Learning Objectives
Upon completion of this activity, participants will:
Describe plasma microRNAs (miRNAs) validated with qualitative reverse-transcriptase polymerase chain reaction in patients with type 2 diabetes with or without diabetic retinopathy (DR), and their correlation with neurodegeneration, according to retinal nerve fibre layer (RNFL) thickness measured using spectral-domain optical coherence tomography.
Determine gene targets of miRNA and mechanisms underlying the pathogenic process of DR, according to bioinformatic analysis used to predict potential targets of miRNA associated with RNFL thickness and to investigate the functions of the potential target genes.
Identify clinical implications of bioinformatic analysis of miRNAs in DR and their gene targets.
Continuing Medical Education
In support of improving patient care, this activity has been planned and implemented by Medscape, LLC and Springer Nature. Medscape, LLC is jointly accredited by the Accreditation Council for Continuing Medical Education (ACCME), the Accreditation Council for Pharmacy Education (ACPE), and the American Nurses Credentialing Center (ANCC), to provide continuing education for the healthcare team.
Medscape, LLC designates this Journal-based CME activity for a maximum of 1.0 AMA PRA Category 1 Credit(s)TM. Physicians should claim only the credit commensurate with the extent of their participation in the activity.
Successful completion of this CME activity, which includes participation in the evaluation component, enables the participant to earn up to 1.0 MOC points in the American Board of Internal Medicine’s (ABIM) Maintenance of Certification (MOC) programme. Participants will earn MOC points equivalent to the amount of CME credits claimed for the activity. It is the CME activity provider’s responsibility to submit participant completion information to ACCME for the purpose of granting ABIM MOC credit.
All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test with a 75% minimum passing score and complete the evaluation at www.medscape.org/journal/eye; (4) view/print certificate.
Credit hours
1.0
Release date:
Expiration date:
Post-test link:
https://medscape.org/eye/posttest943521
Authors/Editors disclosure information
SS has disclosed the following relevant financial relationships: Served as an advisor or consultant for: Allergan, Inc.; Apellis; Bayer AG; Boehringer Ingelheim Pharmaceuticals, Inc.; Heidelberg Pharma GmbH; Novartis; Oculis; Optos; Oxurion; Roche; Served as a speaker or a member of a speakers bureau for: Allergan, Inc.; Bayer AG; Novartis Pharmaceuticals Corporation; Optos; Received grants for clinical research from: Allergan, Inc.; Bayer AG; Boehringer Ingelheim Pharmaceuticals, Inc.; Novartis Pharmaceuticals Corporation; Optos. RS, LC, WW, YD, YZL, HZ. RL have disclosed no relevant financial relationships.
Journal CME author disclosure information
Laurie Barclay has disclosed no relevant financial relationships.
Subject terms: Retina, Prognostic markers
Abstract
Purpose
Retinal neurodegeneration is an early pathological change in diabetic retinopathy (DR). Early-stage retinal neurodegeneration is usually asymptomatic. This study aims to identify circulating microRNAs (miRNAs) as sensitive biomarkers for early retinal neurodegeneration.
Methods
We profiled the plasma miRNA expression in three mild nonproliferative diabetic retinopathy (NPDR) cases and three matched non-DR patients using RNA sequencing. The differential miRNAs were validated with qRT-PCR. The retinal nerve fibre layer (RNFL) thickness of the eyes was measured using spectral-domain Optical coherence tomography (SD-OCT). The association between differential miRNAs and RNFL thickness was analysed using the Pearson correlation analysis. Bioinformatics tools were used to predict potential targets of miRNA associated with RNFL thickness and investigate the functions of the potential target genes.
Results
RNA sequencing identified 69 differential miRNAs and eight of them were reported to be associated with DR. The qRT-PCR for these eight miRNAs validated the down-regulation of circulating miR-26a-5p and miR-126-5p in a larger validating cohort. A positive correlation between plasma miR-26a-5p level and the RNFL thickness of the superior quadrant of both eyes was identified in another cohort, including 33 mild NPDR cases, 33 matched non-DR patients and 20 healthy controls. Furthermore, 367 candidate targets of miR-26a-5p were predicted. The functional studies revealed that these target genes are profoundly involved in various cellular functions and signalling pathways.
Conclusions
Circulating miR-26a-5p is a potential biomarker for early-stage retinal neurodegeneration and it may be involved in the development of DR via profoundly influencing the functions of retinal cells.
Introduction
Diabetic retinopathy (DR) is a multifactorial progressive disease of the retina, which has been considered a leading cause of irreversible blindness in patients with diabetes mellitus [1]. The activation of the retinal glial cells, apoptotic cell death, neuroinflammation and gradual neurodegeneration induced by hyperglycaemic environment play important roles in the pathogenesis of DR [2]. Concomitantly with the development of diabetes, the metabolic disorder results in the abnormalities of the microvascular, breakdown of the blood–retinal barrier, impairments in neurovascular interactions, central vision loss, proliferative retinopathy, macular oedema and retinal layer thinning [3, 4]. These changes ultimately lead to progression into advanced DR. Many studies in recent years have paid attention to retinal neurodegeneration, which is the earliest pathology and may precede microvascular changes that are characteristic of DR [5–8]. It has been proven that the retinal nerve fibre layer (RNFL) thinning is a manifestation of diabetic optic neuropathy [9–11]. However, the potential molecular mechanisms underlying these pathologies remain elusive. Besides, although current therapies, such as laser photocoagulation and anti-vascular endothelial growth factor (VEGF) injections, are available for late stages of DR with proliferation and macular oedema, effective strategies still lacked to prevent or reduce vision loss in the future [12]. Therefore, it is imperative to identify novel biomarkers and therapeutic targets to improve the therapeutic effects of early DR.
MicroRNAs (miRNAs) are small non-coding RNA molecules that orchestrate biological networks via modulation of protein expression by interfering with the translation and stability of mRNA. Some miRNAs are released into the circulation and can exist stably in plasma, urine, and other body fluids [13]. The availability of quantitative detection of circulating miRNAs enhances the use of miRNAs as biomarkers for physiological and pathological processes [14, 15]. Numerous clinical and experimental studies have demonstrated the important roles of miRNAs in the development of diabetes and its complications [16]. Additionally, accumulating evidence has indicated that various circulating miRNAs can serve as potential biomarkers or therapeutic targets for DR [17, 18]. However, the relationship between plasma miRNA and diabetic neurodegeneration remains unclear. We speculate that circulating miRNAs may play an important role in the pathogenesis of retinal neurodegeneration in patients with or without DR.
In the present study, we screened the plasma miRNAs in type 2 diabetic patients with or without DR and investigated the neurodegeneration-related miRNAs for correlation analysis. Then we explored the targets of miRNA and elucidated the underlying mechanism in mediating the pathogenic process of DR by bioinformatic analysis.
Materials and methods
Ethics statement
This study was approved by the Institutional Ethics Committee of the Xi’an Jiaotong University, and written informed consent was obtained from each participant. The study adhered to the tenets of the Declaration of Helsinki.
Study design
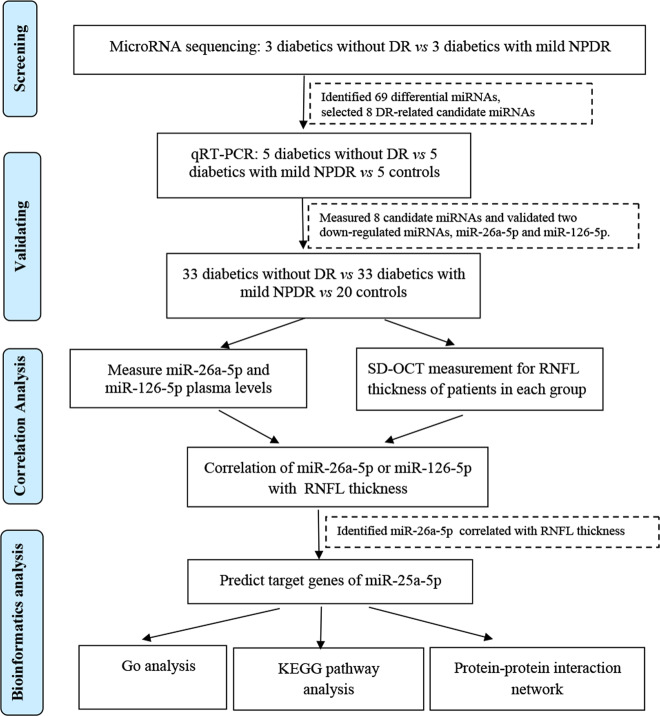
The design of this study is summarized in Fig. 1. In the screening stage, 3 non-DR (NDR) and 3 non-progressive DR (NPDR) plasma samples were subjected for microRNA sequencing to identify the miRNAs that were differentially expressed between type 2 diabetes patients without DR and those with mild NPDR. Thereafter, 33 patients without DR (group 1), 33 patients with mild DR (group 2) and 20 healthy participants (group 3) were included for individual qRT-PCR to further filter signals of the screened miRNAs. The significant differential miRNAs were included in the following experiments. Target genes of the included miRNAs were predicted and the relationship of target miRNAs, target genes and RNFL thickness were assessed with correlation analysis.
Fig. 1. The flow diagram of the study design.
microRNA sequencing was validated with qRT-PCR, and then correlation and bioinformatic analysis was used to identify circulating miRNAs associated with early retinal neurodegeneration. DR: diabetic retinopathy; NPDR: nonproliferative diabetic retinopathy.
Participants and grouping
All patients were recruited from the Department of Ophthalmology at the Shaanxi provincial peoples’ hospital between Jan 1, 2017, and March 31, 2017. The included patients in this study were diagnosed as type 2 diabetes for at least 1 year, without DR or with mild NPDR, and had visual acuity of 6/10 or better. Mild DR was defined as microaneurysms and haemorrhagic spots. The exclusion criteria were as following: diagnosed as any kind of glaucoma or had glaucoma family history; have a cup-to-disc (C/D) ratio of more than 0.5 and a difference in C/D ratio between both eyes that exceeds 0.2; have any kind of coexisting neuro-ophthalmic disease or such a history; with a refractive error of more than the spherical equivalent (SE)+3 or SE−3 dioptres in at least one eye; with axis length (AL) > 25 mm; have a history of refractive surgery or intraocular surgery; have significant media opacity; with a history of uveitis or retinal disease; diagnosed as type 1 diabetes. The recruited patients were divided into three groups based on their DR status: 33 patients without DR were assigned into group 1; 33 patients with mild NPDR were classified into group 2 and 20 healthy people were included as controls (group 3).
Ophthalmological examinations
All patients were identified with careful ophthalmological examinations of both eyes by professional ophthalmologists, including visual acuity, slit-lamp examination, fundus contact lens examination, and spectral domain Optical coherence tomography (SD-OCT) measurement. A detailed review of medical and ocular histories was requested for each subject. The fundus was examined with a handheld lens (90D Volk Optical) and the fundus camera. AL was measured by IOLMaster. OCT imaging system (3D OCT-1 (ver.8.30); Topcon Corporation, Japan) was used to measure peripapillary RNFL thickness with 3D scans (6.0 × 6.0 mm, 512 × 128) after pupillary dilation. Only well-focused, well-centred images without eye movements and with quality strength of 25 or more were selected for further analyses. The total, superior, inferior, nasal and temporal RNFL thickness was calculated by the software and then subjected to analyses.
RNA isolation and microRNA-seq
Up to 5 ml of whole fasting blood was collected into a non-anticoagulant tube from each participant before any therapeutic procedures. Upon collection, each blood sample was immediately centrifuged at 3000 × g for 10 min at room temperature and then centrifuged at 10,000 × g for 5 min at 4 °C. The total RNA was extracted from these samples using TRIzol LS Reagent and then was used to prepare the miRNA sequencing library. The library construction included the following steps: (1) 3′-adaptor ligation; (2) 5′-adaptor ligation; (3) cDNA synthesis; (4) PCR amplification; and (5) size selection for ~135–155 bp PCR-amplified fragments (corresponding to ~15–35 nt small RNAs). The libraries were denatured into single-stranded DNA molecules, which were captured on Illumina flow cells, amplified in situ as clusters and finally sequenced for 50 cycles on Illumina Next Seq per the manufacturer’s instructions.
After sequencing, the Solexa CHASTITY quality-filtered reads were harvested as Clean Reads. The adaptor sequences were trimmed and the adaptor-trimmed-reads (≥15nt) were left. miRDeep2 software was used to predict the novel miRNAs with these trimmed reads. Then, the trimmed reads were aligned to merged pre-miRNA databases (known pre-miRNA from miRBase v21 plus the newly predicted pre-miRNAs) using the Novoalign software (v2.07.11) with at most one mismatch. Reads with counts <2 were discarded when calculating the miRNA expression. To characterize the isomiR variability, sequences that matched the miRNA precursors in the mature miRNAs region ±4 it (no more than 1 mismatch) were accepted as mature miRNA isomiRs, which were grouped according to the 5-prime (5p) or 3-prime (3p) arm of the precursor hairpin. The numbers of mapped tags that were defined as the raw expression levels of that miRNA. To correct for the difference in tag counts between samples, the tag counts were scaled to TPM (the copy number of transcripts per million) based on the total number of tag aligned. Choosing a different isomiR sequence for measuring miRNA expression can affect the ability to detect differential miRNA expression. We use the most abundant isomiR, the mature miRNA annotated in miRBase and all isoforms of miRNA (5p or 3p) to calculate the miRNAs expression. When comparing the differentially expressed miRNA profiles between two groups, fold change and p-value were calculated and used to identify significantly differentially expressed miRNAs (based on ALL_Isoform value). Differentially expressed miRNAs between two groups of samples were filtered through Fold change (based on ALL_Isoform value). Hierarchical clustering was performed.
Measure miRNA levels using quantitative real-time PCR
The expression levels of selected differential miRNAs were measured by qRT-PCR using the SYBR Premix Ex Taq kit (Takara, Tokyo, Japan) according to the manufacturer’s instructions. U6 snRNA was used as the endogenous control. The sequences of primers used for qPCR were presented in Table 1. The PCR amplification protocol was as follows: 95 °C for 2 min, 45 cycles of denaturation at 95 °C for 10 s, followed by 34 s of annealing/extension at 60 °C. Each reaction was performed in triplicate. The relative expression of miRNAs was calculated using the 2−ΔΔCT method.
Table 1.
The sequences of primers used for qRT-PCR.
| miRNA | Primer (5′–3′) |
|---|---|
| hsa-miR-26a-5p |
GSP:5′GGGTTCAAGTAATCCAGG3′ R:5′TGCGTGTCGTGGAGTC3′ |
| hsa-miR-126-3p |
GSP:5′GGGGTCGTACCGTGAGTAAT3′ R:5′GTGCGTGTCGTGGAGTCG3′ |
| hsa-miR-378a-3p |
GSP:5′GGGGTCTGGACTTGGAGTCA3′ R:5′GTGCGTGTCGTGGAGTCG3′ |
| hsa-miR-146b-5p |
GSP:5′GGGGTGAGAACTGAATTCCA3′ R:5′GTGCGTGTCGTGGAGTCG3′ |
| hsa-miR-128-3p |
GSP:5′GGGGAATCACAGTGAACCG3′ R:5′GTGCGTGTCGTGGAGTCG3′ |
| hsa-miR-185-5p |
GSP:5′GGTGGAGAGAAAGGCAGT3′ R:5′TGCGTGTCGTGGAGTC3′ |
| hsa-miR-199a-5p |
GSP:5′GGTGCCCAGTGTTCAGAC3′ R:5′CAGTGCGTGTCGTGGAGT3′ |
| hsa-let-7a-5p |
GSP:5′GGGGGTGAGGTAGTAGGTTGT3′ R:5′GTGCGTGTCGTGGAGTCG3′ |
| hsa-miR-93-5p |
GSP:5′GGCAAAGTGCTGTTCGTG3′ R:5′CAGTGCGTGTCGTGGAGT3′ |
Target gene prediction and gene ontology analysis
TargetScan human 7.1 (http://www.targetscan.org/) and Mirdb V5 (http://www.mirdb.org/) were used to predict the target transcripts of miR-26a-5p. Venny was applied to calculate the intersection of genes. The biological functions of the predicted target genes were evaluated by Gene Ontology (GO) analysis on Enrich (http://amp.pharm.mssm.edu/Enrichr/index.html). For each GO term, the P-value of function clustering and the p-value following multiple detecting corrections, such as Benjamini correction or false discovery rate (FDR) correction, were calculated.
Pathway-enrichment analysis and construction of protein–protein interaction (PPI) network
The Kyoto Encyclopedia of Genes and Genomes (KEGG) is a knowledge base for the systematic analysis of gene functions and links genomic information with higher-order functional information. In this study, pathway-enrichment analysis was performed using Enrichr and the significantly enriched pathways were identified with a P-value of <0.05. The PPI network of proteins was analysed using the Search Tool for the Retrieval of Interacting Genes database (STRING, http://string-db.org/). STRING is an open-access server that provides comprehensive coverage and ease of access to the analysis of gene–gene or PPIs by using four different sources. The list of predicted target genes was uploaded to STRING. Subsequently, PPI networks in STRING were merged and analysed with Cytoscape, an online visualization platform that enables network analysis.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 7 (GraphPad Software Inc., USA). Differences among the groups were assessed using a general linear model, taking into account the eventual variance heterogeneity. Data were presented as mean ± standard deviation (SD) of each group. One-way ANOVA was carried out to compare the means of different groups and multiple comparisons were performed with Bonferroni test. Pearson correlation analysis was used to assess the association between differential miRNAs and RNFL thickness. All tests were two-sided tests and P < 0.05 was considered as statistically significant.
Results
Characteristics of the subjects
A total of 86 participants including 33 diabetic patients without DR (NDR, groups 1), 33 diabetic patients with mild NPDR (group 2) and 20 healthy controls (group 3) were recruited in this study. The patients with mild NPDR had significantly longer diabetic duration than those without DR. Both NDR and NPDR groups had a significantly higher HbA1c level than the healthy controls, while these two groups of patients did not have a significantly different HbA1c level. No significant differences were discovered among the three groups in terms of age, body mass index (BMI), blood lipid profile, axial length and intraocular pressure of the eyes. The basic clinical and laboratory characteristics of each group are summarized in Table 2.
Table 2.
Comparisons of the characteristics of patients according to DR status.
| Group 1 (NDR) |
Group 2 (mild NPDR) |
Group 3 (healthy controls) | F | P | |
|---|---|---|---|---|---|
| Participants (n) | 33 | 33 | 20 | – | – |
| Gender (M/W) | 13/10 | 12/11 | 11/9 | – | – |
| Age (yrs) | 54.03 ± 13.25 | 55.12 ± 14.09 | 52.35 ± 15.77 | 0.237 | 0.789 |
| Diabetes duration (yrs) | 7.62 ± 6.21 | 11.37 ± 4.26* | – | 2.861 | 0.005 |
| BMI (kg/m2) | 24.72 ± 3.01 | 24.83 ± 3.27 | 23.19 ± 2.42 | 2.164 | 0.121 |
| HbA1c (%) | 7.19 ± 2.15*** | 7.94 ± 2.65*** | 4.14 ± 1.36 | 19.27 | 0.0001 |
| Total cholesterol (mmol/l) | 4.16 ± 1.20 | 4.22 ± 1.13 | 4.13 ± 1.12 | 0.042 | 0.958 |
| Triglyceride (mmol/l) | 2.24 ± 1.57 | 2.08 ± 1.26 | 2.15 ± 1.17 | 0.113 | 0.893 |
| LDL-C (mmol/l) | 2.51 ± 0.93 | 2.54 ± 0.85 | 2.48 ± 0.67 | 0.032 | 0.968 |
| HDL-C (mmol/l) | 1.29 ± 0.44 | 1.22 ± 0.31 | 1.26 ± 0.24 | 0.325 | 0.723 |
| AL of right eye (mm) | 23.39 ± 0.61 | 23.36 ± 0.48 | 23.35 ± 0.75 | 0.033 | 0.966 |
| AL of left eye (mm) | 23.48 ± 0.60 | 23.42 ± 0.57 | 23.34 ± 0.58 | 0.359 | 0.699 |
| IOP of right eye (mmHg) | 19.87 ± 3.96 | 18.44 ± 3.70 | 19.02 ± 3.51 | 1.203 | 0.305 |
| IOP of left eye (mmHg) | 19.74 ± 3.68 | 18.99 ± 4.67 | 19.76 ± 4.05 | 0.335 | 0.715 |
Data represented the mean ± standard deviation (SD) of each group. For diabetes duration, * represents comparisons between groups 1 and 2. For HbA1c, * represents the comparisons of group 1 or 2 to group 3. ***P < 0.001.
DR diabetic retinopathy, M/W man/woman, BMI body mass index, HbA1c glycated haemoglobin, HDL-C high-density lipoprotein cholesterol, LDL-C low-density lipoprotein cholesterol, RNFL retinal nerve fibre layer, ACR urinary albumin-to-creatinine ratio, AL axial length, IOP intraocular pressure.
Statistically significant p-values are in bold.
RNA-seq and validation of differential miRNAs
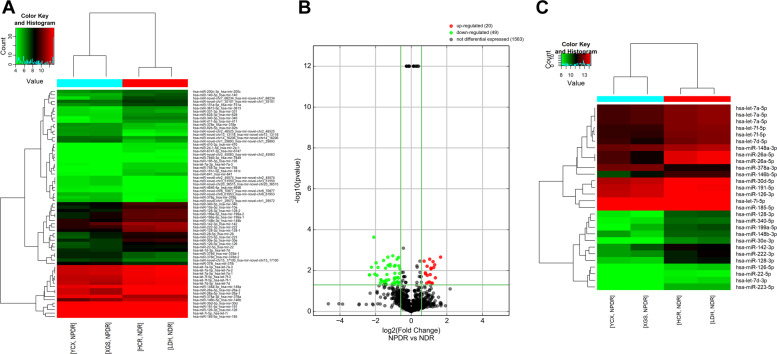
To identify a plasma miRNA signature specific for early-stage DR, a microRNA sequencing assay was performed to identify the differentially expressed miRNAs between three mild NPDR cases and three NDR cases in the initial screening phase. However, because of the sample heterogeneity, only 2 mild NPDR and 2 NDR samples were included for the final analysis. Among the 4158 miRNAs that were analysed, 20 up-regulated and 49 down-regulated miRNAs were identified with the threshold of 1.5 folds change in expression and a P-value < 0.05. The differential miRNAs are hierarchically illustrated by a heatmap (Fig. 2A) and a volcano plot (Fig. 2B). Furthermore, among the differential miRNAs, 28 miRNAs had relatively high abundance in the plasma (RPKM > 200, two up-regulated and 26 down-regulated) (Fig. 2C). Literature search revealed that eight miRNAs among the abundant differential miRNAs are involved in diabetes, inflammation and cell proliferation, including hsa-miR-185-5p, hsa-miR-378a-3p, hsa-miR-199a-5p, hsa-miR-128-3p, hsa-let-7a-5p, hsa-miR-146b-5p, miR-26a-5p, and hsa-miR-126-3p. To validate the results of miRNA sequencing, we measured the expression of the eight candidate miRNAs by qRT-PCR in a validating cohort (5 patients with PDR, 5 patients with NPDR and 5 with healthy controls). The results validated that miR-26a-5p and miR-126-5p were significantly down-regulated in the PDR and NPDR groups compared with healthy controls (P < 0.01 and 0.05, respectively) (Table 3 and Fig. 3).
Fig. 2. Plasma miRNA profiles of NPDR and NDR subjects.
A A heatmap shows the miRNAs with >1.5 fold changes in expression. The miRNAs are hierarchically clustered on the y-axis based on their expression. B A volcano plot showing RNA sequencing results for the comparison of mild NPDR and NDR patients. The line indicates a false discovery rate (FDR)–adjusted P-value of 0.05. Individual microRNAs are displayed by their FDR-adjusted P-value. The up-regulated miRNAs are shown in red, while the down-regulated miRNAs are in green. C A heatmap shows the 28 differential miRNAs that are reported to be related to the development of DR. The expression index is colour-coded with green indicating down-regulation and red indicating up-regulation.
Table 3.
Eight DR-related dysregulated miRNAs detected by miRNA sequencing.
| Dysregulation | miRNA | PRE-ACC | miRNA expression (rpkm) | Fold change | P-value | |
|---|---|---|---|---|---|---|
| NPDR | NDR | |||||
| Up-regulated | hsa-miR-185-5p | MI0000482 | 56,518.5 | 23,555 | 2.399 | 0.0071 |
| Up-regulated | hsa-miR-378a-3p | MI0000786 | 6602 | 2820 | 2.341 | 0.0037 |
| Down-regulated | hsa-miR-199a-5p | MI0000242 | 354.5 | 1151 | 0.308 | 0.0279 |
| Down-regulated | hsa-miR-128-3p | MI0000447 | 405.5 | 1902.5 | 0.213 | 0.0068 |
| Down-regulated | hsa-let-7a-5p | MI0000060 | 4430 | 7599.5 | 0.583 | 0.0361 |
| Down-regulated | hsa-miR-146b-5p | MI0003129 | 1630 | 3919 | 0.416 | 0.0280 |
| Down-regulated | miR-26a-5p | MI0000083 | 3569.5 | 18,071.5 | 0.198 | 0.0162 |
| Down-regulated | hsa-miR-126-3p | MI0000471 | 13,511 | 57,936 | 0.233 | 0.0002 |
PRE-ACC the pre-miRNA ACCESSION, NPDR non-proliferative diabetic retinopathy, NDR no diabetic retinopathy.
Fig. 3. Expression levels of candidate miRNAs in the plasma of patients.
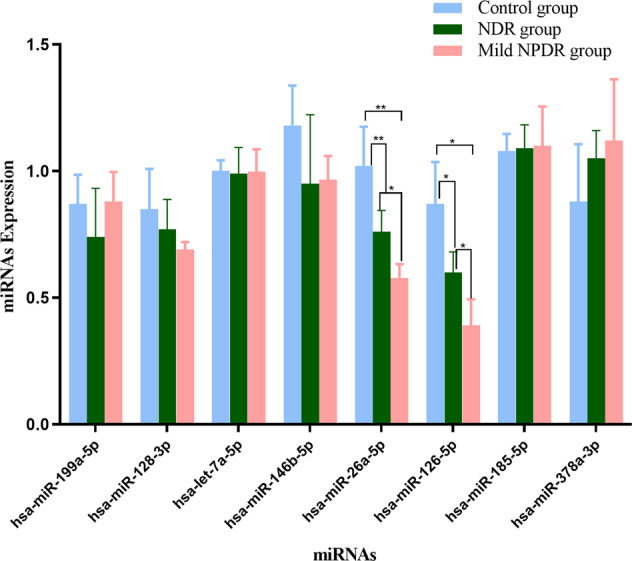
The eight candidate miRNAs were evaluated with qRT-PCR in 15 samples, including 5 patients with PDR, 5 patients with NPDR, and 5 healthy controls. *P < 0.05, **P < 0.01.
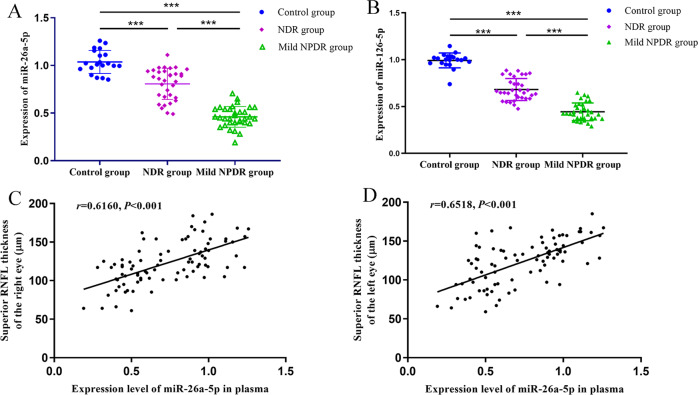
Plasma levels of miR-26a-5p and miR-126-5p
To further validate the down-regulation of miR-26a-5p and miR-126-5p in early-stage DR, qRT-PCR was applied to measure the plasma levels of these two miRNAs in all participants, including 33 cases of NDR, 33 cases of NPDR and 20 healthy controls. As shown in Fig. 4A, the plasma level of miR-26a-5p in NDR and NPDR patients was significantly lower than that of the healthy controls (P < 0.001), and the plasma miR-26a-5p level was further decreased in the NPDR group compared with the NDR group (P < 0.001). Similar results were observed for miR-126-5p, as its plasma level in patients with type 2 diabetes (NDR and NPDR groups) was significantly down-regulated compared with the healthy controls (P < 0.001) and the NPDR patients had further lower circulating miR-126-5p than the NDR patients (P < 0.001) (Fig. 4B). The qRT-PCR results of these two miRNAs are consistent with the results of miRNA sequencing.
Fig. 4. Expression levels of miR-26a-5p and miR-126-5p in patients with or without NPDR.
A miR-26a-5p and B miR-126-5p expressions were measured with qRT-PCR in 86 serum samples (33 patients without DR, 33 patients with NPDR, and 20 healthy controls). C The correlation between miR-26a-5p expression and the RNFL thickness of the right eye. D The association of plasma miR-26a-5p level and RNFL thickness of the left eye was evaluated using the Pearson correlation analysis. A significant positive correlation was observed between plasma miR-26a-5p and RNFL thickness in the upper quadrant of both eyes.***P < 0.001.
Correlation between RNFL thickness and plasma miRNAs
The RNFL thickness of each quadrant of both eyes was then compared among different groups. The results showed that the NPDR but not the NDR patients had significantly thinner RNFL in the superior quadrant of both eyes than the healthy controls, while the superior RNFL thickness of the NDR and the NPDR groups showed no significant difference. However, the RNFL thickness in the inferior, nasal and temporal quadrants of both eyes showed no significant difference among the three groups (Table 4). These data suggest that RNFL thinning in the superior quadrant of the eyes is the earliest change in retinal degeneration in DR.
Table 4.
Peripapillary RNFL thickness of the subjects.
| Eyes | Group 1 (n = 33) |
Group 2 (n = 33) |
Group 3 (n = 20) |
F | P |
|---|---|---|---|---|---|
| Right eye | |||||
| Average | 96.44 ± 10.36 | 97.98 ± 15.61 | 99.83 ± 12.88 | 0.417 | 0.6605 |
| Superior quadrant | 117.24 ± 19.88# | 102.52 ± 21.24** | 128.77 ± 18.69 | 11.140 | 0.0001 |
| Inferior quadrant | 128.54 ± 18.21 | 122.72 ± 25.64 | 131.42 ± 17.46 | 1.190 | 0.3094 |
| Nasal quadrant | 75.33 ± 13.50 | 74.16 ± 17.44 | 76.38 ± 20.91 | 0.111 | 0.8954 |
| Temporal quadrant | 69.56 ± 19.36 | 71.88 ± 25.73 | 73.22 ± 18.40 | 0.199 | 0.8244 |
| Left eye | |||||
| Average | 99.57 ± 19.71 | 95.89 ± 17.46 | 103.88 ± 21.40 | 1.080 | 0.3443 |
| Superior quadrant | 122.51 ± 12.48 | 109.63 ± 27.61* | 127.29 ± 28.39 | 4.336 | 0.0160 |
| Inferior quadrant | 129.28 ± 25.18 | 131.73 ± 19.46 | 130.55 ± 17.20 | 0.108 | 0.8976 |
| Nasal quadrant | 80.62 ± 22.41 | 75.79 ± 29.67 | 79.28 ± 22.05 | 0.313 | 0.7325 |
| Temporal quadrant | 71.50 ± 12.69 | 68.66 ± 20.92 | 74.88 ± 16.33 | 0.834 | 0.4380 |
Data represented mean ± standard deviation (SD) of each group; * represents comparisons of group 1 or 2 to group 3. # represents comparisons between group 1 and 2. *P < 0.05, **P < 0.001, #P < 0.05. P < 0.05 was considered statistically significant.
RNFL retinal nerve fibre layer, DR diabetic retinopathy.
Statistically significant p-values are in bold.
We then investigated the correlation between RNFL thickness and differential miRNAs identified in the miRNA sequencing. A positive correlation between the superior RNFL thickness and plasma miR-26a-5p level was found in both eyes (Fig. 4C, D), but the RNFL thickness of other quadrants of both eyes had no significant association with plasma miR-126-5p level (all P > 0.05, Table 5). Moreover, the RNFL thickness was found to have no significant association with the plasma levels of differential miRNAs (all P > 0.05, Table 5).
Table 5.
Correlation analysis of RNFL thickness with differential miRNA expression.
| Differential miRNAs | Right eye | Left eye | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Superior | Inferior | Temporal | Nasal | Average | Superior | Inferior | Temporal | Nasal | Average | |
| hsa-miR-185-5p | 0.038 | −0.251 | −0.233 | −0.302 | −0.208 | 0.011 | 0.251 | 0.302 | 0.237 | −0.250 |
| P value | 0.713 | 0.300 | 0.336 | 0.208 | 0.059 | 0.966 | 0.300 | 0.208 | 0.339 | 0.062 |
| hsa-miR-378a-3p | −0.233 | −0.069 | 0.072 | −0.324 | 0.190 | −0.274 | 0.070 | 0.120 | −0.103 | 0.009 |
| P value | 0.054 | 0.571 | 0.491 | 0.162 | 0.097 | 0.121 | 0.522 | 0.447 | 0.256 | 0.85 |
| hsa-miR-199a-5p | 0.049 | 0.069 | 0.051 | −0.195 | −0.163 | 0.480 | −0.274 | 0.224 | 0.097 | 0.113 |
| P value | 0.760 | 0.770 | 0.662 | 0.614 | 0.792 | 0.135 | 0.510 | 0.482 | 0.066 | 0.291 |
| hsa-miR-128-3p | 0.178 | −0.092 | −0.113 | 0.019 | 0.274 | 0.075 | −0.018 | −0.196 | 0.019 | 0.120 |
| P value | 0.008 | 0.391 | 0.291 | 0.853 | 0.121 | 0.481 | 0.868 | 0.067 | 0.866 | 0.219 |
| hsa-let-7a-5p | 0.093 | −0.075 | −0.173 | 0.075 | −0.138 | 0.048 | 0.289 | 0.076 | 0.105 | 0.020 |
| P value | 0.383 | 0.485 | 0.110 | 0.490 | 0.195 | 0.651 | 0.865 | 0.479 | 0.323 | 0.846 |
| hsa-miR-146b-5p | 0.072 | 0.104 | 0.004 | −0.143 | −0.131 | 0.099 | 0.010 | 0.188 | 0.015 | 0.102 |
| P value | 0.502 | 0.349 | 0.966 | 0.201 | 0.243 | 0.472 | 0.901 | 0.103 | 0.904 | 0.212 |
| miR-26a-5p | 0.616 | −0.216 | −0.088 | 0.150 | −0.022 | 0.651 | −0.121 | −0.131 | 0.069 | −0.004 |
| P value | *0.000 | 0.050 | 0.422 | 0.181 | 0.834 | *0.000 | 0.205 | 0.243 | 0.517 | 0.967 |
| hsa-miR-126-3p | −0.010 | −0.096 | 0.107 | −0.048 | −0.201 | −0.207 | −0.025 | 0.102 | −0.092 | −0.169 |
| P value | 0.924 | 0.372 | 0.319 | 0.651 | 0.069 | 0.066 | 0.810 | 0.341 | 0.391 | 0.115 |
Pearson correlation analysis was performed to analyse the association between RNFL thinning and differentially expressed miRNAs. *P < 0.05 was defined as statistically significant.
RNFL retinal nerve fibre layer.
Statistically significant p-values are in bold.
Prediction of target genes of miR-26a-5p
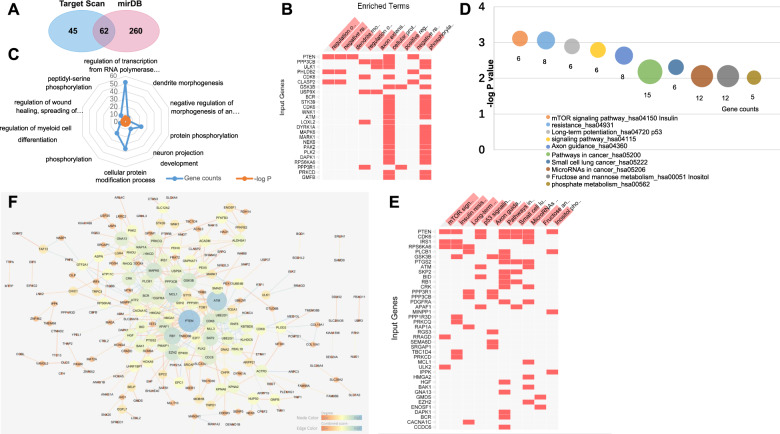
TargetScan and Mirdb servers were then used to predict the potential target genes of miR-26a-5p. The results indicated that 367 genes were candidate targets of miR-26a-5p (107 genes were predicted by TargetScan and 322 genes by mirDB). Among these targets, 62 genes were predicted by both algorisms, including PTEN, CDK8, ATF2, CDC6, EZH2, SMAD1 and ART3 (Fig. 5A, Table S1).
Fig. 5. The bioinformatics analysis of predicted target genes of miR-26a-5p.
A A Venn diagram showing the predicted targets by TargetScan and mirDB is presented, and the intersection indicates the number of genes predicted by both algorisms. B The matrix view of enriched terms in which the overlapping targets are involved. Enriched terms are in the columns, input genes are in the rows, and cells in the matrix indicate if a gene is associated with a term. C The top 10 most enriched biological processes that were analysed by the Enrichr software. D and E KEGG-enrichment analysis of the predicted target genes of miR-26a-5p: D the top10 most enriched KEGG pathways, E the visualization of the KEGG pathway in which the overlapping predicted target genes are involved. F Protein–protein interaction network of predicted target genes of miR-26a-5p. The nodes are proteins that are predicted as the targets of miR-26a-5p, and the edges represent the functional associations.
Functional enrichment analysis of predicted target genes
To further comprehend the functions of predicted target genes of miR-26a-5p, the GO analysis was performed. The GO-enrichment analysis can directly reflect the distribution of target genes for each enriched GO term including biological process (BP), cellular component (CC) and molecular function (MF). As was shown in Fig. 5B, C, the enriched BP include regulation of transcription from RNA polymerase II promoter (GO: 0006357), dendrite morphogenesis (GO: 0048813), negative regulation of morphogenesis of an epithelium (GO: 1905331), regulation of wound healing, spreading of epidermal cells (GO: 1903689), and so forth (all P < 0.05). Moreover, in the GO annotation analysis of these genes, 10 CC items and 57 MF items were enriched (both P < 0.05). Protein serine/threonine kinase activity (GO: 0004674) and kinase activity (GO: 0016301) were the two most enriched subcategories of the MF group; the largest subcategory found in the CC group was actin filament (GO: 0005884). The top 10 items of the CC and MF groups were shown in Table 6.
Table 6.
Top 10 predominant CC and MF terms in GO functional enrichment analysis.
| Category | Term | P-value | Gene counts |
|---|---|---|---|
| GO-CC | Actin filament (GO:0005884) | 0.000533 | 6 |
| Platelet dense granule membrane (GO:0031088) | 0.006565 | 2 | |
| RNA polymerase II transcription factor complex (GO:0090575) | 0.01909 | 7 | |
| Mitochondrial outer membrane (GO:0005741) | 0.019699 | 6 | |
| Actin cytoskeleton (GO:0015629) | 0.020151 | 11 | |
| Polymeric cytoskeletal fibre (GO:0099513) | 0.021144 | 9 | |
| Transcription factor TFIID complex (GO:0005669) | 0.027601 | 3 | |
| Cytoskeleton (GO:0005856) | 0.030491 | 16 | |
| Peroxisomal membrane (GO:0005778) | 0.038572 | 3 | |
| Nucleolus (GO:0005730) | 0.042895 | 19 | |
| GO-MF | Protein serine/threonine kinase activity (GO:0004674) | 4.75E−06 | 21 |
| Kinase activity (GO:0016301) | 1.76E−05 | 17 | |
| Protein kinase activity (GO:0004672) | 0.000209 | 22 | |
| Kinase binding (GO:0019900) | 0.000275 | 19 | |
| Protein kinase binding (GO:0019901) | 0.000848 | 20 | |
| Phosphotransferase activity, alcohol group as acceptor (GO:0016773) | 0.000868 | 13 | |
| Mannosyl-oligosaccharide mannosidase activity (GO:0015924) | 0.000892 | 3 | |
| Ubiquitin-like protein-specific protease activity (GO:0019783) | 0.002478 | 6 | |
| Phosphatidylinositol-4,5-bisphosphate binding (GO:0005546) | 0.005077 | 5 | |
| Signal sequence binding (GO:0005048) | 0.005941 | 4 |
GO gene ontology, CC cellular component, MF molecular function.
KEGG pathway analysis and PPI network constructions for the key target genes of miR-26a-5p
To identify the signalling pathways that the target genes of miR-26a-5p were involved, we mapped the genes to canonical signalling pathways in KEGG. The pathway-enrichment analysis revealed that the targets of miR-26a-5p were involved in 31 statistically enriched categories, including mTOR signalling pathway (hsa04150), insulin resistance (hsa04931), long-term potentiation (hsa04720), and so forth. The top 10 most enriched signalling pathways are summarized in Fig. 5D. Interestingly, PTEN was shown to be associated with 7 of these top 10 pathways (Fig. 5E). The PPI network analysis is a viable tool to understand molecular functions and disease mechanisms. To further understand the role of miR-26a-5p in DR, we performed a PPI analysis for the candidate targets of it. A total of 365 nodes and 391 interactions were involved in the PPI network constructed for the predicted target genes using the STRING tool, and then the genes and interactions were submitted to Cytoscape 3.6.1 software for visualization (Fig. 5F). The network view summarized the network of predicted associations for the predicted target proteins. After degree calculating, a total of 18 genes were identified with the degree of >10 (Table S2). Among these, PTEN (degree = 27), ATM (degree = 23), RB1 (degree = 17), MCL1 (degree = 16), EZH2 (degree = 14), MAPK6 (degree = 14), GSK3B (degree = 13), SKP2 (degree = 13), CDK6 (degree = 13) and GNA13 (degree = 11) were the top 10 genes with the closest connections to other nodes.
Discussion
A growing body of evidence indicated that retinal neurodegeneration plays a significant role in the pathogenesis of early DR [19] and circulating miRNA could be involved in the retinal damage of diabetic subjects [20]. The current study focuses on the role of plasma miRNAs in patients with type 2 diabetes and their association with diabetic neurodegeneration. Using the data of RNA-sequencing assay and genes target prediction, we provide the first evidence that miR-26a-5p as a potential biomarker for early diabetic retinal neurodegeneration in type 2 diabetic patients and the interaction between miR-26a-5p and PTEN might be involved in the diabetes-induced neuroglial degeneration since PTEN has been shown in multiple functional analyses.
DR is characterized by retinal microvascular abnormalities in the eyes, which is a leading cause of blindness in patients with diabetes mellitus. Accumulating clinical and experimental studies indicated that retinal neurodegeneration may precede microvascular changes in DR in diabetes mellitus [6, 7]. Moreover, RNFL loss is considered a manifestation of diabetic optic neuropathy and related to poorly controlled blood glucose and other systemic risk factors [21]. Our previous studies have shown that thinning of RNFL, assessed by SD-OCT, might happen at the earliest stage of DR in type 2 diabetic patients [22, 23]. However, many patients failed to receive the OCT scan at their early-stage DR, because of the limitation of DR screening. Thus, a novel factor for monitoring the progression of the retinal neurodegeneration in early DR is very important for both the prevention of DR and the improvement of the therapeutic effect.
As a potential therapeutic target, miRNA is considered as a useful and easily accessible diagnostic marker for many diseases, which are stable in blood and can be measured by qPCR methodology allowing the multiplexing of several miRNAs in a single experiment [24]. It has been reported that the serum miRNA expression profiles for the progression of DR may serve as fingerprints for disease detection and contributes to the pathogenesis of angiogenesis [25–29]. Recently more and more studies focused on the important roles of neurodegeneration in DR [21], however, there has been limited understanding of the expression profile of total miRNA in circulating serum of DR patients with retinal neurodegeneration, especially RNFL loss.
In the present study, we leveraged the high throughput sequencing technology and investigated the early DR specific circulating miRNA profile that could be of important relevance in the aetiology of retinal neurodegeneration. Finally, we demonstrated a new plasma biomarker to assess the retinal neurodegeneration in patients with type 2 diabetes at the earliest stage.
Although the sample size of our study was small, we were able to identify 20 up-regulated and 49 down-regulated miRNAs with the threshold of 1.5 folds change in expression. The results of qRT-PCR verification indicated that the plasma level of miR-26a-5p was significantly decreased in type 2 diabetic patients with NPDR compared to the ones without DR. Moreover, using the data of correlation between miRNAs and RNFL thinning, we showed that miR-26a-5p was associated with the superior RNFL thickness, which was thinner in NPDR patients but not the NDR ones. To our knowledge, this is the first investigation that demonstrates the circular miRNA profile could be used as a potential biomarker of diabetic neurodegeneration and related to RNFL thickness. However, the genes regulated by miR-26a-5p which may mediate the occurrence of diabetic neurodegeneration are still not clear.
To obtain the target genes of miR-26a-5p, multiple databases were used and 367 target genes were predicted. Some of the miR-26a-5p targets have been reported, such as EZH2, GSK3β, interleukin-6, interleukin-2, SMAD1, CCND2, CCNE2, RB1, and MAP3K2. miR-26a-5p executes functions through target genes and corresponding biological processes. GO analysis was performed using the Enrichr server in this study. Firstly, the top 10 terms of BP functional category were enriched such as regulation of transcription from RNA polymerase II promoter, dendrite morphogenesis, negative regulation of morphogenesis of epithelium, protein phosphorylation, neuron projection development, cellular protein modification process, phosphorylation, regulation of myeloid cell differentiation, regulation of wound healing, spreading of epidermal cells, peptidyl-serine phosphorylation. Additionally, regulation of transcription from RNA polymerase II promoter, dendrite morphogenesis, negative regulation of morphogenesis of epithelium, protein phosphorylation and neuron projection development biological process are connected to DR. To achieve a comprehensive analysis of target genes, MF and CC analyses were also applied, and the analysis showed that target genes were also significantly enriched in the protein serine/threonine kinase activity and kinase activity. The largest subcategory found in the CC group was actin filament, which is important in the pathogenesis of DR. The results of GO analysis were similar to the studies that miR-26a-5p serves different functions in various diseases. For instance, miR-26a-5p has been proved to regulate cardiac fibroblasts collagen, promote metastasis of lung cancer cells, and inhibit neuropathic pain in rats.
Our results of KEGG pathway-enrichment analysis showed that predicted target genes of miR-26a-5p were mainly enriched in the mTOR signalling pathway, insulin resistance, long-term potentiation, p53 signalling pathway, axon guidance, pathways in cancer, small cell lung cancer, miRNAs in cancer, fructose and mannose metabolism and inositol phosphate metabolism. A previous study has indicated that the mTOR signalling pathway is an important pathway encompassing a number of consecutive steps indispensable for the development of DR, which means that the miR-26a-5p may participate in the process of DR by regulating mTOR pathway. Particularly, a great number of studies suggest a mutual connection between the Insulin resistance pathway and the development of retinal neurodegenerative. The PPI is a key feature of numerous cellular processes, involved in gene expression and signalling. In the present study, a representative PPI network of predicted target genes was constructed by STRING and visualized by Cytoscape. After degree calculating, PTEN, ATM, RB1, MCL1, EZH2, MAPK6, GSK3B, SKP2, CDK6, and GNA13 were identified as the top 10 genes with the closest connections to other nodes. Our results demonstrated that PTEN was a key node in the PPI network, as the degree of PTEN was the highest in the whole predicted targets. Besides, the results of GO BP and KEGG annotations also provided reliable evidence that PTEN plays an important role in the effect of miR-26a-5p. Our combining results suggest that PTEN may be the key target gene of miR-26a-5p. PTEN is found as one of the most mutated and deleted tumour suppressors in human cancer, and it has been proved to regulate various disease processes. Consistent with our bioinformatics analysis, a previous study demonstrated that miR-26a-5p protects against myocardial ischaemia/reperfusion injury potential via suppression of PTEN and activation of the AKT pathway [30]. Thus, we hypothesize that miR-26a-5p may mediate early diabetic retinal neurodegeneration with type 2 diabetes through PTEN.
Overall, our results indicated that the plasma level of miR-26a-5p, which was related to the superior RNFL thickness, was significantly decreased in type 2 diabetic patients with NPDR compared to the patients without DR. Then we predicted the target genes of miR-26a-5p and revealed the underlying mechanism by GO, KEGG and PPI analyses. The bioinformatic analyses highlight the functions of a key target gene PTEN, which suggested that miR-26a-5p might mediate retinal neurodegeneration by regulating PTEN expression. Our results demonstrated that miR-26a-5p might serve as a new plasma biomarker to assessing retinal neurodegeneration. Further studies need to be designed to determine the functional roles and underlying mechanisms before miR-26a-5p becomes a biomarker and a target for the treatment of diabetic retinal neurodegeneration.
Summary
What was known before
Retinal neurodegeneration is an early pathological change in diabetic retinopathy (DR).
The availability of quantitative detection of circulating miRNAs enhances the use of miRNAs as biomarkers for physiological and pathological processes.
What this study adds
Circulating miR-26a-5p is a potential biomarker for early-stage retinal neurodegeneration and it may be involved in the development of DR via profoundly influencing the functions of retinal cells.
Supplementary information
Author contributions
RS and RL designed the study; LC, HZ and YZL collected data; WW and YD performed the analyses; RS drafted the manuscript; RS contributed to the discussion and revised the manuscript.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (81600720) and the Natural Science Foundation of Shaanxi province (2017JQ8012). The funders had no role in study design, data collection, and analysis, decision to publish, or preparation of the manuscript.
Data availability
The datasets generated during and/or analysed during the current study are not publicly available due to the risk of violating patient privacy but are available from the corresponding author on reasonable request.
Compliance with ethical standards
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41433-021-01393-5.
References
- 1.Stitt AW, Curtis TM, Chen M, Medina RJ, McKay GJ, Jenkins A, et al. The progress in understanding and treatment of diabetic retinopathy. Prog Retin Eye Res. 2016;51:156–86. doi: 10.1016/j.preteyeres.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Liu F, Saul AB, Pichavaram P, Xu Z, Rudraraju M, Somanath PR, et al. Pharmacological inhibition of spermine oxidase reduces neurodegeneration and improves retinal function in diabetic mice. J Clin Med 2020;9:340. [DOI] [PMC free article] [PubMed]
- 3.Sohn EH, van Dijk HW, Jiao C, Kok PH, Jeong W, Demirkaya N, et al. Retinal neurodegeneration may precede microvascular changes characteristic of diabetic retinopathy in diabetes mellitus. Proc Natl Acad Sci USA. 2016;113:E2655–E2664. doi: 10.1073/pnas.1522014113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simo R, Hernandez C. Novel approaches for treating diabetic retinopathy based on recent pathogenic evidence. Prog Retin Eye Res. 2015;48:160–80. doi: 10.1016/j.preteyeres.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 5.Simo R, Stitt AW, Gardner TW. Neurodegeneration in diabetic retinopathy: does it really matter? Diabetologia. 2018;61:1902–12. doi: 10.1007/s00125-018-4692-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sampedro J, Bogdanov P, Ramos H, Sola-Adell C, Turch M, Valeri M, et al. New insights into the mechanisms of action of topical administration of GLP-1 in an experimental model of diabetic retinopathy. J Clin Med 2019;8:339. [DOI] [PMC free article] [PubMed]
- 7.Hafner J, Zadrazil M, Grisold A, Ricken G, Krenn M, Kitzmantl D, et al. Retinal and corneal neurodegeneration and their association with systemic signs of peripheral neuropathy in type 2 diabetes. Am J Ophthalmol. 2020;209:197–205. doi: 10.1016/j.ajo.2019.09.010. [DOI] [PubMed] [Google Scholar]
- 8.Bogdanov P, Simo-Servat O, Sampedro J, Sola-Adell C, Garcia-Ramirez M, Ramos H, et al. Topical administration of bosentan prevents retinal neurodegeneration in experimental diabetes. Int J Mol Sci 2018;19:3578. [DOI] [PMC free article] [PubMed]
- 9.Choi JA, Ko SH, Park YR, Jee DH, Ko SH, Park CK. Retinal nerve fiber layer loss is associated with urinary albumin excretion in patients with type 2 diabetes. Ophthalmolology. 2015;122:976–81. doi: 10.1016/j.ophtha.2015.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Nadri G, Saxena S, Stefanickova J, Ziak P, Benacka J, Gilhotra JS, et al. Disorganization of retinal inner layers correlates with ellipsoid zone disruption and retinal nerve fiber layer thinning in diabetic retinopathy. J Diabetes Complicat. 2019;33:550–3. doi: 10.1016/j.jdiacomp.2019.05.006. [DOI] [PubMed] [Google Scholar]
- 11.Liu L, Wang Y, Liu HX, Gao J. Peripapillary region perfusion and retinal nerve fiber layer thickness abnormalities in diabetic retinopathy assessed by OCT angiography. Transl Vis Sci Technol. 2019;8:14. doi: 10.1167/tvst.8.4.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obeid A, Su D, Patel SN, Uhr JH, Borkar D, Gao X, et al. Outcomes of eyes lost to follow-up with proliferative diabetic retinopathy that received panretinal photocoagulation versus intravitreal anti-vascular endothelial growth factor. Ophthalmolology. 2019;126:407–13. doi: 10.1016/j.ophtha.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 13.Shafabakhsh R, Aghadavod E, Mobini M, Heidari-Soureshjani R, Asemi Z. Association between microRNAs expression and signaling pathways of inflammatory markers in diabetic retinopathy. J Cell Physiol. 2019;234:7781–7. doi: 10.1002/jcp.27685. [DOI] [PubMed] [Google Scholar]
- 14.Willeit P, Skroblin P, Moschen AR, Yin X, Kaudewitz D, Zampetaki A, et al. Circulating MicroRNA-122 is associated with the risk of new-onset metabolic syndrome and type 2 diabetes. Diabetes. 2017;66:347–57. doi: 10.2337/db16-0731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gandhi R, Healy B, Gholipour T, Egorova S, Musallam A, Hussain MS, et al. Circulating microRNAs as biomarkers for disease staging in multiple sclerosis. Ann Neurol. 2013;73:729–40. doi: 10.1002/ana.23880. [DOI] [PubMed] [Google Scholar]
- 16.Platania C, Maisto R, Trotta MC, D’Amico M, Rossi S, Gesualdo C, et al. Retinal and circulating miRNA expression patterns in diabetic retinopathy: an in silico and in vivo approach. Br J Pharmacol. 2019;176:2179–94. doi: 10.1111/bph.14665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mammadzada P, Bayle J, Gudmundsson J, Kvanta A, Andre H. Identification of diagnostic and prognostic microRNAs for recurrent vitreous hemorrhage in patients with proliferative diabetic retinopathy. J Clin Med 2019;8:2217. [DOI] [PMC free article] [PubMed]
- 18.Liu HN, Cao NJ, Li X, Qian W, Chen XL. Serum microRNA-211 as a biomarker for diabetic retinopathy via modulating Sirtuin 1. Biochem Biophys Res Commun. 2018;505:1236–43. doi: 10.1016/j.bbrc.2018.10.052. [DOI] [PubMed] [Google Scholar]
- 19.Singh RP, Elman MJ, Singh SK, Fung AE, Stoilov I. Advances in the treatment of diabetic retinopathy. J Diabetes Complicat. 2019;33:107417. doi: 10.1016/j.jdiacomp.2019.107417. [DOI] [PubMed] [Google Scholar]
- 20.Zampetaki A, Willeit P, Burr S, Yin X, Langley SR, Kiechl S, et al. Angiogenic microRNAs linked to incidence and progression of diabetic retinopathy in Type 1 diabetes. Diabetes. 2016;65:216–27. doi: 10.2337/db15-0389. [DOI] [PubMed] [Google Scholar]
- 21.Jeon SJ, Park HL, Lee JH, Park CK. Relationship between systemic vascular characteristics and retinal nerve fiber layer loss in patients with type 2 diabetes. Sci Rep. 2018;8:10510. doi: 10.1038/s41598-018-28985-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shi R, Guo Z, Wang F, Li R, Zhao L, Lin R. Alterations in retinal nerve fiber layer thickness in early stages of diabetic retinopathy and potential risk factors. Curr Eye Res. 2018;43:244–53. doi: 10.1080/02713683.2017.1387669. [DOI] [PubMed] [Google Scholar]
- 23.Shi R, Zhao L, Qi Y. The effect of fenofibrate on early retinal nerve fiber layer loss in type 2 diabetic patients: a case-control study. BMC Ophthalmol. 2018;18:100. doi: 10.1186/s12886-018-0769-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joglekar MV, Januszewski AS, Jenkins AJ, Hardikar AA. Circulating microRNA biomarkers of diabetic retinopathy. Diabetes. 2016;65:22–24. doi: 10.2337/dbi15-0028. [DOI] [PubMed] [Google Scholar]
- 25.Farr RJ, Januszewski AS, Joglekar MV, Liang H, McAulley AK, Hewitt AW, et al. A comparative analysis of high-throughput platforms for validation of a circulating microRNA signature in diabetic retinopathy. Sci Rep. 2015;5:10375. doi: 10.1038/srep10375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pusparajah P, Lee LH, Abdul KK. Molecular markers of diabetic retinopathy: potential screening tool of the future? Front Physiol. 2016;7:200. doi: 10.3389/fphys.2016.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen Q, Qiu F, Zhou K, Matlock HG, Takahashi Y, Rajala R, et al. Pathogenic role of microRNA-21 in diabetic retinopathy through downregulation of PPARalpha. Diabetes. 2017;66:1671–82. doi: 10.2337/db16-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han N, Tian W, Yu N, Yu L. YAP1 is required for the angiogenesis in retinal microvascular endothelial cells via the inhibition of MALAT1-mediated miR-200b-3p in high glucose-induced diabetic retinopathy. J Cell Physiol. 2020;235:1309–20. doi: 10.1002/jcp.29047. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Dong Y, He C, Pan X, Liu D, Yang J, et al. RNA-Seq revealed novel non-proliferative retinopathy specific circulating MiRNAs in T2DM patients. Front Cenet. 2019;10:531. doi: 10.3389/fgene.2019.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xing X, Guo S, Zhang G, Liu Y, Bi S, Wang X, et al. miR-26a-5p protects against myocardial ischemia/reperfusion injury by regulating the PTEN/PI3K/AKT signaling pathway. Braz J Med Biol Res. 2020;53:9106. doi: 10.1590/1414-431x20199106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are not publicly available due to the risk of violating patient privacy but are available from the corresponding author on reasonable request.