Abstract
The intricate arrangement of numerous and closely placed chromophores on nanoscale scaffolds can lead to key photonic applications ranging from optical waveguides and antennas to signal-enhanced fluorescent sensors. In this regard, the self-assembly of dye-appended DNA sequences into programmed photonic architectures is promising. However, the dense-packing of dyes can result in not only compromised DNA assembly (leading to ill-defined structures and precipitates) but also to essentially non-fluorescent systems (due to π-π aggregation). Here, we introduce a two-step “tether and mask” strategy wherein large porphyrin dyes are first attached to short G-quadruplex forming sequences and then reacted with per-O-methylated β-cyclodextrin (PMβCD) caps, to form supramolecular synthons featuring the porphyrin fluor fixed into a masked porphyrin lantern (PL) state, due to intramolecular host-guest interactions in water. The PL-DNA sequences can then be self-assembled into cyclic architectures or unprecedented G-wires tethered with hundreds of porphyrin dyes. Importantly, despite the closely arrayed PL units (~2 nm), the dyes behave as bright chromophores (up to 180-fold brighter than the analogues lacking the PMβCD masks). Since other self-assembling scaffolds, dyes, and host molecules can be used in this modular approach, this manuscript lays a general strategy for the bottom-up aqueous self-assembly of bright nanomaterials containing densely packed dyes.
Graphical Abstract
INTRODUCTION
The use of DNA-based assembly approaches to construct 2D and 3D nanoscale structures with photonic properties is currently an intensive area of research.1 The advantage of this strategy is that chromophores can be addressed at defined positions, onto programmable DNA platforms, by using specific dye-appended DNA sequences. The resultant multi-chromophoric DNA assemblies are expected to have breakthrough applications, including in nanoscale optical waveguides for photonic circuits, artificial light-harvesting constructs for solar energy funneling, signal enhanced fluorescent devices for (bio)sensing, and components for super-resolution imaging and optical antennas.2-11 However, a long-standing hurdle that needs to be overcome to fully develop DNA-based assemblies composed of densely arranged chromophores is the minimization of dye-dye aggregation.12 Such dye contact can significantly attenuate the photophysical properties of many chromophores (e.g., quenching of excited states) thereby precluding the use of the excited state for intended processes.12-16 As importantly, increasing dye density can substantially compromise the DNA assembly mode due to competing hydrophobic interactions.
Porphyrins are large aromatic chromophores with attractive photophysical—as well as electronic and catalytic—properties.17-22 These tetrapyrrolic macrocycles exhibit intense absorption and emission bands that can be fine-tuned by meta-lation state and peripheral substitution. Due to these attributes, considerable effort has been spent appending the functional porphyrin dyes onto DNA with the goal of harnessing the DNA domain for the programmed placement of the porphyrins into specific supramolecular architectures in water.23-31 Indeed, porphyrins have been attached to DNA at the 3’ and/or 5’ termini as well as on the phosphate backbone and nucleobase positions. While relatively few attached porphyrin units do not significantly compromise DNA assembly, for photonic nanomaterials with multiple porphyrins, the systems tend to form ill-defined aggregates and precipitates because the assembly is dominated by hydrophobic porphyrin aggregation rather than by DNA hybridization. As importantly, this aggregation leads to attenuated absorption and strongly quenched porphyrin emission, resulting in poor photonic systems.23,27,32
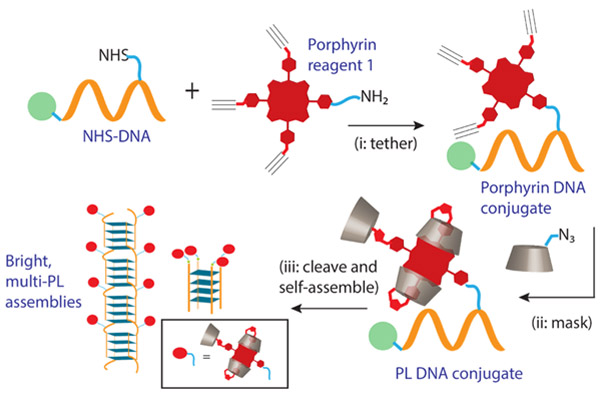
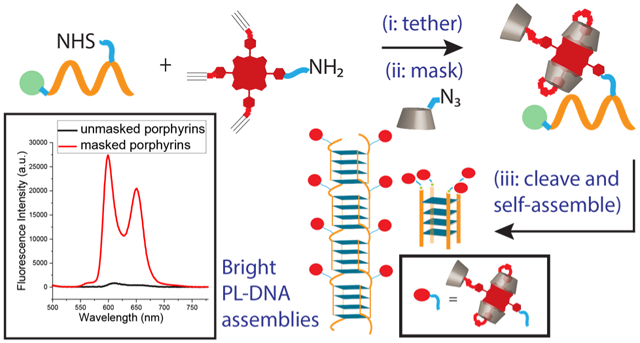
With this manuscript, we show how a large (in the hundreds) population of closely spaced dyes (attached to a DNA scaffold at ~2 nm spacing), exemplified using porphyrins, can be decorated onto DNA quadruplex-based nanostructures wherein (a) the nanostructure integrity is intact and (b) the dyes behave as bright chromophores. Our approach is to “tether and mask” the dyes prior to DNA self-assembly (Scheme 1). The masked dyes access a porphyrin lantern (PL) state via β-cyclodextrin self-encapsulation that precludes the dyes from detrimental non-covalent interactions and provides a low-dielectric microenvironment. Specifically, we demonstrate that bright PL decorated guanine-rich sequences are versatile and can be used to self-assemble (a) cyclic architectures containing 4 porphyrins and (b) unprecedented guanine (G)-quadruplex wires appended with hundreds of porphyrins. Additionally, while the focus here is on DNA assembly, our simple post-synthesis ‘tether and mask’ strategy is expected to be readily extended to other assembling molecules (such as peptides, foldamers, and polymers). Hence, this manuscript lays a general foundation for the bottom-up aqueous self-assembly of bright nanomaterials containing densely packed dyes.
Scheme 1.
A two-step approach to obtain porphyrin lantern (PL) tethered DNA sequences. Subsequent programmed G-quadruplex assembly leads to bright fluorescent architectures with densely packed PLs.
RESULTS AND DISCUSSION
Design of a Porphyrin Conjugation Reagent.
A commonly used and effective conjugation technique involves the coupling of amines with activated N-hydroxy succinimidyl (NHS) esters. Indeed, a variety of functional self-assembling molecules (including DNA, peptides, proteins, and polymers) can be functionalized with NHS esters. Thus, we decided to develop a porphyrin conjugation reagent 1 that contains a primary amino group, on one of its meso-phenyl positions, for conjugation with NHS esters. The design also includes three alkyne arms on porphyrin 1 since these latter groups can be subsequently (i.e., after the initial DNA conjugation step) used to attach masking hosts, per-O-methylated β-cyclodextrins (PMβCDs), via copper-catalyzed alkyne-azide click chemistry (CuAAC).
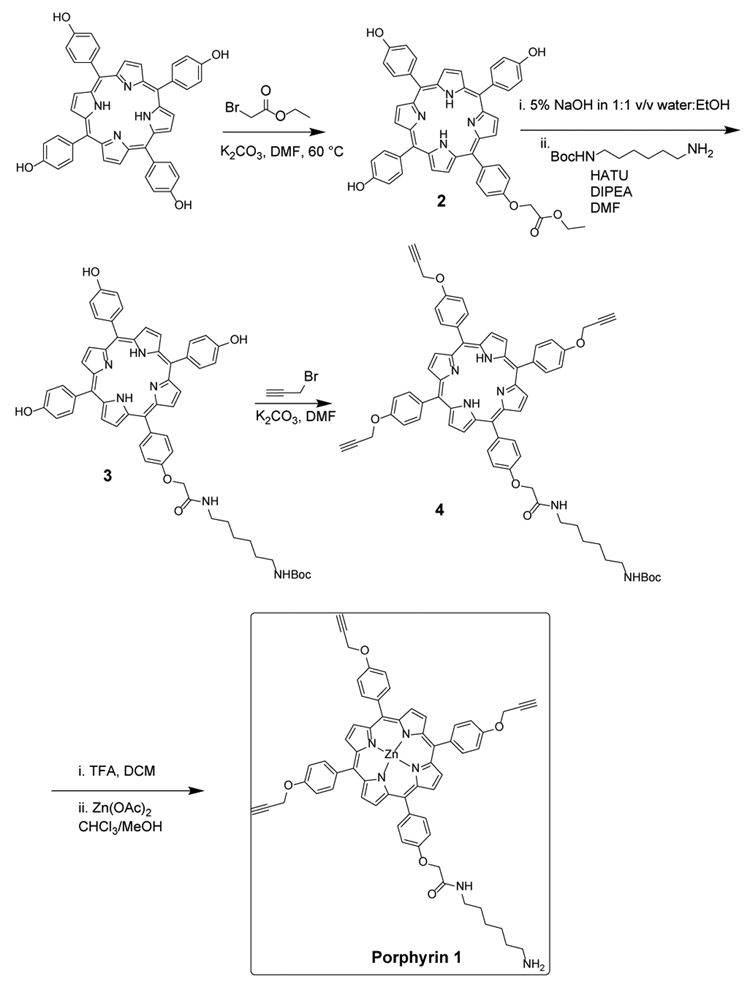
The synthesis of porphyrin 1 is illustrated in Scheme 2. Briefly, 5, 10, 15, 20–tetrakis(4-hydroxyphenyl) porphyrin was monofunctionalized via a nucleophilic displacement reaction with ethyl 2-bromoacetate to afford porphyrin monoester 2. The base catalyzed hydrolysis of 2 followed by amide coupling with mono-boc-protected hexamethylenediamine yielded precursor 3. Propargylation of the remaining three phenolic OH groups on the porphyrin followed by deprotection of the boc moiety and zinc-metallation offered final porphyrin reagent 1.
Scheme 2.
Synthesis of Porphyrin 1.
Tethering of Porphyrins onto Guanine-rich DNA Sequences.
With porphyrin reagent 1 in hand, we next focused on the NHS functionalized DNA partner for conjugation. In particular, our group is interested in the self-assembly properties of synthetically functionalized G-quadruplexes. From a nanomaterials standpoint G-quadruplex based DNA assemblies are attractive because they have high mechanical and thermal stability, can array a large number of functional groups (such as dyes), and have increased nuclease stability.33 Additionally, a single and short G-rich sequence can form programmed multimeric self-assemblies and hence provides an opportunity to pursue a DNA-minimal strategy to construct robust functional nanostructures.34-36
Two G-rich sequences were chosen for this study; the first is a sequence (5’-TG4T2-3’) that forms a discrete quadruplex cylinder composed of 4 parallel strands,37 and the second (5’-G4T2G4-3’) is derived from the Tetrahymena telomere region and is known to assemble into long nano wires.38-40 Further, in order to show the versatility of our approach, the former sequence was functionalized with an NHS ester at its 5’ end (i.e., NHS-DNA 1: 5’-NHS-TG4T2-3’), while the latter sequence, NHS-DNA 2 (5’-G4T(NHS)TG4-3’), incorporates the NHS ester off carbon-5 on an interior thymidine residue.
As shown in Scheme 1(i) and discussed in the experimental section the precursor porphyrin-DNA conjugates were prepared by reacting resin-bound DNA with porphyrin 1 using standard NHS-ester/amine coupling. Cleavage of the porphyrin-DNA conjugate from the solid support afforded Porph-DNA 1 and Porph-DNA 2 (see Table 1). In these control sequences the porphyrin dyes are not masked with PMβCDs and thus should undergo dye-dye interactions. In order to probe the scope of our “tether and mask” strategy, porphyrin 1 was also conjugated to NHS-functionalized polyethylene glycol (PEG; with Mn = 750 Da) to afford Porph-PEG. Porph-PEG also serves as a water-soluble control that lacks the complexity of the DNA domain. All porphyrin tethered (bio)polymers were purified via RP HPLC and analyzed using MALDI-TOF.
Table 1.
List of various conjugates studied and the schematics of their corresponding assemblies.
| Name | Structure | Sequence (5’-3’) | Assembly fonned |
|---|---|---|---|
| PL-DNA 1 | PMβCD3-Porph-TG4T2 |  |
|
| Porph-DNA 1 | Porph-TG4T2 |
|
|
| PL-DNA 2 | G4T(PMβCD3-Porph)TG4 | 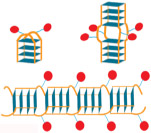 |
|
| Porph-DNA 2 | G4T(Porph)TG4 | Aggregates not mobile on PAGE | |
| PL-PEG | PMβCD3-Porph-PEG750 | — | |
| Porph-PEG | Porph-PEG750 | — |
Masking and Formation of Porphyrin Lantern Complexes.
The pronounced tendency of hydrophobic dye molecules to non-covalently interact with other dyes, attached biomolecules, and the solvent is a major reason for their unfavorable photophysical properties in water. Such interactions can be prevented using compatible hosts that provide a hydrophobic microenvironment and serve to encapsulate and protect the chromophores. Indeed, fluorescent proteins and natural light-harvesting complexes use peptide scaffolds to constrain and protect the chromophores from unwanted interactions.14,41-44 We, and others, have shown that βCDs can bind to the porphyrin dipyrro-methene and meso-phenyl units and enhance the photophysical properties of the porphyrins.45-48 Here, we were eager to use a tandem porphyrin conjugation and masking approach to prepare PL containing templates that can be used for programmed nanomaterial self-assembly.
As shown in Scheme 1(ii) resin-bound porphyrin DNA sequences were subjected to CuAAC reaction conditions49 with PMβCD monoazide. This afforded DNA sequences wherein the Porphyrin is attached to three PMβCDs. Cleavage from the solid support and subsequent RP HPLC purification yielded masked conjugates PL-DNA 1 and PL-DNA 2 (Table 1). In addition, the PL-PEG conjugate was also prepared.
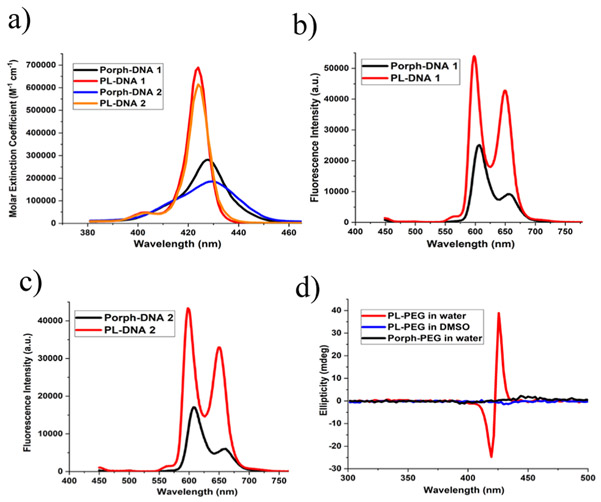
For these masked porphyrin (bio)polymers in aqueous media, the PL state of the porphyrin is proposed to occur via a 360° rotation of the two trans-PMβCD units, facilitating host-guest interactions of the porphyrin moiety in a self-inclusion phenomenon. Hence, even prior to any targeted self-assembly, in water, the CD-capped conjugates were expected to exhibit superior photophysical properties than that of the uncapped species. As illustrated in Fig.1a the UV-Vis profiles for the PL state of the DNA sequences clearly show a sharper porphyrin Soret band accompanied by a higher molar extinction coefficient (2-3-fold higher) compared to the unmasked systems. Furthermore, a 3-5-fold enhancement of the integrated emission was also observed (Fig. 1b, c).
Figure 1.
Optical spectra of non-annealed porphyrin containing DNA or PEG conjugates in water. a) UV-Vis absorption profiles of 1 μM solutions of porphyrin-DNA conjugates showing the porphyrin Soret bands. b) Emission profiles of 1 μM solutions of Porph-DNA 1 and PL-DNA 1. Excitation was absorbance matched at 429 nm. c) Emission profiles of 1 μM solutions of Porph-DNA 2 and PL-DNA 2. Excitation was absorbance matched at 430 nm. d) CD spectra of 30 μM solutions of Porph-PEG and PL-PEG. Also shown is the CD spectra of 30 μM PL-PEG in DMSO.
While the above results suggest the formation of intramolecular βCD-porphyrin interactions, we performed similar studies with PL-PEG, where the absence of nucleotides ensures the difference in the spectral characteristics solely springs from the self-inclusion phenomenon. Here the PL-PEG species showed a 2-fold increase in the molar extinction coefficient and a 5-fold superior fluorescence over Porph-PEG (SI, Fig. S-7). Moreover, when circular dichroism experiments were performed, a distinct chiral signature (positive maximum at 426 nm and negative peak at 420 nm) was observed in the Soret region for PL-PEG (Fig. 1d). Importantly, this bisignate curve is absent in Porph-PEG as there is no chiral environment in this latter compound. Further, when PL-PEG was exposed to a less polar solvent environment (DMSO), the induced circular dichroism band disappears. This result, when taken in conjunction with the UV-Vis and fluorescence studies, makes us suggest that an intramolecular PL state is indeed formed via hydrophobically driven host-guest interactions between PMβCD and porphyrin units. The circular dichroism spectra of PL-DNA sequences in water also show the induced peaks, which are notably absent in the Porph-DNA sequences, indicating negligible contribution of the DNA domain to the induced circular dichroism feature (SI, Fig S-12a).
Photophysical Properties of Porphyrin Lantern Containing G-Quadruplexes.
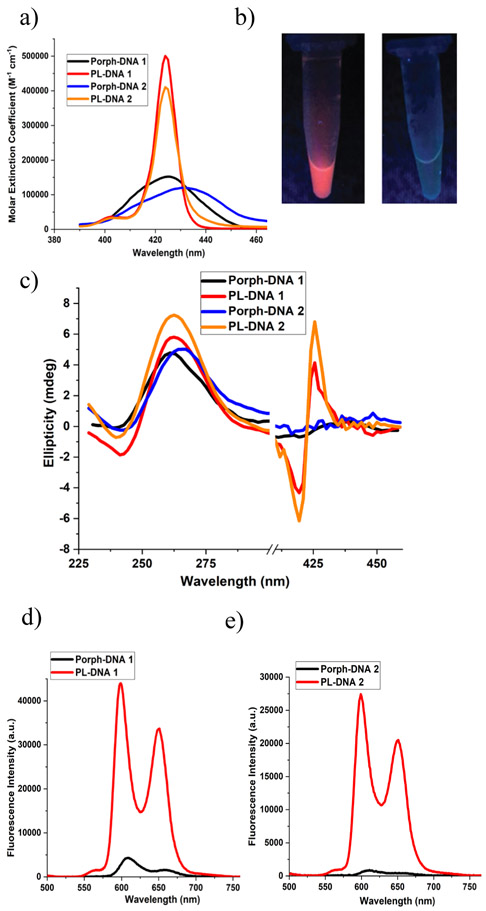
Given the substantial enhancement in optical properties of the encapsulated porphyrin chromophore when the DNA sequences are not annealed, we were eager to investigate the effects after self-assembly, where the dyes will be placed in proximity. The desired G-quadruplex assembly of the PL containing sequences was induced by incubating the samples in appropriate buffers, and their photophysical properties were compared to that of the unmasked porphyrin-DNA conjugates. Specifically, Porph-DNA 1 and PL-DNA 1 were annealed in the presence of K+ to induce the formation of a discrete tetramolecular parallel G-quadruplex structure with four porphyrins arranged in a cyclical fashion off the 5’-terminus. This quadruplex exhibited a minimum at 241 nm and a maximum at 263 nm in the circular dichroism spectra (Fig. 2c) confirming the formation of parallel G-quadruplexes.50-52 In the case of Porph-DNA 2 and PL-DNA 2, annealing was conducted in the presence of K+ and Mg++—conditions that favor growth of aligned G-quadruplexes into higher-order G-wires. The formation of a parallel G-quadruplex structure is the first requirement to grow the G-wires and this was confirmed by the circular dichroism spectra (Fig. 2c). We also synthesized PL-polyT (5’-PMβCD3-Porph-T10-3’) that served (in conjunction with the PL-PEG) as a non self-assembling control. Not surprisingly, even after incubation under quadruplex forming conditions, this control does not exhibit the G-quadruplex signature at 200-300 nm, yet does show the presence of a sharp bisignate peak at the porphyrin Soret band indicating the presence of host-guest interactions (SI, Fig. S-12b).
Figure 2.
Optical spectra of porphyrin containing DNA sequences after quadruplex assembly. a) UV-Vis absorption profiles of 1 μM annealed solutions of conjugates showing the Soret bands. b) 30 μM annealed solutions of PL-DNA 2 (left) and Porph-DNA 2 (right) illuminated under UV lamp (broadband 365 nm). c) CD spectra of 30 μM annealed solutions of porphyrin-DNA conjugates. d) Emission profiles of 1 μM annealed solutions of Porph-DNA 1 and PL-DNA 1. Excitation was absorbance matched at 431 nm. e) Emission profiles of 1 μM annealed solutions of Porph-DNA 2 and PL-DNA 2. Excitation was absorbance matched at 432 nm.
The UV-Vis profiles for the annealed G-quadruplex solutions of all porphyrin-DNA conjugates are shown in Fig. 2a and SI, S-9b. The molar extinction coefficient of both PL-DNA G-quadruplex species (at the Soret band) is 3-fold higher than that of the uncapped Porph-DNA G-quadruplexes. Further, the full-width half-maximum (fwhm) for the G-quadruplex Porph-DNA systems is larger (30 nm for Porph-DNA 1, and 40 nm for Porph-DNA 2) than those for the single-stranded precursors (17 nm for Porph-DNA 1, and 29 nm for Porph-DNA 2). However, the fwhm is not significantly affected by the assembly state of the PL-DNA sequences. These results indicate that the ground state π-π aggregation of the porphyrin dyes when attached to self-assembling DNA is attenuated by the masking PMβCD units.
More striking results came from fluorescence studies (Fig. 2d), where the masked porphyrin unit showed a 12-fold fluorescence enhancement for G-quadruplex PL-DNA 1 when compared to the emission from G-quadruplex Porph-DNA 1 (under absorbance-matched excitation). The increase in fluorescence was even greater, exhibiting 56-fold higher integrated emission, for G-quadruplex PL-DNA 2 when compared to the G-quadruplex Porph-DNA 2 (Fig. 2e). We attribute these large differences in the fluorescence of the assembled quadruplexes to the caging effect of PMβCD moieties that prevents porphyrin aggregation and therefore attenuates fluorescence quenching pathways. In this context, the reason that the fluorescence enhancement is more pronounced for the G-quadruplex PL-DNA 2 case versus PL-DNA 1 is because PL-DNA 2 quadruplexes form higher-order porphyrin arrays with a larger population of closely spaced porphyrin units. Such an increase in the local concentration of unmasked porphyrins would dramatically increase the extent of concentration quenching for the G-quadruplex Porph-DNA 2 species.
The trend observed in the integrated fluorescence intensity measurements were also verified by fluorescence quantum yields (Φ) of the G-quadruplex species (Porph-DNA 1 (3.0 x 10−3), PL-DNA 1 (2.7 × 10−2), Porph-DNA 2 (3.0 x 10−4), PL-DNA 2 (1.6 x 10−2)). What is more, from quantum yield and extinction coefficient measurements, the relative (theoretical) brightness increased 30-fold for the porphyrin units on G-quadruplex PL-DNA 1 vs G-quadruplex Porph-DNA 1, and 180-fold for G-quadruplex PL-DNA 2 vs G-quadruplex Porph-DNA 2. Indeed, whilst the fluorescence from PL-DNA 2 assembly can be readily observed by the naked eye, the Porph-DNA 2 assembly is essentially non-fluorescent (Fig. 2b).
Since the PMβCD driven intramolecular host-guest complexation is pivotal for the enhancement in photophysical properties of the porphyrin G-quadruplex systems, we were eager to determine how robust these interactions are. Hence, a well-known guest of PMβCD, 1-adamantanecarboxylic acid (Ad-COOH), was incubated with an annealed solution of PL-DNA 1. Even at 500 eq. of Ad-COOH, the fluorescence emission intensity was not attenuated to an appreciable extent (SI, Fig. S-13), illustrating the robustness of the intramolecular self-inclusion complex that leads to the PL state. Additional control circular dichroism studies wherein PL-PEG was incubated with 500 eq. of Ad-COOH verified that Ad-COOH is not capable of displacing the intramolecular PMβCD-porphyrin complex at room temperature (see SI, Fig. S-24).
Characterization of Porphyrin Lantern Containing G-quadruplex Nanostructures.
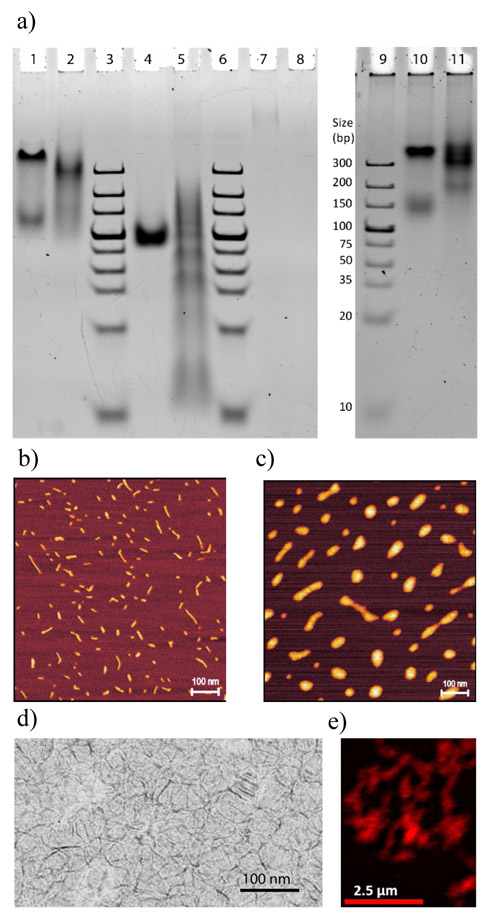
The marked enhancement in brightness of the PL appended G-quadruplexes gave impetus for analyzing their nanostructures. First, we probed the G-quadruplex self-assemblies using non-denaturing PAGE (120 V) at room temperature (Fig. 3a left). PL-DNA 1 annealed under quadruplex forming conditions (lane 1), showed two distinct bands with the faster moving band running similar to single stranded PL-DNA control (PL-polyT, lane 4). Thus the slow-moving band was ascribed to the tetramolecular G-quadruplex that projects four PL units in a cyclical fashion (see Table 1). Interestingly, when unmasked Porph-DNA 1 is annealed and introduced into the gel (lane 7), only a very small quantity migrates on the gel, whilst most of the assembly does not enter the gel, suggesting a porphyrin interaction driven DNA aggregation.
Figure 3.
Characterization of quadruplex nanostructures. a, left) Non-denaturing PAGE (120 V at RT). Control lane 3 (and 6) is double stranded DNA ladder and lane 4 is single stranded PL-polyT. All G-rich sequences were first annealed to form G-quadruplex assembly: PL-DNA 1 (lane 1), PL-DNA 2 (lane 2), parent DNA 2 (G4T2G4) (lane 5), Porph-DNA 1 (lane 7), and Porph-DNA 2 (lane 8).. (a, right) Non-denaturing PAGE (60 V at 4 °C). Control lane 9 is double stranded DNA ladder. The G-rich sequences were first annealed to form G-quadruplex assembly: PL-DNA 1 (lane 10), and PL-DNA 2 (lane 11). b) AFM image of G-wires of parent DNA 2 (not conjugated with porphyrin). c) AFM image of annealed PL-DNA 2. d) Cry-oTEM image of annealed PL-DNA 2. e) Confocal microscopy image of G-wires of PL-DNA 2. Exciting laser was 561 nm.
When PL-DNA 2 is incubated to form a G-quadruplex assembly (lane 2), a streaking band is observed—suggesting multiple assemblies. The less intense portion of the streak that migrates faster than the more intense portion is ascribed to smaller bimolecular quadruplexes (2 PLs, see Table 1). On the other hand, the most intense region (that migrates slightly faster than tetramolecular PL-DNA 1, cf. lane 1) possibly corresponds to tetramolecular quadruplex species of PL-DNA 2 (4 PL units) and short G-wires (e.g., hexamers). Indeed, when a PAGE under milder conditions was employed (60 V at 4°C, Fig 3a right) multiple closely migrating bands are observed in this region (see lane 11). In both gels, the reason we do not observe long G-wires is likely that the 20% PAGE conditions induce the breakup of the longer wires into short wires. Moreover, for the Porph-DNA 2 G-quadruplex system (lane 8), no migrating bands are observed. This indicates that G-quadruplex assembly of Porph-DNA 2 likely forms an ill-defined large aggregate, due to inter alia extensive porphyrin self-stacking, that cannot readily dissociate any smaller DNA species for migration on PAGE. As a control, we also annealed parent DNA 2 (G4T2G4) that lacks the PL units and the linkers (lane 5). DNA 2 shows a long streak with multiple bands corresponding to bimolecular and tetramolecular species as well as G-wires of variable length.
While the above gel-based migration studies provide good evidence of the discrete tetramolecular assembly formed by PL-DNA 1, they do not furnish conclusive evidence for the formation of long G-wires for PL-DNA 2. In order to gather direct evidence for the higher-order structures formed by G-quadruplex PL-DNA 2, AFM was conducted. First, we annealed parent strand DNA 2 in K+ and Mg++ containing buffer. The AFM micrograph, imaged on mica (in air), is shown in Fig. 3b and the average peak height was found to be 2.3 ± 0.14 nm (SI, S-16a). Although the heights of G-wires significantly depend on the buffer composition and imaging conditions, this value matches the previously determined height (2.3 nm)39,40,53 for G-wires formed by the same sequence. Further, these heights are larger than those typically observed for single strands and DNA duplexes.
Next, we performed the same experiments with annealed porphyrin containing DNA 2 sequences. While the AFM imaging of annealed Porph-DNA 2 is characterized by ill-defined aggregates (SI, Fig. S-16d), for the case of PL-DNA 2, defined wires were observed with the height of 3.5 ± 0.22 nm, which is 1.2 nm larger than the parent unfunctionalized G-wires (Fig. 3c and SI, S-16b). This difference is ascribed to the PL units and linkers. Furthermore, while we observed longer G-wires (~400 nm, see SI, Fig. S-16c) the majority were below 100 nm Based on an established model of the parent G-wire, we estimate that an 8 nm length PL-DNA 2 G-wire contains an array of 14 PLs. Thus, the long PL-DNA 2 G-wires are thought to contain hundreds of bright, non-aggregated, porphyrin chromophores. Further, from the model,40 the average nearest PL-containing dT to dT distance is 2.03 ± 0.37 nm, and the minimum nearest distance is 1.40 nm (SI, Fig. S-23). Given the flexibility and the length of the linkers this would readily lead to porphyrin π-stacking in the non-masked Porph-DNA 2 G-wires. However, the PL units are precluded from such interactions due to the intramolecular host-guest interactions for the PL-DNA 2 G-wires. Similarly, the terminal 5’-dT residues are separated by an average nearest distance of 1.23 ± 0.02 nm in a parallel G-quadruplex crystal structure (for a sequence TG4T) that is nearly identical to the sequence used in PL-DNA 1 (see SI, Fig. S-23).
To further confirm the presence of G-wires in a more solution-like native state, the annealed PL-DNA 2 was also probed via cryoTEM. The micrograph in Fig. 3d. clearly shows the presence of nano wires of an average thickness of 3.7 nm, corroborating the AFM studies. In addition, larger networks of fluorescent PL-DNA 2 G-wires were directly observed under a confocal microscope (Fig. 3e). Additionally, DLS measurements were used to probe the G-quadruplex assemblies in solution. From these experiments the estimated hydrodynamic diameter for PL-DNA 1 (assuming a spherical model) was 4.4 nm and the average length of the PL-DNA 2 G-wires (assuming a rod-shape model) was 40-45 nm (see SI, Fig. S-25).
CONCLUSIONS
In summary, we have introduced a simple two-step “tether and mask” strategy wherein large porphyrin dyes are first reacted with short G-rich sequences and then chased with per-O-methylated β-cyclodextrin (PMβCD) caps, to form single stranded DNA sequences containing masked porphyrin lanterns. The masking not only enables the dyes to function as bright chromophores but also does not hinder the programmability and self-assembly capacity of the DNA sequences, even when placed in a closely packed manner (~2 nm). Additionally, this work discloses, for the first time, the construction of defined and bright G-wires covalently appended with hundreds of porphyrin dyes in the nanometer regime. Previous studies have focused on the electronic properties of G-wires54 but the construction of robust and fluorescent G-wires described in this work paves the way towards exciting photonic applications.
Here, it is important to note that elegant research has been conducted on the so-called aggregation-induced emission (AIE) dyes that exhibit enhanced fluorescence in the aggregated state as a result of, inter alia, restriction of intramolecular rotation (RIR).55 AIE luminogens (AIEgens) tethered to DNA sequences have been used to monitor the formation of DNA self-assemblies.56 In fact, recently AIEgens have been attached to G-rich sequences that form a discrete tetramolecular G-quadruplex leading to dye-dye aggregation with enhanced and programmable dye emission.57 AIEgens are particularly promising for applications where a turn-ON response is needed (such as sensing nucleic acid sequences).58 However, in terms of the formation of defined nanowires (and other architectures) appended to large numbers of dyes, hydrophobically-driven AIEgen aggregation is still a limitation due to the possible formation of ill-defined structures and precipitates. Further, AIEgens are often non-innocent and can bind to DNA assemblies.59,60 Interestingly, however, βCD can also serve as a host for AIEgens with dye inclusion leading to RIR-based enhanced fluorescence.61 Thus, the “tether and mask” strategy outlined in this manuscript is also expected to be applicable for AIEgens with the salient advantage that adverse dye-dye aggregation and dye-DNA interactions can be circumvented.
Furthermore, the field of supramolecular chemistry has designed hosts that are tailored to sequester various fluorophores such as DASPMI (calixarene host), ANS (βCD host), thioflavin (cucurbituril 7 host), and squaraine (cyclic tetralactam host).62, 63 Thus, with compatible functional group handles, and appropriate choice of self-assembling template scaffolds, the strategy outlined in this manuscript can be applied in a general manner to develop bright nanostructures—composed of densely-decorated dye-lanterns.
EXPERIMENTAL PROCEDURES
1. General Experimental.
Unless otherwise noted, all chemicals were purchased from Millipore Sigma. 5, 10, 15, 20–tetrakis(4-hydroxyphenyl) porphyrin was purchased from Frontier Scientific and the solvents were acquired from Fischer Scientific. PMβCD-N3 and porphyrin monoester 2 were prepared following procedures described in literature.64,65 The precursor oligodeoxynucleotide (ODN) sequences were synthesized by the W. M. Keck Foundation Biotechnology Resource Laboratory located at Yale University, using standard automated solid phase synthesis. All the phosphoramidites used to prepare these ODNs were purchased from Glen Research. NMR spectra were recorded on a Bruker 500 or 300 MHz spectrometer. The concentration of purified ODNs were quantified based on their UV absorption at 260 nm and their molar extinction coefficients were obtained by nearest neighbor calculations. High Resolution (ESI) Mass Spectra were recorded on Theormo Electron Corp. MAT 95XP spectrometer. MALDI-TOF spectra were recorded on a Bruker Daltonics Autoflex III matrix assisted laser desorption ionization-time of flight mass spectrometer (MALDI-TOF MS). The matrix used was 3-hydroxypicolinic acid for ODNs and α-cyano-4-hydroxycinnamic acid for the small organic molecules. UV–Vis studies were undertaken using a Hewlett Packard 8452A Diode Array Spectrophotometer. Fluorescence spectra were recorded on a PerkinElmer EnSpire Multimode Plate Reader. Circular dichroism spectra were obtained on an Olis RSM 1000 CD using a cylindrical cuvette with 1 mm path length.
2. Preparation of Porphyrin Conjugation Reagent.
Synthesis of tert-butyl (6-(2-(4-(10,15,20-tris(4-hydroxy-phenyl)porphyrin-5-yl)phenoxy)acetamido)hexyl)carbamate (3)
Porphyrin 2 (185 mg, 0.24 mmol) was suspended in 5 mL 5% NaOH solution in aqueous ethanol (1:1 v/v) and stirred for 24 h at room temperature (RT). The reaction mixture was diluted with 45 mL water followed by slow addition of aq. HCl (0.1 M) to reduce the pH of the solution to 6.0. The hydrolyzed product, carboxylic acid derivative, was then extracted with ethyl acetate. The organic phase was dried over anhydrous Na2SO4 and concentrated in a rotary evaporator. To the resulting crude residue, HATU (106 mg, 0.28 mmol, 1.2 eq), N-Boc-1,6-hexanedi-amine hydrochloride (70 mg, 0.28 mmol, 1.2 eq), and DIPEA (150 mg, 1.16 mmol, 5.0 eq) were added and the reaction mixture was stirred in 8 mL DMF for 18 h at RT. The crude product was then dissolved in 50 mL ethyl acetate, washed with water (2 x 100 mL) and brine (50 mL). The organic phase was isolated, dried with anhydrous Na2SO4. The solvent was evaporated under reduced pressure and the crude residue was purified by silica gel column chromatography (0.5-3% MeOH in DCM, eluent) to furnish porphyrin 3 (175 mg, 82% yield). 1H NMR (500 MHz, DMSO-d6) δ 9.98 (s, 3H), 8.90 (d, J = 5.0 Hz, 6H), 8.84 (d, J = 5.0 Hz, 2H), 8.31 (t, J = 5.8Hz, 1H), 8.15 (d, J = 8.3 Hz, 2H), 8.02 (d, J = 7.7 Hz, 6H), 7.42 (d, J = 8.3 Hz, 2H), 7.23 (d, J = 7.7 Hz, 6H), 6.76 (t, J = 5.8 Hz, 1H), 4.79 (s, 2H), 3.26 (q, 2H), 2.92 (q, 2H), 1.54 (p, J = 7.1 Hz, 2H), 1.41 (m, 2H), 1.34 (br, s, 13H), −2.86 (s, 2H). 13C NMR (126 MHz, DMSO-d6) δ 168.0, 158.2, 157.9, 156.1, 136.0, 135.8, 134.7, 132.38, 132.36, 120.7, 120.6, 119.7, 114.4, 113.8, 77.7, 67.8, 38.8, 30.0, 29.7, 28.7, 26.6, 26.5. ESI-HRMS calcd for C57H55O7N6 [(M+H)+]: 935.4127, found: 935.4129
Synthesis of tert-butyl (6-(2-(4-(10,15,20-tris(4-(prop-2-yn-1-yloxy)phenyl)porphyrin-5-yl)phenoxy)acetam-ido)hexyl)carbamate (4)
Compound 3 (150 mg, 0.16 mmol) was dissolved in 5 mL DMF and was charged with potassium carbonate (221 mg, 1.6 mmol). Propargyl bromide 80 wt. % in toluene (190 mg, 3.2 mmol) was added to the reaction mixture. After stirring for 24 h at RT, the reaction content was dissolved in 40 mL ethyl acetate and the organic phase was extracted with water (2 x 80 mL) and brine (50 mL). The solvent was evaporated and the crude residue was purified via silica gel column chromatography (0.5-1.0% MeOH in DCM, eluent) to yield porphyrin 4 (140 mg, 83%). 1H NMR (500 MHz, DMSO-d6) δ 8.86 (br, s, 8H), 8.29 (t, J = 5.8 Hz, 1H), 8.17 – 8.14 (m, 8H), 7.45 – 7.41 (m, 8H), 6.74 (br, s, 1H), 5.11 (br, s, 6H), 4.78 (s, 2H), 3.77 – 3.76 (m, 3H), 3.25 (q, 2H), 2.92 (q, 2H), 1.54 (p, J = 6.8 Hz, 2H), 1.42 – 1.32 (m, 13H), 1.24 – 1.21 (m, 2H), −2.88 (s, 2H). 13C NMR (75 MHz, Chloroform-d) δ 168.1, 157.5, 157.1, 156.0, 136.1, 135.8, 135.6, 135.5, 135.4, 131.1, 119.74, 119.67, 119.1, 113.2, 113.0, 79.1, 78.7, 75.9, 67.7, 56.2, 40.4, 39.0, 30.1, 29.6, 28.5, 26.5, 26.4. ESI-HRMS calcd for C66H61O7N6 [(M+H)+]: 1049.4596, found: 1049.4603.
Synthesis of N-(6-aminohexyl(2-(4-(10,15,20-tris(4-(prop-2-yn-1-yloxy)phenyl)porphyrin-5-yl)phenoxy)acetamide (1)
Porphyrin 4 (120 mg, 0.11 mmol) was dissolved in 5 mL DCM. Trifluoroacetic acid (3 mL) was then added to the solution and the reaction mixture was stirred for 2 h at RT. The solvent was evaporated with rotary evaporator and the residue was neutralized with a saturated sodium bicarbonate solution. Subsequently, the crude product was extracted with ethyl acetate, concentrated, and directly transferred to next step without further purification. The boc-deprotected crude product was dissolved in 10 mL chloroform and to this solution was added zinc acetate (404 mg, 2.2 mmol) in 3 mL methanol. The reaction mixture was stirred for 5 h at RT. After 5 h the reaction mixture was diluted with 30 mL chloroform and washed with water (40 mL) and brine (40 mL). The solvent was evaporated, and the residue was purified via silica gel column chromatography (2-7% MeOH in DCM with 0.1-1.0% aqueous ammonia, eluent) to yield porphyrin reagent 1 (91 mg, 80%). 1H NMR (500 MHz, DMSO-d6) δ 8.80 (d, J = 3.9 Hz, 8H), 8.26 (t, J = 5.8 Hz, 1H), 8.12 – 8.08 (m, 8H), 7.42 – 7.37 (m, 8H), 5.11 – 5.07 (m, 6H), 4.76 (s, 2H), 3.76 – 3.74 (m, 3H), 3.21 (q, 2H), 1.74 (br, s, 2H),1.45 (p, J = 7.2 Hz, 2H), 1.18 (p, J = 7.4 Hz, 2H), 1.05 (p, J = 7.5 Hz, 2H), 0.87 – 0.84 (m, 2H). 13C NMR (126 MHz, DMSO-d6) δ 168.0, 157.7, 157.3, 150.0, 136.2, 135.6, 131.9, 120.2, 113.4, 80.0, 79.0, 67.8, 56.2, 41.0, 38.8, 32.1, 29.6, 26.7, 26.4. ESI-HRMS calcd for C61H51O5N6Zn [(M+H)+]: 1011.3207, found: 1011.3212.
3. Conjugation and Masking of Porphyrin 1 onto (Bio)polymers.
Synthesis of Porph-DNA 1 and Porph-DNA 2
Porphyrin 1 (5mg, 4.95 μmol) was dissolved in 600 μL anhydrous DMF and added to NHS modified DNA 1 or DNA 2 (DNA synthesis scale: 1 μmol) on controlled pore glass (CPG) beads. 35 μL DIPEA was added to the mixture and the reaction was stirred in the dark for 6 h in a 1.5 mL microcentrifuge tube at RT. Subsequently, the reaction mixture was centrifuged at 10000 g for 1.5 min and the supernatant solution was discarded. The beads were washed with 3 x DMSO (1 mL), 2 x water (1 mL) and 1 x MeCN (1 mL). The product conjugates were cleaved from the beads and deprotected with 1:1 solution (500 μL each) of 30% aq. ammonia and 40% aq. methylamine at 65 °C for 15 min. The content after deprotection was cooled to RT and centrifuged. The supernatant was collected and the solvent was evaporated using a speedvac at 65 °C to reduce the volume to ~150 μL. The solution then was desalted with a G-25 microspin column, centrifuged to remove any particles, and was purified via reverse phase HPLC column at 65 °C. 65% (Porph-DNA 1) and 60% (Porph-DNA 2) product yields were estimated from HPLC traces of the reaction mixtures by using the integrated peak areas of the starting materials, the products, and ODN side products. MALDI-TOF analysis of Porph-DNA 1, MW calcd for C141H155N32O52P7Zn [(M+H)+]: 3413.17 Da, found: 3412.19 Da. MALDI-TOF analysis of Porph-DNA 2, MW calcd for C163H171N50O66P9Zn [(M+H)+]: 4231.60 Da, found: 4231.58 Da.
Synthesis of PL-DNA 1, PL-DNA 2, and PL-polyT
PL-DNA conjugates were prepared on beads from the Porph-DNA conjugates. Therefore, the protocol was identical as described above until the Porph-DNA conjugates were washed with DMSO, water, and MeCN. To the washed beads was added a 400 μL DMSO:H2O (1:1) solution of PMβCD-N3 (16 mg, 11 μmol). 60 μL 0.2 M TBTA and 30 μL 0.2 M freshly prepared CuBr solutions in DMSO/t-butanol (3:1) were mixed together and then added to the reaction mixture containing DNA beads. The solution was degassed and stirred at 40 °C for 1 h. After 1 h, the beads were washed with 3 x DMSO (1 mL), 2 x water (1 mL) and 1 x MeCN (1 mL). The product conjugates were cleaved from the beads and deprotected with 1:1 solution (500 μL each) of 30% aq. ammonia and 40% aq. methylamine at 65 °C for 15 min. The content after deprotection was cooled to RT and centrifuged. The supernatant was collected and evaporated at 65 °C to reduce the volume to ~150 μL in a speedvac. The solution then was desalted with a G-25 microspin column, centrifuged to remove any particles, and was purified with reverse phase HPLC column at 65 °C. 45% (PL-DNA 1), 40% (PL-DNA 2), 49% (PL-PolyT) product yields were estimated from HPLC traces of the reaction mixtures by using the integrated peak areas of the starting materials, the products, and ODN side products. MALDI-TOF analysis of PL-DNA 1, MW calcd for C327H482N41O154P7Zn [(M+H)+]: 7734.79 Da, found: 7735.70 Da. MALDI-TOF analysis of PL-DNA 2, MW calcd for C349H498N59O168P9Zn [(M+Na)+]: 8575.20 Da, found: 8574.85 Da. MALDI-TOF analysis of PL-polyT, MW calcd for C357H525N35O179P10Zn [(M+Na)+]: 8569.30 Da, found: 8568.96 Da.
Synthesis of Porph-PEG
Porphyrin 1 (2 mg, 1.98 μmol) and O-Methyl-O′-[(N-sucein-imidyl)succinyl-aminoethyl]polyethylene glycol 750 (10 mg, 10.22 μmol) were dissolved in 300 μL anhydrous DMF. 25 μL DIPEA was added and the reaction was stirred for 6 h at RT. The solvent was completely dried in a speedvac and the content was dissolved in 500 μL ultrapure water with 10% MeCN. The insoluble particles were removed by centrifuging the crude mixture at 10000 g for 8 min. The remaining solution was desalted with G-25 microspin columns and finally purified with RP HPLC. (3 mg, 81% yield) MALDI-TOF analysis of Porph-PEG, number average MW observed, Mn = 1830 Da.
Synthesis of PL-PEG
Porph-PEG (2 mg, 1.07 μmol) and 200 μL DMSO solution of PMβCD-N3 (16 mg, 11 μmol) were mixed together. 60 μL 0.2 M TBTA and 30 μL 0.2 M freshly prepared CuBr solutions in DMSO/t-butanol (3:1) were mixed together and then added to the reaction mixture. The solution was degassed and stirred at 40 °C for 1 h. After 1 h, the solvent was completely evaporated in a speedvac and the content was redissolved in ultrapure water. The insoluble particles were removed by centrifuging the reaction content at 10000 g for 8 min. This step was repeated one more time to make sure the water-insoluble unreacted porphyrin is removed. The remaining solution was desalted with G-25 microspin columns and finally purified with RP HPLC. (4 mg, 65% yield) MALDI-TOF analysis of PL-PEG, number average MW observed, Mn = 6170 Da.
HPLC conditions.
RP-HPLC purification was achieved using a Varian Prostar HPLC system, equipped with a Polymer Laboratories 100 Å 5 μm 4.6 × 250 mm PLRP-S reverse phase column. The column was maintained at 65 °C for all runs. The flow rate was set at 1 mL/min. A gradient composed of two solvents (solvent A is 0.1 M TEAA in 5% acetonitrile and solvent B is 100% acetonitrile) was used.
PAGE Studies.
Nondenaturing polyacrylamide gel electrophoresis was conducted using an Invitrogen XCell SureLock Mini-Cell electrophoresis system. Precast TBE gels (20%) purchased from Invitrogen was used for the gel studies. The gel-electro-phoresis was run at i) RT for 1.5 h at 120 V, ii) 4 °C for 5 h at 60 V with 1 x TBE buffer containing 100 mM KCl. 10 μL 15 μM solution of ODNs (single strand concentration) were applied to the gel along with 2 μL loading dye after 48 h incubation under the standard quadruplex forming condition. After electrophoresis, the gel was visualized by SYBR Safe staining in a Bio-Rad ChemiDoc Imaging system.
Annealing conditions for quadruplex assembly and G-wire growth.
Porph-DNA 1 and PL-DNA 1 were annealed in 100 mM potassium phosphate buffer, pH-7.5 whereas Porph-DNA 2 and PL-DNA 2 were annealed in 50 mM Tris buffer, pH-7.5 containing 50 mM KCl and 10 mM MgCl2. Annealing was done by heating 30 μM solution of each sample at 95 °C for 5 min and then slowly cooling to the room temperature. The samples were kept at 4 °C for at least 48 h before studying assembly related properties.
Microscopy Protocols
a. Cryo-TEM experiments were performed on a FEI Tecnai G2 F30 Twin Transmission Electron Microscope Instrument (accelerating voltage = 200 kV). Sample was prepared on 200 mesh copper grids with lacey carbon film (purchased from Electron Microscopy Science). The samples were diluted to 2 μM concentration in the G-wire growing buffer (50 mM Tris, 50 mM KCl, 10 mM MgCl2, pH-7.5) prior to imaging. Briefly, 5 μL of the sample solution was applied to the copper grid, blotted for 2 s, and directly plunged into liquid ethane to get thin firm layer. The sample was kept immersed in liq. N2 until inserted in the TEM machine for imaging.
b. AFM experiments were carried out on a Bruker Dimension ICON Atomic Force Microscope under scanasyst mode in air. Bruker SCANASYST-AIR AFM probes with nominal frequency, tip radius, and spring constants of 70 kHz, 2 nm, and 0.4 N/m, respectively were used. Mica (highest grade V1 Mica disc, 10 mm diameter, was purchased from TED PELLA, Inc.) was used as the substrate and the mica plate was freshly cleaved via scotch tape to achieve a flat surface before use. Prior to sample introduction, a 1 mM MgCl2 aqueous solution was applied for 5 minutes. The excess salt was rinsed off with 50 μL ultrapure water twice and then dried using an argon gas flow. All G-wire forming samples were diluted to the final ODN concentration of 2 μM with a buffer containing 10 mM Tris and 1 mM MgCl2 and were subsequently applied on the mica substrate for 15 minutes, then washed 5 times with 100 μL aliquots of deionized water. After drying with argon gas flow, the sample was imaged. The images were processed using gwyddion software. Normal flattening steps were performed before measuring the heights of the wires. Average peak height was determined by cross sectioning a number of regions of interest and averaging the data.
c. Confocal microscopy images were collected on a Nikon A1RSi equipped with a 32-channel multianode photomultiplier detector. 5 μL of 1 μM solution of the sample was dried on a glass slide and mounted with a glass cover slip in Eukitt quickhardening mounting medium (45% acrylic resin and 55% xylenes). 561 nm laser was chosen for the excitation as it falls within one of the porphyrin Q-bands.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful for support from the NSF (Grant CHE 1609603 to JJ). We thank Anthony Wishard for help with DLS.
Footnotes
Supporting Information.
Detailed synthetic scheme, characterization of synthetic compounds, oligonucleotides, and PEG polymer (NMR, MALDI-TOF, RP-HPLC, optical spectroscopies) and characterization of porphy-rin-DNA assemblies (CD spectroscopy, AFM, DLS)
The authors declare no competing financial interest.
REFERENCES
- (1).Kuzyk A; Jungmann R; Acuna GP; Liu N DNA Origami Route for Nanophotonics. ACS Photonics 2018, 5, 1151–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Vyawahare S; Eyal S; Mathews KD; Quake SR Nanometer-Scale Fluorescence Resonance Optical Waveguides. Nano Lett. 2004, 4, 1035–1039. [Google Scholar]
- (3).Nicoli F; Barth A; Bae W; Neukirchinger F; Crevenna AH; Lamb DC; Liedl T Directional Photonic Wire Mediated by Homo-Förster Resonance Energy Transfer on a DNA Origami Platform. ACS Nano 2017, 11, 11264–11272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Choi Y; Kotthoff L; Olejko L; Resch-Genger U; Bald I DNA Origami-Based Förster Resonance Energy-Transfer Nanoarrays and Their Application as Ratiometric Sensors. ACS Appl. Mater. Interfaces 2018, 10, 23295–23302. [DOI] [PubMed] [Google Scholar]
- (5).Kownacki M; Langenegger SM; Liu SX; Haner R Integrating DNA Photonic Wires into Light-Harvesting Supramolecular Polymers. Angew. Chemie - Int. Ed 2018, 58, 751–755. [DOI] [PubMed] [Google Scholar]
- (6).Albinsson B; Hannestad JK; Borjesson K Functionalized DNA Nanostructures for Light Harvesting and Charge Separation. Coord. Chem. Rev 2012, 256, 2399–2413. [Google Scholar]
- (7).Klein WP; Díaz SA; Buckhout-White S; Melinger JS; Cunningham PD; Goldman ER; Ancona MG; Kuang W; Medintz IL Utilizing HomoFRET to Extend DNA-Scaffolded Photonic Networks and Increase Light-Harvesting Capability. Adv. Opt. Mater 2018, 6, 1700679. [Google Scholar]
- (8).Nieves DJ; Gaus K; Baker MAB DNA-Based Super-Resolution Microscopy: DNA-PAINT. Genes. 2018, 9, 621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hannestad JK; Sandin P; Albinsson B Self-Assembled DNA Photonic Wire for Long-Range Energy Transfer. J. Am. Chem. Soc 2008, 130, 15889–15895. [DOI] [PubMed] [Google Scholar]
- (10).Selnihhin D; Sparvath SM; Preus S; Birkedal V; Andersen ES Multifluorophore DNA Origami Beacon as a Biosensing Platform. ACS Nano 2018, 12, 5699–5708. [DOI] [PubMed] [Google Scholar]
- (11).Jungmann R; Steinhauer C; Scheible M; Kuzyk A; Tinnefeld P; Simmel FC Single-Molecule Kinetics and Super-Resolution Microscopy by Fluorescence Imaging of Transient Binding on DNA Origami. Nano Lett. 2010, 10, 4756–4761. [DOI] [PubMed] [Google Scholar]
- (12).Schröder T; Scheible MB; Steiner F; Vogelsang J; Tinnefeld P Interchromophoric Interactions Determine the Maximum Brightness Density in DNA Origami Structures. Nano Lett. 2019, 19, 1275–1281. [DOI] [PubMed] [Google Scholar]
- (13).Schwartz E; Le Gac S; Cornelissen JJLM; Nolte RJM; Rowan AE Macromolecular Multi-Chromophoric Scaffolding. Chem. Soc. Rev 2010, 39, 1576–1599. [DOI] [PubMed] [Google Scholar]
- (14).Croce R; Van Amerongen H Natural Strategies for Photosynthetic Light Harvesting. Nat. Chem. Biol 2014, 10, 492–501. [DOI] [PubMed] [Google Scholar]
- (15).Teo YN; Kool ET DNA-Multichromophore Systems. Chem. Rev 2012, 112, 4221–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Peng H-Q; Niu L-Y; Chen Y-Z; Wu L-Z; Tung C-H; Yang Q-Z Biological Applications of Supramolecular Assemblies Designed for Excitation Energy Transfer. Chem. Rev 2015, 115, 7502–7542. [DOI] [PubMed] [Google Scholar]
- (17).Sessler JL; Jayawickramarajah J; Gouloumis A; Torres T; Guldi DM; Maldonado S; Stevenson KJ Synthesis and Photophysics of a Porphyrin-Fullerene Dyad Assembled through Watson-Crick Hydrogen Bonding. Chem. Commun 2005, 1892–1894. [DOI] [PubMed] [Google Scholar]
- (18).Aratani N; Kim D; Osuka A Discrete Cyclic Porphyrin Arrays as Artificial Light-Harvesting Antenna. Acc. Chem. Res 2009, 42, 1922–1934. [DOI] [PubMed] [Google Scholar]
- (19).Hsiao JS; Krueger BP; Wagner RW; Johnson TE; Delaney JK; Mauzerall DC; Fleming GR; Lindsey JS; Bocian DR; Donohoe RJ Soluble Synthetic Multiporphyrin Arrays. 2. Photodynamics of Energy-Transfer Processes. J. Am. Chem. Soc 1996, 118, 11181–11193. [Google Scholar]
- (20).Zhang Y; Ying JY Main-Chain Organic Frameworks with Advanced Catalytic Functionalities. ACS Catal. 2015, 5, 2681–2691. [Google Scholar]
- (21).Beletskaya I; Tyurin VS; Tsivadze AY; Guilard R; Stem C Supramolecular Chemistry of Metalloporphyrins. 2009, 1659–1713. [DOI] [PubMed] [Google Scholar]
- (22).Tanaka T; Osuka A Conjugated Porphyrin Arrays: Synthesis, Properties and Applications for Functional Materials. Chem. Soc. Rev 2015, 44, 943–969. [DOI] [PubMed] [Google Scholar]
- (23).Fendt LA; Bouamaied I; Thöni S; Amiot N; Stulz E DNA as Supramolecular Scaffold for Porphyrin Arrays on the Nanometer Scale. J. Am. Chem. Soc 2007, 129, 15319–15329. [DOI] [PubMed] [Google Scholar]
- (24).Wellner C; Wagenknecht HA Synthesis of DNA Conjugates with Metalated Tetracationic Porphyrins by Postsynthetic Cycloadditions. Org. Lett 2014, 16, 1692–1695. [DOI] [PubMed] [Google Scholar]
- (25).Anderson NT; Dinolfo PH; Wang X Synthesis and Characterization of Porphyrin-DNA Constructs for the Self-Assembly of Modular Energy Transfer Arrays. J. Mater. Chem. C 2018, 6, 2452–2459. [Google Scholar]
- (26).Morales-Rojas H; Kool ET A Porphyrin C-Nucleoside Incorporated into DNA. Org. Lett 2002, 4, 4377–4380. [DOI] [PubMed] [Google Scholar]
- (27).Vybornyi M; Nussbaumer AL; Langenegger SM; Häner R Assembling Multiporphyrin Stacks inside the DNA Double Helix. Bioconjug. Chem 2014, 25, 1785–1793. [DOI] [PubMed] [Google Scholar]
- (28).Balaz M; Holmes AE; Benedetti M; Rodriguez PC; Berova N; Nakanishi K; Proni G Synthesis and Circular Dichroism of Tetraarylporphyrin-Oligonucleotide Conjugates. J. Am. Chem. Soc 2005, 127,4172–4173. [DOI] [PubMed] [Google Scholar]
- (29).Balaz M; Li BC; Jockusch S; Ellestad GA; Berova N Tetraarylporphyrin as a Selective Molecular Cap for Non-Watson-Crick Guanine-Adenine Base-Pair Sequences. Angew. Chemie - Int. Ed 2006, 45, 3530–3533. [DOI] [PubMed] [Google Scholar]
- (30).Jayawickramarajah J; Tagore DM; Tsou LK; Hamilton AD Allosteric Control of Self-Assembly: Modulating the Formation of Guanine Quadruplexes through Orthogonal Aromatic Interactions. Angew. Chemie - Int. Ed 2007, 46, 7583–7586. [DOI] [PubMed] [Google Scholar]
- (31).Woller JG; Hannestad JK; Albinsson B Self-Assembled Nanoscale DNA-Porphyrin Complex for Artificial Light Harvesting. J. Am. Chem. Soc 2013, 135, 2759–2768. [DOI] [PubMed] [Google Scholar]
- (32).Stulz E Nanoarchitectonics with Porphyrin Functionalized DNA. Acc. Chem. Res 2017, 50, 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Tran PLT; De Cian A; Gros J; Moriyama R; Mergny JL. (2012) Tetramolecular Quadruplex Stability and Assembly. In: Chaires J, Graves D (eds) Quadruplex Nucleic Acids. Topics in Current Chemistry, vol 330. Springer, Berlin, Heidelberg, pp 243–273. [DOI] [PubMed] [Google Scholar]
- (34).Aldaye FA; Palmer AL; Sleiman HF Assembling Materials with DNA as the Guide. Science. 2008, 321, 1795–1799. [DOI] [PubMed] [Google Scholar]
- (35).McLaughlin CK; Hamblin GD; Hanni KD; Conway JW; Nayak MK; Cameiro KMM; Bazzi HS; Sleiman HF Three-Dimensional Organization of Block Copolymers on “DNA-Minimal” Scaffolds. J. Am. Chem. Soc 2012, 134, 4280–4286. [DOI] [PubMed] [Google Scholar]
- (36).Zhang N; Chu X; Fathalla M; Jayawickramarajah J Photonic DNA-Chromophore Nanowire Networks: Harnessing Multiple Supramolecular Assembly Modes. Langmuir 2013, 29, 10796–10806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Zhou J; Yuan G; Liu J; Zhan C-G Formation and Stability of G-Quadruplexes Self-Assembled from Guanine-Rich Strands. Chem. Eur. J 2007, 13, 945–949. [DOI] [PubMed] [Google Scholar]
- (38).Greider CW; Blackburn EH Telomeric Sequence in the RNA of Required for Repeat Synthesis. Nature 1989, 337, 331–337. [DOI] [PubMed] [Google Scholar]
- (39).Marsh TC; Vesenka J; Henderson E A New DNA Nanostructure, the G-Wire, Imaged by Scanning Probe Microscopy. Nucleic Acids Res. 1995, 23, 696–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Bose K; Lech CJ; Heddi B; Phan AT High-Resolution AFM Structure of DNA G-Wires in Aqueous Solution. Nat. Commun 2018, 9, 1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Delor M; Dai J; Roberts TD; Rogers JR; Hamed SM; Neaton JB; Geissler PL; Francis MB; Ginsberg NS Exploiting Chromophore-Protein Interactions through Linker Engineering to Tune Photoinduced Dynamics in a Biomimetic Light-Harvesting Platform. J. Am. Chem. Soc 2018, 140, 6278–6287. [DOI] [PubMed] [Google Scholar]
- (42).Miller RA; Presley AD; Francis MB Self-Assembling Light-Harvesting Systems from Synthetically Modified Tobacco Mosaic Virus Coat Proteins. J. Am. Chem. Soc 2007, 129, 3104–3109. [DOI] [PubMed] [Google Scholar]
- (43).Springer JW; Parkes-Loach PS; Reddy KR; Krayer M; Jiao J; Lee GM; Niedzwiedzki DM; Harris MA; Kirmaier C; Bocian DF; Lindsey JS; Holten D; Loach PA Biohybrid Photosynthetic Antenna Complexes for Enhanced Light-Harvesting. J. Am. Chem. Soc 2012, 134 (10), 4589–599. [DOI] [PubMed] [Google Scholar]
- (44).Hu X; Damjanovic A; Ritz T; Schulten K Architecture and Mechanism of the Light-HarvestingApparatus of Purple Bacteria. Proc. Natl. Acad. Sci 1998, 95, 5935–5941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Nishiyabu R; Kano K Double Self-Inclusion by Rotating Glucopyranose Units in per-O-Methylated β-Cyclodextrin Moieties Attached to a Porphyrin in Aqueous Solution. Eur. J. Org. Chem 2004, 4985–4988. [Google Scholar]
- (46).Liu Y; Ke CR; Zhang HY; Cui J; Ding F Complexation-Induced Transition of Nanorod to Network Aggregates: Alternate Porphyrin and Cyclodextrin Arrays. J. Am. Chem. Soc 2008, 130, 600–605. [DOI] [PubMed] [Google Scholar]
- (47).Zhang H; Zhang B; Zhu M; Grayson SM; Schmehl R; Jayawickramarajah J Water-Soluble Porphyrin Nanospheres: Enhanced Photo-Physical Properties Achieved via Cyclodextrin Driven Double Self-Inclusion. Chem. Commun 2014, 50, 4853–4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Tsuchiya Y; Shiraki T; Matsumoto T; Sugikawa K; Sada K; Yamano A; Shinkai S Supramolecular Dye Inclusion Single Crystals Created from 2,3,6-Trimethyl-β-Cyclodextrin and Porphyrins. Chem. Eur. J 2012, 18, 456–465. [DOI] [PubMed] [Google Scholar]
- (49).Zhou X; Pathak P; Jayawickramarajah J Design, Synthesis, and Applications of DNA-Macrocyclic Host Conjugates. Chem. Commun 2018, 54, 11668–11680. [DOI] [PubMed] [Google Scholar]
- (50).Pierce SE; Wang J; Jayawickramarajah J; Hamilton AD; Brodbelt JS Examination of the Effect of the Annealing Cation on Higher Order Structures Containing Guanine or Isoguanine Repeats. Chem. Eur. J 2009, 15, 11244–11255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Burge S; Parkinson GN; Hazel P; Todd AK; Neidle S Quadruplex DNA: Sequence, Topology and Structure. Nucleic Acids Res. 2006, 34, 5402–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Răsădean DM; Sheng B; Dash J; Pantoş, G. D. Amino-Acid-Derived Naphthalenediimides as Versatile G-Quadruplex Binders. Chem. Eur. J 2017, 23, 8491–8499. [DOI] [PubMed] [Google Scholar]
- (53).Troha T; Drevenšek-Olenik I; Webba Da Silva M; Spindler L Surface-Adsorbed Long G-Quadruplex Nanowires Formed by G:C Linkages. Langmuir 2016, 32, 7056–7063. [DOI] [PubMed] [Google Scholar]
- (54).Thazhathveetil AK; Harris MA; Young RM; Wasielewski MR; Lewis FD Efficient Charge Transport via DNA G-Quadruplexes. J. Am. Chem. Soc 2017, 139, 1730–1733. [DOI] [PubMed] [Google Scholar]
- (55).Mei J; Leung NLC; Kwok RTK; Lam JWY; Tang BZ Aggregation-Induced Emission: To-gether We Shine, United We Soar! Chem. Rev. 2015, 115, 11718–11940. [DOI] [PubMed] [Google Scholar]
- (56).Li S; Langenegger SM; Haner R Control of Aggregation-Induced Emission by DNA Hybridization. Chem. Commun. 2013, 49, 5835–5837. [DOI] [PubMed] [Google Scholar]
- (57).Zhu L; Zhou J; Xu G; Li C; Ling P; Liu B; Ju H; Lei J DNA Quadruplexes as Molecular Scaffolds for Controlled Assembly of Fluorogens with Aggregation-Induced Emission. Chem. Sci 2018, 9, 2559–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Li Y; Kwok RTK; Tang BZ; Liu B Specific Nucleic Acid Detection Based on Fluorescent Light-up Probe from Fluorogens with Aggregation-Induced Emission Characteristics. RSC Adv. 2013, 3, 10135–10138. [Google Scholar]
- (59).Hong Y; Chen S; Leung CWT; Lam JWY; Tang BZ Water-Soluble Tetraphenylethene Derivatives as Fluorescent “Light-Up” Probes for Nucleic Acid Detection and Their Applications in Cell Imaging. Chem. – An Asian J 2013, 8, 1806–1812. [DOI] [PubMed] [Google Scholar]
- (60).Hong Y; Häußler M; Lam JWY; Li Z; Sin KK; Dong Y; Tong H; Liu J; Qin A; Renneberg R; et al. Label-Free Fluorescent Probing of G-Quadruplex Formation and Real-Time Monitoring of DNA Folding by a Quaternized Tetraphenylethene Salt with Aggregation-Induced Emission Characteristics. Chem. – A Eur. J 2008, 14, 6428–6437. [DOI] [PubMed] [Google Scholar]
- (61).Liang G; Lam JWY; Qin W; Li J; Xie N; Tang BZ Molecular Luminogens Based on Restriction of Intramolecular Motions through Host–Guest Inclusion for Cell Imaging. Chem. Commun 2014, 50, 1725–1727. [DOI] [PubMed] [Google Scholar]
- (62).Dsouza RN; Pischel U; Nau WM Fluorescent Dyes and Their Supramolecular Host/Guest Complexes with Macrocycles in Aqueous Solution. Chem. Rev 2011, 111, 7941–7980. [DOI] [PubMed] [Google Scholar]
- (63).Dempsey JM; Zhang Q-W; Oliver AG; Smith BD New Tetralactam Hosts for Squaraine Dyes. Org. Biomol. Chem 2018, 16, 8976–8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Hocquelet C; Blu J; Jankowski CK; Arseneau S; Buisson D; Mauclaire L Synthesis of Calixarene-Cyclodextrin Coupling Products. Tetrahedron 2006, 62, 11963–11971. [Google Scholar]
- (65).Cao X; Lin W; Yu Q A Ratiometric Fluorescent Probe for Thiols Based on a Tetrakis(4-Hydroxyphenyl)Porphyrin-Coumarin Scaffold. J. Org. Chem 2011, 76, 7423–7430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.