Abstract
The southernmost region of earth, Antarctica, has world's most challenging environments. Those who live for long time and work in Antarctic stations are subjected to environmental stresses such as cold weather, photoperiod variations leading to disrupted sleep cycles, constrained living spaces, dry air, non-availability of fresh food items, and high electromagnetic radiations, psychological factors, such as geographical and social isolation, etc. All these factors have a significant impact on the human body. The present study investigated the impact of Antarctica harsh environment on human physiology and its metabolic processes by evaluating urine metabolome, using 1H NMR spectroscopy and analyzing certain physiological and clinical parameters for correlation with physiological expression data and metabolite results. Two study groups - before Antarctic exposure (B) and after Antarctic exposure (E), consisting of 11 subjects, exposed to one-month summer expedition, were compared. 35 metabolites in urine samples were identified from the 700 MHz 1H NMR spectra from where integral intensity of 22 important metabolites was determined. Univariate analysis indicated significant decrease in the levels of citrate and creatinine in samples collected post-expedition. Multivariate analysis was also performed using 1H NMR spectroscopy, because independent metabolite abundances may complement each other in predicting the dependent variables. 10 metabolites were identified among the groups; the OPLS-DA and VIP score indicated variation in appearance of metabolites over different time periods with insignificant change in the intensities. Metabolite results illustrate the impact of environmental stress or altered life style including the diet with absence of fresh fruits and vegetables, on the pathophysiology of the human health. Metabolic adaptation to Antarctic environmental stressors may help to highlight the effect of short-term physiological status and provide important information during Antarctic expeditions to formulate management programmes.
Keywords: Expedition, Antarctic stress, Metabolite, NMR spectroscopy, Metabolism
Expedition; Antarctic stress; Metabolite; NMR spectroscopy; Metabolism
1. Introduction
The Antarctica situated at south pole, is with most extreme environmental conditions on earth as the coldest (-25 °C to -80 °C), windiest (Katabatic winds 15–50 km/h) and driest (RH 0.03%) continent, leading to series of physiological, psychological and metabolic perturbations in expedition members. Antarctic milieu comes with monthly mean temperatures falling below −60 °C from March to October, intense magnetic fields, and extreme UV radiations as well as disrupted dark/light cycles and psychological stressors make conditions difficult to live [Lugg and Shepanek 1999]. Under Antarctic treaty the member countries conduct various expeditions for different scientific studies. Expedition members stay in extreme conditions for summer and winter weathers for conducting research. These environmental conditions may cause disease exacerbation, which can be fatal [McMichael et al., 2016; Storey and Storey 2005]. The metabolic responses to intense environmental conditions have not been thoroughly studied so far. However, there is also a deep dependence on metabolic processes for non-shivering thermogenesis, with acclimatization. Of special importance are those changes, which occur inside adipose tissue in mitochondria. Oxidative phosphorylation uncoupling is thought to play an important role in cold-induced thermogenesis, a reaction expected to be regulated by the cold-sensing receptor TRPM8 expressed in both white and brown adipose tissue [Frontini and Cinti 2010; Rossato et al., 2014]. A clear association was found between basal metabolic rate (BMR) and climate. For example, the BMR of indigenous Siberian populations was found to be 5 % higher than the body mass values expected [Roberts 1952]. As previously believed that, these elevations tended to be due to environmental stress rather than elevated dietary protein intakes [Leonard et al., 2015].
Bio-fluid metabolites are in complex balance with cells and tissues, and consequently any alteration related to irregular cellular processes could result in altered bio-fluid metabolites composition [Lindon et al., 2000]. Urine is versatile bio-fluid formed by kidneys from blood and comprises both endogenous as well as exogenous compounds such as amino acids, fatty acids, lipids, carbohydrates, hormones, peptides, xenobiotics and metabolism end products, such as glucuronides and sulfo-conjugates [Bouatra et al., 2013]. The urine has many benefits among the bio-fluids widely used in epidemiology: it is abundant, clean, and convenient to collect [Emwas et al., 2016]. It indicates many body functions through various metabolites from many primary biochemical processes in relation to (patho-) physiology and cardio-metabolic factors, gut microbial metabolism and short-term dietary consumption [Holmes et al., 2008; Bouatra et al., 2013; Elliott et al., 2015; Van Duynhoven and Jacobs, 2016]. Therefore, urine samples provide sufficient and underused information for epidemiology and future translational applications [Nicholson et al., 2011].
Metabolomics, in the last decade has become a powerful experimental tool for generating biomedical data. 1H Nuclear Magnetic Resonance (NMR) spectroscopy offers a detailed quantitative methodology [Bouatra et al., 2013; Emwas et al., 2016; Van Duynhoven and Jacobs, 2016], giving snap shot of the large number of metabolites present in biofluids. It also has the ability to provide cost-effective, automated high-throughput procedures, which would be necessary for large-scale systems epidemiology [Soininen et al., 2009; Soininen et al., 2015; Würtz et al., 2017]. There are various software programs used in urine NMR data analysis, but only few of them provide comprehensive automated metabolic information quantification [Weljie et al., 2006; Zheng et al., 2011; Da Silva et al., 2013; Hao et al., 2014]. Taking clue from those methods, we investigated the metabolome of urine during summer Antarctic expedition using NMR approach and multivariate analysis methods for determining the complex metabolic changes that occur during Antarctic expedition. A panel of metabolic biomarkers in urine samples has potential to differentiate metabolism of expedition members and may complement traditional tests to improve health status during Antarctic Expedition. The alteration in metabolic processes of body to allow survival at the extremes of Antarctic environmental conditions, means the metabolic profile undergoes drastic changes under extremes hence warrants deep study.
2. Materials and methods
2.1. Study subjects
Eleven male members (n = 11); age group: 22–60, of the 34th Indian Scientific Summer Expedition to Antarctica in January to April 2015, volunteered to participate in this study. The team had undergone pre-departure clinical, psychological, and laboratory examinations to ensure a healthy population at the Antarctic stations. None of the subjects used any drugs that could significantly affect the body metabolism, or had symptoms indicative of any infection.
2.2. Urine sample collection
Urine sample 120ml (first pass in the morning), was collected in sterile urine vials as base line at Cape Town before starting of the expedition (B) in the month of January and after the completion of summer expedition of one month (E) in the month of March. After collection (usually within 1 h of collection), all the samples were immediately treated with 2.5 mM sodium azide, centrifuged at 4000 rpm for 10 min for particle removal and immediately distributed in aliquots of 2ml each, and stored at -20 °C for further analysis. At the end of expedition, samples were transported by ship to Cape Town and then by air to Delhi, India, strictly maintaining the cold chain till they reached to our laboratory in Delhi. Before each analysis, the samples were thawed at room temperature for 30 min and filtered second time by centrifugation for metabolite analysis by 1H NMR spectroscopy in the Department of NMR & MRI Facility, All India Institute of Medical Sciences (AIIMS), New Delhi.
2.3. Ethics statement and volunteer information
All expedition participants understood the nature of the study and gave their written consent. The Ethics Committee of the Defence Institute of Physiology and Allied Sciences, DRDO, New Delhi, India, approved all the relevant parameters of the study. The study protocols were in accordance with the approved guidelines.
2.4. Sample preparation and NMR measurement
Frozen urine samples were thawed, 400 μl of urine was diluted with 170 μl of sodium phosphate buffer 200mM (pH 7.4) containing sodium azide prepared in D2O (Sigma, Aldrich). Sodium phosphate buffer was added to the samples in order to minimize pH variation among samples and. 30 μl of TSP (0.5mM) (Sigma- Aldrich, St. Louis, USA), was added to serve as a reference standard for any chemical shift for the 1H NMR spectroscopy. The mixture was transferred to 5mm NMR tubes. The experiment was carried out on a narrow bore NMR spectrometer operating at 700 MHz (Agilent, U.S.A.). 1D spectrum was acquired with a single 90° pulse with water suppression (using a presaturation RF pulse). The parameters used for 1D experiment were as follow: spectral width = 9124.1 Hz, data points = 32 K, number of scans = 64 and relaxation delay = 5s. The 1D NMR data was processed on a Dell 390N, PC, Red Hat Enterprise Linux workstation using Vnmrj 2.3 A. For processing of 1D NMR data, the free induction decays were multiplied by an exponentially decaying function prior to Fourier transformation leading to a line broadening of 0.3Hz. Spectra were manually phase corrected using zero and first order corrections. In addition, selected metabolite detection was also cross-checked from the Human Metabolome Database (HMDB) [Wishart et al., 2012] and the published literature [Bouatra et al., 2013]. The integral intensity of metabolites that showed well-resolved resonances was determined by comparing the integrated intensity of isolated resonance of compounds of interest with the intensity of the reference compound TSP.
2.5. Statistical analysis
All univariate statistical analyses were carried out using the Graphpad Prism software V 5.0. Integral intensity of the metabolites was presented as median with Inter Quartile Range (IQR). Student ‘t’ test was used for the comparison of integral intensity of metabolites between groups, p ≤ 0.05 was considered significant.
Targeted metabolomics approach was applied to understand the classification and clustering pattern of the samples. Multivariate data analyses were performed on the integral intensity data, using Metabo-Analyst 4.0 web server [Chong et al., 2019]. Prior to multivariate analysis, the data was normalized using log transformation and auto scaling. First, the unsupervised principal component analysis (PCA) was applied for initial exploratory analysis for outlier detection. Followed by, supervised orthogonal partial least squares discriminant analysis (OPLS-DA) method to improve the separation between groups of samples and to minimize other biological analytical variations. The OPLS-DA model were cross-validated by a permutation analysis (n = 20), and the resulting goodness of fit parameter (R2Y) and Q2 (predictive ability) were calculated. The variable importance in projection (VIP) scores of the metabolites was analyzed to identify discriminating metabolites. The metabolites with VIP (>1.0) were considered relevant for group differentiation.
2.6. Functional analysis
Using Metabo-Analyst 4.0, metabolic pathway-impact analysis, which incorporates pathway enrichment analysis and topology analysis, was conducted to classify the most important pathways altered during summer Antarctic Expedition. On the basis of p value (<0.05) and effect value (>0.1), the possible target metabolic pathways were filtered out. From quantitative enrichment analysis, the p value was estimated, while from pathway topology analysis, the effect value of the altered metabolic pathway was estimated. Pathway research includes the name or code (identifier) of dataset metabolites searched in multiple databases of metabolites, such as the Human Metabolomic Database (HMDB) and the Kyoto Encyclopedia of Genes and Genomes (KEGG).
3. Results
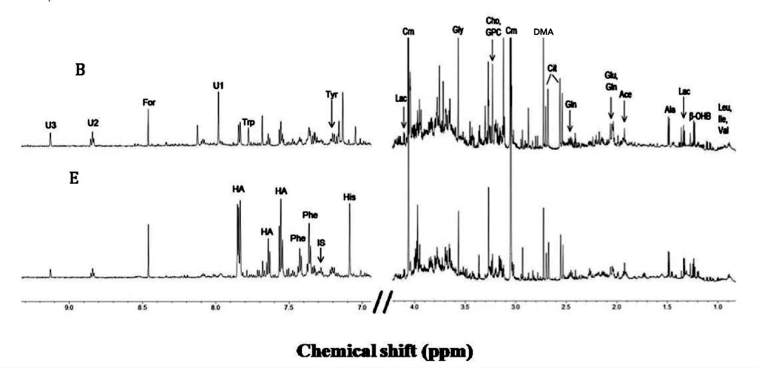
In the present study urine samples of 11 summer expedition members before proceeding to Antarctica (B) in the month of January and after being one month at maître station (E) in the month of February, were analyzed for alteration in metabolites by 700MHz 1HNMR spectrum. The representative one-dimensional proton 1HNMR spectroscopy of - urine sample of an individual before and after the expedition is demonstrated in Figure 1. A total of 35 metabolites were unambiguously identified based on characteristic chemical shift values and multiplicities. The peak areas were integrated relative to reference signals from TSP for quantification. The identified metabolites were compared to the metabolites reported in literature [Cassiède et al., 2017]. Resonances from various metabolites like amino acids such as leucine, isoleucine, valine, alanine, glutamate, glutamine, glycine, histidine, tryptophan; sugars like myo-inositol, organic acids such as lactate, acetate, and other metabolites like choline, glycerophosphocholine, dimethyamine, creatinine, indoxyl sulfate, hippuarte, allantoin were identified (Figure 1). Of these 35 metabolites, the integral intensity of 22 metabolites was determined.
Figure 1.
Representative one dimensional 1H NMR of urine obtained from summer expedition member before (B) and after (E) groups, acquired at 700 MHz in D2O at 25 °C. Abbreviations: β-OHB- β hydroxybutyrate; Ace: Acetate; Ala: Alanine; Cho: Choline; Cit: Citrate; Crn: Creatinine; For: Formate; Glu: Glutamate; Gln: Glutamine; GPC: Glycerophosphocholine; Gly: Glycine; HA: Hippurrate; His: Histidine; IS: Indoxyl sulfate; Ile: Isoleucine; Lac: Lactate; Leu: Leucine; Phe: Phenylalanine, DMA: Dimethylamine; Trp: Tryptophan; Tyr: tyrosine; U1, U2, U3: Unknown, Val: Valine.
The Univariate analysis indicated significantly lower levels of citrate 3.36 (1.76–4.79) AU and creatinine 15.4 (6.24–18.6) AU after one months stay (E) as compared to baseline (B) control sample values (Table 1).
Table 1.
Comparison of intensity of metabolites, VIP score and intensities of metabolites present in urine samples of summer expedition members.
| Metabolites | Chemical Shift (δ, ppm) | Intensity |
p-value | VIP Score | |
|---|---|---|---|---|---|
| Before (B) (n = 11) Median (IQR) AU |
After (E) (n = 11) Median (IQR) |
||||
| Leucine (Leu) | 0.96(δ-CH3), 1.69(γ –CH2), 1.72(β-CH2) | 0.714 (0.524–0.977) | 0.632 (0.379–0.734) | 0.181 | 0.989 |
| Isoleucine (Ile) | 0.94(δ-CH3), 1.26(γ –CH2), 1.48(γ –CH2), 1.97(β-CH), 3.62 (α-CH) |
0.282 (0.227–0.363) | 0.228 (0.214–0.300) | 0.334 | 0.842 |
| Valine (Val) | 1.00(γ –CH2), 1.04(γ –CH3), 2.21(β-CH), 3.52 (α-CH) |
0.349 (0.313–0.737) | 0.391 (0.244–0.445) | 0.473 | 0.367 |
| Alanine (Ala) | 1.46(β-CH3), 3.76(α-CH) | 1.93 (1.89–2.92) | 1.75 (0.682–2.26) | 0.284 | 0.938 |
| Glycine (Gly) | 3.55(CH2) | 2.37 (1.75–4.27) | 2.00 (0.993–3.21) | 0.583 | 0.673 |
| Histidine (His) |
3.14(β-CH2), 7.06 (H4), 7.83(H2) |
0.439 (0.402–0.730) | 0.419 (0.170–0.455) | 0.292 | 1.035 |
| Tryptophan (Try) |
3.26(β-CH), 3.43(β-CH), 4.06(α-CH), 7.55 (H7), 7.69(H4) |
0.360 (0.272–0.471) | 0.173 (0.113–0.322) | 0.087 | 1.612 |
| Dimethylamine (DMA) | 2.73 (CH3) | 2.45 (2.10–4.81) | 2.31 (0.894–2.96) | 0.174 | 1.128 |
| Lactate | 1.33 (β-CH3), 4.12 (α-CH) | 1.69 (1.34–2.30) | 1.38 (0.651–1.83) | 0.186 | 1.145 |
| β-hydroxybutyrate (β –OBH) | 1.21 (γ –CH3), 4.16 (β-CH) |
0.990 (0.760–1.82) | 0.829 (0.361–1.87) | 0.741 | 0.825 |
| Citrate (Cit) |
2.52 (CH2), 2.64 (CH2) |
6.13 (2.83–10.7) | 3.36 (1.76–4.79) | 0.042∗ | 1.414 |
| Formate (For) | 8.46 (CH) | 0.286 (0.221–0.416) | 0.286 (0.136–0.346) | 0.405 | 0.718 |
| Creatine (Cr) |
3.04 (NCH3), 3.93 (CH2) |
42.1 (34.7–66.9) | 35.9 (12.5–45.5) | 0.143 | 1.219 |
| Creatinine (Crn) | 4.06 (CH2) | 24.8 (22.3–34.4) | 15.4 (6.24–18.6) | 0.006∗ | 1.870 |
| Myo-inositol (MI) | 3.28 (H5′), 3.53 (H1′, H3′), 3.63 (H4′, H6′), 4.07 (H2′) |
2.13 (1.64–9.36) | 5.21 (1.56–6.23) | 0.771 | 0.115 |
| Indoxyl Sulphate (IS) |
7.17 (H5′), 7.25 (H6′) |
0.973 (0.704–1.36) | 0.681 (0.283–1.12) | 0.133 | 1.258 |
| Hippurate (HA) | 7.53 (H3′/H5′), 7.61 (H4′), 7.81 (H2′/H6′) |
1.53 (1.35–2.03) | 1.32 (0.524–2.48) | 0.927 | 0.332 |
| Aminohippurate (AHA) | 6.83 (H3/H6) | 0.662 (0.391–0.822) | 0.424 (0.245–0.531) | 0.271 | 1.144 |
| Allantoin (Alln) | 5.42 (CH) | 0.231 (0.171–0.359) | 0.183 (0.128–0.251) | 0.190 | 1.157 |
| U1 | - | 0.244 (0.177–0.319) | 0.282 (0.194–0.359) | 0.778 | 0.139 |
| U2 | - | 0.503 (0.382–0.675) | 0.524 (0.374–0.705) | 0.655 | 0.086 |
| U3 | - | 0.740 (0.236–1.02) | 0.874 (0.129–1.39) | 0.717 | 0.138 |
Bold Indicates those metabolites which, have VIP score greater than 1.
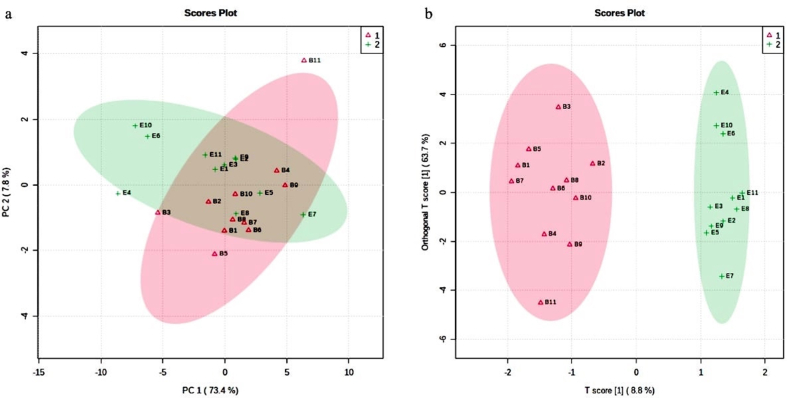
3.1. The PCA analysis of urine samples
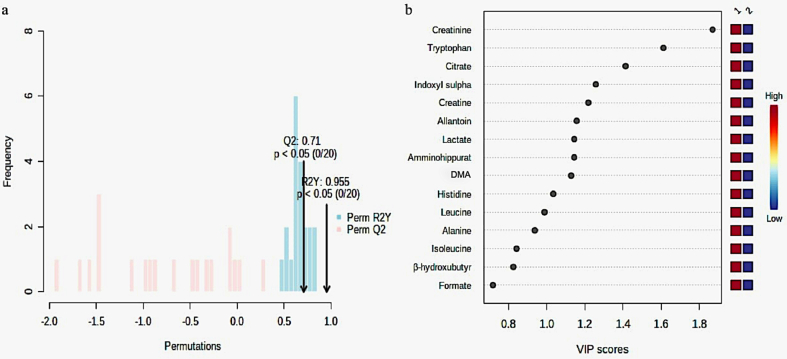
The systematic representation of metabolic responses to Antarctic stressful environment was performed by generating a PCA map for analysis of urine samples by 1HNMR from 11 individuals at two time points during expedition. The PCA score plot illustrated a trend of group clustering allowing visualization of differentiation of each data point, representing the metabolic spectrum of every individual between groups B and E (Figure 2a). The OPLS-DA model was built using the 22 variables. The OPLS-DA score plot revealed clear distinction between the baseline sample and the samples collected from the same individulas after the stay (Figure 2b). The values of R2Y and Q2 for this model were 0.95 and 0.71 respectively, suggesting that the model was robust (Figure 3a) VIP score plot indicated that the separation in each data point is attributable to the relative decrease in the concentration of metabolites. It revealed that metabolites such as histidine, lactate, citrate, DMA, creatine, creatinine, allantoin, amminohippurate, indoxylsulphate and tryptophan contributed for the separation between two groups (Table 1, Figure 3b). A higher level of all these metabolites was observed before the start of expedition (B) as compared to the levels after completion of the summer expedition (E).
Figure 2.
‘a’ and ‘b’ represents principal component analysis (PCA) score plot and Loading score plots of orthogonal projection to latent structure discriminant analysis (OPLSDA) score plot of metabolites present in the urine of expedition member before proceeding to Antarctica (B, Δ) and after completion of summer expedition (E, +).
Figure 3.
Model validation by the permutation test based on 20 permutations of metabolites obtained by 1H NMR analysis (a) and variable importance in Projection (VIP) plot (b) based on the intensity of metabolites present in the urine samples of the two groups under study. Abbreviation: DMA; Dimethylamine.
3.2. Metabolic pathway analysis based on changes in metabolic profile
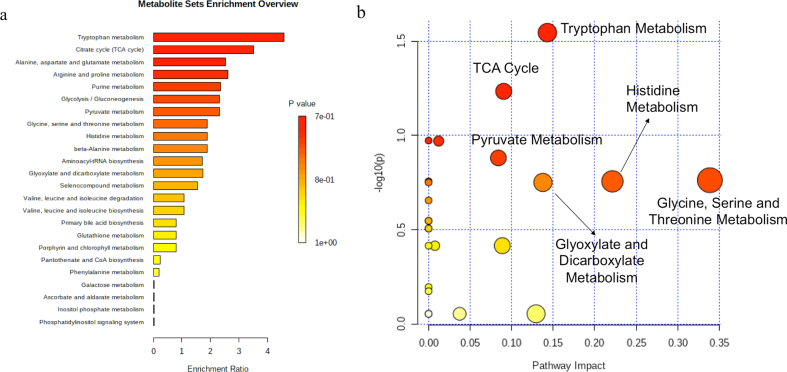
The metabolic pathways that are enriched in urine samples of Antarctic summer expedition members are shown in Figure 4a. Figure 4b represents the highly altered metabolic pathways based on topology analysis. The pathway impact analysis (p < 0.05 and impact value >0.1), indicated six potential target metabolic pathways during Antarctic expedition (Table 2). Total number of compounds involved in each pathway and metabolites actually matched from the uploaded data on metaboanalyst 4.0, p-value calculated from the enrichment analysis; the impact is the pathway impact value calculated from pathway topology analysis. Tryptophan metabolism led pathways were most affected. Other affected pathways are the TCA cycle; Pyruvate, Histidine, Glycine, Serine, Threonine and Glyoxylate and Dicarboxylate Metabolism.
Figure 4.
Urine metabolic pathway analysis: a) metabolic functions correlated with distinguishing urine-identified metabolites of expedition members using the MetaboAnalyst 4.0 quantitative enrichment analysis tool. Pathways are shown from top to bottom in order of decreasing significance; with bars representing their approximate fold enrichment. b) Altered metabolic functions of Antarctic Expedition members, based on the distinguishing metabolites found in urine, using pathway topology analysis. One bubble represents each metabolic pathway. The size of the bubble is proportional to the influence of each path, with the significance of the colour from the lowest (yellow) to the highest (red).
Table 2.
Metabolic pathways most affected during summer Antarctic Expedition determined from pathway-impact analysis with MetaboAnalyst 4.0.
| Pathway Name | Matched Metabolites | p-value | Impact |
|---|---|---|---|
| Tryptophan Metabolism | 1/41 | 0.028 | 0.143 |
| TCA cycle | 1/20 | 0.058 | 0.090 |
| Histidine Metabolism | 1/16 | 0.175 | 0.221 |
| Pyruvate Metabolism | 1/22 | 0.131 | 0.083 |
| Glycine, Serine Threonine Metabolism | 3/33 | 0.172 | 0.338 |
| Glyoxylate and Dicarboxylate Metabolism | 3/32 | 0.177 | 0.137 |
4. Discussion
In characterizing a pathological condition, the biochemical composition of urine has become a central element in clinical relevance. To the best of our knowledge, this is the first study to provide a holistic view of changes in the metabolism of members of the Antarctic expedition by deciphering the effects of the harsh Antarctic climate on the metabolic composition of the excretory fluid (urine) in the members of summer Antarctic expedition.
The metabolomics method is intended to quantify metabolites in body fluids. It was proposed that environmental physiology would provide insight into the perception of the human body at the system level [Edwards and Thiele, 2013], and into a variety of pathological conditions. In line with that, the extreme environment of Antarctica as a natural laboratory has provided a major expansion of knowledge of human physiology and psychology under extreme stress [Carrère et al., 1991; Strike and Steptoe 2004].
There are categories of factors, which affect metabolite concentration levels in human urine. Including uncontrollable natural factors such as age, gender, microflora and gut alterations, while others which could be regulated, such as diet, methods of sample collection, handling and processing should be focused on. Most people reside or visit in extreme heat or cold. Each of these environmental stresses can cause disease exacerbation and are potentially lethal [Storey and Storey 2005; McMichael et al., 2006]. However, such variables are not entirely distinct. They can influence each other considerably, which makes studies more complicated. Physical activities can impact urine metabolic signature over both short and long periods of stress. The primary path for the disposal of metabolic waste from the body is urine. Changes in urinary metabolites may indicate not only the characteristics of the overall metabolism of the body, but also irregular tissue or organ function.
This study highlighted the temporal role of metabolite variability in urine after exposure to physiological, environmental, and psychological stress in Antarctica. The novelty of the present research lies in the inclusion of data obtained during the 34th ISEA summer months and the stay at the station in Antarctica. 1H NMR-based metabolomics methods were used to study urinary metabolomics changes after various forms and levels of physical activity [Enea et al., 2010].
The study is a first of its kind to document the overall effect of the Antarctic environment on the metabolic profile of the urine samples of eleven healthy Antarctic expedition members. The NMR spectral data of urine samples was grouped as before (B) and after (E) expedition time-points allowing simple detection of changes in metabolites, associated with physiological and psychological stresses. Univariate analysis shows significant difference in the intensity of two metabolites, citrate and creatinine only, whereas multivariate analysis significantly differentiated 10 metabolites. This helped to reduce the dimensionality of data and helped to distinguish samples on temporal scales.
Most of the altered metabolites have marginal increments thus could not be finely separated between the two groups. Consequently, a substantial decrease in majority of metabolites was observed. Creatinine, citrate and lactate are major metabolites, which were decreased after summer expedition. Creatinine is the end product of the catabolism of creatine phosphate. Researchers have found a strong association between the concentrations of urinary creatinine and muscle mass [Edwards and Whyte, 1959; Fuller and Rich 1982]. It is enhanced by protein intake and altered by changes in the balance of salt and water, physical activity or even the psychological state [Camara 1951; Brod 1953], very common factors at Antarctica, contributing to both, excessive fluid loss and reduced fluid intake due to cold, or the relative contribution of each, as well as the sum of their effect on body fluid balance. Furthermore, earlier studies have reported that cold stress is also linked to a decreased creatinine level in urine [Gandhi et al., 2012], which is consistent with our finding.
In urine, low citrate excretion elevates the risk of formation of calcium stone in the kidney [Menon and Mahle, 1983]. The proximal tubules reabsorb more than 50 percent of the filtrate citrate from glomeruli [Hamm 1990]. Citrate and hippurate are derived from the citrate cycle and the synthesis of phenylalanine [Zhao et al., 2016]. It is reported that the reduced level of hippurate in urine under chronic cold stress induces disturbance within the activity of gut microflora [Yang et al., 2015], which has also been observed during the same expedition (data not published yet). Glycine levels, participating in hippurate synthesis [Beyoğlu and Idle, 2012], decreased in model rats, which revealed the potential overuse of glycine for hippurate synthesis. On the other hand, decreased citrate excretion could be due to less fluid intake. Minor decrease in lactate level was observed, which could be the result of protein catabolism, stimulated by corticosterone. Though it is a major raw material for gluconeogenesis, produced by active skeletal muscles. The decrease in lactate could therefore suggest a stimulation of gluconeogenic pathways after the summer expedition. Similar findings were observed in sera samples during 28th ISEA [Yadav et al., 2014]. Study on rats also reported decreasing lactate under acute psychological stress [Teague et al., 2007].
Incidentally, metabolic pathway analysis in the present study revealed an alteration in Tryptophan metabolism. As stated earlier, the concentrations of amino acid tryptophan was lower in the urine after completion of the expedition. A key centre for the metabolic regulation of immunological and neuropsychological mechanisms is tryptophan metabolism. Multiple downstream metabolites of kynurenine are neuroactive and some are also capable of breaching the blood-brain barrier [Fukui et al., 1991]. A deeper insight into the link between the availability of tryptophan and systemic responses could be useful not only for disease surveillance or clinical treatments but the presence of tryptophan is a major factor in the regulation of protein biosynthesis. This may be one significant explanation why tryptophan starvation is used by the immune system to limit the unwanted proliferation of pathogens and malignant cells [Pfefferkorn 1984]. Microbiota may be a source of metabolites derived from tryptophan, some of which may be transported via membranes [Zelante et al., 2013], as well as neurotransmitters and precursors such as noradrenaline, gamma-aminobutyric acid (GABA), and other neuroactive agents [Cryan and Dinan 2012].
The evaluated changes in the urinary metabolic profiles were confirmed by PCA score plot, which indicated the tendency to separate the expression of different metabolites before and after expedition with different stress intensities. The current study was established to explore the differential metabolites expressed by internal metabolic changes under stressful environment.
5. Conclusion
We have presented the first human urine metabolomics in response to exposure to extreme environmental conditions in the Antarctic summer expedition. It has highlighted key areas of interest for future investigations, particularly in the context of acute exposure and prolonged expeditions of a similar nature. The preliminary findings reveal the alterations in metabolic pathways, demonstrating stable physical status of expedition members, possibly due to metabolic adaptation to different environmental stressors. Thus, providing useful information for formulating management strategies for future Antarctic expeditions. As a result, further studies on a larger sample size is needed before any conclusions can be drawn.
Declarations
Author contribution statement
Lilly Ganju: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Uma Sharma and Naranamangalam Jagannathan: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Shashi Bala Singh: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Brij Bhushan and Deepti Upadhyay: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Data availability statement
Data included in article/supplementary material/referenced in article.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Acknowledgements
Authors thank National Centre for Polar and Ocean Research (NCPOR) for their logistic support during expedition and all the participants of the study. Brij Bhushan would like to recognise Defence Research and Development organisation (DRDO) for the research fellowship.
References
- Beyoğlu D., Idle J.R. The glycine deportation system and its pharmacological consequences. Pharmacol. Therapeut. 2012 Aug 1;135(2):151–167. doi: 10.1016/j.pharmthera.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouatra S., Aziat F., Mandal R., Guo A.C., Wilson M.R., Knox C., Bjorndahl T.C., Krishnamurthy R., Saleem F., Liu P., Dame Z.T. The human urine metabolome. PloS One. 2013;8(9) doi: 10.1371/journal.pone.0073076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brod J. Regulation of renal function. Acta Med. Acad. Sci. Hungar. 1953;4(3-4):369–396. [PubMed] [Google Scholar]
- Camara A.A. The twenty-four hourly endogenous creatinine clearance as a clinical measure of the functional state of the kidneys. J. Lab. Clin. Med. 1951;37:743–763. [PubMed] [Google Scholar]
- Carrère S., Evans G.W., Stokols D. InFrom Antarctica to Outer Space. Springer; New York, NY: 1991. Winter-over stress: physiological and psychological adaptation to an Antarctic isolated and confined environment; pp. 229–237. [Google Scholar]
- Cassiède M., Nair S., Dueck M., Mino J., McKay R., Mercier P., Quémerais B., Lacy P. Dataset of urinary metabolites measured by 1H NMR analysis of normal human urine. Data in brief. 2017 Feb;10:227. doi: 10.1016/j.dib.2016.11.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Wishart D.S., Xia J. Using metaboanalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr. Protocols Bioinf. 2019 Dec;68(1):e86. doi: 10.1002/cpbi.86. [DOI] [PubMed] [Google Scholar]
- Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat. Rev. Neurosci. 2012 Oct;13(10):701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- Da Silva L., Godejohann M., Martin F.P., Collino S., Bürkle A., Moreno-Villanueva M., Bernhardt J., Toussaint O., Grubeck-Loebenstein B., Gonos E.S., Sikora E. High-resolution quantitative metabolome analysis of urine by automated flow injection NMR. Anal. Chem. 2013 Jun 18;85(12):5801–5809. doi: 10.1021/ac4004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K.D., Whyte H.M. Creatinine excretion and body composition. Clin. Sci. 1959;18:361–366. [PubMed] [Google Scholar]
- Edwards L.M., Thiele I. Applying systems biology methods to the study of human physiology in extreme environments. Extreme Physiol. Med. 2013 Dec;2(1):8. doi: 10.1186/2046-7648-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott P., Posma J.M., Chan Q., Garcia-Perez I., Wijeyesekera A., Bictash M., Ebbels T.M., Ueshima H., Zhao L., Van Horn L., Daviglus M. Urinary metabolic signatures of human adiposity. Sci. Transl. Med. 2015 Apr 29;7(285):285ra62. doi: 10.1126/scitranslmed.aaa5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emwas A.H., Roy R., McKay R.T., Ryan D., Brennan L., Tenori L., Luchinat C., Gao X., Zeri A.C., Gowda G.N., Raftery D. Recommendations and standardization of biomarker quantification using NMR-based metabolomics with particular focus on urinary analysis. J. Proteome Res. 2016 Feb 5;15(2):360–373. doi: 10.1021/acs.jproteome.5b00885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enea C., Seguin F., Petitpas-Mulliez J., Boildieu N., Boisseau N., Delpech N., Diaz V., Eugene M., Dugue B. 1 H NMR-based metabolomics approach for exploring urinary metabolome modifications after acute and chronic physical exercise. Anal. Bioanal. Chem. 2010 Feb 1;396(3):1167–1176. doi: 10.1007/s00216-009-3289-4. [DOI] [PubMed] [Google Scholar]
- Frontini A., Cinti S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell Metabol. 2010 Apr 7;11(4):253–256. doi: 10.1016/j.cmet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- Fukui S., Schwarcz R., Rapoport S.I., Takada Y., Smith Q.R. Blood–brain barrier transport of kynurenines: implications for brain synthesis and metabolism. J. Neurochem. 1991 Jun;56(6):2007–2017. doi: 10.1111/j.1471-4159.1991.tb03460.x. [DOI] [PubMed] [Google Scholar]
- Fuller L, Rich AJ. An index of lean body-mass from 24-h urinary creatinine excretion. Inproceedings of the Nutrition Society 1982 Jan 1 (vol. 41, no. 3, pp. A104-a104). C/o publishing division, wallingford, oxon, england ox10 8de: cab international.
- Gandhi S., Devi M.M., Pal S., Tripathi R.P., Khushu S. Metabolic regulatory variations in rats due to acute cold stress & Tinospora Cordifolia intervention: high resolution 1 H NMR approach. Metabolomics. 2012;8(3):444–453. [Google Scholar]
- Hamm L.L. Renal handling of citrate. Kidney Int. 1990 Oct 1;38(4):728–735. doi: 10.1038/ki.1990.265. [DOI] [PubMed] [Google Scholar]
- Hao J., Liebeke M., Astle W., De Iorio M., Bundy J.G., Ebbels T.M. Bayesian deconvolution and quantification of metabolites in complex 1D NMR spectra using BATMAN. Nat. Protoc. 2014 Jun;9(6):1416. doi: 10.1038/nprot.2014.090. [DOI] [PubMed] [Google Scholar]
- Holmes E., Loo R.L., Stamler J., Bictash M., Yap I.K., Chan Q., Ebbels T., De Iorio M., Brown I.J., Veselkov K.A., Daviglus M.L. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008 May;453(7193):396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard W.R., Snodgrass J.J., Sorensen M.V. Metabolic adaptation in indigenous Siberian populations. Annu. Rev. Anthropol. 2005 Oct 21;34:451–471. [Google Scholar]
- Lindon J.C., Nicholson J.K., Holmes E., Everett J.R. Metabonomics: metabolic processes studied by NMR spectroscopy of biofluids. Concepts Magn. Reson.: Educ. J. 2000;12(5):289–320. [Google Scholar]
- Lugg D., Shepanek M. Space analogue studies in Antarctica. Acta Astronaut. 1999 Apr 1;44(7-12):693–699. doi: 10.1016/s0094-5765(99)00068-5. [DOI] [PubMed] [Google Scholar]
- McMichael A.J., Woodruff R.E., Hales S. Climate change and human health: present and future risks. Lancet. 2006 Mar 11;367(9513):859–869. doi: 10.1016/S0140-6736(06)68079-3. [DOI] [PubMed] [Google Scholar]
- Menon M., Mahle C.J. Urinary citrate excretion in patients with renal calculi. J. Urol. 1983 Jun 1;129(6):1158–1160. doi: 10.1016/s0022-5347(17)52618-x. [DOI] [PubMed] [Google Scholar]
- Nicholson G., Rantalainen M., Maher A.D., Li J.V., Malmodin D., Ahmadi K.R., Faber J.H., Hallgrímsdóttir I.B., Barrett A., Toft H., Krestyaninova M. Human metabolic profiles are stably controlled by genetic and environmental variation. Mol. Syst. Biol. 2011 Jan 1;7(1) doi: 10.1038/msb.2011.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfefferkorn E.R. Interferon gamma blocks the growth of Toxoplasma gondii in human fibroblasts by inducing the host cells to degrade tryptophan. Proc. Natl. Acad. Sci. Unit. States Am. 1984 Feb 1;81(3):908–912. doi: 10.1073/pnas.81.3.908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts D.F. Basal metabolism, race and climate. J. Roy. Anthropol. Inst. G. B. Ireland. 1952 Jul 1;82(2):169–183. [Google Scholar]
- Rossato M., Granzotto M., Macchi V., Porzionato A., Petrelli L., Calcagno A., Vencato J., De Stefani D., Silvestrin V., Rizzuto R., Bassetto F. Human white adipocytes express the cold receptor TRPM8 which activation induces UCP1 expression, mitochondrial activation and heat production. Mol. Cell. Endocrinol. 2014 Mar 5;383(1-2):137–146. doi: 10.1016/j.mce.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Soininen P., Kangas A.J., Würtz P., Suna T., Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in cardiovascular epidemiology and genetics. Circulation: Cardiovascular Genetics. 2015 Feb;8(1):192–206. doi: 10.1161/CIRCGENETICS.114.000216. [DOI] [PubMed] [Google Scholar]
- Soininen P., Kangas A.J., Würtz P., Tukiainen T., Tynkkynen T., Laatikainen R., Järvelin M.R., Kähönen M., Lehtimäki T., Viikari J., Raitakari O.T. High-throughput serum NMR metabonomics for cost-effective holistic studies on systemic metabolism. Analyst. 2009;134(9):1781–1785. doi: 10.1039/b910205a. [DOI] [PubMed] [Google Scholar]
- Storey J.M., Storey K.B. 2005 Feb 25. Cold Hardiness and Freeze Tolerance. Functional Metabolism: Regulation and Adaptation; pp. 473–503. [Google Scholar]
- Strike P.C., Steptoe A. Psychosocial factors in the development of coronary artery disease. Prog. Cardiovasc. Dis. 2004 Jan 1;46(4):337–347. doi: 10.1016/j.pcad.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Teague C.R., Dhabhar F.S., Barton R.H., Beckwith-Hall B., Powell J., Cobain M., Singer B., McEwen B.S., Lindon J.C., Nicholson J.K., Holmes E. Metabonomic studies on the physiological effects of acute and chronic psychological stress in Sprague− Dawley rats. J. Proteome Res. 2007 Jun 1;6(6):2080–2093. doi: 10.1021/pr060412s. [DOI] [PubMed] [Google Scholar]
- Van Duynhoven J.P., Jacobs D.M. Assessment of dietary exposure and effect in humans: the role of NMR. Prog. Nucl. Magn. Reson. Spectrosc. 2016 Aug 1;96:58–72. doi: 10.1016/j.pnmrs.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Weljie A.M., Newton J., Mercier P., Carlson E., Slupsky C.M. Targeted profiling: quantitative analysis of 1H NMR metabolomics data. Anal. Chem. 2006 Jul 1;78(13):4430–4442. doi: 10.1021/ac060209g. [DOI] [PubMed] [Google Scholar]
- Wishart D.S., Jewison T., Guo A.C., Wilson M., Knox C., Liu Y., Djoumbou Y., Mandal R., Aziat F., Dong E., Bouatra S. HMDB 3.0—the human metabolome database in 2013. Nucleic Acids Res. 2012 Nov 17;41(D1):D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtz P., Kangas A.J., Soininen P., Lawlor D.A., Davey Smith G., Ala-Korpela M. Quantitative serum nuclear magnetic resonance metabolomics in large-scale epidemiology: a primer on-omic technologies. Am. J. Epidemiol. 2017 Nov 1;186(9):1084–1096. doi: 10.1093/aje/kwx016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav A.P., Chaturvedi S., Mishra K.P., Pal S., Ganju L., Singh S.B. Evidence for altered metabolic pathways during environmental stress: 1H-NMR spectroscopy based metabolomics and clinical studies on subjects of sea-voyage and Antarctic-stay. Physiol. Behav. 2014 Aug 1;135:81–90. doi: 10.1016/j.physbeh.2014.05.045. [DOI] [PubMed] [Google Scholar]
- Yang Y., Wang Y., Zhang J., Han Z., Chen A., Pan S., Liang S., Wang S. System responses to chronic cold stress probed via 1 H NMR spectroscopy in plasma and urine matrices. Mol. Biosyst. 2015;11(5):1425–1433. doi: 10.1039/c5mb00033e. [DOI] [PubMed] [Google Scholar]
- Zelante T., Iannitti R.G., Cunha C., De Luca A., Giovannini G., Pieraccini G., Zecchi R., D’Angelo C., Massi-Benedetti C., Fallarino F., Carvalho A. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013 Aug 22;39(2):372–385. doi: 10.1016/j.immuni.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Zhao L., Dong M., Liao S., Du Y., Zhou Q., Zheng H., Chen M., Ji J., Gao H. Identification of key metabolic changes in renal interstitial fibrosis rats using metabonomics and pharmacology. Sci. Rep. 2016 Jun 3;6:27194. doi: 10.1038/srep27194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Zhang S., Ragg S., Raftery D., Vitek O. Identification and quantification of metabolites in 1H NMR spectra by Bayesian model selection. Bioinformatics. 2011 Jun 15;27(12):1637–1644. doi: 10.1093/bioinformatics/btr118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supplementary material/referenced in article.