Abstract
The devastating outbreak of COVID-19 has spread all over the world and has become a global health concern. There is no specific therapeutics to encounter the COVID-19. Small interfering RNA (siRNA)-based therapy is an efficient strategy to control human viral infections employing post-transcriptional gene silencing (PTGS) through neutralizing target complementary mRNA. RNA-dependent RNA polymerase (RdRp) encoded by the viral RdRp gene as a part of the replication-transcription complex can be adopted as an acceptable target for controlling SARS-CoV-2 mediated infection. Therefore, in the current study, accessible siRNA designing tools, including significant algorithms and parameters, were rationally used to design the candidate siRNAs against SARS-COV-2 encoded RdRp. The designed siRNA molecules possessed adequate nucleotide-based and other features for potent gene silencing. The targets of the designed siRNAs revealed no significant matches within the whole human genome, ruling out any possibilities for off-target silencing by the siRNAs. Characterization with different potential parameters of efficacy allowed selecting the finest siRNA among all the designed siRNA molecules. Further, validation assessment and target site accessibility prediction also rationalized the suitability of this siRNA molecule. Molecular docking study between the selected siRNA molecule and component of RNA interference (RNAi) pathway gave an excellent outcome. Molecular dynamics of two complexes: siRNA and argonaute complex, guide RNA, and target protein complex, have shown structural stability of these proteins. Therefore, the designed siRNA molecule might act as an effective therapeutic agent against the SARS-CoV-2 at the genome level and can prevent further outbreaks of COVID-19 in humans.
Keywords: COVID-19, siRNA, Post-transcriptional gene silencing, SARS-CoV-2, RNA-dependent RNA polymerase
1. Introduction
The pandemic state of COVID-19 has created chaos worldwide. As a result, the WHO has declared COVID-19 as a global health emergency (Chakraborty et al., 2020a; Chakraborty et al., 2020b; Dong et al., 2020). It is causing high morbidity and mortality across the world since its outbreak noted in China in December 2019 (Rosenbaum, 2020; Sohrabi et al., 2020). The pathological progression of this disease follows well-characterized stages. There is an asymptomatic state at the initial one or two days of infection when the inhaled SARS-CoV-2 possibly binds to the epithelial cells of the nasal cavity, and replication in the cells leads to local propagation of the virus with the exertion of restricted innate immune response (Mason, 2020). The second stage of the infection becomes clinically vivid. The propagation and migration of the virus down to the respiratory tract generate a profound immune response (Harapan et al., 2020; Mason, 2020). About 20% of the infected patients enter into stage 3 of the disease and develop pulmonary infiltrates, causing acute respiratory distress syndrome (i.e., ARDS) (Harapan et al., 2020; Mason, 2020). Massive damage of the alveolar epithelial cells and progressive respiratory failure may lead to the death of the patients in severe cases (Chen et al., 2020; Huang et al., 2020; Zhou et al., 2020).
SARS-CoV-2 is an enveloped single-stranded RNA (ssRNA) virus (Dhama et al., 2020), and it comprises four structural proteins: spike (S), membrane (M), envelope (E), and nucleocapsid (N). Transmembrane trimetric spike glycoprotein is protruded from the viral surface and consists of two functional subunits (i.e., S1 and S2) (Harapan et al., 2020; Yuki et al., 2020). S1 subunit contributes to the interaction of SARS-CoV-2 to the angiotensin-converting enzyme 2 (ACE2) receptor of the host cell, while the S2 subunit plays a role in the fusion of viral and host cell membranes (Harapan et al., 2020; Yuki et al., 2020). Viral replication is the crucial step after the integration of the viral genome into the host cell, which is mediated by a multi-subunit replication/transcription complex factors of viral non-structural proteins (Yin et al., 2020). The essential component of this complex is the RNA-dependent RNA polymerase (RdRp) encoded by the RdRp gene (Lung et al., 2020; Yin et al., 2020).
To date, there exists no specific therapeutics against the fatal COVID-19. Several strategies have been adopted or are underway to prevent or control emerging infections of COVID-19, including the development of vaccines, oligonucleotide-based therapies, small-molecule drugs, monoclonal antibodies, interferon therapies, and peptides (Li and De Clercq, 2020). Promising results have been achieved so far with several repurposed drugs such as remdesivir, dexamethasone, favipiravir, tocilizumab, sarilumab, etc.(Saha et al., 2020a; Saha et al., 2020b; Saha et al., 2020c). However, so far, these approaches have not met the optimum level to improve the clinical outcomes. Thus, identifying more potent and specific therapeutics is urgently required for the containment of the COVID-19 outbreak.
Small interfering RNA (siRNA) therapy, considered as a potential tool of the RNA interference (RNAi) pathway (Chakraborty et al., 2017; Setten et al., 2019), might be an excellent option to curb SARS-CoV-2-mediated infection. The siRNA is a 19–25 base pairs long RNA duplex bearing overhangs of two nucleotides on the 3′ end. It brings about post-transcriptional gene silencing (PTGS) by binding to target complementary mRNA leading to its enzymatic degradation (Chakraborty, 2017; Elbashir et al., 2001; Zamore et al., 2000). siRNA-induced inhibition of gene expression is a highly complex event. siRNA duplex, after its entrance within the cell, is cleaved by dicer (RNase III-like enzyme) and incorporated into RNA-induced silencing complex (RISC), which is a multi-protein component complex (Bernstein et al., 2001; Dana et al., 2017; Hammond et al., 2000). Within RISC, the strands of RNA get separated by the ATP-dependent RNA helicase domain of RISC (Dana et al., 2017). The passenger (sense) strand is degraded within RISC, while the antisense single-stranded RNA (guide RNA) facilitates the alignment of RISC on the target mRNA, enabling its catalytic RISC protein (i.e., an argonaute family member) to cleave the target mRNA (Dana et al., 2017). Subsequent to the revelation of its mechanism, this process has become a powerful experimental gene silencing tool in basic research (Setten et al., 2019; Jana et al., 2004a). It holds promising therapeutic potentials as well. A sufficient number of siRNA-based clinical trials have been conducted for treating diseases such as hepatitis B, ebola, malignancy, diabetic macular edema, hypercholesterolemia, etc. (Hu et al., 2019; Setten et al., 2019). Patisiran (Onpattro; Alnylam Pharmaceuticals) is the first siRNA drug approved by the United States Food and Drug Administration (USFDA) for the treatment of polyneuropathy in patients with hereditary transthyretin amyloidosis (hATTR) (Hu et al., 2019; Setten et al., 2019).
Considering the fact of being the part of the replication/transcription complex, the RdRp enzyme encoded by the viral RdRp gene can be utilized as a suitable target for controlling SARS-CoV-2 mediated infection. In the present manuscript, siRNA molecules for the SARS-CoV-2 RdRp gene have been rationally designed by using various computational tools. Designed siRNAs might contribute to the development of effective therapeutic agents against this deadly virus.
2. Materials and methods
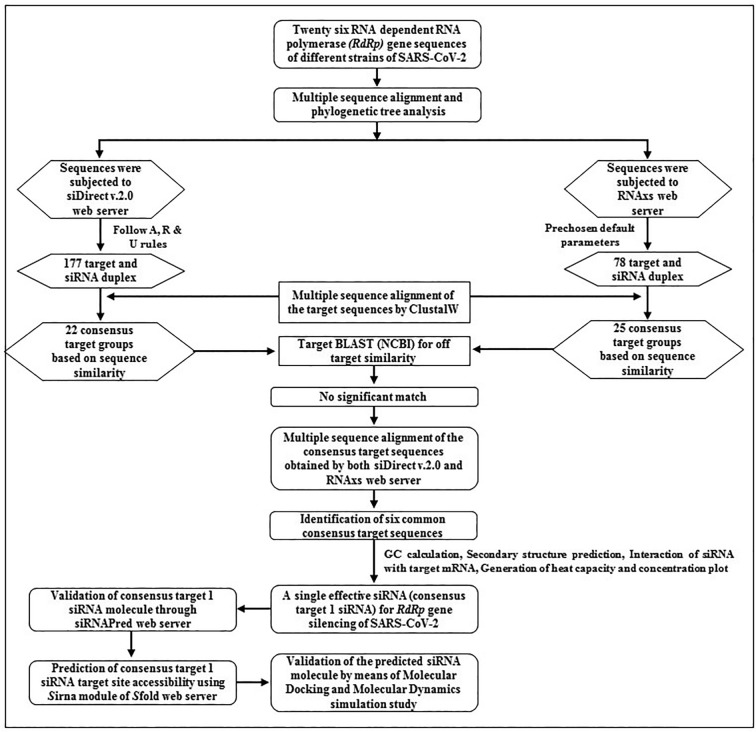
The complete adopted methodology for the design and/or prediction of effective siRNA molecules against the RdRp gene sequence of SARS-CoV-2 is given in Fig. 1 .
Fig. 1.
The complete methodology to design and predict effective siRNA molecules against RdRp gene sequences of SARS-CoV-2.
2.1. Retrieval of RdRp gene sequences
Twenty six RdRp gene sequences of different SARS-COV-2 strains were retrieved from the GenBank available at the National Center for Biotechnology Information (NCBI) (Singh et al., 2012) (Supplementary Table 1).
2.2. Target recognition and designing of siRNA molecules
Two siRNA design tools (i.e., siDirect version 2.0 and RNAxs webserver) were used to design effective and target-specific siRNA molecules against SARS-COV-2 RdRp gene sequences (Naito et al., 2009; Ok-Seon Kwon et al., 2018). The retrieved RdRp gene sequences were subjected to siDirect 2.0 in FASTA format (Naito et al., 2009). Whereas the siDirect 2.0 tool utilizes some rules such as Ui-Tei, Amarzguioui, and Renold algorithms (Table 1 ) for designing siRNAs, it keeps the melting temperature (Tm) of the seed-target duplex below 21.5 °C as a default parameter. It selects siRNA sequences that have at least two mismatches to any other non-targeted transcripts to avoid off-target silencing. The desired siRNA molecules generated through siDirect 2.0, fulfilling all these criteria, were selected for subsequent analysis. Retrieved 26 RdRp gene sequences were also subjected to RNAxs webserver to design functional siRNA and reveal the relevant target sequences (Ok-Seon Kwon et al., 2018). RNAxs web server utilizes prechosen reasonable default values (i.e., accessibility threshold value of 8 nt seed region of siRNA: 0.01157, accessibility threshold value of 16 nt of siRNA: 0.001002, self-folding energy: 0.9022, sequence and energy asymmetry: 0.5 and 0.4655, respectively, and free end: 0.625) for siRNA design, which gives optimal separation of functional and non-functional siRNAs along with displaying the relevant target sequences. ClustalW was used to execute multiple sequence alignment of the siRNA target sequences obtained by siDirect 2.0 and RNAxs webserver (Nur et al., 2013).
Table 1.
Algorithms/rules for rational designing of siRNA molecules.
| Ui-Tei rules | Amarzguioui rules | Reynolds rules |
|---|---|---|
|
|
|
2.3. Investigation of parameters for siRNA refinement
The standard nucleotide BLAST at the NCBI database was used against the whole GenBank database by applying the expected threshold value of 10 to detect any off-target nucleotide sequence similarity (Altschul et al., 1990). The GC calculator was used to calculate the GC content of the selected siRNA sequences (Nur et al., 2013). Prediction of the secondary structure of the siRNA guide strand is considered an essential parameter to determine the efficacy of siRNA. The secondary structure of siRNAs was predicted as per the computed free energy of folding by using the RNAstructure webserver (Bellaousov et al., 2013). Moreover, RNAstructure webserver was even used to analyze the thermodynamic interaction between predicted siRNA (guide strand) and target gene (Bellaousov et al., 2013). The ‘Predict a Bimolecular Secondary Structure’ is one of the essential web-based tools of RNAstructure web server, which predicts free energy of RNA-RNA interaction. It folds two sequences, either RNA or DNA, into their lowest hybrid free energy conformation. Subsequently, the DINAMelt webserver was used to generate heat capacity and concentration plots for the consensus siRNA target strand (Shawan et al., 2015).
2.4. Validation of the predicted siRNA molecule
The anticipated siRNA molecule was validated by using siRNAPred webserver from IMTECH (Kumar and Raghava, 2009). This server predicts the actual efficacy of a query siRNA sequence (both 21 and 19 mer) based on the SVM (support vector machine) algorithm with high accuracy level. In the current study, the efficacy of consensus target 1 siRNA was assessed under homogenous experimental conditions against the main 21 mer dataset (consisting of 2182 siRNA (21 mer) types) using a binary pattern (Kumar and Raghava, 2009).
2.5. Prediction of siRNA target site accessibility within target mRNA
Specifically, the siRNA module inside the Sfold webserver was adopted to determine the target site accessibility for a particular siRNA molecule (Ding et al., 2004). The algorithm within the siRNA module considers a target mRNA as an input sequence and assesses the accessibility of a target site (for a specific siRNA) inside the mRNA by employing different important parameters (i.e., probability profile, loop specific profiles, and siRNA internal stability profile) for distinct target position (Ding et al., 2004). Herein, a single RdRp gene sequence (accession no. LC522350.1) was randomly picked for the analysis purpose, and the mRNA sequence of the selected gene was achieved using the standard BioModels transcription and translation tool (Ebenezer, 2020).
2.6. RNA modeling and interaction pattern analysis
Triggering of a suitable antiviral response employing RNAi-mediated viral gene silencing involves the appropriate interaction between siRNA duplex (mainly guide strand) and RISC complex along with guide siRNA strand and target mRNA within RISC complex (Jana et al., 2004b; Majumdar et al., 2017). Herein, for interaction pattern analysis, computer-assisted molecular docking and molecular dynamics simulation was employed between siRNA (guide strand) and argonaute protein. The 3D modeled structure (PDB file) of guide siRNA was generated by an intuitive molecular editor and visualization tool, i.e., Avogadro 1.2.0 software program. It was followed by structure optimization with the Universal Force Field (UFF) and molecular dynamics (300K) using the same program (Hanwell et al., 2012). The X-ray crystallographic structures of the argonaute protein of human were retrieved from RCSB Protein Data Bank (PDB ID: 4F3T) (Burley et al., 2019).
2.7. Molecular docking of siRNA (guide strand) and argonaute protein
The molecular docking between siRNA (guide strand) and argonaute protein was done by using the HDOCK server. This server uses HDOCK (HDOCK algorithm) lite program, a hierarchical Fast Fourier Transform (FFT) -related docking program. It is a hybrid docking algorithm that uses free docking and template-based modeling and shows high efficacy and robustness (Yan et al., 2017). 2D representation and analysis of protein-siRNA complex was done using UCSF Chimera software (Pettersen et al., 2004; Rodriguez-Guerra Pedregal and Marechal, 2018).
2.8. Molecular dynamics (MD) simulation of siRNA (guide strand)‑argonaute protein docked complex and guide-target complex
MD simulation is an appropriate and robust tool for studying the stability of macromolecular complexes (Boehr et al., 2009). The simulation was performed with the NAMD 2.14 package with the selection of the appropriate CHARMM force field topology (Kalé et al., 1999; MacKerell Jr et al., 2000). Here, NAMD 2.14 software calculates frames step trajectory files (each frame step corresponds to every time steps). The protein solvation was executed with VMD1.9.3 software, where we set a cubic box (5 × 5 × 5 nm3) water sphere area around the selected molecule (Humphrey et al., 1996). The solvated system of the docked complex was processed for energy minimization. In this work, we set the NVT thermodynamic parameter at 310 K, and 1.013 atm pressure to perform the MD simulation on siRNA‑argonaute docked complex. Finally, the simulations were evaluated through Root Mean Square Deviation (RMSD) and Root Mean Square Fluctuation (RMSF), calculated for a complete episode of simulations.
3. Results
3.1. Target specific designing of potential siRNA molecules and revelation of consensus targets
This study included 26 SARS-CoV-2 RdRp gene sequences available in the GenBank database (Supplementary Table 1). For designing target-directed siRNA by siDirect version 2.0, algorithms of Ui-Tei, Amarzguioui, and Reynolds were followed, which demonstrate sufficient nucleotide-based features required for effective gene silencing. Moreover, several default parameters were chosen for minimization of seed dependent off-target hits, such as the melting temperature (Tm) of the seed target duplex was kept below 21.5 °C (seed duplex stability: Max Tm 21. 5 °C) and only those siRNAs were selected that had at least two mismatches to any other non-targeted transcripts (i.e. number of off-target hits within three mismatches). Using siDirect 2.0 tool, a total of 177 putative siRNAs were obtained against 177 target sequences maintaining all rules and algorithms of Ui-Tei, Amarzguioui, and Reynolds (Supplementary Table 2). Multiple sequence alignment of these 177 siRNA targets revealed twenty-two target groups on the basis of their sequence similarity index (Table 2 and Supplementary Table 3). The retrieved 26 RdRp gene sequences were also subjected to RNAxs program to design functional siRNA and to reveal the relevant targets. RNAxs web server generated a total of 78 siRNA molecules from the 26 RdRp gene sequences input (Supplementary Table 4). Multiple sequence alignment of the siRNA targets obtained by RNAxs showed 25 consensus targets (Table 3 and Supplementary Table 5).
Table 2.
Consensus target revealed through multiple sequence alignment of the targets obtained by siDirect 2.0.
| Consensus target sequence no. | Consensus target sequences |
|---|---|
| 1 | GTGTCTCTATCTGTAGTACTATG |
| 2 | GACTTTATGAGTGTCTCTATAGA |
| 3 | TAGTACTATGAC-CAATAGACAGT |
| 4 | GACCAATAGACAGTTTCATCAAA |
| 5 | ATGAAGTATTTTGTGAAAATAGG |
| 6 | TTGGAATGTAGTGCGTATAAAGA |
| 7 | GCGTATAAAGATTGTACAAATGT |
| 8 | TGCTAATAGTGTTTTTAACATTT |
| 9 | ATCTACTGATGGTAACAAAATTG |
| 10 | ATCAAAAATTATTGAAATCAATA |
| 11 | ATGCCATTAGTGCAAAGAATAGA |
| 12 | CCCTACTATAACTCAAATGAATC |
| 13 | GGCTAGCATAAAGAACTTTAAGT |
| 14 | ATGTTAAGTGACACACTTAAAAA |
| 15 | TGGCTTTGAGTTGACATCTATGA |
| 16 | TTGGATTTGATTACGTCTATAAT |
| 17 | TGGCATCATTCTATTGGATTTGA |
| 18 | TGCCTAACATGCTTAGAATTATG |
| 19 | GCCTAACATGCTTAGAATTATGG |
| 20 | TGCGTAAACATTTCTCAATGATG |
| 21 | ATGCACTTTTCGCATATACAAAA |
| 22 | GCCAATGTTAATGCACTTTTATC |
Table 3.
Consensus target sequences of SARS-CoV-2 RdRp gene revealed through multiple sequence alignment of the targets obtained by RNAxs web server.
| Consensus target sequence no. | Consensus target sequences |
|---|---|
| A | GGCCAATGTTAATGCACTT |
| B | GCCAATGTTAATGCACTTT |
| C | GCTAGCATAAAGAACTTTA |
| D | TCCACTGCTTCAGACACTT |
| E | CCCTACTATAACTCAAATG |
| F | TCCCTACTATAACTCAAAT |
| G | GACATCTATGAAGTATTTT |
| H | TGACATCTATGAAGTATTT |
| I | CACACCGTTTCTATAGATT |
| J | GCTTGTCACACCGTTTCTA |
| K | CGGTTCACTATATGTTAAA |
| L | CGGTTCACTATATGTTAAA |
| M | TCAGTTCTTTATTATCAAA |
| N | TCTCTATCTGTAGTACTAT |
| O | AGACAGTTTCATCAAAAAT |
| P | GACAGTTTCATCAAAAATT |
| Q | ACAGTTTCATCAAAAATTA |
| R | CAGTTTCATCAAAAATTAT |
| S | TCTTTATTATCAAAACAAT |
| T | AGTAATTGGAACAAGCAAA |
| U | CCTAACATGCTTAGAATTA |
| V | AGGACTTCCTTGGAATGTA |
| W | CGCATATACAAAACGTAAT |
| X | GCATATACAAAACGTAATG |
| Y | GGATTATCCTAAATGTGAT |
3.2. Analysis of target and off-target similarity
The consensus targets obtained by both siDirect 2.0 and RNAxs webserver (i.e., twenty-two consensus targets from siDirect 2.0 and twenty-five consensus targets from RNAxs webserver) were subjected to NCBI-BLAST for searching similarity against the whole human genome, where no significant matches were detected (data not shown). Therefore, these targets ruled out any possibilities for the off-target nucleotide silencing.
3.3. Multiple sequence alignment to determine the common consensus target sequences
Afterward, another multiple sequence alignment was conducted with the consensus target sequences which were obtained by siDirect 2.0 and RNAxs web server. Six consensus target sequences (i.e. consensus target-1: 5′-GTGTCTCTATCTGTAGTACTATG-3′, consensus target 12: 5′-CCCTACTATAACTCAAATGAATC-3′, consensus target 13: 5′-GGCTAGCATAAAGAACTTTAAGT-3′, consensus target 18: 5′-TGCCTAACATGCTTAGAATTATG-3′, consensus target-19: 5′-GCCTAACATGCTTAGAATTATGG-3′, consensus target 22: 5′GCCAATGTTAATGCACTTTTATC3′) were obtained that are shared by the both servers (i.e. siDirect 2.0 and RNAxs web server) (Table 4 ). The siRNA molecules against these sequences were selected for subsequent analysis with different critical parameters (i.e. calculation of siRNA, GC content, secondary structure determination of siRNA guide strand, guide target interaction, and heat capacity) to determine their specification.
Table 4.
Multiple sequence alignment of targets obtained by siDirect 2.0 and RNAxs server.
| Consensus target sequences no. | Consensus target sequences obtained by siDirect 2.0 and RNAxs server |
|---|---|
| 12 | ----------CCCTACTATAACTCAAATGAATC-- |
| E | ----------CCCTACTATAACTCAAATG---------- |
| F | -----------TCCCTACTATAACTCAAAT--------- |
| 19 | ----------GCCTAACATGCTTAGAATTATGG-- |
| U | -------------CCTAACATGCTTAGAATTA-------- |
| 18 | --------TGCCTAACATGCTTAGAATTATG---- |
| 3 | ------TAGTACTATGACCAATAGACAGT------ |
| T | ---------AGTAATTGGAACAAGCAAA----------- |
| 13 | -----ATGTTAAGTGACACACTTAAAAA------- |
| O | ------------AGACAGTTTCATCAAAAAT--------- |
| P | -------------GACAGTTTCATCAAAAATT-------- |
| Q | --------------ACAGTTTCATCAAAAATTA------- |
| R | ---------------CAGTTTCATCAAAAATTAT------ |
| 4 | --GACCAATAGACAGTTTCATCAAA---------- |
| 8 | --------TGCTAATAGTGTTTTTAACATTT------ |
| D | --------------TCCACTGCTTCAGACACTT------- |
| 13 | ----------GGCTAGCATAAAGAACTTTAAGT-- |
| C | ------------GCTAGCATAAAGAACTTTA-------- |
| 22 | -----------GCCAATGTTAATGCACTTTTATC-- |
| B | -----------GCCAATGTTAATGCACTTT---------- |
| A | ------------GGCCAATGTTAATGCACTT--------- |
| 9 | --------- ATCTACTGATGGTAACAAAATTG --- |
| G | -------------GACATCTATGAAGTATTTT-------- |
| H | ------------TGACATCTATGAAGTATTT--------- |
| 5 | --------ATGAAGTATTTTGTGAAAATAGG---- |
| 10 | --------ATCAAAAATTATTGAAATCAATA ---- |
| V | -----------AGGACTTCCTTGGAATGTA---------- |
| 17 | ---------TGGCATCATTCTATTGGATTTGA----- |
| Y | -------------GGATTATCCTAAATGTGAT--------- |
| W | ------------CGCATATACAAAACGTAAT--------- |
| X | --------------GCATATACAAAACGTAATG------- |
| 21 | ATGCACTTTTCGCATATACAAAA------------- |
| 7 | -----------GCGTATAAAGATTGTACAAATGT- |
| 20 | -----------TGCGTAAACATTTCTCAATGATG-- |
| 1 | -------------GTGTCTCTATCTGTAGTACTATG |
| N | -------------------TCTCTATCTGTAGTACTAT--- |
| K | -------------CGGTTCACTATATGTTAAA--------- |
| I | --------------------CACACCGTTTCTATAGATT-- |
| J | -------------GCTTGTCACACCGTTTCTA--------- |
| 2 | -----------GACTTTATGAGTGTCTCTATAGA--- |
| 15 | ---------- TGGCTTTGAGTTGACATCTATGA --- |
| 16 | ---------- TTGGATTTGATTACGTCTATAAT--- |
| 6 | ----------TTGGAATGTAGTGCGTATAAAGA-- |
| 14 | ----------- ATGTTAAGTGACACACTTAAAAA ---- |
| L | --------------- CGGTTCACTATATGTTAAA -------- |
| M | ---------------TCAGTTCTTTATTATCAAA--------- |
| S | --------------------TCTTTATTATCAAAACAAT--- |
3.4. Calculation of siRNA GC content
GC content of siRNA is inversely related to its RNAi activity (Amarzguioui and Prydz, 2004; Chan et al., 2009). siRNA sequences with low GC content are recommended (between 31.6% and 57.9%) (Chan et al., 2009). siRNAs picked against the selected common consensus targets obtained by both siDirect 2.0 and RNAxs web server were checked in their guide strands for the GC content using the GC calculator program (Nur et al., 2013). As shown in Table 5 , GC content in the siRNAs selected against consensus targets- 1, 18, 19, and 22 was in the range of 31.6% and 57.9%.
Table 5.
Proposed siRNA molecules and their consensus target of RdRp gene sequence from SARS-CoV-2 with GC%, free energy of folding, free energy of binding with target and heat capacity.
| Target | siRNA consensus target sequence 21 nt target + 2 nt overhang (5′ → 3′) | Predicted siRNA duplex at 37 °C 21 nt guide (5′ → 3′) 21 nt passenger (5′ → 3′) |
GC% | ΔG of folding (kcal/mol) | ΔG of binding (kcal/mol) | TmCp (°C) | TmConc. (°C) |
|---|---|---|---|---|---|---|---|
| Consensus Target-1 | GTGTCTCTATCTGTAGTACTATG | UAGUACUACAGAUAGAGACAC | 38.09 | 0.1 | −35.2 | 79.3 | 80.6 |
| GUCUCUAUCUGUAGUACUAUG | |||||||
| Consensus Target-12 | CCCTACTATAACTCAAATGAATC | UUCAUUUGAGUUAUAGUAGGG | 30.95 | – | −32.0 | 72.6 | 73.8 |
| CUACUAUAACUCAAAUGAAUC | |||||||
| Consensus Target-13 | GGCTAGCATAAAGAACTTTAAGT | UUAAAGUUCUUUAUGCUAGCC | 30.95 | – | −31.8 | 73.7 | 75.1 |
| CUAGCAUAAAGAACUUUAAGU | |||||||
| Consensus Target-18 | TGCCTAACATGCTTAGAATTATG | UAAUUCUAAGCAUGUUAGGCA | 33.33 | −1.4 | −32.5 | 76.2 | 77.4 |
| CCUAACAUGCUUAGAAUUAUG | |||||||
| Consensus Target-19 | GCCTAACATGCTTAGAATTATGG | AUAAUUCUAAGCAUGUUAGGC | 33.33 | −1.4 | −32.1 | 74.1 | 75.3 |
| CUAACAUGCUUAGAAUUAUGG | |||||||
| Consensus Target-22 | GCCAATGTTAATGCACTTTTATC | UAAAAGUGCAUUAACAUUGGC | 33.33 | −0.4 | −31.6 | 79.2 | 80.5 |
| CAAUGUUAAUGCACUUUUAUC |
3.5. Secondary structure determination of siRNA guide strand
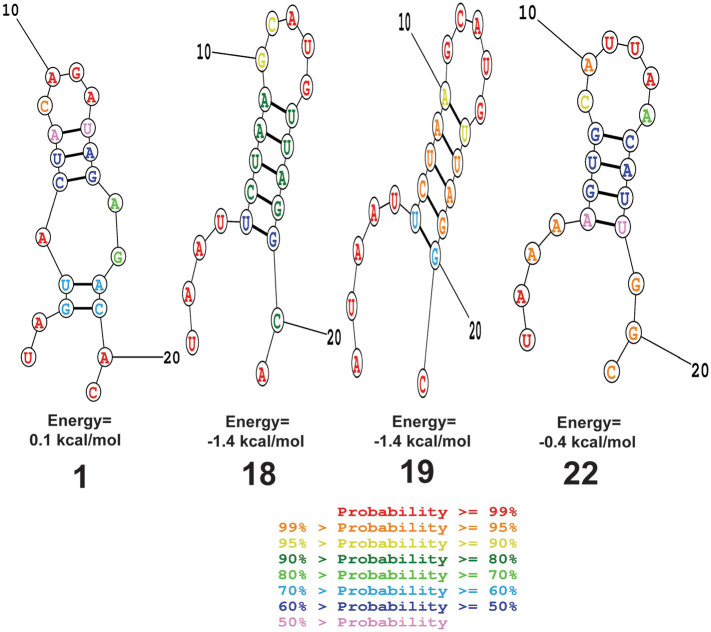
The probable folding and minimum free energy (MFE) of folding of the predicted siRNA guide strands were computed with the online RNA structure web server, given the assumption that siRNA with the positive free energy of folding might facilitate more permissibility to the target leading to better gene silencing (Singh et al., 2012). RNA structure web server follows the most commonly used algorithms for RNA secondary structure prediction based on a search for the minimal free energy state calculation (Bellaousov et al., 2013). This server, thus, can explore the effective lowest free energy structure of the siRNA guide strands. Out of the six selected siRNA, only one siRNA (i.e., consensus target 1 siRNA) showed positive free energy of folding (i.e., 0.1 (kcal/mol)), while other siRNA molecules displayed less than zero kcal/mol free energy of folding (Table 5 and Fig. 2 ).
Fig. 2.
The possible folding and minimum free energy of the guide strands of the predicted siRNA molecules computed using with the online RNA structure web server. A) consensus target 1 siRNA; B) consensus target 18 siRNA; C) consensus target 19 siRNA; D) consensus target 22 siRNA. Secondary structure of siRNA 11 and siRNA 12 could not be predicted.
3.6. RNA-RNA interaction analysis
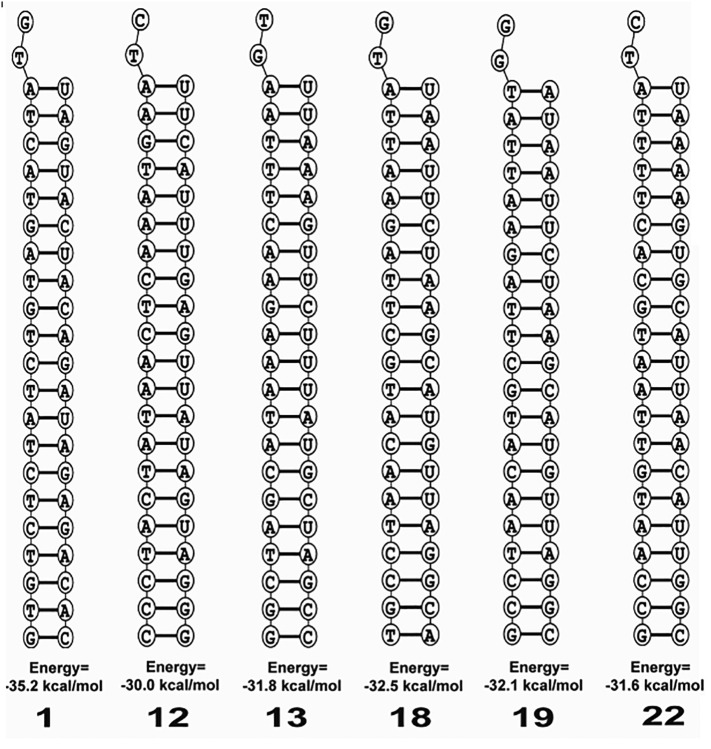
Binding energies of the siRNAs to the respective mRNA target affect the efficiency of siRNAs (Schubert et al., 2005). The thermodynamics of the RNA-RNA interaction of siRNAs with the target was predicted with the aid of the RNA structure web server. As shown in Fig. 3 , the free energy of binding for the consensus target 1, 12, 13, 18, 19 and 22 siRNAs with their targets was −35.2 kcal/mol, −32.0 kcal/mol, −31.8 kcal/mol, −32.5 kcal/mol, −32.1 kcal/mol and − 31.6 kcal/mol, respectively. Optimum free energy favors siRNA binding exclusively to the intended target site of nucleotide strands (Schubert et al., 2005). Thus, consensus target 1 siRNA (i.e., guide strand: 5′- UAGUACUACAGAUAGAGACAC -3′; passenger strand: 5′-GUCUCUAUCUGUAGUACUAUG-3′) with the optimum free energy of binding (i.e., −35.2 kcal/mol) is likely to bind more favorably with the target site (Table 5 and Fig. 3) than other siRNAs.
Fig. 3.
Predicted lowest free energy structure for binding of target RNA with the guide strand of A) consensus target 1 siRNA, B) consensus target 11 siRNA, C) consensus target 12 siRNA, D) consensus target 18 siRNA, E) consensus target 19 siRNA and F) consensus target 22 siRNA.
3.7. Determination of heat capacity and duplex concentration plot
In this study, the DINAMelt webserver was employed to estimate the melting temperature of hybridized nucleic acid pairs and determine the melting profiles of the entire equilibrium. As a function of temperature, the ensemble heat capacity (Cp) was plotted with the indication of melting temperature (Tm) (Supplementary Fig. 1). The comprehensive heat capacity graph displays the involvement of each species within the ensemble heat capacity. Within concentration plot-Tm (Conc), the melting temperature Tm is defined by the concentration of a double-stranded molecule of one-half of its maximum value. The higher values of heat capacity and duplex concentration plot are an indication for highly effective siRNA species. As shown in Table 5, consensus target 1 siRNA showed the highest Tm (Cp) and Tm (Conc) value (i.e.79.3 °C and 80.6 °C, respectively) among all the selected siRNAs (Supplementary Fig. 1).
3.8. Verification of the most efficient siRNA molecule against its consensus target
The siRNA (i.e., guide strand: 5′- UAGUACUACAGAUAGAGACAC -3′; passenger strand: 5′-GUCUCUAUCUGUAGUACUAUG-3′) against consensus target 1 (i.e., 5′-GTGTCTCTATCTGTAGTACTATG-3′) was revealed as an efficient siRNA molecule as observed from the analysis of different parameters (i.e., siRNA GC content, secondary structure determination of siRNA guide strand, guide-target interaction, heat capacity, etc.). To further validate the consensus target 1 siRNA molecule, siRNAPred webserver was utilized (Kumar and Raghava, 2009), which delivered 10 top-ranked siRNAs with great accuracy based on a high siRNAPred score. siRNAPred algorithm score greater than or equal to 1.0 predicts a siRNA with very high efficacy, while score ranging from 0.8–0.9 and 0.7–0.8 predicts with high and moderate efficacy, respectively. In this experiment, a prediction score of 0.857 was found for our query siRNA molecule, which lies within the range of high efficacy prediction score for siRNAPred webserver.
3.9. Target site accessibility prediction
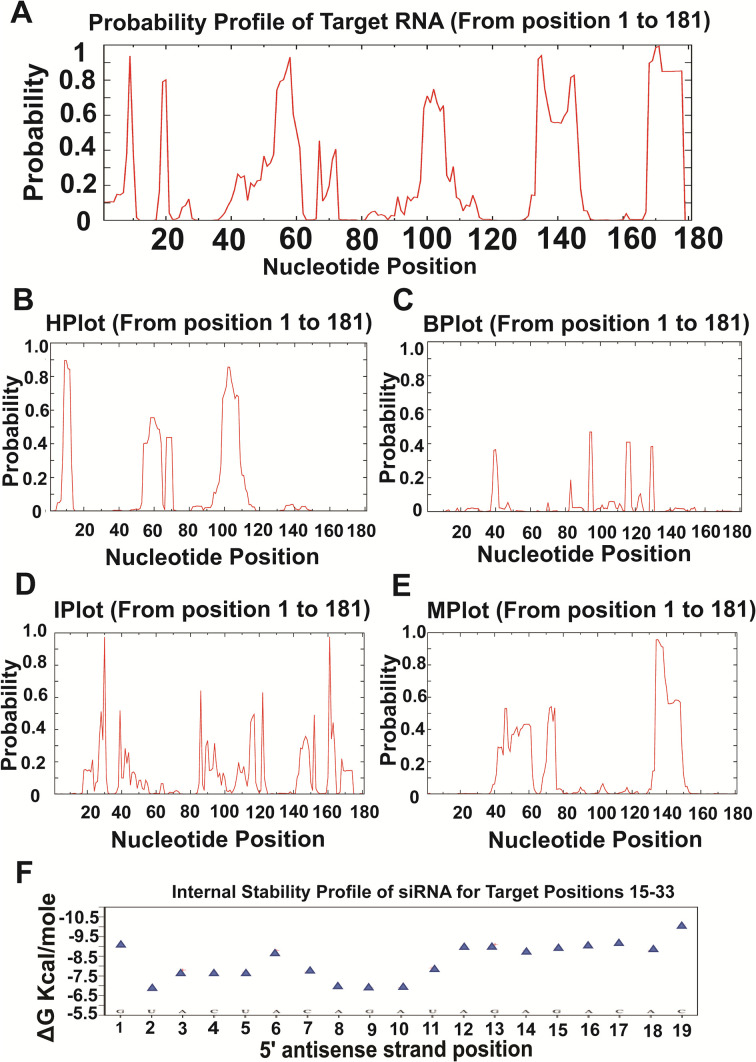
siRNA-mediated gene silencing efficacy also depends on the siRNA target site accessibility (Gatta et al., 2018), which makes the necessity to predict target site accessibility of the siRNA molecule. In order to evaluate target site accessibility of consensus target 1 siRNA molecule, an RdRp gene sequence (Accession no. LC522350.1) having 181 nucleotides was selected in which the siRNA attachment site was within position 15 and 37. After that, the siRNA section within Sfold webserver was employed for an exhaustive analysis of target site accessibility (Ding et al., 2004), which accurately predicted the binding of consensus target 1 siRNA into that specific region (Fig. 4 ). The probability of target site availability for designed siRNA (consensus target 1 siRNA) binding was executed by a siRNA algorithm that provides a probability profile for a single-stranded specific region within the target mRNA. The probability profile for consensus target 1 siRNA target site availability evidently demonstrated the possibility of a location between 15 and 33 within mRNA as highly accessible for siRNA binding (Fig. 4A). Besides, loop specific probability profiles are vital for assessing target site accessibility that reports on the folding patterns of a target binding site. Loop specific probability profiles for consensus target 1 siRNA target site accessibility showed various folding patterns (Fig. 4B–E) in the form of hairpin loop (Hplot), interior loop (Iplot), bulge loop (Bplot), and multi-branched loop (Mplot). From Fig. 4B, C, and E, it is evident that consensus target 1 siRNA binding site (positioned between 15 and 33 in mRNA) was probably not involved in the hairpin, bulge, and multi-branched loop/fold formation and thus, available for siRNA binding. Additionally, the formation of an interior loop (separates the double-stranded RNA due to the absence of Watson-Crick base pairing) within the target site (Fig. 4D) also supports the notion. The internal stabilities of siRNA for specific target sites were also computed using the Srna algorithm of the Sfold server. The free energy of the antisense strand of the consensus target 1 siRNA ranged from −6.5 to −9.5 kcal/mol (within −10 kcal/mol), indicating its efficacy (Fig. 4F).
Fig. 4.
Considerable profiles of consensus target 1 siRNA target site accessibility prediction. A) Probability profile, B) Hairpin loop (Hplot) profile, C) Bulge loop (Bplot) profile, D) Interior loop (Iplot) profile, E) Multi branched loop (Mplot) profile of target mRNA, and F) Internal stability profile of consensus target 1 siRNA for target position 15–33.
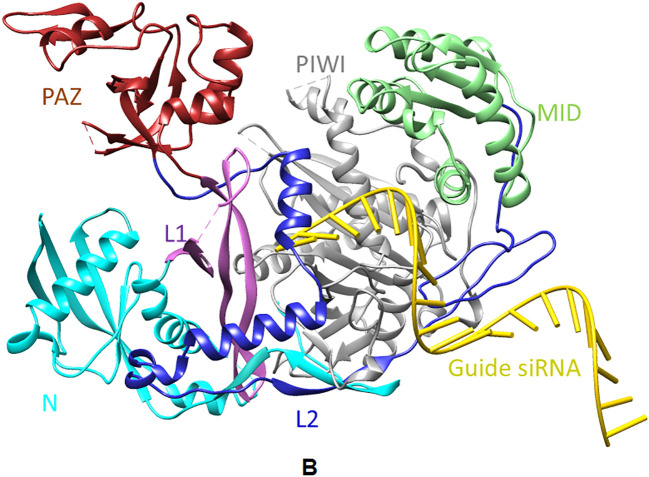
3.10. Molecular docking of siRNA (guide strand) and argonaute protein
The best model was chosen from the top 10 docked models of siRNA (guide) and argonaute. The chosen model has shown the following two results. The docking score (minimal free energy) was noted as −344.12, and the ligand RMSD was observed as 94.65 Angstrom (Fig. 5 ). Afterward, the siRNA‑argonaute docked complex having the lowest energy conformation was chosen for the MD simulation study.
Fig. 5.
3D interaction diagram of different docked complex. A) guide (red)-passenger (blue), B) siRNA‑argonaute, C) siRNA‑argonaute (extended view) and D) guide (red)-target (blue).
3.11. Molecular dynamics (MD) simulation of siRNA (guide strand)‑argonaute protein docked complex and guide-target complex
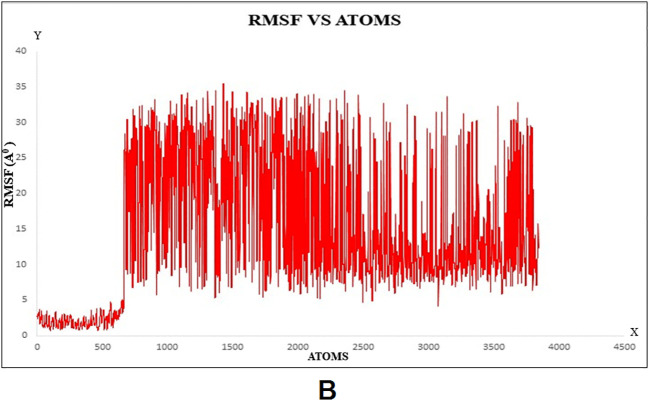
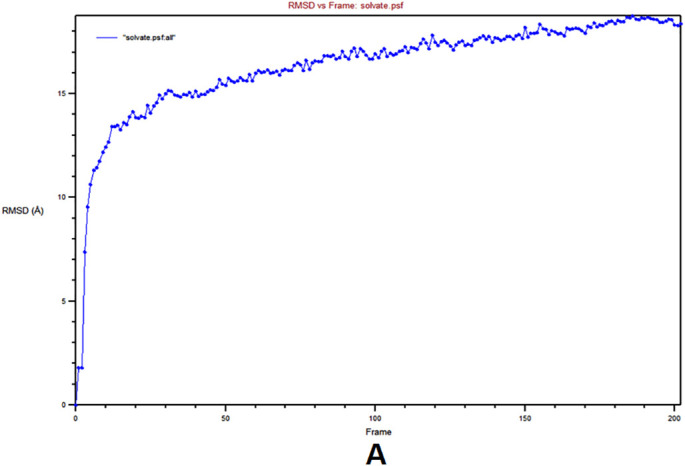
MD simulation was used to determine the durability of the siRNA‑argonaute docked complex by inspecting the physical movements of atoms and molecules within the complex. At the time of MD simulation, energy components, density, pressure, volume, temperature as well as stability of the docked complex within a system were examined. The structural stability of the desired protein-protein complex was assessed via changes in RMSD and RMSF of proteins backbone and all side-chain residues of the docked structure with respect to the initial conformation. The complex was energy minimized using the CHARMM force-field parameter built-in NAMD 2.14 software. RMSF is the time-average of RMSD for all residues that were also calculated for siRNA docked with argonaute. The RMSD and RMSF graphs are shown in Fig. 6A–B, respectively. Stated confirmation was almost stable throughout the simulation. Here, it produced 2000 frames (2000 ps) trajectory run of the docking molecule. RMSF was calculated for Cα atoms, and in RMSD, the entire complex was selected against the backbone of Cα. In order to evaluate, guide siRNA-target protein residue flexibility during MD simulation, RMSF and RMSD of all residues of the molecule were also calculated under 200 frame step simulation run (200 ps = 2 ns) as shown in Fig. 7A–B, respectively. The value of guide-target residue fluctuation excluding 0–1400 atom; they form stable conformation.
Fig. 6.
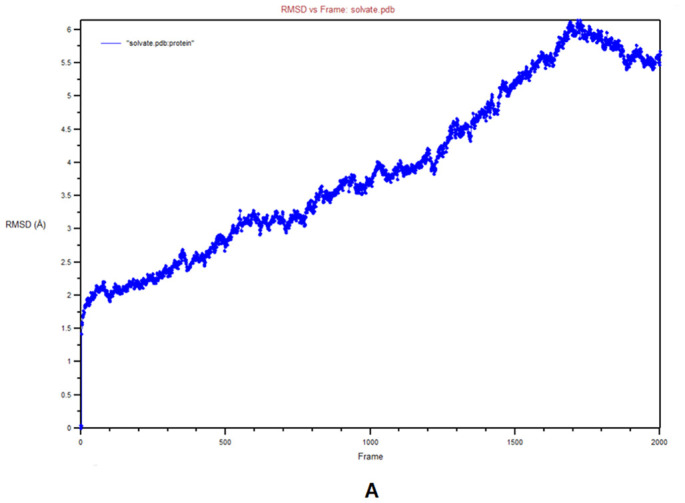
Molecular Dynamics (MD) simulation of guide siRNA‑argonaute complex A) Root Mean Square deviations (RMSD) plot, B) Root mean square Fluctuation (RMSF) plot.
Fig. 7.
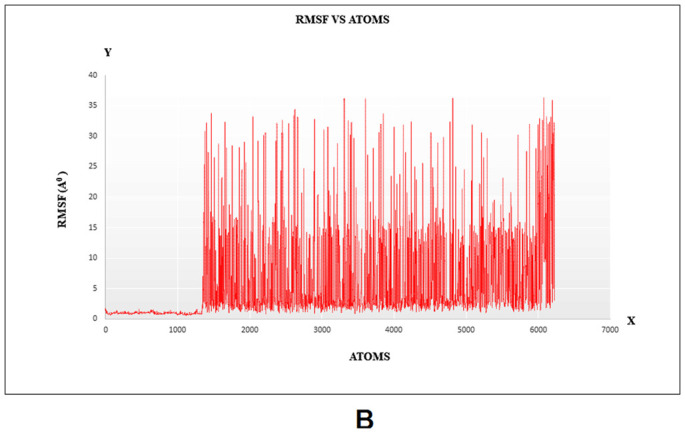
Molecular Dynamics (MD) simulation of target protein-siRNA docked complex A) Root Mean Square Fluctuation (RMSF) plot, B) Root mean square deviations (RMSD) plot.
4. Discussion
COVID-19 pandemic has hit the whole globe aggressively. A confrontation of this pandemic requires promising diverse approaches like gene therapy along with other therapeutic strategies, such as the development of vaccines, drugs, monoclonal antibodies, peptides, etc. Given the facts, RNA-dependent RNA polymerase enzymes encoded by the RdRp gene are the core component of the multi-subunit replication/transcription complex mediating viral replication, siRNA-mediated silencing of RdRP gene with a view to retard SARS-CoV-2 growth might be a promising option. Herein, cogent and extensive bioinformatics-based steps were undertaken to design siRNA against SARS-CoV-2 RdRp (Fig. 1).
Effective and rational designing of the siRNA candidate is necessitated by the unwanted effects produced by the non-specific siRNA (Ebenezer, 2020). The difference in the activities is displayed when the same mRNA is targeted at a different position with randomly designed siRNA (Amarzguioui and Prydz, 2004). Characteristics of nucleotide sequences strongly influence the siRNA silencing efficacy (Ui-Tei et al., 2004). siRNA silencing efficiency is improved by nucleotide sequence preference as target recognition, and cleavage depends on the bases at a particular position. In this regard, an excellent feature of siDirect 2.0 tool is that it selects highly functional siRNA sequences using Ui-Tei (Ui-Tei et al., 2004), Amarzguioui (Amarzguioui and Prydz, 2004), and Reynolds (Reynolds et al., 2004) algorithms. Ui-Tei algorithm (Ui-Tei et al., 2004) follows specific rules, such as i) 5′ end of the sense/passenger strand must contain G/C nucleotide, ii) 5′ terminus of the antisense/guide strand has to include A/U nucleotide, iii) 5′ terminal 7 base pairs of sense/passenger strand has to contain at least 4 A/U nucleotides, and iv) GC stretch should not be longer than 9 nucleotides (Table 1). A highly effective RNA-interference to firefly luciferase activity and chick embryo was reported with the siRNAs, satisfying the Ui-Tei guidelines (Ui-Tei et al., 2004). Meanwhile, siRNAs obeying the nucleotide features of Amarzguioui guidelines are regarded as efficient (Amarzguioui and Prydz, 2004). Amarzguioui rules include the parameters such as i) the A/U differential of the duplex end should be more than zero, ii) robust binding of 5′ sense/passenger strand, iii) position 1 must contain any base except U, iv) position 6 should always contain A, v) weak binding of 3′ sense/passenger strand and vi) position 19 must contain any base except G (Amarzguioui and Prydz, 2004) (Table 1). Reynolds algorithm also follows several criteria, such as i) maintenance of GC content in the designed siRNA between 30% to 52%, ii) the sense/passenger strand must maintain ≥3 base pairs at the position between 15 and 19, iii) internal stability has to be low at a target site, iv) position 19 and 3 of the sense/passenger strand must contain A, v) sense/passenger strand should contain U at position 10, vi) position 13 of the sense/passenger strands must contain any bases other than G. Incorporation of these criteria of Reynolds significantly improves potent siRNA selection (Reynolds et al., 2004) (Table 1). siRNA molecules following all the rules and algorithms of Ui-Tei, Amarzguioui, and Reynolds are considered to be the most operative (Taxman et al., 2006). Another important feature of siDirect 2.0 involves the significant reduction of off-target silencing with the maintenance of seed target duplex Tm values for all predicted siRNAs less than 21.5 °C (Naito et al., 2009). Designing features of siRNA by siDirect 2.0 also includes the elimination of siRNAs that have near-perfect matches to any other non-targeted transcripts (Naito et al., 2009). The siRNAs obtained by siDirect 2.0 targeting RdRp, therefore, are rationally functional considering the aforementioned criteria followed by this tool (Table 2, and Supplementary Tables 2 & 3). RNAxs another webserver, considers several critical parameters such as accessibility threshold, self-folding energy, sequence asymmetry, energy asymmetry for designing effective siRNAs (Table 3, and Supplementary Tables 4 & 5).
Though the major purpose of designing siRNAs is to knock down specific targets, there remain possibilities of silencing an unknown number of unintended genes (Ozcan et al., 2015). RISCs have the potentiality to inhibit the expression of any mRNAs with perfect complementary base-pairing to the guide strand seed region (bases 2–8 from 5′ end) (Song et al., 2004). There is a possibility for a large number of potential off-target matches as the seed region consists of only seven bases. However, human argonaute 2 requires more extensive base pairing for the efficient catalytic degradation of target mRNAs (Setten et al., 2019; Song et al., 2004). Two mechanisms can explain the off-target effect. First, tolerance of siRNA to several mismatches of the target sequences of mRNA retains their capacity to execute silencing of those targets bearing imperfect complementarity (Ozcan et al., 2015; Snove Jr. and Holen, 2004). Second, siRNAs and their related class of miRNAs are nearly identical. The promiscuous entrance of siRNAs into endogenous miRNA machinery allows them to recognize the nucleotide sequences of the ‘seed regions' (i.e., 2–8 nucleotides) of targets with perfect complementarity. In contrast, the complementarity of remaining nucleotides (i.e., the region excluding the seed sequence) for recognition possesses less significance (Doench et al., 2003; Ozcan et al., 2015), leading to the downregulation of the unpredictable number of mRNAs. Therefore, siRNAs designed by different algorithms might still induce off-target effects (Ozcan et al., 2015). Hence, siRNA design processes are typically followed by a BLAST search for cross-reactive 21-bp siRNA sequences to ensure siRNA target specificity. In this analysis, NCBI-BLAST revealed the absence of significant matches of the SARS-CoV-2 RdRp target sequences against the whole genome ruling out the possibilities for off-target silencing (data not shown).
The siRNAs shared by both siDirect 2.0 and RNAxs, in this study, possessed all the characteristics (i.e., nucleotide-based and other characteristics) for being effective siRNAs (i.e., following of the UAR algorithms, bearing melting temperature (Tm) of seed-target duplex below 21.5 °C, possessing of at minimum two mismatches to any other non-targeted transcripts to avoid off-target silencing, potential accessibility threshold, self-folding energy of 0.9022, devoid of off-target silencing, etc.) (Table 4). Determination of several other parameters like siRNA GC content, the secondary structure of the siRNA guide strand, target guide interaction, etc., in the subsequent steps addressed queries related to the effectiveness of siRNAs. GC content is inversely related to the siRNA functionality (Amarzguioui and Prydz, 2004; Chan et al., 2009). The unwinding of the siRNA duplex may be slower by RISC complex-associated helicase when the content of GC is too high (Amarzguioui and Prydz, 2004). The efficiency of target mRNA recognition and hybridization, on the other hand, might be reduced by the lower GC content (Amarzguioui and Prydz, 2004). It is, thus, recommended to pick siRNA sequences with low GC content (between 31.6% and 57.9%) (Chan et al., 2009). GC content in the range of 31.6%–57.9% in the selected siRNAs against the consensus target 1, 18, 19, and 22 further strengthens the feasibility of these siRNAs (Table 5). Previous studies have suggested that the stability of an RNA molecule should possess the minimum free energy of folding (Vickers et al., 2000). Therefore, the siRNA molecules with the positive free energy of folding of the guide strands might get more entrance to the target and have higher potential to interact with the target, causing effective gene silencing (Shawan et al., 2015; Singh et al., 2012). The positive free energy of folding (i.e., ΔG of folding: 0.1 kcal/mol) by consensus target 1 siRNA (i.e., siRNA against consensus target sequence 1) rationalizes the general assumption for its better entrance to target for effective gene silencing (Table 5 and Fig. 2). However, a previous study did not provide a noteworthy contribution of intramolecular siRNA sequence secondary structure to siRNA efficiency (Amarzguioui and Prydz, 2004). To investigate the efficiency of the predicted siRNA, the free energy of binding with the target (i.e., computational RNA-RNA interaction) is another important parameter since RNAi efficiency is well correlated with the binding energies of siRNAs to their respective target mRNA (Schubert et al., 2005), and thus, lies at the core of target prediction algorithm. Given that optimum free energy favors siRNA-target binding, the highest free energy (i.e., −35.2 kcal/mol) of binding of the consensus target sequence 1 siRNA with its target indicate its strong binding with the target site among all the siRNAs (Table 5 and Fig. 3). The better effectiveness of the siRNAs is also indicated by the higher values of Tm (Cp) and Tm (Conc). Consensus target 1 siRNA showed an indication of higher effectiveness than other siRNAs by demonstrating higher values of Tm (Cp) and Tm (Conc) (Table 5 and Supplementary Fig. 1).
The consensus target 1 siRNA appears to be the potential siRNA targeting RdRp among all the siRNAs. siRNAPred, a prediction web server for the prediction of the efficacy of a query siRNA, further verified this siRNA molecule with a prediction score of 0.857, indicating it as a high efficacy siRNA molecule.
The gene silencing efficiency of a selected siRNA candidate depends not only on the degree of its potentiality but also on the availability of the target site within the mRNA molecule. Hence, siRNA target site accessibility is ultimately a crucial benchmark for establishing siRNA-mediated gene silencing efficacy (Gatta et al., 2018). A comprehensive analysis of the target site of the RdRp gene sequence, having 181 nucleotides, predicting the availability for siRNA binding, was performed using the Sfold web server. The probability profile and loop-specific probability profile of siRNA for specific target sites indicated the accessibility of the consensus target 1 siRNA to its target site (Fig. 4). The thermodynamics of siRNA duplex ends defines its efficacy (Khvorova et al., 2003). An effective siRNA should possess free energy in its antisense strand, not exceeding beyond −10 kcal/mol (Ui-Tei et al., 2004). The free energy of the antisense strand of the consensus target 1 siRNA observed in this study was −6.5 to −9.5 kcal/mol, indicating its gene silencing potential (Fig. 4).
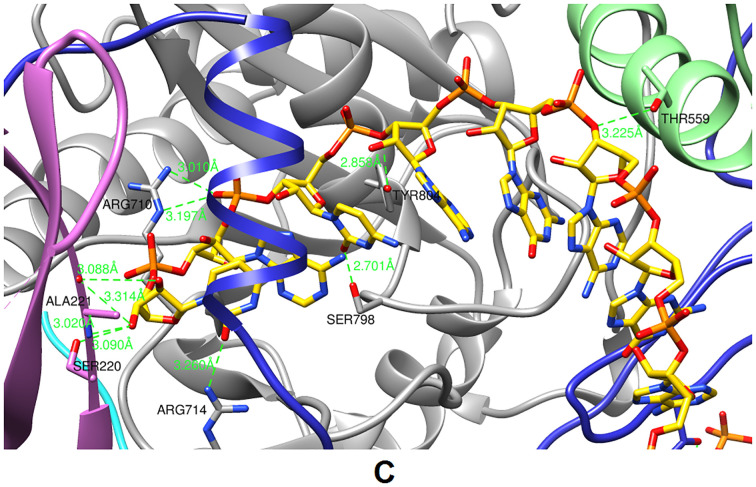
The in silico molecular docking simulation successfully docked our proposed siRNA molecule against argonaute protein with a docking score of −344.12 and ligand RMSD of 94.65 Å (Fig. 5). This docking score showed impressive binding between the designed siRNA and argonaute protein within the docked complex, facilitated by the formation of nine-strong H-bonds (length is in between 2.70 Å–3.2 Å) involving SER220, ALA221, THR559, ARG710, ARG714, SER798, and TYR864 residues (Fig. 5).
The studies of MD simulation assess the stability of the siRNA (guide strand)‑argonaute and target protein-siRNA docked complex (Fig. 6, Fig. 7). The molecular simulation of guide-target molecules has a prospect to acquire the best conformations and calculates the best modes of interactions relative to each other. MD simulation results established the proper stability of the guide siRNA‑argonaute protein complex signifying perdurable interactions among target protein-siRNA complex.
Therapeutic applications of siRNAs for desired gene silencing might face difficulties due to several factors such as instability of siRNAs, reduced cellular uptake, and the absence of a competent delivery method (Tanaka et al., 2010). For effective gene therapy, a suitable promoter-controlled vector can facilitate the targeting of therapeutic genes to the desired cell/location (Glorioso and DeLuca, 1995). To assess the potentiality of a newly designed siRNA, vector-based siRNA in plasmid form can also be used to target desired genes within a specific cell culture/cell line (ElHefnawi et al., 2016). Furthermore, plasmid bearing siRNA can directly be injected into desired organs (Giladi et al., 2003).
5. Conclusion
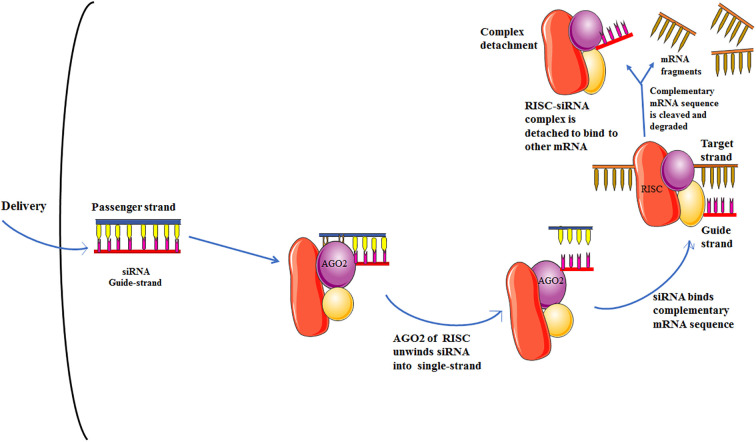
siRNA therapy might be a potential tool of the RNAi pathway to control human viral infections efficiently. In the present pandemic situation of COVID-19, this study was undertaken to design the siRNA molecule rationally as a therapeutic agent using various computational tools against RdRp of SARS-CoV-2. The investigation led to the selection of a single effective siRNA molecule (i.e., guide strand: 5′- UAGUACUACAGAUAGAGACAC -3′; passenger strand: 5′-GUCUCUAUCUGUAGUACUAUG-3′). The selected siRNA molecule was subjected to molecular docking and molecular dynamics simulation, which gave a promising outcome. We have proposed a general antiviral mechanism (Fig. 8 ) for this siRNA molecule that may be effective against SARS-CoV-2 RdRp. However, our work will require further validations in vitro and in vivo animal models for future applications.
Fig. 8.
Proposed mechanism of action of the designed siRNA showing the PTGS of RdRp gene from SARS-CoV-2.
Author's contribution
Mohammad Mahfuz Ali Khan Shawan and Ashish Ranjan Sharma conceptualized, conceived the research, and wrote the manuscript. Manojit Bhattacharya, Bidyut Mallik, Farhana Akhter, Md. Salman Shakil, Md. Mozammel Hossain and Subrata Banik interpreted data, edited, and reviewed the final manuscript. Sang-Soo Lee, Chiranjib Chakraborty, and Md. Ashraful Hasan supervised all the work.
Ethics approval and consent to participate
Ethical approval or individual consent is not applicable.
Availability of data and materials
All data and materials are included within the manuscript.
Conflicts of interest
The authors declare no conflict of interest.
Acknowledgments
This study was supported by Hallym University Research Fund and by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF - 2020R1C1C1008694 & NRF - 2020R1I1A3074575).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.meegid.2021.104951.
Appendix A. Supplementary data
Supplementary material
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/s0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amarzguioui M., Prydz H. An algorithm for selection of functional siRNA sequences. Biochem. Biophys. Res. Commun. 2004;316:1050–1058. doi: 10.1016/j.bbrc.2004.02.157. [DOI] [PubMed] [Google Scholar]
- Bellaousov S., Reuter J.S., Seetin M.G., Mathews D.H. RNAstructure: Web servers for RNA secondary structure prediction and analysis. Nucleic Acids Res. 2013;41:W471–W474. doi: 10.1093/nar/gkt290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Boehr D.D., Nussinov R., Wright P.E. The role of dynamic conformational ensembles in biomolecular recognition. Nat. Chem. Biol. 2009;5:789–796. doi: 10.1038/nchembio.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burley S.K., et al. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019;47:D464–d474. doi: 10.1093/nar/gky1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C. Potentiality of small interfering RNAs (siRNA) as recent therapeutic targets for gene-silencing. Curr. Drug Targets. 2017;3:469–482. doi: 10.2174/138945007780058988. [DOI] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Sharma G., Doss C.G.P., Lee S.-S. Therapeutic miRNA and siRNA: moving from bench to clinic as next generation medicine. Mol. Ther. Nucleic Acids. 2017;8:132–143. doi: 10.1016/j.omtn.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A., Sharma G., Bhattacharya M., Lee S. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4016–4026. doi: 10.26355/eurrev_202004_20871. [DOI] [PubMed] [Google Scholar]
- Chakraborty C., Sharma A.R., Bhattacharya M., Sharma G., Lee S.-S. The 2019 novel coronavirus disease (COVID-19) pandemic: a zoonotic prospective Asian Pacific journal of. Trop. Med. 2020;13:242. [Google Scholar]
- Chan C.Y., Carmack C.S., Long D.D., Maliyekkel A., Shao Y., Roninson I.B., Ding Y. A structural interpretation of the effect of GC-content on efficiency of RNA interference. BMC Bioinformatics. 2009;(10 Suppl 1):S33. doi: 10.1186/1471-2105-10-s1-s33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/s0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dana H., et al. Molecular mechanisms and biological functions of siRNA. Int. J. Biomed. Sci. 2017;13:48–57. [PMC free article] [PubMed] [Google Scholar]
- Dhama K., Sharun K., Tiwari R., Dadar M., Malik Y.S., Singh K.P., Chaicumpa W. COVID-19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum. Vaccin. Immunother. 2020:1–7. doi: 10.1080/21645515.2020.1735227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., Chan C.Y., Lawrence C.E. Sfold web server for statistical folding and rational design of nucleic acids. Nucleic Acids Res. 2004;32:W135–W141. doi: 10.1093/nar/gkh449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen C.P., Sharp P.A. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/s1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebenezer A. Designing effective small interfering RNA for post-transcriptional silencing of human GREM1: a comprehensive. Bioinf. Approach Int. Res. J. Biol. Sci. 2020;2:19–31. [Google Scholar]
- Elbashir S.M., Lendeckel W., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElHefnawi M., et al. In silico design and experimental validation of sirnas targeting conserved regions of multiple hepatitis C virus genotypes. PLoS One. 2016;11 doi: 10.1371/journal.pone.0159211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatta A.K., Hariharapura R.C., Udupa N., Reddy M.S., Josyula V.R. Strategies for improving the specificity of siRNAs for enhanced therapeutic potential. Expert Opin. Drug Discovery. 2018;13:709–725. doi: 10.1080/17460441.2018.1480607. [DOI] [PubMed] [Google Scholar]
- Giladi H., Ketzinel-Gilad M., Rivkin L., Felig Y., Nussbaum O., Galun E. Small interfering RNA inhibits hepatitis B virus replication in mice. Mol. Ther. 2003;8:769–776. doi: 10.1016/s1525-0016(03)00244-2. [DOI] [PubMed] [Google Scholar]
- Glorioso J.C., DeLuca N.A., Fink D.J. Development and application of herpes simplex virus vectors for human gene therapy. Annu. Rev. Microbiol. 1995;49:675–710. doi: 10.1146/annurev.mi.49.100195.003331. [DOI] [PubMed] [Google Scholar]
- Hammond S.M., Bernstein E., Beach D., Hannon G.J. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. s. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- Hanwell M.D., Curtis D.E., Lonie D.C., Vandermeersch T., Zurek E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. Aust. J. Chem. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harapan H., et al. Coronavirus disease 2019 (COVID-19): a literature review. J. Infect. Public Health. 2020;13:667–673. doi: 10.1016/j.jiph.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Weng Y., Xia X.H., Liang X.J., Huang Y. Clinical advances of siRNA therapeutics. J. Gene Med. 2019;21 doi: 10.1002/jgm.3097. [DOI] [PubMed] [Google Scholar]
- Huang C., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/s0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Dalke A., Schulten K.J., Jomg . Vol. 14. 1996. VMD: visual molecular dynamics; pp. 33–38. [DOI] [PubMed] [Google Scholar]
- Jana S., Chakraborty C., Nandi S. Mechanisms and roles of the RNA-based gene silencing. Electron. J. Biotechnol. 2004;7(3):15–16. [Google Scholar]
- Jana S., Chakraborty C., Nandi S. Mechanisms and roles of the RNA-based gene silencing. Electron. J. Biotechnol. 2004;7(3):15–16. [Google Scholar]
- Kalé L., et al. Vol. 151. 1999. NAMD2: greater scalability for parallel molecular dynamics; pp. 283–312. [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kumar M.L.S., Raghava G.P.S. International Conference on Open Source for Computer Aided Drug Discovery, IMTECH, Chandigarh. 2009. siRNApred: SVM based method for predicting efficacy value of siRNA. Proceedings of the OSCADD-2009. [Google Scholar]
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Lung J., et al. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020 doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKerell A.D., Jr., Banavali N., Foloppe N.J. Vol. 56. 2000. Development and current status of the CHARMM force field for nucleic acids; pp. 257–265. [DOI] [PubMed] [Google Scholar]
- Majumdar R., Rajasekaran K., Cary J.W. RNA interference (RNAi) as a potential tool for control of mycotoxin contamination in crop plants: concepts and considerations. Front. Plant Sci. 2017;8:200. doi: 10.3389/fpls.2017.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020;55 doi: 10.1183/13993003.00607-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naito Y., Yoshimura J., Morishita S., Ui-Tei K. siDirect 2.0: updated software for designing functional siRNA with reduced seed-dependent off-target effect. BMC Bioinformatics. 2009;10:392. doi: 10.1186/1471-2105-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nur S.M., Al Amin M., Alam R., Hasan M.A., Hossain M.A., Mannan A. An in silico approach to design potential siRNA molecules for ICP22 (US1) gene silencing of different strains of human herpes simplex 1. J. Young Pharm. 2013;5:46–49. doi: 10.1016/j.jyp.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ok-Seon Kwon S.-J.K., Kim Jin Sang, Lee Gunbong, Maeng Han-Joo, Lee Jeongmi, Hwang Gwi Seo, Cha Hyuk-Jin, Chun Kwang-Hoon. Designing tyrosinase siRNAs by multiple prediction algorithms and evaluation of their anti-melanogenic effects vol 26. Biomol. Ther. 2018;26(3):282–289. doi: 10.4062/biomolther.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan G., Ozpolat B., Coleman R.L., Sood A.K., Lopez-Berestein G. Preclinical and clinical development of siRNA-based therapeutics. Adv. Drug Deliv. Rev. 2015;87:108–119. doi: 10.1016/j.addr.2015.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Reynolds A., Leake D., Boese Q., Scaringe S., Marshall W.S., Khvorova A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Guerra Pedregal J., Marechal J.D. PyChimera: use UCSF chimera modules in any Python 2.7 project. Bioinformatics. 2018;34:1784–1785. doi: 10.1093/bioinformatics/bty021. [DOI] [PubMed] [Google Scholar]
- Rosenbaum L. Facing covid-19 in Italy - ethics, logistics, and therapeutics on the epidemic’s front line. N. Engl. J. Med. 2020;382:1873–1875. doi: 10.1056/NEJMp2005492. [DOI] [PubMed] [Google Scholar]
- Saha A., Sharma A.R., Bhattacharya M., Sharma G., Lee S.-S., Chakraborty C. Probable molecular mechanism of remdesivir for the treatment of covid-19: need to know more. Arch. Med. Res. 2020;51(6):585–586. doi: 10.1016/j.arcmed.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha A., Sharma A.R., Bhattacharya M., Sharma G., Lee S.-S., Chakraborty C. Tocilizumab: a therapeutic option for the treatment of cytokine storm syndrome in COVID-19. Arch. Med. Res. 2020;51(6):595–597. doi: 10.1016/j.arcmed.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R.P., et al. Repurposing drugs, ongoing vaccine, and new therapeutic development initiatives against COVID-19. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.01258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert S., Grünweller A., Erdmann V.A., Kurreck J. Local RNA target structure influences siRNA efficacy: systematic analysis of intentionally designed binding regions. J. Mol. Biol. 2005;348:883–893. doi: 10.1016/j.jmb.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Setten R.L., Rossi J.J., Han S.P. The current state and future directions of RNAi-based therapeutics. Nat. Rev. Drug Discov. 2019;18:421–446. doi: 10.1038/s41573-019-0017-4. [DOI] [PubMed] [Google Scholar]
- Shawan M., et al. Design and prediction of potential RNAi (siRNA) molecules for 3’UTR PTGS of different strains of zika virus: a computational approach. Nat. Sci. 2015;13:37–50. [Google Scholar]
- Singh S., Gupta S.K., Nischal A., Khattri S., Nath R., Pant K.K., Seth P.K. Design of potential siRNA molecules for hepatitis delta virus gene silencing. Bioinformation. 2012;8:749–757. doi: 10.6026/97320630008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snove O., Jr., Holen T. Many commonly used siRNAs risk off-target activity. Biochem. Biophys. Res. Commun. 2004;319:256–263. doi: 10.1016/j.bbrc.2004.04.175. [DOI] [PubMed] [Google Scholar]
- Sohrabi C., et al. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J.J., Smith S.K., Hannon G.J., Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434–1437. doi: 10.1126/science.1102514. [DOI] [PubMed] [Google Scholar]
- Tanaka K., Kanazawa T., Ogawa T., Takashima Y., Fukuda T., Okada H. Disulfide crosslinked stearoyl carrier peptides containing arginine and histidine enhance siRNA uptake and gene silencing. Int. J. Pharm. 2010;398:219–224. doi: 10.1016/j.ijpharm.2010.07.038. [DOI] [PubMed] [Google Scholar]
- Taxman D.J., et al. Criteria for effective design, construction, and gene knockdown by shRNA vectors. BMC Biotechnol. 2006;6:7. doi: 10.1186/1472-6750-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei K., et al. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickers T.A., Wyatt J.R., Freier S.M. Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res. 2000;28:1340–1347. doi: 10.1093/nar/28.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y., Zhang D., Zhou P., Li B., Huang S.Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45:W365–w373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., et al. Structural basis for inhibition of the RNA-dependent RNA polymerase from SARS-CoV-2 by remdesivir. Science. 2020 doi: 10.1126/science.abc1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore P.D., Tuschl T., Sharp P.A., Bartel D.P. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/s0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhou P., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
All data and materials are included within the manuscript.