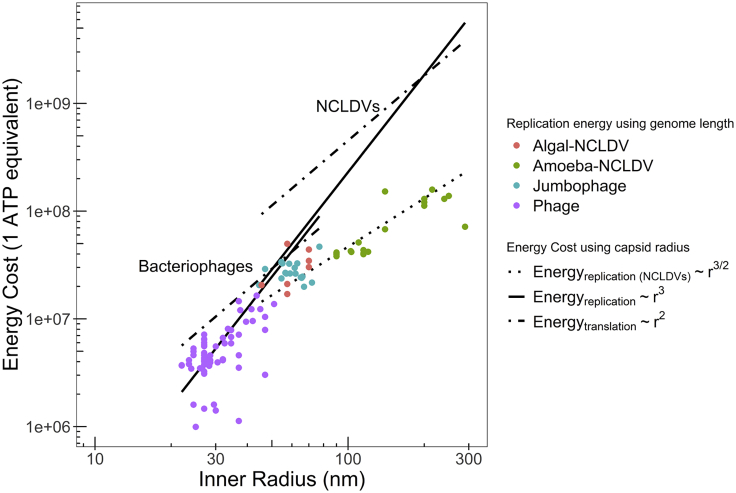
Figure 3.
The log-log plot of translation and replication energy cost of icosahedral dsDNA bacteriophages and NCLDVs as a function of their inner radius
The replication cost is obtained using genome length (Equation 1) and capsid radius (Equation 2). The translation cost for capsid protein molecules is obtained from Equation 3. The gap in the energy costs of NCLDVs and bacteriophages is because of the difference in the thickness t of their viral capsids (Equation 3). Energy costs are reported in terms of the number of ATP hydrolysis events (Mahmoudabadi et al., 2017). As discussed in Mahmoudabadi et al. (2017) translation () and replication ( ) rates dominate at lower and higher capsid sizes, respectively. This trend works well for bacteriophages (data points for replication cost using actual length). For NCLDVs, however, the translation cost always dominates because . See text for more detail. All logs are to the base 10. The entire data are represented in the Table S1.