Abstract
MicroRNAs (miRNAs/miRs) are sensitive biomarkers and endogenous repressors of gene expression by decreasing mRNA stability and interfering with mRNA translation. Despite a number of investigations revealing the dysregulation of miRNA expression associated with cardiotoxicity induced by doxorubicin (Dox), perturbation of miRNAs directly resulting from Dox at early stage in cardiomyocytes and the target gene interaction remain largely unknown. In the present study, high-throughput deep-sequencing was used to analyze changes in global miRNA expression in H9c2 cardiomyocytes exposed to 5 µg/ml Dox for 0, 12 or 24 h. Compared with the 0-h time point, the expression levels of 386 unique miRNAs were altered. Based on miRNA expression and fold-change, the target genes of 76 selected miRNAs were further analyzed using gene interaction networks and pathway enrichment analysis. These miRNAs were involved in the regulation of different pathways, whose functions included apoptosis, cell proliferation, extracellular matrix remodeling, oxidative stress and lipid metabolism. These differentially expressed miRNAs included let-7 family, miR-29b-3p, miR-378-3/5p, miR-351-3p, miR-664-3p, miR-455-3p, miR-298-3p, miR-702-5p, miR-128-1-5p, miR-671 and miR-421-5p. The present data indicated that global wide miRNA profiling in Dox-induced cardiomyocytes may provide a novel mechanistic insight into understanding Dox-induced heart failure and cardiotoxicity, as well as novel biomarkers and therapeutic targets.
Keywords: doxorubicin, microRNA, cardiomyocyte, cardiotoxicity
Introduction
Doxorubicin (Dox) belongs to the anthracycline family of antibiotics and is widely used for the treatment of several types of cancer, including gastric cancer, breast cancer, leukemia, sarcoma and lymphoma (1). Dox efficiently induces apoptosis of tumor cells and prevents cell proliferation by intercalating into DNA and stabilizing a ternary complex with topoisomerase II to interfere with DNA replication (1). The use of Dox is limited due to the development of cardiotoxicity; the risk of heart disease is 3–5% in patients who receive a cumulative dose of 400 mg/m2 and up to 48% in patients receiving 700 mg/m2 (2). A number of studies have demonstrated that Dox is a mitochondrial toxin (1,2). Dox induces over-production of reactive oxygen species (ROS) and ATP imbalance through electron transport chain uncoupling, resulting in cardiac injury (1).
MicroRNAs (miRNAs/miRs) are a class of small non-coding single-stranded RNAs (~22 nucleotides in length) that bind to complementary sequences found in the 3′-untranslated region of mRNAs and regulate post-transcriptional gene expression by inducing mRNA degradation or inhibiting mRNA translation (3). miRNAs are ubiquitously involved in developmental and pathological processes, such as cell differentiation (4), organogenesis (5), tissue injury and remodeling (6), and tumorigenesis. Increasing data have revealed that miRNAs serve an important role in cardiac functions and several cardiovascular diseases, such as ischemic heart diseases and arrhythmia (7,8). Several biological and pathological functions of miRNAs in the regulation of cardiomyocyte survival or apoptosis have been directly confirmed in animal or cell models (9,10). In recent years, miRNA expression profiling of heart tissues in mouse or rat models of Dox-induced cardiotoxicity have been investigated by miRNA array or quantitative PCR assay, and numerous changes in miRNA expression were reported 2–8 weeks after Dox treatment (11,12). It is well known that the transcription of miRNAs is sensitive to cellular stress and triggers a number of downstream biological processes (13). Changes in miRNA expression profiling in cardiomyocytes resulting from Dox at the early stage (12 or 24 h post Dox treatment) remain unknown. In the present study, the rat cardiomyocyte H9c2 cell line was used to investigate changes in miRNA expression at the early stage of Dox treatment and the contribution of miRNAs in the initial phase of Dox-induced cardiomyocytes dysfunction.
Materials and methods
Cell culture and treatment
Rat H9c2 cardiomyocytes were purchased from The Cell Bank of Type Culture Collection of the Chinese Academy of Sciences. Cells were maintained in DMEM containing 10% FBS (both Gibco; Thermo Fisher Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml streptomycin (Thermo Fisher Scientific, Inc.) at 37°C in a humidified incubator containing 5% CO2. Dox (Zhejiang Hisun Chemical Co., Ltd.) was obtained from Tongren Hospital (Shanghai, China). Cells were seeded in a 15-cm culture dish at a density of 1×106. Dox was dissolved in normal saline. Cells were treated with a final concentration of 5 µg/ml Dox for 0, 12 or 24 h at 37°C in a humidified incubator containing 5% CO2, and then samples were collected. The morphology of H9c2 cells was observed at the aforementioned time points under a light microscope (Nikon Eclipse Ti-S; Nikon Corporation; magnification, ×10) (Fig. S1).
RNA isolation, RNA-sequencing library construction and sequencing
Total RNA was extracted from cells using TRIzol® according to the manufacturer's instructions (cat. no. 15596026; Thermo Fisher Scientific, Inc.). A total of 3 µg total RNA per sample was used to generate the small RNA library. Sequencing libraries were prepared using TruSeq® Small RNA Sample Prep kit following the manufacturer's protocol (cat. no. RS-200-0012; Illumina, Inc.). The library quality was examined using Agilent Bioanalyzer 2100 (Agilent Technologies, Inc.). NEBNext Multiplex Small RNA Library Prep Set for Illumina was used for sequencing (cat. no. E7300S; New England BioLabs, Inc.). Deep sequencing of the library was performed on Illumina NextSeq platform using single-end 1×75 sequencing mode.
Quality control and alignment of sequencing data
Raw reads were filtered to obtain high-quality clean reads by removing sequencing adapters, short reads (<35 bp in length) and low-quality reads using Cutadapt v1.9.1 (14) and Trimmomatic v0.35 (14). Subsequently, FastQC v0.11.5 (15) was used for ensuring high quality reads. The resulting clean reads were mapped to rat genome (assembly Rnor_6.0) using bowtie2 v2.2.9 with default parameters (16). Using miRBase as the reference database (http://www.mirbase.org), reads of each miRNA were determined, and reads per million (RPM) was adopted as the normalization method to quantify the abundance of each miRNA.
Analysis of differential expression
Differential miRNA expression analyses were conducted using DESeq2 v1.20.0 (17). The false discovery rate control method was used to calculate the adjusted P-values in multiple testing, in order to determine the significance of the differences. Genes with an adjusted P<0.05, regression to the mean value of 10 at the 0-h time point and an expression level varying ≥1.5-fold at 24 vs. 0 h or 12 vs. 0 h were used for subsequent analysis. The cluster analysis of differentially expressed miRNAs was performed using the Hierarchical Clustering Explorer v3.5 software (http://www.cs.umd.edu/hcil/hce/hce3.html).
Functional enrichment and target genes interaction network analysis
The target genes of rat miRNAs were downloaded from mirTarBase v7.0 (10), the online miRNA reference database that stores manually curated collections of experimentally validated miRNA targets. Enrichment analysis based on Gene Ontology (GO; http://geneontology.org/) annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/pathway.html) pathways for the target genes of differentially expressed miRNAs was implemented using the ClusterProfiler package v3.11.0 (7). A pathway (or GO term) with an adjusted P<0.05 was considered to be a significantly enriched pathway (or GO term). Based on the lists of differentially expressed miRNAs, information on the target gene interactions was obtained from the Search Tool for the Retrieval of Interacting Genes/Proteins (www.string-db.com). Target gene interaction networks were mapped using Cytoscape v3.7 software (https://cytoscape.org/index.html).
Reverse transcription-quantitative (q)PCR analysis for miRNA expression
Total RNA was extracted from cells using TRIzol® (cat. no. 15596026; Thermo Fisher Scientific, Inc.). miRNA first-strand cDNA was synthesized from 1 µg total RNA from cells using the PrimeScript™ II 1st Strand cDNA Synthesis kit (Takara Bio, Inc.). The 20-µl volume reaction system contained 1 µg total RNA, 5 nM reverse transcription primer (Stem-loop RT primer from Shanghai GenePharma Co., Ltd.), 20 U reverse transcriptase, 20 U RNase inhibitor and 0.2 mM dNTPs. The mixture was incubated at 42°C for 15 min and at 85°C for 5 min. qPCR was performed with the SYBR Green Hairpin-it MicroRNA Quantitation PCR kit (cat. no. E01006; Shanghai GenePharma Co., Ltd.) according to the manufacturer's instructions. cDNA (2 µl) was used as the template for the qPCR reaction system. PCR was performed using the StepOnePlus PCR system (Thermo Fisher Scientific, Inc.), and the PCR parameters were as follows: 95°C for 3 min, followed by 40 cycles at 95°C for 12 sec and 62°C for 40 sec. Rox was used as the reference dye in PCR. The expression levels of rno-miR-29b-3p, rno-miR-145-5p and rno-miR-378a-3p were analyzed using the 7500 real-time PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.). The primers were included in the aforementioned SYBR Green Hairpin-it MicroRNA Quantitation PCR kit. Primers used were as follows: Rno-mir-29b-3p forward, 5′-ACAGCAATTAGCACCATTTGAA-3′ and reverse, 5′-TATGCTTCTTCTCGTCTCTGTGTC-3′; rno-mir-145-5p forward, 5′-CAGTCTTGTCCAGTTTTCCCAG-3′ and reverse, 5′-TATGCTTGTTCTCGTCTCTGTGTC-3′; rno-mir-378a-3p forward, 5′-ATGGTGGACTGGACTTGGAGT-3′, and reverse, 5′-GTGCAGGGTCCGAGGT-3′; rat U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′. Relative miRNA expression was normalized to U6 expression and calculated using the relative 2−ΔΔCq method (18).
Statistical analysis
Statistical analysis was performed using SPSS v21 (IBM Corp.) and data were analyzed using one-way ANOVA followed by Tukey's HSD test. All data were presented as the mean ± SD. All experiments were repeated at least three times. P<0.05 was considered to indicate a statistically significant difference.
Results
Dynamic changes in miRNA expression profiling in cardiomyocytes treated with Dox
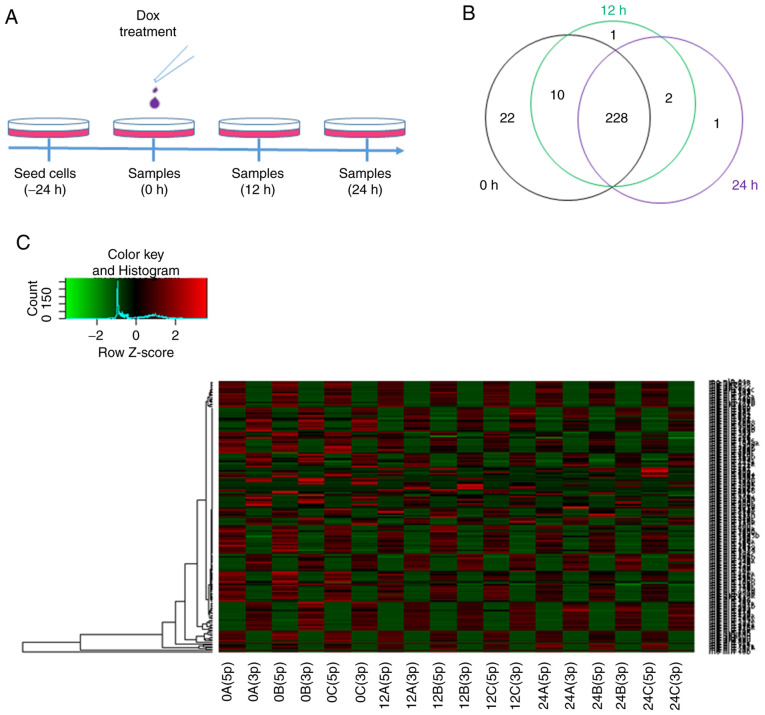
In order to reveal the dynamic changes in miRNAs at the early stage of Dox treatment, deep miRNA sequencing was performed to determine miRNA expression at the 0-, 12- and 24-h time points (Fig. 1A). Read counts of each miRNA were normalized to RPM, and the expression levels of 386 rat miRNAs were identified to be altered after 12 or 24 h of Dox treatment compared with the 0-h time point (Tables SI and SII). In the present study, 228 miRNAs were expressed continuously at the 0-, 12- and 24-h time points by intersecting miRNAs (Fig. 1B). Mature miRNAs can be generated from the 5p- or 3p-arm of miRNA precursors, and the majority of miRNA isoform accumulation is tissue-dependent (19). A total of 198 pairs of miRNAs were analyzed, and 5p- or 3p-biased miRNA expression was presented at different time points (Fig. 1C). The results revealed that Dox decreased the expression levels of most miRNAs; however, the efficiency of regulation was different between the 5p and 3p isoforms in each pair of miRNAs. The mechanism of 5p and 3p selectivity and accumulated expression remains unknown.
Figure 1.
Characteristics of miRNAs in Dox-induced H9c2 cells. (A) Procedure of Dox treatment in H9c2 cells. (B) Venn diagram of miRNA expression at each time point. (C) Heatmap of the 198 pairs of miRNAs with-3p and −5p mature forms in each sample. Normalized reads per million values were used. 0A/B/C, 12A/B/C and 24A/B/C represent samples collected at 0, 12 and 24 h of Dox treatment, respectively. miRNA, microRNA; Dox, doxorubicin.
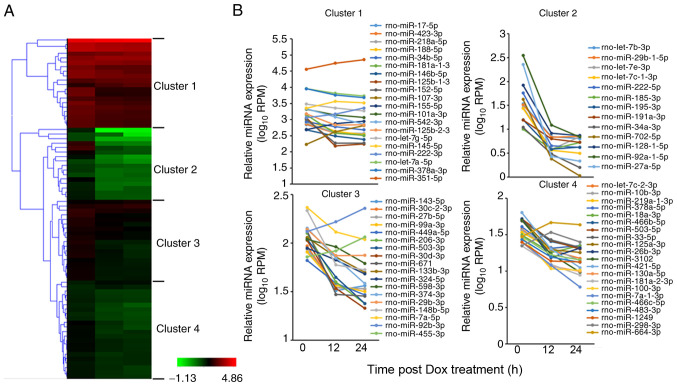
In order to narrow down the target miRNAs, 76 miRNAs whose expression levels changed by ≥1.5-fold compared with the 0-h time point were selected, including 67 downregulated miRNAs and 9 upregulated miRNAs (Table SIII), and were further classified into 4 clusters using the Hierarchical Clustering Explorer v3.5 software (Fig. 2A). Changes in miRNA expression patterns for 4 typical clusters are shown in Fig. 2B. miRNAs in cluster 1 had high expression levels, with average RPM values >100 at the 0-h time point in H9c2 cells. Notably, miR-351-5p exhibited the highest RPM value in cluster 1. The expression levels of miRNAs of cluster 2 rapidly decreased at both 12 and 24 h after Dox treatment. The expression levels of miRNAs of cluster 3 and cluster 4 tended to decrease following Dox treatment, although the range of change was smaller compared with that in cluster 2. The expression levels of most miRNAs in the 4 clusters were downregulated at 12 and 24 h after Dox administration. Among the selected miRNAs, some miRNAs are known to function in Dox-induced heart diseases, such as members of the miR-30 family or let-7 family, miR-133b, miR-143, miR-298, miR-29 and miR-34a (6–13). However, the functions of most of the selected miRNAs in Dox-induced cardiotoxicity or heart injury remain unclear.
Figure 2.
Dynamics of miRNAs in H9c2 cells during Dox treatment. Cluster analysis of miRNAs with altered miRNA expression, determined using high-throughput sequencing, were analyzed at 0, 12 and 24 h after Dox treatment. (A) A total of 76 differentially expressed miRNAs (RPM >10) were identified, whose expression levels changed ≥1.5-fold compared with the 0-h time point. The miRNA RPM values were normalized with log10-transformation before using cluster analysis. The 76 miRNAs were classified into 4 clusters (clusters 1–4) using the Hierarchical Clustering Explorer v3.5 software. (B) Dynamic miRNA expression curves for each cluster. miR/miRNA, microRNA; RPM, reads per million; Dox, doxorubicin.
Contribution of Dox-mediated miRNA expression to different biological processes and pathways
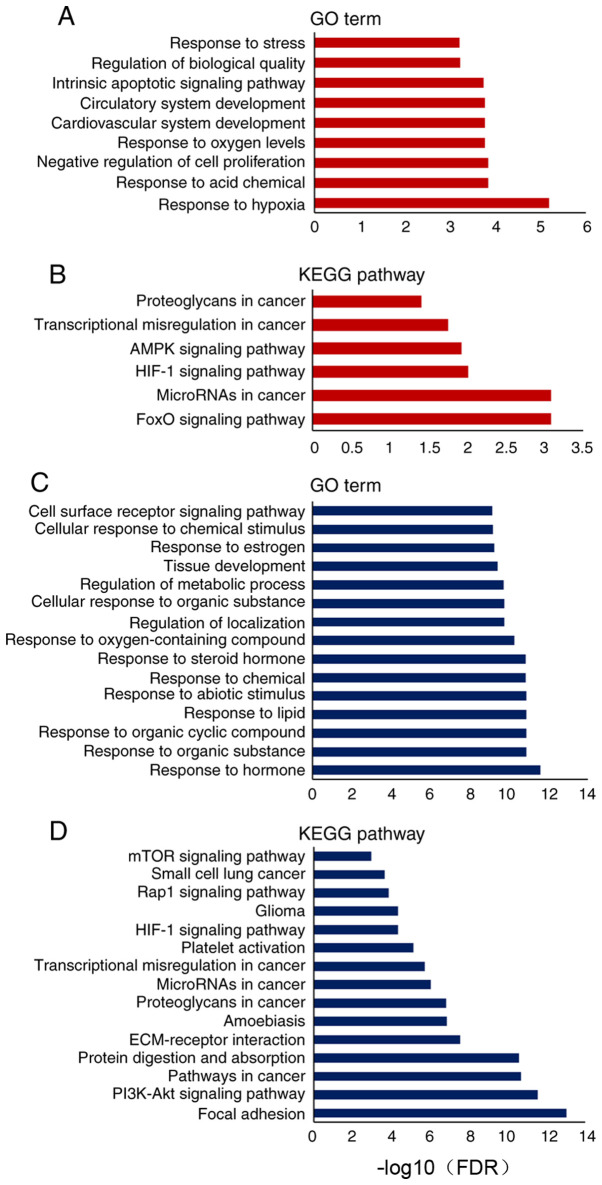
miRNAs directly bind to target mRNAs to induce mRNA degradation or inhibit mRNA translation in cells. Based on the mirTarBase database of differentially expressed miRNAs, information on the downstream targets of upregulated or downregulated miRNAs was collected. A total of 87 genes targeted by the downregulated miRNAs, and 20 genes targeted by the upregulated miRNAs were analyzed according to GO terms and KEGG pathways (Tables SIV–SVII). The upregulated miRNAs contributed to the biological processes of ‘response to hypoxia’, ‘negative regulation of cell proliferation’ and ‘cardiovascular system development’ (Fig. 3A). KEGG pathway analysis revealed that the upregulated miRNAs were involved in the regulation of cell survival and apoptotic pathways, such as the ‘hypoxia-inducible factor 1 (HIF-1) signaling pathway’, ‘AMPK signaling pathway’ and ‘FoxO signaling pathway’ (Fig. 3B). Notably, the analysis of downregulated miRNAs revealed that several processes of cell response to chemicals were disturbed, such as ‘response to organic substance’, ‘response to lipid’ and ‘response to steroid hormone’ (Fig. 3C). Additionally, these downregulated miRNAs were involved in ‘extracellular matrix (ECM)-receptor interaction’, ‘focal adhesion’ and ‘PI3K-Akt signaling pathway’ (Fig. 3D).
Figure 3.
Representative GO and KEGG pathway enrichment analysis of differentially expressed miRNAs. (A) GO and (B) KEGG pathway analysis of upregulated miRNAs after Dox treatment, with the top 9 biological processes and 6 pathways presented, respectively. (C) GO and (D) KEGG pathway analysis of downregulated miRNAs after Dox treatment, with the top 15 biological processes or the top 15 pathways presented. GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; miRNA, microRNA; Dox, doxorubicin.
Regulation of gene networks by miRNAs in H9c2 cells
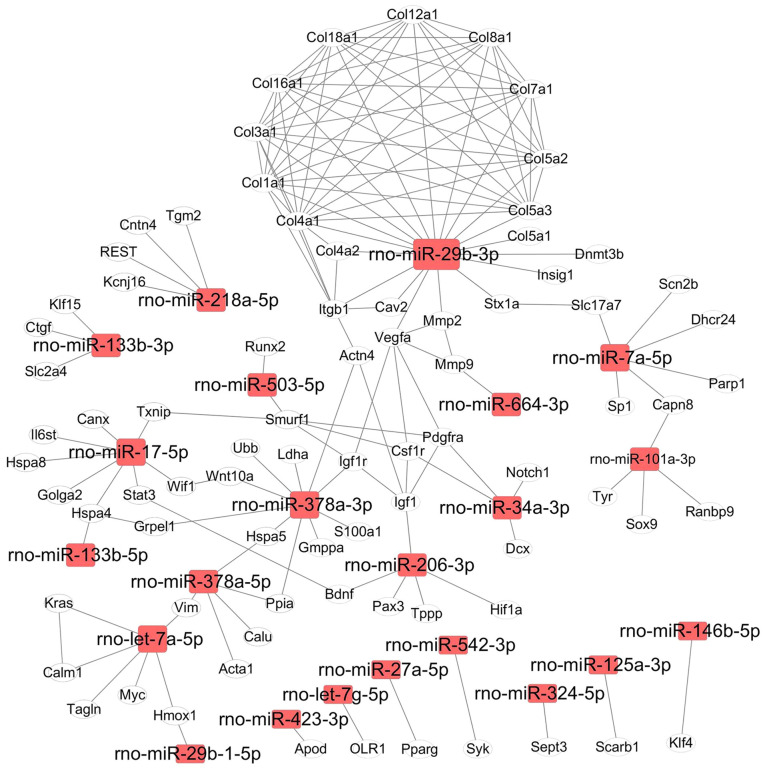
Proteins, as products of gene translation, execute biological functions in cells. Most proteins need be associated with their partners to form a protein complex, which then has the capability to be involved in biological functions (20). Differentially expressed miRNAs induced by Dox in the present study regulated a number of target genes, and the proteins translated from these genes were used for mapping protein-protein interaction networks. For the upregulated miRNAs (miR-455-3p, −92b-3p, −222-3p, −155-5p, −298-3p, −351-5p, 664-3p, −145p-5p and −107-3p), the interaction network consisted of 20 genes (Fig. 4), which were involved in the regulation of cell fate and stress response pathways (Table SIV). For example, rno-miR-145-5p targeted 7 genes in rats (Fig. 4), and these genes were involved in the regulation of autophagy, lipid metabolism and mitochondrial homeostasis (Table SIV). The function of rno-miR-222-3p in heart disease or cardiomyocytes has been previously reported (13); however, its expression profiling during Dox treatment was newly discovered (Fig. 4). rno-miR-92b-3p expression was significantly increased at 12 and 24 h after Dox treatment in H9c2 cells, and it targeted NADPH oxidase 4 (Nox4), whose expression levels and mRNA alternative splicing induce the development of heart failure by increasing ROS production (3).
Figure 4.
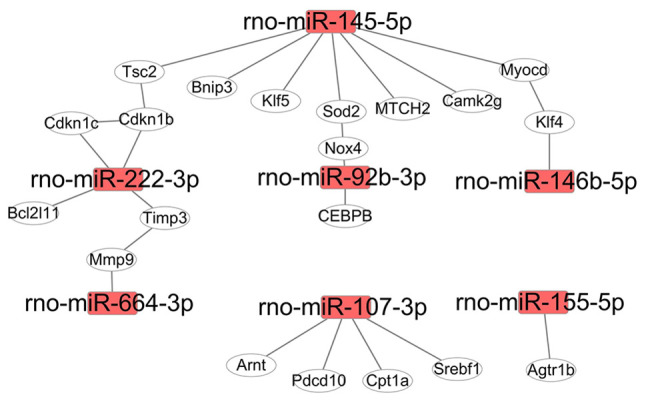
Interaction network of targeted genes downstream of upregulated miRNAs. A total of 7 miRNAs and their targets were included in the interaction network. The interaction network was visualized using the Cytoscape software. Rectangle nodes denote miRNAs, and oval nodes denote genes. miR/miRNA, microRNA.
In the downregulated miRNA group, the range of fold-change was 1.76–384.67 at 12 or 24 h vs. the 0-h time point (Table SII), and the network comprised a total of 178 interactions between genes (Fig. 5). According to the number of targets genes and biological functions, four miRNAs (rno-miR-29b-3p, rno-miR-378a-3p, rno-let-7a-5p and rno-miR-17-5p) were localized on critical network nodes and regulated over 40 genes (Fig. 5). It is well know that heat shock proteins (Hsp) are important for regulating cell survival and protein homeostasis in cells. Notably, rno-miR-378a-5p targeted Hspa5, and rno-miR-17-5p targeted Hspa4 and Hspa8 (Fig. 5). In the current map, rno-let-7a-5p regulated the expression of 6 genes, whose functions are associated with maintaining cell phenotype (such as Myc, Kras and Tagln) and functions (such as Hmox1, Calm1 and Vim) (21). rno-miR-29b-3p expression was significantly downregulated by Dox in H9c2 cells, and it interfered with the expression levels of 10 genes in the collagen protein family, which are important proteins in the ECM. It is well know that the ECM serves a critical role in signal transduction and material exchange between the environment and cells. Three miRNAs, namely rno-miR-145-5p (upregulated), rno-miR-29b-3p and rno-miR-378a-3p (both downregulated), served as important nodes in the gene networks, and their expression levels were therefore further analyzed using qPCR. The expression levels of these three miRNAs were consistent with the results from deep sequencing analysis, with the expression levels of rno-miR-145-5p being significantly upregulated and those of rno-miR-29b-3p and rno-miR-378a-3p being significantly downregulated following Dox treatment for 12 and 24 h (Fig. S2).
Figure 5.
Interaction networks of targeted genes downstream of downregulated miRNAs. A total of 22 miRNAs and their targets were included in the interaction network. The interaction network was visualized using the Cytoscape software. Rectangle nodes denote miRNAs, and oval nodes denote genes. miR/miRNA, microRNA.
Discussion
Cardiotoxicity is the main side effect of Dox treatment that restricts its clinical application in cancer therapy (1). Specific biomarkers at the early stage of cardiotoxicity resulting from Dox treatment are very important to predict and decrease the risk of heart failure in the clinic. Previous studies have revealed that circulating miRNAs (8,9), as potential biomarkers of heart failure, were very sensitive in patients with heart failure (13). In the present study, 76 miRNAs were identified to be dysregulated by Dox treatment in H9c2 myoblast cells. In the upregulated miRNA group, the expression levels of 9 miRNAs (miR-455-3p, −92b-3p, −222-3p, −155-5p, −298-3p, −351-5p, 664-3p, −145p-5p and −107-3p) were significantly upregulated at 12 or 24 h after Dox treatment. Altered expression levels of miR-145-5p, miR-155-5p and miR-222-3p have been previously observed in animal models of Dox administration (22–24). In particular, miR-145-5p has been previously reported as a cardioprotective molecule in myocardial ischemic injury (25) and as a biomarker in LaminA/C-related dilated cardiomyopathy (26). miR-351-5p exhibited the highest RPM value in this group, and has been shown to inhibit Pten expression in H9c2 cells (27). It is well known that PTEN serves an important role in the development of heart failure by regulating multiple signaling pathways (28,29). Additionally, miR-351-5p expression is upregulated by ROS and functions as a proinflammatory and proapoptotic factor by targeting the MAPK signaling pathway in both ICE-6 and H9c2 cells (30,31). Furthermore, several studies have identified that MAPK/NF-κB, a classical proinflammatory and proapoptotic signaling pathway, is involved in cardiotoxicity induced by Dox (32,33).
In the downregulated miRNA group, two out of the ten most altered miRNAs, miR-133b (34) and let-7e-3p (23), have been reported in previous studies, revealing a minor increase in circulating miR-133b expression and a dominant contribution from the skeletal muscle toxicity induced by Dox (34,35). In the present study, miR-133b-5p expression in H9c2 cells was inhibited by Dox and markedly decreased. The main side effect of Dox are both acute or chronic heart failure, whose progression begins from myocardial injury that develops into left ventricular dysfunction (36). These pathological changes are very important for the progression to heart failure. miR-133 expression in the heart functions as a suppressor of cardiac fibrosis and cardiac remodeling (10).
miR-29b-3p expression in the present study was markedly downregulated after Dox administration, in accordance with previous studies (13,22,23). miR-29b-3p targeted 12 collagen genes, whose protein levels have been detected and used as prognostic biomarkers in patients with chronic heart failure (37,38). Additionally, inhibition of miR-29b-3p expression directly results in apoptosis of myoblast cells in vitro (39). Recent studies have indicated that miR-29b-3p serves a central role in the processes of cardiac fibrosis formation and adverse ventricular remodeling (40,41). Both cell proliferation and apoptosis are involved in the process of tissue remodeling. In the present study, miR-702-5p and miR-128-1-5p expression was inhibited by Dox in H9c2 cells. In previous studies, miR-702 has been shown to inhibit apoptosis and improve survival of different types of cells, such as NIH3T3, 293T and embryonic stem cells (42,43). miR-128 serves a critical role in promoting cardiomyocyte proliferation and heart regeneration by targeting Suz12, a component of polycomb repressive complex 2, and downregulating Suz12 protein expression in cardiomyocytes (44).
The members of the let-7 miRNA family are involved in the regulation of cardiomyocyte differentiation and metabolism, as well as myocardial infarction (45). In the present study, the expression levels of seven members of let-7 (rno-let-7a-1-3p, -a-5p, -b-3p, -c-1-3p, -c-2-3p, -e-3p and -g-5p) were inhibited by Dox in H9c2 cells, and, to the best of our knowledge, the expression profiles of six let-7 members in Dox induced cardiomyocytes have not been previously reported, except for let-7g (9). In previous studies, both let-7a and −7e downregulated β1-adrenoceptors expression and modulated their downstream signaling pathways in cardiomyocytes of rat heart failure models (46,47). Inhibition of let-7a and −7e expression significantly results in deterioration of cardiac fibrosis and heart failure (46). However, data on the let-7 miRNA family in regulating heart failure are not consistent across different studies and their biological functions in cardiomyocytes need to be further investigated (48,49). miR-378 improves cardiomyocyte survival (50) and protects cardiomyocytes against myocardial fibrosis in vivo (51). In the present study, Dox repressed miR-378-3p and −5p expression in H9c2 cells at the 12- and 24-h time points. In vivo and in vitro studies have confirmed that miR-378 directly targets caspase-3 (50) and four key components of the MAPK signaling pathway (Mapk1, Igfr1, Gfrbp 2 and Ksr1) (52).
H9c2 rat cardiac cells, as immortalized cardiomyocytes, are widely used in establishing cell models of cardiomyocyte injury induced by different compounds in the medium with or without the addition of serum (32,53). Previous studies have indicated that serum withdrawal can significantly induce changes in miRNA expression in HUVECs and A549 cells (54,55). In the present study, cells were treated with Dox in complete medium with serum, since serum deprivation could have induced disturbances of miRNA expression associated with apoptosis. There is a limitation in the present study, as the effects of cell proliferation and cell cycle on miRNA expression in H9c2 cells could not be avoided. To further investigate the impacts of serum on miRNA expression, miRNA expression profiles should be analyzed after treating H9c2 cells with Dox in medium with or without serum in future studies.
In conclusion, 76 miRNAs were identified to have altered expression levels directly induced by Dox in a cardiomyocyte cell line. Notably, to the best of our knowledge, the present study described for the first time the altered expression levels of miRNAs in cardiomyocytes treated with Dox, including let-7 family, miR-664-3p, −455-3p, −298-3p, −702-5p, −128-1-5p, −671, −421-5p, −378-3/5p and −351-3p. Pathway analysis revealed that these miRNAs were involved in the dysregulation of multiple signaling pathways, including the FoxO, HIF-1, AMPK, focal adhesion, PI3K-Akt, ECM-receptor and mTOR signaling pathways. The expression levels of these miRNAs served an important role in the regulation of biological processes that support cell metabolism and cell stability. Accordingly, previous studies have reported that the MAPK and PI3K-Akt signaling pathways serve an important role in Dox-induced cardiotoxicity (53,56). However, the present study presents some limitations, since the direct cardiotoxicity of Dox was only investigated in vitro. Dox and products of Dox metabolism may serve a role in these biological processes in vivo. The control group always serves an important role in experiments, and the 0-h time point group (untreated group) was chosen as a control in the present study. However, H9c2 cells treated with a vehicle for 0, 12 and 24 h could also be used as experiment controls. Additionally, using different doses of Dox on H9c2 cells may exclude effects of vehicle on miRNA expression. These strategies may be useful for future research in Dox-induced cardiotoxicity in vitro. In future studies, lower doses of Dox and longer treatment times in H9c2 cells may more efficiently mimic the progression of chronic cardiotoxicity induced by Dox. In the future, animal models of spontaneous tumors or primary tumors should be used to study Dox cardiotoxicity and analyze miRNA expression profiles in cardiomyocytes. Analysis of tissues from patients with Dox-induced heart failure may provide further insight into miRNA expression profiles, but samples cannot be collected at the early stage of Dox-induced cardiotoxicity. The use of induced pluripotent stem cell-derived human heart organoids may replace some in vivo experiments and partly overcome the limitations of cell models in the study of Dox cardiotoxicity. The present study provided new insights for understanding the dysregulation of miRNA expression in H9c2 cardiomyocytes induced by Dox at an early stage. Some of these miRNAs may have potential as biomarkers or therapeutic targets.
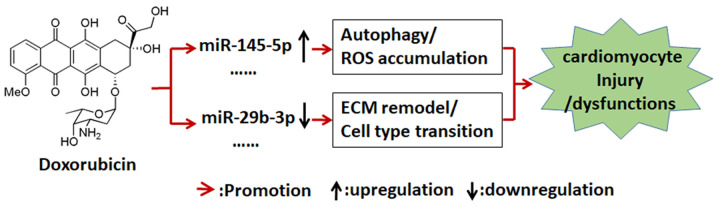
Overall, the present study focused on investigating alterations of miRNA expression in rat cardiomyocyte within 24 h (12- and 24-h time points) of Dox treatment. The expression levels of 76 miRNAs were significantly altered in cardiomyocytes treated with Dox, and had >1.5-fold-changes compared with the 0-h time point group. Only 9 of these miRNAs have been reported in previous studies of Dox-induced cardiotoxicity (22–24,34,35). These differentially expressed miRNAs were ubiquitously involved in numerous biological processes and functional pathways involved in the regulation of apoptosis, ECM remodeling, cell differentiation and cell metabolism. Based on the present data and biological information database, the interactions with the target genes of these miRNAs were analyzed and visual gene interaction networks were mapped. These interaction networks may help to further understand the roles of miRNA dysregulation in the development of Dox-induced cardiotoxicity at the early stage, as well as provide further insight in the involved molecular mechanisms (Fig. 6).
Figure 6.
Model of Dox-induced cardiotoxicity through profiles of miRNA expression in cardiomyocyte cells. In cardiomyocyte cells, Dox may directly disturb the homeostasis of miRNA expression. Differentially expressed miRNAs, such as miR-145-5p and miR-29b-3p, may be enriched in several biological processes associated with autophagy, ROS accumulation and ECM remodeling. miR/miRNA, microRNA; ROS, reactive oxygen species; ECM, extracellular matrix; Dox, doxorubicin.
Supplementary Material
Acknowledgements
The authors would like to thank Dr Yali Kang and Dr Lingyi Li (Shanghai Jiao Tong University School of Biomedicine Engineering, Shanghai, China) for helping to analyze the microRNA sequencing data and data normalization, and Dr Xiaohong Ma (Huadong Hospital Affiliated to Fudan University, Shanghai, China) for his advice on this project and for reviewing the manuscript.
Funding Statement
The present study was supported by the Shanghai Municipal Commission of Health and Family planning (grant no. 201540302), the Science and Technology Commission of Shanghai Changning (grant no. CNKW2016Y03) and Shanghai Jiao Tong University Medicine-Engineering Joint Fund (grant no. YG2015MS64).
Funding
The present study was supported by the Shanghai Municipal Commission of Health and Family planning (grant no. 201540302), the Science and Technology Commission of Shanghai Changning (grant no. CNKW2016Y03) and Shanghai Jiao Tong University Medicine-Engineering Joint Fund (grant no. YG2015MS64).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the National Genomics Data Center repository (https://bigd.big.ac.cn/; accession no. CRA003714).
Authors' contributions
LJ and WJ designed the experiments. YC and YX performed the tissue culture and established the cell models. ZD and YW prepared the microRNA library. YC, YZ and YJ analyzed the data of the miRNA expression profiles and the biological information. LJ and WJ wrote the manuscript. YC, YX, LJ and WJ confirm the authenticity of all the raw data and the processed data in the present study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Mitry MA, Edwards JG. Doxorubicin induced heart failure: Phenotype and molecular mechanisms. Int J Cardiol Heart Vasc. 2016;10:17–24. doi: 10.1016/j.ijcha.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koleini N, Kardami E. Autophagy and mitophagy in the context of doxorubicin-induced cardiotoxicity. Oncotarget. 2017;8:46663–46680. doi: 10.18632/oncotarget.16944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu HJ, Han ZY, He SF, Jin SY, Xu SJ, Fang XD, Zhang Y. Specific MicroRNAs comparisons in hypoxia and morphine preconditioning against hypoxia-reoxgenation injury with and without heart failure. Life Sci. 2017;170:82–92. doi: 10.1016/j.lfs.2016.11.028. [DOI] [PubMed] [Google Scholar]
- 4.Li N, Wang WB, Bao H, Shi Q, Jiang ZL, Qi YX, Han Y. MicroRNA-129-1-3p regulates cyclic stretch-induced endothelial progenitor cell differentiation by targeting Runx2. J Cell Biochem. 2019;120:5256–5267. doi: 10.1002/jcb.27800. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, Yuan Y, He X, Xia X, Mo X. MicroRNA-1 upregulation promotes myocardiocyte proliferation and suppresses apoptosis during heart development. Mol Med Rep. 2017;15:2837–2842. doi: 10.3892/mmr.2017.6282. [DOI] [PubMed] [Google Scholar]
- 6.Ren N, Wang M. microRNA-212-induced protection of the heart against myocardial infarction occurs via the interplay between AQP9 and PI3K/Akt signaling pathway. Exp Cell Res. 2018;370:531–541. doi: 10.1016/j.yexcr.2018.07.018. [DOI] [PubMed] [Google Scholar]
- 7.Guo L, Zheng X, Wang E, Jia X, Wang G, Wen J. Irigenin treatment alleviates doxorubicin (DOX)-induced cardiotoxicity by suppressing apoptosis, inflammation and oxidative stress via the increase of miR-425. Biomed Pharmacother. 2020;125:109784. doi: 10.1016/j.biopha.2019.109784. [DOI] [PubMed] [Google Scholar]
- 8.Ji X, Ding W, Xu T, Zheng X, Zhang J, Liu M, Liu G, Wang J. MicroRNA-31-5p attenuates doxorubicin-induced cardiotoxicity via quaking and circular RNA Pan3. J Mol Cell Cardiol. 2020;140:56–67. doi: 10.1016/j.yjmcc.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Fu J, Peng C, Wang W, Jin H, Tang Q, Wei X. Let-7 g is involved in doxorubicin induced myocardial injury. Environ Toxicol Pharmacol. 2012;33:312–317. doi: 10.1016/j.etap.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 10.Li N, Zhou H, Tang Q. miR-133: A suppressor of cardiac remodeling? Front Pharmacol. 2018;9:903. doi: 10.3389/fphar.2018.00903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu JN, Fu YH, Hu ZQ, Li WY, Tang CM, Fei HW, Yang H, Lin QX, Gou DM, Wu SL, Shan ZX. Activation of miR-34a-5p/Sirt1/p66shc pathway contributes to doxorubicin-induced cardiotoxicity. Sci Rep. 2017;7:11879. doi: 10.1038/s41598-017-12192-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao L, Qi Y, Xu L, Tao X, Han X, Yin L, Peng J. MicroRNA-140-5p aggravates doxorubicin-induced cardiotoxicity by promoting myocardial oxidative stress via targeting Nrf2 and Sirt2. Redox Biol. 2018;15:284–296. doi: 10.1016/j.redox.2017.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruggeri C, Gioffré S, Achilli F, Colombo GI, D'Alessandra Y. Role of microRNAs in doxorubicin-induced cardiotoxicity: An overview of preclinical models and cancer patients. Heart Fail Rev. 2018;23:109–122. doi: 10.1007/s10741-017-9653-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andrews S. FastQC: A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Apr 26;2010 ];2010 [Google Scholar]
- 16.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Buermans HP, Ariyurek Y, van Ommen G, den Dunnen JT't, Hoen PA. New methods for next generation sequencing based microRNA expression profiling. BMC Genomics. 2010;11:716. doi: 10.1186/1471-2164-11-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Legrain P, Rain JC. Twenty years of protein interaction studies for biological function deciphering. J Proteomics. 2014;107:93–97. doi: 10.1016/j.jprot.2014.03.038. [DOI] [PubMed] [Google Scholar]
- 21.Lee H, Han S, Kwon CS, Lee D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein Cell. 2016;7:100–113. doi: 10.1007/s13238-015-0212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desai VG, C Kwekel J, Vijay V, Moland CL, Herman EH, Lee T, Han T, Lewis SM, Davis KJ, Muskhelishvili L, et al. Early biomarkers of doxorubicin-induced heart injury in a mouse model. Toxicol Appl Pharmacol. 2014;281:221–229. doi: 10.1016/j.taap.2014.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Vacchi-Suzzi C, Bauer Y, Berridge BR, Bongiovanni S, Gerrish K, Hamadeh HK, Letzkus M, Lyon J, Moggs J, Paules RS, et al. Perturbation of microRNAs in rat heart during chronic doxorubicin treatment. PLoS One. 2012;7:e40395. doi: 10.1371/journal.pone.0040395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roca-Alonso L, Castellano L, Mills A, Dabrowska AF, Sikkel MB, Pellegrino L, Jacob J, Frampton AE, Krell J, Coombes RC, et al. Myocardial MiR-30 downregulation triggered by doxorubicin drives alterations in β-adrenergic signaling and enhances apoptosis. Cell Death Dis. 2015;6:e1754. doi: 10.1038/cddis.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan M, Zhang L, You F, Zhou J, Ma Y, Yang F, Tao L. MiR-145-5p regulates hypoxia-induced inflammatory response and apoptosis in cardiomyocytes by targeting CD40. Mol Cell Biochem. 2017;431:123–131. doi: 10.1007/s11010-017-2982-4. [DOI] [PubMed] [Google Scholar]
- 26.Toro R, Blasco-Turrión S, Morales-Ponce FJ, Gonzalez P, Martínez-Camblor P, López-Granados A, Brugada R, Campuzano O, Pérez-Serra A, Rosa Longobardo F, et al. Plasma microRNAs as biomarkers for Lamin A/C-related dilated cardiomyopathy. J Mol Med (Berl) 2018;96:845–856. doi: 10.1007/s00109-018-1666-1. [DOI] [PubMed] [Google Scholar]
- 27.da Silva W, dos Santos RA, Moraes KC. Mir-351-5p contributes to the establishment of a pro-inflammatory environment in the H9c2 cell line by repressing PTEN expression. Mol Cell Biochem. 2016;411:363–371. doi: 10.1007/s11010-015-2598-5. [DOI] [PubMed] [Google Scholar]
- 28.Billia F, Hauck L, Konecny F, Rao V, Shen J, Mak TW. PTEN-inducible kinase 1 (PINK1)/Park6 is indispensable for normal heart function. Proc Natl Acad Sci USA. 2011;108:9572–9577. doi: 10.1073/pnas.1106291108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gao Y, Chu M, Hong J, Shang J, Xu D. Hypoxia induces cardiac fibroblast proliferation and phenotypic switch: A role for caveolae and caveolin-1/PTEN mediated pathway. J Thorac Dis. 2014;6:1458–1468. doi: 10.3978/j.issn.2072-1439.2014.08.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng L, Han X, Hu Y, Zhao X, Yin L, Xu L, Qi Y, Xu Y, Han X, Liu K, Peng J. Dioscin ameliorates intestinal ischemia/reperfusion injury via adjusting miR-351-5p/MAPK13-mediated inflammation and apoptosis. Pharmacol Res. 2019;139:431–439. doi: 10.1016/j.phrs.2018.11.040. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y, Mao Z, Xu L, Yin L, Tao X, Tang Z, Qi Y, Sun P, Peng J. Protective effect of dioscin against intestinal ischemia/reperfusion injury via adjusting miR-351-5p-mediated oxidative stress. Pharmacol Res. 2018;137:56–63. doi: 10.1016/j.phrs.2018.09.016. [DOI] [PubMed] [Google Scholar]
- 32.Venkatakrishnan CD, Tewari AK, Moldovan L, Cardounel AJ, Zweier JL, Kuppusamy P, Ilangovan G. Heat shock protects cardiac cells from doxorubicin-induced toxicity by activating p38 MAPK and phosphorylation of small heat shock protein 27. Am J Physiol Heart Circ Physiol. 2006;291:H2680–H2691. doi: 10.1152/ajpheart.00395.2006. [DOI] [PubMed] [Google Scholar]
- 33.Guo R, Wu K, Chen J, Mo L, Hua X, Zheng D, Chen P, Chen G, Xu W, Feng J. Exogenous hydrogen sulfide protects against doxorubicin-induced inflammation and cytotoxicity by inhibiting p38MAPK/NFκB pathway in H9c2 cardiac cells. Cell Physiol Biochem. 2013;32:1668–1680. doi: 10.1159/000356602. [DOI] [PubMed] [Google Scholar]
- 34.Nishimura Y, Kondo C, Morikawa Y, Tonomura Y, Torii M, Yamate J, Uehara T. Plasma miR-208 as a useful biomarker for drug-induced cardiotoxicity in rats. J Appl Toxicol. 2015;35:173–180. doi: 10.1002/jat.3044. [DOI] [PubMed] [Google Scholar]
- 35.Rigaud VO, Ferreira LR, Ayub-Ferreira SM, Ávila MS, Brandão SM, Cruz FD, Santos MH, Cruz CB, Alves MS, Issa VS, et al. Circulating miR-1 as a potential biomarker of doxorubicin-induced cardiotoxicity in breast cancer patients. Oncotarget. 2017;8:6994–7002. doi: 10.18632/oncotarget.14355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cappetta D, Rossi F, Piegari E, Quaini F, Berrino L, Urbanek K, De Angelis A. Doxorubicin targets multiple players: A new view of an old problem. Pharmacol Res. 2018;127:4–14. doi: 10.1016/j.phrs.2017.03.016. [DOI] [PubMed] [Google Scholar]
- 37.Dupuy AM, Kuster N, Curinier C, Huet F, Plawecki M, Solecki K, Roubille F, Cristol JP. Exploring collagen remodeling and regulation as prognosis biomarkers in stable heart failure. Clin Chim Acta. 2019;490:167–171. doi: 10.1016/j.cca.2018.08.042. [DOI] [PubMed] [Google Scholar]
- 38.Nagao K, Inada T, Tamura A, Kajitani K, Shimamura K, Yukawa H, Aida K, Sowa N, Nishiga M, Horie T, et al. Circulating markers of collagen types I, III, and IV in patients with dilated cardiomyopathy: Relationships with myocardial collagen expression. ESC Heart Fail. 2018;5:1044–1051. doi: 10.1002/ehf2.12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou S, Lei D, Bu F, Han H, Zhao S, Wang Y. MicroRNA-29b-3p targets SPARC gene to protect cardiocytes against autophagy and apoptosis in hypoxic-induced H9c2 cells. J Cardiovasc Transl Res. 2019;12:358–365. doi: 10.1007/s12265-018-9858-1. [DOI] [PubMed] [Google Scholar]
- 40.Han Z, Zhang T, He Y, Li G, Li G, Jin X. Inhibition of prostaglandin E2 protects abdominal aortic aneurysm from expansion through regulating miR-29b-mediated fibrotic ECM expression. Exp Ther Med. 2018;16:155–160. doi: 10.3892/etm.2018.6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drummond CA, Fan X, Haller ST, Kennedy DJ, Liu J, Tian J. Na/K-ATPase signaling mediates miR-29b-3p regulation and cardiac fibrosis formation in mice with chronic kidney disease. PLoS One. 2018;13:e0197688. doi: 10.1371/journal.pone.0197688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang WG, Chen L, Dong Q, He J, Zhao HD, Li FL, Li H. Mmu-miR-702 functions as an anti-apoptotic mirtron by mediating ATF6 inhibition in mice. Gene. 2013;531:235–242. doi: 10.1016/j.gene.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Kim BM, Choi MY. Non-canonical microRNAs miR-320 and miR-702 promote proliferation in Dgcr8-deficient embryonic stem cells. Biochem Biophys Res Commun. 2012;426:183–189. doi: 10.1016/j.bbrc.2012.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang W, Feng Y, Liang J, Yu H, Wang C, Wang B, Wang M, Jiang L, Meng W, Cai W, et al. Loss of microRNA-128 promotes cardiomyocyte proliferation and heart regeneration. Nat Commun. 2018;9:700. doi: 10.1038/s41467-018-03019-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kuppusamy KT, Jones DC, Sperber H, Madan A, Fischer KA, Rodriguez ML, Pabon L, Zhu WZ, Tulloch NL, Yang X, et al. Let-7 family of microRNA is required for maturation and adult-like metabolism in stem cell-derived cardiomyocytes. Proc Natl Acad Sci USA. 2015;112:E2785–E2794. doi: 10.1073/pnas.1424042112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du Y, Zhang M, Zhao W, Shu Y, Gao M, Zhuang Y, Yang T, Mu W, Li T, Li X, et al. Let-7a regulates expression of β1-adrenoceptors and forms a negative feedback circuit with the β1-adrenoceptor signaling pathway in chronic ischemic heart failure. Oncotarget. 2017;8:8752–8764. doi: 10.18632/oncotarget.14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Wang B, Cui H, Du Y, Song Y, Yang L, Zhang Q, Sun F, Luo D, Xu C, et al. let-7e replacement yields potent anti-arrhythmic efficacy via targeting beta 1-adrenergic receptor in rat heart. J Cell Mol Med. 2014;18:1334–1343. doi: 10.1111/jcmm.12288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bao MH, Feng X, Zhang YW, Lou XY, Cheng Y, Zhou HH. Let-7 in cardiovascular diseases, heart development and cardiovascular differentiation from stem cells. Int J Mol Sci. 2013;14:23086–23102. doi: 10.3390/ijms141123086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tolonen AM, Magga J, Szabó Z, Viitala P, Gao E, Moilanen AM, Ohukainen P, Vainio L, Koch WJ, Kerkelä R, et al. Inhibition of Let-7 microRNA attenuates myocardial remodeling and improves cardiac function postinfarction in mice. Pharmacol Res Perspect. 2014;2:e00056. doi: 10.1002/prp2.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fang J, Song XW, Tian J, Chen HY, Li DF, Wang JF, Ren AJ, Yuan WJ, Lin L. Overexpression of microRNA-378 attenuates ischemia-induced apoptosis by inhibiting caspase-3 expression in cardiac myocytes. Apoptosis. 2012;17:410–423. doi: 10.1007/s10495-011-0683-0. [DOI] [PubMed] [Google Scholar]
- 51.Yuan J, Liu H, Gao W, Zhang L, Ye Y, Yuan L, Ding Z, Wu J, Kang L, Zhang X, et al. MicroRNA-378 suppresses myocardial fibrosis through a paracrine mechanism at the early stage of cardiac hypertrophy following mechanical stress. Theranostics. 2018;8:2565–2582. doi: 10.7150/thno.22878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ganesan J, Ramanujam D, Sassi Y, Ahles A, Jentzsch C, Werfel S, Leierseder S, Loyer X, Giacca M, Zentilin L, et al. MiR-378 controls cardiac hypertrophy by combined repression of mitogen-activated protein kinase pathway factors. Circulation. 2013;127:2097–2106. doi: 10.1161/CIRCULATIONAHA.112.000882. [DOI] [PubMed] [Google Scholar]
- 53.Kobashigawa LC, Xu YC, Padbury JF, Tseng YT, Yano N. Metformin protects cardiomyocyte from doxorubicin induced cytotoxicity through an AMP-activated protein kinase dependent signaling pathway: An in vitro study. PLoS One. 2014;9:e104888. doi: 10.1371/journal.pone.0104888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X, Wei J, Ma Z, He Y. Rapamycin- and starvation-induced autophagy are associated with miRNA dysregulation in A549 cells. Acta Biochim Biophys Sin (Shanghai) 2019;51:393–401. doi: 10.1093/abbs/gmz022. [DOI] [PubMed] [Google Scholar]
- 55.Kim JH, Lee DK, Kim J, Choi S, Park W, Ha KS, Kim TH, Choe J, Won MH, Kwon YG, Kim YM. A miRNA-101-3p/Bim axis as a determinant of serum deprivation-induced endothelial cell apoptosis. Cell Death Dis. 2017;8:e2808. doi: 10.1038/cddis.2017.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu J, Sun C, Wang R, Li J, Zhou M, Yan M, Xue X, Wang C. Cardioprotective effect of paeonol against epirubicin-induced heart injury via regulating miR-1 and PI3K/AKT pathway. Chem Biol Interact. 2018;286:17–25. doi: 10.1016/j.cbi.2018.02.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the National Genomics Data Center repository (https://bigd.big.ac.cn/; accession no. CRA003714).