Abstract
Background/Aims
XRCC1 (X-ray repair cross-complementing protein 1) expression and its single nucleotide polymorphism XRCC1 rs25487 (G>A) may be related to radiotherapy-related cancer prognosis or radiation-induced side effects. However, this association is controversial. We performed a bioinformatic analysis and a meta-analysis to obtain comprehensive results.
Methods
TCGA data sets and eligible publications published before November 31, 2020 were retrieved by searching the PubMed, Web of Science and CNKI (China National Knowledge Infrastructure) databases. ORs (odds ratios) and HRs (hazard ratios) with their corresponding 95% CIs (confidence intervals) were calculated to evaluate associations. For XRCC1 single nucleotide polymorphisms, we employed three types of comparisons: GA vs GG, AA vs GG and GA+AA vs GG.
Results
Sixty nine articles with 10232 patients and 17 TCGA data sets with 2705 patients were included in the analysis. We observed that high XRCC1 expression was associated with an increased risk of minor treatment response and poor overall survival, XRCC1 rs25487 was associated with reduced risk of minor treatment response in esophageal cancer and an increased risk of high-grade side effects in head and neck cancer.
Conclusion
The results suggest that XRCC1 expression and rs25487 polymorphism are prognostic factors for patients receiving radiotherapy-related treatment. Considering the insufficient treatment parameters provided and the various sample sizes in most of the studies, we suggest that genetic association studies related to radiation-based treatment should include more cancer types with sufficient statistical power and more detailed clinical parameters.
Keywords: radiation, side-effects, treatment response, overall survival, rs25487, XRCC1
Introduction
Radiotherapy using ionizing radiation is among the main treatments used to control or kill malignant neoplasms. Ionizing radiation functions by creating double-strand breaks (DSBs) or by damaging cell membranes, which can lead to cell death. However, the responses to radiotherapy vary between different cancer types, or even between cancer cells inside a tumor (1). Several factors are related to the responses of a specific tumor to radiotherapy: dose and fraction of radiation, and clinicopathologic characteristics including TNM stage (which is a notation system that employs alphanumeric codes to describes the stage of cancer that originates from a solid tumor with.), tumor size (2), and biological characteristics, such as pathological type, hypoxic state, DNA repair capacity, gene expression level and functional gene mutations. Normal tissue surrounding the tumor region or in the path of the radiation beams can be temporally injured during radiation treatment, lasting for a period after treatment, or even irrevocably damaged in some cases. Moreover, radiation might also have an impact on remote normal tissue. Patients treated with radiotherapy, even under the same treatment procedures, may experience a significant difference in radiation-induced early or late side effects, in terms of incidence and severity (3), which is a major challenge for radiotherapy practice. For patients, the major treatment response and slight/no side effects are important factors for long-term survival (4) and quality of life.
Since 2002, more than two-hundred published studies have reported an association between single nucleotide polymorphisms (SNPs) and cancer prognosis in patients receiving radiotherapy-related treatment. In the meantime, many studies have focused on the gene expression level, radiotherapy-related treatment response and cancer prognosis. According to our statistics, the key factors in evaluating prognosis mainly include survival (e.g. overall survival, progression-free survival), treatment response (e.g. complete remission, partial remission), and radiotherapy-related side effects (e.g. pneumonitis, esophagitis). One of the important goals of these studies is to identify genetic factors that can be used to predict radiotherapy-related cancer prognosis (5). Among these published studies, X-ray repair cross-complementing protein 1 (XRCC1) is one of the most studied genes and has been investigated for its possible association with cancer prognosis. The protein encoded by XRCC1, is involved in DNA repair with DNA ligase III and DNA polymerase (6). Preclinically, XRCC1 deficiency delays single-strand break rejoining, induces mutations and results in elevated levels of sister chromatid exchanges, a hallmark of genomic instability. XRCC1 deficiency results in hypersensitivity to ionizing radiation (7). Some clinical studies have indicated that a high XRCC1 expression level is associated with poor overall survival and treatment response in patients treated with radiotherapy (8, 9). Another study concludes that the XRCC1 expression level has no impact on overall survival or treatment response in rectal cancer patients treated with radiotherapy (10). The SNP rs25487 [Arg399Gln, G>A substitution at position 28152, exon 10, Arg to Gln (11)] is among the most widely researched XRCC1 SNPs, and yet researchers still do not agree on its impact. A number of studies with small sample sizes have investigated the association between this SNP and response to chemoradiotherapy (12–15). However, the results are not consistent. A previous meta-analysis of four studies has stated that this SNP does not predict response to chemoradiotherapy (n = 511) in patients with rectal cancer (16). However, the sample size in genetic association studies is still too small to obtain sufficient power to a robust conclusion.
To address the issues of inconsistent conclusions and limited sample size, we performed meta-analyses that included the most comprehensive literature and obtained the largest sample size for this topic to date. To minimize the effect of publication bias, we collected data from published articles (including published Ph.D. and master’s theses) on the association between XRCC1 expression level and radiotherapy-related treatment response/cancer prognosis; we also explored the relationship between XRCC1 rs25487 polymorphism and radiotherapy-related treatment response/radiation-induced side effects.
Methods
Literature Search
Eligible publications were retrieved by searching the PubMed, Web of Science and Chinese National Knowledge Infrastructure (CNKI) databases up to November 31, 2020. The search strategy was based on the following keywords: [(X-ray Repair Cross Complementing Protein 1(MeSH Terms)] AND Radiotherapy [MeSH Terms]) AND Neoplasms [MeSH Terms]. Studies focusing on the association between XRCC1 expression/XRCC1 rs25487 and radiotherapy-related cancer prognosis were screened for further analysis.
Inclusion Criteria and Exclusion Criteria
Two independent reviewers performed the inclusion assessment. The inclusion criteria were as follows: (1) an independent case-control or cohort study; (2) evaluation of the association between the XRCC1 expression level and radiotherapy-related treatment response/cancer prognosis; (3)evaluation of the association between XRCC1 rs25487 and radiotherapy-related treatment response/radiation-induced side effects in patients receiving radiotherapy-related treatment; (4) sufficient data on the relationship between XRCC1 expression and overall survival, with estimated hazard ratios (HRs) and 95% confidence intervals(CIs) determined by multivariate analysis and reported in the articles or available to be indirectly calculated via Kaplan Meier (K-M) curves; and (5) provision of the number of patients in the high/low XRCC1 expression group or with different genotypes in the case-control group.
Exclusion criteria: (1) studies without sufficient data associated with radiotherapy-related treatment response, overall survival (OS), or radiation-induced side effects, (2) duplicated publications, (3) studies based on animal models or cell lines, (4) other literature types: reviews, letters, abstracts, meta-analysis, case reports, etc.
Data Extraction and Quality Assessment
Two independent reviewers collected data from the studies included in the meta-analysis. The following data were collected: first author, publication year, cancer type, treatment, side effects, acute/late degree, evaluation criteria, cutoff value, follow-up period, survival outcome, HR (95% CI), number of patients in the high/positive XRCC1 expression group and low/negative XRCC1 expression group associated with treatment response, and number of patients in the case-control group and the association between XRCC1 rs25487 and treatment response/side effects. If survival rates were not obtained from multivariate analysis, the survival HR (95% CI) was indirectly retrieved from K-M curves using Engauge Digitizer software. This method was described in detail by Tierney et al. (17). A calculation spreadsheet was prepared in Microsoft Excel to obtain the observed minus expected events (O-E), the variance (V), the HR, the log (HR), and its standard error (SE) for each of the individual trials. Newcastle-Ottawa Scale (NOS) criteria were used to assess the quality of studies. NOS score ≥ 6 were considered high-quality studies, otherwise, the studies were considered as low-quality.
Validation of TCGA Database
The Cancer Genome Atlas (TCGA) database was searched to further verify the relationship between XRCC1 expression and radiotherapy-related cancer prognosis. The patients were filtered out if they had not undergone radiotherapy and/or had no survival information. Overall survival was assessed using R software (version 4.0.3).
Subgroup Analysis
To reduce the effect of specific parameters (e.g. cancer type, side effects, treatment, acute/late degree and cutoff value) on the association between rs25487 and treatment/side effects, we performed subgroup analysis if there were five or more studies. When we performed subgroup analysis for cancer, we classified by the type of tissue and the primary site. In treatment response, the patients in each study were divided into two groups according to their treatment response: minor treatment response (case group) and major treatment response (control group). Major treatment response refers to a complete/partial response (complete response: disappearance of all known disease, confirmed at 4 weeks; partial response ≥50% decrease, confirmed at 4 weeks.) or grade <3 regression grade (grade 1: absence of residual cancer and extensive fibrosis, grade 2: rare residual cancer cells scattered through the fibrosis), and minor treatment response refers to stable/progressive disease (stable disease: neither partial response nor complete response criteria meet, progressive disease: ≥25% increase, no partial response or complete response, stable disease documented before increased disease, new lesion(s), or a ≥25% increase in one lesion) or a ≥3 regression grade (grade 3: increased residual cancer cells but fibrosis still predominating; grade 4: residual cancer outgrowing fibrosis, grade 5: an absence of regressive changes). We classified side effects by organ system and clinical dissection/irradiation region. Cystitis and proctitis were classified as bladder and/or rectal toxicity. Digestive system toxicity included dysphagia and radiation esophagitis. Anemia, leukocytopenia, neutropenia and thrombocytopenia were classified as hematological toxicity. Skin toxicity included radiation dermatitis and erythema. Telangiectasia, fibrosis or fat necrosis were classified as soft tissue injury.
Statistical Analysis
HRs and ORs, with their corresponding 95% CIs, were utilized to analyze the association between XRCC1 expression and prognostic indicators (OS) and treatment response, respectively. ORs and 95%CIs were also calculated to evaluate the association between XRCC1 rs25487 and treatment response/side effects. For XRCC1 rs25487 analysis, we used three genetic models to combine data: heterozygote model (GA vs GG), homozygote model (AA vs GG), and dominant model (GA+AA vs GG). A chi-squared test and Higgins’s (I2) test were used to assess heterogeneity. If I2 <50%, the fixed effect model was chosen to combine data, otherwise the random effect model would be adopted. In the sensitivity analysis, the “metaninf” module was used to investigate the influence of each individual study on the overall meta-analysis summary estimate by omitting each study in turn. Begg’s rank correlation test and Egger’s linear regression test were chosen to assess publication bias and P<0.05 was considered statistically significant. All statistical analyses were performed using STATA 14.0 software (StataCorp. 2015. Stata Statistical Software: Release 14. College Station, TX: StataCorp LP.).
Results
Literature Search and Study Characteristics
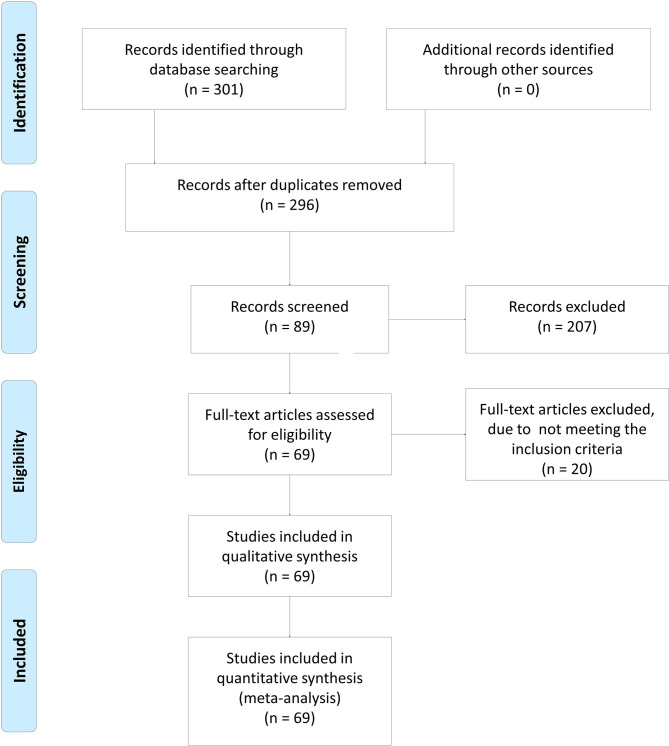
Figure 1 shows the flow diagram of the study selection process. In total, 301 articles were identified using the search strategy. Of these, 207 articles were excluded due to irrelevancy. Another five articles were excluded due to duplication. Finally, 69 articles were included in the meta-analysis (8–10, 13–15, 19–79). The articles were allocated into two parts: nine articles included XRCC1 expression and treatment response/OS data, of which seven articles reported treatment response data, six articles reported OS data, and four articles reported both treatment response and OS data. Patients in these studies were divided into high/positive and low/negative groups. 60 articles included XRCC1 rs25487 and treatment response/side effects data, in which 24 articles reported treatment response data, 40 articles reported side effects data, and four articles reported both treatment response and side effects data. Patients in these studies were divided into case and control groups. All studies scored ≥6 on the NOS, which indicated that all studies were of high quality. Tables 1 and 2 , a partial list of the characteristics of the included articles is presented. In total, 10,232 patients were included in the meta-analysis. Supplementary Table 1 shows the full characteristics of these articles.
Figure 1.
The flow diagram of study inclusion. In total, 301 articles were identified using the searching strategy. Of these, 207 articles were excluded because they do not report the association. Then, five articles were excluded due to duplication. Finally, 69 articles were included in meta-analysis. The flow diagram of study inclusion was cited from Moher et al. (18).
Table 1.
Characteristics of studies associated with XRCC1 expression and prognosis.
| Author (Ref) | Cancer | Treatment | Prognosis type | Sample size | Cutoff value | HR (95% CI) | OR (95% CI) | NOS |
|---|---|---|---|---|---|---|---|---|
| Liu et al. (58) | ESCC | RT | Treatment response | 59 | 10% | / | 0.32 (0.06–1.74) | 8 |
| Zhao and Yu (9) | NSCLC | RT | Treatment response | 62 | 50% | / | 0.34 (0.10–1.14) | 6 |
| Ang et al. (8) | HNSC | RCT+Surgery | OS | 68 | 8 score | 6.02 (2.36–15.37) | / | 8 |
| Sakano et al. (59) | Bladder cancer | RCT | Treatment response | 142 | H-score ≥1.0 | / | 0.97 (0.48–1.96) | 8 |
| Ge et al. (60) | ESCC | RCT+Surgery | OS | 44 | 2–6 scores | 1.09 (0.43–2.77) | / | 8 |
| Zheng (61) | ESCC | RT | Treatment response and OS | 76 | >4 scores | 1.48 (0.84–2.60) | 0.53 (0.19–1.47) | 7 |
| Geng (62) | Gastric cancer | RT | Treatment response and OS | 46 | NM | 1.45 (0.36–6.96) | 0.11 (0.02–0.57) | 8 |
| Huang et al. (10) | Rectal cancer | RCT+Surgery | Treatment response and OS | 86 | 50% | 1.80 (0.48–6.82) | 0.54 (0.21–1.38) | 8 |
| Zhang et al. (63) | ESCC | RT | Treatment response and OS | 76 | 2–6 scores | 1.08 (0.59–1.99) | 0.47 (0.18–1.23) | 8 |
RT, Radiotherapy; RCT, Radio-chemotherapy; OS, Overall survival; NM, Not mentioned; ESCC, Esophageal squamous cell carcinoma; NSCLC, Non-small cell lung cancer; HNSC, Head and neck squamous cancer.
Table 2.
Characteristics of studies associated with XRCC1 rs25487 and prognosis.
| Author (Ref) | Cancer | Treatment 1 | Prognosis type 2 | Number of Patients | NOS |
|---|---|---|---|---|---|
| Sakano et al. (54) | Bladder cancer | RCT | Treatment response | 72 | 8 |
| Qing-hua et al. (55) | NSCLC | RCT | Treatment response | 120 | 6 |
| Warnecke-Eberz et al. (56) | ESCA | RCT | Treatment response | 50 | 8 |
| Xu-sheng et al. (57) | ESCC | RT | Treatment response | 94 | 7 |
| Grimminger et al. (14) | Rectal cancer | RCT | Treatment response | 81 | 7 |
| Lamas et al. (15) | Rectal cancer | RCT | Treatment response | 93 | 8 |
| Balboa et al. (13) | Rectal cancer | RCT | Treatment response | 65 | 8 |
| Cecchin et al. (12) | Rectal cancer | RCT | Treatment response | 235 | 8 |
| Yoon et al. (41) | EAC | RCT | Treatment response | 60 | 6 |
| Paez et al. (72) | Rectal cancer | RCT | Treatment response | 126 | 8 |
| Zha et al. (73) | NSCLC | RCT | Treatment response | 52 | 6 |
| Huang et al. (74) | ESCC | RT | Treatment response | 150 | 8 |
| Fan et al. (75) | CESC | RT | Treatment response | 73 | 7 |
| Chen et al. (76) | NSCLC | RT | Treatment response | 60 | 8 |
| Yu et al. (77) | ESCC | RCT | Treatment response | 73 | 8 |
| Wu (49) | NPC | RT+RCT | Treatment response and side effects | 114 | 7 |
| Zhai et al. (39) | NPC | RCT | Treatment response and side effects | 60 | 8 |
| Huang et al. (78) | ESCC | RCT | Treatment response | 50 | 8 |
| Sun et al. (79) | ESCC | RT+RCT | Treatment response | 97 | 6 |
| Wang et al. (65) | NPC | RT+RCT | Treatment response and side effects | 174 | 7 |
| Zhang et al. (66) | NPC | RCT | Treatment response | 100 | 8 |
| Zhang (69) | Rectal cancer | RCT | Treatment response | 55 | 8 |
| Nicosia et al. (68) | Rectal cancer | RCT | Treatment response | 80 | 8 |
| Yang and Liu (71) | NSCLC | RT | Treatment response and side effects | 486 | 6 |
| Moullan et al. (19) | Breast cancer | RT | Side effects | 254 | 6 |
| Chang-Claude et al. (20) | Breast cancer | RT | Side effects | 446 | 7 |
| Giotopoulos et al. (21) | Breast cancer | RCT | Side effects | 82 | 6 |
| Suga et al. (22) | Breast cancer | RT | Side effects | 389 | 6 |
| Alsbeih et al. (23) | NPC | RT+RCT | Side effects | 50 | 7 |
| Burri et al. (24) | Prostate adenocarcinoma | RT+(RT+HT) | Side effects | 135 | 7 |
| Falvo et al. (25) | Breast adenocarcinoma | RCT+(RT+HT) | Side effects | 403 | 7 |
| Chang-Claude et al. (26) | Breast cancer | RT | Side effects | 403 | 7 |
| Popanda et al. (27) | Prostate cancer | RT | Side effects | 405 | 6 |
| Zschenker et al. (28) | Breast cancer | RT+RCT+(RT+HT)+(RCT+HT) | Side effects | 69 | 7 |
| Alsbeih et al. (29) | NPC | RT+RCT | Side effects | 60 | 8 |
| Mangoni et al. (30) | Breast cancer | RT+RCT | Side effects | 87 | 6 |
| Zhou et al. (31) | Breast cancer | RT | Side effects | 171 | 6 |
| Sakano et al. (32) | Bladder cancer | RCT | Side effects | 95 | 6 |
| Ishikawa et al. (33) | Cervical cancer | RT | Side effects | 208 | 8 |
| Yin et al. (34) | NPC | RT+RCT+(RT+Other) | Side effects | 165 | 8 |
| Pratesi et al. (35) | HNSC | RCT | Side effects | 101 | 6 |
| Langsenlehner et al. (36) | Prostate cancer | RT+(RT+HT) | Side effects | 575 | 8 |
| Terrazzino et al. (37) | Breast cancer | RT | Side effects | 237 | 7 |
| Yoon et al. (41) | EAC | RCT | Side effects | 60 | 7 |
| Raabe et al. (40) | Breast cancer | RT+(RCT+HT) | Side effects | 83 | 6 |
| Terrazzino et al. (38) | Breast cancer | RT+RCT+(RT+HT) +(RCT+HT) | Side effects | 285 | 6 |
| Li et al. (42) | NPC | RT+RCT | Side effects | 114 | 8 |
| Duldulao et al. (43) | Rectal cancer | RCT | Side effects | 347 | 6 |
| Tucker et al. (44) | NSCLC | RT+RCT | Side effects | 169 | 6 |
| Zhu (45) | ESCC | RT+RCT | Side effects | 182 | 7 |
| Cheuk et al. (46) | NPC | RT+RCT | Side effects | 120 | 6 |
| Venkatesh et al. (47) | Head and neck cancer | RT+RCT | Side effects | 166 | 6 |
| Alsbeih et al. (48) | NPC | RT+RCT | Side effects | 155 | 7 |
| Chen et al. (50) | NSCLC | RT | Side effects | 60 | 7 |
| Lan et al. (51) | Cervical cancer | RT | Side effects | 152 | 8 |
| Mumbrekar et al. (52) | Breast cancer | RT+RCT | Side effects | 119 | 8 |
| Smith et al. (53) | Rectal cancer | RCT | Side effects | 165 | 7 |
| Chen et al. (64) | NPC | RCT | Side effects | 114 | 7 |
| Du et al. (67) | Lung cancer | RT | Side effects | 149 | 8 |
| Xie et al. (70) | NSCLC | RCT | Side effects | 178 | 8 |
RT, radiotherapy; RCT, radio-chemotherapy; HT, hormonal therapy; NSCLC, non-small cell lung cancer; ESCA, esophageal carcinoma; ESCC, esophageal squamous cell carcinoma; EAC, esophageal adenocarcinoma; CESC, cervical squamous cell carcinoma; NPC, nasopharyngeal cancer; HNSC, head and neck squamous cell carcinoma.
Association Between XRCC1 Expression and Treatment Response/OS
Overall Analysis
Seven studies reported data showing patient treatment response. The pooled OR indicated that high XRCC1 expression was closely related to an increased risk of minor treatment response (OR = 0.52, 95% CI: 0.36–0.76, P = 0.001). A heterogeneity test showed low heterogeneity among these studies (I2 = 17.9%, P = 0.294). The association between XRCC1 and OS was assayed using data from six studies, and we found that high XRCC1 expression was significantly related to poor OS (HR = 1.67, 95% CI: 1.00–2.78, P = 0.048), However, heterogeneity was found to be relatively large (I2 = 50.2%, P = 0.074). Supplementary Files 1 and 2 show the forest plots of overall analysis of the association between XRCC1 expression and treatment response and OS, respectively.
Subgroup Analysis
In treatment response analysis, high XRCC1 expression was associated with increased risk of minor treatment response in esophageal cancer (OR = 0.46, 95% CI: 0.24–0.88, P = 0.019) and in patients receiving radiotherapy only (OR = 0.36, 95% CI: 0.21–0.62, P = 0.000). In OS analysis, we found no relation between XRCC1 expression and OS in the cancer subgroup or treatment subgroup. Supplement Files 1 and 2 show the forest plots of subgroups with regard to treatment response and OS, respectively.
Sensitivity Analysis and Publication Bias
To assess whether the combined results were affected by a single study, sensitivity analysis was performed by a single study by calculating the results when individual studies were omitted and determining if the result was within the CI. The results are shown in Supplementary Files 5 , indicating that the results were robust and reliable. To evaluate publication bias, Begg’s test and Egger’s test were conducted, and we found publication bias in the analysis between XRCC1 expression and treatment response (Begg’s test P = 0.035, Egger’s test P = 0.004). No significant publication bias was detected for OS (Begg’s test P = 0.452, Egger’s test P = 0.554). Supplementary File 8 show the funnel plots of treatment response and OS.
Validation of TCGA Data Set Results
To further explore the prognostic value of XRCC1 expression in cancer patients who received radiotherapy, we retrieved expression data for radiotherapy-related cancer prognosis in all cancer types from TCGA data set. A total of 2705 patients with 17 cancer types, consisting of digestive, respiratory, urinary, female reproductive, head and neck, neurological, urinary, and soft tissue system cancers, were included in the analysis. The patients were divided into high- and low- XRCC1 expression groups according to the median XRCC1 expression value. Meta-analysis of all the studies indicated that high XRCC1 expression may be related to poor OS; however, the result was not statistically significant (HR = 1.08, 95% CI: 0.93–1.25, P = 0.329). Supplement File 2 show the forest plots of the result. We also explored the prognostic value of XRCC1 expression in cancer patients regardless of treatment. Surprisingly, the results showed that high XRCC1 expression was significantly associated with better OS (HR = 0.88, 95% CI: 0.82–0.94, P = 0.00038), which was contrary to our results related to radiotherapy ( Figure 2 ).
Figure 2.

Overall survival of XRCC1 expression in TCAG data set.
Association Between XRCC1 rs25487 and Treatment Response/Side Effects
Overall Analysis
No significant association was found between rs25487 and treatment response ( Table 3 ). In the side effects analysis, increased risks were found to be associated with the GA genotype (OR = 1.67, 95% CI: 1.05–1.29, P = 0.004) and GA+AA genotype (OR = 1.18, 95% CI: 1.07–1.30, P = 0.001) compared with GG genotype ( Table 4 ). Supplementary Files 3 and 4 show the forest plots of overall and subgroup analyses for treatment response and side effects, respectively.
Table 3.
Overall and subgroup analysis for relation between XRCC1 rs25487 and treatment response.
| Comparison | Group | Subgroup | No. Patients | No. Study | OR | 95% CI | P value | Heterogeneity | Effect model |
|---|---|---|---|---|---|---|---|---|---|
| AA vs GG | Overall | 1396 | 19 | 0.89 | 0.53–1.49 | 0.658 | 61.0% | Random | |
| Cancer | Esophageal cancer | 345 | 6 | 0.65 | 0.15–2.73 | 0.554 | 80.9% | Random | |
| Rectal cancer | 383 | 6 | 0.63 | 0.39–1.01 | 0.056 | 0.0% | Fixed | ||
| Cutoff | SD+PD | 737 | 8 | 0.97 | 0.41–2.30 | 0.626 | 67.2% | Random | |
| Grade ≥3 | 325 | 5 | 0.52 | 0.31–0.86 | 0.012 | 0.0% | Fixed | ||
| Treatment | RCT | 688 | 12 | 0.69 | 0.47–1.01 | 0.059 | 20.2% | Fixed | |
| RT | 557 | 5 | 1.30 | 0.36–4.63 | 0.687 | 84.7% | Random | ||
| GA vs GG | Overall | 1843 | 19 | 1.14 | 0.93–1.40 | 0.221 | 19.1% | Fixed | |
| Cancer | Esophageal cancer | 375 | 6 | 0.50 | 0.30–0.83 | 0.008 | 15.5% | Fixed | |
| Rectal cancer | 537 | 6 | 1.41 | 0.99–2.01 | 0.059 | 0.0% | Fixed | ||
| Cutoff | SD+PD | 983 | 8 | 1.06 | 0.81–1.38 | 0.673 | 48.3% | Fixed | |
| Grade ≥3 | 388 | 5 | 1.20 | 0.78–1.86 | 0.407 | 6.1% | Fixed | ||
| Treatment | RCT | 945 | 12 | 1.19 | 0.90–1.59 | 0.222 | 0.0% | Fixed | |
| RT | 683 | 5 | 0.92 | 0.46–1.86 | 0.819 | 60.7% | Random | ||
| GA+AA vs GG | Overall | 2580 | 24 | 1.08 | 0.90–1.29 | 0.401 | 48.8% | Fixed | |
| Cancer | Esophageal cancer | 573 | 7 | 0.71 | 0.29–1.77 | 0.464 | 76.6% | Random | |
| Rectal cancer | 742 | 7 | 1.05 | 0.77–1.44 | 0.743 | 0.0% | Fixed | ||
| Cutoff | SD+PD | 1223 | 9 | 0.85 | 0.51–1.43 | 0.536 | 68.4% | Random | |
| Grade ≥3 | 586 | 6 | 0.85 | 0.59–1.21 | 0.360 | 0.0% | Fixed | ||
| Treatment | RCT | 1393 | 16 | 1.03 | 0.81–1.30 | 0.817 | 14.0% | Fixed | |
| RT | 963 | 6 | 0.99 | 0.46–2.14 | 0.975 | 78.4% | Random |
RCT, radio-chemotherapy; RT, radiotherapy; SD, stable disease; PD, progressive disease. The bold values mean that they are statistically significant.
Table 4.
Overall and subgroup analysis for relation between XRCC1 rs25487 and side effects.
| Comparison | Group | Subgroup | No. Patients | No. Study | OR | 95% CI | P value | Heterogeneity | Effect model |
|---|---|---|---|---|---|---|---|---|---|
| AA vs GG | Overall | 5333 | 57 | 1.05 | 0.90–1.23 | 0.525 | 44.5% | Fixed | |
| Acute/Late | Late | 1795 | 22 | 0.81 | 0.62–1.07 | 0.139 | 3.6% | Fixed | |
| Acute | 2950 | 31 | 1.29 | 0.90–1.85 | 0.168 | 53.0% | Random | ||
| Cancer | Breast cancer | 1538 | 11 | 0.85 | 0.55–1.31 | 0.462 | 53.9% | Random | |
| Head and neck cancer | 1642 | 26 | 1.04 | 0.77–1.42 | 0.780 | 16.8% | Fixed | ||
| Prostate cancer | 688 | 5 | 0.88 | 0.51–1.52 | 0.656 | 0.0% | Fixed | ||
| Non-small cell lung cancer | 799 | 9 | 1.07 | 0.57–2.00 | 0.833 | 44.5% | Fixed | ||
| Cutoff | Grade ≥2 | 3777 | 42 | 1.06 | 0.88–1.28 | 0.523 | 46.1% | Fixed | |
| Grade ≥3 | 820 | 8 | 0.98 | 0.59–1.62 | 0.943 | 46.5% | Random | ||
| Side effects | Skin toxicity | 1558 | 17 | 0.84 | 0.63–1.13 | 0.259 | 46.9% | Fixed | |
| Soft tissue injury | 803 | 8 | 0.75 | 0.51–1.11 | 0.151 | 47.4% | Fixed | ||
| Mucositis | 579 | 9 | 1.30 | 0.78–2.16 | 0.310 | 0.0% | Fixed | ||
| Treatment | RT | 2164 | 18 | 1.03 | 0.63–1.68 | 0.905 | 66.9% | Random | |
| RCT | 746 | 14 | 1.86 | 1.25–2.77 | 0.002 | 0.0% | Fixed | ||
| RT+RCT | 1383 | 16 | 0.87 | 0.60–1.26 | 0.463 | 22.1% | Fixed | ||
| GA vs GG | Overall | 7732 | 57 | 1.17 | 1.05–1.30 | 0.004 | 40.4% | Fixed | |
| Acute/Late | Late | 2660 | 22 | 1.00 | 0.83–1.20 | 0.985 | 22.4% | Fixed | |
| Acute | 4248 | 31 | 1.29 | 1.12–1.48 | 0.001 | 47.3% | Fixed | ||
| Cancer | Breast cancer | 2369 | 11 | 1.07 | 0.89–1.28 | 0.470 | 13.9% | Fixed | |
| Head and neck cancer | 2194 | 26 | 1.25 | 1.03–1.52 | 0.025 | 45.2% | Fixed | ||
| Prostate cancer | 1161 | 5 | 1.03 | 0.73–1.44 | 0.885 | 0.0% | Fixed | ||
| Non-small cell lung cancer | 1066 | 9 | 1.09 | 0.84–1.43 | 0.526 | 10.7% | Fixed | ||
| GA vs GG | Cutoff | Grade ≥2 | 5472 | 42 | 1.19 | 1.05–1.35 | 0.006 | 29.2% | Fixed |
| Grade ≥3 | 1208 | 8 | 1.47 | 0.86–2.50 | 0.162 | 63.1% | Random | ||
| Side effects | Skin toxicity | 2249 | 17 | 1.10 | 0.91–1.33 | 0.347 | 19.0% | Fixed | |
| Soft tissue injury | 1171 | 8 | 1.07 | 0.66–1.72 | 0.796 | 57.9% | Random | ||
| Mucositis | 781 | 9 | 1.70 | 1.13–2.55 | 0.011 | 31.6% | Fixed | ||
| Treatment | RT | 3198 | 18 | 1.27 | 1.08–1.49 | 0.003 | 50.0% | Fixed | |
| RCT | 1036 | 14 | 1.23 | 0.94–1.61 | 0.134 | 47.6% | Fixed | ||
| RT+RCT | 1853 | 16 | 1.11 | 0.88–1.39 | 0.390 | 43.2% | Fixed | ||
| GA+AA vs GG | Overall | 10200 | 67 | 1.18 | 1.07–1.30 | 0.001 | 48.8% | Fixed | |
| Acute/Late | Late | 3278 | 23 | 0.93 | 0.79–1.10 | 0.401 | 23.7% | Fixed | |
| Acute | 5953 | 40 | 1.42 | 1.17–1.73 | 0.000 | 50.6% | Random | ||
| Cancer | Breast cancer | 3469 | 18 | 1.03 | 0.88–1.20 | 0.738 | 9.6% | Fixed | |
| Head and neck cancer | 2526 | 26 | 1.19 | 0.99–1.43 | 0.068 | 49.9% | Fixed | ||
| Prostate cancer | 1310 | 5 | 1.00 | 0.72–1.38 | 0.985 | 0.0% | Fixed | ||
| Non-small cell lung cancer | 1298 | 9 | 1.13 | 0.89–1.45 | 0.319 | 25.8% | Fixed | ||
| Cutoff | Grade ≥2 | 6566 | 45 | 1.15 | 1.03–1.29 | 0.013 | 47.3% | Fixed | |
| Grade ≥3 | 2311 | 15 | 1.57 | 1.07–2.30 | 0.020 | 52.2% | Random | ||
| Side effects | Skin toxicity | 2719 | 19 | 1.06 | 0.89–1.27 | 0.498 | 26.7% | Fixed | |
| Soft tissue injury | 1579 | 9 | 0.92 | 0.60–1.41 | 0.706 | 61.4% | Random | ||
| Mucositis | 902 | 9 | 1.52 | 1.13–2.06 | 0.006 | 27.2% | Fixed | ||
| Treatment | RT | 3913 | 20 | 1.23 | 0.95–1.61 | 0.119 | 62.5% | Random | |
| RCT | 2163 | 21 | 1.55 | 1.25–1.92 | 0.000 | 39.7% | Fixed | ||
| RT+RCT | 2214 | 17 | 1.07 | 0.86–1.32 | 0.548 | 45.3% | Fixed |
RCT, radio-chemotherapy; RT, radiotherapy.
The bold values mean that they are statistically significant.
Meta-Analysis of Adjusted Data
Among all the included studies focusing on association between XRCC1 rs25487 and treatment response/side effects, adjusted data of side effects were available in seven studies, adjusted data of treatment response were available in two studies. Adjusted factors included age, tumor size, body mass index, adjuvant treatment, dose of radiotherapy, smoking status and so on. When we used adjusted data to analyze the association between rs25487 and side effects/treatment response, no significant association were found for both side effects analysis and treatment response analysis, which were consistent with our results when we used the crude data in corresponding studies. Supplementary Files 11 and 12 show characteristics of studies with adjusted data and the forest plots of adjusted data and crude data of overall analyses for treatment response/side effects, respectively.
Subgroup Analysis
In the treatment response analysis, the GA genotype (OR = 0.50, 95% CI: 0.30–0.83, P = 0.008) was associated with a reduced risk of minor treatment response in esophageal cancer. The AA genotype (OR = 0.52, 95% CI: 0.31–0.86, P = 0.012) was associated with a reduced risk of minor treatment response when using grade ≥3 as the cutoff value ( Table 3 ). We found that this SNP does not predict treatment response in rectal cancer, which is consistent with previous studies (16).
In the side effects analysis, an increased risk was found to be associated with the GA genotype (OR = 1.25, 95% CI: 1.03–1.52, P = 0.025) in head and neck cancer. The results further validated the results for the GA genotype (OR = 1.70, 95% CI: 1.13–2.55, P = 0.011) and GA+AA genotype (OR = 1.52, 95% CI: 1.13–2.06, P = 0.006), in terms of mucositis. Genotypes with variant alleles were associated with increased risks of side effects in patients receiving radiotherapy (GA: OR = 1.27, 95% CI: 1.08–1.49, P = 0.003) or radio-chemotherapy (AA: OR 1.86, 95% CI: 1.25–2.77, P = 0.002; GA+AA: OR = 1.55, 95% CI: 1.25–1.92, P = 0.000). GA (OR = 1.29, 95% CI: 1.12–1.48, P = 0.001) and the GA+AA genotype (OR = 1.42, 95% CI: 1.17–1.73, P = 0.000) was found to be associated with an increased risk of acute side effects. Heterozygous and mutant homozygotes were also associated with an increased risk of side effects when using Grade ≥2 or ≥3 as the cutoff value ( Table 4 ).
Sensitivity Analysis and Publication Bias
In the sensitivity analysis, we removed the studies one by one to clarify their influence on the results. The confidence intervals for all the studies indicated that the results are stable. Table 5 shows the P value for Begg’s rank correlation test and Egger’s linear response test. In addition, we found publication bias in the heterozygote and dominant models of side effects analysis ( Table 5 ). Supplementary Files 6 and 7 show the sensitivity analysis of treatment response and side effects, respectively. Supplementary Files 9 and 10 show the publication bias in treatment response and side effects, respectively.
Table 5.
P value in publication bias.
| Group | Test | GA vs GG | AA vs GG | GA+AA vs GG |
|---|---|---|---|---|
| Treatment response | Begg | 0.263 | 0.529 | 0.535 |
| Egger | 0.116 | 0.542 | 0.161 | |
| Side effects | Begg | 0.042 | 0.660 | 0.006 |
| Egger | 0.095 | 0.673 | 0.024 |
P<0.05 means publication bias is statistically significant.
The bold values mean that they are statistically significant.
Discussion
Encoded by XRCC1 (X-ray repair cross complementing 1) gene, XRCC1 plays a crucial role in the oxidative DNA damage repair through the base excision repair (BER) pathway and single-stranded break repair (SSBR) processes, after exposure to ionizing irradiation or alkylating agents (80). XRCC1 functions as a scaffold protein to assemble a complex with polymerase beta (polβ), DNA ligase III (lig III), and poly (ADP-ribose) polymerase (PARP) (81). Changes in the expression of XRCC1 can affect the ability of cells to repair DNA damage, which may influence cell radiosensitivity. Previous studies have revealed that high XRCC1 expression levels are associated with resistance to radiotherapy in patients with lung, head, and neck cancer (82). However, conclusions about the relationship between XRCC1 expression and treatment efficacy are not consistent. On the other hand, XRCC1 SNPs are also associated with radiotherapy-related outcomes (65), and one of the most common polymorphisms of XRCC1 (rs25487; Arg399Gln) can impair the repair process through a missense mutation in exon 10 (codon 399) which results in dysfunction of the binding domain of PARP or polynucleotide kinase (PNK) (83). Studies have demonstrated that the XRCC1 rs25487 polymorphism is associated with an increased risks for several malignancy types. However, its association with RT-based treatment response and RT-induced normal tissue damage has not been to consistently confirmed.
To the best of our knowledge, this is the first comprehensive meta-analysis with the largest sample size investigating both the relationship between XRCC1 expression and radiotherapy-related treatment response/overall survival and the association between the XRCC1 r25487 polymorphism and radiotherapy-related treatment response/radiation-induced side effects. The results demonstrated that high XRCC1 expression is correlated with poor treatment response. For OS, the pooled HRs showed a strong correlation between high XRCC1 expression and poor OS, indicating that high XRCC1 expression could be considered a risk factor in cancer patients receiving radiotherapy. To further validate the radiotherapy-related prognostic value of XRCC1, we performed a TCGA data review. The pooled HRs indicated that high XRCC1 expression tended to be correlated with poor OS, although the result was not statistically significant. However, interestingly, when we performed a TCGA analysis in different tumor types without considering therapeutic modalities, we found that high XRCC1 expression was significantly related to better OS, indicating that high XRCC1 expression is a protective factor in the undifferentiated treatment cancer population. Studies have demonstrated that XRCC1 facilitates efficient DNA damage processing, which is pertinent in patients undergoing radiochemotherapy. This is mitigated to a certain extent by non−specific DNA repair systems; therefore, high XRCC1 expression levels may increase the DNA repair capacity of tumor cells, leading to an increased tolerance to DNA damage induced by radiochemotherapy (7). On the other hand, studies have also suggested that XRCC1 deficiency can sensitize cells to irradiation, and this enhanced sensitivity could be attributed to increased DNA damage and increased cell cycle arrest, which might be related to an increase in DNA-PKcs and gadd153 mRNA expression (84). All these evidences may explain why high XRCC1 expression is associated with poor radiotherapy-related treatment response and OS, which was identified in our meta-analysis results. Regarding TCGA results without considering therapeutic modalities, some studies concluded that high XRCC1 expression was significantly associated with early clinical stages and nodal status (85, 86). These results suggest that XRCC1 may play its normal role and act to protect individuals. However, one of our included studies (8) did not indicate an association between these parameters. One of the reasons may be that we did not consider the cancer type, and the impact of XRCC1 may differ from cancer to cancer, which may lead to conflicting results, further investigations are needed.
No high-throughput studies (such as GWAS) were found that reported a correlation between XRCC1 rs25487 and radiotherapy-related prognosis. However, given that many single variable studies with different sample sizes that drew different conclusions about the relationship between XRCC1 r25487 and radiotherapy-related prognosis, this meta-analysis is of special importance to obtain a more robust conclusion with higher statistical power. Overall, we found an increased risk of side effects (RT-induced normal tissue damage) associated with the GA genotype and GA + AA genotype, compared to the GG genotype. Furthermore, the GA + AA genotype was found to be associated with acute and severe side effects, especially in terms of mucositis in head and neck cancer (HNC). PARP and PNK are two important enzymes in the BER pathway. The variant alleles of XRCC1 rs25487 might significantly change the affinity of the binding domain of PARR or PNK (83), which could result in a reduced efficiency of DNA damage repair processes in the acute damage phase. However, in the late damage phase, which usually takes a period of months to years for late damage to manifest itself after the end of radiotherapy. It is a far more complicated event than the acute damage phase and has still not been fully elucidated. Radiobiological studies have demonstrated that irradiation initiates a network of pro- and anti-inflammatory cytokine cascades, which are crucial for tissue regeneration and healing. However, the balance of network changes with time and space during the late damage phase. With depletion of target cells and a lack of stem cells, it ends up a failure to regenerate functional tissue, which is replaced by fibrogenesis. Our study found that the XRCC1 SNP is associated with radiation-induced acute side effects instead of late side effects in HNCs. Further research is needed to determine whether XRCC1 plays a role in the late damage phase.
The era of advanced multimodality radiotherapy requires the development of approaches for tailoring treatments to the individual. Irradiation-related genomic biomarkers can contribute by identifying sufficient genetic variants associated with a patient’s risks of radiotherapy-related toxicity and cancer prognosis. Increasing clinical trials (ClinicalTrials.gov identifier: NCT00122239; NCT02573636; NCT00099112; NCT02112162; NCT03296124) are focusing on the relationship between genes and radiotherapy-related side effects, most of the results have not been released. One of our included studies is registered on the ClinicalTrials database (identifier: NCT01316328), which indicates no relationship between XRCC1 rs25487 and radiotherapy-induced fibrosis or fat necrosis in breast cancer patients. Our meta-analysis shows no relationship between XRCC1 rs25487 and radiotherapy-induced soft tissue injury, which confirmed the clinical trial results from a broader perspective with a larger sample size. The evidence above shows that it will be valuable that referencing our meta-analysis results when using the XRCC1 gene and XRCC1 rs25487 polymorphisms as biomarkers for patients receiving radiotherapy-related treatments in clinical practice.
According to statistics, nearly 90% of patients suffer from different grades of mucositis during radiotherapy (87), which has a great detrimental impact on the radiation effect and local control rate, as well as on long-term survival. However, in the subgroup analysis, we found that the XRCC1 rs25487 GA genotype was associated with a reduced risk of minor treatment response in esophageal cancer, which indicates that it is a prognostic factor of better treatment response for people with esophageal cancer under radiotherapy-related treatment. For more than a decade, studies (88) have shown that patients with variant alleles (GA + AA) of XRCC1 rs25487 have a lower pathological complete response (pCR) rate after neoadjuvant chemoradiotherapy. The meta-analysis in the present study further confirmed this result. According to our analysis, patients with the XRCC1 GA genotype and XRCC1 AA genotype are more likely to suffer from grade II or higher acute side effects, especially mucositis. The XRCC1 SNP might be a potential biomarker to tailor individual radiotherapy. In treatment response analysis, we found that the XRCC1 rs25487 GA genotype was associated with reduced risk of minor treatment response in esophageal cancer. In addition, there was no difference in treatment-related toxicity between the RT and RCT groups.
However, some methodological issues should be taken into consideration. First, the sample size among the included studies ranged from 50 to 575, which is relatively small. Thirty-three studies had fewer than 100 patients and only five studies had more than 400 patients. Andreassen et al. reported that studies with small sample sizes were underpowered to detect SNPs with a modest impact on complication risk (89). Therefore, both the conclusions about XRCC1 expression and cancer prognosis and the association between rs25487 and treatment response/side effects should be carefully assessed. Second, most included studies about XRCC1 expression and cancer prognosis are from the Chinese population, and most included XRCC1 SNP analysis studies did not provide information on ethnicity; thus, we could not conduct further research on the association in different populations. Therefore, further studies should specify the ethnicity of the patients involved. Third, in some articles, the HR value was not provided, and we had to extract the HR value from the K-M curve, a process that may introduce errors. Furthermore, the therapeutic regimen, evaluation criteria, and cutoff values adopted in these studies were not uniform, especially the cutoff for XRCC1 expression. Only two studies classified the cutoff according to medians, and we extracted survival data in TCGA by selecting the median as the cutoff value, which may lead to our conclusions being less persuasive. To solve this problem, a unified XRCC1 cutoff value and a more detailed subgroup analysis (such as age, cancer, ethnicity etc.) are necessary. Finally, although our meta-analysis indicates a strong association between XRCC1 expression and rs25487 and radiotherapy-related side effects/treatment response, it does not necessarily mean that XRCC1 expression and rs25487 are the only factors that influenced these outcomes. The genotype-phenotype relationship shows that a gene alone can neither cause an observable phenotypic trait, nor can it be necessary and sufficient to the emergence of observable characteristics. Genes need a cellular environment, the combined action of multiple other genes, as well as certain physico-chemical conditions to have an observable effect on organisms (90). It is important to remember that phenotypes are equally, or even sometimes more greatly influenced by environmental effects than genetic effects. So, a phenotype can be directly related to a genotype, but not necessarily. Our results should be carefully interpreted, other multiple genes and SNPs, environmental effects (like age, radiation dose, radiation quality, adjuvant treatment and so on) can all be the factors that influenced side effects and treatment response. Therefore, further studies should pay greater attention to detailed treatment parameters. These efforts might contribute to bringing radiation therapy from physical precision to biological precision.
Conclusion
Our meta-analysis suggested that high XRCC1 expression is associated with increased risk of minor treatment response and poor OS. XRCC1 rs25487 is associated with reduced risk of minor radiotherapy-related/radiation-induced treatment response in esophageal cancer and increased risk of mucositis in head and neck cancer. Considering the insufficient reporting of treatment parameters and the various sample sizes for different cancer types, we suggest that genetic association studies related to radiation-based treatment should include more cancer types and patients with sufficient statistical power and more detailed clinical parameters.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.
Author Contributions
ZL designed the study. LG, ML, and RS reviewed articles and collected data. LG performed statistical analysis. LG, ML, and RS wrote the manuscript. LQ and CC revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (No.81802997), the HuBei Provincial Department of Science and Technology Innovation Group Programme (No.2019CFA034), The Free Exploration Foundation of the Hubei University of Medicine (No.FDFR201802).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2021.654784/full#supplementary-material
References
- 1. Cosh A, Carslaw H. What does a GP need to know about radiotherapy? Innovait (2014) 7(6):349–55. 10.1177/1755738013484908 [DOI] [Google Scholar]
- 2. Al-Sukhni E, Attwood K, Mattson DM, Gabriel E, Nurkin SJ. Predictors of Pathologic Complete Response Following Neoadjuvant Chemoradiotherapy for Rectal Cancer. Ann Surg Oncol (2016) 23(4):1177–86. 10.1245/s10434-015-5017-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bentzen SM, Overgaard J. Patient-to-Patient Variability in the Expression of Radiation-Induced Normal Tissue Injury. Semin Radiat Oncol (1994) 4(2):68–80. 10.1016/S1053-4296(05)80034-7 [DOI] [PubMed] [Google Scholar]
- 4. Wang M, Delasalle K, Feng L, Thomas S, Giralt S, Qazilbash M, et al. CR represents an early index of potential long survival in multiple myeloma. Bone Marrow Transplant (2010) 45(3):498–504. 10.1038/bmt.2009.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kerns SL, West CM, Andreassen CN, Barnett GC, Bentzen SM, Burnet NG, et al. Radiogenomics: the search for genetic predictors of radiotherapy response. Future Oncol (London England) (2014) 10(15):2391–406. 10.2217/fon.14.173 [DOI] [PubMed] [Google Scholar]
- 6. Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol (1998) 18(6):3563–71. 10.1128/MCB.18.6.3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horton JK, Watson M, Stefanick DF, Shaughnessy DT, Taylor JA, Wilson SH. XRCC1 and DNA polymerase beta in cellular protection against cytotoxic DNA single-strand breaks. Cell Res (2008) 18(1):48–63. 10.1038/cr.2008.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ang MK, Patel MR, Yin XY, Sundaram S, Fritchie K, Zhao N, et al. High XRCC1 protein expression is associated with poorer survival in patients with head and neck squamous cell carcinoma. Clin Cancer Res (2011) 17(20):6542–52. 10.1158/1078-0432.CCR-10-1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao Y-y, Yu Z. Association between XRCC1 , DNA-PKcs, and PCNA and radiotherapeutic effect in non-small cell lung cancer. Chin Clin Oncol (2010) 15(6):514–7. [Google Scholar]
- 10. Huang M-Y, Huang J-J, Huang C-M, Lin C-H, Tsai H-L, Huang C-W, et al. Relationship Between Expression of Proteins ERCC1, ERCC2, and XRCC1 and Clinical Outcomes in Patients with Rectal Cancer Treated with FOLFOX-Based Preoperative Chemoradiotherapy. World J Surg (2017) 41(11):2884–97. 10.1007/s00268-017-4070-z [DOI] [PubMed] [Google Scholar]
- 11. Nedooshan JJ, Yazdi MF, Neamatzadeh H, Shehneh MZ, Kargar S, Seddighi N. Genetic Association of XRCC1 Gene rs1799782, rs25487 and rs25489 Polymorphisms with Risk of Thyroid Cancer: a Systematic Review and Meta-Analysis. Asian Pac J Cancer Prev (2017) 18(1):263. 10.22034/APJCP.2017.18.1.263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cecchin E, Agostini M, Pucciarelli S, De Paoli A, Canzonieri V, Sigon R, et al. Tumor response is predicted by patient genetic profile in rectal cancer patients treated with neo-adjuvant chemo-radiotherapy. Pharmacogenomics J (2011) 11(3):214–26. 10.1038/tpj.2010.25 [DOI] [PubMed] [Google Scholar]
- 13. Balboa E, Duran G, Lamas MJ, Gomez-Caamano A, Celeiro-Munoz C, Lopez R, et al. Pharmacogenetic analysis in neoadjuvant chemoradiation for rectal cancer: high incidence of somatic mutations and their relation with response. Pharmacogenomics (2010) 11(6):747–61. 10.2217/pgs.10.51 [DOI] [PubMed] [Google Scholar]
- 14. Grimminger PP, Brabender J, Warnecke-Eberz U, Narumiya K, Wandhofer C, Drebber U, et al. XRCC1 gene polymorphism for prediction of response and prognosis in the multimodality therapy of patients with locally advanced rectal cancer. J Surg Res (2010) 164(1):e61–6. 10.1016/j.jss.2010.08.002 [DOI] [PubMed] [Google Scholar]
- 15. Lamas MJ, Duran G, Gomez A, Balboa E, Anido U, Bernardez B, et al. X-ray cross-complementing group 1 and thymidylate synthase polymorphisms might predict response to chemoradiotherapy in rectal cancer patients. Int J Radiat Oncol Biol Phys (2012) 82(1):138–44. 10.1016/j.ijrobp.2010.09.053 [DOI] [PubMed] [Google Scholar]
- 16. Guo CX, Yang GP, Pei Q, Yin JY, Tan HY, Yuan H. DNA repair gene polymorphisms do not predict response to radiotherapy-based multimodality treatment of patients with rectal cancer: a meta-analysis. Asian Pac J Cancer Prev (2015) 16(2):713–8. 10.7314/APJCP.2015.16.2.713 [DOI] [PubMed] [Google Scholar]
- 17. Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials (2007) 8:16. 10.1186/1745-6215-8-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moher D, Liberati A, Tetzlaff J, Altman DG, The PRISMA Group . Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med (2009) 6(7):e1000097. 10.1371/journal.pmed1000097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Moullan N, Cox DG, Angele S, Romestaing P, Gerard JP, Hall J. Polymorphisms in the DNA repair gene XRCC1, breast cancer risk, and response to radiotherapy. Cancer Epidemiol Biomarkers Prev (2003) 12(11 Pt 1):1168–74. 10.1023/B:CACO.0000003854.34221.a8 [DOI] [PubMed] [Google Scholar]
- 20. Chang-Claude J, Popanda O, Tan XL, Kropp S, Helmbold I, von Fournier D, et al. Association between polymorphisms in the DNA repair genes, XRCC1, APE1, and XPD and acute side effects of radiotherapy in breast cancer patients. Clin Cancer Res (2005) 11(13):4802–9. 10.1158/1078-0432.CCR-04-2657 [DOI] [PubMed] [Google Scholar]
- 21. Giotopoulos G, Symonds RP, Foweraker K, Griffin M, Peat I, Osman A, et al. The late radiotherapy normal tissue injury phenotypes of telangiectasia, fibrosis and atrophy in breast cancer patients have distinct genotype-dependent causes. Br J Cancer (2007) 96(6):1001–7. 10.1038/sj.bjc.6603637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Suga T, Ishikawa A, Kohda M, Otsuka Y, Yamada S, Yamamoto N, et al. Haplotype-based analysis of genes associated with risk of adverse skin reactions after radiotherapy in breast cancer patients. Int J Radiat Oncol Biol Phys (2007) 69(3):685–93. 10.1016/j.ijrobp.2007.06.021 [DOI] [PubMed] [Google Scholar]
- 23. Alsbeih GA, El-Sebaie MM, Al-Rajhi NM, Al-Harbi NM, Al-Hadyan KS, Al-Buhairi MH, et al. Association between XRCC1 G399A Polymorphism and Late Complications to Radiotherapy in Saudi Head and Neck Cancer Patients. J Egypt Natl Canc Inst (2008) 20(3):302–8. 10.1667/RR1219.1 [DOI] [PubMed] [Google Scholar]
- 24. Burri RJ, Stock RG, Cesaretti JA, Atencio DP, Peters S, Peters CA, et al. Association of single nucleotide polymorphisms in SOD2, XRCC1 and XRCC3 with susceptibility for the development of adverse effects resulting from radiotherapy for prostate cancer. Radiat Res (2008) 170(1):49–59. 10.1667/RR1219.1 [DOI] [PubMed] [Google Scholar]
- 25. Falvo E, Strigari L, Citro G, Giordano C, Boboc G, Fabretti F, et al. SNPs in DNA repair or oxidative stress genes and late subcutaneous fibrosis in patients following single shot partial breast irradiation. J Exp Clin Cancer Res (2012) 31(1):7. 10.1186/1756-9966-31-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chang-Claude J, Ambrosone CB, Lilla C, Kropp S, Helmbold I, von Fournier D, et al. Genetic polymorphisms in DNA repair and damage response genes and late normal tissue complications of radiotherapy for breast cancer. Br J Cancer (2009) 100(10):1680–6. 10.1038/sj.bjc.6605036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Popanda O, Marquardt JU, Chang-Claude J, Schmezer P. Genetic variation in normal tissue toxicity induced by ionizing radiation. Mutat Res (2009) 667(1-2):58–69. 10.1016/j.mrfmmm.2008.10.014 [DOI] [PubMed] [Google Scholar]
- 28. Zschenker O, Raabe A, Boeckelmann IK, Borstelmann S, Szymczak S, Wellek S, et al. Association of single nucleotide polymorphisms in ATM, GSTP1, SOD2, TGFB1, XPD and XRCC1 with clinical and cellular radiosensitivity. Radiother Oncol (2010) 97(1):26–32. 10.1016/j.radonc.2010.01.016 [DOI] [PubMed] [Google Scholar]
- 29. Alsbeih G, Al-Harbi N, Al-Hadyan K, El-Sebaie M, Al-Rajhi N. Association between normal tissue complications after radiotherapy and polymorphic variations in TGFB1 and XRCC1 genes. Radiat Res (2010) 173(4):505–11. 10.1667/RR1769.1 [DOI] [PubMed] [Google Scholar]
- 30. Mangoni M, Bisanzi S, Carozzi F, Sani C, Biti G, Livi L, et al. Association between genetic polymorphisms in the XRCC1, XRCC3, XPD, GSTM1, GSTT1, MSH2, MLH1, MSH3, and MGMT genes and radiosensitivity in breast cancer patients. Int J Radiat Oncol Biol Phys (2011) 81(1):52–8. 10.1016/j.ijrobp.2010.04.023 [DOI] [PubMed] [Google Scholar]
- 31. Zhou L, Xia J, Li H, Dai J, Hu Y. Association of XRCC1 variants with acute skin reaction after radiotherapy in breast cancer patients. Cancer Biother Radiopharm (2010) 25(6):681–5. 10.1089/cbr.2010.0811 [DOI] [PubMed] [Google Scholar]
- 32. Sakano S, Hinoda Y, Sasaki M, Wada T, Matsumoto H, Eguchi S, et al. Nucleotide excision repair gene polymorphisms may predict acute toxicity in patients treated with chemoradiotherapy for bladder cancer. Pharmacogenomics (2010) 11(10):1377–87. 10.2217/pgs.10.106 [DOI] [PubMed] [Google Scholar]
- 33. Ishikawa A, Suga T, Shoji Y, Kato S, Ohno T, Ishikawa H, et al. Genetic variants of NPAT-ATM and AURKA are associated with an early adverse reaction in the gastrointestinal tract of patients with cervical cancer treated with pelvic radiation therapy. Int J Radiat Oncol Biol Phys (2011) 81(4):1144–52. 10.1016/j.ijrobp.2010.09.012 [DOI] [PubMed] [Google Scholar]
- 34. Yin M, Liao Z, Liu Z, Wang LE, Gomez D, Komaki R, et al. Functional polymorphisms of base excision repair genes XRCC1 and APEX1 predict risk of radiation pneumonitis in patients with non-small cell lung cancer treated with definitive radiation therapy. Int J Radiat Oncol Biol Phys (2011) 81(3):e67–73. 10.1016/j.ijrobp.2010.11.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pratesi N, Mangoni M, Mancini I, Paiar F, Simi L, Livi L, et al. Association between single nucleotide polymorphisms in the XRCC1 and RAD51 genes and clinical radiosensitivity in head and neck cancer. Radiother Oncol (2011) 99(3):356–61. 10.1016/j.radonc.2011.05.062 [DOI] [PubMed] [Google Scholar]
- 36. Langsenlehner T, Renner W, Gerger A, Hofmann G, Thurner EM, Kapp KS, et al. Association between single nucleotide polymorphisms in the gene for XRCC1 and radiation-induced late toxicity in prostate cancer patients. Radiother Oncol (2011) 98(3):387–93. 10.1016/j.radonc.2011.01.021 [DOI] [PubMed] [Google Scholar]
- 37. Terrazzino S, La Mattina P, Masini L, Caltavuturo T, Gambaro G, Canonico PL, et al. Common variants of eNOS and XRCC1 genes may predict acute skin toxicity in breast cancer patients receiving radiotherapy after breast conserving surgery. Radiother Oncol (2012) 103(2):199–205. 10.1016/j.radonc.2011.12.002 [DOI] [PubMed] [Google Scholar]
- 38. Terrazzino S, La Mattina P, Gambaro G, Masini L, Franco P, Canonico PL, et al. Common variants of GSTP1, GSTA1, and TGFbeta1 are associated with the risk of radiation-induced fibrosis in breast cancer patients. Int J Radiat Oncol Biol Phys (2012) 83(2):504–11. 10.1016/j.ijrobp.2011.06.2012 [DOI] [PubMed] [Google Scholar]
- 39. Zhai XM, Hu QC, Gu K, Wang JP, Zhang JN, Wu YW. Significance of XRCC1 Codon399 polymorphisms in Chinese patients with locally advanced nasopharyngeal carcinoma treated with radiation therapy. Asia Pac J Clin Oncol (2016) 12(1):e125–32. 10.1111/ajco.12117 [DOI] [PubMed] [Google Scholar]
- 40. Raabe A, Derda K, Reuther S, Szymczak S, Borgmann K, Hoeller U, et al. Association of single nucleotide polymorphisms in the genes ATM, GSTP1, SOD2, TGFB1, XPD and XRCC1 with risk of severe erythema after breast conserving radiotherapy. Radiat Oncol (London England) (2012) 7:65. 10.1186/1748-717X-7-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yoon HH, Catalano P, Gibson MK, Skaar TC, Philips S, Montgomery EA, et al. Genetic variation in radiation and platinum pathways predicts severe acute radiation toxicity in patients with esophageal adenocarcinoma treated with cisplatin-based preoperative radiochemotherapy: results from the Eastern Cooperative Oncology Group. Cancer Chemother Pharmacol (2011) 68(4):863–70. 10.1007/s00280-011-1556-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li H, You Y, Lin C, Zheng M, Hong C, Chen J, et al. XRCC1 codon 399Gln polymorphism is associated with radiotherapy-induced acute dermatitis and mucositis in nasopharyngeal carcinoma patients. Radiat Oncol (London England) (2013) 8:31. 10.1186/1748-717X-8-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Duldulao MP, Lee W, Nelson RA, Ho J, Le M, Chen Z, et al. Gene polymorphisms predict toxicity to neoadjuvant therapy in patients with rectal cancer. Cancer (2013) 119(5):1106–12. 10.1002/cncr.27862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tucker SL, Li M, Xu T, Gomez D, Yuan X, Yu J, et al. Incorporating single-nucleotide polymorphisms into the Lyman model to improve prediction of radiation pneumonitis. Int J Radiat Oncol Biol Phys (2013) 85(1):251–7. 10.1016/j.ijrobp.2012.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu M. Associations of Functional Single Nucleotide Polymorphisms in DNA Repair Genes with Susceptibility, Radiation-induced Injury and Survival in Esophageal Cancer Patients. (2013) pp. 1–104. [Google Scholar]
- 46. Cheuk IW, Yip SP, Kwong DL, Wu VW. Association of XRCC1 and XRCC3 gene haplotypes with the development of radiation-induced fibrosis in patients with nasopharyngeal carcinoma. Mol Clin Oncol (2014) 2(4):553–8. 10.3892/mco.2014.276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Venkatesh GH, Manjunath VB, Mumbrekar KD, Negi H, Fernandes DJ, Sharan K, et al. Polymorphisms in radio-responsive genes and its association with acute toxicity among head and neck cancer patients. PloS One (2014) 9(3):e89079. 10.1371/journal.pone.0089079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Alsbeih G, El-Sebaie M, Al-Rajhi N, Al-Harbi N, Al-Hadyan K, Al-Qahtani S, et al. Among 45 variants in 11 genes, HDM2 promoter polymorphisms emerge as new candidate biomarker associated with radiation toxicity. 3 Biotech (2014) 4(2):137–48. 10.1007/s13205-013-0135-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wu M-x. Effect of the XRCC1 single nucleotide polymorphisms of radiotherapy in patients locally advanced nasopharyngeal carcinoma. (2015) pp. 1–52. [Google Scholar]
- 50. Chen W, Feng H-s, Wen J-y, Meng J-g, Zhang Y. Analysis of the Correlation Between Single Nucleotide Polymorphisms of XRCC1 and Irradia-tion-induced Injury of Patients with Non-small Cell Lung Cancer. J Clin Res (2015) 0(2):331–4 10.3969/j.issn.1671-7171.2015.02.043 [DOI] [Google Scholar]
- 51. Lan M-l, Xiao H, Yu X, Zhang Z-m, Zhang H, He X, et al. Correlation of XRCC1 Arg 399Gln polymorphism with acute irradiation injury of cervical cancer: report of 152 cases. J Third Mil Med Univ (2016) 38(9):1010–4. 10.16016/j.1000-5404.201510068 [DOI] [Google Scholar]
- 52. Mumbrekar KD, Bola Sadashiva SR, Kabekkodu SP, Fernandes DJ, Vadhiraja BM, Suga T, et al. Genetic Variants in CD44 and MAT1A Confer Susceptibility to Acute Skin Reaction in Breast Cancer Patients Undergoing Radiation Therapy. Int J Radiat Oncol Biol Phys (2017) 97(1):118–27. 10.1016/j.ijrobp.2016.09.017 [DOI] [PubMed] [Google Scholar]
- 53. Smith JJ, Wasserman I, Milgrom SA, Chow OS, Chen CT, Patil S, et al. Single Nucleotide Polymorphism TGFbeta1 R25P Correlates with Acute Toxicity during Neoadjuvant Chemoradiotherapy in Rectal Cancer Patients. Int J Radiat Oncol Biol Phys (2017) 97(5):924–30. 10.1016/j.ijrobp.2016.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sakano S, Wada T, Matsumoto H, Sugiyama S, Inoue R, Eguchi S, et al. Single nucleotide polymorphisms in DNA repair genes might be prognostic factors in muscle-invasive bladder cancer patients treated with chemoradiotherapy. Br J Cancer (2006) 95(5):561–70. 10.1038/sj.bjc.6603290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Deng Q-h, Su D, Ma S-l, Liu P, Zhang Y-m, Jiang Z-m, et al. Single nucleotide polymorphisms involved in base excision repair and treatment response in the patients with advanced non-small cell lung cancer. Cancer Res Clin (2007) 19(7):457–60. 10.3760/cma.j.issn.1006-9801.2007.07.008 [DOI] [Google Scholar]
- 56. Warnecke-Eberz U, Vallbohmer D, Alakus H, Kutting F, Lurje G, Bollschweiler E, et al. ERCC1 and XRCC1 gene polymorphisms predict response to neoadjuvant radiochemotherapy in esophageal cancer. J Gastrointest Surg (2009) 13(8):1411–21. 10.1007/s11605-009-0881-z [DOI] [PubMed] [Google Scholar]
- 57. Zhang X-s, Nasiroula A, Zhang J-r, Lv Y, GF. Association study on single nucleotide polymorphism in hOGG1, XRCC1,XRCC3 and radiosensitivity in esophageal cancer. J Xinjiang Med Univ (2010) 33(5):473–81. 10.3969/j.issn.1009-5551.2010.05.002 [DOI] [Google Scholar]
- 58. Liu S-s, Yiliyasi M, Zhang J-r, Zeng M, Ge F. A correlation study of the expression of XRCC1 and ERCC1 to the effect of radiotherapy and prognosis in esophageal squamous cell carcinoma. J Xinjiang Med Univ (2010) 33(5):477–81. 10.3969/j.issn.1009-5551.2010.05.003 [DOI] [Google Scholar]
- 59. Sakano S, Ogawa S, Yamamoto Y, Nishijima J, Miyachika Y, Matsumoto H, et al. ERCC1 and XRCC1 expression predicts survival in bladder cancer patients receiving combined trimodality therapy. Mol Clin Oncol (2013) 1(3):403–10. 10.3892/mco.2013.85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ge H, Lu Y, Chen Y, Zheng X, Wang W, Yu J. ERCC1 expression and tumor regression predict survival in esophageal squamous cell carcinoma patients receiving combined trimodality therapy. Pathol Res Pract (2014) 210(10):656–61. 10.1016/j.prp.2014.06.013 [DOI] [PubMed] [Google Scholar]
- 61. Zheng A-p. To study the correlation of the expression of COX2, XRCC1, RASSF1 and radiosensitivity in esophageal squamous-cell carcinoma. (2015) pp. 1–50. [Google Scholar]
- 62. Geng W. The molecular basis and clinical significance of CK2 inhibitor CX-4945 reversing radiation resistance in cisplatin resistant gastric cancer cells. (2016) pp. 1–102. [Google Scholar]
- 63. Zhang Y, Dong S, Xu R, Yang Y, Zheng Z, Wang X, et al. Prognostic and predictive role of COX-2, XRCC1 and RASSF1 expression in patients with esophageal squamous cell carcinoma receiving radiotherapy. Oncol Lett (2017) 13(4):2549–56. 10.3892/ol.2017.5780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Chen H, Wu M, Li G, Hua L, Chen S, Huang H. Association between XRCC1 single-nucleotide polymorphism and acute radiation reaction in patients with nasopharyngeal carcinoma. Medicine (2017) 96(44):e8202. 10.1097/MD.0000000000008202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang J, Guo C, Gong X, Ao F, Huang Y, Huang L, et al. The impacts of genetic polymorphisms in genes of base excision repair pathway on the efficacy and acute toxicities of (chemo)radiotherapy in patients with nasopharyngeal carcinoma. Oncotarget (2017) 8(45):78633–41. 10.18632/oncotarget.20203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhang H-p, Zhang D, Xu E-c, Liu Y-p, Zhou J. Relationship Between Single Nucleotide Polymorphisms of ERCC1, XRCC1 and Efficacy of Radiochemotherapy in Patients with Nasopharyngeal Cancer. J Chin Oncol (2017) 23(1):40–4. 10.11735/j.issn.1671-170X.2017.01.B008 [DOI] [Google Scholar]
- 67. Du L, Yu W, Huang X, Zhao N, Liu F, tong F, et al. GSTP1 Ile105Val polymorphism might be associated with the risk of radiation pneumonitis among lung cancer patients in Chinese population: A prospective study. J Cancer (2018) 9(4):726–35. 10.7150/jca.20643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nicosia L, Gentile G, Reverberi C, Minniti G, Valeriani M, de Sanctis V, et al. Single nucleotide polymorphism of GSTP1 and pathological complete response in locally advanced rectal cancer patients treated with neoadjuvant concomitant radiochemotherapy. Radiat Oncol J (2018) 36(3):218–26. 10.3857/roj.2018.00094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhang Y-n. Clinical analysis of XPC, ERCC1, and XRCC1 gene polymorphisms and sensitivity of neoadjuvant radiochemotherapy for rectal cancer. (2018) pp. 1–38. [Google Scholar]
- 70. Xie X, Lin SH, Welsh JW, Wei X, Jin H, Mohan R, et al. Radiation-induced lymphopenia during chemoradiation therapy for non-small cell lung cancer is linked with age, lung V5, and XRCC1 rs25487 genotypes in lymphocytes. Radiother Oncol (2020) 154:187–93. 10.1016/j.radonc.2020.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yang Z, Liu Z. Potential Functional Variants in DNA Repair Genes Are Associated with Efficacy and Toxicity of Radiotherapy in Patients with Non-Small-Cell Lung Cancer. J Oncol (2020) 2020:1–7. 10.1155/2020/3132786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Paez D, Salazar J, Pare L, Pertriz L, Targarona E, del Rio E, et al. Pharmacogenetic study in rectal cancer patients treated with preoperative chemoradiotherapy: polymorphisms in thymidylate synthase, epidermal growth factor receptor, GSTP1, and DNA repair genes. Int J Radiat Oncol Biol Phys (2011) 81(5):1319–27. 10.1016/j.ijrobp.2011.01.025 [DOI] [PubMed] [Google Scholar]
- 73. Zha Y-y, Zhang J-n, Nan X-l, Peng Q, Wang S-c, Liu K-l, et al. Correlation Between the Short-term Effect of NP Chemotherapy Regiment Combined with Radiotherapy in the Treatment of Stage III NSCLC and Single Nucleotide Polymorphism of XRCC1 Gene Codon399. Pract J Cancer (2012) 27(5):472–4. 10.3969/j.issn.1001-5930.2012.05.010 [DOI] [Google Scholar]
- 74. Huang H, Han G-h, Xu Q-m, Wang S-h, Guo T, G-c C. Association between XRCC1 and APE1 single nucleotide polymorphism and radiosensitivity in esophageal cancer. J Shanxi Med Univ (2014) 45(10):916–9. 10.13753/j.issn.1007-6611.2014.10.003 [DOI] [Google Scholar]
- 75. Fan X-m, Li K-x, Niu S-h, Fang Z-h, Jin G. Correlation of XRCC1 Polymorphism with Radiotherapy Response in Squamous Cell Carcinoma of Cervix. Tianjin Med J (2014) 42(6) 588–90. 10.3969/j.issn.0253-9896.2014.06.022 [DOI] [Google Scholar]
- 76. Chen W, Feng H-s, Wen J-y, Meng J-g, YZ. Relevance between radiation effect of patients with advanced NSCLC and single nucleotide polymorphisms of XRCC1. J Clin Pulm Med (2014) 19(11):2042–6. 10.3969/j.issn.1009-6663.2014.011.034 [DOI] [Google Scholar]
- 77. Yu X, Xiao H, Zhao B, Zhang X, Wang G. DNA repair gene ERCC1 C118T polymorphism predicts sensitivity of recurrent esophageal cancer to radiochemotherapy in a Chinese population. Thorac Cancer (2015) 6(6):741–8. 10.1111/1759-7714.12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Huang X, Liu C, Cui Y, Zhang H, Liu Y, Zhou X, et al. Association between XRCC1 and ERCC1 single-nucleotide polymorphisms and the efficacy of concurrent radiochemotherapy in patients with esophageal squamous cell carcinoma. Oncol Lett (2017) 13(2):704–14. 10.3892/ol.2016.5496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sun X-j, Zhang Q, Yu H, Ding N, Wu L, Liu Y, et al. XRCC1 polymorphisms rs25487 and rs1799782 and radiotherapy sensitivity in esophageal squamous cell carcinoma patients. Int J Clin Exp Pathol (2017) 10(6):7082–7. [Google Scholar]
- 80. Wong HK, Kim D, Hogue BA, Mcneill DR. DNA Damage Levels and Biochemical Repair Capacities Associated with XRCC1 Deficiency. Biochemistry (2005) 44(43):14335–43. 10.1021/bi051161o [DOI] [PubMed] [Google Scholar]
- 81. Petermann E, Keil C, Oei SL. Roles of DNA ligase III and XRCC1 in regulating the switch between short patch and long patch BER. DNA Repair (2006) 5(5):544–55. 10.1016/j.dnarep.2005.12.008 [DOI] [PubMed] [Google Scholar]
- 82. Vaezi A, Feldman CH, Niedernhofer LJ. ERCC1 and XRCC1 as biomarkers for lung and head and neck cancer. Pharmgenomics Pers Med (2011) 4:47–63. 10.2147/PGPM.S20317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kirk GD, Turner PC, Gong Y, Lesi OA, Mendy M, Goedert JJ, et al. Hepatocellular carcinoma and polymorphisms in carcinogen-metabolizing and DNA repair enzymes in a population with aflatoxin exposure and hepatitis B virus endemicity. Cancer Epidemiol Biomarkers Prev (2005) 14(2):373–9. 10.1158/1055-9965.EPI-04-0161 [DOI] [PubMed] [Google Scholar]
- 84. Niu Y, Zhang X, Zheng Y, Zhang R. XRCC1 deficiency increased the DNA damage induced by γ-ray in HepG2 cell: Involvement of DSB repair and cell cycle arrest. Environ Toxicol Pharmacol (2013) 36(2):311–9. 10.1016/j.etap.2013.04.009 [DOI] [PubMed] [Google Scholar]
- 85. Santana T, Sá MC, de Moura Santos E, Galvão HC, Coletta RD, Freitas RA. DNA base excision repair proteins APE-1 and XRCC-1 are overexpressed in oral tongue squamous cell carcinoma. J Oral Pathol Med (2017) 46(7):496–503. 10.1111/jop.12529 [DOI] [PubMed] [Google Scholar]
- 86. Mahjabeen I, Ali K, Zhou X, Kayani MA. Deregulation of base excision repair gene expression and enhanced proliferation in head and neck squamous cell carcinoma. Tumour Biol (2014) 35(6):5971–83. 10.1007/s13277-014-1792-5 [DOI] [PubMed] [Google Scholar]
- 87. Million RR, Parsons JT, Mendenhall WM. Effect of radiation on normal tissues in the head and neck. Bone, cartilage, and soft tissue. Front Radiat Ther Oncol (1989) 23:221–37; discussion 51-4. 10.1159/000416586 [DOI] [PubMed] [Google Scholar]
- 88. Xifeng W, Jian G, Tsung-Teh W, Swisher SG, Zhongxin L, Correa AM, et al. Genetic variations in radiation and chemotherapy drug action pathways predict clinical outcomes in esophageal cancer. J Clin Oncol (2006) 24(23):3789–98. 10.1200/JCO.2005.03.6640 [DOI] [PubMed] [Google Scholar]
- 89. Andreassen CN, Alsner J. Genetic variants and normal tissue toxicity after radiotherapy: a systematic review. Radiother Oncol (2009) 92(3):299–309. 10.1016/j.radonc.2009.06.015 [DOI] [PubMed] [Google Scholar]
- 90. Orgogozo V, Morizot B, Martin A. The differential view of genotype-phenotype relationships. Front Genet (2015) 6:179. 10.3389/fgene.2015.00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its additional files.