Abstract
Background
Pancreatic cancer is the fourth leading cause of cancer deaths in the United States both in females and in males, and is projected to become the second deadliest cancer by 2030. The overall 5-year survival rate remains at around 10%. Cancer metabolism and specifically lipid metabolism plays an important role in pancreatic cancer progression and metastasis. Lipid droplets can not only store and transfer lipids, but also act as molecular messengers, and signaling factors. As lipid droplets are implicated in reprogramming tumor cell metabolism and in invasion and migration of pancreatic cancer cells, we aimed to identify lipid droplet-associated genes as prognostic markers in pancreatic cancer.
Methods
We performed a literature search on review articles related to lipid droplet-associated proteins. To select relevant lipid droplet-associated factors, bioinformatics analysis on the GEPIA platform (data are publicly available) was carried out for selected genes to identify differential expression in pancreatic cancer versus healthy pancreatic tissues. Differentially expressed genes were further analyzed regarding overall survival of pancreatic cancer patients.
Results
65 factors were identified as lipid droplet-associated factors. Bioinformatics analysis of 179 pancreatic cancer samples and 171 normal pancreatic tissue samples on the GEPIA platform identified 39 deferentially expressed genes in pancreatic cancer with 36 up-regulated genes (ACSL3, ACSL4, AGPAT2, BSCL2, CAV1, CAV2, CAVIN1, CES1, CIDEC, DGAT1, DGAT2, FAF2, G0S2, HILPDA, HSD17B11, ICE2, LDAH, LIPE, LPCAT1, LPCAT2, LPIN1, MGLL, NAPA, NCEH1, PCYT1A, PLIN2, PLIN3, RAB5A, RAB7A, RAB8A, RAB18, SNAP23, SQLE, VAPA, VCP, VMP1) and 3 down-regulated genes (FITM1, PLIN4, PLIN5). Among 39 differentially expressed factors, seven up-regulated genes (CAV2, CIDEC, HILPDA, HSD17B11, NCEH1, RAB5A, and SQLE) and two down-regulation genes (BSCL2 and FITM1) were significantly associated with overall survival of pancreatic cancer patients. Multivariate Cox regression analysis identified CAV2 as the only independent prognostic factor.
Conclusions
Through bioinformatics analysis, we identified nine prognostic relevant differentially expressed genes highlighting the role of lipid droplet-associated factors in pancreatic cancer.
Keywords: Bioinformatics, GEPIA, Lipid droplet-associated genes, Pancreatic cancer, Lipid metabolism
Background
Pancreatic cancer is the fourth leading cause of cancer deaths in the United States both in females and males [1], and is predicted to be the second most common cancer by 2030 [2]. Although recent therapeutic advances such as more effective adjuvant and neo-adjuvant chemotherapies, together with more radical and safer surgery, pancreatic cancer prognosis is very poor and the overall 5-year survival rate is still 10% [1, 3]. Metabolic reprogramming has been recognized as a hallmark of cancer [4]. Aberrant lipid synthesis and reprogrammed lipid metabolism are both associated with the development and progression of pancreatic cancer [5]. This can be because lipids such as phospholipid bilayers are fundamental structural components enabling cellular proliferation [5]. Lipid droplets (LDs) are ubiquitous intracellular organelles that store neutral lipids, such as, triacylglycerides, and cholesterol esters [6, 7]. The storage of neutral lipids in LDs is important for protecting cells from lipotoxicity due to the buildup of excess lipids in cell membranes [7]. Some cancer cells accumulate massive amount of LDs [7]. LDs are further implicated to mediate the proliferation, invasion, metastasis, as well as chemotherapy resistance in several types of cancer [8]. Oncogenic KRAS, which is the most important driver for pancreatic cancer development, controls the storage and utilization of LD supporting reprogramming of tumor cell metabolism, invasion and migration [9]. LDs are composed of a monolayer of phospholipids together with a variety of proteins such as structural proteins, membrane transport proteins, and enzymes [8]. LDs can associate with most other cellular organelles through membrane contact sites mediated by a set of proteins [10]. Since LD-associated proteins play an important role in dynamics of LD, we hypothesized that LD-associated factors may be associated with the outcome of pancreatic cancer patients.
Methods
Literature search for selecting lipid droplet-associated factors
To identify lipid droplet-associated factors, we performed a literature search in the PubMed database related to lipid droplet-associated proteins (last search date: July 2020). All publications with keywords “lipid droplet-associated protein” and the category “review” were collected. There was no restriction on the publication period. We analyzed all retrieved articles [11–39], and selected factors that were described to localize on lipid droplet including factors on contact sites to other organelles, such as endoplasmic reticulum and mitochondria.
Gene expression profiling interactive analysis (GEPIA) bioinformatics analysis
For gene expression profiling and overall survival analysis, we conducted bioinformatics analysis on the GEPIA platform (http://gepia.cancer-pku.cn/) [40]. GEPIA is an online analysis tool for processing high-throughput RNA sequencing expression data of bulk tumorous and normal samples based on the Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and the Genotype-Tissue Expression (GTEx, https://www.gtexportal.org/) databases. Our analysis included 179 pancreatic cancer samples and 171 normal pancreatic tissue samples. Dot maps of selected genes were generated. Furthermore, the GEPIA differential analysis module was used for analyzing gene expression profiles of pancreatic cancer and paired normal samples, and for screening differentially expressed genes (DEGs) between tumor and normal tissues. DEGs were further analyzed for overall survival (OS) on the GEPIA platform. Median expression was used as the threshold between high expression and low expression cohorts. The Human Protein Atlas version 20.1 (https://www.proteinatlas.org) [41] was used to analyze DEGs at the protein level by immunohistochemistry.
Statistical analysis
Statistical analysis was performed through the plug-in units of GEPIA. Analysis of variance (ANOVA) and Limma package plug-in were performed for screening DEGs. The data analysis function of Limma package is for the construction of linear models and differential expression for RNA-seq data. Genes with log2 (fold change) > 1 or < − 1, and a P value < 0.05 were considered DEGs. P-values were calculated as false discovery rate (FDR)-adjusted P-values with the Limma packages. OS analysis was performed using the Kaplan-Meier survival plots tool in the GEPIA platform. Via Log-rank test, log-rank P-values, hazards ratios (HR), and Cox P-values were obtained. Univariate and multivariate Cox regression analysis was carried out via Cox Proportional-Hazards (CoxPH) function in R. P < 0.05 was considered statistically significant.
Results
Selection of lipid droplet-associated factors for the bioinformatics analysis
Following the literature search and analysis, 65 factors were retrieved as LD-associated factors. The selecting criteria included factors not only directly implicated as LD factors, but also factors localized on contact sites of LD with other organelles such as endoplasmic reticulum and mitochondria. The majority of publications indicated perilipin family members PLIN1–5 (Table 1). The selected factors further included: Abhydrolase 5 (ABHD5, also known as Comparative gene identification CGI-58), Acetyl-CoA acetyltransferase 1 (ACAT1), Acyl-CoA synthetase long chain family member (ACSL) 3–4, 1-acylglycerol-3-phosphate O-acyltransferase 2 (AGPAT2, also known as Berardinelli-Seip congenital lipodystrophy BSCL1), Angiopoietin like 8 (ANGPTL8, also known as C19orf80), Apolipoprotein APOA4, APOB, Ancient ubiquitous protein 1 (AUP1), BSCL2 (Seipin), Caveolin 1 (CAV1, also known as BSCL3), CAV2, Caveolae-associated protein 1 (CAVIN1), Carboxyesterase 1 (CES1), Cell death-inducing DNA fragmentation factor-like effector (Cide) family members (CIDEA, CIDEB, CIDEC), Diacylglycerol O-Acyltransferase (DGAT) 1–2, Fas-associated factor 2 (FAF2, also known as UBX domain UBXD8), Fat storage inducing transmembrane protein (FITM) 1–2, G0/G1 switch protein (G0S2), Glycerol-3-phosphate acyltransferase 4 (GPAT4), Hypoxia inducible LD associated (HILPDA), Hydroxysteroid HSD17B11, HSD17B13, Interactor of little elongation complex ELL ICE2, LD-associated hydrolase (LDAH), Lipase E (LIPE, also known as Hormone sensitive lipase HSL), Lysophosphatidylcholine Acyltransferase (LPCAT) 1–2, phosphatidic acid phosphohydrolase Lipin 1 (LPIN1, PAP1), Lanosterol synthase (LSS), Methyltransferase like 7A (METTL7A, also known as AAMB), METTL7B (also known as Associated with LD protein I, ALDI), Monoglyceride lipase (MGLL), Microsomal triglyceride transfer protein (MTTP), N-ethylmaleimide-sensitive factor (NSF)-attachment protein α (NAPA, also known as α-SNAP), Neutral cholesterol ester hydrolase 1 (NCEH1), NSF, Oxysterol binding protein like 2 (OSBPL2, also known as ORP2), Phosphate Cytidylyl transferase PCYT1A (also known as CTP:phosphocholine cytidyltransferase CCTA), Phosphatidylethanolamine N-methytransferase (PEMT), Phospholipase 1 (PLD1), Patatin-like phospholipase domain containing 2 (PNPLA2), PNPLA3, PNPLA5, RAS oncogene family RAB5A, RAB7A, RAB8A, RAB18, SNAP23, Squalene epoxidase (SQLE), Synthaxin 5 (STX5), Ubiquitin conjugating enzyme UBE2G2, Vesicle associated membrane protein 4 (VAMP4), Vesicle-associated membrane protein-associated protein (VAPA), Valosin containing protein (VCP, also known as p97), and Vesicular membrane protein 1 (VMP1) (Table 1).
Table 1.
Lipid droplet-associated factors retrieved by literature search. Sixty-five factors were identified as lipid droplet-associated factors by literature search and analysis.
| Factor name | References | Factor name | References | Factor name | References |
|---|---|---|---|---|---|
| ABHD5 (CGI-58) | [11, 12, 15, 17–20, 22–29, 34, 35, 37, 38] | ACAT1 | [28, 33] | ACSL3 | [14, 15, 18, 35] |
| ACSL4 | [18, 35] | AGPAT2 (BSCL1) | [18, 21] | ANGPTL8 (C19orf80) | [21] |
| APOA4 | [18] | APOB | [18] | AUP1 | [15, 18] |
| BSCL2 (Seipin) | [14, 18, 20] | CAV1 (BSCL3) | [18, 20, 29] | CAV2 | [18, 29] |
| CAVIN1 | [18] | CES1 | [18, 32] | CIDEA | [13, 18–20, 22–24, 28–31, 39] |
| CIDEB | [13, 18, 19, 22–24, 31, 39] | CIDEC (FSP27) | [13, 17–20, 22–25, 28–31, 39] | DGAT1 | [11, 15, 18, 33, 38] |
| DGAT2 | [11, 14, 15, 18, 20, 21, 33, 35, 38] | FAF2 (UBXD8) | [15, 18] | FITM1 | [19] |
| FITM2 | [14, 19, 20] | G0S2 | [11, 17, 19, 20, 25, 28] | GPAT4 | [15, 18, 21, 38] |
| HILPDA | [11, 17, 19] | HSD17B11 | [15, 18, 35] | HSD17B13 | [15, 18] |
| ICE2 | [14] | LDAH | [18] | LIPE (HSL) | [11, 15, 17, 18, 20, 22, 23, 28, 29, 31, 32, 34, 35, 37, 38] |
| LPCAT1 | [18, 21] | LPCAT2 | [18, 21] | LPIN1 | [38] |
| LSS | [35] | METTL7A (AAMB) | [15] | METTL7B (ALDI) | [15] |
| MGLL | [11] | MTTP | [20] | NAPA | [38] |
| NCEH1 | [18] | NSF | [38] | OSBPL2 (ORP2) | [21] |
| PCYT1A (CCTA) | [18, 20] | PEMT | [21] | PLD1 | [38] |
| PLIN1 | [11, 14–20, 22–25, 28–33, 35, 36, 38] | PLIN2 (ADRP) | [11, 14–20, 22–25, 28, 30–33, 35, 36, 38] | PLIN3 (TIP47) | [11, 14, 16–20, 22–25, 28, 31–36, 38] |
| PLIN4 (S3–12) | [11, 14, 16–20, 22–24, 28, 33, 35] | PLIN5 (OXPAT/MLDP/LSDP5) | [11, 14, 16–20, 22–25, 28, 31, 33–35] | PNPLA2 (ATGL) | [11, 12, 15, 17–25, 28, 29, 31–38] |
| PNPLA3 (ADPN) | [12, 18, 27] | PNPLA5 | [21] | RAB5A | [18, 36, 38] |
| RAB7A | [21, 36] | RAB8A | [18] | RAB18 | [14, 15, 18, 20, 24, 38] |
| SNAP23 | [38] | SQLE | [35] | STX5 | [38] |
| UBE2G2 | [15] | VAMP4 | [38] | VAPA | [21] |
| VCP (p97) | [15, 18] | VMP1 | [14] |
Expression of a large number of lipid droplet-associated genes is significantly altered in pancreatic cancer patient specimens
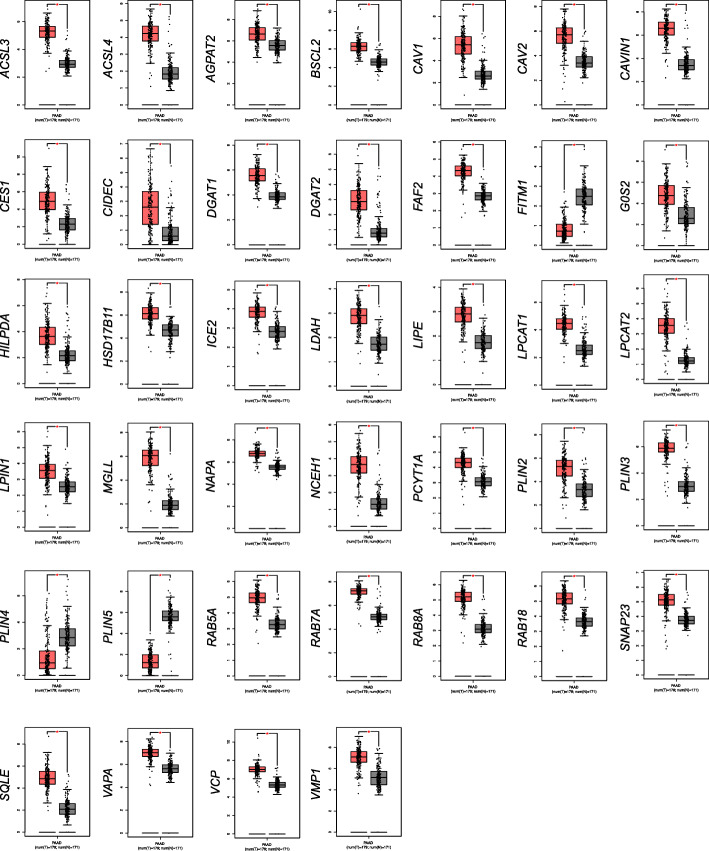
We analyzed the RNA-seq expression data of 65 genes related to lipid metabolism in 179 PDAC and 171 normal samples. Thirty-nine differentially expressed genes (DEGs), with 36 up-regulated (ACSL3, ACSL4, AGPAT2, BSCL2, CAV1, CAV2, CAVIN1, CES1, CIDEC, DGAT1, DGAT2, FAF2, G0S2, HILPDA, HSD17B11, ICE2, LDAH, LIPE, LPCAT1, LPCAT2, LPIN1, MGLL, NAPA, NCEH1, PCYT1A, PLIN3, RAB5A, RAB7A, RAB8A, RAB18, SNAP23, SQLE, VAPA, VCP, VMP1) and 3 down-regulated genes (FITM1, PLIN4, PLIN5) were identified in bulk pancreatic cancer as compared to normal pancreatic tissues (Fig. 1). Statistical data are summarized in Table 2.
Fig. 1.
The mRNA expression levels of 39 differentially expressed genes in pancreatic cancer. Gene mRNA expression level of lipid droplet-associated factors was analyzed with the GEPIA platform. The 39 differentially expressed genes (ACSL3, ACSL4, AGPAT2, BSCL2, CAV1, CAV2, CAVIN1, CES1, CIDEC, DGAT1, DGAT2, FAF2, FITM1, G0S2, HILPDA, HSD17B11, ICE2, LDAH, LIPE, LPCAT1, LPCAT2, LPIN1, MGLL, NAPA, NCEH1, PCYT1A, PLIN2, PLIN3, PLIN4, PLIN5, RAB5A, RAB7A, RAB8A, RAB18, SNAP23, SQLE, VAPA, VCP, VMP1) were depicted via bar plots between pancreatic cancer versus pancreas normal tissue (T: pancreatic cancer tissue, N: pancreas normal tissue)
Table 2.
Statistical data of lipid droplet-associated factors identified as DEGs. Differential expression analysis of lipid droplet-associated genes was performed on the GEPIA platform. The statistical data of 39 DEGs between pancreatic cancer versus normal pancreatic tissues are shown
| Gene Symbol | Gene ID | Median (Tumor) | Median (Normal) | Log2(Fold Change) | Adj. P-value |
|---|---|---|---|---|---|
| ACSL3 | ENSG00000123983.13 | 38.869 | 6.610 | 2.389 | 9.39e-76 |
| ACSL4 | ENSG00000068366.19 | 17.629 | 2.540 | 2.396 | 4.33e-66 |
| AGPAT2 | ENSG00000169692.12 | 100.071 | 46.240 | 1.097 | 6.24e-17 |
| BSCL2 | ENSG00000168000.14 | 77.772 | 23.070 | 1.710 | 9.80e-44 |
| CAV1 | ENSG00000105974.11 | 42.261 | 5.060 | 2.836 | 6.07e-52 |
| CAV2 | ENSG00000105971.14 | 51.282 | 9.600 | 2.302 | 2.89e-39 |
| CAVIN1 | ENSG00000177469.12 | 95.510 | 9.460 | 3.879 | 1.89e-40 |
| CES1 | ENSG00000198848.12 | 28.929 | 3.970 | 2.590 | 4.39e-31 |
| CIDEC | ENSG00000187288.10 | 5.060 | 0.490 | 2.024 | 1.74e-20 |
| DGAT1 | ENSG00000185000.9 | 45.730 | 13.480 | 1.690 | 3.74e-42 |
| DGAT2 | ENSG00000062282.14 | 6.230 | 0.710 | 2.080 | 5.63e-36 |
| FAF2 | ENSG00000113194.12 | 19.030 | 6.270 | 1.462 | 1.49e-59 |
| FITM1 | ENSG00000139914.6 | 0.640 | 4.620 | −1.777 | 7.21e-56 |
| G0S2 | ENSG00000123689.5 | 25.751 | 5.000 | 2.157 | 9.55e-22 |
| HILPDA | ENSG00000135245.9 | 11.370 | 3.420 | 1.485 | 8.02e-28 |
| HSD17B11 | ENSG00000198189.10 | 67.930 | 24.891 | 1.413 | 7.59e-32 |
| ICE2 | ENSG00000128915.11 | 13.590 | 6.080 | 1.043 | 5.07e-38 |
| LDAH | ENSG00000118961.14 | 6.480 | 2.310 | 1.176 | 2.26e-45 |
| LIPE | ENSG00000079435.9 | 5.960 | 1.870 | 1.278 | 2.58e-21 |
| LPCAT1 | ENSG00000153395.9 | 21.251 | 4.670 | 1.972 | 4.51e-53 |
| LPCAT2 | ENSG00000087253.11 | 10.760 | 1.330 | 2.335 | 5.24e-57 |
| LPIN1 | ENSG00000134324.11 | 10.770 | 4.790 | 1.024 | 1.04e-23 |
| MGLL | ENSG00000074416.13 | 65.639 | 2.700 | 4.171 | 6.12e-80 |
| NAPA | ENSG00000105402.7 | 108.774 | 45.581 | 1.237 | 7.56e-36 |
| NCEH1 | ENSG00000144959.9 | 11.570 | 1.480 | 2.342 | 3.75e-57 |
| PCYT1A | ENSG00000161217.11 | 18.930 | 7.370 | 1.252 | 9.92e-42 |
| PLIN2 | ENSG00000147872.9 | 37.980 | 8.970 | 1.967 | 2.64e-29 |
| PLIN3 | ENSG00000105355.8 | 58.351 | 6.890 | 2.911 | 1.13e-74 |
| PLIN4 | ENSG00000167676.4 | 0.900 | 6.180 | −1.918 | 5.30e-13 |
| PLIN5 | ENSG00000214456.8 | 1.340 | 46.610 | −4.347 | 3.06e-86 |
| RAB5A | ENSG00000144566.10 | 30.130 | 8.570 | 1.702 | 1.41e-58 |
| RAB7A | ENSG00000075785.12 | 148.024 | 31.851 | 2.182 | 1.25e-66 |
| RAB8A | ENSG00000167461.11 | 35.579 | 7.510 | 2.104 | 3.17e-70 |
| RAB18 | ENSG00000099246.16 | 34.750 | 11.450 | 1.522 | 4.92e-49 |
| SNAP23 | ENSG00000092531.9 | 33.981 | 12.280 | 1.397 | 1.09e-34 |
| SQLE | ENSG00000104549.11 | 28.019 | 3.220 | 2.782 | 4.34e-62 |
| VAPA | ENSG00000101558.13 | 128.952 | 48.600 | 1.390 | 1.38e-39 |
| VCP | ENSG00000165280.15 | 128.470 | 39.180 | 1.688 | 1.08e-50 |
| VMP1 | ENSG00000062716.10 | 136.901 | 34.789 | 1.946 | 7.73e-33 |
Lipid droplet-associated gene expression of CAV2, CIDEC, HILPDA, HSD17B11, NCEH1, RAB5A, and SQLE is associated with poor survival while expression of BSCL2 and FITM1 is associated with longer overall survival of pancreatic cancer patients
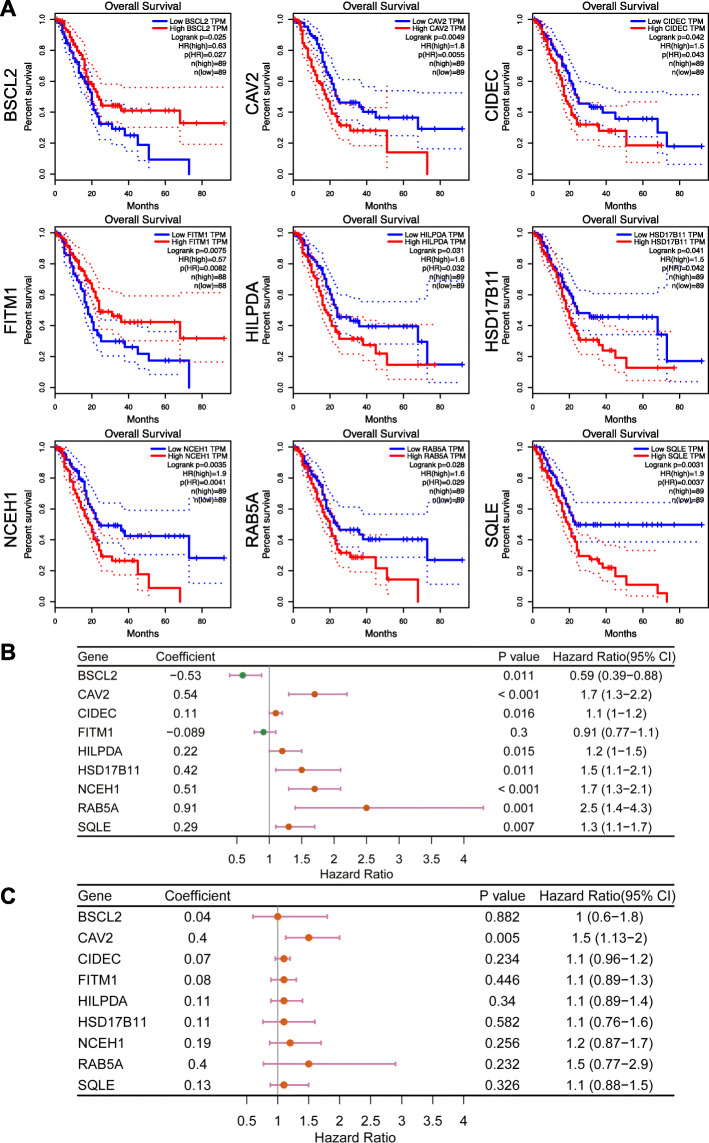
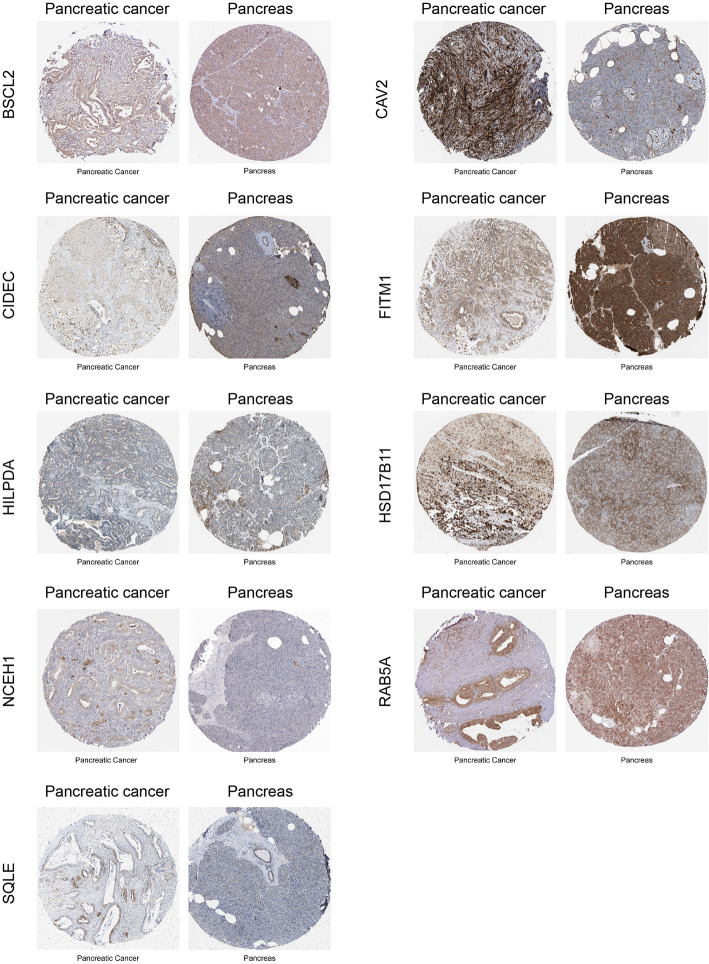
Next, overall survival (OS) analysis of 39 DEGs was performed on the GEPIA platform. The results revealed that up-regulation of seven genes (CAV2, CIDEC, HILPDA, HSD17B11, NCEH1, RAB5A, and SQLE) and down-regulation of FITM1 were significantly associated with a decrease in OS. Interestingly, up-regulation of BSCL2 was associated with favorable OS. Hence, through the above analysis, nine prognostic DEGs of lipid droplet-associated factors were identified in pancreatic cancer (Fig. 2, 3a). To further verify, whether these nine genes have prognostic power, we performed univariate Cox proportional hazards regression analysis and calculated hazard ratios (HRs) and 95% confidence intervals (CIs). Among nine prognostic DEGs, FITM1 was not a significant prognostic factor in the univariate Cox regression analysis (coefficient: -0.089, HR: 0.91 (0.77–1.1), P = 0.3), whereas the other eight genes were confirmed as prognostic factors (BSCL2 coefficient: -0.53, HR: 0.59 (0.39–0.88), P = 0.011; CAV2 coefficient: 0.54, HR: 1.7 (1.3–2.2), P < 0.001; CIDEC coefficient: 0.11, HR: 1.1 (1–1.2), P = 0.016; HILPDA coefficient: 0.22, HR: 1.2 (1–1.5), P = 0.015; HSD17B11 coefficient: 0.42, HR: 1.5 (1.1–2.1), P = 0.011; NCEH1 coefficient: 0.51, HR: 1.7 (1.3–2.1), P < 0.001; RAB5A coefficient: 0.91, HR: 2.5 (1.4–4.3), P = 0.001; SQLE coefficient: 0.29, HR: 1.3 (1.1–1.7), P = 0.007) (Fig. 3b). To identify, whether these genes were independent prognostic factors, we performed multivariate Cox proportional hazards regression analysis. Among the prognostic DEGs, only CAV2 was an independent prognostic factor (coefficient: 0.4, HR: 1.5, P = 0.005) (Fig. 3c). We further analyzed protein expression of the nine DEGs on the Human Protein Atlas database. Representative immunohistochemistry pictures of the DEGs are shown in Fig. 4.
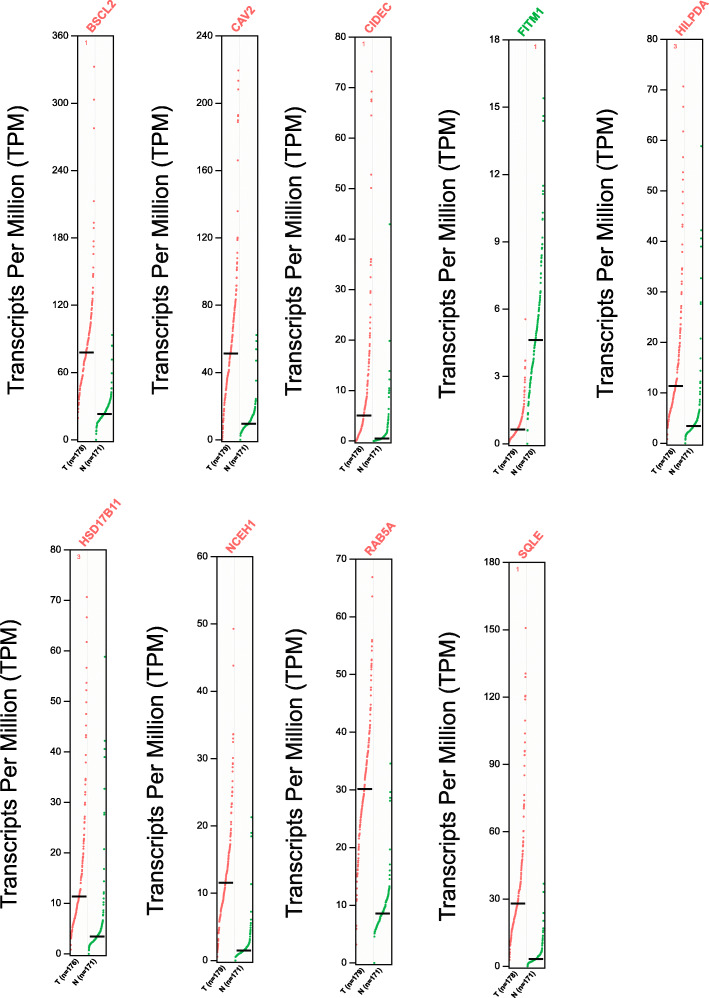
Fig. 2.
Prognostic gene expression profiling. The dot plots profiling of 9 prognostic gene expression (BSCL2, CAV2, CIDEC, FITM1, HILPDA, HSD17B11, NCEH1, RAB5A, and SQLE) were generated across pancreatic cancer and paired pancreas samples. Each dot represents an independent cancer or normal samples
Fig. 3.
Analysis of 9 genes at pancreatic cancer overall survival. a. Overall survival analyses of 9 prognostic genes (BSCL2, CAV2, CIDEC, FITM1, HILPDA, HSD17B11, NCEH1, RAB5A, and SQLE) at pancreatic cancer based on the GEPIA database. b. Univariate Cox regression analysis of nine prognostic genes. c. Multivariate Cox regression analysis of nine prognostic genes
Fig. 4.
Representative immunohistochemistry of nine lipid droplet-associated genes between pancreatic cancer and normal pancreas tissues in the Human Protein Atlas database
Taken together, LD-associated factors seem to be relevant in pancreatic cancer since there are linked by bioinformatics analyses to overall survival of pancreatic cancer patients.
Discussion
LD accumulation in non-adipose tissues has been recognized as a new hallmark of cancer cells [8]. Higher LD content has been reported in colorectal, breast, prostate cancer, hepatocellular carcinoma, renal cell carcinoma and glioblastoma [8]. LDs have been implicated to mediate aspects of proliferation, invasion, metastasis, as well as chemotherapy resistance in several types of cancer [8]. Increased storage of lipids in LDs has been suggested to be beneficial for the survival of cancer cells. Increased LD contents may expand the energy source for the metabolic need of proliferative cancer cells [8]. Storage of excess FAs and cholesterol in LDs can prevent lipotoxicity and endoplasmic reticulum (ER) stress [8]. Crucial regulators in LD homeostasis include structural proteins, membrane transport proteins and enzymes [8]. We hypothesized that expression of LD-associated factors is relevant in pancreatic cancer, because LD-associated factors support the storage of neutral lipids in lipid droplets providing energy source for cancer cells and potentially protecting cancer cells from lipotoxicity. Furthermore, LDs can associate with most other cellular organelles such as ER, nucleus, mitochondria, peroxisomes, and lysosomes through membrane contact sites [10]. It is not known whether LDs can be transferred between cells, or can be secreted into the blood stream. It has been shown that tumors signal over long distances to sites of future metastases to promote formation of a pre-metastatic niche that potentially supports growth of disseminated tumor cells upon their arrival [42]. If lipid droplets act as “transporters” of lipids and signaling molecules between cells, LDs and LD-associated factors might be involved in metastasis. The molecular mechanisms, however, need to be clarified in future.
In the current study, we identified 36 upregulated and 3 downregulated genes characterized as LD-associated factors in pancreatic cancer (Table 2). We further identified that enhanced expression of 7 genes namely CAV2, CIDEC, HILPDA, HSD17B11, NCEH1, RAB5A, and SQLE are associated with significantly shorter overall survival whereas elevated expression of BSCL2 and FITM1 correlates with longer overall survival of pancreatic cancer patients (Fig. 3a). To verify those findings also on the protein level, normal pancreatic tissue samples, in addition to cancerous and para-cancerous tissues, would be required. However, we confirmed protein expression of the DEGs by using the Human Protein Atlas database. Although gene transcripts levels and protein concentrations do not always correspond to each other, we identified prognostic LD-associated factors at the transcriptome level, providing clues for future proteomics, metabolomics and downstream functional analysis.
CAV-2 is a caveolin family member and CAV-2 expression is increased during the accumulation of intracellular LDs and the adipogenic differentiation [43]. CAV-2 plays a key role in intracellular cell transport and higher level of CAV-2 is associated with different types of cancer progression [44]. It has been demonstrated that higher expression of CAV-2 and its upstream regulator bromodomain containing 4 (BRD4) is associated with shorter overall survival of 76 pancreatic cancer patients [44]. In our study, both CAV1 and CAV2 were identified as DEGs, but only CAV2 was a prognostic DEG associated with shorter OS in pancreatic cancer patients.
CIDEC (also known as fat-specific protein of 27 kDa, FSP27) has been suggested to mediate LD-LD contact for promoting LD fusion [6, 10]. CIDE proteins are enriched at LD-LD contact sites and physically chaining the adjacent organelles [10]. So far it has been demonstrated that CIDEC promotes development of hepatic steatosis and steatohepatitis [45, 46]. In our study, only CIDEC but not CIDEA or CIDEB was identified as a prognostic DEG. The precise and specific role of CIDEC in cancer needs to be clarified.
In various cancer cells including renal cell carcinoma, ovarian clear cell carcinoma, colorectal adenoma and carcinoma, upregulation of HILPDA has been observed [47–49]. Hypoxia-inducible factor 1 (HIF1) regulates the expression of HILPDA [11]. HILPDA preferentially accumulates in LDs undergoing remodeling (e.g. expansion). HILPDA has been shown to co-localize with the lipogenic enzymes DGAT1 and DGAT2 [11], which were both identified as DEGs in our study (Table 2). DGATs catalyze the final rate-limiting step in the formation of triglycerides. In line with this, HILPDA has been shown to promote intracellular lipid accumulation by enhancing triglyceride synthesis [11]. Overexpression of HILPDA, but not DGAT1/2, is associated with shorter overall survival in pancreatic cancer patients (Fig. 3a), suggesting that additional roles of HILPDA, other than regulating triacylglyceride (TAG) synthesis, may influence the outcome of pancreatic cancer patients. Indeed, HILPDA inhibits the rate-limiting enzyme of TAG hydrolysis PNPLA2 (ATGL), leading to inhibition of lipolysis, attenuated fatty acid oxidation and ROS production [11].
HSD17B11 is known to convert 5α-androstan-3α, 17β-diol (3α-diol) to androsterone [50]. HSD17B11 regulates size of LDs, LD distribution and TAG content [51]. The role of HSD17B11 in pancreatic cancer has not been elucidated. It has been shown that HSD17B13 variants are associated with nonalcoholic fatty liver disease (NAFLD) and HSD17B13 expression is elevated in nonalcoholic steatohepatitis (NASH) patients, or with risk of cirrhosis and hepatocellular carcinoma (HCC), while a HSD17B13 variant has been demonstrated to protect from HCC development [52–54]. In our study, HSD17B13 was not identified as DEG. Regarding NCEH1, which hydrolyzes 2-acetyl monoalkylglycerol in the metabolism of ether lipids, it has been suggested as a prognostic marker for pancreatic cancer [55], supporting our analysis.
Rab family protein RAB5A belongs to the Ras family of G-proteins, which regulate membrane vesicle trafficking. In our study, several Rab family members namely RAB5A, RAB7A, RAB8A, and RAB18 were identified as DEGs (Table 2). Among these four genes, expression of RAB5A was associated with shorter OS of pancreatic cancer patients. Expression of RAB5A correlated with pancreatic tumor progression in another study of 111 patients as well [56]. Increased expression of RAB5A predicts metastasis and shorter OS in colorectal cancer patients [57]. It has been suggested that RAB5A regulates Wnt signaling, proliferation, invasion and 5-FU drug resistance [56]. Further, RAB5A activates IRS1, Akt and mTOR signaling [58], suggesting that LD-associated factors are also involved in regulating signaling pathways. SQLE is a key enzyme in the cholesterol synthesis pathway and converts squalene to 2,3-epoxysqualene [59]. SQLE increase epigenetic silencing of PTEN, leading to activation of the Akt-mTOR pathway and NAFLD-induced HCC growth. High expression of SQLE was associated with shorter OS of HCC patients [60]. On the other hand, it has been demonstrated that reduction of SQLE mRNA and protein expression is associated with shortened survival of colorectal cancer patients. SQLE reduction aggravates colorectal cancer progression via the activation of the β-catenin pathway and deactivation of p53 pathway [61]. In our study with pancreatic cancer patient databases, SQLE was identified as a DEG and higher expression of SQLE was associated with shorter OS of pancreatic cancer patients. The precise and cancer type-specific role of SQLE has to be further clarified.
Study strengths and limitations
Bioinformatics analysis revealed that expression of LD-associated factors is associated with overall survival in pancreatic cancer patients. Although aberrant lipid synthesis and reprogrammed lipid metabolism are both associated with the development and progression of pancreatic cancer, LD and LD-associated factors have not been considered in this disease. In the current study, we identified 65 factors as LD-associated factors. We identified 39 DEGs with 36 up-regulated and 3 down-regulated genes. Among 39 DEGs, 7 up-regulated genes and 2 down-regulated genes were significantly associated with overall survival of pancreatic cancer patients. Cox regression analysis further validated 8 factors as prognostic factors.
There are also several limitations of the study. Cofounding factors such as body-mass index (BMI) and others could not be considered, since the data of normal tissue from the GTEx database do not include these information. Furthermore, findings were not validated on the protein level. Further studies must include functional analysis using knockout animal model of the prognostic candidate genes to analyze whether deletion/ inhibition of the lipid droplet-associated factors can change the metabolic profile of the cells, proliferation, and the ability to metastasize. It would also be of interest to clarify whether lipid droplets can be detected in liquid biopsies (blood), and whether deletion/ inhibition of LD-associated factors reduce lipid droplet contents in the blood.
Conclusions
LDs are ubiquitous cellular organelles, involved not only in lipid metabolism but also in diverse biological functions such as regulating signaling pathways. LDs mediate proliferation, invasion, metastasis, as well as chemotherapy resistance in several types of cancer. LD-associated proteins play an important role in dynamics of LD and it is now evident that expression of several LD-associated genes are associated with overall survival in pancreatic cancer patients. The current study identified prognostic LD-associated factors at the transcriptome level, providing clues for future proteomics and downstream functional and pathway analysis. It is important to increase our understanding of cancer type-specific roles of LD-associated factors, which may help to develop more specific and personalized therapies for pancreatic cancer patients in the future.
Acknowledgments
Not applicable.
Abbreviations
- ABHD
Abhydrolase
- ACAT
Acetyl-CoA acetyltransferase
- ACSL
Acyl-CoA synthetase long chain family member
- ADRP
Adipocyte differentiation-related protein
- AGPAT
1-Acylglycerol-3-phosphate O-acyltransferase
- ALDI
Associated with LD protein I
- ANGPTL
Angiopoietin like
- APO
Apolipoprotein
- ATGL
Adipose triglyceride lipase
- AUP
Ancient ubiquitous protein
- BSCL
Berardinelli-seip congenital lipodystrophy
- CAV
Caveolin
- CAVIN
Caveolae-associated protein
- CCT
CTP:phosphocholine cytidyltransferase
- CES
Carboxyesterase
- CGI
Comparative gene identification
- CI
Confidence interval
- CIDE
Cell death-inducing DNA fragmentation factor-like effector
- DEG
Differentially expressed gene
- DFF
Death-inducing DNA fragmentation factor
- DGAT
Diacylglycerol O-acyltransferase
- ER
Endoplasmic reticulum
- FAF
Fas-associated factor
- FITM
Fat storage inducing transmembrane protein
- FSP
Fat-specific protein
- G0S2
G0/G1 switch protein
- GEPIA
Gene expression profiling interactive analysis
- GPAT
Glycerol-3-phosphate acyltransferase
- HCC
Hepatocellular carcinoma
- HIF
Hypoxia-inducible factor
- HILPDA
Hypoxia inducible LD associated
- HR
Hazard ratio
- HSD
Hydroxysteroid
- HSL
Hormone sensitive lipase
- ICE
Interactor of little elongation complex ELL
- LD
Lipid droplet
- LDAH
LD-associated hydrolase
- LIPE
Lipase E
- LPCAT
Lysophosphatidylcholine acyltransferase
- LPIN
Lipin
- LSDP
Lipid storage droplet protein
- LSS
Lanosterol synthase
- METTL
Methyltransferase like
- MGLL
Monoglyceride lipase
- MLDP
Myocardial LD protein
- MTTP
Microsomal triglyceride transfer protein
- NAFLD
Nonalcoholic fatty liver disease
- NAP
N-ethylmaleimide-sensitive factor attachment protein
- NASH
Nonalcoholic steatohepatitis
- NCEH
Neutral cholesterol ester hydrolase
- NSF
N-ethylmaleimide-sensitive factor
- OSBPL
Oxysterol binding protein like
- ORP
Oxysterol-binding protein-related protein
- PAP
Phosphatidic acid phosphohydrolase
- PAT
Perilipin, ADRP, TIP47
- PCYT
Phosphate cytidylyl transferase
- PEMT
Phosphatidylethanolamine N-methytransferase
- PLD
Phospholipase
- PLIN
Perilipin
- PNPLA
Patatin-like phospholipase
- PTRF
Polymerase I and transcript release factor
- SNAP
Synaptosome associated protein
- SQLE
Squalene epoxidase
- STX
Syntaxin
- TIP
Tail-interacting protein
- UBE
Ubiquitin conjugating enzyme
- UBXD
UBX domain
- VAMP
Vesicle associated membrane protein
- VAPA
Vesicle-associated membrane protein-associated protein
- VCP
Valosin containing protein
- VMP
Vesicular membrane protein
Authors’ contributions
RB performed the gene bioinformatics analysis and wrote the manuscript. AR gave administrative support. JK supervised the study. YS designed the study, performed the gene selection analysis, and wrote the manuscript. The author(s) read and approved the final manuscript.
Funding
There is no funding obtained for the current manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
The Human Protein Atlas version 20.1 (https://www.proteinatlas.org) was used to analyze differentially expressed genes at the protein level by immunohistochemistry. Image credit: Human Protein Atlas.
Competing interests
The authors have no conflict of interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka M, Mihaljevic AL, Probst P, Heckler M, Klaiber U, Heger U, Büchler MW, Hackert T. Meta-analysis of recurrence pattern after resection for pancreatic cancer. Br J Surg. 2019;106(12):1590–1601. doi: 10.1002/bjs.11295. [DOI] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Sunami Y, Rebelo A, Kleeff J. Lipid metabolism and lipid droplets in pancreatic cancer and stellate cells. Cancers (Basel). 2017;10(1). pii: E3. doi: 10.3390/cancers10010003. Review. [DOI] [PMC free article] [PubMed]
- 6.Walther TC, Farese RV., Jr Lipid droplets and cellular lipid metabolism. Annu Rev Biochem. 2012;81(1):687–714. doi: 10.1146/annurev-biochem-061009-102430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walther TC, Chung J, Farese RV., Jr Lipid Droplet Biogenesis. Annu Rev Cell Dev Biol. 2017;33(1):491–510. doi: 10.1146/annurev-cellbio-100616-060608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Liu H, Luo X. Lipid droplet and its implication in cancer progression. Am J Cancer Res. 2020;10(12):4112–4122. [PMC free article] [PubMed] [Google Scholar]
- 9.Rozeveld CN, Johnson KM, Zhang L, Razidlo GL. KRAS controls pancreatic Cancer cell lipid metabolism and invasive potential through the lipase HSL. Cancer Res. 2020;80(22):4932–4945. doi: 10.1158/0008-5472.CAN-20-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olzmann JA, Carvalho P. Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol. 2019;20(3):137–155. doi: 10.1038/s41580-018-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de la Rosa Rodriguez MA, Kersten S. Regulation of lipid droplet homeostasis by hypoxia inducible lipid droplet associated HILPDA. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865(9):158738. doi: 10.1016/j.bbalip.2020.158738. [DOI] [PubMed] [Google Scholar]
- 12.Dong XC. PNPLA3-A potential therapeutic target for personalized treatment of chronic liver disease. Front Med (Lausanne) 2019;6:304. doi: 10.3389/fmed.2019.00304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Slayton M, Gupta A, Balakrishnan B, Puri V. CIDE proteins in human health and disease. Cells. 2019;8(3):238. doi: 10.3390/cells8030238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salo VT, Ikonen E. Moving out but keeping in touch: contacts between endoplasmic reticulum and lipid droplets. Curr Opin Cell Biol. 2019;57:64–70. doi: 10.1016/j.ceb.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Zhang C, Liu P. The new face of the lipid droplet: lipid droplet proteins. Proteomics. 2019;19(10):e1700223. doi: 10.1002/pmic.201700223. [DOI] [PubMed] [Google Scholar]
- 16.Zhang P, Meng L, Song L, Du J, Du S, Cui W, et al. Roles of Perilipins in diseases and cancers. Curr Genomics. 2018;19(4):247–257. doi: 10.2174/1389202918666170915155948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerk IK, Wechselberger L, Oberer M. Adipose triglyceride lipase regulation: an overview. Curr Protein Pept Sci. 2018;19(2):221–233. doi: 10.2174/1389203718666170918160110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu S, Zhang X, Liu P. Lipid droplet proteins and metabolic diseases. Biochim Biophys Acta Mol Basis Dis. 2018;1864(5 Pt B):1968–1983. doi: 10.1016/j.bbadis.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 19.de la Rosa Rodriguez MA, Kersten S. Regulation of lipid droplet-associated proteins by peroxisome proliferator-activated receptors. Biochim Biophys Acta Mol Cell Biol Lipids. 2017;1862(10 Pt B):1212–1220. doi: 10.1016/j.bbalip.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Engin A. Fat cell and fatty acid turnover in obesity. Adv Exp Med Biol. 2017;960:135–160. doi: 10.1007/978-3-319-48382-5_6. [DOI] [PubMed] [Google Scholar]
- 21.Barbosa AD, Siniossoglou S. Function of lipid droplet-organelle interactions in lipid homeostasis. Biochim Biophys Acta Mol Cell Res. 2017;1864(9):1459–1468. doi: 10.1016/j.bbamcr.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 22.Kimmel AR, Sztalryd C. The Perilipins: major cytosolic lipid droplet-associated proteins and their roles in cellular lipid storage, mobilization, and systemic homeostasis. Annu Rev Nutr. 2016;36:471–509. doi: 10.1146/annurev-nutr-071813-105410. [DOI] [PubMed] [Google Scholar]
- 23.Plakkal Ayyappan J, Paul A, Goo YH. Lipid droplet-associated proteins in atherosclerosis (review) Mol Med Rep. 2016;13(6):4527–4534. doi: 10.3892/mmr.2016.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikura Y, Caldwell SH. Lipid droplet-associated proteins in alcoholic liver disease: a potential linkage with hepatocellular damage. Int J Clin Exp Pathol 2015;8(8):8699–8708. eCollection 2015. PMID: 26464614 [PMC free article] [PubMed]
- 25.D'Andrea S. Lipid droplet mobilization: the different ways to loosen the purse strings. Biochimie. 2016;120:17–27. doi: 10.1016/j.biochi.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Zierler KA, Zechner R, Haemmerle G. Comparative gene identification-58/α/β hydrolase domain 5: more than just an adipose triglyceride lipase activator? Curr Opin Lipidol. 2014;25(2):102–109. doi: 10.1097/MOL.0000000000000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krawczyk M, Portincasa P, Lammert F. PNPLA3-associated steatohepatitis: toward a gene-based classification of fatty liver disease. Semin Liver Dis. 2013;33(4):369–379. doi: 10.1055/s-0033-1358525. [DOI] [PubMed] [Google Scholar]
- 28.Christian M. Nuclear receptor-mediated regulation of lipid droplet-associated protein gene expression in adipose tissue. Horm Mol Biol Clin Investig. 2013;14(3):87–97. doi: 10.1515/hmbci-2013-0028. [DOI] [PubMed] [Google Scholar]
- 29.Konige M, Wang H, Sztalryd C. Role of adipose specific lipid droplet proteins in maintaining whole body energy homeostasis. Biochim Biophys Acta. 2014;1842(3):393–401. doi: 10.1016/j.bbadis.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan Y, Li P, Ye J. Lipid homeostasis and the formation of macrophage-derived foam cells in atherosclerosis. Protein Cell. 2012;3(3):173–181. doi: 10.1007/s13238-012-2025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Girousse A, Langin D. Adipocyte lipases and lipid droplet-associated proteins: insight from transgenic mouse models. Int J Obes. 2012;36(4):581–594. doi: 10.1038/ijo.2011.113. [DOI] [PubMed] [Google Scholar]
- 32.Quiroga AD, Lehner R. Role of endoplasmic reticulum neutral lipid hydrolases. Trends Endocrinol Metab. 2011;22(6):218–225. doi: 10.1016/j.tem.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 33.Buers I, Hofnagel O, Ruebel A, Severs NJ, Robenek H. Lipid droplet associated proteins: an emerging role in atherogenesis. Histol Histopathol. 2011;26(5):631–642. doi: 10.14670/HH-26.631. [DOI] [PubMed] [Google Scholar]
- 34.Lass A, Zimmermann R, Oberer M, Zechner R. Lipolysis - a highly regulated multi-enzyme complex mediates the catabolism of cellular fat stores. Prog Lipid Res. 2011;50(1):14–27. doi: 10.1016/j.plipres.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodman JM. Demonstrated and inferred metabolism associated with cytosolic lipid droplets. J Lipid Res 2009. 50(11):2148–56. 10.1194/jlr.R001446 Epub 2009 Aug 20. PMID: 19696439. [DOI] [PMC free article] [PubMed]
- 36.Zehmer JK, Huang Y, Peng G, Pu J, Anderson RG, Liu P. A role for lipid droplets in inter-membrane lipid traffic. Proteomics. 2009;9(4):914–921. doi: 10.1002/pmic.200800584. PMID: 19160396 [DOI] [PMC free article] [PubMed]
- 37.Zechner R, Kienesberger PC, Haemmerle G, Zimmermann R, Lass A. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J Lipid Res. 2009;50(1):3–21. doi: 10.1194/jlr.R800031-JLR200. [DOI] [PubMed] [Google Scholar]
- 38.Olofsson SO, Boström P, Andersson L, Rutberg M, Levin M, Perman J, Borén J. Triglyceride containing lipid droplets and lipid droplet-associated proteins. Curr Opin Lipidol. 2008;19(5):441–447. doi: 10.1097/MOL.0b013e32830dd09b. [DOI] [PubMed] [Google Scholar]
- 39.Puri V, Virbasius JV, Guilherme A, Czech MP. RNAi screens reveal novel metabolic regulators: RIP140, MAP 4k4 and the lipid droplet associated fat specific protein (FSP) 27. Acta Physiol (Oxf) 2008;192(1):103–115. doi: 10.1111/j.1748-1716.2007.01786.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 42.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar C, Nitadori-Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J, Lyden D. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18(6):883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Codenotti S, Vezzoli M, Poliani PL, Cominelli M, Bono F, Kabbout H, Faggi F, Chiarelli N, Colombi M, Zanella I, Biasiotto G, Montanelli A, Caimi L, Monti E, Fanzani A. Caveolin-1, Caveolin-2 and Cavin-1 are strong predictors of adipogenic differentiation in human tumors and cell lines of liposarcoma. Eur J Cell Biol. 2016;95(8):252–264. doi: 10.1016/j.ejcb.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Jiao F, Han T, Yuan C, Liang Y, Cui J, Zhuo M, Wang L. Caveolin-2 is regulated by BRD4 and contributes to cell growth in pancreatic cancer. Cancer Cell Int. 2020;20(1):55. doi: 10.1186/s12935-020-1135-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsusue K, Kusakabe T, Noguchi T, Takiguchi S, Suzuki T, Yamano S, Gonzalez FJ. Hepatic steatosis in leptin-deficient mice is promoted by the PPARgamma target gene Fsp27. Cell Metab. 2008;7(4):302–311. doi: 10.1016/j.cmet.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu MJ, Cai Y, Wang H, Altamirano J, Chang B, Bertola A, et al. Fat-specific protein 27/CIDEC promotes development of alcoholic steatohepatitis in mice and humans. Gastroenterology. 2015;149(4):1030–41.e6. doi: 10.1053/j.gastro.2015.06.009. Epub 2015 Jun 20. PMID: 26099526 [DOI] [PMC free article] [PubMed]
- 47.Nishimura S, Tsuda H, Ito K, Takano M, Terai Y, Jobo T, Kigawa J, Sugiyama T, Yaegashi N, Aoki D. Differential expression of hypoxia-inducible protein 2 among different histological types of epithelial ovarian cancer and in clear cell adenocarcinomas. Int J Gynecol Cancer. 2010;20(2):220–226. doi: 10.1111/IGC.0b013e3181ca1e16. [DOI] [PubMed] [Google Scholar]
- 48.Kim SH, Wang D, Park YY, Katoh H, Margalit O, Sheffer M, Wu H, Holla VR, Lee JS, DuBois RN. HIG2 promotes colorectal cancer progression via hypoxia-dependent and independent pathways. Cancer Lett. 2013;341(2):159–165. doi: 10.1016/j.canlet.2013.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Saarinen AM, Hitosugi T, Wang Z, Wang L, Ho TH, Liu J. Inhibition of intracellular lipolysis promotes human cancer cell adaptation to hypoxia. Elife. 2017;6:e31132. doi: 10.7554/eLife.31132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brereton P, Suzuki T, Sasano H, Li K, Duarte C, Obeyesekere V, Haeseleer F, Palczewski K, Smith I, Komesaroff P, Krozowski Z. Pan1b (17betaHSD11)-enzymatic activity and distribution in the lung. Mol Cell Endocrinol. 2001;171(1–2):111–117. doi: 10.1016/s0303-7207(00)00417-2. [DOI] [PubMed] [Google Scholar]
- 51.Liu Y, Xu S, Zhang C, Zhu X, Hammad MA, Zhang X, Christian M, Zhang H, Liu P. Hydroxysteroid dehydrogenase family proteins on lipid droplets through bacteria, C. elegans, and mammals. Biochim Biophys Acta Mol Cell Biol Lipids. 2018;1863(8):881–894. doi: 10.1016/j.bbalip.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 52.Ma Y, Belyaeva OV, Brown PM, Fujita K, Valles K, Karki S, de Boer YS, Koh C, Chen Y, du X, Handelman SK, Chen V, Speliotes EK, Nestlerode C, Thomas E, Kleiner DE, Zmuda JM, Sanyal AJ, (for the Nonalcoholic Steatohepatitis Clinical Research Network) Kedishvili NY, Liang TJ, Rotman Y. 17-Beta Hydroxysteroid dehydrogenase 13 is a hepatic retinol dehydrogenase associated with histological features of nonalcoholic fatty liver disease. Hepatology. 2019;69(4):1504–1519. doi: 10.1002/hep.30350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gellert-Kristensen H, Richardson TG, Davey Smith G, Nordestgaard BG, Tybjaerg-Hansen A, Stender S. Combined effect of PNPLA3, TM6SF2, and HSD17B13 variants on risk of cirrhosis and hepatocellular carcinoma in the general population. Hepatology. 2020;72(3):845–856. doi: 10.1002/hep.31238. [DOI] [PubMed] [Google Scholar]
- 54.Yang J, Trépo E, Nahon P, Cao Q, Moreno C, Letouzé E, Imbeaud S, Bayard Q, Gustot T, Deviere J, Bioulac-Sage P, Calderaro J, Ganne-Carrié N, Laurent A, Blanc JF, Guyot E, Sutton A, Ziol M, Zucman-Rossi J, Nault JC. A 17-Beta-Hydroxysteroid dehydrogenase 13 variant protects from hepatocellular carcinoma development in alcoholic liver disease. Hepatology. 2019;70(1):231–240. doi: 10.1002/hep.30623. [DOI] [PubMed] [Google Scholar]
- 55.Lu Y, Zhang L, Chen X, Zhang Q. NCEH1 may be a prognostic biomarker for pancreatic cancer. Int J Clin Exp Pathol. 2020;13(11):2746–2752. [PMC free article] [PubMed] [Google Scholar]
- 56.Li Y, Sun X, Ji D, Kong X, Liu D, Zhao Z, Yan J, Chen S. Expression of Rab5a correlates with tumor progression in pancreatic carcinoma. Virchows Arch. 2017;470(5):527–536. doi: 10.1007/s00428-017-2098-y. [DOI] [PubMed] [Google Scholar]
- 57.Yu MH, Luo Y, Qin SL, Zhong M. Increased expression of Rab5A predicts metastasis and poor prognosis in colorectal cancer patients. Int J Clin Exp Pathol. 2015;8(6):6974–6980. [PMC free article] [PubMed] [Google Scholar]
- 58.Cong XX, Gao XK, Rao XS, Wen J, Liu XC, Shi YP, He MY, Shen WL, Shen Y, Ouyang H, Hu P, Low BC, Meng ZX, Ke YH, Zheng MZ, Lu LR, Liang YH, Zheng LL, Zhou YT. Rab5a activates IRS1 to coordinate IGF-AKT-mTOR signaling and myoblast differentiation during muscle regeneration. Cell Death Differ. 2020;27(8):2344–2362. doi: 10.1038/s41418-020-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu H, Zhou S, Tang Q, Xia H, Bi F. Cholesterol metabolism: new functions and therapeutic approaches in cancer. Biochim Biophys Acta Rev Cancer. 2020;1874(1):188394. doi: 10.1016/j.bbcan.2020.188394. [DOI] [PubMed] [Google Scholar]
- 60.Liu D, Wong CC, Fu L, Chen H, Zhao L, Li C, et al. Squalene epoxidase drives NAFLD-induced hepatocellular carcinoma and is a pharmaceutical target. Sci Transl Med. 2018;10(437):eaap9840. doi: 10.1126/scitranslmed.aap9840. [DOI] [PubMed] [Google Scholar]
- 61.Jun SY, Brown AJ, Chua NK, Yoon JY, Lee JJ, Yang JO, et al. Reduction of squalene epoxidase by cholesterol accumulation accelerates colorectal cancer progression and metastasis. Gastroenterology. 2020;S0016-5085(20):35146–35145. doi: 10.1053/j.gastro.2020.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.