Abstract
Differential metabolites between tuberous roots from cultivated variety (ZP) and wild variety (YS) of Rehmannia glutinosa were analyzed by widely targeted metabolomics, and annotated to KEGG pathways. 228 secondary metabolites (SM) in ZP and YS were detected, of which 58 were differential metabolites (DM), including 41 flavonoids, 10 phenolic acids, 3 terpenoids, 2 alkaloids and 2 others, and 170 were unchanged; Among 58 DMs, 44 (75.9%) were up-regulated in YS, of which 30 were unique to YS, while 14 (24.1%) were down-regulated in YS, of which 10 were unique to ZP; Among flavonoids, 33 (80.5%) were more highly expressed in YS than in ZP; Among phenolic acids, 7 (70%) were more highly expressed in YS than in ZP; 12 of 58 DMs were annotated into 17 types of KEGG pathways. Among them, benzoic acid and p-Coumaryl alcohol were up-regulated in YS, and annotated into 10 pathways (58.8%) and 4 pathways (23.5%), respectively. In addition, much of DMs possess various pharmacological effects. These results indicated better quality of YS than ZP and the necessity of YS domestication. Taken together, this study will provide a reference for the scientific introduction, comprehensive development and utilization of wild Rehmannia glutinosa.
Subject terms: Genetics, Plant sciences
Introduction
Rehmannia glutinosa Libosch. (R. glutinosa) is a perennial herb belonging to the genus Rehmannia (Scrophulariaceae), and has important economic value as medicinal and food materials. Its tuberous root is a commonly used bulk Chinese herbal medicine. The pharmaceutical effects of R. glutinosa are closely related to its secondary metabolites. Many metabolites in its tuberous root have been identified up to date. For example, 1049 metabolites were identified from the developing tuberous root of its variety Jinjiu1. They are composed of catalpol, acteoside, saccharides, terpene glycosides, amino acids, trace elements and other components, which possesses some bioactivities such as anti-cardiovascular diseases, nerve protection, hypertension resistance, immunity enhancement and so on2–4. R. glutinosa is mainly distributed in some provinces, such as Henan, Shanxi, Shandong and others, in China. Among them, better Rehmanniae Radix is from cultivated R. glutinosa in Huaiqing region including Wen County, Wuzhi County, Boai County and Jiaozuo city, Henan, which has higher contents of bioactive components such as catalpol and verbascoside, and lower clinical dosage than that from other places5. Its commonly cultivated varieties are Beijing No.3, Wen 85-5, Jinjiu, etc. In recent years, with its increasing consumption and loss of its farmland, its resources become increasingly exhausted. Moreover, its variety complexity and long-term vegetative propagation make its varieties degenerated, which lead to its poor quality, low yield and narrow genetic basis6. Therefore, it urgently requires new germplasms. To our knowledge, one practical solution to its new germplasms is the introduction, comprehensive development and utilization of wild R. glutinosa.
Wild R. glutinosa resources with many good traits and genes are very important and significant for enriching R. glutinosa germplasm resources and improving the yield and quality of cultivated R. glutinosa varieties, but their qualities vary. As a result, they need be identified and evaluated before their development and utilization. SNP-based R. glutinosa germplasms analysis indicated that there were more significant differences of its cultivated varieties from its wild resources7. Moreover, compared with wild Rehmannia germplasms, the qualities of cultivated R. glutinosa varieties decrease in that their some genes were lost via long-term artificial selection. Accurate chemical composition analysis of R. glutinosa varieties is crucial to their quality evaluation. Therefore, the study of chemical composition of wild and cultivated R. glutinosa will help to solve the lack of its new germplasms. Metabolomics, an ongoing and practical technology, has been widely used in medicine development, medicine toxicity and mechanism research, medicine screening and efficacy evaluation, mining new secondary metabolites, plant metabolism and response mechanisms, microbial interactions, gene function elucidation, effective metabolic pathways and related regulatory mechanisms1,8–14. At present, metabolomic technologies include targeted metabolomics, untargeted metabolomics and widely targeted metabolomics. So far, the first two metabolomic technologies have widely been used in plants, including R. glutinosa15–20, but both have advantages and disadvantages. Widely targeted metabolomics is next generation metabolomics, combining the advantages of untargeted metabolomics and targeted metabolomics, and possesses some advantages such as qualitative and quantitative accuracy, high throughput, high sensitivity and wide coverage. Using Q-TRAP mass spectrometry based on MRM mode, hundreds of known metabolites and nearly a thousand unknown metabolites can be simultaneously quantified, and the detection and identification of highly sensitive and widely targeted metabolites come true21,22. In recent years, this technology has been successfully applied in Sesame23, Rice22,24,25, Vanilla26 and Chrysanthemum morifolium27, respectively.
In our study, widely targeted metabolomics was used to compare the metabolic profiles between ZP and YS of R. glutinosa for the first time in order to qualitatively and quantitatively identify differential metabolites between ZP and YS. The results of this study will provide a reference for the scientific introduction, comprehensive development and utilization of wild R. glutinosa.
Results
Phenotypic analysis
There were some significant differences between ZP and YS in phenotype before the propagation of wild R. glutinosa (Table 1). However, the metabolite profiles of these two varieties were unknown and needed further analysis in this study.
Table 1.
Phenotypic analysis of two varieties.
| Variety | Wild type | Beijing No.3 |
|---|---|---|
| Elevation | 184 m | 97.10 m |
| Root number per plant | 2–4 | 4.00 |
| Fresh weight of the heaviest root per plant (g) | 2.10 | 100 |
| Growing period (year) | Natural growth for many years | Vegetative reproduction for many years |
| Growing environment | Natural mountain soil | Cultivated soil |
| Tuberous root color | Dark yellow | Bright yellow |
Multivariate analysis
Multivariate analysis is the analysis of three or more variables. It is used to deal with the relationship between variables, including different methods such as Principal components analysis (PCA). Orthogonal Partial Least Squares-Discriminant Analysis (OPLS-DA) and so on. PCA and OPLS-DA were used in this study.
PCA
PCA was used to reveal the internal structure of several variables by a few principal components. In the PCA plot (Fig. 1), three mix samples as QC were grouped together, suggesting QC samples with similar metabolic profiles and the entire analysis with stability and repeatability. Meanwhile, it could be seen from Fig. 1 that YS samples were clustered together, located to the left side of the QC samples, and that ZP samples gathered together, located on the right side to the QC samples. These results indicated that our analysis was stable and repeatable.
Figure 1.
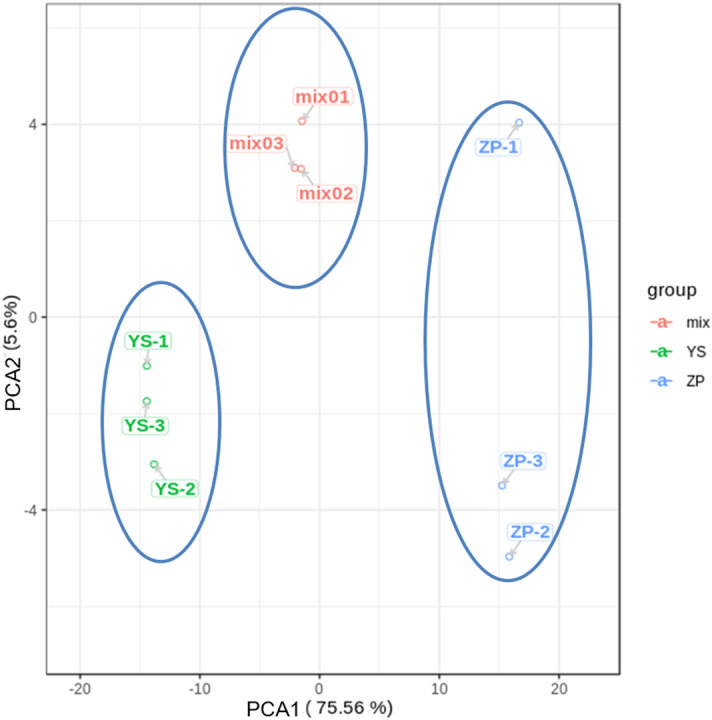
PCA score chart based on mass spectrum data of ZP, YS and QC samples. Ordinate: the second principal component, Abscissa: the first principal component. PCA Plot visualized in ggplot2 toolkit of R v3.5.0 (https://www.r-project.org/).
OPLS-DA
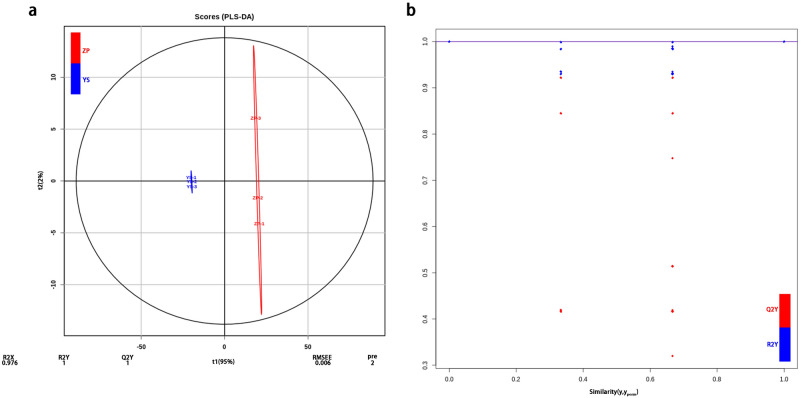
OPLS-DA is a multivariate statistical analysis method with supervised pattern recognition, and can solve the problem that PCA is not sensitive to the variables with little correlation. According to the differential variables, the score plot of every component (Fig. 2a) was formed to further show the differences between the components28. It was seen from Fig. 2a that R2Y and Q2 Y(Q2) were 1, while R2X equals 0.976, suggesting that OPLS-DA should be stable and reliable. Because Q2 equals 1, more than 0.9, OPLS-DA is excellent. The OPLS-DA model is verified using 200 alignment experiments. The horizontal line corresponds to the R2Y and Q2 of the original model, while the red and blue dots represent R2Y and Q2 after replacement, respectively. R2Y (0.81) and Q2 (0.52) in Fig. 2b were smaller than R2Y (1) and Q2 (1) of the original model, suggesting that the corresponding points should not exceed the corresponding lines. Therefore, OPLS-DA is meaningful, from which variable infuence in projection (VIP) values are obtained. VIP value is used to screen differential metabolites.
Figure 2.
(a) OPLS-DA score Plot. t1: Predicted principal components-score value of main components and difference between observation groups, t01: Orthogonal principal component-score value of orthogonal components and difference in observation group, R2Y: Percentage of Y matrix information that can be explained by OPLS-DA, Q2Y (Q2): Prediction ability of OPLS-DA, Pre: Predicted principal component number, RMSEE: Root mean squared error, Red: ZP, Blue: YS. (b) OPLS-DA permutation verification plot. Red: Q2Y, Blue: R2Y. OPLS-DA score Plot visualized in ggplot2 toolkit of R v1.0.1 (https://www.r-project.org/).
Identification of differential metabolites
A total of 228 SMs between YS and ZP were detected by HMDB, MWDB and METLIN databases (see Supplementary Table S1 online). Using both FC ≥ 2 or ≤ 0.5 and VIP ≥ 1 as the screening standards, 58 DMs were screened and identified (Table 2).
Table 2.
DMs from 228 SMs.
| No | Formula | Compounds | VIP | Fold change | p-value | Type |
|---|---|---|---|---|---|---|
| 1 | C29H34O16 | Limocitrin-O-rhamnoside-O-rhamnoside | 1.329 | 102.477 | 0.002 | Up |
| 2 | C31H48O7 | Phytolaccagenin | 1.929 | 0.000 | 0.000 | Down |
| 3 | C14H19NO7 | Ehretioside | 1.895 | 12,259.259 | 0.000 | Up |
| 4 | C16H12O6 | Aracarpene 1 | 1.952 | 21,777.778 | 0.004 | Up |
| 5 | C27H28O17 | Kaempferol 3-glucuronide-7-glucoside | 1.857 | 8529.630 | 0.008 | Up |
| 6 | C27H26O17 | Apigenin-7-O-diglucuronide | 2.074 | 79,666.667 | 0.005 | Up |
| 7 | C17H14O7 | Tricin | 1.681 | 1644.444 | 0.001 | Up |
| 8 | C27H30O15 | Kaempferol glc-rha | 1.926 | 16,814.815 | 0.007 | Up |
| 9 | C16H12O6 | 6,7,8-Tetrahydroxy-5-methoxyflavone | 1.852 | 8066.667 | 0.004 | Up |
| 10 | C21H18O11 | Baicalin | 1.166 | 34.653 | 0.004 | Up |
| 11 | C16H12O6 | Diosmetin | 1.950 | 21,444.444 | 0.002 | Up |
| 12 | C27H31ClO15 | Pelargonin chloride | 1.804 | 0.000 | 0.007 | Down |
| 13 | C9H10O2 | p-Coumaryl alcohol | 1.028 | 15.936 | 0.000 | Up |
| 14 | C20H39NO2 | N-Oleoylethanolamine | 1.035 | 16.587 | 0.000 | Up |
| 15 | C33H40O19 | Robinin(kaempferol-3-O-gal-rham-7-O-rham) | 1.832 | 0.000 | 0.170 | Down |
| 16 | C31H38O16 | 2′-Acetylacteoside | 1.195 | 0.023 | 0.029 | Down |
| 17 | C16H22O10 | Geniposidic acid | 1.329 | 0.010 | 0.001 | Down |
| 18 | C28H32O15 | Diosmin | 1.234 | 47.068 | 0.000 | Up |
| 19 | C9H12O3 | Homovanillic alcohol | 1.683 | 1718.519 | 0.017 | Up |
| 20 | C22H23O11 + | Peonidin O-hexoside | 1.804 | 5107.407 | 0.006 | Up |
| 21 | C23H24O13 | Syringetin 3-O-hexoside | 1.125 | 0.036 | 0.002 | Down |
| 22 | C28H32O16 | Chrysoeriol O-hexosyl-O-hexoside | 1.836 | 6937.037 | 0.004 | Up |
| 23 | C26H26O15 | Tricin O-malonylhexoside | 1.896 | 12,407.407 | 0.000 | Up |
| 24 | C23H24O12 | Tricin 7-O-hexoside | 1.164 | 33.997 | 0.001 | Up |
| 25 | C17H22O10 | 1-O-β-d-Glucopyranosyl sinapate | 1.910 | 0.000 | 0.012 | Down |
| 26 | C28H32O15 | Chrysoeriol 7-O-rutinoside | 1.902 | 13,111.111 | 0.002 | Up |
| 27 | C22H20O12 | Chrysoeriol O-glucuronic acid | 1.440 | 228.916 | 0.005 | Up |
| 28 | C23H22O13 | Tricin O-glucuronic acid | 2.081 | 85,814.815 | 0.006 | Up |
| 29 | C22H28O13 | 3-O-p-coumaroyl quinic acid O-hexoside | 1.604 | 859.259 | 0.006 | Up |
| 30 | C22H26O12 | 5-O-p-Coumaroyl shikimic acid O-hexoside | 1.749 | 3048.148 | 0.005 | Up |
| 31 | C15H10O6 | Luteolin | 1.521 | 452.222 | 0.037 | Up |
| 32 | C7H6O2 | Benzoic acid | 1.937 | 18,703.704 | 0.002 | Up |
| 33 | C21H20O10 | Kaempferol 7-O-rhamnoside | 1.708 | 0.000 | 0.196 | Down |
| 34 | C7H6O3 | Protocatechuic aldehyde | 1.918 | 15,407.407 | 0.003 | Up |
| 35 | C27H30O14 | Kaempferol 3,7-dirhamnoside (kaempferitrin) | 2.059 | 0.000 | 0.151 | Down |
| 36 | C22H23O11 + | Peonidin 3-O-glucoside | 1.797 | 4762.963 | 0.003 | Up |
| 37 | C27H30O16 | Bioquercetin | 1.571 | 0.001 | 0.046 | Down |
| 38 | C27H30O14 | Kaempferol-3,7-O-α-l-rhamnoside | 1.872 | 0.000 | 0.108 | Down |
| 39 | C23H32O15 | β-d-Furanofructosyl-α-d-(3-mustard cyl)glucoside | 1.029 | 15.965 | 0.000 | UP |
| 40 | C16H24O7 | 3-Hydroxy-4-isopropylbenzylalcohol 3-glucoside | 1.986 | 31,074.074 | 0.001 | Up |
| 41 | C21H18O11 | Apigenin-7-O-β-d-glucuronide | 1.220 | 49.046 | 0.006 | Up |
| 42 | C21H18O12 | Tetahydroxy-flavone-7-O-β-d-glucuronide | 2.006 | 38,074.074 | 0.000 | Up |
| 43 | C16H12O6 | Hispidulin | 1.854 | 8237.037 | 0.003 | Up |
| 44 | C17H14O7 | Jaceosidin | 1.632 | 1081.111 | 0.002 | Up |
| 45 | C21H18O12 | Scutellarin | 2.008 | 39,037.037 | 0.001 | Up |
| 46 | C24H22O13 | Malonyglygenistin | 1.505 | 40.364 | 0.000 | Up |
| 47 | C16H12O6 | Pratensein | 1.856 | 8407.407 | 0.004 | Up |
| 48 | C25H18O9 | Luteolin-7-O-glucuronide | 1.608 | 60.303 | 0.003 | Up |
| 49 | C22H22O11 | Diosmetin-7-O-galactoside | 1.016 | 14.960 | 0.001 | Up |
| 50 | C22H20O12 | Diosmetin-7-O-glucuronide | 1.388 | 156.084 | 0.004 | Up |
| 51 | C25H24O14 | Diosmetin-7-O-(6′-O-malonyl)-β-d-glucoside | 1.672 | 1533.333 | 0.003 | Up |
| 52 | C27H30O15 | Luteolin-7-O-rutinoside | 1.873 | 0.000 | 0.165 | Down |
| 53 | C22H30O14 | 3′-O-d-glucosylgentiopicroside | 1.694 | 1862.963 | 0.003 | Up |
| 54 | C15H22O8 | Bartsioside | 1.977 | 28,333.333 | 0.002 | Up |
| 55 | C30H38O15 | Cistanoside C | 1.044 | 0.056 | 0.038 | Down |
| 56 | C22H34NO10 + | Sinapine glucoside | 1.828 | 0.000 | 0.315 | Down |
| 57 | C28H28O18 | Chrysoeriol-7-O-[β-d-glucuronopyranosyl- (1 → 2)-O-β-d-glucuronopyranoside] | 2.067 | 74,185.185 | 0.013 | Up |
| 58 | C27H26O17 | Apigenin-7-O-[β-d-glucuronopyranosyl(1 → 2)-O-β-d-glucuronopyranoside) | 2.109 | 116,407.407 | 0.002 | Up |
Up: compared with ZP, the corresponding metabolite was up-regulated in YS. Down: compared with ZP, the corresponding metabolite was down-regulated in YS.
Display of the difference data between groups by Volcano plot
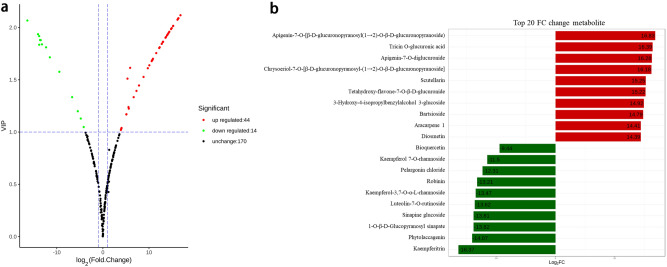
Based on log2 FC value and p-value, DMs were displayed in Volcano plot (Fig. 3a). In Fig. 3a, there were 44 up-regulated metabolites indicating that their expression contents in YS were higher than that in ZP, 14 down-regulated metabolites indicating that their expression contents in ZP were higher than that in YS, and 170 unchanged metabolites indicating that their expression contents did not vary between YS and ZP. Meanwhile, Top 20 FC change metabolites were presented in Fig. 3b. This result was consistent with that based on the VIP values and FCs (Table 2).
Figure 3.
(a) Volcano Plot of differential metabolites. Abscissa: FC value, Ordinate: VIP value, The dots: differential metabolites, Green dots: down regulated metabolites, Red dots: up regulated metabolites, Black dots: detected metabolites without significant difference. Volcano Plot visualized in ggplot2 toolkit of R v3.5.0 (https://www.r-project.org/). (b) Top 20 FC change metabolites. Ordinate: metabolite, Blackish green color: down regulated metabolites, Red: up regulated metabolites.
Heatmap clustering
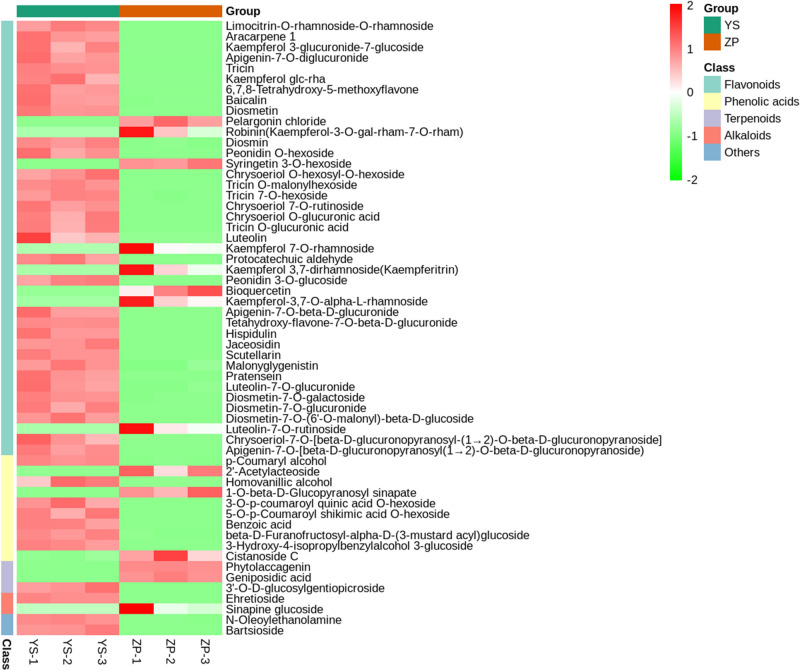
In order to show the varying law of 58 DMs with significant differences, their heat map was drawn (Fig. 4). The results showed that three YS repeats were grouped into one category and three ZP repeats were grouped into the other category. Because different metabolites had different accumulation trends in different samples, and the closer the accumulation trends, the closer the distances, 58 DMs had obvious expression differences between ZP and YS. 58 DMs were grouped into 5 clusters: (1) flavonoids including 41 ones, which were dominant in the SM of R. glutinosa tuberous roots. Among them, 33 (80.5%) metabolites were expressed higher in YS than in ZP. (2) 10 phenolic acids, of which 7 (70%) metabolites were expressed higher in YS than in ZP. (3) 3 terpenoids, 1 (33.33%) of which was expressed higher in YS than in ZP. (4) 2 alkaloids, half of which was higher expressed in YS than in ZP. (5) 2 metabolites that were higher expressed in YS than in ZP. In total, 44 differential metabolites (75.9%) were higher expressed in YS than in ZP, which was consistent with the screening of DMs based on VIP and FC as well as by Volcano plot (Table 2, Fig. 3b).
Figure 4.
Heat map of 58 DMs. Ordinate: clustering after normalization of relative contents of different metabolites, 5 kinds of colors stand for 5 classes of metabolites, Abscissa: grouping of samples, 2 kinds of colors stand for 2 groups of samples, Color scale from green to red indicated the contents of differentially expressed metabolites vary from low to high. Heatmap visualized in ggplot2 toolkit of R v1.0.12 (https://www.r-project.org/).
Pharmaceutical activities of DMs suggesting quality changes
It was seen from Fig. 4 that DMs were grouped into 5 types such as flavonoids, phenolic acids, terpenoids, alkaloids and and others. Among them, flavonoids including luteolin, protocatechuic aldehyde and tricin, were a large family of plant secondary metabolites and a subdivision of polyphenols, a versatile class of natural compounds that represented secondary metabolites from higher plants. Flavonoids were an effective ingredient of many Chinese herbal medicines and had various medicinal effect, including antibacterial, anti-inflammatory, anti-oxidant29,30. Phenolic acids including benzoic acid and p-Coumaryl alcohol, had been proved to have a variety of pharmacological activities, such as cardiovascular and cerebrovascular effects, anti-tumor, anti-oxidation, anti-inflammation, anti-fibrosis, etc31. Terpenoids including phytolaccagenin, geniposidic acid, were also a kind of important compounds in Chinese herbal medicine and played an important role in plant growth and development, resistance and defense, etc32. Alkaloids including ehretioside were a kind of nitrogen-containing basic organic compounds existing in nature (mainly plants). Alkaloids had many pharmacological activities, such as analgesia, spasmolysis, anti-inflammation, anti-tumor, etc. In addition, some compounds in alkaloids could interact with chemical components such as saponins and terpenoids, which could achieve the pharmacological effects of relieving cough, eliminating phlegm and relieving asthma33.
Differential metabolic pathways
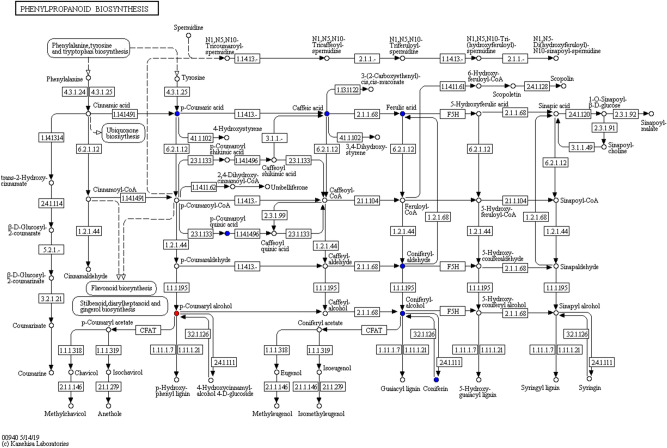
When 228 SMs were annotated to KEGG pathways by KEGG database (https://www.kegg.jp/kegg/kegg1.html)34, 38 SMs were annotated to KEGG pathways 202 times. Their annotation times ranged from 1 times per metabolite to 33 times per metabolite (succinic acid). After some duplicate KEGG pathways were removed, there were still 77 KEGG pathways (see Supplementary Table S2 online). Take Phenylpropanoid biosynthesis of KEGG pathway as an example (Fig. 5). In Fig. 5, p-Coumaric acid, p-coumaryl alcohol, caffeic acid, ferulic acid, coniferyl aldehyde, coniferyl alcohol, p-coumaroyl quinic acid and coniferin were annotated in Phenylpropanoid biosynthesis, of which p-coumaryl alcohol was a differential metabolite up-regulated in YS, compared with ZP, but p-Coumaric acid, caffeic acid, ferulic acid, coniferyl aldehyde, coniferyl alcohol, p-coumaroyl quinic acid and coniferin were unchanged.
Figure 5.
Phenylpropanoid biosynthesis. The red dots: the differentially expressed metabolites that are increased, the blue dots: the detected metabolites, but there is no significant difference.
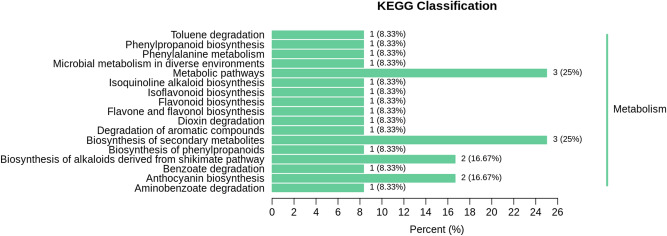
Among 58 DMs, 12 were annotated to KEGG database, 7 of which were annotated to KEGG pathways 23 times (see Supplementary Table S3 online). After some duplicate KEGG pathways were removed, there were still 17 KEGG pathways (Fig. 5). The seven metabolites were pelargonin chloride, peonidin 3-O-glucoside, p-Coumaryl alcohol, luteolin, benzoic acid, protocatechuic aldehyde and pratensein, respectively. These 17 KEGG pathways were categories anthocyanin biosynthesis, phenylpropanoid biosynthesis, biosynthesis of phenylpropanoids, metabolic pathways, biosynthesis of secondary metabolites, flavonoid biosynthesis, flavone and flavonol biosynthesis, phenylalanine metabolism, benzoate degradation, dioxin degradation, toluene degradation, aminobenzoate degradation, biosynthesis of alkaloids derived from shikimate pathway, microbial metabolism in diverse environments, degradation of aromatic compounds, isoquinoline alkaloid biosynthesis and isoflavonoid biosynthesis. Among them, phenylpropanoid biosynthesis and biosynthesis of phenylpropanoids were different metabolic pathway names in KEGG, which had similar names, but corresponded to two different pathway IDs (Table 3).
Table 3.
The categories of 12 DMs-annotated KEGG pathways.
| KEGG pathway | Ko-ID | Number | DM | Cpd-ID |
|---|---|---|---|---|
| Anthocyanin biosynthesis | ko00942 | 2 | Pelargonin chloride; peonidin 3-O-glucoside | C08725 + C12141 |
| Phenylpropanoid biosynthesis | ko00940 | 1 | p-Coumaryl alcohol | C02646 |
| Biosynthesis of phenylpropanoids | ko01061 | 1 | p-Coumaryl alcohol | C02646 |
| Metabolic pathways | ko01100 | 3 | Luteolin; p-coumaryl alcohol; benzoic acid | C01514 + C02646 + C00180 |
| Biosynthesis of secondary metabolites | ko01110 | 3 | Benzoic acid; luteolin; p-coumaryl alcohol | C00180 + C01514 + C02646 |
| Flavonoid biosynthesis | ko00941 | 1 | Luteolin | C01514 |
| Flavone and flavonol biosynthesis | ko00944 | 1 | Luteolin | C01514 |
| Phenylalanine metabolism | ko00360 | 1 | Benzoic acid | C00180 |
| Benzoate degradation | ko00362 | 1 | Benzoic acid | C00180 |
| Dioxin degradation | ko00621 | 1 | Benzoic acid | C00180 |
| Toluene degradation | ko00623 | 1 | Benzoic acid | C00180 |
| Aminobenzoate degradation | ko00627 | 1 | Benzoic acid | C00180 |
| Biosynthesis of alkaloids derived from shikimate pathway | ko01063 | 2 | Benzoic acid; protocatechuic aldehyde | C00180 + C16700 |
| Microbial metabolism in diverse environments | ko01120 | 1 | Benzoic acid | C00180 |
| Degradation of aromatic compounds | ko01220 | 1 | Benzoic acid | C00180 |
| Isoquinoline alkaloid biosynthesis | ko00950 | 1 | Protocatechuic aldehyde | C16700 |
| Isoflavonoid biosynthesis | ko00943 | 1 | Pratensein | C10520 |
Ko-ID = ID of KEGG pathway, Number = the number of metabolites that can be annotated to the corresponding KEGG pathways, Cpd-ID = Number of compound in KEGG.
Discussion
Rehmannia glutinosa is an important perennial herb, and its tuberous roots is clinically used to treat fever, nervous conditions, diabetes, and hypertension, and to increase liver function, hematopoietic function and immune defense and so on in different ways1. The great medicinal market demand for R. glutinosa leads to its overexploitation and germplasm resource scarcity, and long-term vegetative propagation makes the variety degenerate, quality poor, yield low and genetic basis narrow, so it is urgent to enrich and improve R. glutinosa by using candidate germplasm resources. Wild R. glutinosa is a kind of important candidate resources for this objective. Before we make good use of its wild resources, we used widely targeted metabolomics to compare the SM between its cultivated and wild varieties in that its SM are vital for its important pharmacological activities.
On the one hand, we compared the phenotypic characteristics of ZP and YS. It was found that both differences were very obvious, especially in both sizes and weights (Table 1). This was consistent with that of Zuo et al.35. On the other hand, 228 SMs were detected in both YS and ZP using widely targeted metabolomics technology coupled with MRM and public databases (see Supplementary Table S1 online). They were divided into 170 unchanged metabolites and 58 DMs (44 upregulated, 14 downregulated) (Table 2, Fig. 3b). 58 DMs were divided into 5 types such as flavonoids (41), phenolic acids (10), terpenoids (3), alkaloids (2) and others (2) (Table 2).
It is known that plant metabolomes are composed of over 200,000 metabolites that control plant development, and even Arabidopsis contains some 5000 metabolites1, so R. glutinosa should contain more than these 228 SMs in YS and ZP of R. glutinosa Libosch. Moreover, 1049 metabolites were ever identified in the developing tuberous roots of R. glutinosa variety Jinjiu36. Therefore, the high throughout identification of widely targeted metabolomics used in this study was limited, compared with untargeted metabolomics36 and others37,38. Its limitation should be attributed to the lack of large herbal medicine public metabolite databases.
The SMs of medicinal plants are very important substances in their life activities, closely related to their defense against diseases and insect pests and environmental stress, and important pharmacological activities. Based on these, our DMs may reflect the differences between wild and cultivated varieties of R. glutinosa. In this study, (1) the number of upregulated metabolites (44) were bigger than that of downregulated metabolites (14) in YS, compared to ZP (Table 2, Fig. 3b), Among 41 flavonoids, 33 (80.5%) were more highly expressed in YS than ZP, while among 10 phenolic acids, 7 (70%) were more highly expressed in YS than ZP. The number of unique metabolites to YS (30) was bigger than that of unique metabolites to ZP (10). Because these DMs possess important effects, these results suggested that the quality of YS is better than that of ZP. For example, flavonol had antioxidation; luteolin had a variety of pharmacological effects such as anti-tumor, antibacterial, anti-inflammatory, antiviral and analgesic effects39. Anthocyanin, a water-soluble flavonoid compound, had a variety of biological functions such as protecting plants from UV damage, scavenging reactive oxygen species, resisting adversity and changing the color of plants, as well as health efficacy such as anti-aging, anti-obesity and prevention of cardiovascular diseases40. According to investigation and research, flavonoids were synthesized by using intermediate products from phenylalanine converted via phenylpropane route as synthetic precursors, which were formed by different synthetic routes. Due to their various pharmacological effects such as cardiovascular protection, anti-cancer, anti-oxidation, anti-inflammation, liver protection and anti-tumor etc. flavonoids had become hot spots in the development and research of natural medicine at home and abroad27,41,42. According to our results, the expression of flavonoids and phenolic acids in YS was much higher; (2) Among quality control standards of R. glutinosa such as catalpol and rehmannioside D (now) or verbascoside (ever) in the Pharmacopoeia of the People’s Republic of China43, rehmannioside D, catalpol and verbascoside were all contained in unchanged. These result showed that YS could be used to modify ZP, of which catalpol′ result is consistent with a previous report44; (3) In this study, as the autotoxic metabolites such as ferulic acid, benzoic acid, protocatechuic aldehyde, 4-Hydroxybenzoic acid and so on, their the contents in YS were also much higher than that in ZP. Among them, benzoic acid had antibacterial and antiseptic effects45, ferulic acid could inhibit obesity and improve the steady state of blood lipid and blood sugar46,47, and protocatechuic aldehyde had cardiovascular and cerebrovascular protective effects48. Moreover, 30 kinds of DMs such as protocatechuic aldehyde, luteolin, tricin, diosmetin, homovanillic alcohol, jaceosidin, pratensein, hispidulin, malonyglygenistin, bartsioside, scutellarin and other compounds were unique to YS and reported for the first time in R. glutinosa. These results indicate ZP and YS contained unique DMs, unique ones to YS were much more than that to ZP, and that artificial breeding increased the contents of main active metabolites in cultivated variety of R. glutinosa and selected out many metabolite in its wild variety.
In addition, KEGG database helps researchers to study genes, expression information and metabolite content as a whole network, and provides integrated metabolic pathways involved in such as the pathways of carbohydrate, nucleoside and amino acids as well as biodegradation of organic compounds with enzymes. Therefore, it is a powerful tool for metabolism analysis and metabolic network research in vivo1,49. In the present study, 12 of 58 DMs between ZP and YS were annotated to 17 non-repetitive KEGG pathways (Fig. 6, Table 3). The main differential metabolic pathways between ZP and YS included metabolic pathways and biosynthesis of secondary metabolites. Among them, benzoic acid and p-Coumaryl alcohol up-regulated in YS were annotated into 10 (58.8%) and 4 (23.5%), respectively (Table 3). These results provided a clue for analyzing the metabolism of these metabolites and DMs, and their metabolic networks in R. glutinosa. In conclusion, based on the phenotypic differences between YS and ZP, we detected 228 SMs of YS and ZP by using widely targeted metabolomics, and then identified 58 DMs between ZP and YS via multivariate analysis. It was found that the metabolites of YS were more unique, and some of them were quality control metabolites in YS instead of ZP. Our results indicated better quality of YS than that of ZP and the necessity of YS domestication, and will provide a reference for the scientific introduction, comprehensive development and utilization of wild R. glutinosa. In addition, the related metabolic pathways will provide a theoretical basis for the subsequent exploration of biosynthesis of related metabolites in R. glutinosa.
Figure 6.
KEGG pathway classification map of DMs. Ordinate: KEGG metabolic pathway types. Abscissa: the number of DMs annotated to the KEGG pathway type and the proportion of this number to the total number of all the DMs annotated to all the KEGG pathways.
Materials and methods
Plant materials
The plant materials of R. glutinosa were provided by Four huaiyao research institutes in Wuzhi county, Jiaozuo city, Henan province on November 10, 2019. Among materials, the cultivated variety (ZP) of R. glutinosa was collected from germplasm resource nursery of Four huaiyao research institutes and the wild variety (YS) of R. glutinosa was collected from Sumen mountain located in Wuzhi county, Jiaozuo city, Henan province. Relevant Permissions had been provided. Then, two varieties were planted in experimental field of Henan Normal University. All samples were collected with approval and permission. Both tuberous roots were collected and freeze–dried for metabolites profiling. Three samples were used, of which each was composed of 6 different tuberous roots from 6 different mature plants in their harvest time. In each sample, they were pooled in equal parts after collection. These samples were metabolically profiled at Wuhan Maitville Biotechnology Co., Ltd (http://www.metware.cn).
Statement about the wild plant collected
Collection of Rehmannia glutinosa in this research material conforms to and complies with the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. In addition, according to the List of National Key Protected Wild Plants issued by the State Forestry and Grassland Bureau of China, Rehmannia glutinosa, the experimental material of this study, is neither a national key protected wild plant nor an endangered plant species.
Sample preparation and extraction
Related experiments such as sample preparation and extraction of metabolites were conducted by Wuhan Maitville Biotechnology Co., Ltd. Gradient grades of methanol, acetonitrile, and acetic acid were purchased from Merck Company, Germany (http://www.merck-chemicals.com). The internal standard lidocaine was bought from Shanghai New Asiatic Pharmaceuticals Co., Ltd (http://www.xinyapharm.com/). Authentic standards of phenylpropanoids were purchased from BioBioPha Co., Ltd (http://www.biobiopha.com/) and all other standards were from Sigma-Aldrich, USA (http://www.sigmaaldrich.com/united-states.html). The vacuum freeze- dried samples (three biological replicates) were ground into powder using a mixer mill (MM 400, Retsch) with a zirconia bead for 1.5 min at 30 Hz. 100 mg powder was weighed and extracted overnight at 4 °C with 1.0 ml pure methanol (or 70% aqueous methanol) containing 0.1 mg/L lidocaine for lipid-solubility metabolites (or water-solubility metabolites). After centrifuged at 10,000g for 10 min, the supernatants were absorbed, and filtrated by millipore filter (SCAA-104, 0.22 μm pore size; ANPEL, Shanghai, China, http://www.anpel.com.cn/) and stored in vials for UPLC-MS/MS analysis38.
Widely targeted metabolomics conditions
Data acquisition instrument system: Ultra Performance Liquid Chromatography (UPLC) (Shim-pack UFLC SHIMADZU CBM30A, http://www.shimadzu.com.cn/) and Tandem mass spectrometry, MS/MS (Applied Biosystems 4500 QTRAP, http://www.appliedbiosystems.com.cn/). Liquid chromatographic conditions: Column- Waters ACQUITY UPLC HSS T3 C18 (1.8 µm, 2.1 mm * 100 mm); solvent system, water (0.04% acetic acid): acetonitrile (0.04% acetic acid); gradient program, 100:0 V/V at 0 min, 5:95 V/V at 11.0 min, 5:95 V/V at 12.0 min, 95:5 V/V at 12.1 min, 95:5 V/V at 15.0 min; flow rate, 0.40 mL/min50; temperature, 40 °C; injection volume: 5 μL. The effluent was alternatively connected to an ESI-triple quadrupole-linear ion trap (Q TRAP)-MS.
LIT and triple quadrupole (QQQ) scans were acquired on a triple quadrupole-linear ion trap mass spectrometer (Q TRAP), API 4500 Q TRAP LC/MS/MS System, equipped with an ESI Turbo Ion-Spray interface, operating in a positive ion mode and controlled by Analyst 1.6.3 software (AB Sciex). The ESI source operation parameters were as follows: ion source, turbo spray; source temperature 550 °C; ion spray voltage (IS) 5500 V; ion source gas I (GSI), gas II(GSII), curtain gas (CUR) were set at 55, 60, and 25.0 psi, respectively; the collision gas(CAD) was high. Instrument tuning and mass calibration were performed with 10 and 100 μmol/L polypropylene glycol solutions in QQQ and LIT modes, respectively. QQQ scans were acquired as MRM experiments with collision gas (nitrogen) set to 5 psi. DP and CE for individual MRM transitions were done with further DP and CE optimization. A specific set of MRM transitions were monitored for each period according to the metabolites eluted within this period. Similar but inconsistent experimental procedures had been successfully applied and implemented by Zhang et al.51 before.
Qualitative and quantitative analyses of metabolites
Metabolite structure analysis referred to existing mass spectrum public databases such as MassBank (http://www.massbank.jp/), KNAPSAcK (http://kanaya.naist.jp/KNApSAcK/), HMDB (http://www.hmdb.ca/)52, MoTo DB (http://www.ab.wur.nl/moto/) and METLIN (http://metlin.scripps.edu/index.php)53 and others. The primary and secondary spectra detected by mass spectrometry were analyzed qualitatively, and isotopic signals were removed during the analysis of some substances, including repeated signals of K+ ions, Na+ ions, NH4+ ions, and fragment ions that were themselves other larger molecular weight substances repeating signal; Metabolites were quantified using MRM mode for mass spectrum peaks of metabolites, a peak per metabolite (mass spectrum file). Mass spectrum file was processed by MultiaQuant software for integration and correction of chromatographic peaks, a chromatographic peak per metabolite54.
Statistical data analysis
Metabolite data were log2-transformed for statistical analysis to improve normality and normalized. In order to explore the metabolites of cultivated and wild R. glutinosa, the 228 SMs had been used for cluster analysis by R v3.5.0 (http://www.r-project.org/). The cluster heat map were obtained using the agglomeration method of ‘complete linkage’ based on the Euclidean distances of 228 SMs between accessions. The color scale indicates the intensity of the metabolites (log2-transformed). Peak areas were integrated using the IntelliQuan algorithm50. Differences in the metabolites of root tissue between ZP and YS were determined using Welch’s t-test (P < 0.01). The significantly changed (P < 0.01) metabolites were used for subsequent PCA. In parallel, unsupervised PCA was carried out by R v3.5.0 (https://www.r-project.org/). The supervised OPLS-DA was carried out by R v1.0.1, MetaboAnalystR (https://www.r-project.org/)51. Data were processed using Analyst 1.6.3 software.
KEGG function annotation
DMs were annotated to KEGG pathways by KEGG database34. These KEGG pathways were classified according to their types.
Compliance with ethical standards
The conducted experiment complies with the laws of China.
Supplementary Information
Acknowledgements
This research was funded by National Natural Science Foundation of China (31870312), Natural Science Foundation of Henan province (No. 182300410018), Key R & D and promotion projects in Henan Province((No. 212102110405) and Fund of Henan Normal University (No. 2019JQ01). The authors would like to thank Kanehisa Laboratories for providing the image of Phenylpropanoid biosynthesis, and thank Director Huojin Li, Four HuaiyaoInstitute of Qinhuai, Wuzhi County, Henan, China for his provision of Rehmannia glutinosa materials.
Author contributions
Z.Y.Q. conceptualized the study, carried out the conceptualization, validation, data curation, project administration, reviewed and editd manuscript. S.L.Y. and Z.J.L. performed the formal analysis, data curation and original draft preparation. L.H.M. and D.H.Y. helped improve the content of the manuscript. All authors gave final approval for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Luying Shao and Jialin Zhu.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-90961-6.
References
- 1.Zhou YQ, et al. Metabolite accumulation and metabolic network in developing roots of Rehmannia glutinosa reveals its root developmental mechanism and quality. Sci. Rep. 2018;8:14127–14137. doi: 10.1038/s41598-018-32447-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li HW, Meng XL. Research progress on chemical constituents and pharmacological activities of Rehmannia glutinosa. Drug Eval. Res. 2015;38:218–228. [Google Scholar]
- 3.Li HX, Wei SJ. Studies on chemical constituents and pharmacology of Rehmannia glutinosa. Chin. Med. Mod. Distance Educ. China. 2012;10:116–117. [Google Scholar]
- 4.Alipieva K, Korkina L, Orhan IE, Georqiev MI. Verbascoside—A review of its occurrence, (bio)synthesis and pharmacological significance. Biotechnol. Adv. 2014;32:1065–1076. doi: 10.1016/j.biotechadv.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Quan YP, Wang YS, Xin ZH, Ji SD. The genuine medicinal materials of four-major Huai-yao and its developmental prospects. Hubei Agric. Sci. 2011;50:3097–3101. [Google Scholar]
- 6.Wang FQ, Xie CX, Sun RB, Zhang ZY. Progress on germplasm enhancement and breeding of Rehmannia glutinosa. China J. Chin. Materia Med. 2018;43:4203–4209. doi: 10.19540/j.cnki.cjcmm.20180820.012. [DOI] [PubMed] [Google Scholar]
- 7.Guo MM, Zhou YQ, Duan HY, Yang K, Shao LY. Development of SNP marker based on the Rehmannia glutinosa transcriptome database and construction of DNA fingerprint in Rehmannia. Biotechnol. Bull. 2019;35:224–230. [Google Scholar]
- 8.Lea G, Vessela AP, Sylvain C, Florence RF. Metabolomics to decipher the chemical defense of cereals against Fusarium graminearum and deoxynivalenol accumulation. Int. J. Mol. Sci. 2015;16:24839–24872. doi: 10.3390/ijms161024839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steinmeyer S, Lee K, Jayaraman A, Alaniz RC. Microbiota metabolite regulation of host immune homeostasis: A mechanistic missing link. Curr. Allergy Asthma Rep. 2015;15:24. doi: 10.1007/s11882-015-0524-2. [DOI] [PubMed] [Google Scholar]
- 10.Chen ZM, et al. Overview on application of metabonomics in study on processing mechanism of traditional Chinese materia medica. Chin. Arch. Tradit. Chin. Med. 2018;36:275–279. [Google Scholar]
- 11.Wang CC, et al. Thoughts of omics research on quality formation in Dao-di herbs. China J. Chin. Materia Med. 2018;43:2307–2412. doi: 10.19540/j.cnki.cjcmm.20180307.010. [DOI] [PubMed] [Google Scholar]
- 12.Lin S, Wu HY, Li KP. Advances in metabonomics of medicinal plants based on XC-MS and NMR. J. Chin. Med. Mater. 2015;38:2651–2654. [Google Scholar]
- 13.Liu X, Wang YH. The application of NMR-based metabolomics technique to medicinal plants. Acta Pharmaceutica Sinica. 2017;52:541–549. [Google Scholar]
- 14.Xue S, Chen SQ. Analysis on change rule of carbohydrates in Rehmanniae radix praeparata with different processing times based on metabolomics. Chin. J. Exp. Tradit. Med. Formulae. 2018;24:1–5. [Google Scholar]
- 15.Chen, W. K. Study of targeted metabolome and transcriptome in Grape berries grown under double cropping viticulture system. Doctoral thesis, China Agricultural University, Beijing (2018).
- 16.Tian, W. J. Targeted metabolomics profiling of Loquat during fruit development and postharvest ripening. Master thesis, Zhejiang University, Zhejiang (2017).
- 17.Dai WD, Hu ZY, Xie DC, Tan JF, Lin Z. A novel spatial-resolution targeted metabolomics method in a single leaf of the tea plant (Camellia sinensis) Food Chem. 2020;311:1–4. doi: 10.1016/j.foodchem.2019.126007. [DOI] [PubMed] [Google Scholar]
- 18.Li PL, et al. Metabolomic analysis reveals the composition differences in 13 Chinese tea cultivars of different manufacturing suitabilities. J. Sci. Food Agric. 2018;98:1153–1161. doi: 10.1002/jsfa.8566. [DOI] [PubMed] [Google Scholar]
- 19.Tan JF, et al. Study of the dynamic changes in the non-volatile chemical constituents of black tea during fermentation processing by a non-targeted metabolomics approach. Food Res. Int. 2016;79:106–113. doi: 10.1016/j.foodres.2015.11.018. [DOI] [Google Scholar]
- 20.Li J, et al. Phytochemical comparison of different tea (Camellia sinensis) cultivars and its association with sensory quality of finished tea. LWT. 2020;117:108595. doi: 10.1016/j.lwt.2019.108595. [DOI] [Google Scholar]
- 21.Sawada Y, et al. Widely targeted metabolomics based on large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol. 2009;50:37–47. doi: 10.1093/pcp/pcn183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen W, et al. A novel integrated method for large-scale detection, identification, and quantification of widely targeted metabolites: Application in the study of rice metabolomics. Mol. Plant. 2013;6:1769–1780. doi: 10.1093/mp/sst080. [DOI] [PubMed] [Google Scholar]
- 23.Wang DD, et al. Identification of nutritional components in black sesame determined by widely targeted metabolomics and traditional Chinese medicines. Molecules. 2018;23:1180–1192. doi: 10.3390/molecules23051180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen W, et al. Comparative and parallel genome-wide association studies for metabolic and agronomic traits in cereals. Nat. Commun. 2016;7:12767. doi: 10.1038/ncomms12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang CY, et al. Control of leaf senescence by an MeOH-jasmonates cascade that is epigenetically regulated by OsSRT1 in rice. Mol. Plant. 2016;9:1366–1378. doi: 10.1016/j.molp.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Cai YY, Chen XX, Gu FL, Xu F. Vanilla beans forming different processing stages with widely targeted metabolomic technique. Chin. J. Trop. Crops. 2019;40:1325–1335. [Google Scholar]
- 27.Wang T, et al. Widely targeted metabolomics analysis reveals the effect of flooding stress on the synthesis of flavonoids in Chrysanthemum morifolium. Molecules. 2019;24:3695. doi: 10.3390/molecules24203695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thévenot EA, Roux A, Xu Y, Ezan E, Junot C. Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. J. Proteome Res. 2015;14:3322–3335. doi: 10.1021/acs.jproteome.5b00354. [DOI] [PubMed] [Google Scholar]
- 29.Iker H, Leonor A, Frank VB, Sergi MB. How relevant are flavonoids as antioxidants in plants? Trends Plant Sci. 2009;14:125–132. doi: 10.1016/j.tplants.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Dalia S, Vanessa P, Ramiro MM. Review of flavonoids: A diverse group of natural compounds with anti-Candida albicans activity in vitro. Arch. Oral Biol. 2017;76:76–83. doi: 10.1016/j.archoralbio.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 31.Liang WY, et al. Research progress on salvianlic acids of Salvia miltiorrhiza. China J. Chin. Mater. Med. 2016;41:806–812. doi: 10.4268/cjcmm20160508. [DOI] [PubMed] [Google Scholar]
- 32.Zhao HY, et al. Cloning and bioinformatics analysis of promoters of three terpene synthase genes and their terpenoid regulation in Amomum villosum. Chin. Tradit. Herb. Drugs. 2021;52:1117–1127. [Google Scholar]
- 33.Zhang QL, Zhao HB, Wang J, Cui ZJ. Research progress on chemical constituents and pharmacological activities of Aconitum gymnandodrum. West China J. Pharm. Sci. 2021;36:99–102. [Google Scholar]
- 34.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2020;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuo M, Zuo JJ. Domestication, cultivation and application of wild Rehmannia glutinosa. Agric. Technol. 2019;39:93–94. [Google Scholar]
- 36.Yang, K. Metabolite accumulation and metabolic network in developing roots of Rehmannia glutinosa reveal its root developmental mechanism, quality and verbascoside biosynthesis pathway. Master thesis, Henan Normal University, Xinxiang (2019).
- 37.Maldini M, et al. Metabolomic study of wild and cultivated caper (Capparis spinosa L.) from different areas of Sardinia and their comparative evaluation. J. Mass Spectrom. 2016;51:716–728. doi: 10.1002/jms.3830. [DOI] [PubMed] [Google Scholar]
- 38.Zhu G, et al. Rewiring of the fruit metabolome in tomato breeding. Cell. 2018;172:249–261. doi: 10.1016/j.cell.2017.12.019. [DOI] [PubMed] [Google Scholar]
- 39.Huang S, Cai T. Research progress of flavonoid eutectic compound. J. Mod. Med. Health. 2019;35:3791–3795. [Google Scholar]
- 40.Qiu ZM, et al. Research progress on anthocyanins in Lettuce. China Veg. 2019;12:25–30. [Google Scholar]
- 41.Zou LQ, Wang CX, Kuang XJ, Li Y, Sun C. Advance in flavonoids biosynthetic pathway and synthetic biology. China J. Chin. Mater. Med. 2016;41:4124–4128. doi: 10.4268/cjcmm20162207. [DOI] [PubMed] [Google Scholar]
- 42.Kang YL, et al. Research progress on flavonoid metabolic synthesis pathway and related function genes in medicinal plants. Chin. Tradit. Herb. Drugs. 2014;45:1336–1341. [Google Scholar]
- 43.Pharmacopoeia of the People's Republic of China: Part I [S]. 2015 Edition 156–157 (China Medical Science Press, 2015).
- 44.Li, X. E. Physiological characterization of Rehmannia glutinosa cultivar and wild type varieties and its relationship with the tuberous root development. Doctoral thesis, Nanjing Agricultural University, Jiangsu (2013).
- 45.Xie Q, et al. Research progress on chemical constituents and pharmacological effects of benzoin plants. J. Chin. Med. Mater. 2020;1:243–248. [Google Scholar]
- 46.Wang W, et al. Ferulic acid suppresses obesity and obesity-related metabolic syndromes in high fat diet-induced obese C57BL/6J mice. Food Agric. Immunol. 2018;29:1116–1125. doi: 10.1080/09540105.2018.1516739. [DOI] [Google Scholar]
- 47.Naowaboot J, et al. Ferulic acid improves lipid and glucose homeostasis in high-fat diet induced obese mice. Clin. Exp. Pharmacol. Physiol. 2016;43:242–250. doi: 10.1111/1440-1681.12514. [DOI] [PubMed] [Google Scholar]
- 48.Ren Y, et al. Study on components efficacy of Salviae Miltiorrhizae Radix et Rhizoma based on systematic traditional Chinese medicine. China J. Chin. Mater. Med. 2020;45:3251–3258. doi: 10.19540/j.cnki.cjcmm.20200210.401. [DOI] [PubMed] [Google Scholar]
- 49.Zhou YQ, Wang XN, Wang WS, Duan HYD. novo transcriptome sequencing-based discovery and expression analyses of verbascoside biosynthesis-associated genes in Rehmannia glutinosa tuberous roots. Mol. Breed. 2016;36:139–149. doi: 10.1007/s11032-016-0548-x. [DOI] [Google Scholar]
- 50.Qian R, Ren XY, Liu YX, Jiang XL. Widely-targeted metabolomics analysis of Shiraia bambusicola bioactive components. Res. Pap. 2019;38:631–645. [Google Scholar]
- 51.Zhang, S. S. et al. Identification of potential metabolites mediating bird’s selective feeding on Prunus mira flowers. BioMed Res. Int. 1–8 (2019). [DOI] [PMC free article] [PubMed]
- 52.Wishart DS, et al. HMDB 3.0—The human metabolome database in 2013. Nucleic Acids Res. 2013;41:801–807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu ZJ, et al. Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database. Nat. Protoc. 2013;8:451–460. doi: 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fraga CG, Clowers BH, Moore RJ, Zink EM. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography−mass spectrometry, XCMS, and chemometrics. Anal. Chem. 2010;82:4165–4173. doi: 10.1021/ac1003568. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.