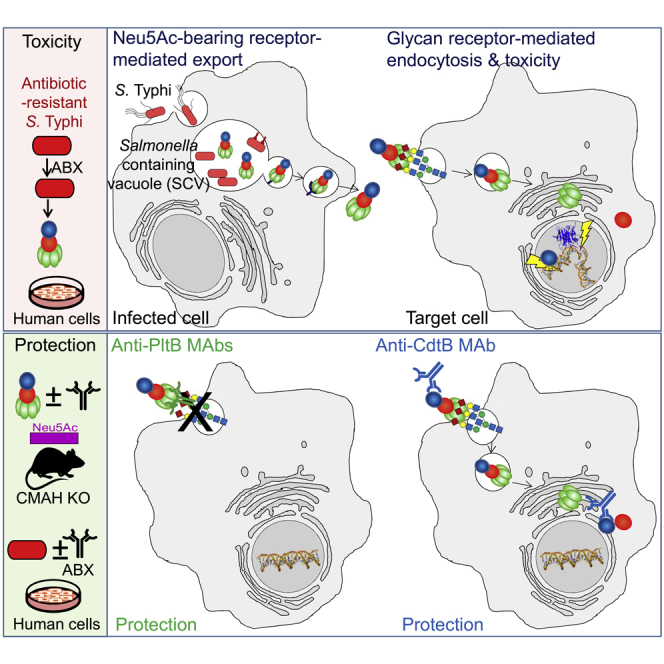
Summary
Nearly all clinical isolates of Salmonella Typhi, the cause of typhoid fever, are antibiotic resistant. All S. Typhi isolates secrete an A2B5 exotoxin called typhoid toxin to benefit the pathogen during infection. Here, we demonstrate that antibiotic-resistant S. Typhi secretes typhoid toxin continuously during infection regardless of antibiotic treatment. We characterize typhoid toxin antibodies targeting glycan-receptor-binding PltB or nuclease CdtB, which neutralize typhoid toxin in vitro and in vivo, as demonstrated by using typhoid toxin secreted by antibiotic-resistant S. Typhi during human cell infection and lethal dose typhoid toxin challenge to mice. TyTx11 generated in this study neutralizes typhoid toxin effectively, comparable to TyTx4 that binds to all PltB subunits available per holotoxin. Cryoelectron microscopy explains that the binding of TyTx11 to CdtB makes this subunit inactive through CdtB catalytic-site conformational change. The identified toxin-neutralizing epitopes are conserved across all S. Typhi clinical isolates, offering critical insights into typhoid toxin-neutralizing strategies.
Graphical abstract
Highlights
-
•
Antibiotic-resistant S. Typhi secretes typhoid toxin despite antibiotic treatment
-
•
MAb targeting the receptor-binding or nuclease subunits neutralizes typhoid toxin
-
•
TyTx11 makes nuclease CdtB inactive by causing catalytic-site conformational change
-
•
Toxin-neutralizing epitopes identified are conserved across S. Typhi clinical isolates
Introduction
Salmonella enterica serovar Typhi (S. Typhi), the causative agent of typhoid fever, sickens an estimated 22 million people and claims over 0.2 million people's lives every year (Crump and Mintz, 2010; Wain et al., 2015). The global spread of multidrug-resistant (MDR) and extensively drug-resistant (XDR) S. Typhi makes typhoid fever a major worldwide public health challenge (Baker et al., 2011; Feasey et al., 2015; Holt et al., 2011; Ingle et al., 2019; Klemm et al., 2018; Yan et al., 2016). Antibiotic-resistant S. Typhi infection is more common among children; more than 90% of the XDR typhoid cases are currently from children younger than 15 years (Feasey et al., 2015; Klemm et al., 2018; Qamar et al., 2018). The typhoid mortality in the preantibiotic era was approximately 25% (Levine and Simon, 2018). Typhoid fever is only partly preventable by vaccines, and two types of typhoid fever vaccines, the live-attenuated Ty21a and Vi and its variant subunit vaccines, are currently available (Yang et al., 2018a). These vaccines exhibit an efficacy of approximately 55%–85%, with the Vi-protein conjugate subunit vaccine being the most efficacious (Voysey and Pollard, 2018; WHO, 2018; Yang et al., 2018a). Nonetheless, there are no vaccines for early-life populations younger than six months available (WHO, 2018).
S. Typhi can induce acute life-threatening conditions, with children being the most vulnerable group. Disease timelines can vary, but generally there is an initial 1- to 2-week incubation period, a symptomatic disease period for 2–3 weeks, followed by a long-term colonization and transmission period in some individuals known as asymptomatic healthy carriers (CDC, 2017). This dynamic infectious cycle of S. Typhi is coordinated by many virulence factors, including typhoid toxin, Vi capsular polysaccharides, and Type III secretion system Salmonella Pathogenicity Island (SPI)-1 and SPI-2 effector toxins (Abrahams and Hensel, 2006; Galán, 2009; Monack, 2012; Nikolaus et al., 2001; Song et al., 2010, 2013; Srikanth et al., 2011; Wangdi et al., 2014). Most of these virulence factors remain on the bacterial surface or in the cytoplasm of S. Typhi-infected host cells, altering host responses primarily at the site of infection and in infected cells. Unlike others, however, typhoid toxin is an exotoxin, which targets host cells located locally and remotely to benefit the pathogen primarily by altering host immune cell numbers and functions (Chong et al., 2017; Galan, 2016; Gibani et al., 2019; Lee et al., 2020; Song et al., 2013; Yang et al., 2018b).
Typhoid toxin is a heptameric A2B5 complex consisting of one each of two enzymatic “A” subunits CdtB and PltA and a glycan receptor-binding “B” homopentamer PltB (Song et al., 2013). In the pyramid-shaped three-dimensional typhoid toxin structure (PDB:4K6L), CdtB is located in the vertex, with PltA in the middle linking the CdtB to the PltB homopentamer situated in the base (Song et al., 2013). S. Typhi secretes typhoid toxin only when the bacteria are inside the host cells (Haghjoo and Galán, 2004; Spano et al., 2008). Receptor-binding PltB recognizes a specific set of host glycan receptors to deliver the toxin's A subunits into target host cells (Lee et al., 2020; Song et al., 2013; Yang et al., 2018b). PltA's role in target host cell intoxication is not prominent, whereas CdtB's nuclease activity is essential for typhoid toxin-mediated cellular and in vivo toxicities (Lee et al., 2020; Song et al., 2013; Yang et al., 2018b). We can evaluate typhoid toxin-induced cellular toxicities through quantitative fluorescent microscopy by measuring host cell DNA damage repair responses and by quantitative flow cytometry measuring host cell-cycle arrest in G2/M (Song et al., 2013; Spano et al., 2008; Yang et al., 2018b). Similarly, we can also evaluate typhoid toxin-mediated in vivo toxicities by analyzing toxin binding to target host cells, immune cell population changes/leukopenia, upper motor function defects, body weights, and survival (Yang et al., 2018b). These in vitro and in vivo virulence phenotypes were observed only when both PltB and CdtB functions are intact, because the assembled toxin variants containing a single amino acid point mutation associated with either glycan-receptor-binding or nuclease functions did not induce the toxicities (Lee et al., 2020; Song et al., 2013; Yang et al., 2018b). Therefore, we exploited these in vitro and in vivo toxicity readouts in this study when evaluating antibody-mediated toxin neutralization.
Here, we demonstrate that antibiotic-resistant S. Typhi secretes typhoid toxin continuously during infection regardless of antibiotic treatment, using several in vitro and in vivo infection models and multiple quantitative approaches. We also show that the identified four toxin-neutralizing epitopes located on PltB and CdtB are conserved across all S. Typhi clinical isolates, including all antibiotic-resistant strains.
Results
Antibiotic-resistant S. Typhi secretes typhoid toxin continuously during human cell infection regardless of antibiotic treatment
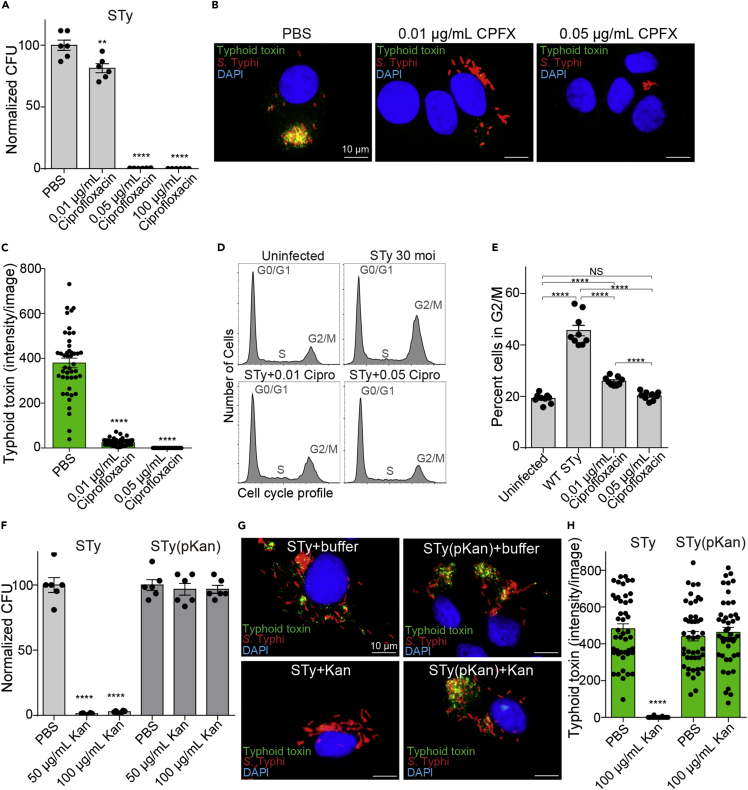
To examine whether antibiotic-resistant S. Typhi produces typhoid toxin continuously during infection even when S. Typhi-infected host cells are treated with antibiotics, we established a human cell infection system through careful verification of the system using both antibiotic-susceptible and antibiotic-resistant S. Typhi strains. As antibiotic ciprofloxacin is one of the standard treatment options for patients with typhoid, S. Typhi-infected human cells (Henle-407 infected with 30 MOI of S. Typhi) were treated with ciprofloxacin (CPFX) when indicated (Figures 1A–1E). As two doses of 500 mg ciprofloxacin/dose are used in human patients (∼5 L blood volume in adults), where one 500 mg dose is estimated to be equivalent to 100 μg/mL, we started the experiments with 100 μg/mL ciprofloxacin treatment. When this dose was used, nearly zero colonies of S. Typhi were recovered from S. Typhi-infected host cells (Figure 1A). To establish an experimental condition leading to the partial killing of S. Typhi, we titrated down the ciprofloxacin concentrations and found that 0.05 and 0.01 μg/mL ciprofloxacin treatment resulted in 0.37% and 81% S. Typhi colony-forming unit (CFU), relative to S. Typhi CFU recovered from PBS-treated control samples (Figure 1A). We then carried out fluorescent microscopy to monitor and quantify typhoid toxin production with these two ciprofloxacin concentrations. The 3xFLAG epitope tag was conjugated to the C-terminal end of CdtB in the chromosome of S. Typhi (Spano et al., 2008), allowing for an anti-FLAG M2 antibody-mediated specific detection of typhoid toxin in host cells during infection. Based on previous time course studies, we quantified typhoid toxin 24 h post-infection (Figure 1B). Strong typhoid toxin signals (green) were observed in ciprofloxacin-untreated, S. Typhi-infected human cells. In contrast, little-to-no typhoid toxin signals were observed in the counterpart cells treated with 0.01 and 0.05 μg/mL ciprofloxacin, respectively (Figures 1B and 1C). Consistently, we observed the background level of host cells in G2/M in uninfected host cells and 0.05 μg/mL ciprofloxacin-treated, S. Typhi-infected host cells, which is markedly different from ciprofloxacin-untreated, S. Typhi-infected host cells (Figures 1D and 1E). Also consistent with the small amount of typhoid toxin produced (Figures 1B and 1C), 0.01 μg/mL ciprofloxacin-treated, S. Typhi-infected host cells exhibited a mild increase of host cells in G2/M (Figures 1D and 1E).
Figure 1.
Antibiotic-resistant S. Typhi secretes typhoid toxin continuously during human cell infection regardless of antibiotic treatment
(A–C) Evaluation of the ciprofloxacin effects on intracellular S. Typhi colony-forming unit (CFU) (A) and typhoid toxin secretion (B and C). Henle-407 cells were infected with 30 MOI. S. Typhi for 24 h in the presence and absence of indicated ciprofloxacin concentrations. Bars represent average ±standard error of the mean (SEM). ∗∗p < 0.01, ∗∗∗∗p < 0.0001, relative to PBS. n = 6 per group. Two-tailed unpaired t tests. (B) Representative fluorescent microscope images showing typhoid toxin (green), S. Typhi (red), and host cell DNA (blue). Scale bar, 10 μm. (C) All microscopy results reflecting the typhoid toxin signal intensity obtained from three independent experiments. Bars represent average ±SEM. ∗∗∗∗p < 0.0001, relative to the PBS group. n = 45 per group. Two-tailed unpaired t tests.
(D and E) Representative flow cytometry histograms reflecting cell cycle profiles of Henle-407 cells left uninfected or infected with 30 MOI S. Typhi for 72 h in the presence and absence of indicated ciprofloxacin concentrations (D) and corresponding percent of cells in the G2/M cell cycle obtained from three independent experiments (E). Bars represent average ±SEM. ∗∗∗∗p < 0.0001, NS, not significant. n = 9 per group. Two-tailed unpaired t tests.
(F) Evaluation of the kanamycin effects on intracellular S. Typhi CFU. Henle-407 cells were infected with 30 MOI S. Typhi for 24 h in the presence and absence of indicated kanamycin concentrations. Bars represent average ±SEM. ∗∗p < 0.01,∗∗∗∗p < 0.0001, relative to PBS. n = 6 per group. Two-tailed unpaired t tests.
(G) Representative fluorescent microscope images showing typhoid toxin (green), S. Typhi (red), and host cell DNA (blue). STy, antibiotic-susceptible S. Typhi; STy(pKan), S. Typhi carrying the pKan plasmid. Kan, 100 μg/mL kanamycin treatment. Scale bar, 10 μm.
(H) Typhoid toxin signal intensity per image obtained from three independent experiments. Bars represent average ±SEM. ∗∗∗∗p < 0.0001, relative to the PBS group. n = 45 per group. Two-tailed unpaired t tests.
After verifying that this system can faithfully quantify the effect of antibiotic treatment on typhoid toxin production during infection, to establish a similar approach for antibiotic-resistant S. Typhi, we next prepared an antibiotic-resistant S. Typhi strain by transforming a low-copy plasmid carrying the kanamycin-resistant gene named pSB4004 (Spano et al., 2008) into kanamycin-susceptible S. Typhi, here referred to as S. Typhi(pKan). Note that this S. Typhi strain harboring the antibiotic-resistant plasmid was used for conducting proof-of-principle studies to avoid creating drug-resistant S. Typhi strains resembling the clinical isolates currently posing a significant public health threat in humans. We do not have live MDR and XDR S. Typhi clinical isolates in the laboratory, which are common in the endemic region, and exhibit resistance to ciprofloxacin. To verify the antibiotic-resistant S. Typhi system, we infected Henle-407 cells with 30 MOI of either kanamycin-susceptible S. Typhi or resistant S. Typhi(pKan) and treated them with PBS, 50 μg/mL, or 100 μg/mL kanamycin as indicated (Figure 1F). As expected, kanamycin treatment almost completely cleared S. Typhi but not S. Typhi(pKan) (Figure 1F). Consistently, S. Typhi(pKan) located inside human cells expressed and secreted typhoid toxin regardless of kanamycin treatment (Figures 1G and 1H). These results indicate that antibiotic-resistant S. Typhi clinical isolates widespread in the endemic regions will likely secrete typhoid toxin continuously during human infection irrespective of antibiotic treatment.
Generation and characterization of neutralizing antibodies targeting the receptor-binding PltB and the nuclease CdtB subunits of typhoid toxin
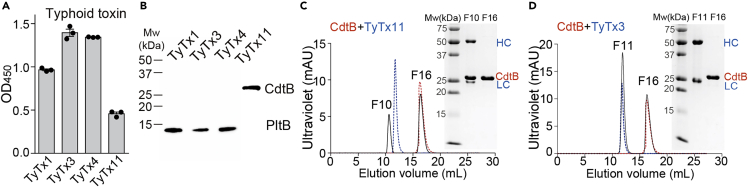
To obtain a panel of neutralizing antibodies (nAbs) targeting PltB and CdtB, we immunized a group of female BALB/c mice with recombinant typhoid toxoid, generated B cell hybridomas using splenic B cells (Van Slyke et al., 2018), and successfully established three B cell hybridomas referred to as TyTx11, TyTx12, and TyTx13. Western blot analyses of these hybridoma culture supernatants indicated that TyTx11 produces a monoclonal antibody targeting CdtB, whereas TyTx12 and 13 produce antibodies specific for PltB (Figure S1). Based on our recently generated antibody collection containing three unique and well-characterized anti-PltB nAbs TyTx1, 3, and 4, whose epitopes overlap with glycan receptor-binding sites on PltB subunits (Nguyen et al., 2021), we proceeded with TyTx11, along with TyTx1, 3, and 4, by purifying a milligram scale of these antibodies via Protein G agarose columns.
TyTx11 recognizes the CdtB subunit of typhoid holotoxin as verified by ELISA, western blot, and size-exclusion chromatography (SEC) (Figures 2A–2C). Specifically, when we incubated TyTx11 and TyTx3 (as a control to demonstrate the specificity) with purified CdtB and subjected them to Superdex 200, it was apparent that TyTx11 is a bona fide anti-CdtB monoclonal antibody (MAb) (Figures 2C, 2D, and S2). Consistently, the variable regions of the light (kappa) and heavy chains of TyTx11 are distinct from the sequences of TyTx1, 3, and 4, indicating that we have four unique hybridoma clones producing antibodies targeting the different epitopes on typhoid toxin (Figures S3 and S4). The apparent binding affinity of TyTx11 to the CdtB subunit was evaluated by surface plasmon resonance (SPR) assays using recombinant typhoid toxin immobilized on the sensor chip, resulting in ∼1.61 nM (Table S1). This nanomolar binding affinity is in line with the binding affinities of TyTx1 and 3, ones that bind to PltB subunits generally via a monovalent interaction due to the toxin's A-subunit-mediated interference with antibody-binding to laterally located epitopes on the PltB pentamer (Nguyen et al., 2021). This binding affinity is markedly different from the overall binding avidity of TyTx4 that can bind to all PltB subunits available per holotoxin (Nguyen et al., 2021) (Table S1).
Figure 2.
Generation and characterization of antibodies targeting the receptor-binding PltB and the nuclease CdtB subunits of typhoid toxin
(A) ELISAs were performed to determine their specificities to typhoid toxin. Bars represent the mean values of three independent experiments ±SEM, which were obtained by measuring the absorbance at 450 nm.
(B) Typhoid toxin (200 ng) was separated using 15% SDS-PAGE, followed by western blot analysis for each MAb to determine their toxin subunit specificities. Representative blot results from three independent experiments are shown.
(C and D) Overlays of Superdex 200 chromatograms of CdtB only (red), MAb only (blue, TyTx3 or TyTx11), and CdtB + MAb together (black). Fractions 10 and 16 for the TyTx11 mixture and fractions 11 and 16 for the TyTx3 mixture were analyzed using 15% SDS-PAGE (insets in C and D, respectively). SEC standards were run on the same Superdex 200 Increase column (Figure S2).
See also Figures S1–S4 and Table S1.
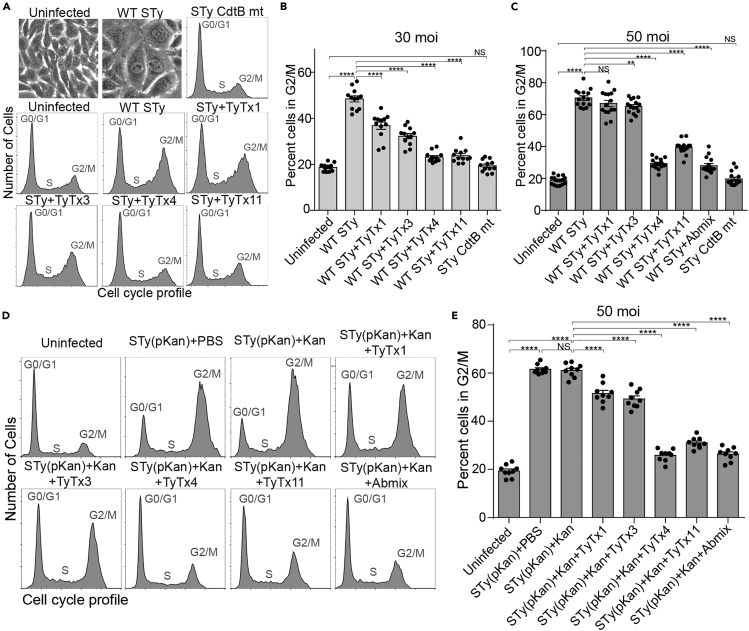
All antibodies neutralize typhoid toxin produced by antibiotic-susceptible and resistant S. Typhi during infection yet with different neutralizing capabilities
Based on the essential roles of PltB and CdtB subunits in typhoid toxin-mediated in vitro and in vivo toxicities, we hypothesized that all antibodies neutralize typhoid toxin. To test the hypothesis, each antibody or antibody mixture was added to culture media for Henle-407 cells infected with S. Typhi (Figures 3A–3C) and S. Typhi(pKan) (Figures 3D and 3E). Consistent with previous observations, typhoid toxin produced by wild-type S. Typhi caused cell distension and cell-cycle arrest in G2/M in a toxin dose-dependent manner (Figures 3A–3C), whose specificity was verified using an isogenic mutant S. Typhi-producing typhoid toxin carrying the catalytic-mutant CdtBH160Q subunit (STy CdtB mt) whose cell-cycle arrest profiles in G2/M was comparable with that of uninfected cells (Figures 3A–3C).
Figure 3.
All antibodies neutralize typhoid toxin produced by antibiotic-susceptible and -resistant S. Typhi during infection yet with different neutralizing capabilities
(A–C) Note that data for TyTx1, 3, and 4 (anti-PltB antibodies) in figure panels (A−C) are adapted from Nguyen et al. (2021) to compare their toxin neutralizing outcomes to TyTx11-mediated toxin neutralization outcomes (anti-CdtB antibody). Measurements of nAb-mediated toxin neutralization against typhoid toxin continuously produced by S. Typhi during human cell infection. Henle-407 cells were left uninfected or infected with S. Typhi (STy) at an MOI 30 (A and B) or 50 (C) for 3 days in the presence and absence of indicated monoclonal antibodies (MAbs) (10 nM). Abmix was prepared by mixing 2.5 nM each of four MAbs. STy CdtB mt, an isogenic S. Typhi carrying CdtB catalytic-mutant CdtBH160Q, was used to demonstrate the specificity of typhoid toxin-induced intoxication. Representative bright-field images of uninfected and WT S. Typhi-infected Henle-407 cells reflecting typhoid toxin-mediated intoxication phenotypes are shown in the first two panels in (A). Representative flow cytometry histograms reflecting cell cycle profiles are shown in the remaining panels in (A), and percent cells in the G2/M cell cycle summarizing all results are shown in (B and C). Bars represent average ±SEM. ∗∗p < 0.01, ∗∗∗∗p < 0.0001, NS, not significant. n = 12–15 per group. Two-tailed unpaired t tests. At least three independent experiments were performed.
(D) Representative flow cytometry histograms reflecting cell cycle profiles of Henle-407 cells left uninfected or infected with 50 MOI. STy(pKan) for 3 days ±50 μg/mL Kan ± indicated MAbs (10 nM). Abmix was prepared by mixing 2.5 nM each of four MAbs.
(E) Percent of cells in the G2/M cell cycle obtained from three independent experiments using antibiotic-resistant S. Typhi. Bars represent average ±SEM. ∗∗∗∗p < 0.0001, NS, not significant. n = 9 per group. Two-tailed unpaired t tests.
All MAbs neutralized typhoid toxin, although two antibodies TyTx4 and 11 were more productive than the other (Figures 3A–3C). At an MOI of 30, 52% of host cells infected with wild-type (WT) S. Typhi were arrested in G2/M (Figures 3A and 3B). When we added 10 nM MAbs to the culture media during a 3-day-infection period, we observed that MAbs protected host cells from toxicity (Figures 3A and 3B). TyTx4 and TyTx11 offered complete protection resulting in the background level of host cells in G2/M observed in uninfected host cells, whereas TyTx1 and TyTx3 exhibited partial protection (Figures 3A and 3B). In the 50 MOI infection, approximately 70% of host cells infected with WT S. Typhi were arrested in G2/M, indicating a higher concentration of typhoid toxin present in this condition than 30 MOI infection (Figure 3C). When the same 10 nM MAbs were used, TyTx4 and 11 still neutralized this increased dose of typhoid toxin effectively, but TyTx1 and 3 neutralized typhoid toxin only at a modest level (Figure 3C).
Similarly, S. Typhi(pKan)-infected host cells exhibited a marked increase in G2/M arrest regardless of kanamycin treatment (Figures 3D and 3E). These results support the suitability of this system for testing MAb efficacy in antibiotic-treatment conditions. Kanamycin and indicated MAbs were present in the media for the 3-day infection period. All four MAbs were able to neutralize typhoid toxin, as shown by a significant reduction in the percentage of host cells arrested in G2/M, even though TyTx4 and 11 were more efficacious in neutralizing typhoid toxin than TyTx1 and 3 (Figures 3D and 3E). These results indicate that all MAbs neutralize typhoid toxin regardless of antibiotic treatment.
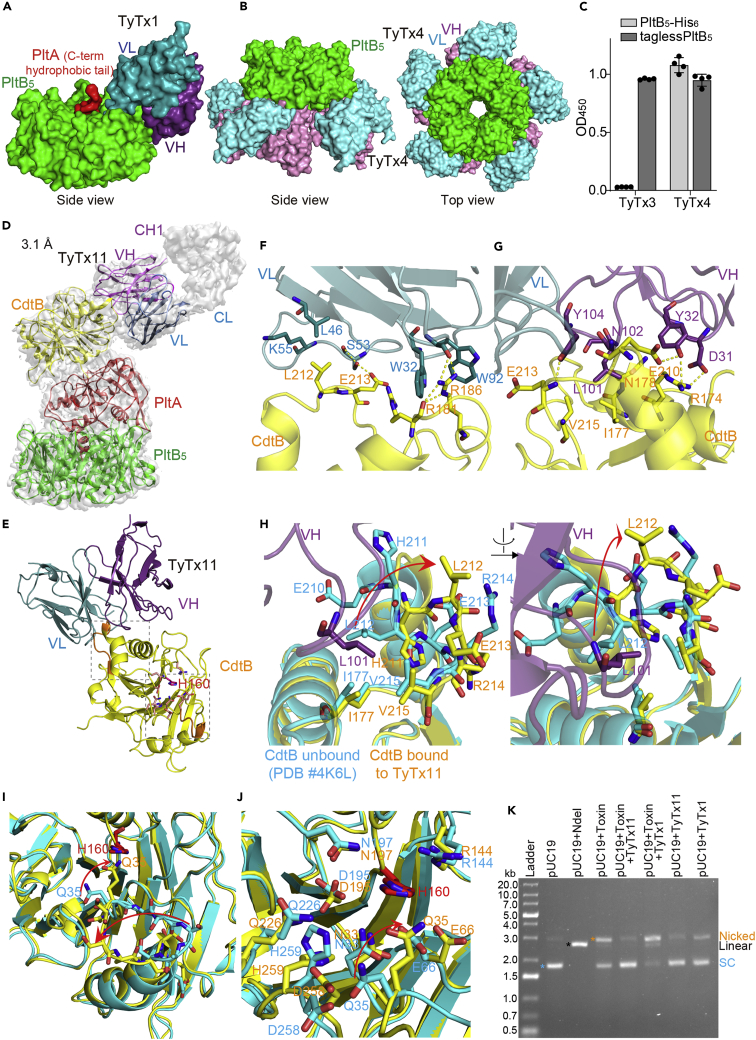
TyTx4 binds to all PltB subunits available per holotoxin, whereas TyTx11 makes CdtB inactive through CdtB catalytic-site conformational change
We have recently mapped toxin-neutralizing epitopes recognized by TyTx1 and 4 via cryoelectron microscopy (cryo-EM) (Nguyen et al., 2021) (summary is shown in Figures 4A and 4B) and TyTx3 via modified ELISA using two types of PltB preparations, PltB containing a histidine hexamer on the predicted epitope site on the C-terminal end and tagless PltB (Figures 4C and S5). In the study, we found that TyTx1 and 3 recognize amino acid (a.a.) residues (a.k.a., epitopes) that are laterally located on PltB, whereas TyTx4 recognizes a.a. located on PltB far from the PltA subunit (Nguyen et al., 2021). Quantitative cryo-EM analyses of the antibody-toxin complexes led us to find that antibodies recognizing the laterally located epitopes on PltB, such as TyTx1 and 3, can occupy up to two PltB subunits among five available per holotoxin due to the interference by the toxin's A subunit PltA (Nguyen et al., 2021) (Figures 4A and S5). This A-subunit-mediated antibody binding interference is not applied to antibodies that recognize the bottom-located epitopes on PltB, such as TyTx4 (Nguyen et al., 2021) (Figure 4B). This molecular mechanism demonstrated through cryo-EM analyses (Nguyen et al., 2021) explains the superior neutralizing capability of TyTx4 among anti-PltB antibodies. The structural studies of the interfaces between the toxin and anti-PltB antibodies also revealed that the epitopes recognized by TyTx1 and TyTx4 overlap with glycan-receptor-binding sites on PltB subunits that are required for the initial toxin membrane trafficking process (Nguyen et al., 2021). Consistently, in the same study, we also demonstrated that anti-PltB antibodies inhibited typhoid toxin binding to cell surface glycan receptors, which subsequently resulted in neutralization of typhoid toxin-mediated cellular toxicity (Nguyen et al., 2021).
Figure 4.
TyTx4 binds to all PltB subunits available per holotoxin, whereas TyTx11 makes CdtB inactive through CdtB catalytic-site conformational change
(A) Note that figure (A and B) are adapted from Nguyen et al. (2021) to compare neutralizing epitopes recognized by anti-PltB antibodies to the epitope recognized by TyTx11. Close-up view of the TyTx1 Fab-typhoid toxin complex structure solved via cryo-EM (EMDB-22699, PDB:7K7H). TyTx1 variable region light chain (VL) and heavy chain (VH), dark cyan and dark purple, respectively; PltB pentamer, green; PltA C-term hydrophobic tail, red. PltA and CdtB subunits are not shown.
(B) Side and top views of the TyTx4 Fab-typhoid toxin complex structure solved via cryo-EM (EMDB-22700, PDB:7K7I). TyTx4 variable region light chain (VL) and heavy chain (VH), cyan and purple, respectively; PltB pentamer, green. PltA and CdtB subunits are not shown.
(C) TyTx3 epitope mapping by modified ELISA. TyTx3 bound to tagless PltB but not to PltB-His6 at the C-terminal end that is located on the lateral side of PltB pentamer in 3-dimensional structure (PDB:4K6L). Binding sites on PltB that are predicted to be recognized by TyTx3 are shown in Figure S5.
(D) Sharpened cryo-EM density map (gray) of typhoid toxin complexed with TyTx11 MAb with ribbon diagram of the refined structure of typhoid toxin (CdtB, yellow; PltA, red; PltB pentamer, green) bound to variable regions of the light chain (VL, blue) and the heavy chain (VH, purple).
(E) Ribbon diagram of the interface between CdtB subunit (yellow) and TyTx11 VL and VH. The flexible loop recognized by TyTx11 and the CdtB catalytic residue H160 are highlighted in orange and red, respectively. Two locations on CdtB where the conformational changes occurred are highlighted in dotted boxes. The box near the interface is highlighted in Figure 4H, and the box on the bottom is zoomed in in Figures 4I and 4J).
(F and G) Close-up views of the interactions between CdtB subunit (yellow) and TyTx11 VL (blue) and VH (purple) (F and G, respectively). Arg174, Arg174, Ile177, Asn178∗, Arg181, Arg186, Arg186, Glu210, Leu212, Leu212, Glu213, Glu213, and Val215 in the CdtB subunit interact with Asp31 (VH, H-bond), Tyr32 (VH, H-bond), Leu101 (VH, hydrophobic), Asn102 (VH, H-bond), Trp32 (VL, π stacks), Trp32 (VL, π stacks), Trp92 (VL, H-bond and π stacks), Tyr32 (VH, H-bond), Leu46 (VL, hydrophobic), Lys55 (VL, hydrophobic), Tyr104 (VH, H-bond), Ser53 (VL, H-bond), and Leu101 (VH, hydrophobic), respectively. Asterisk indicates H-bond via their main chains.
(H) Structure alignment of CdtB bound to TyTx11 (CdtB, yellow; TyTx11 VH, purple) and CdtB unbound (PDB:4K6L, cyan).
(I and J) Two different close-up views of the region near CdtB H160. (I) Close-up view of residues in the catalytic site. (J) Further close-up view of residues near CdtB H160.
(K) DNA agarose gel image assessing antibody-mediated protection of typhoid toxin (toxin)-induced cleavage of pUC19 plasmid. SC, supercoiled.
See also Figures S5–S7 and Table S2.
With this information in hand, to explain and compare the mechanism by which TyTx11 is also superior in neutralizing typhoid toxin, we solved the atomic structure of TyTx11 complexed with typhoid toxin via cryo-EM. In contrast to other cryo-EM approaches that use Fab fragments, here, we directly used TyTx11 IgG and successfully obtained the complex structure at a resolution of 3.1 Å (Figures 4D–4J, and S6, Table S2). It is conceivable that this new approach will make similar structural analyses much more straightforward in other antibody studies.
We found that TyTx11 bound to the CdtB subunit of typhoid toxin through the flexible region adjacent to the CdtB catalytic site containing the key residue His160 (Figures 4D and 4E). Significant inter-molecular interactions between CdtB and TyTx11 are Arg174, Arg174, Ile177, Asn178∗, Arg181, Arg186, Arg186, Glu210, Leu212, Leu212, Glu213, Glu213, and Val215 of the CdtB subunit forming H-bonds, hydrophobic interactions, and/or π stacks with Asp31 (VH, H-bond), Tyr32 (VH, H-bond), Leu101 (VH, hydrophobic), Asn102 (VH, H-bond), Trp32 (VL, π stacks), Trp32 (VL, π stacks), Trp92 (VL, H-bond & π stacks), Tyr32 (VH, H-bond), Leu46 (VL, hydrophobic), Lys55 (VL, hydrophobic), Tyr104 (VH, H-bond), Ser53 (VL, H-bond), and Leu101 (VH, hydrophobic), respectively (Figures 4F, 4G, and S7). Asterisk indicates H-bond via their main chains. VH and VL stand for the variable regions of the heavy and light chains, respectively. The kappa light chain was used in TyTx11 because the PCR primer sets recognizing the kappa chain, but not the counterpart primer sets for the lambda chain, yielded the PCR amplicons (Figure S3).
To understand the mechanism of how TyTx11 can neutralize typhoid toxin without directly masking the CdtB catalytic site, the region near H160, we compared CdtB bound to TyTx11 with CdtB unbound (PDB:4K6L) to investigate whether a conformational change occurred (Figures 4E–4J). We found that notable conformational changes occurred in two locations on CdtB: the flexible loop near the interface between CdtB and TyTx11 (gray box near the interface in Figures 4E and 4H) and the region near CdtB's H160 (gray box on the bottom in Figures 4E, 4I, and 4J). The position of CdtB Leu212 in the flexible loop was drastically changed in the complex, primarily due to the repulsion between CdtB Leu212 and TyTx11 Leu101 (Figure 4H). This, in turn, resulted in the position changes of not only several other residues in this flexible loop, including CdtB Ile177, His211, Glu213, Arg214, and Val215 (Figure 4H), but also several residues located near CdtB H160 (Figures 4I and 4J).
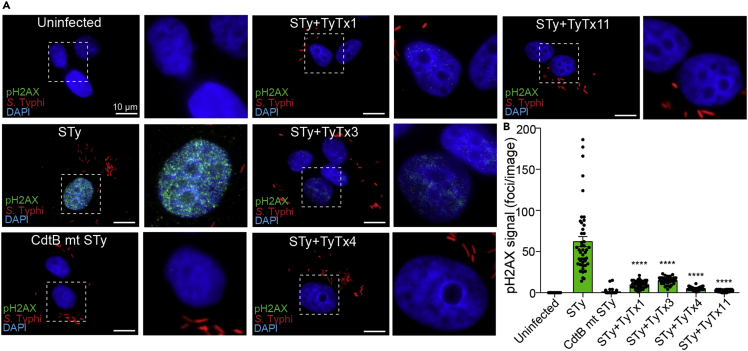
Based on the observed toxin-neutralizing activity of TyTx11, we then hypothesized that these drastic conformational changes of the flexible loop and the region adjacent to the CdtB catalytic site inhibit the nuclease activity of CdtB. To test this hypothesis, we conducted two experiments: standard plasmid cleavage assay evaluating the nicking of the plasmid by bacterial genotoxins and more sensitive quantitative fluorescent microscopy that uses a phospho-γ-H2AX (Ser139) MAb in quantifying the host cell DNA damage repair response. Through pUC19 plasmid cleavage assay, we found that TyTx11 effectively inhibited typhoid toxin-mediated nicking of the plasmid pUC19, but anti-PltB antibody TyTx1 did not (Figure 4K). To quantify the results using a more sensitive readout, we carried out fluorescence microscopy to detect host cell DNA damage repair responses (Figure 5). The strong positive phospho-γ-H2AX signal (green) in the nucleus of host cells reflects the host cell DNA damage induced by the CdtB subunit of typhoid toxin, which is strikingly different from no detectable signal of phospho-γ-H2AX observed in uninfected cells or host cells infected with S. Typhi harboring catalytic-mutant CdtBH160Q (Figure 5A). With this system, we then treated S. Typhi-infected host cells with TyTx11 or other indicated antibodies. The phospho-γ-H2AX signal in the nucleus of host cells infected with S. Typhi was significantly lower in the presence of TyTx11 than the signal observed in S. Typh-infected host cells (Figures 5A and 5B), indicating that the TyTx11-induced conformational changes of the flexible loop and CdtB catalytic site indeed inhibits the CdtB activity. Moreover, consistent with the neutralizing activities of anti-PltB MAbs by inhibiting toxin-glycan receptor interactions (Nguyen et al., 2020), an essential initial step in the toxin endocytosis process in host cells, TyTx1, 3, and 4 also reduced the phospho-γ-H2AX signal, compared with S. Typhi-infected host cells (Figures 5A and 5B). Consistent with other neutralizing results, this microscopy-based quantification also indicates that TyTx4 and 11 are more productive in neutralizing typhoid toxin (Figure 5B).
Figure 5.
The CdtB catalytic-site conformational change induced by TyTx11 makes CdtB an inactive form
(A) Henle-407 cells were left uninfected or infected with 30 MOI S. Typhi(pKan) for 24 h. Host cells were incubated with 30 MOI bacteria for 1 h, followed by 100 μg/mL gentamicin treatment for 30 min to eliminate extracellular bacteria, washing, and incubation in complete culture media containing 10 μg/mL gentamicin for 24 hr. S. Typhi CdtB catalytic mutant (pKan) was included as a control. When indicated, 10 nM antibody was added to cell culture media during the 24-h incubation period. Representative fluorescent microscope images showing pH2AX (green, reflecting host cell DNA damage repair response), S. Typhi (red), and host cell DNA (blue). Zoom-in images (right panels) corresponding to the dotted boxes in the overall images (left panels) are also shown. Scale bar, 10 μm.
(B) pH2AX signal quantification in microscopic images obtained from three independent experiments. Bars represent average ±SEM. ∗∗∗∗p < 0.0001, relative to the STy group. n = 45 per group. Two-tailed unpaired t tests.
All nAbs neutralize typhoid toxin in vivo, as demonstrated by using lethal dose typhoid toxin challenge to mice
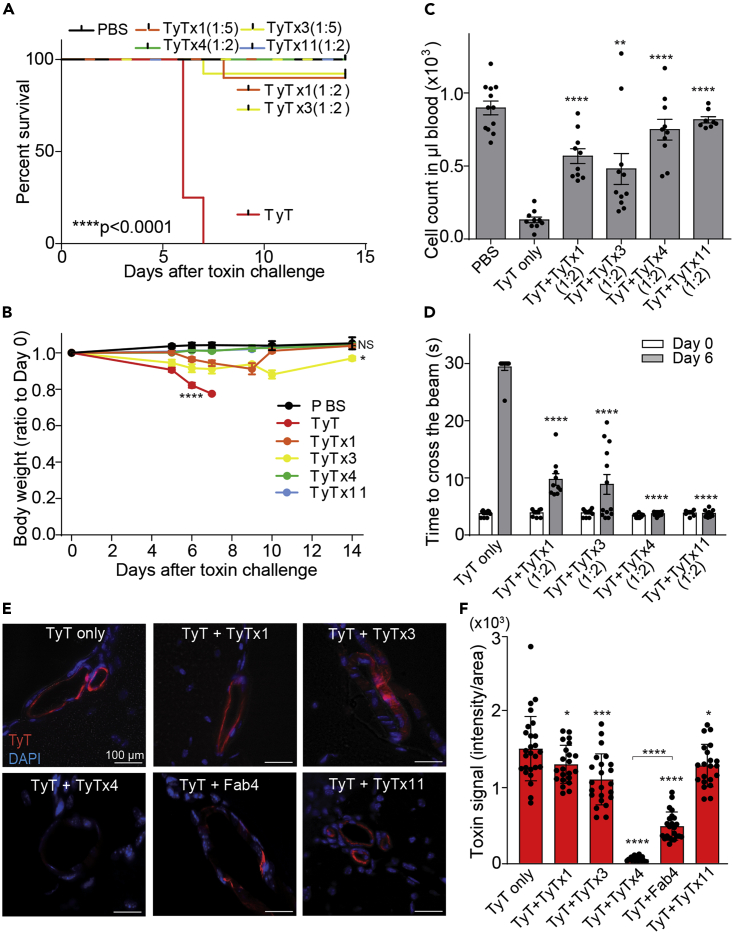
We next investigated whether these newly identified neutralizing epitopes could also neutralize typhoid toxin in vivo. As described in previous studies (Yang et al., 2018b), we intravenously administered mice with a mixture of the lethal dose of typhoid toxin (LD100) and the indicated nAb at an approximate molar ratio of 1:2 (typhoid toxin: nAb) or 1:5. TyTx1 (1:5), TyTx3 (1:5), TyTx4 (1:2), and TyTx11 (1:2) each afforded mice protection against a lethal dose typhoid toxin challenge, as measured by survival, body weight changes, neutrophil counts, and beam balance walking tests (Figures 6A–6D). Consistent with an increased dose of TyTx1 (1:5) and TyTx3 (1:5) exhibiting the complete protection against a lethal dose typhoid toxin challenge, mild signs of typhoid toxin-mediated toxicities were observed in some mice when a lower dose of TyTx1 and 3 (1:2) was used (Figures 6A–6D), which is also consistent with in vitro neutralizing assay results.
Figure 6.
All nAbs neutralize typhoid toxin in vivo
(A–D) Groups of Cmah null mice were administered with LD100 of typhoid toxin with or without nAbs (1:2 molar ratio between toxin and nAb; when indicated, 1:5 mixtures for TyTx1 and TyTx3). Survival (A), body weight changes of mice (B), circulating neutrophil counts in mouse peripheral blood on Day 6 (C), and balance beam walking results of the mice on Day 0 and Day 6 (D) after receiving toxin with or without indicated nAbs. Bars represent the mean ± SEM. n = 8–12. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001, NS, not significant, relative to the PBS group for (B) and the TyT only group for (C and D). The low-rank test was performed for (A) and two-tailed unpaired t tests for (B, C, and D).
(E) Representative images of brain tissue sections showing toxin signals in brain endothelial cells that were evaluated 2 h after administration of AF555-conjugated typhoid toxin (red) and nAbs (1:2) into Cmah null mice. Scale bar, 100 µm.
(F) Quantification of the toxin signals in brain tissue sections.
To better understand the mechanism of antibody-mediated toxin binding inhibition to target host cells and tissues, we administered mice with a mixture of Alexa Fluor 555-conjugated typhoid toxin and indicated nAbs and quantified the toxin binding to brain endothelial cells, one of typhoid toxin's target host cells. Consistent with the observed antibody-mediated protection of upper motor function defects induced by typhoid toxin treatment (Figure 6D), TyTx4 almost completely inhibited typhoid toxin binding to brain endothelial cells (Figures 6E and 6F). In contrast, TyTx1 and 3 only partially inhibited typhoid toxin binding to these cells (Figures 6E and 6F). Unlike some host cells, such as intestinal epithelial cells that predominantly express α2-3 sialosides, brain endothelial cells predominantly express α2-6 sialosides (Lee et al., 2020). Typhoid toxin can use both types of glycans as receptors for toxin endocytosis (Lee et al., 2020; Nguyen et al., 2020; Song et al., 2013; Yang et al., 2018b). We previously demonstrated that the laterally located glycan-binding pocket called BS1 on PltB is used when typhoid toxin uses α2-6 sialosides as a glycan receptor for endocytosis (Lee et al., 2020). Given these previous observations, TyTx4's superior inhibition of typhoid toxin binding to brain endothelial cells was a surprising observation, because TyTx4 recognizes a.a. residues located on the bottom side of PltB pentamer. Based on these findings, we hypothesized that the constant regions of IgG inhibit the interaction of the laterally located glycan-binding pockets on PltB with α2-6 sialosides. To test this hypothesis, we prepared TyTx4-Fab fragments by papain digestion and column purification and compared the effects between TyTx4 Fab fragment (Fab4) and TyTx4. Fab4 indeed exhibited a reduced toxin binding inhibition compared with TyTx4 IgG (Figures 6E and 6F). Also, in support of the hypothesis, we observed a similar difference in antibody-mediated toxin binding inhibition between TyTx4 and Fab4 in another cell type that also predominantly expresses α2-6 sialosides, such as murine primary lymphocytes as demonstrated by SNA (a lectin specific for α2-6 sialosides) (Figures S8A and S8B). In contrast, this phenotype was not observed when we used a cell type that predominantly expresses α2-3 sialosides such as murine primary granulocytes (Lee et al., 2020; Nguyen et al., 2020) (Figures S8C and S8D). These results indicate that both the epitope-binding variable region and the remaining part of nAbs contribute to the toxin-neutralizing outcomes, providing valuable insights into typhoid toxin-neutralizing strategies for use in humans.
Last, through BLAST, we found that all newly identified neutralizing epitopes are highly conserved across MDR and XDR S. Typhi clinical isolates because all clinical isolates identified thus far encode identical typhoid toxin genes, including pltB and cdtB (Lee et al., 2020) (Tables S3 and S4). These results demonstrating the high conservation of neutralizing epitopes on PltB and CdtB identified in this study also provide critical insights into typhoid toxin-neutralizing strategies for future therapeutic use and subunit vaccine development.
Discussion
We demonstrated that typhoid toxin is continuously secreted by antibiotic-resistant S. Typhi even when S. Typhi-infected host cells are treated with antibiotics. Given the global spread of antibiotic-resistant S. Typhi, this is a significant finding of public health relevance. Using the four unique hybridoma clones generated, we identified toxin-neutralizing epitopes located on glycan receptor-binding PltB and nuclease CdtB subunits. Using several quantitative in vitro and in vivo assays, some of which have been newly established as part of this study, we demonstrated that two nAbs are more effective at neutralizing typhoid toxin. The other nAbs were also able to neutralize typhoid toxin when excessive amounts of antibodies were administered. The neutralizing epitopes newly identified are highly conserved across all clinical isolates of S. Typhi, because pltB and cdtB genes found in all S. Typhi clinical isolates are identical (Tables S3 and S4). The remarkable high conservation of typhoid toxin genes throughout the evolution process and across all clinical isolates indicates (1) the anticipated significant contribution of typhoid toxin to infection of S. Typhi and (2) the expected high effectiveness of neutralizing epitopes identified in this study as anti-toxin strategies. Therefore, we envision that the development of anti-toxin strategies against typhoid toxin is valuable, and that anti-toxin strategies developed based on the findings made in this study may be sufficient against typhoid toxin produced by S. Typhi (and S. Paratyphi A, the cause of paratyphoid fever that also secretes the typhoid toxin identical to S. Typhi's typhoid toxin) during actual human infection.
We found that both direct and indirect mechanisms contribute to the outcomes of typhoid toxin neutralization by anti-PltB nAbs. The variable regions of anti-PltB nAbs can directly mask the glycan receptor-binding pockets located on the lateral and bottom sides of PltB subunits. However, we found that none of the anti-PltB nAbs can directly mask all 15 glycan receptor-binding pockets available per holotoxin (Nguyen et al., 2021 and this study). Of the 15 glycan-binding sites, five glycan receptor-binding pockets are located on the lateral side of PltB pentamer, which plays an essential role in binding to target host cells predominantly expressing α2-6 sialosides (e.g., brain endothelial cells and lymphocytes) and α2-3 sialosides (e.g., epithelial cells and neutrophils) (Lee et al., 2020). The remaining 10 glycan receptor-binding pockets are located on the bottom side of the PltB pentamer, which interacts exclusively with α2-3 sialosides (Lee et al., 2020). Intriguingly, the constant regions of anti-PltB nAbs can hinder glycan receptor-binding pockets that are not directly masked by the variable regions of the antibodies from glycan receptor binding, as demonstrated by using TyTx4 as an example. It is conceivable that multivalent anti-PltB antibodies targeting multiple neutralizing epitopes located on the lateral and bottom sides of PltB pentamer would be better at directly masking most of the glycan receptor-binding pockets available per holotoxin. However, it is essential to note that the direct masking of all five laterally located glycan receptor-binding pockets is not straightforward because of the “A” subunit-mediated interference with antibody binding to the laterally located epitopes (Nguyen et al., 2021).
We identified that the primary mechanism of protection for anti-CdtB nAb TyTx11 was by inducing CdtB conformational changes in the flexible loop and the region adjacent to H160, thereby making CdtB an inactive form. TyTx11 binds to the flexible loop and causes the repulsion between CdtB Leu212 and TyTx11 Leu101, leading to the position changes of several other residues in this flexible loop and the catalytic site (Figures 4E–4J). Follow-up plasmid cleavage assays and quantitative fluorescent microscopy demonstrated that the CdtB conformational change induced by TyTx11 resulted in typhoid toxin neutralization.
In cryo-EM studies, we used TyTx11 IgG rather than the Fab fragments and successfully solved the antibody-toxin complex's atomic structure. This is the first report using such a simple procedure and therefore serves as an important example for similar antibody structure studies. Structure and function studies of the antibody-toxin complexes helped determine the molecular protection mechanisms of the neutralizing epitopes. We also successfully mapped B-cell epitope using modified ELISAs with two antigen preparations with unmodified and modified epitopes. Furthermore, several quantitative approaches have been established as part of this study to demonstrate typhoid toxin production by antibiotic-resistant S. Typhi and to objectively evaluate typhoid toxin neutralization capabilities by different nAbs, including quantitative fluorescent microscopy and flow cytometry measuring typhoid toxin production and host responses to typhoid-toxin-induced toxicities. These approaches will help other studies concerning bacterial toxins and toxin-neutralizing strategies.
In conclusion, we report the first map of neutralizing B cell epitopes on typhoid toxin that is highly conserved across all S. Typhi clinical isolates, with the molecular mechanisms of antibody-mediated toxin neutralization. Given that typhoid toxin is continuously produced by antibiotic-resistant S. Typhi irrespective of antibiotic treatment, the identified neutralizing epitopes would serve as a basis for future typhoid toxin-neutralizing strategies. We also predict that this strategy will be most effective when anti-toxin strategies are combined with anti-bacterial strategies (e.g., anti-LPS, anti-Vi, anti-outer membrane proteins), allowing for the elimination of both the bacteria and the toxin that is expected to be secreted until complete clearance of bacteria in infected patients is obtained.
Limitations of the study
Primarily associated with the strict human adaption of S. Typhi, in this study, we used human cell infection models with antibiotic-susceptible and resistant S. Typhi and in vivo mouse models challenged with lethal dose typhoid toxin when evaluating typhoid toxin-neutralizing capabilities of antibodies. It is important to note that non-human primates (NHP) such as chimpanzees are not appropriate for assessing typhoid toxin neutralization because NHP do not express specific glycan receptors recognized by typhoid toxin, required for the first step of the toxin endocytosis process (Deng et al., 2014). Further studies in human volunteer trials with antibiotic-resistant S. Typhi (or antibiotic-susceptible S. Typhi without antibiotic-mediated early intervention) can help verify whether the identified neutralizing epitopes are also neutralizing typhoid toxin in people.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-typhoid toxin MAbs and Fabs | This manuscript | See methods details |
| Anti-Flag M2 antibody | Sigma #F1804 | RRID:AB_262044 |
| Anti-phospho-histone H2AX antibody | ThermoFisher LF-PA0025 | RRID:AB_1958836 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Typhoid Toxin, Toxoid, & PltB subunits | This manuscript | See methods details |
| Critical Commercial Assays | ||
| Alexa Fluor 555 Protein Labeling Kit | ThermoFisher | A20174 |
| RNeasy mini kit | Qiagen | 74104 |
| iScript cDNA synthesis kit | Bio-Rad Laboratories | 1708891 |
| Herculase II Fusion DNA polymerase | Agilent | 600677 |
| Phusion DNA polymerase | New England BioLabs | M0530 |
| QIAquick Gel Extraction Kit | Qiagen | 28706 |
| Deposited Data | ||
| Typhoid toxin bound to TyTx11 Mab | RCSB Protein Data Bank | PDB:6VX4 (see Table S2) |
| Experimental Models: Cell Lines | ||
| Henle-407 Cells | Originally obtained from ATCC | Validated using intestinal epithelial markers |
| Jurkat Cells | Originally obtained from ATCC | TIB-152 |
| Sp2/0-Ag14 | Originally obtained from ATCC | CRL-1581 |
| Experimental Models: Organisms/Strains | ||
| B6.129X1-Cmah tm1Avrk/J Mice (a.k.a. Cmah KO) | Jackson Lab | 017588 |
| Salmonella enterica serovar Typhi | See methods details | See methods details |
| Salmonella Typhi CdtBH160Q | See methods details | See methods details |
| Oligonucleotides | ||
| See Table S5 | IDT | See Table S5 |
| Software and Algorithms | ||
| GraphPad Prism 6 or 8.3 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| LI-COR Image Studio | LI_COR Image Studio Software | https://www.licor.com/bio/image-studio/ |
| Excel | Microsoft | Office 365 |
| FlowJo V10 | TreeStar | https://www.flowjo.com/ |
| ImageJ | National Institutes of Health | https://imagej.nih.gov/ij/ |
| Unicorn 6.3 | GE Healthcare Life Sciences | https://www.gelifesciences.com/en/us/shop/unicorn-6-3-p-01118 |
| BLASTp | National Institutes of Health | https://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins |
| Biacore T200 Evaluation Software | GE Healthcare | https://www.cytivalifesciences.com |
| CryoSparc 2 | Cryosparc | https://cryosparc.com |
| Relion 3 | Relion | https://www3.mrc-lmb.cam.ac.uk/relion/index.php/Main_Page |
| PyMol | Schrodinger | https://pymol.org/2/ |
| Chimera | UCSF Chimera | https://www.cgl.ucsf.edu/chimera/ |
| PHENIX | PHENIX | https://www.phenix-online.org/ |
| Coot | Coot | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| Other Recombinant Proteins | ||
| Thyroglobulin | Sigma | T9145 |
| Ferritin | Sigma | F4503 |
| Adolase | Sigma | A2714 |
| Conalbumin | GE Healthcare Life Sciences | Part of 28403841 |
| Ovalbumin | GE Healthcare Life Sciences | Part of 28403841 |
| Carbonic Anhydrase | GE Healthcare Life Sciences | Part of 28403841 |
| Ribonuclease A | GE Healthcare Life Sciences | Part of 28403841 |
Resource availability
Lead contact
Further requests for resources and materials should be directed to and will be fulfilled by the lead contact, Jeongmin Song (jeongmin.song@cornell.edu).
Materials availability
Antibodies generated as part of this study are available upon request via an appropriate material transfer agreement.
Data and code availability
The published article includes all datasets generated during this study. The atomic structure of TyTx11 bound to typhoid toxin has been deposited in the RCSB Protein Data Bank under PDB:6VX4 (Table S2).
Experimental models and subject details
Bacterial strains
WT and CdtB catalytic mutant Salmonella enterica serovar Typhi ISP2825 have been described previously (Song et al., 2013). For infection experiments, S. Typhi strains were grown at 37°C in 2 mL LB broth containing 0.3 M NaCl to an OD600nm of ∼0.9 after inoculation from an overnight culture in 2 mL LB broth at a dilution of 1:50.
Mammalian cell culture conditions
Human intestinal epithelial Henle-407 cells and human peripheral blood T lymphocyte Jurkat cells were cultured in DMEM high glucose (Invitrogen) and RPMI-1640 (Invitrogen) supplemented with 10% FBS (Hyclone cat# SH30396.03, Lot# AD14962284), respectively. Sialic acid contents of the FBS used were validated, which was ∼99% Neu5Ac and less than 1% Neu5Gc. Cells were kept at 37°C in a cell culture incubator with 5% CO2. Mycoplasma testing was conducted on a regular basis as part of the cell maintenance practice.
Mouse intoxication model
All mouse experiments were conducted following protocols approved by the Cornell University's institutional Animal Care and Use Committee (IACUC; protocol # 2014-0084) for mouse neutralization experiments or the Wadsworth Center's IACUC (protocol # 17-428) for immunization experiments. The experiments were followed by IACUC and AAALAC guidelines. Age- and sex-matched 5- to 8- week-old CMAH null mice were randomly allocated to each group. The mice used in this study were originally purchased from the Jackson Laboratory and bred in a vivarium in the animal facility at Cornell University. All the knockout mice used were genotyped regularly.
Methods details
Expression and purification of recombinant typhoid toxin, typhoid toxoid, and PltB pentamer
Overexpression and purification of typhoid toxin, typhoid toxoid, and PltB homopentamer were carried out as previously described (Deng et al., 2014; Song et al., 2013; Yang et al., 2018b). Purified toxin preparations were further subjected to high-capacity endotoxin removal resin (Pierce, Thermo Fisher Scientific). LPS levels of all the toxin preparations were determined using a ToxinSensor chromogenic LAL endotoxin assay kit (GenScript; <0.2 EU/ml). When indicated, purified toxins were fluorescently labeled with Alexa Fluor-555 dye or (Molecular Probes, Thermo Fisher Scientific) according to the vendor’s recommendation.
Hybridoma generation
Female BALB/c mice approximately 5-7 weeks of age were purchased from Taconic Biosciences (Albany, NY) and housed under conventional, specific pathogen-free conditions. Mice were immunized via intraperitoneal injections with 2 μg of ultrapure endotoxin-free recombinant typhoid toxoid without adjuvant at two-week intervals. Genetically engineered inactive forms of typhoid toxin were used in the study. Specifically, the toxoid carries point mutations in PltAE133A and CdtBH160Q, D195S. The toxoid has the same A2B5 stoichiometry as WT typhoid toxin, as determined via size exclusion chromatography. Serum reciprocal endpoint titers were >100,000 following three immunizations. B cell hybridomas were then generated by performing Hybri-Max PEG fusion procedures using the Sp2/0-Ag14 (ATCC® CRL-1581) myeloma. Similar to the procedure established previously (Van Slyke et al., 2018), we seeded the resulting hybridomas onto wells of 96-well cell culture-treated microtiter plates and hybridomas were selected in DMEM media supplemented with 10% UltraCruz® Hybridoma Cloning Supplement (HCS) (Santa Cruz Biotechnology, Dallas, TX), 10% fetal calf serum, oxaloacetate, pyruvate, and insulin (OPI), hypoxanthine/aminopterin/thymidine (HAT), and penicillin/streptomycin. During the procedures, HAT was gradually replaced with hypoxanthine-thymidine (HT) and surviving hybridomas secreting antibodies of interest were cloned by limiting dilution and expanded in DMEM media without HT, before being transitioned to either CD Hybridoma AGT (Invitrogen) or DMEM supplemented with 4 mM L-glutamine, 4.5 g/L glucose, 1 mM sodium pyruvate, 1.5 g/L sodium bicarbonate, 10% HCS, 20% Ultra Low IgG Fetal Bovine Serum (Invitrogen), and penicillin/streptomycin. B cell hybridomas were cloned by limiting dilution three times to ensure clonality.
MAb purification
Hybridoma clones were selected upon a positive reaction against typhoid toxoid in a direct enzyme-linked immunosorbent assay (ELISA). Murine MAbs were purified from hybridoma supernatants via a standard procedure using protein G chromatography and subjected for epitope characterization using standard ELISAs, Western blots, and Size-exclusion chromatography. In brief, hybridoma cell culture supernatants were cleared via a two-stage process, centrifugation for 15 min at 5,600 rpm at 4°C, followed by filtration through 0.2 μm filters, to remove cell debris. Protein G resins were packed in a 25 ml gravity plastic column and equilibrated by flowing 20 ml binding buffer (20 mM sodium phosphate buffer, pH 7.0). The cleared hybridoma supernatants were submitted to the prepared Protein G resin three times. The bound antibodies were eluted by applying 10 ml elution buffer (0.1 M glycine-HCl, pH 2.7), which was harvested in a falcon tube containing 1 ml equilibration buffer (1 M Tris-HCl, pH 8.0) for immediate neutralization. The purity and yield of purified antibodies were monitored via SDS-PAGE. Purified antibodies were aliquoted and stored at -80°C until use.
MAb affinity determination: Surface Plasmon Resonance assay
TyTx11 binding kinetics and affinity were determined by performing SPR assays using the Biacore T200 (GE Healthcare), as previously described (Rudolph et al., 2020). Series S CM5 chips were immobilized with WT typhoid toxin using amine coupling in 10 mM acetate buffer at pH 4.5. Amine coupling was performed at ambient temperature using a concentration of 5 μg/mL of typhoid toxin, with a contact time of 30 seconds at a flow rate of 10 μl/min. Immobilization procedures were run in 0.1 M Hepes buffer containing 1.5 M NaCl at pH 7.4 to obtain a target response bound of ∼100-120 RU. Pilot experiments to optimize experimental parameters were then performed using 10-fold serial dilutions of each MAb injected at a flow rate of 50 μL/min with a contact time of 120 sec and dissociation time of 600 sec, which were conditions deemed appropriate by the Biacore T200 Evaluation software (GE Healthcare) for the affinity determination of each MAb. Following optimization of the concentration range and conditions, 4-fold serial dilutions of each MAb were input for replicate experiments. Sensograms for each replicate were analyzed to fit a bivalent binding model using the Biacore T200 Evaluation software. Data obtained from the Biacore T200 was imported into GraphPad Prism 8.3 for the generation of final sensograms.
TyTx11 variable region sequencing
Antibody sequencing was conducted based on a recently published method (Meyer et al., 2019). In brief, total RNA was extracted from the hybridoma cells using the RNeasy mini kit (Qiagen). The cDNA synthesis and PCR amplification of antibody variable regions were performed. The iScript Select cDNA Synthesis Kit (Bio-Rad) and mouse IgG reverse transcription primers were used for reverse transcription. A touch-down/step-down PCR was occurred with universal forward primer and reverse PCR primer based on the antibody chain to amplify the antibody variable regions (Table S5). The amplicon appeared between 550-600 base pairs on 1% agarose gel. DNA was extracted from the gel using the QIAquick Gel Extraction Kit (Qiagen). The extracted DNA was Sanger-sequenced using the IgG PCR reverse primers.
In vitro intoxication assay
Henle-407 cell intoxication assays were performed to evaluate MAb-mediated neutralization in the context of S. Typhi infection. In brief, Henle-407 human intestinal epithelial cells were cultured in DMEM high glucose +10% FBS (HyClone) and kept at 37°C in a cell culture incubator with 5% CO2. Cells were seeded at 3x104/well into 12-well culture plates and incubated overnight. The next day, cells were infected with WT, WT carrying pKan, or CdtB catalytic mutant Salmonella Typhi carrying the cdtBH160Q gene (replaced the WT cdtB gene) at a multiplicity of infection of 30 or 50 as indicated in the figure legend for 1 hour in HBSS (Invitrogen), treated with 100 μg/mL gentamicin to kill all extracellular bacteria for 45 min, and washed with PBS. Infected cells were then incubated in complete cell culture medium containing 10 μg/mL gentamicin for 72 hours in the presence and absence of 10 nM MAbs. When indicated, 50 μg/mL kanamycin, 0.01 μg/mL ciprofloxacin, or 0.05 μg/mL ciprofloxacin was also added in cell culture media during this 72 hr-incubation period. Fresh media containing 10 μg/mL gentamicin ± 10 nM MAb ± ciprofloxacin or kanamycin were provided to the cells every 24 hrs during incubation. Abmix in 3C-E was prepared by mixing 2.5 nM each of TyTx1, TyTx3, TyTx4, and TyTx11. After incubation for 72 hrs, cells were collected, washed, and fixed for 2 hrs in 70% ethanol/PBS at -20°C. Fixed cells were washed with PBS and resuspended in 500 μL of PBS containing 50 μg/mL propidium iodide, 0.1 mg/mL DNase-free RNase A, and 0.05% Triton X-100. After incubation for 40 min at 37°C, stained cells were washed with PBS, resuspended in 150 μL PBS, filtered, and read using BD Accuri C6 Plus (BD Biosciences), followed by cell-cycle arrest profile analysis using FlowJo software (Treestar Inc).
Plasmid cleavage assay
Standard plasmid cleavage assays were carried out to assess TyTx11-mediated inhibition of the CdtB nuclease activity with some modifications. In brief, 2.5 μl of 25 ng/μl of supercoiled wild-type pUC19 plasmid was mixed with 2 μl of 10x digestion buffer (10x buffer composition: 2.5 M HEPES, pH 7.0, 0.5 M MgCl2, 0.5 M CaCl2), 11 μl of dH2O, and 4.5 μl of the typhoid toxin and antibody premix (1 μM toxin and 1.5 μM antibody mixtures were prepared in PCR tubes and incubated for 30 min at room temperature before adding the remaining ingredient). The final mix was incubated at 37°C for 2 hrs and quenched by the addition of 6x DNA loading dye (NEB). Ten microliters of each reaction were loaded onto 1% agarose gel, and DNA electrophoresis was carried out at 65 V for 1.5 hr. After gel electrophoresis was completed, DNA was stained for 30 min in DNA electrophoresis running buffer containing dye and imaged.
Colony-forming unit (c.f.u.) assay
Henle cells infected with 30 m.o.i. S. Typhi as indicated above. Infected cells were incubated for 24 hrs with antibiotics indicated in Figures 2A and 3A. As described elsewhere, we then lysed the cells in 1 ml sterile PBS containing 0.1% sodium deoxycholate. Bacterial loads were determined by plating 10-fold serial dilutions of homogenates on LB agar plates and incubated overnight at 37°C. Colonies were counted and the number of total c.f.u. recovered was calculated.
Immunofluorescent Staining
Cells seeded on slides were fixed with 1% paraformaldehyde (PFA) for 10 min, washed with PBS, and permeabilized in PBS containing 50 mM NH4Cl, 0.2% Triton X-100 and 3% BSA for 30 min. The slides were stained with indicated antibodies (anti-S. Typh antibody, 1:2000, anti-Flag M2 antibody, Sigma #F1804, 1:2000, anti-phospho-histone H2AX antibody, Thermo Fisher #LF-PA0025, 1:1000) for 2 hrs at room temperature, counterstained with 4′,6-diamidino-2-phenylindole (DAPI), mounted using ProLong antifade mounting solution (Molecular Probes, ThermoFisher Scientific), and viewed. Immunofluorescent images were acquired with a Leica DMI6000B/DFC340 FX fluorescence microscope system. For image acquisition, 1,600 × 1,200-pixel full-frame pictures of various channels were recorded as 16-bit TIFF files with ×20 (numerical aperture (NA) 0.5) or ×40 (NA 0.75) objectives. The filter wavelengths were as follows: Alexa Fluor 488 (for Flag and Phospho-Histon H2AX) – excitation filter 470/40 nm, emission filter 525/50 nm; Alexa Fluor 594 (for Salmonella) and mCherry – excitation filter 545/30 nm, emission filter 610/75 nm; DAPI – excitation filter 340∼380 nm, emission filter 425 nm. The fluorescent signal intensity of images was quantified using the measure function of ImageJ (National Institutes of Health, USA) after subtracting the background. Recorded images were merged using the ImageJ merge channels function and processed further with Adobe Photoshop to adjust the brightness and contrast equally for all recordings.
In vivo mouse experiments
For toxin neutralization experiments, age- and sex-matched 5- to 8- week-old CMAH null mice were randomly allocated to each group. The mice used in this study were originally purchased from the Jackson Laboratory and bred in a vivarium in the animal facility at Cornell University. All the knockout mice used were genotyped regularly. Groups of mice were injected with 100 μL solutions containing 2 μg of the purified typhoid toxin (endotoxin-free) only or a mixture containing typhoid toxin and each MAb (4 μg TyTx1, 3, 4, and 11) via retro-orbital injections. When indicated as 1:5, 10 μg TyTx1 and TyTx3 were used. Changes in the behavior, weight, and survival of the toxin-injected mice were closely monitored as previously described (Deng et al., 2014; Song et al., 2013; Yang et al., 2018b). Balance beam tests were performed to evaluate toxin-mediated motor function deficits, as previously described (Yang et al., 2018b). To assess toxin-mediated damage on immune cells, blood samples were collected by submandibular bleeding in Microtainer tubes coated with EDTA as an anticoagulant (BD Biosciences), kept at room temperature, and analyzed within 2 hrs after blood collection using a Hemavet 950FS hematology analyzer (Drew Scientific).
Cryo-EM, data collection and structure determination
Preparation of MAb and typhoid toxin complex
TyTx11 MAbs bound to wild-type purified typhoid toxin (PltB, PltA, CdtB) were prepared by overnight incubation of the 1:1 mixture of antibody and toxin (250 μl each of ∼1.2 mg/ml proteins) at 4°C, followed by a submission to a Superdex 200 10/300 increase column (GE Healthcare) using a constant flow of 0.5 mL/min of a buffer containing 15 mM Tris-HCl, pH 7.5 and 150 mM NaCl. Purified TyTx11 MAb bound to typhoid toxin was then directly used for Cryo-EM grid/sample preparations using a buffer containing 15 mM Tris-HCl, pH 7.5 and 150 mM NaCl.
Cryo-EM grid/sample preparation and data acquisition
Mab-toxin complex samples were diluted with a buffer containing 15 mM Tris-HCl, pH 7.5 and 150 mM NaCl to a final concentration of 0.1 and 0.2 mg/mL. The initial screen indicated that a concentration of 0.1 mg/ml was suitable for imaging. Samples (3.5 μl) were applied to glow-discharged copper Quantifoil R1.2/1.3 300 Mesh Cryo-EM grid (EMS) mounted, blotted, and plunge frozen in liquid ethane using a Vitribot (ThermoFisher) at 100% humidity at 4°C. Data were acquired at a nominal magnification of 63,000 x for TyTx11 using a Talos Artica (Thermofisher) operating at 200 kV equipped with a Gatan energy filter set to a slit width of 20 eV and K3 detector operating in super-resolution counting mode using a defocus range of -0.8 to -1.8 μm. Fifty frame movies with a super-resolution pixel size of 0.83 were collected over a 4-second exposure for both datasets. Both datasets were collected using similar exposure and dose rates resulting in a total dose of 54.7 e- Å-2 (1.06 e- Å-2/frame) for the TyTx11/Toxin complex.
Image processing
Motion correction of each image stacks was performed using MotionCor2 function in RELION (Nakane et al., 2018; Zheng et al., 2017).
TyTx11/Toxin complex
(Figure S6): Following CTF estimation, a total of 935 movies collected during data acquisition were subjected to Laplacian-of-Gaussian particle picking in RELION 3. Approximately 1000 particles were manually picked, extracted, and 2D classified to serve as initial templates for automated particle picking in RELION 3. The resulting 1,009,626 particles were extracted at 3.0457 Å/pixel to reduce the memory constrain. The 2D classification was used to remove incorrectly picked particles. CryoSparc 2 was used to generate an ab-initio model from the cleaned particle set. This ab-initio model was used as a reference map for subsequent 3D classifications and refinement in RELION 3. Initial 3D classification revealed three out of five classes contain particle resemble the structure of Fab. The particles from those classes were combined and submitted to another round of 3D classification, which revealed one of four classes contained high-quality particles resulting in a high-resolution estimate for the model. Particles from this class were then used for subsequent per-particle CTF refinements and 3D refinement. When no further improvement can be done, the particles were then unbinned and reextracted at 1.23 Å/pixel. After subsequent CTF refinements and 3D refinements, a Bayesian polishing step followed by a final 3D classification was used to remove any final junk particles. Final reconstruction using 61,478 particles were generated. The final 3D reconstruction has a 0.143 FSC cutoff resolution of 3.12 Å according to RELION.
Model building and refinement
Chimera (www.cgl.ucsf.edu) was used to place the crystal structures of typhoid toxin (PDB:4K6L) into the sharpened reconstructions as preliminary atomic models. The preliminary atomic models for TyTx11 Fab variable region were obtained from the following models: the light chain from PDB:6GFF and the heavy chain from PDB:1F11. Constant regions of the TyTx11 are too flexible for the proper density fitting and thus left unbuilt in the final deposited model. Coot (https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/) was then used to manually rebuild these preliminary models. The flexibility of the antibody constant regions, CdtB subunit, and the majority of the PltA subunit prevented us from confidently building these portions of the maps, so they were left unbuilt. Models were refined in Phenix using the real-space refinement module (phenix-online.org). The quality of the final models was validated using MolProbity (molprobity.biochem.duke.edu). Figures were generated using Chimera and Pymol (Schrodinger, Inc). Cryo-EM density maps and refined complex structures were deposited under EMD-21429 and PDB:6VX4 for the TyTx11 MAb-toxin complex (Table S2).
Quantification and statistical analysis
Data were tested for statistical significance with GraphPad Prism software. The number of replicates for each experiment and the statistical test performed are indicated in the figure legends. Image analysis and quantification and cell cycle profile analysis and quantification were performed using ImageJ and FlowJo software, respectively.
Acknowledgments
We thank Greta Van Slyke for her comments on hybridoma establishment and maintenance and the Wadsworth Center's Media and Tissue Culture Core facility for preparing media and assistance with hybridoma scale-up. We thank J. Ryan Feathers for his comments on cryo-EM data analysis and Mariena S. Ramos and Katie Spoth of the Cornell Center for Material Research (CCMR) for their assistance in using FEI Vitrobot and Talos Artica TEM/SEM. This work was supported in part by NIH R03 AI135767 to J.S. and N.J.M.; NIH R01 AI137345, AI139625, and AI141514 to J.S.; and NSF DMR-1719875 to the CCMR. The funders had no role in the study design, data collection, analysis, decision to publish, or manuscript preparation.
Author contributions
C.A. conducted experiments shown in Figures 1A–1C, 1F-H, 4C, 5, and 6. Y.-A.Y. conducted or contributed to experiments shown in Figures 2A–2D, and 6. D.P.N. conducted experiments shown in Figures 1D, 1E, and 3A–3E. T.N. conducted antibody sequencing and experiments shown in Figures 4A, 4B, and 4D–4K. A.F.R. generated the TyTx11 hybridoma clone and conducted SPR experiments, and N.J.M. supervised this work. J.H.S. conducted experiments shown in Figures 2A–2D. J.S. supervised this study and prepared the manuscript with input from all authors.
Declaration of interests
The authors declare no competing interests.
Published: May 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102454.
Supplemental information
References
- Abrahams G.L., Hensel M. Manipulating cellular transport and immune responses: dynamic interactions between intracellular Salmonella enterica and its host cells. Cell Microbiol. 2006;8:728–737. doi: 10.1111/j.1462-5822.2006.00706.x. [DOI] [PubMed] [Google Scholar]
- Baker S., Holt K.E., Clements A.C., Karkey A., Arjyal A., Boni M.F., Dongol S., Hammond N., Koirala S., Duy P.T. Combined high-resolution genotyping and geospatial analysis reveals modes of endemic urban typhoid fever transmission. Open Biol. 2011;1:110008. doi: 10.1098/rsob.110008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CDC . Oxford University Press; 2017. CDC Yellow Book 2020: Health Information for International Travel. Chapter 4. Travel-related infectious diseases. [Google Scholar]
- Chong A., Lee S., Yang Y.A., Song J. The role of typhoid toxin in Salmonella typhi virulence. Yale J. Biol. Med. 2017;90:283–290. [PMC free article] [PubMed] [Google Scholar]
- Crump J.A., Mintz E.D. Global trends in typhoid and paratyphoid Fever. Clin. Infect. Dis. 2010;50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng L., Song J., Gao X., Wang J., Yu H., Chen X., Varki N., Naito-Matsui Y., Galan J.E., Varki A. Host adaptation of a bacterial toxin from the human pathogen salmonella typhi. Cell. 2014;159:1290–1299. doi: 10.1016/j.cell.2014.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feasey N.A., Gaskell K., Wong V., Msefula C., Selemani G., Kumwenda S., Allain T.J., Mallewa J., Kennedy N., Bennett A. Rapid emergence of multidrug resistant, H58-lineage Salmonella typhi in Blantyre, Malawi. PLoS Negl. Trop. Dis. 2015;9:e0003748. doi: 10.1371/journal.pntd.0003748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan J.E. Typhoid toxin provides a window into typhoid fever and the biology of Salmonella Typhi. Proc. Natl. Acad. Sci. U S A. 2016;113:6338–6344. doi: 10.1073/pnas.1606335113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galán J.E. Common themes in the design and function of bacterial effectors. Cell Host Microbe. 2009;5:571–579. doi: 10.1016/j.chom.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibani M.M., Jones E., Barton A., Jin C., Meek J., Camara S., Galal U., Heinz E., Rosenberg-Hasson Y., Obermoser G. Investigation of the role of typhoid toxin in acute typhoid fever in a human challenge model. Nat. Med. 2019;25:1082–1088. doi: 10.1038/s41591-019-0505-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haghjoo E., Galán J.E. Salmonella typhi encodes a functional cytolethal distending toxin that is delivered into host cells by a bacterial-internalization pathway. Proc. Natl. Acad. Sci. U S A. 2004;101:4614–4619. doi: 10.1073/pnas.0400932101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K.E., Dolecek C., Chau T.T., Duy P.T., La T.T., Hoang N.V., Nga T.V., Campbell J.I., Manh B.H., Vinh Chau N.V. Temporal fluctuation of multidrug resistant salmonella typhi haplotypes in the mekong river delta region of Vietnam. PLoS Negl. Trop. Dis. 2011;5:e929. doi: 10.1371/journal.pntd.0000929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle D.J., Nair S., Hartman H., Ashton P.M., Dyson Z.A., Day M., Freedman J., Chattaway M.A., Holt K.E., Dallman T.J. Informal genomic surveillance of regional distribution of Salmonella Typhi genotypes and antimicrobial resistance via returning travellers. PLoS Negl. Trop. Dis. 2019;13:e0007620. doi: 10.1371/journal.pntd.0007620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm E.J., Shakoor S., Page A.J., Qamar F.N., Judge K., Saeed D.K., Wong V.K., Dallman T.J., Nair S., Baker S. Emergence of an extensively drug-resistant Salmonella enterica serovar typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. mBio. 2018;9:e00105–e00118. doi: 10.1128/mBio.00105-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Yang Y.A., Milano S.K., Nguyen T., Ahn C., Sim J.H., Thompson A.J., Hillpot E.C., Yoo G., Paulson J.C. Salmonella typhoid toxin PltB subunit and its non-typhoidal Salmonella ortholog confer differential host adaptation and virulence. Cell Host Microbe. 2020;27:937–949.e6. doi: 10.1016/j.chom.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M.M., Simon R. The gathering storm: is untreatable typhoid fever on the way? mBio. 2018;9 doi: 10.1128/mBio.00482-18. e00482–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer L., Lopez T., Espinosa R., Arias C.F., Vollmers C., DuBois R.M. A simplified workflow for monoclonal antibody sequencing. PLoS One. 2019;14:e0218717. doi: 10.1371/journal.pone.0218717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack D.M. Salmonella persistence and transmission strategies. Curr. Opin. Microbiol. 2012;15:100–107. doi: 10.1016/j.mib.2011.10.013. [DOI] [PubMed] [Google Scholar]
- Nakane T., Kimanius D., Lindahl E., Scheres S.H. Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. Elife. 2018;7:e36861. doi: 10.7554/eLife.36861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Lee S., Yang Y.A., Ahn C., Sim J.H., Kei T.G., Barnard K.N., Yu H., Millano S.K., Chen X. The role of 9-O-acetylated glycan receptor moieties in the typhoid toxin binding and intoxication. PLoS Pathog. 2020;16:e1008336. doi: 10.1371/journal.ppat.1008336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Richards A., Neupane D., Feathers R., Yang Y.-A., Sim J.H., Byun H., Lee S., Ahn C., Van Slyke G. The structureal basis of Salmonella A2B5 toxin neutralization by antibodies targeting the glycan-receptor binding subunits. 2021. https://ssrncom/abstract=3745287http://dxdoiorg/102139/ssrn3745287 [DOI] [PMC free article] [PubMed]
- Nikolaus T., Deiwick J., Rappl C., Freeman J.A., Schröder W., Miller S.I., Hensel M. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J. Bacteriol. 2001;183:6036–6045. doi: 10.1128/JB.183.20.6036-6045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qamar F.N., Yousafzai M.T., Sultana S., Baig A., Shakoor S., Hirani F., Wassay A., Khushboo S., Mehmood J., Freeman A. A retrospective study of laboratory-based enteric fever surveillance, Pakistan, 2012-2014. J. Infect. Dis. 2018;218:S201–S205. doi: 10.1093/infdis/jiy205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph M.J., Czajka T.F., Davis S.A., Thi Nguyen C.M., Li X.P., Tumer N.E., Vance D.J., Mantis N.J. Intracellular neutralization of ricin toxin by single-domain antibodies targeting the active site. J. Mol. Biol. 2020;432:1109–1125. doi: 10.1016/j.jmb.2020.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Gao X., Galan J.E. Structure and function of the Salmonella Typhi chimaeric A(2)B(5) typhoid toxin. Nature. 2013;499:350–354. doi: 10.1038/nature12377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J., Willinger T., Rongvaux A., Eynon E.E., Stevens S., Manz M.G., Flavell R.A., Galan J.E. A mouse model for the human pathogen Salmonella typhi. Cell Host Microbe. 2010;8:369–376. doi: 10.1016/j.chom.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spano S., Ugalde J.E., Galan J.E. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe. 2008;3:30–38. doi: 10.1016/j.chom.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Srikanth C.V., Mercado-Lubo R., Hallstrom K., McCormick B.A. Salmonella effector proteins and host-cell responses. Cell. Mol. Life Sci. 2011;68:3687–3697. doi: 10.1007/s00018-011-0841-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Slyke G., Angalakurthi S.K., Toth R.T.t., Vance D.J., Rong Y., Ehrbar D., Shi Y., Middaugh C.R., Volkin D.B., Weis D.D. Fine-specificity epitope analysis identifies contact points on ricin toxin recognized by protective monoclonal antibodies. Immunohorizons. 2018;2:262–273. doi: 10.4049/immunohorizons.1800042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Pollard A.J. Seroefficacy of Vi polysaccharide-tetanus toxoid typhoid conjugate vaccine (typbar TCV) Clin. Infect. Dis. 2018;67:18–24. doi: 10.1093/cid/cix1145. [DOI] [PubMed] [Google Scholar]
- Wain J., Hendriksen R.S., Mikoleit M.L., Keddy K.H., Ochiai R.L. Typhoid fever. Lancet. 2015;385:1136–1145. doi: 10.1016/S0140-6736(13)62708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangdi T., Lee C.Y., Spees A.M., Yu C., Kingsbury D.D., Winter S.E., Hastey C.J., Wilson R.P., Heinrich V., Baumler A.J. The Vi capsular polysaccharide enables Salmonella enterica serovar typhi to evade microbe-guided neutrophil chemotaxis. PLoS Pathog. 2014;10:e1004306. doi: 10.1371/journal.ppat.1004306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Typhoid vaccines: WHO position paper, March 2018 - Recommendations. Vaccine. 2018;37:214–216. doi: 10.1016/j.vaccine.2018.04.022. [DOI] [PubMed] [Google Scholar]
- Yan M., Li X., Liao Q., Li F., Zhang J., Kan B. The emergence and outbreak of multidrug-resistant typhoid fever in China. Emerg. Microbes Infect. 2016;5:e62. doi: 10.1038/emi.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.A., Chong A., Song J. Why is eradicating typhoid fever so challenging: implications for vaccine and therapeutic design. Vaccines (Basel) 2018;6:E45. doi: 10.3390/vaccines6030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.A., Lee S., Zhao J., Thompson A.J., McBride R., Tsogtbaatar B., Paulson J.C., Nussinov R., Deng L., Song J. In vivo tropism of Salmonella Typhi toxin to cells expressing a multiantennal glycan receptor. Nat. Microbiol. 2018;3:155–163. doi: 10.1038/s41564-017-0076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., Agard D.A. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The published article includes all datasets generated during this study. The atomic structure of TyTx11 bound to typhoid toxin has been deposited in the RCSB Protein Data Bank under PDB:6VX4 (Table S2).