Abstract
Background
The difference between an individual's chronological and DNA methylation predicted age (DNAmAge), termed DNAmAge acceleration (DNAmAA), can capture life-long environmental exposures and age-related physiological changes reflected in methylation status. Several studies have linked DNAmAA to morbidity and mortality, yet its relationship with kidney function has not been assessed. We evaluated the associations between seven DNAm aging and lifespan predictors (as well as GrimAge components) and five kidney traits (estimated glomerular filtration rate [eGFR], urine albumin-to-creatinine ratio [uACR], serum urate, microalbuminuria and chronic kidney disease [CKD]) in up to 9688 European, African American and Hispanic/Latino individuals from seven population-based studies.
Results
We identified 23 significant associations in our large trans-ethnic meta-analysis (p < 1.43E−03 and consistent direction of effect across studies). Age acceleration measured by the Extrinsic and PhenoAge estimators, as well as Zhang’s 10-CpG epigenetic mortality risk score (MRS), were associated with all parameters of poor kidney health (lower eGFR, prevalent CKD, higher uACR, microalbuminuria and higher serum urate). Six of these associations were independently observed in European and African American populations. MRS in particular was consistently associated with eGFR (β = − 0.12, 95% CI = [− 0.16, − 0.08] change in log-transformed eGFR per unit increase in MRS, p = 4.39E−08), prevalent CKD (odds ratio (OR) = 1.78 [1.47, 2.16], p = 2.71E-09) and higher serum urate levels (β = 0.12 [0.07, 0.16], p = 2.08E−06). The “first-generation” clocks (Hannum, Horvath) and GrimAge showed different patterns of association with the kidney traits. Three of the DNAm-estimated components of GrimAge, namely adrenomedullin, plasminogen-activation inhibition 1 and pack years, were positively associated with higher uACR, serum urate and microalbuminuria.
Conclusion
DNAmAge acceleration and DNAm mortality predictors estimated in whole blood were associated with multiple kidney traits, including eGFR and CKD, in this multi-ethnic study. Epigenetic biomarkers which reflect the systemic effects of age-related mechanisms such as immunosenescence, inflammaging and oxidative stress may have important mechanistic or prognostic roles in kidney disease. Our study highlights new findings linking kidney disease to biological aging, and opportunities warranting future investigation into DNA methylation biomarkers for prognostic or risk stratification in kidney disease.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13148-021-01082-w.
Keywords: Aging, Kidney function, Epigenetic age acceleration, DNAm age, Glomerular filtration rate, UACR, Serum urate
Background
The kidneys are responsible for maintenance of homeostasis and blood filtration, and their function is most commonly clinically assessed by measuring serum creatinine levels to estimate glomerular filtration rate (eGFR) [1, 2]. Chronic kidney disease (CKD), defined by low eGFR (< 60 ml/min/m2) and/or presence of protein in urine, is an increasingly prevalent non-communicable disease with a considerable burden worldwide [3–5]. Increased urinary albumin-to-creatinine ratio (uACR) is a marker of kidney injury, measured to identify early kidney damage which can precede eGFR decline (for example, diabetic nephropathy) [6]. Albuminuria is a predictor of CKD progression and mortality [6, 7], whereas high serum levels of urate, a molecule of purine nucleotide metabolism excreted by the kidney, is a risk factor for incident cardiovascular and kidney disease, and also a biomarker of low eGFR [8].
DNA methylation (DNAm), defined as the covalent addition of a methyl group to a DNA nucleotide (usually the cytosine of a cytosine-guanine dinucleotide [CpG]), is the most extensively studied epigenetic mechanism, and its role in numerous conditions and diseases has been demonstrated [9]. Age-predicting algorithms based on the percentages of DNAm observed at sets of CpGs, such as the ones proposed by Hannum [10] and Horvath [11], have been used to predict an individual’s age (DNAmAge) and assess biological aging by calculating the difference between an individual’s predicted and chronological age—a concept known as DNAmAge acceleration (DNAmAA) [12, 13]. Other measures have been derived to assess specific aspects of aging mechanisms, such as intrinsic epigenetic age acceleration (IEAA), which assesses aging independent of blood immune system changes [14], or extrinsic epigenetic age acceleration (EEAA) [15, 16], which specifically estimates aging as related to the immune system and reflected in changes in blood immune cell-type proportions. A “second generation” of DNAm-based aging signatures incorporated physiological markers to better capture changes in traditional biological aging biomarkers [12]. PhenoAge was developed as a marker meant to mirror physiological dysregulation as reflected in changes in age and 9 additional age-related features, such as C-reactive protein and serum glucose [17]. GrimAge is a mortality predictor based on mortality-related DNAm-estimated traits [18]. Another DNAm-based lifespan predictor, the 10-CpG epigenetic mortality risk score (MRS), stands out for its simplicity and its recent validation [19, 20].
Multiple studies have shown, although with varying findings, a positive relationship between DNAmAge measured in blood and aging-related diseases and mortality [12, 13, 21]. DNAmAge is associated with all-cause mortality [22, 23], frailty [24], cognitive function and physical fitness [25], body mass index (BMI) [26] and obesity [27], lifetime stress [28] and a number of other age-related conditions [12]. The available evidence points to DNAmAge as a potential global biomarker of biological aging and health, though potential for publication bias must be considered [21]. Although some of the “second-generation” DNAm-based aging measures include proteins or markers known to be associated with kidney function [17, 18], whether these and other DNAm-based predictors are correlated with different parameters of kidney aging and low function has not been investigated [13, 29, 30].
We evaluated the association between five kidney traits (eGFR, prevalent CKD, uACR, microalbuminuria and serum urate) and seven DNAm-based age and/or lifespan predictors (HannumAA, HorvathAA, EEAA, IEAA, PhenoAA, GrimAA and MRS) in up to seven population-based studies in a large trans-ethnic meta-analysis. We also evaluated kidney trait associations with secondary DNAm-based predictors: categorical epigenetic mortality risk score (MRS) variables, and eight DNAm-estimated traits underlying the GrimAge mortality predictor. We additionally performed ethnic-specific meta-analyses to identify robust associations across cohorts of different ethnicities.
Results
Study characteristics
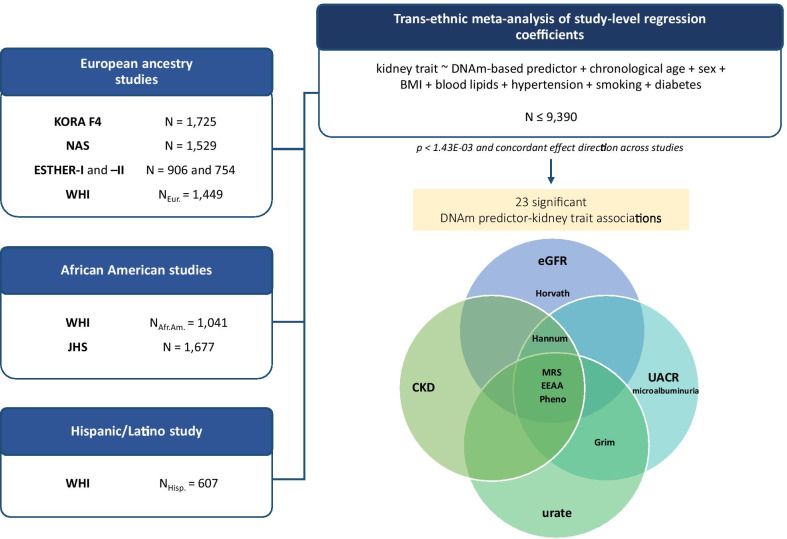
We conducted a trans-ethnic meta-analysis of seven DNAm-based age/lifespan predictors and five kidney traits using data from seven population-based cohorts (Fig. 1, population characteristics in Table 1). Serum creatinine-based traits (eGFR and prevalent CKD) were available for all European ancestry (k = 5), African American (k = 2) and one additional Hispanic/Latino study (N = 9688). Sample sizes for the other traits were smaller: serum urate was available in four studies with European ancestry and one study with African American participants (N = 5903), while uACR and microalbuminuria were available in three of the European ancestry studies and one African American study (N = 4110). Additional details on the cohorts are provided in Additional file 1: Table S1.
Fig. 1.
Study design. Regression analyses were conducted in each study by modeling DNAm-based predictors as independent variables and kidney traits as dependent variables, adjusting for confounders. Results from the fully adjusted model (with chronological age, sex, BMI, log-transformed triglycerides, HDL, hypertension, smoking status, diabetes as covariates and baseline eGFR for serum urate analyses) were meta-analyzed using inverse-variance weighted fixed-effects and random-effects models. We based our main interpretations on the fixed-effects results; if heterogeneity was large (I2 > 0.50 and Cochran’s Q phet < 0.05), we based our interpretations on the random-effects results. The Venn diagram shows the set of 23 statistically significant associations between DNAm-based predictors and kidney traits identified in the trans-ethnic meta-analysis (p < 1.43E−03 and consistent direction of effect across studies)
Table 1.
Population characteristics
| Traits | KORA | ESTHER-I | ESTHER-II | NAS | WHI | JHS | ||
|---|---|---|---|---|---|---|---|---|
| Ethnic background | Eur | Eur | Eur | Eur | Afr. Am | His | Eur | Afr. Am |
| N | 1725 | 906 | 754 | 1529 | 1041 | 607 | 1449 | 1677 |
| Age | 60.98 (8.88) | 62.00 (6.53) | 62.76 (6.75) | 74.61 (7.05) | 61.83 (6.67) | 60.93 (6.64) | 66.32 (6.73) | 56.23 (12.31) |
| Male | 843 (48.9) | 435 (48.0) | 316 (41.9) | 1529 (100) | 0 (0) | 0 (0) | 0 (0) | 649 (37.17) |
| BMI | 28.11 (4.78) | 27.75 (4.25) | 27.47 (4.76) | 28.00 (4.13) | 31.61 (6.43) | 29.21 (5.22) | 28.84 (5.83) | 32.02 (7.37) |
| Smoking status | ||||||||
| Never smoker | 720 (41.7) | 443 (48.9) | 354 (47.0) | 478 (31.3) | 496 (48.06) | 376 (62.46) | 766 (53.34) | 1490 (85.88) |
| Ever smoker | 1003 (58.2) | 463 (51.1) | 400 (53.0) | 1051 (68.7) | 536 (51.94) | 226 (37.54) | 670 (46.66) | 245 (14.12) |
| Serum creatinine | 0.91 (0.27) | 0.69 (0.31) | 0.85 (0.31) | 1.11 (0.45) | 0.82 (0.21) | 0.73 (0.23) | 0.74 (0.14) | 0.96 (0.59) |
| eGFR | 86.77 (16.02) | 99.77 (21.15) | 86.63 (18.94) | 69.32 (16.14) | 92.31 (19.65) | 88.64 (15.39) | 83.69 (13.33) | 93.57 (22.37) |
| CKD | 99 (5.7) | 54 (5.9) | 72 (9.5) | 407 (26.6) | 59 (5.67) | 31 (5.11) | 82 (5.66) | 115 (6.59) |
| uACR ** | 6.15 (3.85, 11.97) | 9.14 (5.49, 17.98) | 8.92 (5.34, 16.53) | NA | NA | NA | NA | 5.95 (3.95,13.23) |
| Microalbuminuria | 150 (8.7) | 125 (13.8) | 104 (13.8) | NA | NA | NA | NA | 106 (13.75) |
| Serum urate | 5.37 (1.46) | 4.22 (1.49) | 4.85 (1.47) | 6.13 (1.51) | NA | NA | NA | 5.64 (1.70) |
| Diabetes | 158 (9.2) | 141 (15.56) | 150 (19.89) | 235 (15.4) | 166 (15.95) | 71 (11.70) | 96 (6.64) | 433 (24.81) |
| Hypertension | 788 (45.7) | 516 (55.95) | 446 (59.15) | 1130 (73.9) | 561 (56.21) | 207 (35.94) | 471 (35.33) | 1027 (58.82) |
| HDL cholesterol | 56.47 (14.64) | 51.70 (15.79) | 53.21 (15.45) | 48.81 (12.85) | 54.76 (13.78) | 51.05 (13.02) | 51.38 (11.92) | 51.35 (14.73) |
| Triglycerides ** | 110 (77, 158) | 89.70 (58.30, 140.50) | 111.85 (76.50, 116.30) | 114 (83, 158) | 104 (75, 141) | 140 (105, 188) | 133 (95, 184) | 92 (64, 129) |
| C-reactive protein ** | 1.27 (0.63, 2.655) | 1.62 (0.83, 3.46) | 2.40 (1.06, 4.72) | 1.47 (0.75, 3.04) | NA | NA | NA | 2.71 (1.18, 5.96) |
Population characteristics of all participating studies. The means and standards deviation (SD) are shown for continuous traits, and N (%) for categorical traits. **Skewed variables, for which median and (1st, 3rd quartile) are shown. The sample sizes presented here for each of the studies correspond to the number of observations with information on DNAm-predictors, serum-based creatinine kidney traits, chronological age and sex. Age was measured in years at time of participation in study; BMI in kg/m2; serum creatinine in mg/dL; eGFR, serum-creatinine-based estimated glomerular filtration rate in mL/min/1.73 m2; CKD: prevalent chronic kidney disease, defined as eGFR < 60 ml/min/1.73 m2; uACR in mg/g; microalbuminuria was defined as uACR ≥ 30 mg/g; serum urate in mg/dl; prevalent diabetes was defined based on use of glucose lowering drugs or fasting plasma glucose ≥ 126 mg/dl; hypertension defined using the Joint National Committee (JNC) VII definition (blood pressure > 140/90 mm Hg or use of anti-hypertensive medications); HDL cholesterol and triglycerides in mg/dl; C-reactive protein in mg/L; NA denotes the trait was not available. Abbreviations used in ethnic background row: Eur., European ancestry; Afr.Am.., African American; His., Hispanic/Latino. An extended version of this table is shown in Additional file 1: Table S1
DNAm-based age and lifespan predictors were used as independent variables and kidney traits as dependent variables in covariate-adjusted regression models. Study-level results showed associations were slightly attenuated after the inclusion of additional covariates in model 2 (namely BMI, log-transformed triglycerides, HDL, hypertension, smoking status and diabetes) in comparison with model 1 (basic model adjusting for chronological age and sex; Additional file 1: Table S2). Sensitivity analyses showed that “crude” bivariate correlations were largely attenuated after adjustment for chronological age; the introduction of additional variables did not significantly alter the observed effects, despite a slight increase in the coefficient after the introduction of smoking in models with MRS and GrimAge (Additional file 3: Note S1). Estimates from the fully adjusted model were meta-analyzed using inverse-variance weighted fixed-effects and random-effects models. We based our main interpretations on the fixed-effects results; if heterogeneity was large (I2 > 0.50 and Cochran’s Q phet < 0.05), we based our interpretations on the random-effects results (Methods).
Meta-analysis of associations between DNAm-based predictors and kidney traits
We identified 23 significant DNAm-based predictor-kidney trait associations (p < 1.43E−03 and concordant direction of effect across studies; Table 2). Three interesting groups worth further discussion are: (1) PhenoAA, MRS and EEAA were associated with all parameters of poor kidney health (lower eGFR, prevalent CKD, higher uACR or microalbuminuria and higher serum urate); (2) the “first-generation” epigenetic aging markers, where HannumAA was associated with all kidney traits but serum urate, and HorvathAA was only associated with lower eGFR; and (3) an analogous measure to age acceleration in GrimAge was associated with uACR, microalbuminuria and serum urate (Fig. 1). Six associations between DNAm-based predictors and kidney traits were replicated across ethnic groups, and ethnic-specific replication was observed for 16 associations in total.
Table 2.
Trans-ethnic meta-analyses of associations between kidney traits and DNAm-based age and lifespan predictors
| Clock | eGFR | CKD | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | 95% CI | p | I2 | phet | OR | 95% CI | p | I2 | phet | |
| HorvathAA | − 0.006 | − 0.01, − 0.003 | 5.15E−04 | 38.696 | 0.121 | 1.019 | 1.003, 1.034 | 0.016 | 13.663 | 0.323 |
| HannumAA | − 0.007 | − 0.011, − 0.004 | 1.05E−04 | 0 | 0.816 | 1.033 | 1.016, 1.05 | 8.55E−05 | 44.898 | 0.08 |
| GrimAA | − 0.006 | − 0.01, − 0.002 | 1.94E−03 | 63.474 | 0.008 | 1.027 | 1.01, 1.044 | 1.27E−03 | 77.714 | 5.2E−05 |
| PhenoAA | − 0.005 | − 0.008, − 0.002 | 2.62E−04 | 0 | 0.564 | 1.031 | 1.018, 1.044 | 3.19E−06 | 33.885 | 0.158 |
| EEAA | − 0.008 | − 0.012, − 0.005 | 2.09E−06 | 43.328 | 0.09 | 1.038 | 1.022, 1.055 | 3.41E−06 | 49.636 | 0.053 |
| MRS | − 0.117 | − 0.158, − 0.075 | 4.39E−08 | 27.67 | 0.208 | 1.784 | 1.474, 2.159 | 2.71E−09 | 68.295 | 0.002a |
| IEAA | − 0.004 | − 0.008, 0 | 0.051 | 16.08 | 0.303 | 1.007 | 0.99, 1.025 | 0.427 | 2.365 | 0.411 |
| Clock | uACR | Microalbuminuria | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| β | 95% CI | p | I2 | phet | OR | 95% CI | p | I2 | phet | |
| HorvathAA | 0.002 | − 0.004, 0.008 | 0.606 | 0 | 0.632 | 1.014 | 0.992, 1.036 | 0.223 | 0 | 0.703 |
| HannumAA | 0.014 | 0.009, 0.02 | 2.04E−06 | 0 | 0.967 | 1.054 | 1.032, 1.076 | 1.08E−06 | 0 | 0.898 |
| GrimAA | 0.029 | 0.021, 0.037 | 1.07E−12 | 7.22 | 0.357 | 1.106 | 1.074, 1.138 | 7.58E−12 | 0 | 0.59 |
| PhenoAA | 0.01 | 0.005, 0.015 | 2.71E−05 | 0 | 0.787 | 1.035 | 1.017, 1.053 | 8.96E−05 | 0 | 0.612 |
| EEAA | 0.013 | 0.008, 0.017 | 4.62E−07 | 0 | 0.989 | 1.048 | 1.03, 1.066 | 1.42E−07 | 0 | 0.86 |
| MRS | 0.252 | 0.179, 0.324 | 1.01E−11 | 76.128 | 0.006a | 2.238 | 1.734, 2.889 | 6.14E−10 | 67.72 | 0.026a |
| IEAA | 0.002 | − 0.004, 0.008 | 0.529 | 0 | 0.847 | 1.015 | 0.991, 1.04 | 0.214 | 0 | 0.769 |
| Clock | Urate | ||||
|---|---|---|---|---|---|
| β | 95% CI | p | I2 | phet | |
| HorvathAA | 0.003 | − 0.001, 0.007 | 0.12 | 0 | 0.453 |
| HannumAA | 0.005 | 0.001, 0.009 | 0.011 | 0 | 0.919 |
| GrimAA | 0.009 | 0.004, 0.013 | 1.17E−04 | 56.31 | 0.057 |
| PhenoAA | 0.009 | 0.006, 0.012 | 4.71E−08 | 0 | 0.432 |
| EEAA | 0.007 | 0.004, 0.011 | 4.37E−05 | 41.60 | 0.144 |
| MRS | 0.115 | 0.067, 0.162 | 2.08E−06 | 12.16 | 0.336 |
| IEAA | − 0.001 | − 0.005, 0.003 | 0.675 | 0 | 0.797 |
Results from trans-ethnic meta-analyses of associations between kidney traits and DNAm-based age and lifespan predictors in up to seven population-based studies. Study-level associations were adjusted for chronological age, sex, BMI, blood lipids, hypertension, smoking and diabetes. Beta coefficients are given as changes in one standard deviation (SD) of the continuous kidney trait. Fully adjusted associations of serum-creatinine-based traits (eGFR, CKD) are based on N ≤ 9390 observations, whereas the sample size for urinary albumin-based traits (uACR, microalbuminuria) is N ≤ 4406 and for urate N ≤ 5769. I2 is the heterogeneity statistic, and (Q) phet corresponds to Cochran’s Q heterogeneity statistic
Shown in bold are statistically significant associations (p < 1.43E-03 and consistent direction of effect across studies) with either no evidence of heterogeneity in the fixed-effects model or supporting findings from the random-effects model
aphet of MRS with CKD, uACR and microalbuminuria < 0.05, therefore reported association based on significant random-effects models: MRS-uACR: β = 0.248 [0.1, 0.397], p = 1.061E−03; MRS-CKD: OR = 1.915 [1.316, 2.786], p = 6.89E−04; and MRS-microalbuminuria: OR = 2.197 [1.403, 3.439], p = 5.82E−04 (Additional file 1: Table S3)
PhenoAA, EEAA and MRS universally associated with poor kidney health
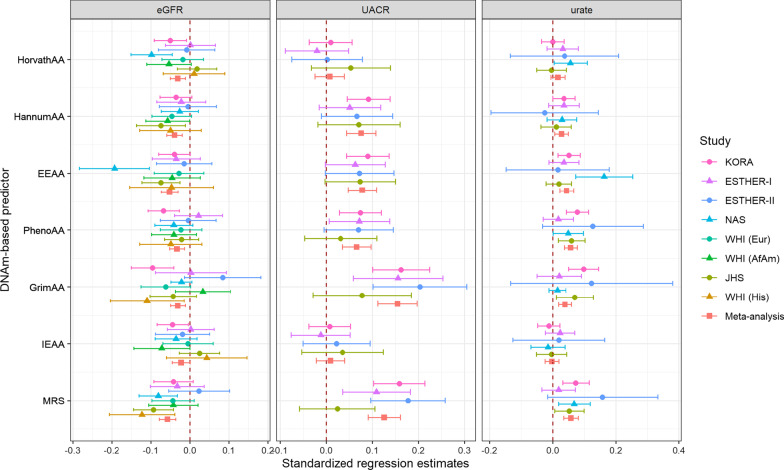
From these three DNAm-based predictors, associations with MRS had the smallest p-values: one MRS unit increase had a − 0.12 (95% CI = [− 0.16, − 0.08]) change in one standard deviation (SD) of log-transformed eGFR (p = 4.39E−08) and was associated with 78% [47–116%] increased odds of prevalent CKD (p = 2.71E−09) (Table 2). Although high heterogeneity was identified in associations between MRS and CKD, uACR and microalbuminuria, the results from random-effects models (Additional file 1: Table S3) were consistent with the estimates produced by the fixed-effect model: for example, one MRS unit increase was associated with a 0.25 [0.10, 0.40] change in one SD of log-transformed UACR (p = 1.06E−03). Figure 2 shows the study-level regression coefficients of the association between the continuous kidney traits and DNAm-based predictors standardized to one SD deviation in both terms to allow for their comparison. While the strength of association with serum urate of these three DNAm-based predictors was similar, MRS and EEAA had the largest effects on eGFR (Fig. 2, standardized effects in Additional file 1: Table S4). Similar observations were done for the binary kidney traits (Additional file 2: Fig. S1).
Fig. 2.
Standardized effect estimates from DNAm-based predictors and kidney traits. Scatter plot showing the effect estimates from the DNAmAge and lifespan predictors across continuous kidney traits for individual studies and trans-ethnic fixed-effects meta-analysis. Effect estimates have been standardized to one SD in both variables to allow for comparison of effect sizes. The legend shows the combination of shape and color coding assigned for the studies
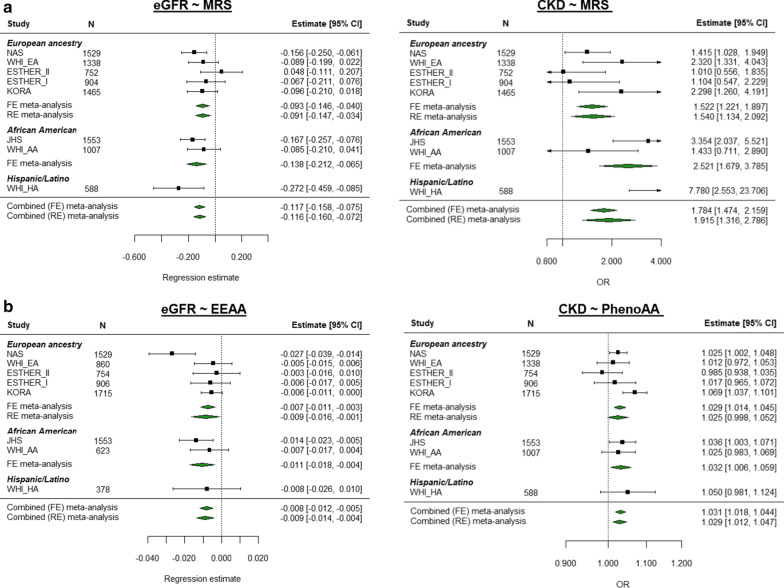
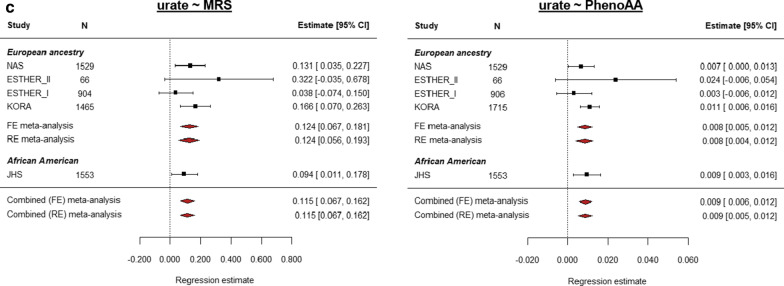
Figure 3 shows results from ethnic-specific meta-analyses significant in both European ancestry and African American meta-analyses (p < 1.43E−03, Additional file 1: Table S3). MRS stands out for its replication across all subgroups, including the small Hispanic/Latino cohort (Fig. 3a, study-level results in Additional file 1: Table S2). The eGFR-EEAA and CKD-PhenoAA effects were also replicated at the Bonferroni-corrected significance level across ethnic-specific meta-analyses (Fig. 3b), whereas their “complementary” associations (CKD-EEAA and eGFR-PhenoAA) were nominally significant (Additional file 2: Fig. S2A). Further effects observed in both European ancestry meta-analysis and the African American cohort were the associations of higher serum urate with MRS and PhenoAA (Fig. 3c). On the other hand, the associations between EEAA, PhenoAA and MRS with higher uACR (and prevalent microalbuminuria) identified in the trans-ethnic meta-analysis were mostly driven by the effects from the European ancestry cohorts (Additional file 2: Figs. S3 and S4), as well as that of serum urate and EEAA (Additional file 2: Fig. S5A). Most notably, the association between uACR and MRS was replicated at the Bonferroni-corrected level in two of the European ancestry cohorts (KORA and ESTHER-II), and nominally significant in the third one (Additional file 1: Table S2).
Fig. 3.
Multivariate regression meta-analysis of association between kidney traits and MRS, EEAA and PhenoAA. Multivariate regression models were used to assess the relationship between kidney traits and age acceleration as measured by seven DNAm-based predictors of age and/or lifespan, including Zhang’s 10-CpG mortality risk score (MRS), PhenoAge and extrinsic epigenetic age acceleration (EEAA). Results from the fully adjusted model included chronological age, sex, BMI, log-transformed triglycerides, HDL, hypertension, smoking status and diabetes as covariates, and baseline eGFR for serum urate analyses. Inverse-variance weighted fixed-effects models meta-analysis was conducted, where if heterogeneity was observed ([Q] phet < 0.05), a random-effects model was further interpreted. Individual panels show forest plots with the study-level and meta-analytic results for each association between kidney trait and DNAm-based predictor from the fully adjusted model. The rows correspond to the different studies and the sample size for each analysis (N). For eGFR and urate, the regression estimates represent the change in one standard deviation of the kidney trait per unit change in the MRS or per one year of age acceleration for PhenoAA and EEA. For CKD, the estimate column corresponds to the odds ratio (OR). The x-axis shows the estimates obtained from either the regression model (for single studies, data point shape is a black square) or the meta-analytic estimate (data point shape is a green diamond for serum creatinine-based traits and a red diamond for urate) with their 95% CI. Estimate: regression coefficient for the continuous traits and OR for CKD, 95% CI: 95% confidence interval of the estimate
Associations with “first-generation” DNAm clocks and GrimAge
HorvathAA, age acceleration measured by the “first-generation” DNAm-based predictor HorvathAge, was exclusively associated with low eGFR: a one year difference between DNAm-estimated age and chronological age was associated with a − 0.006 [− 0.01, − 0.003] change in log-transformed eGFR (p = 5.15E−04, Table 2). HannumAA, another “first-generation” DNAm-based age predictor, was also associated with eGFR; the strength of the association with both “first-generation” DNAm-based predictors was similar (Fig. 2). HannumAA was additionally associated with CKD, uACR and microalbuminuria (Additional file 2: Figs. S2–S4B). While the effects observed for HannumAA with eGFR and CKD were robustly replicated by cohorts with African American participants (Additional file 2: Fig. S2B), the association between HorvathAA and eGFR was mostly driven by studies with European ancestry (Additional file 2: Fig. S2C).
A year of GrimAge acceleration was associated with an increase of 0.03 SD of log-transformed uACR and a 10.6% increase in the odds of having microalbuminuria, both early markers of renal damage (Additional file 2: Figs S3–S4C). Likewise, a one-year difference in GrimAge was associated with higher serum urate levels, a risk factor for cardiorenal disease (Additional file 2: Fig. S5B). The GrimAA effects on uACR were the largest across DNAm-based predictors and kidney traits (Fig. 2), and a similar effect was observed for microalbuminuria (Additional file 2: Fig. S1).
Associations with categorical MRS and GrimAge components
In the categorical MRS, where risk groups were defined based on the number of CpGs with methylation levels beyond pre-defined high-risk thresholds, a 0.431 (95% CI = [0.29, 0.57]) SD increase in log-uACR was observed for individuals with > 5 “aberrantly” methylated CpGs (high risk MRS) compared to those with 0–1 aberrantly methylated loci (low risk MRS) (p = 4.01E-09, Additional file 2: Fig. S6A). Similar associations were observed for microalbuminuria (OR = 2.075 [1.582, 2.722], p = 1.36E-07; Additional file 1: Table S3). The moderate MRS category (defined as those with 2–5 “aberrantly” methylated loci) was also associated with serum urate (Additional file 2: Fig. S6B). These effects were mostly driven by European ancestry studies, where effects had a similar strength of association and replicated within this group (Additional file 2: Fig. S7). None of these effects replicated in the African American cohort, perhaps due to the smaller sample size and reduced statistical power.
In regard to the secondary analyses with the eight DNAm-based components that constitute GrimAge, we found associations for three of them. DNAm-estimated adrenomedullin (DNAmADM), plasminogen-activation inhibition 1 (DNAmPAI1) and pack years (DNAmPACKYRS) were positively associated with higher uACR, higher serum urate levels and microalbuminuria (Additional file 1: Table S5). Of note, the effects identified for serum urate replicated in both the European-ancestry analyses and in the African American cohort (Additional file 1: Table S2), whereas those in uACR and microalbuminuria were mostly replicated within the European studies.
Discussion
We identified 23 associations between kidney traits and DNAm-based predictors of aging and/or mortality in a large trans-ethnic meta-analysis from up to seven multi-ethnic population-based cohorts. PhenoAA, MRS and EEAA were associated with all parameters of poor kidney health (lower eGFR, prevalent CKD, higher uACR or microalbuminuria and higher serum urate). Distinct patterns of association were observed with age acceleration in the “first-generation” clocks (Hannum and Horvath) and an analogous measure in GrimAge: HorvathAA was only associated with lower eGFR, while HannumAA was associated with all kidney traits but serum urate. Finally, GrimAA was associated with uACR, microalbuminuria and serum urate.
Sensitivity analyses showed that the inclusion of chronological age in a “crude” model lead to large attenuation of the correlation between DNAm-based predictors and kidney traits, as expected when adjusting for variables confounding the exposure-outcome association. The posterior introduction of additional variables did not further alter the observed effects. A slight increase in the coefficient after the introduction of smoking was observed for MRS and GrimAge, which may be explained in general by the strong correlation between smoking and DNAm/DNAmAge acceleration in blood [31–34], and specifically in relation to the these two markers as they either directly incorporate cigarette smoking into its formulation [19] or capture the effects of cigarette smoking [20].
From the three “universally” associated DNAm-based predictors, MRS was robustly associated with multiple kidney phenotypes, some of which replicated across ethnic-specific analyses (i.e., eGFR, prevalent CKD and urate). Together with EEAA and GrimAA, MRS showed the largest effects in comparison with the other DNAm-based predictors. Moreover, MRS moderate and high risk categories [20] were also associated with eGFR and markers of kidney injury in the secondary analyses. Recent evidence suggests the MRS is a DNAm-based biomarker which reflects mortality risks by capturing the effects of oxidative stress and systemic inflammation, as well as inflammation-driven changes in immune cell counts—all mechanisms shared by numerous chronic diseases [35–38]. Particularly in kidney aging, mitochondrial dysfunction, uremia-induced epigenetic changes and the production of reactive oxygen species in the glomeruli that lead to barrier function impairment and albuminuria are all mechanisms promoting oxidative stress [8, 39–41]. These factors may thus explain the associations between kidney traits and MRS here identified. The grounding of the MRS in inflammation and oxidative stress mechanisms may also explain the predictive power of this predictor in regard to outcomes from cancer to cardiovascular disease mortality [20, 21].
EEAA and PhenoAA, also associated with all kidney traits, are both extrinsic aging measures (i.e., tracking changes in blood cell composition) that also capture (intrinsic) aging-related physiological dysregulation [12, 18]. EEAA, identified as a measure of immune system aging, is considered better at predicting age-related decline of tissue performance [12, 14, 18]. Increased allostatic load, activation of stress and pro-aging pathways, impairment of protective pathways as well as exogenous lifestyle and environmental factors are all factors driving premature aging [17, 39]. Immunosenescence [42, 43], systemic low-grade inflammation ('inflammaging') [44] and oxidative stress [37, 39] contribute to an increased allostatic load and are mechanisms present in kidney aging [17, 45]. In particular, the immune system plays an important role in the incidence, development and resolution of renal disease [43]. A signature of 447 genes involved in renal aging further confirmed the relevance of cellular pathways common to immune function and renal physiological decline [46]. Likewise, CKD patients show signs of premature immunological aging (such as poor naïve T-cell frequency and reduced thymic output), a status induced by ureamia (high concentrations of serum urate in blood) that is associated with poor clinical outcomes [47]. At the cellular level, telomere attrition [39, 48] and the cytokine secretory profile of senescent cells promote inflammation and lead to fibrotic damage [37, 39, 43, 45], further linking inflammaging to renal dysfunction [9]. Our findings, in line with the positive association between PhenoAge and albumin excretion rate identified in 499 subjects with type 1 diabetes [49], may be thus explained by the relationship between these DNAm-based predictors and the aging-related changes to the immune system, low-grade chronic inflammation and oxidative stress that impact renal disease [37, 47]. Moreover, EEAA, PhenoAge and the MRS have shown a stronger predictive association with time to death than HannumAA, HorvathAA and IEAA, suggesting they better reflect mortality risks associated with biological aging [16, 20]—and based on our findings, also better reflect immune system changes associated with kidney aging. The correlation between these DNAm-based predictors and poor kidney health may also explain their strong associations with mortality, as the ensuing contribution of renal disease to physiological dysregulation may increase mortality risk [6, 7, 39, 50].
Distinct patterns of association were identified with two of the “first-generation” clocks, HorvathAA and HannumAA. HorvathAA, thought to track cell-intrinsic aging (e.g., epigenetic stability mechanisms, cell growth and survival as well as organismal development) [11, 12, 16, 18], was exclusively associated with eGFR in our study. The statistical power derived from the larger sample size in our study likely explains this positive association, unlike prior studies reporting null findings with eGFR [49, 51]. Considering tissue from individuals with renal disease was included in the development of this pan-tissue marker [11], this epigenetic marker reflects alterations in ubiquitous cell-intrinsic pathways that our findings suggest may be relevant for renal (glomerular) function. Genome-wide association studies of the Hannum and Horvath DNAm-based predictors have shown that, even though they capture different aspects of aging, both markers are influenced by genes associated with metabolic and immune system pathways [52, 53]. Consequently, these DNAm-based predictors may also somewhat be reflective of the immune molecular mechanisms previously described. An additional factor of potential relevance for the eGFR-HorvathAA association may be aberrant glucocorticoid signaling, given its potentially pathogenic role in renal function [54] and the enrichment of glucocorticoid response elements in this DNAm marker [29].
HannumAA, associated with lower renal function and markers of early renal damage (uACR and microalbuminuria) in our study, is also strongly correlated with blood cell counts [16, 23] and is sensitive to environmental influences [15, 53]. Moreover, it has also been associated with higher levels of inflammatory biomarkers, creatinine and certain lipid classes in individuals of European ancestry [14, 15, 55]. Like other extrinsic measures, Hannum seems to be a better marker for later-life diseases and mortality than Horvath [22] and has even been proposed as a prognostic marker of pathological metabolic processes [56]. HannumAA in cancerous kidney tissue vs normal samples has also been reported [10], thus offering further evidence on the relevance of our findings in blood to renal disease.
The association between one-year difference in the GrimAge predictor and higher uACR, serum urate and microalbuminuria—but not eGFR or prevalent CKD—suggests this DNAm-based predictor might be more sensitive to systemic inflammation and early renal damage. Albuminuria changes can occur before eGFR decline in early kidney disease [6]: early structural glomerular lesions in patients with normal eGFR are better correlated with changes in uACR than with GFR decline, where the latter might not be present yet [57]. Moreover, albuminuria is a predictor of CKD progression and mortality independently from eGFR changes, which suggests they represent two independent mechanisms underlying renal disease progression [6, 7]. Our findings are in line with prior reports of GrimAge association with albumin excretion in T1D patients and non-diabetic subjects [19, 49], where lower power may explain the null findings reported in [51]. Asymptomatic hyperuricaemia, or high serum urate levels, is involved in pro-inflammatory mechanisms and is associated with a high risk of cardiovascular and renal disease [8]. Serum urate, both in its crystal and soluble forms, activates innate immunity and triggers DNAm epigenetic mechanisms (e.g., promoting cytokine secretion, pro-inflammatory pathways including oxidative stress) leading to persisting inflammation and an increased allostatic load [8, 39]. These effects may, in turn, explain our findings in relation to GrimAge. A high degree of heterogeneity in the eGFR- and CKD-GrimAA analyses was observed, similarly to reports in prior studies [58, 59].
Overall, the observed associations did not show a clear pattern across kidney traits, thus supporting the proposed notion that the existing DNAm-based predictors might reflect different aspects of biological aging. This is in line with their differential association with risk factors, intermediate phenotypes and diseases [12, 58, 60, 61], and their inclusion of non-overlapping CpGs sets [19, 20, 60, 62]. The CpG overlap between DNAm-based predictors was assessed in detail by Liu et.al, where the lack of CpG overlap may be explained by the redundancy of the methylome: CpGs selected in the construction of different DNAm predictors may represent different aging hallmarks or pathways, despite the potential biological similarities of their genomic regions [62]. Our findings are also consistent with the many associations observed with EEAA and other blood immune system correlated DNAm-based predictors [12], and with the lack of associations with IEAA observed in prior studies [52, 61].
DNAm-estimated adrenomedullin (DNAmADM), plasminogen activator inhibitor-1 (DNAmPA1) and smoking pack years (DNAmPackYears) were positively associated with decreased renal function. Consistent with our findings, patients with chronic cardiorenal diseases have higher blood levels of ADM [63, 64] and PAI-1 [65–67], where the first is a potential biomarker of CKD progression [64, 68] and the latter a risk factor for cardiorenal disease [66, 69]. Several factors involved in kidney disease pathogenesis (e.g., oxidative stress, inflammation) induce PAI-1 expression (66, 70, 71), which in turn has been linked to fibrosis, glomeruli damage and other pathogenic mechanisms in renal disease [65, 67], as well as to thrombosis and an increased hypercoagulable state—a shared phenotype of inflammaging [37] and renal disease [72]. Moreover, the effects of smoking in DNAm [31, 32] and their association to DNAmAge acceleration in blood are well known [33, 34]. PhenoAge and MRS capture effects of cigarette smoking [18, 20, 33, 58, 60], whereas GrimAge specifically incorporates cigarette smoking into its formulation [19]. Cigarette smoking has been associated with renal function decline and increased inflammation [73], where several mechanisms explaining the negative effects of smoking on renal function (e.g., oxidative stress, endothelial dysfunction, immune function modulation) contribute to renal disease progression [74]. Overall, our findings are in line with the roles described in the literature for the studied DNAm-estimated markers, and further support the notion that associations between epigenetic aging and health outcomes may be mediated by age-related pro-inflammatory mechanisms [75]. Moreover, they suggest DNAm-based estimates might prove to be valuable proxies in settings where such variables are not available (e.g., limitations in the clinical use of ADM [68] and self-reported smoking [34]).
Strengths of this work are the large sample size and inclusion of multiple independent studies involving multi-ethnic populations. We comprehensively addressed biological aging and lifespan as predicted by DNAm and assessed multiple kidney traits reflecting different aspects of renal health. Our results in regard to PhenoAge and GrimAge represent confirmatory findings to some extent, given that renal function variables were included in the derivation of these algorithms [18, 19]. Of note, although the MRS was derived using data of two of the cohorts included in this study, it has been independently validated [20, 21] and the replication of its associations across multiple cohorts suggest our findings are not a product of data overfitting. All in all, our study meets the considerations proposed by a recent literature review and meta-analysis on the topic [22].
Limitations of this study include the estimation of DNAm markers in blood samples rather than renal tissue, although there is currently no epigenetic age predictor derived in kidney tissue. Age-related methylation changes can be tissue-specific [76] and show inter-individual variation [46], yet associations between eGFR and DNAm in blood have been demonstrated to be relevant to kidney traits [46, 77]. Moreover, this remains the only viable approach for research conducted in population-based studies, where taking renal biopsies from participants is not done due to practical and ethical considerations. Despite the bias inherent to the calculation of eGFR using equations that systematically produce higher values for individuals identified as black [2, 78, 79], associations with trans-ethnic replication in our study featured lower eGFR (CKD). Nevertheless, future kidney research would benefit from the development and use of methods relying on filtration markers independent from muscle mass and/or moving beyond race as a variable [78]. The lack of trans-ethnic replication of all associations may be explained by multiple factors, most notably the smaller sample sizes from non-European studies, or ethnic biases in DNAm-based predictors (as those reported for PhenoAge in [80]). Future studies using larger, homogeneous sample sizes from diverse ethnic groups are needed to address the generalizability of our findings, and to interrogate the contributions of environmental and social determinants of health disparities in epigenetic aging. Our models assumed a linear relationship in the age range studied here, and residual confounding after adjustment for the covariates included in our regression analyses is a possibility. Another potential limitation is that the cross-sectional nature of the study does not allow to draw conclusions on temporal relationships between DNAm and renal phenotypes, although DNAm patterns reflect lifetime environmental exposures and genetic factors. Our findings do not provide a mechanistic or causal explanation for renal aging and blood epigenetic aging markers, but should be considered hypothesis-generating research. Preliminary results from the largest genome-wide association (GWAS) study of DNAm-based aging and lifespan predictors found no evidence of causal effects on renal outcomes (uACR, eGFR, albuminuria and serum urate, among 150 studied traits) [81]. Nevertheless, DNAm-based aging/lifespan signatures could still be a valuable biomarker of kidney disease prognosis, risk stratification or kidney-related outcomes. Future research should aim to expand our understanding of epigenetic aging in chronic diseases and on the clinical utility of the DNAm-based predictors.
Conclusion
In this study of multi-ethnic population-based cohorts, kidney traits were robustly associated with DNAm-based aging and lifespan predictors measured in whole blood, as well as with some secondary DNAm-estimated markers. Our findings are consistent with a body of literature on the role immunosenescence, inflammaging and oxidative stress play in renal function and damage, as well as offer evidence on the relevance of cell-intrinsic aging mechanisms. DNAm age and lifespan predictors seem to capture the contribution of multiple CpGs to pathological changes common to systemic inflammation and renal disease, highlighting the systemic nature of age-related physiological functional decline. Future research in longitudinal studies is required to evaluate the translational value of our findings as either prognostic biomarkers for disease progression and mortality, or as means to enhance risk stratification; functional studies to explore the complex physiological interplay between epigenetic mechanisms and biological aging are also warranted.
Methods
Study design
The association between kidney traits as dependent variables and DNAm aging/lifespan predictors as independent variables was modeled using linear regression following a meta-analytic approach (Fig. 1). Study-level results from up to seven studies were included in the meta-analyses: four studies with participants of European ancestry, one study of African American participants and three substudies from the WHI with European American, African American and Hispanic/Latino participants. The studies were KORA (Kooperative Gesundheitsforschung in der Region Augsburg), NAS (Normative Aging Study), ESTHER (Epidemiologische Studie zu Chancen der Verhütung, Früherkennung und optimierten THerapie chronischer ERkrankungen in der älteren Bevölkerung), WHI (Women's Health Initiative) and the Jackson Heart Study (JSH). The ESTHER, NAS and WHI studies contributed multiple sets of data that were analyzed separately: data sets from ESTHER corresponded to two surveys with non-overlapping sets of participants, whereas data from NAS were longitudinal and collected over consecutive examinations and analyzed taking into account these repeated measures. Further details on the data collection and methods used in each study are available in Additional file 3.
Outcome definition
Serum creatinine values obtained with a Jaffé assay before 2009 were calibrated by multiplying by 0.95 [82] and used to calculate estimated glomerular filtration rate (eGFR) as per the CKD-EPI equation [2] in its implementation in the R package nephro [83]. Prevalent chronic kidney disease (CKD) was defined as eGFR < 60 ml/min/1.73 m2 [84]. eGFR and urinary albumin-creatinine ratio (UACR) were log transformed prior to statistical analysis. Microalbuminuria was defined as uACR ≥ 30 mg/g. Serum urate was also studied.
DNAmAge assessment
Methylation was measured using the Illumina Infinium HumanMethylation450K or EPIC array in whole blood and used to estimate measures of DNAmAge and mortality (additional details on each predictor are described in Additional file 3: Note S2). Five DNAmAge and lifespan predictors were calculated using the online DNAm Age calculator (https://dnamage.genetics.ucla.edu/) [11]: Hannum’s estimate (HannumAge), ExtrinsicAge (EEAA) [10], Horvath’s estimate (HorvathAge) [11], PhenoAge [18] and GrimAge [19]. IEAA, a marker capturing cell-intrinsic aging properties that are independent of blood cell types, was derived by regressing HorvathAge on cell counts [10]. Quality control was conducted as in previous meta-analyses of epigenetic measures [16], with exclusion of individuals with mismatching predicted and reported sex data. Age acceleration (AA) measures were calculated in each study as the difference between the predicted DNAmAge and chronological age, with chronological age included in all models as an adjustment for known chronological age effects across the lifespan. Defining age acceleration as the difference, rather than the residual of chronological age regressed on epigenetic age, has advantages as it is an individual measure as opposed to a population measure, and is not defined to have mean 0 in each population as is the case for the residual measure. A sixth measure, the 10-CpG-based epigenetic mortality risk score (MRS) in its continuous form was calculated as the sum of the methylation β values multiplied by the regression coefficients of each of the ten CpGs for all-cause mortality, as described in [20].
Further measures of epigenetic aging were included in the secondary analysis: the risk-level MRS variable was built based on the cumulative number of “aberrantly” methylated CpG sites, defined by the cut-offs derived from the 4th quartile of the CpG positively correlated with mortality (cg08362785) and the 1st quartile of the other nine loci defined in [20]. Participants were then assigned to one of three risk levels based on the total number of “aberrantly” methylated CpGs: low risk, MRS = 0–1; moderate risk, MRS = 2–5; and high risk, MRS > 5. Finally, to better understand GrimAge, we also included in the analysis its eight underlying traits (smoking pack-years, adrenomedullin, beta-2 microglobulin, cystatin C, growth differentiation factor 15, leptin, plasminogen activation inhibitor 1, tissue inhibitor metalloproteinase 1) [19] if a renal phenotype was associated with GrimAge.
Statistical analysis
Linear and logistic regression models were run with kidney traits as outcomes and measures of DNAm-based age acceleration and lifespan as predictors, including covariates to adjust for potential confounding by biological and technical factors. Chronological age and sex were included in a basic model, whereas additional adjustment for BMI (kg/m2), log transformed triglycerides, HDL, hypertension, smoking status (current/ever, never) and diabetes was done in the fully adjusted model. Linear regression models for serum urate additionally adjusted for baseline eGFR. Details on the study-specific definition or exclusion of variables, as well as additional information on all of the cohorts, are given in Additional file 3: Note S3. Leukocyte count (either measured or estimated by the Houseman approach [85]) was additionally included in the regression models for Horvath’s estimate as to obtain the intrinsic age acceleration measure (IEAA). In the secondary analyses, the aforementioned covariates from the basic and the fully adjusted models were used, with the exception of no smoking adjustment for DNAm-predicted pack years. All measures of association between epigenetic markers and continuous renal traits (eGFR, uACR, urate) were standardized to the standard deviation of the given renal trait as to obtain estimates comparable across renal traits.
All outcomes were available in at least one cohort of European ancestry and African American studies (Additional file 1: Table S1), although eGFR and CKD were the only outcomes reported by all participating studies. Each cohort provided regression estimates and standard errors, which were pooled using inverse-variance fixed-effects and random-effects models. Between-study heterogeneity was assessed using Cochran’s Q and I2 statistics. High heterogeneity was defined as I2 > 0.50 and (Q) phet < 0.05. If high heterogeneity was detected in the fixed-effects model, the random-effects model was interpreted. All statistical analyses were conducted using R version 3.5.3 [86], where meta-analyses were conducted using the metafor package v2.0 [87]. Multiple testing was addressed by correcting the significance level for the total number of statistical tests (i.e., Bonferroni correction, 0.05/7 epigenetic markers * 5 renal traits). Associations were considered significant if p < 1.43 E−03 in the trans-ethnic meta-analysis and if they had consistent direction of effect across studies. Ethnic-specific replication was defined as associations with consistent direction of effect reaching nominal statistical significance (p < 0.05) in two or more studies.
Supplementary Information
Additional file 1: Tables S1–S5 showing the results from all conducted analyses across renal parameters and DNAm-based predictors, providing both estimates from individual studies and meta-analyses.
Additional file 2: Figures S1–S7 showing forest plots for all significant associations not presented in the main figures, as well as secondary traits (categorical MRS), comparison of MRS and DNAmAA effect sizes across renal traits.
Additional file 3: Notes including additional information on sensitivity analyses, the derivation of the DNAm-based predictors, cohort-specific details (background and study design, as well as data collection and pre-processing), and all legends to supplemental tables and figures.
Acknowledgements
We are grateful to all study participants of ESTHER, JHS, KORA, NAS and WHI for their invaluable contributions to these studies, as well as all members of field staff conducting the studies.
Abbreviations
- AA
Age acceleration defined as the difference between chronological and DNAmAge
- HorvathAA
Age acceleration calculated with Horvath’s DNAmAge predictor
- HannumAA
Age acceleration calculated with Hannum’s DNAmAge predictor
- CKD
Chronic kidney disease
- CpG
Cytosine-phosphate-guanine dinucleotide
- DNAm
DNA methylation
- DNAmAge
Estimated “biological” age using DNA methylation information, also known in the literature as epigenetic age
- EEAA
“Extrinsic epigenetic age acceleration,” calculated with Hannum’s DNAmAge predictor
- eGFR
Estimated glomerular filtration rate
- ESTHER
Epidemiologische Studie zu Chancen der Verhütung, Früherkennung und optimierten THerapie chronischer ERkrankungen in der älteren Bevölkerung
- GrimAA
Measure analogous to other age acceleration markers calculated with DNAm GrimAge predictor
- JHS
Jackson Heart Study
- KORA
Kooperative Gesundheitsforschung in der Region Augsburg
- IEAA
Intrinsic epigenetic age acceleration
- MRS
Epigenetic mortality risk score
- NAS
US Department of Veterans Affairs’ Normative Ageing Study
- PhenoAA
Age acceleration calculated with DNAm PhenoAge predictor
- uACR
Urinary albumin-to-creatinine ratio
- WHI
Women’s Health Initiative
Authors' contributions
PRMG, CKWC, LMR, RW, XuG, JA, JW, AP, CG, NF and MW contributed to the design of the study. PRMG, LMR, XuG, JS and YaZ conducted the study-level data processing and/or analyses. PRMG carried out meta-analyses and was the major contributor in writing the manuscript. CKWC, LMR, XuG, YaZ, RW, XiG, JN, EC, BC, LH, ACJ, SK, XL, JCNE, JiS, BS, PV, YiZ, JoS, EAW, WK, JA, JW, HB, AAB, CG, NF, MW oversaw additional input and revisions to manuscript drafts. CKWC, LMR, XuG, YaZ, RW, XiG, JA, JW, CG, NF and MW provided statistical guidance throughout the study, as well as actively took part in the interpretation of data. AB, EC, AC, BC, CE, LH, ACJ, SK, LL, EL, SL, AR, JiS, BY, JoS, SH, AL, EAW, SH made contributions in the acquisition of data in their respective studies. All authors read and approved the final manuscript.
Funding
The KORA study was initiated and financed by the Helmholtz Zentrum München–German Research Center for Environmental Health, which is funded by the German Federal Ministry of Education and Research (BMBF) and by the State of Bavaria. Furthermore, KORA research has been supported within the Munich Center of Health Sciences (MC-Health), Ludwig-Maximilians-Universität, as part of LMUinnovativ. The ESTHER study was supported by the Baden-Württemberg State Ministry of Science, Research and Arts (Stuttgart, Germany), the Federal Ministry of Education and Research (Berlin, Germany), and the Federal Ministry of Family Affairs, Senior Citizens, Women and Youth (Berlin, Germany). The sponsors had no role in the study design, in the collection, analysis, and interpretation of data and preparation, review, or approval of the manuscript. The Normative Aging Study is supported by the National Institute of Environmental Health Sciences (grants P30ES009089, R01ES021733, R01ES025225, and R01ES027747). The VA Normative Aging Study is supported by the Cooperative Studies Program/Epidemiology Research and Information Center of the U.S. Department of Veterans Affairs and is a component of the Massachusetts Veterans Epidemiology Research and Information Center, Boston, Massachusetts. The WHI program is funded by the National Heart, Lung and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C and HHSN268201600004C. The authors thank the WHI investigators and staff for their dedication, and the study participants for making the program possible. A full listing of WHI investigators can be found at: http://www.whi.org/researchers/Documents%20%20Write%20a%20Paper/WHI%20Investigator%20Long%20List.pdf. This study was supported by the National Institutes of Health R01- MD012765, R01-DK117445 and R21-HL140385 to NF; by NIH/NIEHS R01-ES020836 to LH, AB and EAW; and by NIH/NHLBI contract 60442456 BAA23 to SH. The Jackson Heart Study (JHS) is supported and conducted in collaboration with Jackson State University (HHSN268201800013I), Tougaloo College (HHSN268201800014I), the Mississippi State Department of Health (HHSN268201800015I) and the University of Mississippi Medical Center (HHSN268201800010I, HHSN268201800011I and HHSN268201800012I) contracts from the National Heart, Lung and Blood Institute (NHLBI) and the National Institute on Minority Health and Health Disparities (NIMHD). The authors also wish to thank the staffs and participants of the JHS. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung and Blood Institute; the National Institutes of Health; or the US Department of Health and Human Services. The funders had no role in the design and conduct of the study, in the collection, analysis and interpretation of the data, and in the preparation, review or approval of the manuscript. The project described was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant KL2TR002490 (LMR). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. LMR was also funded by T32 HL129982 and R21-HL140385.
Availability of data and materials
The dataset(s) supporting the conclusions of this article is(are) included within the article (and its additional file(s)). The informed consent given by the study participants does not cover posting of participant level phenotype data in public databases. Pre-existing data access policies for each of the five studies state research data requests can be submitted to each steering committee. Study-specific details regarding such requests are described in the next points: KORA data are available upon request from KORA Project Application Self-Service Tool (https://epi.helmholtz-muenchen.de/); data requests can be submitted online and are subject to approval by the KORA Board. ESTHER data are not allowed to be publicly available due to restrictions of informed consent. However, the use of the data for collaboration projects has been and will remain the approach for data sharing. Data used in this analysis were produced and used in accordance with the policies of the Jackson Heart Study under contracts from the National, Heart, Lung and Blood Institute and are not the domain of the authors but that of the Jackson Heart Study. These data are available to other researchers for purposes of reproducing the results or replicating the procedures by submitting a manuscript proposal to the Jackson Heart Study at jhspub@umc.edu, as described at https://www.jacksonheartstudy.org/Research/Publications#submitmanuscript. Data updates for the Jackson Heart Study are also deposited regularly in the National Institutes of Health data repositories, dbGaP (https://www.ncbi.nlm.nih.gov/gap/) and BioLincc (https://biolincc.nhlbi.nih.gov/home/). NAS data are available on request due to privacy/ethical restrictions. Data that support the findings of this study are available from AAB upon reasonable request. The DNA methylation datasets for WHI are publicly available under dbGAP access number phs000200.v10.p3 or upon request to www.whi.org. WHI datasets are also available through BioLINCC https://biolincc.nhlbi.nih.gov/studies/whi_ctos/. Scripts used in data processing and statistical analyses have been made publicly accessible at https://ascgitlab.helmholtz-muenchen.de/pamela.matias/dnam-aging-in-kora-f4 and https://ascgitlab.helmholtz-muenchen.de/pamela.matias/kidney-dnamage.
Declarations
Ethics approval and consent to participate
All participants provided written consent, and study-specific details are described next: ESTHER: The study was approved by the ethics committees of the University of Heidelberg and of the Medical Association of Saarland. All participants provided written informed consent. JHS: All participants included in this analysis provided written, informed consent for use of genetic data, and all study protocols conform to the 1975 Declaration of Helsinki guidelines. The study was approved by the Institutional Review Boards of the participating institutions (University of Mississippi Medical Center, Jackson State University and Tougaloo College). KORA: The KORA cohort ethical approval was granted by the ethics committee of the Bavarian Medical Association (REC reference numbers: F4: #06068) and all were carried out in accordance with the principles of the Declaration of Helsinki. All research participants have signed informed consent prior to taking part in any research activities. The KORA data protection procedures were approved by the responsible data protection officer of the Helmholtz Zentrum München. NAS: The NAS was approved by the Department of Veterans Affairs Boston Healthcare System and written informed consent was obtained from each subject before participation. WHI: All study participants have provided written consent to participate in genetic studies.
Consent for publication
Not applicable.
Competing interests
The authors declare the following competing interests: WK reports personal fees from AstraZeneca, Novartis, Pfizer, The Medicines Company, DalCor, Kowa, Amgen, Corvidia, Daiichi-Sankyo, Berlin-Chemie, Sanofi, Genentech, and Bristol-Myers Squibb, grants and non-financial support from Singulex, Abbott, Roche Diagnostics, Beckmann, all outside the submitted work.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Nora Franceschini and Melanie Waldenberger shared senior authorship
Contributor Information
Pamela R. Matías-García, Email: pamela.matias@helmholtz-muenchen.de
Melanie Waldenberger, Email: waldenberger@helmholtz-muenchen.de.
References
- 1.Levey AS, Inker LA, Coresh J. GFR estimation: from physiology to public health. Am J Kidney Dis Off J Natl Kidney Found. 2014;63(5):820–834. doi: 10.1053/j.ajkd.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Levey AS, Stevens LA, Schmid CH, Zhang YL, Castro AF, 3rd, Feldman HI, et al. A new equation to estimate glomerular filtration rate. Ann Internal Med. 2009;150(9):604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckardt KU, Coresh J, Devuyst O, Johnson RJ, Kottgen A, Levey AS, et al. Evolving importance of kidney disease: from subspecialty to global health burden. Lancet (London, England) 2013;382(9887):158–169. doi: 10.1016/S0140-6736(13)60439-0. [DOI] [PubMed] [Google Scholar]
- 4.Hill NR, Fatoba ST, Oke JL, Hirst JA, O'Callaghan CA, Lasserson DS, et al. Global prevalence of chronic kidney disease: a systematic review and meta-analysis. PLoS ONE. 2016;11(7):e0158765. doi: 10.1371/journal.pone.0158765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levey AS, Atkins R, Coresh J, Cohen EP, Collins AJ, Eckardt KU, et al. Chronic kidney disease as a global public health problem: approaches and initiatives: a position statement from Kidney Disease Improving Global Outcomes. Kidney Int. 2007;72(3):247–259. doi: 10.1038/sj.ki.5002343. [DOI] [PubMed] [Google Scholar]
- 6.Matsushita K, van der Velde M, Astor BC, Woodward M, Levey AS, de Jong PE, et al. Association of estimated glomerular filtration rate and albuminuria with all-cause and cardiovascular mortality in general population cohorts: a collaborative meta-analysis. Lancet (London, England) 2010;375(9731):2073–2081. doi: 10.1016/S0140-6736(10)60674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Astor BC, Matsushita K, Gansevoort RT, van der Velde M, Woodward M, Levey AS, et al. Lower estimated glomerular filtration rate and higher albuminuria are associated with mortality and end-stage renal disease. A collaborative meta-analysis of kidney disease population cohorts. Kidney Int. 2011;79(12):1331–1340. doi: 10.1038/ki.2010.550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joosten LAB, Crişan TO, Bjornstad P, Johnson RJ. Asymptomatic hyperuricaemia: a silent activator of the innate immune system. Nat Rev Rheumatol. 2020;16(2):75–86. doi: 10.1038/s41584-019-0334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salameh Y, Bejaoui Y, El Hajj N. DNA methylation biomarkers in aging and age-related diseases. Front Genetics. 2020;11:171. doi: 10.3389/fgene.2020.00171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49(2):359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14(10):R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horvath S, Raj K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genetics. 2018;19(6):371–384. doi: 10.1038/s41576-018-0004-3. [DOI] [PubMed] [Google Scholar]
- 13.Dhingra R, Nwanaji-Enwerem JC, Samet M, Ward-Caviness CK. DNA methylation age—environmental influences, health impacts, and its role in environmental epidemiology. Curr Environ Health Rep. 2018;5(3):317–327. doi: 10.1007/s40572-018-0203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, et al. An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 2016;17(1):171. doi: 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quach A, Levine ME, Tanaka T, Lu AT, Chen BH, Ferrucci L, et al. Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging. 2017;9(2):419–446. doi: 10.18632/aging.101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging. 2016;8(9):1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 2018;10(4):573–591. doi: 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging. 2019;11(2):303–327. doi: 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Wilson R, Heiss J, Breitling LP, Saum K-U, Schöttker B, et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat Commun. 2017;8:14617. doi: 10.1038/ncomms14617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao X, Colicino E, Shen J, Just AC, Nwanaji-Enwerem JC, Wang C, et al. Comparative validation of an epigenetic mortality risk score with three aging biomarkers for predicting mortality risks among older adult males. Int J Epidemiol. 2019;48(6):1958–1971. doi: 10.1093/ije/dyz082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fransquet PD, Wrigglesworth J, Woods RL, Ernst ME, Ryan J. The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clin Epigenetics. 2019;11(1):62. doi: 10.1186/s13148-019-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H. Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics. 2016;8:64. doi: 10.1186/s13148-016-0228-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Breitling LP, Saum KU, Perna L, Schottker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics. 2016;8:21. doi: 10.1186/s13148-016-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44(4):1388–1396. doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ryan J, Wrigglesworth J, Loong J, Fransquet PD, Woods RL. A systematic review and meta-analysis of environmental, lifestyle, and health factors associated with DNA methylation age. J Gerontol Ser A Biol Sci Med Sci. 2020;75(3):481–494. doi: 10.1093/gerona/glz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schönfels W, Ahrens M, et al. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci USA. 2014;111(43):15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Röh S, Ressler KJ, et al. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 2015;16:266. doi: 10.1186/s13059-015-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiels PG, McGuinness D, Eriksson M, Kooman JP, Stenvinkel P. The role of epigenetics in renal ageing. Nat Rev Nephrol. 2017;13(8):471–482. doi: 10.1038/nrneph.2017.78. [DOI] [PubMed] [Google Scholar]
- 30.Jung M, Pfeifer GP. Aging and DNA methylation. BMC Biol. 2015;13:7. doi: 10.1186/s12915-015-0118-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joehanes R, Just AC, Marioni RE, Pilling LC, Reynolds LM, Mandaviya PR, et al. Epigenetic signatures of cigarette smoking. Circ Cardiovas Genetics. 2016;9(5):436–447. doi: 10.1161/CIRCGENETICS.116.001506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson R, Wahl S, Pfeiffer L, Ward-Caviness CK, Kunze S, Kretschmer A, et al. The dynamics of smoking-related disturbed methylation: a two time-point study of methylation change in smokers, non-smokers and former smokers. BMC Genomics. 2017;18(1):805. doi: 10.1186/s12864-017-4198-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang Y, Gao X, Just AC, Colicino E, Wang C, Coull BA, et al. Smoking-related DNA methylation is associated with DNA methylation phenotypic age acceleration: the veterans affairs normative aging study. Int J Environ Res Public Health. 2019;16(13):2356. doi: 10.3390/ijerph16132356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao X, Zhang Y, Breitling LP, Brenner H. Relationship of tobacco smoking and smoking-related DNA methylation with epigenetic age acceleration. Oncotarget. 2016;7(30):46878–46889. doi: 10.18632/oncotarget.9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao X, Gào X, Zhang Y, Holleczek B, Schöttker B, Brenner H. Oxidative stress and epigenetic mortality risk score: associations with all-cause mortality among elderly people. Eur J Epidemiol. 2019;34(5):451–462. doi: 10.1007/s10654-019-00493-7. [DOI] [PubMed] [Google Scholar]
- 36.Ward-Caviness CK, Pu S, Martin CL, Galea S, Uddin M, Wildman DE, et al. Epigenetic predictors of all-cause mortality are associated with objective measures of neighborhood disadvantage in an urban population. Clin Epigenetics. 2020;12(1):44. doi: 10.1186/s13148-020-00830-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Franceschi C, Campisi J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J Gerontol Ser A Biol Sci Med Sci. 2014;69(Suppl 1):S4–9. doi: 10.1093/gerona/glu057. [DOI] [PubMed] [Google Scholar]
- 38.Wang C, Just A, Heiss J, Coull BA, Hou L, Zheng Y, et al. Biomarkers of aging and lung function in the normative aging study. Aging. 2020;12(12):11942–11966. doi: 10.18632/aging.103363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kooman JP, Kotanko P, Schols AM, Shiels PG, Stenvinkel P. Chronic kidney disease and premature ageing. Nat Rev Nephrol. 2014;10(12):732–742. doi: 10.1038/nrneph.2014.185. [DOI] [PubMed] [Google Scholar]
- 40.Young GH, Wu VC. KLOTHO methylation is linked to uremic toxins and chronic kidney disease. Kidney Int. 2012;81(7):611–612. doi: 10.1038/ki.2011.461. [DOI] [PubMed] [Google Scholar]
- 41.McCarthy ET, Sharma R, Sharma M, Li JZ, Ge XL, Dileepan KN, et al. TNF-alpha increases albumin permeability of isolated rat glomeruli through the generation of superoxide. J Am Soc Nephrol JASN. 1998;9(3):433–438. doi: 10.1681/ASN.V93433. [DOI] [PubMed] [Google Scholar]
- 42.Mueller AL, McNamara MS, Sinclair DA. Why does COVID-19 disproportionately affect older people? Aging. 2020;12(10):9959–9981. doi: 10.18632/aging.103344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tecklenborg J, Clayton D, Siebert S, Coley SM. The role of the immune system in kidney disease. Clin Exp Immunol. 2018;192(2):142–150. doi: 10.1111/cei.13119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Franceschi C, Bonafè M, Valensin S, Olivieri F, De Luca M, Ottaviani E, et al. Inflamm-aging: an evolutionary perspective on immunosenescence. Ann N Y Acad Sci. 2000;908:244–254. doi: 10.1111/j.1749-6632.2000.tb06651.x. [DOI] [PubMed] [Google Scholar]
- 45.Rowland J, Akbarov A, Maan A, Eales J, Dormer J, Tomaszewski M. Tick-tock chimes the kidney clock—from biology of renal ageing to clinical applications. Kidney Blood Press Res. 2018;43(1):55–67. doi: 10.1159/000486907. [DOI] [PubMed] [Google Scholar]
- 46.Rodwell GE, Sonu R, Zahn JM, Lund J, Wilhelmy J, Wang L, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2(12):e427. doi: 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Crépin T, Legendre M, Carron C, Vachey C, Courivaud C, Rebibou JM, et al. Uraemia-induced immune senescence and clinical outcomes in chronic kidney disease patients. Nephrol Dial Transplant Off Publ Eur Dial Transplant Assoc Eur Renal Assoc. 2020;35(4):624–632. doi: 10.1093/ndt/gfy276. [DOI] [PubMed] [Google Scholar]
- 48.Carrero JJ, Stenvinkel P, Fellström B, Qureshi AR, Lamb K, Heimbürger O, et al. Telomere attrition is associated with inflammation, low fetuin-A levels and high mortality in prevalent haemodialysis patients. J Internal Med. 2008;263(3):302–312. doi: 10.1111/j.1365-2796.2007.01890.x. [DOI] [PubMed] [Google Scholar]
- 49.Roshandel D, Chen Z, Canty AJ, Bull SB, Natarajan R, Paterson AD, et al. DNA methylation age calculators reveal association with diabetic neuropathy in type 1 diabetes. Clin Epigenetics. 2020;12(1):52. doi: 10.1186/s13148-020-00840-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Hare AM, Choi AI, Bertenthal D, Bacchetti P, Garg AX, Kaufman JS, et al. Age affects outcomes in chronic kidney disease. J Am Soc Nephrol JASN. 2007;18(10):2758–2765. doi: 10.1681/ASN.2007040422. [DOI] [PubMed] [Google Scholar]
- 51.Smith JA, Raisky J, Ratliff SM, Liu J, Kardia SLR, Turner ST, et al. Intrinsic and extrinsic epigenetic age acceleration are associated with hypertensive target organ damage in older African Americans. BMC Med Genomics. 2019;12(1):141. doi: 10.1186/s12920-019-0585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu AT, Xue L, Salfati EL, Chen BH, Ferrucci L, Levy D, et al. GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat Commun. 2018;9(1):387. doi: 10.1038/s41467-017-02697-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibson J, Russ TC, Clarke TK, Howard DM, Hillary RF, Evans KL, et al. A meta-analysis of genome-wide association studies of epigenetic age acceleration. PLoS Genet. 2019;15(11):e1008104. doi: 10.1371/journal.pgen.1008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hunter RW, Ivy JR, Bailey MA. Glucocorticoids and renal Na(+) transport: implications for hypertension and salt sensitivity. J Physiol. 2014;592(8):1731–1744. doi: 10.1113/jphysiol.2013.267609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irvin MR, Aslibekyan S, Do A, Zhi D, Hidalgo B, Claas SA, et al. Metabolic and inflammatory biomarkers are associated with epigenetic aging acceleration estimates in the GOLDN study. Clin Epigenetics. 2018;10:56. doi: 10.1186/s13148-018-0481-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morrison FG, Logue MW, Guetta R, Maniates H, Stone A, Schichman SA, et al. Investigation of bidirectional longitudinal associations between advanced epigenetic age and peripheral biomarkers of inflammation and metabolic syndrome. Aging. 2019;11(11):3487–3504. doi: 10.18632/aging.101992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Looker HC, Mauer M, Saulnier PJ, Harder JL, Nair V, Boustany-Kari CM, et al. Changes in albuminuria but not GFR are associated with early changes in kidney structure in type 2 diabetes. J Am Soc Nephrol JASN. 2019;30(6):1049–1059. doi: 10.1681/ASN.2018111166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fiorito G, McCrory C, Robinson O, Carmeli C, Rosales CO, Zhang Y, et al. Socioeconomic position, lifestyle habits and biomarkers of epigenetic aging: a multi-cohort analysis. Aging. 2019;11(7):2045–2070. doi: 10.18632/aging.101900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rezwan FI, Imboden M, Amaral AFS, Wielscher M, Jeong A, Triebner K, et al. Association of adult lung function with accelerated biological aging. Aging. 2020;12(1):518–542. doi: 10.18632/aging.102639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao W, Ammous F, Ratliff S, Liu J, Yu M, Mosley TH, et al. Education and lifestyle factors are associated with dna methylation clocks in older African Americans. Int J Environ Res Public Health. 2019;16(17):3141. doi: 10.3390/ijerph16173141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Z, Chen BH, Assimes TL, Ferrucci L, Horvath S, Levine ME. The role of epigenetic aging in education and racial/ethnic mortality disparities among older U.S. Women. Psychoneuroendocrinology. 2019;104:18–24. doi: 10.1016/j.psyneuen.2019.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Z, Leung D, Thrush K, Zhao W, Ratliff S, Tanaka T, et al. Underlying features of epigenetic aging clocks in vivo and in vitro. Aging Cell. 2020;19(10):e13229. doi: 10.1111/acel.13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jougasaki M, Burnett JC., Jr Adrenomedullin as a renal regulator peptide. Nephrol Dial Transplant. 2000;15(3):293–295. doi: 10.1093/ndt/15.3.293. [DOI] [PubMed] [Google Scholar]
- 64.Kronenberg F. Emerging risk factors and markers of chronic kidney disease progression. Nat Rev Nephrol. 2009;5(12):677–689. doi: 10.1038/nrneph.2009.173. [DOI] [PubMed] [Google Scholar]
- 65.Ma L-J, Fogo AB. PAI-1 and kidney fibrosis. Front Biosci (Landmark Ed) 2009;14:2028–2041. doi: 10.2741/3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaughan DE. PAI-1 and atherothrombosis. J Thromb Haemost JTH. 2005;3(8):1879–1883. doi: 10.1111/j.1538-7836.2005.01420.x. [DOI] [PubMed] [Google Scholar]
- 67.Eddy AA, Fogo AB. Plasminogen activator inhibitor-1 in chronic kidney disease: evidence and mechanisms of action. J Am Soc Nephrol JASN. 2006;17(11):2999–3012. doi: 10.1681/ASN.2006050503. [DOI] [PubMed] [Google Scholar]
- 68.Dieplinger B, Mueller T, Kollerits B, Struck J, Ritz E, von Eckardstein A, et al. Pro-A-type natriuretic peptide and pro-adrenomedullin predict progression of chronic kidney disease: the MMKD study. Kidney Int. 2009;75(4):408–414. doi: 10.1038/ki.2008.560. [DOI] [PubMed] [Google Scholar]
- 69.Yamamoto K, Takeshita K, Kojima T, Takamatsu J, Saito H. Aging and plasminogen activator inhibitor-1 (PAI-1) regulation: implication in the pathogenesis of thrombotic disorders in the elderly. Cardiovasc Res. 2005;66(2):276–285. doi: 10.1016/j.cardiores.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 70.Eddy AA. Plasminogen activator inhibitor-1 and the kidney. Am J Physiol Renal Physiol. 2002;283(2):F209–F220. doi: 10.1152/ajprenal.00032.2002. [DOI] [PubMed] [Google Scholar]
- 71.D'Elia JA, Bayliss G, Gleason RE, Weinrauch LA. Cardiovascular-renal complications and the possible role of plasminogen activator inhibitor: a review. Clin Kidney J. 2016;9(5):705–712. doi: 10.1093/ckj/sfw080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lutz J, Menke J, Sollinger D, Schinzel H, Thürmel K. Haemostasis in chronic kidney disease. Nephrol Dial Transplant. 2013;29(1):29–40. doi: 10.1093/ndt/gft209. [DOI] [PubMed] [Google Scholar]
- 73.Hall ME, Wang W, Okhomina V, Agarwal M, Hall JE, Dreisbach AW, et al. Cigarette smoking and chronic kidney disease in African Americans in the Jackson heart study. J Am Heart Assoc. 2016;5(6):e003280. doi: 10.1161/JAHA.116.003280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orth SR, Hallan SI. Smoking: a risk factor for progression of chronic kidney disease and for cardiovascular morbidity and mortality in renal patients–absence of evidence or evidence of absence? Clin J Am Soc Nephrol CJASN. 2008;3(1):226–236. doi: 10.2215/CJN.03740907. [DOI] [PubMed] [Google Scholar]
- 75.Ward-Caviness CK, Huffman JE, Evertt K, Germain M, Van Dongen J, Hill WD, et al. DNA methylation age is associated with an altered hemostatic profile in a multi-ethnic meta-analysis. Blood. 2018;132:1842–1850. doi: 10.1182/blood-2018-02-831347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Slieker RC, Relton CL, Gaunt TR, Slagboom PE, Heijmans BT. Age-related DNA methylation changes are tissue-specific with ELOVL2 promoter methylation as exception. Epigenetics Chromatin. 2018;11(1):25. doi: 10.1186/s13072-018-0191-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chu AY, Tin A, Schlosser P, Ko Y-A, Qiu C, Yao C, et al. Epigenome-wide association studies identify DNA methylation associated with kidney function. Nat Commun. 2017;8(1):1286. doi: 10.1038/s41467-017-01297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eneanya ND, Yang W, Reese PP. Reconsidering the consequences of using race to estimate kidney function. JAMA. 2019;322(2):113–114. doi: 10.1001/jama.2019.5774. [DOI] [PubMed] [Google Scholar]
- 79.Vyas DA, Eisenstein LG, Jones DS. Hidden in plain sight—reconsidering the use of race correction in clinical algorithms. N Engl J Med. 2020;383(9):874–882. doi: 10.1056/NEJMms2004740. [DOI] [PubMed] [Google Scholar]
- 80.Philibert R, Beach SRH, Lei MK, Gibbons FX, Gerrard M, Simons RL, et al. Array-based epigenetic aging indices may be racially biased. Genes. 2020;11(6):685. doi: 10.3390/genes11060685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCartney DL, Min JL, Richmond RC, Lu AT, Sobczyk MK, Davies G, et al. Genome-wide association studies identify 137 loci for DNA methylation biomarkers of ageing. bioRxiv. 2020; 2020.06.29.133702. [DOI] [PMC free article] [PubMed]
- 82.Coresh J, Turin TC, Matsushita K, Sang Y, Ballew SH, Appel LJ, et al. Decline in estimated glomerular filtration rate and subsequent risk of end-stage renal disease and mortalityrisk of end-stage renal disease and mortalityrisk of end-stage renal disease and mortality. JAMA. 2014;311(24):2518–2531. doi: 10.1001/jama.2014.6634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pattaro C, Riegler P, Stifter G, Modenese M, Minelli C, Pramstaller PP. Estimating the glomerular filtration rate in the general population using different equations: effects on classification and association. Nephron Clin Pract. 2013;123(1–2):102–111. doi: 10.1159/000351043. [DOI] [PubMed] [Google Scholar]
- 84.Jha V, Garcia-Garcia G, Iseki K, Li Z, Naicker S, Plattner B, et al. Chronic kidney disease: global dimension and perspectives. Lancet (London, England) 2013;382(9888):260–272. doi: 10.1016/S0140-6736(13)60687-X. [DOI] [PubMed] [Google Scholar]
- 85.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinform. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.R Core Team. R: a language and environment for statistical computing. 2015.
- 87.Viechtbauer W. Conducting meta-analyses in R with the metafor package. J Stat Softw. 2010;36(3):48. doi: 10.18637/jss.v036.i03. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Tables S1–S5 showing the results from all conducted analyses across renal parameters and DNAm-based predictors, providing both estimates from individual studies and meta-analyses.
Additional file 2: Figures S1–S7 showing forest plots for all significant associations not presented in the main figures, as well as secondary traits (categorical MRS), comparison of MRS and DNAmAA effect sizes across renal traits.
Additional file 3: Notes including additional information on sensitivity analyses, the derivation of the DNAm-based predictors, cohort-specific details (background and study design, as well as data collection and pre-processing), and all legends to supplemental tables and figures.
Data Availability Statement
The dataset(s) supporting the conclusions of this article is(are) included within the article (and its additional file(s)). The informed consent given by the study participants does not cover posting of participant level phenotype data in public databases. Pre-existing data access policies for each of the five studies state research data requests can be submitted to each steering committee. Study-specific details regarding such requests are described in the next points: KORA data are available upon request from KORA Project Application Self-Service Tool (https://epi.helmholtz-muenchen.de/); data requests can be submitted online and are subject to approval by the KORA Board. ESTHER data are not allowed to be publicly available due to restrictions of informed consent. However, the use of the data for collaboration projects has been and will remain the approach for data sharing. Data used in this analysis were produced and used in accordance with the policies of the Jackson Heart Study under contracts from the National, Heart, Lung and Blood Institute and are not the domain of the authors but that of the Jackson Heart Study. These data are available to other researchers for purposes of reproducing the results or replicating the procedures by submitting a manuscript proposal to the Jackson Heart Study at jhspub@umc.edu, as described at https://www.jacksonheartstudy.org/Research/Publications#submitmanuscript. Data updates for the Jackson Heart Study are also deposited regularly in the National Institutes of Health data repositories, dbGaP (https://www.ncbi.nlm.nih.gov/gap/) and BioLincc (https://biolincc.nhlbi.nih.gov/home/). NAS data are available on request due to privacy/ethical restrictions. Data that support the findings of this study are available from AAB upon reasonable request. The DNA methylation datasets for WHI are publicly available under dbGAP access number phs000200.v10.p3 or upon request to www.whi.org. WHI datasets are also available through BioLINCC https://biolincc.nhlbi.nih.gov/studies/whi_ctos/. Scripts used in data processing and statistical analyses have been made publicly accessible at https://ascgitlab.helmholtz-muenchen.de/pamela.matias/dnam-aging-in-kora-f4 and https://ascgitlab.helmholtz-muenchen.de/pamela.matias/kidney-dnamage.