Abstract
Alterations in the deformability of red blood cells (RBCs), occurring in hemolytic blood disorders such as sickle cell disease (SCD), contribute to vaso-occlusion and disease pathophysiology. There are few functional in vitro assays for standardized assessment of RBC-mediated microvascular occlusion. Here, we present the design, fabrication, and clinical testing of the Microfluidic Impedance Red Cell Assay (MIRCA) with embedded capillary network-based micropillar arrays and integrated electrical impedance measurement electrodes to address this need. The micropillar arrays consist of microcapillaries ranging from 12 μm to 3 μm, with each array paired with two sputtered gold electrodes to measure the impedance change of the array before and after sample perfusion through the microfluidic device. We define RBC Occlusion Index (ROI) and RBC Electrical Impedance Index (REI), which represent the cumulative percentage occlusion and cumulative percentage impedance change, respectively. We demonstrate the promise of MIRCA in two common red cell disorders, SCD and hereditary spherocytosis. We show that the electrical impedance measurement reflects the microvascular occlusion, where REI significantly correlates with ROI that is obtained via high resolution microscopy imaging of the microcapillary arrays. Further, we show that RBC-mediated microvascular occlusion, represented by ROI and REI, associates with clinical treatment outcomes and correlates with in vivo hemolytic biomarkers, lactate dehydrogenase (LDH) level and absolute reticulocyte count (ARC) in SCD. Impedance measurement obviates the need for high-resolution imaging enabling future translation of this technology for widespread access, portable and point-of-care use. Our findings suggest that the presented microfluidic design and the integrated electrical impedance measurement provide a reproducible functional test for standardized assessment of RBC-mediated microvascular occlusion. MIRCA and the newly defined REI may serve as an in vitro therapeutic efficacy benchmark for assessing clinical outcome of emerging RBC modifying targeted and curative therapies.
Keywords: Microfluidics, Impedance, Red blood cell deformability, Microvascular occlusion, Microcirculation, Inherited red cell disorders, Rheology
INTRODUCTION
Red blood cells (RBCs) are inherently flexible, which allows them to swiftly pass through microcapillaries, facilitating oxygen transport to tissues and removal of damaged or infected red cells, respectively 1. The exceptional deformational capacity of RBCs is due to their large surface area-to-volume ratio, the integrity and organization of the membrane cytoskeletal protein network, and the low cytoplasmic viscosity 2, 3. Pathological changes may cause significant alterations in one or more of a combination of these factors, and consequently lead to decreased RBC deformability or increased stiffness 4, 5. For example, Plasmodium falciparum, an infectious parasite associated with lifelong morbidity and early mortality, significantly reduces host RBC deformability by producing cytoadherence-related antigens and by impairing the shear elastic moduli of the membrane 6, 7. In sickle cell disease (SCD), abnormal intracellular polymerization of sickle hemoglobin causes distorted membrane morphology, increased adhesiveness, and decreased cellular deformability 8–15. Finally, in hereditary spherocytosis (HS), the molecular defect in the genes encoding RBC cytoskeletal proteins (e.g., spectrin, ankyrin, Band 3, and protein 4.2) result in a number of abnormalities, including loss of membrane surface area, decreased cellular deformability, and defective membrane mechanical stability 16. RBCs with abnormal deformability and membrane properties in the microcirculation may hemolyze and/or disrupt normal blood perfusion, resulting in a range of abnormalities, including anemia, reticulocytosis, vascular inflammation, and microcirculatory occlusion 17–20.
Conventional techniques for RBC deformability measurement, including atomic force microscopy (AFM) 21, optical tweezers 22, and micropipette aspiration 23, 24, measure cellular deformability at the single-cell level. These methods involve technically challenging procedures that require skilled personnel and specialized equipment. Bulk-cell approaches include shear flow-based and deformation-based techniques. Shear flow-based techniques, including ektacytometry 25–28 and related microfluidic techniques 29–36, measure RBC deformability by stretching RBCs under shear flow to obtain their elongation or deformation index via laser diffraction or high-speed camera. Deformation-based techniques, including cell transit analyzer 37, 38 and other microscale techniques 13, 39–50, infer RBC deformability from the cell’s transit time, transit velocity, transit pressure, wedging, or cell retention rate through capillary networks that are narrower than the RBC. Even though these techniques allow RBC deformability measurements, they can only describe rheological properties of cellular membranes rather than measure the capacity of RBCs to mechanically deform and pass through microcapillaries. We explored measuring this functional property in this work, which is the vital characteristic of RBCs for traversing the intricate capillary networks to deliver oxygen to the cells and tissues.
The development of a standardized, functional in vitro assay for objective and quantitative assessment of RBC-mediated microvascular occlusion would contribute to a better understanding of the pathophysiological impact of abnormal RBC deformability on the microcirculation. A physiologically relevant device to assess RBC-mediated microvascular occlusion should mimic key dimensions of microcapillaries (3 μm to 10 μm) observed in the capillary bed 51. Microfluidic designs with microstructures of several micrometers molded in polydimethylsiloxane (PDMS) elastomers have been designed to partially mimic the capillary bed to investigate the impact of abnormal RBC deformability in malaria and other pathological conditions 52–54. To measure microcapillary occlusion or RBC retention, these earlier techniques involve an experimental procedure that relies on high-resolution optical imaging, which significantly limits their potential for widespread and point-of-care use. The concept of measuring cellular obstruction of capillary networks due to abnormal RBC deformability through electrical impedance readout was recently introduced 55, 99. The microfluidic design in this study allowed relatively low-throughput processing volume and thus offered limited sampling of RBCs in a blood sample. Pathological RBCs often constitute a small fraction of the entire blood cell population; thus, processing a large number of RBCs is critical in acquiring meaningful results from clinical blood samples, as demonstrated in this study.
Here, we present a standardized, functional in vitro microfluidic assay that allowed objective and quantitative assessments of RBC mediated microvascular occlusion. To that end, we designed the Microfluidic Impedance Red Cell Assay (MIRCA) with embedded micropillar arrays comprising gradient-narrow microcapillaries from 12 μm down to 3 μm along the flow direction mimicking the non-uniform small blood vessels in the capillary bed. Such a design enabled stiffer RBCs to be retained in the upstream arrays representing larger microcapillaries, while less stiff RBCs were retained in the downstream arrays representing smaller microcapillaries. The near-inlet array with 12-μm microcapillaries, which does not represent the typical microcapillary dimension in the capillary bed (which are typically less than 10 μm), was included to trap potential large cells and cell aggregates. Moreover, the micropillar arrays in MIRCA were coupled with two 40-μm-wide side pathways that mimic arteriovenous anastomoses. Anastomoses are bypass passageways around capillary beds, which provide alternative flow paths in the event of blockages in the microcapillaries 56, 57. These anastomosis-mimicking pathways helped regulate the blood flow such that when the upstream portion of an array was fully saturated, the upcoming RBCs could still flow around and into the downstream portion of the array, which prevented congestion of the microchannel. These features enabled testing of clinical samples at near-hematocrit levels and full utilization of the microcapillary domain. Finally, each micropillar array was paired with two sputtered gold electrodes on the channel bottom surface for electrical impedance measurement. The impedance of each array across the paired electrodes was obtained before and after sample perfusion. We used healthy RBCs, glutaraldehyde-stiffened RBCs, and RBCs from two common red cell disorders, SCD and HS, to validate MIRCA and to demonstrate its clinical relevance. We introduced two new parameters, RBC Occlusion Index (ROI) and RBC Electrical Impedance Index (REI), which represent the cumulative percentage occlusion and cumulative percentage impedance change, respectively. We showed that the REI significantly correlates with the ROI, and both the ROI and REI associate with in vivo hemolytic biomarkers, serum lactate dehydrogenase (LDH) levels and absolute reticulocyte counts (ARCs) as well as treatment outcomes in subjects with SCD. MIRCA and its electrical impedance-based readout obviates the need for high-resolution microscopy and enables future translation for widespread access and point-of-care use.
METHODS
Concept and design
The microfluidic design of MIRCA comprises capillary network-inspired micropillar arrays and sputtered electrodes paired with each arrays. As RBCs flow through the microchannel, deformable RBCs are able to clear through, while stiff RBCs are retained within the microcapillaries (Fig. 1A). As a result, the electrical impedance of the micropillar array across the paired electrodes increases in accordance with the resultant occlusion due to the retained RBCs. The microchannel geometry features two key aspects of the human capillary bed: the small capillaries (less than 10 μm) and the arteriovenous anastomoses 56, 57. To mimic small capillaries, micropillar arrays were embedded into the microchannel comprising microcapillaries from 12 μm down to 3 μm along the flow direction (Fig. 1B). The micropillars were designed to be 20-μm long, 10-μm wide, and 12-μm tall (Fig. 1B inset), with a column-to-column spacing of 20 μm and array-to-array spacing of 1 mm. This feature enabled less stiff RBCs to be retained by downstream finer microcapillaries while stiffer RBCs were retained by upstream coarse microcapillaries. The near-inlet 12 μm micropillar array was designed to retain large-cell aggregates or contamination that may be present in the blood flow. To mimic the anastomoses around capillary beds, the micropillar arrays were coupled with two 40-μm-wide side pathways (Fig. 1B inset). This feature helped regulate blood flow around the obstructed area to prevent clogging of the microchannel. Moreover, each micropillar array was flanked by a pair of planar gold electrodes for sensing variation of electrical impedance due to retained RBCs (Fig. 1B). Overall, the microchannel is 24-mm long, 4-mm wide, and 12-μm thick.
Fig. 1.
Microfluidic electrical impedance assessment of RBC-mediated microvascular occlusion. (A) Electrical impedance across two electrodes placed on either side of the micropillar array is measured before and after sample perfusion. The resultant impedance change depends on microcapillary occlusion in the array. (B) The Microfluidic Impedance Red Cell Assay (MIRCA) consists of six micropillar arrays comprising microcapillaries ranging from 12 μm down to 3 μm, which mimic key dimensions of small blood vessels observed in the capillary bed. The 12 μm array is designed to trap potential large-cell aggregates that may cause microchannel clogging. Inset: schematic of the capillary-inspired micropillar array. The 40 μm-wide side pathway is designed to mimic arteriovenous anastomoses to help regulate flow and to prevent upstream blockage. Schematics are not drawn to scale, and all dimensions are in microns. (C) Photograph of MIRCA is shown. The arrow indicates flow direction. Inset: close-up views of the microcapillary occlusion within the 3 μm array induced by glutaraldehyde-stiffened RBCs or healthy RBCs. (D) Temporal variation in electrical impedance of the 3 μm array observed at 10 kHz is shown for a sample with 2% stiff RBCs and 98% healthy RBCs, a sample with 100% healthy RBCs, and PBS. Each sample was perfused for 20 min, which was followed by post-perfusion wash with PBS for another 20 min. The initial impedance reading was taken at the start point of each test (time = 0 min), and the second impedance reading was taken at the endpoint (time = 40 min).
Device fabrication
MIRCA was fabricated using standard soft photolithography protocols. Initially, a master silicon wafer was fabricated by micropatterning a uniform SU8–2010 (Microchem, Newton, MA) photoresist layer. Briefly, the SU8–2010 layer was spin-coated at 2500 rpm over the silicon wafer and soft-baked at 95 °C for 4 min. Next, the wafer was exposed to UV light and post-exposure baked at 95 °C for 4 min. Thereafter, the wafer was developed in 1-methoxy-2-propanol acetate (Sigma Aldrich, St. Louis, MO), and hard-baked at 110 °C overnight. The master wafer was then used for PDMS casting (10:1 ratio, 80 °C overnight). The PDMS blocks were peeled-off and punched with 0.5 mm-diameter ports for inlets and outlets. Substrate fabrication process started with sonicating standard microscope glass slides with isopropanol for 15 min at room temperature. After drying, multiple gold electrodes with dimensions of 10.5 mm × 0.5 mm with a spacing of 2.5 mm and contact pads with dimensions of 4.5 mm × 4.5 mm were sputter-deposited (150 Å/2000 Å Ti/Au) under a laser-micromachined Kapton tape mask. Thereafter, a layer of amorphous silica (5000 Å SiO2) was sputter-deposited under a secondary laser-micromachined Kapton tape mask to ensure proper sealing. All radiofrequency (RF) sputtering processes were performed in Denton Vacuum Explorer 14 System (Moorestown, NJ). Finally, the PDMS block was covalently bonded to the substrate using oxygen plasma. The obtained device was incubated at 60 °C for 30 min on a hotplate to increase the bonding strength. Detailed fabrication process of the substrate is illustrated in Fig. S1. The cross marks were designed to achieve electrode and micropillar array alignment.
Blood sample acquisition and processing
De-identified blood samples were collected in ethylenediaminetetraacetic acid (EDTA)-containing vacutainers from healthy donors or subjects with homozygous (HbSS) SCD or HS at University Hospitals Cleveland Medical Center under Institutional Review Board (IRB)-approved protocols. The obtained blood samples were stored at 4 °C until tested. Signed informed consent forms were obtained from all study participants. RBCs were isolated from the obtained whole blood samples by centrifuging at 500× g for 5 min at room temperature. Plasma, buffy coat, and the near-plasma portion of the RBC pellet were carefully removed. The RBC pellet was then washed twice in phosphate-buffered saline (PBS). Unless otherwise stated, the isolated RBCs were re-suspended in PBS at 20% hematocrit and tested. All experiments were completed within 48 hours of venipuncture in this study. Clinical variables of the study population with SCD are summarized in Table S1.
Glutaraldehyde stiffening of healthy RBCs
In order to validate the microfluidic device functionality of assessing RBC mediated microvascular occlusion, healthy RBCs were isolated and treated with 0.08% w/v glutaraldehyde (Sigma Aldrich) for 10 min at room temperature. The glutaraldehyde-stiffened RBCs were then washed with PBS, mixed with untreated healthy RBCs from the same donor at either 1% or 2% v/v ratio, and re-suspended in PBS at 20% hematocrit for testing. Healthy RBCs from the same healthy donor were included in the study design as control.
Test procedure
Microchannels were initially washed with absolute ethanol (100%) and PBS (1×), which was followed by incubation with 2% bovine serum albumin (BSA) overnight to prevent non-specific binding of RBCs to the channel walls. A Flow-EZ™ microfluidic flow control system (Fluigent, Lowell, MA) was used to regulate the flow (Fig. S2). An impedance analyzer (Agilent 4294A, Santa Clara, CA) coupled with a custom printed-circuit board was used to perform electrical impedance measurements. Briefly, impedance magnitude across each of the 3-μm to 10-μm micropillar arrays was recorded over the frequency range of 40 Hz – 1 MHz before introducing blood into the microchannel and after completing the washing step. Fig. S3 shows the raw impedance data of the 3-μm micropillar array measured for a typical clinical blood sample. For all impedance analyses, a spot frequency of 10 kHz was chosen to minimize potential electrode polarization effects at lower frequencies and avoid parasitic inductances associated with higher frequencies. Hence, data are reported as a percentage change of impedance at 10 kHz in this study.
Prior to the experiment, PBS was perfused through the microchannel at 100 mBar for 15 min to allow for any volumetric changes of the microchannel. Thereafter, the baseline impedance reading was obtained for each micropillar array without stopping the PBS flow. Next, the RBC sample was loaded into the sample reservoir and perfused for 20 min, followed by post-perfusion PBS washing for 20 min. A second impedance reading was then obtained for each micropillar array upon the conclusion of the post-perfusion wash step. The 12-μm micropillar array was excluded from the measurement since it was designed to prevent large-cell aggregates or contamination, as stated earlier. System background noise was characterized by repeatedly testing PBS (background electrolyte) using five different devices. For select experiments, the electrical impedance of the 3-μm micropillar array was continuously monitored over the entire duration of the experiment.
To determine the association between the electrical impedance change and microcapillary occlusion in the microchannel, an Olympus IX83 inverted motorized microscope with Olympus CellSense live-cell imaging and analysis software were used to obtain high-resolution microscopic images. The images were further processed by Adobe Photoshop software (San Jose, CA), in which the obstructed microcapillaries were manually counted. Typically, it takes less than 45 min to complete the microfluidic processing and electrical impedance data acquisition/analysis for testing one clinical blood sample. MIRCA has been designed to be single-use and disposable to prevent any cross-contamination between the samples tested. Each data point in this manuscript was generated with a single use of a newly made device.
RBC Occlusion Index and RBC Electrical Impedance Index
To effectively compare the overall microcapillary occlusion and the overall impedance change caused by different RBC samples, we defined RBC Occlusion Index (ROI) and RBC Electrical Impedance Index (REI) using the following equations,
| (1) |
| (2) |
Where,
Therefore, ROI represents the cumulative percentage occlusion of the capillary networks and REI represents the cumulative percentage impedance change of the capillary networks across the device. The area of interest in our device contains the 3-μm to 10-μm micropillar arrays. The upstream 12-μm array was included to trap potential large-cell aggregates. The concepts of ROI (i.e., the cumulative percentage occlusion of the capillary networks) and REI (i.e., the cumulative percentage impedance change of the capillary networks) are translatable to any microfluidic device employing capillary networks and electrical impedance measurement to assess occlusion/impedance change due to abnormal cellular deformability.
Statistical methods
Data were reported as mean ± standard deviation (mean ± SD) in this study. Data were initially analyzed for normality followed by appropriate comparison methods: paired t-test for paired data, one-way ANOVA for normally distributed data, or Mann-Whitney for non-normally distributed data. Linear regression was used to assess the relationship between two variables, and the Pearson correlation coefficient (PCC) was reported. Mean-shift clustering technique 58, 59 was used to categorize the study population with SCD based on their ROI and REI results. A custom-written code in MATLAB (MathWorks, Natick, MA) was utilized for the clustering analysis. Statistical significance was defined with p-value less than 0.05 (p<0.05). All statistical analyses were carried out using Minitab 19 software (Minitab Inc., State College, PA).
RESULTS
Characterization of system background noise
We tested PBS (background electrolyte) using five different devices and found that the resultant electrical impedance changes of individual micropillar arrays ranged from −0.57% to 0.95% (Fig. S4, mean ± SD = 0.14% ± 0.33%). The 95% confidence interval was calculated as 0.01–0.27%. Hence, any impedance changes less than 0.27% could be attributed to the system background noise, and were thereby rounded to zero in blood tests.
Validation of the micropillar arrays and the integrated electrical impedance measurement using glutaraldehyde-stiffened RBCs
Glutaraldehyde is a non-specific protein cross-linker, which is commonly used to stiffen RBCs in order to mimic pathologically abnormal RBC deformability 60–62. Mild glutaraldehyde stiffening (0.08% w/v in PBS) was applied to RBCs in order to validate the micropillar arrays and the integrated electrical impedance measurement (Fig. 1C and insets). Of note, continuous monitoring of the impedance of the 3-μm micropillar array with four different samples, including PBS, a sample with 100% healthy RBCs, and a sample with 98% healthy RBCs and 2% stiff RBCs, revealed a unique feature of the presented device: the profile of the impedance change is significantly affected by the presence of healthy RBCs and stiff RBCs in the blood flow (Fig. 1D). In addition, we found that the level of microcapillary occlusion increased as the fraction of the stiff RBCs in the tested RBC samples increased (Fig. 2A). Further, the ROI of samples with 98% healthy RBCs and 2% stiff RBCs was significantly higher compared to that of samples with 99% healthy RBCs and 1% stiff RBCs or 100% healthy RBCs (Fig. 2B, mean ± SD = 36.16% ± 4.44% vs. 17.15% ± 1.78% or 7.61% ± 1.67% for 2% stiff RBCs vs. 1% stiff RBCs or healthy, p=0.003 or p=0.001, paired t-test), and the ROI of samples with 99% healthy RBCs and 1% stiff RBCs was significantly higher compared to that of 100% healthy RBCs (Fig. 2B, p=0.010, paired t-test). Notably, we observed that magnitude of impedance variation also increased as the fraction of the stiff RBCs in the tested RBC samples increased (Fig. 2C). Moreover, the REI of samples with 98% healthy RBCs and 2% stiff RBCs was significantly higher compared to that of samples with 99% healthy RBCs and 1% stiff RBCs or 100% healthy RBCs (Fig. 2D, mean ± SD = 20.99% ± 1.42% vs. 9.40% ± 1.61% or 4.35% ± 1.44% for 2% stiff RBCs vs. 1% stiff RBCs or healthy, p=0.002 or p<0.001, paired t-test), and the REI of samples with 99% healthy RBCs and 1% stiff RBCs was significantly higher compared to that of 100% healthy RBCs (Fig. 2D, p=0.013, paired t-test). Importantly, our results reveal that REI significantly correlates with ROI in these tests (Fig. 2E, PCC=0.987, p<0.0001, N=12).
Fig. 2.
MIRCA system characterization using glutaraldehyde-stiffened RBCs (stiff RBCs) and analysis of the microcapillary occlusion and impedance data. (A) Profiles of microcapillary occlusion in the 3 μm to 10 μm arrays shown as histograms for the tested RBC samples with 100% healthy RBCs, with 99% healthy RBCs and 1% stiff RBCs, or with 98% healthy RBCs and 2% stiff RBCs. (B) The ROI of samples with 98% healthy RBCs and 2% stiff RBCs was significantly higher compared to that of samples with 99% healthy RBCs and 1% stiff RBCs or 100% healthy RBCs, and the ROI of samples with 99% healthy RBCs and 1% stiff RBCs was significantly higher compared to that of 100% healthy RBCs (p < 0.05, paired t-test). (C) Profiles of impedance change in those arrays shown as histograms for the tested RBC samples. (D) The REI of samples with 98% healthy RBCs and 2% stiff RBCs was significantly higher compared to that of samples with 99% healthy RBCs and 1% stiff RBCs or 100% healthy RBCs, and the REI of samples with 99% healthy RBCs and 1% stiff RBCs was significantly higher compared to that of 100% healthy RBCs (p < 0.05, paired t-test). (E) The REI significantly correlates with the ROI in the tested samples (PCC = 0.987, p < 0.0001, N = 12). ROI: RBC occlusion index. REI: RBC electrical impedance index. PCC: Pearson correlation coefficient. Error bars represent standard deviation. N = 4 for each group.
Validation of the process repeatability and reproducibility of results
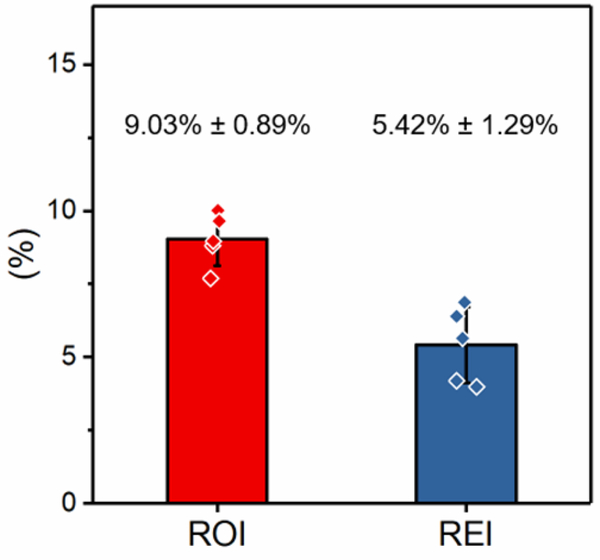
To validate the process repeatability and reproducibility of results, we tested one RBC sample obtained from a single healthy donor using five MIRCA devices manufactured from different batches. The results were highly consistent, where the ROI of the sample is 9.03% ± 0.89%, and the REI of the sample is 5.42% ± 1.29% (Fig. 3, mean ± SD).
Fig. 3.
Measurement reproducibility was assessed by repeatedly testing one RBC sample from a single healthy donor using five different devices. Shown are the ROI and REI results of the five repeats (mean ± standard deviation). ROI: RBC occlusion index. REI: RBC electrical impedance index. Error bars represent standard deviation.
Assessments of RBC-mediated microvascular occlusion in sickle cell disease and hereditary spherocytosis
To demonstrate the clinical relevance of MIRCA, we tested clinical samples from 12 subjects with homozygous (HbSS) SCD and 2 subjects with HS and compared the results with samples from 5 health donors. We found that the level of microcapillary occlusion increased when comparing RBCs from subjects with SCD or HS to RBCs from healthy donors (Fig. 4A). Further, the ROI of RBCs from subjects with SCD or HS is significantly higher compared to that of RBCs from healthy donors (Fig. 4B, mean ± SD = 34.65% ± 21.99% or 23.46% ± 2.55% vs. 8.02% ± 1.71% for SCD or HS vs. healthy, p=0.018 or p<0.001, one-way ANOVA). Notably, magnitude of impedance variation also increased when comparing RBCs from subjects with SCD or HS to RBCs from healthy donors (Fig. 4C). Moreover, the REI of RBCs from subjects with SCD or HS is significantly higher compared to that of RBCs from healthy donors (Fig. 4D, mean ± SD = 14.93% ± 10.48% or 11.17% ± 1.14% vs. 4.31% ± 1.25% for SCD or HS vs. healthy, p=0.043 or p=0.001, one-way ANOVA). Further, our results indicate that REI significantly correlates with ROI in these tests (Fig. 4E, PCC=0.946, p<0.0001, N=19).
Fig. 4.
Assessment of RBC deformability and microvascular occlusion in two common red cell disorders, sickle cell disease (SCD) and hereditary spherocytosis (HS). (A) Histograms of microcapillary occlusion for the tested RBC samples from healthy donors and subjects with SCD or HS. (B) The ROI of RBCs from subjects with SCD or HS is significantly higher compared to that from healthy donors (p < 0.05, one-way ANOVA). (C) Histograms of impedance change for the tested RBC samples from healthy donors and subjects with SCD or HS. (D) The REI of RBCs from subjects with SCD or HS is also significantly higher compared to that from healthy donors (p < 0.05, one-way ANOVA). (E) The REI significantly correlates with the ROI in the tested samples (PCC = 0.946, p < 0.0001, N = 19). ROI: RBC occlusion index. REI: RBC electrical impedance index. PCC: Pearson correlation coefficient. Error bars represent standard deviation. N = 5 for healthy, N = 12 for SCD, and N = 2 for HS.
RBC-mediated microvascular occlusion and the resultant ROI and REI correlate with clinical hemolytic biomarkers in sickle cell disease
We next explored whether the clinical phenotypes of the study population with SCD affected microvascular occlusion. We found that ROI and REI significantly associated with in vivo biomarkers of hemolysis, including serum LDH levels (Fig. S5A, PCC=0.814, p=0.001) and ARCs (Fig. S5B, PCC=0.582, p=0.047) in the study population with SCD. Next, we assessed whether the electrical impedance change is associated with these biomarkers. Our results indicate that the REI significantly correlates with serum LDH levels (Fig. S5C, PCC=0.698, p=0.012) and ARCs (Fig. S5D, PCC=0.731, p=0.007) in the study subjects with SCD.
ROI and REI as an in vitro therapeutic efficacy benchmark to assess clinical outcome of treatments in sickle cell disease
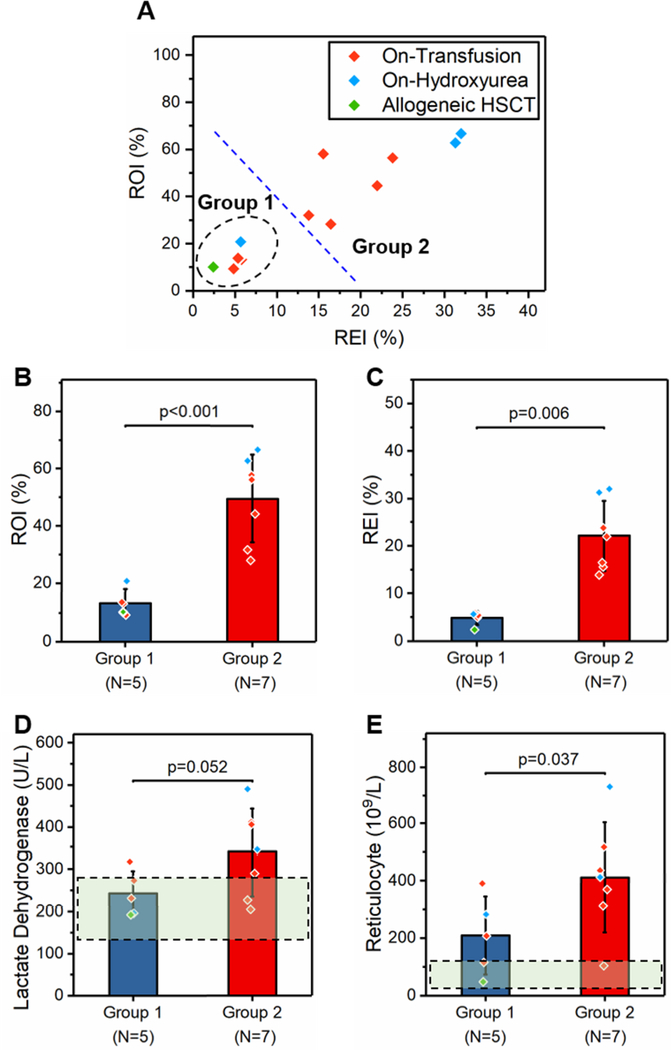
We further assessed whether treatments of the study population with SCD affected microvascular occlusion. We performed mean-shift clustering analysis and identified a sub-group of subjects (Group 1, N=5) with distinct ROI and REI profiles compared to the rest (Group 2, N=7, Fig. 5A). We found that Group 1 subjects had significantly lower levels of ROI (Fig. 5B, mean ± SD = 13.42 ± 4.57% vs. 49.81 ± 15.13%, p<0.001, one-way ANOVA) and REI (Fig. 5C, mean ± SD = 4.84 ± 1.39% vs. 22.13 ± 7.40%, p=0.006, Mann-Whitney) compared to Group 2 subjects. We then determined that among the five Group 1 subjects who had less severe microvascular occlusion, one received allogeneic hematopoietic stem-cell transplantation (HSCT), one was on-hydroxyurea (HU), and the other three were on-transfusion (Fig. 5A). We next explored whether the two groups of subjects differ in terms of other clinical variables. Accordingly, we found that Group 1 subjects had relatively lower serum LDH levels (Fig. 5D, 242 ± 53 vs. 339 ± 105 U/L, p=0.052, Mann-Whitney) and significantly lower ARC levels (Fig. 5E, mean ± SD = 209 ± 136 vs. 412 ± 192 109/L, p=0.037, Mann-Whitney) compared to Group 2 subjects. Comparison of clinical variables between the two groups is summarized in Table S2.
Fig. 5.
MIRCA assessment of RBC-mediated microvascular occlusion as an in vitro therapeutic efficacy benchmark to assess the clinical outcome of treatments in SCD. (A) A subpopulation (group 1, N = 5) with distinct ROI and REI profiles compared to the rest (group 2, N = 7) was identified. Group 1 subjects had significantly lower (B) ROI (p < 0.001, one-way ANOVA) and (C) REI (p = 0.006, Mann–Whitney) compared to group 2 subjects. Moreover, group 1 subjects had relatively lower (D) serum lactate dehydrogenase (LDH) levels (p = 0.052, Mann–Whitney) and (E) absolute reticulocyte counts (ARCs) (p = 0.037, Mann–Whitney) compared to group 2 subjects. The dashed rectangular regions represent typical normal ranges for the given clinical parameters. HSCT: hematopoietic stem-cell transplantation. ROI: RBC occlusion index. REI: RBC electrical impedance index. Error bars represent standard deviation.
DISCUSSION
Results reported in this study suggest that MIRCA, with its integrated electrical impedance-based readout, provides a functional, reproducible in vitro approach for assessing microvascular occlusion associated with abnormal RBC deformability. Glutaraldehyde is a non-specific protein cross-linker, which is commonly used to verify new RBC deformability measurement techniques 62. Our results on glutaraldehyde-stiffened RBCs suggest that MIRCA is deformability-based and is able to discriminate different RBC samples with 1% variation in the fraction of stiff RBCs over the entire RBC population (Fig. 2B&D). However, the RBC deformability resulting from glutaraldehyde stiffening is not comparable to that of pathological RBCs in red cell disorders. Therefore, we further validated MIRCA with samples from people with RBC disorders, namely, SCD and HS. HS, mostly prevalent among northern Europeans, results in a fragile RBC membrane 53, 63. We found that RBCs in two subjects with HS were less deformable, as reflected by a higher ROI and REI compared to healthy RBCs (Fig. 4B&D). SCD, prevalent in the African diaspora, is one of the most common inherited blood disorders worldwide and affects millions of people with considerable morbidity and mortality 64–67. Both the abnormal RBC deformability and the molecular basis of SCD are well established in SCD 68, 69. In accordance with previous studies, we found that the ROI and REI of RBCs from people with SCD were significantly higher, therefore less deformable, compared to RBCs from control subjects (Fig. 4B&D). Interestingly, our results show that RBCs from people with SCD had relatively higher ROI and REI compared to those from the two subjects with HS (Fig. 4B&D). An early study using isotonic ektacytometry revealed that reduction in isotonic RBC deformability is higher in SCD than in HS 70. We postulate that these observations are due to the fact that pathological RBCs from different diseases are affected to different extents.
Electrical measurement is widely adopted in numerous microfluidic designs for biological sample testing, largely due to its simplicity, efficiency, and its potential for adaptation for point-of-care tests 64, 71–78. We have recently developed a novel microfluidic platform, termed OcclusionChip, for assessing RBC-mediated microvascular occlusion in one of our previous studies 79. Compared to the OcclusionChip, MIRCA design has a micropillar array in the downstream with narrower microcapillaries (3 μm compared to 4 μm). This feature may improve the sensitivity of this test for disease in which a subtle fraction of RBCs are abnormal. Flow rate is a key parameter that could affect the measurement. The flow rate here is mediated by inlet pressure, medium viscosity (dominated by hematocrit 67), and microchannel flow resistance. For standardized microfluidic assessment, we employed a digital microfluidic pressure pump to allow a constant inlet pressure, and also carefully adjusted the hematocrit value of RBC suspension at 20% for all the tested samples. Further, the microchannel is 12-μm thick, which is larger than the thickness of RBCs (~ 2 μm). Hence, when RBCs are retained in the microcapillaries, other cells are still able to transit through the microcapillaries (as evidenced in Video S1). This feature, along with the large scale of the micropillar arrays and the two 40-μm-wide side passageways (anastomoses), makes the MIRCA system hard to saturate, largely preventing a potential build-up of flow resistance in the microchannel due to RBC retention. Moreover, the total volume of the blood sample processed within one testing cycle was measured as approximately 40 μL, which translates to a processing rate of 4 million RBCs per min (with an estimated blood density of 1.06 g/mL 80 and 2 million RBCs per μL of blood at 20% hematocrit 81). Such high volume-processing throughput of MIRCA ensures meaningful test results with clinical blood samples.
MIRCA is integrated with surface electrodes for electrical impedance measurement capabilities, and the REI measurements significantly correlate with ROI measurements (Fig. 2E and Fig. 3E), suggesting that REI could serve as a robust indicator of the overall microcapillary occlusion across the device, and that the integrated electrical impedance measurement could replace high-resolution imaging when applying the device in primary healthcare settings. MIRCA provides a finer and more rapid detection compared to previous devices, an alternative functional metric for standardized assessments of RBC mediated microvascular occlusion with no need for high-resolution imaging, with the potential to be developed as a truly small-sized, portable device providing real-time results at the point-of-care.
SCD is a clinically heterogeneous disease, as the clinical phenotypes vary considerably from subject to subject 79, 82, 83. Here, our results show that both the ROI and REI significantly correlate with two in vivo hemolytic biomarkers, serum LDH levels and ARCs (Fig. S5), suggesting that subjects with a more severe intravascular hemolysis are more likely to have RBC-driven microvascular occlusion in SCD. We postulate that two factors may have contributed to these results. Firstly, sickle RBCs have been shown to be vulnerable to mechanical stress and microvesicles shedding-off 84–86, 100, which leads to increased hemolysis, reduced membrane surface area-to-volume ratio, and decreased deformability. Secondly, reticulocytes are known to be less deformable compared to mature RBCs, due to their spherical shape with smaller surface area-to-volume ratio, less optimal organization of membrane lipids and proteins, and more viscous cytoplasm with a mass of chromatin granules 87. The presence of reticulocytosis in a subset of people with SCD, due to hemolytic stress, may contribute to the elevated microcapillary occlusion seen in these studies.
Of note, we observed a cluster of subjects who had significantly lower levels of ROI and REI compared to the rest (Fig. 5A–C) over the study population with SCD. Among these five subjects, one received Allogeneic HSCT, one was on-HU, and the other three were on exchange transfusion (Fig. 5A). We determined that these subjects had less severe RBC-mediated microvascular occlusion as measured by MIRCA, which was consistent with their relatively lower serum LDH levels and significantly lower ARCs (Fig. 5D&E). Exchange transfusion is one of the main therapeutic treatments in SCD, in which normal RBCs from healthy donors are exchanged with sickle RBCs to dilute the concentration of sickle hemoglobin and sickle RBCs in circulation 88. However, vaso-occlusive events can still occur in transfused patients due to the remaining and newly made sickle, non-deformable RBCs. Accordingly, we found that the 8 subjects on transfusion therapy among the study population with SCD had significantly higher ROI and REI compared to healthy donors (Fig. S6). Notably, we did not notice any significant difference when comparing ROI and REI between females and males over the study population (Fig. S7). Together, these analyses demonstrate the promise of MIRCA and the REI as an in vitro therapeutic efficacy benchmark to assess clinical outcomes. MIRCA is likely to significantly benefit the development and assessment of targeted and curative therapies for SCD.
Therapeutic ex vivo gene transfer into autologous hematopoietic stem cells, also known as gene therapy, is currently the most promising approach that can repair the fundamental cause of SCD and provide long-term curative treatment for the patients 89, 90. The National Heart, Lung, and Blood Institute recently launched the ‘Cure Sickle Cell Initiative’ to keep nourishing the collaborative, patient-focused research environment and to support the further development of gene therapy for treating SCD. Close monitoring of the ability of RBCs to clear microcapillaries in MIRCA before and after a curative therapy would provide valuable insights into the patient clinical outcome. In particular, discrepant RBC populations may arise in the patient after receiving a curative therapy, at which time it is crucial to discern the heterogeneity in the entire cell population and its effects 91. In this study, we found that one patient with SCD, who received Allogeneic HSCT (Fig. 5A), had similar levels of ROI and REI compared to healthy donors (ROI: 10.08% vs. 8.02 ± 1.71%; REI: 2.44% vs. 4.31 ± 1.25%). Based on our results, we will test whether, following curative therapy, we will see a transition from high to low microcapillary occlusion and electrical impedance as non-sickling RBCs replace sickle RBCs in vivo.
SARS-CoV-2, a new RNA coronavirus leading to a global pandemic, is the etiological driver of the clinical syndrome coronavirus disease 2019 (COVID-19) 92, 93. The disease is characterized by a number of manifestations including persistent dry cough, shortness of breath, hypoxemia, and fever 94, 95. In SCD, COVID-19 may trigger severe acute chest syndrome (ACS) and vaso-occlusive crisis 96. Since RBCs play an important role in oxygen delivery, it is plausible to suspect that their biophysical properties, such as deformability, are deleteriously altered and thus contribute to congestions of microvessels in COVID-19. Importantly, a recent study showed that COVID-19 is linked with significantly less deformable RBCs 97, even though the underlying mechanism is yet to be uncovered. Within the context of COVID-19, we envision that MIRCA and the REI may provide a functional biomarker to supplement the current diagnostic tools for disease assessment and monitoring.
A limitation of this study is that when applying the ROI and REI in other microfluidic capillary networks for assessment of pathological RBC deformability, their absolute values may be affected by experimental parameters such as inlet pressure, duration of sample perfusion, and design of the microfluidic capillary networks. Future studies may prospectively focus on characterizing the intrinsic electrical properties of pathological RBCs from a wider range of red cell disorders, including malaria and thalassemia, which will provide more insights into disease pathophysiology and assist the assessment of RBC mediated microvascular occlusion through electrical impedance measurement.
CONCLUSIONS
In this study, a novel microfluidic device integrated with capillary network-inspired micropillar matrices and electrodes for electrical impedance measurement (MIRCA) is presented for standardized assessment of RBC-mediated microvascular occlusion. Healthy RBCs, glutaraldehyde-stiffened RBCs, and RBCs from subjects with two red cell disorders, SCD and HS, were used to validate MIRCA. Two new highly correlated parameters, ROI and REI, have been defined, which represent the cumulative percentage occlusion and the cumulative percentage impedance change, respectively, across the device. Unique features of MIRCA include processing of clinical blood samples at near-physiologic hematocrit (20%), rapid (less than 5 min) data acquisition and analysis, and REI as an alternative functional metric for occlusion assessment in the absence of high-resolution imaging and as a functional test for microvascular health and disease status monitoring. Future work will focus on adapting the microfluidic device for point-of-care use in resource-limited settings and improving the repeatability of portable analyses when performed by minimally trained personnel in the field. The clinical utility of MIRCA and the REI as an in vitro therapeutic efficacy benchmark in a larger patient population with a wider range of treatments, including emerging therapies with curative intent, warrants future investigation.
Supplementary Material
ACKNOWLEDGEMENTS
This work is supported by National Science Foundation (NSF) Career Award 1552782, National Heart, Lung, and Blood Institute (NHLBI) awards: R01HL133574, U01HL117659, T32HL134622, the Cure Sickle Cell Initiative (OT2HL152643), and American Heart Association 17GRNT33661005 grants. Authors acknowledge with gratitude the contributions of patients and clinicians at University Hospitals Cleveland Medical Center. The authors gratefully acknowledge Laurie Dudik’s help on the device substrate fabrication at the Electronics Design Center, Case Western Reserve University.
Footnotes
CONFLICT OF INTEREST
A patent application has been filed by Case Western Reserve University for this technology.
REFERENCES
- 1.Diez-Silva M, Dao M, Han JY, Lim CT and Suresh S, Mrs Bull, 2010, 35, 382–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tomaiuolo G, Biomicrofluidics, 2014, 8, 051501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo P and Fu BM, Journal of biomechanical engineering, 2012, 134, 041003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lacelle PL, Semin Hematol, 1970, 7, 355–371. [PubMed] [Google Scholar]
- 5.Mohandas N, Clark MR, Jacobs MS and Shohet SB, J Clin Invest, 1980, 66, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sisquella X, Nebl T, Thompson JK, Whitehead L, Malpede BM, Salinas ND, Rogers K, Tolia NH, Fleig A, O’Neill J, Tham WH, David Horgen F and Cowman AF, eLife, 2017, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dondorp AM, Kager PA, Vreeken J and White NJ, Parasitol Today, 2000, 16, 228–232. [DOI] [PubMed] [Google Scholar]
- 8.Alapan Y, Matsuyama Y, Little JA and Gurkan UA, Technology, 2016, 4, 71–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huisjes R, Bogdanova A, van Solinge WW, Schiffelers RM, Kaestner L and van Wijk R, Front Physiol, 2018, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alapan Y, Kim C, Adhikari A, Gray KE, Gurkan-Cavusoglu E, Little JA and Gurkan UA, Translational research : the journal of laboratory and clinical medicine, 2016, 173, 74–91 e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kucukal E, Little JA and Gurkan UA, Integr Biol-Uk, 2018, 10, 194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim M, Alapan Y, Adhikari A, Little JA and Gurkan UA, Microcirculation, 2017, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carden MA, Fay ME, Lu X, Mannino RG, Sakurai Y, Ciciliano JC, Hansen CE, Chonat S, Joiner CH, Wood DK and Lam WA, Blood, 2017, 130, 2654–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu Y, Ahn B, Sakurai Y, Hansen CE, Tran R, Mimche PN, Mannino RG, Ciciliano JC, Lamb TJ, Joiner CH, Ofori-Acquah SF and Lam WA, Nature biomedical engineering, 2018, 2, 453–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Praljak N, Iram S, Goreke U, Singh G, Hill A, Gurkan UA and Hinczewski M. J. b., bioRxiv, 2020, DOI: 10.1101/2020.07.01.181545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perrotta S, Gallagher PG and Mohandas N, Lancet, 2008, 372, 1411–1426. [DOI] [PubMed] [Google Scholar]
- 17.Frei AC, Guo Y, Jones DW, Pritchard KA Jr., Fagan KA, Hogg N and Wandersee NJ, Blood, 2008, 112, 398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Da Costa L, Galimand J, Fenneteau O and Mohandas N, Blood reviews, 2013, 27, 167–178. [DOI] [PubMed] [Google Scholar]
- 19.Manwani D and Frenette PS, Blood, 2013, 122, 3892–3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beare NA, Harding SP, Taylor TE, Lewallen S and Molyneux ME, The Journal of infectious diseases, 2009, 199, 263–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lekka M, Fornal M, Pyka-Fosciak G, Lebed K, Wizner B, Grodzicki T and Styczen J, Biorheology, 2005, 42, 307–317. [PubMed] [Google Scholar]
- 22.Henon S, Lenormand G, Richert A and Gallet F, Biophys J, 1999, 76, 1145–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuzman D, Svetina S, Waugh RE and Zeks B, Eur Biophys J Biophy, 2004, 33, 1–15. [DOI] [PubMed] [Google Scholar]
- 24.Evans EA, Waugh R and Melnik L, Biophys J, 1976, 16, A167–A167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Streekstra GJ, Dobbe JGG and Hoekstra AG, Opt Express, 2010, 18, 14173–14182. [DOI] [PubMed] [Google Scholar]
- 26.Baskurt OK, Hardeman MR, Uyuklu M, Ulker P, Cengiz M, Nemeth N, Shin S, Alexy T and Meiselman HJ, Biorheology, 2009, 46, 251–264. [DOI] [PubMed] [Google Scholar]
- 27.Llaudet-Planas E, Vives-Corrons JL, Rizzuto V, Gomez-Ramirez P, Sevilla Navarro J, Coll Sibina MT, Garcia-Bernal M, Ruiz Llobet A, Badell I, Velasco-Puyo P, Dapena JL and Manu-Pereira MM, International journal of laboratory hematology, 2018, 40, 94–102. [DOI] [PubMed] [Google Scholar]
- 28.Rab MA, Kanne CK, van Oirschot BA, Bos JF, Houwing ME, Cnossen MH, Schutgens R, Pasterkamp G, van Wijk R and Sheehan VA, Blood, 2018, 132, 2360–2360. [Google Scholar]
- 29.Tomaiuolo G, Barra M, Preziosi V, Cassinese A, Rotoli B and Guido S, Lab Chip, 2011, 11, 449–454. [DOI] [PubMed] [Google Scholar]
- 30.Lee SS, Yim Y, Ahn KH and Lee SJ, Biomed Microdevices, 2009, 11, 1021–1027. [DOI] [PubMed] [Google Scholar]
- 31.Guruprasad P, Mannino RG, Caruso C, Zhang HQ, Josephson CD, Roback JD and Lam WA, Am J Hematol, 2019, 94, 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zheng Y, Chen J, Cui T, Shehata N, Wang C and Sun Y, Lab Chip, 2014, 14, 577–583. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Y, Cachia MA, Ge J, Xu ZS, Wang C and Sun Y, Lab Chip, 2015, 15, 3138–3146. [DOI] [PubMed] [Google Scholar]
- 34.Cluitmans JC, Chokkalingam V, Janssen AM, Brock R, Huck WT and Bosman GJ, Biomed Res Int, 2014, 764268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu Z, Zheng Y, Wang X, Shehata N, Wang C and Sun Y, Microsyst Nanoeng, 2018, 4, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sakuma S, Kuroda K, Tsai C-HD, Fukui W, Arai F and Kaneko M, Lab Chip, 2014, 14, 1135–1141. [DOI] [PubMed] [Google Scholar]
- 37.Rendell M, Luu T, Quinlan E, Knox S, Fox M, Kelly S and Kahler K, Biochim Biophys Acta, 1992, 1133, 293–300. [DOI] [PubMed] [Google Scholar]
- 38.Rendell M, Fox M, Knox S, Lastovica J, Kirchain W and Meiselman HJ, J Lab Clin Med, 1991, 117, 500–504. [PubMed] [Google Scholar]
- 39.Zheng Y, Shojaei-Baghini E, Azad A, Wang C and Sun Y, Lab Chip, 2012, 12, 2560–2567. [DOI] [PubMed] [Google Scholar]
- 40.Adamo A, Sharei A, Adamo L, Lee B, Mao S and Jensen KF, Anal Chem, 2012, 84, 6438–6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bow H, Pivkin IV, Diez-Silva M, Goldfless SJ, Dao M, Niles JC, Suresh S and Han J, Lab Chip, 2011, 11, 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Islamzada E, Matthews K, Guo Q, Santoso AT, Duffy SP, Scott MD and Ma H, Lab Chip, 2019, 20, 226–235. [DOI] [PubMed] [Google Scholar]
- 43.Ciciliano JC, Abbaspour R, Woodall J, Wu C, Bakir MS and Lam WA, Lab Chip, 2017, 17, 3804–3816. [DOI] [PubMed] [Google Scholar]
- 44.Guo Q, Duffy SP, Matthews K, Deng X, Santoso AT, Islamzada E and Ma H, Lab Chip, 2016, 16, 645–654. [DOI] [PubMed] [Google Scholar]
- 45.Santoso AT, Deng XY, Lee JH, Matthews K, Duffy SP, Islamzada E, McFaul SM, Myrand-Lapierre ME and Ma HS, Lab Chip, 2015, 15, 4451–4460. [DOI] [PubMed] [Google Scholar]
- 46.Guo Q, Park S and Ma H, Lab Chip, 2012, 12, 2687–2695. [DOI] [PubMed] [Google Scholar]
- 47.Duez J, Holleran JP, Ndour PA, Loganathan S, Amireault P, Francais O, El Nemer W, Le Pioufle B, Amado IF, Garcia S, Chartrel N, Le Van Kim C, Lavazec C, Avery VM and Buffet PA, Antimicrobial agents and chemotherapy, 2015, 59, 4206–4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diakite SAS, Ndour PA, Brousse V, Gay F, Roussel C, Biligui S, Dussiot M, Prendki V, Lopera-Mesa TM, Traore K, Konate D, Doumbia S, Cros J, Dokmak S, Fairhurst RM, Diakite M and Buffet PA, Malaria J, 2016, 15, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Man Y, Kucukal E, An R, Bode A, Little JA and Gurkan UA, Microcirculation, 2020, e12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Q, Reiling SJ, Rohrbach P and Ma H, Lab Chip, 2012, 12, 1143–1150. [DOI] [PubMed] [Google Scholar]
- 51.Buffet PA, Safeukui I, Deplaine G, Brousse V, Prendki V, Thellier M, Turner GD and Mercereau-Puijalon O, Blood, 2011, 117, 381–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shelby JP, White J, Ganesan K, Rathod PK and Chiu DT, P Natl Acad Sci USA, 2003, 100, 14618–14622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Picot J, Ndour PA, Lefevre SD, El Nemer W, Tawfik H, Galimand J, Da Costa L, Ribeil JA, de Montalembert M, Brousse V, Le Pioufle B, Buffet P, Le Van Kim C and Francais O, Am J Hematol, 2015, 90, 339–345. [DOI] [PubMed] [Google Scholar]
- 54.Du E, Diez-Silva M, Kato GJ, Dao M and Suresh S, Proceedings of the National Academy of Sciences of the United States of America, 2015, 112, 1422–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiang Y, Liu J, Dieujuste D and Du E, bioRxiv, 2020. [Google Scholar]
- 56.Hudetz AG, Microcirculation, 1997, 4, 233–252. [DOI] [PubMed] [Google Scholar]
- 57.Delaney JP, Am J Physiol, 1969, 216, 1556–1561. [DOI] [PubMed] [Google Scholar]
- 58.Derpanis KG, Lecture Notes, 2005, 32. [Google Scholar]
- 59.Comaniciu D and Meer P, IEEE Transactions on Pattern Analysis Machine Intelligence, 2002, 24, 603–619. [Google Scholar]
- 60.Pantaleo A, Ferru E, Giribaldi G, Mannu F, Carta F, Matte A, De Franceschi L and Turrini F, Biochem J, 2009, 418, 359–367. [DOI] [PubMed] [Google Scholar]
- 61.Forsyth AM, Wan JD, Ristenpart WD and Stone HA, Microvasc Res, 2010, 80, 37–43. [DOI] [PubMed] [Google Scholar]
- 62.Sosa JM, Nielsen ND, Vignes SM, Chen TG and Shevkoplyas SS, Clin Hemorheol Micro, 2014, 57, 275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eber S and Lux SE, 2004. [Google Scholar]
- 64.Alapan Y, Little JA and Gurkan UA, Scientific reports, 2014, 4, 7173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Platt OS, Brambilla DJ, Rosse WF, Milner PF, Castro O, Steinberg MH and Klug PP, The New England journal of medicine, 1994, 330, 1639–1644. [DOI] [PubMed] [Google Scholar]
- 66.Man Y, Goreke U, Kucukal E, Hill A, An R, Liu S, Bode A, Solis-Fuentes A, Nayak LV, Little JA and Gurkan UA, Blood cells, molecules & diseases, 2020, 83, 102424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kucukal E, Man Y, Hill A, Liu S, Bode A, An R, Kadambi J, Little JA and Gurkan UA, Am J Hematol, 2020, 1246–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Noomuna P, Risinger M, Zhou S, Seu K, Man Y, An R, Sheik DA, Wan J, Little JA, Gurkan UA, Turrini FM, Kalfa T and Low PS, British journal of haematology, 2020, DOI: 10.1111/bjh.16671, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pace BS, Ofori-Acquah SF and Peterson KR, Anemia, 2012, 2012, 143594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Renoux C, Faivre M, Bessaa A, Da Costa L, Joly P, Gauthier A and Connes P, Sci Rep, 2019, 9, 6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maji D, Suster MA, Kucukal E, Sekhon UDS, Gupta AS, Gurkan UA, Stavrou EX and Mohseni P, IEEE transactions on biomedical circuits and systems, 2017, 11, 1459–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maji D, Nayak L, Martin J, Sekhon UDS, Sen Gupta A, Mohseni P, Suster MA and Ahuja SP, Haemophilia : the official journal of the World Federation of Hemophilia, 2019, 25, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maji D, De La Fuente M, Kucukal E, Sekhon UDS, Schmaier AH, Sen Gupta A, Gurkan UA, Nieman MT, Stavrou EX, Mohseni P and Suster MA, Journal of thrombosis and haemostasis : JTH, 2018, 16, 2050–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu J, Qiang Y, Alvarez O and Du E, Sensors and actuators. B, Chemical, 2018, 255, 2392–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zheng Y, Nguyen J, Wang C and Sun Y, Lab Chip, 2013, 13, 3275–3283. [DOI] [PubMed] [Google Scholar]
- 76.Dou W, Wang L, Malhi M, Liu H, Zhao Q, Plakhotnik J, Xu Z, Huang Z, Simmons CA, Maynes JT and Sun Y, Biosensors & bioelectronics, 2021, 175, 112875. [DOI] [PubMed] [Google Scholar]
- 77.Dou W, Zhao Q, Malhi M, Liu X, Zhang Z, Wang L, Masse S, Nanthakumar K, Hamilton R, Maynes JT and Sun Y, Biosensors & bioelectronics, 2020, 167, 112468. [DOI] [PubMed] [Google Scholar]
- 78.An R, Wipf DO and Minerick AR, Biomicrofluidics, 2014, 8, 021803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Man Y, Kucukal E, An R, Watson Q, Bosch J, Zimmerman PA, Little JA and Gurkan UA, Lab Chip, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vitello DJ, Ripper RM, Fettiplace MR, Weinberg GL and Vitello J. M. J. J. o. v. m., 2015, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Judy HE and Price NB, Journal of the American Medical Association, 1958, 167, 563–566. [DOI] [PubMed] [Google Scholar]
- 82.Habara A and Steinberg MH, Exp Biol Med (Maywood), 2016, 241, 689–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kucukal E, Man Y, Quinn E, Tewari N, An R, Ilich A, Key NS, Little JA and Gurkan UA, Blood Adv, 2020, 4, 3688–3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lizarralde Iragorri MA, El Hoss S, Brousse V, Lefevre SD, Dussiot M, Xu T, Ferreira AR, Lamarre Y, Silva Pinto AC, Kashima S, Lapoumeroulie C, Covas DT, Le Van Kim C, Colin Y, Elion J, Francais O, Le Pioufle B and El Nemer W, Lab Chip, 2018, 18, 2975–2984. [DOI] [PubMed] [Google Scholar]
- 85.Ferru E, Giger K, Pantaleo A, Campanella E, Grey J, Ritchie K, Vono R, Turrini F and Low PS, Blood, 2011, 117, 5998–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nader E, Romana M and Connes P, Frontiers in immunology, 2020, 11, 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xie L, Jiang Y, Yao W, Gu L, Sun D, Ka W, Wen Z and Chien S, J Biomech, 2006, 39, 530–535. [DOI] [PubMed] [Google Scholar]
- 88.Josephson CD, Su LL, Hillyer KL and Hillyer CD, Transfus Med Rev, 2007, 21, 118–133. [DOI] [PubMed] [Google Scholar]
- 89.Ribeil JA, Hacein-Bey-Abina S, Payen E, Magnani A, Semeraro M, Magrin E, Caccavelli L, Neven B, Bourget P, El Nemer W, Bartolucci P, Weber L, Puy H, Meritet JF, Grevent D, Beuzard Y, Chretien S, Lefebvre T, Ross RW, Negre O, Veres G, Sandler L, Soni S, de Montalembert M, Blanche S, Leboulch P and Cavazzana M, The New England journal of medicine, 2017, 376, 848–855. [DOI] [PubMed] [Google Scholar]
- 90.Esrick EB and Bauer DE, Semin Hematol, 2018, 55, 76–86. [DOI] [PubMed] [Google Scholar]
- 91.Demirci S, Uchida N and Tisdale JF, Cytotherapy, 2018, 20, 899–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC and Zhang YZ, Nature, 2020, 579, 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yuki K, Fujiogi M and Koutsogiannaki S, Clin Immunol, 2020, 215, 108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Herrmann J, Mori V, Bates JHT and Suki B, Nat Commun, 2020, 11, 4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thomas T, Stefanoni D, Dzieciatkowska M, Issaian A, Nemkov T, Hill RC, Francis RO, Hudson KE, Buehler PW and Zimring JC, Journal of Proteome Research, 2020, 19, 4455–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nur E, Gaartman AE, van Tuijn CF, Tang MW and Biemond BJ, Am J Hematol, 2020, 95, 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kubankova M, Hohberger B, Hoffmanns J, Fuerst J, Hermann M, Guck J and Krater M, bioRxiv, 2021, DOI: 10.1101/2021.02.12.429482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Man Y, Maji D, An R, Ahuja SP, Little JA, Mohseni P, Suster MA and Gurkan UA, Blood, 2020, 136, 10. [Google Scholar]
- 100.An R, Man Y, Kucukal E, Cheng K, Wulftange WJ, Little JA and Gurkan UA, Blood, 2020, 136, 13–14. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.