ABSTRACT
Objective: To compare the accuracy parameters of seven commercial molecular in vitro diagnostic tests for detecting SARS-CoV-2.
Methods: Studies evaluating the accuracy of seven different commercial molecular diagnostic tests for detecting SARS-CoV-2 (Cepheid Xpert Xpress SARS-CoV-2 test, Simplexa COVID-19 Direct, Abbott ID NOW COVID-19, Cobas SARS-CoV-2, Allplex 2019-nCoV Assay, Panther Fusion SARS-CoV-2, and BioFire COVID-19 Test) were included. The quality of the included studies was assessed using the QUADAS-2 checklist. A bivariate random-effects regression model was implemented..
Results: Meta-analysis of 12 included studies showed that the performances of commercial COVID-19 molecular in vitro diagnostic tests were high, with a summary sensitivity of 95.9% (95% CI 93.9–97.2%, I2 = 60.22%) and specificity of 97.2% (95% CI 95.5–98.3%, I2 = 56.66%). Among seven evaluated tests, the Abbott ID NOW COVID-19 and Simplexa COVID-19 Direct displayed lower sensitivity (91.6%, 95% CI 80.5–96.6% and 92%, 95% CI 86.2–95.5, respectively).
Conclusion: All evaluated tests showed good accuracy. However, the slightly lower sensitivity observed in the Abbott ID Now COVID-19 and Simplexa COVID-19 Direct should be considered when deciding on a test platform. Moreover, the diagnostic accuracy of COVID-19 commercial diagnostic tests should be weighed against their ease of use and speed.
KEYWORDS: SARS-CoV-2, COVID-19, diagnostic accuracy, diagnostic performance, molecular in vitro diagnostic tests
1. Introduction
To date, the coronavirus disease 2019 (COVID-19) pandemic remains a major burden worldwide [1–6]. Accurate diagnosis of COVID-19 still relies on reverse transcription-polymerase chain reaction (RT-PCR) of the SARS-CoV-2 viral RNA as the gold standard [7]. To contain and help stop the spread of the disease, rapid, precise, and large-scale detection of COVID-19 is crucial, and the need for a sensitive, user-friendly, and rapid diagnostic test becomes increasingly urgent.
Currently, many commercial molecular in vitro diagnostic tests for COVID-19 have become available to fulfill this demand. However, accurate and validated data on the diagnostic tests are still needed, as manufacturer-independent evaluation data are scarce. Several early reports have shown the higher rate of false-negative findings [8] as a flaw of currently available tests. This emphasizes the fact that many factors can influence the sensitivity and specificity of a test [9], such as the core amplification technology, variations in the performance of the tests, and sampling method. Hence, in this current study, we aimed to compare the performance of seven readily available COVID-19 molecular in vitro diagnostic tests from different manufacturers through a meta-analysis.
2. Methods
2.1. Search strategy and eligibility criteria
This study was performed according to Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies (PRISMA-DTA) statement [10]. First, a literature search was conducted in PubMed and Scopus without limits of time frame or language (dated up to December 2020), with the following terms used individually or in combination: ‘diagnostic,’ ‘sensitivity,’ ‘specificity,’ ‘commercial,’ ‘molecular in vitro diagnostic tests,’ ‘nucleic acid amplification test (NAAT),’ ‘COVID-19,’ and ‘SARS-CoV-2.’ An additional literature search approach was implemented using the brand name of US FDA-approved molecular in vitro diagnostic tests for SARS-CoV-2 detection as the descriptors.
Initially, studies were included if they meet the following criteria: (1) Evaluation of any FDA-approved, commercially available molecular in vitro diagnostic tests for COVID-19; (2) utilizing human clinical sample; (3) reporting accuracy data; and (4) using either composite standard reference, modified CDC SARS-CoV-2 assay, or consensus standard as the study reference standard. Any commercial kits reported in a single study were excluded, leaving only seven kits (Xpert Xpress SARS-CoV-2 test (Cepheid), Simplexa COVID-19 Direct (DiaSorin Molecular LLC), ID NOW COVID-19 (Abbott Diagnostics Scarborough, Inc.), Cobas SARS-CoV-2 (Roche Molecular Systems, Inc.), Allplex 2019-nCoV Assay (Seegene Inc.), Panther Fusion SARS-CoV-2 (Hologic, Inc.), and BioFire COVID-19 Test (BioFire Defense, LLC)) were then further analyzed. The manufacturer’s specifications are summarized in Table 1.
Table 1.
Comparison of seven different commercial molecular in vitro diagnostic tests for the detection of SARS-CoV-2
| Brand name | Method | Sample type | Assay run time (min) | Sample volume required (μl) | .Extraction required | Analytical sensitivity per claim | Target |
|---|---|---|---|---|---|---|---|
| Xpert Xpress SARS-CoV-2 test (Cepheid) | Real-time RT-PCR | Nasopharyngeal, oropharyngeal, nasal, or mid-turbinate swab and/or nasal wash/aspirate | ∼45 | 300 | Yes (automated) | 250 copies/mL | N2 and E genes |
| Simplexa COVID-19 Direct (DiaSorin Molecular LLC) | Real-time RT-PCR | Bronchoalveolar lavage, nasal swab, nasal wash/aspirate, nasopharyngeal swab, and saliva specimens | ∼60 | 50 | No | 242 copies/mL | ORF1ab and S genes |
| Cobas SARS-CoV-2 (Roche Molecular Systems, Inc.) | Real-time RT-PCR | Nasal, nasopharyngeal, or oropharyngeal swabs | ∼210 | 600 | Yes (automated) | 46 copies/mL | ORF1ab, a non-structural region that is unique to SARS-CoV-2, and E genes. |
| ID NOW COVID-19 (Abbott Diagnostics Scarborough, Inc.) | Isothermal nucleic acid amplification | Nasal, nasopharyngeal, or throat swabs | <15 | 200 | No | 100 copies/mL | RdRp gene |
| Allplex 2019-nCoV Assay (Seegene Inc.) | Real-time RT-PCR | Nasopharyngeal swab, oropharyngeal swab, anterior nasal swab, and midturbinate and sputum specimens | 75 | 300 | Yes (automated) | 100 copies/ml | E, RdRP, and N genes |
| Panther Fusion SARS-CoV-2 (Hologic, Inc.) | Real-time RT-PCR | Nasopharyngeal and oropharyngeal swabs | ∼145 | 250–500 | Yes (automated) | 62.5 copies/ml | Two conserved regions of ORF1ab gene |
| BioFire COVID-19 Test (BioFire Defense, LLC) | Nested multiplexed RT-PCR test | Nasopharyngeal swab | ∼50 | 300 | Yes (automated) | 165 copies/mL | ORF1ab and ORF8, N1, N2, and N3 genes |
N2: nucleocapsid gene; E: envelope gene; RdRp: RNA-dependent RNA polymerase; ORF: Open reading frame, S: spike glycoprotein gene.
2.2. Data extraction and statistical analysis
The following data were extracted: First author, year of publication, type of sample, type of reference standard, brand name, type of sample, sample size, and data of the diagnostic value for each test [number of true positive (TP), false positive (FP), true negative (TN), and false negative (FN)].
The risk bias in each study was evaluated using the Diagnostic Precision Study Quality Assessment Tool (QUADAS-2). The meta-analyses were performed according to the brand name and type of reference standard. The bivariate (random effect) model was implemented, with a minimum of two studies. Forest plots and summary receiver operating characteristic (SROC) curves were performed using Review Manager (RevMan) Version 5.3 and OpenMeta-Analyst [11–14].
3. Results
A total of 188 articles were identified after duplicate removal, of which 176 were excluded during the screening phase and further evaluation, leaving 12 records being fully examined (Figure 1) [15–26]. The main characteristics of the included study are summarized in Supplementary Table 1. The overall sensitivity and specificity of all included studies on commercial molecular in vitro diagnostic tests for COVID-19 were examined, reaching 95.9% (95% CI 93.9–97.2%, I2 = 60.22%) and 97.2% (95% CI 95.5–98.3%, I2 = 56.66%), respectively (Supplementary Figure 1(a,b)), with SROC curves are shown in Supplementary Figure 2.
Figure 1.
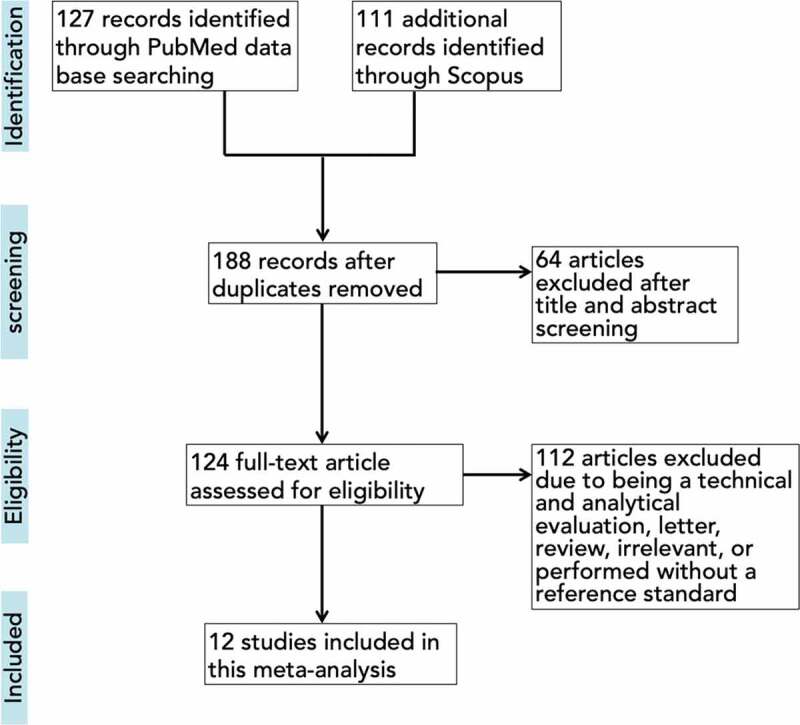
Flowchart of included studies
We then further analyzed the parameters of accuracy (sensitivity and specificity) from seven commercially available molecular in vitro diagnostic tests for COVID-19, with results depicted in Figure 2 and Table 2. Regardless of the reference standard and sample types used in this study, sensitivity and specificity from five tests were comparable (sensitivity ranging from 95.6% to 99.4%; specificity ranging from 96.4% to 99.8%; Table 2). However, studies utilizing ID NOW COVID-19 and the Simplexa COVID-19 Direct exhibited lower sensitivity 91.6% (95% CI 80.5–96.6%, I2 = 65.42) and 92% (95% CI 86.2–95.5, I2 = 42.13%), respectively (Table 2). Although the specificity of ID NOW COVID-19 was slightly lower compared to other kits, it is notable that substantial heterogeneity existed (I2 = 79.63%; Table 2), and thus, this should be interpreted with caution. The SROC plot and overview of seven molecular in vitro diagnostic tests for COVID-19 with their summary sensitivity and specificity are shown in Figure 3.
Figure 2.
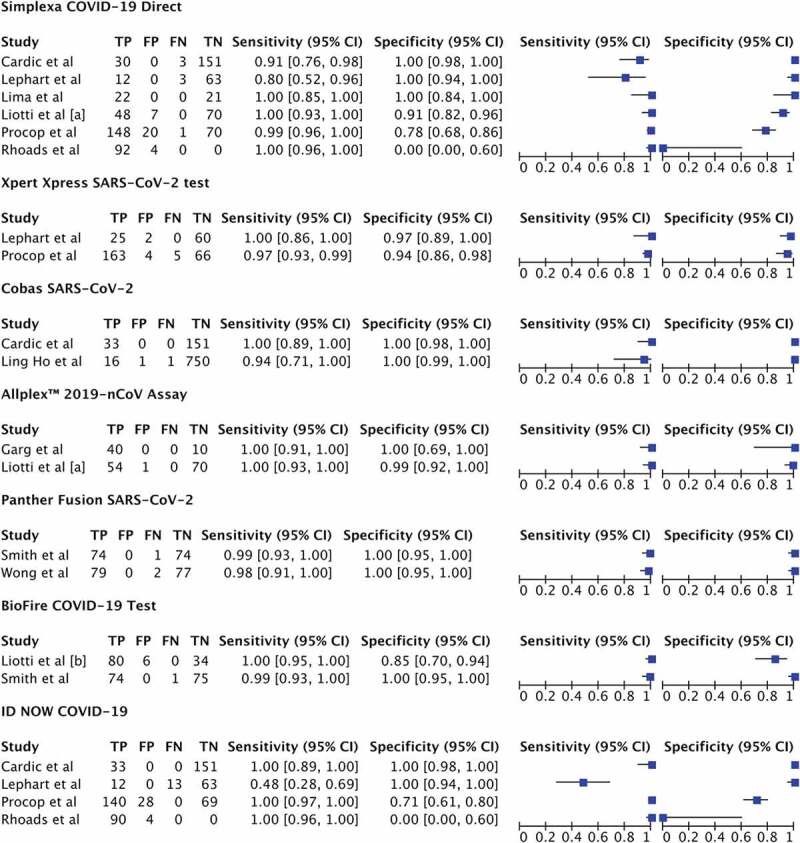
Forest plot of pairs of sensitivity and specificity in each study included stratified by brand name. TP: true positive; FP: false positive; FN: false negative; TN: true negative
Table 2.
Meta-analysis of the parameters of accuracy in different commercial molecular in vitro diagnostic tests for the detection of SARS-CoV-2 stratified by brand name
| Brand name | Sample | No. of studies | Pooled sensitivity (95% CI) | Pooled specificity (95% CI) |
|---|---|---|---|---|
| Xpert Xpress SARS-CoV-2 test (Cepheid) | Nasopharyngeal and nasal swab | 2 | 0.956 (0.849–0.988) I2 = 63.75% |
0.964 (0.779–0.995) I2 = 54.54% |
| Simplexa COVID-19 Direct (DiaSorin Molecular LLC) | Nasopharyngeal, oropharyngeal, and nasal swab | 6 | 0.920 (0.862–0.955) I2 = 42.13% |
0.970 (0.937–0.986) I2 = 18.32% |
| Cobas SARS-CoV-2 (Roche Molecular Systems, Inc.) | Nasopharyngeal, throat, sputum, saliva, stool, aspiration, and serum | 2 | 0.963 (0.836–0.993) I2 = 0% |
0.998 (0.991–1.000) I2 = 0% |
| ID NOW COVID-19 (Abbott Diagnostics Scarborough, Inc.) | Nasopharyngeal and nasal swab | 4 | 0.916 (0.805–0.966) I2 = 65.42% |
0.942 (0.708–0.991) I2 = 79.63% |
| Allplex 2019-nCoV Assay (Seegene Inc.) | Nasopharyngeal, oropharyngeal, and nasal swab | 2 | 0.978 (0.916–0.995) I2 = 0% |
0.982 (0.884–0.998) I2 = 0% |
| Panther Fusion SARS-CoV-2 (Hologic, Inc.) | Nasopharyngeal swabs, deep throat saliva, and lower respiratory tract | 2 | 0.994 (0.956–0.999) I2 = 0% |
0.982 (0.931–0.995) I2 = 0% |
| BioFire COVID-19 Test (BioFire Defense, LLC) | Nasopharyngeal, oropharyngeal, and nasal swab | 2 | 0.967 (0.743–0.997) I2 = 64.77% |
0.982 (0.931–0.995) I2 = 0% |
Figure 3.
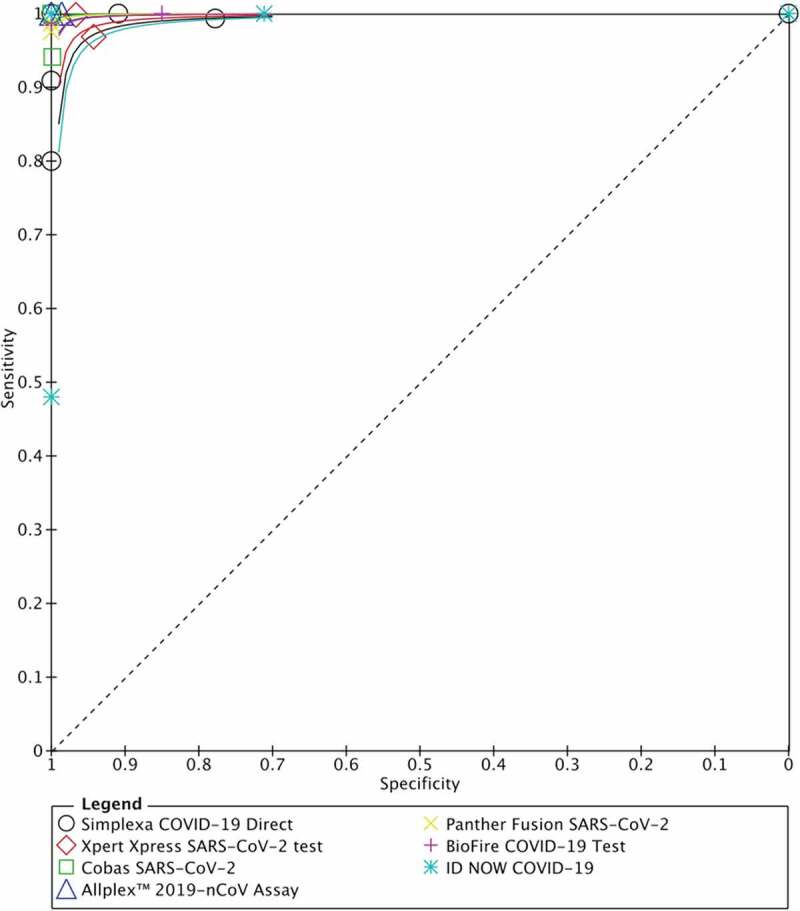
Summary of ROC curves from seven commercial molecular in vitro diagnostic tests for detecting SARS-CoV-2
The quality assessment of the included studies is presented in Figure 4(a,b). None of the studies had a low risk of bias in all four domains of QUADAS-2. Of the total 12 included studies, 7 (58.3%) had unclear risk of bias due to the lack of information regarding selection and randomization of patients/samples, whereas one study (8.3%) exhibited high risk of bias due to the study utilizing convenience sampling method. All studies (100%) scored unclear risk of bias for the reference standard domain because gold standard (culture or sequencing) was not employed. Two studies (16.7%) had high risk of bias for the flow and timing domain, mainly because of the lack of information on the interval time between index test and reference standard.
Figure 4.

Methodological quality of the included studies. (a) Individual assessment and (b) summary
4. Discussion
Accurate diagnostic confirmation of COVID-19 followed by subsequent isolation and tracing is the core approach for mitigating the current spread of SARS-CoV-2 infection. Because molecular diagnostic testing for SARS-CoV-2 is crucial and urgently needed during this challenging period, accelerated development of SARS-CoV-2 nucleic acid amplification test (NAAT) as well as Emergency Use Authorization (EUA) from FDA has been implemented [18,27]. However, recent studies have highlighted the potential problems of diagnostic accuracy from several platforms [15,19].
In this study, we performed a meta-analysis on the performance of seven FDA-approved and commercially available SARS-CoV-2 molecular diagnostic tests. We found that the overall performance of commercial COVID-19 molecular in vitro diagnostic tests was high, with a summary sensitivity of 95.9% (95% CI 93.9–97.2%, I2 = 60.22%) and specificity of 97.2% (95% CI 95.5–98.3%, I2 = 56.66%). However, our study revealed that the ID NOW COVID-19 (Abbott) and the Simplexa COVID-19 Direct exhibited lower sensitivity relative to other platforms, consistent with previously reported studies [8,15,18,19,28–30]. Previously, several studies have also shown reduced sensitivity of both ID NOW COVID-19 and the Simplexa COVID-19 Direct in samples with higher CT values (lower viral load) [8,19]. Since both platforms utilize extraction-free approaches for amplification, a plausible reason for the reduced sensitivity may be due to the potential presence of multiple inhibitory substances or contaminants in the raw sample matrix [31]. The inhibitory effect of the raw samples not only is observable in RT-PCR assays but has also been shown to occur in isothermal amplification assays such as loop-mediated isothermal amplification [32,33].
Other factors, such as LoD (limit of detection), also contribute to differences in comparative performance between kits. Despite ID NOW COVID-19 demonstrating comparable analytical LoD (Table 1), Zhan et al. [28] observed that ID NOW COVID-19 had much higher LoD (20,000 copies/mL) than that claimed (100 copies/mL). Therefore, caution should be considered when using ID NOW COVID-19 for patients with lower viral load, despite having shorter turnaround time.
Limitations of our analysis include variations in the reference standard used (due to lack of concrete gold standard diagnostics), patient characteristics, sampling method and medium, specimen variations, and small sample size of each test. Additionally, the low-quality score reported in some studies may also influence the accuracy of our analyses. Therefore, these findings should be interpreted with caution.
In summary, the lower sensitivity found in the ID NOW COVID-19 (Abbott) and the Simplexa COVID-19 Direct should be taken into consideration by decision makers when deciding on a testing platform, particularly in community setting. Appropriate sample specimens as well as confirmatory testing need to be comprehensively evaluated prior to clinical use. In the end, diagnostic accuracy of COVID-19 commercial diagnostic tests should be weighed against their ease of use and speed.
5. Expert opinion
Early detection of SARS-CoV-2 is crucial in mitigating the COVID-19 pandemic. Several commercial molecular in vitro SARS-CoV-2 detection tests have been introduced with the Emergency Use Authorization (EUA) from the U.S. Food and Drug Administration (FDA). Since the rapid approval process was necessary as a quick response towards demands for diagnostic modalities during the pandemic, post-market surveillance of diagnostics performance becomes even more crucial to ensure optimal field implementation. Therefore, evaluations on the diagnostic performance of commercial molecular in vitro test for SARS-CoV-2 is urgently needed as a guide in choosing the right testing platform for clinical implementation.
Supplementary Material
Funding Statement
This paper was not funded.
Declaration of interest
The authors have no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
Supplementary material
Supplemental data for this article can be accessed here.
References
Papers of special note have been highlighted as either of interest (•) or of considerable interest (••) to readers.
- 1.Ulhaq ZS, Soraya GV.. Anti-IL-6 receptor antibody treatment for severe COVID-19 and the potential implication of IL-6 gene polymorphisms in novel coronavirus pneumonia. Med Clin (Barc) [Internet]. 2020. [cited 2020 August7];155:548–556. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7351402/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ulhaq ZS, Soraya GV. Interleukin-6 as a potential biomarker of COVID-19 progression. Med Mal Infect. 2020;50:382–383. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Elucidating the role of IL-6 as a discriminat for COVID-19 severity.
- 3.Soraya GV, Ulhaq ZS. Interleukin-6 levels in children developing SARS-CoV-2 infection. Pediatr Neonatol. 2020;61:253–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ulhaq ZS, Soraya GV, Fauziah FA. Recurrent positive SARS-CoV-2 RNA tests in recovered and discharged patients. Rev Clin Esp. 2020;220:524–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ulhaq ZS, Soraya GV. The prevalence of ophthalmic manifestations in COVID-19 and the diagnostic value of ocular tissue/fluid. Graefes Arch Clin Exp Ophthalmol Albrecht Von Graefes Arch Klin Exp Ophthalmol. 2020;258:1351–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Soraya GV, Ulhaq ZS. Crucial laboratory parameters in COVID-19 diagnosis and prognosis: an updated meta-analysis. Med Clin (Barc). 2020;155:143–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wirden M, Feghoul L, Bertine M, et al. Multicenter comparison of the Cobas 6800 system with the RealStar RT-PCR kit for the detection of SARS-CoV-2. J Clin Virol Off Publ Pan Am Soc Clin Virol. 2020;130:104573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Basu A, Zinger T, Inglima K, et al. Performance of Abbott ID NOW COVID-19 rapid nucleic acid amplification test using nasopharyngeal swabs transported in viral transport media and dry nasal swabs in a New York City academic institution. J Clin Microbiol [Internet]. 2020. [cited 2020 December14];58. Available from: https://jcm.asm.org/content/58/8/e01136-20. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Describe a lower sensitivity of the Abbott ID NOW COVID-19 in detecting SARS-CoV-2.
- 9.Zhang Y, Wang C, Han M, et al. Discrimination of false negative results in RT-PCR detection of SARS-CoV-2 RNAs in clinical specimens by using an internal reference. Virol Sin [Internet]. 2020. [cited 2020 December14];35:885–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salameh J-P, Bossuyt PM, McGrath TA, et al. Preferred reporting items for systematic review and meta-analysis of diagnostic test accuracy studies (PRISMA-DTA): explanation, elaboration, and checklist. BMJ. 2020;370:m2632. [DOI] [PubMed] [Google Scholar]
- 11.Ulhaq ZS, Soraya GV. Aqueous humor interleukin-6 levels in primary open-angle glaucoma (POAG): a systematic review and meta-analysis. Arch Soc Esp Oftalmol. 2020;95:315–321. [DOI] [PubMed] [Google Scholar]
- 12.Ulhaq ZS. Chemokine IL-8 level in aqueous humor of open-angle glaucoma: a meta-analysis. Arch Soc Esp Oftalmol. 2020;95:114–119. [DOI] [PubMed] [Google Scholar]
- 13.Ulhaq ZS, Soraya GV. Roles of IL-8-251A/T and +781C/T polymorphisms, IL-8 level, and the risk of age-related macular degeneration. Arch Soc Esp Oftalmol. 2021. [DOI] [PubMed] [Google Scholar]
- 14.Ulhaq ZS, Soraya GV, Budu, et al. The role of IL-6-174 G/C polymorphism and intraocular IL-6 levels in the pathogenesis of ocular diseases: a systematic review and meta-analysis. Sci Rep. 2020;10:17453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Procop GW, Brock JE, Reineks EZ, et al. A comparison of five SARS-CoV-2 molecular assays with clinical correlations. Am J Clin Pathol [Internet]. 2020. [cited 2020 December15];153:328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith E, Zhen W, Manji R, et al. Analytical and clinical comparison of three nucleic acid amplification tests for SARS-CoV-2 detection. J Clin Microbiol [Internet]. 2020. [cited 2020 December15];58. Available from: https://jcm.asm.org/content/58/9/e01134-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cradic K, Lockhart M, Ozbolt P, et al. Clinical evaluation and utilization of multiple molecular in vitro diagnostic assays for the detection of SARS-CoV-2. Am J Clin Pathol. 2020;154:201–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lephart PR, Bachman MA, LeBar W, et al. Comparative study of four SARS-CoV-2 nucleic acid amplification test (NAAT) platforms demonstrates that ID NOW performance is impaired substantially by patient and specimen type. Diagn Microbiol Infect Dis. 2021;99:115200. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Describe a lower sensitivity of the Abbott ID NOW COVID-19 compare with other NAAT platforms.
- 19.Rhoads DD, Cherian SS, Roman K, et al. Comparison of Abbott ID NOW, DiaSorin Simplexa, and CDC FDA Emergency Use Authorization methods for the detection of SARS-CoV-2 from nasopharyngeal and nasal swabs from individuals diagnosed with COVID-19. J Clin Microbiol [Internet]. 2020. [cited 2020 December15];58. Available from: https://jcm.asm.org/content/58/8/e00760-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ho H-L, Lin -Y-Y, Wang F-Y, et al. Establishing diagnostic algorithms for SARS-CoV-2 nucleic acid testing in clinical practice. J Chin Med Assoc JCMA. 2020. DOI: 10.1097/JCMA.0000000000000456 [DOI] [PubMed] [Google Scholar]
- 21.Liotti FM, Menchinelli G, Marchetti S, et al. Evaluating the newly developed BioFire COVID-19 test for SARS-CoV-2 molecular detection. Clin Microbiol Infect. 2020;26:1699–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garg A, Ghoshal U, Patel SS, et al. Evaluation of seven commercial RT-PCR kits for COVID-19 testing in pooled clinical specimens. J Med Virol [Internet]. [cited 2020 December15];93:2281–2286. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/jmv.26691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lima A, Healer V, Vendrone E, et al. Validation of a modified CDC assay and performance comparison with the NeuMoDxTM and DiaSorin® automated assays for rapid detection of SARS-CoV-2 in respiratory specimens. J Clin Virol Off Publ Pan Am Soc Clin Virol. 2020;133:104688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wong RC-W, Wong AH, Ho Y-I-I, et al. Performance evaluation of panther fusion SARS-CoV-2 assay for detection of SARS-CoV-2 from deep throat saliva, nasopharyngeal, and lower-respiratory-tract specimens. J Med Virol. 2020;93:1226–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liotti FM, Menchinelli G, Marchetti S, et al. Evaluation of three commercial assays for SARS-CoV-2 molecular detection in upper respiratory tract samples. Eur J Clin Microbiol Infect Dis. 2020;40:269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JH-K, Yip CC-Y, Chan JF-W, et al. Clinical performance of the Luminex NxTAG CoV extended panel for SARS-CoV-2 detection in nasopharyngeal specimens from COVID-19 patients in Hong Kong. J Clin Microbiol. 2020;58. DOI: 10.1128/JCM.00936-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smithgall MC, Scherberkova I, Whittier S, et al. Comparison of Cepheid Xpert Xpress and Abbott ID NOW to Roche Cobas for the rapid detection of SARS-CoV-2. J Clin Virol. 2020;128:104428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhen W, Manji R, Smith E, et al. Comparison of four molecular in vitro diagnostic assays for the detection of SARS-CoV-2 in nasopharyngeal specimens. J Clin Microbiol. 2020;58. DOI: 10.1128/JCM.00743-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thwe PM, Ren P. How many are we missing with ID NOW COVID-19 assay using direct nasopharyngeal swabs? Findings from a mid-sized academic hospital clinical microbiology laboratory. Diagn Microbiol Infect Dis. 2020;98:115123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moore NM, Li H, Schejbal D, et al. Comparison of two commercial molecular tests and a laboratory-developed modification of the CDC 2019-nCoV reverse transcriptase PCR assay for the detection of SARS-CoV-2. J Clin Microbiol [Internet]. 2020. [cited 2020 December16];58. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7383545/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker FM, Hsieh K. Advances in directly amplifying nucleic acids from complex samples. Biosensors (Basel) [Internet]. 2019. [cited 2020 December19];9:117. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6955841/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schellenberg JJ, Ormond M, Keynan Y. Is the glass half full? Extraction-free RT-LAMP to detect SARS-CoV-2 is less sensitive but highly specific compared to standard RT-PCR in 101 samples. medRxiv. 2020;2020.12.07.20239558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dudley DM, Newman CM, Weiler AM, et al. Optimizing direct RT-LAMP to detect transmissible SARS-CoV-2 from primary nasopharyngeal swab and saliva patient samples. medRxiv. 2020;2020.08.30.20184796. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


