Abstract
Background
Chemotherapy and targeted agent anti-cancer efficacy is largely dependent on the proliferative state of tumours, as exemplified by agents that target DNA synthesis/replication or mitosis. As a result, cell cycle specificities of a number of cancer drugs are well known. However, they are yet to be described in a quantifiable manner.
Methods
A scalable cell synchronisation protocol used to screen a library of 235 anti-cancer compounds exposed over six hours in G1 or S/G2 accumulated AsPC-1 cells to generate a cell cycle specificity (CCS) score.
Findings
The synchronisation method was associated with reduced method-related cytotoxicity compared to nocodazole, delivering sufficient cell cycle purity and cell numbers to run high-throughput drug library screens. Compounds were identified with G1 and S/G2-associated specificities that, overall, functionally matched with a compound's target/mechanism of action. This annotation was used to describe a synergistic schedule using the CDK4/6 inhibitor, palbociclib, prior to gemcitabine/AZD6738 as well as describe the correlation between the CCS score and published synergistic/antagonistic drug schedules.
Interpretation
This is the first highly quantitative description of cell cycle-dependent drug sensitivities that utilised a tractable and tolerated method with potential uses outside the present study. Drug treatments such as those shown to be G1 or S/G2 associated may benefit from scheduling considerations such as after CDK4/6 inhibitors and being first in drug sequences respectively.
Funding
Cancer Research UK (CRUK) Institute core grants C14303/A17197 and C9545/A29580. The Li Ka Shing Centre where this work was performed was generously funded by CK Hutchison Holdings Limited, the University of Cambridge, CRUK, The Atlantic Philanthropies and others.
Keywords: Cancer, cell cycle, synchronisation, high throughput screening, combination treatment
Research in Context.
Evidence before this study
Drug combinations and drug scheduling are increasingly being employed to combat the lack of efficacy or development of drug resistance often observed in cancer treatment. The cell cycle is well known to not only play a role in tumour formation but also in treatment response where previous attempts at combining cell cycle inhibitors with standard chemotherapies have given mixed outcomes.
Added value of this study
This study describes a better tolerated, scalable synchronisation method used to interrogate cell cycle dependencies of anti-cancer compounds. This phenotypic annotation was utilised to describe a novel and productive combination of sequential cell cycle treatments as well as implicate cell cycle specificity with scheduling antagonism using an external dataset.
Implications of all the available evidence
Understanding cell cycle specificities of anti-cancer compounds may aid selection of productive combinations and schedules and further exploration as a routine metric used in pre-clinical drug development may be warranted.
Alt-text: Unlabelled box
Introduction
The interplay between cancer and the cell cycle is well known, with amplification of mitogenic signals through oncogenes such as c-Myc and phosphoinositide 3-kinases (PI3Ks) and loss of tumour suppressors like p53, p21 and pRb converging to promote uncontrolled proliferation [1]. Yet, in the context of cancer therapy, exploitation of this connection to improve the therapeutic index of treatments outside the basic anti-proliferative mechanism of chemotherapy is somewhat limited. Such methods, broadly termed “cyclotherapy”, have been of interest for some time but have yet to break into significant use likely due to both biological and clinical complexities [2]. However, the interplay between cell cycle and drug activity, particularly in combination therapy, has important consequences when considering drug synergy or antagonism [3]. The most prototypical example of cell cycle dependency is the scheduling effects seen with microtubule poisons such as paclitaxel that are most potent during mitosis. Many agents including platinum-based chemotherapies as well as non-specific and specific cyclin-dependent kinase (CDK) inhibitors have all been shown to antagonise paclitaxel activity when given concurrently or as the first drug in a schedule due to the negative impact of these drugs on progression through M phase [4], [5], [6].
Despite promising pre-clinical data, early efforts to utilise pan-CDK inhibitors such as flavopiridol in combinations were typically hampered by a low therapeutic index due to low anti-tumour efficacies and major off-target toxicities, particularly neutropenia and thrombocytopenia [7]. However, there has been a resurgence in interest in this class of agents with the advent of third generation CDKi's, such as palbociclib, ribociclib and abemaciclib, that target non-conserved ATP binding pockets and exhibit higher selectivity alongside fewer dose-limiting toxicities [8,9]. Even so, combination as well as scheduling of these inhibitors with traditional chemotherapies has produced mixed cellular responses with both synergistic and antagonistic interactions being observed [10], [11], [12]. This contrasts with the promising combination of CDK4/6 compounds with inhibitors whose targets are associated with mitogenic signalling in G1 phase, such as mammalian target of rapamycin (mTOR) inhibitors [13], [14], [15], [16], where the cell cycle-dependent potencies of each drug may be mechanistically complementary. Given the importance of combination therapy and drug scheduling in cancer treatment, alongside these complex cell cycle interactions, the ability to quantify these broad G1 and S/G2/M cell cycle phase dependencies may better inform treatment strategies.
Many key cell cycle processes have been uncovered using methods that induce synchronous cell cycle progression and as such have helped form the basis of our understanding of this complex system. However, there are both biological and technical caveats associated with these methods. Chemical synchronisation approaches are often associated with a negative impact on proliferation/survival and increased DNA damage such as that seen with microtubule poisons (e.g. nocodazole) [17,18], as well as those that disrupt DNA replication including hydroxyurea and thymidine [19,20]. Use of fluorescent cell cycle reporters, such as those used in the FUCCI system [21], has bypassed these issues to some extent by avoiding synchronisation but its use is generally limited to low-throughput assays due to intensive live cell imaging and low cell numbers. Based on these technical limitations, use of a tolerated and high-yield method was paramount in order to tackle the question of cell cycle-dependent drug sensitivities. Agents used in G2/M synchronisation methods result in subsequent G1/S/G2 populations that occur in the absence of the arresting agent, better mimicking unperturbed cell cycle progression. Despite its limitations, nocodazole is widely used to induce prometaphase mitotic arrest and is often coupled with a mechanical detachment (a mitotic shake-off) to produce highly pure mitotic fractions [19]. The CDK1 inhibitor RO-3306, was first described as a reversible inducer of G2/M arrest with synchronous entry into M phase upon inhibitor withdrawal and demonstrated potential to be combined with a shake-off method [22].
In this study, we describe a better tolerated, efficient and scalable synchronisation method using the reversible CDK1 inhibitor RO-3306. This method was used to screen a library of over 200 anti-cancer compounds to generate a cell cycle specificity score based on relative drug activity between G1 and S/G2 accumulated states. This cell cycle specificity was explored further in the context of a published dataset describing effects of sequential drug treatment, revealing correlations with synergistic and antagonistic combinations.
Methods
All experiments were performed using established human cancer cell lines in vitro using techniques described in the following sections.
Cell culture & chemicals
All cell lines were cultured at 37°C, 5% CO2 for a maximum of 20 passages after thawing and were cultured in DMEM (Thermo Fisher #41966029) (MIA PaCa-2, PANC-1, HeLa) or RPMI-1640 (Thermo Fisher #21875034) (AsPC-1) and supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher #10270106). Murine cell line K8484 were previously established from KPC mice of 129/SvJae/C57Bl/6 background and were grown in DMEM supplemented with 5% FBS [23]. Each cell line passed mycoplasma and single tandem repeat (STR) genotyping tests performed internally by the CRUK-CI Cell Services core facility. RO-3306 (Merck, #217699), nocodazole (Sigma-Aldrich, #M1404), gemcitabine (Tocris, #3259), CB-5083 (BioVision via Cambridge Bioscience, #B1032-5), palbociclib (Sigma, PZ0199) and SN-38 (Merck, H0165) were dissolved in DMSO, aliquoted and stored at -20°C. A customised Cambridge Cancer Compound Library (L2300) was purchased from Selleckchem pre-dissolved in DMSO at 10 mM. AZD6738 was provided by AstraZeneca.
Cell cycle synchronisation
For nocodazole-based synchronisation, cells were incubated with 100 nM nocodazole for 14 hours before a shake off was performed using physical agitation of the dish to collect mitotic cells. For the RO-3306-based method, cells were incubated with 6 μM RO-3306 for 20 hours before being washed three times with 1xPBS, replenished with media and incubated at 37°C/5% CO2 for a further hour. A shake off was then performed in a similar manner to nocodazole-treated cells. Cells collected after the shake off were then washed in 1xPBS, spun using a benchtop centrifuge and the pellet suspended in culture media before re-seeding for downstream analysis. All centrifugation steps were performed at 1000 rpm.
Flow cytometry
Cells were collected via trypsinisation at the indicated times, washed with 1xPBS and fixed in ice-cold 70% ethanol overnight at -20°C. Fixed cells were then washed once with 1xPBS before being incubated in 500 μL blocking buffer (1xPBS +2% BSA + 0.1% TritonX-100/Tween20) for 1 hour at room temperature (RT), primary antibody solution (phospho-MPM-2, 1:500, Merck Millipore #05-368; phospho-histone H2AX S139, 1:500, Merck Millipore #05-636) in blocking buffer for 2 hours at RT and secondary antibody solution (Alexa Fluor 488 Goat anti-mouse, 1:500, Thermo Fisher #A11017) in blocking buffer for 1 hour at RT in the dark with three washes using blocking buffer between each step. Fixed cells were then incubated with blocking solution containing the DNA dye FxCycle Violet (1 μg/mL, Thermo Fisher #F10347) for at least 1 hour at RT in the dark before samples were run using the BD Biosciences LSRFortessa™ flow cytometer and analysed using FlowJo® and GraphPad Prism software. Gating strategies are outlined in the supplemental material.
Proliferation assays
For colony forming and clonocidal assays, 300 or 120 cells (MIA PaCa-2, HeLa, PANC-1, MCF7, K8484) and 1000 or 400 cells (AsPC-1) were seeded in 6 or 12 well plates respectively and grown for 11-14 days depending on the cell line with the final four days including drug exposure where indicated. Cells were fixed with 3% trichloroacetic acid (TCA) for 30 minutes at 4°C before staining with the protein dye sulforhodamine B (SRB). Images were then taken and colonies counted using a GelCount™ (Oxford Optronix) before dissolving in 10 mM Tris pH 8.0 solution for fluorescence quantification using a spectrophotometer. For colony forming assays, plating efficiency was calculated by dividing the number of colonies by the number of cells seeded, multiplied by 100. SRB and live cell IncuCyte time lapse experiments using the YOYO-3 viability dye (Thermo Fisher, #Y3606) were performed in 96-well plates as described previously [24].
Compound library screen
AsPC-1 cells were synchronised using RO-3306 as described above and isolated mitotic cells were seeded at 500 cells per well into 384-well plates using the Multidrop™ Combi dispenser (Thermo Fisher). Each plate consisted of 12 DMSO control wells alongside an 11-point dose response (from 0.3 nM to 30 μM) for seven compounds with two replicates for each condition. Four wells were treated with high dose SN-38 (10 μM) as a positive control to ensure no technical issues with dosing was observed. Compound was added using the LabCyte Echo 555 acoustic dispenser in a randomised well layout at 4 hours post-seeding for G1max and 18 hours post-seeding for S/G2max. For pulse treatment, compound-containing media was removed from the wells, washed with sterile, pre-warmed PBS and fresh media added using a combination of the Integra Viaflo 384 benchtop electronic pipette system and the Multidrop™ Combi dispenser. Plates were then fixed with 4% formaldehyde at 96 hours post-drug addition, stained with 1 μg/mL Hoechst 33342 (Thermo Fisher, #H3570) and nuclei counted using the CellInsight NXT (Thermo Fisher). Each condition was normalised to the mean DMSO value within the same plate and non-linear regression fitted to the dose response (log[inhibitor] vs response – variable slope, GraphPad Prism 8). The area under the curve (AUC) was then calculated for each dose response and compared between cell cycle conditions. By modifying the relative change/fractional difference formula [B/A]-1 we calculated cell cycle specificity score (CCS) as CCS = 1-(AUC[MIN]/AUC[MAX]) to generate a consistent fold change metric with a value of 0 denoting no change. If the AUC[MIN] was in the G1-accumulated condition the score was transformed by multiplying by -1 such that a compound with a score >0 suggested more S/G2 specificity and <0 suggested more G1 specificity.
Statistics
Data is reported as mean with ± SEM or SD, detailed in the figure legend as applicable, of at least three independent biological replicates except for Figure 2 and 5 describing the compound library screen and scheduling comparison respectively where sample size is detailed in the figure legends. Statistical analysis of two parameters was performed using either a two-tailed, unpaired t test (Figure 1d), or two-tailed, unpaired Mann-Whitney test (Figure 4d/e) based on data distribution. Standard deviations were not assumed to be equal. For multiple comparisons involving more than two parameters, a multiple unpaired t test with Holm-Sidak multiple comparisons correction (Figure 1b) or a one-way or two-way ANOVA was performed with a Bonferroni correction (Figure 3b, 4d/f) were performed. A Gaussian non-linear regression was used with a least-squares fit for frequency distributions. P values that were <0.05 (two-sided) were considered statistically significant for all statistical calculations. Comparisons with non-significant p-values >0.05 were not displayed on all figures for better clarity. PRISM 8.4.0 (GraphPad Software, San Diego, CA, USA) was used for data analysis.
Figure 2.
Compound library screen in G1 or S/G2 accumulated AsPC-1 cells. a. Frequency distribution of CCS scores for each compound tested with a Gaussian curve fitted to describe the mean (μ), standard deviation (σ) and goodness of fit (R2). b. CCS score plotted against the minimum AUC value across both conditions (AUCmin). c. Average CCS scores and AUCmin values when compounds are grouped by pathway as provided by Selleckchem. d-h. Waterfall plots of CCS score by pathway with average DMSO control scores plotted as the first column. Compound names added numerical order if score was above 1.5σ with colour shading representing the CCS score being above 1.5, 2 or 3σ.
Figure 5.
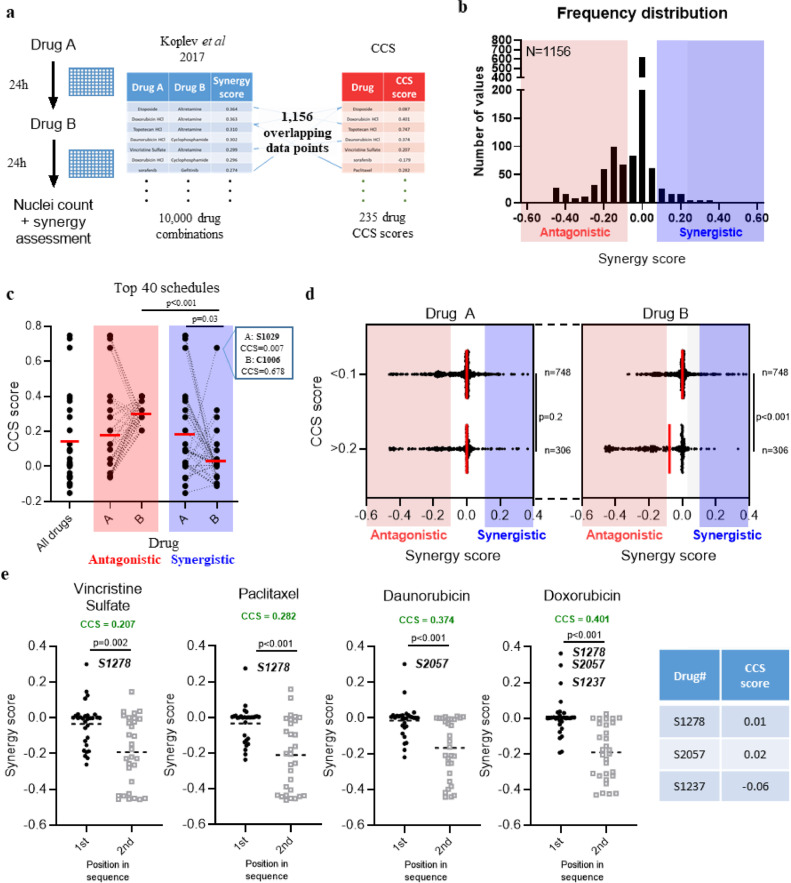
Correlation between CCS score and drug scheduling synergy/antagonism. a. General scheme showing how the synergy/CCS score combined dataset was made using synergy data generated by Koplev et al26. b. A frequency distribution of synergy scores for the included set of schedules where both compounds have an associated CSS score. c. Summaries showing median cell cycle score for all drugs alongside the top 40 antagonistic or synergistic schedules. Each point represents a single compound, n=40 and dashed lines connect two compounds within the schedule. Box inset shows the CCS scores of the synergitic schedule of lenalidomide (S1029) and gemcitabine (C1006) d. Drug schedules were split into distinct clusters based on whether the second drug in the sequence (Drug B) has a CCS score of <0.1 (not S/G2 specific) or >0.2 (likely to be S/G2 specific) and plotted against synergy score. Each point represents a single schedule alongside the median value; statistical significance tested using a two-tailed Mann-Whitney t-test. e. Four of the most common compounds in antagonistic schedules were selected. The synergy score of schedules where each drug is first or second in the sequence was compared. Each data point is a single schedule of a total (n) of 30 and the dashed line represents the mean; statistical significance tested using a two-tailed Mann-Whitney test. The CCS scores of notable compounds are added for reference. Sample size (n) shown on b and d is the number of independent schedules in that group. Statistical significance tested using either a One-way ANOVA with a Bonferroni's multiple comparison correction (c) or two-tailed Mann-Whitney test (d and e).
Figure 1.
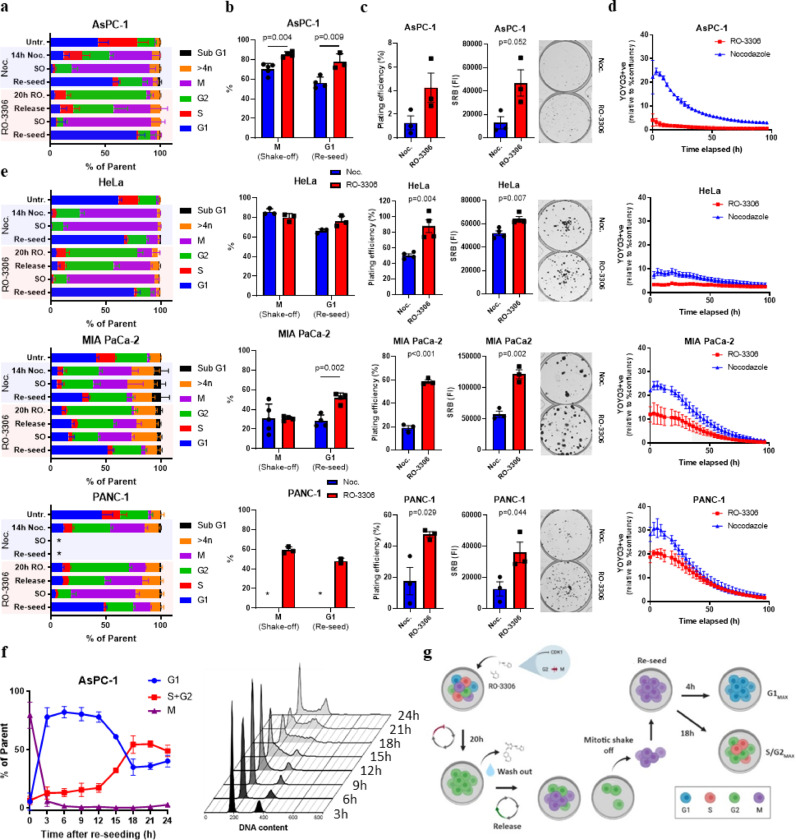
Characterisation of the RO-3306 shake off method. a. Cell cycle quantification using DNA content and a mitotic marker, phospho-MPM2, after flow cytometry analysis of cells treated with DMSO or after a nocodazole (noc.) or RO-3306-based mitotic shake off. SO, shake-off. Release = 1 hour after RO-3306 washout. Re-seed condition used isolated mitotic cells harvested 3h post-seeding. b. Same quantification as in a highlighting mitotic or G1 content of shake-off or re-seeded cells respectively after nocodazole or RO-3306 treatment. c. Plating efficiency and Sulforhodamine B (SRB) quantification alongside representative images of cell colonies grown after either a nocodazole or RO-3306-based mitotic shake off. d. Live cell imaging quantification of YOYO-3+ve objects normalised to total growth area. e. Repeat of a-d in three other cell lines, HeLa, MIA PaCa-2 and PANC-1. f. Cell cycle quantification of G1, S+G2 and M phases using DNA content and the mitotic marker, phospho-MPM2, at given times post-RO-3306 mitotic shake off. Representative DNA content histograms for each time point are shown on the right. g. Schematic summarising the method. *= Insufficient cell numbers for analysis. All data shown are means from at least three independent experiments, with error shown either as SD (a, b, d-f) or SEM (c). Students t tests (c) or multiple t tests (b) with a Holm-SIdak multiple comparisons correction was performed for comparisons between two groups or more respectively.
Figure 4.
Transient G1 accumulation increases the cytotoxicity associated with the gemcitabine/AZD6738 combination. a. Dose response curves of gemcitabine, AZD6738 and combinations where AZD6738 is fixed at either 0.3 or 1 μM in G1 or S/G2 accumulated AsPC-1 cells. Gemcitabine was added as a 6h pulse whereas all AZD6738 doses were continuous. A non-linear regression was fitted to each dose response and a GI50 value calculated where appropriate. Cell cycle specificity (CCS) score was calculated for each biological replicate and the mean/standard error included. Scores of >0.2 or <-0.2 are denoted S/G2 or G1-specific respectively. b. Schematic outlining the “clonocidal” assay, a modified clonogenic assay where treatment of established cell colonies allows identification of truly cytotoxic drug treatments. c. Representative images of MIA PaCa-2 cells after performing the clonocidal assay with 22h pre-treatment with either DMSO or 1 μM palbociclib followed by 6h of 100 nM gemcitabine and continuous 1 μM AZD6738. Colonies were fixed and stained with sulforhodamine B (SRB) dye before visualisation. d. Quantification of SRB intensity compared to DMSO after assessing the schedule in a clonocidal assay using MIA PaCa-2, K8484, MCF7 and HeLa cells e. Representative DNA content histograms (top) and γH2AX vs DNA content (bottom) of MIA PaCa-2 cells after respective treatments. f. Quantification of γH2AX positive cells across each treatment. Palbo. = palbociclib, gem. = gemcitabine. All data are shown as means ± SEM (a, d) or SD (f) from three independent experiments.
Figure 3.
Compound dose responses in G1 or S/G2 accumulated AsPC-1 cells. a. Dose response curves of selected compounds with biological triplicates. A non-linear regression was fitted to each dose response and a GI50 value calculated and reported where appropriate. *= Extrapolated value, where GI50 > 30 μM max concentration tested. b. Average CCS score of selected compounds as well as the DMSO control. All data are shown as mean ± SEM from three independent experiments; for b comparisons between control and each compound was assessed using a one-way ANOVA with a Bonferroni multiple comparison correction.
Role of funding source
The funders of this study had no role in study design, data collection, data analyses, interpretation, or writing of the report.
Results
An RO-3306 based mitotic shake off protocol produces pure fractions with improved viability
To assess the utility of an RO-3306 shake off protocol in producing synchronised cell fractions we first exposed the pancreatic adenocarcinoma (PDAC) cell line, AsPC-1, to increasing concentrations of RO-3306. After flow cytometry-based cell cycle analysis using DNA content and a mitotic marker, the minimum concentration needed for maximal G2 arrest was 6 μM after 20 hours (Figure S1). Subsequent analysis of mitotic entry after RO-3306 release, mitotic shake-off and subsequent G1 entry post re-seeding indicated that this method produced highly pure mitotic and G1 fractions with minimal contamination from other phases (Figure 1a). The increase in the mitotic fraction between RO-3306 released cells and the shake off demonstrates the added benefit of including this mechanical detachment step within the method. Both mitotic accumulation after shake-off and subsequent G1 entry improved using the RO-3306-based method compared to a standard nocodazole-based mitotic shake off protocol (Figure 1b). Importantly, mitotic cells isolated using the RO-3306 based method demonstrated improved long-term proliferative capacity compared to nocodazole as assessed by clonogenic assay (Figure 1c), as well as improved viability after re-seeding post shake-off as demonstrated by the increase in a cell death marker, YOYO-3 staining, in live cell imaging assays (Figure 1d). This method was also tested after optimisation in three other cell lines; HeLa, MIA PaCa-2 and Panc-1 cells (Figure 1e). The RO-3306 based method consistently increased G1 purity post-shake off compared to nocodazole across all tested cell lines. Given the purity of the mitotic population, AsPC-1 cells were taken forward to characterise further. To assess downstream synchronisation post-mitosis, cells were re-seeded into dishes and collected at 3-hour intervals. Using DNA content and a mitotic marker, we found AsPC-1 cells exhibited a moderately static 2N population from 4-12 hours post-seeding suggesting minimal DNA replication during these time points (Figure 1f). From 12 hours onwards, we could detect a time dependent increase in >2N DNA content that was maximal by 18 hours post-seeding (Figure 1f). As expected, the percentage of mitotic cells remained low after re-seeding with some evidence of mitotic entry by 24h (Figure 1f). A summary of the final method is shown in Figure 1g. Additionally, as HeLa cells are commonly used in synchronisation assays, downstream synchronisation of HeLa cells after the same method was also tested (Figure S2) demonstrating the potential utility of this method outside the present study.
The synchronisation method is amenable to higher throughput drug screening approaches
The described method was ideally suited to address the question of cell cycle dependent drug sensitivities based on the purity of the cell cycle fractions and reduced method-induced cytotoxicity. Given the kinetics of DNA content changes we observed, we selected appropriate time points to transiently expose synchronised cells to anti-cancer compounds for 6 hours between 4-10 hours and 18-24 hours post-seeding after mitotic shake-off to minimise potential overlap between cell cycle phases, with the caveat that this short-term exposure could compromise drug potency. Continuous drug exposure was also included for comparison and plates were fixed 96h after drug addition. An 11-point dose response from 0.3 nM to 30 μM was chosen to cover a broad spectrum of drug potencies with each concentration in duplicate, with well randomisation across the plate and both negative and positive control wells. Without direct access to large-scale robotic automation, we optimised an up-scaled method that utilised semi-automated machinery that permitted the assessment of cell cycle drug-sensitivities in a medium throughput fashion (Figure S3). Although each aspect of this optimisation was integral, drug washout using the Integra Viaflo benchtop pipette system was particularly crucial as we demonstrated this method was the least disruptive to cells but maintained throughput with minimal variability, compared to manual multichannel pipetting or using a plate washer (Figure S3c). Considering the scale of through-put we could achieve, we used a custom commercial drug library containing 231 well-known anti-cancer compounds covering 15 different pathways (Selleckchem, Table S1). An additional 4 drugs obtained separately to the commercial library were added to give a total of 235 compounds. Comparison of area under the curve (AUC) using a fitted dose response curve was chosen as the most effective assessment of cell cycle-dependent effects, given that it did not rely on reaching 50% growth inhibition as needed for GI50 although the two measures are linked. To compare AUC values between the conditions, we used a fractional difference metric to calculate a cell cycle specificity (CCS) score and was defined as 1-(AUC[MIN]/AUC[MAX]) for each compound such that the metric was not biased by order. Finally, if the AUC[MIN] value was associated with the G1-accumulated condition, the score was multiplied by minus one to achieve a score where <0 indicated G1 sensitivity and >0 indicated S/G2 sensitivity. The potency of each drug was also described as the minimum AUC value of the two cell cycle conditions (AUCmin), allowing assessment of drug activity over 6 hours of drug exposure.
High throughput compound library screen identifies cell cycle dependencies
The drug library was tested across 44 separate 384-well plates where mean DMSO nuclei counts across plates treated during 4-10h and 18-24h each exhibited plate-to-plate variability but with coefficient of variations (CV) of less than 10% (Figure S4a). CCS was calculated for all 235 compounds and plotted as a frequency distribution where the data followed a Gaussian distribution with R2=0.98 and mean/SD values of -0.004 and 0.099 respectively (Figure 2a). Across all compounds tested we observed 54, 39 and 25 candidates that fell outside 1.5, 2 and 3 times the standard deviation (σ) respectively indicating “hits”, drugs with cell cycle phase-dependency, at all levels of stringency (Table S1). Given this distribution of the data, we can confidently define G1 and S/G2 specificity at CCS scores less than -0.153 or greater than 0.153 respectively. AUCmin also followed a Gaussian distribution (R2=0.79) where almost half of all compounds (112/235) had a value within +/- 10% of 3000, indicating either that 6h exposure at any concentration was insufficient to induce obvious anti-proliferative effects, or a lack of efficacy in this cellular model (Figure S4b). Plotting CCS against AUCmin identified compounds that possessed both high potency as a pulse and cell cycle phase specificity (Figure 2b). A correlation was observed between AUCmin and CCS score although the correlation coefficient was low and reduced further when only considering compounds with an AUCmin of less than 2000 (Figure S4c, R2= 0.49 & 0.22). This suggested that, although there was a correlation between these two factors, AUCmin (i.e. potency) was a poor predictor of CCS score. This is exemplified by drugs such as ispinesib, a kinesin spindle protein inhibitor, that displayed high potency (AUCmin = 229) but no cell cycle specificity (CCS = 0.15), contrary to other mitotic drugs such as paclitaxel (CCS = 0.28, AUCmin = 266; Table S1). When drugs were clustered based on the pathway annotation provided by the drug library provider, pathway dependent cell cycle-based potencies were observed that seemed to match the broad mechanistic annotation for that class of compounds (Figure 2c), given they are an average score they are typically lower than those seen for individual compounds. For instance, the “DNA damage” and “Cell cycle” pathways contain classical chemotherapies and checkpoint kinase inhibitors respectively, consistent with their average CCS scores of 0.16 and 0.17 as being S/G2 specific. Further breakdown of these two pathways identifies the main drivers behind these scores such as Checkpoint kinase (Chk) 1/2 and Wee1 kinase inhibitors (Cell cycle) as well as topoisomerase and DNA/RNA synthesis inhibitors (DNA damage, Figure 2d/f). Despite the “Angiogenesis” pathway having an S/G2-related CCS score, closer examination of hits identified ENMD-2076, a pan-kinase inhibitor known to inhibit Aurora A (AurA) kinase, as significantly contributing to the overall score (Figure 2e). On the opposite end of the spectrum, both “PI3K/Akt/mTOR” and “MAPK” pathways displayed lower average CCS scores of -0.06 and -0.10 respectively demonstrating more G1-associated sensitivity, with mTOR complex 1/2 (mTORC1/2) inhibitors AZD8055 and sapanisertib having top CCS scores (Figure 2g/h). Dissection of cell cycle specificity by target identified a spectrum of CCS and AUCmin values indicating, at least in this cell line, that there may be differences in cell cycle specificity between molecules that have the same target. For example, cladribine and clofarabine are both purine analogues that broadly disrupt DNA/RNA synthesis but have different CCS scores (0.17 and 0.73, Table S1). Similar observations could be made when comparing mTORC1 inhibitors temsirolimus (CCS -0.11) and everolimus (-0.06) versus dual mTORC1/2 inhibitors AZD8055 (-0.37) and sapanisertib (-0.30), with the latter exhibiting higher specificity to G1 accumulated cells (Table S1). Visual inspection of dose response curves generated for selected G1 and S/G2 associated compounds matched the CCS annotation (Figure 3a). Furthermore, in cases where GI50 calculations were possible, a number of S/G2-associated compounds demonstrated log fold changes in GI50 concentrations between cell cycle phases. Consistent with lower potency scores, changes with G1-associated compounds were less striking although given the duration of drug exposure and proposed mechanisms of action this is perhaps not surprising. CCS scores averaged over three biological triplicates were found to be statistically significant, showing the robustness of the method and analysis pipeline (Figure 3b). Comparison of GI50 and CCS scores in the continuous treatment condition highlighted the impact of drug washout on potency and also the ability to identify cell cycle specific hits at higher degrees of confidence (Figure S4c/d). Only 5 compounds were outside the 3σ threshold and each of these displayed low AUC values that did not correspond to a significant shift in GI50 values (Table S1). Surprisingly, we did find compounds which were just as potent in either pulse or continuous treatment after synchronisation (e.g. rabusertib) suggesting these compounds are active with short exposure times. Importantly, similar levels of cell cycle synchronisation were observed between the independent biological replicates (Figure S5). Overall, these results demonstrated that cell cycle-dependent drug sensitivities could be quantified, broadly matched known cell cycle-dependent mechanisms but also highlighted differences in sensitivities of compounds with the same target that could not be explained by drug potency.
Transient G1 phase accumulation accentuates gemcitabine/AZD6738 efficacy
Previous work from our lab identified synergy between gemcitabine and an ATR inhibitor, AZD6738, in human and mouse models of pancreatic cancer in vitro and in vivo [24,25]. Both gemcitabine and AZD6738 had high CCS scores within the drug library screen (0.53 and 0.60 respectively) indicating high S/G2 specificity, consistent with their known modes of action. Given the ability of these compounds to synergise and induce replication catastrophe [24,25], we investigated whether exposure to both compounds would affect the CCS score as part of a schedule. To match a typical dosing schedule given for this combination in vivo [24,25], we added AZD6738 continuously at two fixed doses (0.3 and 1 μM) after a 6h gemcitabine pulse in G1 or S/G2 accumulated cells. Given the limited gap between these two compounds, as well as the DNA and ribonucleotide reductase large subunit (RRM) 1 binding mechanisms of gemcitabine [26], [27], [28], [29], this drug schedule, for the purposes of this work, is referred more generally as a combination although given sequentially. As a single agent, continuous AZD6738 exhibited a CCS score of -0.04 with a GI50 of 1.99 and 2.76 μM in G1 and S/G2-accumulated cells respectively indicating that continuous exposure did not demonstrate any particular cell cycle phase-dependent sensitivity (Figure 4a). Given AZD6738 did show S/G2-associated sensitivity as a pulse (Figure 3a), this is consistent with the fact that cells will lose cell cycle synchronicity over time which renders the initial cell cycle phase irrelevant. However, we were surprised to observe that the addition of continuous 0.3 μM AZD6738 reduced the concentration of gemcitabine needed to reach 50% growth inhibition in G1 (1.46 to 0.88 μM) but not S/G2 (0.39 to 0.34 μM) accumulated cells (Figure 4a). The extent of growth inhibition across the full range of gemcitabine doses in G1 versus S/G2 was boosted further with 1 μM AZD6738, although this schedule was highly toxic in both cell cycle conditions (Figure 4a). Given that AZD6738 was used after a gemcitabine pulse at concentrations that did not appear to reduce cell proliferation as a single agent, these data supported previous reports of a synergistic relationship between these two compounds. These data suggested that exposure to both compounds may be more effective in transiently G1-accumulated cells with increasing concentrations of AZD6738, even though both compounds have S/G2 specific mechanisms.
Given this finding, we hypothesised that palbociclib, a reversible CDK4/6 inhibitor, used as a pre-treatment would mimic the transient G1 accumulation seen after synchronisation and potentiate the effects of the gemcitabine/AZD6738 combination. MIA PaCa-2 cells have been shown to be sensitive to the combination in vitro and in vivo [24,25], and also demonstrated G1 accumulation after 22 hours of exposure to palbociclib, with associated decreases in S, G2 and M phase populations consistent with inhibited S-phase entry (Figure S6) and positive Rb status (data not shown) and were therefore an appropriate model to test further.
Given the potency of the combination and the fact that 96-well assays with low cell numbers are inherently sensitive to both cytotoxic and cytostatic effects, we wanted to design an in vitro assay able to distinguish between these two phenotypes. To achieve this, we seeded 6-well plates at low density analogous to a colony forming assay but allowed the cells to form visible colonies before treatment. This would mean that only large-scale but not small-scale cell death or transient arrest would be observable post-treatment (Figure 4b) and so was subsequently referred to as a “clonocidal” assay. Colonies were visible roughly 8 days after seeding whereupon they were pre-treated with 1 μM palbociclib or DMSO for 22h before treatment with gemcitabine (6h pulse) or AZD6738 (continuous). Significant colony death was observed after the gemcitabine/AZD6738 combination but only after palbociclib pre-treatment (Figure 4c) suggesting G1 accumulation increases the cell death associated with this combination. In addition, when palbociclib was not removed prior to exposure to gemcitabine/AZD6738 this affect was completely abrogated and, alongside additional evidence of effective S phase entry after palbociclib release, suggests the G1 synchronisation effect elicited by palbociclib is critical for the observed cytotoxicity (Figure S6). We then tested this schedule in the same assay across three other cancer cell lines of various cancer types including HeLa cells which are positive for the E6/E7 human papillomavirus proteins rendering them insensitive to CDK4/6 inhibition. All cell lines demonstrated increased colony death after the triple schedule compared to the gemcitabine/AZD6738 combination alone, apart from HeLa cells confirming that palbociclib-induced G1 accumulation appears to be responsible for this increased efficacy (Figure 4d). Treatment with palbociclib alone had no significant effect on colony number and palbociclib pre-treatment did increase single agent efficacy of gemcitabine in K8484 and MCF7 cells as well as increase single agent AZD6738 potency in K8484 cells. Although the differential potency between DMSO and palbociclib pre-treatment was lost as expected in HeLa cells due to inactivation of Rb, these cells demonstrated high colony death with the combination alone suggesting that Rb pathway inactivation may be increasing sensitivity to this combination.
The gemcitabine/AZD6738 combination has been previously shown to potently induce markers of DNA damage including those associated with S phase and replication-associated damage [24,25]. To ascertain the effect of palbociclib pre-treatment prior to this combination on cell cycle and DNA damage we analysed treated MIA PaCa-2 cells by flow cytometry. DNA content analysis highlighted an increase in intra-S accumulated cells 24h after the triple schedule compared to the combination without palbociclib pre-treatment (Figure 4e). Furthermore, we observed an increase in cells positive for phospho-H2AX (γH2AX), a broad marker of DNA damage, that were pre-treated with palbociclib and persisted after 48h suggesting a lack of DNA repair (Figure 4f). Together, these data suggest that palbociclib pre-treatment leads to an increase in DNA damage and cell death associated with combined gemcitabine and AZD6738 exposure, reflecting the changes that we observed when assessing cell cycle sensitivity scores in synchronised AsPC-1 cells.
Higher CCS scores correlate with drug scheduling antagonism
Although we had investigated utility of the CCS score by candidate approach using gemcitabine/AZD6738, we also wanted to evaluate the broader utility of this phenotype. To utilise our CCS score dataset and assess potential uses for this annotation, we investigated whether there was any correlation between CCS score and drug scheduling synergy/antagonism as assessed by Koplev and colleagues [30]. In their study, the effect of drug sequence (drug A followed by drug B, and vice versa) within some 10,000 different drug schedules was assessed in pancreatic cancer cells in a high throughput assay. Our CCS score was compared with their synergy scores derived using the PANC-1 cell line, as their AsPC-1 dataset included a much smaller subset of compounds (Figure 5a). The authors did however note a good correlation between PANC1 vs AsPC-1 lines, justifying the comparison. A total of 1,156 schedules contained compounds that overlapped with our library and the synergy score of these (where >0 = synergistic, <0 = antagonistic) were plotted as a histogram to determine their distribution (Figure 5b). This compiled dataset contained both antagonistic and synergistic interactions, although the majority (765/1,156) of these schedules were clustered around a neutral synergy score between -0.05 and 0.05 with neither synergy or antagonism being observed. To investigate the effect of cell cycle specificities at the extremes of synergy/antagonism, we interrogated CCS score using the top 40 antagonistic and synergistic schedules from the Koplev dataset (Figure 5c). Although no difference was observed between CCS scores of the first drug (drug A) in antagonistic versus synergistic schedules, the second drug (drug B) displayed a significantly higher CCS score (p=0.001, one-way ANOVA) within antagonistic schedules, suggesting that when an S/G2 specific compound is used as the second drug in a sequence it is more likely to be antagonistic (Figure 5c). This association was also true when all the schedules were clustered based on having S/G2 (CCS>0.2) or non-S/G2 (CCS<0.1) scores for drug B (Figure 5d), with average synergy scores of -0.129 and -0.012 respectively across a total of 1,054 schedules. To see whether particular drugs were driving this association, we selected the four most frequent compounds in schedules with a synergy score of <-0.1 (i.e. antagonistic), doxorubicin, vincristine sulphate, paclitaxel and daunorubicin, and directly compared synergy scores between matched schedules (A->B vs. B->A). They were all found to have a significantly lower synergy score when used as the second drug in a schedule (i.e. B->A) and all had been assigned a CCS score of >0.2 (Figure 5e). In cases where synergy was observed when these drugs were used as the first compound in the sequence, the associated second compound had a neutral CCS score (S1237, temozolomide, CCS=-0.06; S2057, cyclophosphamide, CCS=0.02; S1278, altretamine, CCS=0.01). A notable exception to this correlation included a synergistic schedule involving the S/G2 associated gemcitabine as the second compound (Figure 5c). This effect was observed when cells were exposed to gemcitabine after, but not before, the compound lenalidomide (after, synergy score = 0.19; before, synergy score = 0). These compounds showed promise as a combination in vitro but failed to offer significant benefits in a clinical trial [31,32].
Overall, these significant associations between drug scheduling antagonism and CCS score may suggest this annotation has utility in optimising effective drug combinations.
Discussion
The significant limitations surrounding current synchronisation methods, namely toxicity, cell yield, time required and cell line variability, are well known. The benefits of an RO-3306-based method described in this work could be of value when optimising such techniques in other cell lines. The reduction in toxicity associated with the RO-3306 shake-off method was critical to ensure unbiased downstream analysis within our proliferation-based drug library screen but may also have important implications for the wider cell cycle field by potentially reducing spurious off-target effects. The limitation of this method appears to be the variable level of synchronisation between different cell lines, a common feature of synchronisation techniques utilising cell cycle checkpoints.
Overall, the CCS score of hits we identified from the compound library screen generally matched their known targets such as topoisomerase inhibitors for S/G2 and PI3K/mTOR inhibitors for G1, validating the synchronisation method and providing a platform to investigate compounds with less well-known mechanisms. We were also able to identify cases where the CCS score varied despite having the same molecular target (e.g. TOP1; topotecan = 0.70, SN-38 = 0.38) as well as identifying a number of compounds that had no apparent cell cycle specificity despite having high levels of potency (e.g. ispinesib = 0.15, kinesin spindle protein inhibitor). Drug potency was one factor related to generation of CCS scores, although the two were not significantly correlated. It is possible that use of longer incubation times may have uncovered more cell cycle dependencies, however significant overlap with cell cycle transitions could impact on the reliability of the CCS score. The comparison of CCS scores in pulse versus continuous exposure can also provide insights into drug mode of action. AZD6738, for example, demonstrated S/G2 associated activity that was not dramatically shifted by continuous exposure (GI50: pulse = 5 μM, cont. = 2.4 μM), suggesting transiently inhibiting ATR in S/G2 cells accounts for a significant proportion of its associated cytotoxicity. With previous reports of hypersensitivity of ATM-null cell lines to ATR inhibition [25,33,34], it is possible this effect could potentially be boosted in cells without functional ATM. This may explain why others have found AZD6738-induced bone marrow toxicity with such tissue types having an abundance of highly proliferative cells [35]. The inherent lack of AZD6738-induced G1-associated toxicity, however, may also be exploited in schedules that use transient CDK4/6 inhibition to protect the bone marrow in patients with Rb negative tumours [36,37].
Exposure to both gemcitabine and AZD6738 was more productive in G1-accumulated cells, suggesting a pre-replicative mechanism and could be exploited using transient CDK4/6 inhibition in a schedule that appeared Rb-dependent. Of note was the extent of cell death following treatment in the “clonocidal” assay, an adaptation of the colony forming assay, which assesses the extent of treatment cell death after large, visible colonies have been allowed to form. This ensures that the measured effect is less influenced by low seeding densities but also by compounds that induce cytostatic rather than cytotoxic effects. Taken together with the increase in intra-S phase of the cell cycle post-treatment alongside increased γH2AX, a marker of DNA damage, further work should characterise any replication-mediated effect of G1 accumulation prior to gemcitabine/AZD6738 treatment to elucidate the mechanism of action. Given the effect of gemcitabine and ATR inhibition on ribonucleotide reductase it is possible that the dual activities of these compounds directly impact on pre-replication events prior to S phase thereby increasing the affected population after palbociclib pre-treatment [29,38,39]. Interestingly, the sensitivity of HeLa cells to the combination without pre-treatment with palbociclib may suggest that Rb pathway loss is a predictive biomarker of enhanced sensitivity to gemcitabine/AZD6738 and warrants further investigation. Overall, we demonstrated that a drug treatment with a G1-associated CCS score showed increased efficacy when combined with a drug that induces G1 accumulation, suggesting that other drugs with these attributes may also be viable candidates. Indeed, the utility of combining mTOR inhibitors with palbociclib has been well validated in vitro and in vivo indicating that CCS score may be useful in finding effective drug combinations [13], [14], [15], [16].
Despite the limitations with respect to cell lines, our correlation between CCS score in AsPC-1 cells and scheduling data in PANC-1 cells produced by Koplev et al demonstrated that CCS score may also inform scheduling strategies [30]. Of the 1,156 drug schedules used in our analysis, the majority (765/1,156) showed neither antagonism nor synergy (scores between -0.05 and 0.05). Even so, multiple comparative analyses suggested that if the second drug in the schedule had a higher, S/G2-associated CCS score then the schedule is more likely to be antagonistic. This seemed to be particularly true for microtubule poisons such as paclitaxel and vincristine where drug sequence is known to impact anti-tumour efficacy, with this cell cycle-dependent antagonism also noted by Koplev and colleagues [30]. Although these analyses are by no means exhaustive, along with existing published data it contributes to the growing body of evidence that timing and cell cycle specificity are important aspects of pre-clinical, in vitro compound profiling studies that could be incorporated into early stage drug development pipelines.
As a final remark, the clinical complexities of schedules such as those described here with palbociclib are significant and subsequent work in vivo should aim to address how this could be implemented in humans. The importance of these scheduling studies is likely to increase given the significant interest in DDR inhibitors with recent evidence highlighting how scheduling of these inhibitors with other compounds can impact tolerability, a significant issue with this class of compounds [40]. As the response in HeLa cells demonstrated, the key consideration for this approach is genetic context and as such patient stratification and biomarker development will be of paramount importance in translating such treatment schedules to clinical setting.
Contributors
Conceptualization, T.I.J.; Methodology, T.I.J, C.J.M., D.K.; Investigation, T.I.J, C.J.M., D.K.; Writing – Original Draft, T.I.J.; Writing – Review & Editing, T.I.J., F.M.R., D.I.J., C.R.D., L.S.C., Y.W., D.K., C.J.M, S.BQF., A.L.; Funding Acquisition, D.I.J.; Resources; S.BQF., Y.W., A.L., L.S.C.; Supervision, F.M.R., D.I.J.
T.I.J, C.J.M. and D.K. all verify the underlying data is correct. All authors have read and approved the final version of the manuscript.
Data Sharing Statement
Data supporting this study, including the compound library screen results, are included in the supplementary data files. Data used from the Koplev et al study is available online at http://dx.doi.org/10.17632/wgybvcvjwf.7 (Data Table 2).
Declaration of Competing Interest
D.K., C.J.M. and S.BQF. have nothing to disclose. T.I.J. reports other from Artios Parma Ltd, outside the submitted work; and Artios Pharma employee and shareholder. C.R.D, L.S.C., Y.W. and A.L. reports other from AstraZeneca PLC, outside the submitted work; and AstraZeneca employee and shareholder. F.M.R. reports grants from Cancer Research UK, during the conduct of the study. D.I.J. reports grants from Cancer Research UK, non-financial support from AstraZeneca, during the conduct of the study.
Acknowledgements
We would like to acknowledge and thank Lorna Hopcroft (AZ), Chrysiis Michaloglou (AZ), Christopher Way (AZ) and Abigail Shea (CRUK CI) for help with training and use of lab equipment. We thank the Core Facilities at the Cancer Research UK Cambridge Institute, including the Biological Resources Unit, Research Instrumentation & Cell Services, Light Microscopy and Flow Cytometry for technical provision, as well as members of the Pharmacology & Drug Development Group for discussions. Thanks to AstraZeneca for provision of AZD6738 and our funders: Cancer Research UK (CRUK) and the Li Ka Shing Centre where this work was performed was generously funded by CK Hutchison Holdings Limited, the University of Cambridge, CRUK, The Atlantic Philanthropies and others.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103396.
Contributor Information
Timothy I. Johnson, Email: Isaac.Johnson@cantab.net.
Duncan I. Jodrell, Email: dij21@cam.ac.uk.
Appendix. Supplementary materials
Figure S1: RO-3306 optimisation. Flow cytometry-based quantification of cell cycle phases using DNA content and a mitotic marker after treatment with increasing concentrations of RO-3306 (0.5-8 μM) or DMSO for 20 hours prior to harvesting. Cell lines indicated for each graph, data displayed as mean ± SD after three independent experiments.
Figure S2: RO-3306 synchronisation time-course in HeLa cells. a. Gating strategy for quantifying cell cycle stages. b. Cell cycle quantification of HeLa cells synchronised using the RO-3306 shake-off method with a time-course taken over 21 hours. c. Same as in B with S and G2 phase percentages added together with the 0h time point taken from isolated mitotic cells (pre-seeding). Data generated from a single experiment.
Figure S3: Optimised, medium-throughput sample processing for compound library screens. a. Coefficient of variation (CV) of SRB values after seeding either 96-well plates by hand or 384-well plates using the Multidrop Combi dispenser. Line represents median with each point representing an individual biological replicate, comparison analysed using a two-tailed unpaired t-test. b. Time taken to dose 96/384 well plates by hand or by using the Labcyte Echo acoustic dispenser. c. Normalised SRB values of 108 DMSO treated wells in a 384-well plate where 2x PBS washes were performed by hand (red), using an automated plate washer (blue) or the Integra Viaflo electronic pipetting station.
Figure S4: Additional parameters identified from the drug library screen. a. Mean nuclei count of DMSO-treated wells for all assay plates with those used for early (4-10h) and late (18-24h) dosing identified alongside CV values for each set. b. Frequency distribution with a Gaussian least squares fit of the minimum AUC value found in either G1 or S/G2 accumulated AsPC-1 cells. Standard deviation is displayed. c. Correlation between CCS score and AUCmin values of all drugs tested (left) and drugs with an AUCmin value <2000 (right) with a simple linear regression. Number of observations and R2 values are displayed in each panel. d. Comparison of GI50 values generated after continuous or pulse treatment in G1 (n=57) or S/G2 (n=70) accumulated AsPC-1 cells. Each point represents a single compound.
Figure S5: Cell cycle distributions of AsPC-1 cells used to generate CCS scores in biological triplicate. Cell cycle quantification of AsPC-1 cells at various stages through an RO-3306-based mitotic shake off assay in three independent replicates used to generate cell cycle specificity scores (CCS).
Figure S6: Cell cycle distribution after palbociclib treatment and subsequent release. a. DNA content histograms of MIA PaCa-2 cells with and without 1 μM palbociclib treatment for 22 hours and quantification of cell cycle phases using DNA content and a mitotic marker, phospho-mpm2, across three biological replicates. Error bars represent ± SD b. Clonocidal assay in MIA PaCa-2 cells treated either with DMSO or gem./AZD6738 after pre-treatment with DMSO, continuous (C) palbociclib or palbociclib wash out (WO). Data is shown as mean ± SEM of three independent experiments. One-way ANOVA with Bonferroni multiple comparisons test was used for comparisons. c. Cell cycle analysis using DNA content in MIA PaCa-2 cells treated with 1 μM palbociclib for 22 hours before release into fresh medium and subsequently collected at the indicated times. Error bars represent ± SD of two independent experiments. d. Live cell confocal imaging of MIA PaCa-2 cells expressing a CDT1 and geminin fluorescent cell cycle reporter. Cells were treated with 1 μM palbociclib for 22 hours before wash out and resumption of imaging. Data is expressed as number of nuclei counted as red-fluorescent G1 cells, green-fluorescent S/G2/M cells or double positive (early S) from a single experiment. e. Live cell imaging using the IncuCyte Zoom microscope and MIA PaCa-2 cells grown as colonies and expressing the fluorescent cell cycle reporter. Due to the fluorescent proteins used, only Ggreen fluorescence could be captured on this system. Cells were treated with 1 μM palbociclib for 22 hours before release into fresh medium and imaged at the indicated times. Yellow lines indicate phase contrast detection of colony borders. Data shown is from a single experiment.
References
- 1.Williams G.H., Stoeber K. The cell cycle and cancer. J Pathol. 2012;226:352–364. doi: 10.1002/path.3022. [DOI] [PubMed] [Google Scholar]
- 2.Blagosklonny M.V., Darzynkiewicz Z. Cyclotherapy: Protection of Normal Cells and Unshielding of Cancer Cells. Cell Cycle. 2002;1(6):375–382. doi: 10.4161/cc.1.6.259. [DOI] [PubMed] [Google Scholar]
- 3.Mills C.C., Kolb E.A., Sampson V.B. Development of Chemotherapy with Cell-Cycle Inhibitors for Adult and Pediatric Cancer Therapy. Cancer Res. 2018;78(2) doi: 10.1158/0008-5472.CAN-17-2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong X., Sui M., Fan W., Kraft A. Cell-cycle dependent antagonistic interactions between paclitaxel and carboplatin in combination therapy. Cancer biology & therapy. 2007;6:1067–1073. doi: 10.4161/cbt.6.7.4323. [DOI] [PubMed] [Google Scholar]
- 5.Smorenburg C.H., Sparreboom A., Bontebal M., Verweij J. Combination chemotherapy of the taxanes and antimetabolites: its use and limitations. Eur J Cancer. 2001;37:2310–2323. doi: 10.1016/s0959-8049(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 6.Shah M.A., Schwartz G.K. Cell cycle-mediated drug resistance. Clin Cancer Res. 2001;7(8):2168–2181. [PubMed] [Google Scholar]
- 7.Schwartz G.K., Ilson D., Saltz L., O'Reilly E., Tong Masalak. Phase II study of the cyclin-dependent kinase inhibitor flavopiridol administered to patients with advanced gastric carcinoma. J Clin Oncol. 2001;19(7):1985–1992. doi: 10.1200/JCO.2001.19.7.1985. Apr 1. [DOI] [PubMed] [Google Scholar]
- 8.Chen P., Lee N.V., Hu W., Xu M., Ferre R.A., Lam H. Spectrum and Degree of CDK Drug Interactions Predicts Clinical Performance. Molecular cancer therapeutics. 2016;15(10):2273–2281. doi: 10.1158/1535-7163.MCT-16-0300. pmid27496135. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton E., Infante J.R. Targeting CDK4/6 in patients with cancer. Cancer Treat Rev. 2016;45:129–138. doi: 10.1016/j.ctrv.2016.03.002. pmid:27017286. [DOI] [PubMed] [Google Scholar]
- 10.Jin D., Tran N., Thomas N., Tran D.D. CombiningCDK4/6 inhibitors ribociclib and palbociclib with cytotoxic agents does not enhance cytotoxicity. PLoS ONE. 2019;14(10) doi: 10.1371/journal.pone.0223555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pikman Y., Alexe G., Roti G., Conway A.S., Furman A., Lee E.S. Synergistic Drug Combinations with a CDK4/6 Inhibitor in T-cell Acute Lymphoblastic Leukemia. Clin. Cancer Res. 2017;23(4):1012–1024. doi: 10.1158/1078-0432.CCR-15-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McClendon A.K., Dean J.L., Rivadeneira D.B., Yu J.E., Reed C.A., Gao E. CDK4/6 inhibition antagonizes the cytotoxic response to anthracycline therapy. Cell Cycle. 2012;11:2747–2755. doi: 10.4161/cc.21127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Michaloglou C., Crafter C., Siersbaek R., Delpuech O., Curwen J.O., Carnevalli L.S. Combined Inhibition of mTOR and CDK4/6 Is Required for Optimal Blockade of E2F Function and Long-term Growth Inhibition in Estrogen Receptor–positive Breast Cancer. Mol. Cancer Ther. 2018;17(5):908–920. doi: 10.1158/1535-7163.MCT-17-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto T., Kanaya N., Somlo G., Chen S. Synergistic anti-cancer activity of CDK4/6 inhibitor palbociclib and dual mTOR kinase inhibitor MLN0128 in pRb-expressing ERnegative breast cancer. Breast Cancer Res Treat. 2019;174(3):615–625. doi: 10.1007/s10549-018-05104-9. April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cretella D., Ravelli A., Fumarola C., La Monica S., Digiacomo G., Cavazzoni A. The anti-tumor efficacy of CDK4/6 inhibition is enhanced by the combination with PI3K/AKT/mTOR inhibitors through impairment of glucose metabolism in TNBC cells. Journal of Experimental & Clinical Cancer Research. 2018;37:72. doi: 10.1186/s13046-018-0741-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bonelli M.A., Digiacomo G., Fumarola C., Alfieri R., Quaini F., Falco A. Combined Inhibition of CDK4/6 and PI3K/AKT/mTOR Pathways Induces a Synergistic Anti-Tumor Effect in Malignant Pleural Mesothelioma Cells. Neoplasia. 2017;19:637–648. doi: 10.1016/j.neo.2017.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elhajouji A., Cunha M., Kirsch-Volders M. Spindle poisons can induce polyploidy by mitotic slippage and micronucleate mononucleates in the cytokinesis-block assay. Mutagenesis. 1998;13(2):193–198. doi: 10.1093/mutage/13.2.193. [DOI] [PubMed] [Google Scholar]
- 18.Brito D.A., Rieder C.L. The Ability to Survive Mitosis in the Presence of Microtubule Poisons Differs Significantly Between Human Nontransformed (RPE-1) and Cancer (U2OS, HeLa) Cells. Cell Motil Cytoskeleton. 2009:437–447. doi: 10.1002/cm.20316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banfalvi, G. Overview of Cell Synchronization, in: Banfalvi, G. (Ed.), Cell Cycle Synchronization: Methods and Protocols, Methods in Molecular Biology 2017. Springer New York, New York, NY, pp. 3–27.
- 20.Johnson T.I, Costa A.S.H., Ferguson A.N., Frezza C. Fumarate hydratase loss promotes mitotic entry in the presence of DNA damage after ionising radiation. Cell Death Dis. 2018;9(9):913. doi: 10.1038/s41419-018-0912-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008;132(3):487–498. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 22.Vassilev L.T., Tovar C., Chen S., Knezevic D., Zhao X., Sun H. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. PNAS. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Olive K.P., Jacobetz M.A., Davidson C.J., Gopinathan A., McIntyre D., Honess D., Madhu B. Inhibition of Hedgehog Signaling Enhances Delivery of Chemotherapy in a Mouse Model of Pancreatic Cancer. Science. 2009;324(5933):1457–1461. doi: 10.1126/science.1171362. Jun 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallez Y., Dunlop C.R., Johnson T.I., Koh SB., Fornari C., Yates J.W.T. The ATR Inhibitor AZD6738 Synergizes with Gemcitabine In Vitro and In Vivo to Induce Pancreatic Ductal Adenocarcinoma Regression. Mol Cancer Ther. 2018;17(8):1670–1682. doi: 10.1158/1535-7163.MCT-18-0010. Aug 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunlop C.R., Wallez Y., Johnson T.I., Fernandez S.B.Q., Durant S.T., Cadogan E.B. Complete loss of ATM function augments replication catastrophe induced by ATR inhibition and gemcitabine in pancreatic cancer models. Br J Cancer. 2020 doi: 10.1038/s41416-020-1016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pereira S, Fernandes PA, Ramos MJ. Mechanism for ribonucleotide reductase inactivation by the anticancer drug gemcitabine. J Comput Chem. 2004;25:1286–1294. doi: 10.1002/jcc.20054. [DOI] [PubMed] [Google Scholar]
- 27.Plunkett W, Huang P, Gandhi V. Preclinical characteristics of gemcitabine. Anticancer Drugs. 1995;6:7–13. doi: 10.1097/00001813-199512006-00002. Suppl 6. [DOI] [PubMed] [Google Scholar]
- 28.Chen Z, Zhou J, Zhang Y, Bepler G. Modulation of the ribonucleotide reductase M1-gemcitabine interaction in vivo by N-ethylmaleimide. Biochem Biophys Res Commun. 2011;413:383–388. doi: 10.1016/j.bbrc.2011.08.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerqueira N.M., Fernandes P.A., Ramos M.J. Understanding ribonucleotide reductase inactivation by gemcitabine. Chemistry. 2007;13(30):8507–8515. doi: 10.1002/chem.200700260. [DOI] [PubMed] [Google Scholar]
- 30.Koplev S., Longden J., Ferkinghoff-Borg J., Bjerregard M.B., Cox T.R., Erler J.T. Dynamic Rearrangement of Cell States Detected by Systematic Screening of Sequential Anticancer Treatments. Cell Reports. 2017;20:2784–2791. doi: 10.1016/j.celrep.2017.08.095. [DOI] [PubMed] [Google Scholar]
- 31.Fryer RA, Barlett B, Galustian C, Dalgleish AG. Mechanisms underlying gemcitabine resistance in pancreatic cancer and sensitisation by the iMiD™ lenalidomide. Anticancer Res. 2011 Nov;31(11):3747–3756. [PubMed] [Google Scholar]
- 32.Ullenhag GJ, Mozaffari F, Broberg M, Mellstedt H, Liljefors M. Clinical and Immune Effects of Lenalidomide in Combination with Gemcitabine in Patients with Advanced Pancreatic Cancer. PLoS One. 2017;12(1) doi: 10.1371/journal.pone.0169736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt A., Knittel G., Welcker D., Yang TP., Geroge J., Nowak M. ATM Deficiency Is Associated with Sensitivity to PARP1- and ATR Inhibitors in Lung Adenocarcinoma. Cancer research. 2017:77. doi: 10.1158/0008-5472.CAN-16-3398. [DOI] [PubMed] [Google Scholar]
- 34.Jette N.R., Kumar M., Radhamani S., Arthur G., Goutam S., Yip S. ATM-Deficient Cancers Provide New Opportunities for Precision Oncology. Cancers. 2020;12:687. doi: 10.3390/cancers12030687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vendetti F.P., Lau A., Schamus S., Conrads T.P., O'Connor M.J., Bakkenist C.J. The orally active and bioavailable ATR kinase inhibitor AZD6738 potentiates the anti-tumor effects of cisplatin to resolve ATM-deficient non-small cell lung cancer in vivo. Oncotarget. 2015;6(42):44289–44305. doi: 10.18632/oncotarget.6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bisi J.E., Sorrentino J.A., Roberts P.J., Tavares F.X., Strum J.C. Preclinical Characterization of G1T28: A Novel CDK4/6 Inhibitor for Reduction of Chemotherapy-Induced Myelosuppression. Mol. Cancer Therap. 2016 doi: 10.1158/1535-7163.MCT-15-0775. [DOI] [PubMed] [Google Scholar]
- 37.He S., Roberts P.J., Sorrentino J.A., Bisi J.E., Storrie-White H., Tiessen R.G. Transient CDK4/6 inhibition protects hematopoietic stem cells from chemotherapy-induced exhaustion. Sci. Trans. Med. 2017;9:387. doi: 10.1126/scitranslmed.aal3986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X., Muller E.G., Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 39.Buisson R., Boisvert J.L., Benes C.H., Zou L. Distinct but concerted roles of ATR, DNA-PK, and Chk1 in countering replication stress during S phase. Mol Cell. 2015;59:1011–1024. doi: 10.1016/j.molcel.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang, Y., McGrail, DJ., Sun, C., Labrie, Marilyne., Chen, X., Zhang, D., et al. Sequential Therapy with PARP and WEE1 Inhibitors Minimizes Toxicity while Maintaining Efficacy. Cancer Cell. 2019 Jun 10;35(6):851-867.e7 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: RO-3306 optimisation. Flow cytometry-based quantification of cell cycle phases using DNA content and a mitotic marker after treatment with increasing concentrations of RO-3306 (0.5-8 μM) or DMSO for 20 hours prior to harvesting. Cell lines indicated for each graph, data displayed as mean ± SD after three independent experiments.
Figure S2: RO-3306 synchronisation time-course in HeLa cells. a. Gating strategy for quantifying cell cycle stages. b. Cell cycle quantification of HeLa cells synchronised using the RO-3306 shake-off method with a time-course taken over 21 hours. c. Same as in B with S and G2 phase percentages added together with the 0h time point taken from isolated mitotic cells (pre-seeding). Data generated from a single experiment.
Figure S3: Optimised, medium-throughput sample processing for compound library screens. a. Coefficient of variation (CV) of SRB values after seeding either 96-well plates by hand or 384-well plates using the Multidrop Combi dispenser. Line represents median with each point representing an individual biological replicate, comparison analysed using a two-tailed unpaired t-test. b. Time taken to dose 96/384 well plates by hand or by using the Labcyte Echo acoustic dispenser. c. Normalised SRB values of 108 DMSO treated wells in a 384-well plate where 2x PBS washes were performed by hand (red), using an automated plate washer (blue) or the Integra Viaflo electronic pipetting station.
Figure S4: Additional parameters identified from the drug library screen. a. Mean nuclei count of DMSO-treated wells for all assay plates with those used for early (4-10h) and late (18-24h) dosing identified alongside CV values for each set. b. Frequency distribution with a Gaussian least squares fit of the minimum AUC value found in either G1 or S/G2 accumulated AsPC-1 cells. Standard deviation is displayed. c. Correlation between CCS score and AUCmin values of all drugs tested (left) and drugs with an AUCmin value <2000 (right) with a simple linear regression. Number of observations and R2 values are displayed in each panel. d. Comparison of GI50 values generated after continuous or pulse treatment in G1 (n=57) or S/G2 (n=70) accumulated AsPC-1 cells. Each point represents a single compound.
Figure S5: Cell cycle distributions of AsPC-1 cells used to generate CCS scores in biological triplicate. Cell cycle quantification of AsPC-1 cells at various stages through an RO-3306-based mitotic shake off assay in three independent replicates used to generate cell cycle specificity scores (CCS).
Figure S6: Cell cycle distribution after palbociclib treatment and subsequent release. a. DNA content histograms of MIA PaCa-2 cells with and without 1 μM palbociclib treatment for 22 hours and quantification of cell cycle phases using DNA content and a mitotic marker, phospho-mpm2, across three biological replicates. Error bars represent ± SD b. Clonocidal assay in MIA PaCa-2 cells treated either with DMSO or gem./AZD6738 after pre-treatment with DMSO, continuous (C) palbociclib or palbociclib wash out (WO). Data is shown as mean ± SEM of three independent experiments. One-way ANOVA with Bonferroni multiple comparisons test was used for comparisons. c. Cell cycle analysis using DNA content in MIA PaCa-2 cells treated with 1 μM palbociclib for 22 hours before release into fresh medium and subsequently collected at the indicated times. Error bars represent ± SD of two independent experiments. d. Live cell confocal imaging of MIA PaCa-2 cells expressing a CDT1 and geminin fluorescent cell cycle reporter. Cells were treated with 1 μM palbociclib for 22 hours before wash out and resumption of imaging. Data is expressed as number of nuclei counted as red-fluorescent G1 cells, green-fluorescent S/G2/M cells or double positive (early S) from a single experiment. e. Live cell imaging using the IncuCyte Zoom microscope and MIA PaCa-2 cells grown as colonies and expressing the fluorescent cell cycle reporter. Due to the fluorescent proteins used, only Ggreen fluorescence could be captured on this system. Cells were treated with 1 μM palbociclib for 22 hours before release into fresh medium and imaged at the indicated times. Yellow lines indicate phase contrast detection of colony borders. Data shown is from a single experiment.