Abstract
Despite studies providing insight into the neurobiology of chronic stress, depression and anxiety, long noncoding RNA (lncRNA)-mediated mechanisms underlying the common and distinct pathophysiology of these stress-induced disorders remain nonconclusive. In a previous study, we used the chronic mild stress paradigm to separate depression-susceptible, anxiety-susceptible and insusceptible rat subpopulations. In the current study, lncRNA and messenger RNA (mRNA) expression was comparatively profiled in the hippocampus of the three stress groups using microarray technology. Groupwise comparisons identified distinct sets of lncRNAs and mRNAs associated with the three different behavioral phenotypes of the stressed rats. To investigate the regulatory roles of the dysregulated lncRNAs upon mRNA expression, correlations between the differential lncRNAs and mRNAs were first analyzed by combined use of weighted gene coexpression network analysis and ceRNA theory-based methods. Subsequent functional analysis of strongly correlated mRNAs indicated that the dysregulated lncRNAs were involved in various biological pathways and processes to specifically induce rat susceptibility or resiliency to depression or anxiety. Further intersectional analysis of phenotype-associated and drug-associated lncRNA-mRNA networks and subnetworks assisted in identifying 16 hub lncRNAs as potential targets of anti-depression/anxiety drugs. Collectively, our study established the molecular basis for understanding the similarities and differences in pathophysiological mechanisms underlying stress-induced depression or anxiety and stress resiliency, revealing several important lncRNAs that represent potentially new therapeutic drug targets for depression and anxiety disorders.
Keywords: Depression, Anxiety, Chronic mild stress, lncRNA, Drug target
Highlights
-
•
LncRNAs/mRNAs and pathways related to three CMS-induced phenotypes were identified.
-
•
Specific and common molecular basis of three CMS-induced phenotypes was provided.
-
•
Phenotype- and drug-associated lncRNA-mRNA networks were intersectionally applied.
-
•
16 hub lncRNAs as potential targets of anti-depression/anxiety drugs were revealed.
1. Introduction
Depression and anxiety are two common and chronic psychiatric diseases, having a significant impact on the socio-occupational well-being of patients, family, and society (Almeida et al., 2012; Hamilton et al., 2015; Larson et al., 2007). The core symptoms of depression and anxiety disorders are differentially exhibited but frequently coexist clinically (Brodbeck et al., 2011; Melton et al., 2016). Due to their considerable comorbidity and pathophysiological overlap, results in most basic and clinical studies are frequently mixed (Chiba et al., 2012; Yun et al., 2016), potentially leading to our obscure understanding of the factors moderating these two disorders. Common and specific features in the neural system are still largely unknown, even though some recent studies have slowly begun to separately investigate noncomorbid subjects with these two disorders (Chen et al., 2018; Frick, 2017; Lotan et al., 2014; Zhao et al., 2017).
Mounting evidence indicates that depression and anxiety disorders share several putative risk factors, such as life event stress and chronic stress (Leuner and Shors, 2013; Mathew et al., 2011; Pittenger and Duman, 2008). In a given society, chronic stressful life events have been proposed as adverse environmental factors underlying the etiologies of depressive and anxiety disorders (Chang and Grace, 2014; Yun et al., 2016). However, many individuals exposed to chronic stressful events do not display depressive or anxious symptoms (Henningsen et al., 2012; Krishnan et al., 2007; Russo et al., 2012; Uchida et al., 2011). To model some of the environmental factors affecting humans, chronic mild stress (CMS) is commonly employed in rodents (Chang and Grace, 2014; Henningsen et al., 2012). To explore gene-environment interactions in these disorders, genetic and epigenetic dysregulations in adult rats exposed to CMS is generally investigated to gain insight into the biological basis of stress-induced behavioral variations (Argentieri et al., 2017; Golden et al., 2013; Kang et al., 2012; Krishnan et al., 2007; Pena and Nestler, 2018).
At the epigenetic level, mRNA transcription, mRNA post-transcription, and long noncoding RNAs (lncRNAs) are critical players in regulating coding gene expression (Yang et al., 2016). Although lncRNAs do not code for proteins, they are annotated due to the presence of a cryptic open reading frame. With the advancement of biomedical research on lncRNAs, increasing evidence indicates that lncRNAs participate in various critical events, such as allosterically mediating enzymatic activity, impacting chromosome conformation, and genomic imprinting (Quinn and Chang, 2016). Moreover, mutations and dysregulation of lncRNA expression have been linked with various developmental processes and disease pathogenesis (Huang et al., 2017; Sunwoo et al., 2016; Yang et al., 2016). Interestingly, lncRNAs are highly expressed in the brain and contribute to key neuronal functions, including neurogenesis, brain patterning, synaptic efficiency, and neural plasticity (Yang et al., 2016). In this vein, lncRNAs may play powerful regulatory roles in CMS-associated depression and anxiety pathologies (Huang et al., 2017; Spadaro et al., 2015), potentially representing a new class of therapeutic targets that can be exploited for disease treatment (Qureshi and Mehler, 2013). Therefore, investigating lncRNA-directed regulatory networks should contribute to our understanding of the mechanisms underlying susceptibility and resilience to depression or anxiety and to drug target discovery (Matsui and Corey, 2017).
The hippocampus is well-known as a malleable brain region with respect to stress stimulation and is closely related with learning and memory, as well as emotions. Chronic stress has been shown to detrimentally affect hippocampal neurogenesis and neuroplasticity, consequently influencing learning and memory abilities in depression and anxiety (Bannerman et al., 2014; Malykhin and Coupland, 2015). Previously, we performed quantitative proteomics analysis to explore common and distinct hippocampal proteins associated with depressive and anxiety phenotypes in the CMS rat model (Tang et al., 2019). This model distinguishes between depression-susceptible, anxiety-susceptible, and insusceptible subpopulations, representing the three different responses to CMS (Tang et al., 2019). In the present study, hippocampal tissues from the same batch of CMS rats from the previous publication were used to explore dysregulated lncRNAs that may contribute to chronic stress-induced depression or anxiety. Hippocampal lncRNA and mRNA profiles were comparatively assessed in depression-susceptible, anxiety-susceptible and insusceptible phenotypes, providing a unique chance to decipher the molecular profiles associated with susceptibility or resilience to CMS. To predict the potential functions of these dysregulated lncRNAs, strongly correlated differentially expressed mRNAs were subjected to gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis using the OmicsBean tool. By coexpression analysis, phenotype-associated and drug-associated lncRNA-mRNA networks and subnetworks were constructed to provide new insight into disease pathophysiologies and drug targets. Further integrated analysis of the subnetworks revealed that sixteen hub lncRNAs may represent important new therapeutic drug targets for depression and anxiety disorders.
2. Materials and methods
2.1. Animals
Animals used for collecting the brain samples represented three different behavioral phenotypes: (1) CMS-induced depression-susceptible (assessed in the sucrose preference test (SPT) and the forced swimming test (FST)); (2) CMS-induced anxiety-susceptible (assessed in the elevated plus maze test (EMT)); and (3) CMS-introduced but stress-insusceptible. Additional non-CMS-introduced group served as a negative control. Finally the four groups were obtained with five rats in each group. For a more detailed description, please refer to our previous work (Tang et al., 2019). The animal protocols of this study were approved by the Ethics Committee of Chongqing Medical University, and animals were cared for in accordance with the National Institutes of Health protocols for the use and care of laboratory animals (2017013).
2.2. Collection of tissues and extraction of RNA
After the tests, animals were decapitated, and brains were removed and dissected on ice. Hippocampal tissues were separated, weighed, rapidly frozen in liquid N2 and stored at a temperature of −80 °C. From each group, the tissues of four rats were randomly selected for the following microarray analysis. Total RNA was extracted from tissues with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. Moreover, we measured RNA quality and quantity with a NanoDrop ND-1000. Then, we evaluated RNA integrity using standard electrophoresis.
2.3. Microarray analysis
Arraystar profiling (Rockville, USA) was employed for expression profiling of protein-coding mRNAs and lncRNAs. We selected approximately 9000 lncRNAs from the NCBI RefSeq databases. We performed microarray analysis and sample labeling in accordance with the Agilent One-Color Microarray-Based Gene Expression Analysis protocol. Briefly, after removing rRNA, we performed purification of mRNA using mRNA-ONLY™. Afterward, we transcribed and amplified the samples into fluorescent cDNAs (Arraystar Flash RNA Labeling Kit, Arraystar). A random priming method was used to evaluate transcript length without 3’ bias. Using the Qiagen RNeasy Mini Kit, we purified the labeled cDNAs. With the NanoDrop ND-1000 tool, we examined the specific activity and the concentration of labeled cDNAs (pmol Cy3/μg cDNA). By adding 1 μl of 25 × fragmentation buffer, as well as 5 μl of 10 × blocking agents, we fragmented 1 μg of each labeled cDNA. Moreover, heating of the mixture was performed at 60 °C for 30 min. Afterward, dilution was performed by mixing the labeled cDNAs with 25 μl H2O, which was then mixed with GE hybridization buffer. Then, we dispensed the hybridization solution (50 μl) into the gasket slide, which was later assembled onto lncRNA expression microarray slides. The slides were subjected to 17-h incubation at 65 °C in a hybridization oven (Agilent Technologies). Using the Agilent DNA Microarray Scanner (G2505C), hybridized arrays were scanned, washed, and fixed.
2.4. Microarray data analysis
Extraction of raw data and analysis of the array images was performed using Feature Extraction software (Agilent version 11.0.1.1). Moreover, we performed quantile normalization using the GeneSpring GX v12.1 software package (Agilent Technologies). Differential expression analysis of normalized signal data was performed using the Limma software package. A log10-fold-change of unity and a false discovery rate (FDR) of less than 0.05 was used as a threshold. For differentially expressed genes, we performed a hierarchical cluster analysis to visualize the data (TreeView).
2.5. Correlation analysis of lncRNAs and mRNAs
Weighted correlation network analysis (WGCNA) assists in building a gene network based on coexpression relationships that helps to identify gene modules that are tightly coordinated across entire datasets (Alaei et al., 2018; Ren et al., 2015). We computed pairwise correlations between mRNA and lncRNA. The power function was used to analyze the topological overlap between lncRNAs and mRNAs, serving as a measure to evaluate neighborhood sharing or connection strength.
Meanwhile, ceRNA analysis was employed to identify mRNAs and lncRNAs that share biologically meaningful correlations (Zhang et al., 2018). Moreover, we searched for possible miRNA response elements in the mRNAs and lncRNAs. To predict an interaction between lncRNA, miRNA, and mRNA, we overlapped miRNA binding sites on both mRNAs and lncRNAs. Interactions of lncRNAs and mRNAs with miRNAs were predicted using miRNA target prediction software based on RNAhybird and miRanda. Finally, data obtained from the two methods were combined, and the overlapping sets revealed the relatively stronger correlations between lncRNAs and mRNAs.
2.6. Analysis of anti-depression/anxiety drug targets
To identify anti-depression and anti-anxiety drugs and their target genes, we searched the DrugBank database (Version 5.1.4) (https://www.drugbank.ca/). Information, including drug name, DrugBank ID and target genes, was collected in this study. The identified genes were considered primary targets of the drugs. For further analysis, species type for these genes was converted from homo sapiens to rattus norvegicus. Subsequently, interactions of the primary target genes were determined from six interaction databases: I2D, APID, Mentha, IntAct, BioGRID and MatrixDB. For each gene, interactions from all six databases were then combined to derive a universal gene set. To obtain reliably interacting genes, the network link detection Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) was used to analyze interactions between primary target genes and their interacting genes in a further step. Network analysis was conducted under the guidance of the medium confidence (STRING score = 0.4) and seven linkage criteria. In the generated network, genes directly interacting with primary target genes were considered secondary targets of the drugs.
2.7. Functional and network analysis
For bioinformatics analysis, the multiomics tool OmicsBean (http://www.omicsbean.cn) was used to identify enrichment of cellular component (CC), biological process (BP), and molecular function (MF) based on Gene Ontology (GO) categories (Tang et al., 2019; Xie et al., 2018). KEGG analysis was used to identify significantly enriched pathways, where p < 0.01 was considered significant. Furthermore, lncRNAs and their strongly correlated mRNAs were selected to develop gene expression networks. Pearson's correlation coefficients of greater than 0.99 were adopted for network construction. Calculating the degree of connectivity as previously described (Wang et al., 2019b), hub genes in the networks were identified using CytoHubba, a plugin in Cytoscape software (Version 3.7.1). According to node degree, the top 30 hub genes were displayed in the Cytoscape software.
3. Results
3.1. Identification of differentially expressed lncRNAs and mRNAs of the CMS model rats
In the present study, samples used were from the same batch of CMS rats in our previous study (Tang et al., 2019). Based on the behavioral assessments of SPT, FST and EMT, a subset of control, depression-susceptible, anxiety-susceptible, and insusceptible mice was obtained. The CMS paradigm may represent an effective tool for analyzing unique and common neural characteristics of noncomorbid depression and anxiety.
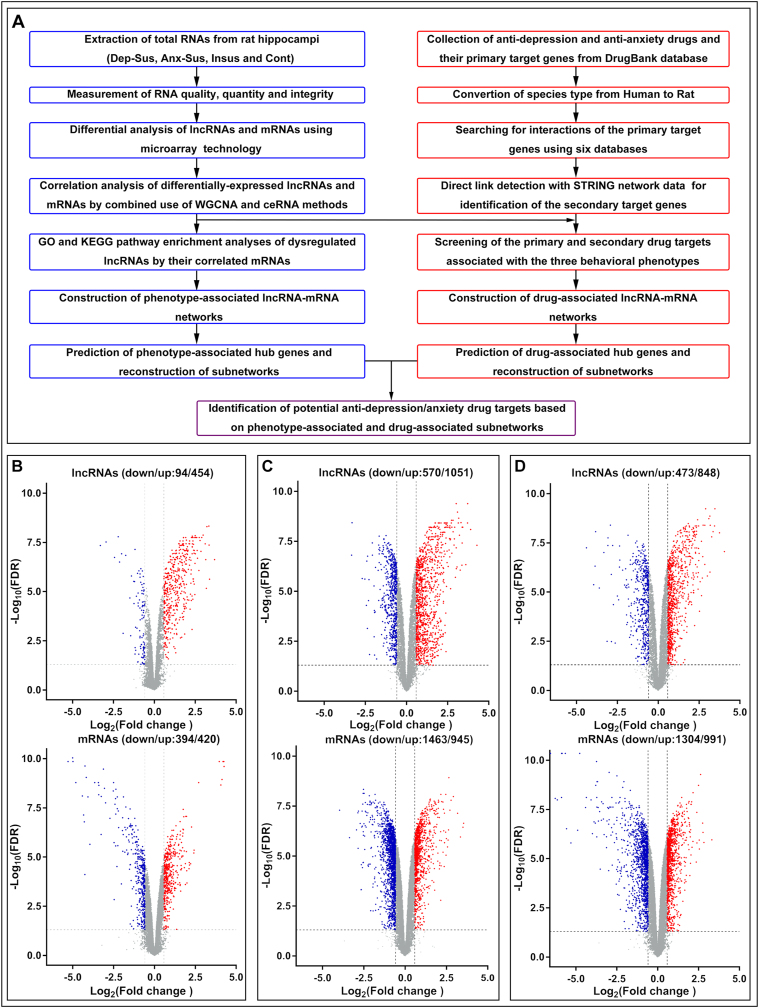
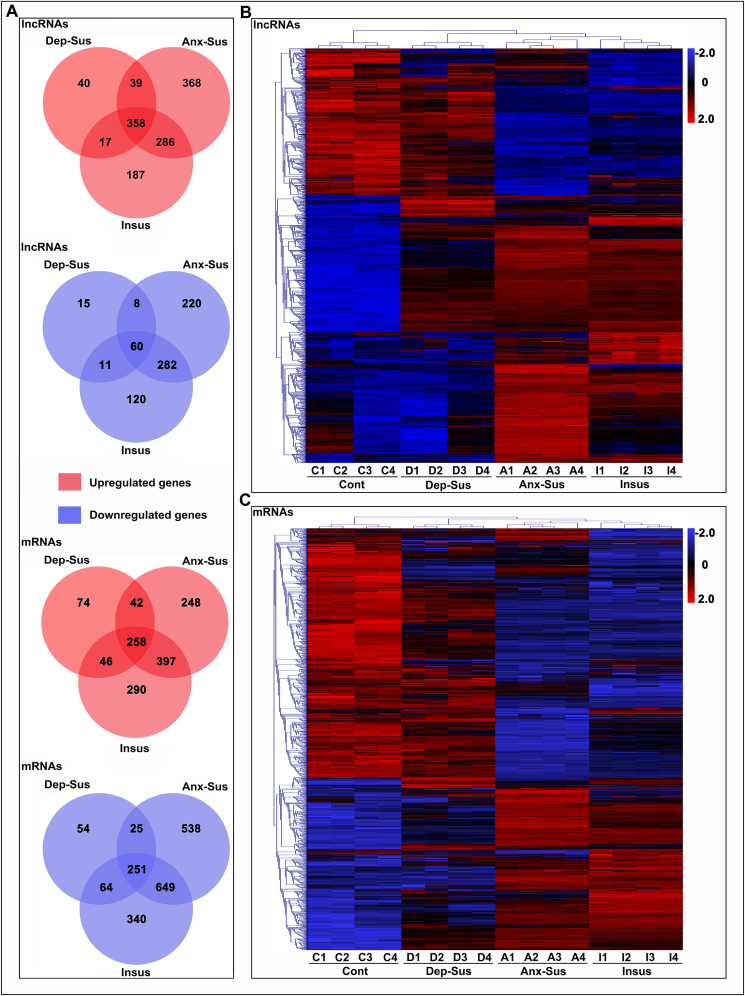
Next, we conducted the microarray screening to identify the lncRNA and mRNA profiles in hippocampal tissues from control, depression-susceptible, anxiety-susceptible and insusceptible rats, as shown in Fig. 1A. Using volcano plot filtering, differentially expressed lncRNAs and mRNAs with an FDR of less than 0.05 and greater than 1.5-fold change in both pairwise comparisons were identified (Fig. 1B–D; Supplementary Table S1). In the depression-susceptible group, 454 lncRNAs and 420 mRNAs were upregulated, and 94 lncRNAs and 394 mRNAs were downregulated (Fig. 1B). In the anxiety-susceptible group, 1051 lncRNAs and 945 mRNAs were upregulated, and 570 lncRNAs and 1463 mRNAs were downregulated (Fig. 1C). In the insusceptible group, 848 lncRNAs and 991 mRNAs were upregulated, and 473 lncRNAs and 1304 mRNAs were downregulated (Fig. 1D). Further analysis by Venn diagrams indicated that, among the insusceptible and the two susceptible groups, a set of lncRNAs (n = 1014) and a set of mRNAs (n = 1665) were regulated similarly as a result of exposure to CMS or potential intermediate variants (Fig. 2A). Interestingly, we also observed 465 lncRNAs and 576 mRNAs with similar regulation between the two susceptible (depression and anxiety) groups, which may indicate some common abnormal components in these two CMS-induced disorders. Despite the similarity in the expression changes in the three groups, a substantial percentage (47%) of lncRNAs and mRNAs were unique to the three CMS groups, suggesting that these groups have different molecular mechanisms underlying responses to stress. In addition, according to unsupervised hierarchical cluster analysis, gene expression profiles were categorized into three distinct groups representing the three different responses to CMS (Fig. 2B–C).
Fig. 1.
Analysis of differentially expressed long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs). (A) Overview of the workflow of the intersectional network analysis. (B, C and D) Identification of differentially expressed lncRNAs and mRNAs in the hippocampus of depression-susceptible (B), anxiety-susceptible (C) and insusceptible (D) groups. Volcano plot showing variations in gene expression. The fold change log (base 2) is on the x-axis, and the negative false log discovery rate (FDR) (base 10) is on the y-axis. Red signifies high relative expression, and blue signifies low relative expression. Dep-Sus, depression-susceptible; Anx-Sus, anxiety-susceptible; Insus, insusceptible; Cont, control. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Fig. 2.
Intergroup analysis of differentially expressed long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs). (A) Venn diagram illustrating the number of altered lncRNAs and mRNAs in the three groups. (B, C and D) Heatmap of differentially expressed lncRNAs and mRNAs based on hierarchical clustering in the depression-susceptible (D1-4), anxiety-susceptible (A1-4) and Insusceptible (I1-4) groups with respect to control (C1-4) (n = 4 per group). Red signifies high relative expression, and blue signifies low relative expression. Color intensity represents expression level, which is noted in the key bar with a log2-scale. Dep-Sus, depression-susceptible; Anx-Sus, anxiety-susceptible; Insus, insusceptible; Cont, control. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.2. Correlations between differentially expressed lncRNAs and mRNAs
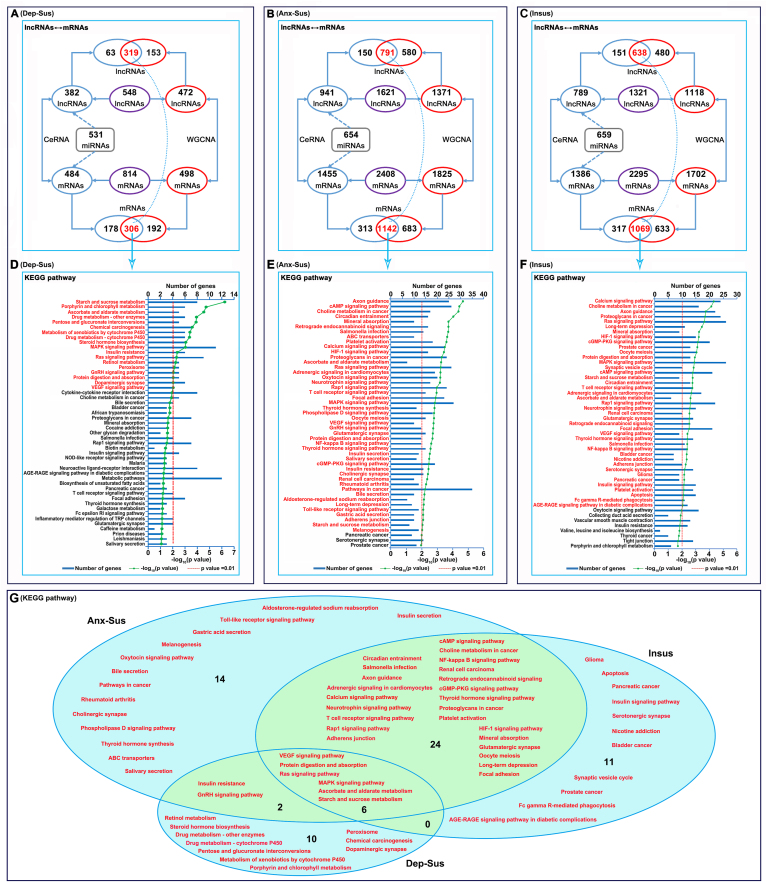
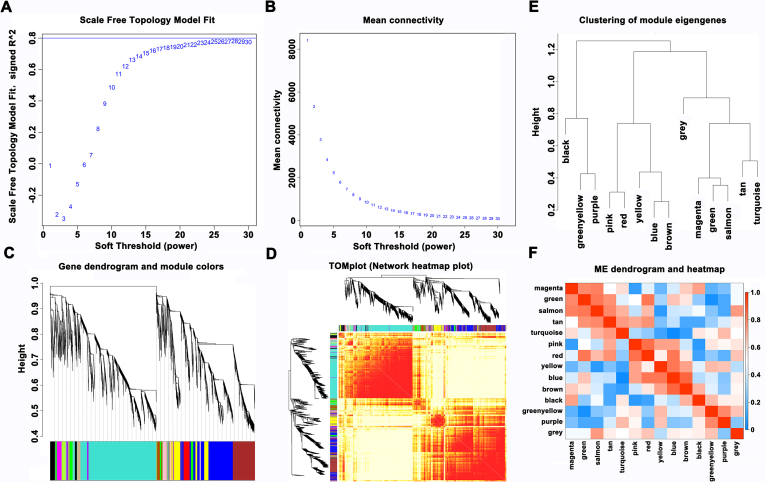
WGCNA was performed to explore the relationship between differentially expressed lncRNAs and mRNAs (Supplementary Fig. S1) (Alaei et al., 2018; Ren et al., 2015). In the depression-susceptible group, 472 lncRNAs were identified as positively associated with 498 mRNAs (Fig. 3A). In the anxiety-susceptible group, 1371 lncRNAs were positively associated with 1825 mRNAs (Fig. 3B). In the insusceptible group, 1118 lncRNAs were positively associated with 1702 mRNAs (Fig. 3C). Meanwhile, a ceRNA theory-based method was also implemented to ascertain the regulatory relationships between dysregulated lncRNAs, miRNAs, and mRNAs (Zhang et al., 2018). A lncRNA-mRNA competing interaction pair was selected based on the lncRNAs and mRNAs sharing common miRNAs. As a result, a total of 85,269 lncRNA-mRNA pairs were identified in the depression-susceptible groups, and a total of 699,622 dysregulated lncRNA-mRNA pairs were identified in the anxiety-susceptible groups. In addition, a total of 520,899 dysregulated lncRNA-mRNA pairs were identified in the insusceptible groups. By merging these dysregulated interactions, 382 lncRNAs were found to be associated with 484 mRNAs in the depression-susceptible group, 941 lncRNAs were associated with 1455 mRNAs in the anxiety-susceptible group, and 789 lncRNAs were associated with 1386 mRNAs in the insusceptible group. Notably, the vast majority of information collected from the ceRNA model was included in the WGCNA model. Finally, the combined data indicated that 319 lncRNAs were strongly correlated with 306 mRNAs in the depression-susceptible group, 791 lncRNAs were correlated with 1142 mRNAs in the anxiety-susceptible group and 638 lncRNAs were correlated with 1069 mRNAs in the insusceptible group (Fig. 3A–C; Supplementary Table S2).
Fig. 3.
Gene correlation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses. Correlation of differentially expressed long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) in the depression-susceptible (Dep-Sus, A), anxiety-susceptible (Anx-Sus, B) and insusceptible (Insus, C) groups was analyzed using the weighted correlation network analysis (WGCNA) and ceRNA theory-based methods. Then, KEGG pathway enrichment analysis of differentially expressed mRNAs correlated with differentially expressed lncRNAs was performed. The top enriched terms from the depression-susceptible (D), anxiety-susceptible (E) and insusceptible (F) groups were identified, and titles in red indicate significantly enriched KEGG pathways. The p-value negative log (base 10) is on the x-axis. (G) Venn diagram showing the common and unique significantly-enriched KEGG pathways in the three groups. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Functional characterization of dysregulated lncRNAs by their correlated mRNAs
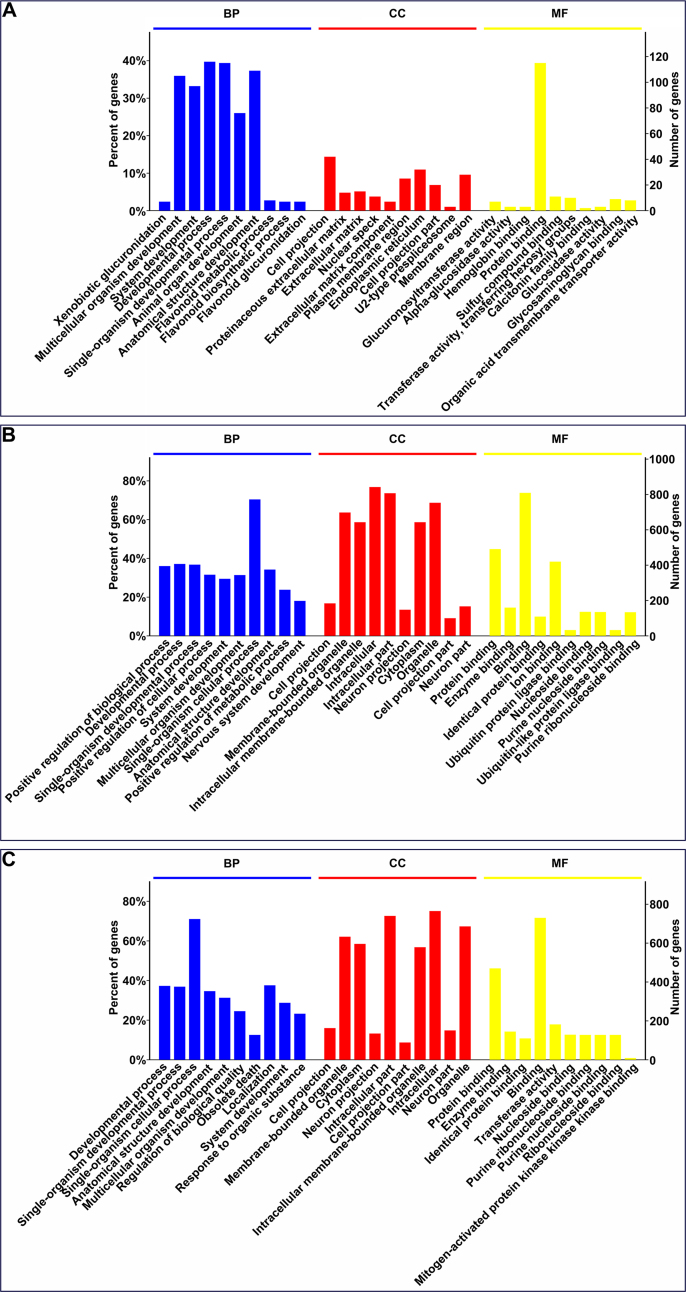
To predict lncRNA function, we conducted functional enrichment of mRNAs correlated with each differentially expressed lncRNA (Yang et al., 2016). The enriched terms were used as the predicted functional terms for each given lncRNA. Concretely, GO and KEGG pathway analysis was performed on the 306 correlated mRNAs in the depression-susceptible group (Supplementary Table S3). Overall, 471, 40, 74, and 18 terms were significantly enriched in the BP, CC, MF, and KEGG pathways, respectively (p < 0.01). Supplementary Fig. S2A depicts the top 10 enriched GO terms. BP gene analysis revealed significant enrichment for organism and system development. Moreover, CC analysis revealed significant enrichment for extracellular matrix and membrane components. MF analysis revealed significant enrichment for binding, enzyme and transporter activities. KEGG pathway enrichment analysis revealed dysregulation in small molecular and drug metabolism, as well as MAPK, Ras, GnRH and VEGF signaling pathways (Fig. 3D).
GO and KEGG pathway analysis was performed on the 1142 correlated mRNAs in the anxiety-susceptible group. Overall, 1174, 179, 149 and 46 terms were significantly enriched in BP, CC, MF and KEGG pathways, respectively (p < 0.01). Supplementary Fig. S2B shows the top 10 enriched GO terms. BP analysis revealed significant enrichment for developmental process, and positive regulation of biological and cellular processes. CC analysis revealed significant enrichment for membrane-bound organelle and intracellular part. MF classification revealed significant enrichment for various molecule binding. KEGG pathway enrichment analysis revealed dysregulation in axon guidance and various signaling and secretion pathways (Fig. 3E).
GO and KEGG pathway analysis was performed on the 1069 correlated mRNAs in the insusceptible group. Overall, 1047, 161, 158 and 41 terms were significantly enriched in the BP, CC, MF and KEGG pathways, respectively (p < 0.01). Supplementary Fig. S2C shows the top 10 enriched GO terms. BP analysis revealed significant enrichment for developmental and cellular processes. CC analysis revealed significant enrichment for intracellular part and membrane-bound organelle. MF classification revealed significant enrichment for molecule binding and enzyme activity. KEGG pathway enrichment analysis revealed dysregulation in many signaling, synapse and metabolic pathways (Fig. 3F).
Overall, of these significantly-enriched KEGG pathways, six were observed to be common among the three groups (Fig. 3G). Between the two susceptible groups, the eight pathways were shared. Importantly, the 10, 14 and 11 pathways were found to be uniquely associated to the depression-susceptible, anxiety-susceptible and insusceptible groups, respectively, potentially suggesting the three different biological processes as a response to CMS.
3.4. Construction of phenotype-associated lncRNA-mRNA networks and subnetworks
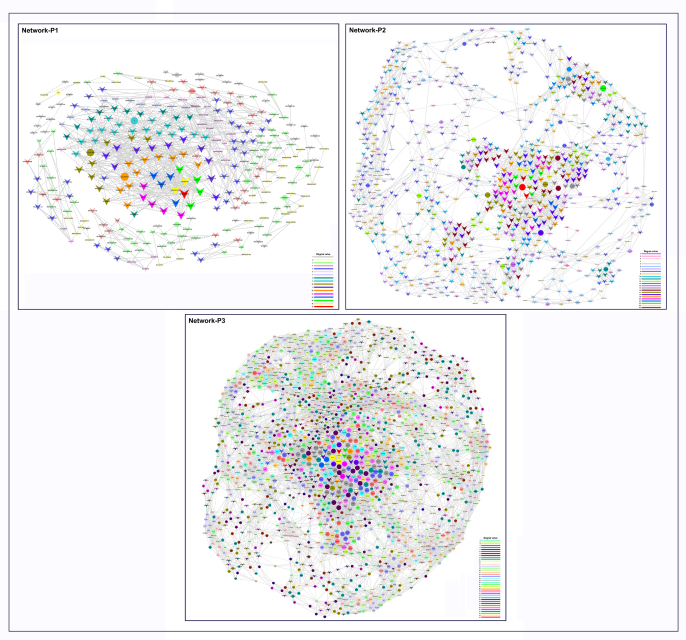
To explore putative lncRNA regulatory mechanisms on mRNA expression, differentially expressed lncRNAs were correlated with dysregulated mRNAs using coexpression network analysis. The coexpression network in the depression-susceptible group was comprised of 307 network nodes (network-P1 in Supplementary Fig. S3). There were 804 connections (405 negative and 399 positive interactions) between 10 differentially expressed mRNAs and 297 differentially expressed lncRNAs (Supplementary Table S4). The coexpression network in the anxiety-susceptible group was comprised of 840 network nodes, with 4058 connections (2045 positive and 2013 negative interactions) among 49 differentially expressed mRNAs and 791 differentially expressed lncRNAs (network-P2 in Supplementary Fig. S3). The coexpression network in the insusceptible group was comprised of 1707 network nodes, with 15875 connections (8071 positive and 7804 negative interactions) among 1069 differentially expressed mRNAs and 638 differentially expressed lncRNAs (network-P3 in Supplementary Fig. S3). In these networks, one dysregulated mRNA and one dysregulated lncRNA are typically correlated with 1–18 lncRNAs and 1–26 mRNAs, respectively. Further, some hub genes were identified according to node degree in these networks. For better visualization, subnetworks of the top 30 hub genes were rebuilt using Cytoscape software for further analyses (subnetwork-P1, -P2 and -P3).
3.5. Construction of drug-associated lncRNA-mRNA networks and subnetworks
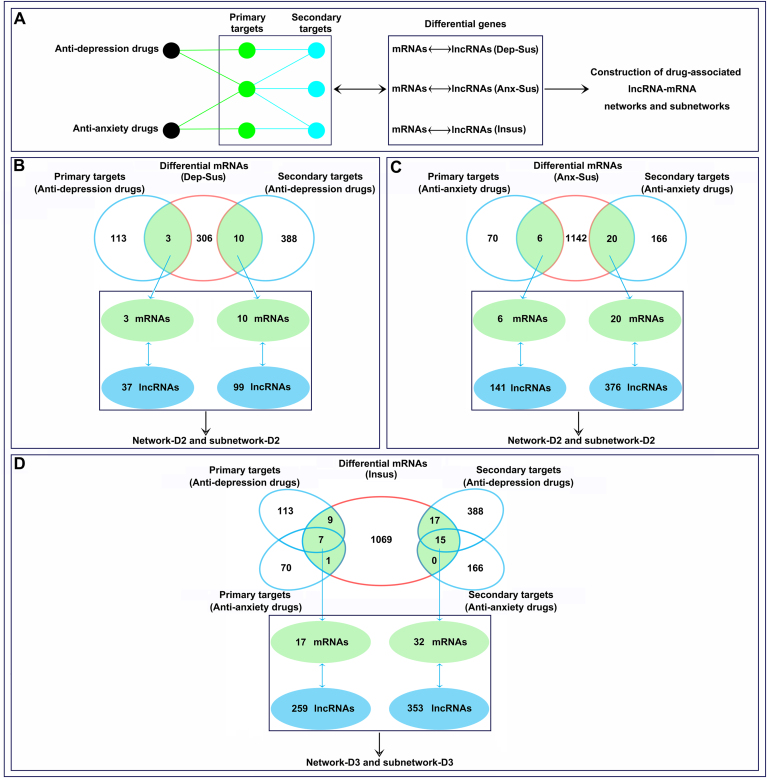
In the present study, we first retrieved a total of 38 anti-depression and 13 anti-anxiety drugs approved by the US FDA by searching DrugBank (Fig. 4A). Details of these drugs, including drug name, DrugBank ID, and target genes, are displayed in Supplementary Table S5. A total of 113 target genes were obtained from 38 anti-depression drugs, and a total of 70 target genes were obtained from 13 anti-anxiety drugs. These collected genes were considered the primary targets of the drugs.
Fig. 4.
Analysis of drug-associated messenger RNAs (mRNAs) and long noncoding RNAs (lncRNAs). According to the designed workflow (A), differential mRNAs and correlated lncRNAs from the depression-susceptible (Dep-Sus, B), anxiety-susceptible (Anx-Sus, C) and insusceptible (Insus, D) groups were screened as primary and secondary targets of anti-depression/anxiety drugs. The obtained mRNAs and lncRNAs were then used to construct drug-associated networks and subnetworks.
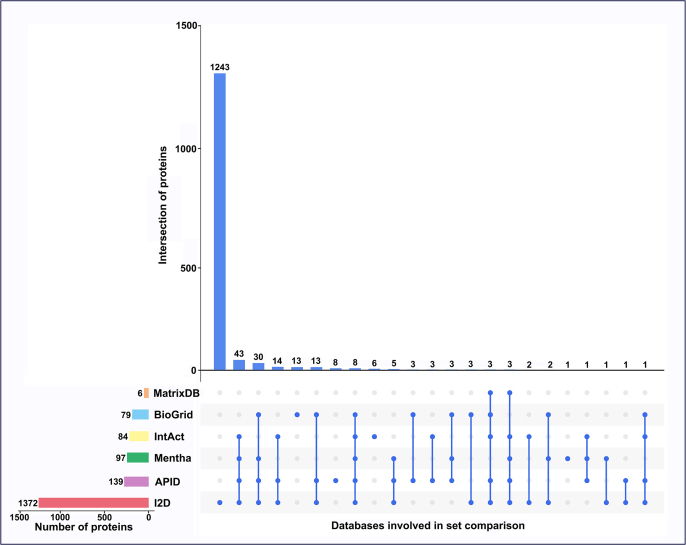
After species type was converted from human to rat, interactions of these primary target genes were determined from the six databases. Comparative analysis of the six databases revealed a unique curated set of 1410 genes that interacted with 120 primary target genes. Individual results from I2D, APID, Mentha, IntAct, BioGRID, and MatrixDB revealed 1372, 139, 97, 84, 79, and 6 genes, respectively, suggesting that none of the databases entirely represented the interaction data of the primary target genes (Supplementary Fig. S4). For each primary target gene, the interactions from all six databases were then combined to derive a universal gene set. Further, interactions between primary target genes and their interacting genes were analyzed by network link detection using STRING data. Those genes that were linked to the primary target genes directly and with medium-confidence were subsequently selected. As a result, a total of 392 genes interacting with 120 primary target genes were obtained and thereby considered secondary target genes.
To explore the relationship between drug targets and behavioral phenotypes, primary and secondary target genes were compared with the differential mRNAs identified in the depression-susceptible, anxiety-susceptible and insusceptible groups. As shown in Fig. 4B, 3 primary and 10 secondary target genes of anti-depression drugs were found to be differentially expressed in the depression-susceptible group, which were associated with 37 and 99 differentially expressed lncRNAs, respectively. Likewise, there were 6 primary and 20 secondary target genes of anti-anxiety drugs that were differentially expressed in the anxiety-susceptible group, which were associated with 141 and 376 differentially expressed lncRNAs, respectively (Fig. 4C). In addition, 17 primary and 32 secondary target genes of anti-depression/anxiety drugs were also found to be differentially expressed in the insusceptible groups, which were associated with 259 and 353 differentially expressed lncRNAs, respectively (Fig. 4D). Together, these differentially expressed lncRNAs and mRNAs from the three groups were considered target genes of anti-depression/anxiety drugs.
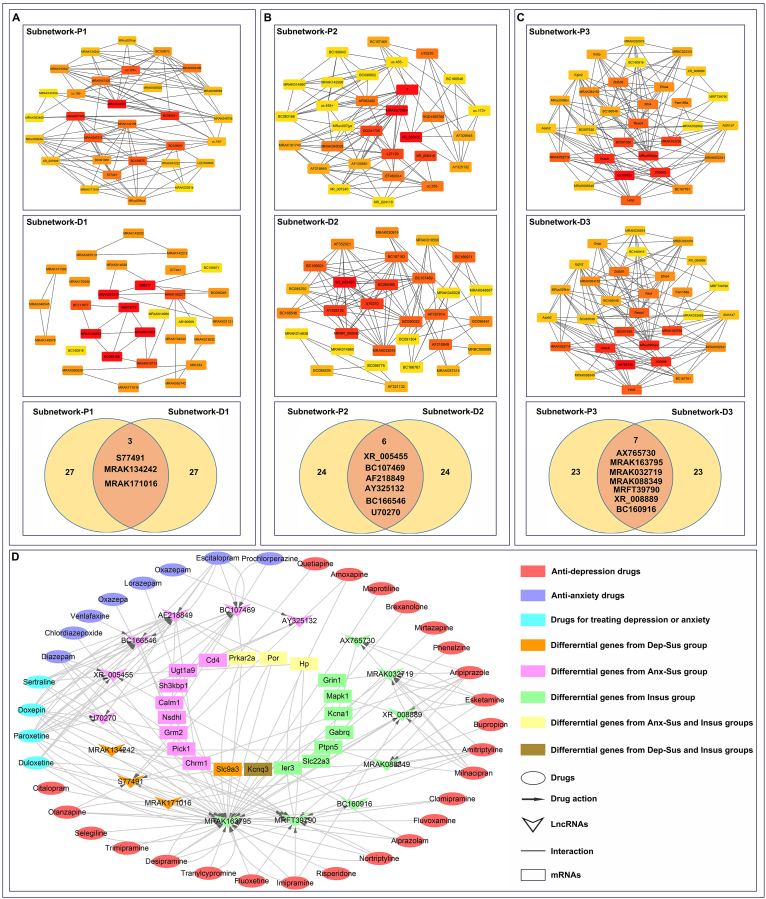
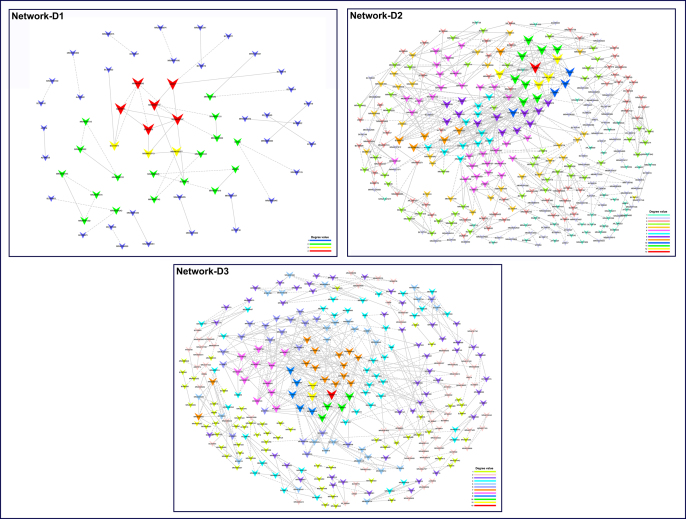
Afterward, the coexpression networks between these drug-correlated lncRNAs and mRNAs were constructed. As shown in Supplementary Fig. S5, network D1 was comprised of 61 network nodes. There were 51 connections (25 negative and 26 positive interactions) among 61 lncRNAs (Supplementary Table S6). Network D2 was comprised of 320 network nodes, with 642 connections (326 positive and 316 negative interactions) among 320 lncRNAs. Network D3 was comprised of 293 network nodes, with 526 connections (268 positive and 258 negative interactions) among 293 lncRNAs. Further, some hub genes were identified according to node degree. For better visualization, the subnetworks of the top 30 hub genes were rebuilt using Cytoscape software for the following analyses (subnetwork-D1, -D2 and -D3 in Fig. 5A–C).
Fig. 5.
Subnetwork analysis. (A, B and C) Phenotype-associated (subnetwork-P1, -P2 and -P3) and drug-associated (subnetwork-D1, -D2 and -D3) lncRNA-mRNA subnetworks were compared and analyzed, respectively. Subnetworks were constructed using the CytoHubba plugin. Advanced ranking of hub genes is represented by redder color. (D) Potential network of drug-lncRNA-mRNA-phenotype. Thirty-six anti-depression/anxiety drugs potentially act on 16 target lncRNAs to affect expression of the correlated 20 mRNAs. These lncRNAs and mRNAs were differentially expressed in the depression-susceptible (Dep-Sus), anxiety-susceptible (Anx-Sus) and insusceptible (Insus) groups. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.6. Integrated analysis of phenotype-associated and drug-associated lncRNA-mRNA subnetworks
To further identify hub lncRNAs and mRNAs correlated with both phenotype and drug categories, we focused on and comparatively analyzed nodes in the reconstructed phenotype-associated and drug-associated subnetworks. In comparison, three hub lncRNAs (S77491, MRAK134242, and MRAK171016) were common genes between subnetwork-P1 and subnetwork-D1 and potentially correlated with anti-depression drugs. By comparing subnetwork-P2 and subnetwork-D2, six hub lncRNAs (XR_005455, U70270, BC107469, AF218849, AY325132, and BC166546) as common genes were correlated with anti-anxiety drugs. By comparing subnetwork-P3 and subnetwork-D3, seven hub lncRNAs (AX765730, MRAK163795, MRAK032719, MRAK088349, XR_008889, BC160916, and MRFT39790) as common genes were identified to be correlated with anti-depression and/or anti-anxiety drugs. Of note, these identified hub lncRNAs potentially mediated the 20 differential mRNAs that were potential target genes of anti-depression/anxiety drugs. By means of this functional link, the hub lncRNAs could be predicted to be potential targets of the corresponding drugs. Meanwhile, it could be observed from subnetwork-P1, -P2 and -P3 that these hub lncRNAs might also represent some novel core regulatory factors associated with behavioral phenotypes. It can be assumed that the drugs may act on the potential target lncRNAs to affect expression of correlated mRNAs, thereby ameliorating depression and anxiety symptoms in stress-susceptible subjects or enabling them to resist stress (Fig. 5D). Therefore, it is tempting to speculate that the 16 hub lncRNAs identified in the present study may represent important new therapeutic drug targets and are worthy of further research for drug development in depression and anxiety disorders.
4. Discussion
4.1. Phenotype-correlated differential lncRNAs/mRNAs and significant pathways could be specific and common molecular basis of susceptibility and resiliency to stress-induced depression or anxiety
As chronic stress is a risk factor in depression and anxiety, CMS induces depression and anxiety related behaviors in laboratory rats (Chang and Grace, 2014; Henningsen et al., 2012). In previous publications (Tang et al., 2019), there are three different stress response phenotypes that we identified in the CMS rat model, which primarily include depression-susceptible, anxiety-susceptible, and insusceptible groups. This classification offers a valuable approach for identifying common and distinct molecular bases of vulnerability to stress-induced depression or anxiety and stress resiliency. Moreover, this CMS model also provides valuable information in the field of translational research. Analyzing the expression profiles of lncRNAs linked to the three CMS-induced behavioral phenotypes may provide new insights into our understanding of clinical depression and anxiety. In some previous studies, a large number of lncRNAs have been found to be differentially regulated in the hippocampus of learned helpless Holtzman rat model of depression (Wang et al., 2019a) and the prefrontal cortex of repeated social defeat stress mouse model of anxiety (Wang et al., 2019c). Yet, these studies only from a single perspective demonstrated a strong association between lncRNA expression and depressive or anxious behaviors (Huang et al., 2017, Spadaro et al., 2015, Wang et al., 2019a, Wang et al., 2019c). Thus, there is an urgent need to explore the common and distinct lncRNA characteristics that underlie susceptibility and resilience to depression or anxiety.
Therefore, we investigated stress-induced lncRNA expression and mRNA-lncRNA coexpression in the hippocampi of CMS rats exhibiting the three differential stress responses. Microarray data indicated that a large set of differentially expressed lncRNAs and mRNAs were recognized from the hippocampi of depression-susceptible, anxiety-susceptible, and insusceptible groups compared to nonstressed controls. The lncRNAs and mRNAs were that were altered similarly between the depression/anxiety-susceptible and insusceptible groups might represent general functional components or intermediate variants involved in the response to CMS. The common patterns of these two different disorders might contribute to those overlapping lncRNAs and mRNAs between depression-susceptible and anxiety-susceptible groups. Specifically, the unique gene regulation patterns in the three groups may underlie the observed behavioral differences. Heatmap analysis also suggested that the three stressed groups displayed differential lncRNA and mRNA functional expression profiles. These results may help other researchers identify genes and pathways associated with stress-induced depression or anxiety and stress resilience.
To interrogate the functions of the differentially expressed lncRNAs, we screened their correlated differentially expressed mRNAs using WGCNA and ceRNA theory-based methods (Alaei et al., 2018; Ren et al., 2015; Zhang et al., 2018). Results obtained from the two methods were combined, with the overlapping results providing more robust correlations between the differentially expressed lncRNAs and mRNAs. Next, GO and KEGG pathway analyses on the correlated differentially expressed mRNAs were conducted for functional prediction of these differentially expressed lncRNAs. GO and pathway enrichment analyses revealed that CMS-induced mRNAs were enriched for signaling pathways previously implicated in neurological stress and mood disorders (Leuner and Shors, 2013; Pittenger and Duman, 2008). Of note, the pathway prediction algorithms indicated that mRNAs correlated with the dysfunctional lncRNAs were significantly enriched for metabolism dysfunction in the depression-susceptible group, signaling and secretion dysregulations in the anxiety-susceptible group, and signaling and synapse aberrations in the insusceptible group. Intriguingly, many significant KEGG pathways were observed to be uniquely correlated with the three phenotypes. This might represent differences in important protein dysregulation systems and active biological pathways that occurred in these stressed groups. These bioinformatics data implicated lncRNA-mediated gene regulatory mechanisms specifically associated with stress-induced behaviors of depression, anxiety, and resilience in the hippocampus.
4.2. Intersectional analysis of phenotype-associated and drug-associated lncRNA-mRNA interaction networks revealed sixteen hub lncRNAs as potential targets of anti-depression/anxiety drugs
To systemically investigate these functional relationships, we further used lncRNA-mRNA coexpression analysis to ascertain putative interactions between differentially expressed lncRNAs and mRNAs from the three different phenotypes. The phenotype-associated networks suggested that lncRNA and mRNA interregulation was associated with susceptibility to CMS, implicating lncRNA-related regulatory machinery in determining stress-induced behavioral phenotypes. It has been widely acknowledged that genes with more node degrees in the network usually play more roles. Therefore, we screened hub genes in the three networks according to node degree. Accordingly, the top 30 hub genes were selected as key genes associated with susceptibility and resilience to depression or anxiety, and then the corresponding subnetworks were reconstructed for better visualization. Of note, hub lncRNAs identified in the networks may serve as key regulatory factors in abnormal expressions of mRNAs.
In addition, to understand disease pathophysiologies, these differentially expressed lncRNAs and mRNAs examined in the preclinical disease model, including depression-susceptible and anxiety-susceptible groups, may provide interesting targets for anti-depression and anti-anxiety drugs. Of note, recent studies demonstrated that molecules associated with stress resilience also had the potential to be useful drug targets (Henningsen et al., 2012; Krishnan et al., 2007; Russo et al., 2012; Uchida et al., 2011). Accordingly, differential genes from the insusceptible group were considered for the following analysis. In this study, we first retrieved all anti-depression and anti-anxiety drugs and their corresponding primary target genes from DrugBank database. Then, the secondary target genes of the drugs were also obtained by combined searching of all interactions for the primary target genes from the six databases followed by identification of direct interactions using the STRING tool. By detailed comparison, we subsequently found that 13 and 26 differentially expressed mRNAs identified from the depression-susceptible and anxiety-susceptible groups, respectively, were predicted to be the targets (including primary and secondary targets) of the anti-depression and anti-anxiety drugs. Meanwhile, it was also discovered that 49 differentially expressed mRNAs from the insusceptible groups were predicted to be the anti-depression/anxiety drug targets. In theory, expression of these mRNAs as potential targets might be subjected to regulation by their correlated lncRNAs. Based on these functional correlations, a total of 610 lncRNAs as upstream regulatory factors could be considered potential drug targets. To further ascertain putative interactions, drug-associated lncRNA-mRNA networks were constructed in the present study. Likewise, the top 30 hub genes were selected as key genes associated with anti-depression/anxiety drugs, and corresponding subnetworks were subsequently rebuilt for better visualization.
Furthermore, we comparatively analyzed hub genes in the reconstructed phenotype-associated and drug-associated subnetworks. It can be speculated that common hub lncRNAs and mRNAs between two types of subnetworks might be correlated with both phenotypes and drugs. Pairwise comparison analyses identified 16 hub lncRNAs. These hub lncRNAs may not only play key regulatory roles in expression of corresponding protein-coding genes (mRNAs) associated with CMS-induced disorders, but may also represent key targets of anti-depression/anxiety drugs. In the scope of our knowledge, this is the first study that effectively combined phenotype-associated and drug-associated subnetworks for screening important drug targets. Excitingly, 16 hub lncRNAs were identified in this study and may represent new important therapeutic drug targets that are worthy of further research for drug development in depression and anxiety disorders. Certainly, although attractive findings have been obtained by a series of bioinformatics analyses in our present study, additional laboratory experiments such as experimental proof-of-principle studies need to be conducted in the future.
5. Conclusion
This study employed a microarray-based analysis to investigate hippocampal lncRNA and mRNA expression patterns that profoundly affect rodent susceptibility to CMS-induced depression or anxiety and stress resilience. These findings provide significant and fresh new insights into the regulatory mechanisms underlying stress in CMS-exposed rats, which may serve as the molecular basis for future research. This research facilitates a better understanding of the similarities and differences in pathophysiological mechanisms underlying stress-induced depression or anxiety and stress resiliency. By intersectional analysis of phenotype-associated and drug-associated lncRNA-mRNA networks and subnetworks, we identified sixteen hub lncRNAs with potential as important new therapeutic drug targets of depression and anxiety disorders.
Data availability
The microarray data reported in this paper have been deposited in NCBI Gene Expression Omnibus (GEO: GSE156580), and are publicly accessible at https://www.ncbi.nlm.nih.gov/geo/.
CRediT authorship contribution statement
Wei Liao: Methodology, Investigation, Formal analysis, Writing – original draft. Yanchen Liu: Methodology, Investigation, Formal analysis, Writing – original draft. Haojun Huang: Methodology, Investigation, Formal analysis, Writing – original draft. Hong Xie: Methodology, Formal analysis, Funding acquisition. Weibo Gong: Methodology, Investigation. Dan Liu: Methodology, Investigation. Fenfang Tian: Methodology, Investigation. Rongzhong Huang: Methodology, Investigation, Formal analysis. Faping Yi: Conceptualization, Methodology, Formal analysis, Writing – review & editing. Jian Zhou: Conceptualization, Supervision, Methodology, Formal analysis, Writing – review & editing, Funding acquisition.
Declaration of competing interest
The authors have declared no competing interests.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grants No. 31570826, 31770890) and the Health and Family Planning Commission of Chongqing Nanan District (Grant No. 2018–03).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ynstr.2021.100347.
Contributor Information
Faping Yi, Email: 100506@cqmu.edu.cn.
Jian Zhou, Email: zhoujian@cqmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
A complete list of differentially expressed long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) in the depression-susceptible (Dep-Sus), anxiety-susceptible (Anx-Sus) and insusceptible (Insus) groups.
Correlation data of differentially expressed long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) in the depression-susceptible, anxiety-susceptible and insusceptible groups obtained by weighted correlation network analysis (WGCNA) and ceRNA theory-based methods.
Complete list of Gene Ontology (GO) biological process (BP), cellular component (CC), molecular function (MF) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway terms for differentially expressed messenger RNAs (mRNAs) correlated with differential long noncoding RNAs (lncRNAs) in the depression-susceptible (Dep-Sus), anxiety-susceptible (Anx-Sus) and insusceptible (Insus) groups. Terms with p-value < 0.01 are highlighted in gray.
Negative and positive correlations between long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) in the network-P1, -P2 and -P3.
Primary and secondary target genes of FDA-approved anti-depression and anti-anxiety drugs.
Negative and positive correlations between long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) in the network-D1, -D2 and -D3.
Supplementary Figure S1.
Correlation analysis of long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) in the depression-susceptible, anxiety-susceptible and insusceptible groups with weighted correlation network analysis (WGCNA). (A) The effect of different power values on the scale independence degree of coexpression modules of lncRNAs and mRNAs. (B) The effect of different power value on the average connectivity degree of coexpression modules of lncRNAs and mRNAs. (C) The constructed coexpression modules of lncRNAs and mRNAs by WGCNA method. Each branch above represented each gene and the different color below represents each coexpression module. (D) Interaction analyses of coexpression modules. Different colors on the horizontal axis and vertical axis represent different modules. The brightness of the color yellow in the middle represents the relative strength among these modules. (E) Cluster analysis of eigengenes in the module. (F) Adjacency heatmap of eigengenes in the module. The gradient change of color from blue (0) to red (1) in the heatmap represents the adjacency of eigengenes in different modules from weak to strong, respectively.
Supplementary Figure S2.
Enrichment analysis of Gene Ontology (GO) pathway terms for differentially expressed messenger RNAs (mRNAs) correlated with differentially expressed long noncoding RNAs (lncRNAs). The top 10 enriched terms from depression-susceptible (A), anxiety-susceptible (B) and insusceptible (C) groups in biological process (BP), cellular component (CC), and molecular function (MF) were identified.
Supplementary Figure S3.
Construction of phenotype-associated lncRNA-mRNA networks (network-P1, -P2 and -P3). The arrows and circles represent dysregulated lncRNAs and mRNAs, respectively. Different colors and sizes represent the corresponding node degrees. The dashed and solid lines between genes indicate negative and positive correlations, respectively.
Supplementary Figure S4.
Analysis of interactions of primary targets of anti-depression and anti-anxiety drugs. Interactions from the six databases (I2D, APID, Mentha, IntAct, BioGRID, and MatrixDB) were comparatively analyzed.
Supplementary Figure S5.
Construction of drug-associated lncRNA-mRNA networks (network-D1, -D2 and -D3). The arrows and circles represent dysregulated lncRNAs and mRNAs, respectively. Different colors and sizes represent the corresponding node degrees. The dashed and solid lines between genes indicate negative and positive correlations, respectively.
References
- Alaei S., Sadeghi B., Najafi A., Masoudi-Nejad A. LncRNA and mRNA integration network reconstruction reveals novel key regulators in esophageal squamous-cell carcinoma. Genomics. 2018;111(1):76–89. doi: 10.1016/j.ygeno.2018.01.003. [DOI] [PubMed] [Google Scholar]
- Almeida O.P., Draper B., Pirkis J., Snowdon J., Lautenschlager N.T., Byrne G., Sim M., Stocks N., Kerse N., Flicker L., Pfaff J.J. Anxiety, depression, and comorbid anxiety and depression: risk factors and outcome over two years. Int. Psychogeriatr. 2012;24:1622–1632. doi: 10.1017/S104161021200107X. [DOI] [PubMed] [Google Scholar]
- Argentieri M.A., Nagarajan S., Seddighzadeh B., Baccarelli A.A., Shields A.E. Epigenetic pathways in human disease: the impact of DNA methylation on stress-related pathogenesis and current challenges in biomarker development. EBioMedicine. 2017;18:327–350. doi: 10.1016/j.ebiom.2017.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannerman D.M., Sprengel R., Sanderson D.J., McHugh S.B., Rawlins J.N., Monyer H., Seeburg P.H. Hippocampal synaptic plasticity, spatial memory and anxiety. Nat. Rev. Neurosci. 2014;15:181–192. doi: 10.1038/nrn3677. [DOI] [PubMed] [Google Scholar]
- Brodbeck J., Abbott R.A., Goodyer I.M., Croudace T.J. General and specific components of depression and anxiety in an adolescent population. BMC Psychiatr. 2011;11:191. doi: 10.1186/1471-244X-11-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.H., Grace A.A. Amygdala-ventral pallidum pathway decreases dopamine activity after chronic mild stress in rats. Biol. Psychiatr. 2014;76:223–230. doi: 10.1016/j.biopsych.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.J., Bai S.J., Li W.W., Zhou C.J., Zheng P., Fang L., Wang H.Y., Liu Y.Y., Xie P. Urinary biomarker panel for diagnosing patients with depression and anxiety disorders. Transl. Psychiatry. 2018;8:192. doi: 10.1038/s41398-018-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba S., Numakawa T., Ninomiya M., Richards M.C., Wakabayashi C., Kunugi H. Chronic restraint stress causes anxiety- and depression-like behaviors, downregulates glucocorticoid receptor expression, and attenuates glutamate release induced by brain-derived neurotrophic factor in the prefrontal cortex. Prog. Neuro-Psychopharmacol. Biol. Psychiatry. 2012;39:112–119. doi: 10.1016/j.pnpbp.2012.05.018. [DOI] [PubMed] [Google Scholar]
- Frick A. Common and distinct gray matter alterations in social anxiety disorder and major depressive disorder. EBioMedicine. 2017;21:53–54. doi: 10.1016/j.ebiom.2017.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden S.A., Christoffel D.J., Heshmati M., Hodes G.E., Magida J., Davis K., Cahill M.E., Dias C., Ribeiro E., Ables J.L., Kennedy P.J., Robison A.J., Gonzalez-Maeso J., Neve R.L., Turecki G., Ghose S., Tamminga C.A., Russo S.J. Epigenetic regulation of RAC1 induces synaptic remodeling in stress disorders and depression. Nat. Med. 2013;19:337–344. doi: 10.1038/nm.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton J.P., Chen M.C., Waugh C.E., Joormann J., Gotlib I.H. Distinctive and common neural underpinnings of major depression, social anxiety, and their comorbidity. Soc. Cognit. Affect Neurosci. 2015;10:552–560. doi: 10.1093/scan/nsu084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningsen K., Palmfeldt J., Christiansen S., Baiges I., Bak S., Jensen O.N., Gregersen N., Wiborg O. Candidate hippocampal biomarkers of susceptibility and resilience to stress in a rat model of depression. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Luo Y.-l., Mao Y.-s., Ji J.-l. The link between long noncoding RNAs and depression. Prog. NeuroPsychopharmacol. Biol. Psychiatr. 2017;73:73–78. doi: 10.1016/j.pnpbp.2016.06.004. [DOI] [PubMed] [Google Scholar]
- Kang H.J., Voleti B., Hajszan T., Rajkowska G., Stockmeier C.A., Licznerski P., Lepack A., Majik M.S., Jeong L.S., Banasr M., Son H., Duman R.S. Decreased expression of synapse-related genes and loss of synapses in major depressive disorder. Nat. Med. 2012;18:1413–1417. doi: 10.1038/nm.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V., Han M.H., Graham D.L., Berton O., Renthal W., Russo S.J., Laplant Q., Graham A., Lutter M., Lagace D.C., Ghose S., Reister R., Tannous P., Green T.A., Neve R.L., Chakravarty S., Kumar A., Eisch A.J., Self D.W., Lee F.S., Tamminga C.A., Cooper D.C., Gershenfeld H.K., Nestler E.J. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Larson C.L., Nitschke J.B., Davidson R.J. Common and distinct patterns of affective response in dimensions of anxiety and depression. Emotion. 2007;7:182–191. doi: 10.1037/1528-3542.7.1.182. [DOI] [PubMed] [Google Scholar]
- Leuner B., Shors T.J. Stress, anxiety, and dendritic spines: what are the connections? Neuroscience. 2013;251:108–119. doi: 10.1016/j.neuroscience.2012.04.021. [DOI] [PubMed] [Google Scholar]
- Lotan A., Fenckova M., Bralten J., Alttoa A., Dixson L., Williams R.W., van der Voet M. Neuroinformatic analyses of common and distinct genetic components associated with major neuropsychiatric disorders. Front. Neurosci. 2014;8:331. doi: 10.3389/fnins.2014.00331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malykhin N.V., Coupland N.J. Hippocampal neuroplasticity in major depressive disorder. Neuroscience. 2015;309:200–213. doi: 10.1016/j.neuroscience.2015.04.047. [DOI] [PubMed] [Google Scholar]
- Mathew A.R., Pettit J.W., Lewinsohn P.M., Seeley J.R., Roberts R.E. Co-morbidity between major depressive disorder and anxiety disorders: shared etiology or direct causation? Psychol. Med. 2011;41:2023–2034. doi: 10.1017/S0033291711000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui M., Corey D.R. Non-coding RNAs as drug targets. Nat. Rev. Drug Discov. 2017;16:167–179. doi: 10.1038/nrd.2016.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton T.H., Croarkin P.E., Strawn J.R., McClintock S.M. Comorbid anxiety and depressive symptoms in children and adolescents: a systematic review and analysis. J. Psychiatr. Pract. 2016;22:84–98. doi: 10.1097/PRA.0000000000000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pena C.J., Nestler E.J. Progress in epigenetics of depression. Prog. Mol. Biol. Transl. Sci. 2018;157:41–66. doi: 10.1016/bs.pmbts.2017.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger C., Duman R.S. Stress, depression, and neuroplasticity: a convergence of mechanisms. Neuropsychopharmacology. 2008;33:88–109. doi: 10.1038/sj.npp.1301574. [DOI] [PubMed] [Google Scholar]
- Quinn J.J., Chang H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016;17:47–62. doi: 10.1038/nrg.2015.10. [DOI] [PubMed] [Google Scholar]
- Qureshi I.A., Mehler M.F. Long non-coding RNAs: novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics. 2013;10:632–646. doi: 10.1007/s13311-013-0199-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Cui Y., Li X., Wang B., Na L., Shi J., Wang L., Qiu L., Zhang K., Liu G., Xu Y. A co-expression network analysis reveals lncRNA abnormalities in peripheral blood in early-onset schizophrenia. Prog. NeuroPsychopharmacol. Biol. Psychiatr. 2015;63:1–5. doi: 10.1016/j.pnpbp.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Russo S.J., Murrough J.W., Han M.H., Charney D.S., Nestler E.J. Neurobiology of resilience. Nat. Neurosci. 2012;15:1475–1484. doi: 10.1038/nn.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spadaro P.A., Flavell C.R., Widagdo J., Ratnu V.S., Troup M., Ragan C., Mattick J.S., Bredy T.W. Long noncoding RNA-directed epigenetic regulation of gene expression is associated with anxiety-like behavior in mice. Biol. Psychiatr. 2015;78:848–859. doi: 10.1016/j.biopsych.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo J.-S., Lee S.-T., Im W., Lee M., Byun J.-I., Jung K.-H., Park K.-I., Jung K.-Y., Lee S.K., Chu K., Kim M. Altered expression of the long noncoding RNA NEAT1 in huntington's disease. Mol. Neurobiol. 2016;54:1577–1586. doi: 10.1007/s12035-016-9928-9. [DOI] [PubMed] [Google Scholar]
- Tang M., Huang H., Li S., Zhou M., Liu Z., Huang R., Liao W., Xie P., Zhou J. Hippocampal proteomic changes of susceptibility and resilience to depression or anxiety in a rat model of chronic mild stress. Transl. Psychiatry. 2019;9:260. doi: 10.1038/s41398-019-0605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S., Hara K., Kobayashi A., Otsuki K., Yamagata H., Hobara T., Suzuki T., Miyata N., Watanabe Y. Epigenetic status of Gdnf in the ventral striatum determines susceptibility and adaptation to daily stressful events. Neuron. 2011;69:359–372. doi: 10.1016/j.neuron.2010.12.023. [DOI] [PubMed] [Google Scholar]
- Wang Q., Roy B., Dwivedi Y. Co-expression network modeling identifies key long non-coding RNA and mRNA modules in altering molecular phenotype to develop stress-induced depression in rats. Transl. Psychiatry. 2019;9:125. doi: 10.1038/s41398-019-0448-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Lou W., Ding B., Yang B., Lu H., Kong Q., Fan W. A novel mRNA-miRNA-lncRNA competing endogenous RNA triple sub-network associated with prognosis of pancreatic cancer. Aging (Albany NY) 2019;11:2610–2627. doi: 10.18632/aging.101933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Ma S., Mao M., Li C., Shen X., Xu S., Yang J. RNA-sequencing and bioinformatics analysis of long noncoding RNAs and mRNAs in the prefrontal cortex of mice following repeated social defeat stress. BioMed Res. Int. 2019 doi: 10.1155/2019/7505260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Huang H., Tang M., Wu Y., Huang R., Liu Z., Zhou M., Liao W., Zhou J. iTRAQ-based quantitative proteomics suggests synaptic mitochondrial dysfunction in the Hippocampus of rats susceptible to chronic mild stress. Neurochem. Res. 2018;43:2372–2383. doi: 10.1007/s11064-018-2664-y. [DOI] [PubMed] [Google Scholar]
- Yang B., Xia Z.-a., Zhong B., Xiong X., Sheng C., Wang Y., Gong W., Cao Y., Wang Z., Peng W. Distinct hippocampal expression profiles of long non-coding RNAs in an alzheimer's disease model. Mol. Neurobiol. 2016;54:4833–4846. doi: 10.1007/s12035-016-0038-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun S., Donovan M.H., Ross M.N., Richardson D.R., Reister R., Farnbauch L.A., Fischer S.J., Riethmacher D., Gershenfeld H.K., Lagace D.C., Eisch A.J. Stress-induced anxiety- and depressive-like phenotype Associated with transient Reduction in neurogenesis in adult nestin-CreERT2/diphtheria toxin fragment A transgenic mice. PloS One. 2016;11 doi: 10.1371/journal.pone.0147256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Sun H., Zhang Y., Zhao H., Fan W., Li J., Lv Y., Song Q., Li J., Zhang M., Shi H. Characterization of dysregulated lncRNA-mRNA network based on ceRNA hypothesis to reveal the occurrence and recurrence of myocardial infarction. Cell Death Dis. 2018;4:35. doi: 10.1038/s41420-018-0036-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chen L., Zhang W., Xiao Y., Shah C., Zhu H., Yuan M., Sun H., Yue Q., Jia Z., Zhang W., Kuang W., Gong Q., Lui S. Gray matter abnormalities in non-comorbid medication-naive patients with major depressive disorder or social anxiety disorder. EBioMedicine. 2017;21:228–235. doi: 10.1016/j.ebiom.2017.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A complete list of differentially expressed long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) in the depression-susceptible (Dep-Sus), anxiety-susceptible (Anx-Sus) and insusceptible (Insus) groups.
Correlation data of differentially expressed long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) in the depression-susceptible, anxiety-susceptible and insusceptible groups obtained by weighted correlation network analysis (WGCNA) and ceRNA theory-based methods.
Complete list of Gene Ontology (GO) biological process (BP), cellular component (CC), molecular function (MF) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway terms for differentially expressed messenger RNAs (mRNAs) correlated with differential long noncoding RNAs (lncRNAs) in the depression-susceptible (Dep-Sus), anxiety-susceptible (Anx-Sus) and insusceptible (Insus) groups. Terms with p-value < 0.01 are highlighted in gray.
Negative and positive correlations between long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) in the network-P1, -P2 and -P3.
Primary and secondary target genes of FDA-approved anti-depression and anti-anxiety drugs.
Negative and positive correlations between long noncoding RNAs (lncRNAs) and messenger RNAs (mRNAs) in the network-D1, -D2 and -D3.
Data Availability Statement
The microarray data reported in this paper have been deposited in NCBI Gene Expression Omnibus (GEO: GSE156580), and are publicly accessible at https://www.ncbi.nlm.nih.gov/geo/.