.05). No Q344X larvae was classified as Strong with or without carvedilol treatment.
Higher doses of carvedilol were tested at 31.6 μM and 100 μM to determine if a larger treatment dose would improve rod number, but these concentrations were toxic to the zebrafish larvae. Thus, further rod number improvement was evaluated with a longer carvedilol treatment period. Q344X larvae were treated with 10 μM carvedilol beginning at 3 dpf. The drug and media were refreshed daily to maintain the health of the larvae. Larval treatment beginning at 3 dpf was compared to treatment beginning at 5 dpf to determine if earlier carvedilol treatment is more effective. There was no difference in rod number between any of the Q344X and WT groups at 3 dpf and 4 dpf indicating that Q344X rod degeneration is not significant at these stages (Fig. 4a). Q344X rod degeneration does become significant at 5 dpf, and the earlier carvedilol treatment beginning at 3 dpf significantly increased the rod number at 5 dpf (Fig. 4a). Carvedilol treatment beginning at 5 dpf with daily refreshment still improved rod number in the Q344X zebrafish at 6 dpf and 7 dpf, however carvedilol treatment beginning at 3 dpf resulted in significantly higher rod numbers than the later 5 dpf treatment (Fig. 4a.). Correlating with increased rod number, the VMR of Q344X larvae treated with carvedilol beginning at 3 dpf displayed a significantly more rapid light-off VMR (Fig. 4b) compared with the VMR of larvae treated with carvedilol treatment at 5 dpf (Hotellings T-squared test, N = 3 replicates of 24 larvae, T = 397, df = 30, p value < 0.0001). Carvedilol treatment beginning at 3 dpf did not have a significant effect on the photopic VMR of Q344X larvae (Supplementary Fig. S4). These results suggest that earlier carvedilol drug treatment improves the number of Q344X rods better than later treatment, and that the carvedilol effect primarily acts on the rod photoreceptors.
Figure 4.
Carvedilol treatment beginning at 3 dpf increased rod numbers in the Q344X larvae greater than the treatment beginning at 5 dpf. (a) Quantification of rod number in WT, Q344X treated with DMSO beginning at 3 dpf, and Q344X treated with carvedilol beginning at 3 dpf or 5 dpf. Rods were quantified from their retinal cryosections beginning at 3–7 dpf. There was no statistically significant difference in rod number between groups at 3 dpf and 4 dpf determined by one-way ANOVA (3 dpf; N = 10; F(3,36) = 0.1, p value = 0.95); (4 dpf; N = 10; F(3,36) = 0.5, p value = 0.69). There was a statistically significant difference in rod number between groups at 5 dpf through 7 dpf determined by one-way ANOVA (5 dpf, N = 10; F(3,36) = 0.1, p value < 0.0001), (6 dpf, N = 10; F(3,36) = 0.1, p value < 0.0001), (7 dpf, N = 10; F(3,36) = 0.1, p value < 0.0001). The effect of Q344X rod degeneration and carvedilol treatment on rod number was assessed post hoc by pairwise t-test with false discovery rate correction at 5 dpf (WT − Q344X, p value < 0.0001; Q344X − Q344X + car3dpf, p value < 0.001; Q344X − Q344X + car5dpf, p value = 0.36, Q344X + car3dpf − Q344X + car5dpf, p value < 0.0001), at 6 dpf (WT − Q344X, p value < 0.0001; Q344X − Q344X + car3dpf, p value < 0.0001; Q344X − Q344X + car5dpf, p value < 0.05; Q344X + car3dpf − Q344X + car5dpf, p value < 0.05), and at 7 dpf (WT − Q344X, p value < 0.0001; Q344X − Q344X + car3dpf, p value < 0.0001; Q344X − Q344X + car5dpf, p value < 0.05; Q344X + car3dpf − Q344X + car5dpf, p value < 0.05). (b) Carvedilol treatment of Q344X larvae beginning at 3 dpf (purple trace) displayed a significant scotopic light-off VMR when compared to Q344X larvae treated with DMSO (red trace) (Hotellings T-squared test, N = 3 replicates of 24 larvae, T = 397, df = 30, p value < 0.0001). Each trace shows the average displacement of each replicate, and the color ribbons indicate µ ± s.e.m.
Carvedilol can inhibit β-adrenergic signaling in Y79 retinoblastoma cells
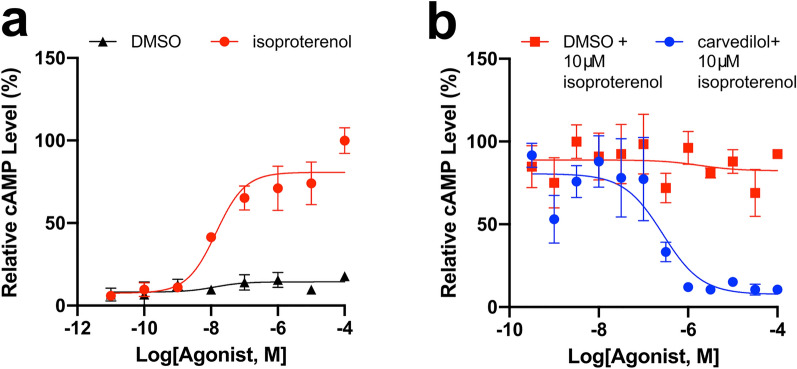
Carvedilol is a β-blocker that that binds to β1-adrenergic receptors, β2-adrenergic receptors, α1-adrenergic receptors and inhibits adrenergic signaling. However, its retinal target is unknown, and it may act directly on rods. To evaluate this possibility, we examined the effect of carvedilol treatment on the Y79 human retinoblastoma line which uniquely expresses rod-specific genes59. The Y79 cell line exists as a photoreceptor-like precursor that shows differentiation potential for the rod lineage60. Activin treatment of the Y79 line increases the expression of the transcription factor Nrl which induces progenitor differentiation into rods60. Previous work has leveraged this line to conduct expression studies in a photoreceptor-like cellular environment biased towards the rod lineage61. The level of adrenergic signaling was determined by GPCR-modulated changes in cAMP levels as measured by a cAMP-sensitive luciferase. First, the Y79 cells were transfected with the luciferase reporter, and then they were exposed to half-log dilutions of isoproterenol, a β-adrenergic receptor agonist. Isoproterenol was capable of inducing cAMP signaling in the transfected Y79 cells with a pEC50 of 7.5 ± 1.1 (Fig. 5a). The cAMP level was not increased in controls treated with matching DMSO percentage to dissolve isoproterenol. The relative cAMP level did not increase much above 10 μM isoproterenol. To determine if carvedilol treatment can inhibit this isoproterenol-mediated cAMP increase, the transfected Y79 cells were pretreated with half-log dilutions of carvedilol and then challenged with a dose of 10 μM isoproterenol that would induce saturating relative cAMP level according to Fig. 5a. Carvedilol pretreatment was able to prevent isoproterenol-mediated cAMP signaling with a pIC50 of 6.5 ± 0.7 (Fig. 5b).
Figure 5.
Carvedilol treatment might directly act on rods cells. To determine the extent to which carvedilol act directly on rods, we conducted a GloSensor cAMP assay with human Y79 cells. (a) Representative dose–response curves of GloSensor-transfected Y79 cells treated with half-log concentrations of isoproterenol (red trace; N = 4) or percentage-matched DMSO (black trace; N = 4). These plots were normalized to the maximum average luminescent level recorded per experiment. Error bars show ± 1 s.e.m. Isoproterenol was capable of increasing cAMP signaling through β-adrenergic receptor binding with an pEC50 of 7.49 ± 1.07. (b) Representative dose–response curves of GloSensor-transfected Y79 cells pretreated with half-log doses of carvedilol (blue trace; N = 4) or percentage-matched DMSO (red trace; N = 4). Cells were then challenged with a 10 μM isoproterenol that could induce maximal cAMP response, as shown in (a). Carvedilol pretreatment was capable of preventing isoproterenol-mediated cAMP increases with an pIC50 of 6.51 ± 0.67.
Discussion
There are no approved cures to treat the majority of RP subtypes. To address this unmet need, we have utilized the Q344X adRP zebrafish model to establish a drug screening platform that can be expanded into other subtypes of RP. We found that rod degeneration in the Q344X zebrafish resulted in a deficient scotopic light-off VMR, a locomotor response displayed during drastic light offset. This behavior was driven by rods, as it was diminished by chemical ablation of the rods. We leveraged this behavior as a functional assay to screen for beneficial drugs that could enhance the response in the Q344X model. We found that both the scotopic VMR and retinal histology of the Q344X model were improved by carvedilol. Despite showing a clear behavioral response to the scotopic light offset, the carvedilol-treated Q344X did not display a similar scotopic light-off VMR profile compared with WT. This is likely because carvedilol treatment did not restore the rod number or distribution to the WT levels. Because eyeless chokh larvae and enucleated Q344X larvae treated with carvedilol did not display an improved scotopic light-off VMR, it is likely that carvedilol acted at the eye level. Since carvedilol treatment of Q344X larvae shows efficacy and is approved by the FDA, studying its mechanism in the retina can potentially expedite the development of a new treatment for adRP patients.
Carvedilol has several known modes of action. It is primarily classified as a β-blocker; however, it has also been demonstrated to act as an α1-blocker, a calcium channel agonist at high concentration, and a free radical scavenger62,63. Carvedilol may mediate its visual benefit through some of these pathways. Traditionally, β-blockers are seen only as antagonists that prevent epinephrine from binding β-adrenergic receptors. Epinephrine is present in the mouse subretinal space and increases with light exposure64. Blocking epinephrine signaling can potentially lower cAMP levels in the Q344X rods by preventing endogenous ADCY signaling. Interestingly, carvedilol also acts as an atypical β-blocker which is capable of inducing biased signaling65. Specifically, carvedilol can promote β-arrestin signaling while acting as an inverse agonist towards G protein signaling66. This type of β-arrestin signaling has been shown to have anti-apoptotic effects67–69 that may prevent Q344X rod death. This pathway is feasible in the Q344X zebrafish as RNA-seq of adult zebrafish rods detected the expression of β2-adrenergic receptors70. Carvedilol may also exert protective effects on Q344X rods through α1 blockade since selectively blocking Gq-coupled α1 adrenergic receptors can prevent photoreceptor degeneration in a Stargardt Disease model71,72. In our study, it is unlikely that carvedilol is acting as a calcium channel blocker at the tested dose since it would likely stop the larval heart beating and kill the larva before its VMR could be measured73,74. Since our study did not identify any other beneficial compounds from this REDOX library, carvedilol probably did not exert its visual benefit on the Q344X model as a radical-scavenging antioxidant.
More evidence is available which suggests that β-blockers may be able to treat RP. A recent study has identified that another β-blocker, metipranolol, is capable improving rod survival and electroretinogram in the rd10 mouse75. Another study has found that the β-blocker metoprolol can provide protection against bright light-induced retinal degeneration, and metoprolol protection can be increased by co-treatment with other GPCR agonists and antagonists72. Carvedilol has already been shown to have beneficial effects with treating other eye-disease models. Carvedilol can lower intraocular pressure (IOP) in the eye of rabbits76. Also, carvedilol has neuroprotective effects on retinal ganglion cells in an optic nerve injury mouse model77. However, it is unknown if β-blockers such as carvedilol can work directly on rods, so we employed the human Y79 line to determine this. We were able to demonstrate that carvedilol can inhibit isoproterenol-mediated activation of β receptors in the Y79 cells. Therefore, carvedilol likely bound to the β-adrenergic receptors and directly elicited its beneficial effects on rods. In addition, one of carvedilol’s target receptors, the β1-adrenergic receptor, is expressed in mouse rods78. These results suggest that carvedilol may be able to elicit its therapeutic effects directly on the rods, and it targets adrenergic signaling in the eye that may treat Q344X adRP.
Targeting GPCR signaling through adrenergic receptors is an attractive method for the treating Q344X adRP. While the full disease mechanism is unknown, it is believed that mislocalized activation of rhodopsin in the inner segment of Q344X rods induces ADCY activation resulting in cAMP increase and apoptosis15,21,23,24. This highlights ADCY as a potential drug target. Previous work with the Q344X zebrafish, as well as the Stargardt Disease mouse, has shown that inhibition of ADCY with the inhibitor SQ 22,536 improved photoreceptor survival15,71. SQ 22,536 treatment did improve the VMR displayed by Q344X larvae, however the overall resulting VMR was smaller than that of both carvedilol treatment conditions. Therefore, the improved Q344X VMR from carvedilol treatment may be due to other chemical properties of the drug or more efficient uptake of the drug.
We have performed the first functional drug screen for RP and have discovered carvedilol as a positive hit. Drugs identified through the presented screening method may provide a beneficial lead, but the screening parameters may not be the optimal treatment conditions for that particular drug. In the case of carvedilol, we tested a treatment period beginning at 3 dpf and determined that earlier treatment further improved rod survival. Utilizing the behavioral drug screen at 5 dpf and further investigating hits with earlier treatment is an efficient method for identifying the best hits for further translation. Investigating the VMR and rod survival at stages later than 7 dpf becomes more complicated because larval zebrafish deplete their yolk at around 9 dpf and require feeding to survive. Larval feeding and foraging introduce extra variability in the behavioral characterizations of drug effects. It may also be possible to evaluate the effect of positive drug hits by utilizing the OKR behavior of zebrafish larvae. However, it is possible for a mutant larval zebrafish to be light-sensitive and display a VMR while being incapable of displaying an OKR38. Emran et. al.38 found that the nrc mutant zebrafish is light sensitive while failing to produce an OKR due to disruption of ON retinal pathway. Future research will elucidate the mechanism through which these carvedilol-regulated pathways increase rod numbers, and will test carvedilol’s efficacy on the Q344X mouse model79 to validate the translational value of carvedilol for adRP treatment and its capability to improve vision. Positive findings in the Q344X mouse would pave the way for further screening and testing of drugs that modulate the adrenergic system to treat adRP. Our phenotypic drug screen with the Q344X zebrafish lays the foundation for drug screening with fish modeling different classes of RP mutations.
Materials and methods
Animals
Zebrafish of the AB background were utilized for all experiments https://zfin.org/ZDB-GENO-960809-7. The chokh/rx3 zebrafish line (chks399) was utilized in this study58. Adult and larval zebrafish were maintained and bred using standard procedure80. Adult fish were placed in breeding tanks the night before breeding after receiving all meals. Adult fish began spawning at 9:00am, and embryos were collected before 10:30am. Larval zebrafish were reared until 7 days post-fertilization (dpf) in E3 medium in an incubator at 28 °C. The fish incubator was kept on a 14 hr light and 10 hr dark cycle. E3 medium was changed daily, and healthy embryos were kept for experiments. All protocols were approved by the Purdue University Institutional Animal Care and Use Committee. This study was completed in compliance to the ARRIVE guidelines.
Transgenic animals
Tg(rho:Hsa.RH1_Q344X) transgenic animals were generated previously15 and are referred to in this study as Q344X. Q344X larvae were identified on 2 dpf through the expression of EGFP under the control of 1.1 kb promoter of olfactory marker protein (omp) contained in the transgenic cassette. Their genotype was verified via PCR with the following primers: 5′-CCAGCGTGGCATTCTACATC-3′ and 5′-AACGCTTACAATTTACGCCT-3′. The rods in the Q344X line were labeled with the Tg(-3.7rho:EGFP) transgene81 and are referred to in this study as rho:EGFP. Zebrafish expressing nitrodreductase in rod photoreceptors, Tg(-3.7rho:YFP-NfsB)gmc500, were generated previously46 and referred to in this study as rho:NTR. To chemically ablate rods, we used the zebrafish line, Tg(-3.7rho:YFP-NfsB)gmc500, expressing nitroreductase (NTR) under the control of the rhodopsin promotor46 (rho:NTR).
Drug treatment
The ENZO SCREEN-WELL REDOX library was used for drug screening (ENZO Life Sciences, BML-2835-0100). Carvedilol was also ordered from ENZO Life Sciences (BML-AR112-0100) for further experiments and from MilliporeSigma (C3993-50MG) for confirming the positive effects observed in specific behavioral experiments (data not shown). SQ 22,536 was purchased from Sigma (S153-5MG). All drugs tested were dissolved in DMSO. The DMSO percentage that zebrafish larvae were exposed during experiments to was 0.1% except for SQ 22,536 treatment where DMSO exposure was 1%. WT larvae were only exposed to 0.1% DMSO to control for behavioral effects in the Fig. 2a WT dataset. Thirty larvae were exposed per drug dissolved in 15 mL E3 media in a 100 × 15 mm petri dish. The treatment began on 3 dpf or 5 dpf as stated. The drug-containing media were not refreshed during experiment unless otherwise stated. The treated larvae were directly transferred into the 96-well plate with their corresponding E3 medium with drugs to ensure consistent drug dosing throughout the treatment period.
The common starting concentration of 10 μM was chosen to minimize toxic effects while maximizing the chances of finding an effective dose55. Drug screening with the Q344X zebrafish model begun at 5 dpf due to the onset of rod degeneration and the display of a variety of visual behaviors including the VMR and OKR37–40. This stage also was chosen to allow the larvae to developmentally mature to a stage as far as possible to minimize potential toxicity from drugs.
Rod photoreceptor ablation
Treatment with the prodrug metronidazole (MTZ) specifically ablates the rod photoreceptors of the rho:NTR zebrafish line. Specifically, NTR-expressing larvae were treated with 2.5 mM MTZ from 5 to 7 dpf. Their VMR was compared with the untreated larvae on 7 dpf.
Retinal histology and imaging
All larvae were fixed in 4% paraformaldehyde (PFA) overnight at 4 °C. For retinal cryosections, fixed larvae were infiltrated with 30% sucrose overnight at 4 °C prior to imbedding in Tissue Freezing Medium (GeneralData, TFM). Ten micrometers-thick cryosections were collected on Fisherbrand Superfrost Plus Microscope Slides (Thermo Fisher Scientific, 12-550-15). The sections containing the optic nerve were analyzed for anatomical reference. Rod photoreceptors were identified and quantified by utilizing the rho:EGFP transgene as a marker. Since the rods in the ventral retina are present in a high density, high exposure and low exposure images were captured of every cryosection to view the rods in all areas of the retina. A rod was defined as the presence of an identifiable, single soma expressing EGFP. Cryosection images presented in this study are high-exposure to visualize all of the rod signal present in the retinal slice.
Whole-animal preparation
To visualize rod distribution in the retina, PFA-fixed larvae were bleached with 1% KOH/3% H2O2 for 40 min at room temperature to bleach the black pigment from the retinal pigment epithelium. The bleached embryos were imbedded a 3% methyl cellulose solution for observation.
Microscope and camera
All samples were imaged with an Olympus BX51 microscope (Olympus) and a SPOT RT3 Color Slider camera (SPOT Imaging).
Visual motor response assay
A ZebraBox system from ViewPoint Life Sciences was utilized for the Visual Motor Response (VMR) assay. Individual zebrafish larvae were placed in 96-well plate format using Whatman UNIPlate square 96-well plates (VWR, 13503-152). In order to produce a scotopic stimulus, the ZebraBox was modified to attenuate the light intensity beyond its lowest limit by fitting neutral-density filters in the light path. Seven neutral density filters (BarnDoor Film and Video Lighting, E209R), each allowing approximately 40% transmittance, were stacked between the light source and the plate holder until a final intensity of 0.01 lx was attained. In our scotopic experiments, the machine was also powered at 5% in order to prevent instability from the LED light source. The larval displacement was collected by the tracking mode which binned the activity every second.
To conduct the scotopic VMR assay, larvae were sorted and grown in 100 × 15 mm petri dishes (VWR, 25384-088) with 15 mL E3 media in a density of 30 larvae from 2 to 5 dpf. Larvae were transferred to 96-well plates with one larva per well on the morning of 6 dpf and dark adapted overnight. On 7 dpf, the dark-adapted larvae were placed in the ZebraBox and their scotopic VMR was measured. For drug screening with larvae, this procedure was the same except larvae were exposed to drugs in petri dishes on 5 dpf. In this study, the following protocol was used: 30 min in the dark followed by a 60-min scotopic light illumination at 0.01 lx, and then a light offset for 5 min (Fig. 1a). All VMR experiments were conducted on 7 dpf between 9 am and 6 pm to minimize the effect of circadian rhythm on vision82.
Light stimulus intensity
Light intensity of the ZebraBox LED spectrum was measured with a SpectriLight ILT950 Spectroradiometer (International Light Technologies). The total irradiance of the LED stimulus over the entire visible spectrum at 5% power output was 3.2 µW cm−2 (0.0063 µW cm−2 at 500 nm wavelength). The corresponding illuminance was 4.5 lx. The light intensity was further reduced by fitting neutral-density filters in the light path as described above. These neutral-density filters did not alter the color spectrum of the LED light (Supplementary Fig. S1). The light intensity with neutral-density filters was calculated by multiplying the light intensity emitted by the machine with the transmittance of each neutral-density filter. The irradiance of the final scotopic stimulus used in this study was 0.005 µW cm−2 (1.80e−5 µW cm−2 at 500 nm). The corresponding illuminance was 0.01 lx.
Y79 cell culture and cAMP assays
The human Y79 cell line was obtained from American Type Culture Collection (ATCC HTB-18). These cells were cultured in RPMI-1640 Medium (ATCC, 30-2001) with 15% Fetal Bovine Serum (ATCC, 30-2020) at 37 °C with 5% CO2. To measure cAMP levels in the cells, the GloSensor Technology − 22F cAMP plasmid (Promega, E2301) was used with GloSensor Assay Reagent (Promega, E1290). Four million cells were seeded into 10-cm dishes with 10 mL of Opti-MEM Reduced Serum Medium (ThermoFisher, 31,985,062) for transfection. The cells were transfected with 20 µg of GloSensor plasmid utilizing X-tremeGENE HP DNA Transfection Reagent (MilliporeSigma, 6,366,244,001) at a 2:1 ratio of plasmid to X-tremeGENE reagent, according to manufacturer’s instructions. These cells were transfected for 24 h, and then they were transferred back into RPMI for another 24 h. Twenty-five thousand cells were then seeded into a low-volume white 384-well plate per well (Greiner Bio-one, 784,080). The cAMP assay was carried out according to the GloSensor protocol for suspension cells. Cells were either treated with DMSO or carvedilol for 20 min at room temperature prior to treatment with isoproterenol. Luminosity was recorded 20 min after drug or DMSO vehicle addition with a FlexStation 3 Multi-Mode Microplate Reader (Molecular Devices).
Data visualization and statistical analysis
General data and statistics
All standard statistical analyses were performed with R version 3.6.083.
VMR data
Raw data from the VMR assay was processed and extracted by Data Workshop (ViewPoint Life Sciences). Data figures were created using ggplot2 package in R84. The VMR data were normalized for baseline activity, light intensity variation per well, and batch effect (i.e. biological replicate) by linear-regression models as previously described56. Additionally, offset values were added to the normalized activity to prevent negative values in displacement.
To determine if each VMR replicate from drug-treated Q344X larvae during drug screening was consistent with the other replicate, a high-dimensional nonparametric multivariate test57 was performed. This test was chosen because the number of observations (i.e. sample size) for each VMR is less than the dimension of the dataset. The dimension is the length of the time period used in the analysis and the sample size is the number of drug-treated larvae. The High-Dimensional Hypothesis test was implemented in the R package HDtest.
The Hotelling’s T-squared test85 was used to test significant changes in zebrafish displacement from 1 to 30 s after the light change. This test is the multivariate version of the T-test which follows the F-distribution. The test statistic for the Hotelling’s T-squared test is calculated as: where and are the sample size. ρ is the dimension which is the time interval used in the analysis. The Hotelling’s T-squared test was used for VMR analysis due to a number of advantages: 1. The Type I error rate is controlled. 2. The relationship between multiple variables is considered. 3. It can generate an overall conclusion even if multiple (single) t-tests are inconsistent. The null hypothesis for the experiment is the group means for all response variables are equal which means the mean vector of the distance travelled for the two chosen groups are the same . The Hotelling T-squared test analysis was performed on the R package Hotelling with some reshape of the dataset.
Y79 cAMP data
Luminosity data obtained from the Y79 cell line was analyzed and plotted using Graphpad Prism (version 8, GraphPad Software). Data were plotted with the non-linear fit method under “log(agonist) vs. response—Variable slope (three parameters)”. pEC50 (negative log of half maximal effective concentration) and pIC50 (negative log of half maximal inhibitory concentration) were calculated through the above-mentioned non-linear fit.
Supplementary Information
Author contributions
Y.F.L., C.P.P., M. Z. and M. T. conceptualized this study. Y.F.L. and L.G. designed the experiments. M.T. generated and provided the Tg(rho:Hsa.RH1_Q344X) zebrafish line. L.G. performed drug screening with VMR. L.G. and R.J. performed rod imaging through cryosections and whole mount. J.M. and L.Z. contributed the Tg(-3.7rho:YFP-NfsB)gmc500 zebrafish line and performed metronidazole treatment. M.Z., R.X., Y.C., and W.Z. analyzed the VMR data. L.G., M.J.K. and R.M.vR. performed cAMP assays on the Y79 cell line and interpreted the data. L.G., M.T. and Y.F.L. interpreted the results. L.G., and Y.F.L. drafted the manuscript. All authors contributed to the critical revision of the manuscript and approved the final version.
Funding
Logan Ganzen was supported by Grant Numbers TL1 TR001107 and UL1 TR001108 (A. Shekhar, PI) from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award. Rebecca James was partially supported by a Grant-in-Aid from Sigma Xi. Mengrui Zhang, Rui Xie, Yongkai Chen, and Wenxuan Zhong would like to acknowledge the partial support from the National Science Foundation under grants DMS-1903226, DMS-1925066, and the National Institute of Health under grant R01GM122080. Liyun Zhang and Jeff Mumm recieved support from the Wynn-Gund TRAP award from the Foundation Fighting Blindness. Chi Pui Pang was partially supported by a Research Grant (Grant No. 06170896) from the Health and Medical Research Fund, Hong Kong. Mingzhi Zhang is partially supported by Science and Technology Innovation Strategy Special Fund Project of Guangdong Province (project code: [2018]157-46), the Grant for Key Disciplinary Project of Clinical Medicine under the Guangdong High-level University Development Program, China (002-18120304), and LKSF cross-disciplinary research grants (2020LKSFG16B). Richard van Rijn was supported by grants from NIH (R01AA025368, R21AA026949, R21AA026675 R03DA045897) and the Purdue Institute for Drug Discovery. Motokazu Tsujikawa was supported by AMED under Grant Number 19gm1210004 and JSPS KAKENHI Grant Numbers JP 17K11448 and JP 21H03093. Yuk Fai Leung was partially supported by research grants from the Purdue Research Foundation and the International Retinal Research Foundation.
Data availability
The raw zebrafish behavioral data is available on the Harvard Dataverse https://doi.org/10.7910/DVN/JYLWH1. The R scripts to reproduce the analyses and plots reported in this paper are available in GitHub https://github.com/zhanzmr/Zebrafish_Model.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mingzhi Zhang, Email: zmz@jsiec.org.
Motokazu Tsujikawa, Email: moto@ophthal.med.osaka-u.ac.jp.
Yuk Fai Leung, Email: yfleung@purdue.edu.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-89482-z.
References
- 1.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 2.O’Neal TB, Luther EE. Retinitis Pigmentosa. StatPearls. StatPearls Publishing; 2020. [PubMed] [Google Scholar]
- 3.Hamel C. Retinitis pigmentosa. Orphanet J. Rare Dis. 2006;1:40. doi: 10.1186/1750-1172-1-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin. Genet. 2013;84:132–141. doi: 10.1111/cge.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daiger SP, Bowne SJ, Sullivan LS. Genes and mutations causing autosomal dominant retinitis pigmentosa. Cold Spring Harb. Perspect. Med. 2014;5:a017129. doi: 10.1101/cshperspect.a017129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sorrentino FS, Gallenga CE, Bonifazzi C, Perri P. A challenge to the striking genotypic heterogeneity of retinitis pigmentosa: a better understanding of the pathophysiology using the newest genetic strategies. Eye. 2016;30:1542–1548. doi: 10.1038/eye.2016.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frick KD, Roebuck MC, Feldstein JI, McCarty CA, Grover LL. Health services utilization and cost of retinitis pigmentosa. Arch. Ophthalmol. 2012;130:629–634. doi: 10.1001/archophthalmol.2011.2820. [DOI] [PubMed] [Google Scholar]
- 8.National Academies of Sciences Engineering and Medicine . Making Eye Health a Population Health Imperative. National Academies Press; 2016. [PubMed] [Google Scholar]
- 9.Ganzen L, Venkatraman P, Pang CP, Leung YF, Zhang M. Utilizing zebrafish visual behaviors in drug screening for retinal degeneration. Int. J. Mol. Sci. 2017;18:1185. doi: 10.3390/ijms18061185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodrigues GA, et al. Pharmaceutical development of AAV-based gene therapy products for the eye. Pharm. Res. 2019;36:29. doi: 10.1007/s11095-018-2554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brockerhoff SE, Fadool JM. Genetics of photoreceptor degeneration and regeneration in zebrafish. Cell. Mol. Life Sci. 2011;68:651–659. doi: 10.1007/s00018-010-0563-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris AC. The genetics of ocular disorders: insights from the zebrafish. Br. Defects Res. Part C Embryo Today Rev. 2011;93:215–228. doi: 10.1002/bdrc.20211. [DOI] [PubMed] [Google Scholar]
- 13.Link BA, Collery RF. Zebrafish models of retinal disease. Annu. Rev. Vis. Sci. 2015;1:125–153. doi: 10.1146/annurev-vision-082114-035717. [DOI] [PubMed] [Google Scholar]
- 14.Gross JM, Perkins BD. Zebrafish mutants as models for congenital ocular disorders in humans. Mol. Reprod. Dev. 2008;75:547–555. doi: 10.1002/mrd.20831. [DOI] [PubMed] [Google Scholar]
- 15.Nakao T, Tsujikawa M, Notomi S, Ikeda Y, Nishida K. The role of mislocalized phototransduction in photoreceptor cell death of retinitis pigmentosa. PLoS ONE. 2012;7:e32472. doi: 10.1371/journal.pone.0032472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasamoto Y, Nakao T, Matsumura N, Tsujikawa M. Generation of transgenic zebrafish lines with human rhodopsin mutations. Investig. Ophthalmol. Vis. Sci. 2010;51:1354. [Google Scholar]
- 17.Athanasiou D, et al. The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog. Retin. Eye Res. 2018;62:1–23. doi: 10.1016/j.preteyeres.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rossmiller B, Mao H, Lewin AS. Gene therapy in animal models of autosomal dominant retinitis pigmentosa. Mol. Vis. 2012;18:2479–2496. [PMC free article] [PubMed] [Google Scholar]
- 19.Sung CH, et al. Rhodopsin mutations in autosomal dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 1991;88:6481–6485. doi: 10.1073/pnas.88.15.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobson SG, Kemp CM, Sung CH, Nathans J. Retinal function and rhodopsin levels in autosomal dominant retinitis pigmentosa with rhodopsin mutations. Am. J. Ophthalmol. 1991;112:256–271. doi: 10.1016/S0002-9394(14)76726-1. [DOI] [PubMed] [Google Scholar]
- 21.Sung CH, Makino C, Baylor D, Nathans J. A rhodopsin gene mutation responsible for autosomal dominant retinitis pigmentosa results in a protein that is defective in localization to the photoreceptor outer segment. J. Neurosci. 1994;14:5818–5833. doi: 10.1523/JNEUROSCI.14-10-05818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kremmer S, et al. Ocular findings in patients with autosomal dominant retinitis pigmentosa and Cys110Phe, Arg135Gly, and Gln344stop mutations of rhodopsin. Graefes Arch. Clin. Exp. Ophthalmol. 1997;235:575–583. doi: 10.1007/BF00947087. [DOI] [PubMed] [Google Scholar]
- 23.Concepcion F, Chen J. Q344ter mutation causes mislocalization of rhodopsin molecules that are catalytically active: a mouse model of q344ter-induced retinal degeneration. PLoS ONE. 2010;5:e10904. doi: 10.1371/journal.pone.0010904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portera-Cailliau C, Sung CH, Nathans J, Adler R. Apoptotic photoreceptor cell death in mouse models of retinitis pigmentosa. Proc. Natl. Acad. Sci. U.S.A. 1994;91:974–978. doi: 10.1073/pnas.91.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alfinito PD, Townes-Anderson E. Activation of mislocalized opsin kills rod cells: a novel mechanism for rod cell death in retinal disease. Proc. Natl. Acad. Sci. 2002;99:5655–5660. doi: 10.1073/pnas.072557799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tam BM, Xie G, Oprian DD, Moritz OL. Mislocalized rhodopsin does not require activation to cause retinal degeneration and neurite outgrowth in Xenopus laevis. J. Neurosci. 2006;26:203–209. doi: 10.1523/JNEUROSCI.3849-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patton EE, Zon LI. The art and design of genetic screens: zebrafish. Nat. Rev. Genet. 2001;2:956–966. doi: 10.1038/35103567. [DOI] [PubMed] [Google Scholar]
- 28.Fadool JM, Dowling JE. Zebrafish: a model system for the study of eye genetics. Prog. Retin. Eye Res. 2008;27:89–110. doi: 10.1016/j.preteyeres.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dowling JE. The Retina : An Approachable Part of the Brain. Belknap Press of Harvard University Press; 2012. [Google Scholar]
- 30.Morris AC, Fadool JM. Studying rod photoreceptor development in zebrafish. Physiol. Behav. 2005;86:306–313. doi: 10.1016/j.physbeh.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hensley MR, et al. Cellular expression of Smarca4 (Brg1)-regulated genes in zebrafish retinas. BMC Dev. Biol. 2011;11:45. doi: 10.1186/1471-213X-11-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitt EA, Dowling JE. Early retinal development in the zebrafish, Danio rerio: light and electron microscopic analyses. J. Comp. Neurol. 1999;404:515–536. doi: 10.1002/(SICI)1096-9861(19990222)404:4<515::AID-CNE8>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 33.Branchek T, Bremiller R. The development of photoreceptors in the zebrafish, Brachydanio rerio. I. Structure. J. Comp. Neurol. 1984;224:107–115. doi: 10.1002/cne.902240109. [DOI] [PubMed] [Google Scholar]
- 34.Fadool JM. Development of a rod photoreceptor mosaic revealed in transgenic zebrafish. Dev. Biol. 2003;258:277–290. doi: 10.1016/S0012-1606(03)00125-8. [DOI] [PubMed] [Google Scholar]
- 35.Schmitt EA, Dowling JE. Early-eye morphogenesis in the zebrafish, Brachydanio rerio. J. Comp. Neurol. 1994;344:532–542. doi: 10.1002/cne.903440404. [DOI] [PubMed] [Google Scholar]
- 36.Easter SS, Jr, Nicola GN. The development of vision in the zebrafish (Danio rerio) Dev. Biol. 1996;180:646–663. doi: 10.1006/dbio.1996.0335. [DOI] [PubMed] [Google Scholar]
- 37.Easter SS, Nicola GN. The development of eye movements in the zebrafish (Danio rerio) Dev. Psychobiol. 1997;31:267–276. doi: 10.1002/(SICI)1098-2302(199712)31:4<267::AID-DEV4>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 38.Emran F, et al. OFF ganglion cells cannot drive the optokinetic reflex in zebrafish. Proc. Natl. Acad. Sci. 2007;104:19126–19131. doi: 10.1073/pnas.0709337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chhetri J, Jacobson G, Gueven N. Zebrafish–on the move towards ophthalmological research. Eye (Lond). 2014;28:367–380. doi: 10.1038/eye.2014.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y, et al. Statistical analysis of zebrafish locomotor response. PLoS ONE. 2015;10:e0139521. doi: 10.1371/journal.pone.0139521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emran F, Rihel J, Dowling JE. A behavioral assay to measure responsiveness of zebrafish to changes in light intensities. J. Vis. Exp. 2008;20:923. doi: 10.3791/923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y, et al. A high-throughput zebrafish screening method for visual mutants by light-induced locomotor response. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014;11:693–701. doi: 10.1109/TCBB.2014.2306829. [DOI] [PubMed] [Google Scholar]
- 43.Gao Y, et al. Computational classification of different wild-type zebrafish strains based on their variation in light-induced locomotor response. Comput. Biol. Med. 2016;69:1–9. doi: 10.1016/j.compbiomed.2015.11.012. [DOI] [PubMed] [Google Scholar]
- 44.Deeti S, O’Farrell S, Kennedy BN. Early safety assessment of human oculotoxic drugs using the zebrafish visualmotor response. J. Pharmacol. Toxicol. Methods. 2014;69:1–8. doi: 10.1016/j.vascn.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 45.Zhang L, et al. A naturally-derived compound schisandrin b enhanced light sensation in the pde6c zebrafish model of retinal degeneration. PLoS ONE. 2016;11:e0149663. doi: 10.1371/journal.pone.0149663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walker SL, et al. Automated reporter quantification in vivo: high-throughput screening method for reporter-based assays in zebrafish. PLoS ONE. 2012;7:e29916. doi: 10.1371/journal.pone.0029916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White DT, et al. ARQiv-HTS, a versatile whole-organism screening platform enabling in vivo drug discovery at high-throughput rates. Nat. Protoc. 2016;11:2432–2453. doi: 10.1038/nprot.2016.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bilotta J, Saszik S, Sutherland SE. Rod contributions to the electroretinogram of the dark-adapted developing zebrafish. Dev. Dyn. 2001;222:564–570. doi: 10.1002/dvdy.1188. [DOI] [PubMed] [Google Scholar]
- 49.Saade CJ, Alvarez-Delfin K, Fadool JM. Rod photoreceptors protect from cone degeneration-induced retinal remodeling and restore visual responses in zebrafish. J. Neurosci. 2013;33:1804–1814. doi: 10.1523/JNEUROSCI.2910-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moyano M, Porteros Á, Dowling JE. The effects of nicotine on cone and rod b-wave responses in larval zebrafish. Vis. Neurosci. 2013;30:141–145. doi: 10.1017/S0952523813000187. [DOI] [PubMed] [Google Scholar]
- 51.Venkatraman P, et al. Rods contribute to visual behavior in larval zebrafish. Investig. Ophthalmol. Vis. Sci. 2020;61:11. doi: 10.1167/iovs.61.12.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Punzo C, Xiong W, Cepko CL. Loss of daylight vision in retinal degeneration: Are oxidative stress and metabolic dysregulation to blame? J. Biol. Chem. 2012;287:1642–1648. doi: 10.1074/jbc.R111.304428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Titialii K, Morris A. Embryonic hyperglycemia causes a reduction in photoreceptor cells and increased oxidative stress in the developing retina. FASEB J. 2020;34:1. doi: 10.1096/fasebj.2020.34.s1.00336. [DOI] [Google Scholar]
- 54.Li C, et al. CERKL interacts with mitochondrial TRX2 and protects retinal cells from oxidative stress-induced apoptosis. Biochim. Biophys. Acta Mol. Basis Dis. 2014;1842:1121–1129. doi: 10.1016/j.bbadis.2014.04.009. [DOI] [PubMed] [Google Scholar]
- 55.Wiley DS, Redfield SE, Zon LI. Chemical screening in zebrafish for novel biological and therapeutic discovery. Methods Cell Biol. 2017;138:651–679. doi: 10.1016/bs.mcb.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xie R, et al. Normalization of large-scale behavioural data collected from zebrafish. PLoS ONE. 2019;14:e0212234. doi: 10.1371/journal.pone.0212234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang J, Zheng C, Zhou WX, Zhou W. Simulation-based hypothesis testing of high dimensional means under covariance heterogeneity. Biometrics. 2017;73:1300–1310. doi: 10.1111/biom.12695. [DOI] [PubMed] [Google Scholar]
- 58.Loosli F, et al. Loss of eyes in zebrafish caused by mutation of chokh/rx3. EMBO Rep. 2003;4:894–899. doi: 10.1038/sj.embor.embor919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Polo A, Farber DB. Rod photoreceptor-specific gene expression in human retinoblastoma cells. Proc. Natl. Acad. Sci. 1995;92:4016–4020. doi: 10.1073/pnas.92.9.4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanno C, et al. Activin inhibits cell growth and induces differentiation in human retinoblastoma Y79 cells. Curr. Eye Res. 2009;34:652–659. doi: 10.1080/02713680903007147. [DOI] [PubMed] [Google Scholar]
- 61.Oura Y, et al. High-temperature requirement A 1 causes photoreceptor cell death in zebrafish disease models. Am. J. Pathol. 2018;188:2729–2744. doi: 10.1016/j.ajpath.2018.08.012. [DOI] [PubMed] [Google Scholar]
- 62.Giannattasio C, et al. α1-Blocking properties of carvedilol during acute and chronic administration. J. Cardiovasc. Pharmacol. 1992;19:S18–S22. doi: 10.1097/00005344-199219001-00005. [DOI] [PubMed] [Google Scholar]
- 63.McTavish D, Campoli-Richards D, Sorkin EM. Carvedilol. A review of its pharmacodynamic and pharmacokinetic properties, and therapeutic efficacy. Drugs. 1993;45:232–258. doi: 10.2165/00003495-199345020-00006. [DOI] [PubMed] [Google Scholar]
- 64.Hadjiconstantinou M, Cohen J, Neff NH. Epinephrine: a potential neurotransmitter in retina. J. Neurochem. 1983;41:1440–1444. doi: 10.1111/j.1471-4159.1983.tb00843.x. [DOI] [PubMed] [Google Scholar]
- 65.Wisler JW, et al. A unique mechanism of beta-blocker action: carvedilol stimulates beta-arrestin signaling. Proc. Natl. Acad. Sci. 2007;104:16657–16662. doi: 10.1073/pnas.0707936104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, et al. Gαi is required for carvedilol-induced β1 adrenergic receptor β-arrestin biased signaling. Nat. Commun. 2017;8:1706. doi: 10.1038/s41467-017-01855-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.DeFea KA, et al. beta-arrestin-dependent endocytosis of proteinase-activated receptor 2 is required for intracellular targeting of activated ERK1/2. J. Cell Biol. 2000;148:1267–1281. doi: 10.1083/jcb.148.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Povsic TJ, Kohout TA, Lefkowitz RJ. β-Arrestin1 mediates insulin-like growth factor 1 (IGF-1) activation of phosphatidylinositol 3-kinase (PI3K) and anti-apoptosis. J. Biol. Chem. 2003;278:51334–51339. doi: 10.1074/jbc.M309968200. [DOI] [PubMed] [Google Scholar]
- 69.Ahn S, Kim J, Hara MR, Ren X-R, Lefkowitz RJ. β-Arrestin-2 mediates anti-apoptotic signaling through regulation of BAD phosphorylation. J. Biol. Chem. 2009;284:8855–8865. doi: 10.1074/jbc.M808463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun C, Galicia C, Stenkamp DL. Transcripts within rod photoreceptors of the Zebrafish retina. BMC Genom. 2018;19:127. doi: 10.1186/s12864-018-4499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen Y, et al. Systems pharmacology identifies drug targets for stargardt disease–associated retinal degeneration. J. Clin. Investig. 2013;123:5119–5134. doi: 10.1172/JCI69076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen Y, et al. Synergistically acting agonists and antagonists of G protein–coupled receptors prevent photoreceptor cell degeneration. Sci. Signal. 2016;9:ra74. doi: 10.1126/scisignal.aag0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chopra SS, et al. Voltage-gated sodium channels are required for heart development in zebrafish. Circ. Res. 2010;106:1342–1350. doi: 10.1161/CIRCRESAHA.109.213132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stainier DYR. Zebrafish genetics and vertebrate heart formation. Nat. Rev. Genet. 2001;2:39–48. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- 75.Kanan Y, et al. Metipranolol promotes structure and function of retinal photoreceptors in the rd10 mouse model of human retinitis pigmentosa. J. Neurochem. 2019;148:307–318. doi: 10.1111/jnc.14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakaya M, et al. Induction of cardiac fibrosis by β-blocker in G protein-independent and G protein-coupled receptor kinase 5/β-arrestin2-dependent signaling pathways. J. Biol. Chem. 2012;287:35669–35677. doi: 10.1074/jbc.M112.357871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu B, Liu Y-J. Carvedilol promotes retinal ganglion cell survival following optic nerve injury via ASK1-p38 MAPK pathway. CNS Neurol. Disord. Drug Targets. 2019;18:695–704. doi: 10.2174/1871527318666191002095456. [DOI] [PubMed] [Google Scholar]
- 78.Siegert S, et al. Transcriptional code and disease map for adult retinal cell types. Nat. Neurosci. 2012;15:487–495. doi: 10.1038/nn.3032. [DOI] [PubMed] [Google Scholar]
- 79.Sandoval IM, et al. Abrupt onset of mutations in a developmentally regulated gene during terminal differentiation of post-mitotic photoreceptor neurons in mice. PLoS ONE. 2014;9:e108135. doi: 10.1371/journal.pone.0108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Westerfield M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio rerio) 5. University of Oregon Press; 2007. [Google Scholar]
- 81.Hamaoka T, Takechi M, Chinen A, Nishiwaki Y, Kawamura S. Visualization of rod photoreceptor development using GFP-transgenic zebrafish. Genesis. 2002;34:215–220. doi: 10.1002/gene.10155. [DOI] [PubMed] [Google Scholar]
- 82.Emran F, Rihel J, Adolph AR, Dowling JE. Zebrafish larvae lose vision at night. Proc. Natl. Acad. Sci. 2010;107:6034–6039. doi: 10.1073/pnas.0914718107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.R Core Team (2020). R: A language and environment for statistical computing. R: A language and environment for statistical computing. (R Foundation for Statistical Computing, 2020).
- 84.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; 2016. [Google Scholar]
- 85.Hotelling H. The Generalization of Student’s Ratio. Ann. Math. Stat. 1931;2:360–378. doi: 10.1214/aoms/1177732979. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw zebrafish behavioral data is available on the Harvard Dataverse https://doi.org/10.7910/DVN/JYLWH1. The R scripts to reproduce the analyses and plots reported in this paper are available in GitHub https://github.com/zhanzmr/Zebrafish_Model.