Abstract
In view of many European countries and the USA leading to the second wave of COVID-19 pandemic, winter season, the evolution of new mutations in the spike protein, and no registered drugs and vaccines for COVID-19 treatment, the discovery of effective and novel therapeutic agents is urgently required. The degrees and frequencies of COVID-19 clinical complications are related to uncontrolled immune responses, secondary bacterial infections, diabetes, cardiovascular disease, hypertension, and chronic pulmonary diseases. It is essential to recognize that the drug repurposing strategy so far remains the only means to manage the disease burden of COVID-19. Despite some success of using single-target drugs in treating the disease, it is beyond suspicion that the virus will acquire drug resistance by acquiring mutations in the drug target. The possible synergistic inhibition of drug efficacy due to drug-drug interaction cannot be avoided while treating COVID-19 and allied clinical complications. Hence, to avoid the unintended development drug resistance and loss of efficacy due to drug-drug interaction, multi-target drugs can be promising tools for the most challenging disease. In the present work, we have carried out molecular docking studies of compounds from the FDA approved drug library, and the FDA approved and passed phase −1 drug libraries with ten therapeutic targets of COVID-19. Results showed that known drugs, including nine anti-inflammatory compounds, four antibiotics, six antidiabetic compounds, and one cardioprotective compound, could effectively inhibit multiple therapeutic targets of COVID-19. Further in-vitro, in vivo, and clinical studies will guide these drugs' proper allocation to treat COVID-19.
Communicated by Ramaswamy H. Sarma
Keywords: FDA/passed phase-1 inhibitors, COVID-19, multi-targeting compounds, molecular docking, molecular dynamics
Introduction
On March 11, 2020, WHO has announced coronavirus disease 2019 (COVID-19) as a "global pandemic". The disease, caused by SARS-CoV-2, has been unceasingly spreading and becoming the most devastating health crisis of the century (Akriti et al., 2021; Ozkendir et al., 2020). As of November 14 2020, COVID-19 claimed 53,164,803 infections and 1,300,576 deaths worldwide. It has been predicted that the SARS-CoV-2 spread could gain further momentum, in low temperatures and dry weather conditions, during the coming winter months (Mandal & Panwar, 2020; Sarkodie & Owusu, 2020). Instantaneously, recent UK and USA studies have identified a new mutation, known as D614G, in the virus's spike protein that could make it more transmissible with increasing viral load (B Korber et al.; Bette Korber et al., 2020). Altogether, it can be assumed that the second wave of COVID-19 will be riskier than the earlier one. Like all other viruses, it is well within the realm of possibility that SARS-CoV-2 will acquire some new mutations in the drug targets to get around our existing therapeutics. Dealing with the evolutionary trend of drug resistance, drugs interfering with multiple viral proteins' functions all together will have substantial implications in controlling the disease.
Accumulating evidence suggests that COVID-19 patients have varied clinical manifestations, extending from no symptoms and mild symptoms to severe respiratory failure, septic shock, and multiple organ dysfunction (Di Gennaro et al., 2020). The damages caused by uncontrolled immune responses are majorly responsible for the disease's clinical deterioration (Felsenstein et al., 2020). Moreover, some of the severe COVID-19 patients admitted to ICU have been reported to have bacterial co-infections caused by Staphylococcus aureus, Haemophilus influenza, Enterobacteriaceae, and Streptococcus pneumonia (Contou et al., 2020). A meta-analysis of literature studies showed that comorbidities, including hypertension, diabetes, cardiovascular disease, chronic pulmonary disease, and chronic kidney disease, are major risk factors for severe patients (Yang et al., 2020).
As of now, we do not have any clinically approved and targeted therapy for treating COVID-19 (Kheirandish, 2021). However, many repurposed drugs, including antivirals, antimalarials, antibacterial, and immunomodulators, are in different stages of clinical trials for treatment (Lam et al., 2020). Many computational efforts by employing various in silico tools have been conducted searching for drug candidates against COVID-19 (Harismah & Mirzaei, 2020; Khalid et al., 2020; Mirzaei et al., 2020; Mohamed et al., 2021). We also have done a high throughput virtual screening of 3963 natural compounds (NPASS database- http://bidd.group/NPASS/index.php) to identify possible drug candidates for COVID-19, which might inhibit multiple crucial proteins of SARS-CoV-2 (Biswajit Naik et al., 2020).
The current interdisciplinary chemical system biology approach connects chemical biology and system biology (Networking Chemical Biology, 2008) is focused on identifying polypharmacological compounds from FDA approved drug library and FDA approved and passed phase −1 drug library to expedite the discovery of specific drugs for the treatment of COVID-19 and associated health complications. We have selected essential viral and host proteins for our high-throughput virtual screening approach. The proteins include spike protein, spike protein complex with ACE2, PL protein (PLpro), 3 C-like proteinase (3CLpro), RNA dependent RNA polymerase (RDRP), helicase, endoRNAse, 2′-O-ribo methyltransferase (Methyltransferase), 3′-5′ exoribonuclease, and nucleocapsid, which have been reported as prime targets for developing drugs against COVID-19 (Saxena, 2020). High throughput virtual screening of the above-mentioned libraries resulted in twenty best-docked multi-targeting molecules with known anti-inflammatory, antibacterial, cardioprotective, or antidiabetic properties were reported in this work. Further, obtained results were analyzed, and only five potential compounds were selected based on multi-target inhibitory activity. Later, molecular simulation dynamics also confirmed the microscopic interaction between identified SARS-CoV-2 inhibitors and their target proteins. Thus, five specific inhibitors identified under this investigation could be used to treat COVID-19 in combination with associated clinical complications.
Material and methods
Proteins as therapeutic targets, homology modeling, refinement and validation
Ten proteins of SARS-CoV-2 were studied as therapeutic targets for the identification of antagonists. At the time of the experiment, the crystal structure was available for the eight proteins, namely 3 C-like proteinase (PDB ID:6LU7), RNA dependent RNA polymerase (PDB ID: 6M71), EndoRNase (PDB ID: 6VWW), Methyltransferase (PDB ID: 6W61), Spike glycoprotein (PDB ID: 6VXX), Spikeglycoprotein-ACE2 complex (PDB ID: 6LZG), Papain like proteinase (PDB ID: 6W9C) and Nucleocapsid (PDB ID: 6M3M). However, for two target proteins, i.e. helicase (accession no: YP_009725308.1) and exoribonuclease (accession no: YP_009725309.1), homology structures were modeled using SWISS-model server (https://swissmodel.expasy.org/). These models were taken from the previous report published by Naik et al. (2020). In brief, the templates for the homology modeling were selected on two criteria, their sequence identity and sequence coverage, which is estimated by Global Model Quality Estimate (GMQE) and Quaternary Structure Quality Estimate (QSQE) score (Cardoso et al., 2018). Although homology modeling is a prevalent technique to model the tertiary structure of a protein sequence, an accurate estimation of each atom's three-dimensional coordinates in a protein sequence is a challenge (Cheng, 2008). Therefore, the obtained models are needed to be refined to attain their nearest native structure. Here, the modelled proteins were refined using a 3 D-refine server (http://sysbio.rnet.missouri.edu/3Drefine/). Further, a Ramachandran plot was developed for both refined protein models to estimate the number of amino acids present in the allowed, disallowed, and favorable region (Lovell et al., 2003) using RAMPAGE server (http://mordred.bioc.cam.ac.uk/∼rapper/rampage.php).
Selection of antagonist library (ligand selection)
Two libraries, namely FDA approved drug library (https://www.selleckchem.com/screening/fda-approved-drug-library.html) and FDA approved and passed Phase I library (https://www.selleckchem.com/screening/fda-approved-passed-phase-i-drug-library.html) from a Bioactive compounds expert company, Sellekchem were used as ligands.
Molecular docking
Protein preparation
Crystallographic protein structures of eight targets were retrieved using their PDBIDs from the RCSB Protein Data Bank (https://www.rcsb.org/). However, in the case of helicase and exoribonucleases targets, refined homology models were used for docking. All the structures were pre-processed and optimized at neutral physiological condition (pH 7) for the proper protonations of the residues in protein preparation wizard of maestro v11.9 module of Schrodinger suite 2019-1 (Release, 2016) to correct the bond information by adding hydrogen bonds, filling in missing side chains, and deleting water molecules and hetero groups to escape from hindrance in the binding zone.
Ligand preparation
A set of 2,683 compounds from FDA approved drug library, and 2820 compounds from FDA approved and passed phase I library were retrieved in .sdf format for their screening and identification of antagonists against SARS-CoV-2 targets. The compounds were processed by removing salts, adding hydrogens, deprotonation, and neutralization at the physiologically relevant pH 7 to obtain their possible isomers, tautomer, and stereoisomeric forms of three-dimensional conformation. All these actions were performed by the Ligprep v4.7 tool of Schrodinger 2019-1.
Grid generation
Generation of the grid around a receptor/protein is a very important step in a molecular docking study as it allows the ligands to bind with the specific binding region of the receptor or the target proteins. For this study, a three-dimensional grid box was generated of size 20 Å around the active site amino acid residues of each target protein using the glide module v8.2 of Schrodinger. The active site residues were retrieved from the previously reported literature studies (Supplementary Table 1).
Table 1A.
List of selected compounds (along with their role) which showed interaction with therapeutics targets of SARS-Cov-2.
| Name of the Compound | Drug used for the purpose | References | Therapeutic targets of SARS-CoV-2 | Docking Score | MMGBSA Score |
|---|---|---|---|---|---|
| Anti-inflammatory | |||||
| Rutin Hydrate | Anti-inflammatory, Antioxidant, Inhibitor of platelet aggregation, reduced heart risks |
(Ojha et al., 2016) | RDRP | −11.37 | −65.34 |
| Helicase | −9.57 | −87.32 | |||
| Spike Protein | −9.83 | −71.83 | |||
| PLprotein | −11.44 | −72.29 | |||
| 3CLpro | −12.66 | −101.17 | |||
| EndoRNAse | −10.13 | −92.21 | |||
| Methyltransferase | −9.07 | −70.4 | |||
| Nucleocapsid | −13.76 | −100.03 | |||
| Silymarin | Anti-inflammatory Hepatoprotective, antioxidant, anticancer, and cardioprotective activities | (Baliga et al., 2014) | Exoribonucleases | −6.14 | −77.75 |
| Spike Protein | −5.88 | −58.7 | |||
| Spike protein complex with ACE2 | −5.61 | −64.41 | |||
| RDRP | −5.9 | −41.01 | |||
| EndoRNAse | −7.08 | −62.4 | |||
| Helicase | −7.82 | −113.86 | |||
| Nucleocapsid | −10.02 | −60.65 | |||
| Quercetin | Antioxidant and anti-aging agent and anti-carcinogenic | (Li et al., 2016) | 3CLpro | −9.07 | −70.4 |
| Helicase | −13.76 | −100.03 | |||
| Spike Protein | −12.66 | −101.17 | |||
| Spike protein complex with ACE2 | −9.57 | −87.32 | |||
| RDRP | −5.35 | −48.75 | |||
| EndoRNAse | −6.7 | −50.99 | |||
| Exoribonucleases | −6.55 | −40.73 | |||
| Mitoxantrone 2Hcl | Antineoplastic agent used to treat multiple sclerosis, Immunosuppressant | (Fox, 2004) | Helicase | −8.35 | −98.48 |
| Spike Protein | −6.15 | −48.46 | |||
| Spike protein complex with ACE2 | −8.11 | −68.28 | |||
| RDRP | −8.13 | −77.04 | |||
| EndoRNAse | −6.66 | −71.5 | |||
| Methyltransferase | −6.69 | −65.44 | |||
| Nucleocapsid | −9.5 | −63.22 | |||
| Luteolin | Anti-inflammatory and Neuroprotective agent | (Nabavi et al., 2015) | Helicase | −4.52 | −73.99 |
| 3CLpro | −7.50 | −61.63 | |||
| Spike protein complex with ACE2 | −5.84 | −46.31 | |||
| PLprotein | −4.80 | −33.54 | |||
| EndoRNAse | −7.18 | −61.06 | |||
| Methyltransferase | −3.87 | −42.55 | |||
| Aloin | Anti-inflammatory used to treat acute Influenza. Antiarrhythmic, Anti-carcinogenic | (Huang et al., 2019) | Spike Protein | −7.52 | −46.36 |
| Spike protein complex with ACE2 | −6.19 | −52.55 | |||
| RDRP | −7.47 | −43.78 | |||
| EndoRNAse | −10.25 | −50.69 | |||
| Methyltransferase | −7.13 | −27.61 | |||
| Nucleocapsid | −8.24 | −50.03 | |||
| Morin Hydrate | Anti-inflammatory, antiproliferative | (Choudhury et al., 2017) | 3CLpro | −6.92 | −58.7 |
| Helicase | −4.95 | −75.72 | |||
| Spike Protein | −3.13 | −37.39 | |||
| Spike protein complex with ACE2 | −6.15 | −45.3 | |||
| EndoRNAse | −6.32 | −44.79 | |||
| Exoribonucleases | −6.56 | −52 | |||
| Kaempferol | Anti-colorectal cancer (CRC) drug | (Riahi-Chebbi et al., 2019) | 3CLpro | −7.48 | −43.88 |
| Spike Protein | −3.58 | −40.92 | |||
| Spike protein complex with ACE2 | −5.49 | −43.44 | |||
| Exoribonucleases | −5.23 | −41.17 | |||
| Nucleocapsid | −6.1 | −34.75 | |||
| Madecassoside | Anti-inflammatory, wound healing, and antioxidant | (Li et al., 2007) | Helicase | −9.62 | −131.31 |
| RDRP | −10.05 | −36.38 | |||
| Antibiotics | |||||
| Amikacin Hydrate | Antibiotic | (Torres et al., 2018) | Helicase | −7.71 | −53.43 |
| Spike Protein | −9.28 | −66.72 | |||
| Spike protein complex with ACE2 | −13.08 | −71.28 | |||
| EndoRNAse | −10.37 | −82.08 | |||
| Methyltransferase | −7.73 | −71.19 | |||
| Exoribonucleases | −9.61 | −88.19 | |||
| Geneticin | Aminoglycoside antibiotic | (Zufferey et al., 2012) | Methyltransferase | −5.37 | −59.72 |
| Spike protein complex with ACE2 | −7.19 | −63.84 | |||
| RDRP | −10.25 | −61.95 | |||
| EndoRNAse | −6.91 | −76.69 | |||
| Spike Protein | −4.4 | −44.03 | |||
| Netilmicin Sulfate | Antibiotic | (Craig et al., 1983) | Spike Protein | −2.96 | −50.37 |
| Spike protein complex with ACE2 | −9.85 | −86.57 | |||
| Methyltransferase | −6.86 | −71.81 | |||
| Hygromycin B | Antibiotic (kills bacteria, fungi and higher eukaryotic cells) | (Kirst & Allen, 2007) | EndoRNAse | −8.12 | −94.09 |
| Spike protein complex with ACE2 | −10.97 | −68.83 | |||
| RDRP | −9.98 | −63.5 | |||
| Spike Protein | −3.71 | −37.87 | |||
| Nucleocapsid | −10.1 | −55.66 | |||
| Antidiabetics | |||||
| Acarbose | Antidiabetic (Type 2 Diabetes Mellitus) | (Chiasson et al., 2002) | Spike protein complex with ACE2 | −11.07 | −74.88 |
| PLprotein | −9.74 | −55.2 | |||
| RDRP | −10.99 | −60.78 | |||
| EndoRNAse | −10.67 | −73.16 | |||
| Exoribonucleases | −10.67 | −104.11 | |||
| Nucleocapsid | −14.58 | −81.08 | |||
| Apigenin | Anti-diabetes, amnesia and Alzheimer's disease, depression and insomnia, Anti-cancer, etc. | (Salehi et al., 2019) | 3CLpro | −6.86 | −49.99 |
| Spike protein complex with ACE2 | −4.38 | −34.62 | |||
| Methyltransferase | −3.98 | −31.76 | |||
| Exoribonucleases | −4.07 | −48.02 | |||
| Nucleocapsid | −5.37 | −40.02 | |||
| Hesperidin | Antihyperlipidemic, Cardioprotective, Antihypertensive, Antidiabetic a | (Zanwar et al., 2014) | 3CLpro | −10.68 | −91.32 |
| Helicase | −7.71 | −67.17 | |||
| PL protein | −8.47 | −52.6 | |||
| Nucleocapsid | −11.68 | −84.31 | |||
| Neohesperidin | Diabetes Mellitus | (Wu et al., 2017) | Spike Protein | −6.83 | −65.7 |
| RDRP | −9.95 | −87.01 | |||
| EndoRNAse | −8.69 | −70.83 | |||
| Nucleocapsid | −10.84 | −75.55 | |||
| Troxerutin | Antidiabetic, Chronic Venous Insufficiency | (Zhang et al., 2013) | RDRP | −10.01 | −72.04 |
| EndoRNAse | −10.31 | −88.41 | |||
| Exoribonucleases | −10.75 | −71 | |||
| Nucleocapsid | −13.0 | −91.87 | |||
| Notoginsenoside R1 | Diabetic nephropathy, cardiovascular disease, cerebral vascular disease, and liver dysfunction | (Zhang et al., 2018) | RDRP | −9.73 | −56.44 |
| EndoRNAse | −11.27 | −93.95 | |||
| Cardiovascular Disease | |||||
| Salvianolic Acid B | Anti-diabetic, Angina pectoris, and other heart diseases, anti-inflammatory, antioxidant, hepatoprotective, neuroprotective, antidepressant | (Ma et al., 2019; Zhuang et al., 2012) | Helicase | −6.6 | −119.88 |
| RDRP | −8.83 | −61.9 | |||
| EndoRNAse | −10.08 | −78.14 | |||
| Nucleocapsid | −11.24 | −86.25 | |||
Virtual screening
All the ligands were docked against each target protein for the virtual screening purpose using the Glide tool v8.2 of Schrodinger 2019-1 (Friesner et al., 2004). A total of 6647 ligands and positive controls of respective proteins were screened in a 3-step rotational workflow for virtual screening i.e. High Throughput Virtual Screening (HTVS) to Standard Precision (SP) to Extra Precision (XP). For instance, the ligands were first screened with HTVS, output screened ligands from HTVS filter were put into the filter of SP and again the successfully screened ligands from SP filter were put forwarded to XP docking. The HTVS and SP mode of screening utilize the same scoring function to eliminate false positives with a difference of speed and accuracy, whereas XP utilizes extensive sampling protocols and advanced scoring to give higher enrichment (Pandey et al., 2015).
MM-GBSA: Estimation of binding free energy
Prime Molecular Mechanics Generalized Born model and Solvent Accessibility (MM-GBSA) module of Schrodinger 2019-1 was used to calculate the binding free energy (ΔG bind) between the target proteins and ligands. For this action, glide pose viewer complexes are taken as input files with the SVGB solvation model, and the OPLS3 force field was set in the setting tab of prime MM-GBSA module of Schrodinger suite (Lyne et al., 2006).
Pharmacokinetics and physiochemical properties of ligands
To study the druggable characteristics of ligands, we used the Qikprop v5.9 tool of Schrodinger, which reveals the absorption, distribution, metabolism, and excretion properties of individual ligands for the evaluation of suitable drugs. Furthermore, we have also analyzed the toxicity profile of compounds using the most reliable webserver pkCSM (http://biosig.unimelb.edu.au/pkcsm/).
ROC plot analysis
For the validation of molecular docking protocol, here we performed the enrichment study that estimates the Receiver Operating Characteristics (ROC), Area Under Curve (AUC) and Boltzmann-Enhanced Discrimination of Receiver Operating Characteristic (BEDROC) in the form of different metric values (Truchon & Bayly, 2007). In this study, the effectiveness of SP and XP mode of virtual screening protocol was observed through a statistical receiver operating characteristics curve that shows a significant difference between the active compounds and the decoy (false positive/inactive) molecules (Fawcett, 2006). The ROC plot was represented by 1-specificity on the X-axis representing the false-positive molecules and sensitivity on Y-axis representing the active compounds. For this action, an active-decoy docked file in pose viewer (pv. maegz) format for SP and XP mode of docking was given as input, and all these processes were done by the enrichment calculator tool provided by Schrodinger suite. Furthermore, AUC and BEDROC values were also calculated for enrichment study of the SP and XP docking.
Molecular dynamics simulation
We performed Molecular dynamics simulation for the five complexes A) RDRP-Rutinhydrate B) Exo-Silymarin C) Helicase-Luteolin D) Methyltransferase-Amikacin hydrate E) Methyltransferase-Geneticin using the Particle Mesh Ewald Molecular Dynamics (PMEMD) (Kaus et al., 2013) module of AMBER12 (Case et al., 2010) software package. The partial charges and force field parameters for the ligands were generated automatically using the antechamber program (Wang et al., 2006) in AMBER12. Force field parameters were given for the ligand and the protein using the xleap module in AMBER12. General Amber force field (GAFF) (Wang et al., 2004) and AM1-BCC (Jakalian et al., 2002) charges were used for ligands, while AMBERff99SB force field was used for the protein. Using xleap and antechamber module, we obtained the initial coordinate and the topology files for all the complex systems. The resultant initial structure of the individual complex system was solvated using TIP3P (Jorgensen et al., 1983) water model, the water box size of 10 Å from the solute in the x, y, and z coordinates, and then counter-ions were added to neutralize the complex. The potential energy of initial structures was minimized using the 1000 steps of steepest descents followed by 2000 steps of the conjugate gradient method. We fixed the complex molecule using harmonic constraints (excluding the water molecules) with force constant of 30 kcal mol−1Å2 to overcome the bad contacts between water molecules and the complex. Again minimization was carried out without any constraints for 3000 cycles (1000 cycles of steepest descents and 2000 cycles of the conjugate gradient). The complex systems were then slowly heated from 0 to 300 K with weak 20 kcal mol−1Å−2 constraints at 40 ps in NVT condition. This allows the structure to undergo slow relaxation. In MD simulation, using geometrical tolerances of 5 × 10-4 Å, the SHAKE constraints were imposed on all covalent bonds involving hydrogen atoms, with a time set to 2 fs. Then, equilibration and long production steps were performed in NPT condition (T = 300 K and P = 1atm) and temperature regulation was achieved with the help of Berendsen weak coupling method (0.5 ps time constant for heat bath coupling and 0.2 ps pressure relaxation time) (Berendsen et al., 1984) (Ryckaert & Bellemans, 1978). Finally, we carried out 100 ns long NPT MD simulation using a heat bath coupling time constant of 1 ns to analyze the conformational dynamics of five complexes. The Analysis of structural convergence properties such as Root Mean Square Deviation (RMSD), Root Mean Square Fluctuation (RMSF), the radius of gyration (Rg) were carried out using cpptraj (Roe & Cheatham III, 2013) in AMBER12. For inspecting the 3 D structure of the molecule, we used UCSF Chimera (Pettersen et al., 2004) and VMD (Humphrey et al., 1996). And for the solvation free energy calculations of the complexes, we have used 3 D RISM theory (Hirata, 2003).
Results
SARS-CoV-2 therapeutic target modelling & retrieval of crystal structure and antagonists library compounds
The models for two therapeutic targets, i.e. helicase and exoribonuclease of SARS-CoV-2, were modelled using template sequences of 6jyt.2.A & 5nfy.1.A respectively. The targets were sharing 97.95% and 94.07% sequence identity with the chosen templates respectively. Indeed, it is known that the homology model is considered to be excellent and reliable if the template is chosen, and the target shares minimum of 30% sequence identity (Xiang, 2006).
Further, the homology model's quality was estimated by the QMEAN score (Benkert et al., 2009). The GMQE score ranges between 0 to 1, which means the higher the score, the higher the model's reliability and accuracy for the used template. In our case, the value for GMQE score was 0.98 for both targets, which denotes higher accuracy. Next, the models for both target enzymes were validated before and after refinement using the Ramachandran plot. Where Ramachandran plot confirmed that models in .pdb format after refinement are stable and better suited for docking with ligands, i.e. for helicase (non-refined % of favored residues 84.34% whereas refined % of favored residues 88.60%) and exoribonuclease (non-refined % of favored residues 89.44% whereas refined % of favored residues 93.10%) both, rest eight therapeutic targets were retrieved using their PDBIDs from the RCSB Protein Data Bank (https://www.rcsb.org/). Structural based drug screening is a method based upon the libraries of small compounds and targets active residues. Selleckchem has a collection of numerous libraries, which also includes FDA approved drug library (2683 compounds) and FDA approved and passed Phase I library (2820 compounds). Repurposing or already FDA approved drugs is the only secure and rapid way to meet the urgent requirement for ongoing pandemic treatment. We selected both FDA approved drug library, and FDA approved and passed Phase I library for our experiments from Selleckchem.
Computational virtual screening of FDA compounds against potential therapeutic targets of SARS-CoV-2
To find out molecules of therapeutic effects for inflammation, bacterial infection, diabetes, cardiovascular diseases, and kidney-related disorders and that can hit multiple therapeutic targets of SARS-CoV-2, molecular docking studies of compounds from FDA approved drug library, and FDA approved and passed phase -I drug library was performed against the target proteins including spike protein, spike protein complex with ACE2, PL protein, 3CLpro, RdRp, helicase, endoRNAse, methyltransferase, exoribonucleases, and nucleocapsid. The top 20 molecules showing better binding interactions with multiple proteins of SARS-CoV-2 compared to control compounds depicted in Table 1B and are listed in Table 1A. These compounds include nine anti-inflammatory compounds, four antibiotics, six antidiabetic compounds, and one cardioprotective compound. Among those compounds, four anti-inflammatory compounds, two antibacterial compounds, four antidiabetic compounds, and one therapeutic compound related to cardiovascular diseases are discussed in detail. Moreover, based on their docking scores with multiple therapeutic targets, three anti-inflammatory agents and two antibiotics are selected for molecular dynamic analysis.
Table 1B.
List of compounds used as positive control against the therapeutic targets of SARS-CoV-2.
| Name of the compound used as control | References | Therapeutic targets of SARS-CoV-2 | Docking Score | MMGBSA Score |
|---|---|---|---|---|
| TG-0205221-102285029 | (Jin et al., 2020) | 3CLpro | −6.918 | −71.39 |
| Tideglusib | −4.349 | −50.28 | ||
| Iodobananin | (Tanner et al., 2005) | Helicase | −4.877 | −40.25 |
| MLS001181552 | −2.248 | −38.09 | ||
| Zorubicin | (Vincent et al., 2005) | Spike Protein | −5.339 | −67 |
| SSAA09E2 | −2.718 | −47.49 | ||
| Chloroquine | −1.179 | −31.2 | ||
| Hydroxychloroquine | (Samarth & Kirk, 2020) | Spike protein complex with ACE2 | −7.693 | −67.56 |
| Azithromycin | −6.808 | −62.72 | ||
| Cinacalcet | (Rimanshee et al., 2020) | PL protein | −2.007 | −36.63 |
| Biltricide | −1.572 | −41.41 | ||
| IDX-144 | (Elfiky, 2020) | RDRP | −8.225 | −63.65 |
| Remdesivir | −5.068 | −41.77 | ||
| Setrobuvir | −2.85 | −45.81 | ||
| Idarubicin | (Chandra et al., 2020) | EndoRNAse | −4.9 | −71.14 |
| Glisoxepide | −1.79 | −56.6 | ||
| Raltegravir | (Khan et al., 2020) | Methyltransferase | −3.14 | −39.82 |
| Dolutegravir | −3.117 | −46.74 | ||
| Ritonavir | (Narayanan & Nair) | Exoribonucleases | −4.98 | −67.52 |
| Favipiravir | −4.062 | −17.85 | ||
| Gallocatechin | (Roh, 2012) | Nucleocapsid | −9.302 | −77.23 |
| Catechin | −8.933 | −74.46 |
Anti-inflammatory compounds against multiple therapeutic targets of SARS-CoV-2
Rutin hydrate
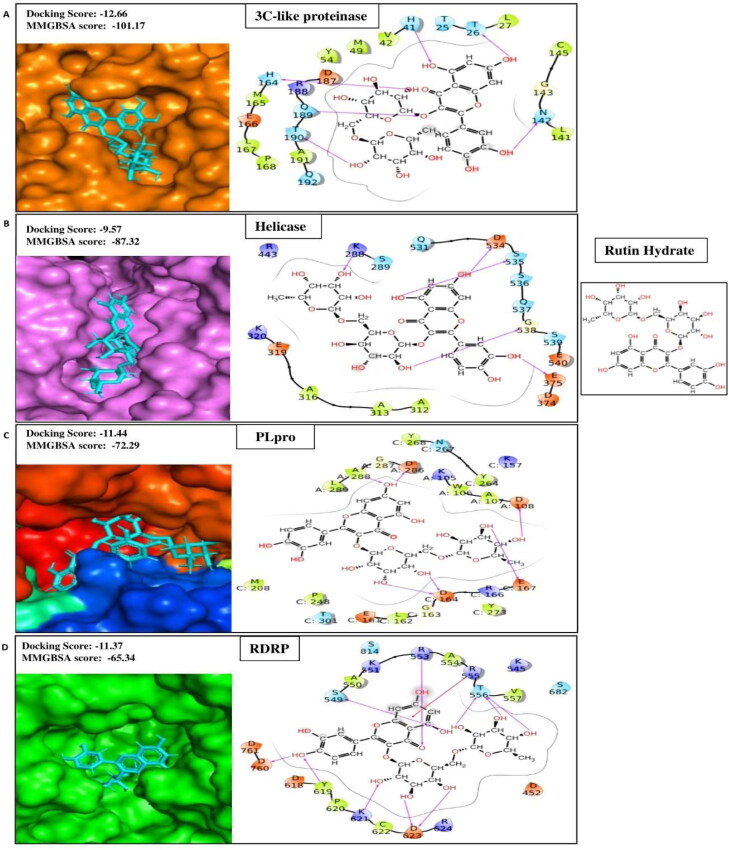
Rutin is a water-soluble flavone glycoside converted into quercetin in the bloodstream (Morand et al., 2000). It is widely found in buckwheat, apple skin, red wine, tea, and coffee (Kim et al., 2005; Kreft et al., 1999). It is reputed for its potent antioxidant capacity and has been reported effective against oxidative-stress-mediated diseases (López-Revuelta et al., 2006). Besides, rutin possesses several pharmacological properties, including analgesic, anticarcinogenic, cardioprotective, neuroprotective, vasoprotective, and cytoprotective activities (Ganeshpurkar & Saluja, 2017). Rutin works as a potential anti-inflammatory agent in many ways. It increases prostaglandins production, decreases histamine synthesis (Hussain et al., 2009; Kaithwas & Majumdar, 2010), inhibits the migration and/or degranulation of neutrophils (Selloum et al., 2003), blocks lipoxygenase pathway (Kurisawa et al., 2003), and suppresses proinflammatory cytokines production (S.-w. Wang et al., 2012). The antimicrobial and antiviral activities of rutin have been extensively studied against various bacteria, fungi, and viruses (Ganeshpurkar & Saluja, 2017). Computational studies of different research groups have revealed helicase (Wu et al., 2020), spike protein (Kadioglu et al., 2021), main protease (Mpro) (Das et al., 2020; Huynh et al., 2020), RdRp (Altayeb et al., 2020), envelope protein (E) (Bhowmik et al., 2020), methyltransferase (Kadioglu et al., 2021) as the therapeutic targets of rutin for SARS-CoV-2. In our molecular docking studies with ten therapeutic targets of SARS-CoV-2, rutin occupied the first rank by displaying better docking scores such as −11.37, −9.57, −9.83, −11.44, −12.66, −10.13, −9.07 and −13.76 for RdRp, Helicase, Spike protein, PL protein, 3CLpro, EndoRNAse, Methyltransferase, and Nucleocapsid respectively (Table 1A) than the reported positive inhibitors (Table 1B). The ligand-binding patterns of the docked complexes are displayed in Figure 1A, showing Rutin Hydrate strongly binds with the active site pocket of each target protein. For instance, it interacts through strong hydrogen bonds with the active site pocket residues of 3CLpro (Thr26, His41, Asn142, His164, Gln189 and Thr190), helicase (Lys288, Glu375, Asp534, Ser535 and Gly538), PLpro (chainA:Asp108, Asp286 and Ala288; chainC:Asp164 and Glu167), RDRP (Ser549, Arg553, Thr556, Tyr619, Lys621, Asp624 and Asp760) and Pi-cation inetractin with Arg555 of RDRP. Our findings have shown strong agreement with molecular docking studies reported by various research groups, besides, our studies have revealed the binding affinity of rutin with two more therapeutic targets including Nucleocapsid and EndoRNAse of SARS-CoV-2. In this sense, it can be predicted that the multi targeting property of rutin will eventually potentiate its synergistic antiviral effect against SARS-CoV-2.
Figure 1A.
Representation of protein-ligand interacting residues of compound Rutin hydrate with its best four protein targets i.e. (A) 3 C-like proteinase, (B) Helicase, (C) Papine-like proteinase (PLpro) and (D) RNA Dependent RNA Polymerase (RDRP).
Silymarin
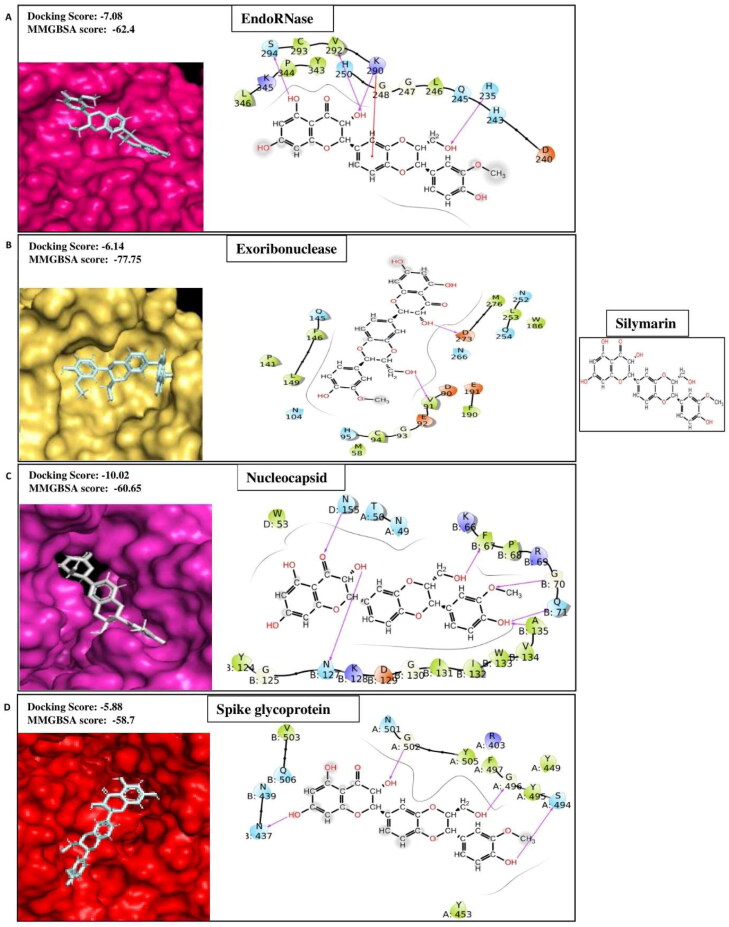
Silymarin is a flavonolignan, commonly extracted from milk thistle fruits and seeds (Silybum marianum) (Shibano et al., 2007). It possesses widespread therapeutic applications for diseases including cancers, cardiotoxicity, nephrotoxicity, diabetes, Parkinson's disease, Alzheimer's disease, hepatic and lung disease, and prostate disease (Karimi et al., 2011; Milić et al., 2013). The anti-inflammatory effect of silymarin is mediated in several ways, including suppression of the signaling pathway of nuclear factor-kB (NF-kB) (Kang et al., 2004), cytokine production, and T cell proliferation (Gharagozloo et al., 2013). The potent antiviral activities against a wide range of viruses, including hepatitis B virus, hepatitis C virus, influenza virus, dengue virus, Chikungunya virus, and human immunodeficiency virus, have been reported (Liu et al., 2019). Recently, in silico studies have revealed that the compound has an inhibitory effect on S-glycoprotein and Mpro of SARS-CoV-2 (Ubani et al., 2020). Consequently, in our search of compounds having the multi-target ability for SARS-CoV-2, we have found that silymarin possesses better docking scores such as −7.82, −5.9, −5.88, −7.08, −6.14, and −10.02 for Helicase, RdRp, Spike protein, EndoRNAse, Exoribonucleases, and Nucleocapsid respectively (Table 1A) compared to the reported positive inhibitors (Table 1B). The ligand-binding patterns of the docked complexes are displayed in Figure 1B. Silymarin interacts by making hydrogen bond with the active site residues of EndoRNAse (His235, Lys290, Val292 and Ser 294), Exoribnuclease (Val91 and Asp273), nucleocapsid (chainB:Phe67, Gly70, Gln71, Asn127 and Ala135; chainD:Asn155), Spike glycoprotein (chainA:Ser494, Gly496 and Gly502; chainB:Asn437) and through Pi-cation interaction with Lys290 of EndoRNAse.
Figure 1B.
Showing protein-ligand contacts between the compound Silymarin and its best four protein targets i.e. (A) EndoRNase, (B) Exoribonuclease, (C) Nucleocapsid and (D) Spike glycoprotein.
This study suggests the antiviral activity of silymarin against multiple essential viral proteins of SARS-CoV-2. It further supports the recently reviewed hypothesis of silymarin's dual targeting ability, i.e. against both essential viral proteins and host cytokine storm (Bosch-Barrera et al., 2020).
Luteolin
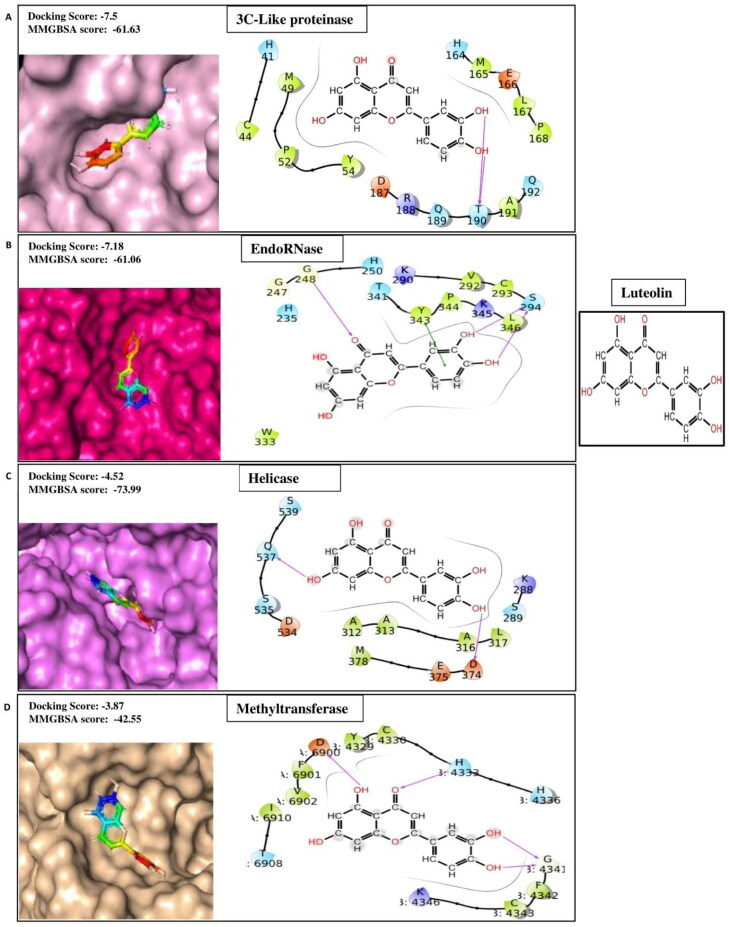
Luteolin is a flavone compound present as glycosides in many vegetables and fruits, including celery, pepper, thyme, and broccoli (Lopez-Lazaro, 2009). It possesses a broad range of pharmacological properties such as anticancer, neuroprotective, antioxidant, and immunomodulatory capacities ( Imran et al., 2019; Park & Song, 2019; Xagorari et al., 2001 ). The current advancements of in silico, in vitro, and in vivo studies reveal that luteolin exhibit strong anti-inflammatory activities via regulation of MAPK in the AP-1 (activator protein −1) pathway, Src in the NF-κB pathway, and SOCS3 in the STAT3 (signal transducer and activator of transcription 3) pathway (Aziz et al., 2018). Because of these well-studied profiles of actions, it has been proposed that luteolin may have a therapeutic role in COVID-19 treatment by attenuating the secretion of proinflammatory cytokine and chemokines from pulmonary mast cells (Theoharides, 2020). In addition to that, luteolin has been shown to have potent antiviral activity against the Japanese encephalitis virus (Fan et al., 2016), influenza A virus (Yan et al., 2019), and HIV-1 (Mehla et al., 2011). Relevant computational docking studies on the binding affinity of luteolin against therapeutic targets of SARS-CoV-2, including 3CL protease, PLpro, RdRp, and spike proteins, have been reported (Pandey et al., 2020; Yu et al., 2020 ). In our SARS-CoV-2 multi-target based drug discovery approach, luteolin has shown binding affinities with 3CLpro, Helicase, Spike protein complex with ACE2, PLprotein, EndoRNAse, and Methyltransferase with better docking scores −7.50, −4.52, −5.84, −4.80, −7.18, and −3.87 respectively (Table 1A) as compared to the control molecules (Table 1B). The ligand-binding patterns of the docked complexes are displayed in Figure 1C. For example, Luteolin get binds with the active site pocket residue Thr190 of 3CLPro; Gly248 and Ser294 of EndoRNAse; D374 and Gln537 of Helicase; chainA:Asp6900, chainB:His4333 and Gly4341 of methyltransferase through strong hydrogen bonds and by making Pi-Pi skacking with Tyr343 of EndoRNAse.
Figure 1C.
2 D-interaction diagram representing protein-ligand contacts of Luteolin with its best four targets i.e. (A)3c-like proteinase (B) EndoRNase (C) Helicase (D) Methyltransferase.
Aloin
Aloin is abundantly found in the leaf exudates of various Aloe species such as Aloe vera. Aloin and its semisynthetic derivatives have been reported to possess invaluable medicinal characteristics, including anti-inflammatory (Park et al., 2011), antiproliferative (Nićiforović et al., 2007), antioxidant (Tian & Hua, 2005), and anticancer properties (Kumar et al., 2010). Nevertheless, it is an anthraquinone glycoside, and its antiviral activity has recently been documented (Huang et al., 2019). In the ongoing COVID-19 pandemic situation, the binding affinity of aloin for therapeutic targets such as transmembrane serine protease 2 (Tmprss2)(Coban, 2020), ACE2(da Silva Antonio et al., 2020), Mpro (Das & Singha Roy, 2020) is evaluated. Here, our results yielded the higher binding affinity represented by docking scores of aloin as −7.52, −6.19, −7.47, −10.25, −7.13, and −8.24 for Spike Protein, Spike protein complex with ACE2, RDRP, EndoRNAse, Methyltransferase, and Nucleocapsid respectively (Table 1A) compared to the positive controls (Table 1B).
Antibiotics against multiple therapeutic targets of SARS-CoV-2
Amikacin hydrate
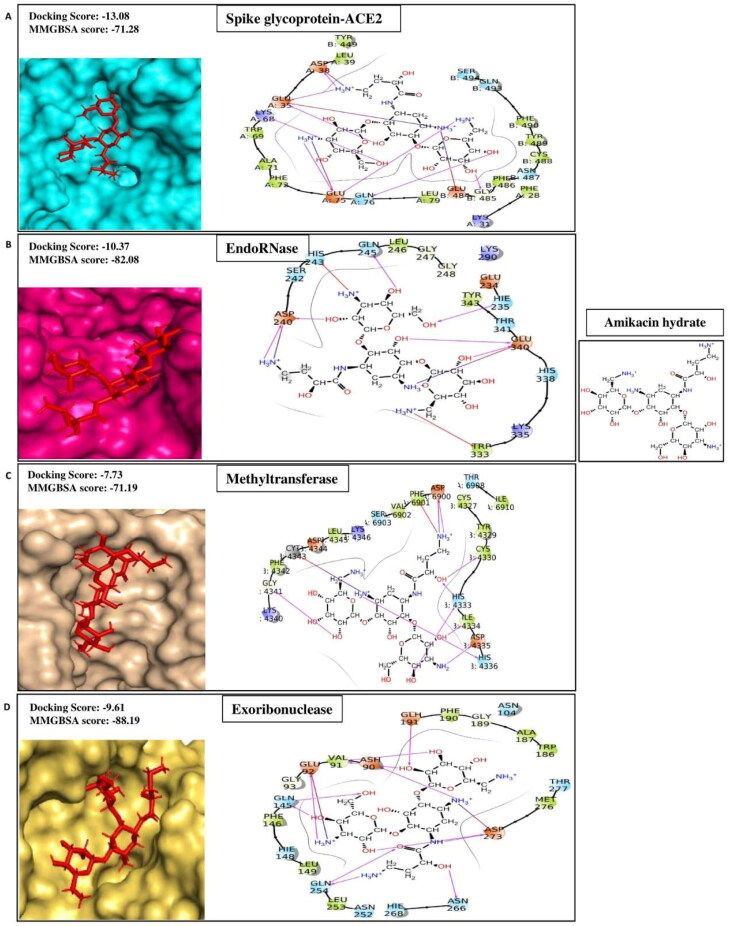
Amikacin hydrate is a semisynthetic aminoglycoside antibiotic derived from Kanamycin A. It is broad specific, and effectively used in treating a wide range of infections caused by gram-negative bacteria, including pneumonia, meningitis, sepsis, intra-abdominal infections, and urinary tract infections. It is also used to treat mycobacterial, Nocardial, and neonatal infections (Ramirez & Tolmasky, 2017). It inhibits the translocation process of bacterial translation by binding to the A site of the16S RNA of the 30S ribosomal subunits (Kondo et al., 2006). In our high throughput virtual screening experiment against multiple therapeutic targets of SARS-CoV2, we have identified amikacin hydrate has higher affinities signified by docking scores −7.71, −9.28, −13.08, −10.37, −7.73, and −9.61 for Helicase, Spike Protein, Spike protein complex with ACE2, EndoRNAse, Methyltransferase, and Exoribonucleases respectively (Table 1A) than the positive controls (Table 1B). The ligand-binding patterns of the docked complexes are displayed in Figure 1D, in which Amikacin hydrate docked in the binding pocket of target proteins. It interacts through strong hydrogen bonds with Spike-ACE2 (chainA:Glu35, Asp38, Lys68, Glu75, Gln76; chainB:Gly485), EndoRNase (His235, Asp240, Gln245 and Glu340), methyltransferase (chainA:Asp6900; chainB:Cys4330, His4333, Ile4334, Asp4335, His4336 and Gly4341), Exoribinuclease (Asp90, Glu92, Glu191 and Asp273); by making salt bridge with Spike-ACE2 (chainA:Glu35, Asp38, Glu75; chainB:Glu484), EndoRNAse (Asp240 and Glu340), methyltransferase (chianA:Asp6900; chainB:Cys4343), Exoribonuclease (Glu92 and Asp273); and through Pi-cation interaction with His243, Trp333 of EndoRNAse and chainA:Phe6901 of methyltransferase. Our results agree with the studies of Vijayan et al., 2021 and Shaankar et al., 2020 where they have shown the significant affinity of amikacin hydrate with EndoRNAse and Methyltransferase, respectively (Shankar et al., 2020; Vijayan & Gourinath, 2021).
Figure 1D.
Representing protein-ligand contacts of Amikacin hydrate with its best four protein targets i.e. (A) Spike glycoprotein-ACE2 (B) EndoRNase (C) Methyltransferase (D) Exoribonuclease.
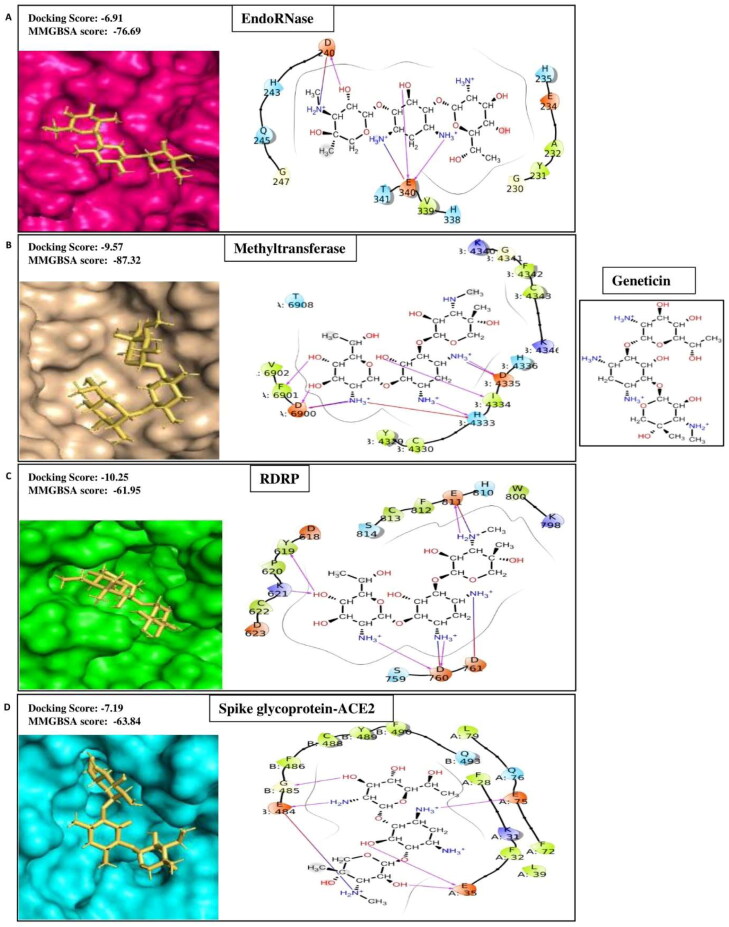
Geneticin
Geneticin, also known as gentamycin, is another aminoglycoside antibiotic. It induces non-functional or truncated proteins in bacteria by binding to 16 s rRNA of 30S ribosomal subunit and misreading genetic code (Beganovic et al., 2018). It is inexpensive and successfully used to treat a wide range of infections, including neonatal sepsis, gynecological infection, nasal infection, ear diseases, eye diseases, diabetic foot infections, and surgical site infection (Chen et al., 2014). Moreover, inhalational gentamycin therapy has been shown to be effective in treating lung infections such as pneumonic plague in the mouse model (Gur et al., 2018). Here, we have demonstrated that geneticin, along with its antibacterial property, retains binding affinities signified by docking scores −5.37, −7.19, −10.25, −6.91, and −4.4 for Methyltransferase, Spike protein complex with ACE2, RdRp, EndoRNAse and Spike Protein of SARS-CoV2 respectively (Table 1A). The ligand-binding patterns of the docked complexes are displayed in Figure 1E. Geneticin interacts within the binding pocket of EndoRNAse through strong hydrogen bond and salt bridge with Asp240 and Glu340. In the active site pocket of methyltransferase, geneticin binds with chianA:Asp6900, Phe6901 and chainB:His4333, Ile4334 and Asp4335 through hydrogen bond; chainA:Asp6900 and chainB:Asp4335 through salt bridges; and with chainB:His4333 by forming Pi-cation interaction. In RDRP binding site, geneticin forms hydrogen bonds with Tyr619, Lys621, Asp760, Glu811 and salt bridges with Asp761 and Glu811. Geneticin occupied inside the active site pocket of spike-ACE2 by forming hydrogen bonds with amino acids of chainA: Glu35 and Glu75, chainB: Glu484 and Gly485. Thus, our data suggest that aerosolized geneticin's inhalation will be clinically relevant in treating COVID-19 infection and associated respiratory bacterial infections.
Figure 1E.
Displaying protein-ligand contact residues of Geneticin with its best four protein targets i.e. (A) EndoRNase, (B) Methyltransferase, (C) RNA dependent RNA polymerase (RDRP) and (D) Spike glycoprotein-ACE2.
Netilmicin sulfate
Netilmicin sulfate is another semisynthetic aminoglycoside antibiotic derived from sisomicin. It is used for the treatment of P. aeruginosa infections in cystic fibrosis (Prayle & Smyth, 2010), neonatal sepsis (Bacopoulou et al., 2009), and Septic Burn Patients (Munster, 1979). It has also been demonstrated that the entrapment of netilmicin sulfate in poly(DL-lactide-co-glycolide) (PLGA) hydrophobic matrix enhanced its bioavailability, reduce dosing frequency, and side effects (Kolate et al., 2015). In our molecular docking study designed to identify antibiotics with inhibitory actions against multiple therapeutic targets of SAR-CoV-2, we found that netilmicin sulfate has better docking scores such as −2.96, −9.85, and −6.86 for Spike Protein, Spike protein complex with ACE2, and Methyltransferase, respectively (Table 1A) as compared to the positive control (Table 1B).
Antidiabetic compounds against multiple therapeutic targets of SARS-CoV-2
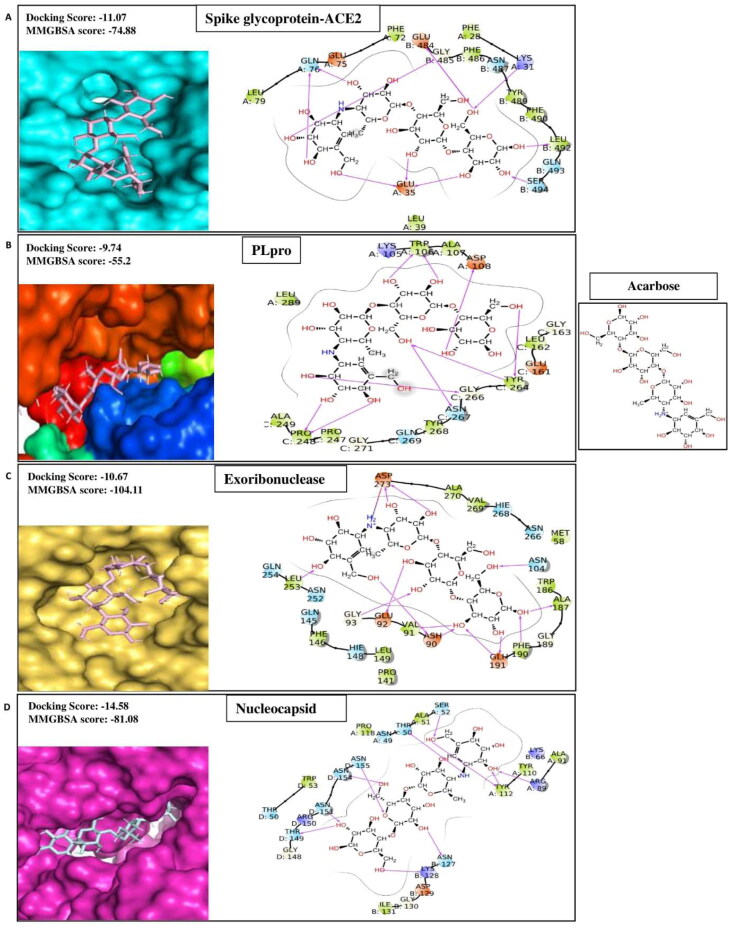
Acarbose
Acarbose is the most commonly recommended oral drug for impaired glucose tolerance (IGT) treatment and is also useful for cardiovascular risk factors (Standl et al., 2014). Besides, it has been observed that acarbose exerted an anti-inflammatory effect in diabetic rats by inhibiting the expression of mRNA of IL-6 and TNF-α (Zhang et al., 2013). In silico studies further shows that the compound binds IL-6 and TNF-α, which could suppress the massive inflammatory response in COVID-19 patients (Xiaoqi et al., 2020). A recent study demonstrated that this compound retains a strong binding affinity for the main protease and spike protein of SARS CoV-2 (Tariq et al., 2020). Our molecular docking analysis showed that acarbose has significantly high binding affinities for six targets of SARS CoV-2 such as Nucleocapsid, Exoribonucleases, EndoRNAse, RDRP, PLprotein, and Spike protein complex with ACE2 with docking scores −14.58, −10.67, −10.67, −10.99, −9.74, and −11.07 respectively (Table 1A). The ligand-binding patterns of the docked complexes are displayed in Figure 1F, showing acarbose strongly binds with the active site pocket of each target protein. For instance, acarbose interacts through hydrogen bond with the active site pocket residues of Spike-ACE2 (chianA:Lys31, Glu35 and Gln76; chainB:Glu484, Gly485, Leu492 and Ser494); PLpro (chainA:Trp106 and Asp108; chainC:Pro248, Tyr262, Gly266 and Asn267); Exoribonuclease (Asp90, Glu92, Asn104, Ala187, Phe190, Glu191, Leu253 and Asp273); and Nucleocapsid (chainA:Thr50, Ser52, Arg89 and Tyr112; chainB:Asn127 and Lys128; chainD:Thr149, Asn151, Asn154 and Asn155).
Figure 1F.
2 D-interaction diagram indicating protein-ligand contacts of acarbose with its best four targets i.e. (A) Spike glycoprotein-ACE2, (B) Papine like proteinase (PLpro), (C) Exoribonuclease and (D) Nucleocapsid.
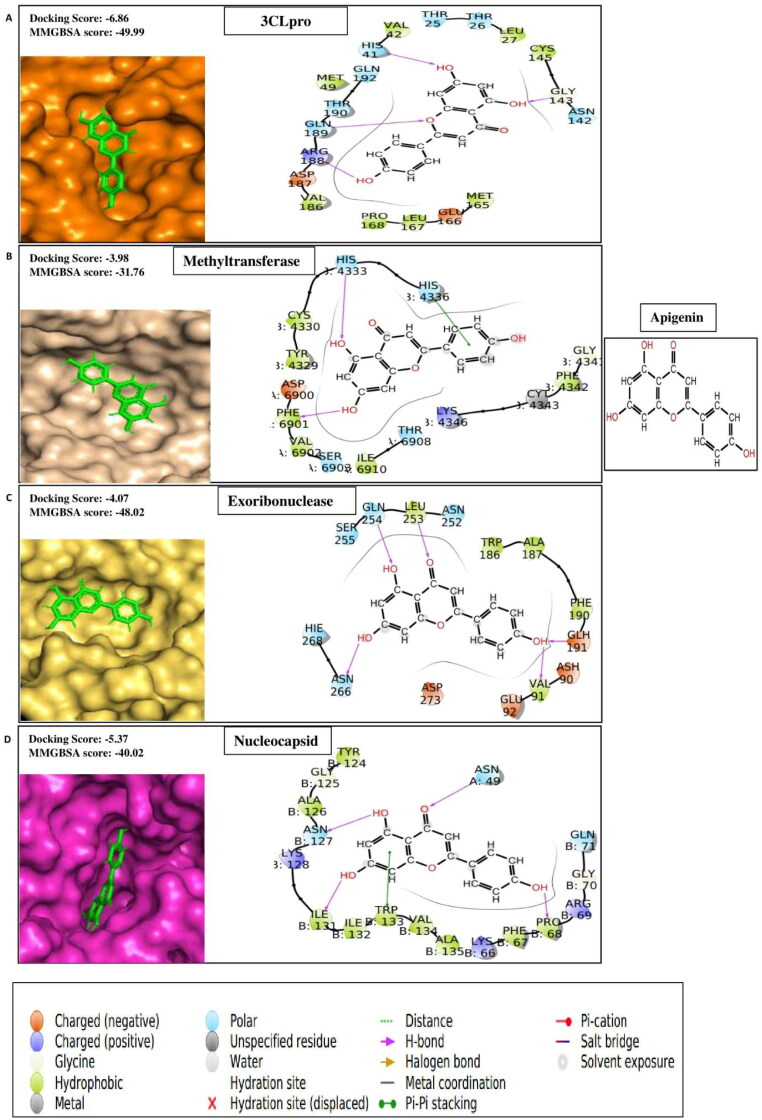
Apigenin
Apigenin, a flavone compound, usually is available in many vegetables, fruits, nuts, and tea. The mechanism of the antidiabetic effects of apigenin is related to inhibition of pancreatic stellate cell activity, oxidative damage of pancreatic β-cells, hyperglycemia condition, and modulation of GLUT4 translocation (Vinayagam & Xu, 2015). This compound has also been reported to possess anti-inflammatory, anti-mutagenic, antioxidant, and antiviral properties (Bahare Salehi et al., 2019). The docking based molecular interaction data of different research groups have indicated that apigenin can inhibit Mpro (Khaerunnisa et al., 2020) and ACE 2 (Preeti Pandey et al., 2020) of SARS CoV-2. Our study reveals that the compound has significantly high binding affinities for five targets of SARS CoV-2, such as 3CLpro, Spike protein complex with ACE2, Methyltransferase, Exoribonucleases, and Nucleocapsid with the docking scores of −6.86, −4.38, −3.98, −4.07, and −5.37 respectively (Table 1A). The ligand-binding patterns of the docked complexes are displayed in Figure 1G. Apigenin forms hydrogen bond and pi-pi stacking interactions with the binding pocket amino acid residues of target proteins. Hydrogen bond formation ocuurs between apigenin and amino acid residue of 3CLpro (His41, Gly143, Arg188 and Gln189); Methyltransferase (chainA:Phe6901; chainB:His4333); Exoribonuclease (Val91, Glu191, Leu253, Gln254, Asn266); and Nucleocapsid (chainA:Asn49; chainB:Pro68, Asn127, Ile131). Apigenin forms pi-pi stacking interaction with the chainB:His4336 and chainB:Trp133 of Methyltransferase and Nucleocapsid respectively.
Figure 1G.
Showing protein-ligand contacts between the compound apigenin and its best four protein targets i.e. (A) 3c-like proteinase (3CLpro), (B) Methyltransferase, (C) Exoribonuclease and (D) Nucleocapsid.
Hesperidin
The antidiabetic properties of hesperidin are documented in many in vitro and in vivo studies (Mahmoud & Hussein, 2016). This compound's antioxidant and anti-inflammatory activities are also documented (Barreca et al., 2020). On the other hand, virtual screening studies have shown the binding affinities of hesperidin for Mpro (Adem et al., 2020), ACE2 receptor (Cheng et al., 2020), and 3CLpro (Chen et al., 2020) of SARS CoV-2. In this context, our studies show that hesperidin has better binding affinities for the key target protein, including 3CLpro, helicase, PL protein, and nucleocapsid, with docking scores −10.68, −7.71, −8.47, and −11.68, respectively (Table 1A).
Neohesperidin
Neohesperidin, flavanone glycoside, exhibited hypolipidemic and antidiabetic effects in diabetic KK-AY mice (Jia et al., 2015). Furthermore, It has been shown that the compound could bind with high affinity with ACE2 (Cheng et al., 2020), Tmprss2 (Coban, 2020), and 3CLpro (Ghosh et al., 2020; Jannat et al., 2020). In our present study, the compound's strong binding affinities have been observed for four proteins, including Spike Protein, RDRP, EndoRNAse, and Nucleocapsid, with docking scores −6.83, −9.95, −8.69, and −10.84, respectively (Table 1A).
Cardio-protective compound against multiple therapeutic targets of SARS-CoV-2
Salvianolic acid B
Salvianolic acid B (SalB), the major component of Salviae miltiorrhizae, plays significant roles in cardio-protection in various ways, including antithrombotic and anticoagulant effects, inhibiting apoptosis of myocardial cells, inhibiting myocardial injury, and promoting cardiac angiogenesis (Jie Wang et al., 2013). In the virtual screening campaign against different drug targets of SARS CoV-2, SalB has been reported to have a significant binding affinity with 3CLpro (Contini, 2020) and Mpro (Ibrahim et al., 2020). Our result indicated that SalB has a potential binding affinity for nucleocapsid, endoRNAse, RDRP, and helicase with docking scores −11.24, −10.08, −8.83 and −6.6, respectively (Table 1A).
QikProp analysis of ADME and principal descriptor values
Lipinski's rule of five is beneficial for determining essential pharmacokinetics such as absorption, distribution, metabolism, and excretion (ADME) of drug molecules (Lipinski, 2004). Therefore, ADME studies of selected 20 compounds using Schrödinger QikProp module have been performed. Among them Rutin hydrate, Silymarin, Quercetin, Luteolin, Aloin, Morin hydrate, Kaempferol and Apigenin were successfully passed through the maximum number of the physiochemical filters, including QPlogPo/w (Predicted octanol/water partition coefficient), QPlogS (Predicted aqueous solubility), QPlogHERG (Predicted IC50 value for the blockage of HERG K + channels), QPPCaco (Predicted apparent Caco-2 cell permeability for the gut blood barrier), QPlogBB (predicted brain/blood partition coefficient), QPPMDCK (Predicted apparent MDCK cell permeability in nm/s), QPlogKp (Predicted skin permeability) and QPlogKhsa (prediction of binding to human serum albumin) as shown in Table 2A. The drug-like properties of those compounds were further analyzed through the principal descriptor values, including Accept HB (Estimated number of hydrogen bonds that would be accepted by the solute from water molecules in an aqueous solution), DonorHB (Estimated number of hydrogen bonds that would be donated by the solute to water molecules in an aqueous solution), PHOA (Percent human oral absorption), SASA (Total solvent accessible surface area), FOSA (Hydrophobic component of the SASA), FISA (Hydrophilic component of the SASA), PISA (π (carbon and attached hydrogen) component of the SASA), and MW (molecular weight of molecule). Importantly, we have found that the lead compound's scores lie within the acceptable range of most of the drug-like properties (Table 2B). Toxicity profile including AMES toxicity (prediction of mutagenicity of compounds) and hERG inhibition of selected 20 compounds were evaluated to give better insight for drug likeliness (Aishwarya et al., 2021). All the compounds shows negative value for mutagenicity (supplementary Table 12).
Table 2A.
ADME properties of the selected twenty compounds.
| Drug Name |
QPlogPo/w (-2.5 to 6.5) |
QPlogS (-6.5 to 0 .5) |
QPlogHERG (below -5.0) | QPPCaco (<25%-poor,>500-great) |
QPlogBB (-3 to 1.2) |
QPPMDCK (<25%-poor,>500-great) | QPlogKp (-8.0 to-0.1) |
QPlogKhsa (-1.5 to 1.5) |
|---|---|---|---|---|---|---|---|---|
| Anti-inflammatory | ||||||||
| Rutin Hydrate | −2.476 | −2.624 | −5.864 | 0.895 | −4.915 | 0.251 | −7.269 | −1.358 |
| Silymarin | 1.672 | −4.808 | −5.924 | 26.684 | −2.636 | 9.847 | −4.911 | 0.009 |
| Quercetin | 0.384 | −2.874 | −5.088 | 20.005 | −2.367 | 7.212 | −5.457 | −0.348 |
| Mitoxantrone 2HCl | 0.512 | −1.919 | −7.66 | 0.855 | −3.088 | 0.292 | −9.085 | −0.327 |
| Luteolin | 0.963 | −3.109 | −5.061 | 41.618 | −1.954 | 15.92 | −4.86 | −0.188 |
| Aloin | −0.464 | −2.896 | −5.035 | 9.05 | −3.05 | 3.06 | −5.958 | −0.652 |
| Morin Hydrate | 0.419 | −2.67 | −4.853 | 26.94 | −2.158 | 9.949 | −5.206 | −0.361 |
| Kaempferol | 1.054 | −3.056 | −5.077 | 57.783 | −1.805 | 22.699 | −4.542 | −0.197 |
| Madecassoside | −2.561 | −2.203 | −5.001 | 0.586 | −5.55 | 0.159 | −7.596 | −1.648 |
| Antibiotics | ||||||||
| Amikacin Hydrate | −8.684 | −0.237 | −6.399 | 0.002 | −5.175 | 0.001 | −12.607 | −2.555 |
| Geneticin | −5.151 | 2 | −7.209 | 0.208 | −2.481 | 0.07 | −9.781 | −1.448 |
| Netilmicin Sulfate | −3.598 | 2 | −8.628 | 0.556 | −2.199 | 0.203 | −8.935 | −1.097 |
| Hygromycin B | −5.929 | 1.997 | −6.901 | 0.047 | −3.441 | 0.014 | −10.945 | −1.788 |
| Antidiabetic | ||||||||
| Acarbose | −6.893 | 0.846 | −4.921 | 0.128 | −4.679 | 0.034 | −9.797 | −2.257 |
| Apigenin | 1.632 | −3.341 | −5.126 | 119.76 | −1.428 | 49.902 | −3.942 | −0.035 |
| Hesperidin | −1.437 | −2.798 | −5.425 | 4.802 | −3.926 | 1.543 | −6.139 | −1.172 |
| Neohesperidin | −1.128 | −4.292 | −6.533 | 3.283 | −4.846 | 1.023 | −6.447 | −1.174 |
| Troxerutin | −2.526 | −2.119 | −6.331 | 0.818 | −6.05 | 0.228 | −6.65 | −1.939 |
| Notoginsenoside R1 | −1.195 | −3.123 | −5.482 | 1.138 | −5.704 | 0.325 | −6.847 | −1.423 |
| Cardiovascular Disease | ||||||||
| Salvianolic AcidB | 2.25 | −5.927 | −4.095 | 0.004 | −7.326 | 0.001 | −8.372 | −0.397 |
Table 2B.
Principal drug-like properties of the selected twenty compounds.
| Drug Name | MW (130-725) | SASA (300-1000) | FOSA (0-750) |
FISA (7-330) |
PISA (0-450) |
Volume (500-2000) |
|---|---|---|---|---|---|---|
| Anti-inflammatory | ||||||
| Rutin Hydrate | 610.5 | 778.6 | 199.8 | 368.1 | 210.7 | 1523 |
| Silymarin | 482.4 | 739.7 | 187.4 | 252 | 300.3 | 1343 |
| Quercetin | 302.2 | 516.9 | 0 | 284.2 | 232.8 | 864.3 |
| Mitoxatrone 2HCl | 444.5 | 793.8 | 328.3 | 301.4 | 164.1 | 1392 |
| Luteolin | 286.2 | 504.4 | 0 | 250.6 | 253.8 | 844.8 |
| Aloin | 418.4 | 645.1 | 153 | 320.5 | 171.6 | 1174 |
| Morin Hydrate | 302.24 | 503.217 | 0 | 270.537 | 232.68 | 851.42 |
| Kaempferol | 286.2 | 501.1 | 0 | 235.6 | 265.5 | 839.9 |
| Madecassoside | 975.1 | 1135 | 680.8 | 445.9 | 8.085 | 2514 |
| Antibiotics | ||||||
| Amikacin Hydrate | 585.6 | 835.1 | 342.7 | 492.4 | 0 | 1605 |
| Geneticin | 496.6 | 699.2 | 426.5 | 272.7 | 0 | 1363 |
| Netilmicin Sulfate | 475.6 | 754.9 | 501.5 | 220.4 | 33 | 1421 |
| Hygromycin B | 527.5 | 753.9 | 411.6 | 342.3 | 0 | 1417 |
| Antidiabetic | ||||||
| Acarbose | 645.6 | 803 | 350.5 | 451.9 | 0.589 | 1634 |
| Apigenin | 270.2 | 490.7 | 0 | 202.2 | 288.5 | 819.3 |
| Hesperidin | 610.6 | 928.4 | 375.1 | 377.1 | 176.2 | 1701 |
| Neohesperidin | 612.6 | 904 | 351 | 381.5 | 171.5 | 1697 |
| Troxerutin | 742.7 | 1019 | 445.7 | 430.6 | 142.2 | 1967 |
| Notoginsenoside R1 | 933.1 | 1188 | 765.6 | 415.4 | 7.188 | 2541 |
| Cardiovascular Disease | ||||||
| Salvianolic Acid B | 718.6 | 921.7 | 70.32 | 502.6 | 348.8 | 1871 |
ROC curve plot and enrichment study
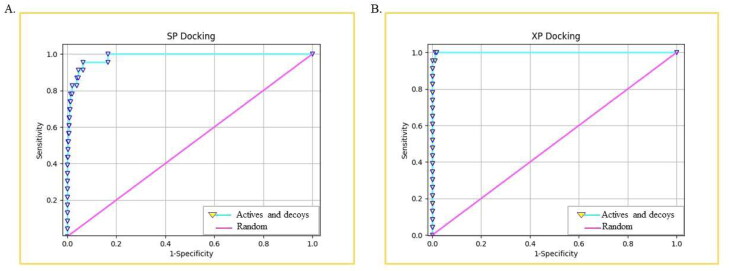
In computer-aided drug designing, the power to identify the true positive docked compounds against the false positive is a major challenge (Awuni & Mu, 2015). Therefore, a method that should have the ability to predict and identify true positive versus false positive in virtual screening mode can be used. In this study, the ROC plot and Area Under Curve (AUC) plot are used for this purpose. The SP and XP molecular docking output file was used for plotting the graphs, and it was observed that the ROC values are 0.94 and 1.00, respectively (Figure 2), revealing the specificity of docking. Further, the docking method's sensitivity and precision were validated by AUC plot, and the obtained values are 0.94 and 0.99 for SP and XP docking, respectively (Table 3). The value closer to 1 represents the higher chances of having true positive docked compounds. The BEDROCK at α = 160.9 is calculated as 0.858 and 0.989 for SP and XP docking, respectively, which specifies the overall sound performance of XP docking (Table 3).
Figure 2.
Protocol validation for SP and XP mode of docking through the depiction of both actives and decoys compounds by mapping the ROC curve.
Table 3.
Table showing the validation of SP and XP docking based upon active counts and ROC.
| Mode of docking | Percentage of results | 1% | 2% | 5% | 10% | 20% | Enrichment Factors | Metric Value | |
|---|---|---|---|---|---|---|---|---|---|
| SP docking | Active counts | 9 | 14 | 18 | 21 | 22 | ROC | 0.94 | |
| % of Actives | 39.1 | 60.9 | 78.3 | 91.3 | 95.7 | AUC | 0.94 | ||
| BEDROC | 160.9 | 0.858 | |||||||
| 20.0 | 0.813 | ||||||||
| 8.0 | 0.870 | ||||||||
| XP Docking | Active counts | 10 | 19 | 23 | 23 | 23 | ROC | 1.00 | |
| % of Actives | 43.5 | 82.6 | 100.0 | 100.0 | 100.0 | AUC | 0.99 | ||
| BEDROC | 160.9 | 0.989 | |||||||
| 20.0 | 0.983 | ||||||||
| 8.0 | 0.992 | ||||||||
MD simulation of top drug-target protein complexes
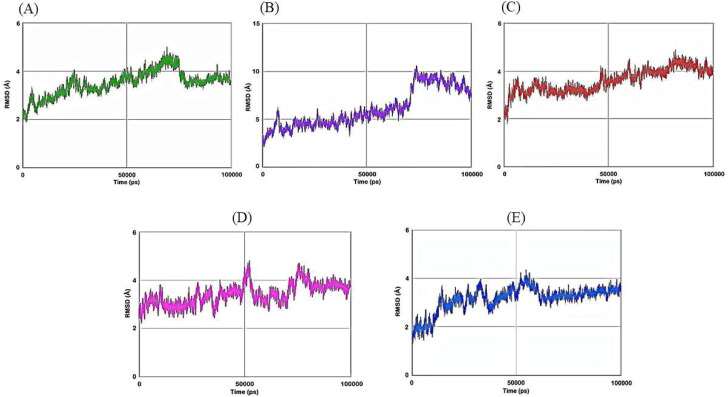
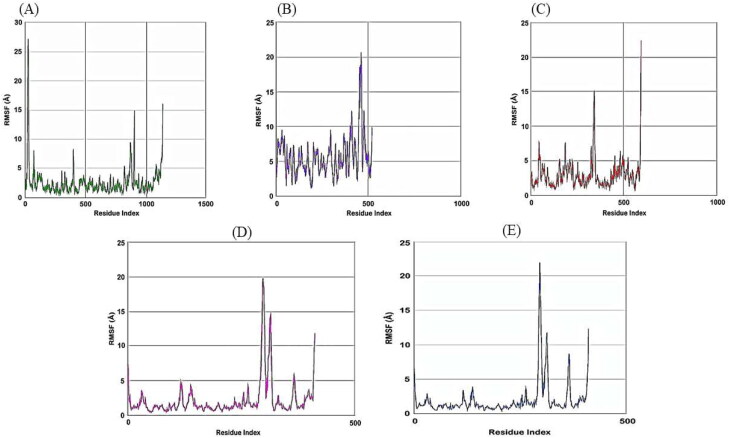
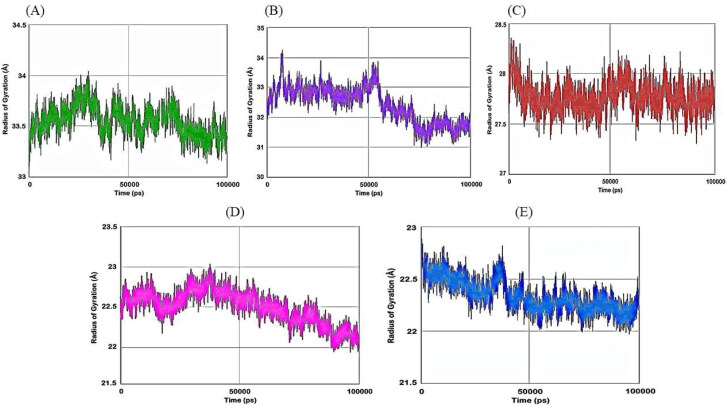
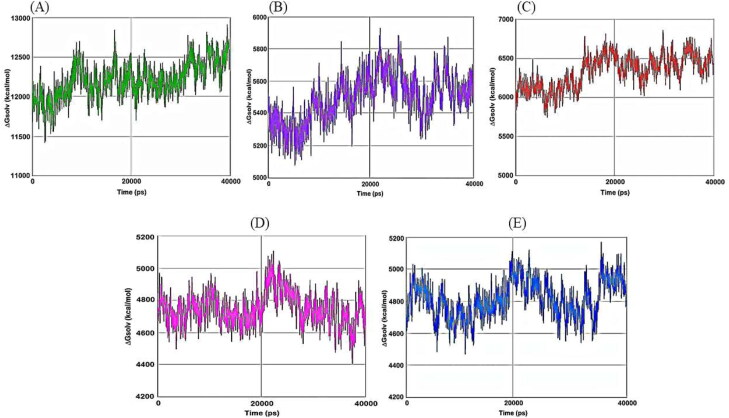
Then we have carried out molecular dynamics simulation studies on the top five drug target protein complexes (RDRP-Rutinhydrate, Exo-Silymarin, Helicase-Luteolin, Methyltransferase-Amikacin Hydrate, and Methyltransferase-Geneticin). For the study of the top five complexes' conformational dynamics, Root Mean Square Deviation (RMSD) analysis with respect to their corresponding initial reference structure was carried out. Figure 3 shows the RMSD for Cα atoms of the five complex systems as a function of time in pico-seconds. The RMSD values for the five complex systems were observed to be in the range 2 Å to 10 Å, and the values perhaps indicate the structural stability of all the complexes. The degree of flexibility of chain in the five complexes was depicted in terms of Root Mean Square Fluctuation (RMSF) of each residue from its time – average position in Figure 4. The RMSF plots show that the fluctuations were more in the Exo-Silymarin and Helicase-Luteolin complexes among the five complexes. We have also carried out the radius of gyration analysis to check the compactness of five complexes in a dynamic system (as shown in Figure 5). From Figure 5, we can infer that the five complexes' overall size remains altered, but subtle change we noticed in the Exo-Silymarin and Helicase-Luteolin complexes. In Figure 6, we have depicted the distribution of water molecules around the five complexes in terms of solvation free energy in kcal/mol. The five complexes' solvation-free energy shows different profiles because of the difference in the size of the protein molecule, which affects the protein-water interaction energy and solvent reorganization energy. We have also analyzed the different intermolecular interactions between the protein and the ligand in the five complexes. The intermolecular interactions observed in the five complexes have been summarized in supplementary Tables 2–6. We see that the ligands display an appropriate number of hydrogen bonds in the dynamic system and are correlated with the molecular docking results from the intermolecular interactions. MMGBSA thermodynamics calculation for every 10000ps intervals was estimated for each protein-ligand complex during the course of simulation and standard deviations of binding free energy were tabulated in supplementary Tables 7–11.
Figure 3.
Plot of RMSD of Cα atoms of A) RDRP-Rutinhydrate B) Exo-Silymarin C) Helicase-Luteolin D) Methyltransferase-Amikacin hydrate E) Methyltransferase-Geneticin in angstroms as a function of simulation time in pico seconds.
Figure 4.
Plot of RMSF of Cα atoms of A) RDRP-Rutinhydrate B) Exo-Silymarin C) Helicase-Luteolin D) Methyltransferase-Amikacin hydrate E) Methyltransferase-Geneticin in angstroms as a function of Residue Index.
Figure 5.
Plot of Radius of gyration of A) RDRP-Rutinhydrate B) Exo-Silymarin C) Helicase-Luteolin D) Methyltransferase-Amikacin hydrate E) Methyltransferase-Geneticin in angstroms as a function of simulation time in pico seconds.
Figure 6.
Plot of solvation free energy of A) RDRP-Rutinhydrate B) Exo-Silymarin C) Helicase-Luteolin D) Methyltransferase-Amikacin hydrate E) Methyltransferase-Geneticin in kcal/mol as a function of simulation time in pico seconds.
Discussion
The global COVID-19 load has mounted to 53,164,803 confirmed cases and 1,300,576 deaths as of November 14, 2020. To break the rising trend of COVID-19 and related detrimental effects, drug repurposing therapeutic strategies have widely opted on a health emergency basis (Drożdżal et al., 2020). The unique advantages of using repurposed drugs in treating human diseases, including cancer, have already been well documented (Pushpakom et al., 2019). Numerous in silico incisive investigations of FDA approved drugs against the druggable targets of SARS-CoV-2 have been undertaken to boost the COVID-19 treatment strategies. Conventionally, compounds are selected to act on a single drug target with great potency. These compounds may turn to be unsuccessful in the long run, as the virus is evolving with new mutations. COVID-19 treatment difficulties are also deeply rooted in its complex association with uncontrolled immune response, secondary bacterial infection, diabetes, and cardiovascular diseases. The development of new inhibitors with known targets for different pathways of COVID-19 associated diseases and acting multiple targets of SARS CoV-2 will bring a paradigm change for COVID-19 therapy. Moreover, these polypharmacological compounds can avoid the possible adverse synergistic effects of drug-drug interactions during the treatment of COVID-19 and accompanying diseases.
Therefore, we investigated the capacity of compounds from FDA approved drug library, and FDA approved and passed phase −1 drug library towards polypharmacological drug identification through high throughput virtual screening. Our molecular docking studies assess how the two libraries' compounds can inhibit multiple SARS-CoV-2 essential proteins or host proteins that interact with the virus component to stop the virus's entry and proliferation. We found nine anti-inflammatory compounds such as Rutin Hydrate, Silymarin, Luteolin, Aloin, Madecassoside, Mitoxantrone 2Hcl, Kaempferol, Quercetin, and Morin hydrate; four antibacterial such as Amikacin Hydrate, Geneticin, Netilmicin Sulfate, and Hygromycin B; six antidiabetic compounds such as Hesperidin, Apigenin, Acarbose, Notoginsenoside R1, Neohesperidin, and troxerutin; and one cardioprotective compound such as Salvianolic Acid B, that could effectively interact with multiple therapeutic targets of SARS CoV-2. Some of these compounds are extensively discussed in the result section. For further validation of ligand-protein interactions, molecular dynamic studies for five compounds, including Rutin Hydrate, Silymarin, Luteolin, Amikacin Hydrate, and Geneticin, have also been performed.
As the two libraries have been thoroughly investigated for COVID-19 therapy, we found that these compounds' binding affinities with different therapeutic targets of SARS CoV-2 have been reported in many studies. However, our screening approach against ten validated drug targets of SARS CoV-2 together rather than individual targets provides us a unique opportunity to discover multi-target inhibitors. Remarkably, in many cases, our data agree with the earlier in silico studies of many computational groups. In some cases, we observe the differences in selecting best-docked molecules with earlier studies due to the difference in reference molecules used by various groups in their docking analysis.
Our studies are limited with a lack of experimental evidence that the compounds identified here can directly inhibit their target proteins or/and inhibit SARS-CoV-2 proliferation in patients. However, as per the list of clinical trials (clinicaltrial.gov website) on COVID-19 treatment to date, five of our best-docked compounds, including Quercetin (NCT04468139, phase-4), Silymarin (NCT04394208, phase-3), and Morin hydrate (NCT04348656, phase-3), Luteolin (NCT04404218, phase-2), and Apigenin (NCT04404218, phase-2) are in different phases of clinical studies. Hence, it is reasonable to assume that the best-scored compounds obtained by the present docking analysis may also retain the therapeutic capacity for COVID-19 treatment. Researchers can also focus prospectively on research and clinical trials of the remaining 15 best-docked molecules for developing multi-target drugs against the COVID-19 pandemic.
Finally, this drug repositioning method using the in silico screening method can be a novel computational strategy to explore single, multi-targeting, and multi-purposing therapeutic molecules. These drugs can treat COVID-19 and frequently associated co-morbid disorders such as uncontrolled immune response, secondary bacterial infection, diabetes, and cardiovascular diseases and can overcome the limitations of probable patient compliance out of drug interactions associated with combination therapy.
Supplementary Material
Acknowledgements
BN is thankful to the Central University of Rajasthan for providing a research fellowship. DP is thankful to the Central University of Rajasthan for providing the computational facility.
Disclosure statement
No potential conflict of interest was reported by the authors.
Competing financial interests
The authors have declared no competing interest.
References
- Adem, S., Eyupoglu, V., Sarfraz, I., Rasul, A., & Ali, M. (2020). Identification of potent COVID-19 main protease (Mpro) inhibitors from natural polyphenols: An in silico strategy unveils a hope against CORONA. Preprints, 2020, 2020030333. 10.20944/preprints202003.0333.v1. [DOI] [PMC free article] [PubMed]
- Aishwarya, S., Gunasekaran, K., Sagaya Jansi, R., & Sangeetha, G. (2021). From genomes to molecular dynamics- A bottom up approach in extrication of SARS CoV-2 main protease inhibitors. Computational Toxicology (Amsterdam, Netherlands), 18, 100156. https://doi.org/ 10.1016/j.comtox.2021.100156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akriti, K., Satpathy, I., & Patnaik, B. (2021). Covid-19 and its impact on livelihood: An Indian perspective. Eurasian Chemical Communications, 3(2), 81–87. [Google Scholar]
- Altayeb, H., Bouslama, L., Abdulhakimc, J. A., Chaieb, K., Baothman, O. A., & Zamzami, M. A. (2020). Potential activity of a selected natural compounds on SARS-CoV-2 RNA-dependent-RNA polymerase, and binding affinity of the receptor-binding domain (RBD).
- Awuni, Y., & Mu, Y. (2015). Reduction of false positives in structure-based virtual screening when receptor plasticity is considered. Molecules, 20(3), 5152–5164. 10.3390/molecules20035152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz, N., Kim, M. Y., & Cho, J. Y. (2018). Anti-inflammatory effects of luteolin: A review of in vitro, in vivo, and in silico studies. Journal of Ethnopharmacology, 225, 342–358. 10.1016/j.jep.2018.05.019 [DOI] [PubMed] [Google Scholar]
- Bacopoulou, F., Skouroliakou, M., & Markantonis, S. L. (2009). Netilmicin in the neonate: Pharmacokinetic analysis and influence of parenteral nutrition. Pharmacy World & Science, 31(3), 365–368. 10.1007/s11096-009-9278-z [DOI] [PubMed] [Google Scholar]
- Baliga, M. S., Saxena, A., Kaur, K., Kalekhan, F., Chacko, A., Venkatesh, P., & Fayad, R. (2014). Chapter 50 - Polyphenols in the prevention of ulcerative colitis: Past, present and future. In Watson R. R., Preedy V. R., & Zibadi S. (Eds.), Polyphenols in human health and disease (pp. 655–663).Academic Press. [Google Scholar]
- Barreca, D., Mandalari, G., Calderaro, A., Smeriglio, A., Trombetta, D., Felice, M. R., & Gattuso, G. (2020). Citrus flavones: An update on sources, biological functions, and health promoting properties. Plants, 9(3), 288. 10.3390/plants9030288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beganovic, M., Luther, M. K., Rice, L. B., Arias, C. A., Rybak, M. J., & LaPlante, K. L. (2018). A review of combination antimicrobial therapy for Enterococcus faecalis bloodstream infections and infective endocarditis. Clinical Infectious Diseases, 67(2), 303–309. 10.1093/cid/ciy064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert, P., Künzli, M., & Schwede, T. (2009). QMEAN server for protein model quality estimation. Nucleic Acids Research, 37(suppl_2), W510–W514. 10.1093/nar/gkp322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen, H. J., Postma, J. v., van Gunsteren, W. F., DiNola, A. R. H. J., & Haak, J. R. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690. 10.1063/1.448118 [DOI] [Google Scholar]
- Bhowmik, D., Nandi, R., Jagadeesan, R., Kumar, N., Prakash, A., & Kumar, D. (2020). Identification of potential inhibitors against SARS-CoV-2 by targeting proteins responsible for envelope formation and virion assembly using docking based virtual screening, and pharmacokinetics approaches. Infection, Genetics and Evolution, 84, 104451. 10.1016/j.meegid.2020.104451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch-Barrera, J., Martin-Castillo, B., Buxó, M., Brunet, J., Encinar, J. A., & Menendez, J. A. (2020). Silibinin and SARS-CoV-2: Dual targeting of host cytokine storm and virus replication machinery for clinical management of COVID-19 patients. Journal of Clinical Medicine, 9(6), 1770. 10.3390/jcm9061770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso, J. M. S., Fonseca, L., Egas, C., & Abrantes, I. (2018). Cysteine proteases secreted by the pinewood nematode, Bursaphelenchus xylophilus: In silico analysis. Computational Biology and Chemistry, 77, 291–296. https://doi.org/ 10.1016/j.compbiolchem.2018.10.011 [DOI] [PubMed] [Google Scholar]
- Case, D. A., Darden, T. A., Cheatham, T. E. III, Simmerling, C. L., Wang, J., Duke, R. E., Luo, R., Walker, R. C., Zhang, W., Merz, K. M., Roberts, B., Hayik, S., Roitberg, A., Seabra, G., Swails, J., Goetz, A. W., Kolossváry, I., Wong, K. F., Paesani, F., … Kollman, P. A . (2010). AMBER 12. University of California, San Francisco. [Google Scholar]
- Chandra, A., Gurjar, V., Qamar, I., & Singh, N. (2020). Identification of potential inhibitors of SARS-COV-2 endoribonuclease (EndoU) from FDA approved drugs: A drug repurposing approach to find therapeutics for COVID-19. Journal of Biomolecular Structure and Dynamics, 1–11. 10.1080/07391102.2020.1775127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Chen, Y., Wu, P., & Chen, B. (2014). Update on new medicinal applications of gentamicin: Evidence-based review. Journal of the Formosan Medical Association, 113(2), 72–82. 10.1016/j.jfma.2013.10.002 [DOI] [PubMed] [Google Scholar]
- Chen, Y. W., Yiu, C.-P B., & Wong, K.-Y. (2020). Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL pro) structure: Virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Research, 9, 129. 10.12688/f1000research.22457.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, J. J. B. s b. (2008). A multi-template combination algorithm for protein comparative modeling. BMC Structural Biology, 8(1), 18. 10.1186/1472-6807-8-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, L., Zheng, W., Li, M., Huang, J., Bao, S., Xu, Q., & Ma, Z. (2020). Citrus fruits are rich in flavonoids for immunoregulation and potential targeting ACE2. Preprints, 2020, 2020020313. [DOI] [PMC free article] [PubMed]
- Chiasson, J. L., Josse, R. G., Gomis, R., Hanefeld, M., Karasik, A., & Laakso, M. (2002). Acarbose for prevention of type 2 diabetes mellitus: The STOP-NIDDM randomised trial. The Lancet, 359(9323), 2072–2077. 10.1016/S0140-6736(02)08905-5 [DOI] [PubMed] [Google Scholar]
- Choudhury, A., Chakraborty, I., Banerjee, T. S., Vana, D. R., & Adapa, D. (2017). Efficacy of morin as a potential therapeutic phytocomponent: Insights into the mechanism of action. International Journal of Medical Research & Health Sciences, 6(11), 175–194. [Google Scholar]
- Coban, M. (2020). Attacking COVID-19 progression using multi-drug therapy for synergetic target engagement. arXiv Preprint arXiv:2007.02557. [DOI] [PMC free article] [PubMed]
- Contini, A. (2020). Virtual screening of an FDA approved drugs database on two COVID-19 coronavirus proteins. ChemRxiv. Preprint. 10.26434/chemrxiv.11847381.v1 [DOI]
- Contou, D., Claudinon, A., Pajot, O., Micaëlo, M., Longuet Flandre, P., Dubert, M., Cally, R., Logre, E., Fraissé, M., Mentec, H., & Plantefève, G. (2020). Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Annals of Intensive Care, 10(1), 1–9. 10.1186/s13613-020-00736-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, W. A., Gudmundsson, S., & Reich, R. M. (1983). Netilmicin sulfate: A comparative evaluation of antimicrobial activity, pharmacokinetics, adverse reactions and clinical efficacy. Pharmacotherapy, 3(6), 305–315. 10.1002/j.1875-9114.1983.tb03283.x [DOI] [PubMed] [Google Scholar]
- da Silva Antonio, A., Wiedemann, L. S. M., & Veiga-Junior, V. F. (2020). Natural products' role against COVID-19. RSC Advances, 10(39), 23379–23393. 10.1039/D0RA03774E [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, S., & Singha Roy, A. (2020). Naturally occurring anthraquinones as potential inhibitors of SARS-CoV-2 main protease: A molecular docking study. ChemRxiv. Preprint. 10.26434/chemrxiv.12245270.v1 [DOI] [PMC free article] [PubMed]
- Das, S., Sarmah, S., Lyndem, S., & Singha Roy, A. (2020). An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics, 39, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gennaro, F., Pizzol, D., Marotta, C., Antunes, M., Racalbuto, V., Veronese, N., & Smith, L. (2020). Coronavirus diseases (COVID-19) current status and future perspectives: A narrative review. International Journal of Environmental Research and Public Health, 17(8), 2690. 10.3390/ijerph17082690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drożdżal, S., Rosik, J., Lechowicz, K., Machaj, F., Kotfis, K., Ghavami, S., & Łos, M. J. (2020). FDA approved drugs with pharmacotherapeutic potential for SARS-CoV-2 (COVID-19) therapy. Drug Resistance Updates, 53, 100719. 10.1016/j.drup.2020.100719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elfiky, A. A. (2020). SARS-CoV-2 RNA dependent RNA polymerase (RdRp) targeting: An in silico perspective. Journal of Biomolecular Structure and Dynamics, 39, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan, W., Qian, S., Qian, P., & Li, X. (2016). Antiviral activity of luteolin against Japanese encephalitis virus. Virus Research, 220, 112–116. 10.1016/j.virusres.2016.04.021 [DOI] [PubMed] [Google Scholar]
- Fawcett, T. J. P. r l. (2006). An introduction to ROC analysis. Pattern Recognition Letters, 27(8), 861–874. 10.1016/j.patrec.2005.10.010 [DOI] [Google Scholar]
- Felsenstein, S., Herbert, J. A., McNamara, P. S., & Hedrich, C. M. (2020). COVID-19: Immunology and treatment options. Clinical Immunology, 215, 108448. 10.1016/j.clim.2020.108448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, E. J. (2004). Mechanism of action of mitoxantrone. Neurology, 63(12 Suppl 6), S15–S18. 10.1212/wnl.63.12_suppl_6.s15 [DOI] [PubMed] [Google Scholar]
- Friesner, R. A., Banks, J. L., Murphy, R. B., Halgren, T. A., Klicic, J. J., Mainz, D. T., Repasky, M. P., Knoll, E. H., Shelley, M., Perry, J. K., Shaw, D. E., Francis, P., & Shenkin, P. S. (2004). Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry, 47(7), 1739–1749. https://doi.org/ 10.1021/jm0306430 [DOI] [PubMed] [Google Scholar]
- Ganeshpurkar, A., & Saluja, A. K. (2017). The pharmacological potential of rutin. Saudi Pharmaceutical Journal, 25(2), 149–164. 10.1016/j.jsps.2016.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharagozloo, M., Jafari, S., Esmaeil, N., Javid, E. N., Bagherpour, B., & Rezaei, A. (2013). Immunosuppressive effect of silymarin on mitogen‐activated protein kinase signalling pathway: The impact on T cell proliferation and cytokine production. Basic & Clinical Pharmacology & Toxicology, 113(3), 209–214. 10.1111/bcpt.12088 [DOI] [PubMed] [Google Scholar]
- Ghosh, K., Amin, S. A., Gayen, S., & Jha, T. (2020). Chemical-informatics approach to COVID-19 drug discovery: Exploration of important fragments and data mining based prediction of some hits from natural origins as main protease (Mpro) inhibitors. Journal of Molecular Structure, 1224, 129026. 10.1016/j.molstruc.2020.129026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gur, D., Glinert, I., Aftalion, M., Vagima, Y., Levy, Y., Rotem, S., Zauberman, A., Tidhar, A., Tal, A., Maoz, S., Ber, R., Pass, A., & Mamroud, E. (2018). Inhalational gentamicin treatment is effective against pneumonic plague in a mouse model. Frontiers in Microbiology, 9, 741. 10.3389/fmicb.2018.00741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harismah, K., & Mirzaei, M. (2020). Favipiravir: Structural analysis and activity against COVID-19. Advanced Journal of Chemistry, Section B: Natural Products and Medical Chemistry, 2(2), 55–60. [Google Scholar]
- Hirata, F. (2003). Molecular theory of solvation (Vol. 24). Springer Science & Business Media. http://biosig.unimelb.edu.au/pkcsm/.). [Google Scholar]
- Huang, C.-T., Hung, C.-Y., Hseih, Y.-C., Chang, C.-S., Velu, A. B., He, Y.-C., Huang, Y.-L., Chen, T.-A., Chen, T.-C., Lin, C.-Y., Lin, Y.-C., Shih, S.-R., & Dutta, A. (2019). Effect of aloin on viral neuraminidase and hemagglutinin-specific T cell immunity in acute influenza. Phytomedicine : international Journal of Phytotherapy and Phytopharmacology, 64, 152904. https://doi.org/ 10.1016/j.phymed.2019.152904 [DOI] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Hussain, M. T., Verma, A. R., Vijayakumar, M., Sharma, A., Mathela, C., & Rao, C. V. (2009). Rutin, a natural flavonoid, protects against gastric mucosal damage in experimental animals. 亚洲传统医药, 4(5), 188–197. [Google Scholar]
- Huynh, T., Wang, H., Cornell, W., & Luan, B. (2020). In silico exploration of repurposing and optimizing traditional Chinese medicine rutin for possibly inhibiting SARS-CoV-2's main protease. chemrxiv.org
- Ibrahim, M. A. A., Abdelrahman, A. H. M., Hussien, T. A., Badr, E. A. A., Mohamed, T. A., El-Seedi, H. R., Pare, P. W., Efferth, T., & Hegazy, M.-E F. (2020). In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Computers in Biology and Medicine, 126, 104046. 10.1016/j.compbiomed.2020.104046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imran, M., Rauf, A., Abu-Izneid, T., Nadeem, M., Shariati, M. A., Khan, I. A., Imran, A., Orhan, I. E., Rizwan, M., Atif, M., Gondal, T. A., & Mubarak, M. S. (2019). Luteolin, a flavonoid, as an anticancer agent: A review. Biomedicine & Pharmacotherapy, 112, 108612. https://doi.org/ 10.1016/j.biopha.2019.108612 [DOI] [PubMed] [Google Scholar]
- Jakalian, A., Jack, D. B., & Bayly, C. I. (2002). Fast, efficient generation of high‐quality atomic charges. AM1‐BCC model: I. Method. Journal of Computational Chemistry, 23(16), 1623–1641. 10.1002/jcc.10128 [DOI] [PubMed] [Google Scholar]
- Jannat, K., Hasan, A., Mahamud, R., Jahan, R., Bondhon, T. A., & Farzana, B-n. (2020). In silico screening of Vigna radiata and Vigna mungo phytochemicals for their binding affinity to SARS-CoV-2 (COVID-19) main protease (3CLpro). Journal of Medicinal Plants, 8(4), 89–95. [Google Scholar]
- Jia, S., Hu, Y., Zhang, W., Zhao, X., Chen, Y., Sun, C., Li, X., & Chen, K. (2015). Hypoglycemic and hypolipidemic effects of neohesperidin derived from Citrus aurantium L. in diabetic KK-A(y) mice. Food & Function, 6(3), 878–886. https://doi.org/ 10.1039/c4fo00993b [DOI] [PubMed] [Google Scholar]
- Jin, Z., Du, X., Xu, Y., Deng, Y., Liu, M., Zhao, Y., Zhang, B., Li, X., Zhang, L., Peng, C., Duan, Y., Yu, J., Wang, L., Yang, K., Liu, F., … Yang, H. (2020). Structure of Mpro from COVID-19 virus and discovery of its inhibitors. Nature, 582(7811), 289–293. [DOI] [PubMed]
- Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W., & Klein, M. L. (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics, 79(2), 926–935. 10.1063/1.445869 [DOI] [Google Scholar]
- Kadioglu, O., Saeed, M., Johannes Greten, H., & Efferth, T. (2021). Identification of novel compounds against three targets of SARS CoV-2 coronavirus by combined virtual screening and supervised machine learning. Computers in biology and medicine, 133, 104359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaithwas, G., & Majumdar, D. K. (2010). Evaluation of antiulcer and antisecretory potential of Linum usitatissimum fixed oil and possible mechanism of action. Inflammopharmacology, 18(3), 137–145. 10.1007/s10787-010-0037-5 [DOI] [PubMed] [Google Scholar]
- Kang, J. S., Jeon, Y. J., Park, S.-K., Yang, K.-H., & Kim, H. M. (2004). Protection against lipopolysaccharide-induced sepsis and inhibition of interleukin-1β and prostaglandin E2 synthesis by silymarin. Biochemical Pharmacology, 67(1), 175–181. 10.1016/j.bcp.2003.08.032 [DOI] [PubMed] [Google Scholar]
- Karimi, G., Vahabzadeh, M., Lari, P., Rashedinia, M., & Moshiri, M. (2011). “Silymarin”, a promising pharmacological agent for treatment of diseases. Iranian Journal of Basic Medical Sciences, 14(4), 308. [PMC free article] [PubMed] [Google Scholar]
- Kaus, J. W., Pierce, L. T., Walker, R. C., & McCammon, J. A. (2013). Improving the efficiency of free energy calculations in the amber molecular dynamics package. Journal of Chemical Theory and Computation, 9(9), 4131–4139. 10.1021/ct400340s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khaerunnisa, S., Kurniawan, H., Awaluddin, R., Suhartati, S., & Soetjipto, S. (2020). Potential inhibitor of COVID-19 main protease (Mpro) from several medicinal plant compounds by molecular docking study. 1–14. preprints.org
- Khalid, H., Hussain, R., & Hafeez, A. (2020). Virtual screening of Piperidine based small molecules against COVID-19. Lab-in-Silico, 1(2), 50–55. [Google Scholar]
- Khan, R. J., Jha, R. K., Amera, G. M., Jain, M., Singh, E., Pathak, A., Singh, R. P., Muthukumaran, J., & Singh, A. K. (2020). Targeting SARS-CoV-2: A systematic drug repurposing approach to identify promising inhibitors against 3C-like proteinase and 2′-O-ribose methyltransferase. Journal of Biomolecular Structure and Dynamics, 39(8), 2679–2614. 10.1080/071102.2020.1753577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheirandish, H. (2021). Corona Virus and Salt Intake. Journal of Medicinal and Chemical Sciences, 4(1), 1–7. [Google Scholar]
- Kim, H., Kong, H., Choi, B., Yang, Y., Kim, Y., Lim, M. J., Neckers, L., & Jung, Y. (2005). Metabolic and pharmacological properties of rutin, a dietary quercetin glycoside, for treatment of inflammatory bowel disease. Pharmaceutical Research, 22(9), 1499–1509. https://doi.org/ 10.1007/s11095-005-6250-z [DOI] [PubMed] [Google Scholar]
- Kirst, H. A., & Allen, N. E. (2007). 7.21 - Aminoglycosides antibiotics. In Taylor J. B. & Triggle D. J. (Eds.), Comprehensive medicinal chemistry II (pp. 629–652). Elsevier. [Google Scholar]
- Kolate, A., Kore, G., Lesimple, P., Baradia, D., Patil, S., Hanrahan, J. W., & Misra, A. (2015). Polymer assisted entrapment of netilmicin in PLGA nanoparticles for sustained antibacterial activity. Journal of Microencapsulation, 32(1), 61–74. 10.3109/02652048.2014.944951 [DOI] [PubMed] [Google Scholar]
- Kondo, J., François, B., Russell, R. J., Murray, J. B., & Westhof, E. (2006). Crystal structure of the bacterial ribosomal decoding site complexed with amikacin containing the γ-amino-α-hydroxybutyryl (haba) group. Biochimie, 88(8), 1027–1031. 10.1016/j.biochi.2006.05.017 [DOI] [PubMed] [Google Scholar]
- Korber, B., Fischer, W. M., Gnanakaran, S., Yoon, H., Theiler, J., Abfalterer, W., Hengartner, N., Giorgi, E. E., Bhattacharya, T., Foley, B., Hastie, K. M., Parker, M. D., Partridge, D. G., Evans, C. M., Freeman, T. M., de Silva, T. I., McDanal, C., Perez, L. G., Tang, H., … Wyles, M. D. (2020). Tracking changes in SARS-CoV-2 Spike: Evidence that D614G increases infectivity of the COVID-19 virus. Cell, 182(4), 812–827. 10.1016/j.cell.2020.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber, B., Fischer, W., Gnanakaran, S., Yoon, H., Theiler, J., & Abfalterer, W., & Partridge, D. G. (2020). Spike mutation pipeline reveals the emergence of a more transmissible form of SARS-CoV-2, BioRxiv.
- Kreft, S., Knapp, M., & Kreft, I. (1999). Extraction of rutin from buckwheat (Fagopyrum esculentum Moench) seeds and determination by capillary electrophoresis. Journal of Agricultural and Food Chemistry, 47(11), 4649–4652. 10.1021/jf990186p [DOI] [PubMed] [Google Scholar]
- Kumar, S., Priya Matharasi, D., Gopi, S., Sivakumar, S., & Narasimhan, S. (2010). Synthesis of cytotoxic and antioxidant Schiff's base analogs of aloin. Journal of Asian Natural Products Research, 12(5), 360–370. 10.1080/10286021003775327 [DOI] [PubMed] [Google Scholar]
- Kurisawa, M., Chung, J. E., Uyama, H., & Kobayashi, S. (2003). Enzymatic synthesis and antioxidant properties of poly (rutin). Biomacromolecules, 4(5), 1394–1399. 10.1021/bm034136b [DOI] [PubMed] [Google Scholar]
- Lam, S., Lombardi, A., & Ouanounou, A. (2020). COVID-19: A review of the proposed pharmacological treatments. European Journal of Pharmacology, 886, 173451. 10.1016/j.ejphar.2020.173451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, G. G., Bian, G. X., Ren, J. P., Wen, L. Q., Zhang, M., & Lü, Q. J. (2007). Protective effect of madecassoside against reperfusion injury after regional ischemia in rabbit heart in vivo. Yao xue xue bao = Acta Pharmaceutica Sinica, 42(5), 475–480 [PubMed] [Google Scholar]
- Li, Y., Yao, J., Han, C., Yang, J., Chaudhry, M. T., Wang, S., Liu, H., & Yin, Y. (2016). Quercetin, Inflammation and Immunity. Nutrients, 8(3), 167. https://doi.org/ 10.3390/nu8030167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinski, C. A. (2004). Lead-and drug-like compounds: The rule-of-five revolution. Drug Discovery Today: Technologies, 1(4), 337–341. 10.1016/j.ddtec.2004.11.007 [DOI] [PubMed] [Google Scholar]
- Liu, C.-H., Jassey, A., Hsu, H.-Y., & Lin, L.-T. (2019). Antiviral activities of silymarin and derivatives. Molecules, 24(8), 1552. 10.3390/molecules24081552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Lazaro, M. (2009). Distribution and biological activities of the flavonoid luteolin. Mini-Reviews in Medicinal Chemistry, 9(1), 31–59. 10.2174/138955709787001712 [DOI] [PubMed] [Google Scholar]
- López-Revuelta, A., Sánchez-Gallego, J. I., Hernández-Hernández, A., Sánchez-Yagüe, J., & Llanillo, M. (2006). Membrane cholesterol contents influence the protective effects of quercetin and rutin in erythrocytes damaged by oxidative stress. Chemico-Biological Interactions, 161(1), 79–91. 10.1016/j.cbi.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Lovell, S. C., Davis, I. W., Arendall, W. B., de Bakker, P. I. W., Word, J. M., Prisant, M. G., Richardson, J. S., & Richardson, D. C. (2003). Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins: Structure, Function, and Bioinformatics, 50(3), 437–450. 10.1002/prot.10286 [DOI] [PubMed] [Google Scholar]
- Lyne, P. D., Lamb, M. L., & Saeh, J. C. (2006). Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. Journal of Medicinal Chemistry, 49(16), 4805–4808. https://doi.org/ 10.1021/jm060522a [DOI] [PubMed] [Google Scholar]
- Ma, L., Tang, L., & Yi, Q. (2019). Salvianolic acids: Potential source of natural drugs for the treatment of fibrosis disease and cancer. Frontiers in Pharmacology, 10, 97. 10.3389/fphar.2019.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmoud, A. M., & Hussein, O. (2016). Hesperidin as a promising anti-diabetic flavonoid: The underlying molecular mechanism. International Journal of Food and Nutritional Science, 3, 1–2. [Google Scholar]
- Mandal, C. C., & Panwar, M. (2020). Can the summer temperature drop COVID-19 cases? Public Health, 185, 72–79. 10.1016/j.puhe.2020.05.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehla, R., Bivalkar-Mehla, S., & Chauhan, A. (2011). A flavonoid, luteolin, cripples HIV-1 by abrogation of tat function. PLoS One, 6(11), e27915. 10.1371/journal.pone.0027915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milić, N., Milošević, N., Suvajdžić, L., Žarkov, M., & Abenavoli, L. (2013). New therapeutic potentials of milk thistle (Silybum marianum). Natural Product Communications, 8(12), 193451300801236. 10.1177/1934578X1300801236 [DOI] [PubMed] [Google Scholar]
- Mirzaei, M., Harismah, K., Da'i, M., Salarrezaei, E., & Roshandel, Z. (2020). Screening efficacy of available HIV protease inhibitors on COVID-19 protease. Journal Mil Med, 22(2), 100–107. [Google Scholar]
- Mohamed, K., Yazdanpanah, N., Saghazadeh, A., & Rezaei, N. (2021). Computational drug discovery and repurposing for the treatment of Covid-19: A systematic review. Bioorganic chemistry, 106, 104490. [DOI] [PMC free article] [PubMed]
- Morand, C., Manach, C., Crespy, V., & Remesy, C. (2000). Respective bioavailability of quercetin aglycone and its glycosides in a rat model. Biofactors, 12(1–4), 169–174. 10.1002/biof.5520120127 [DOI] [PubMed] [Google Scholar]
- Munster, A. M. (1979). Netilmicin: A new aminoglycoside in the treatment of septic burn patients. Archives of Surgery, 114(1), 28–30. 10.1001/archsurg.1979.01370250030005 [DOI] [PubMed] [Google Scholar]
- Nabavi, S. F., Braidy, N., Gortzi, O., Sobarzo-Sanchez, E., Daglia, M., Skalicka-Woźniak, K., & Nabavi, S. M. (2015). Luteolin as an anti-inflammatory and neuroprotective agent: A brief review. Brain Research Bulletin, 119(Pt A), 1–11. 10.1016/j.brainresbull.2015.09.002 [DOI] [PubMed] [Google Scholar]
- Naik, B., Gupta, N., Ojha, R., Singh, S., Prajapati, V. K., & Prusty, D. (2020). High throughput virtual screening reveals SARS-CoV-2 multi-target binding natural compounds to lead instant therapy for COVID-19 treatment. International Journal of Biological Macromolecules, 160, 1–17. 10.1016/j.ijbiomac.2020.05.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan, N., & Nair, D. T. Ritonavir may inhibit exoribonuclease activity of nsp14 from the SARS-CoV-2 virus and potentiate the activity of chain terminating drugs, Laboratory of Genomic Integrity and Evolution, Regional Centre for Biotechnology, NCR Biotech Science Cluster, 3rd Milestone, Faridabad-Gurgaon Expressway, Faridabad- 121001. Haryana. 2Manipal Academy of Higher Education, Manipal 576104, Karnataka. India. chemrxiv.org
- Networking Chemical Biology. (2008). Nat Chem Biol, 4, 633. 10.1038/nchembio1108-633 [DOI] [PubMed]
- Nićiforović, A., Adžić, M., Zarić, B., & Radojčić, M. (2007). Adjuvant antiproliferative and cytotoxic effect of aloin in irradiated HeLaS3 cells. Russian Journal of Physical Chemistry A, 81(9), 1463–1466. 10.1134/S0036024407090221 [DOI] [Google Scholar]
- Ojha, H., Sharma, K., Kallepalli, S., Raina, S., & Agrawala, P. K. (2016). In-vitro evaluation of rutin and rutin hydrate as potential radiation countermeasure agents. Internatuinal Journal of Radiation Research, 14(1), 9–16. 10.18869/acadpub.ijrr.14.1.9 [DOI] [Google Scholar]
- Ozkendir, O. M., Askar, M., & Kocer, N. E. (2020). Influence of the epidemic COVID-19: An outlook on health, business and scientific studies. Lab-in-Silico, 1(1), 26–30. [Google Scholar]
- Pandey, P., Rane, J. S., Chatterjee, A., Kumar, A., Khan, R., Prakash, A., & Ray, S. (2020). Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: An in silico study for drug development. Journal of Biomolecular Structure and Dynamics, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, R. K., Sharma, D., Bhatt, T. K., Sundar, S., & Prajapati, V. K. (2015). Developing imidazole analogues as potential inhibitor for Leishmania donovani trypanothione reductase: Virtual screening, molecular docking, dynamics and ADMET approach. Journal of Biomolecular Structure and Dynamics, 33(12), 2541–2553. 10.1080/07391102.2015.1085904 [DOI] [PubMed] [Google Scholar]
- Park, C. M., & Song, Y. S. (2019). Luteolin and luteolin-7-O-glucoside protect against acute liver injury through regulation of inflammatory mediators and antioxidative enzymes in GalN/LPS-induced hepatitic ICR mice. Nutrition Research and Practice, 13(6), 473–479. 10.4162/nrp.2019.13.6.473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, M.-Y., Kwon, H.-J., & Sung, M.-K. (2011). Dietary aloin, aloesin, or aloe-gel exerts anti-inflammatory activity in a rat colitis model. Life Sciences, 88(11–12), 486–492. 10.1016/j.lfs.2011.01.010 [DOI] [PubMed] [Google Scholar]
- Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF Chimera-a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612. https://doi.org/ 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Prayle, A., & Smyth, A. R. (2010). Aminoglycoside use in cystic fibrosis: Therapeutic strategies and toxicity. Current Opinion in Pulmonary Medicine, 16(6), 604–610. 10.1097/MCP.0b013e32833eebfd [DOI] [PubMed] [Google Scholar]
- Pushpakom, S., Iorio, F., Eyers, P. A., Escott, K. J., Hopper, S., Wells, A., Doig, A., Guilliams, T., Latimer, J., McNamee, C., Norris, A., Sanseau, P., Cavalla, D., & Pirmohamed, M. (2019). Drug repurposing: progress, challenges and recommendations. Nature Reviews. Drug Discovery, 18(1), 41–58. https://doi.org/ 10.1038/nrd.2018.168 [DOI] [PubMed] [Google Scholar]
- Ramirez, M. S., & Tolmasky, M. E. (2017). Amikacin: Uses, resistance, and prospects for inhibition. Molecules, 22(12), 2267. 10.3390/molecules22122267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Release, S. J. E. Schrödinger, LLC, New York, NY, Impact, Schrödinger, LLC, New York, NY. (2016). 1: Schrödinger Suite 2017-1 Protein Preparation Wizard.
- Riahi-Chebbi, I., Souid, S., Othman, H., Haoues, M., Karoui, H., Morel, A., Srairi-Abid, N., Essafi, M., & Essafi-Benkhadir, K. (2019). The Phenolic compound Kaempferol overcomes 5-fluorouracil resistance in human resistant LS174 colon cancer cells. Scientific Reports, 9(1), 195. 10.1038/s41598-018-36808-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimanshee, A., Amit, D., Vishal, P., & Mukesh, K. (2020). Potential inhibitors against papain-like protease of novel coronavirus (SARS-CoV-2) from FDA approved drugs.
- Roe, D. R., & Cheatham, T. E. (2013). PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. Journal of Chemical Theory and Computation, 9(7), 3084–3095. https://doi.org/ 10.1021/ct400341p [DOI] [PubMed] [Google Scholar]
- Roh, C. (2012). A facile inhibitor screening of SARS Coronavirus N protein using nanoparticle-based RNA oligonucleotide. International journal of nanomedicine, 7, 2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckaert, J.-P., & Bellemans, A. (1978). Molecular dynamics of liquid alkanes. Faraday Discussions of the Chemical Society, 66, 95–106. 10.1039/dc9786600095 [DOI] [Google Scholar]
- Salehi, B., Venditti, A., Sharifi-Rad, M., Kręgiel, D., Sharifi-Rad, J., Durazzo, A., Lucarini, M., Santini, A., Souto, E., Novellino, E., Antolak, H., Azzini, E., Setzer, W., & Martins, N. (2019). The therapeutic potential of apigenin. International Journal of Molecular Sciences, 20(6), 1305. 10.3390/ijms20061305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarth, S., & Kirk, M. (2020). Energetics based modeling of hydroxychloroquine and azithromycin binding to the SARS-CoV-2 spike (S)Protein - ACE2 complex.
- Sarkodie, S. A., & Owusu, P. A. (2020). Impact of meteorological factors on COVID-19 pandemic: Evidence from top 20 countries with confirmed cases. Environmental Research, 191, 110101. 10.1016/j.envres.2020.110101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, A. (2020). Drug targets for COVID-19 therapeutics: Ongoing global efforts. Journal of Biosciences, 45(1), 1–24. 10.1007/s12038-020-00067-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selloum, L., Bouriche, H., Tigrine, C., & Boudoukha, C. (2003). Anti-inflammatory effect of rutin on rat paw oedema, and on neutrophils chemotaxis and degranulation. Experimental and Toxicologic Pathology, 54(4), 313–318. 10.1078/0940-2993-00260 [DOI] [PubMed] [Google Scholar]
- Shankar, U., Jain, N., Majee, P., Mishra, S. K., Rathi, B., & Kumar, A. (2020). Potential drugs targeting Nsp16 protein may corroborates a promising approach to combat SARS-CoV-2 virus.
- Shibano, M., Lin, A.-S., Itokawa, H., & Lee, K.-H. (2007). Separation and characterization of active flavonolignans of Silybum marianum by Liquid Chromatography connected with Hybrid Ion-Trap and Time-of-Flight Mass Spectrometry (LC–MS/IT-TOF). Journal of Natural Products, 70(9), 1424–1428. 10.1021/np070136b [DOI] [PubMed] [Google Scholar]
- Standl, E., Theodorakis, M. J., Erbach, M., Schnell, O., & Tuomilehto, J. (2014). On the potential of acarbose to reduce cardiovascular disease. Cardiovascular Diabetology, 13(1), 81. 10.1186/1475-2840-13-81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner, J. A., Zheng, B.-J., Zhou, J., Watt, R. M., Jiang, J.-Q., Wong, K.-L., Lin, Y.-P., Lu, L.-Y., He, M.-L., Kung, H.-F., Kesel, A. J., & Huang, J.-D. (2005). The adamantane-derived bananins are potent inhibitors of the helicase activities and replication of SARS coronavirus. Chemistry & Biology, 12(3), 303–311. https://doi.org/ 10.1016/j.chembiol.2005.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tariq, A., Mateen, R., Sohail Afzal, S., & Saleem, M. (2020). Paromomycin: A potential dual targeted drug effectively inhibits both spike (S1) and main protease of COVID-19. International Journal of Infectious Diseases, 98, 166–175. [DOI] [PMC free article] [PubMed]
- Theoharides, T. C. (2020). COVID-19, pulmonary mast cells, cytokine storms, and beneficial actions of luteolin. Biofactors, 46(3), 306–308. 10.1002/biof.1633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, B., & Hua, Y. (2005). Concentration-dependence of prooxidant and antioxidant effects of aloin and aloe-emodin on DNA. Food Chemistry, 91(3), 413–418. 10.1016/j.foodchem.2004.06.018 [DOI] [Google Scholar]
- Torres, A., Motos, A., Battaglini, D., & Li Bassi, G. (2018). Inhaled amikacin for severe Gram-negative pulmonary infections in the intensive care unit: Current status and future prospects. Crit Care, 22(1), 343. 10.1186/s13054-018-1958-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truchon, J.-F., & Bayly, C. I. (2007). Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. Journal of Chemical Information and Modeling, 47(2), 488–508. https://doi.org/ 10.1021/ci600426e [DOI] [PubMed] [Google Scholar]
- Ubani, A., Agwom, F., Morenikeji, O. R., Shehu, N. Y., Luka, P., Umera, E. A., Umar, U., Omale, S., Aguiyi, J. C., Nnadi, N. E., & Luka, P. D. (2020). Molecular docking analysis of some phytochemicals on two SARS-CoV-2 targets. F1000Research, 9(1157), 1157. [Google Scholar]
- Vijayan, R., & Gourinath, S. (2021). Structure-based inhibitor screening of natural products against NSP15 of SARS-CoV-2 revealed Thymopentin and Oleuropein as potent inhibitors. Journal of proteins and proteomics, 1–10. [DOI] [PMC free article] [PubMed]
- Vinayagam, R., & Xu, B. (2015). Antidiabetic properties of dietary flavonoids: A cellular mechanism review. Nutrition & Metabolism, 12(1), 60. 10.1186/s12986-015-0057-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent, M. J., Bergeron, E., Benjannet, S., Erickson, B. R., Rollin, P. E., Ksiazek, T. G., Seidah, N. G., & Nichol, S. T. (2005). Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virology Journal, 2, 69. https://doi.org/ 10.1186/1743-422X-2-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J., Wang, W., Kollman, P. A., & Case, D. A. (2006). Automatic atom type and bond type perception in molecular mechanical calculations. Journal of Molecular Graphics & Modelling, 25(2), 247–260. https://doi.org/ 10.1016/j.jmgm.2005.12.005 [DOI] [PubMed] [Google Scholar]
- Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A., & Case, D. A. (2004). Development and testing of a general amber force field. Journal of Computational Chemistry, 25(9), 1157–1174. 10.1002/jcc.20035 [DOI] [PubMed] [Google Scholar]
- Wang, J., Xiong, X., & Feng, B. (2013). Cardiovascular effects of salvianolic acid B. Evidence-Based Complementary and Alternative Medicine, 2013, 1–16. 10.1155/2013/247948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S-w., Wang, Y.-J., Su, Y-j., Zhou, W-w., Yang, S-g., Zhang, R., Zhao, M., Li, Y-n., Zhang, Z-p., Zhan, D-w., & Liu, R-t. (2012). Rutin inhibits β-amyloid aggregation and cytotoxicity, attenuates oxidative stress, and decreases the production of nitric oxide and proinflammatory cytokines. Neurotoxicology, 33(3), 482–490. https://doi.org/ 10.1016/j.neuro.2012.03.003 [DOI] [PubMed] [Google Scholar]
- Wu, C., Liu, Y., Yang, Y., Zhang, P., Zhong, W., Wang, Y., Wang, Q., Xu, Y., Li, M., Li, X., Zheng, M., Chen, L., & Li, H. (2020). Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharmaceutica Sinica B, 10(5), 766–788. 10.1016/j.apsb.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, H., Liu, Y., Chen, X., Zhu, D., Ma, J., Yan, Y., Si, M., Li, X., Sun, C., Yang, B., He, Q., & Chen, K. (2017). Neohesperidin Exerts Lipid-Regulating Effects in vitro and in vivo via Fibroblast Growth Factor 21 and AMP-Activated Protein Kinase/Sirtuin Type 1/Peroxisome Proliferator-Activated Receptor Gamma Coactivator 1α Signaling Axis. Pharmacology, 100(3–4), 115–126. https://doi.org/ 10.1159/000452492 [DOI] [PubMed] [Google Scholar]
- Xagorari, A., Papapetropoulos, A., Mauromatis, A., Economou, M., Fotsis, T., & Roussos, C. (2001). Luteolin inhibits an endotoxin-stimulated phosphorylation cascade and proinflammatory cytokine production in macrophages. Journal of Pharmacology and Experimental Therapeutics, 296(1), 181–187. [PubMed] [Google Scholar]
- Xiang, Z. (2006). Advances in homology protein structure modeling. Current Protein and Peptide Science, 7(3), 217–227. 10.2174/138920306777452312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaoqi, W., Xin, B., Xu, Z., Li, K., Li, F., Zhong, W., … Peng, S. (2020). Network representation learning-based drug mechanism discovery and anti-inflammatory response against COVID-19.
- Yan, H., Ma, L., Wang, H., Wu, S., Huang, H., Gu, Z., Jiang, J., & Li, Y. (2019). Luteolin decreases the yield of influenza A virus in vitro by interfering with the coat protein I complex expression. Journal of Natural Medicines, 73(3), 487–496. https://doi.org/ 10.1007/s11418-019-01287-7 [DOI] [PubMed] [Google Scholar]
- Yang, J., Zheng, Y., Gou, X., Pu, K., Chen, Z., Guo, Q., Ji, R., Wang, H., Wang, Y., & Zhou, Y. (2020). Prevalence of comorbidities and its effects in patients infected with SARS-CoV-2: a systematic review and meta-analysis. International Journal of Infectious Diseases, 94, 91–95. https://doi.org/ 10.1016/j.ijid.2020.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, R., Chen, L., Lan, R., Shen, R., & Li, P. (2020). Computational screening of antagonists against the SARS-CoV-2 (COVID-19) coronavirus by molecular docking. International Journal of Antimicrobial Agents, 56(2), 106012. 10.1016/j.ijantimicag.2020.106012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanwar, A. A., Badole, S. L., Shende, P. S., Hegde, M. V., & Bodhankar, S. L. (2014). Chapter 76 - Cardiovascular effects of hesperidin: A flavanone glycoside. In Watson R. R., Preedy V. R., & Zibadi S. (Eds.), Polyphenols in human health and disease (pp. 989–992). Academic Press. [Google Scholar]
- Zhang, B., Zhang, J., Zhang, C., Zhang, X., Ye, J., Kuang, S., Sun, G., & Sun, X. (2018). Notoginsenoside R1 protects against diabetic cardiomyopathy through activating estrogen receptor α and its downstream signaling, Frontiers in pharmacology, 9, 1227. 10.3389/fphar.2018.01227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q., Xiao, X., Li, M., Li, W., Yu, M., Zhang, H., Wang, Z., & Xiang, H. (2013). Acarbose reduces blood glucose by activating miR-10a-5p and miR-664 in diabetic rats. PloS One, 8(11), p.e79697. 10.1371/annotation/330eea00-9c09-4982-a458-3add994bab9d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z. F., Fan, S. H., Zheng, Y. L., Lu, J., Wu, D. M., Shan, Q., & Hu, B. (2014). Troxerutin improves hepatic lipid homeostasis by restoring NAD(+)-depletion-mediated dysfunction of lipin 1 signaling in high-fat diet-treated mice. Biochemical Pharmacology, 91(1), 74–86. 10.1016/j.bcp.2014.07.002 [DOI] [PubMed] [Google Scholar]
- Zhuang, P., Zhang, Y., Cui, G., Bian, Y., Zhang, M., Zhang, J., Liu, Y., Yang, X., Isaiah, A. O., Lin, Y., & Jiang, Y. (2012). Direct stimulation of adult neural stem/progenitor cells in vitro and neurogenesis in vivo by salvianolic acid B. PloS One, 7(4), e35636. https://doi.org/ 10.1371/journal.pone.0035636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey, R., Bibis, S. S., Zhu, T., & Dhalladoo, S. (2012). Characterization of a compensatory mutant of Leishmania major that lacks ether lipids but exhibits normal growth, and G418 and hygromycin resistance. Experimental Parasitology, 130(3), 200–204. 10.1016/j.exppara.2012.01.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.