Abstract
Camel chymosin can be efficiently employed to produce cheese. Traditionally the rennet enzyme produced by the glands of the fourth stomach of ruminant animals (abomassum) is used in cheese making. Full-length Camelus bactrianus (Bactrian camel) prochymosin gene was synthesized and constitutively expressed in Pichia pastoris cells under glyceraldehydes-3-phosphate dehydrogenase (GAP) promoter. It was purified by sequential anion and cation exchange chromatography. SDS-PAGE analysis resulted in two bands, approximately 42 and 35 kDa. The 42 kDa band vanished when the sample was treated with endoglycosidase H, indicating that the recombinant protein is partially glycosylated. Optimal pH for the activity of the highest-purity recombinant chymosin was pH 4.5 for cow's milk and pH 4.0 for mare's milk. The range 45–50 °C and 70 °C for cow's and mare's milk types, respectively, was found to be the most appropriate for maximal relative milk-clotting activity. Concentration of CaCl2 that ensured the stability of the chymosin milk-clotting activity was between 20 and 50 mM with an optimum at 30 mM. Milk-clotting activity of camel recombinant chymosin and ability to make curd was successfully tested on fresh mare's milk. Pichia pastoris strain with integrated camel chymosin gene showed high productivity of submerged fermentation in bioreactor with milk-clotting activity 1412 U/mL and 80 mg/L enzyme yield. These results suggest that the constitutive expression of the camel chymosin Camelus bactrianus in the yeast Pichia pastoris has good prospects for practical applications.
Keywords: Recombinant DNA, Protein purification, Camel chymosin, Constitutive expression, Milk coagulation, Glycosylation, Rennet
Recombinant DNA; Protein purification; Camel chymosin; Constitutive expression; Milk coagulation; Glycosylation; Rennet.
1. Introduction
One of the earliest biotechnological applications of enzymes is cheese production [1]. The proteolytic enzymes pepsin and chymosin [2], previously known as "rennet," [3] have been identified as active ingredients in this process. Chymosin (EC 3.4.23.4) is an aspartic peptidase that belongs to the pepsin-like family [4]. Chymosin particularly targets the κ-casein peptide link between Phe105 and Met106 (Figure 1A), triggering the formation of destabilized casein micelles, milk coagulation, and separation of the milk into curds and whey [5]. Although plants [6, 7, 8, 9, 10, 11] and microorganisms [12, 13, 14, 15, 16, 17] can be used as different sources of milk-coagulating enzymes, plant and microbial rennet are characterized by nonspecific proteolytic activity against casein, and animal rennet is more frequently used in the dairy sector for cheese making. Because of the high specific milk-clotting activity, chymosin is considered the most efficient protease for the cheese-making industry. Today, fermentation-produced chymosin accounts for more than 90% of rennet utilized, and it has the added benefit of being both kosher and halal acceptable [18]. There are chymosins, obtained from different animals, that arouse interest, for example the yak (Bos grunniens) [19] and Altai maral (Cervus elaphus) [20].
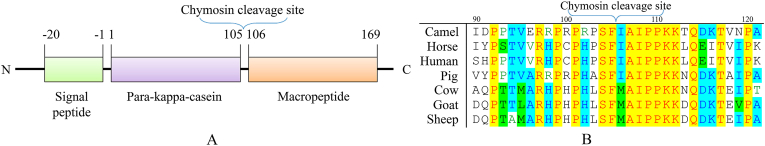
Figure 1.
A - Domain structure of kappa-casein of cow's milk; B - Comparative sequence of chymosin sensitive region of kappa-casein molecules of some mammals.
Chymosin refers to a specific action aspartic proteases [21]. Analysis of the amino acid sequences of the milk for camel, horse, human, pig, cow, goat, and sheep showed that the cleavage site of κ-casein by chymosin is the sequence SFXAIPPKK, where X is methionine in the bovids and isoleucine in other mammals (Figure 1B). Highly conserved region 97–116 of κ-casein between species [22], that κ-casein proteolysis and subsequent milk coagulation are important biological processes. Since chymosin possesses high substrate specificity, this difference may explain why camel's milk cannot be coagulated with bovine chymosin [23, 24]. At the same time, it is interesting to test the activity of camel's chymosin on mare's milk.
Mare's κ-casein contains 165 amino acids residues. Isoelectric point of equine and human κ-casein, 8.03 and 8.68 respectively, are higher than bovine κ-casein at 5.93, and they have a net positive charge at physiological pH, whereas bovine κ-casein has a net negative charge [25]. In terms of hydropathy dispersion along the polypeptide chain, human κ-casein appears to be more comparable to horse than bovine κ-casein [26].
Camel chymosin can be employed to produce cheese from cow's milk [27, 28, 29, 30] and camel milk [24, 31]. In comparison, camel chymosin has 70 % more milk-clotting activity than bovine chymosin and is more thermostable than bovine chymosin [23, 29], making it useful and appealing for commercial cheese manufacture. Chymosin from Camelus dromedarius is characterized well [23, 29, 32], and its recombinant version is available as commercial milk-clotting product Chy-MAX M (Ch. Hansen). The recombinant chymosin of C. dromedarius has been successfully expressed in the filamentous fungus Aspergillus niger [23] and in the yeast Pichia pastoris [33]. Chymosin from the Central Asian camel Camelus bactrianus is not characterized, but its full nucleotide sequence is known [34]. Analysis of the nucleotide sequence of the two camel chymosins shows that the C. bactrianus chymosin gene differs from the dromedary chymosin gene at three positions, C685G, A880T, A881C, resulting in two amino acid substitutions, Val229Leu and Ser294Asn.
In compare to Dromedary, Bactrian camels are found in considerably colder climates [35]. The Bactrian camel may be found all throughout the Siberian steppe as well as in the harsh, dry deserts of Central and Eastern Asia. In the winter, temperatures in their native habitat, for example in Trans-Altai region of Gobi desert, can drop as low as –35 °C [36]. This arises interest to obtain and study C. bactrianus chymosin.
The aim of this work was secretory expression of C. bactrianus chymosin B in the yeast P. pastoris and biochemical characterization of the resultant enzyme. P. pastoris (also known as Komagataella phaffi), an eukaryotic expression system, is a methylotrophic yeast suitable as a host organism for heterologous expression of proteins owing to simplicity of its genetic manipulation, rapid growth on an inexpensive medium to high cell densities, and capability for complex post-translational modifications [37, 38]. The P. pastoris yeast expression system is a good choice for obtaining a heterologous recombinant protein with prospects for large-scale microbial production [39], and C. bactrianus chymosin holds promise for the cheese-making industry.
At the best of our knowledge, a comprehensive research on dairy products from minor species (equine, camel, yak) is not yet available in the scientific literature, due to a lack of economic relevance. The most well-known dairy product made from mare's milk is koumiss. It is a fermented alcoholic milk beverage that has been created since around the year 2000 BC. It is most well-known and commonly consumed in Central Asian regions. It is usually obtained from fresh raw milk fermented by starters (mainly lactic acid bacteria and yeast) at 20–30% dose and stored in a suitable container or in a fermentation bag, usually at 20–30 °C [40]. The production of cheese from mare's milk is considered inadvisable due to rennet coagulation problems [26]. However, several recent studies have shown that cheese can be made from donkey milk by following special technological approaches such as using certain types of rennet, strong coagulation conditions, fortification of other types of milk, and the addition of transglutaminase to better crosslink the milk proteins [41]. The collateral aim of the present work was to show ability of camel chymosin to coagulate mare's milk, and partially characterize its' clotting features.
2. Materials and methods
2.1. Vectors, strains, cloning enzymes and chemicals
Vector pGAPZαA (Invitrogen, USA) was used to construct the expression cassette. Restriction enzymes EcoRI and NotI, phosphatase FastAP, T4 DNA ligase, and Phusion High-Fidelity DNA Polymerase (Thermo Fisher, USA) were employed for the amplification and cloning of the target gene. Escherichia coli strain DH5α was acquired from Thermo Fisher Scientific. P. pastoris GS115 cells were purchased from Invitrogen (USA). The chemical reagents used in this study were of molecular biology or pure analytical grade and purchased from Sigma-Aldrich (St. Louis, USA) and AppliChem (Darmstadt, Germany). The vectors and enzyme were stored at -20 °C, the strains were stored at -80 °C and chemicals were stored accordingly manufactured recommendations.
2.2. Synthesis of the gene and vector construction
The amino acid sequence of the C. bactrianus prochymosin gene was retrieved from GenBank (accession No. JARL00000000.1) [34]. The nucleotide sequence was codon optimized for expression in yeast. The gene was synthesized by the Macrogen Company (Korea) and was shipped in the pTOP_Blunt_V2 vector. The prochymosin gene was amplified from pTOP_Blunt_V2/ProchymCB with PCR primers ProchymCB-EcoRI (5′-CCGGAATTCTCTGGAATTACTAGAATCCCATTG-3′) and ProchymCB-NotI (5′-ATAGTTTAGCGGCCGCTTAGATAGCTTTAGCCAATCCAACTCTGTTGTTAGC-3′) and was cloned into the pGAPZαA vector at the EcoRI/NotI sites, resulting in shuttle plasmid pGAPZαA/ProchymCB. The encoded prochymosin protein carries an N-terminal α-factor signal peptide for secretion in yeast culture.
2.3. Transformation of P. pastoris cells and producer strain preparation
The pGAPZαA/ProchymCB vector was linearized with endonuclease PagI (Thermo Fisher, USA) in Buffer O (Thermo Fisher, USA). The linearized vector was purified by phenol/chloroform extraction followed by ethanol precipitation. P. pastoris GS115 cells were electroporated with the linearized vector as follows. Fresh competent P. pastoris cells were prepared and transformed according to the instructions included with the EasyselectTM Pichia Expression Kit manual (Invitrogen). To 80 μL of the cells, 3 μg of purified linearized plasmid DNA was added, and then the suspension was pulsed in a 0.2 cm electroporation cuvette at 2 kV for 4.8 ms with electoporator (MicroPulser™, Bio-Rad, USA). Clones of P. pastoris GS115/pGAPZαA/ProchymCB were selected on an agar plate with zeocin (200 μg/mL). After that, the clones were screened for the presence of the insert by PCR with primers GAPfw (5′-GTCCCTATTTCAATCAATTGAA-3′) and AOX1rv (5′-GCAAATGGCATTCTGACATCC-3′). The clones positive for pGAPZαA/ProchymCB insert were analyzed for chymosin expression by a milk-clotting assay. Clone with the highest chymosin expression was preserved with glycerol at −80 °C and served as a producer strain.
2.4. Determination of stability of the gene integration in the genome of P. pastoris
Determination of stability of the gene integration in the yeast genome was performed accordingly [42], the strain was cultivated on the YPD medium without any antibiotic for 20 generations. Colonies were then plated to YPD agar containing zeocin and grown for 3 days. The viable colonies on each plate were then counted. The genomic DNA of the transformants was analyzed by PCR to confirm integration of the cassette in the genome of P. pastoris.
2.5. Determination of an optimal medium for prochymosin expression
Cultivation of the Pichia pastoris GS115/pGAPZαA/ProchymCB strain was evaluated in eight media (Table 1). The cultivation was carried out according to the EasySelect™ Pichia pastoris Kit (Invitrogen, USA) recommendations in 250 mL flasks with 50 mL of a medium at 28°С and 250 rpm for 72 h. Then, 1 mL the culture was collected every 24 h and centrifuged at 13200 × g, for 20 min, at 4 °C (Eppendorf Centrifuge 5415R, Hamburg, Germany). The supernatants were stored at 4 °C and were subjected to the milk-clotting assay.
Table 1.
Composition of the media tested in this work.
| Medium name | Composition |
|---|---|
| YPD | 1% yeast extract, 2% peptone, 3% dextrose, pH 7.0 |
| YPD + BMz(5g/L) | 1% yeast extract, 2% peptone, 3% dextrose, 5 g/L beet molasses, pH 7.0 |
| YPD + BMz(10g/L) | 1% yeast extract, 2% peptone, 3% dextrose, 10 g/L beet molasses, pH 7.0 |
| YPD + BMz(20g/L) | 1% yeast extract, 2% peptone, 3% dextrose, 20 g/L beet molasses, pH 7.0 |
| YP + BMz(10g/L) | 1% yeast extract, 2% peptone, 10 g/L beet molasses, pH 7.0 |
| YPD + CAS + BMz(10g/L) | 1% yeast extract, 2% peptone, 3% dextrose, 100 mМ citrate-phosphate buffer рН 4.0, 10 mМ ascorbic acid, 5% sorbitol, 10 g/L beet molasses |
| YPD + CAS + CrMz(10g/L) | 1% yeast extract, 2% peptone, 3% dextrose, 100 mМ citrate-phosphate buffer рН 4.0, 10 mМ ascorbic acid, 5% sorbitol, 10 g/L corn molasses |
| D + CAS | 3% dextrose, 100 mМ citrate-phosphate buffer рН 4.0,10 mМ ascorbic acid, 5% sorbitol |
YP – yeast extract and peptone; D – dextrose; CAS – citrate-phosphate buffer with ascorbic acid and sorbitol; CrMz – corn molasses; BMz – beet molasses.
2.6. Enzyme production in shake flasks
Pichia pastoris GS115/pGAPZαA/ProchymCB strain cells were inoculated into a 50 mL flask containing 5 mL of YPD with zeocin (100 μg/mL) and were cultured overnight at 30 °C and 250 rpm in a shaking incubator (KS 4000 i control, IKA, Germany). The overnight culture was inoculated into 50 mL of YPD in a 500 mL shake flask and was incubated at 30 °C and 250 rpm for overnight. The overnight culture was inoculated into 500 mL of YPD + CAS + BMz (10g/L) in a 5 L shake flask and was incubated in a shaking incubator (Climo-Shaker ISF1-X, Kuhner, Switzerland) at 28 °C and 250 rpm for 120 h with daily addition of 40% glucose (0.8 % final) and citrate-phosphate buffer рН 4.0 (100 mМ final). The cells were collected by centrifugation at 3500×g, for 20 min, at 4 °C (Avanti J-26SXP with rotor JA-10, Beckman Coulter, USA) and discarded. The supernatant was used for enzyme purification.
2.7. Deglycosylation
Deglycosylation was performed accordingly [43]. The Pichia pastoris GS115/pGAPZαA/ProchymCB culture of was grown in 200 mL of the YPD + CAS + BMz (10g/L) medium in a flask for 144 h at 28 °C and 250 rpm. The cells were collected by centrifugation (3500 × g, 4 °C, 15 min) (Avanti J-26SXP with rotor JA-10, Beckman Coulter, USA) and discarded. The supernatant was clarified by centrifugation at 40000 × g, for 1 h at 4 °C (Avanti J-26SXP with rotor JA-20, Beckman Coulter, USA) and filtered through a 0.22 μm filter, and 20 mL of the filtrate was loaded onto 10 kDa MWCO (molecular-weight cutoff) protein concentrators (Thermo Fisher, USA). After concentration the chymosin (1 mg/mL) was denatured at 95 °C for 5 min and deglycosylated with endoglycosidase H (New England Biolabs, USA) at 37 °C for 16 h. The reaction was stopped by heating at 65 °C. The result was analyzed by western blotting.
2.8. Western blotting with an anti-prochymosin antibody
For western blotting, we applied polyclonal antibodies raised in rabbits against calf prochymosin. Western blotting was performed according to the standard protocol [44]. Briefly, protein samples were separated by SDS-PAGE in a 12% (w/v) gel and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was stained with Ponceau S dye (0.1% (w/v) of Ponceau S in 5% (v/v) acetic acid) to confirm protein transfer. Membrane was blocked by 5% (w/v) skim milk powder in Tween 20/Tris-buffered saline (TBST: 50 mM Tris-HCl pH 7.6, 150 mM NaCl, 0.1% (w/v) Tween 20). The C. bactrianus prochymosin protein was detected with the rabbit polyclonal antibody against calf prochymosin (with 1:5000 dilution) as a primary antibody and a goat anti-rabbit IgG antibody conjugated with horseradish peroxidase (Sigma-Aldrich Chimie S.a.r.l., Lyon, France) (with 1:10,000 dilution) as a secondary antibody. The bands were detected by the Enhanced Chemi-Luminescence Detection Kit (Applichem GmbH, Darmstadt, Germany), and an X-ray film was then exposed to the membrane (AgfaPhoto GmbH, Germany).
2.9. Purification of recombinant camel chymosin
Ion exchange chromatography was used to purify recombinant camel chymosin from yeast culture [45]. The supernatant of GS115/pGAPZαA/ProchymCB culture was passed through a 0.22 μm filter and pH was changed to 4.5 with 25 mM sodium acetate, followed by incubation at room temperature for 24 h with stirring to activate all produced chymosin. Activated culture supernatant was diluted 3 times with 25 mM sodium acetate, and pH was lowered to 3.0 with 1M HCl to make chymosin net charge positive. The mixture was loaded onto anion-exchange column, using peristaltic pump P1 (Pharmacia, Sweden), with DEAE-Sepharose FF (GE Healthcare, USA) equilibrated with 25 mM NaCl in 50 mM sodium citrate buffer (pH 3.0); this step was defined as clarification. The flow through the column was collected and loaded onto cation-exchange column, using peristaltic pump P1, with SP-Sepharose FF (Sigma-Aldrich, USA) equilibrated with 25 mM NaCl in 50 mM sodium citrate buffer (pH 3.0). The column was next washed with 5 mL of 25 mM NaCl in 50 mM sodium citrate buffer (pH 3.0) and 15 mL of 50 mM NaCl in 25 mM sodium acetate buffer (pH 5.5) to change the net charge of chymosin to negative. Then, the protein was eluted with 750 mM NaCl in 25 mM sodium acetate buffer (pH 5.5), and was collected by 1 mL fractions. The fractions with milk-clotting activity were combined, and the concentration of NaCl was lowered to 25 mM by dilution with 25 mM sodium acetate buffer (pH 5.5) and loaded, using peristaltic pump P1, onto the strong cation-exchange column with Q-Sepharose FF (Sigma-Aldrich, USA) equilibrated with 25 mM NaCl in 25 mM sodium acetate buffer (pH 5.5). The column was washed with 25 mM NaCl in 25 mM sodium acetate buffer (pH 5.5), and chymosin was eluted by means of a 50–2000 mM gradient of NaCl in 25 mM sodium acetate buffer (pH 5.5). The fractions were analyzed by the milk-clotting assay and the most active fractions were applied to SDS-PAGE. The SDS-PAGE was conducted according to the Laemmli method [46] in a Mini-PROTEAN Tetra cell (Bio-Rad Laboratories Inc., USA).
2.10. Determination of protein concentration
Protein concentration was determined by Bradford method [47] using Bio-Rad assay reagent (Bio-Rad Protein Assay Day, Munich, Germany) and bovine serum albumin as the standard. The measurements were carried out in three repetitions, with the average of the three repetitions being reported as the specified result.
2.11. The milk-clotting assay
The milk-clotting assay was carried out in accordance with ref. [48]. This assay was performed with powdered cow's skim milk reconstituted at 12% (w/v) in 0.025 M sodium acetate buffer, pH 6.0, as a substrate. The enzymatic reactions for clone selection were carried out at least in triplicate at 37 °C in test tubes with 20 μL of an enzyme solution and 1 mL of the substrate. The milk clots were visualized by turning the tubes upside down. One unit of milk-clotting activity was defined as the quantity of the enzyme required for clotting 1 mL of cow's skim milk in 40 min at 35 °C. Eq. (1) below shows how chymosin activity units (A) were calculated:
| (Equation 1) |
there Vmilk is milk volume (mL), Vchymosin is volume of added chymosin (mL), Tmc is milk-clotting time (sec).
The milk-clotting assay on mare's milk was carried out by using freeze-dried mare's milk (SaumalBioTech LLP, Kazakhstan.). Powder milk was reconstituted at 20% in deionized water, preheated to 40 °C, defatted by centrifugation at 3000 × g for 15 min (Eppendorf Centrifuge 5415R, Hamburg, Germany). Final composition of the substrate solution was brought to next: 10% mare milk, 0.2M buffer solution with an appropriate pH, 30 mM CaCl2. Purified recombinant camel chymosin was diluted to 50 ng/μl for cow's milk and to 2 μg/μl for mare's milk with 10 mM sodium acetate buffer (pH 5.5) and 10 μl of enzyme was incubated with 1 mL of the milk substrate at 37 °C. For cow's milk clotting time in each tube was measured by time first flakes observed by inversion of tube. For mare's milk reaction at each time point (15 s interval) was stopped by addition of pepstatin A in a final concentration in tube 50 μg. Stopped reaction was then centrifuged at 3500 × g for 3 min to divide precipitated casein. And time of the most transparent supernatant was recorded as clotting time.
2.12. Measurement of optimal substrate pH for the milk-clotting activity
To this end, skim milk powder was dissolved in either 50 mM sodium acetate buffer (pH 4.5–5.5), 50 mM imidazole buffer (pH 6.0–6.5), or 50 mM Tris-HCl buffer (pH 7.0–8.0). The milk solutions were placed as a substrate in 1.5 mL tubes. Maximum enzyme activity was defined as 100% activity, and the other samples were evaluated based on that. The tests were carried out in three repetitions, and the outcome was determined as the average of the three repetitions.
2.13. Determination of optimal temperature for the milk-clotting activity
The milk-clotting activity was measured over the temperature range of at 0–70 °C (with 5 °C intervals) at 25 mM sodium acetate buffer (pH 6.0) for cow's milk and at 200 mM sodium acetate buffer (pH 5.0) for mare's milk. Substrate and enzyme solutions were preheated to the reaction temperature for 5 min, and the solutions were mixed. Maximum enzyme activity was defined as 100% activity, and the other samples were evaluated based on that. The tests were carried out in three repetitions, and the outcome was determined as the average of the three repetitions.
2.14. Effect of CaCl2 on the milk-clotting activity
The effect of calcium on the milk-clotting activity was performed as described by [23]. Cow's milk-clotting activity were examined in an assay at 37 °C in the presence of CaCl2 at a concentration from 0 to 160 mM, dissolved in 25 mM sodium acetate buffer (pH 6.0). Enzyme activity in the absence of metal ions was assumed to be 100%, and the other samples with varied metal ions were tested based on that. The tests were carried out in three repetitions, and the outcome was determined as the average of the three repetitions.
2.15. Effects of metal ions on the milk-clotting activity
The effects of various metal ions on the milk-clotting activity were examined in an assay at 37 °C in the presence of each of these 11 chlorides: CaCl2, CoCl2, NiCl2, FeCl2, BaCl2, ZnCl2, MgCl2, MnCl2, LiCl, KCl, and NaCl at a concentration of 10 mM, dissolved in 25 mM sodium acetate buffer (pH 6.0). Enzyme activity in the absence of metal ions was assumed to be 100%, and the other samples with varied metal ions were tested based on that. The tests were carried out in three repetitions, and the outcome was determined as the average of the three repetitions.
2.16. Enzyme production in a bioreactor
A 10 L fermenter (Biostat, Sartorius, Germany) was utilized to determine the ability of the yeast strain to produce C. bactrianus recombinant chymosin on a large scale. A single colony was inoculated into 5 mL of YPD broth and grown at 28 °C and 250 rpm. After 24 h, the culture was transferred to 30 mL of YPD broth and grown in the same condition for 24 h. The culture was inoculated into 300 mL of the YPD + CAS + BMz (10g/L) medium and grown at 28 °C and 250 rpm for 24 h. Then, the culture was inoculated into 3 L of the YPD + CAS + BMz (10g/L) medium in the 10 L bioreactor. Standard procedures were utilized to operate the fermenter under the following cultivation conditions: 28 °C, 400 rpm, aeration 2–4 L/min, pH 4.0, fed carbon sources: 3% glucose and 5% 100 mM citrate-phosphate buffer (pH 4.0) (added each 24 h), and cultivation duration: 144 h.
Using Eqs. (2) and (3), the biomass and enzyme production rates (ΔBiomassmax and ΔEnzymemax) were computed as the ratio of the increase in wet cells or activity to the cultivation period during which the rise in biomass/activity occurred.
| (Equation 2) |
| (Equation 3) |
where ΔWCW is weight of produced wet cells (g) in period of time, ΔEC as enzyme activity of produced cells (U) in period of time, T is cultivation period (h). ΔWCW was calculated as a difference of wet cells weight before and after cultivation period. ΔEC was calculated as an activity unit yield difference before and after cultivation period.
2.17. Software and statistical analysis
All tests were carried out in triplicate. In the quantitative study, the mean values and standard deviations were obtained using the GraphPad Prism Version 8.0.1 software in this work. The mean value of enzyme activity was shown, while other values were shown as mean ± standard deviations (SD; n = 3). Calculation of protein molecular mass, isoelectic point, nucleotide sequence reading and primers design and other manipulations was performed using Vector NTI Advance 11 and SnapGene Viewer 5.2.4 software.
3. Results and discussion
The prochymosin gene of C. bactrianus was inserted into a yeast expression vector pGAPZαA under the control of the constitutive GAP promoter to construct plasmid pGAPZαA/ProchymCB. Twenty transformants were checked by PCR for the presence of the chymosin gene in genomic DNA. Ten transformants with the most intense glow of the PCR product under UV light were chosen and grown in YPD + CAS + BMz (10g/L) broth. The supernatants were analyzed by the milk-clotting assay, the results are indicated in Figure 2, milk-clotting activity units were calculated using Eq. (1). Six clones expressed chymosin with a good milk-clotting activity (Figure 2, Supplementary materials: Figure S1). Clone N8 showed maximum milk-clotting activity (1379 U/mL) and was chosen as a producer strain of camel chymosin.
Figure 2.
Results of milk-clotting screening of culture supernatants of the GS115/pGAPZαA/ProchymCB clones.
A basal salt medium (BSM) proposed through Invitrogen is usually hired to acquire the excessive-density cultures, however it isn't always the proper complement because of its excessive ionic strength, medium additives precipitation, and imbalanced nutrient concentration [49]. The media is troublesome to organize for big scale uses and needs the addition of vitamin H that is pricey vitamin, and of mixture of micronutrients, several of which can inhibit recombinant protein production [50]. Well-known medium for the cultivation of Pichia pastoris yeast is YPD, containing peptone and yeast extract as a nitrogen source and glucose is added as a carbon source. The addition of beet molasses to YPD at a concentration of 0.5% and 1% increases the level of chymosin expression by 2.6 and 11.3%, respectively (Table 2). A further increase in the molasses content up to 2% strongly inhibited the expression level of the recombinant chymosin. The absence of glucose in the medium also had a negative effect, which is explained by the very low content of glucose in molasses - about 0.28 % of dry matter [51], which is needed to activate the P. pastoris GAP promoter [52]. Biotin is the sole vitamin in a commonly used rich growth medium for P. pastoris [53] to increase growth rate and biomass yield [54], but other complex vitamin sources such as yeast extract and molasses may achieve the same effect [55]. The pH of the medium has a significant impact on chymosin production by the recombinant yeast strain [56]. Optimal pH of medium for chymosin production is pH 4.0 [57,58] and it was maintained by citrate-phosphate buffer. Multicomponent medium with beet (YPD + CAS + BMz (10g/L)) or corn molasses (YPD + CAS + CrMz(10g/L)) increase chymosin production for 26 and 14%, respectively (Table 1). The lack of yeast extract and peptone in the medium (D + CAS) leads to the absence of strain growth, demonstrating the importance of both carbon and nitrogen [55].
Table 2.
The milk-clotting activity of the camel recombinant chymosin depending on the growth medium.
| Medium | Milk-clotting activity, U/mL |
|---|---|
| YPD | 1043 ± 18 |
| YPD + BMz(5g/L) | 1071 ± 33 |
| YPD + BMz(10g/L) | 1176 ± 37 |
| YPD + BMz(20g/L) | - |
| YP + BMz(10g/L) | - |
| YPD + CAS + BMz(10g/L) | 1411 ± 45 |
| YPD + CAS + CrMz(10g/L) | 1212 ± 27 |
| D + CAS | - |
Yeasts have a strong ability to glycosylate proteins. Pichia may have an advantage in the glycosylation of secreted proteins over Saccharomyces cerevisiae because it does not hyperglycosylate (Figure 3, Supplementary materials: Figure S2). The average length of the oligosaccharide chains added post-translationally to Pichia proteins is 8–14 mannose residues per side chain [59]. From amino acid sequence of the C. bactrianus prochymosin, it follows that the protein has two potential sites for N-glycosylation: Asp100-Gln101-Thr102 and Asp291-Asp292-Ser293. Furthermore, very little O-linked glycosylation has been detected in Pichia [59]. Figure 3 (Supplementary materials: Figure S2) depicts the results of western blotting before (three bands on lane 8) and after (one band on lane 9) treatment with Endo H and indicates that two well-defined bands in the culture supernatant are forms of the same protein with one or two glycosylated sites and without glycosylation.
Figure 3.
Western blotting analysis of hourly accumulation of the camel recombinant chymosin in culture media, and analysis of the culture supernatant of strain GS115/pGAPZαA/ProchymCB before (lane 8) and after deglycosylation (lane 9). 1: bovine chymosin; 2: negative control; 3: 24 h; 4: 48 h; 5: 72 h; 6: 96 h; 7: 120 h; 8: 144 h. 9: 144 h supernatant treated with Endo H.
The calculated molecular mass of C. bactrianus unglycosylated chymosin is 35.6 kDa. It follows from Figure 3 that the N-glycosylation increases the molecular mass by approximately 7 kDa. Using a mannose oligosaccharide linked to asparagine by two N-acetylglucosamines, P. pastoris may N-glycosylate proteins [60]. The most popular and significant form of post-translational modification is N-glycosylation [61]. Glycosylation of various chymosins expressed in yeast has been observed for: buffalo (Bubalus arnee bubalis) [62], camel (Camelus dromedaries) [33], cow (Bos Taurus) [50], goat (Capra hircus) [63], yak (Bos grunniens) [19] and this is not unusual for yeast-produced proteins [62].
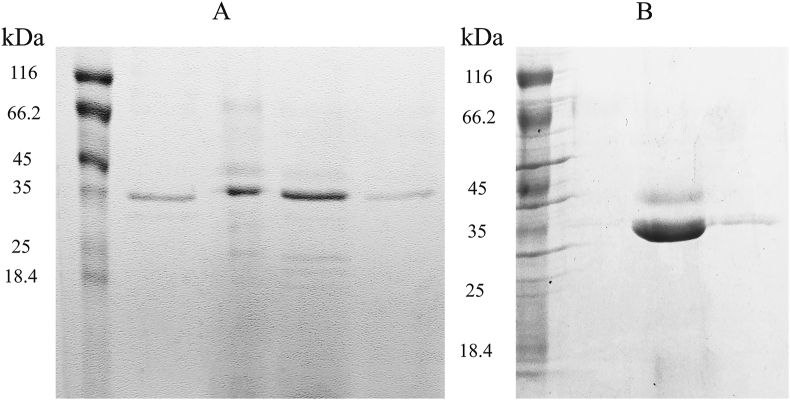
The combination of anion exchange and cation exchange chromatography made it possible to purify chymosin from the medium and to concentrate the recombinant protein. By calculation, was established that bactrian camels’ mature chymosin has isoelectric point = 4.87 and was found to not bind to DEAE-Sepharose and binds to SP-Sepharose at pH 3.0 and to Q-Sepharose at pH 5.5 (Figure 4, Supplementary materials: Figure S3). As shown in Figure 4A recombinant chymosin purified on SP-Sepharose column migrated in SDS-PAGE as 2 protein bands (with molecular weights of 42 and 35 kDa). This reflects the glycosylation pattern. As we see in Figure 4B Q-Sepharose allowed to concentrate the chymosin and to get rid of other proteins.
Figure 4.
Purification of the camel recombinant chymosin on SP-Sepharose FF (A) and Q-Sepharose FF (B).
Optimal pH for the activity of highest-purity C. bactrianus recombinant chymosin was pH 4.5 and 4.0 for cow's and mare's milk, respectively (Figure 5). With rising pH, the relative clotting activity decreased. At pH 6.0, the relative milk-clotting activity on cow's milk reduced to 47%, and at pH 7.8 and 8.0, it reduced to 36% and 12%, respectively. In mare's milk the chymosin activity pattern observed only at pH 3.0–5.5 range. Lower pH values for both milk types due to its acid coagulation occurrence [25] couldn't be achieved, and at higher pH values no coagulation was observed. These data indicated that the camel chymosin just as other chymosins has the highest activity under acidic conditions. For bovine recombinant chymosin, the highest activity is detected at pH 5.5 [56], whereas for dromedary recombinant chymosin expressed in the filamentous fungus A. niger or yeast P. pastoris, the highest activity is seen at pH 6.0 [23] and 5.04 [33], respectively. These data imply that such factors as the origin of the enzyme, an expression system, and the degree of its purification influence chymosin activity.
Figure 5.
Optimal substrate pH for the milk-clotting activity of the camel recombinant chymosin.
The milk-clotting activity of recombinant chymosin is strongly affected by temperature [23, 33, 56]. In our study, the optimal temperature for relative milk-clotting activity was found to be in the range 45–50 °C for cow's milk and 70 °C for mare's milk (Figure 6). Similar data are reported in the literature [23, 33]. For calf recombinant chymosin, the optimum temperature is 37 °C [50], and for lamb chymosin 44 °C [64]. Here, the reaction rate at 45 °C on cow's milk and at 70 °C on mare's milk increased two-fold as compared with the physiological temperature of 37 °C. At 70 °C, the activity of chymosin on cow's milk decreases to zero, while it reaches a maximum on mare's milk. Different values of the optimum temperature for two substrates (cow's and mare's milk) we associate with different mechanisms of action of camel chymosin. In cow's milk, the main substrate for the chymosin is a kappa-casein, which ensures the stability of casein micelles [65], and for mare's milk, which is albumin milk, kappa-casein does not play such a key role [25] and in this case, all milk components undergo proteolysis. Mare's milk has a chymosin-induced clotting system that probably differs from bovine milk.
Figure 6.
Optimal temperature for the milk-clotting activity of the camel recombinant chymosin.
The activity of aspartic proteases is influenced by the presence of chloride and calcium metal ions in a substrate solution. Here, the dependence of camel recombinant chymosin activity on the concentration of calcium ions in the range of 0–150 mM and the influence of 10 kinds of metal ions at 10 mM on the milk-clotting activity were studied too. The results indicated that concentration of CaCl2 that ensured the milk-clotting activity stability of the camel recombinant chymosin was in the range of 20 and 50 mM with an optimum at 30 mM (Figure 7).
Figure 7.
The impact of CaCl2 concentration on the cow's milk-clotting activity of the camel recombinant chymosin.
Relative clotting activity was 24% when CaCl2 was absent and less than 35% when the concentration of CaCl2 was 150 mM. These results are consistent with similar data on bovine and dromedary chymosins [33, 50].
The cow's milk-clotting activity of C. bactrianus recombinant chymosin increased twofold when 10 mM CoCl2 and MgCl2 were added (Table 3). After the addition of 10 mM FeCl2 and BaCl2, the activity increased 3.5- to 3.7-fold and increased more than fivefold when 10 mM MnCl2 was provided. In contrast, 10 mM NiCl2 strongly suppressed the cow's milk-clotting activity. Our data on C. bactrianus recombinant chymosin are similar to findings about calf recombinant chymosin [50]. Unlike cow's milk, no dependence of chymosin activity in the presence of metal ions was observed in mare's milk. We assume that the mare's milk clotting occurs due to the proteolytic activity of chymosin, which is weakly impacted by metal ions [23].
Table 3.
Effects of metal ions on the milk-clotting activity of the camel recombinant chymosin.
| Metal ion | Concentration, mM | Relative activity, % (±st.d.) |
|---|---|---|
| Control | - | 100 ± 3.95 |
| Ca2+ (CaCl2) | 10 mM | 298.8 ± 16.04 |
| Co2+ (CoCl2) | 10 mM | 182.7 ± 8.21 |
| Ni2+ (NiCl2) | 10 mM | 19.9 ± 0.39 |
| Fe2+ (FeCl2) | 10 mM | 338.7 ± 12.99 |
| Ba2+ (BaCl2) | 10 mM | 373.5 ± 20.06 |
| Zn2+ (ZnCl2) | 10 mM | 98.5 ± 4.54 |
| Mg2+ (MgCl2) | 10 mM | 188.2 ± 5.31 |
| Mn2+ (MnCl2) | 10 mM | 518.4 ± 19.02 |
| Li1+ (LiCl) | 10 mM | 102.8 ± 2.73 |
| K1+ (KCl) | 10 mM | 97 ± 4.61 |
| Na1+ (NaCl) | 10 mM | 105 ± 4.06 |
In order to test the possibilities of scaling up the production of recombinant C. bactrianus chymosin, P. pastoris clone 8 was cultivated in a 10 L bioreactor to ferment the yeast strain (Figure 8). Biomass growth was found to correlate well with yield of enzyme activity. This is important because biomass assessment is critical for P. pastoris fermentation process control [66].
Figure 8.
Pilot scale production of C. bactrianus recombinant chymosin by yeast P. pastoris.
Using optimal growth conditions, YPD + CAS + BMz (10g/L) growth medium allowed us reaching a wet cell weight of 382.02 g after 144 h growth of P. pastoris strain GS115/pGAPZαA/ProchymCB (Table 4). Protein production was started from the first 24 h of cultivation and reached 375 U/ml. Total yield of the recombinant enzyme was 80 mg per liter of yeast culture. The resulting supernatant at 144 h cultivation contained 1412 U/mL activity.
Table 4.
Biomass and enzyme production rate.
| Cultivation period, h | Volume, mL | Wet cells weight, g | Activity, U | ΔBiomassmax, g/h | ΔEnzymemax, U/h |
|---|---|---|---|---|---|
| 0–24 | 3000 | 69 | 1 125 000 | 2,88 | 46 875 |
| 24–48 | 3400 | 170 | 2 567 000 | 4,21 | 60 083 |
| 48–72 | 3900 | 273 | 4 067 700 | 4,29 | 62 529 |
| 72–96 | 4500 | 345 | 6 205 500 | 3,00 | 89 075 |
| 96–120 | 5200 | 377 | 7 342 400 | 1,33 | 47 371 |
| 120–144 | 6000 | 382 | 8 472 000 | 0,21 | 47 067 |
According to our calculations, using Eqs. (2) and (3), the maximum of biomass production rate of 4.29 g/h is observed on the third day. Maximum of enzyme production rate of 89 075 U/h was at the fourth day. After 96 h of growth, no further growth in chymosin production was observed, which coincided with the beginning of the stationary phase of growth. In compare, C. dromedarius and Yak chymosins with methanol-induced expression in P. pastoris under AOX1 promoter, maximum enzyme yield was observed at 144 h and 184 h respectively [33, 67]. These results indicate that C. bactrianus chymosin expression in P. pastoris under the control of the constitutive (GAP) promoter is effective.
C. bactrianus chymosin was tested in mare's milk coagulation. It is known that mare's milk differs significantly from that of other dairy mammals [68]. The whey protein fraction represents almost 40 % in mare's milk and less than 20 % in cow's milk. Mare's milk could be defined typically as albuminous milk. Due to this, there is a big problem in processing and clot formation of mare's milk. Previous investigation on rennet-induced coagulation of mare's milk reported that it does not form a gel during renneting, by bovine chymosin and is not suitable for cheese production [26].
Future studies will focus on the kinetic and thermodynamic parameters of the C. bactrianus chymosin, as it was shown for other enzymes [16, 69, 70], optimal parameters for large-scale and industrial fermentation of GS115/pGAPZαA/ProchymCB strain will be find out and cheese making conditions from mare's and camel's milk using C. bactrianus chymosin will be assayed.
4. Conclusions
In this study, a recombinant chymosin from the two-humped camel C. bactrianus was characterized for the first time. The C. bactrianus chymosin gene was cloned and expressed in P. pastoris under the control of the GAP promoter and purified from culture via a combination of cation and anion exchange chromatography. C. bactrianus recombinant chymosin manifested high milk-clotting activity (9605 U/mg) with optimum conditions for cow's milk-clotting activity at 45 °C, pH 4.5, and 30 mM CaCl2 and for clotting mare's milk at 70 °C, pH 4.0 The dependence of this activity on metal ions was investigated too. The activity of cow's milk clotting is increased by Mn2+, Fe3+, Ba2+, Co2+, and Mg2+ and strongly suppressed by Ni2+ and no effect on mare's milk clotting was observed. Recombinant-chymosin production in a 10 L fermenter indicates that the camel chymosin constitutive expression P. pastoris yeasts could be used for large scale production for milk-clotting enzyme manufacturing with 80 mg/L of enzyme yield. The proposed expression system has high efficiency of the production of C. bactrianus recombinant chymosin. The inclusion of the constitutive GAP promoter to control the heterologous expression may be a good solution for the production of a recombinant enzyme for the food industry [71]. Our research on the optimal medium composition allowed us to find out conditions for the recombinant-chymosin manufacture under the control of this constitutive promoter. C. bactrianus recombinant chymosin was used to prepare cheese from bovine milk; this confirms the applicability of the new enzyme to a cheese-making process. C. bactrianus recombinant chymosin appears to be a good alternative to the milk-clotting enzymes currently used in the cheese industry. Milk-clotting activity on a mare's milk has the prospect for processing mare's and donkey's milk.
Declarations
Author contribution statement
Zhiger Akishev: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Assel Kiribayeva, Arman Mussakhmetov, Kairat Baltin: Performed the experiments.
Yerlan Ramankulov: Contributed reagents, materials, analysis tools or data.
Bekbolat Khassenov: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research was funded by the Science Committee of the Ministry of Education and Science of the Republic of Kazakhstan (Grant No. AP09562228 and AP05133470).
Data availability statement
The data that has been used is confidential.
Declaration of interests statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Szecsi P.B. The aspartic proteases. Scand. J. Clin. Labor. Invest. Suppl. 1992;210:5–22. [PubMed] [Google Scholar]
- 2.Fruton J.S. A history of pepsin and related enzymes. Q. Rev. Biol. 2002;77:127–147. doi: 10.1086/340729. [DOI] [PubMed] [Google Scholar]
- 3.Foltmann B. A review on prorennin and rennin. C. R. Trav. Lab. Carlsberg. 1966;35:143–231. [PubMed] [Google Scholar]
- 4.Rawlings N.D., Barrett A.J., Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mohanty A.K., Mukhopadhyay U.K., Grover S., Batish V.K. Bovine chymosin: production by rDNA technology and application in cheese manufacture. Biotechnol. Adv. 1999;17:205–217. doi: 10.1016/s0734-9750(99)00010-5. [DOI] [PubMed] [Google Scholar]
- 6.Abdel-Raouf H.-A., El-Neshwy A., Rabie A., Khalifa S. Evaluation of rennet substitute from artichoke (Cynara scolymus L.) flowers extracts: study the factors affecting the activity of milk clotting. Zagazig J. Agr. Res. 2017;44:2203–2219. [Google Scholar]
- 7.Zhang Y., Wang H., Tao L., Huang A.X. Milk-clotting mechanism of Dregea sinensis Hemsl. protease. J. Dairy Sci. 2015;98:8445–8453. doi: 10.3168/jds.2015-9851. [DOI] [PubMed] [Google Scholar]
- 8.González-Velázquez D.A., Mazorra-Manzano M.A., Martínez-Porchas M., Huerta-Ocampo J.A., Vallejo-Córdoba B., Mora-Cortes W.G. Exploring the milk-clotting and proteolytic activities in different tissues of Vallesia glabra: a new source of plant proteolytic enzymes. Appl. Biochem. Biotechnol. 2021;193:389–404. doi: 10.1007/s12010-020-03432-5. [DOI] [PubMed] [Google Scholar]
- 9.Nasr A.I., Mohamed Ahmed I.A., Hamid O.I. Characterization of partially purified milk-clotting enzyme from sunflower (Helianthus annuus) seeds. Food Sci. Nutr. 2016;4:733–741. doi: 10.1002/fsn3.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Afsharnezhad M., Shahangian S.S., Sariri R. A novel milk-clotting cysteine protease from Ficus johannis: purification and characterization. Int. J. Biol. Macromol. 2019;121:173–182. doi: 10.1016/j.ijbiomac.2018.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Sbhatu D.B., Tekle H.T., Tesfamariam K.H. Ficus palmata ForskåL (BELES ADGI) as a source of milk clotting agent: a preliminary research. BMC Res. Notes. 2020;13:446. doi: 10.1186/s13104-020-05293-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.da Silva R.R., Souto T.B., Gonsales da Rosa N., de Oliveira L.C.G., Juliano M.A., Juliano L. Evaluation of the milk clotting properties of an aspartic peptidase secreted by Rhizopus microsporus. Prep. Biochem. Biotechnol. 2020;50:226–233. doi: 10.1080/10826068.2019.1683861. [DOI] [PubMed] [Google Scholar]
- 13.Ahmed S.A., Wehaidy H.R., Ibrahim O.A., Abd El Ghani S., El-Hofi M.A. Novel milk-clotting enzyme from Bacillus stearothermophilus as a coagulant in UF-white soft cheese. Biocat. Agr. Biotechn. 2016;7:241–249. [Google Scholar]
- 14.Silva B.L., Geraldes F.M., Murari C.S., Gomes E., Da-Silva R. Production and characterization of a milk-clotting protease produced in submerged fermentation by the thermophilic fungus Thermomucor indicae-seudaticae N31. Appl. Biochem. Biotechnol. 2014;172:1999–2011. doi: 10.1007/s12010-013-0655-7. [DOI] [PubMed] [Google Scholar]
- 15.Theron L.W., Divol B. Microbial aspartic proteases: current and potential applications in industry. Appl. Microbiol. Biotechnol. 2014;98:8853–8868. doi: 10.1007/s00253-014-6035-6. [DOI] [PubMed] [Google Scholar]
- 16.Wehaidy H.R., Abdel-Naby M.A., Shousha W.G., Elmallah M.I.Y., Shawky M.M. Improving the catalytic, kinetic and thermodynamic properties of Bacillus subtilis KU710517 milk clotting enzyme via conjugation with polyethylene glycol. Int. J. Biol. Macromol. 2018;111:296–301. doi: 10.1016/j.ijbiomac.2017.12.125. [DOI] [PubMed] [Google Scholar]
- 17.Meng F., Chen R., Zhu X., Lu Y., Nie T., Lu F. Newly effective milk-clotting enzyme from Bacillus subtilis and its application in cheese making. J. Agric. Food Chem. 2018;66:6162–6169. doi: 10.1021/acs.jafc.8b01697. [DOI] [PubMed] [Google Scholar]
- 18.Johnson M.E. A 100-Year Review: cheese production and quality. J. Dairy Sci. 2017;100:9952–9965. doi: 10.3168/jds.2017-12979. [DOI] [PubMed] [Google Scholar]
- 19.Ersöz F., İnan M. Large-scale production of yak (Bos grunniens) chymosin A in Pichia pastoris. Protein Expr. Purif. 2019;154:126–133. doi: 10.1016/j.pep.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 20.Belenkaya S.V., Bondar A.A., Kurgina T.A., Elchaninov V.V., Bakulina A.Y., Rukhlova E.A. Characterization of the Altai maral chymosin gene, production of a chymosin recombinant analog in the prokaryotic expression system, and analysis of its several biochemical properties. Biochemistry Biokhimiia. 2020;85:781–791. doi: 10.1134/S0006297920070068. [DOI] [PubMed] [Google Scholar]
- 21.Visser S., Slangen C.J., van Rooijen P.J. Peptide substrates for chymosin (rennin). Interaction sites in κ-casein-related sequences located outside the (103-108)-hexapeptide region that fits into the enzyme's active-site cleft. Biochem. J. 1987;244:553–558. doi: 10.1042/bj2440553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mercier J.-C., Chobert J.-M. Comparative study of the amino acid sequences of the caseinomacropeptides from seven species. FEBS (Fed. Eur. Biochem. Soc.) Lett. 1976;72:208–214. doi: 10.1016/0014-5793(76)80972-6. [DOI] [PubMed] [Google Scholar]
- 23.Kappeler S.R., van den Brink H.J., Rahbek-Nielsen H., Farah Z., Puhan Z., Hansen E.B. Characterization of recombinant camel chymosin reveals superior properties for the coagulation of bovine and camel milk. Biochem. Biophys. Res. Commun. 2006;342:647–654. doi: 10.1016/j.bbrc.2006.02.014. [DOI] [PubMed] [Google Scholar]
- 24.Al-Zoreky N.S., Almathen F.S. Using recombinant camel chymosin to make white soft cheese from camel milk. Food Chem. 2021;337:127994. doi: 10.1016/j.foodchem.2020.127994. [DOI] [PubMed] [Google Scholar]
- 25.Uniacke-Lowe T., Huppertz T., Fox P.F. Equine milk proteins: chemistry, structure and nutritional significance. Int. Dairy J. 2010;20:609–629. [Google Scholar]
- 26.Uniacke-Lowe T. University College Cork; 2011. Studies on Equine Milk and Comparative Studies on Equine and Bovine Milk Systems. PhD Thesis. [Google Scholar]
- 27.Bansal N., Drake M.A., Piraino P., Broe M.L., Harboe M., Fox P.F. Suitability of recombinant camel (Camelus dromedarius) chymosin as a coagulant for Cheddar cheese. Int. Dairy J. 2009;19:510–517. [Google Scholar]
- 28.Moynihan A.C., Govindasamy-Lucey S., Jaeggi J.J., Johnson M.E., Lucey J.A., McSweeney P.L.H. Effect of camel chymosin on the texture, functionality, and sensory properties of low-moisture, part-skim Mozzarella cheese. J. Dairy Sci. 2014;97:85–96. doi: 10.3168/jds.2013-7081. [DOI] [PubMed] [Google Scholar]
- 29.Langholm Jensen J., Molgaard A., Navarro Poulsen J.C., Harboe M.K., Simonsen J.B., Lorentzen A.M. Camel and bovine chymosin: the relationship between their structures and cheese-making properties. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013;69:901–913. doi: 10.1107/S0907444913003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gumus P., Hayaloglu A.A. Effects of blends of camel and calf chymosin on proteolysis, residual coagulant activity, microstructure, and sensory characteristics of Beyaz peynir. J. Dairy Sci. 2019;102:5945–5956. doi: 10.3168/jds.2018-15671. [DOI] [PubMed] [Google Scholar]
- 31.Bekele B., Hansen E.B., Eshetu M., Ipsen R., Hailu Y. Effect of starter cultures on properties of soft white cheese made from camel (Camelus dromedarius) milk. J. Dairy Sci. 2019;102:1108–1115. doi: 10.3168/jds.2018-15084. [DOI] [PubMed] [Google Scholar]
- 32.Jensen J.L., Jacobsen J., Moss M.L., Rasmussen F., Qvist K.B., Larsen S. The function of the milk-clotting enzymes bovine and camel chymosin studied by a fluorescence resonance energy transfer assay. J. Dairy Sci. 2015;98:2853–2860. doi: 10.3168/jds.2014-8672. [DOI] [PubMed] [Google Scholar]
- 33.Wang N., Wang K.Y., Li G., Guo W., Liu D. Expression and characterization of camel chymosin in Pichia pastoris. Protein Expr. Purif. 2015;111:75–81. doi: 10.1016/j.pep.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 34.Jirimutu Wang Z., Ding G., Chen G., Sun Y., Sun Z. Genome sequences of wild and domestic bactrian camels. Nat. Commun. 2012;3:1202. doi: 10.1038/ncomms2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ming L., Yuan L., Yi L., Ding G., Hasi S., Chen G. Whole-genome sequencing of 128 camels across Asia reveals origin and migration of domestic Bactrian camels. Commun. Biol. 2020;3:1. doi: 10.1038/s42003-019-0734-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaczensky P., Adiya Y., von Wehrden H., Mijiddorj B., Walzer C., Güthlin D. Space and habitat use by wild Bactrian camels in the Transaltai Gobi of southern Mongolia. Biol. Conserv. 2014;169:311–318. doi: 10.1016/j.biocon.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Damasceno L.M., Huang C.J., Batt C.A. Protein secretion in Pichia pastoris and advances in protein production. Appl. Microbiol. Biotechnol. 2012;93:31–39. doi: 10.1007/s00253-011-3654-z. [DOI] [PubMed] [Google Scholar]
- 38.Obst U., Lu T.K., Sieber V. A modular toolkit for generating Pichia pastoris secretion libraries. ACS Synth. Biol. 2017;6:1016–1025. doi: 10.1021/acssynbio.6b00337. [DOI] [PubMed] [Google Scholar]
- 39.Baghban R., Farajnia S., Rajabibazl M., Ghasemi Y., Mafi A., Hoseinpoor R. Yeast expression systems: overview and recent advances. Mol. Biotechnol. 2019;61:365–384. doi: 10.1007/s12033-019-00164-8. [DOI] [PubMed] [Google Scholar]
- 40.Kücükcetin A., Yaygin H., Hinrichs J., Kulozik U. Adaptation of bovine milk towards mares’ milk composition by means of membrane technology for koumiss manufacture. Int. Dairy J. 2003;13:945–951. [Google Scholar]
- 41.Faccia M., D’Alessandro A.G., Summer A., Hailu Y. Milk products from minor dairy species: a review. Animals. 2020;10:1260. doi: 10.3390/ani10081260. https://www.mdpi.com/2076-2615/10/8/1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parashar D., Satyanarayana T. Production of chimeric acidic α-amylase by the recombinant Pichia pastoris and its applications. Front. Microbiol. 2017;8:493. doi: 10.3389/fmicb.2017.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niu C., Han Y., Wang J., Zheng F., Liu C., Li Y. Comparative analysis of the effect of protein Z4 from barley malt and recombinant Pichia pastoris on beer foam stability: role of N-glycosylation and glycation. Int. J. Biol. Macromol. 2018;106:241–247. doi: 10.1016/j.ijbiomac.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Wood E.J. Molecular cloning. A laboratory manual. Biochem. Educ. 1983;11:82. [Google Scholar]
- 45.Jungbauer A., Hahn R. Chapter 22 ion-exchange chromatography. In: Burgess R.R., Deutscher M.P., editors. Methods in Enzymology. Academic Press; 2009. pp. 349–371. [Google Scholar]
- 46.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 47.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 48.Ageitos J.M., Vallejo J.A., Sestelo A.B., Poza M., Villa T.G. Purification and characterization of a milk-clotting protease from Bacillus licheniformis strain USC13. J. Appl. Microbiol. 2007;103:2205–2213. doi: 10.1111/j.1365-2672.2007.03460.x. [DOI] [PubMed] [Google Scholar]
- 49.Cos O., Ramón R., Montesinos J.L., Valero F. Operational strategies, monitoring and control of heterologous protein production in the methylotrophic yeast Pichia pastoris under different promoters: a review. Microb. Cell Factories. 2006;5:17. doi: 10.1186/1475-2859-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang X.P., Yin M.L., Chen P., Yang Q. Constitutive expression, purification and characterization of bovine prochymosin in Pichia pastoris GS115. World J. Microbiol. Biotechnol. 2012;28:2087–2093. doi: 10.1007/s11274-012-1012-7. [DOI] [PubMed] [Google Scholar]
- 51.Palmonari A., Cavallini D., Sniffen C.J., Fernandes L., Holder P., Fagioli L. Short communication: characterization of molasses chemical composition. J. Dairy Sci. 2020;103:6244. doi: 10.3168/jds.2019-17644. 624. [DOI] [PubMed] [Google Scholar]
- 52.Waterham H.R., Digan M.E., Koutz P.J., Lair S.V., Cregg J.M. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene. 1997;186:37–44. doi: 10.1016/s0378-1119(96)00675-0. [DOI] [PubMed] [Google Scholar]
- 53.Gasser B., Dragosits M., Mattanovich D. Engineering of biotin-prototrophy in Pichia pastoris for robust production processes. Metab. Eng. 2010;12:573–580. doi: 10.1016/j.ymben.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 54.Jungo C., Urfer J., Zocchi A., Marison I., von Stockar U. Optimisation of culture conditions with respect to biotin requirement for the production of recombinant avidin in Pichia pastoris. J. Biotechnol. 2007;127:703–715. doi: 10.1016/j.jbiotec.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Kingston D., Novelli G.F., Cerrutti P., Recupero M.N., Blasco M., Galvagno M.A. Use of grape pomaces to produce biomass of a Komagataella pastoris strain expressing a bovine chymosin activity. Food Sci. Nutr. 2014;2:734–743. doi: 10.1002/fsn3.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Noseda D.G., Recupero M.N., Blasco M., Ortiz G.E., Galvagno M.A. Cloning, expression and optimized production in a bioreactor of bovine chymosin B in Pichia (Komagataella) pastoris under AOX1 promoter. Protein Expr. Purif. 2013;92:235–244. doi: 10.1016/j.pep.2013.08.018. [DOI] [PubMed] [Google Scholar]
- 57.Noseda D.G., Recupero M., Blasco M., Bozzo J., Galvagno M.A. Production in stirred-tank bioreactor of recombinant bovine chymosin B by a high-level expression transformant clone of Pichia pastoris. Protein Expr. Purif. 2016;123:112–121. doi: 10.1016/j.pep.2016.03.008. [DOI] [PubMed] [Google Scholar]
- 58.Noseda D.G., Blasco M., Recupero M., Galvagno M.A. Bioprocess and downstream optimization of recombinant bovine chymosin B in Pichia (Komagataella) pastoris under methanol-inducible AOXI promoter. Protein Expr. Purif. 2014;104:85–91. doi: 10.1016/j.pep.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 59.Tschopp J.F., Sverlow G., Kosson R., Craig W., Grinna L. High-level secretion of glycosylated invertase in the methylotrophic yeast. Pichia Pastoris. Bio/Technology. 1987;5:1305–1308. [Google Scholar]
- 60.Pratap J., Rajamohan G., Dikshit K.L. Characteristics of glycosylated streptokinase secreted from Pichia pastoris: enhanced resistance of SK to proteolysis by glycosylation. Appl. Microbiol. Biotechnol. 2000;53:469–475. doi: 10.1007/s002530051643. [DOI] [PubMed] [Google Scholar]
- 61.Parodi A.J. Reglucosylation of glycoproteins and quality control of glycoprotein folding in the endoplasmic reticulum of yeast cells. Biochim. Biophys. Acta Gen. Subj. 1999;1426:287–295. doi: 10.1016/s0304-4165(98)00130-5. [DOI] [PubMed] [Google Scholar]
- 62.Vallejo J.A., Ageitos J.M., Poza M., Villa T.G. Cloning and expression of buffalo active chymosin in Pichia pastoris. J. Agric. Food Chem. 2008;56:10606–10610. doi: 10.1021/jf802339e. [DOI] [PubMed] [Google Scholar]
- 63.Vega-Hernandez M.C., Gomez-Coello A., Villar J., Claverie-Martin F. Molecular cloning and expression in yeast of caprine prochymosin. J. Biotechnol. 2004;114:69–79. doi: 10.1016/j.jbiotec.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 64.Slamani R., Labadi R., Brahim Errahmani M., Bellal M.M. Purification and characterisation of milk-clotting and caseinolytic activities of pepsin isolated from adult sheep abomasa. Int. J. Dai. Techn. 2018;71:764–770. [Google Scholar]
- 65.Carlson A., Hill C.G., Jr., Olson N.F. Kinetics of milk coagulation: I. The kinetics of kappa casein hydrolysis in the presence of enzyme deactivation. Biotechnol. Bioeng. 1987;29:582–589. doi: 10.1002/bit.260290507. [DOI] [PubMed] [Google Scholar]
- 66.Yang Z., Zhang Z. Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: a review. Biotechnol. Adv. 2018;36:182–195. doi: 10.1016/j.biotechadv.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 67.Luo F., Jiang W.H., Yang Y.X., Li J., Jiang M.F. Cloning and expression of yak active chymosin in Pichia pastoris. Asian-Australas. J. Anim. Sci. 2016;29:1363–1370. doi: 10.5713/ajas.16.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Potočnik K., Gantner V., Kuterovac K., Cividini A. Mare’s milk: composition and protein fraction in comparison with different milk species. Mljekarstvo. 2011;61:107–113. https://hrcak.srce.hr/69078 [Google Scholar]
- 69.Germeç M., Turhan I. Thermostability of Aspergillus niger inulinase from sugar-beet molasses in the submerged fermentation and determination of its kinetic and thermodynamic parameters. Biomass Con. Bioref. 2020:1–9. [Google Scholar]
- 70.Germec M., Turhan I. Evaluation of carbon sources for the production of inulinase by Aspergillus niger A42 and its characterization. Bioproc. Biosyst. Eng. 2019;42:1993–2005. doi: 10.1007/s00449-019-02192-9. [DOI] [PubMed] [Google Scholar]
- 71.Prielhofer R., Maurer M., Klein J., Wenger J., Kiziak C., Gasser B. Induction without methanol: novel regulated promoters enable high-level expression in Pichia pastoris. Microb. Cell Factories. 2013;12(5) doi: 10.1186/1475-2859-12-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that has been used is confidential.