Abstract
The heterotrimeric Asi ubiquitin ligase (encoded by ASI1, ASI2, and ASI3) mediates protein degradation in the inner nuclear membrane in Saccharomyces cerevisiae. Asi1p and Asi3p possess catalytic domains, while Asi2p functions as an adaptor for a subset of Asi substrates. We hypothesized the Asi complex is an important mediator of protein quality control, and we predicted that Asi would be required for optimal growth in conditions associated with elevated abundance of aberrant proteins. Loss of Asi1p or Asi3p, but not Asi2p, sensitized yeast to hygromycin B, which promotes translational infidelity by distorting the ribosome A site. Surprisingly, loss of quality control ubiquitin ligase Hul5p did not sensitize yeast to hygromycin B. Our results are consistent with a prominent role for an Asi subcomplex that includes Asi1p and Asi3p (but not Asi2p) in protein quality control.
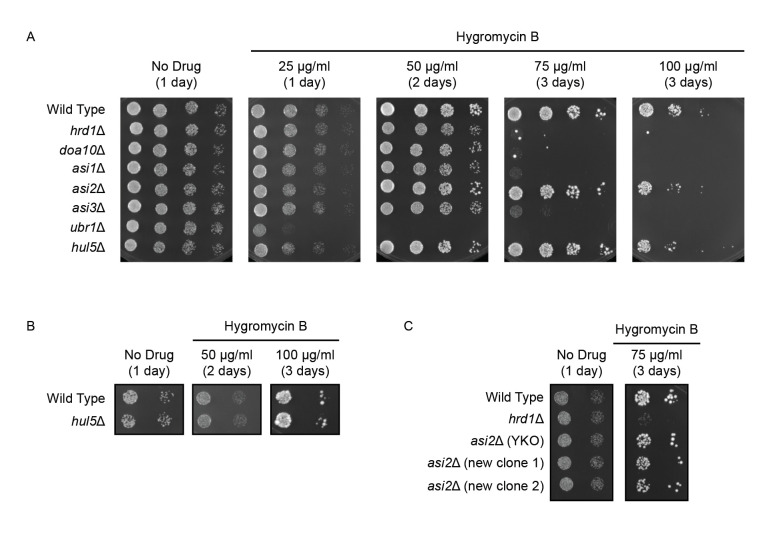
Figure 1. ASI1 and ASI3 confer resistance to hygromycin B.
(A-C) Sixfold serial dilutions of yeast of the indicated genotypes were spotted onto agar plates containing rich growth medium (No Drug) or indicated concentrations of hygromycin B. Plates were incubated at 30°C and imaged after 1-3 days. Experiments were performed in triplicate. (C) “asi2Δ (YKO)” is VJY852 and was obtained from the Yeast Knockout Collection (Tong et al., 2001). “asi2Δ (new clone 1)” and “asi2Δ (new clone 2)” are VJY969 and VJY970, respectively, and were generated for this study.
Description
Organelle proteome maintenance is essential for eukaryotic life. Several dedicated proteolytic systems promote organelle-specific turnover of misfolded or excess proteins. Inner nuclear membrane (INM) proteins are targeted for proteasomal destruction via INM-associated degradation (INMAD). At least three ubiquitin ligases mediate INMAD in the budding yeast Saccharomyces cerevisiae. These include the Asi complex, Doa10p, and the anaphase promoting complex (Deng and Hochstrasser, 2006; Foresti et al., 2014; Khmelinskii et al., 2014; Koch et al., 2019). Asi is composed of three subunits, Asi1p, Asi2p, and Asi3p (Foresti et al., 2014; Khmelinskii et al., 2014). Asi1p and Asi3p possess catalytic Really Interesting New Gene (RING) domains, while Asi2p serves as an adaptor for a subset of Asi substrates (Foresti et al., 2014; Khmelinskii et al., 2014; Natarajan et al., 2020).
Asi contributes to protein quantity control (e.g. degradation of orphan subunits of oligosaccharyl transferase and glycosylphosphatidylinositol transamidase complexes (Natarajan et al., 2020)) and localization control (e.g. degradation of mislocalized ergosterol synthetic enzyme Erg11p (Buchanan et al., 2019; Natarajan et al., 2020) and of transcription factors Stp1p and Stp2p when they inappropriately enter the nucleus (Natarajan et al., 2020; Omnus et al., 2011). The Asi complex and the endoplasmic reticulum-localized ubiquitin ligase Hrd1p redundantly promote the degradation of mutated, hypofunctional translocon subunit sec61-2p (Foresti et al., 2014; Trueman et al., 2011), suggesting Asi may also mediate protein quality control (PQC) of misfolded polypeptides. Only Asi1p and Asi3p (but not Asi2p) promote sec61-2p degradation (Foresti et al., 2014) and mitigate toxicity caused by sec61-2p expression (Flagg et al., 2021), raising the possibility that PQC function of Asi is mediated by Asi1p and Asi3p alone, or in conjunction with unidentified substrate specificity factors.
The aminoglycoside hygromycin B produced by the bacterium Streptomyces hygroscopicus reduces translational fidelity by distorting the ribosome A site, resulting in inaccurately synthesized protein molecules (Brodersen et al., 2000; Ganoza and Kiel, 2001). We previously demonstrated that loss of ER and nuclear PQC ubiquitin ligases Hrd1p, Doa10p, and Ubr1p sensitizes cells to hygromycin B (Crowder et al., 2015; Niekamp et al., 2019; Runnebohm et al., 2020). The extent of Asi’s contribution to PQC relative to these enzymes remains unknown.
We hypothesized that Asi is an important mediator of PQC. We predicted that the Asi complex would be required for resistance to conditions expected to increase the abundance of aberrant proteins. To test this, we cultured wild type yeast, yeast lacking genes encoding each subunit of the Asi complex, and a panel of PQC mutant yeast strains in the absence and presence of increasing concentrations of hygromycin B (Figure 1A). Consistent with previous results, loss of HRD1 or DOA10 sensitized cells to 75 μg/ml hygromycin B, and yeast deleted for UBR1 exhibited sensitivity at concentrations as low as 25 μg/ml. By contrast, deletion in two different genetic backgrounds of the gene encoding PQC ubiquitin ligase Hul5p (Fang et al., 2011; Runnebohm et al., 2020; Sitron and Brandman, 2019) did not sensitize cells to hygromycin B at the concentrations evaluated (Figure 1A, 1B).
Loss of ASI1 and ASI3 sensitized cells to 75 μg/ml hygromycin B to a similar extent as loss of DOA10 or HRD1 (Figure 1A). Intriguingly, loss of ASI2 in multiple independently generated yeast strains did not confer a similar growth disadvantage under these conditions (Figure 1A, 1C). Deletions of ASI genes and HUL5 were validated by PCR. Taken together, our results indicate Asi1p and Asi3p, but not Asi2p, are required for optimal growth in the presence of a compound expected to generate increased numbers of PQC substrates.
The finding that loss of Hul5p does not enhance sensitivity to hygromycin B was surprising, given multiple characterized functions of Hul5p in PQC. Among other roles, Hul5p promotes degradation of substrates that have escaped detection by the ribosome quality control ubiquitin ligase Ltn1p (Sitron and Brandman, 2019) and promotes turnover of misfolded proteins following heat shock (Fang et al., 2011). Loss of Ltn1p sensitizes cells to hygromycin B (Bengtson and Joazeiro, 2010; Crowder et al., 2015). We speculate that a requirement for Hul5p in hygromycin B resistance may become apparent during conditions characterized by elevated cellular dependence on Hul5p, such as compromised Ltn1p function or heat shock.
Multiple lines of evidence suggest that a subcomplex of the Asi ubiquitin ligase including Asi1p and Asi3p (but not Asi2p) mediates PQC degradation of misfolded proteins, potentially in complex with unidentified substrate adaptors. First, as demonstrated here, deletion of ASI1 or ASI3, but not of ASI2, sensitizes cells to conditions expected to increase the abundance of aberrant, mistranslated proteins to an extent similar to that observed following loss of other characterized PQC genes (we note it remains possible that ASI2 is required for optimal growth under different forms of proteotoxic stress, such as elevated temperature). Second, while Asi1p, Asi2p, and Asi3p collaborate to mediate degradation of a host of mislocalized proteins, only Asi1p and Asi3p promote degradation of mutated translocon component sec61-2p (Foresti et al., 2014). Finally, simultaneous deletion of genes encoding Hrd1p, Ire1p (a component of the yeast unfolded protein response), and either Asi1p or Asi3p causes markedly slower growth than concurrent knockout of HRD1, IRE1, and ASI2 (Foresti et al., 2014).
Asi2p function is also dispensable for degradation of some Asi1/3p substrates that do not possess features rendering them predicted PQC substrates (Khmelinskii et al., 2014). Such substrates may expose degradation signals (e.g. when other complex subunits are present in substoichiometric abundance) resembling those of quality control substrates, co-opting a PQC enzyme for regulatory purposes. The precise nature of degradation signal(s) recognized by Asi remains to be resolved.
Methods
ASI2 gene replacement. To generate VJY969 and VJY970, ASI2 was replaced with kanMX4 via homologous recombination. An 1807-bp asi2Δ::kanMX4 cassette was PCR-amplified from VJY852 (Yeast Knockout Collection asi2Δ::kanMX4 strain (Tong et al., 2001)) using primers VJR472 (5’ GACACCGAATCAAACGCATA 3’) and VJR473 (5’ GGAAAGCTTGCAAACAGCTC 3’) and introduced into naïve wild type VJY476 (alias BY4741 (Tong et al., 2001)) by lithium acetate transformation (Guthrie and Fink, 2004). G418-resistant clones were validated by PCR genotyping at the 5’ and 3’ recombination junctions.
Yeast growth assay. Yeast growth analysis was performed as described (Watts et al., 2015). Four μl of sixfold serial dilutions were pipetted onto yeast extract-peptone-dextrose medium (Guthrie and Fink, 2004) in the absence or presence of increasing concentrations of hygromycin B (Gibco). Plates were incubated at 30°C and imaged at the indicated times.
Reagents
| Name | Genotype | Figure | Reference |
| VJY60 (alias W303) | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 | 1B | (Thomas and Rothstein, 1989) |
| VJY85 | MATa ade2-1 ura3-1 his3-11 trp1-1 leu2-3,112 can1-100 hul5Δ::LEU2 | 1B | (Wang et al., 1999) |
| VJY360 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 asi1Δ::kanMX4 | 1A | (Tong et al., 2001) |
| VJY469 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ubr1Δ::kanMX4 | 1A | (Tong et al., 2001) |
| VJY476 (alias BY4741) | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 1A, 1C | (Tong et al., 2001) |
| VJY511 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hrd1Δ::kanMX4 | 1A, 1C | (Tong et al., 2001) |
| VJY662 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 hul5Δ::kanMX4 | 1A | (Tong et al., 2001) |
| VJY667 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 doa10Δ::kanMX4 | 1A | (Tong et al., 2001) |
| VJY696 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 asi3Δ::kanMX4 | 1A | (Tong et al., 2001) |
| VJY852 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 asi2Δ::kanMX4 | 1A, 1C | (Tong et al., 2001) |
| VJY969 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 asi2Δ::kanMX4 | 1C | This study |
| VJY970 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 asi2Δ::kanMX4 | 1C | This study |
Acknowledgments
Acknowledgments
Experiments to determine sensitivity of asi1Δ and hul5Δ yeast to hygromycin B were piloted by undergraduate students in the Spring 2020 Methods in Cell Biology (BIO 315) Course at Ball State University and validated in the research laboratory of EMR. We thank Kamryn Kennedy, Mahmoud Daraghmi, and Seth Horowitz for laboratory assistance. We thank Jon Huibregtse, Christopher Hickey, and Mark Hochstrasser for generously sharing yeast strains. We thank the Ball State University Division of Online and Strategic Learning for supporting an initiative to transform undergraduate laboratory courses into authentic research-based learning experiences. We thank Stefan Kreft and Adrian Mehrtash for thoughtful discussion about this manuscript. We thank our reviewer for thoughtful feedback about this manuscript.
Funding
This work was funded by NIH grant R15 GM111713 (EMR). Work in the lab of PJS is funding by NIH grant R15 G067291 and NIH grant R15 CA252996. KAW and BMV were supported by Ball State University Honors Undergraduate Fellowships. This project was conceived while EMR was supported in part by a Ball State University Excellence in Teaching award (sponsored by the Ball State University Division of Online and Strategic Learning and the Office of the Provost).
References
- Bengtson MH, Joazeiro CA. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature. 2010 Sep 12;467(7314):470–473. doi: 10.1038/nature09371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodersen DE, Clemons WM Jr, Carter AP, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. The structural basis for the action of the antibiotics tetracycline, pactamycin, and hygromycin B on the 30S ribosomal subunit. Cell. 2000 Dec 22;103(7):1143–1154. doi: 10.1016/s0092-8674(00)00216-6. [DOI] [PubMed] [Google Scholar]
- Buchanan BW, Mehrtash AB, Broshar CL, Runnebohm AM, Snow BJ, Scanameo LN, Hochstrasser M, Rubenstein EM. Endoplasmic reticulum stress differentially inhibits endoplasmic reticulum and inner nuclear membrane protein quality control degradation pathways. J Biol Chem. 2019 Nov 13;294(51):19814–19830. doi: 10.1074/jbc.RA119.010295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder JJ, Geigges M, Gibson RT, Fults ES, Buchanan BW, Sachs N, Schink A, Kreft SG, Rubenstein EM. Rkr1/Ltn1 Ubiquitin Ligase-mediated Degradation of Translationally Stalled Endoplasmic Reticulum Proteins. J Biol Chem. 2015 Jun 01;290(30):18454–18466. doi: 10.1074/jbc.M115.663559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M, Hochstrasser M. Spatially regulated ubiquitin ligation by an ER/nuclear membrane ligase. Nature. 2006 Oct 19;443(7113):827–831. doi: 10.1038/nature05170. [DOI] [PubMed] [Google Scholar]
- Fang NN, Ng AH, Measday V, Mayor T. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol. 2011 Oct 01;13(11):1344–1352. doi: 10.1038/ncb2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagg MP, Wangeline MA, Holland SR, Duttke SH, Benner C, Neal S, Hampton RY. Inner-nuclear-membrane-associated degradation employs Dfm1-independent retrotranslocation and alleviates misfolded transmembrane-protein toxicity. Mol Biol Cell. 2021 Feb 10;32(7):521–537. doi: 10.1091/mbc.E20-11-0720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foresti O, Rodriguez-Vaello V, Funaya C, Carvalho P. Quality control of inner nuclear membrane proteins by the Asi complex. Science. 2014 Sep 18;346(6210):751–755. doi: 10.1126/science.1255638. [DOI] [PubMed] [Google Scholar]
- Ganoza MC, Kiel MC. A ribosomal ATPase is a target for hygromycin B inhibition on Escherichia coli ribosomes. Antimicrob Agents Chemother. 2001 Oct 01;45(10):2813–2819. doi: 10.1128/AAC.45.10.2813-2819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C and Fink GR. 2004. Guide to Yeast Genetics and Molecular and Cell Biology, Part B. 1st ed. Elsevier, San Diego. 2002.
- Khmelinskii A, Blaszczak E, Pantazopoulou M, Fischer B, Omnus DJ, Le Dez G, Brossard A, Gunnarsson A, Barry JD, Meurer M, Kirrmaier D, Boone C, Huber W, Rabut G, Ljungdahl PO, Knop M. Protein quality control at the inner nuclear membrane. Nature. 2014 Dec 18;516(7531):410–413. doi: 10.1038/nature14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch BA, Jin H, Tomko RJ Jr, Yu HG. The anaphase-promoting complex regulates the degradation of the inner nuclear membrane protein Mps3. J Cell Biol. 2019 Feb 01;218(3):839–854. doi: 10.1083/jcb.201808024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan N, Foresti O, Wendrich K, Stein A, Carvalho P. Quality Control of Protein Complex Assembly by a Transmembrane Recognition Factor. Mol Cell. 2019 Oct 31;77(1):108–119.e9. doi: 10.1016/j.molcel.2019.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niekamp JM, Evans MD, Scott AR, Smaldino PJ, Rubenstein EM. TOM1 confers resistance to the aminoglycoside hygromycin B in Saccharomyces cerevisiae. MicroPubl Biol. 2019 Dec 01;2019 [PMC free article] [PubMed] [Google Scholar]
- Omnus DJ, Pfirrmann T, Andréasson C, Ljungdahl PO. A phosphodegron controls nutrient-induced proteasomal activation of the signaling protease Ssy5. Mol Biol Cell. 2011 Jun 01;22(15):2754–2765. doi: 10.1091/mbc.E11-04-0282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runnebohm AM, Evans MD, Richardson AE, Turk SM, Olesen JB, Smaldino PJ, Rubenstein EM. Loss of protein quality control gene UBR1 sensitizes Saccharomyces cerevisiae to the aminoglycoside hygromycin B. Fine Focus. 2020 Oct 26;6(1):76–83. doi: 10.33043/FF.6.1.76-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitron CS, Brandman O. CAT tails drive degradation of stalled polypeptides on and off the ribosome. Nat Struct Mol Biol. 2019 May 27;26(6):450–459. doi: 10.1038/s41594-019-0230-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Elevated recombination rates in transcriptionally active DNA. Cell. 1989 Feb 24;56(4):619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001 Dec 14;294(5550):2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- Trueman SF, Mandon EC, Gilmore R. Translocation channel gating kinetics balances protein translocation efficiency with signal sequence recognition fidelity. Mol Biol Cell. 2011 Jul 01;22(17):2983–2993. doi: 10.1091/mbc.E11-01-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Yang J, Huibregtse JM. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol. 1999 Jan 01;19(1):342–352. doi: 10.1128/MCB.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts SG, Crowder JJ, Coffey SZ, Rubenstein EM. Growth-based determination and biochemical confirmation of genetic requirements for protein degradation in Saccharomyces cerevisiae. J Vis Exp. 2015 Feb 16;(96):e52428–e52428. doi: 10.3791/52428. [DOI] [PMC free article] [PubMed] [Google Scholar]