Highlights
-
•
Immunodeficient mice are susceptible to spontaneous tumors which are rarely reported in patient-derived tumor xenografts (PDX) studies.
-
•
Quality control of PDX identity is required at each step on the preclinical assays to avoid erroneous conclusions.
-
•
Description of the approaches used for this PDX check should be clearly detailed in material and methods section.
Keywords: Patient-derived tumor xenograft, Immunodeficient mice, Spontaneous mouse lymphoma, Experimental bias, Reproducibility of experimental results
Abstract
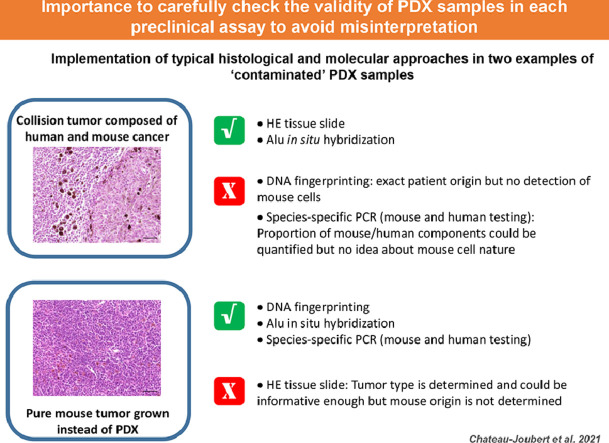
Patient-derived tumor xenograft (PDX) is now largely recognized as a key preclinical model for cancer research, mimicking patient tumor phenotype and genotype. Immunodeficient mice, well-known to develop spontaneous lymphoma, are required for PDX growth. As for all animal models used for further clinical translation, a robust experimental design is strongly required to lead to conclusive results. Here we briefly report unintentional co-engraftment of mouse lymphoma during expansion of well-established PDXs to illustrate the importance of systematic check of the PDX identity to avoid misinterpretation. Besides, this quality control based on complementary approaches deserves a more detailed description in materials and methods section to ensure experimental validity and reproducibility.
Graphical Abstract
Patient-derived tumor xenograft (PDX) model is largely recognized as a powerful model for preclinical anticancer drug testing. Establishment and expansion of patient fresh tumor tissues into mice provide an accurate depiction of human tumor biologic characteristics [1]. Obviously, to avoid immune rejection of human transplants by the mouse host, PDX models have to be generated into immunodeficient mice displaying at least T cell deficiency, as the pioneer nude mice, the SCID mutated or RAG KO mice or more complex multigenic immunodeficient lines [2]. The most appropriate immunodeficient mouse model has to be selected according to several factors: characteristic of immune system components (recipient B cells, NK cells, complement, leakiness with aging), demonstration of higher engraftment rates, propensity to develop PDX metastasis, but also genetic background strains with different disease susceptibility, including spontaneous tumors [2]. The risk of development of spontaneous mouse tumors, most often lymphoma, could be a confounding factor, leading to misinterpretation of preclinical data.
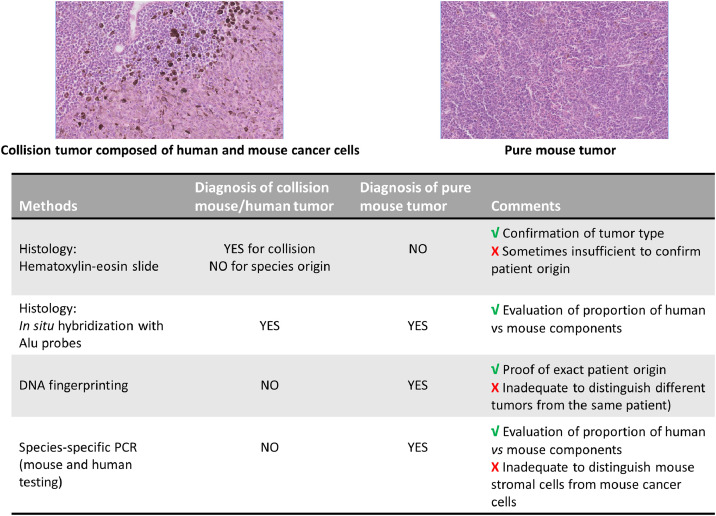
In this context, we illustrate here a potential bias of experimental results through unintentional co-engraftment of mouse lymphoma cells with human tumor xenograft when passages of a well-established human uveal melanoma derived xenograft, emphasizing the absolute need of systematic check of PDX tissue. Indeed, a uveal melanoma PDX was subcutaneously engrafted the same day from 2 Crl:NU(Ico)-Foxn1nu (nude) mice into 28 CB-17/Icr-Prkdcscid/Rj (SCID) for PDX expansion with the aim of anticancer drug efficacy testing. Because of higher robustness, nude mice are routinely used for PDX in vivo banking while SCID mice are selected to promote metastasis development [3]. When growing tumors reached a volume of 40 to 200 mm3, mice were randomly assigned to the control or treatment groups as usually done for drug assay. From 11 to 14 weeks after engraftment, 14 mice, including 10 already included in experimental groups, displayed dehydration, weakness associated with progressive wasting/emaciation. At necropsy, general tissue pallor, large tumors at the engraftment site and splenomegaly were observed, without lymphadenopathy. Histological examination demonstrated that subcutaneous masses were a collision tumor consisting of a melanoma and a lymphoma and that splenomegaly was due to diffuse lymphomatous infiltration. Human origin for this lymphoma was unlikely, as Epstein–Barr virus-associated human lymphomas have been reported in first passages of human solid tumor xenotransplantation in highly immunodeficient mice like NSG [4]. In situ hybridization with Alu probe specific to primate cells confirmed that the SC tumor mass is composed of human melanoma cells but with non-human mouse lymphoma cells. Finally, PCR assay detected Foxn1nu mutation in collision tumor and splenic lymphoma, confirming the nude origin of lymphoma. Thus, mouse lymphoma cells were already present in the ‘donor’ PDX tumor and concomitantly engrafted into recipient mice with the human cancer tissue, grew in parallel with the human cancer cells and disseminated to the spleen. It is particularly noteworthy that the subcutaneous initial growth profile of ‘mixed’ PDX was similar to that of the parent PDX: no statistical difference was noted for the tumor size at the inclusion time compared to previous experiments. Thus, this inclusion time, reflecting tumor growth rate, could not be used here as a warning sign to identify invalid PDXs.
Careful screening of all PDX tumor samples is imperative to identify spontaneous mouse tumors in PDXs to avoid the loss and misuse of valuable PDX models. As spontaneous mouse lymphoma in immunodeficient mouse strains are not rare [5], [6], people involved in PDX bank management are used to facing mouse lymphoma tissue instead of expected PDX during transplantation step. Macroscopic aspect of tumor tissue before engraftment is helpful because of usual gray brittle appearance for mouse lymphoma. Nevertheless, despite attentive visual inspection of tumor sample, this macroscopic examination does not guarantee the absence of mouse tumors. Thus, when maintenance of a bank of colorectal PDXs, which were serially transferred from generation to generation into mice, we had retrospectively observed a ‘mixed’ tumor (i.e. containing tumor cells from both mouse and human origins) at passage P10 which had led to a pure mouse lymphoma at passage P12. More precisely, we have previously reported in a series of 157 samples from 157 different PDX models frozen after routine visual check that the proportion of mouse cells was 100% in 7 samples (5%) [7]. Consequently, when the samples are composed of mixed PDX and mouse tumor tissue, the risk of problematic ‘mixed’ samples would be superior to 5%. Human origin of tissue can be also easily checked by molecular biology using species specific primers for human housekeeping genes or specific for exclusive primate sequence Alu but one more time with no guarantee that samples are mouse tumor-free. In the above-mentioned series of the 150 PDX, we had noted that mouse host cells were found in all specimens with a median proportion of 9%, ranged from 0.5% to 38% according to tumor types and samples, but without confirmation of normal or tumoral mouse components using this approach. In the present case, this PCR approach using species specific primers could have led to erroneous conclusion since human melanoma tissue was present in the engrafted tumor pieces but mixed with highly proliferating mouse lymphoma cells (Fig. 1).
Fig. 1.
Challenging quality control of PDX tissue: example in the case of mouse tumors within or in place of expected human cancer tissue (here mouse lymphoma and human uveal melanoma) Magnification x10.
Consequently, every PDX tissue used for routine tumor passage and drug testing experiments should be carefully checked for its real nature, both at the beginning and at the end of the experiment. Histological examination or molecular analyses (short tandem repeat analysis, presence of Alu sequence, expression of human and mouse housekeeping genes…) are complementary approaches to attest the proper identity of the growing tissue. This verification and the way it is done should be clearly mentioned in Material and methods section but this is never the case today. Regular lack of detail in experimental methods and outcome measurements results in low reproducibility based on provided information, as reported in a review of 145 articles on tumor-graft experiments [8], at a time when lack of reproducibility in scientific and preclinical field is an increasing concern. A checklist of minimal information has been specifically established for PDX use to promote reproducibility in research studies using these models [9], including a quality assurance module. Nevertheless, the criterion ‘Tumor confirmed not to be of mouse/EBV origin’, while considered as an essential attribute in the PDX-MI list, is not recommended to be done neither in every sample nor at every passage. This recurrent validation remains a key point to experimental validity, all the more important when 1 × 1 × 1 (‘one animal per model per treatment’) approach is implemented in newly designed ‘PDX trials’ [10].
Author contributions
Sophie Chateau-Joubert: Formal analysis; Investigation; Methodology.
Miriam Hopfe: Methodology; validation.
Sophie Richon: Data collection and analysis.
Didier Decaudin: Data generation; Writing – review.
Sergio Roman-Roman: expertise and feed back.
Edouard Reyes-Gomez: Expertise, validation.
Ivan Bieche: investigation, expertise and feed back.
Fariba Nemati: Data generation; Writing – review.
Virginie Dangles-Marie: Experimental design, Investigation, Formal analysis, Supervision, Validation, Writing.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Renaud Leclere, Didier Meseure, André Nicolas for tissue sample preparation; and Gérald Massonnet for mouse monitoring. We also thank CReMEC and euroPDX consortium for valuable discussion.
Footnotes
Nonstandard abbreviations: Patient-derived tumor xenograft (PDX).
References
- 1.Byrne A.T., Alférez D.G., Amant F., Annibali D., Arribas J., Biankin A.V., Bruna A., Budinská E., Caldas C., Chang D.K., Clarke R.B., Clevers H., Coukos G., Dangles-Marie V., Eckhardt S.G., Gonzalez-Suarez E., Hermans E., Hidalgo M., Jarzabek M.A., de Jong S., Jonkers J., Kemper K., Lanfrancone L., Mælandsmo G.M., Marangoni E., Marine J.C., Medico E., Norum J.H., Palmer H.G., Peeper D.S., Pelicci P.G., Piris-Gimenez A., Roman-Roman S., Rueda O.M., Seoane J., Serra V., Soucek L., Vanhecke D., Villanueva A., Vinolo E., Bertotti A., Trusolino L. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer. 2017;17(4):254–268. doi: 10.1038/nrc.2016.140. [DOI] [PubMed] [Google Scholar]
- 2.Okada S., Vaeteewoottacharn K., Kariya R. Application of Highly Immunocompromised Mice for the Establishment of Patient-Derived Xenograft (PDX) Models. Cells. 2019;8(8):889–906. doi: 10.3390/cells8080889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khanna C., Hunter K. Modeling metastasis in vivo. Carcinogenesis. 2005;26(3):513–523. doi: 10.1093/carcin/bgh261. [DOI] [PubMed] [Google Scholar]
- 4.Bondarenko G., Ugolkov A., Rohan S., Kulesza P., Dubrovskyi O., Gursel D., Mathews J., O’Halloran T.V., Wei J.J., Mazar A.P. Patient-Derived Tumor Xenografts Are Susceptible to Formation of Human Lymphocytic Tumors. Neoplasia. 2015;17(9):735–741. doi: 10.1016/j.neo.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell M.R., Nation P.N., Andrew S.E. A lack of DNA mismatch repair on an athymic murine background predisposes to hematologic malignancy. Cancer Res. 2005;65(7):2626–2635. doi: 10.1158/0008-5472.CAN-04-3158. [DOI] [PubMed] [Google Scholar]
- 6.Custer R.P., Bosma G.C. Bosma MJ Severe Combined Immunodeficiency (SCID) in the Mouse. Pathology, Reconstitution, Neoplasms. Am. J. Pathol. 1985;120(3):464–477. PMID: 2412448. [PMC free article] [PubMed] [Google Scholar]
- 7.Bieche I., Vacher S., Vallerand D., Richon S., Hatem R., De Plater L., Dahmani A., Némati F., Angevin E., Marangoni E., Roman-Roman S., Decaudin D., Dangles-Marie V. Vasculature analysis of patient derived tumor xenografts using species-specific PCR assays: evidence of tumor endothelial cells and atypical VEGFA-VEGFR1/2 signalings. BMC Cancer. 2014;14:178. doi: 10.1186/1471-2407-14-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugar E., Pascoe A.J., Azad N. Reporting of preclinical tumor-graft cancer therapeutic studies. Cancer Biol. Ther. 2012;13(13):1262–1268. doi: 10.4161/cbt.21782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weroha S.J., Amant F., Pfister S.M., Kool M., Parkinson H., Butte A.J., Bult CJ.PDX-MI. Minimal Information for Patient-Derived Tumor Xenograft Models. Cancer Res. 2017;77(21):e62–e66. doi: 10.1158/0008-5472.CAN-17-0582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao H., Korn J.M., Ferretti S., Monahan J.E., Wang Y., Singh M., Zhang C., Schnell C., Yang G., Zhang Y., Balbin O.A., Barbe S., Cai H., Casey F., Chatterjee S., Chiang D.Y., Chuai S., Cogan S.M., Collins S.D., Dammassa E., Ebel N., Embry M., Green J., Kauffmann A., Kowal C., Leary R.J., Lehar J., Liang Y., Loo A., Lorenzana E., McDonald E 3rd Robert, McLaughlin M.E., Merkin J., Meyer R., Naylor T.L., Patawaran M., Reddy A., Röelli C., Ruddy D.A., Salangsang F., Santacroce F., Singh A.P., Tang Y., Tinetto W., Tobler S., Velazquez R., Venkatesan K., Von Arx F., Wang H.Q., Wang Z., Wiesmann M., Wyss D., Xu F., Bitter H., Atadja P., Lees E., Hofmann F., Li E., Keen N., Cozens R., Jensen M.R., Pryer N.K., Williams J.A., Sellers W.R. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015;21(11):1318–1325. doi: 10.1038/nm.3954. [DOI] [PubMed] [Google Scholar]