When the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic emerged in 2019, researchers rapidly recalibrated epidemiological computer models originally developed for other pandemics to serve as decision support tools for policy-makers and health care professionals planning public health responses. However, no current tools can predict the course of disease and help a doctor decide on the most appropriate treatment for an individual COVID-19 patient. “Digital twins” are software replicas of the dynamic function and failure of engineered products and processes. The medical analog, patient-specific digital twins, could integrate known human physiology and immunology with real-time patient-specific clinical data to produce predictive computer simulations of viral infection and immune response. Such medical digital twins could be a powerful addition to our arsenal of tools to fight future pandemics, combining mechanistic knowledge, observational data, medical histories, and the power of artificial intelligence (AI).
An industrial digital twin of a device, such as a specific jet engine, combines a predictive template computational model calibrated using historical data aggregated from many devices with regularly collected operational data for the particular device. Digital twins allow continual forecasting, and small-scale interventions to prevent problems before they become serious. This greatly reduces the frequency of critical failures. The most sophisticated engineering digital twins are also self-improving—they continuously monitor divergence between predictions and observations and use these divergences to improve their own accuracy.
Although medical digital twins are much more difficult to develop than those for engineered devices, they have begun to find applications in improving human health. Examples include the “artificial pancreas” for type 1 diabetes patients (1, 2). In the artificial pancreas model, a template mathematical model of human glucose metabolism and a closed-loop control algorithm modeling insulin delivery and data from an implanted glucose sensor are customized into a patient-specific digital twin that continuously calculates insulin needs and drives an implanted pump that adjusts blood insulin concentrations. Additionally, pediatric cardiac digital twins combine template models of the heart with patient-derived clinical measurements to optimize some heart surgeries (3) and assess the risk of thrombosis (4). These examples illustrate how current digital twins can operate in real time to maintain health continuously, or they can be used off-line to design personalized medical interventions. The ARCHIMEDES diabetes model expands these technologies by including models not only of the progression of diabetes within individual patients but also of medical diagnosis, treatments, and the functioning of the health care system that is providing the treatment (5). These examples provide a vision of the potential of medical digital twins.
Medical digital twins that combine mechanistic understanding of physiology and viral replication with AI-based models derived from population and individual clinical data are promising as tools for optimizing the treatment of patients infected with a virus. As clinical outcomes of SARS treatment revealed, therapies such as steroidal anti-inflammatories can be lifesaving but also ineffective or even lethal if they are not adjusted carefully to suit individual patient responses (6). Thus, more complex therapies combining antivirals and multiple immune-stimulating and anti-inflammatory drugs would need to be personalized for time of application and dose of each component to be both effective and safe. Validated digital twins could greatly reduce the cost and complexity of such combinatorial clinical applications. Even primitive digital twins could improve diagnosis, prognosis, and treatment by providing a framework to combine patient-specific population data in a consistent framework. Their deployment would enable rapid refinement and improvement, especially if they were designed in a modular fashion to permit the parallel development and optimization of their component submodels.
However, a digital twin that can continuously replicate the complexity of an infection and immune responses sufficiently well to guide individual treatment is currently out of reach. To realize the potential of digital twins in the treatment of viral diseases, there are a number of issues that need to be addressed. The spread of infection within the body and the immune response to viral pathogens are still poorly understood, as are the factors determining if and when specific components of the immune response are beneficial (viral clearance) or harmful (hyperinflammation). Viral infections can be complex, with pathologies developing in organs beyond the sites of primary infection, requiring an understanding of the responses of multiple organs. Therapies are also complex, with combinations integrating phased doses of antivirals, anti-inflammatory drugs, antibodies, and immune-stimulating drugs such as interferon (IFN) or interleukin-7 (IL-7).
Where to start in building viral-infection digital twins? When considering how to build mechanistic model components for a viral-infection digital twin, many of the necessary submodels of relevant pathways and processes already exist (7) or could be developed using existing experimental methodologies (see the figure 1). For example, at the subcellular scale, transcriptomics data analysis of human macrophages can be used to construct dynamic network models of the interactions of highly expressed genes for each macrophage subtype (8), forming the basis for dynamic models of gene regulatory networks. At the multicellular scale, imaging technology reveals spatial aspects of the immune response (9). At the tissue scale, a simulation of an alveolar sac can capture the spatiotemporal variability of the immune response. At the organ scale, computational fluid dynamics models can simulate airflow in the lungs (10). And at the whole-body scale, computational models calibrated with simultaneous data from different organs, such as respiratory contractions and cerebral blood flow, can be used to integrate different organ systems (11). Physiologically based kinetic models (12) are widely used in the development and regulation of pharmaceuticals. Virologists have developed template models that capture aspects of viral spread, immune response, viral replication in individual cells, physiology, and dysfunction of specific organ systems, transport in the blood and lymph, airway transport of viruses, and aerosol therapies (13).
Building a personalized digital twin.
Data from multiple scales are needed to build computational representations of biological processes and body systems that are affected by viral infection. These submodels are integrated and personalized with clinical data from individual patients. The digital twin can then be used to derive predictions about diagnosis, prognosis, and efficacy and optimization of therapeutic interventions.
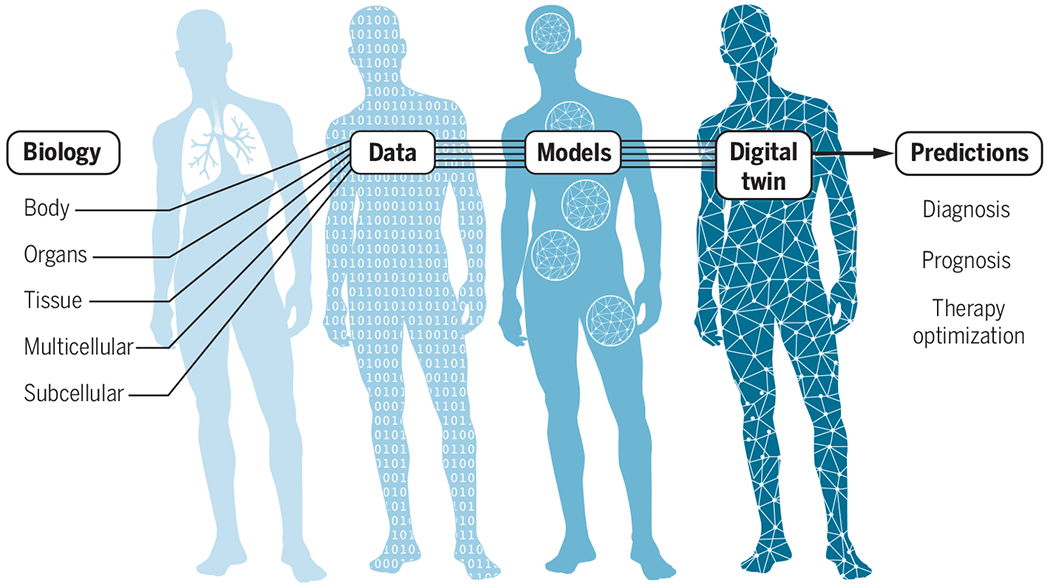
Digital twins describing infection and treatment require the development, validation, and integration of numerous component submodels in the context of a rapidly developing scientific understanding of biological behaviors and continual generation of new experimental and clinical data. Although individual laboratories may construct submodels, the development of comprehensive digital twins will require laboratories and research groups around the world to integrate and validate submodels independently, with only limited central coordination. Enabling such parallel development requires a flexible simulation architecture that uses a multiscale map of all the relevant components of a patient’s response to viral infection, as well as responses to available treatments. Community efforts such as the COVID-19 Disease Map Project and Computational Modeling in Biology Network (COMBINE) are working to build such infrastructure, although much work needs to be done to adapt those for use in digital-twin technology.
Another substantial challenge is the generation of the heterogeneous data required to both calibrate template models to known human biology and physiology and to personalize template models into digital twins. Data from clinical trials are an important resource for model validation and discovery, as are high-resolution time-series data characterizing the immune response in a variety of individuals and settings. Model construction and validation require collection of synchronous measurements at different physiological scales: ‘omics data from tissues and single cells, from diverse experimental systems, including two-dimensional (2D) and 3D cell cultures, in vivo and ex vivo animal models, and patients; at the tissue level, data characterizing immune cell trafficking and patterns of damage and recovery; and biophysical and structural data from tissues and organs, combined with data characterizing transport throughout the body. To personalize a digital twin, the model template must integrate with clinical records and time courses, such as vital signs, immune cell counts, computed tomography scans of infected organs, measured viral loads, and treatment responses. Given the likely complexity of the underlying models, even the abundance of such data will leave considerable challenges in model validation and uncertainty quantification.
Data-driven AI digital twins for prognosis and treatment optimization are often viewed as alternatives to mechanistic modeling. However, AI and mechanistic approaches are most valuable when used together. Mechanistic modeling can provide important constraints for training AI algorithms (14). Conversely, AI can assist mechanistic modeling in a host of ways, including parameter identification, model structure evaluation, simulation acceleration, and continuous model refinement through comparison of model predictions with observations.
Building a useful digital twin requires improved communication between clinicians, experimentalists, and modelers. Relatively few clinical and biological insights are currently translated into computational models that could serve as building blocks for medical digital twins. In many cases, data collected as part of experimental studies are not usable for this purpose in part because simulation was not an explicit aim of these studies.
The many challenges that have arisen as health systems have sought to respond to the current global health crisis highlight the urgent need for a viral-infection digital twin that can serve as an integration platform for heterogeneous high-dimensional data that serve to inform data collection and guide evidence-based approaches to personalized treatment. They could result in a core technology that enables a rapid medical response to future emerging viral pathogens. In addition, they can form the basis for digital twins for other diseases. Initially, repurposing the immune response portion of the viral digital twin to other diseases where the immune system is key, such as cancer, could rapidly leverage the viral models. Longer term, digital twins could be coupled to other disease-specific twins already being developed, resulting in much more comprehensive models. Ultimately, these larger-scale digital twins could increase the resilience of the entire health care system. ■
ACKNOWLEDGMENTS
We thank the members of the NIH Working Group on Multiscale Modeling and Viral Pandemics for many helpful discussions. We thank B. Shapiro and L. Sordo Vieira for help with the figure. R.L. is partially supported by the NIH (NIHR011AI135128, NIHU01EB024501, and NIHR01GM127909). J.P.S. and J.A.G are supported by the NIH (NIHU24EB028887, NIHR01GM122424, and NSF1720625). J.A.G. holds shares in Gilead Pharmaceuticals and is a partner in Apoptocys LLC. He is the sole proprietor of Virtual Tissues for Health LLC. He has a patent pending on the application of virtual tissues to improve therapy for diabetic retinopathy.
REFERENCES AND NOTES
- 1.Kovatchev B, Trends Endocrinol. Metab 30, 432 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Brown SA et al. , N. Engl. J. Med 381, 1707 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shang JK et al. , Ann. Thorac. Surg 107, 1232 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grande Gutierrez N et al. , Int. J. Cardiol 281, 15 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eddy DM, Schlessinger L, Diabetes Care 26, 3102 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Stockman LJ, Bellamy R, Garner P, PLOS Med. 3, e343 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perelson AS, Ke R, Clin. Pharmacol. Ther 10.1002/cpt.2160 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beyer M et al. , PLOS ONE 7, e45466 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martines RB et al. , U.S. Emerg. Infect. Dis 26, 2005 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolanjiyil AV, Kleinstreuer C, Comput. Biol. Med 79, 193 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Cross TJ et al. , PLOS ONE 8, e66950 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goyal A, Cardozo-Ojeda EF, Schiffer JT, Sci. Adv 6, eabc7112 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sego TJ et al. , PLOS Comput. Biol. 16, e1008451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alber M et al. , NPJ Digit. Med 2, 115 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]


