Abstract
Objective
The area postrema (AP) and nucleus tractus solitarius (NTS) located in the hindbrain are key nuclei that sense and integrate peripheral nutritional signals and consequently regulate feeding behaviour. While single-cell transcriptomics have been used in mice to reveal the gene expression profile and heterogeneity of key hypothalamic populations, similar in-depth studies have not yet been performed in the hindbrain.
Methods
Using single-nucleus RNA sequencing, we provide a detailed survey of 16,034 cells within the AP and NTS of mice in the fed and fasted states.
Results
Of these, 8,910 were neurons that group into 30 clusters, with 4,289 from mice fed ad libitum and 4,621 from overnight fasted mice. A total of 7,124 nuclei were from non-neuronal cells, including oligodendrocytes, astrocytes, and microglia. Interestingly, we identified that the oligodendrocyte population was particularly transcriptionally sensitive to an overnight fast. The receptors GLP1R, GIPR, GFRAL, and CALCR, which bind GLP1, GIP, GDF15, and amylin, respectively, are all expressed in the hindbrain and are major targets for anti-obesity therapeutics. We characterise the transcriptomes of these four populations and show that their gene expression profiles are not dramatically altered by an overnight fast. Notably, we find that roughly half of cells that express GIPR are oligodendrocytes. Additionally, we profile POMC-expressing neurons within the hindbrain and demonstrate that 84% of POMC neurons express either PCSK1, PSCK2, or both, implying that melanocortin peptides are likely produced by these neurons.
Conclusion
We provide a detailed single-cell level characterisation of AP and NTS cells expressing receptors for key anti-obesity drugs that are either already approved for human use or in clinical trials. This resource will help delineate the mechanisms underlying the effectiveness of these compounds and also prove useful in the continued search for other novel therapeutic targets.
Keywords: Area postrema, Nucleus tractus solitarius, Gene expression, Obesity, Therapeutics
Highlights
-
•
We provide a survey of 16,034 cells of the murine AP and NTS in the fed and fasted state by single-nucleus RNA sequencing.
-
•
8,910 are grouped into 30 neuronal clusters, and 7,124 are non-neuronal such as oligodendrocytes, astrocytes and microglia.
-
•
We examine the transcriptomes of populations expressing Glp1r, Gipr, Gfral, Calcr, and Pomc.
-
•
The expression profiles for the above populations were not dramatically altered by an overnight fast.
-
•
We provide a detailed single-cell level characterisation of AP and NTS cells expressing anti-obesity drug targets.
1. Introduction
Studies in humans and animal models have highlighted the brain's central role in controlling appetite. Genetic and molecular approaches, for instance, have revealed key circuits within the hypothalamus that sense and integrate peripheral nutritional signals and as a consequence regulate food intake and body weight. A number of nuclei residing within the hindbrain have also been identified as critical centres that regulate feeding behaviour. The area postrema (AP) is a circumventricular organ located within the medulla oblongata below the fourth ventricle, and, similar to the hypothalamus, is able to detect circulating chemical messengers in the blood and transduce them into neural signals and networks, relaying information to other brain areas [1]. Adjacent to the AP is the nucleus tractus solitarius (NTS) consisting of a collection of subnuclei that receive direct inputs from multiple locations including gustatory information from cranial nerves and postprandial signals via vagal afferents [2] and thus acts as a visceral sensory relay station within the brain.
The AP and NTS have been implicated in regulating appetite [3], with a number of therapies for treating obesity that are currently either in clinical trials or already on the market targeting key receptors expressed within one or both nuclei. For example, multiple long-acting GLP1R agonists are indicated for treating type 2 diabetes, with liraglutide further indicated for treating obesity, and semaglutide in submission for regulatory approval for obesity. These have been shown to cause weight loss in humans [4,5] by acting on central GLP1R-expressing neurons in the hypothalamus and NTS to suppress food intake [[6], [7], [8]]. The AP and NTS are also major sites of action for the hormone amylin, where it binds and activates calcitonin receptors (CALCR) coupled to RAMP proteins [9]. GDF15, a sentinel hormone that conveys states of somatic stress to the brain thus influencing appetite, acts exclusively through GFRAL receptors in the AP and NTS [10]. Combinatorial therapies are being studied to assess the efficacy of targeting multiple circuits to enhance obesity treatments. Of interest, GLP1R agonists and dual amylin/calcitonin receptor agonists have been shown to have additive effects on reduction of food intake and body weight in rodents [11]. In addition, with the activation of hypothalamic GIPR neurons resulting in a reduction of food intake [12], dual agonists of GLP1 and GIP receptors are being trialled as treatments for obesity and type 2 diabetes (for a review, see [13]). A population of neurons that play a key role in sensing circulating nutritional signals and in so doing orchestrate appetite and peripheral metabolism express proopiomelanocortin (POMC). Within the brain, while POMC is primarily expressed in the arcuate nucleus of the hypothalamus (ARC), a smaller population exists in the NTS.
Multiple studies using single-cell RNA sequencing have been performed to characterise neurons within the hypothalamus and compare transcript expression across multiple nutritional states [[14], [15], [16], [17]]. The AP's transcriptional landscape upon treatment with amylin has been examined using so-called “bulk” RNA-sequencing studies [9]. More recently, single-nucleus RNA sequencing was performed on 1,848 mouse AP neurons characterising populations involved in nausea-associated behaviours [18]. However, cells from the NTS were not included and only ad libitum fed mice were studied.
Herein, using single-nucleus RNA sequencing (NucSeq), we survey 16,034 cells from the AP and NTS of mice in the fed and fasted states. We report a detailed transcriptional profile of hindbrain neurons expressing GLP1R, GIPR, CALCR, and GFRAL, and that their gene expression profiles are not dramatically altered by an overnight fast. Of note, we find that a large proportion of cells that express GIPR are not actually neurons but oligodendrocytes. We also profile POMC-expressing neurons within the hindbrain and demonstrate that the majority express PCSK1 and/or PCSK2, implying that melanocortin peptides are likely produced by these cells. Thus, we provide a detailed single-cell level characterisation of neurons within the AP and NTS that express receptors for key anti-obesity drugs that are either already approved for human use or in clinical trials.
2. Materials and methods
Mouse studies were performed in accordance with UK Home Office Legislation regulated under the Animals (Scientific Procedures) Act 1986 Amendment, Regulations 2012, following an ethical review by the University of Cambridge Animal Welfare and Ethical Review Body (AWERB). All of the mice were obtained from Charles River Laboratories. Twelve 6- to 8-week-old C57BL/6J male mice were maintained on a 12-h light/12-h dark cycle (lights on 0700–1900) in a temperature-controlled (22 °C) facility with ad libitum access to food (RM3(E) Expanded chow, Special Diets Services, UK) and water. The day before sampling, 6 mice were subjected to an overnight fast for 16 h.
2.1. Nucleus dissociation
The mice were sacrificed by cervical dislocation and a portion of the hindbrain containing the AP and NTS was dissected and snap frozen on dry ice. Samples were stored at −80 °C overnight. Samples from the same nutritional condition were pooled together to yield 1 ad libitum sample and 1 fasted sample. The next day, homogenates were produced using a Dounce homogeniser in 1 ml of homogenate buffer (100 μM of DTT [Sigma–Aldrich, St. Louis, MO, USA], 0.1% Triton X-100 [Sigma–Aldrich], 2X EDTA Protease Inhibitor [Roche, Basel, Switzerland], 0.4 U/μl of RNasin RNase inhibitor [Promega, Madison, WI, USA, 10000 U, 40 U/ml], and 0.2 U/μl of Superase.In RNase Inhibitor [Ambion, Austin, TX, USA, 10000 U, 20 U/μl] in nuclei isolation medium [250 mM of sucrose, 25 mM of KCl (Ambion), 5 mM of MgCl2 (Ambion), and 10 mM of Tris buffer at a pH of 8.0 (Ambion) in nuclease-free water (Ambion)] with 1 μl/ml of DRAQ5 [Biostatus, Loughborough, UK]) on ice. The homogenates were centrifuged at 900×g for 10 min at 4 °C, and the supernatant was discarded. The pellets were then resuspended in homogenate buffer with 25% OptiPrep diluted in iodixanol dilution medium (250 mM of sucrose, 150 mM of KCl, 30 mM of MgCl2, and 60 mM of Tris buffer with a pH of 8 in nuclease-free water). Each suspension was layered on top of separate 29% OptiPrep solutions and centrifuged for 20 min at 13,500×g at 4 °C. For each sample, the pellet was removed and resuspended in 1 ml of wash buffer (1% BSA, 0.4 U/μl of RNasin, and 0.2 of U/μl of Superase.In in PBS [Sigma–Aldrich]). The samples were passed through a 40 μm cell strainer along with 0.7 ml of wash buffer.
2.2. Fluorescent-activated cell sorting
Fluorescent-activated cell sorting (FACS) was performed on both samples using a BD Influx cell sorter (BD Biosciences, Franklin Lakes, NJ, USA). The gating was set according to size and granularity using FSC and SSC to capture singlets and remove debris, and fluorescence at 647/670 nm to detect DraQ5 staining. Each sample was sorted into two separate tubes, with a total of 15,000 particles sorted into each tube containing 10 μl of wash buffer.
2.3. Single-nucleus RNA sequencing
Sequencing libraries for the 4 (2 ad libitum and 2 fasted) single-nuclei suspension samples were generated using 10X Genomics Chromium Single-Cell 3′ Reagent kits (Pleasanton, CA, USA, version 3) according to the standardised protocol. Briefly, nuclear suspensions were loaded onto the chromium chip along with gel beads, partitioning oil, and master mix to generate GEMs containing free RNA. RNA from lysed nuclei was reverse transcribed and cDNA was PCR amplified for 19 cycles. The amplified cDNA was used to generate a barcoded 3′ library according to the manufacturer's protocol, and paired-end sequencing was performed using an Illumina NovaSeq 6000 (San Diego, CA, USA, read 1: 28 bp and read 2: 91 bp). Library preparation and sequencing was performed by the Genomics Core, Cancer Research UK Cambridge Institute.
2.4. Single-cell clustering and differential gene expression analysis
Reads were aligned to mouse genome (GRCm38) with unspliced transcripts being considered during the analysis using the CellRanger package version 4.0 (10X Genomics). A downstream analysis on the raw count matrices was performed using the Seurat package version 3.1.1 [19,20]. Briefly, nuclei expressing less than 200 features or less than 1,000 transcripts were removed as low-quality reads. Nuclei with more than 6,000 different features were removed as these were likely doublets. As the percentage of mitochondrial RNA reads was low, any nuclei expressing more than 0.2% mitochondrial RNA were excluded from the analysis. The data were log-normalised and scaled prior to PCA, followed by unsupervised clustering analysis using the Louvain algorithm and non-linear dimensional reduction via T-distributed stochastic neighbour embedding (tSNE). Any clusters that showed a high expression of more than one cell-type marker (e.g. neuron, astrocyte) were further removed. Marker genes for each cluster were identified using Wilcoxon's rank-sum test and receiver-operating curve (ROC) analyses. Differential expression of genes between nuclei from the ad libitum fed and overnight fasted mice within each cluster was analysed using Wilcoxon's rank-sum test with adjusted P values based on Bonferroni's correction using all of the features within the dataset.
To characterise differential gene expression in the oligodendrocyte population, a subset of all clusters identified as oligodendrocytes was produced and re-clustered using the Seurat pipeline (as previously described). Differential gene expression analysis of the entire population was performed using Wilcoxon's rank-sum test to identify the effects of fasting on transcript expression in oligodendrocytes. To individually characterise populations of GLP1R, GFRAL, CALCR, GIPR, and POMC, subsets of nuclei expressing at least 1 raw UMI count of the gene of interest were created. If the gene was found to be expressed mainly in the neurons, then the subset was obtained from the neuronal dataset (this was the case for GLP1R, CALCR, and GFRAL); however, if there was widespread transcript expression within the non-neuronal clusters, the subset was taken from the entire dataset (GIPR and POMC). Subsets were re-clustered according to the Seurat pipeline (as previously described). Marker genes for each cluster were identified using Wilcoxon's rank-sum test and a ROC analysis. Violin plots were generated using the Seurat package. The differential gene expression cut-off used was unadjusted at P < 0.05 unless otherwise stated.
To characterise receptor and peptide expression profiles of these nuclei subsets, any gene that was expressed in a minimum of 50% of cells in one of the clusters was kept for analysis. The mean relative expression level of each gene was calculated for each cluster to characterise these cells' expression profiles. To determine these subsets’ differential gene expression in response to a fast, the genes that were used for expression profiling were analysed for changes in gene expression within each cluster between fed and fasted nuclei using the EdgeR package [21]. For receptors and transporters, the gene type was determined based on the classification from the Ingenuity pathway analysis (Qiagen). For peptide hormones, genes annotated with the gene ontology (GO) term GO:0005179 (hormone activity) were included.
Genes whose roles in neuronal transmission are well established (Slc17a6, Slc17a7, Slc32a1, Gad1, Gad2, Slc6a5, Ddc, Dbh, Th, Slc6a3, Tph1, Slc6a4, Chat, and Slc18a3) and expressed in a minimum of 10% of cells from one cluster in each subset were also analysed in the same way (average expression level and differential gene expression). The same was done for a number of oligodendrocyte markers for the GIPR and POMC subsets as these had oligodendrocyte clusters (Olig1, Olig2, Olig3, Cldn11, Mbp, Mog, Sox10, Pdgfra, and Cspg4).
2.5. Pathway analysis
The Ingenuity pathway analysis (IPA) was used to identify pathways that were either upregulated or downregulated between the fed and fasted conditions.
2.6. Data availability
The data are available from the NCBI Gene Expression Omnibus, accession GSE168737.
3. Results
3.1. Single-nucleus RNA-sequencing survey of 16,034 nuclei from the mouse AP and NTS
We isolated and performed NucSeq on a total of 16,034 nuclei from the AP/NTS of 12 male mice, six fed ad libitum and six fasted overnight. A median of 2,419 different transcripts were detected per individual nucleus (Figure 1A). Unsupervised clustering analyses (see Methods) separated the nuclei into 41 different clusters on a tSNE plot (Figure 1A), with the cell type of each cluster determined based on the expression of canonical markers (Figure 1C).
Figure 1.
Single-nucleus RNA sequencing of the AP and NTS reveals 41 populations. (A) tSNE plot showing 41 clusters including 8 Oligodendrocyte and Oligodendrocyte Precursor Cells (OPC) clusters, 1 Astrocyte cluster, 1 microglia cluster, 1 epithelial cell cluster and 30 neuronal clusters. (B) Phylogenetic tree showing the hierarchy of the clusters. Similar cell clusters are grouped together. (Abbreviations: O = Oligodendrocytes, A = Astrocytes, E = Epithelial cells, M = Microglia, N = Neurons). (C) Feature plots displaying the relative expression levels of cell type markers for neurons (green, top left), Oligodendrocytes and OPC's (blue, top right), astrocytes (red, bottom left), epithelial cells (pink, bottom centre) and microglia (orange, bottom right). (D) tSNE plot showing the 30 neuronal clusters (8910 nuclei). Cluster numbers remain unchanged from Figure 1A. (E) Scaled expression levels of Slc17a6 (blue) and Slc32a1 (orange) in neuronal nuclei displayed as a combined feature plot.
A total of 11 of the 41 clusters encompassed 7,124 non-neuronal cells (Figure 1B). A large population of oligodendrocytes by marked expression of Mog accounted for seven clusters that were closely adjacent to each other, with the maturity of the oligodendrocytes marked by a corresponding decrease in expression levels of Olig2, increasing from left to right. There was also a cluster of oligodendrocyte precursor cells (OPCs) marked by their expression of Pdgfra. A single cluster of astrocytes was identified by the expression of the excitatory amino acid transporters 1 and 2 (Slc1a3 and Slc1a2) as well as Aldh1l1. There was one cluster of microglia (marked by the expression of Tmem119) and a cluster of epithelial cells (expressing Lum) (Figure 1C).
The remaining 30 clusters were formed from 8,910 neurons, all of which were characterised by their high relative expression of Syt1, Snap25, and Rbfox3 (Figure 1C). We focused on these 30 neuronal clusters (Figure 1D) and performed a differential gene expression analysis to identify their key transcriptional markers (summarised in Figure S1). The primary driver differentiating the transcriptional profile of the neurons was their expression of either Slc17a6 (vGLUT2) and Slc32a1 (vGAT), which are excitatory and inhibitory markers, respectively (Figure 1E). There were two clusters with a low expression of Slc17a6 and Slc32a1: cluster 36, which additionally expressed the cholinergic neuronal markers Slc5a7 (encoding the high-affinity choline transporter) and Chat (choline acetyltransferase) (Figure S1), and cluster 27, which additionally expressed Slc17a7 (vGLUT1), Ngf, and Fam107b (Figure 1E and Figure S1). Other notable populations included cluster 26, which was marked by the expression of Npy; clusters 31 and 34, which both expressed Ddc (DOPA decarboxylase, which converts l-DOPA into dopamine) and Dbh (dopamine-β-hydroxylase, which converts dopamine into noradrenaline), implying that these were noradrenergic neurons; and cluster 40, which was likely to be serotonergic due to the expression of Tph2 (tryptophan hydroxylase 2) and Slc6a4 (a serotonin transporter) (Figure S1).
3.2. The transcriptomic response of AP and NTS cell populations to an overnight fast
We next examined the consequences of an overnight fast on the transcriptional profile of AP and NTS cells, with 3,378 non-neuronal and 4,289 neuronal nuclei (total 7,667) profiled from fed mice, and 3,746 non-neuronal and 4,621 neuronal nuclei (total 8,367) profiled from the fasted mice (Figure 2). Most of the neuronal clusters displayed a relatively equal number of fed:fasted nuclei, the exceptions being clusters 6 (markers Cdh8 and Sox2ot), 25 (Zic4 and Grin2c), and 40 (Slc6a4 and Tph2), which had a disproportionate number of nuclei from the ad libitum fed mice (84%, 67.8%, and 78.3%, respectively), and clusters 13 (Tac1 and Dlk1) and 36 (Slc5a7 and Chat), which had an overrepresentation of nuclei from the fasted mice (66.1% and 69.1% respectively) (Figure 2B).
Figure 2.
The effect of nutritional state on transcriptomic expression in mouse AP/NTS cells. (A) tSNE plot revealing the nutritional status of each cell within the dataset. A total of 7667 nuclei came from mice fed ad libitum (pink), and 8367 cells came from mice who were fasted overnight (blue). (B) Graph showing the proportion of nuclei within each cluster that originated from mice fed ad libitum (pink) or overnight fasted mice (blue). (C) Graph showing the number of genes upregulated (green) and downregulated (blue) in response to an overnight fast in each cluster. Genes included were significantly differentially regulated (P < 0.05). (B–C) Cluster numbers listed down the left-hand side and the cell types of each cluster identified: N = neurons, O = Oligodendrocytes and OPCs, M = Microglia, E = Epithelial cells, A = Astrocytes. (D) A tSNE displaying a subset of oligodendrocyte nuclei (not including the OPC cluster), revealing the nutritional status of each cell (ad libitum in pink, fasted in blue). (E) Relative expression levels of top differentially expressed genes within the oligodendrocyte subset, displayed as feature plots. (F) Top 10 canonical pathways affected by an overnight fast in the oligodendrocyte population. A negative z-score represents an overall downregulation of the pathway, and a positive z score represents an overall upregulation of the pathway in the fasted state. Genes involved in each pathway whose expression was differentially regulated were highlighted for each pathway.
The population of oligodendrocytes was surprisingly transcriptionally responsive to an overnight fast (Figure 2C). Fasting in fact turned out to be the main driver of the oligodendrocyte clustering (Figure 2D). The oligodendrocytes showed consistently high levels of differential gene expression in the fasted state, with 6 out of the 7 clusters exhibiting more upregulated than downregulated genes (Figure 2C). When the entire oligodendrocyte population was considered as a whole, the top differentially regulated genes in response to fasting included upregulation of Sgk1 (serum/glucocorticoid regulated kinase 1), Itgad (integrin alpha-D) encoding the alpha subunit of an integrin glycoprotein, and the GABA transporter 1 gene Slc6a1, which transports GABA into the cell. Additionally, downregulated genes included Eml1 (echinoderm microtubule associated protein-like 1) and two heat shock proteins Hsp90ab1 and Hsp90b1 (Figure 2E). The pathway analysis revealed that neuregulin signalling, important in myelination that involves Hsp90ab1 and Hsp90b1, was downregulated (Figure 2F) [22]. Furthermore, a number of pathways involved in inositol metabolism, thought to regulate oligodendrocyte differentiation and survival [23,24] and PPARα/RXRα signalling, were upregulated in the fasted state (Figure 2F). Interestingly, expression of 25 of the differentially expressed genes in the oligodendrocytes were regulated by the transcription factor Tcf7l2, including Sgk1, Ptpn11, Rap1a, and Ppp1r16b, all of which were implicated in the neuregulin or inositol biosynthesis pathways (Figure 2F).
3.3. Characterisation of neurons expressing key anti-obesity therapeutic targets in the AP and NTS
We next focussed on detailed analyses of four different populations of neurons that express the receptors GLP1R, GIPR, GFRAL, and CALCR, which bind GLP1, GIP, GDF15, and amylin, respectively, and are major targets for anti-obesity therapeutics.
3.3.1. GLP1R
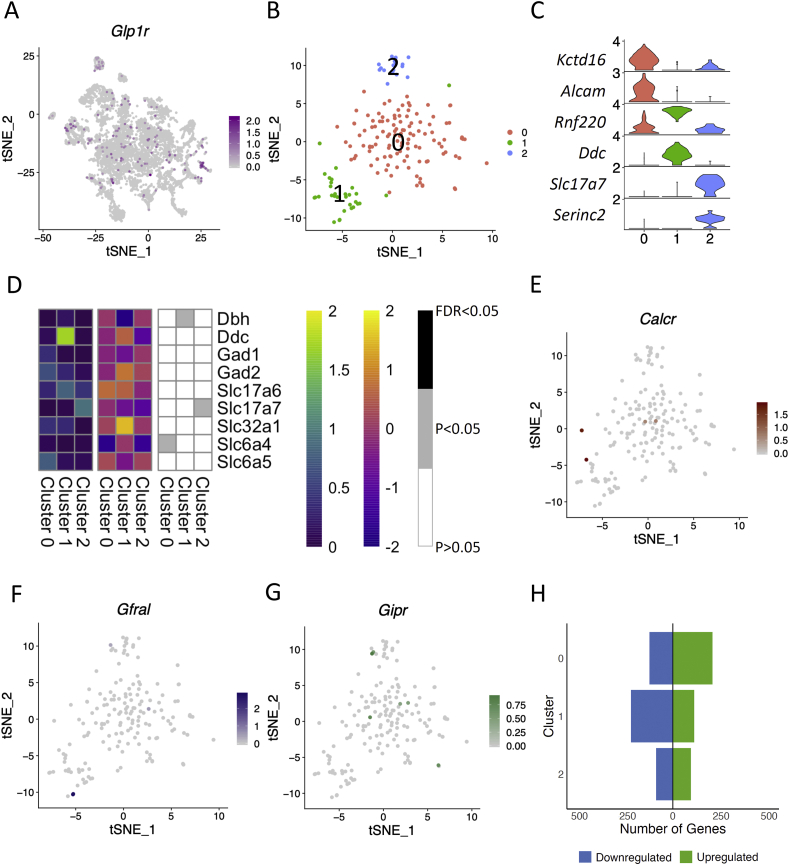
We identified a total of 173 Glp1r-expressing nuclei in 24 of the 30 neuronal clusters (Figure 3A). When we extracted and analysed these neurons in isolation, they fell into three clusters (Figure 3B). Cluster 0, the largest of the three, was characterised by the expression of Kctd16 and Alcam, cluster 1 was marked by Ddc, and cluster 2 was marked by Slc17a7 (Figure 3C).
Figure 3.
Characterisation of GLP1R expression in AP and NTS neurons. (A) Feature plot revealing relative expression levels of Glp1r in the 30 neuronal clusters. (B) tSNE plot of the 173 GLP1R neurons, grouped into 3 clusters. (C) Violin plots showing relative expression levels of 2 marker genes for each cluster of the GLP1R subset. The highest relative expression value for each gene is displayed on the left of the plot. (D) Heatmaps profiling the expression of canonical neuronal markers in each GLP1R cluster. Left: Average relative expression level in each cluster; Middle: Differential expression of each gene per cluster (LogFC values); Right: Determining whether gene expression was significantly changed in the fasted state. (E–G) Feature plots showing relative expression levels of Calcr (E), Gfral (F), and Gipr (G) in the GLP1R subset. (H) Graph showing the number of genes upregulated (green) and downregulated (blue) in response to an overnight fast in each cluster. Genes included were significantly differentially regulated (P < 0.05).
Cluster 1 consisted of both excitatory and inhibitory neurons, with 48.4% expressing Slc17a6 and 22.6% expressing Slc32a1 (Figure 3D). In cluster 2, 83.3% of nuclei expressed Slc17a7 (vGLUT1) and 38.9% expressed Slc17a6 (vGLUT2, Figure 3D). Cluster 0 neurons predominantly expressed inhibitory neuronal markers, with 46% of nuclei expressing Gad2 and Slc6a5 (average relative expression levels: 0.64 and 0.75, respectively). These data showed that GLP1R neurons within the mouse hindbrain can either be excitatory or inhibitory. We next investigated the expression of Gfral, Gipr, and Calcr in the Glp1r subset. These four populations of neurons appear largely distinct, with only 6 Glp1r nuclei co-expressing Gipr, 5 co-expressing Calcr, and 4 co-expressing Gfral (Figure 3E–G), and could suggest that agonists to these receptors act largely independently of each other. However, while this experiment is designed to identify the presence of transcripts, it is entirely possible that cells expressing low levels of transcripts could have been missed. A detailed expression profile of GLP1R neurons is provided in Figure S2.
To determine whether GLP1R neurons are sensitive to an overnight fast, we performed differential gene expression analysis in all 3 clusters (see Methods). Clusters 0 and 1 showed the highest changes in gene expression in the fasted state (326 and 327 differentially expressed genes, respectively; Figure 3H and Table S1). Notably, in cluster 0, Ptgds expression was significantly upregulated (logFC = 1.1566, P = 6.40 × 10−8, and false discovery rate [FDR] = 0.0003), and in cluster 1, Oxr1 (oxidation resistance 1) was significantly downregulated in response to an overnight fast (logFC = −1.9133, P = 3.78 × 10−6, and FDR = 0.0156). The pathway analysis revealed the synaptogenesis signalling pathway to be significantly changed in two of the clusters (downregulated in cluster 0 and upregulated in cluster 2) and calcium signalling to be upregulated in cluster 2 (Table S2).
3.3.2. GIPRs were expressed in both neurons and oligodendrocytes in the mouse hindbrain
GIPR expression within the hypothalamus has been characterised using histological techniques in combination with single-cell RNA sequencing [12]; however, less is known about GIPR-expressing cells within the hindbrain. We identified 436 Gipr-expressing nuclei that were found in all but two clusters within the entire hindbrain dataset (Figure 4A). Neuronal cluster 21 contained the highest proportion of Gipr nuclei, with 11.5% of this cluster expressing Gipr. Marker genes for this cluster included Ccbe1 and Zeb2 (Figure S1). Unexpectedly, a large number of Gipr+ nuclei were oligodendrocytes (Figure 4A), with cluster 9 exhibiting the highest percentage of Gipr nuclei (13.4%).
Figure 4.
GIPR is expressed in neuronal and non-neuronal cells within the mouse AP and NTS. (A) Feature plot displaying the relative expression levels of Gipr in the overall dataset. (B) tSNE plot of the 436 GIPR cells, which grouped into 4 clusters. (C) Violin plots showing relative expression levels of 2 marker genes for each cluster of the GIPR subset. (D–E) Heatmaps profiling the expression of canonical neuronal (D) and oligodendrocyte (E) markers in each GIPR cluster. Left: Average relative expression level in each cluster; Middle: Differential expression of each gene per cluster (LogFC values); Right: Identifying whether gene expression was significantly changed in the fasted state. (F) Graph showing the number of genes upregulated (green) and downregulated (blue) in response to an overnight fast in each cluster. Genes included were significantly differentially regulated (P < 0.05).
When we re-clustered the 436 Gipr-expressing nuclei, they formed 4 clusters, 2 of oligodendrocytes and 2 neuronal (Figure 4B). Both oligodendrocyte clusters expressed Bcas1, a myelin-associated protein, as well as Mbp and Mog (cluster 1: 100% and 99.2% and cluster 2: 99.1% and 91.8%, respectively). Additionally, oligodendrocyte cluster 1 expressed Enpp6, an enzyme involved in choline metabolism that is typically expressed in maturing oligodendrocytes [25]. Both clusters of neurons expressed Scn1a and Lrfn5, with cluster 3 expressing Ccbe1 and Arhgap15 (Figure 4C). Overall, 39.9% of cluster 0 expressed the glutamatergic marker Slc17a6 (average relative expression: 0.37), and 39.9% expressed Slc6a5 (average expression level of 0.45). Cluster 3 was predominantly GABAergic, with 48.7% and 61.5% expressing Gad1 and Gad2 (Figure 4D). Further characterisation of expression profiles of GIPR cells is shown in Figure S3.
Of the 436 nuclei, 190 were derived from the fasted mice. A differential expression analysis in each cluster showed the highest changes in gene expression were within the oligodendrocyte clusters, with 284 and 310 differentially expressed genes in response to a fast in clusters 1 and 2, respectively (Figure 4F and Table S3). Similar to GLP1R neurons (Table S1), Ptgds was also significantly upregulated in clusters 0 and 2 (logFC = 1.4381, P = 3.14 × 10−10, and FDR = 8.69 × 10−7 and logFC = 1.2383, P = 2.69 × 10−7, and FDR = 1.80 × 10−4), respectively. Adipor2 (adiponectin receptor 2) expression was upregulated in cluster 1 (logFC = 0.9853, P = 7.52 × 10−7, and FDR = 1.04 × 10−3) (Table S3). In the neuronal clusters, the pathway analysis revealed that the synaptogenesis signalling pathway was downregulated in cluster 0 in the fasted state (Table S4).
3.3.3. CALCR
We identified 185 Calcr-expressing nuclei that were located in 23 of the 30 neuronal clusters (Figure 5A). When re-clustered, the CALCR neurons grouped into 3 clusters (Figure 5B). Cluster 0 and some of cluster 2 expressed Chrm3 (acetylcholine muscarinic receptor 3), suggesting that they responded to acetylcholine signalling (Figure 5C). Cluster 2 expressed Th (tyrosine hydroxylase), suggesting that these were catecholamine neurons (Figure 5C).
Figure 5.
Profiling CALCR neurons in the AP and NTS. (A) Feature plot revealing relative expression levels of Calcr in the neuronal clusters. (B) tSNE plot of the 185 CALCR neurons, grouped into 3 clusters. (C) Violin plots showing relative expression levels of 2 marker genes for each cluster of the CALCR neurons. (D) Feature plots showing the relative expression levels of Ramp1, Ramp2 and Ramp3 in the CALCR subset. (E) Heatmaps profiling the expression of canonical neuronal markers in each CALCR cluster. Left: Average relative expression level in each cluster; Middle: Differential expression of each gene per cluster (LogFC values); Right: Determining whether gene expression was significantly changed in the fasted state. (F) Graph showing the number of genes upregulated (green) and downregulated (blue) in response to an overnight fast in each cluster. Genes included were significantly differentially regulated (P < 0.05).
Amylin receptors are formed by the heterodimerisation of CALCR with at least one of the 3 RAMP proteins [26]. To identify which of these CALCR neurons were likely expressing the amylin receptor, the expression of RAMP1, RAMP2, and RAMP3 was characterised in CALCR neurons (Figure 5D). RAMP1 and RAMP2 were not widely expressed in the CALCR population, but RAMP3 was expressed in cluster 1 (Figure 5D).
Upon assessing the expression of established neuronal markers in each cluster, we saw that Calcr clusters consisted of mainly glutamatergic neurons (Figure 5E). Cluster 0 was a mix of both excitatory (43.9%) and inhibitory neurons (31.9%). The expression of Slc17a6 in cluster 1 suggested that neurons in this cluster were glutamatergic. Cluster 2 nuclei expressed Slc17a6, Th, Dbh, and Ddc, suggesting that this cluster had catecholamine neurons (Figure 5E). Detailed expression profiles of these clusters are shown in Figure S4.
Cluster 0 exhibited the highest amount of differential gene expression in response to a fast (240 genes; Figure 5F). In both clusters 0 and 2, Meg3 expression was downregulated in response to a fast (logFC = −0.2462, P = 8.13 × 10−15, and FDR = 2.76 × 10−11 and logFC = −0.3476, P = 2.23 × 10−7, and FDR = 3.77 × 10−4, respectively). Scaffold protein Kidins220 was downregulated in cluster 2 in the fasted state (logFC = −2.4617, P = 3.35 × 10−9, and FDR = 1.13 × 10−5; Table S5).
3.3.4. GFRAL
The hormone GDF15 acts to suppress food intake through the GFRAL receptor located on a small population of neurons within the AP and NTS [[27], [28], [29], [30]]. We identified 114 Gfral-expressing nuclei. Cluster 31 contained the highest number, with 20.3% of nuclei expressing Gfral (Figure 6A). A large proportion of GFRAL neurons and GLP1R neurons were located within the same cluster in the overall dataset (cluster 31), indicating the transcriptional similarities between these two populations. However, with our approach, we observed minimal co-expression of these two receptors, although it was possible that we did not detect cells expressing low levels of these transcripts. When re-clustered, they formed 2 clusters (Figure 6B). Cluster 0 was characterised by the expression of Tenm2 and Plcl1, while cluster 1 expressed Ddc, suggesting that these were dopaminergic neurons (Figure 6C). Cluster 1 also expressed Slc17a6, Dbh, and Th, suggesting that these neurons transmitted both glutamatergic and catecholamine signals (Figure 6D).
Figure 6.
Characterising GFRAL neurons in the AP and NTS. (A) Feature plot of Gfral expression in the neuronal dataset. (B) tSNE plot of the 114 GFRAL neurons, grouped into 2 clusters. (C) Violin plots showing relative expression levels of marker genes for both GFRAL clusters. (D) Heatmaps profiling the expression of canonical neuronal markers in both GFRAL clusters. Left: Average relative expression level in each cluster; Middle: Differential expression of each gene per cluster (LogFC values); Right: Determining whether gene expression was significantly changed in the fasted state. (E) Graph showing the number of genes upregulated (green) and downregulated (blue) in response to an overnight fast in both clusters. Genes included were significantly differentially regulated (P < 0.05).
A total of 57 of the 114 Gfral neurons came from the fasted animals. Overall, 177 genes were differentially expressed in the fasted condition in cluster 0, including the inhibitory neuronal markers (Gad1, Slc32a1, and Slc6a5), which were downregulated in the fasted state (logFC = −1.093, P = 0.0454, and FDR = 0.831; logFC = −1.399, P = 0.00563, and FDR = 0.575; and logFC = −1.282, P = 0.0259, and FDR = 0.750, respectively). A total of 118 differentially expressed genes were identified in cluster 1 (Figure 6E and Table S7), including Tenm3 and Pias1, that were both upregulated in the fasted condition (logFC = 2.2753, P = 1.37 × 10−5, and FDR = 0.0372 and logFC = 2.4759, P = 3.33 × 10−5, and P = 0.0450, respectively; Table S7).
3.4. Characterisation of AP/NTS POMC cells
While POMC neurons located within the hypothalamus have been well characterised using single-cell RNA sequencing [16], the NTS POMC neurons have not been similarly studied. Herein, we identified 346 nuclei expressing POMC (Figure 7). Unlike in the hypothalamus, where the expression of Pomc appears to be a main driver of cluster formation [14,15], there was no single cluster that expressed particularly high levels of Pomc (Figure 7A). Instead, its expression was detected in all of the clusters apart from the microglia and epithelial cell clusters, with neuronal clusters 40 (Slc6a4 and Tph2), 36 (Prph, Tbx20, Chat, and Slc5a7), and 22 (Gal and Cartpt) containing the highest percentage of Pomc nuclei (8.7%, 7.2%, and 6.1%, respectively). Interestingly, Pomc expression was also evident throughout the oligodendrocyte clusters. When analysed separately, the 346 nuclei grouped into 3 neuronal and 1 oligodendrocyte clusters (Figure 7B). Clusters 0–2 were neuronal, although their expression profiles were very similar (Figure 7C), with cluster 0 nuclei marked by the expression of Baiap3 and Camk2a, and clusters 1 and 2 expressing Syt2 (synaptotagmin) and Flt3. Cluster 3 was clearly an oligodendrocyte cluster due to its high expression of Mog.
Figure 7.
Characterising POMC cells in the AP and NTS. (A) Feature plot showing relative expression levels of Pomc in the overall dataset. (B) tSNE plot of the 346 POMC nuclei, grouped into 4 clusters. (C) Violin plots showing relative expression levels of marker genes for each cluster of the POMC subset. (D) The relative expression levels of Pcsk1 and Pcsk1 in the POMC subset. (E) Table displaying the number (and percentage) of Pomc neurons expressing Lepr, Cckar, Cckbr, Mc3r and Mc4r in each of the neuronal clusters, and total expression numbers in the neuronal clusters. (F) Graph showing the number of genes upregulated (green) and downregulated (blue) in response to an overnight fast in each cluster. Genes included were significantly differentially regulated (P < 0.05). (G-H) Heatmaps profiling the expression of canonical neuronal (G) and oligodendrocyte (H) markers in each POMC cluster. Left: Average relative expression level in each cluster; Middle: Differential expression of each gene per cluster (LogFC values); Right: Determining whether gene expression was significantly changed in the fasted state.
POMC is processed into its active constituents by prohormone convertases PCSK1 and PCSK2 [31], and a total of 84% of POMC neuronal nuclei expressed either or both enzymes (Figure 7D). In contrast, only four oligodendrocyte nuclei expressed Pcsk2, with none expressing Pcsk1. Only a small proportion of each cluster expressed the leptin receptor Lepr (cluster 0 = 13.1%, cluster 1 = 19.4%, and cluster 2 = 12.3%; Figure 7E). Only 2.5% expressed the Cck receptor (Cckbr) and 5% expressed Mc4r. There was no expression of Cckar and Mc3r in the POMC nuclei.
In the hypothalamus, POMC neurons are very responsive to changes in nutritional state [14]. In the hindbrain, cluster 0 exhibited the highest number of differentially expressed genes, with the majority being downregulated in the fasted state (Figure 7F). This included Ryr2 (logFC = −0.7386, P = 6.51 × 10−6, and FDR = 0.00569; Table S9). A total of 266 genes were differentially regulated in cluster 2. In the oligodendrocyte cluster, Adipor2 expression was upregulated (logFC = 1.4047, P = 7.44 × 10−6, and FDR = 0.00867) and Frmd4a was downregulated (logFC = −3.1281, P = 4.96 × 10−14, and FDR = 1.73 × 10−10). In clusters 0 and 2, the synaptogenesis signalling pathway was the top downregulated pathway (Table S10).
4. Discussion
To date, cells within the AP/NTS have largely been characterised using histological and molecular approaches. In this study, we performed a single-nucleus level survey of 16,034 cells mostly from the mouse AP and NTS in the fed and fasted state. Because of our dissection method, we undoubtedly also captured cells from adjacent nuclei such as the dorsal motor nucleus of the vagus. We provided detailed expression profiles of hindbrain cells expressing the key anti-obesity drug targets GLP1R, GIPR, CALCR, and GFRAL and were able to capture and profile NTS POMC neurons at a single-nucleus level.
A key benefit of NucSeq is that it can be performed on CNS tissue from older animals, as neurons in older animals tend to have longer processes and are likely to be damaged in the single-cell dissociation process [32]. At best, this can result in sampling biases due to cells with long processes being underrepresented in the dataset, while at worst, nothing at all is captured. Thus, NucSeq opens up the possibility of studying the effects of long-term high-fat diets or other diets on changes in gene expression. Furthermore, as the cell nucleus is a fairly robust organelle, NucSeq can be performed on frozen samples, substantially expanding the availability of suitable samples to include those archived in tissue banks. This will certainly be the preferred method to use on human brain samples [33], and thus NucSeq data from mouse samples would be a more appropriate comparator for human single brain cell transcriptomics. While cytosolic RNA is inherently different from the mix of unspliced RNA emerging from the transcriptional machinery within the nucleus, transcriptome data from single nuclei are qualitatively reflective of that from single cells, certainly in terms of different cell types and their unique repertoire of transcripts [34].
A recent study surveying neuronal populations within the AP using NucSeq in relation to nausea-associated behaviours identified seven key AP neuronal cell types [18]. Our current dataset identifies similar populations of neurons expressing GIPR, GFRAL, GLP1R, and CALCR and provides additional novel information on both neuronal and non-neuronal populations located within the NTS. In addition to a census of cells within the AP/NTS, we provide insight into changes in transcriptional expression in hindbrain cells in response to an overnight fast.
Previous transcriptomic analyses of the mouse brain in the fed and fasted states focussed on the hypothalamus, either as a whole, on individually laser captured microdissected regions, or most recently using single-cell/nucleus RNA sequencing [15,35]. From these studies, we know that there are many different populations of hypothalamic neurons that are exquisitely sensitive to changes in the nutritional state [14,15]. In contrast, we showed that hindbrain neuronal populations were not especially responsive to an overnight fast, with the majority exhibiting modest effects on differential gene expression. This is unsurprising and reflects the differences in physiological roles of these two regions in the brain when it comes to appetitive drive, with the hypothalamus regulating hunger and satiety, and the hindbrain mediating the more visceral sensations, ranging from satiety and fullness, to aversion and nausea [3]. Unexpectedly, the one exception was the population of hindbrain oligodendrocytes that we showed to be transcriptionally sensitive to the fasted state. In particular, using pathway analysis, we found that an upstream regulator of differentially expressed genes in oligodendrocytes was the transcription factor Tcf7l2, which was previously shown to be necessary for differentiation of oligodendrocytes [36]. This suggests a possible remodelling of hindbrain oligodendrocytes that occurs in response to an overnight fast, a phenomenon that has previously been seen in oligodendrocytes in the median eminence (ME) of the hypothalamus [37]. As in the ME, the hindbrain oligodendrocytes are positioned near a circumventricular organ, and we suggest they could, in response to nutritional signals, control access of circulating metabolic cues to neurons in the AP.
The hindbrain's role in mediating satiety and aversive responses is part of the reason why many anti-obesity therapies are effective at the hindbrain. For example, once-weekly semaglutide administration over a period of 15 months was recently shown to produce a mean bodyweight reduction of 14.9% in patients with obesity [5]. In addition to signalling to hypothalamic neurons, semaglutide and liraglutide have both been shown to activate GLP1R neurons within the AP and NTS, mediating some of these drugs' weight loss effects [7,38]. The four neuronal populations we focussed on, expressing GLP1R, GIPR, CALCR, and GFRAL appear to be distinct from each other, possibly indicating that each of these receptors signals through independent pathways. However, it is important to note that while NucSeq is effective at identifying the presence of transcripts, it is not designed to demonstrate the absence of a transcript. Thus, it is entirely possible that there are cells co-expressing these receptors at low levels that we did not detect.
Even with dual agonism of GIPR and GLP1R showing an additional effect on weight loss in humans by reduction of food intake [39], the involvement of GIPR in controlling food intake and bodyweight is still not fully understood, with both antagonism of the receptor with an antibody and administration either centrally or peripherally of a GIPR agonist showing efficacious results in weight loss in mice [40,41]. While it is clear that hypothalamic GIPR plays a key role in this effect on weight loss, what is less clear is the role of GIPR in the hindbrain. For example, unlike GLP1R, whose expression in the hindbrain is largely restricted to neurons, around half of the cells expressing GIPR are oligodendrocytes. Similar to the broader population of oligodendrocytes, the subset of GIPR-expressing cells is also transcriptionally sensitive to an overnight fast, although the role GIPR signalling plays in these cells remains unclear.
Another class of compounds that work in concert with GLP1R agonists are amylin mimetics, with dual amylin and calcitonin receptor agonists shown to have complimentary actions on food intake in combination with liraglutide treatment [42]. CALCR neurons within the NTS are a key mediator of food intake reduction from CALCR agonists in comparison with the hypothalamus [43], demonstrating the importance of characterising these hindbrain populations. Of the three distinct populations of CALCR neurons we identified, only one is likely to express the amylin receptor based on its co-expression of Ramp3.
With the approval of the melanocortin agonist setmelanotide for treating rare causes of obesity, there has been renewed interest in the hypothalamic melanocortin pathway. However, the POMC neurons in the hindbrain, which have been studied neurochemically, have never been successfully isolated and characterised using transcriptomic techniques. POMC mRNA expression in the NTS has not been well characterised due to expression levels being so low, so it has been difficult to detect at a single-cell level [44]. Herein, we were able to identify and characterise POMC neurons within the hindbrain and show that the majority of these neurons co-express pro-hormone convertases PCSK1 and/or PCSK2. A number of functional studies have demonstrated that leptin administration activates POMC neurons in the NTS through elevation of c-fos or pSTAT3 immunoreactivity [44,45], indicating these neurons' role in regulating satiety signalling. Indeed, there are contradictory findings on the expression of LepRb on POMC NTS neurons, with studies using POMC-eGFP mice demonstrating that a subpopulation of POMC neurons express LepRb, and a study using a POMC-Cre reporter mouse finding no co-expression of POMC with LepRb [[44], [45], [46]]. In our dataset, we find found that a small subpopulation of POMC neurons (15%) do in fact express Lepr transcripts. A possible explanation for some of the inconsistent findings regarding the properties of POMC neurons within the NTS are the differences in which cells are marked by POMC-eGFP and POMC-Cre mouse lines [47]. Identifying POMC neurons through RNA sequencing will help clarify and strengthen evidence surrounding these neurons’ expression profiles.
To conclude, we report single-nucleus RNA sequencing of 30 neuronal and 11 non-neuronal populations residing within the AP/NTS. We provide extensive profiling of differential gene expression that occurs in response to an overnight fast and identify that oligodendrocytes in fact exhibit the greatest changes in transcriptional expression. We provide detailed profiling of GLP1R, CALCR, GIPR, and GFRAL cells based on their expression of receptors, ion channels, and transporters as well as their neuronal/glial properties, informing future studies that will examine these particular targets. We identify and characterise POMC cells residing within the AP/NTS on the single-cell level. This resource will help delineate the mechanisms underlying these compounds’ effectiveness and also prove useful in the continued search for other novel therapeutic targets for obesity.
Funding
G.K.C.D. is funded by a BBSRC CASE 4-year PhD studentship co-funded by Novo Nordisk. B.Y.H.L. is supported by a BBSRC Project Grant (BB/S017593/1). J.A.T., I.C., D.R., and G.S.H.Y. are supported by the Medical Research Council (MRC Metabolic Diseases Unit (MC_UU_00014/1)). J.A.T. is supported by an NIHR Clinical Lectureship (CL-2019-14-504). Next-generation sequencing was performed by the IMS Genomics and Transcriptomics Core Facility, which is supported by the MRC (MC_UU_00014/5), the Wellcome Trust (208363/Z/17/Z), and the Cancer Research UK Cambridge Institute Genomics Core.
Author contributions
G.K.C.D. collected, analysed, and visualised the data, wrote the original draft, and edited the manuscript. B.Y.H.L. collected, analysed, and visualised the data and edited the manuscript. J.A.T., I.C., and D.R. collected the data and edited the manuscript. A.P.C. provided supervision and edited the manuscript. J.P.W. and L.B.K. conducted the data analysis and edited the manuscript. C.P. was responsible for funding acquisition and supervision and edited the manuscript. G.S.H.Y. provided conceptualisation, funding acquisition, and supervision, wrote the original draft, and reviewed and edited the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101240.
Contributor Information
Georgina K.C. Dowsett, Email: gkcd2@cam.ac.uk.
Brian Y.H. Lam, Email: yhbl2@cam.ac.uk.
John A. Tadross, Email: jt636@medschl.cam.ac.uk.
Irene Cimino, Email: ic326@medschl.cam.ac.uk.
Debra Rimmington, Email: dy222@cam.ac.uk.
Anthony P. Coll, Email: apc36@cam.ac.uk.
Joseph Polex-Wolf, Email: jhpw@novonordisk.com.
Lotte Bjerre Knudsen, Email: lbkn@novonordisk.com.
Charles Pyke, Email: pyke@novonordisk.com.
Giles S.H. Yeo, Email: gshy2@cam.ac.uk.
Conflicts of interest
J.P.W., L.B.K., and C.P. are Novo Nordisk employees and/or shareholders.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Price C.J., Hoyda T.D., Ferguson A.V. The area postrema: a brain monitor and integrator of systemic autonomic state. The Neuroscientist. 2007;14(2):182–194. doi: 10.1177/1073858407311100. [DOI] [PubMed] [Google Scholar]
- 2.Gasparini S., Howland J.M., Thatcher A.J., Geerling J.C. Central afferents to the nucleus of the solitary tract in rats and mice. Journal of Comparative Neurology. 2020;528(16):2708–2728. doi: 10.1002/cne.24927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grill H.J., Hayes M.R. Hindbrain neurons as an essential hub in the neuroanatomically distributed control of energy balance. Cell Metabolism. 2012;16(3):296–309. doi: 10.1016/j.cmet.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pi-Sunyer X., Astrup A., Fujioka K., Greenway F., Halpern A., Krempf M. A randomized, controlled trial of 3.0 mg of liraglutide in weight management. New England Journal of Medicine. 2015;373(1):11–22. doi: 10.1056/NEJMoa1411892. [DOI] [PubMed] [Google Scholar]
- 5.Wilding J.P.H., Batterham R.L., Calanna S., Davies M., Van Gaal L.F., Lingvay I. Once-weekly semaglutide in adults with overweight or obesity. New England Journal of Medicine. 2021 doi: 10.1056/NEJMc2106918. [DOI] [PubMed] [Google Scholar]
- 6.Sisley S., Gutierrez-Aguilar R., Scott M., D'Alessio D.A., Sandoval D.A., Seeley R.J. Neuronal GLP1R mediates liraglutide's anorectic but not glucose-lowering effect. Journal of Clinical Investigation. 2014;124(6):2456–2463. doi: 10.1172/JCI72434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fortin S.M., Lipsky R.K., Lhamo R., Chen J., Kim E., Borner T. GABA neurons in the nucleus tractus solitarius express GLP-1 receptors and mediate anorectic effects of liraglutide in rats. Science Translational Medicine. 2020;12(533) doi: 10.1126/scitranslmed.aay8071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Secher A., Jelsing J., Baquero A.F., Hecksher-Sørensen J., Cowley M.A., Dalbøge L.S. The arcuate nucleus mediates GLP-1 receptor agonist liraglutide-dependent weight loss. Journal of Clinical Investigation. 2014;124(10):4473–4488. doi: 10.1172/JCI75276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liberini C.G., Borner T., Boyle C.N., Lutz T.A. The satiating hormone amylin enhances neurogenesis in the area postrema of adult rats. Molecular Metabolism. 2016;5(10):834–843. doi: 10.1016/j.molmet.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patel S., Alvarez-Guaita A., Melvin A., Rimmington D., Dattilo A., Miedzybrodzka E.L. GDF15 provides an endocrine signal of nutritional stress in mice and humans. Cell Metabolism. 2019;29(3):707–718. doi: 10.1016/j.cmet.2018.12.016. e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liberini C.G., Koch-Laskowski K., Shaulson E., McGrath L.E., Lipsky R.K., Lhamo R. Combined Amylin/GLP-1 pharmacotherapy to promote and sustain long-lasting weight loss. Scientific Reports. 2019;9(1):8447. doi: 10.1038/s41598-019-44591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adriaenssens A.E., Biggs E.K., Darwish T., Tadross J., Sukthankar T., Girish M. Glucose-Dependent insulinotropic polypeptide receptor-expressing cells in the hypothalamus regulate food intake. Cell Metabolism. 2019;30(5):987–996.e6. doi: 10.1016/j.cmet.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathiesen D.S., Bagger J.I., Bergmann N.C., Lund A., Christensen M.B., Vilsbøll T. The effects of dual GLP-1/GIP receptor agonism on glucagon secretion-A review. International Journal of Molecular Sciences. 2019;20(17):4092. doi: 10.3390/ijms20174092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell J.N., Macosko E.Z., Fenselau H., Pers T.H., Lyubetskaya A., Tenen D. A molecular census of arcuate hypothalamus and median eminence cell types. Nature Neuroscience. 2017;20(3):484–496. doi: 10.1038/nn.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen R., Wu X., Jiang L., Zhang Y. Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Reports. 2017;18(13):3227–3241. doi: 10.1016/j.celrep.2017.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lam B.Y.H., Cimino I., Polex-Wolf J., Nicole Kohnke S., Rimmington D., Iyemere V. Heterogeneity of hypothalamic pro-opiomelanocortin-expressing neurons revealed by single-cell RNA sequencing. Molecular Metabolism. 2017;6(5):383–392. doi: 10.1016/j.molmet.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romanov R.A., Zeisel A., Bakker J., Girach F., Hellysaz A., Tomer R. Molecular interrogation of hypothalamic organization reveals distinct dopamine neuronal subtypes. Nature Neuroscience. 2017;20(2):176–188. doi: 10.1038/nn.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang C., Kaye J.A., Cai Z., Wang Y., Prescott S.L., Liberles S.D. Area postrema cell types that mediate nausea-associated behaviors. Neuron. 2020 doi: 10.1016/j.neuron.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology. 2018;36(5):411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., III Comprehensive integration of single-cell data. Cell. 2019;177(7):1888–1902. doi: 10.1016/j.cell.2019.05.031. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Research. 2012;40(10):4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mei L., Nave K.-A. Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron. 2014;83(1):27–49. doi: 10.1016/j.neuron.2014.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mironova Y.A., Lenk G.M., Lin J.-P., Lee S.J., Twiss J.L., Vaccari I. PI(3,5)P2 biosynthesis regulates oligodendrocyte differentiation by intrinsic and extrinsic mechanisms. eLife. 2016;5 doi: 10.7554/eLife.13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vemuri G.S., McMorris F.A. Oligodendrocytes and their precursors require phosphatidylinositol 3-kinase signaling for survival. Development. 1996;122(8):2529. doi: 10.1242/dev.122.8.2529. [DOI] [PubMed] [Google Scholar]
- 25.Morita J., Kano K., Kato K., Takita H., Sakagami H., Yamamoto Y. Structure and biological function of ENPP6, a choline-specific glycerophosphodiester-phosphodiesterase. Scientific Reports. 2016;6(1):20995. doi: 10.1038/srep20995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle C.N., Lutz T.A., Le Foll C. Amylin – its role in the homeostatic and hedonic control of eating and recent developments of amylin analogs to treat obesity. Molecular Metabolism. 2018;8:203–210. doi: 10.1016/j.molmet.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L., Chang C.-C., Sun Z., Madsen D., Zhu H., Padkjær S.B. GFRAL is the receptor for GDF15 and is required for the anti-obesity effects of the ligand. Nature Medicine. 2017;23(10):1158–1166. doi: 10.1038/nm.4394. [DOI] [PubMed] [Google Scholar]
- 28.Emmerson P.J., Wang F., Du Y., Liu Q., Pickard R.T., Gonciarz M.D. The metabolic effects of GDF15 are mediated by the orphan receptor GFRAL. Nature Medicine. 2017;23(10):1215–1219. doi: 10.1038/nm.4393. [DOI] [PubMed] [Google Scholar]
- 29.Hsu J.-Y., Crawley S., Chen M., Ayupova D.A., Lindhout D.A., Higbee J. Non-homeostatic body weight regulation through a brainstem-restricted receptor for GDF15. Nature. 2017;550(7675):255–259. doi: 10.1038/nature24042. [DOI] [PubMed] [Google Scholar]
- 30.Mullican S.E., Lin-Schmidt X., Chin C.-N., Chavez J.A., Furman J.L., Armstrong A.A. GFRAL is the receptor for GDF15 and the ligand promotes weight loss in mice and nonhuman primates. Nature Medicine. 2017;23(10):1150–1157. doi: 10.1038/nm.4392. [DOI] [PubMed] [Google Scholar]
- 31.Harno E., Gali Ramamoorthy T., Coll A.P., White A. POMC: the physiological power of hormone processing. Physiological Reviews. 2018;98(4):2381–2430. doi: 10.1152/physrev.00024.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnaswami S.R., Grindberg R.V., Novotny M., Venepally P., Lacar B., Bhutani K. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nature Protocols. 2016;11(3):499–524. doi: 10.1038/nprot.2016.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu H., Kirita Y., Donnelly E.L., Humphreys B.D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. Journal of the American Society of Nephrology. 2019;30(1):23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bakken T.E., Hodge R.D., Miller J.A., Yao Z., Nguyen T.N., Aevermann B. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PloS One. 2018;13(12) doi: 10.1371/journal.pone.0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deng G., Morselli Lisa L., Wagner Valerie A., Balapattabi K., Sapouckey Sarah A., Knudtson Kevin L. Single-nucleus RNA sequencing of the hypothalamic arcuate nucleus of C57bl/6J mice after prolonged diet-induced obesity. Hypertension. 2020;76(2):589–597. doi: 10.1161/HYPERTENSIONAHA.120.15137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weng C., Ding M., Fan S., Cao Q., Lu Z. Transcription factor 7 like 2 promotes oligodendrocyte differentiation and remyelination. Molecular Medicine Reports. 2017;16(2):1864–1870. doi: 10.3892/mmr.2017.6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kohnke S., Lam B., Buller S., Zhao C., Nuzzaci D., Tadross J. Nutritional signals rapidly activate oligodendrocyte differentiation in the adult hypothalamic median eminence. bioRxiv. 2019:751198. [Google Scholar]
- 38.Gabery S., Salinas C.G., Paulsen S.J., Ahnfelt-Rønne J., Alanentalo T., Baquero A.F. Semaglutide lowers body weight in rodents via distributed neural pathways. JCI insight. 2020;5(6) doi: 10.1172/jci.insight.133429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willard F.S., Douros J.D., Gabe M.B., Showalter A.D., Wainscott D.B., Suter T.M. Tirzepatide is an imbalanced and biased dual GIP and GLP-1 receptor agonist. JCI insight. 2020;5(17) doi: 10.1172/jci.insight.140532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Q., Delessa C.T., Augustin R., Bakhti M., Colldén G., Drucker D.J. The glucose-dependent insulinotropic polypeptide (GIP) regulates body weight and food intake via CNS-GIPR signaling. Cell Metabolism. 2021 doi: 10.1016/j.cmet.2021.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Killion E.A., Wang J., Yie J., Shi S.D.H., Bates D., Min X. Anti-obesity effects of GIPR antagonists alone and in combination with GLP-1R agonists in preclinical models. Science Translational Medicine. 2018;10(472) doi: 10.1126/scitranslmed.aat3392. [DOI] [PubMed] [Google Scholar]
- 42.Larsen A.T., Gydesen S., Sonne N., Karsdal M.A., Henriksen K. The dual amylin and calcitonin receptor agonist KBP-089 and the GLP-1 receptor agonist liraglutide act complimentarily on body weight reduction and metabolic profile. BMC Endocrine Disorders. 2021;21(1):10. doi: 10.1186/s12902-020-00678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cheng W., Gonzalez I., Pan W., Tsang A.H., Adams J., Ndoka E. Calcitonin receptor neurons in the mouse nucleus tractus solitarius control energy balance via the non-aversive suppression of feeding. Cell Metabolism. 2020;31(2):301–312. doi: 10.1016/j.cmet.2019.12.012. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georgescu T., Lyons D., Doslikova B., Garcia A.P., Marston O., Burke L.K. Neurochemical characterization of brainstem pro-opiomelanocortin cells. Endocrinology. 2020;161(4) doi: 10.1210/endocr/bqaa032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellacott K.L.J., Halatchev I.G., Cone R.D. Characterization of leptin-responsive neurons in the caudal brainstem. Endocrinology. 2006;147(7):3190–3195. doi: 10.1210/en.2005-0877. [DOI] [PubMed] [Google Scholar]
- 46.Huo L., Grill H.J., Bjørbæk C. Divergent regulation of proopiomelanocortin neurons by leptin in the nucleus of the solitary tract and in the arcuate hypothalamic nucleus. Diabetes. 2006;55(3):567. doi: 10.2337/diabetes.55.03.06.db05-1143. [DOI] [PubMed] [Google Scholar]
- 47.Rau A.R., Hughes A.R., Hentges S.T. Various transgenic mouse lines to study proopiomelanocortin cells in the brain stem label disparate populations of GABAergic and glutamatergic neurons. American Journal of Physiology. Regulatory, Integrative and Comparative Physiology. 2018;315(1):R144–R152. doi: 10.1152/ajpregu.00047.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available from the NCBI Gene Expression Omnibus, accession GSE168737.