Abstract
Whether and how an acute immune challenge may affect DNA Damage Response (DDR) is unknown. By studying vaccinations against Influenza and SARS-CoV-2 (mRNA-based) we found acute increases of type-I interferon-inducible gene expression, oxidative stress and DNA damage accumulation in blood mononuclear cells of 9 healthy controls, coupled with effective anti-SARS-CoV-2 neutralizing antibody production in all. Increased DNA damage after SARS-CoV-2 vaccine, partly due to increased oxidative stress, was transient, whereas the inherent DNA repair capacity was found intact. In contrast, in 26 patients with Systemic Lupus Erythematosus, who served as controls in the context of chronic immune activation, we validated increased DNA damage accumulation, increased type-I interferon-inducible gene expression and induction of oxidative stress, however aberrant DDR was associated with deficiencies in nucleotide excision repair pathways. These results indicate that acute immune challenge can indeed activate DDR pathways, whereas, contrary to chronic immune challenge, successful repair of DNA lesions occurs.
Keywords: SARS-CoV-2, Influenza, Vaccination, Acute immune activation, Chronic immune activation, Type I interferon, Oxidative stress, DNA damage response
1. Introduction
Vaccination constitutes an essential way to restrain the spread of severe infectious diseases, which impose a potentially serious threat to public health. Life-threatening diseases like smallpox have been eradicated, while others like polio, tetanus, diphtheria, and measles have been significantly restricted, since vaccination implementation [1]. Especially, in the case of influenza, annual vaccination is the most effective protection. When the circulating strain matches the strains included in the Influenza vaccine, vaccination can reduce serious illness probability by 40–60% [2].
At the end of 2019, a new pathogen imposed a major threat to public health [3]. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) caused an outbreak of viral pneumonia, a disease known as Corona Virus Disease-2019 (COVID-19). COVID-19 was declared a pandemic in March 2020 causing more than 2.5 million deaths worldwide and a major socio-economic impact [4,5]. To confront this critical need, a plethora of vaccination technologies are being tested against SARS-CoV-2. The newer mRNA (Pfizer/BioNTech, Moderna) and adenovirus-based (AstraZeneca/Oxford) vaccine platforms have been licensed for use in humans, while other technologies, i.e. inactivated viruses and recombinant protein-based vaccines (Sanofi, Novavax, Sinovac and GSK), are currently being tested [6,7].
Vaccination effectiveness is significantly influenced by the immunological cellular response to vaccine antigens. After vaccination, innate immune response is temporarily activated. Type I interferons (IFN), which are key mediators of antiviral innate immune response, have been shown to be transiently increased after vaccination against viruses like Influenza [8,9]. This acute innate immune activation is thought to be of great importance, since type I interferon induction can activate the adaptive immune response and influence the neutralizing antibody production [10].
Oxidative stress, an imbalance between the oxidant and antioxidant mechanisms after exposure to deleterious stimuli, plays a pivotal role in the pathogenesis of viral infections. Acute immune activation following a viral infection is associated with increased oxidative stress, as a result of viral replication and the consequent inflammation [11,12]. Under these inflammatory conditions, the immune cellular components generate increased levels of reactive oxygen (ROS) and nitrogen species (RNS), two main oxidative representatives, and catalyze the production of oxidative DNA damage and the activation of DNA Damage Response (DDR), indicating an association among acute immune activation, augmented oxidative stress and DNA damage formation [13]. However, to the best of our knowledge, the direct effect of vaccination upon the formation of intracellular oxidative stress and the activation of DDR network has not been adequately studied.
On the other hand, systemic autoimmune diseases are characterized by chronic immune activation, ultimately leading to tissue injury [14]. The prototypic example of chronic autoimmune disorders is Systemic Lupus Erythematosus (SLE), characterized by aberrant immune system activation, production of numerous pathogenic autoantibodies and immune complex deposition [[15], [16], [17], [18]]. Apart from chronic immune activation, SLE patients display excessive production of pro-oxidant species and defective DDR mechanisms [19,20], resulting in cytosolic DNA fragments accumulation which may act as potent immunostimulators [21,22]. Herein, we sought to investigate how transient immune activation triggers the DDR network and whether this differs in chronic immune activation. For this purpose, we evaluated critical DDR parameters and factors leading to DNA damage formation in peripheral blood mononuclear cells (PBMCs) from healthy controls following vaccination against Influenza and SARS-CoV-2 compared to SLE patients with variable disease activity levels.
2. Materials and methods
2.1. Subjects
Nine apparently healthy members of the personnel of our hospital (aged from 27 to 44 years, 7 women) were recruited during the influenza (October 2019, VaxigripTetra, inactivated Influenza vaccine, Sanofi Pasteur) and SARS-CoV2 vaccinations (January–February 2021, Comirnaty, BNT162b2, mRNA vaccine, Pfizer-BioNTech). Peripheral blood samples were collected immediately before and 24 h after the single dose of influenza vaccination, as well as immediately before and 24 h after the first dose of a mRNA SARS-CoV-2 vaccination, and 14 days after both the first and the second dose. Twenty-six patients who fulfilled the American College of Rheumatology criteria for Systemic Lupus Erythematosus [23] (24 women aged from 18 to 70 years) with variable clinical disease activity [Systemic Lupus Erythematosus Disease Activity Index (SLEDAI): mean ± SD (range): 2.88 ± 3.43 (0−12)] served as disease controls in our protocol. All healthy controls and patients gave their informed consent according to the declaration of Helsinki. The study was approved by Laiko Hospital Ethics Committee (Protocol Nr.1110).
2.2. Cell isolation
PBMCs were isolated and purified using Ficoll gradient centrifugation, as previously described [24]. Cells were resuspended in Freezing Medium [90% Fetal Bovine Serum (FBS) -10% Dimethyl sulfoxide (DMSO)] or lysed in TRITidy G (AppliChem, Germany) and stored at −80 °C until further processing.
2.3. RNA extraction, reverse transcription, and type I IFN score calculation
RNA was extracted with the use of TRITidy G Reagent (AppliChem, Germany) according to the manufacturer's instructions and immediately stored at −80 °C. The quantity and quality of RNA samples were spectrophotometrically tested (Biospec Nano, Japan).
One microgram of total RNA was reverse transcribed into cDNA with Superscript III (Thermo Fisher Scientific, USA). Complementary DNA samples were diluted 1:10 with nuclease-free water (Qiagen, Germany) immediately after synthesis and stored at −20 °C.
Quantitative real-time polymerase chain reaction (qRT-PCR) was used to quantify the expression of selected genes using the Bio-Rad IQ5 thermocycler and the KAPA SYBR FAST Mastermix (KAPA Biosystems, South Africa), as previously described [25]. Briefly, genes preferentially induced by type I IFNs according to recent data [20,21] were selected and included the following: IFN-induced protein with tetratricopeptide repeats 1 (IFIT1) and myxovirus (influenza virus) resistance 1 (MX1). As an internal control and normalization gene (housekeeping gene), we used the glyceraldehyde phosphate dehydrogenase (GAPDH). A reference sample was included in each PCR plate to ensure normalization across experiments. Type I IFN score was calculated as previously described [[25], [26], [27]].
2.4. Oxidative stress measurement and abasic sites detection
Basal oxidative stress was quantified using a luminescence-based system that detects and quantifies total glutathione (GSH), oxidized glutathione (GSSG), and the GSH/GSSG ratio according to manufacturer's experimental protocol (GSH/GSSG-GloTM Assay, Promega). The endogenous levels of abasic sites were evaluated using the OxiSelect Oxidative DNA Damage Quantitation Kit (AP-sites) according to the manufacturer's experimental protocol (Cell Signaling Inc., UK).
2.5. DNA damage quantification
Endogenous DNA damage levels in PBMCs were measured by single-cell gel electrophoresis (comet assay) under alkaline conditions, measuring single-strand breaks (SSBs) and/or double-strand breaks (DSBs) as previously described [28].
2.6. Measurement of nucleotide excision repair (NER)
NER capacity was evaluated at cellular and gene-specific level. At cellular level, freshly isolated PBMCs were directly resuspended in PBS and irradiated with UVC with a total dose of 5 J/m2 as previously described [28]. Briefly, PBMCs were centrifuged after UVC irradiation, passed in RPMI medium containing 10% FBS, 100 units/ ml penicillin and 100 mg/ml streptomycin and incubated in a humidified CO2-incubator (37 °C, 99% dH20, 5% CO2) for 1, 2, and 6 h. At each time point, cells were collected in freezing medium (90% FBS, 10% DMSO) and stored at −80 °C until further processing. To evaluate DNA damage, each sample was analyzed by alkaline comet assay.
At gene level, the NER mechanism capacity was evaluated by the monofunctional binding of mono-hydroxy-melphalan to a single site in the DNA molecule (monoadducts) at specific timepoints as previously described [29]. PBMCs were treated with 100 μg/ml of mono-hydroxy-melphalan for 5 min in complete RPMI-1640 medium supplemented with 10% FBS and 100 units/ml penicillin, 100 μg/ml streptomycin and 2 mM l-glutamine. Cells were subsequently incubated in drug-free complete RPMI medium, harvested at specific time-points (15, 30 and 60 min) and DNA monoadducts were measured along the NRAS gene using Southern Blot analysis [28].
2.7. Measurement of neutralizing antibodies against SARS-CoV-2
Neutralizing antibodies (NAbs) against SARS-CoV-2 were quantified using the cPass ™ SARS-CoV-2 Neutralization Antibody detection kit (GenScript, USA), a surrogate virus neutralization assay, which allows the indirect detection of the circulating NAbs against SARS-CoV-2 independently of class in the peripheral blood, by evaluating the antibody-mediated inhibition of SARS-CoV-2 receptor binding domain (RBD) binding to human host receptor angiotensin converting enzyme 2 (ACE2), according to the manufacturer's experimental protocol [30].
2.8. Statistical analysis
The variable distribution was examined by D'Agostino-Pearson and Shapiro-Wilk tests. Continuous variables are presented as mean ± SD. Paired comparisons were performed with the use of Wilcoxon signed-rank test and independent comparisons were performed with the use of Mann-Whiney U test. Results were considered significant when p < 0.05. Statistical analysis was performed in SPSS v.26 and SigmaPlot v.14.5 (IBM, USA) and GraphPad Prism v.9.1.1 (GraphPad, USA).
3. Results
3.1. Type I interferon score in PBMCs upon acute and chronic immune activation in vivo
First, we examined whether the innate immune response is activated after the Influenza and SARS-CoV-2 vaccination. Therefore, we calculated type I IFN score as a composite of two type I IFN-inducible genes (IFIT1 and MX1) and normalized to a housekeeping gene (GAPDH). Significantly increased type I IFN score was detected in healthy individuals 24 h following administration of influenza or SARS-CoV-2 vaccine (p = 0.021 and p = 0.012, respectively), declining thereafter in samples obtained following SARS-CoV-2 vaccination (Fig. 1 ). When we directly compared the induction of type I IFN response after vaccination against Influenza or SARS-CoV-2 vaccines at the individual patient level, we did not observe a significant difference (p = 0.148). Moreover, the type I IFN response in individuals vaccinated against SARS-CoV-2 was comparable between those 2 individuals who showed a systematic reaction (fever, chills), and the rest study subjects who only had pain at the injection site. In addition, compared with healthy individuals before vaccination, SLE patients demonstrated higher type I interferon score (p < 0.05), comparable to those scores detected in 24 h after vaccination.
Fig. 1.
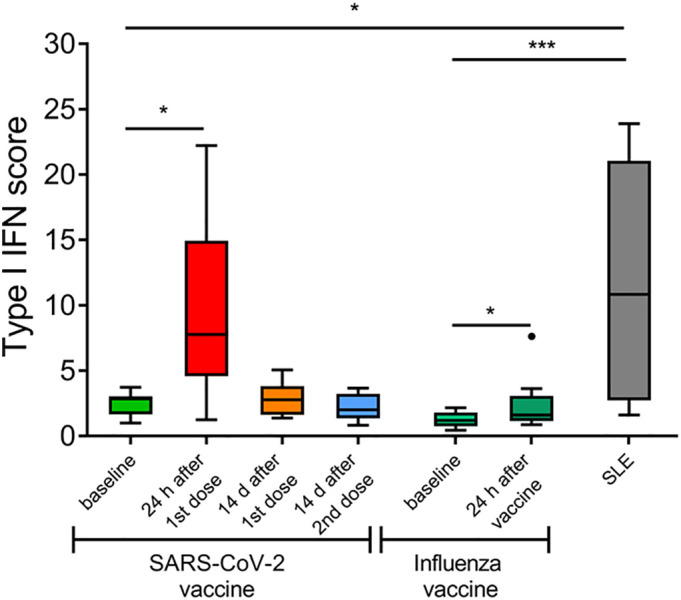
Type I interferon (IFN) signature in peripheral blood mononuclear cells (PBMCs) upon acute and chronic immune activation in vivo. Tukey boxplots showing the type I IFN score, calculated as the total of the relative mRNA expression of two type I IFN-inducible genes (IFIT1 and MX1) in PBMCs of healthy controls (HCs) (n = 8) before and after SARS-CoV-2 and influenza vaccination and SLE patients (n = 10). The relative mRNA expression was measured using RT-qPCR. P-values are derived from Wilcoxon signed-rank test and Mann-Whitney U test. * P < 0.05, *** P < 0.001.
3.2. Successful immunization by SARS-CoV-2 vaccination
Next, we investigated whether SARS-CoV-2 vaccination successfully elicited a strong antibody response. For this purpose, we measured the neutralizing capacity of circulating antibodies 14 days after the second dose of the mRNA SARS-CoV-2 vaccine, as at that timepoint a sufficient antibody response can be observed [31]. Indeed, all vaccinated individuals showed a robust neutralizing antibody production (>90% antibody-mediated inhibition of SARS-CoV-2 RBD binding to human host ACE2), irrespective of changes in DNA damage, oxidative stress levels or type I IFN response.
3.3. Transient induction of oxidative stress by influenza and SARS-CoV-2 vaccination
Moreover, we aimed to investigate whether oxidative stress is induced after acute and chronic immune activation. SARS-CoV-2 and Influenza vaccines generated a transient increase of oxidative stress in the PBMCs of healthy individuals 24 h after vaccination, as indicated by the reduction of GSH to GSSG ratio (p = 0.028 and p = 0.012 vs baseline, respectively; Fig. 2A), and the increased formation of abasic sites, a prototypical oxidative adduct (p = 0.027 and p = 0.011 vs baseline, respectively), which was subsequently resolved in samples obtained following SARS-CoV-2 vaccination (Fig. 2B). Additionally, we found that unstimulated PBMCs from SLE patients exhibited maximal oxidative stress at levels comparable to those of the healthy individuals 24 h after vaccination.
Fig. 2.
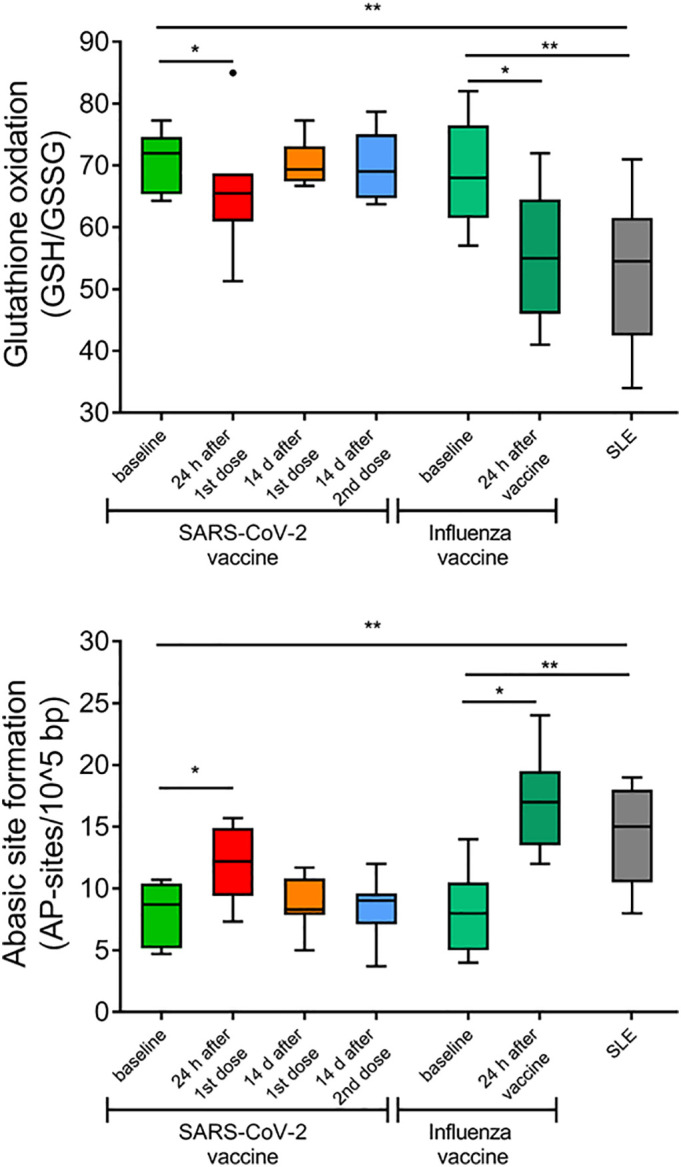
Induction of oxidative stress by SARS-CoV-2 and influenza vaccination in peripheral blood mononuclear cells (PBMCs) of healthy individuals at levels observed in PBMCs of SLE patients. Tukey boxplots representing oxidative stress levels expressed as (Α) the ratio of reduced Glutathione (GSH) to oxidized glutathione (GSSG) and (Β) the amount of abasic sites per 105 base pairs in PBMCs of HCs before and after SARS-CoV-2 (n = 6) and influenza (n = 9) vaccination and SLE patients (n = 10). P-values are derived from Wilcoxon signed-rank test and Mann-Whitney U test. * P < 0.05, ** P < 0.01.
3.4. Oxidative stress fuels DDR activation
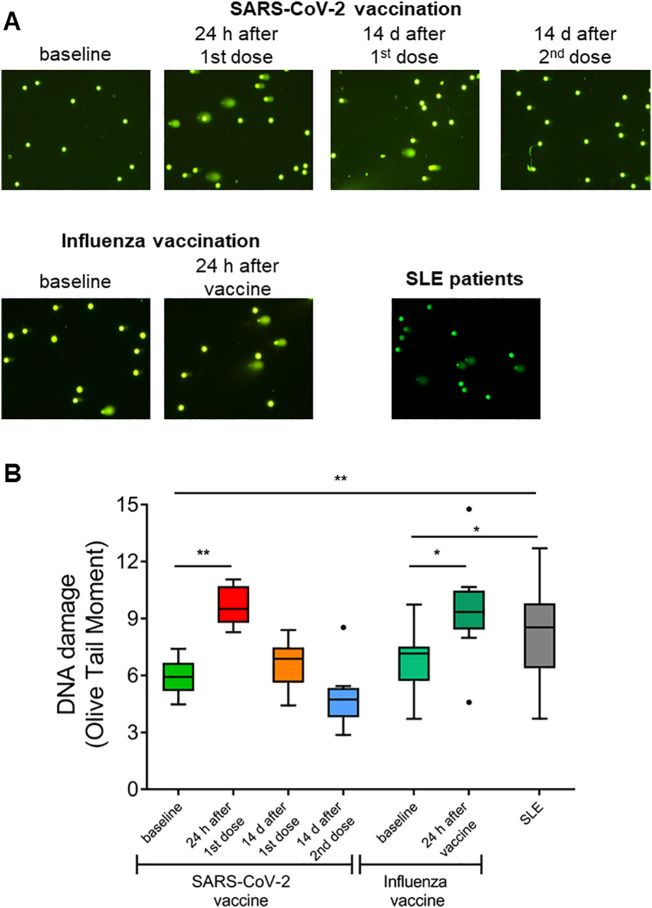
Next, we examined whether the increased oxidative stress, observed in healthy individuals 24 h after vaccination, as well as in SLE patients, resulted in DDR activation, considering that oxidative stress is a major contributor to DNA damage formation [13]. We assessed DNA damage by measuring DSBs and/or SSBs via alkaline comet assay (Fig. 3A). We found that PBMCs from healthy individuals 24 h after SARS-CoV-2 and Influenza vaccination demonstrated significantly higher DNA damage levels, compared to those before vaccination (SARS-CoV-2 vaccine: p = 0.008, Influenza vaccine: p = 0.015), which were successfully repaired 14 days after either the first or the second dose of SARS-CoV-2 vaccination (Fig. 3B). On the other hand, PBMCs from SLE patients showed significantly higher endogenous DNA damage compared with healthy individuals before vaccination (p < 0.05), indicating that SLE patients display a significant DNA damage accumulation without external stimuli and a persistently activated DDR network.
Fig. 3.
DNA damage accumulation and successful repair following SARS-CoV-2 and influenza vaccination in healthy individuals, unlike SLE. Α) Representative alkaline comet assay images of untreated PBMCs from one HC before and at four time-points after SARS-CoV-2 vaccination (1–4), one HC before and after influenza vaccination (5,6) and a SLE patient.Β) Tukey boxplots representing the endogenous DNA damage levels (Olive tail moment arbitrary units) as assessed by alkaline comet assay in peripheral blood mononuclear cells (PBMCs) from healthy controls (HCs) before and after SARS-CoV-2 and influenza vaccination (n = 9) and in SLE patients (n = 26). P-values are derived from Wilcoxon signed-rank test and Mann-Whitney U test. * P < 0.05, ** P < 0.01.
3.5. Effective DNA damage repair in healthy individuals after vaccination opposing to patients with SLE
Next, we investigated whether the aforementioned DNA damage accumulation may be attributed to deficits in the DNA damage repair mechanisms apart from the augmented oxidative stress levels. We examined the efficacy of a central DNA repair pathway, namely NER, both at cellular and gene-specific level.
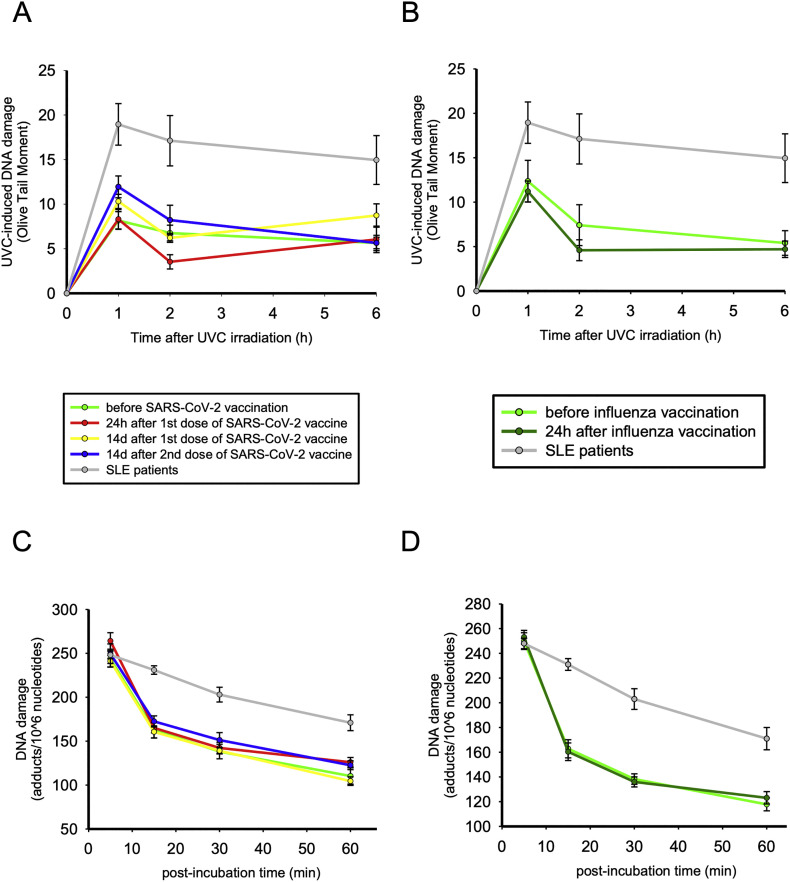
In order to examine the NER capacity at cellular level, we irradiated freshly isolated PBMCs with UVC, which causes 6–4 photoproducts (6–4 PPs) and cyclobutane pyrimidine dimers (CPDs), both of which are exclusively repaired by NER. Peak DNA damage levels were observed 1 h after UVC irradiation and, subsequently, DNA damage levels were reduced. We found that NER capacity remained unaffected after influenza and SARS-CoV-2 vaccination at PBMCs of healthy individuals [AUC of NER kinetics: SARS-CoV-2 vaccine: p = 0.214 vs baseline (Fig. 4A); influenza vaccine: p = 0.678 vs baseline (Fig. 4B)]. However, SLE patients demonstrated significantly reduced NER capacity, as evidenced by the lower rates of DNA damage resolution (AUC of NER kinetics: p = 0.001). Of interest, we observed a trending association between endogenous DNA damage and NER capacity in the SLE patients (r = 0.583, p = 0.099), partly explaining the increased endogenous DNA damage manifested in these patients. However, patients with more severely affected NER mechanism or higher endogenous DNA damage levels do not show signs of higher disease burden, such as higher antinuclear autoantibody titres (ANA < 1/320 vs rest patients p = 0.286 and p = 0.413 respectively) or higher SLE disease activity (SLEDAI > median vs rest patients p = 0.631 and p = 0.556 respectively).
Fig. 4.
Nucleotide excision repair (NER) of DNA damage in healthy individuals differs from patients with SLE. A–B) Line graphs representing NER capacity at cellular level by showing levels of single- and/or double-strand DNA breaks (Olive tail moment ± Standard error) as assessed by alkaline comet assay in peripheral blood mononuclear cells (PBMCs) of HCs before and after SARS CoV-2 and influenza vaccination (n = 9) and SLE patients (n = 10) at baseline and 1, 2, and 6 h after ex vivo UVC irradiation of freshly isolated PBMCs with 5 J/m2. C–D) Line graphs representing NER capacity at gene-specific level (NRAS gene), by showing the removal of monoadducts 0–60 min after treatment of PBMCs derived from HCs before and after SARS-CoV-2 and influenza vaccine, as well as from SLE patients, with mono-hydroxy-melphalan.
Additionally, the efficacy of NER mechanism was examined at the entire NRAS gene by administrating mono-hydroxyl-melphalan, an alkylating agent capable of inducing DNA damage mainly repaired by NER [28]. In all subjects, similar formation of monoadducts was found at the end of the 5-min treatment. Afterwards, we found no differences in the repair capacity of the PBMCs before and after vaccination (SARS-CoV-2 vaccine: p = 0.753, influenza vaccine: p = 0.594; Fig. 4C,D), whereas PBMCs from SLE patients showed significant defects in this mechanism (p < 0.001), which confirms our previous observations in patients with quiescent SLE [20]. Importantly, we found minor individual variability in the NER capacity of the HCs after SARS-CoV-2 and Influenza vaccination, when assessing NER efficiency at both cellular and gene-specific level (Supplementary Fig. 1 A,B).
4. Discussion
Herein we show that vaccination against RNA viruses such as Influenza A and SARS-CoV-2 leads to a transient activation of the DDR network, along with the antiviral innate immune responses, to a comparable level seen in patients with SLE. However, while DNA damage formation via oxidative stress is significantly increased in healthy subjects post-vaccination, the DNA repair capacity of such lesions by the NER mechanism remains intact. On the other hand, SLE patients, where a chronic immune activation is presumably present, display both increased DNA damage formation and significantly impaired repair capacity, which could account for the increased levels of endogenous DNA damage that we and others have previously reported in SLE patients [19,20] or other systemic autoimmune diseases [25,28].
An interplay between DDR and innate immune response has been well-documented in the past years (reviewed in [13,32]). Previous studies have shown that DNA damage accumulation elicits innate immune responses mostly through a cGAS/STING-mediated pathway [21,33,34], while on the other hand chronic activation of the immune system can lead to DNA damage accumulation [35,36]. In our study, we aimed to dissect the effect of an acute immune challenge vs chronic immune activation on the DDR system. For this purpose, we used vaccination as an in vivo model of acute inflammation, since previous studies have shown that vaccines lead to acute systemic inflammatory responses expanding in other tissues beyond the activated lymphocytes, such as the endothelium [37]. More specifically, a previous study examining the transcriptomic alterations in healthy subjects vaccinated against Influenza showed a transient, yet significant upregulation of the type I IFN response in peripheral blood leukocytes [8]. Similarly, we show herein that both Influenza and SARS-CoV-2 vaccination induce a type I IFN response in healthy controls' PBMCs. As a representative chronic inflammatory disease we selected SLE, because type I IFN is integral for its pathogenesis [38], as well as a valuable therapeutic target [39].
Type I IFNs are a critical component of the immune response against viral infections, serving as the link between innate and adaptive immune responses [40]. Among other pleiotropic functions, type I IFNs promote maturation and differentiation of professional antigen presenting cells (APCs), such as dendritic cells [41], which in turn activate B cells for antibody production [42]. Type I IFNs can also directly activate B cells by upregulating the expression of multiple surface antigens or other mediators such as BAFF (nicely reviewed in [43]), while they also promote rapid expansion of antigen-specific B cells, which produce neutralizing antibodies [44]. Moreover, type I IFNs orchestrate antiviral T cell responses by directly activating T cells, enhancing APC-T cell interactions and preventing NK-mediated destruction of clonally expanding viral-specific T cells [45]. It is, thus, not surprising that type I IFNs have been proposed as adjuvants in vaccines to enhance host antiviral immunity [46]. Herein, to the best of our knowledge we show for the first time that SARS-CoV-2 vaccination elicits a potent type I IFN response in healthy young adults, which, however, returns to normal levels 2 weeks after vaccination, in accordance with previous studies on Influenza vaccination [8].
Further, we showed that both Influenza and SARS-CoV-2 vaccination led to increased oxidative stress levels, which however returned to normal after 14 days. In line with our results, previous studies have shown that vaccines can induce a systematic upregulation of oxidative stress [[47], [48], [49]], as evidenced by increased oxidative products in breath [47] and decreased plasma anti-oxidant capacity [48,49]. Of interest, oxidized DNA, either viral or endogenous, can resist degradation by the cytoplasmic exonuclease TREX1, thus accumulating in the cytoplasm and further propagating innate immune activation [34], which is in line with our observation of increased type I IFN in parallel with increased oxidative stress. Although SARS-CoV-2 vaccination induced this transient oxidative stress and DNA damage accumulation, we did not find a direct association with the neutralizing antibody production efficiency, as all participants in our study developed a strong neutralizing antibody response. While oxidative stress levels returned to normal in the healthy vaccinated individuals, we observed a similar increase in oxidative stress levels of SLE patients without an external stimulus. Whether this transient increase in oxidative stress and DNA damage is beneficial (associated with more efficient neutralizing antibody production) or detrimental remains to be elucidated in future studies. Oxidative stress is integrally implicated in the pathogenesis of SLE at multiple levels [50]. Oxidized DNA has been found to co-localize with type I IFN-induced proteins in skin biopsies of SLE patients further supporting the interplay between oxidative stress and innate immune response [34]. Moreover, oxidization of self-antigens renders them more immunogenic enhancing auto-antibody production [51,52], while levels of protein oxidation have been found to strongly correlate with disease activity [53]. T cells of SLE patients show decreased levels of reduced glutathione, while treatment of SLE patients with the potent anti-oxidant N-acetylcysteine improved disease activity [54]. Of note, oxidative stress promotes the expansion of CD4/CD8-double negative T cells, Th2 and Th17 cells (nicely reviewed in [50]), which are central in SLE pathogenesis [[55], [56], [57]]. Whether pre-existing increases in oxidative stress and DNA damage can affect antibody production after vaccination remains unknown. Nevertheless, recent studies examining response of patients with systemic autoimmune diseases to SARS-CoV-2 vaccination show decreased antibody production compared to healthy subjects, which may be attributed either to their underlying disease and/or immunomodulatory therapy [58,59].
Finally, we observed a transient accumulation of DNA damage in PBMCs 24 h post-vaccination, which returned to normal levels a few days later. Previous studies have shown that antigen-activated T lymphocytes show a pronounced DDR [60] and can rapidly proliferate with an initial division time of approximately 2 h [61]. This can lead to genomic stress, which could partially explain the increased DNA damage levels that we observed shortly after vaccination. On the other hand, vaccination did not seem to affect DNA repair capacity of the healthy subjects, which is opposite to the deficiency of central DNA repair mechanisms that we observed in patients with SLE. Of interest, in the acute inflammatory response, DDR seems to have a protective role for the organism; in sepsis, for example, induction of a potent DDR can prevent the development of hyper-inflammatory syndrome and reduce tissue damage [62]. In contrast with the single immune challenge imposed by the vaccination, however, in SLE T cells are constantly stimulated by self-antigens. This chronic antigenic stimulation has been associated with telomere shortening and decreased proliferation of SLE PBMCs [63], a state called replicative senescence [64].
5. Conclusion
To conclude, our study demonstrates that influenza and SARS-CoV-2 vaccines, acting as an acute immune stimulant, successfully activate the DDR network, whereas SLE patients, characterized by chronic immune activation, manifest an aberrant DDR activation. Post-vaccination healthy individuals exhibit a transient increase in type I IFN expression and oxidative stress, inducing resolvable DNA damage. On the other hand, SLE patients display an unresolved persistent increase in type I IFN expression and oxidative stress. Most importantly, the acute immune activation does not influence the DNA repair capacity, in contrast to the deficiencies observed in SLE patients. The aforementioned results can be used to shed light on the cellular pathways involved in the activation of the adaptive immune response and the production of neutralizing antibodies after SARS-CoV-2 and influenza vaccination, as well as analyze the aberrations caused by the chronic immune activation.
The following is the supplementary data related to this article.
Nucleotide excision repair (NER) capacity at cellular- and gene-specific level.
Financial support
This work was supported by educational grants (ELKE: 0974 and Empeirikeion Foundation) to P.P. Sfikakis.
Declaration of Competing Interest
None of the authors has any potential financial conflict of interest related to this manuscript.
References
- 1.Rauch S., Jasny E., Schmidt K.E., Petsch B. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jang H., Ross T.M. Preexisting influenza specific immunity and vaccine effectiveness. Expert Rev. Vaccin. 2019;18:1043–1051. doi: 10.1080/14760584.2019.1675519. [DOI] [PubMed] [Google Scholar]
- 3.Gasmi A., Noor S., Tippairote T., Dadar M., Menzel A., Bjørklund G. Individual risk management strategy and potential therapeutic options for the COVID-19 pandemic. Clin. Immunol. 2020;215:108409. doi: 10.1016/j.clim.2020.108409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hu B., Guo H., Zhou P., Shi Z.-L. Characteristics of SARS-CoV-2 and COVID-19. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yuki K., Fujiogi M., Koutsogiannaki S. COVID-19 pathophysiology: a review. Clin. Immunol. 2020;215:108427. doi: 10.1016/j.clim.2020.108427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grigoryan L., Pulendran B. The immunology of SARS-CoV-2 infections and vaccines. Semin. Immunol. 2020;50:101422. doi: 10.1016/j.smim.2020.101422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Izda V., Jeffries M.A., Sawalha A.H. COVID-19: a review of therapeutic strategies and vaccine candidates. Clin. Immunol. 2021;222:108634. doi: 10.1016/j.clim.2020.108634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obermoser G., Presnell S., Domico K., Xu H., Wang Y., Anguiano E., Thompson-Snipes L., Ranganathan R., Zeitner B., Bjork A., Anderson D., Speake C., Ruchaud E., Skinner J., Alsina L., Sharma M., Dutartre H., Cepika A., Israelsson E., Nguyen P., Nguyen Q.-A., Harrod A.C., Zurawski S.M., Pascual V., Ueno H., Nepom G.T., Quinn C., Blankenship D., Palucka K., Banchereau J., Chaussabel D. Systems scale interactive exploration reveals quantitative and qualitative differences in response to influenza and pneumococcal vaccines. Immunity. 2013;38:831–844. doi: 10.1016/j.immuni.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Athale S., Banchereau R., Thompson-Snipes L., Wang Y., Palucka K., Pascual V., Banchereau J. Influenza vaccines differentially regulate the interferon response in human dendritic cell subsets. Sci. Transl. Med. 2017;9 doi: 10.1126/scitranslmed.aaf9194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aoshi T., Koyama S., Kobiyama K., Akira S., Ishii K.J. Innate and adaptive immune responses to viral infection and vaccination. Curr. Opin. Virol. 2011;1:226–232. doi: 10.1016/j.coviro.2011.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Schwarz K.B. Oxidative stress during viral infection: a review. Free Radic. Biol. Med. 1996;21:641–649. doi: 10.1016/0891-5849(96)00131-1. [DOI] [PubMed] [Google Scholar]
- 12.Liu M., Chen F., Liu T., Chen F., Liu S., Yang J. The role of oxidative stress in influenza virus infection. Microbes Infect. 2017;19:580–586. doi: 10.1016/j.micinf.2017.08.008. [DOI] [PubMed] [Google Scholar]
- 13.Souliotis V.L., Vlachogiannis N.I., Pappa M., Argyriou A., Ntouros P.A., Sfikakis P.P. DNA damage response and oxidative stress in systemic autoimmunity. IJMS. 2019;21:55. doi: 10.3390/ijms21010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doria A., Zen M., Bettio S., Gatto M., Bassi N., Nalotto L., Ghirardello A., Iaccarino L., Punzi L. Autoinflammation and autoimmunity: bridging the divide. Autoimmun. Rev. 2012;12:22–30. doi: 10.1016/j.autrev.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 15.Tsokos G.C. Systemic Lupus Erythematosus. N. Engl. J. Med. 2011;12 doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 16.Pisetsky D.S., Lipsky P.E. New insights into the role of antinuclear antibodies in systemic lupus erythematosus. Nat. Rev. Rheumatol. 2020;16:565–579. doi: 10.1038/s41584-020-0480-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herrada A.A., Escobedo N., Iruretagoyena M., Valenzuela R.A., Burgos P.I., Cuitino L., Llanos C. Innate immune cells’ contribution to systemic lupus erythematosus. Front. Immunol. 2019;10 doi: 10.3389/fimmu.2019.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsokos G.C. Autoimmunity and organ damage in systemic lupus erythematosus. Nat. Immunol. 2020;21:605–614. doi: 10.1038/s41590-020-0677-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Souliotis V.L., Sfikakis P.P. Increased DNA double-strand breaks and enhanced apoptosis in patients with lupus nephritis. Lupus. 2015;24:804–815. doi: 10.1177/0961203314565413. [DOI] [PubMed] [Google Scholar]
- 20.Souliotis V.L., Vougas K., Gorgoulis V.G., Sfikakis P.P. Defective DNA repair and chromatin organization in patients with quiescent systemic lupus erythematosus. Arthrit. Res Ther. 2016;18:182. doi: 10.1186/s13075-016-1081-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Härtlova A., Erttmann S.F., Raffi F.A., Schmalz A.M., Resch U., Anugula S., Lienenklaus S., Nilsson L.M., Kröger A., Nilsson J.A., Ek T., Weiss S., Gekara N.O. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42:332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Shen Y.J., Le Bert N., Chitre A.A., Koo C.X., Nga X.H., Ho S.S.W., Khatoo M., Tan N.Y., Ishii K.J., Gasser S. Genome-derived cytosolic DNA mediates type I interferon-dependent rejection of B cell lymphoma cells. Cell Rep. 2015;11:460–473. doi: 10.1016/j.celrep.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 23.Aringer M., Costenbader K., Daikh D., Brinks R., Mosca M., Ramsey-Goldman R., Smolen J.S., Wofsy D., Boumpas D.T., Kamen D.L., Jayne D., Cervera R., Costedoat-Chalumeau N., Diamond B., Gladman D.D., Hahn B., Hiepe F., Jacobsen S., Khanna D., Lerstrøm K., Massarotti E., McCune J., Ruiz-Irastorza G., Sanchez-Guerrero J., Schneider M., Urowitz M., Bertsias G., Hoyer B.F., Leuchten N., Tani C., Tedeschi S.K., Touma Z., Schmajuk G., Anic B., Assan F., Chan T.M., Clarke A.E., Crow M.K., Czirják L., Doria A., Graninger W., Halda-Kiss B., Hasni S., Izmirly P.M., Jung M., Kumánovics G., Mariette X., Padjen I., Pego-Reigosa J.M., Romero-Diaz J., Fernández Í.R.-F., Seror R., Stummvoll G.H., Tanaka Y., Tektonidou M.G., Vasconcelos C., Vital E.M., Wallace D.J., Yavuz S., Meroni P.L., Fritzler M.J., Naden R., Dörner T., Johnson S.R. 2019 European League Against Rheumatism/American College of Rheumatology classification criteria for systemic lupus erythematosus. Ann. Rheum. Dis. 2019;78:1151–1159. doi: 10.1136/annrheumdis-2018-214819. [DOI] [PubMed] [Google Scholar]
- 24.Vlachogiannis N.I., Gatsiou A., Silvestris D.A., Stamatelopoulos K., Tektonidou M.G., Gallo A., Sfikakis P.P., Stellos K. Increased adenosine-to-inosine RNA editing in rheumatoid arthritis. J. Autoimmun. 2020;106:102329. doi: 10.1016/j.jaut.2019.102329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vlachogiannis N.I., Pappa M., Ntouros P.A., Nezos A., Mavragani C.P., Souliotis V.L., Sfikakis P.P. Association between DNA damage response, fibrosis and type I interferon signature in systemic sclerosis. Front. Immunol. 2020;11:582401. doi: 10.3389/fimmu.2020.582401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nezos A., Gravani F., Tassidou A., Kapsogeorgou E.K., Voulgarelis M., Koutsilieris M., Crow M.K., Mavragani C.P. Type I and II interferon signatures in Sjogren’s syndrome pathogenesis: contributions in distinct clinical phenotypes and Sjogren’s related lymphomagenesis. J. Autoimmun. 2015;63:47–58. doi: 10.1016/j.jaut.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vlachogiannis N.I., Nezos A., Tzioufas A.G., Koutsilieris M., Moutsopoulos H.M., Mavragani C.P. Increased frequency of the PTPN22W* variant in primary Sjogren’s Syndrome: association with low type I IFN scores. Clin. Immunol. 2016;173:157–160. doi: 10.1016/j.clim.2016.10.015. [DOI] [PubMed] [Google Scholar]
- 28.Souliotis V.L., Vlachogiannis N.I., Pappa M., Argyriou A., Sfikakis P.P. DNA damage accumulation, defective chromatin organization and deficient DNA repair capacity in patients with rheumatoid arthritis. Clin. Immunol. 2019;203:28–36. doi: 10.1016/j.clim.2019.03.009. [DOI] [PubMed] [Google Scholar]
- 29.Episkopou H., Kyrtopoulos S.A., Sfikakis P.P., Fousteri M., Dimopoulos M.A., Mullenders L.H.F., Souliotis V.L. Association between transcriptional activity, local chromatin structure, and the efficiencies of both subpathways of nucleotide excision repair of melphalan adducts. Cancer Res. 2009;69:4424–4433. doi: 10.1158/0008-5472.CAN-08-3489. [DOI] [PubMed] [Google Scholar]
- 30.Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.-C., Tiu C., Hu Z., Chen V.C.-W., Young B.E., Sia W.R., Tan Y.-J., Foo R., Yi Y., Lye D.C., Anderson D.E., Wang L.-F. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 31.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., Bailey R., Swanson K.A., Roychoudhury S., Koury K., Li P., Kalina W.V., Cooper D., Frenck R.W., Hammitt L.L., Türeci Ö., Nell H., Schaefer A., Ünal S., Tresnan D.B., Mather S., Dormitzer P.R., Şahin U., Jansen K.U., Gruber W.C., C4591001 Clinical Trial Group Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pateras I.S., Havaki S., Nikitopoulou X., Vougas K., Townsend P.A., Panayiotidis M.I., Georgakilas A.G., Gorgoulis V.G. The DNA damage response and immune signaling alliance: is it good or bad? Nature decides when and where. Pharmacol. Ther. 2015;154:36–56. doi: 10.1016/j.pharmthera.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 33.Günther C., Kind B., Reijns M.A.M., Berndt N., Martinez-Bueno M., Wolf C., Tüngler V., Chara O., Lee Y.A., Hübner N., Bicknell L., Blum S., Krug C., Schmidt F., Kretschmer S., Koss S., Astell K.R., Ramantani G., Bauerfeind A., Morris D.L., Graham D.S. Cunninghame, Bubeck D., Leitch A., Ralston S.H., Blackburn E.A., Gahr M., Witte T., Vyse T.J., Melchers I., Mangold E., Nöthen M.M., Aringer M., Kuhn A., Lüthke K., Unger L., Bley A., Lorenzi A., Isaacs J.D., Alexopoulou D., Conrad K., Dahl A., Roers A., Alarcon-Riquelme M.E., Jackson A.P., Lee-Kirsch M.A. Defective removal of ribonucleotides from DNA promotes systemic autoimmunity. J. Clin. Invest. 2015;125:413–424. doi: 10.1172/JCI78001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gehrke N., Mertens C., Zillinger T., Wenzel J., Bald T., Zahn S., Tüting T., Hartmann G., Barchet W. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity. 2013;39:482–495. doi: 10.1016/j.immuni.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Jaiswal M., LaRusso N.F., Burgart L.J., Gores G.J. Inflammatory cytokines induce DNA damage and inhibit DNA repair in cholangiocarcinoma cells by a nitric oxide-dependent mechanism. Cancer Res. 2000;60:184–190. [PubMed] [Google Scholar]
- 36.Meira L.B., Bugni J.M., Green S.L., Lee C.-W., Pang B., Borenshtein D., Rickman B.H., Rogers A.B., Moroski-Erkul C.A., McFaline J.L., Schauer D.B., Dedon P.C., Fox J.G., Samson L.D. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J. Clin. Invest. 2008;118:2516–2525. doi: 10.1172/JCI35073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vlachopoulos C., Dima I., Aznaouridis K., Vasiliadou C., Ioakeimidis N., Aggeli C., Toutouza M., Stefanadis C. Acute systemic inflammation increases arterial stiffness and decreases wave reflections in healthy individuals. Circulation. 2005;112:2193–2200. doi: 10.1161/CIRCULATIONAHA.105.535435. [DOI] [PubMed] [Google Scholar]
- 38.Crow M.K. Type I interferon in the pathogenesis of lupus. J. Immunol. 2014;192:5459–5468. doi: 10.4049/jimmunol.1002795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morand E.F., Furie R., Tanaka Y., Bruce I.N., Askanase A.D., Richez C., Bae S.-C., Brohawn P.Z., Pineda L., Berglind A., Tummala R. Trial of anifrolumab in active Systemic Lupus erythematosus. N. Engl. J. Med. 2020;382:211–221. doi: 10.1056/NEJMoa1912196. [DOI] [PubMed] [Google Scholar]
- 40.Samuel C.E. Antiviral actions of interferons. Clin. Microbiol. Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gallucci S., Lolkema M., Matzinger P. Natural adjuvants: endogenous activators of dendritic cells. Nat. Med. 1999;5:1249–1255. doi: 10.1038/15200. [DOI] [PubMed] [Google Scholar]
- 42.Jego G., Palucka A.K., Blanck J.-P., Chalouni C., Pascual V., Banchereau J. Plasmacytoid dendritic cells induce plasma cell differentiation through Type I Interferon and Interleukin 6. Immunity. 2003;19:225–234. doi: 10.1016/s1074-7613(03)00208-5. [DOI] [PubMed] [Google Scholar]
- 43.Kiefer K., Oropallo M.A., Cancro M.P., Marshak-Rothstein A. Role of type I interferons in the activation of autoreactive B cells. Immunol. Cell Biol. 2012;90:498–504. doi: 10.1038/icb.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Swanson C.L., Wilson T.J., Strauch P., Colonna M., Pelanda R., Torres R.M. Type I IFN enhances follicular B cell contribution to the T cell-independent antibody response. J. Exp. Med. 2010;207:1485–1500. doi: 10.1084/jem.20092695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crouse J., Kalinke U., Oxenius A. Regulation of antiviral T cell responses by type I interferons. Nat. Rev. Immunol. 2015;15:231–242. doi: 10.1038/nri3806. [DOI] [PubMed] [Google Scholar]
- 46.Bracci L., La Sorsa V., Belardelli F., Proietti E. Type I interferons as vaccine adjuvants against infectious diseases and cancer. Expert Rev. Vaccin. 2008;7:373–381. doi: 10.1586/14760584.7.3.373. [DOI] [PubMed] [Google Scholar]
- 47.Phillips M., Cataneo R.N., Chaturvedi A., Danaher P.J., Devadiga A., Legendre D.A., Nail K.L., Schmitt P., Wai J. Effect of influenza vaccination on oxidative stress products in breath. J. Breath Res. 2010;4 doi: 10.1088/1752-7155/4/2/026001. [DOI] [PubMed] [Google Scholar]
- 48.Vlachopoulos C., Aznaouridis K., Dagre A., Vasiliadou C., Masoura C., Stefanadi E., Skoumas J., Pitsavos C., Stefanadis C. Protective effect of atorvastatin on acute systemic inflammation-induced endothelial dysfunction in hypercholesterolaemic subjects. Eur. Heart J. 2007;28:2102–2109. doi: 10.1093/eurheartj/ehm247. [DOI] [PubMed] [Google Scholar]
- 49.Clapp B.R., Hingorani A.D., Kharbanda R.K., Mohamed-Ali V., Stephens J.W., Vallance P., MacAllister R.J. Inflammation-induced endothelial dysfunction involves reduced nitric oxide bioavailability and increased oxidant stress. Cardiovasc. Res. 2004;64:172–178. doi: 10.1016/j.cardiores.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 50.Perl A. Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2013;9:674–686. doi: 10.1038/nrrheum.2013.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Passam F.H., Giannakopoulos B., Mirarabshahi P., Krilis S.A. Molecular pathophysiology of the antiphospholipid syndrome: the role of oxidative post-translational modification of beta 2 glycoprotein I. J. Thromb. Haemost. 2011;9:275–282. doi: 10.1111/j.1538-7836.2011.04301.x. [DOI] [PubMed] [Google Scholar]
- 52.Otaki N., Chikazawa M., Nagae R., Shimozu Y., Shibata T., Ito S., Takasaki Y., Fujii J., Uchida K. Identification of a lipid peroxidation product as the source of oxidation-specific epitopes recognized by anti-DNA autoantibodies. J. Biol. Chem. 2010;285:33834–33842. doi: 10.1074/jbc.M110.165175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang G., Pierangeli S.S., Papalardo E., Ansari G.A.S., Khan M.F. Markers of oxidative and nitrosative stress in systemic lupus erythematosus: correlation with disease activity. Arthrit. Rheumat. 2010;62:2064–2072. doi: 10.1002/art.27442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lai Z.-W., Hanczko R., Bonilla E., Caza T.N., Clair B., Bartos A., Miklossy G., Jimah J., Doherty E., Tily H., Francis L., Garcia R., Dawood M., Yu J., Ramos I., Coman I., Faraone S.V., Phillips P.E., Perl A. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: a randomized, double-blind, placebo-controlled trial. Arthrit. Rheumat. 2012;64:2937–2946. doi: 10.1002/art.34502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Crispín J.C., Oukka M., Bayliss G., Cohen R.A., Van Beek C.A., Stillman I.E., Kyttaris V.C., Juang Y.-T., Tsokos G.C. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J. Immunol. 2008;181:8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nalbandian A., Crispín J.C., Tsokos G.C. Interleukin-17 and systemic lupus erythematosus: current concepts. Clin. Exp. Immunol. 2009;157:209–215. doi: 10.1111/j.1365-2249.2009.03944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shivakumar S., Tsokos G.C., Datta S.K. T cell receptor alpha/beta expressing double-negative (CD4-/CD8-) and CD4+ T helper cells in humans augment the production of pathogenic anti-DNA autoantibodies associated with lupus nephritis. J. Immunol. 1989;143:103–112. [PubMed] [Google Scholar]
- 58.Geisen U.M., Berner D.K., Tran F., Sümbül M., Vullriede L., Ciripoi M., Reid H.M., Schaffarzyk A., Longardt A.C., Franzenburg J., Hoff P., Schirmer J.H., Zeuner R., Friedrichs A., Steinbach A., Knies C., Markewitz R.D., Morrison P.J., Gerdes S., Schreiber S., Hoyer B.F. Immunogenicity and safety of anti-SARS-CoV-2 mRNA vaccines in patients with chronic inflammatory conditions and immunosuppressive therapy in a monocentric cohort. Ann. Rheum. Dis. 2021 doi: 10.1136/annrheumdis-2021-220272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boyarsky B.J., Ruddy J.A., Connolly C.M., Ou M.T., Werbel W.A., Garonzik-Wang J.M., Segev D.L., Paik J.J. Antibody response to a single dose of SARS-CoV-2 mRNA vaccine in patients with rheumatic and musculoskeletal diseases. Ann. Rheum. Dis. 2021 doi: 10.1136/annrheumdis-2021-220289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McNally J.P., Millen S.H., Chaturvedi V., Lakes N., Terrell C.E., Elfers E.E., Carroll K.R., Hogan S.P., Andreassen P.R., Kanter J., Allen C.E., Henry M.M., Greenberg J.N., Ladisch S., Hermiston M.L., Joyce M., Hildeman D.A., Katz J.D., Jordan M.B. Manipulating DNA damage-response signaling for the treatment of immune-mediated diseases. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E4782–E4791. doi: 10.1073/pnas.1703683114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon H., Kim T.S., Braciale T.J. The cell cycle time of CD8+ T cells responding in vivo is controlled by the type of antigenic stimulus. PLoS One. 2010;5 doi: 10.1371/journal.pone.0015423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Figueiredo N., Chora A., Raquel H., Pejanovic N., Pereira P., Hartleben B., Neves-Costa A., Moita C., Pedroso D., Pinto A., Marques S., Faridi H., Costa P., Gozzelino R., Zhao J.L., Soares M.P., Gama-Carvalho M., Martinez J., Zhang Q., Döring G., Grompe M., Simas J.P., Huber T.B., Baltimore D., Gupta V., Green D.R., Ferreira J.A., Moita L.F. Anthracyclines induce DNA damage response-mediated protection against severe sepsis. Immunity. 2013;39:874–884. doi: 10.1016/j.immuni.2013.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Honda M., Mengesha E., Albano S., Nichols W.S., Wallace D.J., Metzger A., Klinenberg J.R., Linker-Israeli M. Telomere shortening and decreased replicative potential, contrasted by continued proliferation of telomerase-positive CD8+CD28lo T cells in patients with Systemic Lupus Erythematosus. Clin. Immunol. 2001;99:211–221. doi: 10.1006/clim.2001.5023. [DOI] [PubMed] [Google Scholar]
- 64.d’Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer. 2008;8:512–522. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide excision repair (NER) capacity at cellular- and gene-specific level.