Abstract
Background
Clinical management ranges from surveillance or curettage to wide resection for atypical to higher-grade cartilaginous tumours, respectively. Our aim was to investigate the performance of computed tomography (CT) radiomics-based machine learning for classification of atypical cartilaginous tumours and higher-grade chondrosarcomas of long bones.
Methods
One-hundred-twenty patients with histology-proven lesions were retrospectively included. The training cohort consisted of 84 CT scans from centre 1 (n=55 G1 or atypical cartilaginous tumours; n=29 G2-G4 chondrosarcomas). The external test cohort consisted of the CT component of 36 positron emission tomography-CT scans from centre 2 (n=16 G1 or atypical cartilaginous tumours; n=20 G2-G4 chondrosarcomas). Bidimensional segmentation was performed on preoperative CT. Radiomic features were extracted. After dimensionality reduction and class balancing in centre 1, the performance of a machine-learning classifier (LogitBoost) was assessed on the training cohort using 10-fold cross-validation and on the external test cohort. In centre 2, its performance was compared with preoperative biopsy and an experienced radiologist using McNemar's test.
Findings
The classifier had 81% (AUC=0.89) and 75% (AUC=0.78) accuracy in identifying the lesions in the training and external test cohorts, respectively. Specifically, its accuracy in classifying atypical cartilaginous tumours and higher-grade chondrosarcomas was 84% and 78% in the training cohort, and 81% and 70% in the external test cohort, respectively. Preoperative biopsy had 64% (AUC=0.66) accuracy (p=0.29). The radiologist had 81% accuracy (p=0.75).
Interpretation
Machine learning showed good accuracy in classifying atypical and higher-grade cartilaginous tumours of long bones based on preoperative CT radiomic features.
Funding
ESSR Young Researchers Grant.
Keywords: Artificial intelligence, Chondrosarcoma, Machine learning, Multidetector computed tomography
Research in Context.
Evidence before this study
To date, radiomic studies have dealt with MRI of cartilaginous bone lesions with the aim of discriminating among benign enchondroma, atypical cartilaginous tumour and malignant chondrosarcoma, predicting local recurrence of chondrosarcoma and differentiating chondrosarcoma from other entities such as skull chordoma. Machine learning was used in combination with radiomics to address some of these issues. Particularly, an adaptive boosting classifier (AdaBoostM1) was a good predictor of tumour grade based on MRI radiomic features derived from unenhanced T1-weighted sequences, showing 75% accuracy in the test cohort for classification of atypical cartilaginous tumours and chondrosarcomas. This previous study included 58 patients from the same institution and the machine-learning classifier was internally tested using a hold-out set as a test cohort. To our knowledge, no published study has focused on machine learning and CT radiomics of cartilaginous bone lesions, as done in our study.
Added value of this study
We attempted to differentiate atypical cartilaginous tumours from chondrosarcomas of long bones, as this is the most relevant clinical question and orientates towards a conservative approach or aggressive surgery. Our CT radiomics-based machine-learning classifier (boosted [LogitBoost] linear logistic regression classifier) achieved 75% accuracy overall, 81% accuracy in identifying atypical cartilaginous tumours and 70% accuracy in identifying higher-grade chondrosarcomas in the external test cohort, respectively, with no difference in comparison with an experienced radiologist (p=0.75). These results agree with those previously reported for tumour grading based on MRI radiomics. Furthermore, our findings were obtained in a more than twice larger population and validated in an independent test cohort from a second institution, thus ensuring their generalizability in clinical practice. Finally, although statistical significance was not reached (p=0.29), the machine-learning classifier's accuracy was slightly superior compared to preoperative biopsy. We may speculate that this difference could become significant in a larger population.
Implications of all the available evidence
Radiomics-based machine learning may potentially aid in preoperative tumour characterization by integrating the multidisciplinary approach currently based on clinical, conventional imaging and histological assessment.
Alt-text: Unlabelled box
1. Introduction
Chondrosarcoma accounts for 20 to 30% of primary malignant bone lesions [1]. Clinical management primarily depends on tumour grading. Particularly, low-grade (G1) chondrosarcomas of long bones, recently downgraded from malignant to locally aggressive lesions and renamed “atypical cartilaginous tumours” [2], are managed with intralesional curettage or even watchful waiting. Appendicular higher-grade and axial skeleton chondrosarcomas require wide resection with free margins [3]. The 10-year overall survival decreases from 88% for atypical cartilaginous tumour/G1 to 62% and 26% for G2 and G3 chondrosarcoma, respectively [4]. Both imaging and biopsy integrate clinical information before any treatment is started [3]. Magnetic resonance imaging (MRI) is the best imaging modality for local staging [5]. Computed tomography (CT) is used for biopsy guidance [6] and provides additional information, such as matrix mineralization and cortex changes [3]. CT and positron emission tomography-CT (PET-CT) can be both employed for general staging [3]. Biopsy is considered the reference standard for preoperative assessment but suffers from the disadvantages of sampling errors [7] and overlapping histological findings leading to discrepancies even among expert bone pathologists [8]. Thus, the need for new imaging-based tools like radiomics is advocated to better characterize cartilaginous bone lesions preoperatively [9].
Radiomics includes extraction and analysis of large numbers of quantitative characteristics, known as radiomic features, from imaging studies [10]. This research field has gained much attention in oncologic imaging as a potential tool for quantification of tumour heterogeneity, which is hard to capture with conventional imaging assessment or sampling biopsies [11]. Most radiomic studies to date have focused on discriminating tumour grades and types before treatment, monitoring response to therapy and predicting outcome [11]. Due to its high-dimensional nature consisting of numerous radiomic features, radiomics benefits from powerful analytic tools and artificial intelligence with machine learning perfectly addresses this issue [12]. Machine learning algorithms can be trained using subsets of radiomic features creating classification models for the diagnosis of interest [13], [14], [15].
Machine learning has recently shown good accuracy in discriminating between atypical cartilaginous tumours and higher-grade bone chondrosarcomas based on unenhanced MRI radiomic features [16]. The aim of this study is to investigate the diagnostic performance of CT radiomics-based machine learning for classification of atypical cartilaginous tumours and higher-grade chondrosarcomas of long bones.
2. Methods
2.1. Ethics
Our Institutional Review Board approved this retrospective study and waived the need for informed consent (Protocol: “AI tumori MSK”). Our database was anonymized according to the General Data Protection Regulation for Research Hospitals.
2.2. Study design and inclusion criteria
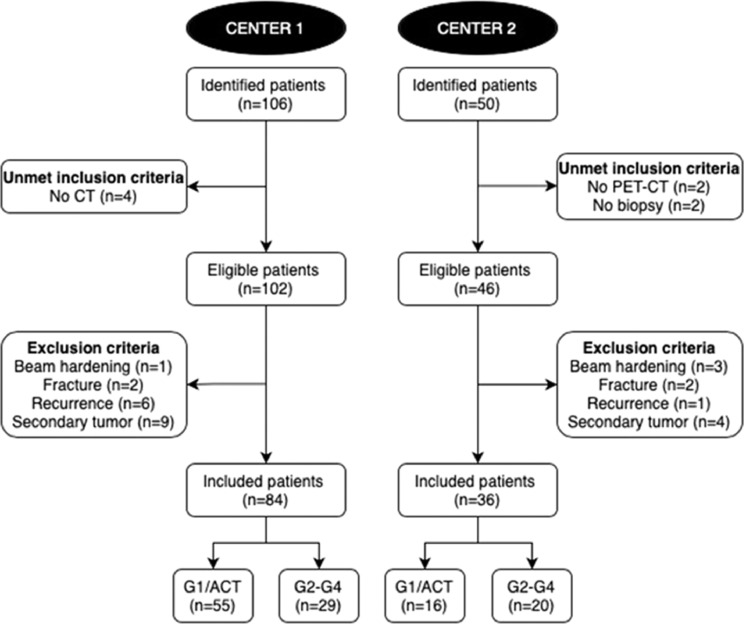
Information was retrieved through electronic records from the orthopaedic surgery and pathology departments. Consecutive patients with an atypical cartilaginous tumour or appendicular chondrosarcoma and CT or PET-CT performed over the last 10 years at one of two tertiary bone tumour centres (centre 1, IRCCS Orthopaedic Institute Galeazzi in Milan, Italy; centre 2, IRCCS Regina Elena National Cancer Institute in Rome, Italy) were considered for inclusion. Inclusion criteria were: (i) atypical cartilaginous tumour or conventional G2-G3-G4 (dedifferentiated) chondrosarcoma of long bones that was surgically treated with intralesional curettage or resection; (ii) definitive histological diagnosis defined on the basis of the surgical specimen assessment; (iii) CT (centre 1) or PET-CT (centre 2) scan performed before biopsy and within 1 month before surgery; and (iv) in centre 2, preoperative biopsy performed within 1 month before surgery. Patients with pathological fractures, secondary tumours arising from pre-existing cartilaginous lesions, recurrent tumours or metal devices resulting in beam hardening artifacts were excluded. A flowchart of patient selection is shown in Fig. 1.
Fig. 1.
Flowchart of patient selection. ACT, atypical cartilaginous tumours.
2.3. Study cohorts
One-hundred-twenty patients were retrospectively included. The training cohort consisted of 84 CT scans by as many patients from centre 1 (n=55 G1 or atypical cartilaginous tumours; n=29 G2-G4 chondrosarcomas). The external test cohort was constituted by the CT component of 36 PET-CT scans by as many patients from centre 2 (n=16 G1 or atypical cartilaginous tumours; n=20 G2-G4 chondrosarcomas). Patients’ demographics and data regarding lesion location, grading and surgical treatment are detailed in Table 1. In centre 1, all examinations were performed using a 64-slice CT unit (Siemens SOMATOM Emotion, Erlangen, Germany). CT specifications were: matrix, 512 × 512; field of view (range), 138-380 mm; slice thickness, 1 mm. In centre 2, all examinations were performed using a 16-slice PET-CT unit (Siemens Biograph, Erlangen, Germany). PET-CT specifications were: matrix, 512 × 512; field of view, 500 mm; slice thickness, 4 mm. All DICOM images were exported and converted to the NiFTI format prior to the analysis [17].
Table 1.
Demographics and clinical data. Age is presented as median and interquartile (1st-3rd) range.
| Centre 1 | Centre 2 | |
|---|---|---|
| Age | 52 (45-65) years | 57 (46-69) years |
| Sex | Men: n=30 Women: n=54 |
Men: n=13 Women: n=23 |
| Lesion location | Femur: n=40 Fibula: n=9 Humerus: n=30 Radius: n=1 Tibia: n=4 |
Femur: n=21 Fibula: n=6 Humerus: n=5 Tibia: n=4 |
| Lesion grading | G1: n=55 G2: n=13 G3: n=9 G4 (dedifferentiated): n=7 |
G1: n=16 G2: n=12 G3: n=3 G4 (dedifferentiated): n=5 |
| Surgery |
G1/Atypical cartilaginous tumours Curettage: n=47 Wide resection: n=8* |
G1/Atypical cartilaginous tumours Wide resection: n=16* |
|
G2-G4 chondrosarcomas Curettage + wide resection: n=5⁎⁎ Wide resection: n=24 |
G2-G4 chondrosarcomas Wide resection: n=20 |
Wide resection was performed in n=8 G1/atypical cartilaginous tumours from centre 1 in case of specific anatomic location (like fibular head) or to prevent the risk of biopsy sampling errors. It was performed in all cases from centre 2 to prevent the risk of biopsy sampling errors, as per routine procedure.
Curettage was initially performed in n=5 G2 chondrosarcomas from centre 2, as preoperative biopsy downgraded the lesions as G1. A second surgery consisting of wide resection was thus required.
2.4. Segmentation
A recently-boarded musculoskeletal radiologist (S.G.) manually performed contour-focused segmentation using a freely available, open-source software, ITK-SNAP (v3.6) [18]. In detail, bidimensional regions of interest were annotated on the axial slice showing the maximum lesion extension. Unenhanced CT scan or CT scan performed as part of PET-CT protocol was used. According to the intraclass correlation coefficient (ICC) guidelines by Koo et al. [19], in a subgroup of 30 patients randomly selected from centre 1, segmentations were additionally performed independently by two radiology residents experienced in musculoskeletal and oncologic imaging (M.A. and A.C.) to meet the requirements of a reliability analysis in terms of patients and readers involved. All the readers knew the study would deal with cartilaginous bone lesions, but they were unaware of tumour grading and disease course, as well as the slice other readers used for segmentation.
2.5. Feature extraction
Image preprocessing and feature extraction were performed using PyRadiomics (v3.0.0) [20]. Regarding preprocessing, image resampling (to an 1 × 1 mm in-plane resolution) was performed to ensure the correct calculation of texture features, following current guidelines [21]. Grey level normalization and discretization followed. For the first, after z-score normalization, grey level values were scaled by a factor of 100. The resulting arrays were shifted by a value of 300 to avoid negative-valued pixels that could cause issues with texture analysis. After this process, the final image grey level range is expected to fall between 0 and 600, excluding outliers. To select the correct bin width for discretization, an exploratory extraction of first order parameters (i.e., grey level range) was performed exclusively on the training set, to avoid any information leak from the external test cohort. In this step, bin widths 2, 3, 4 and 5 were used to analyse grey level range of the normalized images. In addition to the original images, features were also extracted from filtered ones, i.e. after Laplacian of Gaussian (sigma=1, 2, 3, 4, 5) filtering and wavelet decomposition (all combinations of high and low-pass filtering on the x and y axes). All available first-order (histogram analysis), 2D shape-based and texture features were extracted, described in detail in the PyRadiomics official documentation (https://pyradiomics.readthedocs.io/en/latest/features.html).
2.6. Machine learning analysis
Radiomic data processing and machine learning analysis were performed using the Weka data mining platform (v3.8.4), R and scikit-learn Python package [22], [23], [24]. A normalization (min-max range=0-1) scaler was fitted on the training data and applied to both training and external test cohorts prior to the analysis. Feature selection was performed exclusively using the training cohort data and included stability assessment as well as variance and intercorrelation analyses. The first was performed by obtaining feature ICC with a two-way random effect, single rater, absolute agreement model. Features were considered stable if the ICC 95% confidence interval lower bound was ≥0.75. Next, low variance (0.15 threshold) or highly inter-correlated (Pearson correlation coefficient threshold 0.80) features were removed. Finally, features with an information gain ratio >0.35 were selected.
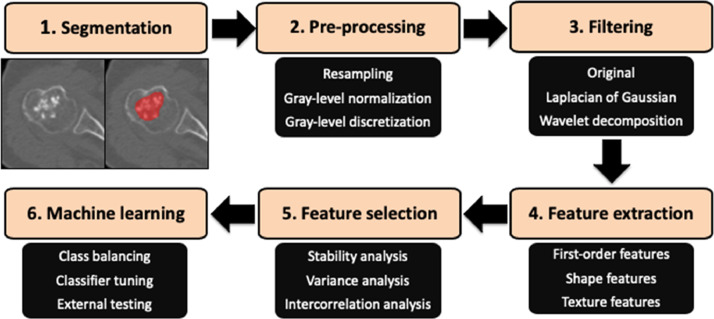
Given the unbalanced nature of the training dataset, the synthetic minority oversampling technique (SMOTE) was used to balance this data by creating new instances from the minority class in centre 1, thus increasing the number of G2-G4 chondrosarcomas to 55 [25]. The test set underwent no oversampling as it was not employed to build the classification model but only to assess its performance. Thereafter, a boosted (LogitBoost) linear logistic regression machine-learning classifier was trained and validated on the training cohort using 10-fold cross validation and tested on the external cohort. The Brier score was obtained, together with calibration curves, for the external test set in order to evaluate prediction and calibration loss. Our radiomics-based machine-learning workflow pipeline is shown in Fig. 2.
Fig. 2.
Radiomics-based machine learning workflow pipeline.
2.7. Qualitative imaging assessment
A musculoskeletal radiologist with 12 years of experience in bone sarcomas (V.A.) read all CT studies from centre 2 blinded to any information regarding tumour grading, disease course and radiomics-based machine learning analysis. G2-G4 chondrosarcomas were differentiated from atypical cartilaginous tumours based on the presence of at least one of the following parameters: medullary cavity expansion with thinner cortex, cortical breakthrough, aggressive periosteal reaction, soft-tissue mass [5,26]. Additionally, maximum lesion diameter was measured.
2.8. Statistical analysis
Continuous data are presented as median and interquartile (1st-3rd) range. Categorical data are presented as value counts and proportions. Data management was performed using the pandas Python software package. The “irr” and “stats” R packages were used for ICC assessment and remaining statistical tests, respectively. In the external test cohort, the classifier's performance was compared with preoperative biopsy and the radiologist's performance using McNemar's test. Mann-Whitney and Fisher's tests were used to assess age and sex differences between the two cohorts. A 2-sided p-value <0.05 indicated statistical significance.
Accuracy measures of the machine-learning classifier performance included, among others:
-
•
F-score, i.e. the harmonic average of the precision (i.e. positive predictive value) and recall (i.e. sensitivity), ranging from 0 to 1 (perfect accuracy)
-
•
Area under the precision-recall curve, i.e. an alternative to the area under the ROC curve, which is more informative for imbalanced classes.
A radiologist with experience in radiomics and artificial intelligence (R.C.) assessed Radiomics Quality Score in the attempt to estimate the methodological rigor of our study, as suggested by Lambin et al. [27].
2.9. Role of funding source
This research was partially funded by the Young Researchers Grant awarded by the European Society of Musculoskeletal Radiology (S.G.). The funding source provided financial support without any influence on the study design; on the collection, analysis, and interpretation of data; and on the writing of the report. The corresponding and senior (last) authors had the final responsibility for the decision to submit the paper for publication.
3. Results
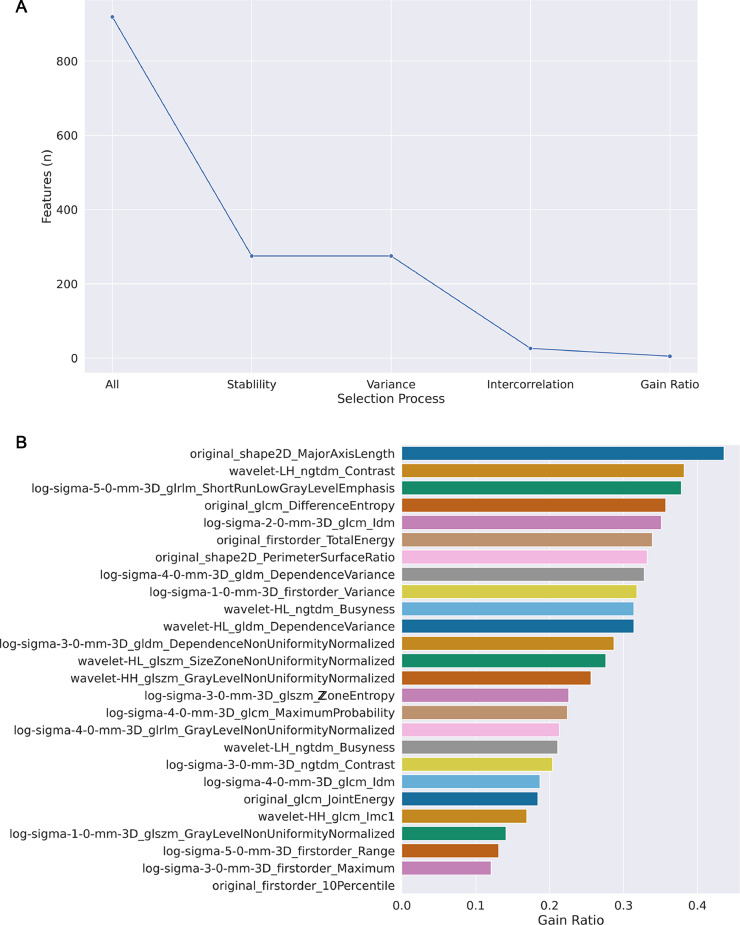
No difference in patients’ age (p=0.25 [Mann-Whitney test]) and sex (p>0.99 [Fisher's test]) was found between the training and the external test cohorts. In our population, a bin width value of 3 presented the best results for feature extraction, with an average of 59 bins (± 30) in the training set. A total of 919 radiomic features were extracted from each segmentation. The rate of stable features was 30% (n=275), none of which had low variance. Removing all inter-correlated features yielded a dataset of 26 non-colinear features. Of these, the five with the highest gain ratio were selected and included: Major Axis Length (2D shape-based) derived from the original images; Contrast (Neighbouring Gray Tone Difference Matrix) derived from wavelet-transformed images (Low-High pass filter); Short Run Low Gray Level Emphasis (Gray Level Run Length Matrix) from LoG-filtered images (sigma=5); Difference Entropy (Gray Level Co-occurrence Matrix) from the original images; Inverse Difference Moment (Gray Level Co-occurrence Matrix) derived from LoG-filtered images (sigma=2). Feature dimensionality reduction is shown in Fig. 3.
Fig. 3.
Feature dimensionality reduction. A Feature selection process was performed exclusively using the training cohort data and included stability assessment as well as variance and intercorrelation analyses. The rate of stable features was 30% (n=275), none of which had low variance. Removing all inter-correlated features yielded a dataset of 26 non-colinear features. B The five features with the highest gain ratio were selected and included.
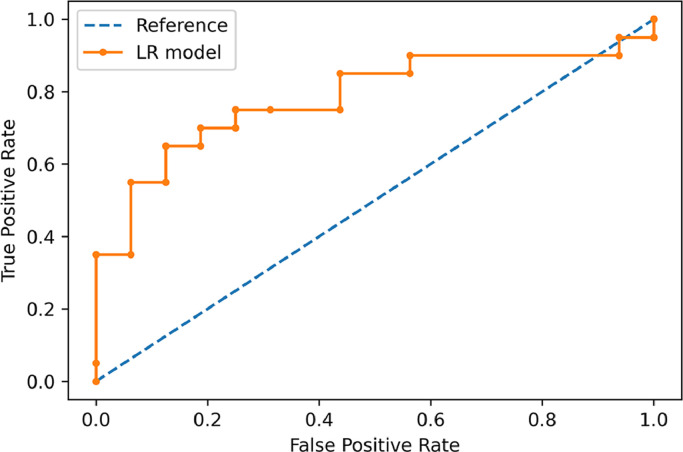
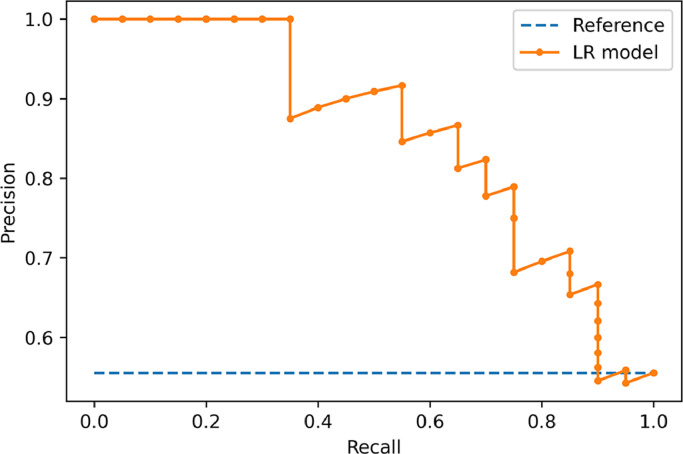
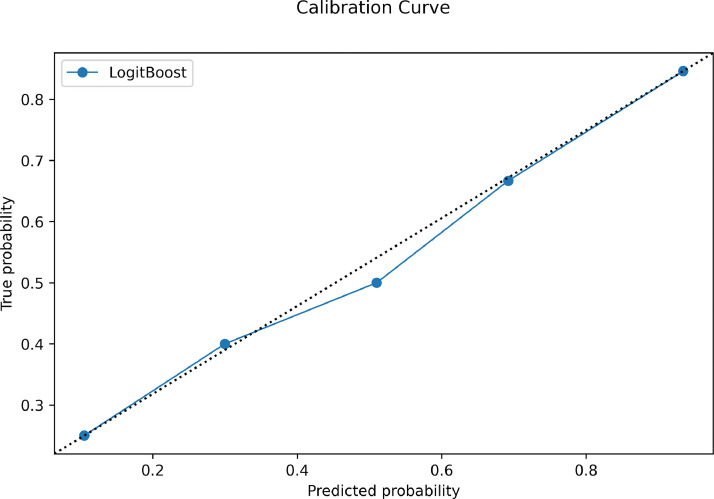
The machine learning classifier had 81% (89/110) and 75% (27/36) accuracy in identifying the cartilaginous bone lesions in the training and external test cohorts, respectively. Area under the ROC curve was, respectively, 0.89 and 0.78. In detail, its accuracy in classifying atypical cartilaginous tumour and higher-grade chondrosarcoma was 84% (46/55) and 78% (43/55) in the training cohort, and 81% (13/16) and 70% (14/20) in the external test cohort, respectively. Other evaluation metrics are derived from confusion matrix in Table 2 and reported in Table 3. Fig. 4 shows the ROC curve illustrating the classifier performance in the external test cohort. Fig. 5 shows the precision-recall curve illustrating the classifier performance for G2-G4 chondrosarcoma identification in the external test cohort. The final model had a Brier score of 0.25, while Fig. 6 depicts its calibration curve in the external test cohort. Our Radiomics Quality Score was 47% (Supplementary material).
Table 2.
Confusion matrix for the training and external test cohorts.
| Predicted class |
||||
|---|---|---|---|---|
| ACT | CS | |||
| Actual class | Training | ACT | 46 | 9 |
| CS | 12 | 43 | ||
| External test | ACT | 13 | 3 | |
| CS | 6 | 14 | ||
ACT, atypical cartilaginous tumour; CS, higher-grade chondrosarcoma.
Table 3.
Classifier accuracy metrics weighted average and by class in both the training and external test cohorts.
| Cohort | Class | TP rate | FP rate | Precision | Recall | F-score | ROC | PRC |
|---|---|---|---|---|---|---|---|---|
| Training | ACT | 0.836 | 0.218 | 0.793 | 0.836 | 0.814 | 0.891 | 0.876 |
| CS | 0.782 | 0.164 | 0.827 | 0.782 | 0.804 | 0.891 | 0.915 | |
| WA | 0.809 | 0.191 | 0.810 | 0.809 | 0.809 | 0.891 | 0.895 | |
| External test | ACT | 0.813 | 0.300 | 0.684 | 0.813 | 0.743 | 0.784 | 0.661 |
| CS | 0.700 | 0.188 | 0.824 | 0.700 | 0.757 | 0.784 | 0.857 | |
| WA | 0.750 | 0.238 | 0.762 | 0.750 | 0.751 | 0.784 | 0.770 |
ACT, atypical cartilaginous tumour; CS, higher-grade chondrosarcoma; FP, false positive; PRC, precision-recall curve; ROC, receiver operator curve; TP, true positive; WA, weighted average.
Fig. 4.
ROC curve showing the classifier performance in the external test cohort.
Fig. 5.
Precision-recall curve illustrating the classifier performance for G2-G4 chondrosarcoma identification in the external test cohort.
Fig. 6.
Calibration curve in the external test cohort. The data is divided into bins, with the y-axis representing the distribution of positive cases in each bin while the x-axis the probability as predicted by the classifier. The closer the resulting calibration curve is to the reference line. the better the model's predictions reflect the actual class distribution in the dataset.
In patients from centre 2, preoperative biopsy had 64% (23/36 correct tumour grade provided) accuracy in grading the cartilaginous bone lesions. Area under the ROC curve was 0.66. Preoperative biopsy provided an inconclusive result (n=5) or downgraded the lesion (n=8) in the remaining patients. Biopsy accuracy was slightly lower in comparison with the machine-learning classifier's accuracy, although this difference was not statistically significant (p=0.29 [McNemar's test]). The experienced radiologist had 81% (29/36 correct diagnosis provided) accuracy in identifying the cartilaginous bone lesions with no statistical difference compared to the classifier (p=0.75 [McNemar's test]). The radiologist's accuracy was 75% (4/16) and 85% (17/20) in classifying atypical cartilaginous tumours and higher-grade chondrosarcomas, respectively, as detailed in Table 4.
Table 4.
Qualitative imaging assessment performed by the experienced radiologist. Lesion diameter is presented as median and interquartile (1st-3rd) range. Other variables are presented as proportions.
| Class | Bone expansion | Cortical breakthrough | Aggressive periostitis | Soft-tissue mass | Maximum diameter | Correct diagnosis |
|---|---|---|---|---|---|---|
| ACT | 1/16 | 3/16 | 1/16 | 0/16 | 45 (31-54) mm | 12/16 |
| CS | 13/20 | 16/20 | 14/20 | 13/20 | 91 (59-124) mm | 17/20 |
| Overall | 14/36 | 19/36 | 15/36 | 13/36 | 60 (42-100) mm | 29/36 |
ACT, atypical cartilaginous tumour; CS, higher-grade chondrosarcoma
4. Discussion
The main finding of this study is that we developed a machine-learning classifier for discrimination between atypical cartilaginous tumours and higher-grade chondrosarcomas of long bones based on preoperative CT radiomic features, which achieved good accuracy in an independent test cohort from an external institution. Its performance did not differ in comparison with both an experienced bone tumour radiologist and preoperative biopsy.
Atypical cartilaginous tumours are locally aggressive lesions of the extremities, relatively indolent as compared with higher-grade tumours, and have a very low metastatic rate [2]. Curettage is the standard of care [3], but its effectiveness in preventing transformation into higher-grade chondrosarcoma has not been demonstrated. Hence, given the similarity to benign enchondroma on both imaging [28] and histology [8], watchful waiting has been proposed as an alternative strategy to prevent overtreatment and morbidity associated with surgery [29], [30], [31]. An accurate differentiation from higher-grade chondrosarcomas requiring wide resection is thus necessary for treatment planning, and currently based on a multidisciplinary approach combining clinical presentation with imaging and biopsy [3]. On imaging, MRI is the method of choice for local staging, while CT and PET-CT are employed for general staging [3]. Both MRI [5] and PET-CT based on standard uptake values [32] are accurate in discriminating between atypical cartilaginous tumours and chondrosarcomas. On the other hand, biopsy may erroneously lead to tumour down-grading in large heterogenous lesions, as only small areas are sampled [7]. Additionally, low reliability in tumour grading has been reported even among specialized bone pathologists [8] and the risk of biopsy-tract contamination also remains a concern. Thus, current imaging techniques may be further equipped to safely grade cartilaginous bone lesions non-invasively, and radiomics looks promising in this regard [9].
To date, radiomic studies have dealt with MRI of cartilaginous bone lesions with the aim of discriminating among benign enchondroma, atypical cartilaginous tumour and malignant chondrosarcoma [16,33,34], predicting local recurrence of chondrosarcoma [35] and differentiating chondrosarcoma from other entities such as skull chordoma [36]. Machine learning was used in combination with radiomics to address some of these issues [16,35,36]. Particularly, machine learning was a good predictor of tumour grade based on MRI radiomic features derived from unenhanced T1-weighted sequences, showing 75% accuracy in the test cohort for classification of atypical cartilaginous tumours and chondrosarcomas [16]. This previous study included 58 patients from the same institution and the machine-learning classifier was internally tested using a hold-out set as a test cohort [16]. To our knowledge, no published study has focused on machine learning and CT radiomics of cartilaginous bone lesions, as done in this study. We attempted to differentiate atypical cartilaginous tumours from chondrosarcomas of long bones, as this is the most relevant clinical question and orientates towards a conservative approach or aggressive surgery. Our machine-learning classifier achieved 75% accuracy overall, 81% accuracy in identifying atypical cartilaginous tumours and 70% accuracy in identifying higher-grade chondrosarcomas in the external test cohort, respectively, with no difference compared to a dedicated radiologist with 12 years of experience in bone sarcomas (p=0.75 [McNemar's test]). These results agree with those previously reported for tumour grading based on MRI radiomics [16]. Furthermore, our findings were obtained in a more than twice larger population and validated in an independent test cohort from a second institution, thus ensuring their generalizability in clinical practice. Finally, although statistical significance was not reached (p=0.29 [McNemar's test]), the machine-learning classifier's accuracy was slightly superior compared to preoperative biopsy. We may speculate that this difference could become significant in a larger population.
Some limitations of our study need to be taken into account. First, our study is retrospective, as this design allowed including relatively large numbers of patients with an uncommon disease, such as chondrosarcoma, and imaging data already available. Additionally, a prospective analysis is not strictly needed for radiomic studies [13]. Second, we performed bidimensional segmentation and chose the image showing the maximum lesion extension. This decision was taken according to a recent study emphasizing that bidimensional segmentation yields better performance than volumetric approach [37], which would also be time-consuming in clinical practice. Third, feature stability was assessed by 3 readers only in a subgroup of 30 patients randomly selected from the training cohort, as 3 observers and 30 samples are the minimum numerical requirements for a reliability analysis according to the ICC guidelines by Koo et al. [19]. Fourth, atypical cartilaginous tumours were twice more numerous than higher-grade chondrosarcomas in the training cohort. However, an imbalance of 2/3 is acceptable in machine-learning studies [38] and SMOTE was used to artificially oversample the minority class in the training cohort [25]. Fifth, the training and external test cohorts respectively included CT scans and the CT portion of combined PET-CT scans with different acquisition parameters. Nonetheless, this is a further point in favour of the reliability of our findings, as the classifier performed well in both cohorts of patients. Sixth, only non-contrast CT was used in this study. However, contrast-enhanced CT was not available in patients from centre 2, as PET-CT was used. It was available only for a limited number of patients from centre 1, where preoperative assessment routinely included both CT and contrast-enhanced MRI; contrast was also administered before CT according to need, mainly to assess tumour-vessel relationships in case of high-grade chondrosarcoma. Our findings open the possibility for future studies to shed light on the value of contrast-enhanced CT radiomics and machine-learning assessment of cartilaginous bone tumours.
In conclusion, our machine-learning classifier showed good accuracy in differentiating atypical cartilaginous tumours from higher-grade chondrosarcomas of long bones based on radiomic features derived from preoperative CT scans. Our large population of study relative to such an uncommon disease, along with the good performance achieved in an independent cohort of patients from an external institution, supports the generalizability of our findings and their transferability into clinical practice. Our method may potentially aid in preoperative tumour characterization by integrating the multidisciplinary approach currently based on clinical, conventional imaging and histological assessment. Future investigations with prospective design are warranted to further validate our findings.
Declaration of Competing Interest
The authors declare no potential conflict of interest related to this work.
Acknowledgments
Contributors
Salvatore Gitto: conceptualization; data curation; funding acquisition; investigation; methodology; project administration; visualization; writing – original draft
Renato Cuocolo: data curation; formal analysis; methodology; software; validation; writing – review and editing
Alessio Annovazzi: data curation; writing – review and editing
Vincenzo Anelli: investigation; writing – review and editing
Marzia Acquasanta: investigation; writing – review and editing
Antonino Cincotta: investigation; writing – review and editing
Domenico Albano: conceptualization; writing – review and editing
Vito Chianca: conceptualization; writing – review and editing
Virginia Ferraresi: resources; writing – review and editing
Carmelo Messina: conceptualization; writing – review and editing
Carmine Zoccali: resources; writing – review and editing
Elisabetta Armiraglio: resources; writing – review and editing
Antonina Parafioriti: resources; writing – review and editing
Rosa Sciuto: resources; writing – review and editing
Alessandro Luzzati: resources; writing – review and editing
Roberto Biagini: resources; writing – review and editing
Massimo Imbriaco: resources; supervision; writing – review and editing
Luca Maria Sconfienza: conceptualization; project administration; resources; supervision; writing – review and editing
Acknowledgements
This research was partially funded by the Young Researchers Grant awarded by the European Society of Musculoskeletal Radiology (S. Gitto). The funding source provided financial support without any influence on the study design; on the collection, analysis, and interpretation of data; and on the writing of the report. The corresponding and senior (last) authors had the final responsibility for the decision to submit the paper for publication.
Data sharing
The trained model and Weka result buffer, containing model weights as well as the entire training, validation and test run, are available in a publicly accessible repository (https://github.com/rcuocolo/csa_ct).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103407.
Appendix. Supplementary materials
References
- 1.Murphey M.D., Walker E.A., Wilson A.J., Kransdorf M.J., Temple H.T., Gannon F.H. From the archives of the AFIP: imaging of primary chondrosarcoma: radiologic-pathologic correlation. Radiographics. 2003;23:1245–1278. doi: 10.1148/rg.235035134. [DOI] [PubMed] [Google Scholar]
- 2.Fletcher C.D.M., Bridge J.A., Hogendoorn P.C.W., Mertens F. International Agency for Research on Cancer Press; Lyon, France: 2013. World Health Organization Classification of Tumours of Soft Tissue and Bone. [Google Scholar]
- 3.Casali P.G., Bielack S., Abecassis N. Bone sarcomas: ESMO–PaedCan–EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv79–iv95. doi: 10.1093/annonc/mdy310. [DOI] [PubMed] [Google Scholar]
- 4.van Praag (Veroniek) V.M., Rueten-Budde A.J., Ho V. Incidence, outcomes and prognostic factors during 25 years of treatment of chondrosarcomas. Surg Oncol. 2018;27:402–408. doi: 10.1016/j.suronc.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Douis H., Singh L., Saifuddin A. MRI differentiation of low-grade from high-grade appendicular chondrosarcoma. Eur Radiol. 2014;24:232–240. doi: 10.1007/s00330-013-3003-y. [DOI] [PubMed] [Google Scholar]
- 6.Cannavò L., Albano D., Messina C. Accuracy of CT and MRI to assess resection margins in primary malignant bone tumours having histology as the reference standard. Clin Radiol. 2019;74:736. doi: 10.1016/j.crad.2019.05.022. e13-736.e21. [DOI] [PubMed] [Google Scholar]
- 7.Hodel S., Laux C., Farei-Campagna J., Götschi T., Bode-Lesniewska B., Müller D.A. The impact of biopsy sampling errors and the quality of surgical margins on local recurrence and survival in chondrosarcoma. Cancer Manag Res. 2018;10:3765–3771. doi: 10.2147/CMAR.S178768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eefting D., Schrage Y.M., Geirnaerdt M.J.A. Assessment of interobserver variability and histologic parameters to improve reliability in classification and grading of central cartilaginous tumours. Am J Surg Pathol. 2009;33:50–57. doi: 10.1097/PAS.0b013e31817eec2b. [DOI] [PubMed] [Google Scholar]
- 9.van de Sande M.A.J., van der Wal R.J.P., Navas Cañete A. Radiologic differentiation of enchondromas, atypical cartilaginous tumours, and high-grade chondrosarcomas—Improving tumour-specific treatment: a paradigm in transit? Cancer. 2019;125:3288–3291. doi: 10.1002/cncr.32404. [DOI] [PubMed] [Google Scholar]
- 10.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lubner M.G., Smith A.D., Sandrasegaran K., Sahani D.V., Pickhardt P.J. CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics. 2017;37:1483–1503. doi: 10.1148/rg.2017170056. [DOI] [PubMed] [Google Scholar]
- 12.Kocak B., Durmaz E.S., Ates E., Kilickesmez O. Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol. 2019;25:485–495. doi: 10.5152/dir.2019.19321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chianca V., Cuocolo R., Gitto S. Radiomic machine learning classifiers in spine bone tumors: a multi-software, multi-scanner study. Eur J Radiol. 2021;137 doi: 10.1016/j.ejrad.2021.109586. [DOI] [PubMed] [Google Scholar]
- 14.Choy G., Khalilzadeh O., Michalski M. Current applications and future impact of machine learning in radiology. Radiology. 2018;288:318–328. doi: 10.1148/radiol.2018171820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cuocolo R., Caruso M., Perillo T., Ugga L., Petretta M. Machine Learning in oncology: a clinical appraisal. Cancer Lett. 2020;481:55–62. doi: 10.1016/j.canlet.2020.03.032. [DOI] [PubMed] [Google Scholar]
- 16.Gitto S., Cuocolo R., Albano D. MRI radiomics-based machine-learning classification of bone chondrosarcoma. Eur J Radiol. 2020;128 doi: 10.1016/j.ejrad.2020.109043. [DOI] [PubMed] [Google Scholar]
- 17.Li X., Morgan P.S., Ashburner J., Smith J., Rorden C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. J Neurosci Methods. 2016;264:47–56. doi: 10.1016/j.jneumeth.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 18.Yushkevich P.A., Piven J., Hazlett H.C. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage. 2006;31:1116–1128. doi: 10.1016/j.neuroimage.2006.01.015. [DOI] [PubMed] [Google Scholar]
- 19.Koo T.K., Li M.Y. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Griethuysen J.J.M., Fedorov A., Parmar C. Computational Radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zwanenburg A., Vallières M., Abdalah M.A. The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology. 2020;295:328–338. doi: 10.1148/radiol.2020191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frank E., Hall M., Trigg L., Holmes G., Witten I.H. Data mining in bioinformatics using Weka. Bioinformatics. 2004;20:2479–2481. doi: 10.1093/bioinformatics/bth261. [DOI] [PubMed] [Google Scholar]
- 23.Pedregosa F., Varoquaux G., Gramfort A. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:825–2830. [Google Scholar]
- 24.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: A language and environment for statistical computing. [Google Scholar]
- 25.Chawla N.V., Bowyer K.W., Hall L.O., Kegelmeyer W.P. SMOTE: synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–357. doi: 10.1613/jair.953. [DOI] [Google Scholar]
- 26.Parlier-Cuau C., Bousson V., Ogilvie C.M., Lackman R.D., Laredo J.-D. When should we biopsy a solitary central cartilaginous tumour of long bones? Literature review and management proposal. Eur J Radiol. 2011;77:6–12. doi: 10.1016/j.ejrad.2010.06.051. [DOI] [PubMed] [Google Scholar]
- 27.Lambin P., Leijenaar R.T.H., Deist T.M. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 28.Crim J., Schmidt R., Layfield L., Hanrahan C., Manaster B.J. Can imaging criteria distinguish enchondroma from grade 1 chondrosarcoma? Eur J Radiol. 2015;84:2222–2230. doi: 10.1016/j.ejrad.2015.06.033. [DOI] [PubMed] [Google Scholar]
- 29.Zoccali C., Baldi J., Attala D. Intralesional vs. extralesional procedures for low-grade central chondrosarcoma: a systematic review of the literature. Arch Orthop Trauma Surg. 2018;138:929–937. doi: 10.1007/s00402-018-2930-0. [DOI] [PubMed] [Google Scholar]
- 30.Deckers C., Schreuder B.H.W., Hannink G., de Rooy J.W.J., van der Geest I.C.M. Radiologic follow-up of untreated enchondroma and atypical cartilaginous tumours in the long bones. J Surg Oncol. 2016;114:987–991. doi: 10.1002/jso.24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Omlor G.W., Lohnherr V., Lange J. Outcome of conservative and surgical treatment of enchondromas and atypical cartilaginous tumours of the long bones: retrospective analysis of 228 patients. BMC Musculoskelet Disord. 2019;20:134. doi: 10.1186/s12891-019-2502-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Annovazzi A., Anelli V., Zoccali C. 18F-FDG PET/CT in the evaluation of cartilaginous bone neoplasms: the added value of tumour grading. Ann Nucl Med. 2019;33:813–821. doi: 10.1007/s12149-019-01392-3. [DOI] [PubMed] [Google Scholar]
- 33.Fritz B., Müller D.A., Sutter R. Magnetic resonance imaging–based grading of cartilaginous bone Tumours. Invest Radiol. 2018;53:663–672. doi: 10.1097/RLI.0000000000000486. [DOI] [PubMed] [Google Scholar]
- 34.Lisson C.S., Lisson C.G., Flosdorf K. Diagnostic value of MRI-based 3D texture analysis for tissue characterisation and discrimination of low-grade chondrosarcoma from enchondroma: a pilot study. Eur Radiol. 2018;28:468–477. doi: 10.1007/s00330-017-5014-6. [DOI] [PubMed] [Google Scholar]
- 35.Yin P., Mao N., Liu X. Can clinical radiomics nomogram based on 3D multiparametric MRI features and clinical characteristics estimate early recurrence of pelvic chondrosarcoma? J Magn Reson Imaging. 2020;51:435–445. doi: 10.1002/jmri.26834. [DOI] [PubMed] [Google Scholar]
- 36.Li L., Wang K., Ma X. Radiomic analysis of multiparametric magnetic resonance imaging for differentiating skull base chordoma and chondrosarcoma. Eur J Radiol. 2019;118:81–87. doi: 10.1016/j.ejrad.2019.07.006. [DOI] [PubMed] [Google Scholar]
- 37.Ren J., Yuan Y., Qi M., Tao X. Machine learning–based CT texture analysis to predict HPV status in oropharyngeal squamous cell carcinoma: comparison of 2D and 3D segmentation. Eur Radiol. 2020;30:6858–6866. doi: 10.1007/s00330-020-07011-4. [DOI] [PubMed] [Google Scholar]
- 38.Sollini M., Antunovic L., Chiti A., Kirienko M. Towards clinical application of image mining: a systematic review on artificial intelligence and radiomics. Eur J Nucl Med Mol Imaging. 2019;46:2656–2672. doi: 10.1007/s00259-019-04372-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.