Abstract
The tumor suppressor function of p14ARF is regulated at a posttranslational level via mechanisms yet to be fully understood. Here, we report the identification of an unconventional p14ARF degradation pathway induced by the chaperone HSP90 in association with the E3 ubiquitin ligase C-terminus of HSP70-interacting protein (CHIP). The ternary complex of HSP90, CHIP, and p14ARF was required to induce the lysosomal degradation of p14ARF by an ubiquitination-independent but LAMP2A-dependent mechanism. Depletion of HSP90 or CHIP induced p14ARF-dependent senescence in human fibroblasts. Premature senescence observed in cells genetically deficient in CHIP was rescued in cells that were doubly deficient in CHIP and p14ARF. Notably, non–small cell lung cancer cells (NSCLC) positive for p14ARF were sensitive to treatment with the HSP90 inhibitor geldanamycin. Furthermore, overexpression of HSP90 and CHIP with a concomitant loss of p14ARF correlated with poor prognosis in patients with NSCLC. Our findings identify a relationship between p14ARF and its chaperones that suggest new therapeutic strategies in cancers that overexpress.
Introduction
Non–small cell lung cancer (NSCLC) constitutes approximately 85% to 90% of lung cancers, with a 5-year survival rate of 15.9% (1, 2). NSCLC is a primary subtype of lung cancer that displays relative insensitivity to chemotherapy and radiotherapy compared with small-cell lung cancer (SCLC), which accounts for the majority of other lung cancers (3–7). NSCLC displays genetic and cellular heterogeneity such as mutation and amplification of oncogenes including HER2, MET, FGFR1, FGFR2, ALK, ROS1, NRG1, NTRK1, and RET (2, 8–10). In contrast, a small number of inhibitors with specific targets such as EGFR mutation or EML4–ALK fusion are currently available for clinical NSCLC treatment. Other clinical trials using inhibitors that target driver mutations including FGFR, DDR2, KRAS, and PI3K are also currently underway (11).
HSP90 is a major factor that maintains the stability of a variety of proteins. Thus, perturbation of the status of HSP90 would disturb cellular homeostasis, leading to cancer cell death. HSP90 is involved in various cellular mechanisms such as protein folding and activation, cell-cycle control, and transcriptional regulation (12–14). In addition to enabling cancer cells to cope with drastic changes in protein homeostasis, HSP90 displays tumorigenic properties via stabilization of the oncogenic proteins such as HER2, AKT, TERT, Raf1, mutant p53, etc (14–27). Because inhibiting HSP90 leads to destabilization of its client oncoproteins, several HSP90 inhibitors were developed as cancer drugs and underwent clinical trials in various human cancers (28). Treatment with geldanamycin (GA) inhibits HSP90 ATPase activity, which is essential for the degradation of HSP90 client oncoproteins and stimulates cell-cycle arrest and apoptosis (29). Recent reports have also suggested that GA treatment induces cellular senescence by an unknown mechanism (30). Although HSP90 may play a role in cellular senescence through TERT stabilization, the accurate mechanism of HSP90 inhibition–induced cellular senescence remains to be identified (26, 31). C-terminus of HSP70-interacting protein (CHIP) is one of the major cochaperones of HSP90 and can regulate the ubiquitylation and degradation of HSP90 target proteins. CHIP contains the N-terminal TPR domain and C-terminal U-Box domain, which are required for the interaction with chaperones and for the recruitment of the E2 enzyme and ubiquitylation, respectively (32, 33). In association with HSP70, CHIP typically induces the degradation of HSP90 client proteins when HSP90 fails to induce their structural maturation into their native forms possibly through a hostile environment or HSP90 inhibition by drugs (33).
p14ARF is an alternative reading frame product of the INK4a/ARF locus and functions as a tumor suppressor through p53-dependent and p53-independent pathways (34). p14ARF is transcriptionally induced by oncogenic signaling such as c-myc and causes oncogene-induced senescence, which results in tumor suppression (35). Although p14ARF has been reported to be regulated by posttranslational modifications (10, 11, 36), the roles of molecular chaperones in directly modulating p14ARF levels and the related cellular senescence have not been identified. Here, we demonstrate that HSP90 plays a powerful oncogenic role by inducing p14ARF degradation, which prevents cellular senescence. The ternary complex of HSP90, CHIP, and p14ARF was required to induce the ubiquitylation-independent and LAMP2A-dependent lysosomal degradation of p14ARF. Furthermore, HSP90 and CHIP overexpression together with a loss of p14ARF expression correlated with poor prognosis in advanced human NSCLC and served as an important prognostic marker. This study is the first to identify molecular chaperone-mediated degradation of the tumor suppressor p14ARF and to demonstrate its clinical importance in NSCLC.
Materials and Methods
Cell culture and transfection
An embryonic kidney cell line (293T), a cervical cancer cell line (HeLa), a human NSCLC cell line (H1299), the IMR90 fibroblast line, the human foreskin fibroblast (HFF) line, and human lung cancer cell lines including A549, NCI-H460, NCI-H522, NCI-H23, and NCI-H226 were provided by ATCC. The human lung cancer cell lines including NCI-H322M, HOP62, and EKVX were provided from the U.S. National Cancer Institute (NCI; MTA no. 2702–09). PC-9 cells were purchased in 2015 from Sigma-Aldrich Corporation. 293T, HeLa, IMR90, and HFF were obtained in 2014. H1299 was purchased in 2012. Rest of the cell lines were obtained in 2009.
All lung cancer cell lines except PC-9 and H1299 were tested and authenticated annually by STR test in National Cancer Center Core Facility, Korea. STR profile was analyzed using Gene Mapper v 5.0 software. All the provided cell lines were aliquoted and frozen within passage 10, and experiments were performed using thawed cell lines from passage 7 to 18. Using e-Myco plus Mycoplasma PCR Detection Kit (Intron), Cell lines were tested for infection of mycoplasma as soon as receipt. Whenever subculture was performed, cells were treated with plamosin (Invivogen) to prevent infection of mycoplasma. 293T and HeLa cells and HFFs were maintained in DMEM (Hyclone) supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco). Human lung cancer cell lines including H1299 and others were grown in RPMI (Gibco), and IMR90 fibroblasts were maintained in MEM (Gibco). RPMI and MEM were also supplemented with 10% FBS and 1% penicillin/streptomycin. All cell lines were grown in 5% CO2 at 37°C. siRNAs were transfected into IMR90 fibroblasts and HFFs as well as in HeLa and lung cancer cell lines using Lipofectamine RNAiMAX. Plasmids expressing p14ARF, CHIP, and HSP90 were transfected into H1299 and 293T cells using Lipofectamine 2000 (Invitrogen).
Biochemical assays
Cell lysates were immunoprecipitated and immunoblotted as described previously (11). SA-β-galactosidase staining, cell viability assays, crystal violet staining assays were performed as described before (11).
Preparation of CHIP−/− MEFs and p19ARF−/−;CHIP−/− MEFs
C57BL/6-Chip+/− mice was kindly provided by RIKEN Bio Resource Center. B6.129 mice with a Cdkn2atm1Cjs/Nci allele (homozygous mice, −/−) were purchased from the National Cancer Institute (Frederick, MD). p19ARF+/− mice were further backcrossed until N6 in the B6 background. Female and male CHIP+/− mice were crossed to produce CHIP+/+ and CHIP−/− MEFs. p19ARF+/− mice and CHIP+/− mice were crossed to generate p19ARF+/−;CHIP+/− mice. p19ARF−/−;CHIP−/− MEFs were produced from crossing female and male p19ARF+/−;CHIP+/− mice. MEFs were generated as described previously (11).
Human tumor samples
In total, 354 NSCLC cases were selected from the pathology case archive of Toyama University Hospital and National Hospital Organization Higashi-Ohmi General Medical Center based on the diagnosis and quality of the available tissue on the paraffin blocks. None of the patients received neoadjuvant treatments and all patients underwent complete resection between 1993 and 2010. The patients who had relapsed were treated with standard chemotherapy. The tumors were staged according to the International Union against Cancer tumor-node-metastasis classification, and histology was defined and graded according to the 2004 WHO guidelines. This study was approved by the ethics committees at Toyama University Hospital and National Hospital Organization Higashi-Ohmi General Medical Center, and informed consent was obtained from each patient. This study was also approved by the Office of Human Subjects Research at the NIH (Bethesda, MD).
Statistical analysis
Student t test was used to determine whether the expression of each protein was associated with clinicopathologic characteristics, with protein expression data expressed as the mean and 95% confidence interval (CI). The cut-off value for discriminating between low and high expression was determined through ROC curve analysis (37). Overall survival was analyzed according to the Kaplan–Meier product-limit method, and the survival curves were compared with the log-rank test. Subsequently, we used a multivariate proportional Cox model and adjusted for the following clinical/pathologic variables: age at diagnosis, gender, smoking history, cancer type, pT factor, and lymph node metastasis. P values were considered significant when less than 0.05. Statistical analysis was performed using SPSS version 21.0 (SPSS) and the R statistical package (Version 3.1.2).
Immunohistochemistry and scoring
Tissue microarrays (TMA) were constructed from 354 formalin-fixed, paraffin-embedded tissue specimens. For each case, areas with the most representative histology were selected from a review of hematoxylin and eosin (H&E)-stained slides. We performed IHC as described previously (38). Images were analyzed using Visiopharm software version 4.5.1.324 (Visiopharm). The thresholds for the size and shape of tumor cells were manually calibrated. Briefly, brown (3,3′-diaminobenzidine, DAB) and blue (hematoxylin) cells were separated by spectrum. A brown staining intensity (0, negative; 1, weak; 2, moderate; and 3, strong) was obtained using a predefined algorithm and optimized settings. The overall immunohistochemical score (histoscore) was expressed as the percentage of positive cells multiplied by their staining intensity (possible range, 0–300).
The information on plasmid construction, sequences of siRNA and shRNA, primer sequences, antibodies, and chemicals are available in Supplementary information.
Results
HSP90 suppresses cellular senescence
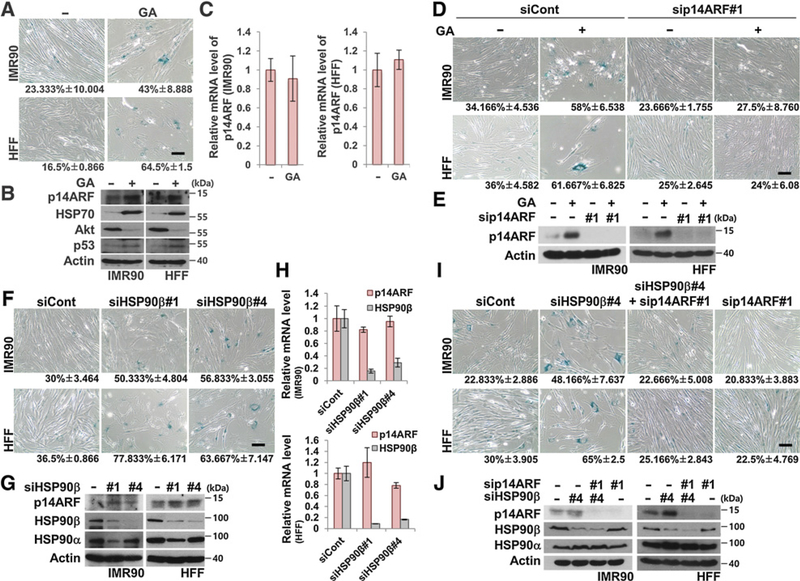
To determine the effect of GA on cellular senescence (30), various concentrations of GA ranging from 0.05 to 1.0 μmol/L were used to treat human fetal lung (IMR90) fibroblasts and HFF. Regardless of the concentration, GA treatment doubled or tripled the amount of β-galactosidase–positive senescent IMR90 fibroblasts and HFFs compared with controls (Fig. 1A and Supplementary Fig. S1A). Interestingly, we observed an increase in p14ARF protein at different concentrations of GA within 24 hours (Supplementary Fig. S1B and S1C). Furthermore, p14ARF protein seemed to be stabilized because there was no change in p14ARF mRNA levels (Fig. 1B and C). Because there was very little cleaved PARP or cleaved caspase-3 detected, IMR90 fibroblasts and HFFs likely did not undergo apoptosis under these conditions (Supplementary Fig. S1B and S1C). The increased and decreased levels of HSP70 and AKT, respectively, suggested that effects of GA treatment (Fig. 1B and Supplementary Fig. S1B). p14ARF, a potent inducer of senescence, was ablated in the GA-treated fibroblasts. Interestingly, GA-induced cellular senescence was suppressed by p14ARF depletion, suggesting possible involvement of p14ARF in GA-induced senescence (Fig. 1D and E). Because GA is a known HSP90 inhibitor, we tested whether HSP90α or β depletion would cause similar effects in human fibroblasts. As expected, HSP90α or β knockdown by isoform-specific HSP90 siRNAs accelerated cellular senescence (Fig. 1F) and increased p14ARF protein levels without altering p14ARF mRNA levels (Fig. 1G and H and Supplementary Fig. S2A and S2B). siHSP90β#1 suppressed expression of both HSP90α and β, which prompted us to exclude this siRNA in later experiments (Fig. 1G). Simultaneous depletion of p14ARF and HSP90 protected fibroblasts against cellular senescence, indicating a possible interaction of HSP90 with p14ARF (Fig. 1I and J and Supplementary Fig. S2C and S2D). Overall, these observations suggest that plausible posttranslational regulation of p14ARF in connection with HSP90 inhibition should affect cellular senescence.
Figure 1.
Inhibition or ablation of HSP90 accelerates cellular senescence via stabilization of p14ARF. A, IMR90 fibroblasts and HFFs were treated with 0.1 μmol/L GA for 24 hours and stained for SA-β-galactosidase activity. Numbers under the images indicate the percentage of senescent cells. Scale bar, 150 μm. B and C, Two normal cell lines were treated with GA under identical conditions as in A. The cell lysates were immunoblotted. Actin was detected as a loading control. The mRNAs were analyzed by qRT-PCR. D, After 48 hours of transfection with control and p14ARF siRNA #1, the cells were treated with 0.1 μmol/L GA for 48 hours, followed by SA-β-galactosidase activity staining. E, Under identical conditions as in Fig. 1D, the cell lysates were immunoblotted. F, After 96 hours of transfection with 40 nmol/L HSP90β siRNAs #1 and #4, two normal cell lines were stained for SA-β-galactosidase activity. G and H, Two normal cell lines were transfected with 40 nmol/L HSP90β siRNAs #1 and #4 for 96 hours. The cell lysates were immunoblotted. mRNA expression was analyzed by qRT-PCR. I, HSP90β and p14ARF siRNAs were transfected as indicated, followed by SA-β-galactosidase staining after 96 hours. J, Under identical conditions as in Fig. 1I, the cell lysates were immunoblotted. Data represent the mean ± SEM; P values by two-tailed Student t test.
CHIP is an essential mediator linking HSP90 to p14ARF for its degradation
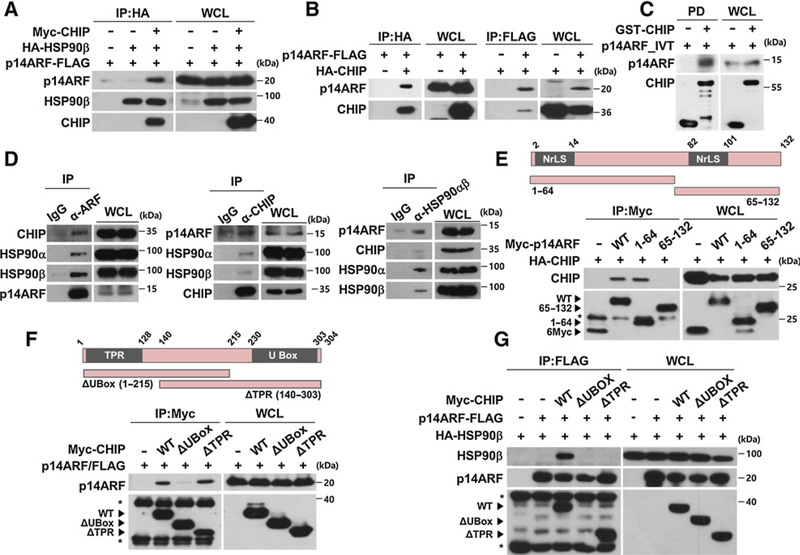
The effects of HSP90 inhibition on the levels of p14ARF were further investigated. GA treatment or HSP90 ablation induced an increase in p14ARF protein levels and prolonged the half-life of the p14ARF protein under cycloheximide treatment in the human cervical cancer cell line HeLa (Supplementary Fig. S3A–S3D). Because HSP90 inhibition induces HSP70 expression (39), we also tested the effects of HSP70 on p14ARF stability. The results showed that the accumulation of p14ARF following GA treatment was not affected by depletion of HSP70, suggesting that HSP70 might not regulate p14ARF stabilization (Supplementary Fig. S3E and S3F). However, no direct interaction between HSP90β and p14ARF was observed, suggesting the existence of plausible connector between the two proteins (Supplementary Fig. S4). Because CHIP is a well-known counterpart of molecular chaperones including HSP90 and HSP70 and delivers their target proteins for degradation, we tested whether CHIP could be associated with HSP90 and p14ARF. Interestingly, HSP90α or β bound p14ARF only when CHIP was coexpressed (Fig. 2A and Supplementary Fig. S5A). In contrast, overexpressed or recombinant CHIP was able to bind to p14ARF without HSP90 overexpression, suggesting a possible interaction between CHIP and p14ARF without the aid of HSP90 (Fig. 2B and C). When endogenous p14ARF protein was immunoprecipitated, it bound both isoforms of HSP90 and CHIP, suggesting that CHIP might mediate the interaction between p14ARF and HSP90α or β (Fig. 2D). Although HSP90 is known to bind to the TPR domain of CHIP (33), p14ARF bound to the U-Box domain of CHIP through its N-terminal region (Fig. 2E and F). Corroborating these findings, p14ARF and ΔTPR mutants formed complexes without HSP90 (Fig. 2F). Because both the TPR and U-Box domains of CHIP were required to form a ternary complex composed of HSP90, CHIP, and p14ARF (Fig. 2G and Supplementary Fig. S5B), CHIP may function as a structural platform that recruits both HSP90 and p14ARF in close proximity.
Figure 2.
HSP90, CHIP, and p14ARF form a ternary complex. A, Plasmids expressing p14ARF-FLAG, Myc-CHIP, and HA-HSP90β were transfected into 293T cells as indicated. After 24 hours, immunoprecipitation assay was performed using cell lysates. HSP90β was immunoprecipitated using anti-HA antibody, and then p14ARF, CHIP, and HSP90 were detected using antibodies as indicated. B, Cells were transfected with plasmids as indicated. After 24 hours, the cell lysates were immunoprecipitated using anti-HA or anti-FLAG antibody. C, GST pull-down assay was performed using in vitro translated p14ARF and purified GST-CHIP. D, HeLa (human cervical cancer) cell lysates were immunoprecipitated using anti-p14ARF, anti-CHIP, and anti-HSP90α/β antibodies. E, Plasmids expressing Myc/full-length p14ARF, Myc/p14ARF deletion mutants, and HA-CHIP were transfected as indicated. The cell lysates were immunoprecipitated using anti-Myc antibody. F, Plasmids expressing Myc/full-length CHIP, Myc/CHIP deletion mutants, and p14ARF-FLAG were transfected as indicated. The cell lysates were immunoprecipitated using anti-Myc antibody. G, Constructs were transfected as indicated. The cell lysates were immunoprecipitated using anti-FLAG antibody. p14ARF protein expression was adjusted for comparing interactions with identical amounts of p14ARF. Asterisks, chains of antibodies.
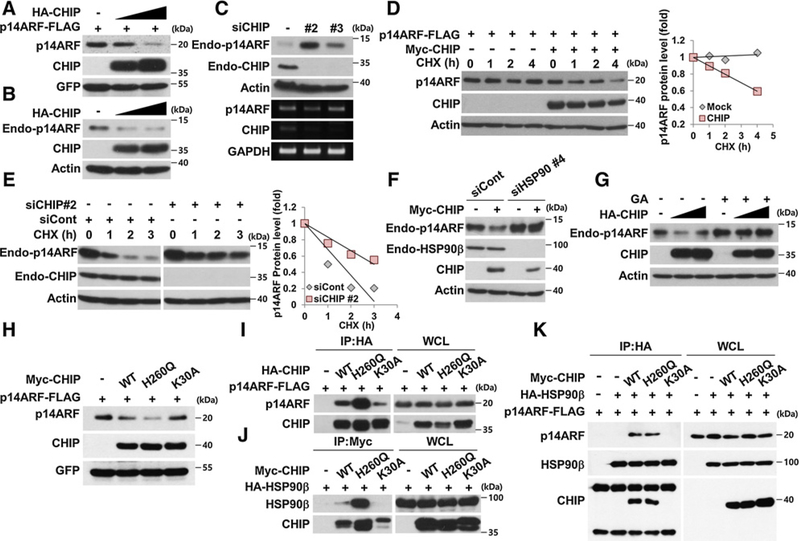
Next, we tested p14ARF degradation under various conditions. CHIP overexpression induced the degradation of exogenously and endogenously expressed p14ARF (Fig. 3A and B). In contrast, CHIP knockdown by siRNA increased endogenous p14ARF protein levels without any change in its mRNA level (Fig. 3C). Upon cycloheximide treatment, CHIP overexpression decreased p14ARF stability while CHIP depletion increased its stability (Fig. 3D and E). We next tested whether HSP90 is required for CHIP-mediated p14ARF degradation. Of note, CHIP could not induce p14ARF degradation when HSP90 was depleted by siRNA or when its function was inhibited by GA treatment (Fig. 3F and G and Supplementary Fig. S6). These results suggest that both HSP90 and CHIP are responsible for p14ARF degradation. To further analyze the action of CHIP in mediating p14ARF degradation in association with HSP90, two mutants of CHIP, K30A and H260Q, were used. As expected, CHIP K30A, a mutant defective in chaperone binding that failed to interact with HSP90 and to form a ternary complex (Fig. 3J and K and Supplementary Fig. S5C), was able to bind to p14ARF but could not induce p14ARF degradation (Fig. 3H and I). In contrast to K30A, CHIP H260Q, a mutant defective in E3 ligase activity, formed a ternary complex with p14ARF and HSP90 (Fig. 3I–K and Supplementary Fig. S5C) and induced p14ARF degradation (Fig. 3H). Overall, these results indicate that CHIP-mediated p14ARF degradation requires HSP90 without ubiquitylation.
Figure 3.
HSP90 is required for CHIP-induced ubiquitylation-independent p14ARF degradation. A and B, H1299 cells were transfected with constructs as indicated. The cell lysates were immunoblotted. C, HeLa cells were transfected with 30 nmol/L CHIP siRNAs for 72 hours. mRNAs were analyzed by RT-PCR using primers specific for p14ARF and CHIP. D, H1299 cells were transfected with constructs for 24 hours, followed by cycloheximide (CHX; 100 μg/mL) treatment. The graph indicates the relative p14ARF protein levels compared with the actin protein levels. E, HeLa cells were transfected with 30 nmol/L CHIP siRNA for 72 hours and treated with cycloheximide. F, H1299 cells were transfected with 40 nmol/L HSP90β siRNA for 72 hours, and then plasmids expressing Myc-CHIP were transfected as indicated. G, HeLa cells were transfected with plasmid expressing HA-CHIP and treated with 0.5 μmol/L GA for 24 hours. H, Constructs expressing Myc-CHIP WT, H260Q, K30A, p14ARF-FLAG, and GFP were transfected into H1299 cells. I, 293T cells were transfected with plasmids as indicated. The cell lysates were immunoprecipitated using anti-HA antibody. J, 293T cells were transfected with plasmids as indicated. The cell lysates were immunoprecipitated using anti-Myc antibody. K, Plasmids were expressed in 293T cells, and the cell lysates were immunoprecipitated using anti-HA antibody as indicated. p14ARF protein expression was adjusted for comparing interactions with identical amounts of p14ARF.
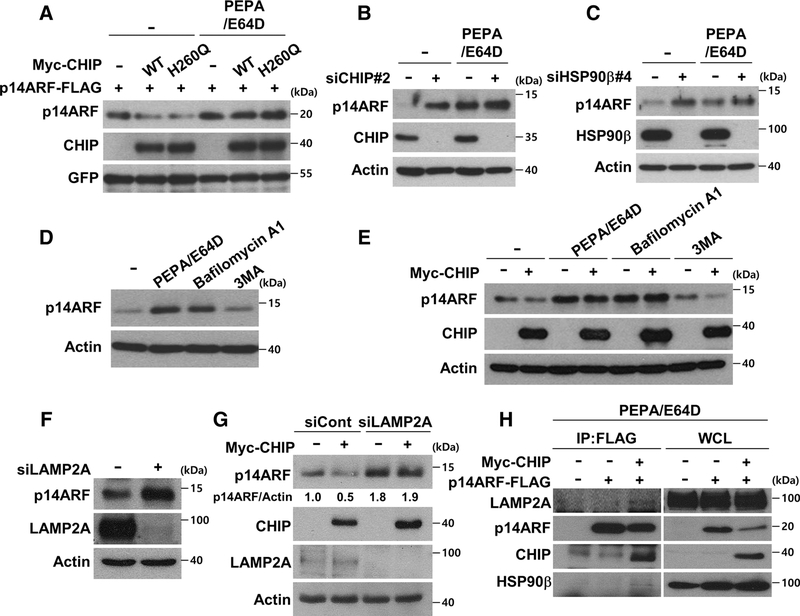
HSP90- and CHIP-dependent p14ARF degradation occurs in an ubiquitin-independent and lysosome-dependent manner
Because CHIP-mediated p14ARF degradation was not rescued by proteasome inhibition (data not shown), the possibility of lysosome-dependent pathways was further examined. CHIP WT- and H260Q mutant-mediated p14ARF degradation was blocked by treatment with the lysosomal inhibitors pepstatin A and E64D (Fig. 4A). p14ARF protein levels that were increased by CHIP or HSP90 knockdown were similar to those stabilized by PEPA/E64D treatment (Fig. 4B and C). Furthermore, no additional stabilization of p14ARF was observed following CHIP or HSP90 ablation when treated with PEPA/E64D, suggesting the involvement of lysosome-dependent degradation (Fig. 4B and C). The autophagy inhibitor 3MA could not stabilize p14ARF excluding involvement of other autophagy processes (Fig. 4D). In addition, although CHIP could not induce p14ARF degradation under lysosomal inhibitor treatment, CHIP induced p14ARF degradation under 3MA treatment (Fig. 4E). Because chaperones are known to mediate the lysosomal degradation of target proteins through their recruitment to LAMP2A, a lysosomal membrane transporter (40), the involvement of LAMP2A in CHIP- and HSP90-mediated p14ARF degradation was tested. LAMP2A ablation increased p14ARF protein levels and blocked CHIP-mediated p14ARF degradation (Fig. 4F and G). Moreover, CHIP increased interactions among p14ARF, HSP90, and LAMP2A upon lysosomal inhibitor treatment, suggesting that CHIP and HSP90 recruit p14ARF to LAMP2A and thus induce lysosomal degradation (Fig. 4H). In conclusion, these data suggest that CHIP and HSP90 cooperatively deliver p14ARF to LAMP2A for lysosomal degradation.
Figure 4.
CHIP and HSP90 induce lysosomal degradation of p14ARF. A, H1299 cells were transfected with constructs as indicated. The transfected cells were treated with 10 μg/mL E64D and 10 μg/mL pepstatin A for 24 hours. B and C, HeLa cells were transfected with 30 nmol/L CHIP siRNA or 40 nmol/L HSP90β siRNA for 72 hours, followed by E64D and pepstatin A treatment for 24 hours. D, HeLa cells were treated with E64D, pepstatin A, 200 nmol/L bafilomycin A1, and 5 mmol/L 3MA for 24 hours as indicated. E, Plasmid expressing Myc-CHIP was transfected into HeLa cells, followed by treatment of inhibitors as indicated. F, After 72 hours transfection with 30 nmol/L LAMP2A siRNA, the cells were immunoblotted. G, HeLa cells were transfected with 30 nmol/L LAMP2A siRNA for 48 hours, followed by overexpression of Myc-CHIP. H, Indicated constructs were expressed in HeLa cells. The transfected cells were treated with E64D and pepstatin A for 24 hours, followed by imunoprecipitation using anti-FLAG antibodies.
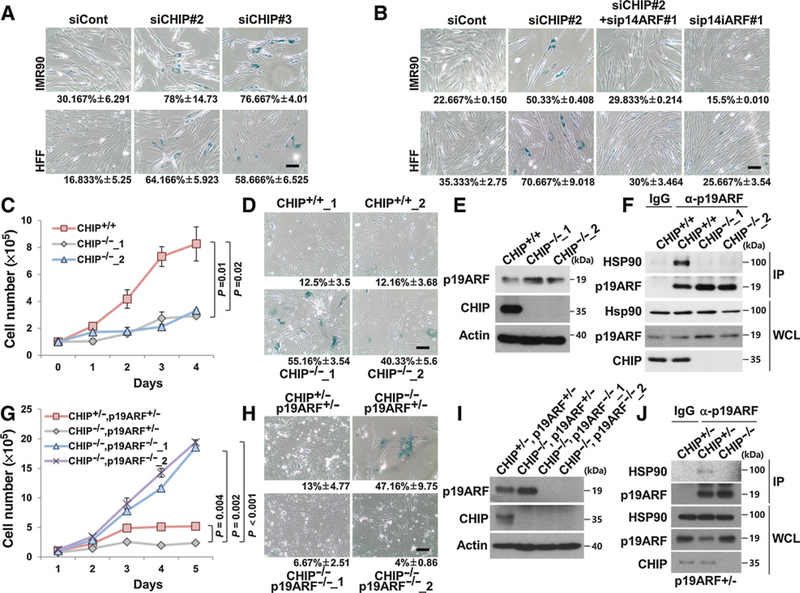
CHIP negatively regulates cellular senescence through p14ARF destabilization
On the basis of the above results, whether CHIP’s ability to induce cellular senescence was dependent on p14ARF was tested. Transient knockdown of CHIP by siRNA accelerated cellular senescence and increased p14ARF protein levels in IMR90 fibroblasts and HFFs (Fig. 5A and Supplementary Fig. S7A and S7B). Simultaneous depletion of p14ARF and CHIP inhibited CHIP knockdown–mediated cellular senescence, suggesting that CHIP prevents cellular senescence via p14ARF destabilization (Fig. 5B and Supplementary Fig. S7C and S7D). To evaluate the physiologic roles of CHIP and p19ARF (mouse homolog of human p14ARF), CHIP KO or CHIP/p19ARF DKO MEFs were generated. CHIP KO MEFs showed growth retardation and premature cellular senescence with increased p19ARF protein levels compared with WT MEFs (Fig. 5C–E). Of note, complex formation between HSP90 and p14ARF disappeared in CHIP KO MEFs consolidating CHIP’s role as a structural mediator (Fig. 5F). The observations of CHIP/p19ARF DKO MEFs displaying significant loss of cellular senescence compared with CHIP KO MEFs and CHIP/p19ARF double hetero MEFs indicate that CHIP KO-induced cellular senescence is p19ARF-dependent (Fig. 5G–I). We further confirmed complex formation between HSP90 and p19ARF in CHIP/p14ARF double hetero MEFs, but not in CHIP-null MEFs, indicating that CHIP was required for HSP90 and p19ARF complex formation (Fig. 5J). CHIP KO MEFs showed cell growth retardation and increased cellular senescence with p19ARF stabilization compared with CHIP/p19ARF double hetero MEFs (Fig. 5G–I). We further investigated the effect of GA treatment on CHIP/p19ARF double hetero, CHIP KO, or CHIP/p19ARF DKO MEFs. The results indicated that the levels of p19ARF in CHIP KO MEFs were not affected by GA treatment, whereas the p19ARF levels in CHIP/p19ARF double hetero MEFs increased. Accordingly, the viability of CHIP KO MEFs was less affected by GA treatment compared with the double hetero MEFs (Supplementary Fig. S8A and S8B). As expected, the DKO MEFs displayed strong resistance to GA treatment. In summary, these results indicate that CHIP prevents cellular senescence by forming ternary complexes with HSP90 and p19ARF, which subsequently induce p19ARF degradation.
Figure 5.
CHIP depletion causes ARF-dependent premature senescence. A, After 72 hours of transfection with 30 nmol/L CHIP siRNAs #2 or #3, the cells were stained for SA-β-galactosidase activity. B, Two normal cell lines were transfected with 20 nmol/L CHIP and p14ARF siRNA for 72 hours, followed by SA-β-galactosidase assay. C, In total, 1 × 105 WT and CHIP KO MEFs were plated equally and counted each day thereafter. This experiment was conducted in triplicate. D, WT and CHIP KO MEFs were stained for SA-β-galactosidase activity at passage 5. E, The levels of p19ARF were detected in CHIP KO MEFs. F, In the same passage as in Fig. 5D, the cell lysates of WT and CHIP KO MEFs were immunoprecipitated using anti-p19ARF antibody. G, In total, 1 × 105 CHIP KO and DKO MEFs were plated equally and counted each day thereafter. H, CHIP KO and DKO MEFs were stained for SA-β-galactosidase activity at passage 8. I, Indicated proteins were detected in DKO MEFs. J, In the same passage, as in Fig. 5H, the cell lysates of littermate double hetero and CHIP KO MEFs were immunoprecipitated using anti-p19ARF antibody. Data represent the mean ± SEM; P values by two-tailed Student t test.
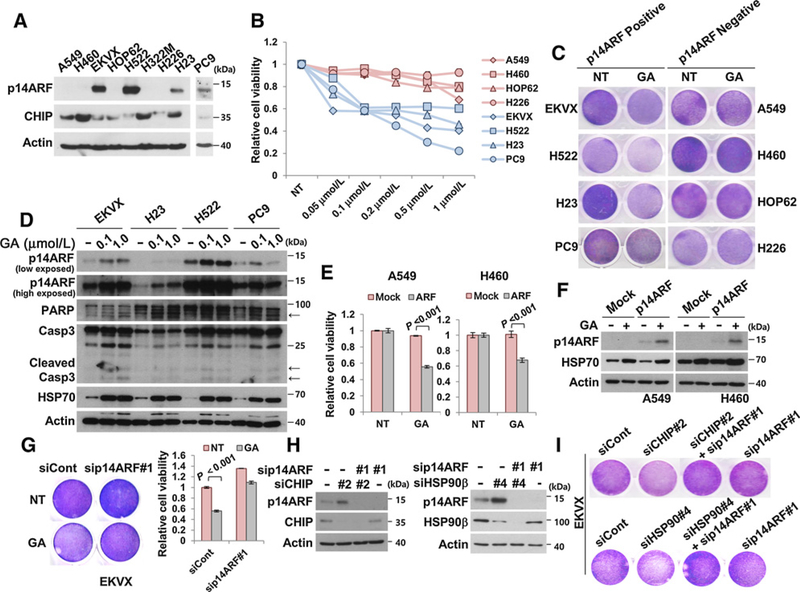
The presence of p14ARF sensitizes lung cancer cells to GA treatment
On the basis of our data that p14ARF depletion prevents GA-induced cellular senescence (Fig. 1D), we examined whether p14ARF could sensitize cancer cells to GA treatment using p14ARF-positive and p14ARF-negative NSCLC cell lines (Fig. 6A). p14ARF-positive NSCLC cell lines, including EKVX, H522, H23, and PC9 were more susceptible to the toxicity of various concentrations of GA treatment compared with p14ARF-negative cell lines (Fig. 6B and C). Furthermore, cell lines containing p14ARF underwent cleavage of PARP and caspase 3, which is a hallmark of apoptosis. This suggested that GA more effectively inhibits growth and induces apoptosis in the presence of p14ARF (Fig. 6D and Supplementary Fig. S9). More importantly, p14ARF expression increased cytotoxicity associated with GA, regardless of ALK fusion or mutations in EGFR or p53, all of which are frequently detected in NSCLC patients (Supplementary Table S2). As expected, GA treatment also increased p14ARF protein levels in NSCLC cell lines, as shown in normal fibroblasts (Fig. 6D). Next, we tested whether p14ARF expression causes cytotoxicity in p14ARF-negative NSCLC cell lines including A549, H460, HOP62, and H226. Under p14ARF overexpression, HOP62 and H226 did not grow (data not shown). In contrast, A549 and H460 survived under stable expression of p14ARF, and they became more susceptible to GA treatment with the elevation of p14ARF proteins (Fig. 6E and F). Finally, the depletion of p14ARF could partially protect EKVX cells from GA-induced cytotoxicity (Fig. 6G). Similar to the normal fibroblasts, CHIP and HSP90 knockdown also induced cell growth retardation with increased p14ARF protein levels. As expected, the effects of CHIP and HSP90 knockdown were blocked by p14ARF ablation (Fig. 6H and I). These suggest that the CHIP- and HSP90-mediated p14ARF regulatory mechanism works in a similar manner in both cancer and normal cells. Importantly, severe sensitization of p14ARF-positive NSCLC cells to GA treatment suggests that p14ARF is suppressed by HSP90.
Figure 6.
p14ARF-positive lung cancer cell lines were more sensitive to GA treatment. A, Cell lysates of indicated cell lines were immunoblotted. B, NSCLC cells were plated in 48-well plates at a density of 4 × 104 cells and treated with the indicated concentrations of GA for 36 hours. C, NSCLC cell lines expressing p14ARF were treated with 0.1 μmol/L GA for 36 hours, followed by crystal violet staining. D, p14ARF-positive NSCLC cell lines were treated with 0.1 or 1.0 μmol/L GA for 36 hours, followed by immunoblotting using antibodies as indicated. Arrows, cleaved proteins. E and F, A549 and H460 were stably transfected with p14ARF. Identical amounts of cells were plated in 48-well plates and treated with 0.1 μmol/L GA for 36 hours. G, EKVX cells were transfected with 30 nmol/L p14ARF siRNA #1. After 24 hours, cells were treated with 0.1 μmol/L GA for 48 hours, followed by crystal violet staining. H, EKVX cells were transfected with 20 nmol/L CHIP siRNA #2, 20 nmol/L HSP90β siRNA #4, or 20 nmol/L p14ARF siRNA #1, as indicated. I, Under the identical condition as in Fig. 6H, cells were stained with crystal violet. Data represent the mean ± SEM. P values were determined using a two-tailed Student t test. Cell viability was measured in triplicate using CellTiter-Glo reagent.
HSP90 overexpression and loss of p14ARF expression correlate with poor prognosis in human NSCLC
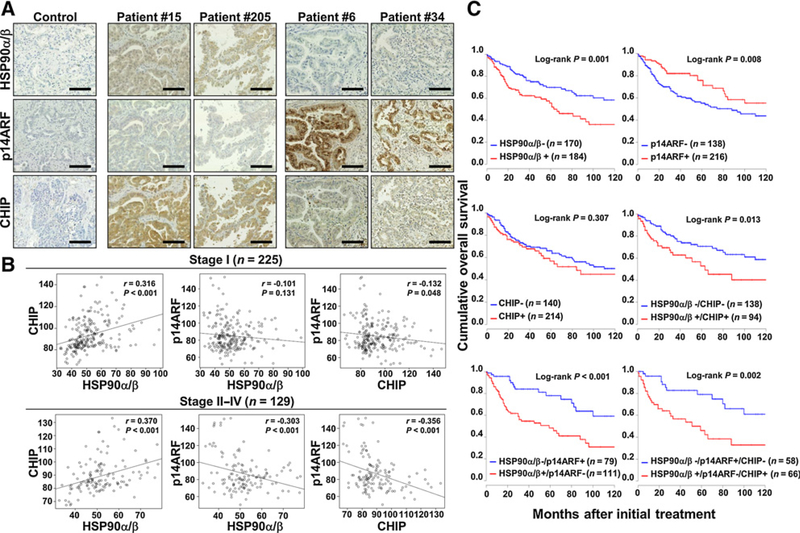
To determine the clinical relevance of HSP90, p14ARF, and CHIP expression in human cancer, we compared their expression levels by IHC in tissues from patients with NSCLC. The clinicopathologic characteristics of the study population are summarized in Supplementary Table S3. CHIP immunoreactivity significantly correlated with ADC (P = 0.007), earlier disease status including stage (P = 0.003) and pN status (P = 0.037). CHIP expression positively correlated with HSP90 expression in both early and advanced stages (both P < 0.001, Spearman ρ = 0.316 and 0.370, respectively), whereas CHIP expression negatively correlated with p14ARF expression in advanced stages (P < 0.001 and Spearman ρ = −0.356). Although the negative correlation between HSP90 and p14ARF was not strong (P = 0.001 and Spearman ρ = −0.175) in all NSCLC cases, significant correlation between both markers was observed in advanced staged specimens (P < 0.001 and Spearman ρ = −0.303; Supplementary Fig. S10 and Fig. 7A and B). Notably, these data suggest that the HSP90–p14ARF–CHIP axis is closely linked in NSCLC.
Figure 7.
HSP90, p14ARF, and CHIP expression in human NSCLC specimens. A, Representative images of immune histochemical staining of HSP90α/β, p14ARF, and CHIP in NSCLC tissues. Scale bar, 50 μm. B, CHIP expression positively correlated with HSP90 expression in both early and advanced stages (both P < 0.001), whereas p14ARF expression showed strong negative correlations with HSP90 and CHIP expression in only the advanced stage (both P < 0.001). C, Kaplan–Meier plots of overall survival for patients with NSCLC were categorized by HSP90, p14ARF, or CHIP expression. HSP90+, histoscore ≥ 50; p14ARF-, histoscore ≥ 92; CHIP+, histoscore ≥ 85.
We next examined the relation of the expression of each protein with patient survival outcome. Kaplan–Meier plots demonstrated that patients with high HSP90 expression and loss of p14ARF expression displayed significantly worse overall survival (median of 63 months vs. 131 months, P = 0.001; median of 74 months vs. 122 months, P = 0.008, respectively). Furthermore, the patients with combined HSP90+/p14ARF−, HSP90+/CHIP+, or HSP90+/p14ARF−/CHIP+ expression showed significantly worse overall survival (median of 51 months vs. 122 months, P < 0.001; median of 59 months vs. 131 months, P = 0.013; and median of 51 months vs. 122 months, P = 0.002, respectively) than patients with HSP90−/p14ARF+, HSP90−/CHIP−, or HSP90−/p14ARF+/CHIP− expression (Fig. 7C). Cox proportional hazards models indicated that high HSP90 expression [HR = 2.13 (95% CI, 1.33–3.70), P < 0.01] or a combination of HSP90+/p14ARF− [HR = 2.63 (95% CI, 1.32–5.25), P < 0.01], HSP90+/CHIP+ [HR = 2.55 (95% CI, 1.29–5.04), P < 0.01], or HSP90+/p14ARF−/CHIP+ [HR = 3.98 (95% CI, 1.43–11.07), P < 0.01] expression was an independent prognostic factor of overall survival (Supplementary Table S1). Altogether, these data indicated that HSP90, p14ARF, and CHIP expression serves as an important prognostic factor in human lung cancer.
Discussion
Unlike previous reports that HSP70 is a major cofactor of CHIP for protein degradation, we found that HSP90, together with CHIP, plays an active role in inducing the degradation of p14ARF, a potent tumor suppressor. CHIP was able to bind to p14ARF but could not induce its degradation in the absence of HSP90. These observations suggest that HSP90 may facilitate the unfolding of p14ARF rather than its maintenance. Further structural analyses are required for a detailed and more insightful interpretation of this mechanism.
Of note, p14ARF degradation occurred regardless of the E3 ubiquitin ligase activity of CHIP and was not prevented by proteasome inhibition but by lysosome inhibition. These results suggest that HSP90- and CHIP-mediated p14ARF degradation is ubiquitylation-independent lysosomal degradation. Although CHIP is known to induce proteasomal degradation of target proteins, recent reports have suggested that CHIP also stimulates degradation through a lysosome-dependent pathway, which is known as chaperone-mediated autophagy (CMA; refs. 41, 42). The mechanism of CMA generally differs from the conventional autophagy system in several ways. The substrates of CMA have KFERQ or similar motifs that are recognized by cytosolic HSC70 or HSP70 and its associated components (43). These substrates are recruited to the lysosome-linked membrane protein, LAMP2A, which is involved in unfolded substrate transport (44). Because substrates are recruited to lysosomes directly in CMA proteolysis, degradation is blocked by lysosome inhibitors but not by autophagy inhibitors such as 3MA, which targets autophagosome formation (40). We observed that HSP90- and CHIP-mediated p14ARF degradation is partly similar to CMA. p14ARF degradation is blocked by lysosome inhibitors but not by autophagy inhibitors. Furthermore, we observed that CHIP increases the interactions among HSP90, p14ARF, and LAMP2A. These results suggest that HSP90 and CHIP stimulate p14ARF degradation through a LAMP2A-dependent lysosomal pathway but not autophagy. However, this mechanism differs from canonical CMA in that HSP90, rather than HSP70, participates in target protein delivery to LAMP2A. No conventional HSP70-interacting KFERQ motif was identified in p14ARF, indicating that lysosome-dependent p14ARF degradation might have a different mode of action compared with conventional degradation. Other factors such as CFTR 508, VHL, p450 2E1, apolipoprotein B, and mutant insulin receptor are also known to be degraded via HSP90 (45–48). Further identifying whether these targets are also similarly affected by HSP90-dependent CMA pathways in future studies would be beneficial.
Because HSP90 inhibition induces the degradation of its oncogenic client proteins, HSP90 has been identified as a therapeutic target for treating cancer. Various types of HSP90 inhibitors such as GA analogs, resorcinol derivatives, and purine analogs were developed as cancer drug (49). Although a HSP90 inhibitor was partially effective to EGFR-mutated NSCLC (50), it was more effective for treatment of ALK-rearranged NSCLC, which accounts for approximately 3% to 5% of lung tumors (9).
Here, we identified that p14ARF expression significantly increased the sensitivity of NSCLC cells to GA treatment regardless of EGFR mutation, ALK fusion, or p53 status. Although HSP90 inhibition has been reported to induce apoptosis through p53 activation, we found that p14ARF-positive NSCLC cell lines showed significant sensitivity to GA treatment despite the presence of mutant p53. Because more than half of patients with NSCLC have p53 mutations, GA treatment that targets p14ARF regardless of p53 activity could be a good therapeutic strategy for patients with NSCLC. Moreover, HSP90+/p14ARF−, HSP90+/CHIP+, and HSP90+/p14ARF−/CHIP+ expression patterns in patients with NSCLC indicated significantly worse overall survival than did HSP90−/p14ARF+, HSP90−/CHIP−, and HSP90−/p14ARF+/CHIP− expression patterns. Moreover, Cox proportional hazards models indicated that high HSP90 expression or a combination of HSP90+/p14ARF−, HSP90+/CHIP+, or HSP90+/p14ARF−/CHIP+ expression was an independent prognostic factor of overall survival.
In summary, HSP90 has powerful oncogenic effects through degrading the tumor suppressor p14ARF followed by prevention of cellular senescence, and based on our results that p14ARF expression increases the cytotoxicity of HSP90 inhibition in NSCLC cell lines, and the HSP90+/p14ARF− expression is an important prognostic marker in NSCLC, we suggest that p14ARF-positive NSCLC may show more marked responses to HSP90 inhibitors than p14ARF-negative NSCLC.
Supplementary Material
Acknowledgments
Grant Support
This work was supported by grants from the Basic Science Research Program of the National Research Foundation of Korea (NRF; 2014R1A1A1002589 to S. Han and A. Ko) and Creative Research Initiative Program of NRF (2015R1A3A2066581 to J. Song) funded by the Ministry of Science, ICT and Future Planning, and from the National Cancer Center, Korea (NCC-1420300 to J. Song).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
References
- 1.Ettinger DS, Akerley W, Borghaei H, Chang AC, Cheney RT, Chirieac LR, et al. Non-small cell lung cancer, version 2.2013. J Natl Compr Canc Netw 2013;11:645–53. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Fillmore CM, Hammerman PS, Kim CF, Wong KK. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 2014;14:535–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wingo PA, Ries LA, Giovino GA, Miller DS, Rosenberg HM, Shopland DR, et al. Annual report to the nation on the status of cancer, 1973–1996, with a special section on lung cancer and tobacco smoking. J Natl Cancer Inst 1999;91:675–90. [DOI] [PubMed] [Google Scholar]
- 4.Travis WD, Linnoila RI, Tsokos MG, Hitchcock CL, Cutler GB Jr., Nieman L, et al. Neuroendocrine tumors of the lung with proposed criteria for large-cell neuroendocrine carcinoma. An ultrastructural, immunohistochemical, and flow cytometric study of 35 cases. Am J Surg Pathol 1991;15:529–53. [DOI] [PubMed] [Google Scholar]
- 5.Subramanian J, Govindan R. Lung cancer in never smokers: a review. J Clin Oncol 2007;25:561–70. [DOI] [PubMed] [Google Scholar]
- 6.Travis WD, Travis LB, Devesa SS. Lung cancer. Cancer 1995;75:191–202. [DOI] [PubMed] [Google Scholar]
- 7.Kenfield SA, Wei EK, Stampfer MJ, Rosner BA, Colditz GA. Comparison of aspects of smoking among the four histological types of lung cancer. Tobacco Control 2008;17:198–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langer CJ, Besse B, Gualberto A, Brambilla E, Soria JC. The evolving role of histology in the management of advanced non-small-cell lung cancer. J Clin Oncol 2010;28:5311–20. [DOI] [PubMed] [Google Scholar]
- 9.Sang J, Acquaviva J, Friedland JC, Smith DL, Sequeira M, Zhang C, et al. Targeted inhibition of the molecular chaperone Hsp90 overcomes ALK inhibitor resistance in non-small cell lung cancer. Cancer Discov 2013;3:430–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D, Shan J, Zhu WG, Qin J, Gu W. Transcription-independent ARF regulation in oncogenic stress-mediated p53 responses. Nature 2010;464:624–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ko A, Shin JY, Seo J, Lee KD, Lee EW, Lee MS, et al. Acceleration of gastric tumorigenesis through MKRN1-mediated posttranslational regulation of p14ARF. J Natl Cancer Inst 2012;104:1660–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol 2010;11:515–28. [DOI] [PubMed] [Google Scholar]
- 13.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer 2005;5:761–72. [DOI] [PubMed] [Google Scholar]
- 14.Kamal A, Boehm MF, Burrows FJ. Therapeutic and diagnostic implications of Hsp90 activation. Trends Mol Med 2004;10:283–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson JL, Brown C. Plasticity of the Hsp90 chaperone machine in divergent eukaryotic organisms. Cell Stress Chaperones 2009;14:83–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basso AD, Solit DB, Munster PN, Rosen N. Ansamycin antibiotics inhibit Akt activation and cyclin D expression in breast cancer cells that overexpress HER2. Oncogene 2002;21:1159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu W, Mimnaugh E, Rosser MF, Nicchitta C, Marcu M, Yarden Y, et al. Sensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J Biol Chem 2001;276:3702–8. [DOI] [PubMed] [Google Scholar]
- 18.Schulte TW, Blagosklonny MV, Ingui C, Neckers L. Disruption of the Raf-1-Hsp90 molecular complex results in destabilization of Raf-1 and loss of Raf-1-Ras association. J Biol Chem 1995;270:24585–8. [DOI] [PubMed] [Google Scholar]
- 19.An WG, Schulte TW, Neckers LM. The heat shock protein 90 antagonist geldanamycin alters chaperone association with p210bcr-abl and v-src proteins before their degradation by the proteasome. Cell Growth Differ 2000;11:355–60. [PubMed] [Google Scholar]
- 20.Yao Q, Nishiuchi R, Li Q, Kumar AR, Hudson WA, Kersey JH. FLT3 expressing leukemias are selectively sensitive to inhibitors of the molecular chaperone heat shock protein 90 through destabilization of signal transduction-associated kinases. Clin Cancer Res 2003;9:4483–93. [PubMed] [Google Scholar]
- 21.Fumo G, Akin C, Metcalfe DD, Neckers L. 17-Allylamino-17-demethoxygeldanamycin (17-AAG) is effective in down-regulating mutated, constitutively activated KIT protein in human mast cells. Blood 2004;103:1078–84. [DOI] [PubMed] [Google Scholar]
- 22.Solit DB, Zheng FF, Drobnjak M, Munster PN, Higgins B, Verbel D, et al. 17-Allylamino-17-demethoxygeldanamycin induces the degradation of androgen receptor and HER-2/neu and inhibits the growth of prostate cancer xenografts. Clin Cancer Res 2002;8:986–93. [PubMed] [Google Scholar]
- 23.Kosano H, Stensgard B, Charlesworth MC, McMahon N, Toft D. The assembly of progesterone receptor-hsp90 complexes using purified proteins. J Biol Chem 1998;273:32973–9. [DOI] [PubMed] [Google Scholar]
- 24.Qing G, Yan P, Xiao G. Hsp90 inhibition results in autophagy-mediated proteasome-independent degradation of IkappaB kinase (IKK). Cell Res 2006;16:895–901. [DOI] [PubMed] [Google Scholar]
- 25.Isaacs JS, Jung YJ, Mimnaugh EG, Martinez A, Cuttitta F, Neckers LM. Hsp90 regulates a von Hippel Lindau-independent hypoxia-inducible factor-1 alpha-degradative pathway. J Biol Chem 2002;277:29936–44. [DOI] [PubMed] [Google Scholar]
- 26.Holt SE, Aisner DL, Baur J, Tesmer VM, Dy M, Ouellette M, et al. Functional requirement of p23 and Hsp90 in telomerase complexes. Genes Dev 1999;13:817–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basso AD, Solit DB, Chiosis G, Giri B, Tsichlis P, Rosen N. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J Biol Chem 2002;277:39858–66. [DOI] [PubMed] [Google Scholar]
- 28.Trepel J, Mollapour M, Giaccone G, Neckers L. Targeting the dynamic HSP90 complex in cancer. Nat Rev Cancer 2010;10:537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ayrault O, Godeny MD, Dillon C, Zindy F, Fitzgerald P, Roussel MF, et al. Inhibition of Hsp90 via 17-DMAG induces apoptosis in a p53-dependent manner to prevent medulloblastoma. Proc Natl Acad Sci U S A 2009;106:17037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Restall IJ, Lorimer IA. Induction of premature senescence by hsp90 inhibition in small cell lung cancer. PLoS ONE 2010;5:e11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soti C, Sreedhar AS, Csermely P. Apoptosis, necrosis and cellular senescence: chaperone occupancy as a potential switch. Aging Cell 2003;2:39–45. [DOI] [PubMed] [Google Scholar]
- 32.Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Hohfeld J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol 2001;3:93–6. [DOI] [PubMed] [Google Scholar]
- 33.Murata S, Chiba T, Tanaka K. CHIP: a quality-control E3 ligase collaborating with molecular chaperones. Int J Biochem Cell Biol 2003;35:572–8. [DOI] [PubMed] [Google Scholar]
- 34.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A 1998;95:8292–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol 2001;2:731–7. [DOI] [PubMed] [Google Scholar]
- 36.Wang X, Zha M, Zhao X, Jiang P, Du W, Tam AY, et al. Siva1 inhibits p53 function by acting as an ARF E3 ubiquitin ligase. Nat Commun 2013;4:1551. [DOI] [PubMed] [Google Scholar]
- 37.Noh KH, Kim BW, Song KH, Cho H, Lee YH, Kim JH, et al. Nanog signaling in cancer promotes stem-like phenotype and immune evasion. J Clin Invest 2012;122:4077–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee MS, Jeong MH, Lee HW, Han HJ, Ko A, Hewitt SM, et al. PI3K/AKT activation induces PTEN ubiquitination and destabilization accelerating tumourigenesis. Nat Commun 2015;6:7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen HY, He JC, Wang Y, Huang QY, Chen JF. Geldanamycin induces heat shock protein 70 and protects against MPTP-induced dopaminergic neurotoxicity in mice. J Biol Chem 2005;280:39962–9. [DOI] [PubMed] [Google Scholar]
- 40.Dice JF. Chaperone-mediated autophagy. Autophagy 2007;3:295–9. [DOI] [PubMed] [Google Scholar]
- 41.Tetzlaff JE, Putcha P, Outeiro TF, Ivanov A, Berezovska O, Hyman BT, et al. CHIP targets toxic alpha-Synuclein oligomers for degradation. J Biol Chem 2008;283:17962–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ferreira JV, Fofo H, Bejarano E, Bento CF, Ramalho JS, Girao H, et al. STUB1/CHIP is required for HIF1A degradation by chaperone-mediated autophagy. Autophagy 2013;9:1349–66. [DOI] [PubMed] [Google Scholar]
- 43.Chiang HL, Terlecky SR, Plant CP, Dice JF. A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 1989;246:382–5. [DOI] [PubMed] [Google Scholar]
- 44.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science 1996;273:501–3. [DOI] [PubMed] [Google Scholar]
- 45.Agarraberes FA, Dice JF. A molecular chaperone complex at the lysosomal membrane is required for protein translocation. J Cell Sci 2001;114:2491–9. [DOI] [PubMed] [Google Scholar]
- 46.Fuller W, Cuthbert AW. Post-translational disruption of the delta F508 cystic fibrosis transmembrane conductance regulator (CFTR)-molecular chaperone complex with geldanamycin stabilizes delta F508 CFTR in the rabbit reticulocyte lysate. J Biol Chem 2000;275:37462–8. [DOI] [PubMed] [Google Scholar]
- 47.Goasduff T, Cederbaum AI. CYP2E1 degradation by in vitro reconstituted systems: role of the molecular chaperone hsp90. Arch Biochem Bioph 2000;379:321–30. [DOI] [PubMed] [Google Scholar]
- 48.McClellan AJ, Scott MD, Frydman J. Folding and quality control of the VHL tumor suppressor proceed through distinct chaperone pathways. Cell 2005;121:739–48. [DOI] [PubMed] [Google Scholar]
- 49.Whitesell L, Lin NU. HSP90 as a platform for the assembly of more effective cancer chemotherapy. Biochim Biophys Acta 2012;1823:756–66. [DOI] [PubMed] [Google Scholar]
- 50.Johnson ML, Yu HA, Hart EM, Weitner BB, Rademaker AW, Patel JD, et al. Phase I/II study of HSP90 inhibitor AUY922 and erlotinib for EGFR-mutant lung cancer with acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors. J Clin Oncol 2015;33:1666–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.