Abstract
TSC1 or TSC2 mutations cause Tuberous Sclerosis Complex (TSC), and lead to mechanistic target of rapamycin (mTOR) hyperactivation evidenced by hyperphosphorylation of ribosomal S6 protein and 4-elongation factor binding protein (4E-BP1). Amino acid (AA) levels modulate mTOR-dependent S6 and 4E-BP1 phosphorylation in non-neural cells, but this has not been investigated in neurons. The effects of AA levels on mTOR signaling and S6 and 4E-BP1 phosphorylation were analyzed in Tsc2 and Depdc5 (a distinct mTOR regulatory gene associated with epilepsy) CRISPR-edited Neuro2a (N2a) cells and differentiated neurons. Tsc2 or Depdc5 knockout (KO) led to S6 and 4E-BP1 hyperphosphorylation and cell soma enlargement, but while Tsc2 KO N2a cells exhibited reduced S6 phosphorylation (Ser240/244) and cell soma size after incubation in AA free (AAF) media, Depdc5 KO cells did not. Using a CFP/YFP FRET-biosensor coupled to 4E-BP1, we assayed 4E-BP1 phosphorylation in living N2a cells and differentiated neurons following Tsc2 or Depdc5 KO. AAF conditions reduced 4E-BP1 phosphorylation in Tsc2 KO N2a cells but had no effect in Depdc5 KO cells. Rapamycin blocked S6 protein phosphorylation but had no effect on 4E-BP1 phosphorylation, following either Tsc2 or Depdc5 KO. Confocal imaging demonstrated that AAF media promoted movement of mTOR off the lysosome, functionally inactivating mTOR, in Tsc2 KO but not Depdc5 KO cells, demonstrating that AA levels modulate lysosomal mTOR localization and account, in part, for differential effects of AAF conditions following Tsc2 versus Depdc5 KO. AA levels and rapamycin differentially modulate S6 and 4E-BP1 phosphorylation and mTOR lysosomal localization in neurons following Tsc2 KO versus Depdc5 KO. Neuronal mTOR signaling in mTOR associated epilepsies may have distinct responses to mTOR inhibitors and to levels of cellular amino acids.
Keywords: tuberous sclerosis complex, cortical malformations, epilepsy, mTORopathies, GATOR1
Introduction
Tuberous Sclerosis Complex (TSC) is a multi-system disorder caused by mutations inTSC1 or TSC2 (Crino et al., 2006) that is highly associated with epilepsy, autism, and intellectual disability. TSC1 or TSC2 mutations cause hyperactivation of the mechanistic target of rapamycin (mTOR) signaling pathway and lead to malformations of cortical development (MCD) linked to the neuropsychiatric phenotypes in TSC; TSC2 variants cause a more severe neurological phenotype than TSC1 (Curatolo, 2015; Curatolo et al., 2015). The association between mTOR complex 1 (mTORC1) hyperactivation, evidenced by enhanced p70S6kinase (S6K) and ribosomal protein S6 (S6) phosphorylation, and abnormalities in neuronal size, axon outgrowth, dendritic arborization, and excitability has been demonstrated in TSC mouse models (Choi et al., 2008; Zeng et al., 2011; Crino, 2013; Lafourcade et al., 2013; Tsai et al., 2014; Crowell et al., 2015). Many of the anatomic and behavioral abnormalities in Tsc1 and Tsc2 conditional KO mice are reduced with the mTOR inhibitor rapamycin (sirolimus).
In addition to TSC1/TSC2, variants in additional mTOR pathway genes have been linked to MCD and epilepsy (so-called “mTORopathies”). Among these, Dep Domain Containing 5 (DEPDC5) is the most common mutation found in focal MCD associated with epilepsy (Baldassari et al., 2019). Like TSC1/TSC2, DEPDC5 mutations cause mTORC1 hyperactivation in neurons and thus treatment with mTOR inhibitors is under consideration for epilepsy associated with DEPDC5 variants. However, clinical trials of mTOR inhibitors in TSC have yielded only modest efficacy for epilepsy (French et al., 2016) suggesting that these compounds may not fully rescue the network consequences of hyperactivated mTORC1 signaling. Furthermore, it is unclear whether mTOR hyperactivation driven by all mTOR pathway gene mutations will respond similarly to mTOR inhibitors.
Apart from S6K and S6, mTORC1 phosphorylates the translational repressor eukaryotic initiation factor 4E binding protein-1 (4E-BP1; Thr 37/46), which in turn releases elongation initiation factor 4E (eIF4E) from inhibition and fosters 5’-cap dependent protein translation associated with cell growth. Recently, a constitutively activated 4E-BP1 construct resistant to mTORC1 phosphorylation rescued cell soma enlargement, altered dendritic complexity, and cortical dyslamination caused by transfection of constitutively activated Rheb (RhebCA), an mTOR activator, into the mouse embryonic cerebral cortex (Lin et al., 2016). Knockdown of S6K did not rescue cortical dyslamination in this strain suggesting independent effects of S6K and 4E-BP1 on brain structure. Similarly, S6K knockdown did not rescue cortical dyslamination in a rat strain with constitutively activated mTOR (Kassai et al., 2014). Interestingly, despite strong mTORC1 kinase activity targeting Thr37/46 of 4E-BP1 in HEK293T cells (Kang et al., 2013), rapamycin has limited effect preventing 4E-BP1 Thr37/46 phosphorylation in vitro. Thus, while rapamycin may block some mTOR signal effects mediated via S6K, unopposed effects of 4E-BP1 phosphorylation may have functional consequences in neurons.
In non-neural cells, cellular amino acid levels regulate mTORC1 kinase activity by modulating the subcellular localization of mTORC1 (Bar-Peled et al., 2013); this effect in neurons has not been fully defined. Optimal kinase activity is achieved when mTORC1 is tethered to the lysosomal membrane. In non-neural cells, several protein complexes e.g., CASTOR, Sestrins, GATOR1, and KICSTOR synergistically modulate the location of mTORC1 on the lysosome, and in turn mTORC1 kinase activity, in response to intracellular amino acid levels (Wolfson and Sabatini, 2017). In HEK293T cells, when amino acids are replete, signaling from GATOR2 to GATOR1 allows active mTORC1 to remain attached to the lysosome (Bar-Peled et al., 2013; Sancak and Sabatini, 2009). Conversely, when intracellular amino acid levels are low, GATOR1 exerts GAP activity on lysosomal RAG proteins thereby preventing mTORC1from contacting the lysosomal surface in non-neural cells. Despite much work in non-neural cells, the movement of mTORC1 onto and off of the lysosome, the role of amino acids on mTORC1 signaling, and the effects of altered amino acid levels on 4E-BP1 phosphorylation has not been comprehensively studied in neurons. Previous work in neurons demonstrated that knockdown of the GATOR1 subunits Depdc5 and Nprl3 leads to mTOR hyperactivation (S6 phosphorylation) even under amino acid depletion conditions (Iffland et al., 2018) suggesting differential effects of nutrient deprivation on mTOR activation. Rapamycin has no effect on the subcellular localization of mTORC1 in non-neural cells (Sancak and Sabatini, 2009) and thus, both unchecked 4E-BP1 phosphorylation and lysosomal mTOR localization persist despite pharmacological mTOR inhibition.
Thus, as a strategy to define how nutrients govern mTOR activation in neurons, we examined phosphorylation of S6 protein (PS6) and 4E-BP1 in the setting of amino acid deprivation following CRISPR editing of Tsc2 or Depdc5. To assess dynamic changes in mTOR activation in living neurons, we implemented a novel FRET-based assay with live-cell confocal microscopy to visualize changes in 4E-BP1 phosphorylation and mTOR subcellular localization in mouse neuroblastoma (N2a) cells and differentiated neurons.
Materials and Methods
CRISPR/Cas9 construct generation and validation
Guide RNA targeting the spCas9 endonuclease to regions in the mouse genome encoding Tsc2 and Depdc5 were calculated in silico using ChopChop software (chopchop.cbu.uib.no). A scramble gRNA (-GACTACCAGAGCTAACTCA-) was used as a transfection and gRNA control. In silico guide RNAs were then assembled into oligonucleotides (Integrated DNA Technologies, Coralville, IA), annealed using ligase buffer (Promega, Madison, WI) at 98°C for 5 min. Annealed gRNA were then sub-cloned into PX330-based plasmid (addgene #48138) using Golden Gate Assembly containing an mCherry reporter linked to Cas9 via a T2a multicistronic element. Plasmid assembly was confirmed by Sanger sequencing (Genewiz, South Plainfield, NJ).
To validate that our gRNA containing CRISPR/Cas9 plasmid created indels in our regions of interest, DNA from Tsc2, Depdc5, and scramble FAC-sorted cells lines (as described below) as well as wildtype (WT) N2aC was assayed for mis-matched DNA pairs (EnGen Mutation Detection Kit; New England Biolabs, Ipswich, MA) with PCR primers targeted towards our genomic region of interest (Integrated DNA technologies, Coralville, IA).
Cell Culture and establishment of cell lines
Neuro2a cells (N2aC; Sigma-Aldrich, St. Louis, MO) were selected for analysis because they express neuronal marker proteins (Figure S10), have a high transfection efficiency, are readily FAC sorted, and can be rapidly differentiated into neurons. N2aC were cultured in complete medium consisting of EMEM (Invitrogen, Carlsbad, CA) supplemented with 10% FBS (Invitrogen, Carlsbad, CA). To create stable CRISPR/Cas9 edited cell lines, N2aC were transfected using Lipofectamine LTX with Plus reagent (ThermoFisher Scientific, Waltham, MA) and 30 μg of plasmid diluted in 300 μl Opti-MEM (Invitrogen, Carlsbad, CA) for 48 hours. Cotransfection experiments were performed by using 30 μg of each plasmid. After 48hrs of transfection, cells were trypsinized (0.25%), centrifuged, washed with ice-cold PBS, passed through a cell strainer into a 5 ml conical tube and assayed by flow cytometry (University of Maryland School of Medicine Flow Cytometry Core) for sorting based on mCherry (Cas9) fluorescence (BD FACSAria II cell sorter; Becton Dickinson and Company, Franklin Lakes, NJ). mCherry+ sorted cells were placed into PBS containing 1% serum until re-plating. Cells were replated in complete media and grown to confluence.
N2a Cell Differentiation
Undifferentiated N2aC, both WT and KO cell lines, were plated onto glass bottom video dishes and allowed to adhere for 24 hrs. After 24hrs, cells were incubated in EMEM media containing 0.5% fetal bovine serum and 20 μM retinoic acid (Millipore Sigma, St Louis, MO) for 24 hrs. After incubation in retinoic acid containing media, N2aC display a change in cell body morphology and extend MAP2 positive neurites, confirmed in live cells using an ZOE Fluorescent Cell Imager (Bio-Rad, Hercules, CA). After confirming neurite extension, cells were transfected with mechanistic target of rapamycin activity reporter (TORCAR) as described above.
Quantitative and Statistical analysis of cell size data
Cell soma diameter was measured in digital images of CRISPR-edited, scramble control, or WT N2aC lines in ImageJ (Schneider et al., 2012). Each measurement was taken using the longest dimension of each soma in 50 total cells (25 cells in 2 replicates). To define the mTOR dependency of cell size changes, cells were incubated with the mTOR inhibitor rapamycin (50 nM, 24hrs, Cell Signaling Technologies, Danvers, MA), torin1, a specific ATP-competitive dual kinase inhibitor of mTORC1 and mTORC2 (50 nM; 24 hrs; Torcris, Bristol, UK), or amino acid free (AAF) media (1 hr., DMEM, US Biological, Salem, MA) using DMSO as a vehicle control. Statistical analysis of cell size differences was assessed in Origin (Northampton, MA) using one-way ANOVA (p < 0.05 considered statistically significant). Measurements were graphed using a box and whisker plot where the box represents the standard error of the mean and the whiskers represent the 5–95% confidence interval.
Analysis of mTORC1 activity using live-cell imaging
N2aC were plated on video dishes coated with poly-L-lysine. Cells were grown to 30–40% confluence and co-transfected with TORCAR (pcDNA3-Lyso-TORCAR was a gift from J. Zhang; Addgene plasmid # 64929; http://n2t.net/addgene:64929; RRID:Addgene_64929) (Zhou et al., 2015; Zhou et al., 2016) and CRISPR-Cas9 Tsc2 or Depdc5 KO plasmids, scramble plasmid or left as wildtype cells. Cells were transfected with Lipofectamine LTX with Plus enhancing reagent (Thermofisher) in optimum and serum-free physiological media. For each chamber slide approximately 30 μg of each plasmid was used. Cells were transfected for 48 hours. After 48 hours, transfection media was removed, and warm complete imaging media was washed on. Cells were cultured for an additional 24 hours in complete DMEM imaging media (FluoroBrite DMEM, ThermoFisher Scientific, Waltham, MA).
Cells were imaged on a Zeiss LSM Duo slit scanning confocal microscope with lasers allowing us to visualize CFP, YFP and RFP channels. Cells were first selected by Cas9 (red) expression followed by determination of TORCAR transfection (green). Images were taken at 40x magnification simultaneously recording fluorescence from the CFP and YFP FRET channels using time-lapse imaging at two-minute intervals for 60 minutes. First, a 10-minute baseline in complete imaging media was established and then experimental media was washed on. Experimental conditions were defined as follows: 1) complete media baseline, 2) AAF imaging media (no phenol red; US Biologicals, Salem, MA) with or without glucose (5.5 mM), 3) complete imaging media with rapamycin (150 nM), 4) complete imaging media with torin1 (100 nM), 5) complete imaging media with rapamycin followed by washing on AAF media with rapamycin (150 nM), 6) complete imaging media with torin1 followed by washing on AAF media with torin1 (100 nM), 7) complete imaging media followed by washing on AAF media and then by washing on complete imaging media again, or 8) complete imaging media containing 150, 250, 350, 450, or 550 nM rapamycin (n=5 cells imaged in each group).
After recording CFP and YFP fluorescence data, the CFP to YFP ratio (C:Y) was calculated (Zhou et al., 2016), averaged across 5 cells per group and a best-fit line was generated in Origin statistical software (Northampton, MA). For experiments with one media change a linear best-fit line was generated. For experiments with multiple media changes a polynomial bet-fit line was generated. For each group, the individual C:Y for each cell is graphed (blue) along with the average C:Y (dashed black line) and best-fit line (red). All data were normalized to account for focal plane changes and differences in transfection efficiency. Percent change was also calculated and is defined as the average of data points recorded during the 10 minute baseline compared to the last data point recorded for each cell. These values were also used to calculate standard error. Statistical differences were determined using a one-way ANOVA with a p value of < 0.05 deemed significant. Statistical significance was determined using the average C:Y ratio during each 10 minute baseline and compared to the overall change in C:Y for each experimental manipulation. A representative video of the time-lapse recording can be found in the supplemental material (Movie S1).
Immunocytochemistry
N2aC were fixed in 4% PFA at room temperature for 20 minutes and then permeabilized in phosphate-buffered saline (PBS) containing 0.3% Triton X-100 (ThermoFisher Scientific, Waltham, MA). Cells were blocked for 2 hours at room temperature (RT) in 5% normal goat serum (Jackson ImmunoResearch, West Grove, PA). Cells were incubated in one of the following primary antibodies in blocking solution containing 5% normal serum at 4°C overnight: F-actin (1:1000; Abcam, Cambridge, UK), mTOR (1:1000; Abcam, Cambridge, UK), lysosome associated membrane protein 2 (LAMP2; 1:1000; Thermofisher, Waltham, MA), microtubule associate protein 2 (MAP2; 1:1000; Abcam, Cambridge, UK), Tau (1:500; Abcam, Cambridge, UK), Nestin (1:1,000; Proteintech, Rosemont, IL), tubulin beta III isoform (TUJ1; 1:500; Millipore, Darmstadt, Germany), or tyrosine hydroxylase (TH; 1:1,000, Abcam, Cambridge, UK). The secondary antibody, containing a fluorochrome (Alexa Fluor 488 or Alexa Fluor 594; all 1:100 dilution; Molecular Probes, Eugene, OR), was incubated with the cells for 2 hours at RT. DAPI counterstain was used to visualize nuclei and was applied along with mounting media during coverslipping. Apoptosis and necrosis assays were performed using WT live N2aC plated in chamber slides using the Annexin V-FITC Early Apoptosis Detection Kit (Cell Signaling Technology, Danvers, MA). Experimental N2aC were incubated in AAF media for 60 minutes prior to imaging. As a positive control for apoptosis and necrosis, 25 nM Etoposide was incubated with WT N2aC for 24 hrs. As a negative control, WT N2aC were used after incubation in complete media.
Western blot analysis
Cells were lysed in RIPA lysis buffer (50 mM Tris HCl, pH 8.0; 150 mM NaCl; 1% NP-40; 0.5% sodium deoxycholate; and 0.1% SDS) with a protease (Sigma-Aldrich, St. Louis, MO) and phosphatase inhibitor (ThermoFisher Scientific, Waltham, MA) followed by brief sonication. Lysates were centrifuged at 4°C for 20 minutes at 14,000 g, and supernatants collected. Protein concentrations (μg/ml) were determined on a nanodrop spectrophotometer (ThermoFisher Scientific, Waltham, MA). A sample mixture containing 30 μg of protein, Nupage Reducing Agent and Nupage Loading Buffer (Invitrogen, Carlsbad, CA) was denatured for 10 minutes at 90°C and loaded onto a Bolt BT Plus 4–12% gel (Invitrogen, Carlsbad, CA). After electrophoresis, proteins were transferred onto PVDF membranes (Immobilon; EMD millipore, Darmstadt, Germany) at 4°C. The membranes were blocked in Odyssey Blocking Buffer (Li-Cor, Lincoln, Nebraska) for 1 hr. at RT. Membranes were probed overnight at 4°C in Odyssey Blocking Buffer with antibodies recognizing PS6 (Ser240/244; rabbit monoclonal; 1:1000; Cell Signaling, Danvers, MA), total S6 ribosomal protein (mouse monoclonal; 1:500; Cell Signaling, Danvers, MA), Tsc2 (1:1,000, Cell Signaling, Danvers, MA). Odyssey secondary antibodies 800CW or 680RD were used to visualize bands on the blot (1:1000; Li-Cor, Lincoln, Nebraska). To ensure equal loading, antibodies recognizing β-actin (1:10,000, Abcam, Cambridge UK) followed by Odyssey secondary (1:1,000, 2 hrs, RT; Li-Cor, Lincoln, Nebraska) were used. Blots were developed using an Odyssey Clx scanner and scanned at 169 μm resolution with exposure for both 700 and 800 nm channels set at 3.5 (relative intensity, no units; Li-Cor, Lincoln, Nebraska). All blots were performed in triplicate and densitometry analysis was performed in Image J (Schneider et al., 2012) normalizing band density to β-actin loading control. Densitometry data were analyzed in Origin software (Northampton, MA). Band density from each blot was averaged and the standard error calculated.
mTOR-lysosome colocalization assay
CRISPR-edited cell lines were plated into chamber slides and allowed to adhere for 24 hours. After 24 hrs, cells were incubated in AAF or complete media for 60 minutes and then PFA fixed. Cells were labeled with mTOR and LAMP2 antibodies. Using a spinning disk confocal microscope (Nikon, Tokyo, Japan), cells were selected and a region of interest (ROI) was isolated. Fluorescence intensity of mTOR and LAMP2 was measured within the ROI using ImageJ (Schneider et al., 2012). A Pearson’s correlation test (Excel, Microsoft, Redmond, WA) was then performed to determine whether or not an increase in fluorescence intensity in mTOR correlated with an increase in fluorescence intensity in LAMP2 - the higher the correlation coefficient (R), the greater the degree of colocalization between mTOR and LAMP2. ROIs were measured for 10 N2aC in Tsc2 KO, Depdc5 KO, scramble control, and wildtype cells each in either AAF or replete amino acid conditions. Statistical significance was determined using a one-way ANOVA of averaged R values, with p < 0.05 deemed significant. To better visualize the degree of colocalization in digital images, each ROI was reconstructed into a 3D fluorescence intensity surface plot. 3D surface plots were then converted into short video clips to better visualize fluorescence colocalization across the entire ROI.
Results
Molecular and Functional Validation of CRISPR/Cas9 constructs
Neuro2a cells (N2aC) were transfected with CRISPR/Cas9 plasmids containing gRNAs targeting murine Tsc2 (exon 29) or Depdc5 (exon 16). After FAC sorting by mCherry fluorescence (Cas9 expression), cell lines were established (Figure S1). To assess whether or not Cas9 cleavage resulted in indels in our region of interest, extracted DNA was analyzed by T7 assay. Upon exposure to T7 endonuclease, heteroduplexes caused by mismatched DNA pairs appear as separate bands when visualized on an agarose gel. In Tsc2 and Depdc5 KO cell lines, a reaction product was observed after T7 digest and in positive control specimens but not in DNA obtained from WT or scramble transfected N2aC (Figure S1A,B) using PCR primers spanning the KO region for Tsc2 and Depdc5. Further, Tsc2 KO cell lines did not express Tsc2 protein compared with WT and scramble control N2aC lines. Published results with antibodies recognizing Depdc5 have been highly variable in Western blot analysis (Iffland et al., 2018; Kim et al., 2015) and thus we did not assay for Depdc5 protein levels.
Increased phosphorylation of ribosomal S6 protein (PS6) and enhanced cell size has been demonstrated in resected MCD tissue specimens from TSC and individuals with DEPDC5 mutations (Baldassari et al., 2019; Crino, 2013; Crino, 2015; Ribierre et al., 2018). An increase in PS6 levels was observed in Tsc2 and Depdc5 KO N2aC compared to WT and scramble N2aC lines (Figure S1E,G; densitometry data in Figure S2) with no commensurate change in non-phosphorylated S6 protein levels. Cell soma size was quantified using fluorescent microscopy in digital images of F-actin-labeled N2aC (Figure S1C) and was significantly increased following Tsc2 KO and Depdc5 KO (p<0.001; Figure S1D). Cell soma size increases were mTOR dependent, as treatment with rapamycin (50 nM for 24 hrs) or torin1 (50 nM for 24hrs) rescued soma size increases in both Tsc2 and Depdc5 KO cells compared with DMSO vehicle (Fig S1D).
FRET Detection of 4E-BP1 Phosphorylation
TORCAR (mTORC1 Activity Reporter) is a CFP/YFP FRET biosensor coupled to 4E-BP1 (Zhou et al., 2015; Zhou et al., 2016) that targets the lysosomal membrane via LAMP1 (lysosomal associated membrane protein-1, similar to LAMP2) and identifies changes in the phosphorylation state of 4E-BP1 at the lysosomal surface (see Figure 1F, and representative video in Movie S1). Live-cell confocal fluorescence microscopy was used to analyze and calculate the ratio of CFP to YFP (C:Y) fluorescence where changes in C:Y ratio indicates a cognate change in 4E-BP1 phosphorylation and serves as a proxy for changes in mTORC1 activity. By convention in each data figure, media change after a 10 minute baseline in complete media for each condition is represented in each data figure by a vertical black line; individual cell C:Y ratios are depicted as blue lines, the average C:Y from each 5- cell cohort is depicted by black dashed lines, and the red line is the best fit line for each cohort.
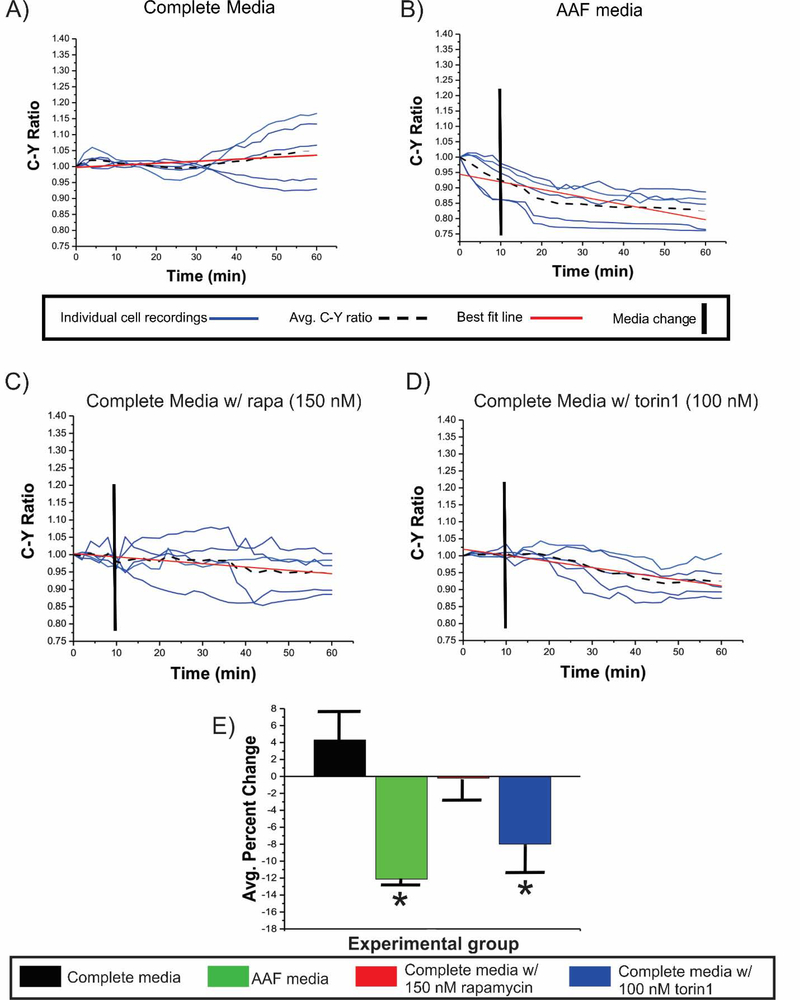
Figure 1: 4E-BP1 Phosphorylation in WT N2aC.
C:Y was measured in WT N2aC transfected with TORCAR alone. Cells were incubated in (A) complete media, (B) AAF media, (C) complete media with rapamycin, or (D) in complete media with torin1 after a 10 minute baseline in complete media for each condition. A non-significant 4.2% increase in C:Y is observed over the 60 minute recording period in cells incubated in complete media while there was a 12.2% decrease in C:Y in WT cells incubated in AAF media (E, p<0.05). WT N2aC incubated with rapamycin (C) showed no change in C:Y, while exposure to torin1 (D, p<0.05) led to a 7.9% decrease in C:Y (E). Only co-transfected cell were used for live-cell imaging experiments. Error bars in (E), standard error of the mean.*p<0.05.
To establish baseline activity of mTORC1, WT N2AC were transfected with TORCAR and live-cell imaging was performed for 60 minutes in complete media. The C:Y ratio (n=5 cells, Figure 1A,E) remained constant over the 60 minute epoch demonstrating little baseline fluctuation in mTOR signaling to 4E-BP1. Then, following a 10 minute baseline incubation in complete media, AAF media, complete media containing rapamycin, or complete media containing torin1, were washed onto cells. While incubation of WT N2aC in AAF media caused a decrease in C:Y (12.2%, p<0.05; Figure 1B,E), and torin1 treatment caused a 7.9% decrease in C:Y (p < 0.05; Figure 1D,E), rapamycin in complete media had no effect on C:Y ratio (Figure 2C,E). Thus, AAF media and torin1 caused reduced 4E-BP1 phosphorylation, while rapamycin did not alter 4E-BP1 phosphorylation in WT N2aC. Next, CRISPR/Cas9 scramble transfected N2aC were assayed under AAF media, rapamycin, or torin1 conditions. After a 10 minute baseline, scramble co-transfected cells incubated in AAF media showed an 8.9% decrease in C:Y (p<0.05; Figure S6A,D), however, no significant change in C:Y (Figure S6B,D) was observed in cells incubated with complete media containing rapamycin. A 7.1% decrease in C:Y was observed in scramble transfected N2aC after incubation with torin1 in complete media (p<0.05; Figure S6C,D). We tested a range of rapamycin concentrations on TORC1 signaling. C:Y ratio was not significantly altered by rapamycin in WT N2aC at doses ranging from 150–550 nM (Figure S8) but all cell types responded consistently to torin1 treatment (Figures 1,2,4). In contrast to our results, previous studies in mouse embryonic fibroblasts and in vivo mouse models showed that loss of Tsc2 resulted in sustained mTORC1 kinase activity during amino acid starvation or protein restriction (Demetriades et al., 2014; Guenther et al., 2014). Thus, our findings suggest differential effects of AAF media on mTOR signaling in neuronal cells compared with non-neural cells. These data demonstrate that WT and CRISPR/Cas9 scramble transfected cells behave similarly when incubated in AAF media or mTOR inhibitors. An important observation was that despite no significant changes in C:Y ratio (4E-BP1 phosphorylation) in N2aC after incubation with rapamycin, ribosomal S6 protein phosphorylation was reduced after incubation with rapamycin (Figure S1F,H) providing evidence that rapamycin appears to differentially effect S6 and 4EBP1 phosphorylation in N2aC (see also, Choo et al., 2008) while torin1 altered both 4E-BP1 and S6 phosphorylation.
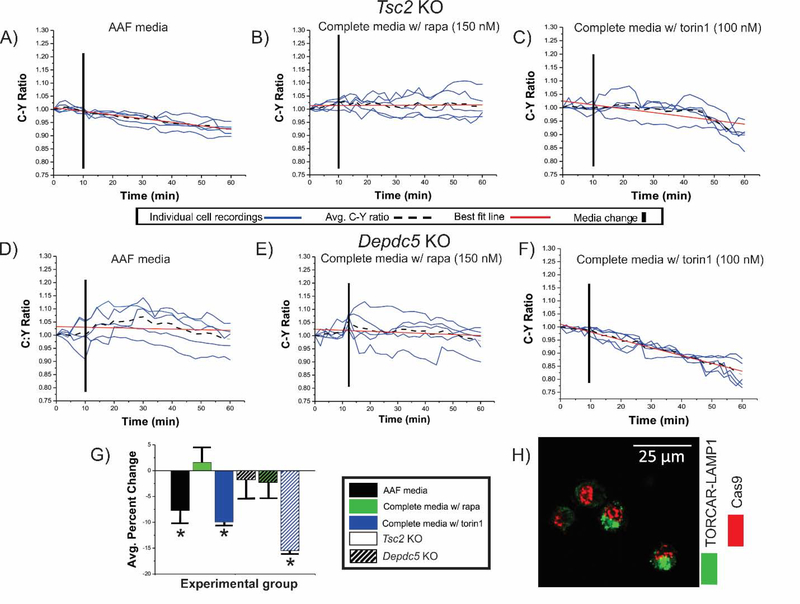
Figure 2: Decrease in C:Y ratio is observed in Tsc2 KO, but not Depdc5 cells after incubation in AAF media.
Tsc2 KO (solid bars in G) and Depdc5 KO (striped bars in G) cells transfected with TORCAR were incubated in AAF media (A, D), in complete media with rapamycin (B,E), or in complete media with torin1 (C,F). After incubation in AAF media (A), C:Y was decreased in Tsc2 KO cells (7.0%, (G); p<0.05). No change in C:Y was observed after incubation in complete media containing rapamycin in Tsc2 KO (B) and Depdc5 KO (E) N2aC, respectively. An average percent decrease in C:Y of 9.9% and 15.4% was observed in both Tsc2 KO and Depdc5 KO N2a cells (p<0.05) after incubation in complete media containing torin1 (C,F), respectively. In (H), representative images of TORCAR and CRISPR/Cas9 plasmid co-transfected cells are shown. AAF= amino acid free, rapa = rapamycin, KO = knockout. Error bars in (D), standard error of the mean. *p<0.05.
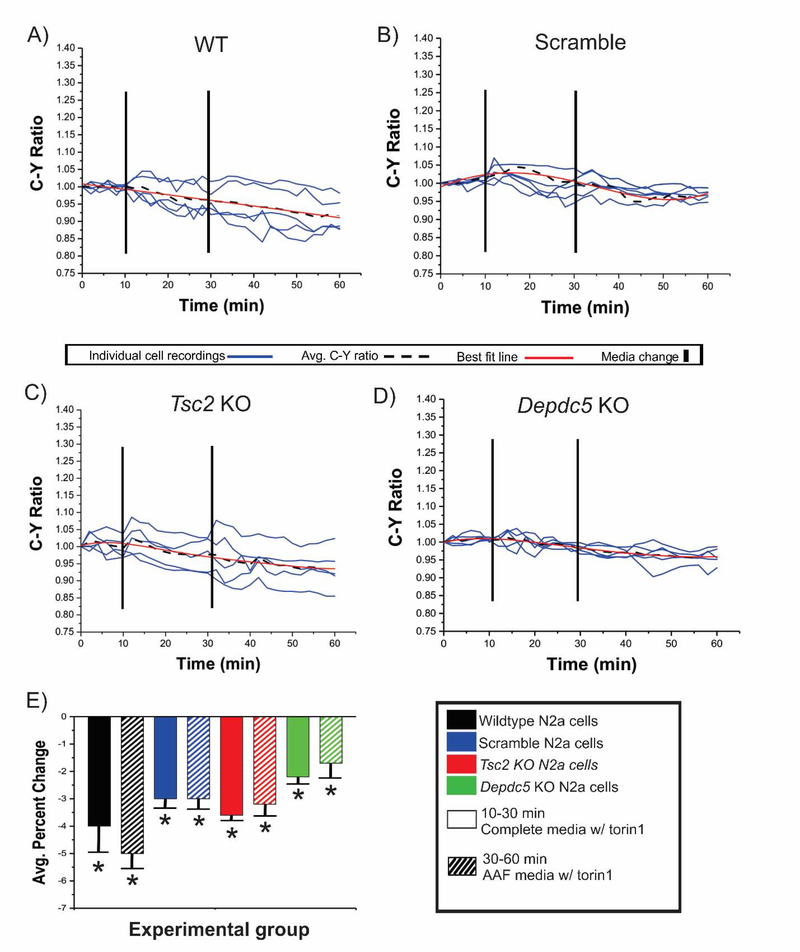
Figure 4: Addition of torin1 to AAF media does not enhance the inhibitory effect of AAF conditions.
After a 10 minute baseline in complete media WT, scramble, Tsc2 KO, and Depdc5 KO cells transfected with TORCAR were incubated in complete media containing torin1 (A-D, 100 nM, mins 10–30, solid bars in E) followed by incubation in AAF media containing torin1 (100 nM, mins 30–60, striped bars in E). During incubation in complete media containing torin1, an average decrease in C:Y was observed across each experimental group: WT = 3.8%, Scram = 3.0%, Tsc2 = 3.7%, Depdc5 = 2.2% (p <0.05). After incubation in AAF media containing torin1, similar percent decreases in C:Y ratio were observed in WT (A; 4.9%), scramble (B; 3.2%), Tsc2 KO (C; 3.4%) and in Depdc5 KO N2aC (D); 1.7%) (p <0.05); Results were statistically significant from baseline but not between groups (e.g., torin1 in complete media vs. torin1 in AAF media).
In parallel experiments, we demonstrate that incubating WT N2aC in AAF media for 60 minutes does not result in apoptosis or necrosis, that alterations in C:Y were not due to extracellular glucose concentrations in the experimental media, and that the effects were not due to photobleaching of the FRET TORCAR sensor (Figures S3–5).
mTOR activation following Tsc2 KO, but not Depdc5 KO, is abolished by amino acid depletion
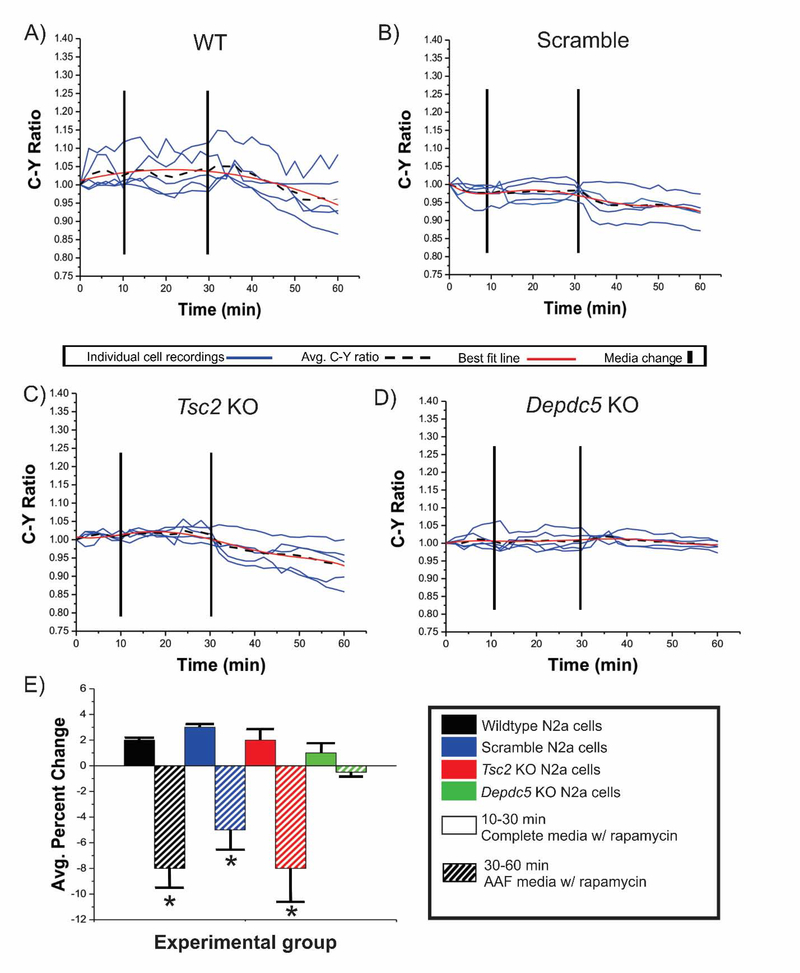
The canonical (via Tsc1/Tsc2) and amino acid signaling (via Depdc5) arms of the mTOR pathway function independently in response to different signaling cues to modulate mTORC1 kinase activity. We hypothesized that an intact amino acid regulatory arm could be leveraged to inhibit Tsc2 KO-induced mTOR pathway hyperactivation in N2aC. To test this, C:Y ratio was assayed in Tsc2 and Depdc5 KO N2aC under AAF media conditions, in complete media containing rapamycin, or in complete media containing torin1 (Figure 2). Incubation in AAF media resulted in a 7.0% decrease in C:Y in Tsc2 KO N2aC (p< 0.05; Figure 2A,G) but no significant change in C:Y in Depdc5 KO cells (Figure 2D,G). We next hypothesized that amino acid deprivation would produce an inhibitory effect that would override, and function independently of, pharmacological mTOR inhibition. We posited that CRISPR/Cas9 Tsc2 KO cells would show a decrease in C:Y after combined rapamycin (150 nM) and AAF media incubation similar to AAF media incubation alone. After a 10 minute baseline, WT, scramble, Tsc2 KO, and Depdc5 KO N2aC were incubated in complete media containing rapamycin for 20 min. During this time period, there was no significant change in C:Y observed across all cell types (Figure 3E). After 20 minutes, complete media containing rapamycin was removed and AAF media containing 150 nM rapamycin was washed on for 30 minutes. During this time period a statistically significant decrease in C:Y (p < 0.05) was observed in Tsc2 KO, and WT and scramble conditions, but not Depdc5 KO cells where no change in C:Y was observed (Figure 3B–E). Thus, rapamycin did not add to the effects of AAF conditions on C:Y. Next, the combined effects of AAF media and torin1 (100 nM) were assayed. As we have demonstrated (Figures 1,2,4), torin1 alone resulted in a decrease in C:Y in each experimental cell type. Therefore, we hypothesized that concomitant AAF media and torin1 incubation would result in a decrease in C:Y similar to what is observed with torin1 media alone. To test this, after a 10 minute baseline, WT, scramble, Tsc2 KO, and Depdc5 KO N2aC were incubated in torin1 containing complete media for 20 min. During this time period a decrease in C:Y across all cell types was observed (Figure 4E). After 20 minutes, complete torin1 containing media was removed and AAF media containing 100 nM torin1 was washed on for 30 minutes. During this time period a decrease in C:Y similar to what was observed with torin1 alone (see Figure 4E). These data suggest that mTOR inhibition caused by amino acid deprivation is independent of and is not augmented by pharmacological inhibition.
Figure 3: Addition of rapamycin to AAF media does not enhance the inhibitory effect of amino acid free conditions.
After a 10 minute baseline in complete media WT, scramble, Tsc2 KO, Depdc5 KO cells transfected with TORCAR were incubated in complete media containing rapamycin (A-D, 150 nM, mins 10–30, solid bars in E) followed by incubation in AAF media containing rapamycin (150 nM, mins 30–60, striped bars in E). During incubation in complete media containing rapamycin, an average increase in C:Y was observed across each experimental group (E, not significant). After incubation in AAF media containing rapamycin, a decrease in C:Y ratio was observed in WT (7.9%; A, E; p<0.05), scramble (B,E; 4.7%; p<0.05), and Tsc2 KO (C,E; 8.1%; p<0.05). No statistically significant changes were observed in Depdc5 KO N2aC (E). *p<0.05.
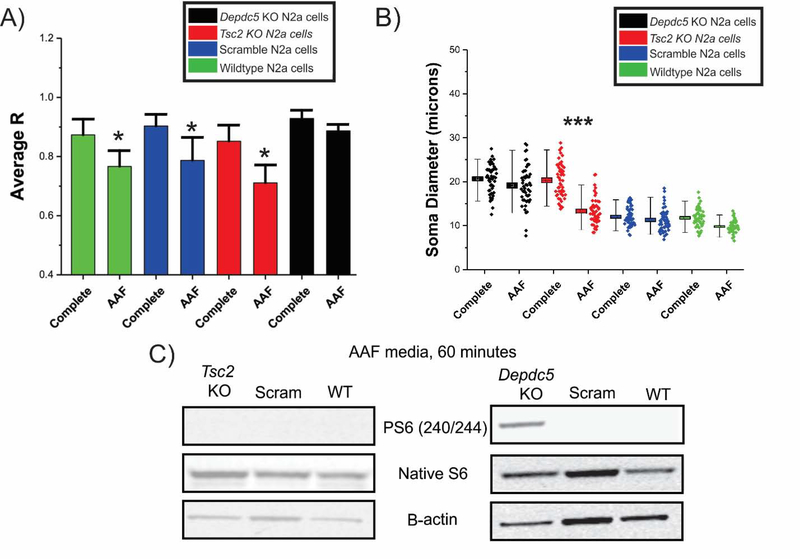
Amino acid levels alter mTOR lysosomal localization following Tsc2 KO but not Depdc5 KO
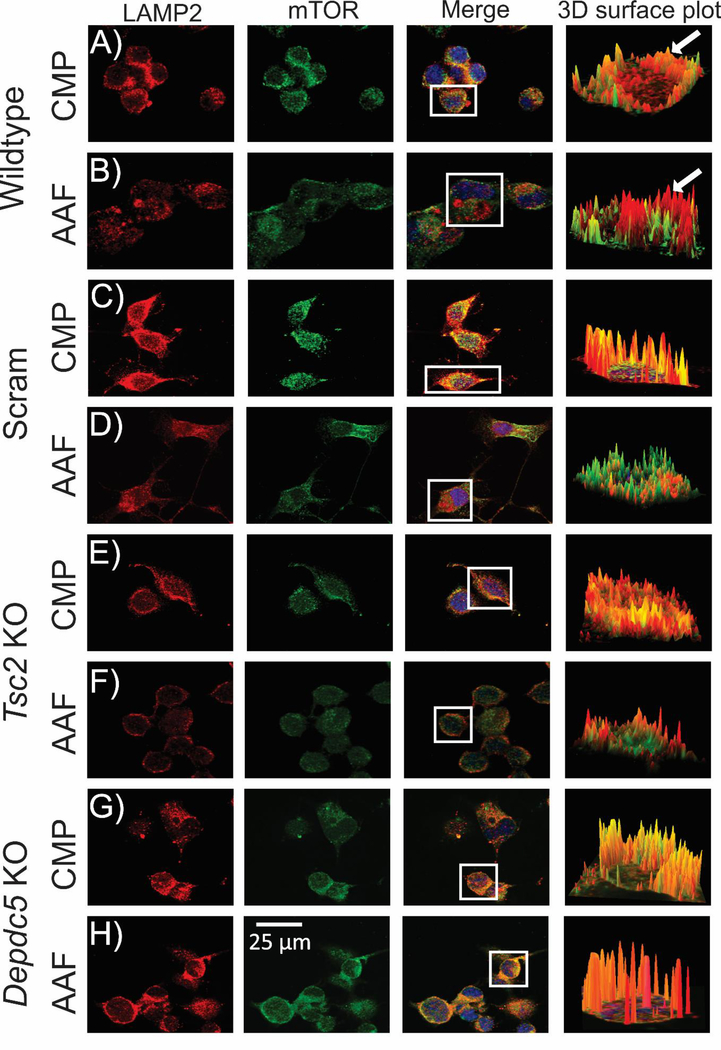
We next hypothesized that AAF conditions would alter localization of mTOR to the lysosomal surface. WT, scramble, Tsc2 KO, and Depdc5 KO cell lines we re incubated in AAF media for 60 minutes, fixed in 4% PFA and probed for mTOR and LAMP2. Images were captured on a spinning disk confocal microscope, ROIs were selected and then reconstructed in 3D to better visualize areas of colocalization defined by a higher correlation between mTOR and LAMP2 (see Methods). Videos of the 3D surface plots yielded dynamic visualization of the colocalization across the cell body (see Movies S2–9). In complete media, WT, scramble, Tsc2 KO, and Depdc5 KO N2aC have a correlation coefficient (R) of 0.87, 0.90, 0.87, and 0.92, respectively between mTOR and LAMP2 labeling (Figure 5 and 6A) demonstrating high colocalization of mTOR to the lysosomal membrane. However, after incubation in AAF media, WT, scramble, and Tsc2 KO N2aC showed a statistically significant decrease in colocalization between mTOR and the lysosome as compared to cells in complete media (Figure 5, 6A) with R’s of 0.76, 0.78, and 0.71, respectively, demonstrating a reduction in mTOR colocalization with the lysosome. No significant change in R level for mTOR-lysosome colocalization was observed in Depdc5 KO cells incubated in AAF media (R=0.88, Figure 6A). After incubation in AAF media, levels of PS6 are abolished in WT, scramble, and Tsc2 KO cells but not in Depdc5 KO cells as measured by Western blot (Figure 6C, densitometry data in Figure S7). Similarly, Tsc2 KO but not Depdc5 KO N2aC displayed a statistically significant decrease in cell size after incubation in AAF media for 1 hr. (Figure 6B).
Figure 5: Reduced colocalization between mTOR and the lysosomal membrane in Tsc2 KO but not Depdc5 KO cells in AAF conditions.
WT, scramble, Tsc2 KO, and Depdc5 KO cell lines incubated in AAF media (60 minutes), fixed in 4% PFA and visualized with antibodies targeting mTOR (red; alexa 594) or LAMP2 (green; alexa 488) on a spinning disk confocal microscope. Regions of interest (ROIs; cell bodies) were selected in digital micrographs and 3D reconstructed (Image J; arrows). ROIs of mTOR and LAMP2 colocalization are orange or yellow (arrow, A) and ROIs where mTOR and LAMP2 are not colocalized appear as distinct red and green areas (arrow, B). After incubation in AAF media WT (B), scramble (D), and Tsc2 KO (F) cell lines display a decrease in colocalization between mTOR and the lysosomal membrane vs. incubation in complete media (A, C, E). However, no statistical difference in colocalization was observed in Depdc5 KO cell lines incubated in AAF media (H) vs. complete media (G). Statistical data for digital micrographs are in Fig. 6. High-definition videos showing 3D surface visualization of colocalization are provided (Movies S2–S9).
Figure 6: AAF conditions alter sub-cellular localization and soma size in Tsc2 KO N2aC.
A) Correlation analysis (R) of fluorescence intensity of mTOR and LAMP2 (see Fig. 5) reveals high correlation between mTOR and the lysosomal membrane with R values of 0.87 (WT), 0.90 (Scram), 0.87 (Tsc2), and 0.92 (Depdc5). A statistically significant decrease in colocalization between mTOR and the lysosomal membrane was observed in WT (0.76), scramble (0.78) and Tsc2 KO (0.71) cells after incubation in AAF media (60 min incubation for each group, n = 10 cells per group, p<0.05). Depdc5 KO cells display an enhanced colocalization in complete media (R= 0.92) vs. WT, scramble and Tsc2 KO cells. Depdc5 KO cells displayed a nonsignificant decrease in (R) after AAF media incubation media (R= 0.88; n =10 cells per group). Incubation in AAF media results in decreased phosphorylated S6 levels in Tsc2 KO, scramble and WT N2aC but not Depdc5 KO N2aC (densitometry in Figure S7).
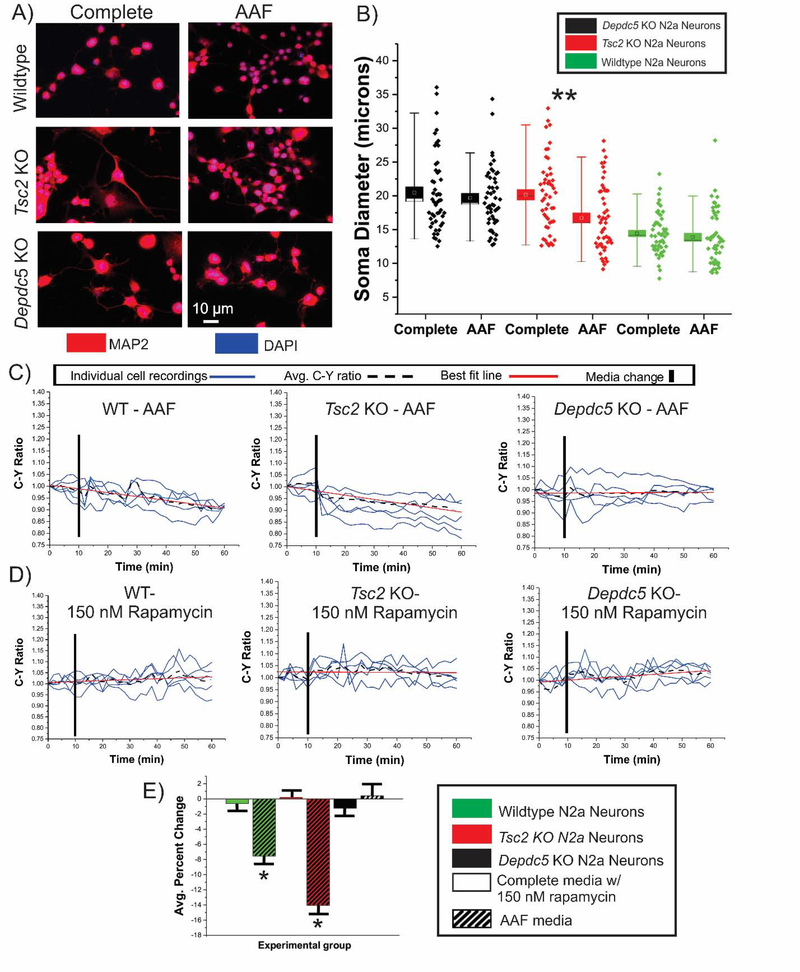
Cell size and 4E-BP1 phosphorylation in differentiated neurons
Our results demonstrate that amino acid deprivation rescues soma diameter and reduces both S6 and 4E-BP1 phosphorylation in undifferentiated N2aC following Tsc2 or Depdc5 KO. We next investigated whether amino acid deprivation would result in a decrease in soma diameter and a decrease in 4E-BP1 phosphorylation in differentiated neurons. To obtain neurons, KO and WT cell lines were differentiated using retinoic acid and then transfected with TORCAR plasmid or incubated in AAF or complete media. After 1 hr. of incubation AAF media, Tsc2 KO but not Depdc5 KO or WT N2a neurons show a decrease in soma diameter (Fig. 7A,B; p<0.05) compared to neurons in complete media, similar to what was observed in undifferentiated N2a cells (Fig 6B). WT, Tsc2 KO, and Depdc5 KO differentiated neurons transfected with TORCAR were incubated in AAF media, complete media containing rapamycin, or complete media containing torin1. Incubation in AAF media led to a decrease in C:Y in WT (7.4%; p<0.05) and Tsc2 KO (14.0%) neurons (Figure 7E). As was observed in undifferentiated N2aC, no change in C:Y was observed when Depdc5 KO neurons were incubated in AAF media (Figure 7C,E). Further, no change in C:Y was observed in any cell type incubated in complete media containing 150 nM rapamycin (Figure 7D,E). WT, Tsc2 KO, and Depdc5 KO neurons incubated in complete media containing 100 nM torin1 displayed a 6.8%, 11.8% and 7.6% decrease in C:Y, respectively (p < 0.05; Figure S9). These data are similar to what was observed in undifferentiated N2a cells (Figure 1,2) demonstrating a consistent effect of AAF conditions on 4E-BP1 phosphorylation in immature and differentiated neurons.
Figure 7: Tsc2 KO neurons exhibit morphological changes and decreased C:Y ratio in AAF media.
Differentiated N2aC were incubated in complete or AAF media for 1 hour, were fixed, and probed with MAP2 antibodies to visualize cell bodies (A). Incubation in AAF media led to a decrease in soma diameter in Tsc2 KO but not Depdc5 KO or WT neurons (N= 50 per group; B; p<0.01). TORCAR transfected Tsc2 KO, Depdc5 KO, and WT neurons were incubated in AAF media (C, striped bars in E) or in complete media with rapamycin (D; solid bars in E). A 7.4% decrease in C:Y was observed in WT neurons incubated in AAF media (C, E; p<0.05). A 14.0% decrease (p<0.05) in C:Y was observed in Tsc2 KO neurons but no change in C:Y was observed in Depdc5 KO neurons (C, E). No significant change in C:Y was observed in WT neurons after rapamycin in complete media (D, E). Data on torin1 treated WT, Tsc2 KO, and Depdc5 KO cells can be found in Figure S9. AAF= amino acid free, rapa = rapamycin, KO = knockout. Error bars in (D), standard error of the mean. *p<0.05, ** p<0.01.
Discussion
We demonstrate that mTOR pathway hyperactivation induced by Tsc2 KO but not Depdc5 KO, in undifferentiated N2aC and differentiated neurons is abolished by amino acid deprivation as measured by S6 phosphorylation in Western assay and by 4E-BP1 phosphorylation using live cell imaging and FRET detection. We specifically implemented FRET in living neurons to show the dynamic changes occurring over minutes to an hour since most prior studies have looked in more static conditions over longer time epochs. Further, we demonstrate that AAF media conditions result in movement of mTOR off of the lysosomal membrane, the key step in functionally inactivating mTORC1 (Sancak and Sabatini, 2009), in Tsc2 KO and WT neurons, but not Depdc5 KO neurons. Rapamycin reduced S6 phosphorylation, but did not alter 4E-BP1 phosphorylation, in undifferentiated and differentiated neurons following Tsc2 KO or Depdc5 KO in N2aC. Finally, while rapamycin decreased cell size in both Tsc2 KO and Depdc5 KO cells, AAF media incubation resulted in decreases in soma diameter in Tsc2 KO, but not Depdc5 KO, cells. These data demonstrate that 4E-BP1 phosphorylation is insensitive to rapamycin and exhibits variable responses to amino acid deprivation following Tsc2 KO versus Depdc5 KO in neurons. Furthermore, movement of mTOR onto and off of the lysosome (a key step in modulation of mTOR signaling) varies in response to AAF conditions following Tsc2 KO versus Depdc5 KO in neurons. Our findings suggest that pharmacological inhibition of mTOR with rapamycin and related compounds may not fully ameliorate all signaling effects of mTOR activation in neurons and that changes in cellular nutrient status in neurons may have distinct effects in neurodevelopmental disorders linked to individual mTOR pathway gene variants i.e., TSC2 versus DEPDC5. Ribosomal S6 and 4E-BP1 are targets of mTORC1 kinase activity responsible for different aspects of cap-dependent protein translation. However, mTOR inhibition by rapamycin in non neural cell types results in differential inhibition of S6 phosphorylation (strongly inhibited), and 4E-BP1 (less inhibited; Choo et al., 2008). Thus, cells can maintain cap-dependent translation even in the presence of rapamycin due to the relative resistance of 4E-BP1 to rapamycin. Indeed, our data demonstrate that S6 phosphorylation is rapidly inhibited by rapamycin but that there is little change in 4E-BP1 phosphorylation in neurons during live-cell imaging even at high rapamycin concentrations. Direct mTOR inhibition via torin1 results in a decrease in the phosphorylation of both ribosomal S6 and 4E-BP1 (Thoreen et al., 2009) and thus compounds like torin1, with direct kinase activity on both mTORC1 and mTORC2 isoforms may have clinical relevance. Similarly, AAF media incubation resulted in a decrease in phosphorylation of both ribosomal S6 and 4E BP1. These findings suggest that mTORC1 inhibition via amino acid modulation may provide a method to prevent phosphorylation of both S6 and 4E-BP in neuronal cells. An intriguing aspect of mTOR inhibition by amino acid deprivation in non-neural cells is the ability of AAF conditions to dissociate mTOR from the lysosomal surface, functionally inactivating mTORC1 and separating it from its binding partners (Bar-Peled et al., 2013). This effect is not achieved by rapamycin or torin1. In fact, rapamycin may enhance the localization of mTORC1 to the lysosomal surface (Ohsaki et al., 2010). Thus, our findings in neurons raise the possibility that the modest clinical efficacy of everolimus (40% seizure reduction in 50% of patients given high dose everolimus on seizures (Franz et al., 2018) might reflect partial inhibition of mTORC1 with residual and unaccounted for effects of 4E-BP1 phosphorylation and cap-dependent translation in neurons. Increased S6 phosphorylation caused by mTOR pathway gene variants results in abnormal morphology, both cell size and process outgrowth, in neurons in vitro (Sokolov et al., 2018) and in human brain tissue specimens from patients with mTORopathies in which PS6 immunolabeled cells are a frequent finding (see Iffland and Crino, 2017). However, recent evidence points toward 4E-BP1 as a driver of pathology in cortical malformations as well. For example, knockdown of 4E BP2, an isoform of 4E-BP1, is sufficient to produce abnormal neuronal migration in vivo (Lin et al., 2016). Further, constitutive expression of 4E-BP2 prevents abnormal neuronal positioning, mTORC1 hyperactivation-associated cytomegaly, and partially rescued dendritic hypertrophy (Lin et al., 2016). Therefore, mechanisms of mTOR inhibition that impact the phosphorylation of both ribosomal S6 protein and 4E-BP isoforms equally may more effectively result in reversal/prevention of TSC-associated neuronal pathology. Interestingly, AAF media conditions result in morphological changes in Tsc2 KO and WT neurons but not in Depdc5 KO neurons. Depdc5 KO cells maintained increased levels of both PS6 and P4E-BP1 during AAF media incubation as well as increased cell size suggesting that our results following Tsc2 KO are dependent on a functional GATOR1 complex, the key complex in the amino acid regulatory arm of the mTOR pathway. GATOR1 proteins are enriched in brain and may thus provide a target to produce enhanced mTOR pathway inhibition in the brain of TSC patients (Dibbens et al., 2013) while impacting mTOR signaling in other organ systems to a lesser degree.
Several amino acids including leucine, methionine, and arginine have been linked to mTOR pathway function and specific amino acids sensors for each of these amino acids including Sestrin 1/2, SAMTOR, and CASTOR, respectively, have been defined in non-neuronal cells (Chantranupong et al., 2014; Chantranupong et al., 2016; Gu et al., 2017). These amino acids will likely provide insights into how specific amino acids effect mTOR signaling in neurons. There are multiple pharmacological targets within the amino acid regulatory arm of the mTOR pathway that could be leveraged to mimic the effect of removal of dietary amino acids. These include compounds that target amino acid transporters (Hafliger et al., 2018) or amino acid sensors (Hasegawa et al., 2019). Indeed, in a phase 1 study using NV-5138 to activate Sestrin, the leucine sensor in the amino acid regulatory arm of the mTOR pathway, an mTORC1-dependent antidepressant effect was observed (Hasegawa et al., 2019). Thus, targeting this arm of the mTOR pathway can indeed have an impact on both mTOR signaling and the pathophysiological consequences associated with aberrant mTOR pathway function.
Conclusions
We demonstrate that amino acid depletion reduces 4E-BP1 phosphorylation and soma size in neuronal cells caused by mTOR pathway hyperactivation following Tsc2 KO. Localization of mTORC1 to the lysosome was diminished under AAF conditions following Tsc2 KO. Torin1, but not rapamycin, mimicked the effects of AAF media following Tsc2 KO. AAF media, but not rapamycin dually affected both S6 and 4E-BP1 phosphorylation. Manipulation of amino acids and rapamycin treatment did not alter the effects of Depdc5 KO on 4E-BP1 phosphorylation or localization of mTORC1 to the lysosome. These findings demonstrate that cellular amino acid levels and rapamycin have distinct effects on mTOR signaling in neurons via S6 versus 4E-BP1 protein phosphorylation, and thus, each may have differential effects in the setting of distinct mTOR pathway gene mutations.
Supplementary Material
Figure S1: CRISPR-Cas9 Tsc2 or Depdc5 KO results in mTOR pathway hyperactivation. CRISPR-Cas9 editing of regions within mouse Tsc2 (A) and Depdc5 (B) were verified using a T7 endonuclease assay. Indels created by CRISPR-Cas9 editing are revealed as multiple bands after exposure to T7 endonuclease (arrows in A and B). To verify a complete digest reaction, a manufacturer provided positive control was used (PC, at 400 and 200 bp respectively, A,B). Wildtype (WT) and Scramble (Scram) control DNA was also assayed by T7 assay using primers covering the same regions as those assayed in CRISPR edited cells. No digested bands were observed in control samples (A,B). A hallmark of mTOR pathway hyperactivation is increased cell body size. Tsc2 and Depdc5 KO cell lines display significantly increased cell body size (n=50 cells per group; two replicates) compared to scramble and WT controls (C and D, p<0.001, ***). Increases in cell size after Tsc2 or Depdc5 KO were rescued with rapamycin (p<0.001) or torin1 application (p<0.001). Size differences between WT and scramble vehicle treated cells were not significant. In rapamycin or torin1 treated N2aC, cell size decreases were significant at p < 0.05 (*). In D, each box represents SEM, whiskers reflect 5–95% confidence intervals. Western assay of Tsc2 KO N2aC reveals a decrease in Tsc2 protein compared to scramble and WT cells as well as an increase in PS6 (240/244) (E). An increase in PS6 (240/244) was also observed in Depdc5 KO N2aC (G). After Tsc2 or Depdc5 KO, increases in PS6 (240/244) were rescued by rapamycin or torin1 application (F and H). Western blot densitometry data are in Figure S2 (n=3 per blot). U = undigested, D= digested, KO = knockout, Rapa= rapamycin, F-actin = filamentous actin.
Figure S2: Densitometry data for figure S1. All densitometry measurements are normalized to loading control (β-actin) and performed in triplicate. Levels of Tsc2 (A) and phosphorylated S6 (240/244) (B-G) were determined using lysates made after incubation in complete media only. All rapamycin and torin1 blots were performed using lysates from cells incubated with rapamycin (150 nM) or torin1 (100 nM) in complete media for 1 hour. Error bars indicate SEM. PS6= phosphorylated S6 (240/244). Densitometry data correspond to blots in Figure S1 as follows: A)= Fig. S1E; B)= Fig. S1E; C)= Fig. S1F; D)= Fig. S1F; E)= Fig. S1G; F)= S1H; G)= Fig. S1H.
Figure S3: Incubation of N2a cells in AAF media for 1 hr. does not result in cell death. Live N2aC were assayed with annexin V and propidium iodide to determine whether or not 1 hr. of incubation in AAF media resulted in induction of apoptosis or necrosis, respectively. As a positive control, the topoisomerase II inhibitor Etoposide was used (A). Etoposide treated cells expressed high levels of annexin V and took up propidium iodide vs. N2a cells in AAF media (B) or WT control (C) cells in complete media.
Figure S4: There is no statistical difference in C:Y between incubation in AAF media with glucose vs. media without glucose. To ensure that the mTOR inhibition observed in AAF media without glucose was not caused by an absence of glucose, WT N2aC were assayed in AAF media without and with glucose (50 mM; “high glucose media”). In (A) WT N2a cell baseline in complete media is shown (same data as in Figure 1). A decrease in C:Y was observed in WT N2a cells in AAF media without glucose (avg. percent decrease of 12.2%, same data as Figure 1; p<0.05) and in AAF media with glucose (avg. percent decrease of 13.6%; p<0.05). The difference between AAF media types was not statistically significant. *p<0.05.
Figure S5: Decreases in C:Y during incubation in AAF media can be rescued by incubation in complete media. (A) A decrease in C:Y during AAF media incubation (striped bars in E) was observed in WT (3.8%), scramble (3.7%), and Tsc2 KO (2.5%) N2aC (all significant, p <0.05). After incubation in complete media (solid bars in E), an increase in C:Y was observed in WT (A, 0.05%), scramble (B, 3.2%) and Tsc2 KO (C, 1.5%) N2aC. In scramble and Tsc2 KO cells, this increase was statistically significant from baseline (p <0.05). No significant change in C:Y was observed during AAF media incubation or in complete media in Depdc5 KO cells (D).
Figure S6: CFP:YFP Ratio in CRISPR scramble and TORCAR co-transfected N2a cells compared to WT N2a cells transfected with TORCAR alone. CRISPR scramble and TORCAR co-transfected cells were incubated in (A) AAF media, (B) in complete media with rapamycin, or (C) in complete media with torin1 after a 10 minute baseline in complete media for each condition. An average 8.9% decrease in C:Y was observed in cells incubated in AAF media (A, D; p < 0.05). Cells incubated in rapamycin (B) or torin1 (C) containing complete media displayed a 1.5% increase and 7.1% decrease in C:Y, respectively (D; p < 0.05). The difference in C:Y between AAF media and torin1 containing complete media was not significant. These trends are similar to what is observed in WT cells in Figure 1. AAF = amino acid free, rapa = rapamycin. Error bars in (D), standard error of the mean. *p<0.05.
Figure S7: Densitometry data for figure 6. All densitometry measurements are normalized to loading control (beta-actin) and performed in triplicate. Levels of PS6 (A,B) were determined using lysates made after incubation in AAF media only for 1 hour. Error bars indicate SEM. Densitometry data correspond to blots in Figure S6 as follows: A)= Fig. 6C left, B)= Fig. 6C right.
Figure S8: Rapamycin dose response curve in WT cells transfected with TORCAR. To ensure that the absence of change in C:Y ratio after rapamycin treatment in N2aC was not due to insufficient concentration of rapamycin, a dose response experiment was performed. N2aC transfected with TORCAR were incubated in complete media containing 150 nM, 250 nM, 350 nM, 450 nM, or 550 nM rapamycin after a 10 minute baseline recording in complete media. During incubation with 150 nM, 250 nM, and 350 nM rapamycin, an increase in average C:Y of 2.6%, 1.0%, and 4.5% was observed, respectively. During incubation with 450 nM and 550 nM rapamycin decreases in C:Y of 2.6% and 4.1% were observed respectively. These concentrations are much higher than was is required to inhibit phosphorylation of S6 (Figure S1) and result in a decrease in C:Y less than torin1 or AAF media (Figure 1). No changes in C:Y were statistically significant from baseline at any rapamycin concentration.
Figure S9: A decrease is C:Y ratio is observed in Tsc2, Depdc5, and WT neurons after incubation in complete media containing torin1. N2a neurons were transfected with TORCAR and imaged in torin1 containing complete media after a 10 minute baseline in complete media only. Similar to what was observed in undifferentiated N2a cells, a statistically significant decrease in C:Y was observed in Tsc2 KO (11.8%), Depdc5 KO (7.6%), and WT neurons (6.8%). *p<0.05.
Figure S10: N2a cells express multiple neuronal makers. N2a cells were used in this study due to their ease of use in transfection, Western blot assays, FAC sorting, and live cell imaging. N2a cells are also easy to maintain in culture and can be rapidly assayed. Further, N2a cells express many typical neuronal markers including tau protein (A), microtubule associated protein 2 (MAP2) and Nestin (B), and Neuron-specific class III beta-tubulin (TUJ-1; C). For these experiments, N2aC were not differentiated using dibutyryl-cAMP and thus they do not express Tyrosine hydroxylase (TH; C). Filamentous actin (F-actin) was used in this study to assess cell soma size (A).
Movie S1: Representative time-lapse of CFP/YFP channels in WT N2a cells treated with torin1
Movie S2: 3D surface plot of a WT N2a cell in complete media. Movie created from surface plot shown in Figure 5.
Movie S3: 3D surface plot of a WT N2a cell in AAF media. Movie created from surface plot shown in Figure 5.
Movie S4: 3D surface plot of a Scramble transfected N2a cell in complete media. Movie created from surface plot shown in Figure 5.
Movie S5: 3D surface plot of a Scramble transfected N2a cell in AAF media. Movie created from surface plot used in Figure 5.
Movie S6: 3D surface plot of a Tsc2 KO N2a cell in complete media. Movie created from surface plot shown in Figure 5.
Movie S7: 3D surface plot of a Tsc2 KO N2a cell in AAF media. Movie created from surface plot shown in Figure 5.
Movie S8: 3D surface plot of a Depdc5 KO N2a cell in complete media. Movie created from surface plot shown in Figure 5.
Movie S9: 3D surface plot of a Depdc5 KO N2a cell in AAF media. Movie created from surface plot shown in Figure 5.
Research Highlights.
TSC is highly associated with epilepsy and autism caused by mutations in TSC1 or TSC2
mTOR pathway hyperactivation is abolished by amino acid deprivation in Tsc2 KO cells
mTOR pathway hyperactivation is not abolished by amino acid deprivation in Depdc5 KO cells
Response of mTOR signaling to nutrients may have relevance for clinical therapeutics
Acknowledgements:
Dr. Joseph Mauban, Manager of the University of Maryland School of Medicine Confocal Microscopy core, for his assistance with both live-cell slit-scanning and spinning disk confocal microscopy. Dr. Xiaoxuan Fan, Director of the Flow Cytometry Shared Service at the University of Maryland for his assistance in establishing CRISPR cell lines. Dr. Alex Poulopoulos at the University of Maryland School of medicine for assistance in generating PX330 CRISPR plasmids. This work was supported by NINDS R01NS099452 and R01NS094596-01A1 (PBC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Baldassari S, Picard F, Verbeek NE, van Kempen M, Brilstra EH, Lesca G, Conti V, Guerrini R, Bisulli F, Licchetta L, Pippucci T, Tinuper P, Hirsch E, de Saint Martin A, Chelly J, Rudolf G, Chipaux M, Ferrand-Sorbets S, Dorfmuller G, Sisodiya S, Balestrini S, Schoeler N, Hernandez-Hernandez L, Krithika S, Oegema R, Hagebeuk E, Gunning B, Deckers C, Berghuis B, Wegner I, Niks E, Jansen FE, Braun K, de Jong D, Rubboli G, Talvik I, Sander V, Uldall P, Jacquemont ML, Nava C, Leguern E, Julia S, Gambardella A, d’Orsi G, Crichiutti G, Faivre L, Darmency V, Benova B, Krsek P, Biraben A, Lebre AS, Jennesson M, Sattar S, Marchal C, Nordli DR Jr., Lindstrom K, Striano P, Lomax LB, Kiss C, Bartolomei F, Lepine AF, Schoonjans AS, Stouffs K, Jansen A, Panagiotakaki E, Ricard-Mousnier B, Thevenon J, de Bellescize J, Catenoix H, Dorn T, Zenker M, Muller-Schluter K, Brandt C, Krey I, Polster T, Wolff M, Balci M, Rostasy K, Achaz G, Zacher P, Becher T, Cloppenborg T, Yuskaitis CJ, Weckhuysen S, Poduri A, Lemke JR, Moller RS, Baulac S, 2019. The landscape of epilepsy related GATOR1 variants. Genet Med 21, 398–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM, 2013. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340, 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L, Scaria SM, Saxton RA, Gygi MP, Shen K, Wyant GA, Wang T, Harper JW, Gygi SP, Sabatini DM, 2016. The CASTOR Proteins Are Arginine Sensors for the mTORC1 Pathway. Cell 165, 153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM, 2014. The Sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep 9, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YJ, Di NA, Kramvis I, Meikle L, Kwiatkowski DJ, Sahin M, He X, 2008. Tuberous sclerosis complex proteins control axon formation. Genes Dev 22, 2485–2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AY, Yoon SO, Kim SG, Roux PP, Blenis J, 2008. Rapamycin differentially inhibits S6Ks and 4E-BP1 to mediate cell-type-specific repression of mRNA translation. Proceedings of the National Academy of Sciences of the United States of America 105, 17414–17419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino PB, 2013. Evolving neurobiology of tuberous sclerosis complex. Acta Neuropathol 125, 317–332. [DOI] [PubMed] [Google Scholar]
- Crino PB, 2015. mTOR signaling in epilepsy: insights from malformations of cortical development. Cold Spring Harb. Perspect. Med 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP, 2006. The tuberous sclerosis complex. N Engl J Med 355, 1345–1356. [DOI] [PubMed] [Google Scholar]
- Crowell B, Lee GH, Nikolaeva I, Dal P,V, D’Arcangelo G, 2015. Complex Neurological Phenotype in Mutant Mice Lacking Tsc2 in Excitatory Neurons of the Developing Forebrain(123). eNeuro 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatolo P, 2015. Mechanistic target of rapamycin (mTOR) in tuberous sclerosis complex associated epilepsy. Pediatr Neurol 52, 281–289. [DOI] [PubMed] [Google Scholar]
- Curatolo P, Moavero R, de Vries PJ, 2015. Neurological and neuropsychiatric aspects of tuberous sclerosis complex. Lancet Neurol 14, 733–745. [DOI] [PubMed] [Google Scholar]
- Demetriades C, Doumpas N, Teleman AA, 2014. Regulation of TORC1 in response to amino acid starvation via lysosomal recruitment of TSC2. Cell 156, 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibbens LM, de Vries B, Donatello S, Heron SE, Hodgson BL, Chintawar S, Crompton DE, Hughes JN, Bellows ST, Klein KM, Callenbach PM, Corbett MA, Gardner AE, Kivity S, Iona X, Regan BM, Weller CM, Crimmins D, O’Brien TJ, Guerrero-Lopez R, Mulley JC, Dubeau F, Licchetta L, Bisulli F, Cossette P, Thomas PQ, Gecz J, Serratosa J, Brouwer OF, Andermann F, Andermann E, van den Maagdenberg AM, Pandolfo M, Berkovic SF, Scheffer IE, 2013. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nature genetics 45, 546–551. [DOI] [PubMed] [Google Scholar]
- Franz DN, Lawson JA, Yapici Z, Brandt C, Kohrman MH, Wong M, Milh M, Wiemer-Kruel A, Voi M, Coello N, Cheung W, Grosch K, French JA, 2018. Everolimus dosing recommendations for tuberous sclerosis complex-associated refractory seizures. Epilepsia 59, 1188–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French JA, Lawson JA, Yapici Z, Ikeda H, Polster T, Nabbout R, Curatolo P, de Vries PJ, Dlugos DJ, Berkowitz N, Voi M, Peyrard S, Pelov D, Franz DN, 2016. Adjunctive everolimus therapy for treatment-resistant focal-onset seizures associated with tuberous sclerosis (EXIST-3): a phase 3, randomised, double-blind, placebo controlled study. Lancet 388, 2153–2163. [DOI] [PubMed] [Google Scholar]
- Gu X, Orozco JM, Saxton RA, Condon KJ, Liu GY, Krawczyk PA, Scaria SM, Harper JW, Gygi SP, Sabatini DM, 2017. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 358, 813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther GG, Liu G, Ramirez MU, McMonigle RJ, Kim SM, McCracken AN, Joo Y, Ushach I, Nguyen NL, Edinger AL, 2014. Loss of TSC2 confers resistance to ceramide and nutrient deprivation. Oncogene 33, 1776–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafliger P, Graff J, Rubin M, Stooss A, Dettmer MS, Altmann KH, Gertsch J, Charles RP, 2018. The LAT1 inhibitor JPH203 reduces growth of thyroid carcinoma in a fully immunocompetent mouse model. J Exp Clin Cancer Res 37, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa Y, Zhu X, Kamiya A, 2019. NV-5138 as a fast-acting antidepressant via direct activation of mTORC1 signaling. J Clin Invest 130, 2207–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iffland PH 2nd, Baybis M, Barnes AE, Leventer RJ, Lockhart PJ, Crino PB, 2018. DEPDC5 and NPRL3 modulate cell size, filopodial outgrowth, and localization of mTOR in neural progenitor cells and neurons. Neurobiol Dis 114, 184–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iffland PH, Crino PB, 2017. Focal Cortical Dysplasia: Gene Mutations, Cell Signaling, and Therapeutic Implications. Annu. Rev. Pathol 12, 547–571. [DOI] [PubMed] [Google Scholar]
- Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM, 2013. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341, 1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassai H, Sugaya Y, Noda S, Nakao K, Maeda T, Kano M, Aiba A, 2014. Selective activation of mTORC1 signaling recapitulates microcephaly, tuberous sclerosis, and neurodegenerative diseases. Cell Rep 7, 1626–1639. [DOI] [PubMed] [Google Scholar]
- Kim JS, Ro SH, Kim M, Park HW, Semple IA, Park H, Cho US, Wang W, Guan KL, Karin M, Lee JH, 2015. Sestrin2 inhibits mTORC1 through modulation of GATOR complexes. Sci Rep 5, 9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafourcade CA, Lin TV, Feliciano DM, Zhang L, Hsieh LS, Bordey A, 2013. Rheb activation in subventricular zone progenitors leads to heterotopia, ectopic neuronal differentiation, and rapamycin-sensitive olfactory micronodules and dendrite hypertrophy of newborn neurons. J. Neurosci 33, 2419–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin TV, Hsieh L, Kimura T, Malone TJ, Bordey A, 2016. Normalizing translation through 4E-BP prevents mTOR-driven cortical mislamination and ameliorates aberrant neuron integration. Proceedings of the National Academy of Sciences of the United States of America 113, 11330–11335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y, Suzuki M, Shinohara Y, Fujimoto T, 2010. Lysosomal accumulation of mTOR is enhanced by rapamycin. Histochem Cell Biol 134, 537–544. [DOI] [PubMed] [Google Scholar]
- Ribierre T, Deleuze C, Bacq A, Baldassari S, Marsan E, Chipaux M, Muraca G, Roussel D, Navarro V, Leguern E, Miles R, Baulac S, 2018. Second-hit mosaic mutation in mTORC1 repressor DEPDC5 causes focal cortical dysplasia-associated epilepsy. J Clin Invest 128, 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Sabatini DM, 2009. Rag proteins regulate amino-acid-induced mTORC1 signalling. Biochem Soc Trans 37, 289–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW, 2012. NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolov AM, Seluzicki CM, Morton MC, Feliciano DM, 2018. Dendrite growth and the effect of ectopic Rheb expression on cortical neurons. Neurosci Lett 671, 140–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS, 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem 284, 8023–8032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai V, Parker WE, Orlova KA, Baybis M, Chi AW, Berg BD, Birnbaum JF, Estevez J, Okochi K, Sarnat HB, Flores-Sarnat L, Aronica E, Crino PB, 2014. Fetal brain mTOR signaling activation in tuberous sclerosis complex. Cereb. Cortex 24, 315–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Sabatini DM, 2017. The Dawn of the Age of Amino Acid Sensors for the mTORC1 Pathway. Cell Metab 26, 301–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng LH, Rensing NR, Zhang B, Gutmann DH, Gambello MJ, Wong M, 2011. Tsc2 gene inactivation causes a more severe epilepsy phenotype than Tsc1 inactivation in a mouse model of tuberous sclerosis complex. Hum. Mol. Genet 20, 445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Clister TL, Lowry PR, Seldin MM, Wong GW, Zhang J, 2015. Dynamic Visualization of mTORC1 Activity in Living Cells. Cell Rep 10, 1767–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li S, Zhang J, 2016. Tracking the Activity of mTORC1 in Living Cells Using Genetically Encoded FRET-based Biosensor TORCAR. Curr Protoc Chem Biol 8, 225–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: CRISPR-Cas9 Tsc2 or Depdc5 KO results in mTOR pathway hyperactivation. CRISPR-Cas9 editing of regions within mouse Tsc2 (A) and Depdc5 (B) were verified using a T7 endonuclease assay. Indels created by CRISPR-Cas9 editing are revealed as multiple bands after exposure to T7 endonuclease (arrows in A and B). To verify a complete digest reaction, a manufacturer provided positive control was used (PC, at 400 and 200 bp respectively, A,B). Wildtype (WT) and Scramble (Scram) control DNA was also assayed by T7 assay using primers covering the same regions as those assayed in CRISPR edited cells. No digested bands were observed in control samples (A,B). A hallmark of mTOR pathway hyperactivation is increased cell body size. Tsc2 and Depdc5 KO cell lines display significantly increased cell body size (n=50 cells per group; two replicates) compared to scramble and WT controls (C and D, p<0.001, ***). Increases in cell size after Tsc2 or Depdc5 KO were rescued with rapamycin (p<0.001) or torin1 application (p<0.001). Size differences between WT and scramble vehicle treated cells were not significant. In rapamycin or torin1 treated N2aC, cell size decreases were significant at p < 0.05 (*). In D, each box represents SEM, whiskers reflect 5–95% confidence intervals. Western assay of Tsc2 KO N2aC reveals a decrease in Tsc2 protein compared to scramble and WT cells as well as an increase in PS6 (240/244) (E). An increase in PS6 (240/244) was also observed in Depdc5 KO N2aC (G). After Tsc2 or Depdc5 KO, increases in PS6 (240/244) were rescued by rapamycin or torin1 application (F and H). Western blot densitometry data are in Figure S2 (n=3 per blot). U = undigested, D= digested, KO = knockout, Rapa= rapamycin, F-actin = filamentous actin.
Figure S2: Densitometry data for figure S1. All densitometry measurements are normalized to loading control (β-actin) and performed in triplicate. Levels of Tsc2 (A) and phosphorylated S6 (240/244) (B-G) were determined using lysates made after incubation in complete media only. All rapamycin and torin1 blots were performed using lysates from cells incubated with rapamycin (150 nM) or torin1 (100 nM) in complete media for 1 hour. Error bars indicate SEM. PS6= phosphorylated S6 (240/244). Densitometry data correspond to blots in Figure S1 as follows: A)= Fig. S1E; B)= Fig. S1E; C)= Fig. S1F; D)= Fig. S1F; E)= Fig. S1G; F)= S1H; G)= Fig. S1H.
Figure S3: Incubation of N2a cells in AAF media for 1 hr. does not result in cell death. Live N2aC were assayed with annexin V and propidium iodide to determine whether or not 1 hr. of incubation in AAF media resulted in induction of apoptosis or necrosis, respectively. As a positive control, the topoisomerase II inhibitor Etoposide was used (A). Etoposide treated cells expressed high levels of annexin V and took up propidium iodide vs. N2a cells in AAF media (B) or WT control (C) cells in complete media.
Figure S4: There is no statistical difference in C:Y between incubation in AAF media with glucose vs. media without glucose. To ensure that the mTOR inhibition observed in AAF media without glucose was not caused by an absence of glucose, WT N2aC were assayed in AAF media without and with glucose (50 mM; “high glucose media”). In (A) WT N2a cell baseline in complete media is shown (same data as in Figure 1). A decrease in C:Y was observed in WT N2a cells in AAF media without glucose (avg. percent decrease of 12.2%, same data as Figure 1; p<0.05) and in AAF media with glucose (avg. percent decrease of 13.6%; p<0.05). The difference between AAF media types was not statistically significant. *p<0.05.
Figure S5: Decreases in C:Y during incubation in AAF media can be rescued by incubation in complete media. (A) A decrease in C:Y during AAF media incubation (striped bars in E) was observed in WT (3.8%), scramble (3.7%), and Tsc2 KO (2.5%) N2aC (all significant, p <0.05). After incubation in complete media (solid bars in E), an increase in C:Y was observed in WT (A, 0.05%), scramble (B, 3.2%) and Tsc2 KO (C, 1.5%) N2aC. In scramble and Tsc2 KO cells, this increase was statistically significant from baseline (p <0.05). No significant change in C:Y was observed during AAF media incubation or in complete media in Depdc5 KO cells (D).
Figure S6: CFP:YFP Ratio in CRISPR scramble and TORCAR co-transfected N2a cells compared to WT N2a cells transfected with TORCAR alone. CRISPR scramble and TORCAR co-transfected cells were incubated in (A) AAF media, (B) in complete media with rapamycin, or (C) in complete media with torin1 after a 10 minute baseline in complete media for each condition. An average 8.9% decrease in C:Y was observed in cells incubated in AAF media (A, D; p < 0.05). Cells incubated in rapamycin (B) or torin1 (C) containing complete media displayed a 1.5% increase and 7.1% decrease in C:Y, respectively (D; p < 0.05). The difference in C:Y between AAF media and torin1 containing complete media was not significant. These trends are similar to what is observed in WT cells in Figure 1. AAF = amino acid free, rapa = rapamycin. Error bars in (D), standard error of the mean. *p<0.05.
Figure S7: Densitometry data for figure 6. All densitometry measurements are normalized to loading control (beta-actin) and performed in triplicate. Levels of PS6 (A,B) were determined using lysates made after incubation in AAF media only for 1 hour. Error bars indicate SEM. Densitometry data correspond to blots in Figure S6 as follows: A)= Fig. 6C left, B)= Fig. 6C right.
Figure S8: Rapamycin dose response curve in WT cells transfected with TORCAR. To ensure that the absence of change in C:Y ratio after rapamycin treatment in N2aC was not due to insufficient concentration of rapamycin, a dose response experiment was performed. N2aC transfected with TORCAR were incubated in complete media containing 150 nM, 250 nM, 350 nM, 450 nM, or 550 nM rapamycin after a 10 minute baseline recording in complete media. During incubation with 150 nM, 250 nM, and 350 nM rapamycin, an increase in average C:Y of 2.6%, 1.0%, and 4.5% was observed, respectively. During incubation with 450 nM and 550 nM rapamycin decreases in C:Y of 2.6% and 4.1% were observed respectively. These concentrations are much higher than was is required to inhibit phosphorylation of S6 (Figure S1) and result in a decrease in C:Y less than torin1 or AAF media (Figure 1). No changes in C:Y were statistically significant from baseline at any rapamycin concentration.
Figure S9: A decrease is C:Y ratio is observed in Tsc2, Depdc5, and WT neurons after incubation in complete media containing torin1. N2a neurons were transfected with TORCAR and imaged in torin1 containing complete media after a 10 minute baseline in complete media only. Similar to what was observed in undifferentiated N2a cells, a statistically significant decrease in C:Y was observed in Tsc2 KO (11.8%), Depdc5 KO (7.6%), and WT neurons (6.8%). *p<0.05.
Figure S10: N2a cells express multiple neuronal makers. N2a cells were used in this study due to their ease of use in transfection, Western blot assays, FAC sorting, and live cell imaging. N2a cells are also easy to maintain in culture and can be rapidly assayed. Further, N2a cells express many typical neuronal markers including tau protein (A), microtubule associated protein 2 (MAP2) and Nestin (B), and Neuron-specific class III beta-tubulin (TUJ-1; C). For these experiments, N2aC were not differentiated using dibutyryl-cAMP and thus they do not express Tyrosine hydroxylase (TH; C). Filamentous actin (F-actin) was used in this study to assess cell soma size (A).
Movie S1: Representative time-lapse of CFP/YFP channels in WT N2a cells treated with torin1
Movie S2: 3D surface plot of a WT N2a cell in complete media. Movie created from surface plot shown in Figure 5.
Movie S3: 3D surface plot of a WT N2a cell in AAF media. Movie created from surface plot shown in Figure 5.
Movie S4: 3D surface plot of a Scramble transfected N2a cell in complete media. Movie created from surface plot shown in Figure 5.
Movie S5: 3D surface plot of a Scramble transfected N2a cell in AAF media. Movie created from surface plot used in Figure 5.
Movie S6: 3D surface plot of a Tsc2 KO N2a cell in complete media. Movie created from surface plot shown in Figure 5.
Movie S7: 3D surface plot of a Tsc2 KO N2a cell in AAF media. Movie created from surface plot shown in Figure 5.
Movie S8: 3D surface plot of a Depdc5 KO N2a cell in complete media. Movie created from surface plot shown in Figure 5.
Movie S9: 3D surface plot of a Depdc5 KO N2a cell in AAF media. Movie created from surface plot shown in Figure 5.