Abstract
Cryptosporidium spp. are common protozoan pathogens in mammals. With pet rodents being integrated into modern life, the potential roles of them in transmitting parasites to humans need assessments. In the present study, we examined the occurrence of Cryptosporidium spp. in pet rodents in Guangdong, south China. A total of 697 fecal samples were collected from 11 species of rodents in seven pet shops, one pet market and one farm. Cryptosporidium spp. were identified by PCR analysis of the small subunit rRNA gene. An overall infection rate of 36.9% (257/697) was obtained, with infection rates varying from 9.3% in chinchillas, 52.3% in guinea pigs, 57.1% in squirrels, to 68.4% in cricetid animals. Nine Cryptosporidium species and genotypes were identified, including C. wrairi (in 129 guinea pigs), C. andersoni (in 34 hamsters), C. homai (in 32 guinea pigs), Cryptosporidium hamster genotype (in 30 hamsters), C. ubiquitum (in 24 chinchillas and squirrels), C. parvum (in 2 chinchillas), Cryptosporidium ferret genotype (in 2 chipmunks), C. muris (in 1 hamster and 1 guinea pig), and Cryptosporidium chipmunk genotype V (in 1 chinchilla and 1 chipmunk). Sequence analysis of the 60 kDa glycoprotein gene identified three subtype families of C. ubiquitum, including family XIId in 15 chinchillas, XIIa in 5 chinchillas, and a new subtype family (XIIi) in 1 squirrel. The identification of C. parvum and C. ubiquitum in pet rodents suggests that these animals, especially chinchillas, could serve as reservoirs of human-pathogenic Cryptosporidium spp. Hygiene should be practiced in the rear and care of these animals, and One Health measures should be developed to reduce the occurrence of zoonotic Cryptosporidium infections due to contact with pet rodents.
Keywords: Cryptosporidium, Pet rodents, Molecular epidemiology, Zoonosis, One health
Graphical abstract
Highlights
-
•
Cryptosporidium spp. were prevalent in pet rodents in Guangdong, China.
-
•
Nine Cryptosporidium species and genotypes were identified.
-
•
Chinchillas were commonly infected with zoonotic C. ubiquitum.
-
•
The XIId subtype family of C. ubiquitum has been imported into China together with chinchillas.
-
•
One Health measures should be developed to control zoonotic cryptosporidiosi.
1. Introduction
Cryptosporidium spp. are causative agents of cryptosporidiosis, leading to a variety of gastrointestinal symptoms in humans, such as nausea, vomiting, moderate-to-severe diarrhea in persons with an intact immune system, and chronic diarrhea-associated wasting and death in neonatal and immunocompromised individuals [1]. Humans can be infected with Cryptosporidium spp. through directly contact with infected individuals or ingesting contaminated water and food [2]. The pathogens in humans can be either human or animal origin, with zoonotic transmission playing an important role in epidemiology of human cryptosporidiosis in some areas [3].
Cryptosporidium spp. have various degrees of host specificity [4]. Some Cryptosporidium species such as C. hominis, C. bovis, C. xiaoi, and C. suis have limited host ranges, being found mostly in humans, bovine animals, ovine animals, and pigs, respectively. Others such as C. parvum and C. ubiquitum are found in multiple species of mammals, including humans [4]. Because of this, each animal species is commonly infected with only a few Cryptosporidium species or genotypes. For example, humans are mostly infected with C. hominis, C. parvum, C. meleagridis, C. felis, C. canis, and C. ubiquitum, with some of them more common than others depending on hygiene levels and intensity of animal farming [4]. Differences in the distribution of Cryptosporidium species reflect variations in the significance of animals in cryptosporidiosis epidemiology [5].
Rodents are common hosts of Cryptosporidium spp. [6]. Because they have high numbers in the ecosystem and live close to the ground, rodents are frequently infected with Cryptosporidium spp. [7]. Thus far, at least 15 known Cryptosporidium species (C. parvum, C. ubiquitum, C. viatorum, C. andersoni, C. muris, C. wrairi, C. homai, C. tyzzeri, C. apodemi, C. ditrichi, C. microti, C. alticolis, C. rubeyi, C. occultus, and C. rati) and 28 genotypes (rat genotypes II-V, ferret genotype, chipmunk genotypes I-V, bamboo rat genotypes I-III, hamster genotype, squirrel genotypes I-III, muskrat genotypes I-II, apodemus genotypes I-II, vole genotypes I-VII and Brandt's vole genotype I) have been identified [[8], [9], [10], [11], [12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24]]. Therefore, rodents are considered reservoirs of some zoonotic Cryptosporidium spp. and play an important role in the ecology of zoonotic Cryptosporidium spp. due to their high numbers and wide distribution [25,26].
With close contact with humans, the role of pet rodents in the transmission of zoonotic pathogens is getting more attention [27]. In modern life, rodents are increasingly used as pets due to their cute appearance, gentle characteristics, and easy care. Some are also farmed extensively as laboratory animals and food for zoo animals, farmed reptiles, and humans. However, few studies have been conducted on the occurrence and genetic identity of Cryptosporidium spp. in pet rodents [19,28,29]. Previous studies have mostly focused on wild rodents, with a few on laboratory rodents. There is a need of data for the formulation of One Health control measures for the prevention of zoonotic cryptosporidiosis associated pet rodents.
In the present study, we have examined the occurrence of Cryptosporidium spp. in 11 species of pet rodents in Guangdong, China, and determined their species and subtype identity through sequence analysis of the small subunit (SSU) rRNA and 60 kDa glycoprotein (gp60) genes. The zoonotic potential of the Cryptosporidium spp. in these animals was assessed based on the genetic identity of the pathogens detected.
2. Material and methods
2.1. Ethics statement
The research protocol was approved by the Research Ethics Committee of the South China Agricultural University. Animal samples were collected following the guidelines of the Chinese Laboratory Animal Administration established in 2017. Fecal pellets were collected from cages with minimum handling of the sampled animals. Permissions were obtained from the animal owners before the fecal sample collection.
2.2. Sample collection
A total of 697 fecal samples were collected during October 2018 to June 2019 from 11 species of rodents in seven pet shops, one pet market, and one farm in Guangdong, southern China. In pet shops, fresh feces were collected from 280 chinchillas (Chinchilla lanigera), 43 guinea pigs (Cavia porcellus), 34 Siberian dwarf hamsters (Phodopus sungorus), 30 Syrian hamsters (Mesocricetus auratus), 3 fancy rats (Rattus norvegicus domestica), 3 Campbell's dwarf hamsters (Phodopus campbelli), 2 common degus (Octodon degus), and 1 Roborovski dwarf hamster (Phodopus roborovskii). Most rodents in these pet shops were kept in separate cages, therefore one sample was collected from each cage. In five pet shops where chinchilla samples were collected, except one of them (Pet Shop 1), the cages were cleaned and sterilized with alcohol spray daily, with fresh food and bedding materials added every morning. The hygiene condition in Pet Shop 1, however, was suboptimal, with infrequent liter replacement (every two weeks) and poor ventilation. In the pet market (Huadiwan Pet Market), fecal samples were collected from 24 Campbell's dwarf hamsters, 4 chipmunks, 3 Siberian dwarf hamsters, 2 Eurasian red squirrels (Sciurus vulgaris), and 1 Siberian flying squirrel (Pteromys volans). The pet market also sold dogs, cats, and birds in addition to pet rodents. On a guinea pig farm in Sihui City (~50 km from Guangzhou), 267 fecal samples were collected from guinea pigs of 3–4 months (n = 221) and > 1 year (n = 46). On the farm, four young animals were kept in each steel cage, while 15–20 adult animals were kept together in fenced pens. To minimize repeated sampling of the same guinea pigs, only one fresh sample was taken from each cage, while five samples of fresh fecal pellets were collected from the four corners and center of each pen. No rodents examined in the study received antiparasitic treatments and had diarrhea or other clinical signs prior to the sample collection.
The samples were collected by hands with disposable gloves, and placed in individual Ziplock bags marked with the collection date, location, animal species, age, sex, colour, and clinical characteristics. The fecal samples were stored at 4 °C in 2.5% potassium dichromate prior to DNA extraction.
2.3. DNA extraction
Fecal samples were washed three times with distilled water by centrifugation to remove potassium dichromate. Genomic DNA was extracted from ~0.5 g of the washed fecal material using the Fast DNA Spin Kit for Soil (MP Biomedical, Santa Ana, CA, USA). The DNA obtained was stored at −20 °C prior to PCR analysis.
2.4. Genotyping and subtyping of Cryptosporidium spp.
Nested PCR analysis of the SSU rRNA gene was used to detect Cryptosporidium spp. in the extract DNA [30]. Cryptosporidium bovis DNA and reagent-grade water were used as positive and negative controls, respectively. The Cryptosporidium species or genotypes present were identified by DNA sequence analysis of positive PCR products. Subtypes of the two human-pathogenic Cryptosporidium spp., C. parvum and C. ubiquitum, were determined by PCR and sequence analysis of the gp60 gene [31,32]. As novel Cryptosporidium genotypes were identified in the study, representative samples of unique Cryptosporidium genotypes identified in the study were further characterized by sequence analysis of the 70 kDa heat-shock protein (hsp70) and actin genes [33,34]. These genes together with the SSU rRNA gene are wildly used in genetic characterization of new Cryptosporidium species and genotypes [35].
2.5. Sequence analysis
PCR products were sequenced bi-directionally on an ABI 3730 instrument (Applied Biosystems, Foster City, CA, USA) by the BioSune Biotechnology Company (Shanghai, China) to determine the species and subtypes of Cryptosporidium spp. The sequences obtained were edited and assembled using ChromasPro 2.1.5.0 (http://technelysium.com.au/ChromasPro.html). Reference sequences were chosen from GenBank (https://www.ncbi.nlm.nih.gov) based on the degree of sequence identity. The DNA sequences obtained and their references were aligned using ClustalX 2.0.11 (http://clustal.org).
2.6. Phylogenetic analysis
To assess the genetic relationships among Cryptosporidium spp., maximum likelihood trees based on substitution rates calculated using the General Time Reversible model were generated using the software Mega 7 (http://www.megasoftware.net/). The reliability of cluster formation in the trees was assessed by bootstrap analysis using 1000 replicates.
2.7. Statistical analysis
The Chi-square test implemented in SPSS v.20.0 (IBM Corp., New York, NY, USA) was used to assess differences in infection rates of Cryptosporidium spp. between sampling locations or age groups. Difference with P value ≤0.05 were considered significant.
2.8. GenBank accession numbers
Representative nucleotide sequences generated in the study were submitted to GenBank under accession numbers MW521241- MW521282.
3. Results
3.1. Prevalence of Cryptosporidium spp. in pet rodents
Of the 697 fecal samples collected from pet rodents, 257 (36.9%) were positive for Cryptosporidium spp. based on PCR analysis of the SSU rRNA gene (Table 1). Syrian hamsters had the highest prevalence (86.7%) of Cryptosporidium spp., followed by Siberian dwarf hamsters (86.5%), sciurids (57.1% in chipmunks, Eurasian red squirrels, and Siberian flying squirrels), and guinea pigs (52.3%). In comparison, the Cryptosporidium infection rates were 22.2% in Campbell's dwarf hamsters and 9.3% in chinchillas. The few fancy rats and common degus sampled were negative for Cryptosporidium spp. All nine sampling locations surveyed were positive for Cryptosporidium spp.
Table 1.
Prevalence of Cryptosporidium species/genotypes in pet rodents in Guangdong Province, China.
| Host | Location | Total no. of samples | No. of positive samples | Prevalence (%) | No. and/or name of Cryptosporidium species/genotype (s) | No. and name of Cryptosporidium subtypes (s) | Zoonotic potential of Cryptosporidium spp. |
|---|---|---|---|---|---|---|---|
| Cavia porcellus (Guinea pig) | Pet shops in Guangzhou, farm in Sihui | 310 | 162 | 52.3 | C. wrairi (129), C. homai (32), C. muris (1) | – | Low |
| Chinchilla lanigera (Chinchilla) | Pet shops in Guangzhou | 280 | 26 | 9.3 | C. ubiquitum (23), C. parvum (2), Cryptosporidium chipmunk genotype V (1) |
C. ubiquitum-XIId (15), C. ubiquitum-XIIa (5) |
High |
| Phodopus sungorus (Siberian dwarf hamster) | Pet shops and market in Guangzhou | 37 | 32 | 86.5 | Cryptosporidium hamster genotype (26),C. andersoni (6) | – | Low |
| Mesocricetus auratus (Syrian hamster) | Pet shops in Guangzhou | 30 | 26 | 86.7 | C. andersoni (26) | – | Low |
| Phodopus campbelli (Campbell's dwarf hamster) | Pet shop and market in Guangzhou | 27 | 6 | 22.2 |
Cryptosporidium hamster genotype (4), C. andersoni (2) |
– | Low |
| Phodopus roborovskii (Roborovski dwarf hamster) | Pet shop in Guangzhou | 1 | 1 | 100.0 | C. muris (1) | – | Medium |
| Tamias (Chipmunk) | Pet market in Guangzhou | 4 | 3 | 75.0 |
Cryptosporidium ferret genotype (2), Cryptosporidium chipmunk genotype V (1) |
– | Low |
| Sciurus vulgaris (Eurasian red squirrel) | Pet market in Guangzhou | 2 | 0 | 0.0 | – | – | – |
| Pteromys volans (Siberian flying squirrel) | Pet market in Guangzhou | 1 | 1 | 100.0 | C. ubiquitum (1) | 66 SNPs from XIIb (1) | Medium |
| Rattus norvegicus domestica (Fancy rat) | Pet shop in Guangzhou | 3 | 0 | 0.0 | – | – | – |
| Octodon degus (Common degu) | Pet shop in Guangzhou | 2 | 0 | 0.0 | – | – | – |
| Total | 697 | 257 | 36.9 | 6 species and 3 genotypes | – |
The infection rates of Cryptosporidium spp. in guinea pigs were 58.8% on the farm examined and 11.6% in pet shops (Table 2; χ2 = 33.0, df = 1, P < 0.0001). There was also a significant difference in prevalence of Cryptosporidium spp. between young (3–4 months old) and adult (> 1 year) animals on the guinea pig farm (χ2 = 5.4, df = 1, P = 0.02). Among the five pet shops providing chinchilla samples, only two were positive for Cryptosporidium spp., with infection rates of 22.3% and 1.0% (Table 3). The overall infection rate in Pet Shop 1 was significantly higher than in Pet Shop 2 (χ2 = 23.5, df = 1, P < 0.0001).
Table 2.
Differences in the occurrence of Cryptosporidium spp. in guinea pigs between pet shops (Pet shop 3, 5, and 7) and one farm.
| Location | Age | Total no. of samples |
Cryptosporidium spp. |
|
|---|---|---|---|---|
| No. of positive | Species/genotypes (no. of samples) | |||
| Pet shops | < 1 year | 14 | 1 (7.1%) | C. homai (1) |
| > 1 year | 24 | 0 (0.0%) | ||
| unknown | 5 | 4 (80.0%) | C. wrairi (2), C. homai (2) | |
| Total | 43 | 5 (11.6%) | C. homai (3), C. wrairi (2) | |
| Farm | 3–4 month | 221 | 137 (62.0%) | C. wrairi (117), C. homai (19), C. muris (1) |
| > 1 year | 46 | 20 (43.5%) | C. wrairi (10), C. homai (10) | |
| Total | 267 | 157 (58.8%) | C. wrairi (127), C. homai (29), C. muris (1) | |
Table 3.
Occurrence of Cryptosporidium spp. in chinchillas from different pet shops in Guangdong Province.
| Location | No. of samples examined |
Cryptosporidium spp. |
|
|---|---|---|---|
| No. of positive | Species/genotypes (no. of samples) | ||
| Pet shop in Zhudao (Pet Shop 1) | 112 | 25 (22.3%) | C. ubiquitum (23), C. parvum (1), Cryptosporidium chipmunk genotype V (1) |
| Pet shop in Huadiwan (Pet Shop 2) | 105 | 1 (1.0%) | C. parvum (1) |
| Pet shop in Tianhe Square (Pet Shop 5) | 39 | 0 | |
| Pet shop in Beijing Road (Pet Shop 6) | 16 | 0 | |
| Pet shop in Guangming Square (Pet Shop 3) | 8 | 0 | |
3.2. Cryptosporidium species and genotypes in pet rodents at the SSU rRNA locus
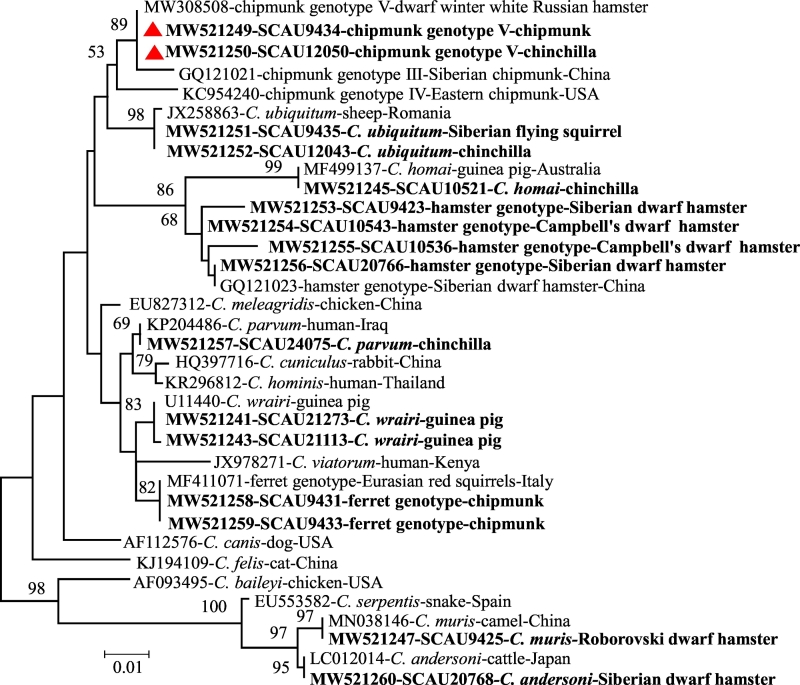
Nine known Cryptosporidium species and genotypes with different zoonotic potential were identified by sequence analysis of the SSU rRNA PCR products and phylogenetic analysis of the sequences obtained (Table 1). They included C. wrairi (129/257), C. andersoni (34/257), C. homai (32/257), Cryptosporidium hamster genotype (30/257), C. ubiquitum (24/257), C. parvum (2/257), C. muris (2/257), Cryptosporidium ferret genotype (2/257), and Cryptosporidium chipmunk genotype V. The latter was erroneously named as Cryptosporidium chipmunk genotype III (MW308508) in GenBank. As it had six nucleotide differences from the reference sequence for Cryptosporidium chipmunk genotype III (GQ121021), it was renamed as Cryptosporidium chipmunk genotype V.
Among these Cryptosporidium species and genotypes, the nucleotide sequences generated from C. andersoni, C. parvum, C. muris, Cryptosporidium ferret genotype, and Cryptosporidium chipmunk genotype V were identical to GenBank sequences LC012014 obtained from cattle, KP204486 from humans, MN038146 from camels, MF411071 from Eurasian red squirrels, and MW308508 from dwarf winter white Russian hamsters (Phodopus sungoris sungoris), respectively. In addition, the nucleotide sequences from two guinea pig-adapted species, C. wrairi and C. homai, had 0–3 and 0–2 nucleotide substitutions compared with the GenBank sequence U11440 and MF499137, respectively. Similarly, the two nucleotide sequences from C. ubiquitum had 0 and 3 nucleotide substitutions compared with the partial SSU rRNA gene sequence JX258863 obtained previously from sheep (Table 1 and Fig. 1). In contrast, among the 30 isolates positive for the hamster genotype, only one isolate generated a SSU rRNA sequence identical to the reference sequence GQ121023 obtained from Siberian dwarf hamsters; 17 generated sequences that had 1–7 nucleotide differences and another 12 generated sequences that had mixed signals in the trace files.
Fig. 1.
Phylogenetic relationships of Cryptosporidium species and genotypes found in the present study based on the maximum likelihood analysis of the partial SSU rRNA gene with substitution rates calculated using the general time reversible model. Bootstrap values above 50% from 1000 replicates are shown at the nodes. The Cryptosporidium species/genotypes identified in this study are indicated in bold, while sequences of the chipmunk genotype V are labeled with red triangles. The scale bar indicates 0.01 nucleotide substitutions per site. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3.3. Characterizations of unique Cryptosporidium genotypes at actin and hsp70 loci
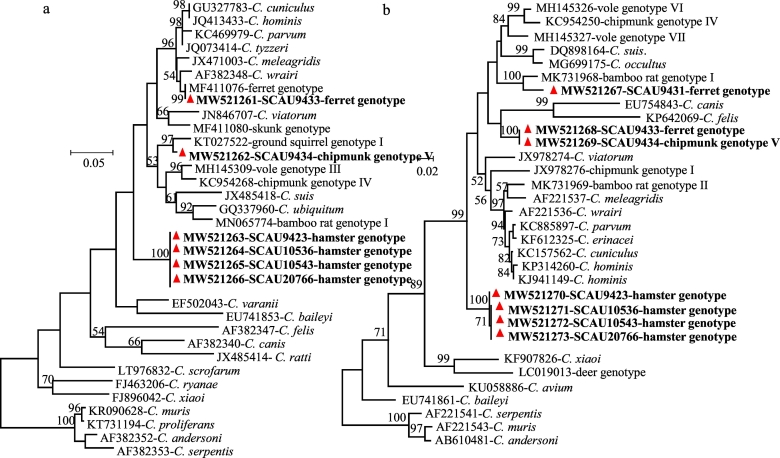
Because of the sequence differences in the Cryptosporidium hamster genotype among isolates and the identification of the chipmunk genotype V at the SSU rRNA locus, these isolates and a few other ones of related Cryptosporidium genotypes were further characterized at the actin and hsp70 loci. Among the two samples (SCAU9431 and SCAU9433) positive for the Cryptosporidium ferret genotype, sample SCAU9433 produced the expected ferret genotype sequence (MF411076) at the actin locus, while SCAU9431 was negative at the locus. These two samples, however produced different sequences at the hsp70 locus; the sequence from SCAU9431 was phylogenetically related to the Cryptosporidium bamboo rat genotype I (MK731968), while the one from SCAU9433 was similar (with 4 nucleotide substitutions) to a sequence generated from SCAU9434, which was positive for the Cryptosporidium chipmunk genotype V at the SSU rRNA locus (Fig. 2). The latter, in contrast, produced a unique nucleotide sequence (with 26 SNPs compared with KT027522 from the phylogenetically related ground squirrel genotype I) at the actin locus, supporting the identification of Cryptosporidium chipmunk genotype V.
Fig. 2.
Phylogeny of Cryptosporidium chipmunk genotype V, ferret genotype, and hamster genotype identified in the study based on the maximum likelihood analyses of actin (a) and hsp70 (b) genes. Bootstrap values greater than 50% from 1000 replicates are shown on the branches. The Cryptosporidium sequences generated in this study are indicated by bold and red triangles. The scale bar indicates 0.05 and 0.02 nucleotide substitutions per site, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Among the samples that generated SSU rRNA sequence similar or identical to the Cryptosporidium hamster genotype (Fig. 1), SCAU9423, SCAU10536, SCAU10543, and SCAU20766 generated actin and hsp70 sequences similar to each other, with 1 nucleotide substitution at the actin locus and up to 3 nucleotide substitutions at the hsp70 locus (Fig. 2).
3.4. Distribution of Cryptosporidium species and genotypes by animal species
Among the nine Cryptosporidium species and genotypes identified in the study, C. wrairi and C. homai, were found in 129 and 32 guinea pigs, respectively. Cryptosporidium andersoni was detected in 26 Syrian hamsters, 6 Siberian dwarf hamsters and 2 Campbell's dwarf hamsters. Cryptosporidium hamster genotype and sequences related to it were found in 26 Siberian dwarf hamsters and 4 Campbell's dwarf hamsters. Cryptosporidium ubiquitum was found in 23 chinchillas and 1 Siberian flying squirrel. Among the less common ones, C. parvum was detected in 2 chinchillas, C. muris in 1 Roborovski dwarf hamster and 1 guinea pig, Cryptosporidium ferret genotype in 2 chipmunks, and chipmunk genotype V in 1 chinchilla and 1 chipmunk. Therefore, most Cryptosporidium spp. were preferentially found in one group of related animals.
By animal species or groups, guinea pigs were infected with C. wrairi (129), C. homai (32), and C. muris (1); chinchillas were infected with C. ubiquitum (23), C. parvum (2), and Cryptosporidium chipmunk genotype V (1); Siberian dwarf hamsters were infected with Cryptosporidium hamster genotype (26) and C. andersoni (6); Syrian hamsters were infected with C. andersoni (26); Campbell's dwarf hamsters were infected with Cryptosporidium hamster genotype (4) and C. andersoni (2); the Roborovski dwarf hamster was infected with C. muris (1); chipmunks were infected with Cryptosporidium ferret genotype (2) and Cryptosporidium chipmunk genotype V (1); and the Siberian flying squirrel was infected with C. ubiquitum (1). Therefore, among the pet rodents sampled, only chinchillas were commonly infected with major zoonotic Cryptosporidium spp.
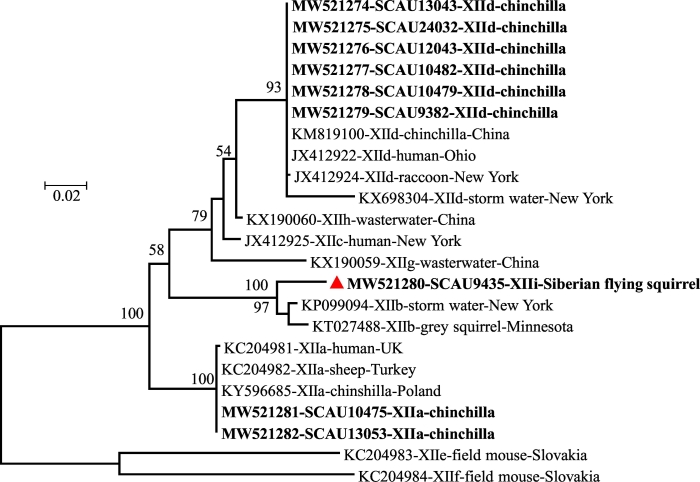
3.5. Subtypes of C. ubiquitum
The C. ubiquitum and C. parvum identified were further characterized at the gp60 locus. Among them, 5 and 15C. ubiquitum isolates from chinchillas produced sequences of the subtype families XIIa and XIId, respectively. One sequence from an isolate from a Siberian flying squirrel generated a nucleotide sequence genetically related to the XIIb subtype family but with 66 nucleotide substitutions (92% sequence identity). In a phylogenetic analysis of gp60 sequences, it clustered with XIIb with full bootstrap support (Fig. 3). This new subtype family was named as XIIi in accordance with established subtype nomenclature [32,36]. The subtype identity of the C. parvum could not be determined, as the gp60 PCR analysis of the two samples was negative, possibly due to low oocyst numbers in the samples.
Fig. 3.
Genetic relationship in the partial the 60 kDa glycoprotein gene among Cryptosporidium ubiquitum subtype families from pet rodents inferred using the maximum likelihood method. Bootstrap values above 50% from 1000 replicates are shown at the nodes. The subtype families identified in this study are indicated in bold, while the novel subtype family is labeled with a red triangle. The scale bar indicates 0.02 nucleotide substitutions per site. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
Results of the study indicate that Cryptosporidium spp. are common in pet rodents in Guangdong, China. The overall infection rate of 36.9% (257/697) is within the range of reported Cryptosporidium infection rates of 1.4% in red-bellied tree squirrels to 85.0% in guinea pigs in China [9,19].The high occurrence of Cryptosporidium spp. in guinea pigs (52.3% or 162/310) and hamsters (68.4% or 65/95) in the present study might be attributed to the poor sanitary conditions in some pet shops and on the guinea pig farm. In comparison, a much lower infection rate of Cryptosporidium spp. was seen in chinchillas (9.3% or 26/280), which were kept in a much cleaner environment. This is similar to the 10.0% (14/140) infection rate in another study in several cities in China [28]. As reported in a previous study of Cryptosporidium spp. in chinchillas [28], the infection rate in young guinea pigs was significantly higher than in old ones on the guinea pig farm in the present study.
Many factors can lead to the differences in the prevalence of Cryptosporidium spp. among animal species. Among them, animal management may be one major factor affecting infection rates; the infection rate of Cryptosporidium spp. in guinea pigs on the sampled farm (58.8% or 157/267) was significantly higher than that in pet shops (11.6% or 5/43). Poor sanitation and housing many susceptible animals together in the same environment probably had led to the high infection rate on the study farm, while the clean environment could have resulted in the lower infection rate in pet shops. This is supported by data from the five pet shops providing chinchilla samples; the infection rate (22.3% or 25/112) in Pet Shop 1, which had relatively poorer sanitary condition, was much higher than infection rates (0.0%–1.0%) in other pet shops.
A high genetic diversity is apparently present in Cryptosporidium spp. from pet rodents. In this study, nine known Cryptosporidium species/genotypes were identified. Among the groups of pet rodents examined in the present study, most appear to have different Cryptosporidium spp. For example, supporting findings in several small-scale studies, C. ubiquitum is the dominant Cryptosporidium species in chinchillas [13,28,37]. Similarly, hamsters are mostly infected with C. andersoni and Cryptosporidium hamster genotype, guinea pigs are mostly infected with C. wrairi and C. homai, while sciurids (squirrels and chipmunks) are infected with several host-adapted Cryptosporidium squirrel and chipmunk genotypes [11,16,19,20,38,39].
Some of the Cryptosporidium species found in pet rodents are known zoonotic pathogens. Cryptosporidium parvum was detected in two of the chinchillas examined. It is one of the two most common Cryptosporidium species causing human cryptosporidiosis. Previously, at least 20 species of rodents, such as rats, mice, voles, and squirrels are known to be positive for C. parvum [[9], [10], [11],19,23,26,[40], [41], [42], [43], [44], [45], [46], [47], [48]]. In China, rodents are frequently infected with this species, and the prevailing subtype family IId in rodents is also commonly found in cattle and other livestock [49].
The C. ubiquitum found in some pet rodents is another major zoonotic Cryptosporidium spp. Like C. parvum, it has a broad host range including primates, carnivores, ruminants, and various rodents such as squirrels, chipmunks, field mice, and brown rats [13,20,28,32,[37], [38], [39],47,[49], [50], [51], [52]]. In some industrialized nations such as the United States, cases of human cryptosporidiosis caused by C. ubiquitum are more than those caused by C. meleagridis, C. felis and C. canis [49]. Among pet rodents, C. ubiquitum appears to be particularly common in chinchillas, as shown in the present and previous studies in China, Japan and Poland [13,28,37].
Results of subtype analysis support the zoonotic potential of C. ubiquitum found in pet rodents. Except a Siberian flying squirrel, all rodents infected with C. ubiquitum were chinchillas. Sequence analysis of the gp60 gene indicated they belonged to XIIa and XIId subtypes. Between them, XIId appears to be common in chinchillas, being already found in several C. ubiquitum isolates examined in China and Japan [13,28,29]. In contrast, XIIa had only been found previously in two chinchillas in Poland [37]. As chinchillas are native animals of South America, the XIId found in them could be brought over from their native land. Beyond chinchillas, most infections with C. ubiquitum XIId subtypes have been found in rodents, other wild mammals, and humans in the United State, supporting the American origin of the subtype family [32]. The XIIa subtype family found in some chinchillas, in contrast, is best known to be the C. ubiquitum subtype family found in sheep, goats and some other small ruminants worldwide, and is the dominant C. ubiquitum subtype family in humans in the United Kingdom [32]. The C. ubiquitum identified in the Siberian flying squirrel, however, belongs to a novel subtype family XIIi, with unknown human-infective potential. Divergent C. ubiquitum subtype families have been found in rodents frequently in recent years [32,36].
Other zoonotic Cryptosporidium spp. identified in pet rodents in the study include C. muris and potentially C. andersoni. The former was identified in one guinea pig and hamster each, while the latter was identified in 32 hamsters. These two Cryptosporidium species are genetically related gastric parasites. Both have been reported in humans, although the human-infectivity of C. andersoni is controversial [[53], [54], [55], [56], [57], [58]]. In the present study, C. andersoni was found at much higher frequency in pet rodents than C. muris. This was probably due to the nature of the animal species examined. C. andersoni appears to be a common Cryptosporidium species in hamsters, as seen in the present and previous studies [19]. In contrast, C. muris is most found in rats, which were poorly sampled in the present study [25,48].
With increasing contact with pet rodents in modern life and increased awareness of zoonotic diseases in the era of COVID-19, residents and consumers should be educated on pet-associated zoonotic diseases, proper handling of animals and their waste, and the need for practicing sanitation and hygiene during the rear and care of these cute animals. This is especially important to immunocompromised persons, as there is no effective treatment against cryptosporidiosis. This needs the adoption of the One Health concept and the participation of veterinarians, physicians as well as pet shop owners, as there is a general lack of awareness of the risks for cryptosporidiosis associated with pet rodents [59]. The human-animal-environment interactions advocated by One Health are especially useful in the formulation of control measures against zoonotic cryptosporidiosis associated with pet rodents, as poor hygiene and over-crowding are known risk factors for Cryptosporidium infection in the present study. Governmental agencies should have better inter-agency collaborations in the inspections of pet farms and retail shops and surveillance and control of zoonotic pathogen transmission. Only the use of the One Health approach would likely reduce transmission of zoonotic Cryptosporidium spp. due to exposure to pet rodents [3,60].
5. Conclusions
Data generated from the study suggest that Cryptosporidium spp. are common in pet rodents in southern China, and some of them are known zoonotic pathogens. This is a public health concern, as rodents infected with Cryptosporidium spp. generally do not have diarrhea and other clinical signs, which might make them look harmless. The zoonotic potential of Cryptosporidium spp. in pet rodents appears to depend on the animal species involved, with different Cryptosporidium species preferentially infecting specific groups of animals, and only some being commonly infected with zoonotic Cryptosporidium species. Chinchillas are of particular concern because of the high occurrence of C. ubiquitum, which is the second most important zoonotic Cryptosporidium species in the United States and has thus far been only occasionally found in humans in developing countries [4]. The introduction of C. ubiquitum XIId subtype family into China, Japan, and European countries could be a public health concern. Europe has already witnessed the introduction of C. ubiquitum XIIb subtype family and the Cryptosporidium skunk genotype as the result of eastern grey squirrel import [39]. With the prevailing trend of keeping rodents as pets, attentions should be paid to the potential public health implications of close contact with them. One Health measures, including the development of guidelines to veterinarians and educational materials on hand wash and hygiene to pet owners, should be developed to reduce the occurrence of zoonotic Cryptosporidium infections due to contact with pet rodents.
Funding source
This work was supported by the National Natural Science Foundation of China (31820103014 and U1901208), the 111 Project (D20008), and Innovation Team Project of Guangdong University (2019KCXTD001).
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgements
We thank the pet shop, pet market and farm owners and staff for their assistance in sample collection during this study.
Contributor Information
Na Li, Email: nli@scau.edu.cn.
Yaqiong Guo, Email: guoyq@scau.edu.cn.
Una Ryan, Email: Una.Ryan@murdoch.edu.au.
Yaoyu Feng, Email: yyfeng@scau.edu.cn.
Lihua Xiao, Email: lxiao1961@gmail.com.
References
- 1.Checkley W., White A.C., Jr., Jaganath D., Arrowood M.J., Chalmers R.M., Chen X., Fayer R., Griffiths J.K., Guerrant R.L., Hedstrom L., Huston C.D., Kotloff K.L., Kang G., Mead J.R., Miller M., Petri W.A., Jr., Priest J.W., Roos D.S., Striepen B., Thompson R.C. Andrew, Ward H.D., Van Voorhis W.A., Xiao L., Zhu G., Houpt E.R. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect. Dis. 2015;15:85–94. doi: 10.1016/S1473-3099(14)70772-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zahedi A., Ryan U. Cryptosporidium - An update with an emphasis on foodborne and waterborne transmission. Res. Vet. Sci. 2020;132:500–512. doi: 10.1016/j.rvsc.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Innes E.A., Chalmers R.M., Wells B., Pawlowic M.C. A one health approach to tackle cryptosporidiosis. Trends Parasitol. 2020;36:290–303. doi: 10.1016/j.pt.2019.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng Y., Ryan U., Xiao L. Genetic diversity and population structure of Cryptosporidium. Trends Parasitol. 2018;34:997–1011. doi: 10.1016/j.pt.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 2010;124:80–89. doi: 10.1016/j.exppara.2009.03.018. [DOI] [PubMed] [Google Scholar]
- 6.Taghipour A., Olfatifar M., Foroutan M., Bahadory S., Malih N., Norouzi M. Global prevalence of Cryptosporidium infection in rodents: a systematic review and meta-analysis. Prev. Vet. Med. 2020;182:105–119. doi: 10.1016/j.prevetmed.2020.105119. [DOI] [PubMed] [Google Scholar]
- 7.Rabiee M.H., Mahmoudi A., Siahsarvie R., Krystufek B., Mostafavi E. Rodent-borne diseases and their public health importance in Iran. PLoS Negl. Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ayinmode A.B., Ogbonna N.F., Widmer G. Detection and molecular identification of Cryptosporidium species in laboratory rats (Rattus norvegicus) in Ibadan, Nigeria. Ann. Parasitol. 2017;63:105–109. doi: 10.17420/ap6302.92. [DOI] [PubMed] [Google Scholar]
- 9.Chai Y., Deng L., Liu H., Yao J., Zhong Z., Xiang L., Fu H., Shen L., Zhou Z., Deng J., Hu Y., Peng G. First detection of Cryptosporidium spp. in red-bellied tree squirrels (Callosciurus erythraeus) in China. Parasite. 2019;26:28. doi: 10.1051/parasite/2019029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Condlova S., Horcickova M., Havrdova N., Sak B., Hlaskova L., Perec-Matysiak A., Kicia M., McEvoy J., Kvac M. Diversity of Cryptosporidium spp. in Apodemus spp. in Europe. Eur. J. Protistol. 2019;69:1–13. doi: 10.1016/j.ejop.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng L., Chai Y., Luo R., Yang L., Yao J., Zhong Z., Wang W., Xiang L., Fu H., Liu H., Zhou Z., Yue C., Chen W., Peng G. Occurrence and genetic characteristics of Cryptosporidium spp. and Enterocytozoon bieneusi in pet red squirrels (Sciurus vulgaris) in China. Sci. Rep. 2016;10:1026. doi: 10.1038/s41598-020-57896-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horcickova M., Condlova S., Holubova N., Sak B., Kvetonova D., Hlaskova L., Konecny R., Sedlacek F., Clark M., Giddings C., McEvoy J., Kvac M. Diversity of Cryptosporidium in common voles and description of Cryptosporidium alticolis sp. n. and Cryptosporidium microti sp. n. Apicomplexa: CryptosporidiidaeParasitology. 2019;146:220–233. doi: 10.1017/S0031182018001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kubota R., Matsubara K., Tamukai K., Ike K., Tokiwa T. Molecular and histopathological features of Cryptosporidium ubiquitum infection in imported chinchillas Chinchilla lanigera in Japan. Parasitol. Int. 2019;68:9–13. doi: 10.1016/j.parint.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Kvac M., McEvoy J., Loudova M., Stenger B., Sak B., Kvetonova D., Ditrich O., Raskova V., Moriarty E., Rost M., Macholan M., Pialek J. Coevolution of Cryptosporidium tyzzeri and the house mouse (Mus musculus) Int. J. Parasitol. 2013;43:805–817. doi: 10.1016/j.ijpara.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kvac M., Vlnata G., Jezkova J., Horcickova M., Konecny R., Hlaskova L., McEvoy J., Sak B. Cryptosporidium occultus sp. n. (Apicomplexa: Cryptosporidiidae) in rats. Eur. J. Protistol. 2018;63:96–104. doi: 10.1016/j.ejop.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Zahedi A., Durmic Z., Gofton A.W., Kueh S., Austen J., Lawson M., Callahan L., Jardine J., Ryan U. Cryptosporidium homai n. sp. (Apicomplexa: Cryptosporidiiae) from the guinea pig (Cavia porcellus) Vet. Parasitol. 2017;245:92–101. doi: 10.1016/j.vetpar.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Zhao W., Zhou H., Huang Y., Xu L., Rao L., Wang S., Wang W., Yi Y., Zhou X., Wu Y., Ma T., Wang G., Hu X., Peng R., Yin F., Lu G. Cryptosporidium spp. in wild rats (Rattus spp.) from the Hainan Province, China: Molecular detection, species/genotype identification and implications for public health. Int. J. Parasitol.-PAR. 2019;9:317–321. doi: 10.1016/j.ijppaw.2019.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F., Zhang Z., Hu S., Zhao W., Zhao J., Kvac M., Guo Y., Li N., Feng Y., Xiao L. Common occurrence of divergent Cryptosporidium species and Cryptosporidium parvum subtypes in farmed bamboo rats (Rhizomys sinensis) Parasit. Vectors. 2020;13:149. doi: 10.1186/s13071-020-04021-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lv C., Zhang L., Wang R., Jian F., Zhang S., Ning C., Wang H., Feng C., Wang X., Ren X., Qi M., Xiao L. Cryptosporidium spp. in wild, laboratory, and pet rodents in China: prevalence and molecular characterization. Appl. Environ. Microbiol. 2009;75:7692–7699. doi: 10.1128/AEM.01386-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stenger B.L.S., Clark M.E., Kvac M., Khan E., Giddings C.W., Prediger J., McEvoy J.M. North American tree squirrels and ground squirrels with overlapping ranges host different Cryptosporidium species and genotypes. Infect. Genet. Evol. 2015;36:287–293. doi: 10.1016/j.meegid.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 21.Feng S., Chang H., Wang Y., Huang C., Han S., He H. Molecular Characterization of Cryptosporidium spp. in Brandt's Vole in China. Front. Vet. Sci. 2020;7:300. doi: 10.3389/fvets.2020.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jezkova J., Prediger J., Holubova N., Sak B., Konecny R., Feng Y., Xiao L., Rost M., McEvoy J., Kvac M. Cryptosporidium ratti n. sp. (Apicomplexa: Cryptosporidiidae) and genetic diversity of Cryptosporidium spp. in brown rats (Rattus norvegicus) in the Czech Republic. Parasitology. 2020;148:1–14. doi: 10.1017/S0031182020001833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q., Li L., Tao W., Jiang Y., Wan Q., Lin Y., Li W. Molecular investigation of Cryptosporidium in small caged pets in Northeast China: host specificity and zoonotic implications. Parasitol. Res. 2016;115:2905–2911. doi: 10.1007/s00436-016-5076-4. [DOI] [PubMed] [Google Scholar]
- 24.Li F., Zhao W., Zhang C., Guo Y., Li N., Xiao L., Feng Y. Cryptosporidium Species and C. parvum Subtypes in Farmed Bamboo Rats. Pathogens. 2020;9:1018. doi: 10.3390/pathogens9121018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng-Hublin J.S., Singleton G.R., Ryan U. Molecular characterization of Cryptosporidium spp. from wild rats and mice from rural communities in the Philippines. Infect. Genet. Evol. 2013;16:5–12. doi: 10.1016/j.meegid.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Danisova O., Valencakova A., Stanko M., Luptakova L., Hatalova E., Canady A. Rodents as a reservoir of infection caused by multiple zoonotic species/genotypes of C. parvum, C. hominis, C. suis, C. scrofarum, and the first evidence of C. muskrat genotypes I and II of rodents in Europe. Acta Trop. 2017;172:29–35. doi: 10.1016/j.actatropica.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Tumova P., Mazanek L., Lecova L., Dluhosova J., Typovska H., Kotrasova V., Tichackova V., Nohynkova E. A natural zoonotic giardiasis: infection of a child via Giardia cysts in pet chinchilla droppings. Parasitol. Int. 2018;67:759–762. doi: 10.1016/j.parint.2018.07.010. [DOI] [PubMed] [Google Scholar]
- 28.Qi M., Luo N., Wang H., Yu F., Wang R., Huang J., Zhang L. Zoonotic Cryptosporidium spp. and Enterocytozoon bieneusi in pet chinchillas (Chinchilla lanigera) in China. Parasitol. Int. 2015;64:339–341. doi: 10.1016/j.parint.2015.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Takaki Y., Takami Y., Watanabe T., Nakaya T., Murakoshi F. Molecular identification of Cryptosporidium isolates from ill exotic pet animals in Japan including a new subtype in Cryptosporidium fayeri. Vet. Parasitol. Reg. Stud. Rep. 2020;21:100430. doi: 10.1016/j.vprsr.2020.100430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiao L., Escalante L., Yang C., Sulaiman I., Escalante A.A., Montali R.J., Fayer R., Lal A.A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999;65:1578–1583. doi: 10.1128/AEM.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alves M., Xiao L., Sulaiman I., Lal A.A., Matos O., Antunes F. Subgenotype analysis of Cryptosporidium isolates from humans, cattle, and zoo ruminants in Portugal. J. Clin. Microbiol. 2003;41:2744–2747. doi: 10.1128/JCM.41.6.2744-2747.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li N., Xiao L., Alderisio K., Elwin K., Cebelinski E., Chalmers R., Santin M., Fayer R., Kvac M., Ryan U., Sak B., Stanko M., Guo Y., Wang L., Zhang L., Cai J., Roellig D., Feng Y. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg. Infect. Dis. 2014;20:217–224. doi: 10.3201/eid2002.121797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sulaiman I.M., Morgan U.M., Thompson R.C., Lal A.A., Xiao L. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microbiol. 2000;66:2385–2391. doi: 10.1128/AEM.66.6.2385-2391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sulaiman I.M., Lal A.A., Xiao L. Molecular phylogeny and evolutionary relationships of Cryptosporidium parasites at the actin locus. J. Parasitol. 2002;88:388–394. doi: 10.1645/0022-3395(2002)088[0388:MPAERO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 35.Xiao L., Fayer R., Ryan U., Upton S.J. Cryptosporidium taxonomy: recent advances and implications for public health. Clin. Microbiol. Rev. 2004;17:72–97. doi: 10.1128/cmr.17.1.72-97.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C., Hu Y., Wang L., Wang Y., Li N., Guo Y., Feng Y., Xiao L. Environmental transport of emerging human-pathogenic Cryptosporidium species and subtypes through combined sewer overflow and wastewater. Appl. Environ. Microbiol. 2017;83 doi: 10.1128/AEM.00682-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kellnerova K., Holubova N., Jandova A., Vejcik A., McEvoy J., Sak B., Kvac M. First description of Cryptosporidium ubiquitum XIIa subtype family in farmed fur animals. Eur. J. Protistol. 2017;59:108–113. doi: 10.1016/j.ejop.2017.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Feng Y., Alderisio K.A., Yang W., Blancero L.A., Kuhne W.G., Nadareski C.A., Reid M., Xiao L. Cryptosporidium genotypes in wildlife from a New York watershed. Appl. Environ. Microbiol. 2007;73:6475–6483. doi: 10.1128/AEM.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prediger J., Horcickova M., Hofmannova L., Sak B., Ferrari N., Mazzamuto M.V., Romeo C., Wauters L.A., McEvoy J., Kvac M. Native and introduced squirrels in Italy host different Cryptosporidium spp. Eur. J. Protistol. 2017;61:64–75. doi: 10.1016/j.ejop.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Condlova S., Horcickova M., Sak B., Kvetonova D., Hlaskova L., Konecny R., Stanko M., McEvoy J., Kvac M. Cryptosporidium apodemi sp. n. and Cryptosporidium ditrichi sp. n. (Apicomplexa: Cryptosporidiidae) in Apodemus spp. Eur. J. Protistol. 2018;63:1–12. doi: 10.1016/j.ejop.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 41.Liu X., Zhou X., Zhong Z., Zuo Z., Shi J., Wang Y., Qing B., Peng G. Occurrence of novel and rare subtype families of Cryptosporidium in bamboo rats (Rhizomys sinensis) in China. Vet. Parasitol. 2015;207:144–148. doi: 10.1016/j.vetpar.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 42.Mohebali M., Zarei Z., Khanaliha K., Kia E.B., Motavalli-Haghi A., Davoodi J., Rezaeian T., Tarighi F., Rezaeian M. Natural intestinal Protozoa in rodents (Rodentia: Gerbillinae, Murinae, Cricetinae) in Northwestern Iran. Iran. J. Parasitol. 2017;12:382–388. [PMC free article] [PubMed] [Google Scholar]
- 43.Montecino-Latorre D., Li X., Xiao C., Atwill E.R. Elevation and vegetation determine Cryptosporidium oocyst shedding by yellow-bellied marmots (Marmota flaviventris) in the Sierra Nevada Mountains. Int. J. Parasitol.-PAR. 2015;4:171–177. doi: 10.1016/j.ijppaw.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perec-Matysiak A., Bunkowska-Gawlik K., Zalesny G., Hildebrand J. Small rodents as reservoirs of Cryptosporidium spp. and Giardia spp. in south-western Poland. Ann. Agric. Environ. Med. 2015;22:1–5. doi: 10.5604/12321966.1141359. [DOI] [PubMed] [Google Scholar]
- 45.Saki J., Foroutan-Rad M., Asadpouri R. Molecular characterization of Cryptosporidium spp. in wild rodents of Southwestern Iran using 18s rRNA gene nested-PCR-RFLP and sequencing techniques. J. Trop. Med. 2016 doi: 10.1155/2016/6834206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei Z., Liu Q., Zhao W., Jiang X., Zhang Y., Zhao A., Jing B., Lu G., Qi M. Prevalence and diversity of Cryptosporidium spp. in bamboo rats (Rhizomys sinensis) in South Central China. Int. J. Parasitol.-PAR. 2019;9:312–316. doi: 10.1016/j.ijppaw.2019.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang X., Jian Y., Li X., Ma L., Karanis G., Karanis P. The first report of Cryptosporidium spp. in Microtus fuscus (Qinghai vole) and Ochotona curzoniae (wild plateau pika) in the Qinghai-Tibetan plateau area, China. Parasitol. Res. 2018;117:1401–1407. doi: 10.1007/s00436-018-5827-5. [DOI] [PubMed] [Google Scholar]
- 48.Zhao Z., Wang R., Zhao W., Qi M., Zhao J., Zhang L., Li J., Liu A. Genotyping and subtyping of Giardia and Cryptosporidium isolates from commensal rodents in China. Parasitology. 2015;142:800–806. doi: 10.1017/S0031182014001929. [DOI] [PubMed] [Google Scholar]
- 49.Feng Y., Xiao L. Molecular epidemiology of cryptosporidiosis in China. Front. Microbiol. 2017;8:1701. doi: 10.3389/fmicb.2017.01701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murakoshi F., Fukuda Y., Matsubara R., Kato Y., Sato R., Sasaki T., Tada C., Nakai Y. Detection and genotyping of Cryptosporidium spp. in large Japanese field mice, Apodemus speciosus. Vet. Parasitol. 2013;196:184–188. doi: 10.1016/j.vetpar.2013.02.011. [DOI] [PubMed] [Google Scholar]
- 51.Song J., Kim C.Y., Chang S.N., Abdelkader T.S., Han J., Kim T.H., Oh H., Lee J.M., Kim D.S., Kim J.T., Oh H.S., Hur M., Suh J.H., Park J.H. Detection and molecular characterization of Cryptosporidium spp. from wild rodents and insectivores in South Korea. Korean J. Parasitol. 2015;53:737–743. doi: 10.3347/kjp.2015.53.6.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao W., Wang J., Ren G., Yang Z., Yang F., Zhang W., Xu Y., Liu A., Ling H. Molecular characterizations of Cryptosporidium spp. and Enterocytozoon bieneusi in brown rats (Rattus norvegicus) from Heilongjiang Province, China. Parasit. Vectors. 2018;11:313. doi: 10.1186/s13071-018-2892-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waldron L.S., Dimeski B., Beggs P.J., Ferrari B.C., Power M.L. Molecular epidemiology, spatiotemporal analysis, and ecology of sporadic human cryptosporidiosis in Australia. Appl. Environ. Microbiol. 2011;77:7757–7765. doi: 10.1128/AEM.00615-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang Y., Ren J., Yuan Z., Liu A., Zhao H., Liu H., Chu L., Pan W., Cao J., Lin Y., Shen Y. Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu Province, China. BMC Infect. Dis. 2014;14:555. doi: 10.1186/s12879-014-0555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hussain G., Roychoudhury S., Singha B., Paul J. Incidence of Cryptosporidium andersoni in diarrheal patients from southern Assam, India: a molecular approach. Eur. J. Clin. Microbiol. Infect. Dis. 2017;36:1023–1032. doi: 10.1007/s10096-016-2887-2. [DOI] [PubMed] [Google Scholar]
- 56.Higuera A., Villamizar X., Herrera G., Giraldo J.C., Vasquez-A L.R., Urbano P., Villalobos O., Tovar C., Ramirez J.D. Molecular detection and genotyping of intestinal protozoa from different biogeographical regions of Colombia. PeerJ. 2020;8 doi: 10.7717/peerj.8554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petrincova A., Valencakova A., Luptakova L., Ondriska F., Kalinova J., Halanova M., Danisova O., Jarcuska P. Molecular characterization and first report of Cryptosporidium genotypes in human population in the Slovak Republic. Electrophoresis. 2015;36:2925–2930. doi: 10.1002/elps.201500230. [DOI] [PubMed] [Google Scholar]
- 58.Chappell C.L., Okhuysen P.C., Langer-Curry R.C., Lupo P.J., Widmer G., Tzipori S. Cryptosporidium muris: infectivity and illness in healthy adult volunteers. Am. J. Trop. Med. Hyg. 2015;92:50–55. doi: 10.4269/ajtmh.14-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abe N., Matsubara K. Molecular identification of Cryptosporidium isolates from exotic pet animals in Japan. Vet. Parasitol. 2015;209:254–257. doi: 10.1016/j.vetpar.2015.02.035. [DOI] [PubMed] [Google Scholar]
- 60.Ryan U., Zahedi A., Paparini A. Cryptosporidium in humans and animals-a one health approach to prophylaxis. Parasite Immunol. 2016;38:535–547. doi: 10.1111/pim.12350. [DOI] [PubMed] [Google Scholar]