Abstract
Background
The recently launched high-throughput assays for detecting antibodies against SARS-CoV-2 has contributed to the managing strategies for the COVID-19 pandemic. This study aimed to investigate the performance of three high-throughput assays and one rapid lateral flow test relative to regulatory authorities' recommended criteria.
Methods
A total of 315 samples, including 150 pre-pandemic samples, 152 samples from SARS-CoV-2 RT-PCR positive individuals and 13 potentially cross-reactive samples were analysed with SARS-CoV-2 IgG (Abbott, Abbott Park, IL), Elecsys Anti-SARS-CoV-2 (Roche, Solna, Sweden), LIAISON SARS-CoV-2 S1/S2 IgG (DiaSorin, Saluggia, Italy) and 2019-nCOV IgG/IgM Rapid Test (Dynamiker Biotechnology Co., Tianjin, China).
Results
All assays performed with a high level of specificity ranging from 96.7% to 99.3%. Sensitivity differed more between the assays, Roche exhibiting the highest sensitivity of 98.7%. The corresponding figures for Abbott, DiaSorin and Dynamiker Biotechnology were 80.9%, 89.0% and 72.4%, respectively.
Conclusions
The results of the evaluated SARS-CoV-2 assays vary considerably, as well as their ability to fulfil the performance criteria proposed by regulatory authorities. Introduction into clinical use in low-prevalent settings, should, therefore, be made with caution.
Keywords: COVID-19, SARS-CoV-2, serology, immunology, antibodies
Introduction
Coronavirus disease 2019 (COVID-19) was first reported in Wuhan, Hubei province, China, in December 2019 [1]. Since then, several rapid tests based on immunochromatographic techniques have been developed. These point-of-care tests usually deliver results within 15–30 min; however, the tests' nature makes large-scale testing inefficient.
Several manufacturers have developed immunoassays based on recognition of recombinant antigens to achieve high-throughput detection of anti-SARS-CoV-2 antibodies in plasma or serum. Some tests claim to detect IgM or IgG, while others also identify IgA or a mixture of different antibody classes. Differences in test design, including choice of antigen, are likely to affect the sensitivity and specificity of tests [2], as well as the characteristics of the SARS-CoV-2 antibody response [3,4]. In this study, the performance of three commercially available high-throughput automated SARS-CoV-2 antibody assays and one rapid immunochromatographic test was investigated and compared to regulatory authorities' recommended criteria.
Materials and methods
Assays
Four commercially available immunoassays and their corresponding platforms were evaluated: (1) Abbott SARS-CoV-2 IgG on the ARCHITECT i2000 (Abbott, Abbott Park, IL), recognizing IgG antibodies binding to the SARS-CoV-2 nucleoprotein in a chemiluminescent microparticle immunoassay (CMIA); (2) Elecsys Anti-SARS-CoV-2 on the Cobas 8000 e801 (Roche Diagnostic Scandinavia AB, Solna, Sweden) detecting antibodies (including IgG) binding to the SARS-CoV-2 nucleocapsid (N) protein; (3) LIAISON SARS-CoV-2 S1/S2 IgG on the LIAISON XL (DiaSorin, Saluggia, Italy) detecting IgG antibodies recognizing the spike glycoprotein of the coronavirus; and (4) the lateral flow test 2019-nCOV IgG/IgM Rapid Test (Dynamiker Biotechnology Co., Tianjin, China) detecting antibodies with unspecified epitope recognition. Kit-recommended positive cut-off values for interpretation were applied: Abbott; ≥1.4 S/CO, Roche; ≥1.0 COI, DiaSorin; ≥15 AU/mL (positive) and 12–15 AU/mL (equivocal). Any positive Dynamiker result, either for IgG, IgM or both, was considered positive.
All automated systems are part of our laboratories' routine operations and are subject to accepted quality assurance procedures. All tests (including calibration and controls) were performed according to manufacturers' instructions.
Sample collections
The samples originated from a Microbiology Department collection obtained after consent to deposit, store and use for research and development. Samples were anonymized before inclusion. Consequently, according to the Swedish Ethical Review Agency's guidelines, the study did not require approval from an ethics committee. All serum samples were stored at −20 °C until analysis.
Specificity was evaluated using 150 pre-pandemic (2018) samples. To challenge the assays, 13 additional serum samples with possible interferences (antinuclear antibodies (n = 2); rheumatoid factor (n = 2); anti-cytomegalovirus IgM (n = 2); anti-Epstein-Barr virus IgM (n = 2) and samples from pregnant donors (n = 5)) were analysed.
Sensitivities were evaluated using 152 outpatient serum samples from 147 individuals that before serum sampling had tested PCR positive for SARS-CoV-2 from nasopharyngeal and/or pharyngeal swabs. Due to a global lack of reagents for RT-PCR diagnostics at the beginning of the pandemic, different protocols and reagents had to be used. Analysis was performed as part of routine using Abbott RealTime SARS-CoV-2 Amplification Reagent Kit on Abbott m2000; Alinity M SARS-CoV-2 AMP Kit or Alinity M Resp-4-Plex AMP Kit on Abbott Alinity M; or Xpert® Xpress SARS-CoV-2 (Cepheid, Sunnyvale, CA) on GeneExpert Dx, or by external laboratories: ABCLabs (Solna, Sweden) using TaqPath™ COVID-19 CE-IVD RT-PCR Kit (Thermo Fisher Scientific, Waltham, MA) or by the National Veterinary Institute (Uppsala, Sweden) using KiCqStart qPCR ReadyMix on ABI 7500 with primers and probes according to Corman et al. [5]. The interval between positive PCR and serum sample collection ranged from 18 to 125 days (median 78 days).
Calculations
Overall per cent agreement, sensitivity (per cent positive agreement) and specificity (per cent negative agreement) were calculated based on a contingency table according to EP12-A2 [6], using Microsoft Excel 2019 (Microsoft Corp., Redmond, WA). The between-test agreement was evaluated using Cohen's kappa calculated with IBM SPSS Statistics for Windows, Version 26.0 (IBM Corp., Armonk, NY). The positive likelihood ratio (LR+) and positive predictive value (PPV) at different prevalence (P) scenarios were calculated as follows:
Further data analysis was performed using GraphPad Prism, version 7.04 for Windows (GraphPad Software, La Jolla, CA).
Clinical performance requirements
The performance of each assay was compared with published guidelines from three regulatory authorities (Table 1): Public Health Agency of Sweden (PHAS); Haute Autorité de Santé, Saint-Denis, France (HAS); and Centres for Disease Control and Prevention, USA (CDC) (Atlanta, GA) [7–9].
Table 1.
Summary of recommended criteria for SARS-CoV-2 serology assays issued by authorities in three different countries.
| Regulatory authority | Sensitivity (%) | Specificity (%) | Additional comment |
|---|---|---|---|
| Public Health Agency of Sweden | 90.0 | 99.5 | Recommendations based on seroprevalence of 5%, rendering a target PPV of >90% |
| Haute Autorité de Santé, France | 95.0 | 98.0 | |
| Centers for Disease Control and Prevention, USA | – | 99.5 | For populations with seroprevalence of ≥5% |
Results
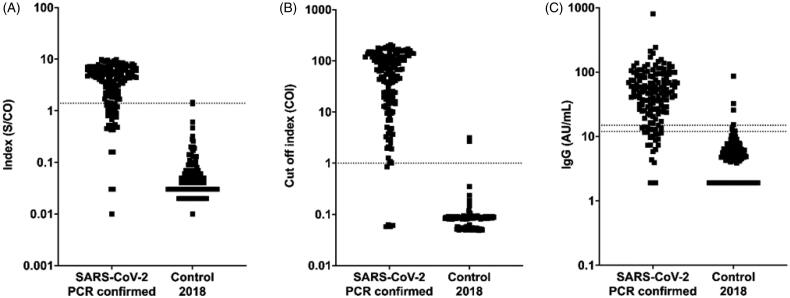
Of the 150 pre-pandemic samples tested, Roche reported two false-positive results, DiaSorin six (including two equivocal results), Dynamiker Biotechnology five and Abbott one (Figure 1). The negative sample collection resulted in median values (range) of 0.08 COI (0.05–3.16), 0.04 S/CO (0.01–1.47) and 4.8 AU/mL (<3.8–85.9) for Roche, Abbott and DiaSorin, respectively.
Figure 1.
Differences in distribution patterns between Abbott, Roche and DiaSorin immunoassays for detection of SARS-CoV-2 antibodies. Measured values from serology testing of 152 positive (SARS-CoV-2 PCR confirmed) and 150 negative samples. Dotted lines represent cut-off values of (A) Abbott: positive result index ≥1.4 S/CO, (B) Roche: positive cut-off index (COI) ≥1.0 and (C) DiaSorin: positive cut-off ≥15 AU/mL and equivocal 12–15 AU/mL. For DiaSorin, negative samples with signals below the detection limit (3.8 AU/mL) were plotted as 1.9 AU/mL and positive signals >400 AU/mL were plotted as 800 AU/mL.
In the panel consisting of 13 potentially cross-reactive pre-pandemic samples, one sample with rheumatoid factor IgM was reported as positive for both SARS-CoV-2 IgM and IgG by Dynamiker. One sample obtained during pregnancy showed an equivocal result on the DiaSorin assay. All samples were negative on the Abbott and Roche assays.
Two RT-PCR positive samples were reported as negative by Roche, 29 by Abbott and 42 by Dynamiker. DiaSorin reported 23 negative results, including seven equivocal results.
The overall agreement was 90.1% for Abbott, 98.6% for Roche, 90.4% for DiaSorin and 84.4% for Dynamiker Biotechnology. The pairwise inter-assay agreements (Cohen’s kappa) were as follows: Roche and Abbott 0.802 (95% CI, 0.735–0.869), p<.0005; Roche and Diasorin 0.863 (95% CI, 0.806–0.920), p<.0005; Roche and Dynamiker Biotechnology 0.689 (95% CI, 0.611–0.767), p<.0005; Abbott and DiaSorin 0.806 (95% CI, 0.737–0.875), p<.0005; Abbott and Dynamiker Biotechnology 0.772 (95% CI, 0.699–0.845), p<.0005; and DiaSorin and Dynamiker Biotechnology 0.721 (95% CI, 0.641–0.801), p<.0005.
Sensitivity and specificity for each assay are presented in Table 2, together with the corresponding data extracted from each manufacturer's test kit insert. Abbott exhibited the highest specificity and Roche the highest sensitivity, 99.3% and 98.7%, respectively. Seven RT-PCR positive samples and two samples from the negative collection were excluded from the DiaSorin sample collection due to equivocal results. LR + were 121.4, 74.0, 32.9 and 21.7 for Abbott, Roche, DiaSorin and Dynamiker Biotechnology, respectively.
Table 2.
Sensitivity and specificity compared to manufacturers' data from samples collected ≥14 days (Abbott, Roche) and >15 days (DiaSorin) post PCR confirmation.
| Study data |
Manufacturers' dataa |
|||||||
|---|---|---|---|---|---|---|---|---|
| n | Sensitivity % (95% CI) | n | Specificity % (95% CI) | n | Sensitivity % (95% CI) | n | Specificity % (95% CI) | |
| Abbott | 152 | 80.9 (74.0–86.4) | 150 | 99.3 (96.3–100) | 88 | 100 (95.9–100) | 997 | 99.6 (99.0–99.9) |
| Roche | 152 | 98.7 (95.3–99.8) | 150 | 98.7 (95.3.99.8) | 185 | 99.5 (97.0–100) | 6305 | 99.8 (99.7–99.9) |
| DiaSorin | 145 | 89.0 (82.8–93.1) | 148 | 97.3 (93.3–98.9) | 48 | 97.9 (89.1–99.6) | 1200 | 98.6 (97.7–99.1) |
| Dynamiker | 152 | 72.4 (64.8–78.9) | 150 | 96.7 (92.4–98.6) | 162 | 93.2 (not reported) | 300 | 95.3 (not reported) |
No information about the time of sampling was available for Dynamiker Biotechnology.
Data retrieved from following versions of product kit inserts: Abbott 6R86 H0791R04, June 2020; Roche V3, 2020-06; DiaSorin 200/007-797, 07, 2020-07; Dynamiker Biotechnology DNK-1419-1.
Benchmarking the sensitivity and specificity data against published guidelines showed that the Abbott and Roche assays met the specificity criteria set by HAS. None of the assays met the specificity criteria by PHAS and CDC. Neither of the Abbott, DiaSorin or Dynamiker Biotechnology assays exhibited a sufficient sensitivity to meet the criteria set by PHAS and HAS (Tables 1 and 2). In contrast, Roche exhibited a sensitivity of 98.7%, thus exceeding the sensitivity criteria set by both these authorities.
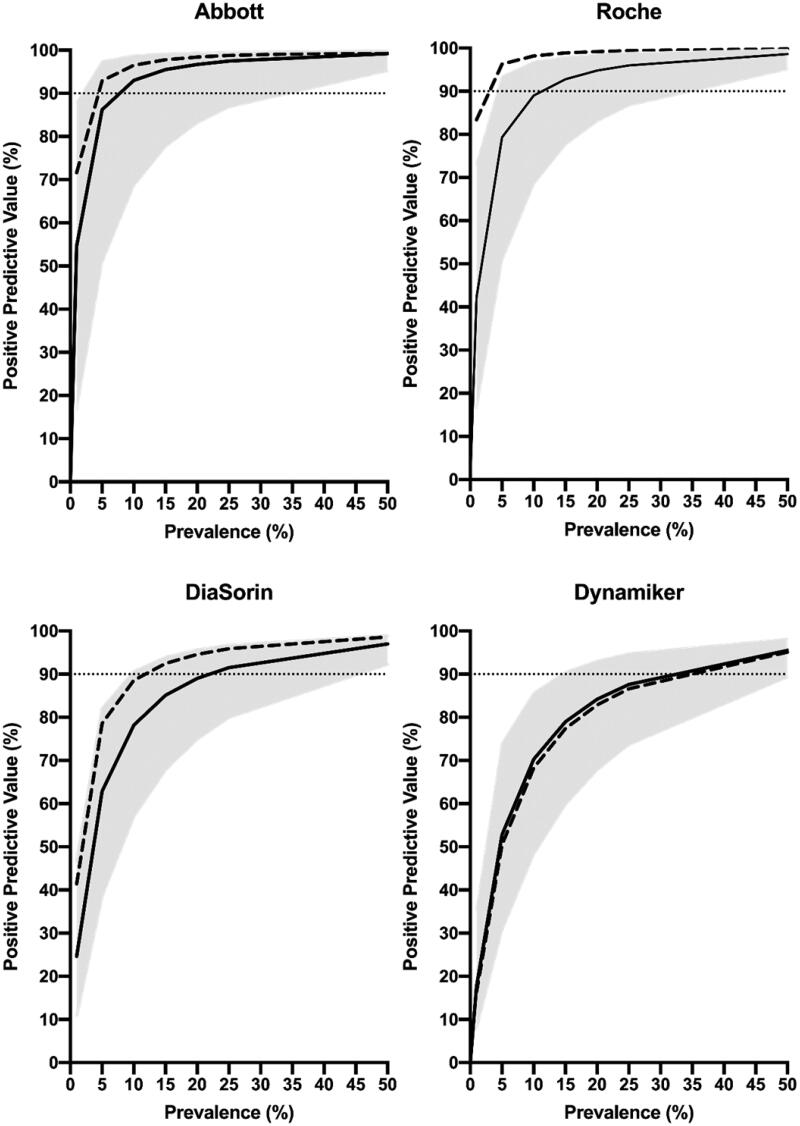
Assay performance recommendations from PHAS are based on a target PPV of ≥90%. Therefore, the seroprevalence required to reach a PPV of ≥90% was calculated (Figure 2).
Figure 2.
Estimated PPV for the assays, calculated at seroprevalences of 1%, 5%, 10%, 15%, 20%, 25% and 50%. Calculations were based on sensitivity, specificity and respective 95% CI limits (solid lines = mean values; grey areas = 95% CI). Dashed lines represent mean sensitivity and specificity data from the manufacturers' kit inserts. Dotted horizontal lines refer to 90% PPV.
Discussion
Although the degree of immunity to SARS-CoV-2 is disputed, it is plausible that individuals undergoing COVID-19 will gain partial or temporary protection against new episodes. The rapidly emerging pandemic has limited the possibility of state-of-the-art validations according to the EP-12 A2 [5], and there is no international consensus on clinical performance requirements for COVID-19 assays. Neither is there an agreed gold standard method for antibody testing [10]. Using the criteria proposed by authorities [7–9], only Roche fulfilled an authority criteria (HAS), which is in accordance with other studies [11,12]. However, in one Austrian study [13], Roche also fulfilled the CDC requirements.
A qualitative diagnostic test's clinical performance is dependent on the prevalence. Both PHAS and CDC accordingly link their respective specificity criteria for SARS-CoV-2 antibody testing to seroprevalence levels where, PHAS recommendations are based on a PPV >90%. Based on our results, a prevalence of approximately 8%, 13%, 29% and 44% would be necessary to reach this goal for Abbott, Roche, Dynamiker Biotechnology and DiaSorin, respectively (Figure 2).
Introducing comprehensive antibody testing for COVID-19 is cumbersome in a low-prevalence setting. In order to reach the proposed performance criteria, two-tier testing could be considered. Alternatively, a high degree of specificity (and thus a high PPV) could be prioritized using modified cut-off values. Our results suggest that this could be conceivable for the Abbott and Roche assays, while the more overlapping results for DiaSorin would make it difficult to find such a specific cut-off with a reasonably preserved degree of sensitivity (Figure 1). As the seroprevalence increases, the need for two-tier testing or modifications to improve specificity will decrease. Due to the disease's novelty and the expedited development of new assays, possible sources of systematic errors, e.g. cross-reacting antibodies against other Coronaviridae, should be carefully considered.
We conclude that the SARS-CoV-2 assays only partly fulfil the criteria proposed by regulatory authorities. Introduction into clinical use must, therefore, be made with careful consideration and well-informed stakeholders.
Acknowledgements
The expert technical assistance of Ola Forsell and Susanna Bergqvist at the Department of Clinical Chemistry and Transfusion Medicine, Växjö Central Hospital, and of Sanna Hjalmarsson, Christina Bojesson and Eline Boesen at the Department of Clinical Microbiology, Region Kronoberg, is greatly appreciated.
Disclosure statement
The authors declare that they have no conflicts of interest relevant to the manuscript submitted to Infectious Diseases. There is no funding to report.
References
- 1.Zhu N, Zhang D, Wang W, et al. . A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zainol Rashid Z, Othman SN, Abdul Samat MN, et al. . Diagnostic performance of COVID-19 serology assays. Malays J Pathol. 2020;42(1):13–21. [PubMed] [Google Scholar]
- 3.Zhou M, Zhong J, Bi L, et al. . Serological characteristics of COVID-19 patients. Infect Dis (Lond). 2020;52(10):749–750. [DOI] [PubMed] [Google Scholar]
- 4.Mo H, Zeng G, Ren X, et al. . Longitudinal profile of antibodies against SARS-coronavirus in SARS patients and their clinical significance. Respirology. 2006;11(1):49–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman VM, Landt O, Kaiser M, et al. . Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrett PE, Lasky FD, Meier KL, et al. . User protocol for evaluation of qualitative test performance: approved guideline. 2nd ed. Wayne (PA): Clinical and Laboratory Standards Institute; 2008. [Google Scholar]
- 7.Folkhälsomyndigheten (Public Health Agency of Sweden) . Vägledning för antikroppspåvisning; 2020; [cited 2020 Jul 3]. Available from: https://www.folkhalsomyndigheten.se/contentassets/2c3d8e40926e4bcc942aa640922bb758/vagledning-antikroppspavisning.pdf
- 8.Haute Authorité de Santé . Specifications setting out the performance assessment methods applicable to serological tests detecting anti-SARS-CoV-2 antibodies; 2020; [cited 2020 Jul 31]. Available from: https://www.has-sante.fr/upload/docs/application/pdf/2020-05/has_serological_tests_covid19_specifications.pdf
- 9.Centers for Disease Control and Prevention . Interim guidelines for COVID-19 antibody testing; interim guidelines for COVID-19 antibody testing in clinical and public health settings; 2020; [cited 2020 Jul 31]. Available from: https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antibody-tests-guidelines.html
- 10.European Commission . Guidelines on COVID-19 in vitro diagnostic tests and their performance (2020/C 122 I/01); 2020; [cited 2020 Aug 4]. Available from: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=OJ:C:2020:122I:FULL&from=SV
- 11.GeurtsvanKessel CH, Okba NMA, Igloi Z, et al. . An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat Commun. 2020;11(1):3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan SS, Saw S, Chew KL, et al. Comparative clinical evaluation of the Roche Elecsys and Abbott SARS-CoV-2 serology assays for COVID-19. Arch Pathol Lab Med. 2020. [DOI] [PubMed]
- 13.Perkmann T, Perkmann-Nagele N, Breyer MK, et al. . Side-by-side comparison of three fully automated SARS-CoV-2 antibody assays with a focus on specificity. Clin Chem. 2020;66(11):1405–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]