Abstract
Background
The availability of thousands of genomes has enabled new advancements in biology. However, many genomes have not been investigated for their quality. Here we examine quality trends in a taxonomically diverse and well-known group, butterflies (Papilionoidea), and provide draft, de novo assemblies for all available butterfly genomes. Owing to massive genome sequencing investment and taxonomic curation, this is an excellent group to explore genome quality.
Findings
We provide de novo assemblies for all 822 available butterfly genomes and interpret their quality in terms of completeness and continuity. We identify the 50 highest quality genomes across butterflies and conclude that the ringlet, Aphantopus hyperantus, has the highest quality genome. Our post-processing of draft genome assemblies identified 118 butterfly genomes that should not be reused owing to contamination or extremely low quality. However, many draft genomes are of high utility, especially because permissibility of low-quality genomes is dependent on the objective of the study. Our assemblies will serve as a key resource for papilionid genomics, especially for researchers without computational resources.
Conclusions
Quality metrics and assemblies are typically presented with annotated genome accessions but rarely with de novo genomes. We recommend that studies presenting genome sequences provide the assembly and some metrics of quality because quality will significantly affect downstream results. Transparency in quality metrics is needed to improve the field of genome science and encourage data reuse.
Keywords: accessibility, genomics, life sciences, open data, Papilionoidea
Introduction
The explosion of available genomes across the Tree of Life has created entirely new fields of science and is changing how we investigate long-standing questions in biology. Studies of gene family evolution and gene mutation have expanded from single genes to mapping the architecture of entire genomes. Macroevolutionary studies using genomic data are now regularly being generated at impressive scales, e.g., complete class [1], continent [2], and spanning up to 500 million years [3]. As the scope of questions addressed with genomic data continues to expand, determining the effect of read length and genome completeness on results is vital. One metric that is often applied to assembled genomes is the N50 score, a weighted median statistic of contig continuity that describes the distribution of contig lengths. The N50 value indicates that half of the assembly is contained in contigs or scaffolds equal to or larger than the value. Assemblies with low N50s are more fragmented and have contigs or scaffolds with less overlap with one another. Completeness of a draft assembly can also be assessed using BUSCO scores [4]. This measure uses a taxonomically informed set of “core” protein-coding orthologs that are theoretically present in a given taxon to evaluate genomic completeness. BUSCO may detect both haplotypes sequenced from diploid tissue with adequate genome coverage. However, high heterozygosity can lead to more fragmented assemblies (low N50), potentially reducing the number of complete protein-coding genes recovered. These scores can be influenced by biological variation through natural variation in chromosome length or lineage-wide loss of core orthologs, but also by systematic error, as in poor sequencing depth [4]. Genomes may be of low quality in terms of continuity, completeness, or a combination of these 2 metrics. Understanding how genomes with low-quality metrics affect downstream analyses is critical.
Here, we provide draft de novo genome assemblies and quality metrics for butterflies that will be useful for studying Lepidoptera evolution, gene discovery, and genomics. To understand how genome quality varies across taxa, we examine genome assembly quality in this exemplar group of organisms that has >935 published genomes. Additionally, we explore potential uses of these data, bearing in mind their draft nature, and discuss the state of butterfly genomics in light of genome quality.
Gene family evolution and mutation holds immense potential in uncovering the mechanisms behind rapid functional adaptation and potential subsequent speciation [5, 6], and significant progress is being made in this area with the inclusion of genomic data [3]. De novo genome assemblies allow for the discovery of novel genes with important ecological implications. For example, genes and gene duplications associated with plant detoxification can be identified [7]. Additionally, expansions of a particular gene copy can be indicative of functional adaptation (e.g., [8, 9]). However, inaccurate assessment of gene copy number will lead to false interpretations. Denton et al. [10] document a pattern of gene misassembly and false gene duplication rates in draft genomes, with gene number either overestimated or underestimated in 40% of all gene families. The mechanism of such error is closely tied to N50, such that when genes are fragmented (low N50), multiple contigs are assembled into non-biological contigs [10]. These types of errors will present as misidentification of gene duplication and loss, as well as non-biological mutations. Gene family evolution and mutation holds immense potential in uncovering the mechanisms behind rapid functional adaptation and potential subsequent speciation [5, 6], and significant progress is being made in this area with the inclusion of genomic data [3]. Including sequences of known identity to identify regions of sequencing artefacts or incorrect annotation and implementing assembly error estimation [11] may mitigate these challenges.
Phylogenetic studies stand to gain enormous taxonomic ground into the 2020s, primarily owing to the explosion of low-coverage genomes that are particularly well suited for phylogenetic studies. Taxonomic coverage in phylogenetic studies is increasing exponentially with the ability to sequence genomes from historical or museum specimens. Advances in both cost and quality of sequencing, as well as the ability to sequence DNA from degraded museum samples [12–15], allow researchers to now produce phylogenies including all extant, and even extinct species in a taxonomic group [16]. Stringency standards for including genomes in phylogenetic studies are not well established, and poor-quality genomes can produce erroneous assemblies of genes of interest [10], as detailed above. Furthermore, quality scores that highlight the completeness of a genome may serve an important quality control step for the inclusion of genomes in phylogenies, and we recommend that researchers prioritize this quality metric for phylogenetic inference. A more complete genome suggests that the sample possesses common and complete protein-coding genes, and thus it is more likely to include the researcher's set of orthologs. By assessing genome completeness, future systematic error due to taxa with low matrix occupancy may be avoided [17].
Here, we provide 822 draft de novo genome assemblies and quality metrics for a taxonomically diverse, well known group, butterflies, that will be useful for studies on their evolution, gene discovery, and genomics. We explore potential uses of these data, bearing in mind their draft nature, and discuss the state of butterfly genomics in light of genome quality.
Methods
We obtained all published genome assemblies and genomic reads of butterflies (Lepidoptera: Papilionoidea) from NCBI [18] and LepBase [19] databases as of 1 July 2020. In the case of NCBI genome assemblies, we searched using the taxonomy database (keywords “Papilionoidea” and “papilionoid”) for the latest assemblies. When multiple were available, we selected the most recently submitted assembly (as of 1 July 2020; see Supplementary Table S1). We also searched the SRA database [18] and published literature for available paired-end, whole-body, whole shotgun genome sequences of papilionoid species [13, 20–31] (search terms butterfly genome; papilionoid genome; butterfly shotgun genome; searches concluded on 1 July 2020).
We trimmed reads using TrimGalore [32] requiring a quality score of 20 and read length of 30. We assembled reads using SPAdes (SPAdes, RRID:SCR_000131) v3.13 [33] using paired reads and allowing values of K to vary based on read length. For the majority of the de novo genomes, 32 CPUs and 128 GB of memory were sufficient. Forty genomes required additional memory; we ran these genomes with 24 threads with 720 GB of memory, potentially due to deeper sequencing or greater genomic complexity.
Following assembly, we performed several post-processing steps to ensure sequence integrity. First, we identified and removed contigs composed of <200 bp using SeqTK (Seqtk, RRID:SCR_018927) [34]. We scanned for evidence of vector contamination using VecScreen (VecScreen, RRID:SCR_016577) [35] and removed affected contigs. Then, we used the NCBI contaminant screening database to identify common contaminants, such as from fungi or bacteria, and removed those contaminant sequences.
To assess assembly quality, we first used assembly-stats [36] to quantify scaffold N50 for each cleaned, contaminant-free assembly. This measure estimates the contiguity of assembly contigs and describes the contig length of half of the genome; i.e., 50% of the genome includes contigs greater than or equal to this length. We also used BUSCO (BUSCO, RRID:SCR_015008) v3.02 [4] to determine the presence of a set of 1,658 core insect single-copy genes (version 9) that are highly conserved across insects and give an approximation of the completeness of the assembly. Herein, we evaluate only the BUSCO Complete score, which requires each of the 1,658 core ortholog genes in the assembly to include both start and stop codons. For the full BUSCO score report, see Supplemental Tables S1–S3. A custom script, filter_seqs_by_NCBI.py (Supplementary File 1), was created to automate NCBI required edits. This script uses the text feedback file from NCBI and will be useful for researchers willing to make their assemblies available on NCBI.
Results
We assembled 873 papilionoid genomes using raw reads from the NCBI SRA database and downloaded 62 pre-assembled genomes from the NCBI Assembly database [18]. These 935 butterfly samples with genomic data represent 665 unique species because some species have multiple subspecies sequenced or have replicate genomes (Supplementary Table S1). We did not attempt to combine genomic reads from multiple conspecific individuals because this will artificially increase heterozygosity and inevitably affect assembly quality [37]. All genomes assembled for this study (Supplementary Table S1) are available for download through the TPA Database (BioProject PRJNA606954) and quality statistics calculated for each genome are listed in Supplementary Table S1.
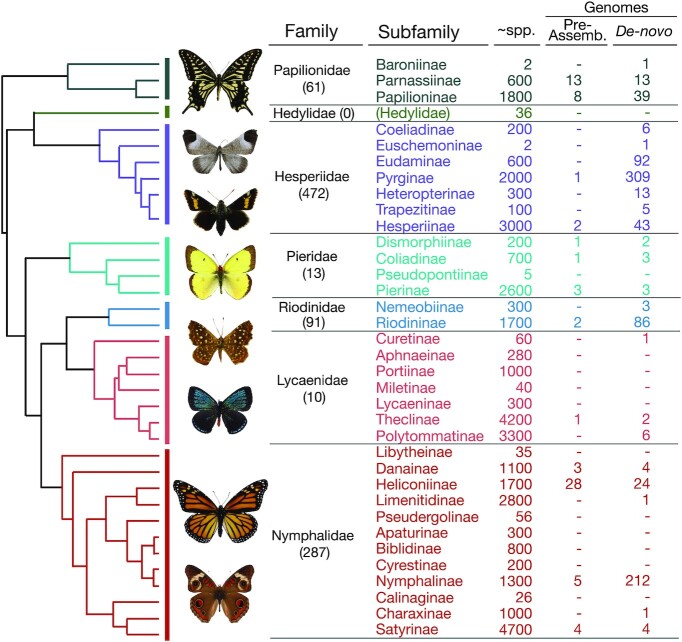
Pre-assembled genomes from NCBI and LepBase span 6 butterfly families and 12 subfamilies; our de novo assembled genomes represent 6 families and 24 subfamilies (Fig. 1). The only family for which no public genomic data are available is the Hedylidae, a family with only 36 described neotropical species [39]. Hesperiidae has the greatest number of species with available genomic data (472), more than half of which are in subfamily Pyrginae (310), largely due to research by Grishin and colleagues [13, 20–27, 30, 31] (Fig. 1). The Nymphalidae, the most species-rich family of butterflies, have 287 genomes available, and 210 of these genomes are in the genus Junonia (Fig. 1). The Lycaenidae have comparatively few genomes available (10), given its high species richness (Fig. 1).
Figure 1:
Pre-assembled and de novo assembled genomes for each butterfly and subfamily shown on a topological sketch of [38]. Species-richness estimates and topology are presented for comparison only.
The metrics that we used revealed large variance in genome assembly quality. N50 and BUSCO scores were often similar, such that the highest quality genomes typically had both high N50 and high BUSCO scores, although this was not always the case (Fig. 2). These quality statistics measure 2 different aspects of quality and should be used in conjunction because length distribution may not be associated with gene content [4]. Pre-assembled genomes downloaded from NCBI and LepBase on average had high quality scores (Supplementary Table S2, Fig. 2) (scaffold N50 = 1,706,589 bp; BUSCO = 81.2%). Of these, 5 Heliconius genomes (H. doris,H. hecuba flava, H. hierax, H. wallacei, and H. xanthocles) have notably lower mean quality scores (N50 = 996.6 bp; BUSCO = 33.66%). The H. hierax genome (GCA_900068475.1) had the lowest quality measures of the pre-assembled genomes that we investigated (N50 = 916 bp; BUSCO = 30.5). The satyrine Aphantopus hyperantus (GCA_902806685.1) had the highest quality scores of all genomes investigated (N50 = 15,230,192 bp; BUSCO = 97.8%).
Figure 2:

Natural log–normalized N50 and BUSCO scores plotted for both pre-assembled (squares) and de novo (circles) genome assemblies. Colors denote taxonomic family designation, as in Fig. 1. Letters correspond to inset images of representative species.
Quality scores varied widely among the draft de novo genome assemblies (Fig. 2). In 51 cases, we found that assemblies comprised only short (<200 bp) fragments and contaminants. In these cases, we removed the assembly and report the N50 score as zero (Supplementary Tables S1 and S3). N50 ranged from 249 bp in Junonia evarete nigrosuffusa (SRR10765819; Nymphalidae) to 43,550 bp in Sertania guttata guttata (Fig. 2E; SRR10158585; Riodinidae). One hundred seven de novo genomes resulted in a BUSCO score of 0% (Supplementary Tables S1 and S3), meaning that these genomes recovered none of the core insect orthologs. Seven had BUSCO scores of ≥90%, with the greatest BUSCO score (96.4%) from Papilio antimachus (Fig. 2D; SRR8954523 [29]). The mean quality scores of the de novo genomes were low (N50 = 15,650 bp; BUSCO = 28.25%; excluding zero values). Proboscis propylea (Fig. 2H) had a greater than average BUSCO score, but low N50 (N50 = 605 bp; BUSCO = 45.3%). In an effort to identify the best exemplar genome for each major butterfly lineage, we present the highest quality genomes per subfamily (Table 1). Table 2 summarizes the 50 highest quality butterfly denovo and preassembled genomes, regardless of taxonomy.
Table 1:
Highest quality genomes by butterfly subfamily, according to N50 and BUSCO scores
| Taxonomy | Organism | Accession ID | N50 (bp) | BUSCO 3 (C%) |
|---|---|---|---|---|
| Hesperiidae; Coeliadinae | Choaspes benjaminii | SRR7174556 | 2,532 | 87.3 |
| Hesperiidae; Eudaminae | Phocides pigmalion | SRR7174453 | 9,497 | 76.7 |
| Hesperiidae; Hesperiinae | Megathymus ursus | GCA_003671415.1 | 4,153,133 | 98.3 |
| Hesperiidae; Heteropterinae | Dalla quadristriga | SRR9330377 | 4,259 | 69.8 |
| Hesperiidae; Pyrginae | Cecropterus lyciades | GCA_002930495.1 | 558,064 | 97.3 |
| Hesperiidae; Trapezitinae | Toxidia parvulus | SRR9330370 | 932 | 21.7 |
| Lycaenidae; Curetinae | Curetis bulis | SRR10158559 | 1,108 | 28.3 |
| Lycaenidae; Polyommatinae | Cyclargus thomasi | SRR6727422 | 13,909 | 91.3 |
| Lycaenidae; Theclinae | Calycopis cecrops | GCA_001625245.1 | 233,537 | 95.5 |
| Nymphalidae; Charaxinae | Charaxes varanes | SRR5175869 | 1,531 | 49.5 |
| Nymphalidae; Danainae | Danaus plexippus | GCA_009731565.1 | 9,209,872 | 93.9 |
| Nymphalidae; Heliconiinae | Heliconius erato | LepBase_Heliconius_erato_demophoon_v1 | 10,688,973 | 97.4 |
| Nymphalidae; Limentidinae | Limenitis arthemis | SRR1504973 | 631 | 12.6 |
| Nymphalidae; Morphinae | Taenaris catops | GCA_009936525.1 | 1,720,500 | 35.2 |
| Nymphalidae; Nymphalinae | Vanessa tameamea | GCA_002938995.1 | 2,988,984 | 98.3 |
| Nymphalidae; Satyrinae | Aphantopus hyperantus | GCA_902806685.1 | 15,230,192 | 97.8 |
| Papilionidae; Baroniinae | Baronia brevicornis | SRR8954515 | 1,886 | 59.0 |
| Papilionidae; Papilioninae | Papilio xuthus | GCA_000836235.1 | 6,198,915 | 97.6 |
| Papilionidae; Parnassiinae | Sericinus montela | SRR8954536 | 3,584 | 59.4 |
| Pieridae; Coliadinae | Zerene cesonia | GCA_012273895.1 | 9,214,832 | 95.6 |
| Pieridae; Dismorphiinae | Leptidea sinapis | GCA_900199415.2 | 857,189 | 97.2 |
| Pieridae; Pierinae | Pieris napi | LepBase_Pieris_napi_v1.1 | 12,597,868 | 94.4 |
| Riodinidae; Nemeobiinae | Euselasia chrysippe | SRR10158562 | 1,806 | 30.3 |
| Riodinidae; Riodininae | Calephelis nemesis | GCA_002245505.1 | 206,312 | 95.6 |
C: complete.
Table 2:
Highest 50 quality papilionoid genome assemblies, regardless of subfamily, ranked using natural log–normalized N50 and BUSCO Complete scores
| Rank | Organism | Accession ID | N50 (bp) | BUSCO 3 (C%) |
|---|---|---|---|---|
| 1 | Aphantopus hyperantus | GCA_902806685.1 | 15,230,192 | 97.8 |
| 2 | Pieris napi | LepBase_Pieris_napi_v1.1 | 12,597,868 | 94.4 |
| 3 | Heliconius erato | LepBase_Heliconius_erato_demophoon_v1 | 10,688,973 | 97.4 |
| 4 | Danaus plexippus | GCA_009731565.1 | 9,209,872 | 98.0 |
| 5 | Zerene cesonia | GCA_012273895.1 | 9,214,832 | 95.6 |
| 6 | Papilio xuthus | GCA_000836235.1 | 6,198,915 | 97.6 |
| 7 | Papilio bianor | GCA_011763625.1 | 13,111,833 | 65.0 |
| 8 | Megathymus ursus | GCA_003671415.1 | 4,153,133 | 98.3 |
| 9 | Papilio memnon | GCA_003118415.2 | 4,560,862 | 92.9 |
| 10 | Papilio polytes | GCA_000836215.1 | 3,672,263 | 91.8 |
| 11 | Vanessa tameamea | GCA_002938995.1 | 2,988,984 | 98.3 |
| 12 | Junonia coenia | LepBase_Junonia_coenia_JC_v1.0 | 1,571,165 | 98.2 |
| 13 | Danaus chryssipus | GCA_004959915.1 | 1,465,393 | 93.9 |
| 14 | Papilio machaon | GCA_001298355.1 | 1,174,287 | 95.5 |
| 15 | Hypolimnas misippus | GCA_008963455.1 | 1,011,763 | 98.1 |
| 16 | Leptidea sinapsis | GCA_900199415.2 | 857,189 | 97.2 |
| 17 | Danaus melanippus | GCA_010014825.1 | 889,656 | 89.4 |
| 18 | Bicyclus anynana | GCA_900239965.1 | 638,282 | 97.6 |
| 19 | Pieris rapae | GCA_001856805.1 | 617,301 | 98.0 |
| 20 | Papilio dardanus | GCA_013186455.1 | 596,599 | 94.3 |
| 21 | Cecropterus lyciades | GCA_002930495.1 | 558,064 | 97.3 |
| 22 | Lerema accius | GCA_001278395.1 | 525,349 | 95.1 |
| 23 | Phoebis sennae | GCA_001586405.1 | 299,140 | 91.1 |
| 24 | Taenaris catops | GCA_009936525.1 | 1,720,500 | 35.2 |
| 25 | Calycopis cecrops | GCA_001625245.1 | 233,537 | 95.5 |
| 26 | Papilio glaucus | GCA_000931545.1 | 230,841 | 95.5 |
| 27 | Calephelis nemesis | GCA_002245505.1 | 206,312 | 95.6 |
| 28 | Delias pasithoe | GCA_010014985.1 | 193,720 | 96.5 |
| 29 | Heliconius melpomene | GCA_000313835.2 | 194,302 | 95.6 |
| 30 | Maniola jurtina | GCA_009667785.1 | 212,945 | 88.3 |
| 31 | Calephelis virginiensis | GCA_002245475.1 | 175,106 | 93.9 |
| 32 | Heliconius burneyi | LepBase_Heliconius_burneyi_helico3 | 106,325 | 96.5 |
| 33 | Melitaea cinxia | GCA_000716385.1 | 119,328 | 83.0 |
| 34 | Colias croceus | GCA_009982905.1 | 95,765 | 92.5 |
| 35 | Heliconius hecalesia | LepBase_Heliconius_hecalesia_helico3 | 68,855 | 96.5 |
| 36 | Heliconius demeter | Lepbase_Heliconius_demeter_helico3 | 67,995 | 96.8 |
| 37 | Heliconius besckei | LepBase_Heliconius_besckei_helico3 | 64,778 | 95.8 |
| 38 | Heliconius himera | LepBase_Heliconius_himera_helico3 | 48,684 | 96.5 |
| 39 | Heliconius sara | LepBase_Heliconius_sara_helico3 | 43,390 | 94.3 |
| 40 | Heliconius telesiphe | LepBase_Heliconius_telesiphe_helico3 | 42,672 | 94.7 |
| 41 | Eueides tales | LepBase_Eueides_tales_helico3 | 32,552 | 94.7 |
| 42 | Megathymus ursus | SRR7174358 | 24,120 | 90.7 |
| 43 | Agraulis vanillae | LepBase_Agraulis_vanillae_helico3 | 21,413 | 94.6 |
| 44 | Dryas iulia | LepBase_Dryas_iulia_helico3 | 21,916 | 92.3 |
| 45 | Delias oraia | SRR4341246 | 18,269 | 92.3 |
| 46 | Pararge aegeria | GCA_900499025.1 | 16,525 | 88.0 |
| 47 | Atrophaneura dixoni | SRR8954516 | 14,618 | 93.5 |
| 48 | Cyclargus thomasi | SRR6727422 | 13,909 | 91.3 |
| 49 | Eumaeus atala | SRR6727440 | 13,611 | 87.0 |
| 50 | Sertania guttata | SRR10158585 | 43,550 | 35.1 |
Discussion
High-quality genomes are required for studies that span the biological sciences, from gene family, mutation research to macroevolutionary phylogenetics and population dynamics. Our results show that available genomes vary widely in quality and taxonomic coverage. The significant variance in N50 and BUSCO scores highlights an important message: in the scientific literature, a “genome” can range from genomic fragments to fully annotated chromosomes. Large-scale genomic studies, especially those that sequence species in an entire clade or geographic region, represent great scientific feats, but if they are based on many low-quality genomes, they may not be useful for subsequent studies. We encourage peer-reviewed journals and public databases to require authors to report genome quality via N50 and BUSCO, which can be accessioned with the assembly on NCBI as Global Statistics. Doing so provides maximum transparency, reproducibility, and a holistic view of future data reuse. In this way, users can easily evaluate whether the quality of the genome is high enough to investigate gene family diversification (prioritize N50) or phylogenetic systematics (prioritize BUSCO).
Our analyses highlight the extensive variation in genome quality. Part of this discrepancy can be alleviated with changes in language. Perhaps we should begin referring to low-quality genomes, such as J. evarete nigrosuffusa (SRR10765819; N50 = 249 bp; BUSCO = 0.1%), as “genomic data,” as opposed to the potentially misleading term, “genome.” Next, accessioning all assemblies would save countless hours of computation time and allow for the validation of results. In addition, assemblies would also allow results (e.g., gene family evolution, sequence identification, ortholog determination) from previous studies to be validated. Accessioning should include low-coverage draft genome assemblies, which can also be deposited in the NCBI's Assembly database. These assemblies have notably lower N50 and BUSCO scores when compared to the average assembly from NCBI and LepBase [19]. Quality metrics of our de novo assembled genomes were, in many cases, comparable to the 5 Heliconius genomes that we investigated, suggesting that even low-quality genome assemblies can and should be accessioned. Including quality scores (as Global Statistics) for each draft assembly via the NCBI Assembly database (in addition to taxon-specific genome databases, such as Lepbase [19]) would provide a transparent overview of available genomes for future studies.
Genome assembly requires considerable computational resources, and assessing genome quality simply from raw file size on GenBank can be misleading. Many studies in the biological and medical sciences rely on existing genomes and their annotations (e.g., [40]). If researchers independently assemble genomes, this can lead to duplicated effort and significant time investment. Furthermore, if raw data quality is poor, assemblies likely will not be useful. In our study, we found that ≥51 of the 873 genomes that we assembled are ultimately unusable, and another 67 should be reused only with caution (Supplementary Table S1). These 118 samples produced assemblies that entirely comprised contamination or contigs <200 bp or were devoid of core insect genes, or a combination of these factors. However, it is possible that alternate assembly methods could produce a better assembly. Low N50 and low BUSCO assemblies are likely composed of fragmented genes, and, most likely, the contigs that are present are the result of very low sequence coverage. This low coverage is indicative of a high error rate and greater likelihood of incorrect sequence frame. As such, while we provide these extremely low-quality genomes, users should exercise caution in mining genes from these samples owing to the high probability of error. Reporting N50 and BUSCO, as well as genome assemblies, in manuscripts and databases promotes transparency and discourages needless computation.
Contamination has been shown to be a pervasive pattern in genome and transcriptome sequencing projects, especially those that use multiplexed sequencing approaches [41–43]. In a recent study, Allio et al. [29] found that cross-contamination accounted for 0.26% of assembly contigs. While contaminants were removed from Allio et al. [29] using CroCo [44] and thus do not affect their results, it remains unknown how much these contaminant sequences will impact future studies that reuse these genomic data. The authors did not accession genome assemblies that had contaminants removed, and contaminants remain in accessioned reads. Furthermore, it is impossible to repeat these necessary decontamination steps without detailed information regarding multiplex strategy [44]. Accessioning decontaminated assemblies to NCBI is a necessary and easy solution.
Our study reveals a significant lack of standardization and reporting across genomic studies because many do not provide genome assemblies and necessary quality metrics. Our main conclusions are that:
We provide draft assemblies and quality metrics for all butterfly genomes available at the time of this study (available through NCBI TPA database) (Supplementary Table S1). We synthesize these data into tables of the 50 highest quality genomes, as well as exemplar genomes for each subfamily.
We found that the ringlet, Aphantopus hyperantus, has the highest quality papilionoid genome, and that ≥51 of 873 genomes that we assembled are ultimately unusable, and another 67 should be reused only with caution. Long and contiguous reads, indicated by high N50 values, are 1 quality metric that should be reported in all studies, especially those of gene mutation, duplication, or genomic architecture.
Quality metrics, such as sequence length, whether sequences are contiguous, and N50 and BUSCO scores, should be reported in all studies. Phylogenetic studies are strengthened when genomes with a high completeness score, such as BUSCO, are used.
Researchers should provide draft assemblies in all genome publications and databases. Accessioning quality scores will enhance transparency and avoid unnecessary use of computational resources. Accessioning assemblies further promotes the FAIR principles of interoperability and reuse by limiting contaminant sequences and allowing confirmation of results.
Data Availability
See Supplementary Tables S1–S3 for genomic read accession numbers used in this study and associated metadata. The 822 viable genome assemblies produced using SPAdes v3.13 are available in the NCBI TPA repository and can be accessed with BioProject PRJNA606954. The sequence assemblies, BUSCO files, scripts, and other supporting data underlying this article are also available via the GigaScience database, GigaDB [45].
Additional Files
Supplementary Table S1. Sample ID, N50, BUSCO, and sequencing metadata for de novo assembled genomes.
Supplementary Table S2. Sample ID, N50, BUSCO, and sequencing metadata for pre-assembled genomes.
Supplementary Table S3. Sample ID, N50, BUSCO, and sequencing metadata for de novo genomes resulting in extremely poor quality assemblies.
Supplementary File S1. Filter_seqs_by_NCBI.py script used to automatically update assemblies with the feedback file from NCBI during the NCBI Accession process.
Abbreviations
bp: base pair; BUSCO: Benchmarking Universal Single-Copy Orthologs; CPU: central processing unit; FAIR: Findability, Accessibility, Interoperability, and Reuse; NCBI: National Center for Biotechnology Information; SPAdes: St. Petersburg genome Assembler; SRA: Sequence Read Archive; TPA: third party database.
Competing Interests
The authors declare that they have no competing interests.
Funding
This work was funded by the National Science Foundation Grants DEB No. 1,541,500 and No. 1,557,007 to A.Y.K.
Authors’ Contributions
A.Y.K. conceived of the study. E.A.E. performed data collection, data analysis, and produced the figures and scripts, with overall guidance from A.Y.K. All authors wrote the manuscript. C.G.S. and E.A.E. deposited the data.
Supplementary Material
Chris Wheat, PhD -- 3/31/2020 Reviewed
Chris Wheat, PhD -- 8/5/2020 Reviewed
Sujai Kumar -- 4/20/2020 Reviewed
ACKNOWLEDGEMENTS
The authors acknowledge the University of Florida Research Computing (https://www.rc.ufl.edu/) for providing computational resources and support that contributed to the research results reported in this study. We are grateful to 2 reviewers, Hans Zauner and Xuankun Li, who provided helpful comments. Other members of the Kawahara Lab participated in thoughtful discussions that greatly improved the quality of this manuscript. We thank Laurel Kaminsky, Anupama Priyadarshini, Victoria Tran, Andrew Warren, and the FLMNH Digitization Team for providing butterfly images.
Contributor Information
Emily A Ellis, McGuire Center for Lepidoptera and Biodiversity, Florida Museum of Natural History, University of Florida, 3215 Hull Road, Gainesville, FL 32611–2710, USA.
Caroline G Storer, McGuire Center for Lepidoptera and Biodiversity, Florida Museum of Natural History, University of Florida, 3215 Hull Road, Gainesville, FL 32611–2710, USA.
Akito Y Kawahara, McGuire Center for Lepidoptera and Biodiversity, Florida Museum of Natural History, University of Florida, 3215 Hull Road, Gainesville, FL 32611–2710, USA.
References
- 1. Prum RO, Berv JS, Dornburg A, et al. A comprehensive phylogeny of birds (Aves) using targeted next-generation DNA sequencing. Nature. 2015;526:569–73. [DOI] [PubMed] [Google Scholar]
- 2. Zhang J, Cong Q, Shen J, et al. Genomics of a complete butterfly continent. bioRxiv 2019, 10.1101/829887. [DOI] [Google Scholar]
- 3. Thomas GWC, Dohmen E, Hughes DST, et al. Gene content evolution in the arthropods. Genome Biol. 2020;21:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Simão FA, Waterhouse RM, Ioannidis P, et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–2. [DOI] [PubMed] [Google Scholar]
- 5. Casacuberta E, González J. The impact of transposable elements in environmental adaptation. Mol Ecol. 2013;22:1503–17. [DOI] [PubMed] [Google Scholar]
- 6. Bennetzen JL. Transposable element contributions to plant gene and genome evolution. Plant Mol Biol. 2000;42:251–69. [PubMed] [Google Scholar]
- 7. Edger PP, Heidel-Fischer HM, Bekaert M, et al. The butterfly plant arms-race escalated by gene and genome duplications. Proc Natl Acad Sci U S A. 2015;112:8362–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown CA, Murray AW, Verstrepen KJ. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol. 2010;20:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gouin A, Bretaudeau A, Nam K, et al. Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci Rep. 2017;7:11816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Denton JF, Lugo-Martinez J, Tucker AE, et al. Extensive error in the number of genes inferred from draft genome assemblies. PLoS Comput Biol. 2014;10:e1003998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Han MV, Thomas GWC, Lugo-Martinez J, et al. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Molecular Biology and Evolution. 2013;30:1987–97. [DOI] [PubMed] [Google Scholar]
- 12. Burrell AS, Disotell TR, Bergey CM. The use of museum specimens with high-throughput DNA sequencers. J Hum Evol. 2015;79:35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang J, Cong Q, Shen J, et al. Genomes reveal drastic and recurrent phenotypic divergence in firetip skipper butterflies (Hesperiidae: Pyrrhopyginae). Proc Biol Sci. 2019;286:20190609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gilbert MTP, Moore W, Melchior L, et al. DNA extraction from dry museum beetles without conferring external morphological damage. PLoS One. 2007;2:e272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. St Laurent RA, Mielke CGC, Herbin D, et al. A new target capture phylogeny elucidates the systematics and evolution of wing coupling in sack-bearer moths. Syst Entomol. 2020;3:17. [Google Scholar]
- 16. Parham JF, Stuart BL, Bour R, et al. Evolutionary distinctiveness of the extinct Yunnan box turtle (Cuora yunnanensis) revealed by DNA from an old museum specimen. Proc Biol Sci. 2004;271(Suppl 6):S391–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sanderson MJ, McMahon MM, Steel M. Phylogenomics with incomplete taxon coverage: the limits to inference. BMC Evol Biol. 2010;10:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration. The Sequence Read Archive. Nucleic Acids Res. 2011;39:D19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Challi RJ, Kumar S, Dasmahapatra KK, et al. Lepbase: the Lepidopteran genome database. bioRxiv 2016, 10.1101/056994. [DOI] [Google Scholar]
- 20. Zhang J, Cong Q, Shen J, et al. Three new subfamilies of skipper butterflies (Lepidoptera, Hesperiidae). Zookeys. 2019;861:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang J, Shen J, Cong Q, et al. Genomic analysis of the tribe Emesidini (Lepidoptera: Riodinidae). Zootaxa. 2019;4668:475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li W, Cong Q, Shen J, et al. Genomes of skipper butterflies reveal extensive convergence of wing patterns. Proc Natl Acad Sci U S A. 2019;116:6232–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cong Q, Shen J, Borek D, et al. Complete genomes of Hairstreak butterflies, their speciation, and nucleo-mitochondrial incongruence. Sci Rep. 2016;6:24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cong Q, Li W, Borek D, et al. The Bear Giant-Skipper genome suggests genetic adaptations to living inside yucca roots. Mol Genet Genomics. 2019;294:211–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cong Q, Shen J, Li W, et al. The first complete genomes of Metalmarks and the classification of butterfly families. Genomics. 2017;109:485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shen J, Cong Q, Borek D, et al. Complete genome of Achalarus lyciades, the first representative of the Eudaminae subfamily of skippers. Curr Genomics. 2017;18:366–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Shen J, Cong Q, Kinch LN, et al. Complete genome of Pieris rapae, a resilient alien, a cabbage pest, and a source of anti-cancer proteins. F1000Res. 2016;5:2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. VanKuren NW, Massardo D, Nallu S, et al. Butterfly mimicry polymorphisms highlight phylogenetic limits of gene reuse in the evolution of diverse adaptations. Mol Biol Evol. 2019;36:2842–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Allio R, Scornavacca C, Benoit N, et al. Whole genome shotgun phylogenomics resolves the pattern and timing of swallowtail butterfly evolution. Syst Biol. 2020;69:38–60. [DOI] [PubMed] [Google Scholar]
- 30. Cong Q, Shen J, Warren AD, et al. Speciation in cloudless sulphurs gleaned from complete genomes. Genome Biol Evol. 2016;8:915–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cong Q, Borek D, Otwinowski Z, et al. Skipper genome sheds light on unique phenotypic traits and phylogeny. BMC Genomics. 2015;16:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. TrimGalore. Retrieved July 12, 2020 from https://github.com/FelixKrueger/TrimGalore. [Google Scholar]
- 33. Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. SeqTK. Retrived July 21, 2020 from https://github.com/lh3/seqtk. [Google Scholar]
- 35. VecScreen. Retrieved May 26, 2020 from https://github.com/aaschaffer/generate_vecscreen_candidates. [Google Scholar]
- 36. assembly-stats. Retrieved November 19 2020 from https://github.com/sanger-pathogens/assembly-stats. [Google Scholar]
- 37. Kajitani R, Toshimoto K, Noguchi H, et al. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24:1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Espeland M, Breinholt J, Willmott KR, et al. A comprehensive and dated phylogenomic analysis of butterflies. Current Biology. 2018;28:770–8. [DOI] [PubMed] [Google Scholar]
- 39. Kawahara AY, Breinholt JW, Espeland M, et al. Phylogenetics of moth-like butterflies (Papilionoidea: Hedylidae) based on a new 13-locus target capture probe set. Mol Phylogenet Evol. 2018;127:600–5. [DOI] [PubMed] [Google Scholar]
- 40. Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–51. [DOI] [PubMed] [Google Scholar]
- 41. Ballenghien M, Faivre N, Galtier N. Patterns of cross-contamination in a multispecies population genomic project: detection, quantification, impact, and solutions. BMC Biol. 2017;15:e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jun G, Flickinger M, Hetrick KN, et al. Detecting and estimating contamination of human DNA samples in sequencing and array-based genotype data. Am J Hum Genet. 2012;91:839–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Merchant S, Wood DE, Salzberg SL. Unexpected cross-species contamination in genome sequencing projects. PeerJ. 2014;2:e675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Simion P, Belkhir K, François C, et al. A software tool “CroCo” detects pervasive cross-species contamination in next generation sequencing data. BMC Biol. 2018;16:e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ellis EA, Storer CG, Kawahara AY. Supporting data for “De novo genome assemblies of butterflies.”. GigaScience Database. 2021. 10.5524/100856. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Chris Wheat, PhD -- 3/31/2020 Reviewed
Chris Wheat, PhD -- 8/5/2020 Reviewed
Sujai Kumar -- 4/20/2020 Reviewed
Data Availability Statement
See Supplementary Tables S1–S3 for genomic read accession numbers used in this study and associated metadata. The 822 viable genome assemblies produced using SPAdes v3.13 are available in the NCBI TPA repository and can be accessed with BioProject PRJNA606954. The sequence assemblies, BUSCO files, scripts, and other supporting data underlying this article are also available via the GigaScience database, GigaDB [45].